Abstract
This study characterizes the differential targeting of recently synthesized immediate early gene (IEG) mRNAs to neuronal cell bodies versus dendrites and tests the hypothesis that this targeting is based on signals in the encoded proteins. A single electroconvulsive seizure induces the expression of a number of IEG mRNAs in granule cells of the dentate gyrus. Most of these IEG mRNAs remain in the cell body, including two that are characterized in the present study (the mRNAs for NGFI-A and COX-2). In contrast, the mRNA for Arcmoved rapidly into dendrites at an apparent rate of ∼300 μm/hr. Inhibiting protein synthesis with cycloheximide did not disrupt the differential mRNA sorting, demonstrating that the differential targeting of mRNAs is not dependent on translation.
Keywords: Arc, dendritic mRNA, mRNA localization, electroconvulsive seizure, mRNA transport, immediate early gene, dendrite
Most of the mRNAs that neurons express are restricted to the region of the cell body and very proximal dendrites; however, a small subset of mRNAs is localized in dendrites at relatively high levels (for a recent review, see Steward, 1997). Such differential sorting of mRNA within the cytoplasm could, in principle, occur in three general ways: (1) Most mRNAs could be restricted to the cell body because of limited diffusion, whereas mRNAs that move into dendrites could be delivered by selective transport processes. (2) mRNAs could be restricted to the the cell body via some selective tethering mechanism, whereas mRNAs that are not tethered would diffuse into dendrites. (3) Both the restriction of certain mRNAs to the cell body and the delivery of other mRNAs into dendrites could be attributable to selective targeting mechanisms.
In terms of these possibilities, any mechanisms that select mRNAs for retention in the cell body or delivery into dendrites presumably require some signals through which the different mRNAs are recognized (what have been termed mRNA zip codes; see Steward and Singer, 1997). These zip codes could be in the nascent protein that the mRNA encodes, so that the mRNA is targeted together with its nascent peptide, or in the nucleotide sequence of the mRNA itself.
A barrier to the analysis of mRNA targeting in neurons is that most differentially sorted mRNAs are expressed constitutively so that the ongoing process of mRNA sorting cannot be evaluated. In the present study we document through nonradioactive in situhybridization the very precise differential sorting of mRNAs encoding different immediate early genes (IEGs)—a phenomenon that provides an opportunity to explore the dynamic regulation of sorting of recently synthesized mRNAs in neurons in vivo. After an appropriate stimulus (for example, a single electroconvulsive seizure, ECS), most IEG mRNAs, like most constituitively expressed mRNAs, remain in the region of the neuronal cell body. However, one IEG has been discovered for which the transcript becomes localized throughout dendrites. Because this gene encodes a protein that becomes associated with the cytoskeleton, it was named Arc [activity-regulated cytoskeleton-associated protein (Lyford et al., 1995); also identified as arg3.1 (Link et al., 1995)].
The differential sorting of IEG mRNAs provides an opportunity to test hypotheses about the nature of the mRNA zip codes. If either the restriction of mRNAs to the cell body or the targeting of mRNAs to dendrites depends on a zip code in the peptide, then proper sorting will require protein synthesis. Thus, the present study evaluates whether the differential sorting of Arc mRNA versus two representatives of the many IEG mRNAs that remain in the cell body (NGFI-A and COX-2) is disrupted by blocking translation with cycloheximide. Our results reveal that cycloheximide treatment, which blocked expression of the IEG proteins, did not disrupt the differential sorting of Arc mRNA versus NGFI-A and COX-2 mRNAs. These results indicate that the zip codes for sorting IEG mRNAs lie in the mRNAs themselves and not in the encoded peptides.
MATERIALS AND METHODS
Electroconvulsive seizures. ECSs were induced in male Sprague Dawley rats 25–40 d of age (108–195 gm). Current was passed transcranially (40 mA for 0.5 sec) via electrodes pressed against the scalp immediately posterior to the orbits or via ear clip electrodes. This treatment resulted in a generalized tonic/clonic seizure. Animals were allowed to survive 15 min (n = 4), 30 min (n = 6), 1 hr (n = 4), 2 hr (n = 2), 4 hr (n = 6), 6 hr (n = 2), 8–9 hr (n = 3), 12 hr (n = 2), and 24 hr (n = 2).
Inhibition of protein synthesis. Cycloheximide (CHX; Sigma, St. Louis, MO) was prepared as a 10 mg/ml solution in sterile saline. Rats prepared for in situ hybridization received an injection of CHX (20 or 50 mg/kg, i.p.). Injection controls (n = 3) were allowed to survive 2 hr and 15 min or 4 hr and 15 min after CHX. Rats in the CHX+ECS group were injected with CHX 15 min before ECS and were allowed to survive 1 (n = 2), 2 (n = 3), or 4 hr (n = 4). In addition, some rats received CHX either immediately after ECS and survived 1–2 hr (n = 2) or received CHX 5 min after ECS and were allowed to survive for 4 hr (n = 2). For immunocytochemistry, rats received CHX (20 mg/kg) 15 min before ECS and were allowed to survive for 2 hr (n = 2).
In situ hybridization. Animals were killed humanely with an overdose of sodium pentobarbital (150 mg/kg) and perfused transcardially with 4% paraformaldehyde in 0.1 m phosphate buffer (PB), pH 7.3. Brains were removed and post-fixed in the perfusate overnight at 4°C and cryoprotected in 30% sucrose in PB for 24 hr. Then they were frozen either in powdered dry ice or over liquid nitrogen vapors. In some cases, hemispheres from different experimental groups were frozen together as hemi- or quadbrains. In this way, sections from different brains could be processed together. Sections were cut at 20 μm, using a cryostat, and thaw-mounted onto poly-l-lysine-coated slides. Sections were stored at −80°C until use.
To detect Arc, NGFI-A, COX-2, and β-tubulin mRNAs, we synthesized cRNA probes. The Arc cRNA probe was transcribed from a full-length clone of the rat Arc gene subcloned into the pBluescript plasmid (Lyford et al., 1995). The probe for NGFI-A was derived from the full-length coding region of rat NGFI-A subcloned in pBluescript vector (Milbrandt, 1987), a gift of Dr. Jeffrey Milbrandt (Washington University School of Medicine, St. Louis, MO). Probe for COX-2 mRNA was synthesized from a nearly full-length clone of rat COX-2 subcloned into the pBluescript vector (Yamagata et al., 1993). Antisense cRNAs for Arc, COX-2, and NGFI-A probes were transcribed from linearized plasmids by using T7 polymerase. The probe for β-tubulin was derived from a 1.25 kb portion of the coding region of chicken β-tubulin mRNA (Cleveland et al., 1980) inserted in pGEM-2. Linearized plasmids were transcribed by the SP6 promoter to yield antisense cRNA. Radiolabeled cRNA probes were prepared byin vitro transcription in the presence of35S-labeled uridine 5′-[α-thio] triphosphate (New England Nuclear, Boston, MA), using the Stratagene (La Jolla, CA) RNA transcription kit. Nonradioactive cRNA probes were synthesized with the Ambion Maxiscript in vitro transcription kit in the presence of digoxygenin-11-uridine-5′-triphosphate (Boehringer Mannheim, Indianapolis, IN).
