Abstract
Neurotrophins activate multiple signaling pathways in neurons. However, the precise roles of these signaling molecules in cell survival are not well understood. In this report, we show that nerve growth factor (NGF) activates the transcription factors NF-κB and AP-1 in cultured sympathetic neurons. Activated NF-κB complexes were shown to consist of heterodimers of p50 and Rel proteins (RelA, as well as c-Rel), and NF-κB activation was found to occur independently ofde novo protein synthesis but in a manner that required the action of the proteasome complex. Treatment with the NF-κB inhibitory peptide SN50 in the continuous presence of NGF resulted in dose-dependent induction of cell death. Under the conditions used, SN50 was shown to selectively inhibit NF-κB activation but not the activation of other cellular transcription factors such as AP-1 and cAMP response element-binding protein. Cells treated with SN50 exhibited morphological and biochemical hallmarks of apoptosis, and the kinetics of cell killing were accelerated relative to death induced by NGF withdrawal. Finally, experiments were conducted to test directly whether NF-κB could act as a survival factor for NGF-deprived neurons. Microinjection of cells with an expression plasmid encoding NF-κB (c-Rel) resulted in enhanced neuronal survival after withdrawal of NGF, whereas cells that were transfected with a vector encoding a mutated derivative of c-Rel lacking the transactivation domain underwent cell death to the same extent as control cells. Together, these findings suggest that the activation of NF-κB/Rel transcription factors may contribute to the survival of NGF-dependent sympathetic neurons.
Keywords: rat, neuron, apoptosis, cell death, NF-κB, transcription factor
Programmed cell death is a naturally occurring process required for nervous system development (Oppenheim, 1991), which has been studied extensively using nerve growth factor (NGF)-dependent sympathetic neurons isolated from rat superior cervical ganglion (SCG) (Deshmukh and Johnson, 1997; Pettmann and Henderson, 1998). When deprived of NGF, these neurons undergo apoptosis (Levi-Montalcini and Booker, 1960; Gorin and Johnson, 1979); addition of exogenous NGF rescues them from death (Hendry and Campbell, 1976). Similarly, newborn mice lacking NGF (Crowley et al., 1994) or its receptor TrkA (Smeyne et al., 1994) have greatly reduced numbers of sympathetic neurons. Binding of NGF to TrkA and the low-affinity neurotrophin receptor p75NTR activates several signaling pathways, including the phosphoinositide-3-kinase (PI3K) and mitogen-activated protein kinase pathways (Bredesen and Rabizadeh, 1997). NGF binding also results in the activation of several transcription factors (Ginty et al., 1994; Bonni et al., 1995; Riccio et al., 1997), including NF-κB (Wood, 1995; Carter et al., 1996). Recently, PI3K has been shown to be important for NGF-mediated survival of rat pheochromocytoma PC12 cells and primary neurons (Yao and Cooper, 1995; Philpott et al., 1997; Crowder and Freeman, 1998). However, it remains unclear whether other NGF-induced events, such as activation of NF-κB, have a role in neuronal survival.
NF-κB plays a key role in the regulation of genes involved in immunity, inflammation (Ghosh et al., 1998), and nervous system function (O’Neill and Kaltschmidt, 1997). The NF-κB/Rel family consists of five major members: p50, RelA, c-Rel, p52, and RelB. NF-κB activation has been shown to protect lymphoid cells and fibroblasts from several apoptotic stimuli (Beg and Baltimore, 1996;Liu et al., 1996; Van Antwerp et al., 1996; Wang et al., 1996; Boothby et al., 1997).
Like NF-κB, AP-1 transcription factors are dimeric molecules comprised of two major protein families: the Jun family and the Fos/activating transcription factor family. The activity of AP-1 components is essential for the proliferation and differentiation of certain cells (Angel and Karin, 1991). However, some AP-1 components may also be involved in apoptosis (Herdegen et al., 1997). Indeed, c-Jun is required for apoptosis in response to withdrawal of NGF from cultured sympathetic neurons (Estus et al., 1994; Ham et al., 1995). C-Jun N-terminal kinase (JNK) activation may also be involved in the induction of apoptosis in neurons by virtue of its ability to induce transcription of the proto-oncogene c-jun(Verheij et al., 1996; Eilers et al., 1998).
Here, we show that NGF activates NF-κB and AP-1 in sympathetic neurons and that NGF-inducible NF-κB is required for the survival of neurons. We also demonstrate that overexpression of NF-κB is sufficient to protect sympathetic neurons from apoptosis caused by NGF deprivation. These results suggest that NF-κB may contribute to the survival of NGF-dependent sympathetic neurons.
MATERIALS AND METHODS
Primary neuronal culture. Primary cultures of sympathetic neurons were obtained from SCG of embryonic day 21 rats as described previously (Crowder and Freeman, 1998). Cultures prepared by this method are >95% postmitotic neurons (Estus et al., 1994; Crowder and Freeman, 1998). Briefly, SCG were dissected, dissociated, and resuspended in media containing 90% MEM (Life Technologies, Gaithersburg, MD), 10% FBS (Sigma, St. Louis, MO), 2 mm glutamine, 20 μm uridine, 20 μm 5-fluoro-2′-deoxyuridine (an antiproliferative agent), 100 U/μl penicillin, 100 μg/ml streptomycin, and 50 ng/ml NGF (Harlan Bioproducts for Science, Madison, WI). The cell suspension was filtered through a Nitex filter (size 3–20/14; Tetko, Briarcliff Manor, NY), and cells were plated onto collagen-coated dishes. For microinjection experiments, dissociated cells were preplated onto plastic tissue culture dishes (Beckton Dickinson, Lincoln Park, NJ) for 1 hr before plating on poly-l-ornithine (Sigma) and laminin-coated (Collaborative Research, Bedford, MA) 35 mm glass-bottomed dishes (MatTek, Ashland, MA). All experimentation was performed on neurons that had been in culture for at least 5 d, but no more than 7 d. To initiate NGF deprivation, culture medium was replaced by rinsing cells once with Leibovitz’s L-15 and then adding medium prepared as described above but lacking NGF and containing excess neutralizing NGF antibody (Harlan, Indianapolis, IN).
Nuclear extract preparation and electrophoresis mobility shift assay. SCG neurons (1 × 105) were collected by centrifugation at 800 × g for 5 min. Nuclear extracts were prepared as described previously (Schreiber et al., 1989). Electrophoresis mobility shift assay (EMSA) was performed by incubating the nuclear extracts with a 32P-radiolabeled probe at room temperature for 10 min, followed by resolution of the DNA–protein complexes on native 4% polyacrylamide gels (Maggirwar et al., 1997). For antibody supershift assays, 2 μg of desired antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) were added to the EMSA reaction 5 min before electrophoresis. For peptide blocking experiments, 0.2 μg of antigenic blocking peptide (Santa Cruz) was added to the EMSA reaction before the addition of corresponding antibodies. The specificity of antibodies was confirmed by performing EMSA in which we used nuclear extracts of COS-7 cells transfected with cDNA expression vectors encoding various NF-κB/Rel and AP-1 transcription factor family members. In addition, the reactivity of the antibodies was confirmed using nuclear extracts of rat cerebellar granule neurons, rat pheochromocytoma cells (PC12), and human monocytic cells (U937).
Double-stranded oligodeoxynucleotide probes used in EMSA were as follows (only the upper strand is indicated): NF-κB, 5′-CAACGGCAGGGGAATTCCCCTCTCCTT-3′; AP-1, 5′-CGCTTGATGAGTCAGCCGGAA; cAMP response element-binding protein (CREB), 5′-AGAGATTGCCTGACGTCAGAGAGCTAG; OCT-1, 5′-TGTCGAATGCAAATCACTAGAA.
Immunoblotting. Whole-cell extracts were prepared from the neurons (1 × 105) by in situ lysis using ELB buffer (50 mm HEPES, pH 7.0, 250 mm NaCl, 0.1% Nonidet P-40, 5 mm EDTA, 1 mm dithiothreitol, and 1 mmphenylmethanesulfonyl fluoride) supplemented with various protease and phosphatase inhibitors. Cell lysates containing equal amounts of total protein were fractionated by reducing 10% SDS-PAGE and were electrophoretically transferred to Hybond ECL nitrocellulose membrane (Amersham, Arlington Heights, IL). The membranes were analyzed for immunoreactivity with antisera (Santa Cruz) recognizing Rel proteins using the ECL detection system (Amersham).
