Abstract
Syntrophins are cytoplasmic peripheral membrane proteins of the dystrophin-associated protein complex (DAPC). Three syntrophin isoforms, α1, β1, and β2, are encoded by distinct genes. Each contains two pleckstrin homology (PH) domains, a syntrophin-unique (SU) domain, and a PDZ domain. The name PDZ comes from the first three proteins found to contain repeats of this domain (PSD-95,Drosophila discs large protein, and the zona occludens protein 1). PDZ domains in other proteins bind to the C termini of ion channels and neurotransmitter receptors containing the consensus sequence (S/T)XV-COOH and mediate the clustering or synaptic localization of these proteins. Two voltage-gated sodium channels (NaChs), SkM1 and SkM2, of skeletal and cardiac muscle, respectively, have this consensus sequence. Because NaChs are sarcolemmal components like syntrophins, we have investigated possible interactions between these proteins. NaChs copurify with syntrophin and dystrophin from extracts of skeletal and cardiac muscle. Peptides corresponding to the C-terminal 10 amino acids of SkM1 and SkM2 are sufficient to bind detergent-solubilized muscle syntrophins, to inhibit the binding of native NaChs to syntrophin PDZ domain fusion proteins, and to bind specifically to PDZ domains from α1-, β1-, and β2-syntrophin. These peptides also inhibit binding of the syntrophin PDZ domain to the PDZ domain of neuronal nitric oxide synthase, an interaction that is not mediated by C-terminal sequences. Brain NaChs, which lack the (S/T)XV consensus sequence, also copurify with syntrophin and dystrophin, an interaction that does not appear to be mediated by the PDZ domain of syntrophin. Collectively, our data suggest that syntrophins link NaChs to the actin cytoskeleton and the extracellular matrix via dystrophin and the DAPC.
Keywords: syntrophin, dystrophin complex, PDZ domain, PH domain, neuromuscular junction, sodium channel, cytoskeleton, surface plasmon resonance
To receive and transduce chemical signals from motor neurons, skeletal muscle cells have developed a complex, highly organized postsynaptic membrane. The hallmark of this specialization is the very high concentrations (∼10,000/μm2) of nicotinic acetylcholine receptors (AChRs) at the crests of the postsynaptic folds (Fertuck and Salpeter, 1974). Voltage-gated sodium channels (NaChs) are also concentrated in the postsynaptic membrane, where they are primarily confined to the deeper portions of the folds, and in the perijunctional membrane (Betz et al., 1984; Beam et al., 1985; Angelides, 1986;Caldwell et al., 1986; Haimovich et al., 1987; Flucher and Daniels, 1989). This uneven distribution may result in part from a gradient of NaCh expression by muscle nuclei along the muscle fiber. Locally, however, it seems most likely to involve NaCh anchoring by cytoskeletal or other elements (Lupa and Caldwell, 1994).
Compared with the wealth of data on AChR clustering, little is known about the mechanism of anchoring of NaChs. The synaptic accumulation of AChRs in skeletal muscle involves the association of AChRs with the postsynaptic cytoskeleton in response to signals initiated by neurally derived agrin (McMahan, 1990; Bowe and Fallon, 1995). The 43 kDa AChR-associated protein rapsyn is believed to play a central role in this process. Rapsyn clusters AChRs in heterologous cells (Froehner et al., 1990; Phillips et al., 1991) and in rapsyn-deficient mice, AChRs are distributed diffusely and do not cluster in response to exogenously applied agrin (Gautam et al., 1995). Recently, NaChs have been shown to aggregate at points of contact between skeletal muscle cells and Chinese hamster ovary (CHO) cells expressing a neural form of agrin, suggesting that similar mechanisms mediate NaCh anchoring (Sharp and Caldwell, 1996). However, a cytoskeletal protein with a function analogous to rapsyn has yet to be identified for NaChs.