In situ hybridization for radioactive cRNA probes was performed as reported previously (Steward et al., 1990). For in situ hybridization with digoxygenin-labeled cRNA probe, the hybridization solution contained 2× SSC, 1% Denhardt’s reagent, 10% dextran sulfate, 0.5 mg/ml heparin, 0.5 mg/ml of yeast tRNA (Sigma), 0.25 mg/ml salmon sperm DNA (Sigma), and 50% deionized formamide. Approximately 0.2–0.5 μg of cRNA probe was added to each section as determined by dot blot, performed by using the manufacturer’s instructions. Digoxygenin-labeled probes were detected with the Genius kit (Boehringer Mannheim).
Autoradiography. Sheet film autoradiograms were prepared by exposing sections to β-Max Hyperfilm (Amersham, Arlington Heights, IL) for 3 d. The slides were dipped in NT2B emulsion (Kodak, Rochester, NY) and exposed for 7–10 d at 4°C and then developed via D19 chemistry (Kodak). The sections were stained with cresyl violet, dehydrated, and coverslipped with DPX (Fluka, Neu-Ulm, Germany).
Digital images of emulsion autoradiograms viewed at 100× under oil were collected by a CCD camera linked to an Olympus Vanox microscope. Grain density was analyzed by using the grain-counting function on the MCID M4 software (Imaging Research, St. Catherine’s, Ontario, Canada). A Wratten 47 B (Kodak) filter was placed in the light path to reduce the interference of the cresyl stain with grain detection. Counts were made by using a 20 × 20 μm sample window superimposed over the following regions of the ventral limb of the dentate gyrus. Three frames were placed randomly over the granule cell layer, and six frames each were placed over the proximal (within ∼50 μm of the granule cell bodies) and distal molecular layer (within ∼50 μm of the pia). Data were collected from three to four sections per animal. The raw counts over the proximal and distal molecular layer were divided by the counts over the cell bodies to generate an index of dendritic labeling normalized to labeling over somata expression.
Western blot analysis. Rats received a single ECS and were killed humanely at varying times postseizure for analysis of the expression of the Arc protein, as previously described (Lyford et al., 1995). Hippocampi were dissected and sonicated into boiling SDS sample buffer. Then homogenates were subjected to Western analysis with a polyclonal antibody raised against a fusion protein that represents the terminal two-thirds of the Arc protein. The experiments describing the specificity of this antibody have been reported previously (Lyford et al., 1995). As a control for equivalency of protein loading, blots also were probed with an antibody specific for the α-subunit of calcium calmodulin-dependent Kinase II (Hendry and Kennedy, 1986).
Immunocytochemistry. Arc protein was detected by using the same antibody as in immunoblots. Egr-1 protein (encoded by the gene also called NGFI-A) was detected by using a commercial rabbit polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Rats were killed humanely with an overdose of sodium pentobarbital (150 mg/kg) and decapitated. Brains were removed quickly, rinsed in ice-cold PBS, pH 7.3, and frozen by immersion in 2-methyl-butane chilled on powdered dry ice. In some cases, hemispheres from two conditions were combined as a single block to control for processing. Frozen blocks were wrapped in foil and stored at −80°C before cryosectioning. Coronal sections (20 μm thickness) were retrieved on gelatin-coated slides and processed immediately for immunocytochemistry.
Immunocytochemistry was performed with reagents provided in the ABC kit (Vector Laboratories, Burlingame, CA). Sections were fixed in 4% paraformaldehyde (10–20 min) in PBS, rinsed in PBS (three times for 10 min), and blocked in 10% normal goat serum (NGS) in blocking buffer (0.5% Triton X-100 and 0.25% λ-carrageenan in 0.1 mphosphate buffer, pH 7.3) for 2 hr. Primary antibody was diluted in blocking buffer with 5% NGS (Arc 1:100–1:250) and incubated overnight at 4°C. Sections were rinsed in PBS and then incubated for 2 hr at room temperature in biotinylated goat anti-rabbit IgG (1:100 in blocking buffer with 5% NGS). Sections were incubated in 2% ABC reagent in blocking buffer without serum for 30 min at room temperature (Vector Laboratories) and rinsed in Tris-imidazole buffer. Controls were processed as above except that the primary antibody was omitted. The antibody was visualized with the ABC kit (Vector Laboratories) with 1 mg/ml DAB (3–3′ diaminobenzidine; Sigma) and 0.02% H202. Sections were dehydrated via ethanol, cleared in xylene, and coverslipped under DPX.
RESULTS
Recently synthesized IEG mRNAs are targeted to different subcellular domains
We evaluated the differential sorting of three IEG mRNAs that are induced by intense neuronal activity: (1) Arc(activity-regulated cytoskeleton-associated protein), (2) NGFI-A (Milbrandt, 1987), and (3) inducible cyclo-oxygenase (COX-2; Yamagata et al., 1993). Arc was chosen because its mRNA and the encoded protein are targeted to dendrites (Lyford et al., 1995). NGFI-A [also known as zif/268 (Christy et al., 1988), egr-1 (Sukhatme et al., 1988), and Krox 24 (Lemaire et al., 1988)] was chosen because it is a classic IEG (Sheng and Greenberg, 1990) that encodes a transcription factor and is strongly induced by ECS in granule cells of the dentate gyrus (Cole et al., 1990). COX-2, like Arc, is a member of a more recently identified class of IEGs that are rapidly induced by neuronal activity but that do not encode transcription factors. COX-2, an enzyme involved in prostaglandin synthesis, was chosen because the COX-2 protein also is localized to dendrites (Kaufmann et al., 1996).
In nonstimulated control animals, the mRNAs for Arc, NGFI-A, and COX-2 were present at low-to-moderate levels in neurons throughout the forebrain. The overall level of labeling in the dentate gyrus was low for all three mRNAs, but in situ hybridization with digoxygenin-labeled riboprobes revealed the presence of a few relatively heavily labeled neurons within the stratum granulosum in the case of both Arc and NGFI-A (Fig.1). In the case of Arc, there was also some labeling of the dendrites of the individual heavily labeled neurons and diffuse labeling in the dendritic layers of the hippocampus proper. Labeling in the dentate molecular layer was usually distinctly lower than in the dendritic lamina of CA1.
Fig. 1.