Pulse–chase studies. Neurons (1 × 105) maintained in NGF-containing media for 5 d were used for these studies. The cells were starved for 1 hr at 37°C by incubating in DMEM medium lacking methionine and cysteine (ICN Biochemicals, Costa Mesa, CA) but containing NGF (100 ng/ml). Subsequently, the cells were metabolically pulse-radiolabeled by addition of 300 μCi/ml [35S]methionine–[35S]cysteine (Dupont NEN, Boston, MA) for 1 hr as described previously (Harhaj et al., 1996). After this, the radiolabeled amino acids were removed, and cells were incubated for 2 hr in normal medium containing NGF (100 ng/ml), in either the presence or absence of the proteasome inhibitor MG132 (50 μm) or the NF-κB-suppressive peptide SN50 (100 μg/ml; Biomol, Plymouth Meeting, PA). Whole-cell extracts were prepared by in situ lysis in ELB buffer. The extracts were precleared with preimmune serum and immobilized Protein A on Trisacryl beads (Pierce, Rockford, IL), and then immunoprecipitated with anti-IκB-α serum (a gift of Dr. Edward M. Schwarz, University of Rochester, Rochester, NY) (Miyamoto et al., 1994) and Protein A–Trisacryl beads. Immunoprecipitates were analyzed by 10% SDS-PAGE and then by autoradiography.
MTT assay. MTT is converted from yellow to a blue formazan crystal by mitochondrial succinate dehydrogenase in viable cells (Mosmann, 1983). After the indicated times and treatments, neurons were washed with PBS and incubated for 20 min at 37°C in 0.5 mg/ml MTT in L-15 culture medium. MTT was removed, and neurons were washed with PBS and lysed in 100 μl of DMSO to extract the formazan crystal. Absorbance was measured at 490 nm on a Victor 1420 microplate reader. At least two wells were treated per experiment, and at least three independent experiments were performed. The data are expressed as a percentage of nontreated neurons that remained in the presence of NGF.
Nissl staining. Equal numbers of neurons plated on collagen-coated two-well chamber slides were treated as indicated and then fixed in 4% paraformaldehyde in PBS. Neurons were rinsed in PBS, stained in 0.1% crystal violet (EM Science, Gibbstown, NJ), dehydrated by immersing in increasing concentrations of ethanol, and mounted using Pro-Texx mounting medium (Baxter, Deerfield, IL). Neurons that were evenly stained and had a discernable nucleus were scored as healthy. Four fields were counted in each well at 20× magnification to obtain an average number of healthy neurons per field. At least two wells were treated and counted for each experiment, and a minimum of three independent experiments were performed. The results are reported as a percentage of nontreated neurons that remained in the presence of NGF.
Terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling assay. Neurons plated on collagen-coated two-well chamber slides were fixed with 4% paraformaldehyde and 0.2% Triton X-100, washed with PBS, and subjected to terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL) assay (Gavrieli et al., 1992). Permeabilized neurons were covered with a reaction mixture containing 1 mmCoCl2, 0.25 U/μl terminal-deoxynucleotidyl–tranferase (Boehringer Mannheim, Indianapolis, IN), and 6 μm digoxigenin-11-dUTP (Boehringer Mannheim) and placed at 37°C for 1 hr. Neurons were then rinsed in PBS, blocked in 5% goat serum and 2% BSA for 2 hr, and labeled with a 1:4 dilution of FITC-conjugated anti-digoxigenin antibody in blocking buffer for 4 hr (Oncor, Gaithersburg, MD). Cells were rinsed in PBS and labeled with 2 μg/ml Hoechst 33,342 (Molecular Probes, Eugene OR) in PBS for 5 min to visualize the nuclei of cells. After two additional rinses in PBS, slides were covered with glass coverslips using mounting solution of 50% glycerol and 0.1% phenylenediamine in PBS and viewed by a Nikon Diaphot 300 inverted microscope equipped for epifluorescence.
Microinjection. Neurons plated on 35 mm glass-bottomed dishes were injected using a Nikon Diaphot 300 inverted microscope equipped with a PLI-100 picoinjector (Medical System, Greensvale, NY) and a Narishige micromanipulator (Nikon-Narishige, Tokyo, Japan). Before injection, cells were placed in serum-free L-15 media (Life Technologies, Gaithersburg, MD) containing penicillin and streptomycin; ∼200–300 neurons were injected for each cDNA. Cells were injected with solutions containing expression vectors at 50 μg/ml in KPi buffer (100 mm KCl and 10 mmpotassium phosphate, pH 7.4) containing 4 mg/ml rhodamine–dextran (10 kDa; Sigma) to permit identification of injected cells. Neurons were returned to NGF-containing media immediately after injections to allow for expression of plasmid cDNA. The number of neurons successfully injected was determined by counting rhodamine-positive cells 18 hr after injection. NGF deprivation was initiated 18 hr after injection and, after an additional 48 hr, neurons were incubated for 5 min with 3 μg/ml Hoechst 33,342 (Molecular Probes) in L-15 to label chromatin; cells were then scored for viability. Injected cells scored as viable had diffuse nuclear Hoechst staining and phase-bright cell bodies with clearly discernable nuclei. Survival was expressed as a ratio of the number of healthy rhodamine-positive cells at 48 hr after NGF withdrawal to the number of rhodamine-positive cells determined at 18 hr. For each cDNA, three independent experiments were performed involving at least 200 injected cells per experiment. In each case, neuronal survival was assessed by an observer who was unaware of the treatment performed.
RESULTS
Effect of NGF on NF-κB, OCT, and AP-1 DNA binding activities in sympathetic neurons
For these studies, neuronal cultures were prepared from SCG of embryonic day 21 rats and were maintained in the presence of NGF and antimitotic agents for 5–7 d. To determine the effects of NGF on the activities of various transcription factors, NGF was withdrawn from the culture for 14 hr, after which NGF was added back to the cultures for various time periods. The 14 hr NGF withdrawal before readdition of NGF permits the downregulation of signaling pathways affected by NGF, including TrkA phosphorylation (Franklin et al., 1995), ERK activation (Creedon et al., 1996), and AKT activation (R. J. Crowder and R. S. Freeman, unpublished observations) but does not result in irreversible consequences of NGF withdrawal (Deckwerth and Johnson, 1993). After NGF withdrawal and readdition, nuclear extracts from these cells were prepared and analyzed by performing EMSA.
To investigate whether NGF induces activation of the NF-κB/Rel family of transcription factors, we used a double-stranded32P-radiolabeled oligonucleotide probe that contained high-affinity palindromic κB binding sequences (Maggirwar et al., 1997). As shown in Figure 1, top panel, EMSA revealed a low level of NF-κB DNA binding activity in cells deprived of NGF for 14 hr. This NF-κB activity was significantly increased when the cells were refed with medium containing NGF for 2 hr. Further incubation of the cells with NGF led to a greater induction of NF-κB, which reached a maximum level at 4 hr and remained elevated after 8 hr of treatment.
Fig. 1.
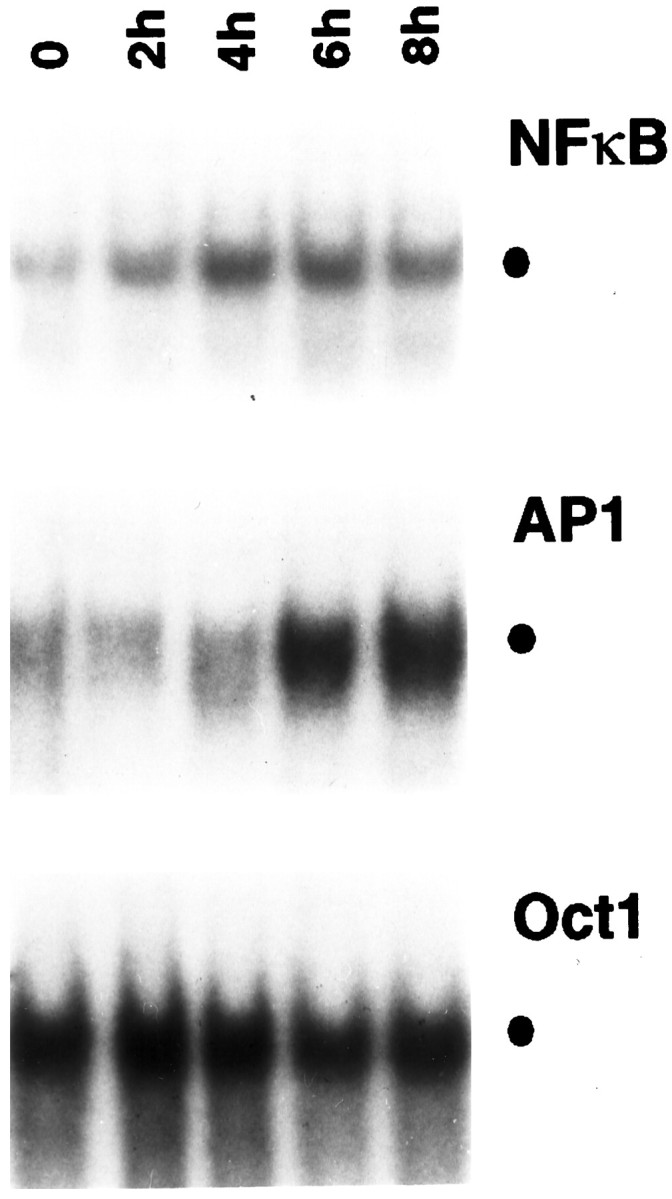
Effect of NGF on various transcription factors in sympathetic neurons. NGF was withdrawn from primary cultures of rat sympathetic neurons for 14 hr, after which NGF (100 ng/ml) was added back to the culture for the indicated time periods. Nuclear extracts were prepared from these cells and were then subjected to EMSA using32P-radiolabeled DNA probes as indicated. The specific binding of protein complexes to the different probes is indicated by the filled circle. The results shown are representative of more than three independent experiments; essentially identical results were also obtained when cells were exposed to 50 ng/ml NGF (data not shown).