The syntrophins are a family of intracellular peripheral membrane proteins that are components of the dystrophin-associated protein complex (DAPC) in skeletal muscle. The three known syntrophin isoforms, α1, β1, and β2, are encoded by separate genes and are differentially expressed. All three isoforms are expressed in skeletal muscle, although α1-syntrophin is the most abundant (Adams et al., 1993, 1995; Ahn et al., 1994; Yang et al., 1994). Each syntrophin has two tandem pleckstrin homology (PH) domains followed by a C-terminal syntrophin-unique (SU) domain (Adams et al., 1995). Inserted in the first PH domain is a single PDZ domain. The name PDZ comes from the first three proteins found to contain repeats of this domain (PSD-95,Drosophila discs large protein, and the zona occludens protein 1) (Cho et al., 1992). In skeletal muscle, α1- and β1-syntrophin are found on the sarcolemma and are relatively concentrated at the neuromuscular junction (NMJ), whereas β2-syntrophin is concentrated at the NMJ but barely detectable on the sarcolemma (Peters et al., 1994, 1997). Syntrophins bind directly to the C-terminal domain of dystrophin and the dystrophin-related proteins utrophin and dystrobrevin (Kramarcy et al., 1994; Ahn and Kunkel, 1995;Dwyer and Froehner, 1995; Yang et al., 1995; Ahn et al., 1996). The absence of dystrophin in Duchenne muscular dystrophy (DMD) and themdx mouse leads to a dramatic reduction in sarcolemmal syntrophin, although the syntrophins remain concentrated at the NMJ (Butler et al., 1992; Peters et al. 1994, 1997; Yang et al., 1995).
The PDZ domain of α1-syntrophin is known to bind to the PDZ-containing N-terminal region of neuronal nitric oxide synthase (nNOS), thereby targeting the enzyme to the sarcolemma (Brenman et al., 1996a). In other proteins, PDZ domains bind to the C termini of proteins containing the consensus sequence (S/T)XV-COOH, an interaction that has been shown to mediate clustering of NMDA receptors andshaker family K+ channels by the PSD-95 family of synapse-associated proteins (Kim et al., 1995, 1996; Kornau et al., 1995). We investigated the possibility that syntrophins bind to ion channels via their PDZ domains. In particular, the α subunits of two skeletal muscle NaChs, SkM1 and SkM2/rH1 (hereafter referred to as SkM2), have C-terminal sequences with the (S/T)XV motif, and NaChs, like syntrophins, are present on the sarcolemma and are concentrated at the NMJ (Froehner et al., 1987; Flucher and Daniels, 1989). We found that syntrophins, dystrophin, and NaChs can be isolated as stable complexes from detergent extracts of skeletal and cardiac muscle, and that the PDZ domains of all three syntrophins bind the C-terminal sequences of SkM1 and SkM2. Brain NaChs, which lack the C-terminal (S/T)XV consensus sequence, can also be isolated in a complex with syntrophins and dystrophin. This interaction does not appear to be mediated by the PDZ domains of syntrophins. Our data suggest that syntrophins link NaChs to the actin cytoskeleton and the extracellular matrix via dystrophin and the DAPC.
MATERIALS AND METHODS
Antisera. Affinity-purified polyclonal antibodies to NaChs were prepared and characterized as described previously (Dugandzija-Novakovic et al., 1995). In brief, antisera were raised against a synthetic 18-mer peptide (TEEQKKYYNAMKKLGSKK) corresponding to a putative intracellular loop connecting homology domains III and IV, a region that is highly conserved in all functional vertebrate NaChs. The preparation and characterization of the anti-syntrophin monoclonal antibody (mAb) 1351 has been described previously (Froehner et al., 1987). mAb 1351 is directed against an epitope within the PDZ domain of syntrophin (S. H. Gee and S. C. Froehner, unpublished data) and recognizes all known mouse syntrophin isoforms (Peters et al., 1994, 1997). mAb 1808 against dystrophin has been described previously (Sealock et al., 1991). Isoform-specific monoclonal antibodies to K+ channel α subunits (Bekele-Arcuri et al., 1996) were a generous gift from Dr. James S. Trimmer (State University of New York at Stony Brook).