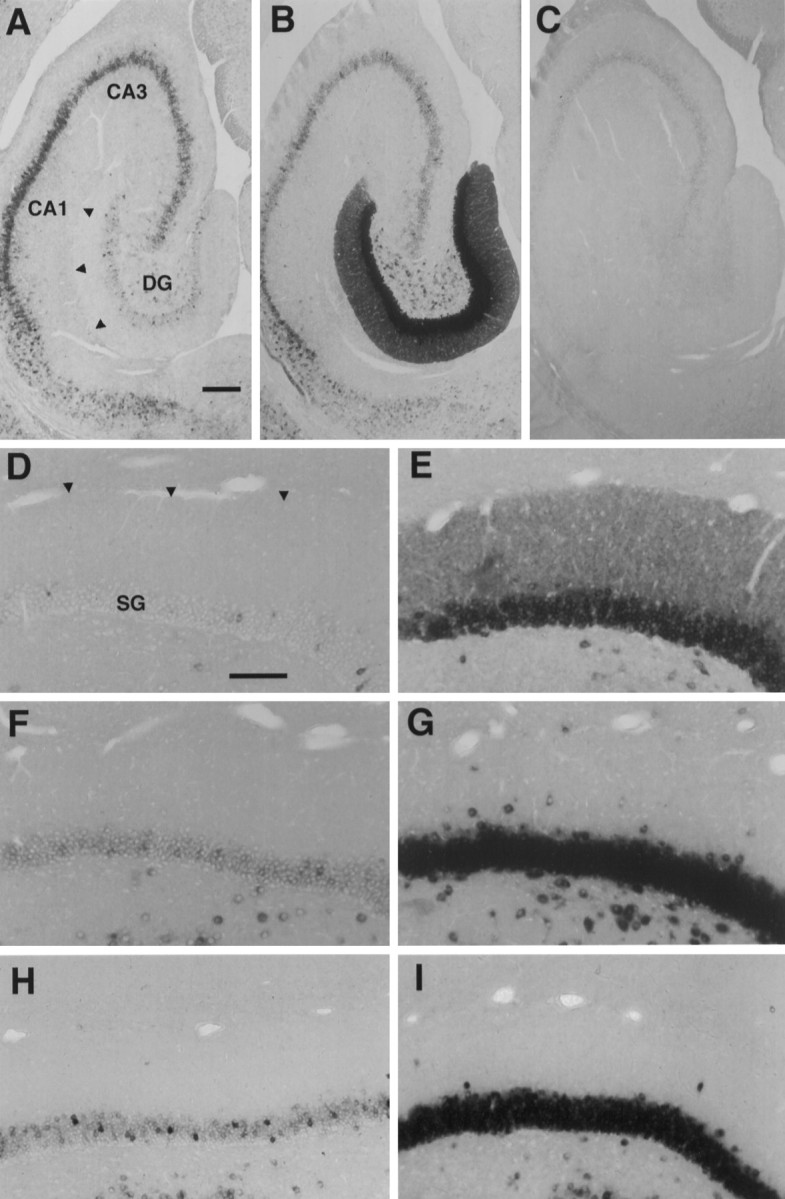
Arc is the first inducible dendritic mRNA. A, B, Views of the rat hippocampal formation showing the distribution of Arc mRNA, using nonradioactive in situ hybridization with digoxygenin-labeled antisense riboprobe. C, Labeling of the rat hippocampal formation, using a digoxygenin-labeled sense probe for Arc mRNA. A, Basal expression ofArc mRNA in the dentate gyrus overall is low, but a small number of neurons that show a high level of expression appear as occasional clusters. Note that basal levels of Arc mRNA are lower in the dentate molecular layer than in the dendritic lamina of CA1. B, This pattern is altered dramatically by ECS when Arc mRNA is induced broadly in dentate granule cells and migrates throughout the dendritic layer. C, Hybridization using an Arc sense probe yielded only background labeling. CA1 and CA3represent the subfields of the hippocampus. DG, Dentate gyrus. Arrowheads indicate the hippocampal fissure, the distal limit of the dentate molecular layer. Scale bar, 250 μm.D–I, Bright-field views of the dorsal limb of the dentate gyrus comparing the localization of Arc mRNA with NGFI-A mRNA and COX-2 mRNA after induction by a single ECS, using digoxygenin-labeled antisense riboprobe. Expression of these mRNAs is low in nonstimulated rats (D, Arc; F, NGFI-A; H, COX-2), and all are elevated substantially by a single ECS (E, Arc;G, NGFI-A; I, COX-2). Note thatArc mRNA was present throughout the dentate molecular layer 1 hr after ECS, but NGFI-A and COX-2 mRNA were confined to the stratum granulosa (SG) despite strong induction. This suggests that the dendritic localization of Arc mRNA is not simply the result of random diffusion but, rather, is the product of selective transport. Arrowheads indicate the distal limit of granule cell dendrites. Scale bar, 100 μm.
The localization of Arc mRNA to dendrites is selective
After a single ECS there was a striking induction of the mRNAs for all three IEGs that was most pronounced in granule cells of the dentate gyrus (Fig. 1). As reported previously,Arc mRNA was distributed rapidly throughout the dendrites of the dentate granule cells. In contrast, the mRNAs for NGFI-A and COX-2 remained tightly localized in the cell body (Fig. 1Evs G and I). The fact that the three IEG mRNAs are sorted differentially demonstrates that the movement ofArc into dendrites is not the result of random diffusion resulting from sudden saturation of the mRNA sorting mechanisms of the neurons.
Kinetics of Arc mRNA translocation to dendritesin vivo
The kinetics of Arc mRNA transport were determined by comparing Arc mRNA expression in the dentate gyrus of naive animals with that at the following times after ECS viain situ hybridization: 15 and 30 min, 1, 2, 4, 6, 8–9, and 12 hr (Fig. 2) and 24 hr (data not shown). At 15 min, increases in Arc mRNA levels were obvious throughout the granule cell layer, indicating the onset of widespread transcriptional activation. Within 30 min, labeling extended well into proximal dendrites and by 1 hr had reached the distal limit of the molecular layer, a distance on average of ∼300 μm. This pattern is consistent with the synchronous delivery of Arc mRNA into the dendrites of a large population of dentate granule cells;Arc mRNA appears first in granule cell somata and then progresses radially to the pia along the major axis of granule cell dendrites. Hence, this observed en masse translocation suggests thatArc mRNA travels along individual dendrites at a rate of at least 300 μm/hr. After labeling filled the molecular layer, it continued to become increasingly dense until reaching peak levels at 2–4 hr.
Fig. 2.