To examine the specificity of this effect of NGF, we analyzed the DNA binding activity of the octamer (OCT) family of transcription factors, which are thought to be steady-state regulators of “housekeeping” genes (Ruvkun and Finney, 1991). We detected a major DNA binding complex in nuclear extracts of neurons deprived of NGF for 14 hr (Fig.1, bottom panel) that remained essentially unaffected by prolonged NGF treatment. Thus, NGF appears to selectively target only certain types of transcription factors within sympathetic neurons.
Recently, Eilers et al. (1998) demonstrated that sympathetic neurons chronically exposed to NGF contain relatively high levels of JNK/SAPK activity, which are further enhanced after NGF deprivation. We asked whether regulation of JNK by NGF might lead to the activation of AP-1 transcription factors in these cells. At 14 hr of NGF deprivation (before readdition of NGF), only very low levels of residual AP-1 activity were detected in the neurons (Fig. 1, middle panel). Incubation of these cells with NGF for up to 4 hr had little effect on AP-1, but prolonged (6 hr) exposure to NGF resulted in a substantial increase in AP-1-specific DNA binding activity, which remained elevated even after 8 hr of NGF treatment. Thus, NGF appears to regulate the activation of both NF-κB and AP-1, although with different kinetics.
NGF-induced NF-κB contains heterodimeric complexes of p50 and Rel proteins
To elucidate the nature of the NF-κB complex activated by NGF, we performed antibody supershift analyses using nuclear extracts from neurons maintained in NGF for 5 d. As shown in Figure2, neither preimmune serum nor p52-specific antiserum exhibited immunoreactivity with the NF-κB–DNA complex. In contrast, antibodies recognizing p50, RelA, or c-Rel proteins abrogated the formation of this complex and led to the appearance of higher molecular weight complexes, indicating that these members of the NF-κB family were activated by NGF. Three pieces of evidence confirmed the specificity of our supershift analyses. First, these antibodies had no effect on the formation of a nonspecific protein–DNA complex (Fig. 2, open circle). Second, incubation of these antisera with the NF-κB probe in the absence of a nuclear extract did not generate a “supershift” (data not shown). Third, the effects of these antibodies could be blocked by cognate antigenic peptides (data not shown). Together, these results suggest NGF-treated neurons contain nuclear NF-κB/Rel complexes predominantly comprised of p50/RelA and p50/c-Rel heterodimers.
Fig. 2.
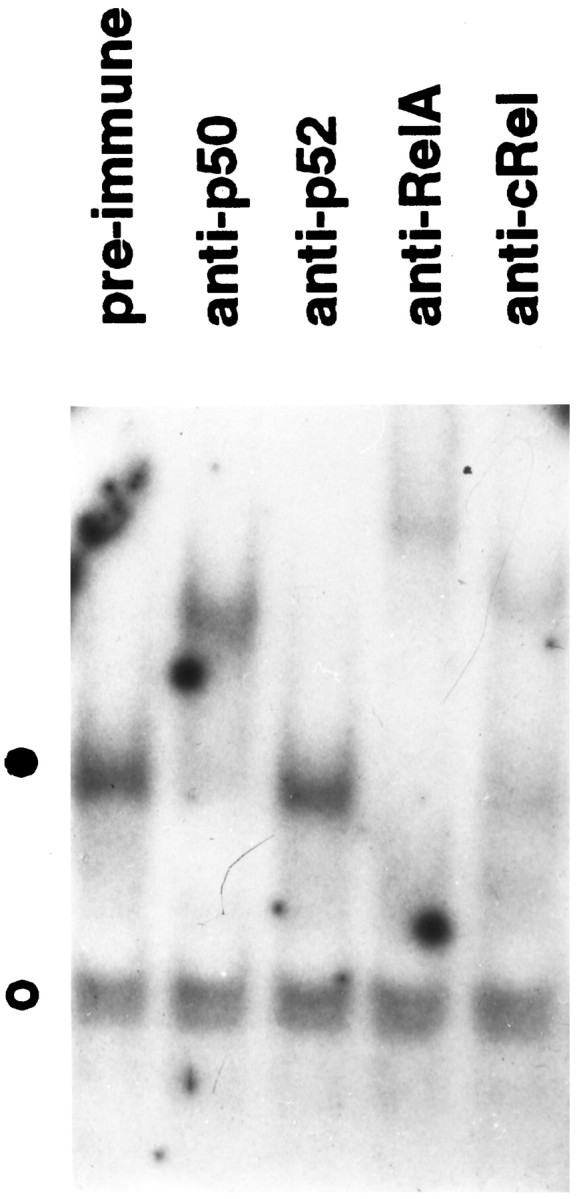
NGF-induced NF-κB contains prototypical complexes. Nuclear extracts from SCG neurons maintained in the presence of NGF were subjected to EMSA in the presence of the indicated antisera. The filled circle indicates the specific NF-κB protein–DNA complex; the open circle shows the formation of a nonspecific protein–DNA complex. The results shown are representative of more than three independent experiments.
Sustained activation of NF-κB by NGF does not require de novo synthesis of RelA
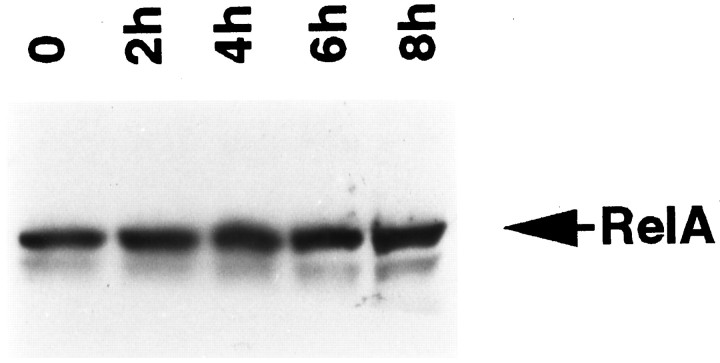
Because total protein synthesis is decreased significantly by 14 hr of NGF withdrawal (Deckwerth and Johnson, 1993), the activation of NF-κB could be the result of increased protein synthesis after the readdition of NGF. To address this possibility, we performed immunoblotting analyses by using RelA-specific antibodies and whole-cell extracts of neurons treated with or without NGF. As shown in Figure 3, we detected a RelA immunoreactive protein in NGF-deprived cells. However, intracellular levels of RelA remained unchanged over an 8 hr period of NGF stimulation. This finding is consistent with the notion that the regulation of NF-κB in sympathetic neurons, as in other cell types, involves cytosolic–nuclear redistribution of protein rather thande novo protein synthesis.
Fig. 3.
NGF-mediated activation of NF-κB is independent of de novo synthesis of RelA. Cultures of SCG neurons were deprived of NGF for 14 hr, followed by incubation with NGF for the indicated periods. Whole-cell extracts were then subjected to immunoblot analysis using antibodies specific for RelA. The results shown are representative of two independent experiments.
Activation of NF-κB by NGF involves proteolysis of IκB-α and is blocked by a proteasome inhibitor
NF-κB is normally sequestered in the cytoplasm by a member of the IκB family of inhibitory proteins (Ghosh et al., 1998). When cells are exposed to inducers of NF-κB, these IκB proteins undergo sequential phosphorylation, ubiquitination, and proteasomal degradation, thereby allowing the nuclear translocation of dimeric complexes of NF-κB (Palombella et al., 1994; Traenckner et al., 1994).
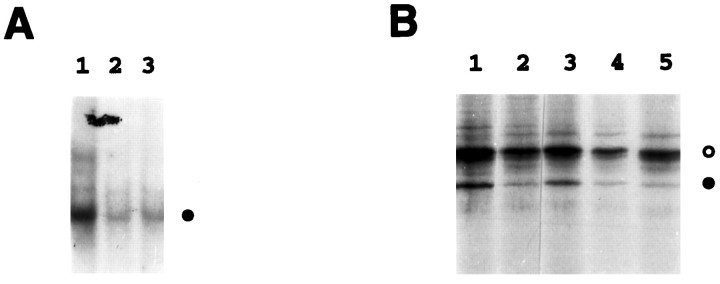
To examine whether activation of NF-κB by NGF also requires the proteasome pathway, we first tested the effect of proteasome inhibitors ALLN and MG132 on NF-κB activities in neurons chronically maintained in NGF. As shown in Figure4A, EMSA revealed a high basal level of activated NF-κB (lane 1), which was significantly inhibited after the incubation of neurons for 2 hr with either ALLN (lane 2) or MG132 (lane 3). No cell death caused by this brief MG132 treatment was observed. These findings suggest that NGF-mediated activation of NF-κB in neurons requires the action of the proteasome complex, as is the case in other cell types.