Fusion proteins. Mouse α1-syntrophin domains were generated by PCR amplification of an α1-syntrophin cDNA clone (GenBank accession number U00677) with oligonucleotide primers flanking nucleotides 111–341 (PH1a), 333–620 (PDZ), 594–923 (PH1b), 948–1328 (PH2), and 1323–1622 (SU) (Adams et al., 1993; Brenman et al., 1996a). For the β2-PDZ domain, the primers flanked nucleotides 320–594 (corresponding to amino acids 90–184) of a cDNA encoding mouse β2-syntrophin (GenBank accession number U00678) (Adams et al., 1993). For the β1-PDZ domain, the primers flanked nucleotides 667–954 (corresponding to amino acids 115–210) of a cDNA encoding mouse β1-syntrophin (GenBank accession number U89997) (Peters et al. 1997). The resultant products were subcloned into the pET-32a vector (Novagen, Madison, WI) that contains sequences encoding thioredoxin, an N-terminal 15 amino acid S-Tag, and a hexahistidine nickel binding motif. The 3′ end of the PH1a PCR product was ligated to the 5′ end of the PH1b PCR product (using engineered XbaI sites) to produce an intact PH1 domain. Positive clones were selected and sequenced. Clones with the correct sequence were electroporated into BL21(λDE3)pLysS competent cells. Overnight cultures were diluted 1:10, grown at 37°C for 2 hr, and then induced for 17 hr at 28°C with 1 mmisopropyl-β-d-thiogalactopyranoside. The expressed fusion proteins were all recognized by the S-Protein HRP conjugate (Novagen) and were purified on nickel-Sepharose columns according to the manufacturer’s instructions. All domains were purified from the soluble fraction. Fusion protein purity was determined by Coomassie blue staining of SDS-polyacrylamide gels. Protein concentration was determined by the method of Bradford (1976). Fusion proteins linking N-terminal domains of nNOS (amino acids 1–150 and 1–299) and glutathione S-transferase (GST) were generated as described (Brenman et al., 1996a,b).
Peptides. Synthetic peptides corresponding to the C-terminal 10 amino acids of the adult skeletal muscle NaCh (SkM1; VRPGVKESLV), the cardiac and embryonic skeletal muscle NaCh (SkM2; SPDRDRESIV), and the NMDA receptor 2B subunit (NR2B; EKLSSIESDV) were purchased from Macromolecular Resources (Colorado State University, Fort Collins, CO). Fas (NFRNEIQLSLV) and adenomatous polyposis coli (APC; HSGSYLVTSV) peptides were a generous gift from Dr. Brian Kay and Stacy Sekely (University of North Carolina at Chapel Hill). All peptides contained an additional four amino acid linker (SGSG) at the N terminus and an N-terminal biotin.
Preparation of detergent-solubilized membrane extracts.Tissues were dissected from C57BL6 mice or Sprague Dawley rats and were frozen in liquid nitrogen or used fresh as needed. Tissue was homogenized in a Waring blender in 10 vol of 50 mm Tris, pH 7.5, 1 mm EDTA, and 1 mm EGTA (TEE). The following protease inhibitors were added immediately before homogenization: 1 mm phenylmethylsulfonyl fluoride and 1 μg/ml each leupeptin, pepstatin, aprotinin, and antipain. The homogenate was centrifuged at 30,000 × g for 10 min, and the supernatant was removed. The pellet was rehomogenized in 10 vol of the same buffer and then centrifuged as above. The final pellet was resuspended in 10 vol of TEE plus 100 mm NaCl plus 1% Triton X-100, extracted for 30 min at 4°C on a rocker platform, and then clarified by centrifuging at 40,000 × g for 20 min. The supernatant was removed from beneath the fat layer, if present.