Kinetics of Arc mRNA translocation into granule cell dendrites. Sheet film autoradiograms show labeling of a 35S-labeled cRNA probe for Arc mRNA in the dentate gyrus at a series of time points after a single ECS. Widespread expression of Arc mRNA in the somata of a large population of dentate granule cells could be seen within 15 min of ECS. By 30 min the labeling extended into the dentate molecule layer, indicating transport of Arc mRNA into dendrites. Within 1 hr, labeling was elevated to the distal extent of granule cell dendrites, a distance of up to ∼300 μm. Levels ofArc mRNA continued to increase, reaching a peak at 2–4 hr. By 6 hr the amount of Arc mRNA over the molecular layer was in decline and fell to basal levels by 12 hr. Scale bar, 500 μm.
Expression of Arc mRNA was decreased at 6–9 hr and returned to basal levels by 12 hr. This decline of Arc mRNA labeling appeared as a general fading of the distribution observed at its peak, with levels always highest over cell bodies.
Time course of protein expression in the hippocampal formation after ECS
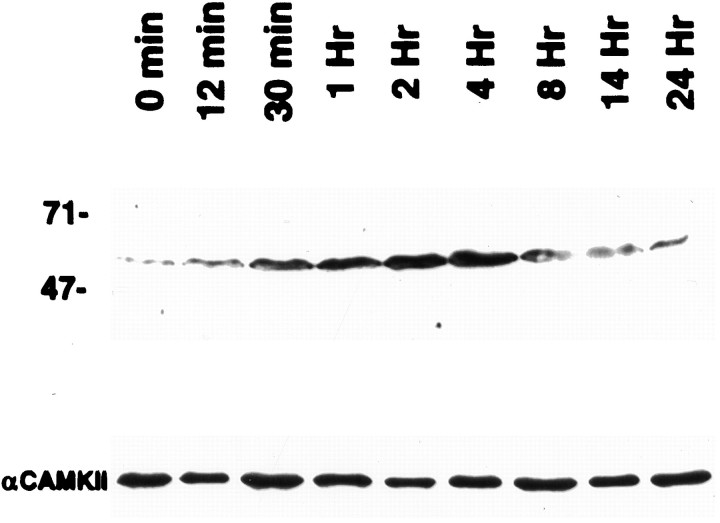
To evaluate whether expression of the Arc protein occurred early enough that the encoded protein could play a role in targeting, we evaluated the time course of protein induction in the hippocampal formation after ECS by Western blot (Fig.3). Increases in protein levels were evident within 30 min after ECS, which is soon enough that the nascentArc protein could provide the signal necessary for dendritic export of the mRNA. The translocation of newly synthesizedArc mRNA to distal dendrites was obvious by 2 hr after ECS, so this interval was chosen to evaluate the possible role of translation in Arc mRNA targeting.
Fig. 3.
Western blot analysis of the induction of theArc protein in the hippocampal formation by ECS. Rats received a single electroconvulsive stimulus and were killed at varying times postseizure. Hippocampi were dissected and sonicated into boiling SDS sample buffer. Homogenates were subjected to Western analysis withArc-specific antibody. A clear increase inArc protein was observed by 30 min, and the highest protein level was seen at 4 hr. Levels of Arc protein returned to the pre-seizure baseline by 24 hr. The blot was probed with an antibody against α-CaMKII to control for equivalency of protein loading.
IEG mRNAs are sorted differentially despite inhibition of translation
To evaluate whether either the selective retention of newly synthesized mRNAs in the cell body or the delivery of mRNAs to dendrites (or both) depends on translation, we evaluated the differential sorting of IEG mRNAs after treatment with CHX, a compound that blocks elongation of the nascent peptide chain. As illustrated in Figure 4, treatment with CHX (20 mg/kg, i.p.) 15 min before the seizure blocked the increases in IEGprotein levels that otherwise are seen after ECS. Using this paradigm, we prepared a separate set of animals to examine IEG mRNA distribution by in situ hybridization.
Fig. 4.
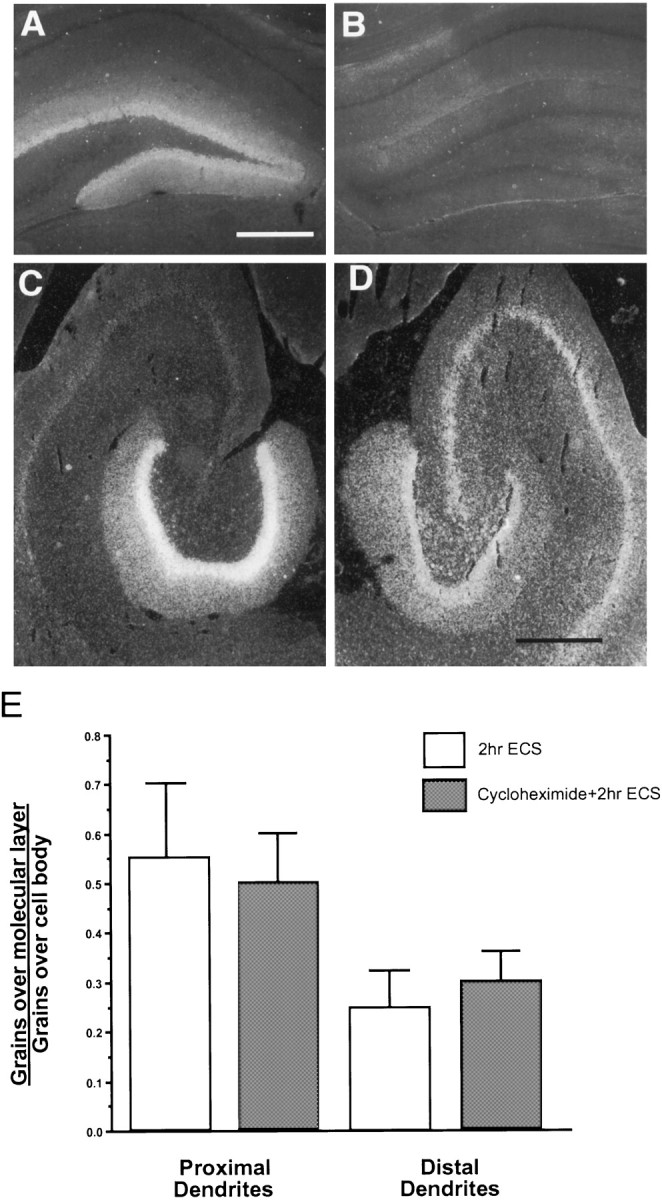
Inhibition of protein synthesis by cycloheximide does not interrupt translocation of ArcmRNA despite blockade of Arc protein expression.A, B, Expression of the Arc protein shown by immunocytochemistry in dark field 2 hr after ECS without cycloheximide (A) and with cycloheximide (B). Scale bar, 500 μm. C, D, Dark-field views of autoradiograms comparing Arc mRNA expression 2 hr after ECS (C) and the same treatment 15 min after cycloheximide injection (D). Scale bar, 500 μm. E, Comparison of the extent of Arc mRNA translocation in the dentate molecular layer in 2 hr ECS and 2 hr CHX+ECS animals. Grain counts of Arc mRNA labeling over the proximal and distal molecular layer were normalized to the counts present over the granule cell layer. Error bars show mean ± SD.