Fig. 4.
Activation of NF-κB by NGF requires the action of the proteasome complex and involves proteolysis of IκB-α.A, SCG neurons maintained in the continuous presence of NGF were treated with the proteasome inhibitors ALLN (50 μm) (lane 2) or MG132 (50 μm) (lane 3), or with medium alone (lane 1), for 2 hr. Nuclear extracts prepared from these cells were subjected to EMSA analysis to detectκB-binding proteins. The filled circle indicates the specific NF-κB–DNA complex.B, SCG neurons were metabolically labeled for 1 hr with 300 μCi/ml [35S]methionine–[35S]cysteine in the presence of NGF (100 ng/ml). The pulse-labeled cells were then lysed and extracted immediately (lane 1) or cold-chased for 2 hr in either the absence (lane 2) or presence of MG132 (50 μm) (lane 3), SN50M (100 μg/ml) (lane 4), or SN50 (100 μg/ml) (lane 5). Whole-cell extracts were isolated and then subjected to immunoprecipitation using IκB-α-specific antiserum as described in Materials and Methods. The filled circleindicates IκB-α, whereas the open circle denotes a nonspecific protein. IκB-α levels were quantitated by densitometric analysis of this autoradiogram (NIH Image software) and normalized to the level of the nonspecific protein (open circle). Based on this analysis, IκB-α levels had declined to 18% of starting levels within 2 hr of NGF withdrawal (compare lanes 1, 2). IκB-α degradation was unaffected by addition of the SN50M or SN50 peptides (IκB-α declined to 19 and 15% of its starting level, respectively) (lanes 4,5), but the protein was protected by addition of the proteasome inhibitor MG132 (IκB-α remained at 62% of its initial level) (lane 3). The results shown are representative of two (B) or three (A) independent experiments.
In view of the effect of the proteasome inhibitors on NGF activation of NF-κB, we next examined whether NGF induces the proteolytic degradation of IκB-α in SCG neurons. For these studies, we initially attempted to perform NGF starvation and stimulation experiments similar to those shown in Figure 1, with detection of IκB-α levels by immunoblot analysis. However, these experiments were inconclusive, in part because of the inherent instability of IκB-α and also because of the overall decline in protein synthesis that was observed in NGF-deprived cells. To circumvent these problems, we used pulse–chase experiments to analyze IκB-α. NGF-maintained neurons were metabolically labeled for 1 hr with [35S]methionine–[35S]cysteine, after which a chase with nonradioactive amino acids was performed for 2 hr, in either the absence or presence of the proteasome inhibitor MG132 (Fig. 4B). Whole-cell extracts were then prepared and subjected to immunoprecipitation using antibodies specific to IκB-α. We detected a radiolabeled band corresponding to IκB-α in neurons before the chase (Fig. 4B, lane 1). After 2 hr of chase in the absence of MG132, the steady-state level of IκB-α dramatically decreased (Fig. 4B,lane 2). These results show that NGF stimulates the signals for proteolytic degradation of IκB-α in sympathetic neurons. When the chase was performed for 2 hr in the presence of MG132, this inhibitor efficiently protected IκB-α from degradation (Fig.4B, lane 3). It is noteworthy that the intensities of radiolabeled bands of nonspecific proteins (for example, Fig. 4B, the band shown asopen circle) were relatively unaltered over the entire period of this experiment, suggesting that the decreased intensity of the IκB-α-specific band is attributable to its selective proteolytic degradation.
SN50 selectively inhibits NGF-dependent NF-κB activity and induces neuronal death
We next sought to examine the relationship between NF-κB activation and survival in NGF-dependent neurons using the NF-κB inhibitory peptide SN50. SN50 is a synthetic oligopeptide that contains a hydrophobic cell-permeable motif, together with nuclear localization sequences (NLS) from the p50 subunit of NF-κB (Lin et al., 1995).
To validate the use of SN50 in this system, we first examined the effect of SN50 on the fate of IκB-α. As expected for an inhibitor of NF-κB translocation, incubation of sympathetic neurons in the presence of either SN50 or an inactive control peptide that is mutated within the NLS motif (SN50M) did not protect IκB-α from degradation (Fig. 4B, lanes 4, 5). This is consistent with previous studies in which the peptide SN50 inhibited nuclear translocation of NF-κB without altering IκB-α degradation (Lin et al., 1995; Kilgore et al., 1997).
We then tested the effects of SN50 on NF-κB activity in neurons maintained in the presence of NGF. After incubating these cells for 4 hr with either SN50 or SN50M, nuclear extracts were prepared and analyzed by performing EMSA. Incubation of cells with the SN50 peptide, but not SN50M, abolished the formation of the specific NF-κB–DNA complex (Fig. 5A, closed circle) but had no effect on a nonspecific protein–DNA complex (Fig. 5A, open circle). Furthermore, the addition of SN50 to in vitro EMSA reactions had no effect on the ability of NF-κB proteins to bind to DNA (data not shown). These results demonstrate that NGF-mediated activation of NF-κB can be blocked by the treatment of neurons with SN50.
Fig. 5.
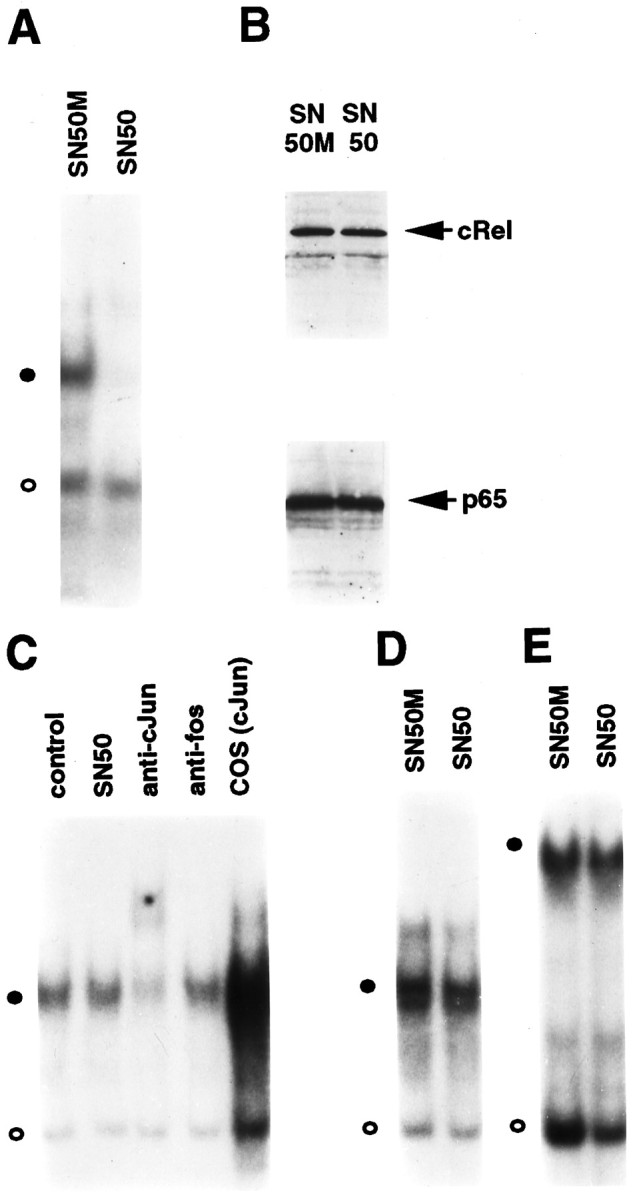
Selective inhibition of NF-κB in sympathetic neurons by synthetic peptide SN50. A, Cultures of SCG neurons maintained in the continuous presence of NGF for 5 d were treated with either SN50M or SN50 at a concentration of 75 μg/ml for 4 hr. Nuclear extracts isolated from these cells were used to perform EMSA for analysis of κB-binding proteins. Thefilled circle indicates the specific NF-κB–DNA complex; the open circle denotes a nonspecific protein–DNA binding complex. B, Whole-cell extracts of SN50M- and SN50-treated neurons maintained in the presence of NGF were subjected to immunoblotting analyses by using anti-RelA and anti-c-RelA antibodies. C, Nuclear extracts of SN50- or SN50M-treated (control) neurons were subjected to EMSA using a 32P-radiolabeled DNA probe corresponding to a consensus AP-1 binding site. The filled circle denotes the AP-1–DNA complex. EMSA was also performed in the presence of anti-c-Jun and anti-c-Fos antibodies (as indicated), and nuclear extracts from c-Jun transfected COS-1 cells were included as a control (COScJun). D, E, EMSA was also performed using nuclear extracts of SN50- and SN50M-treated neurons (as indicated) with radiolabeled oligonucleotide probes specific for CREB (D) and OCT (E) transcription factors. The results shown are representative of a minimum of either two (B) or three (all other panels) experiments.