Peptide affinity chromatography. Biotinylated SkM1, SkM2, and NR2B peptides (200 μg each) were coupled to 0.5 ml of streptavidin–agarose (Sigma, St. Louis, MO) in PBS, pH 7.2, for 5 hr at 4°C on a rocker platform. The beads were then washed extensively with TEE plus 350 mm NaCl plus 0.1% Triton X-100 to remove unbound peptide. Two hundred microliters of a 50% slurry of each agarose-coupled peptide or of uncoupled streptavidin–agarose beads were added to 1 ml of detergent-solubilized cardiac muscle membranes and incubated overnight at 4°C on a rocker platform. After extensive washing with the above buffer, the beads were aspirated to near dryness and heated to 80°C in 100 μl of SDS-PAGE loading buffer.
Immunoaffinity purification. mAb 1351 against syntrophin, mAb 1808 against dystrophin, and control mouse IgG (4 mg each) were coupled to Affi-Gel 10 (Bio-Rad, Hercules, CA) in 20 mmHEPES, pH 7.5, plus 150 mm NaCl according to the manufacturer’s instructions. These antibody resins were incubated with 10 ml of detergent-solubilized membranes for 1 hr at 4°C and then were washed extensively with TEE plus 0.5 m NaCl plus 1% Triton X-100, followed by washing with TEE plus 0.5 m NaCl plus 0.1% Triton X-100 and finally with TEE. Bound proteins were eluted with 2.75 ml of 0.1 m triethylamine, pH 11.5, and were immediately neutralized by the addition of 1.25 ml of 1.5m Tris, pH 6.8. Eluted proteins were precipitated by the addition of 0.5 ml 100% trichloroacetic acid (TCA; final concentration, 12%) and overnight incubation on ice. Proteins were pelleted by centrifugation at 39,000 × g for 1 hr in an SW41Ti rotor (Beckman Instruments) after addition of 12% TCA in TEE to fill the centrifuge tubes. The pellets were rinsed with 99% ethanol, dried, and then resuspended in SDS-PAGE loading buffer.
Fusion protein affinity chromatograpy. Solubilized membranes from mouse cardiac muscle or brain were prepared as described above. Fifty micrograms of purified syntrophin fusion protein were added to 1 ml of extract in 1.4 ml Eppendorf tubes along with 25 μl of S-Protein–agarose beads and incubated overnight at 4°C on a rocker platform. After centrifugation, the S-Protein beads were washed extensively with TEE plus 0.5 m NaCl plus 1% Triton X-100. The final wash was removed completely by aspirating with a flat gel-loading tip. The beads were resuspended in 50 μl of SDS-PAGE loading buffer and heated at ∼80°C for 5 min. For peptide competition experiments, syntrophin PDZ domain fusion proteins were preincubated with 10 μm peptide for 30 min on ice.
Immunoblotting and overlay assays. Proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes (Towbin et al., 1979). The membranes were blocked with 5% skim milk powder in 25 mm Tris, pH 7.5, 150 mm NaCl, and 0.1% Tween 20 (TBS–Tween), and then incubated with primary antibody in the same buffer for 1 hr at room temperature. The blots were washed three times for 10 min each in TBS–Tween, incubated with HRP-conjugated secondary antibody for 30 min, and finally washed another four times for 10 min each. Immunoreactive bands were visualized by enhanced chemiluminescence. Overlays with biotinylated synthetic peptides were carried out as above with the following modifications. Biotinylated peptides were preconjugated to streptavidin–HRP as follows. Streptavidin–HRP (0.66 μg/ml) was added to 0.1 μm peptide in blocking buffer and incubated for 30 min at room temperature on a rocker platform. Blots were incubated with the peptide conjugates for 1 hr and then washed four times for 15 min each with TBS–Tween. Overlays with nNOS were carried out as described previously (Brenman et al., 1996a). The SkM1 and NR2B peptides were added where indicated to a final concentration of 1 μm.