Two hours after animals received an injection of CHX alone (20 or 50 mg/kg, i.p.) the pattern of Arc mRNA expression within the dentate gyrus was comparable to that of controls, with labeling limited to occasional sets of neurons. Expression of Arc mRNA did appear somewhat elevated in neocortex and the CA1 field, however (data not shown). Under protein synthesis inhibition by CHX, ArcmRNA was still induced by ECS in the dentate gyrus and exhibited a clear dendritic translocation similar to that observed in rats receiving ECS alone (compare Fig. 4C and D). Interestingly, levels of Arc mRNA expressed by dentate granule cells were ∼30% lower in CHX-pretreated animals 2 hr after ECS (n = 2) than after ECS alone (n = 2). For this reason, levels of Arc mRNA present in the proximal and distal molecular layer were normalized to the level present over granule cells by using automated grain counting (Fig.4E). If targeting of Arc mRNA into dendrites was blocked by inhibiting protein synthesis, the relative amount of labeling over dendrites should be decreased substantially. However, the ratio of mRNA in dendrites to that in cell bodies was quite similar in CHX-treated and control animals; thus, CHX treatment had no detectable effect on dendritic translocation per se.
Inhibition of protein synthesis also did not disrupt the selective retention of the mRNAs for NGFI-A and COX-2 in neuronal cell bodies (see Fig. 5 for NGFI-A). Moreover, there was no detectable change in the subcellular localization of a noninducible mRNA (β-tubulin mRNA) at 2 or 4 hr after CHX+ECS. These results are important in the light of previous findings that prolonged exposure of cultured hippocampal neurons to protein synthesis inhibitors (6–12 hr) resulted in the drift of mRNAs normally restricted to the cell body into the dendritic compartment (Kleiman et al., 1993). These findings raised the possibility that mRNAs that were not assembled into a translational complex might diffuse nonselectively into dendrites. The failure to see any such delocalization in the present experiments indicates that the mechanisms that underlie the differential sorting of mRNAs in neurons in vivo remain fully operational even when protein synthesis is blocked.
Fig. 5.
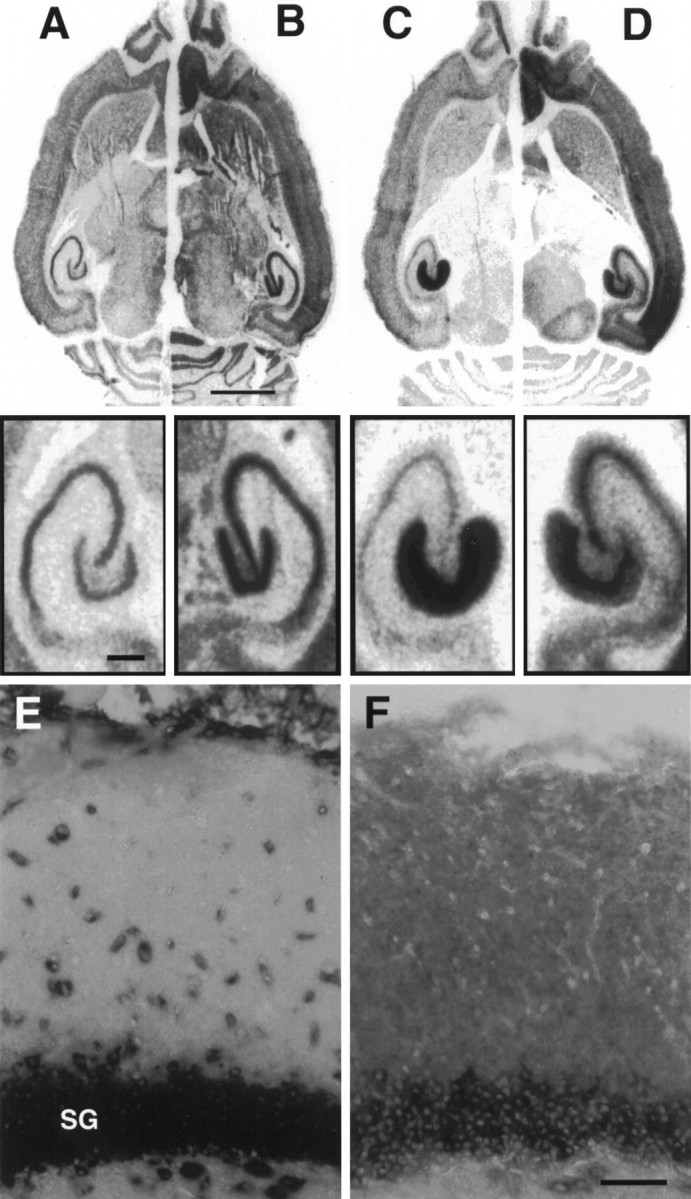
Newly synthesized NGFI-A and ArcmRNA are sorted to their appropriate compartment after inhibition of protein synthesis. A–D, Sheet film autoradiograms of a hemibrain composed of the left hemisphere of an animal 2 hr post-ECS and the right hemisphere of a 2 hr post-ECS animal pretreated with cycloheximide (20 mg/kg) 15 min before ECS. Expression of NGFI-A mRNA was superinduced by ECS in the presence of cycloheximide (B) relative to ECS alone (A) but remained restricted to the granule cell layer. Arc mRNA showed the normal pattern of dendritic translocation (C) despite pretreatment with cycloheximide (D). Scale bar, 2.5 mm.Insets, Higher magnification of the hippocampal formation from A–D. Scale bar, 500 μm. E, F, Nonradioactive in situ hybridization of adjacent sections to compare the cellular distribution of NGFI-A (E) and Arc (F) mRNA 2 hr after ECS in an animal pretreated with cycloheximide. The maintained localization of NGFI-A mRNA over dentate granule cell bodies indicates that mRNA does not diffuse freely in dendrites when it is not part of a translational complex. The migration of ArcmRNA throughout the dendritic lamina therefore is consistent with its selective transport. SG, Stratum granulosa. Scale bar, 50 μm.