To examine whether the inhibition of NF-κB DNA binding activity by SN50 was caused by the depletion of Rel proteins in SCG neurons, we performed immunoblotting analyses. As shown in Figure 5B, intracellular levels of RelA or c-Rel proteins remained unchanged in SN50-treated neurons compared with those treated with SN50M. Together with the findings presented in Figure 4, these results suggest that the observed inhibitory effect of SN50 is attributable to its ability to compete with NGF-induced NF-κB complexes for access to the cellular machinery responsible for nuclear translocation of NF-κB.
To test whether the inhibitory effects of SN50 were specific to NF-κB, EMSA was performed using 32P-radiolabeled probes containing consensus sites for various transcription factors, including AP-1, CREB, and OCT. A high basal level of AP-1 DNA binding activity was detected in NGF-treated neurons (Fig. 5C). This AP-1–DNA complex was supershifted by c-Jun-specific antisera but not by anti-c-Fos antibodies, and it comigrated with the c-Jun homodimer generated with a c-Jun transfected COS-1 cell extract. Treatment with SN50 had no effect on this AP-1–DNA complex (Fig. 5C). Likewise, neither CREB nor OCT DNA binding activities were affected by incubation of neurons with the SN50 peptide (Fig.5D,E). Therefore, the inhibitory effect of SN50 appears to be specific to NF-κB transcription factors under these conditions.
Having determined that SN50 is an inhibitor of NF-κB, but not of other transcription factors, in sympathetic neurons, we proceeded with experiments aimed at examining the role of NF-κB in neuronal survival.
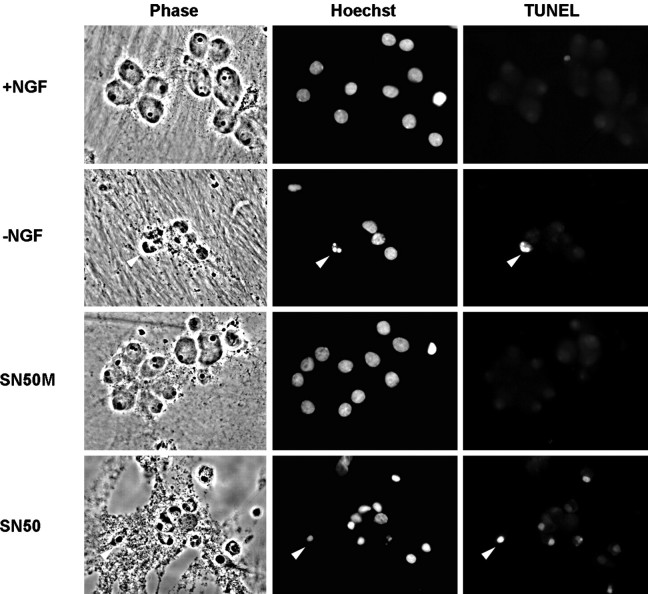
Sympathetic neurons that are deprived of NGF in vitroundergo apoptosis, which first becomes apparent ∼18 hr after NGF withdrawal and is virtually complete by 72 hr (Deckwerth and Johnson, 1993). This process involves atrophy of the cell body, condensation of the nuclear chromatin, DNA fragmentation, and neurite degeneration. In our initial experiments, we noticed that prolonged treatment (>4 hr) with SN50 resulted in significant neuronal death that morphologically resembled the death caused by NGF withdrawal. Like NGF-deprived neurons, neurons treated with 100 μg/ml SN50 in the presence of NGF had severely degenerated neurites and condensed cell bodies (Fig.6). The SN50-treated neurons also exhibited marked chromatin condensation, detected by staining nuclei with Hoechst dye, and DNA fragmentation, as detected by the TUNEL assay. Control NGF-maintained cells and cells treated with SN50M showed no signs of apoptotic morphology or chromatin condensation and were devoid of TUNEL labeling, suggesting that the toxicity of SN50 was dependent on an intact NF-κB NLS.
Fig. 6.
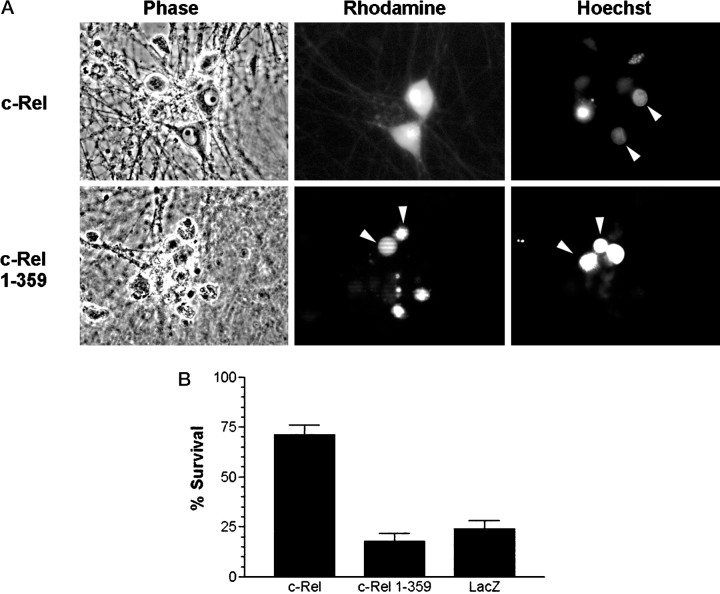
SN50 induces cell death with characteristic features of apoptosis. Cultured neurons were either maintained in the continuous presence of NGF, deprived of NGF, or treated with NGF with SN50 or SN50M peptides (100 μg/ml each). After 24 hr, cells were fixed, analyzed for TUNEL reactivity, and stained with Hoechst dye to label nuclei. Phase-contrast, TUNEL, and Hoechst images of the same field of view are shown for each treatment. Arrowheadsindicate examples of cells deprived of NGF or treated with SN50 that have condensed chromatin and TUNEL reactivity.
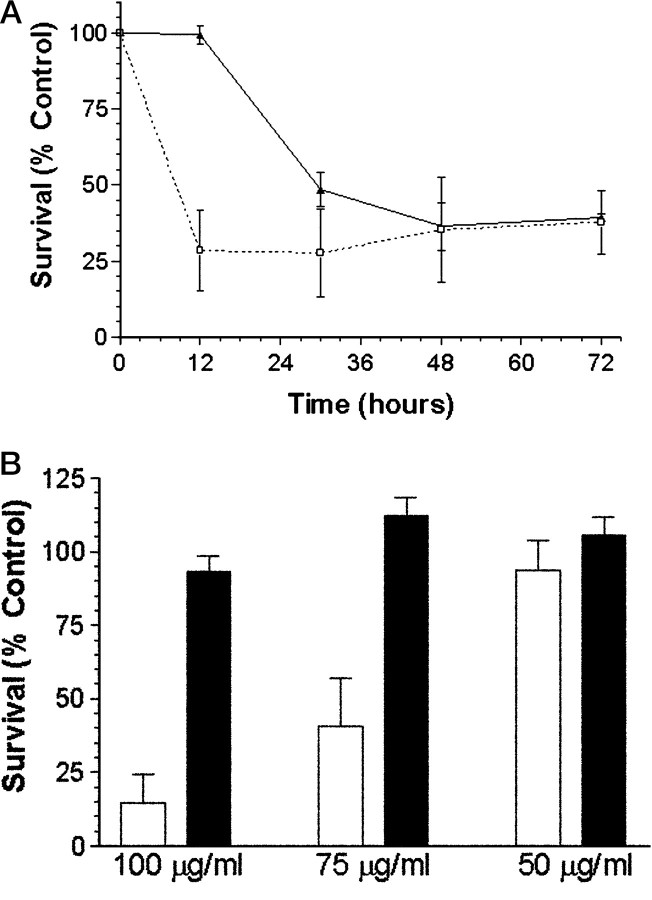
Although SN50-treated neurons exhibited an apoptotic morphology similar to neurons deprived of NGF, the rate at which these cells underwent cell death was accelerated. Using MTT reduction as an indication of mitochondrial function and cell viability, we found that 12 hr of exposure to SN50 resulted in a 70% decrease in MTT reduction, whereas neurons deprived of NGF for 12 hr showed no such decrease (Fig.7A). Only after 30 hr of NGF withdrawal did the decrease in MTT reducing capacity approach the level seen in SN50-treated cells. The reason for the more rapid action of SN50 compared with NGF withdrawal is not known, but it could reflect our observations that SN50 causes more rapid and complete inhibition of NF-κB activation compared with NGF withdrawal (compare Figs. 1 and5A; these figures show NF-κB activity after 14 hr of NGF-deprivation or 4 hr of SN50 treatment, respectively)
Fig. 7.