After CHX, levels of Arc mRNA are induced less strongly and decline more rapidly
Superinduction of mRNA during translation inhibition is a feature characteristic of IEGs in general (Sheng and Greenberg, 1990) and in fact one that was exploited to identify Arc by differential cloning (Link et al., 1995; Lyford et al., 1995). It is noteworthy, however, that Arc mRNA was less strongly induced in dentate granule cells after CHX+ECS despite the fact that it appears to be superinduced in other parts of the hippocampus and in the cortex (Fig.5). Although clearly induced by ECS in the presence of CHX, levels ofArc mRNA in dentate gyrus neurons were actually somewhat lower in CHX/ECS animals than in ECS animals. The finding that full expression of Arc mRNA requires protein synthesis suggests that Arc behaves as both an IEG and a secondary response gene in dentate granule cells. This is similar to other mRNAs induced by ECS (Lauterborn et al., 1996). These differences were relatively subtle during the initial period of translocation but were clear by 4 hr: ECS rats showed peak induction, whereas CHX+ECS showed marked declines in Arc mRNA levels (Fig.6B).
Fig. 6.
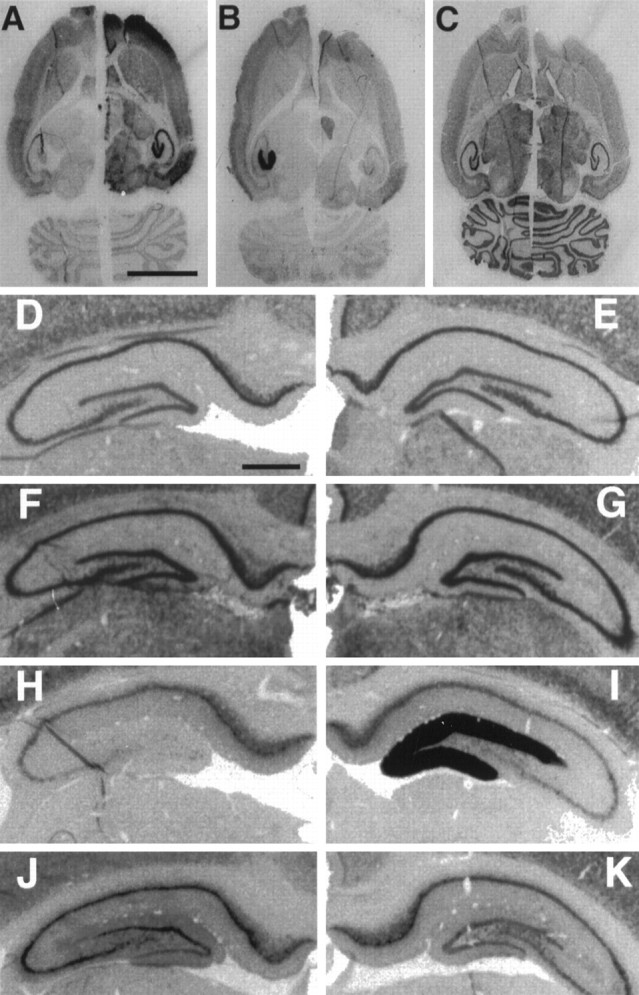
In situ hybridization for NGFI-A,Arc, and β-tubulin mRNA in 4 hr ECS and 4 hr CHX+ECS animals. A–C, Horizontal sections of hemibrains composed of the left hemisphere of a 4 hr ECS animal and the right hemisphere of a 4 hr CHX+ECS animal. Neighboring sections compare the distribution of NGFI-A mRNA (A),Arc mRNA (B), and β-tubulin mRNA (C), using 35S-labeled antisense riboprobes. Scale bar, 0.5 cm. D–G, Expression of NGFI-A mRNA. H–K, Arc mRNA in the dentate gyrus shown by in situ hybridization, using digoxygenin-labeled antisense riboprobes. These views of the hippocampal formation are taken from neighboring sections of the same “quadbrain” composed of hemispheres from four experimental conditions mounted as a single block and sectioned and hybridized together. Shown are basal expression (D,H), 4 hr after ECS (E,I), cycloheximide (20 mg/kg) 15 min before ECS (F, J), and cycloheximide injection (20 mg/kg) 5 min after ECS (G,K). Scale bar, 750 μm.
In contrast to what was seen with Arc, CHX did not diminish the seizure-induced response of NGFI-A in the dentate gyrus. Indeed, inhibition of protein synthesis by CHX, whether administered before ECS (Fig. 6F) or introduced after ECS (Fig.6G), resulted in prolonged elevation of NGFI-A mRNA but a decrease in Arc mRNA (Fig.6J,K). The fact thatArc mRNA decreased even when CHX was given after ECS indicates that CHX does not exert its effect by reducing seizure intensity.
DISCUSSION
The purpose of the present study was to determine whether newly synthesized IEG mRNAs were sorted to cell bodies versus dendrites on the basis of signals in their mRNAs or in the nascent proteins. By massively upregulating IEG mRNAs, a single ECS induced mRNAs localized to both cell bodies (NGFI-A, COX-2) and to dendrites (Arc). These mRNAs were still sorted appropriately when protein synthesis was blocked during the period of induction and targeting, indicating that the targeting of these mRNAs to different subcellular domains cannot depend on a signal or signals within the nascent peptides. Instead, these results point to signals within mRNA itself for both the retention of newly synthesized mRNAs within cell bodies and the targeting to dendrites.
Approaching this question in vivo and using IEG mRNA offer a number of unique advantages over other possible strategies. In the first place, the assessment of IEG mRNAs allows an evaluation of the differential sorting of newly synthesized mRNAs and an analysis of the directionality and kinetics of movement of the mRNAs from their site of synthesis. The examination of the kinetics of movement is not possible with any constituitively expressed mRNA, including mRNA constructs introduced into neurons in vitroby using transfection techniques. Even the observation of the movement of RNA transcripts that have been introduced by microinjection (Muslimov et al., 1997; Prakash et al., 1997) has the potential disadvantage that the introduced mRNAs do not engage the sorting machinery where they normally would (at the nuclear pore just after leaving the nucleus). Another advantage is that the assessment involves native mRNAs rather than genetically modified transcripts; this eliminates any concern about whether the sorting and transport machinery can deal appropriately with an abnormal mRNA or whether the levels of expression of genetically manipulated transcripts may overwhelm the sorting or transport machinery because of the use of unusual promoters. Finally, studies in vivo have revealed a range of mRNA localization patterns (for example, a differential subcellular distribution of the mRNAs for MAP2 and α-CaMKII kinase;Steward and Wallace, 1995). So far, such precise distribution patterns have not been seen in the in vitro setting, raising the concern that localization mechanisms in neurons in vitro may not be fully developed.
All of this is not to discount the advantages of other approaches. In particular, identification of the necessary and sufficient mRNA sequence that represents a particular “zip code” will require the introduction of candidate sequences along with some marker. Consequently, the present approach should be viewed as complementary to others, not a substitute.
There are, however, some technical issues to consider in terms of thein vivo approach. First, the logic of the experiment requires an effective block of the synthesis of the IEG proteins. In this regard, the dose of CHX used was higher than what has been shown previously to block the synthesis of IEG proteins (Hughes et al., 1993). Because CHX inhibits peptide elongation, rather than initiation, it remains a formal possibility that very short polypeptide chains could be formed in the presence of CHX. In this case, however, one still would expect a substantial disruption of mRNA sorting, manifested by either a diminution of the transport of Arc into dendrites or a drift of NGFI-A and COX-2 mRNAs into dendrites. Neither effect was seen, suggesting that the zip codes that determine the localization of IEG mRNAs reside in the nucleotide sequence and not in the nascent protein.
The conclusion that IEG mRNAs are sorted on the basis of signals in the mRNA itself is consistent with a growing body of evidence in other systems. For example, all of the mRNAs that are localized differentially in oocytes and developing embryos apparently are targeted on the basis of signals in the mRNAs themselves, often in the 3′-untranslated region (UTR) (for review, see Wilhelm and Vale, 1993; St. Johnston, 1995). The limited evidence that exists regarding the signals that target other neuronal mRNAs is also consistent with targeting signals in the mRNAs themselves. For example, transgenes consisting of L7 mRNA without translation initiation sites are still properly localized in dendrites of cerebellar Purkinje cells in vivo despite the absence of the nascent L7 peptide (Bian et al., 1996). Similarly, transgenes made up of the 3′-UTR of the α-subunit of calcium calmodulin-dependent Kinase II (α-CaMKII kinase) and bacterial β-galactosidase are properly targeted to dendrites in transgenic mice (Mayford et al., 1996). Vasopressin mRNA also appears to be targeted into dendrites on the basis of signals in the mRNA. Constructs containing labeled fragments of vasopressin mRNA injected into sympathetic neurons are properly sorted into dendrites, although the transcripts are not configured for translation of the cognate peptide (Prakash et al., 1997). It is important to note, however, that there may be exceptions to the rule. It remains possible, for example, that the mRNA for MAP2 is targeted to dendrites on the basis of a signal or signals in the nascent peptide (Marsden et al., 1996).
In many cases, signals necessary and sufficient for mRNA localization are present in the 3′-UTR portion of the mRNA. It is thus noteworthy that there are no regions of obvious homology among the 3′-UTRs ofArc, L7, and α-CaMKII kinase (three mRNAs that are targeted to dendrites), suggesting that localization in dendrites is not determined by a single consensus sequence. Given that the 3′-UTRs of some localized mRNAs contain separate sequences that operate at distinct steps in targeting (for review, see St. Johnston, 1995), the ultimate positioning of mRNAs within dendrites may involve multiple levels of signals and associated factors.
Rate of Arc mRNA transport along dendrites
We interpret the rapid translocation of Arc mRNA into the dendritic laminae of the dentate gyrus as representing an active and selective dendritic transport. The alternative possibility, thatArc mRNA diffuses into dendrites after transcription whereas all other mRNAs are somehow bound within the soma, seems much less likely. In the first place, experimentally induced seizures massively induce the expression of a host of mRNAs. ECS induces a set of IEGs numbering at least 20 (Lanahan et al., 1997), and at least 52 genes are induced by kainic acid administration (Nedivi et al., 1993). Yet, to our knowledge, all of these mRNAs that have been characterized byin situ hybridization are localized to cell bodies exceptArc. It is hard to imagine a selection mechanism that could restrict this heterogeneous population of mRNAs to the cell body and yet somehow spare Arc. This is especially true because association with the translational machinery cannot account for the restriction, because NGFI-A and COX-2 mRNAs remain confined to the cell body despite CHX treatment before ECS. Moreover, there is other evidence that the mRNAs present in the dendrites of neurons in culture are associated with microtubules (Bassell et al., 1994). This is consistent with data in other systems implicating microtubules in the delivery of mRNAs to particular subcellular domains (Macdonald and Struhl, 1988; Mowry and Melton, 1992; Kislauskis et al., 1993, 1994;Wilhelm and Vale, 1993; St. Johnston, 1995).
The translocation of Arc mRNA from granule cell bodies to dendrites after ECS provided the first opportunity to measure the rate at which particular mRNAs migrate in dendrites in vivo. The rate seen (∼300 μm/hr) is ∼10 times faster than the rate of migration previously estimated from pulse–chase studies, using tritiated uridine to label newly synthesized RNA (Davis et al., 1987,1990). Unlike in situ hybridization for a specific mRNA, however, incorporation labels a heterogeneous pool of RNAs predominated by nonmessenger nonmessenger RNAs, with ribosomal RNA accounting for ∼80% of label (Darnell et al., 1986).
The rate we observed is approximately comparable to that reported for the movement of labeled mRNA granules in the dendrites of neurons in culture (360 μm/hr; Knowles et al., 1996) and to the rate of movement of radiolabeled BC1 RNA after microinjection into cultured sympathetic neurons (∼250 μm/hr; Muslimov et al., 1997). Studies of the movement of labeled myelin basic protein mRNA after microinjection into oligodendrocytes indicate a somewhat slower rate of 72 μm/hr (Ainger et al., 1993), however. Our estimates are based on time sampling of the apparent rate of movement from a lamina containing cell bodies into a neuropil region containing a dense network of dendrites, whereas the other studies directly measure the movement of individual packets of mRNA in single processes. Taking this into account, the similarity in rates is more striking than the differences.
The present findings extend our understanding of the mechanisms that lead to a differential localization of mRNAs in vivo and provide new clues about the nature of the transport mechanisms that deliver mRNAs into dendrites. Although it remains to be determined if the trafficking mechanisms that operate for IEG mRNAs are the same or different from the mechanisms that operate for other differentially sorted mRNAs, analysis of the sorting of IEGs provides a currently unique level of access to the dynamic process of mRNA localization.
Footnotes
This work was supported by National Science Foundation Grant IBN92-22120 and National Institutes of Health Grant N512333 (O.S.), National Institutes of Health Grant MH 53603 (P.W.), and National Institutes of Health/National Institute of Neurological Diseases and Stroke Individual National Research Service Award NS0973 (C.S.W.). We thank Leanna Whitmore, Angela Vames, Josh Ajima, and Douglas J. Davis for technical assistance; Ginger Withers for discussion of this manuscript; and Michele Paradies for providing protocols and assistance with in situ hybridization with digoxygenin-labeled cRNA probes.
Correspondence should be addressed to Dr. Oswald Steward, Department of Neuroscience, University of Virginia School of Medicine, Box 5148 MR4, Charlottesville, VA 22908.