Death of sympathetic neurons treated with SN50 is time- and dose-dependent. A, Neuronal cultures were either deprived of NGF (black line, filled triangles) or treated with 100 μg/ml SN50 in the presence of NGF (dashed line, open squares). Survival was assayed by MTT reduction at 12, 30, 48, and 72 hr after a single application of SN50. Data represents the mean ± SEM from three independent experiments. B, Neuronal cultures were treated with a single application of 50, 75, or 100 μg/ml SN50 (open bars) or SN50M (filled bars), both in the presence of NGF. Viability was assayed after 48 hr of treatment by Nissl staining as described in Materials and Methods. The results represent the mean ± SEM of at least three independent experiments. For both A andB, results are expressed as a percentage of the survival of control neurons maintained in the continuous presence of NGF.
In non-neuronal cell lines, SN50 has been shown to inhibit NF-κB activity by ∼85% at a concentration of 100 μg/ml, whereas SN50M has no effect at the same concentration (Lin et al., 1995). Consistent with these observations, SN50 at a comparable concentration completely inhibited NGF-induced NF-κB activity (Fig. 5A). SN50 also elicited a dose-dependent inhibition of NGF-mediated survival, which was maximal at 100 μg/ml and half-maximal (IC50) between 50 and 75 μg/ml (Fig. 7B). Therefore, SN50, but not SN50M, inhibits cell survival over the same concentration range at which it inhibits NF-κB activation. These results suggest that NF-κB may have a role in NGF-mediated neuronal survival.
Overexpression of c-Rel is sufficient to prevent the death of NGF-deprived neurons
To test directly whether NF-κB might inhibit neuronal death after NGF withdrawal, we microinjected sympathetic neurons with plasmid DNAs encoding either full-length c-Rel (pCMV4 c-Rel) or a transactivation-deficient form of c-Rel [pCMV4 c-Rel(1–359)] (Doerre et al., 1993). As a control, cells were injected with an expression vector encoding β-galactosidase (Crowder and Freeman, 1998). NGF deprivation was initiated 18 hr after microinjection, a time when >95% of successfully microinjected cells express exogenous c-Rel (as determined by immunofluorescence staining using an anti-c-Rel antibody; data not shown). After 48 hr of NGF deprivation, neurons injected with the c-Rel expression vector remained phase bright, had a clearly definable nucleus, and had uniformly dispersed chromatin (Fig. 8A). In contrast, uninjected neurons or neurons injected with β-galactosidase or c-Rel(1–359) vectors showed signs of apoptotic cell death, including chromatin condensation and neurite degeneration. When these results were quantitated, >75% of neurons injected with control vectors underwent cell death after NGF withdrawal (Fig.8B). In contrast, ∼70% of neurons injected with the c-Rel expression vector were protected from cell death. These findings demonstrate that the overexpression of c-Rel is sufficient to block apoptosis caused by NGF withdrawal. In addition, our data indicate that the transactivation domain of c-Rel is necessary for the neuroprotective effect of c-Rel.
Fig. 8.
A, Overexpression of c-Rel promotes survival in NGF-deprived neurons. Shown are images of NGF-deprived neurons after injection with either c-Rel or c-Rel(1–359) expression vectors. Neurons were microinjected and then deprived of NGF for 48 hr. Cells were then stained with Hoechst dye and visualized by phase-contrast and fluorescence microscopy. Shown are phase-contrast views, rhodamine-positive cells (injected cells), and Hoechst-stained nuclei in one field of view for each injected DNA.Arrowheads for c-Rel injections indicate healthy looking injected cells with normal nuclear chromatin. Arrowheadsfor c-Rel(1–359) indicate injected cells containing condensed nuclear chromatin. B, Overexpression of c-Rel is sufficient to prevent neuronal cell death caused by NGF deprivation. Cultured neurons were microinjected with plasmids encoding either c-Rel, c-Rel(1–359), or LacZ, and then deprived of NGF for 48 hr as described in Materials and Methods. Survival was assayed on the basis of cellular morphology (as viewed by phase-contrast microscopy) and by the presence or absence of condensed chromatin (detected with Hoechst dye). The results (mean ± SEM) from at least three independent experiments are shown. Survival for c-Rel-injected cells was significantly different from LacZ- or c-Rel(1–359)-injected neurons (two-tailedt test; p < 0.0001 in both cases).
DISCUSSION
We used EMSA to detect the DNA binding activity of various transcription factors in nuclear extracts from primary cultures of rat sympathetic neurons and found that NGF activates NF-κB in these cells. This NF-κB activation pathway appears to be prototypical in that it involves the proteolytic degradation of the inhibitory protein IκB-α via the action of the proteasome complex. Inhibiting NF-κB activity resulted in the death of sympathetic neurons in the presence of NGF, whereas overexpression of c-Rel in the absence of NGF was sufficient to promote survival. These data suggest that activation of NF-κB contributes to the NGF-dependent survival of sympathetic neurons.
NF-κB was found to be constitutively activated in sympathetic neurons maintained in the presence of NGF. Withdrawal of NGF from these cultures resulted in the rapid downregulation of NF-κB activity (data not shown), and readdition of NGF, at a time when the cells could be fully rescued by NGF, resulted in reconstitution of steady-state levels of NF-κB in these cells. NF-κB activity increased gradually over 2–4 hr after NGF addition. These kinetics contrast with the rapid increase of NF-κB activity in Schwann cells detected within 30 min of NGF treatment (Carter et al., 1996) and the relatively delayed activation of NF-κB (>24 hr) observed in NGF-treated PC12 cells (Wood, 1995). In Schwann cells, NGF-induced NF-κB activation is dependent on p75NTR and not TrkA, possibly accounting for the more rapid rate of activation in these cells. Simultaneous expression of p75NTR and TrkA has been shown to repress signal transduction events mediated by either receptor expressed alone (Kaplan and Miller, 1997). For example, the presence of TrkA was shown to suppress p75NTR-mediated ceramide production in PC12 cells (Dobrowsky et al., 1995), and introduction of TrkA into oligodendrocytes caused a reduction in p75NTR-dependent JNK activation (Yoon et al., 1998). On the other hand, activation of NF-κB by NGF in p75NTR-expressing oligodendrocytes was not adversely affected by introducing TrkA into these cells (Yoon et al., 1998). Thus, although p75NTR and TrkA can independently activate downstream pathways, coexpression of both receptors appears to modify certain pathways (ceramide and JNK) but not others (NF-κB). Whether NF-κB activation in sympathetic neurons is mediated by p75NTR, TrkA, or both remains to be determined.
The NGF inducible NF-κB complexes in sympathetic neurons contain p50, RelA, and c-Rel subunits. In its inactive state, NF-κB is sequestered in the cytosol by binding to IκB-α proteins. The phosphorylation and subsequent proteasomal degradation of IκB-α releases NF-κB and permits its translocation into the nucleus in which it can bind DNA and regulate transcription. Translocation of p50-containing NF-κB complexes to the nucleus can be blocked by SN50, a peptide that contains the nuclear localization sequences from p50 and presumably competes with p50 for binding to unknown components of the nuclear transport machinery (Lin et al., 1995). In our experiments, NGF-dependent NF-κB activity was potently inhibited by treatment with SN50. Importantly, SN50 did not inhibit the DNA binding activity of a variety of other transcription factors, including AP-1, CREB, and OCT. Although SN50 was selective for NF-κB, it remains possible that SN50 could inhibit the translocation of other transcription factors or nuclear survival signals emanating from the cytoplasm.
SN50-treated neurons resembled neurons deprived of NGF, suggesting that NF-κB activity may be an important component of a survival pathway mediated by NGF. To test whether ectopic activation of NF-κB could lead to survival in the absence of NGF, we analyzed the effects of overexpressing c-Rel in NGF-deprived neurons. Overexpression of c-Rel was found to be sufficient to protect neurons from apoptosis caused by NGF withdrawal. These results are consistent with previous observations in non-neuronal cells in which ectopic expression of c-Rel provided protection from serum withdrawal-induced apoptosis (Bertrand et al., 1998) and anti-IgM- or TGFβ-1-mediated apoptosis (Arsura et al., 1996; Wu et al., 1996). In contrast to full-length c-Rel, microinjection of c-Rel(1–359), which lacks the C-terminal transactivation domain of the wild-type protein but retains the DNA binding Rel homology domain (Doerre et al., 1993), failed to protect neurons from apoptosis after NGF withdrawal. Therefore, it appears likely that the transcriptional activity of c-Rel is required for its ability to promote survival in the absence of NGF.
Our hypothesis that NF-κB has a role in the survival of NGF-dependent neurons is consistent with several recent studies suggesting a role for NF-κB in protection from apoptosis. Abrogation of NF-κB, either by deletion of RelA or by overexpression of IκB-α, sensitizes immune cells to apoptosis in response to TNFα and DNA-damaging agents (Beg and Baltimore, 1996; Liu et al., 1996; Van Antwerp et al., 1996; Wang et al., 1996). In addition, inhibition of NF-κB in H-Ras-transformed fibroblasts induces apoptosis (Mayo et al., 1997), and in PC12 cells, inhibition of NF-κB blocks the neuroprotection afforded by soluble β-amyloid precursor protein (Guo et al., 1998). NF-κB activity also appears to be required for the ability of NGF to protect undifferentiated PC12 cells from apoptosis after serum withdrawal (Taglialatela et al., 1997). In contrast to these results, NF-κB has been implicated in the death of neurons induced by glutamate and β-amyloid protein (Behl et al., 1994; Grilli et al., 1996), although a causal relationship for NF-κB in these neuronal deaths has not been demonstrated.