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassell GJ, Singer RH, Kosik KS. Association of poly(A) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 3.Bian F, Chu T, Schilling K, Oberdick J. Differential mRNA transport and the regulation of protein synthesis: selective sensitivity of Purkinje cell dendritic mRNAs to translation inhibition. Mol Cell Neurosci. 1996;7:116–133. doi: 10.1006/mcne.1996.0009. [DOI] [PubMed] [Google Scholar]
- 4.Christy BA, Lau LF, Nathans D. A gene activated in mouse 3T3 cells by serum growth factors encodes a protein with “zinc finger” sequences. Proc Natl Acad Sci USA. 1988;85:7857–7861. doi: 10.1073/pnas.85.21.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleveland DW, Lopata MA, MacDonald RJ, Cowan NJ, Rutter WJ, Kirshner MW. Number and evolutionary conservation of alpha and beta cDNA probes. Cell. 1980;20:95–105. doi: 10.1016/0092-8674(80)90238-x. [DOI] [PubMed] [Google Scholar]
- 6.Cole AJ, Abu-Shakra S, Saffen DW, Baraban JM, Worley PF. Rapid rise in transcription factor mRNAs in rat brain after electroshock-induced seizures. J Neurochem. 1990;55:1920–1927. doi: 10.1111/j.1471-4159.1990.tb05777.x. [DOI] [PubMed] [Google Scholar]
- 7.Darnell JE, Lodish H, Baltimore D. Molecular cell biology. Scientific American Books/Freeman; New York: 1986. [Google Scholar]
- 8.Davis L, Banker GA, Steward O. Selective dendritic transport of RNA by hippocampal neurons in culture. Nature. 1987;330:477–479. doi: 10.1038/330477a0. [DOI] [PubMed] [Google Scholar]
- 9.Davis L, Burger B, Banker GA, Steward O. Dendritic transport: quantitative analysis of the time course of somatodendritic transport of recently synthesized RNA. J Neurosci. 1990;10:3056–3068. doi: 10.1523/JNEUROSCI.10-09-03056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendry SHC, Kennedy MB. Immunoreactivity for a calmodulin-dependent protein kinase is selectively increased in macaque striate cortex after monocular deprivation. Proc Natl Acad Sci USA. 1986;83:1536–1540. doi: 10.1073/pnas.83.5.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes P, Beilharz EPG, Dragunow M. Brain-derived neurotrophic factor is induced as an immediate early gene following N-methyl-d-aspartate receptor activation. Neuroscience. 1993;57:319–328. doi: 10.1016/0306-4522(93)90065-n. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann WE, Worley PF, Pegg J, Bremer M, Isackson P. COX-2, a synaptically induced enzyme, is expressed by excitatory neurons at postsynaptic sites in rat cerebral cortex. Proc Natl Acad Sci USA. 1996;93:2317–2321. doi: 10.1073/pnas.93.6.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kislauskis EH, Li Z, Taneja KL, Singer RH. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleiman R, Banker G, Steward O. Inhibition of protein synthesis alters the subcellular distribution of mRNA in neurons but does not prevent dendritic transport of RNA. Proc Natl Acad Sci USA. 1993;90:11192–11196. doi: 10.1073/pnas.90.23.11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knowles RB, Sabry JH, Martone ME, Deerinck DJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanahan A, Lyford G, Stevenson GS, Worley PF, Barnes CA. Selective alteration of long-term potentiation-induced transcriptional response in hippocampus of aged, memory-impaired rats. J Neurosci. 1997;17:2876–2885. doi: 10.1523/JNEUROSCI.17-08-02876.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lauterborn JC, Rivera S, Stinis CT, Hayes VY, Isackson PJ, Gall CM. Differential effects of protein synthesis inhibition on the activity-dependent expression of BDNF transcripts: evidence for immediate-early gene responses from specific promoters. J Neurosci. 1996;16:7428–7436. doi: 10.1523/JNEUROSCI.16-23-07428.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaire P, Revelant O, Bravo R, Charney P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci USA. 1988;85:4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Freu U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyford G, Yamagata K, Kaufmann W, Barnes C, Sanders L, Copeland N, Gilbert D, Jenkins N, Lanahan A, Worley P. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 22.Macdonald PM, Struhl G. Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336:595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 23.Marsden K, Doll T, Ferralli J, Botteri F, Matus A. Transgenic expression of embryonic MAP2 in adult mouse brain: implications for neuronal polarization. J Neurosci. 1996;16:3265–3273. doi: 10.1523/JNEUROSCI.16-10-03265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayford M, Baranes D, Podsypanina K, Kandel E. The 3′-untranslated region of CaMKIIα is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci USA. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 26.Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 27.Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci. 1997;17:4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity genes revealed by differential cDNA cloning. Nature. 1993;263:718–721. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- 29.Prakash N, Fehr S, Mohr E, Richter D. Dendritic localization of rat vasopressin mRNA—ultrastructural analysis and mapping of targeting elements. Eur J Neurosci. 1997;9:523–532. doi: 10.1111/j.1460-9568.1997.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 30.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 31.St Johnston D. The intracellular localization of messenger RNAs. Cell. 1995;81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 32.Steward O. mRNA localization in neurons: a multipurpose mechanism? Neuron. 1997;18:9–12. doi: 10.1016/s0896-6273(01)80041-6. [DOI] [PubMed] [Google Scholar]
- 33.Steward O, Singer RH. The intracellular RNA sorting system: postal zones, zip codes, mail bags, and mail boxes. In: Harford UB, Morris DR, editors. mRNA metabolism and post-transcriptional gene regulation. Wiley–Liss; New York: 1997. pp. 127–146. [Google Scholar]
- 34.Steward O, Wallace CS. mRNA distribution within dendrites: relationship to afferent innervation. J Neurobiol. 1995;26:447–458. doi: 10.1002/neu.480260316. [DOI] [PubMed] [Google Scholar]
- 35.Steward O, Torre ER, Phillips LL, Trimmer PA. The process of reinnervation in the dentate gyrus of adult rats: time course of increases in mRNA for glial fibrillary acidic protein. J Neurosci. 1990;10:2373–2384. doi: 10.1523/JNEUROSCI.10-07-02373.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sukhatme VP, Cao X, Chang LC, Tsai-Morris C-H, Stamenkovich D, Ferriera PCP, Cohen DR, Edwards SA, Shows TB, Curran T, Le Beau MM, Adamson ED. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 37.Wilhelm JE, Vale RD. RNA on the move: the mRNA localization pathway. J Cell Biol. 1993;123:269–274. doi: 10.1083/jcb.123.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. Expression of a mitogen-inducible cyclooxygenase in brain neurons: regulation by synaptic activity and glucocorticoids. Neuron. 1993;11:371–386. doi: 10.1016/0896-6273(93)90192-t. [DOI] [PubMed] [Google Scholar]