Besides NF-κB, the PI3K pathway has recently been implicated in the regulation of NGF-dependent survival. Inhibition of this pathway in PC12 cells blocks the ability of NGF to rescue cells from apoptosis (Yao and Cooper, 1995). A role for PI3K in survival has also been found in primary sympathetic and cerebellar granule neurons (D’Mello et al., 1997; Dudek et al., 1997; Miller et al., 1997; Philpott et al., 1997;Crowder and Freeman, 1998). PI3K and NF-κB may exist in the same or independent survival pathways. Evidence in support of independent pathways comes from experiments in Chinese hamster ovary cells on the antiapoptotic action of insulin (Bertrand et al., 1998). Insulin-dependent survival could be blocked partially by inhibiting either PI3K or NF-κB; inhibiting both proteins was additive and led to complete reversal of the survival promoted by insulin. However, PI3K inhibitors did not impede the ability of insulin to induce NF-κB transcription factor activity. Therefore, at least in these cells, PI3K and NF-κB appear to exist in separate survival pathways.
NGF-treated neurons possess constitutively activated AP-1 transcription factor complexes that contain c-Jun, but not c-Fos, proteins. This observation is consistent with previous immunoblotting and immunofluorescence experiments showing that c-Jun, but not c-Fos, is expressed in NGF-maintained neurons and, in the presence of NGF, these neurons contain high levels of JNK activity (Ham et al., 1995; Eilers et al., 1998). Thus, in the presence of NGF, SCG neurons possess activated NF-κB and AP-1 transcription factors.
c-Jun has recently been implicated in the death of sympathetic neurons after NGF withdrawal. c-Jun mRNA and protein increase after the removal of NGF, as do JNK activity and c-Jun phosphorylation. Inhibition of c-Jun with a dominant negative c-Jun mutant or neutralizing c-Jun antibodies blocks apoptosis caused by NGF deprivation (Estus et al., 1994; Ham et al., 1995). Given the ability of c-Jun to promote neuronal death, our evidence showing that c-Rel can promote survival suggests that NGF withdrawal-induced apoptosis may require both inactivation of NF-κB and activation of AP-1 transcription factors. The observation that NGF-maintained neurons contain both of these activities suggests that the mechanisms that regulate survival and cell death may be coordinately regulated. Along these lines, several recent studies have raised the possibility that both NF-κB and AP-1 may be regulated by a common upstream protein kinase, MEK kinase 1 (Karin and Delhase, 1998).
In summary, the results of our study demonstrate that NGF-dependent sympathetic neurons have constitutive NF-κB activity that can be further induced by treatment with NGF. Because expression of c-Rel can block apoptosis after NGF withdrawal, the activation of this pathway may contribute to the survival mechanisms mediated by NGF. Future studies will be needed to (1) address the relationship of NF-κB to other NGF-dependent survival pathways, such as those involving PI3K and AKT (Yao and Cooper, 1995; Philpott et al., 1997; Crowder and Freeman, 1998); (2) investigate possible interactions between NF-κB and AP-1; and (3) identify the transcriptional targets of NF-κB that are relevant for neuronal survival.
Footnotes
This work was supported by National Institutes of Health Grants PO1 MH57556 (to S.D.) and RO1 NS34400 (to R.S.F.), and Predoctoral Training Grant ES07026 (to P.D.S.). R.S.F. also acknowledges the generous support from the Paul Stark Endowment at the University of Rochester. We thank Drs. W. Greene, E. M. Schwarz, and S. C. Sun for providing research reagents and materials.
Correspondence should be addressed to Dr. Sanjay Maggirwar, Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, NY 14642.
REFERENCES
- 1.Angel P, Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 2.Arsura M, Wu M, Sonenshein GE. TGF β1 inhibits NF-κB/Rel activity inducing apoptosis of B cells: transcriptional activation of IκBα. Immunity. 1996;5:31–40. doi: 10.1016/s1074-7613(00)80307-6. [DOI] [PubMed] [Google Scholar]
- 3.Beg AA, Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 4.Behl C, Davis JB, Lesley R, Schubert D. Hydrogen peroxide mediates amyloid β protein toxicity. Cell. 1994;77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 5.Bertrand F, Atfi A, Cadoret A, L’Allemain G, Robin H, Lascols O, Capeau J, Cherqui G. A role for nuclear factor κB in the antiapoptotic function of insulin. J Biol Chem. 1998;273:2931–2938. doi: 10.1074/jbc.273.5.2931. [DOI] [PubMed] [Google Scholar]
- 6.Bonni A, Ginty DD, Dudek H, Greenberg ME. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol Cell Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- 7.Boothby MR, Mora AL, Scherer DC, Brockman JA, Ballard DW. Perturbation of the T lymphocyte lineage in transgenic mice expressing a constitutive repressor of nuclear factor (NF)-κB. J Exp Med. 1997;185:1897–1907. doi: 10.1084/jem.185.11.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bredesen DE, Rabizadeh S. p75NTR and apoptosis: Trk-dependent and Trk-independent effects. Trends Neurosci. 1997;20:287–290. doi: 10.1016/s0166-2236(96)01049-1. [DOI] [PubMed] [Google Scholar]
- 9.Carter BD, Kaltschmidt C, Kaltschmidt B, Offenhauser N, Bohm-Matthaei R, Baeuerle PA, Barde YA. Selective activation of NF-κB by nerve growth factor through the neurotrophin receptor p75. Science. 1996;272:542–545. doi: 10.1126/science.272.5261.542. [DOI] [PubMed] [Google Scholar]
- 10.Creedon DJ, Johnson EM, Lawrence JC. Mitogen-activated protein kinase-independent pathways mediate the effects of nerve growth factor and cAMP on neuronal survival. J Biol Chem. 1996;271:20713–20718. doi: 10.1074/jbc.271.34.20713. [DOI] [PubMed] [Google Scholar]
- 11.Crowder RJ, Freeman RS. Phosphatidylinositol 3-kinase and Akt protein kinase are necessary and sufficient for the survival of nerve growth factor-dependent sympathetic neurons. J Neurosci. 1998;18:2933–2943. doi: 10.1523/JNEUROSCI.18-08-02933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowley C, Spencer SD, Nishimura MC, Chen KS, Pitts-Meek S, Armanini MP, Ling LH, MacMahon SB, Shelton DL, Levinson AD, Phillips HS. Mice lacking nerve growth factor display perinatal loss of sensory and sympathetic neurons yet develop basal forebrain cholinergic neurons. Cell. 1994;76:1001–1011. doi: 10.1016/0092-8674(94)90378-6. [DOI] [PubMed] [Google Scholar]
- 13.Deckwerth TL, Johnson EM., Jr Temporal analysis of events associated with programmed cell death (apoptosis) of sympathetic neurons deprived of nerve growth factor. J Cell Biol. 1993;123:1207–1222. doi: 10.1083/jcb.123.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshmukh M, Johnson EM., Jr Programmed cell death in neurons: focus on the pathway of nerve growth factor deprivation-induced death of sympathetic neurons. Mol Pharmacol. 1997;51:897–906. doi: 10.1124/mol.51.6.897. [DOI] [PubMed] [Google Scholar]
- 15.D’Mello SR, Borodezt K, Soltoff SP. Insulin-like growth factor and potassium depolarization maintain neuronal survival by distinct pathways: possible involvement of PI 3-kinase in IGF-1 signaling. J Neurosci. 1997;17:1548–1560. doi: 10.1523/JNEUROSCI.17-05-01548.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobrowsky RT, Jenkins GM, Hannun YA. Neurotrophins induce sphingomyelin hydrolysis. Modulation by co-expression of p75NTR with Trk receptors. J Biol Chem. 1995;270:22135–22142. doi: 10.1074/jbc.270.38.22135. [DOI] [PubMed] [Google Scholar]
- 17.Doerre S, Sista P, Sun SC, Ballard DW, Greene WC. The c-rel protooncogene product represses NF-κB p65-mediated transcriptional activation of the long terminal repeat of type 1 human immunodeficiency virus. Proc Natl Acad Sci USA. 1993;90:1023–1027. doi: 10.1073/pnas.90.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. Regulation of neuronal survival by the serine–threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 19.Eilers A, Whitfield J, Babij C, Rubin LL, Ham J. Role of the Jun kinase pathway in the regulation of c-Jun expression and apoptosis in sympathetic neurons. J Neurosci. 1998;18:1713–1724. doi: 10.1523/JNEUROSCI.18-05-01713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estus S, Zaks WJ, Freeman RS, Gruda M, Bravo R, Johnson EM., Jr Altered gene expression in neurons during programmed cell death: identification of c-jun as necessary for neuronal apoptosis. J Cell Biol. 1994;127:1717–1727. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franklin JL, Sanz-Rodriguez C, Juhasz A, Deckwerth TL, Johnson EM., Jr Chronic depolarization prevents programmed death of sympathetic neurons in vitro but does not support growth: requirement for Ca2+ influx but not Trk activation. J Neurosci. 1995;15:643–664. doi: 10.1523/JNEUROSCI.15-01-00643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh S, May MJ, Kopp EB. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 24.Ginty DD, Bonni A, Greenberg ME. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell. 1994;77:713–725. doi: 10.1016/0092-8674(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 25.Gorin PD, Johnson EM. Experimental autoimmune model of nerve growth factor deprivation: effects on developing peripheral sympathetic and sensory neurons. Proc Natl Acad Sci USA. 1979;76:5382–5386. doi: 10.1073/pnas.76.10.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grilli M, Pizzi M, Memo M, Spano P. Neuroprotection by aspirin and sodium salicylate through blockade of NF-κB activation. Science. 1996;274:1383–1385. doi: 10.1126/science.274.5291.1383. [DOI] [PubMed] [Google Scholar]
- 27.Guo Q, Robinson N, Mattson MP. Secreted β-amyloid precursor protein counteracts the proapoptotic action of mutant presenilin-1 by activation of NF-κB and stabilization of calcium homeostasis. J Biol Chem. 1998;273:12341–12351. doi: 10.1074/jbc.273.20.12341. [DOI] [PubMed] [Google Scholar]
- 28.Ham J, Babij C, Whitfield J, Pfarr CM, Lallemand D, Yaniv M, Rubin LL. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 29.Harhaj EW, Maggirwar SB, Sun SC. Inhibition of p105 processing by NF-κB proteins in transiently transfected cells. Oncogene. 1996;12:2385–2392. [PubMed] [Google Scholar]
- 30.Hendry IA, Campbell J. Morphometric analysis of rat superior cervical ganglion after axotomy and nerve growth factor treatment. J Neurocytol. 1976;5:351–360. doi: 10.1007/BF01175120. [DOI] [PubMed] [Google Scholar]
- 31.Herdegen T, Skene P, Bahr M. The c-Jun transcription factor–bipotential mediator of neuronal death, survival and regeneration. Trends Neurosci. 1997;20:227–231. doi: 10.1016/s0166-2236(96)01000-4. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan DR, Miller FD. Signal transduction by the neurotrophin receptors. Curr Opin Cell Biol. 1997;9:213–221. doi: 10.1016/s0955-0674(97)80065-8. [DOI] [PubMed] [Google Scholar]
- 33.Karin M, Delhase M. JNK or IKK, AP-1 or NF-κB, which are the targets for MEK kinase 1 action? Proc Natl Acad Sci USA. 1998;95:9067–9069. doi: 10.1073/pnas.95.16.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilgore KS, Schmid E, Shanley TP, Flory CM, Maheswari V, Tramontini NL, Cohen H, Ward PA, Friedl HP, Warren JS. Sublytic concentrations of the membrane attack complex of complement induce endothelial interleukin-8 and monocyte chemoattractant protein-1 through nuclear factor-κB activation. Am J Pathol. 1997;150:2019–2031. [PMC free article] [PubMed] [Google Scholar]
- 35.Levi-Montalcini R, Booker B. Destruction of the sympathetic ganglia in mammals by an anti-serum to the nerve-growth protein. Proc Natl Acad Sci USA. 1960;46:384–391. doi: 10.1073/pnas.46.3.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin YZ, Yao SY, Veach RA, Torgerson TR, Hawiger J. Inhibition of nuclear translocation of transcription factor NF-κB by a synthetic peptide containing a cell membrane-permeable motif and nuclear localization sequence. J Biol Chem. 1995;270:14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- 37.Liu ZG, Hsu H, Goeddel DV, Karin M. Dissection of TNF receptor 1 effector functions: JNK activation is not linked to apoptosis while NF-κB activation prevents cell death. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 38.Maggirwar SB, Harhaj EW, Sun S-C. Regulation of the IL-2 CD28RE by NF-ATp and various NF-κB/Rel transcription factors. Mol Cell Biol. 1997;17:2605–2614. doi: 10.1128/mcb.17.5.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayo MW, Wang CY, Cogswell PC, Rogers-Graham KS, Lowe SW, Der CJ, Baldwin AS., Jr Requirement of NF-κB activation to suppress p53-independent apoptosis induced by oncogenic Ras. Science. 1997;278:1812–1815. doi: 10.1126/science.278.5344.1812. [DOI] [PubMed] [Google Scholar]
- 40.Miller TM, Tansey MG, Johnson EM, Jr, Creedon DJ. Inhibition of phosphatidylinositol 3-kinase activity blocks depolarization- and insulin-like growth factor I-mediated survival of cerebellar granule cells. J Biol Chem. 1997;272:9847–9853. doi: 10.1074/jbc.272.15.9847. [DOI] [PubMed] [Google Scholar]
- 41.Miyamoto S, Chiao PJ, Verma IM. Enhanced IκBα degradation is responsible for constitutive NF-κB activity in mature murine B-cell lines. Mol Cell Biol. 1994;14:3276–3282. doi: 10.1128/mcb.14.5.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 43.O’Neill LA, Kaltschmidt C. NF-κB: a crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997;20:252–258. doi: 10.1016/s0166-2236(96)01035-1. [DOI] [PubMed] [Google Scholar]
- 44.Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 45.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin–proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 46.Pettmann B, Henderson CE. Neuronal cell death. Neuron. 1998;20:663–674. doi: 10.1016/s0896-6273(00)81004-1. [DOI] [PubMed] [Google Scholar]
- 47.Philpott KL, McCarthy MJ, Klippel A, Rubin LL. Activated phosphatidylinositol 3-kinase and Akt kinase promote survival of superior cervical neurons. J Cell Biol. 1997;139:809–815. doi: 10.1083/jcb.139.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riccio A, Pierchala BA, Ciarallo CL, Ginty DD. An NGF–TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science. 1997;277:1097–1100. doi: 10.1126/science.277.5329.1097. [DOI] [PubMed] [Google Scholar]
- 49.Ruvkun G, Finney M. Regulation of transcription and cell identity by POU domain proteins. Cell. 1991;64:475–478. doi: 10.1016/0092-8674(91)90227-p. [DOI] [PubMed] [Google Scholar]
- 50.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smeyne RJ, Klein R, Schnapp A, Long LK, Bryant S, Lewin A, Lira SA, Barbacid M. Severe sensory and sympathetic neuropathies in mice carrying a disrupted Trk/NGF receptor gene. Nature. 1994;368:246–249. doi: 10.1038/368246a0. [DOI] [PubMed] [Google Scholar]
- 52.Taglialatela G, Robinson R, Perez-Polo JR. Inhibition of nuclear factor κB (NFκB) activity induces nerve growth factor-resistant apoptosis in PC12 cells. J Neurosci Res. 1997;47:155–162. [PubMed] [Google Scholar]
- 53.Traenckner EB, Wilk S, Baeuerle PA. A proteasome inhibitor prevents activation of NF-κB and stabilizes a newly phosphorylated form of IκB-α that is still bound to NF-κB. EMBO J. 1994;13:5433–5441. doi: 10.1002/j.1460-2075.1994.tb06878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM. Suppression of TNF-α-induced apoptosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 55.Verheij M, Bose R, Lin XH, Yao B, Jarvis WD, Grant S, Birrer MJ, Szabo E, Zon LI, Kyriakis JM, Haimovitz-Friedman A, Fuks Z, Kolesnick RN. Requirement for ceramide-initiated SAPK/JNK signalling in stress-induced apoptosis. Nature. 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 56.Wang CY, Mayo MW, Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 57.Wood JN. Regulation of NF-κB activity in rat dorsal root ganglia and PC12 cells by tumour necrosis factor and nerve growth factor. Neurosci Lett. 1995;192:41–44. doi: 10.1016/0304-3940(95)11603-t. [DOI] [PubMed] [Google Scholar]
- 58.Wu M, Lee H, Bellas RE, Schauer SL, Arsura M, Katz D, FitzGerald MJ, Rothstein TL, Sherr DH, Sonenshein GE. Inhibition of NF-κB/Rel induces apoptosis of murine B cells. EMBO J. 1996;15:4682–4690. [PMC free article] [PubMed] [Google Scholar]
- 59.Yao R, Cooper GM. Requirement for phosphatidylinositol-3 kinase in the prevention of apoptosis by nerve growth factor. Science. 1995;267:2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 60.Yoon SO, Casaccia-Bonnefil P, Carter B, Chao MV. Competitive signaling between TrkA and p75 nerve growth factor receptors determines cell survival. J Neurosci. 1998;18:3273–3281. doi: 10.1523/JNEUROSCI.18-09-03273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]