Surface plasmon resonance. The relative binding of syntrophin PDZ domains to SKM1, SKM2, NR2B, APC, and Fas peptides was determined using surface plasmon resonance (Morelock et al., 1995;Myszka et al., 1996). This technique has been discussed in several reviews (Fisher and Fivash, 1994; O’Shannessy, 1994; Myszka, 1997). In brief, ligands are attached to a carboxymethylated dextran gold surface, and analytes are injected over the peptide surface. Binding is measured as an increase in resonance units (RU), which directly reflect changes in refractive index at the surface of the chip. All experiments were performed on a BIAcore 2000 instrument at the University of North Carolina Macromolecular Interactions Facility. Neutravidin (Pierce, Rockford, IL)-coated CM5 sensor chips (BIAcore, Piscataway, NJ) were prepared to capture the biotinylated peptides onto the surface. Neutravidin was dissolved at 1 mg/ml in 20 mm HEPES, pH 7.4, 3.4 mm EDTA, 150 mm NaCl, and 0.005% surfactant P-20 (BIAcore) (HBS). The neutravidin solution was diluted 1:20 in 10 mm sodium acetate, pH 6.0, and then covalently attached to the carboxyl groups on the surface of the CM5 sensor chip activated with 200 mm1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride and 50 mmN-hydroxysuccinimide. This was followed by 10 injections of 100 mm HCl (2 min contact time) to achieve a stable baseline. Biotinylated NaCh peptides were immobilized by one or two injections of 100 nm peptide onto the neutravidin surface. After injection of 10 mm HCl to remove unbound peptide, final surface densities varied between 75 and 125 RU.
To measure relative binding, purified syntrophin PDZ domain fusion proteins in HBS were injected onto the peptide surfaces at a flow rate of 20 μl/min for 2 min. The steady-state binding levels, in RU, were recorded at the end of each injection. Between successive measurements the surfaces were regenerated by treatment with 10 mm HCl (2 min contact time). Binding to a control blank flow cell was also measured and used to subtract nonspecific interactions. To normalize the relative responses to the amount of immobilized peptide, the RU values recorded at the end of the sensorgrams were divided by the amount of each specific peptide immobilized.
To study the inhibition of binding of α1-syntrophin PDZ domain to nNOS by peptides, a monoclonal antibody to GST (Santa Cruz Biotechnology, Santa Cruz, CA) was covalently coupled to a CM5 sensor chip as above. The GST mAb solution was diluted to 5 μg/ml in 10 mm sodium acetate, pH 5.75, and then injected at a flow rate of 20 μl/min for 2 min. After injection of 100 mmNaOH (5 min contact time) to remove unbound antibody, final surface densities varied between 3000 and 10,000 RU. A 100 μg/ml solution (in HBS) of a GST fusion protein encoding the first 299 amino acids of nNOS (Brenman et al., 1996a) was injected onto the chip surface (1 min at a flow rate of 20 μl/min) followed by injection of a 1 μmsolution of α1-syntrophin PDZ domain fusion protein (2 min contact time). Peptides were mixed with the α1-PDZ solution just before injection. Between successive measurements the surface of the chip was regenerated by treatment with 3 m MgCl2 (5 min contact time).
RESULTS
Voltage-gated NaChs are composed of a single transmembrane, channel-forming α subunit (Mr ∼250 kDa) and a variable number of smaller (Mr ∼35 kDa) transmembrane auxiliary β subunits. At least 11 different, but homologous, genes encode the α subunits, each of which is composed of four domains having six transmembrane helices each. The α subunits expressed in adult skeletal muscle (designated SkM1) and in fetal skeletal muscle and cardiac muscle (SkM2) have similar C termini that contain the consensus sequence (Q/E)(S/T)XV-COOH predicted to be a ligand for syntrophin PDZ domains (Songyang et al., 1997).
Muscle and brain NaChs copurify with syntrophins and dystrophin
We first determined whether muscle NaChs exist in a complex with syntrophin and dystrophin. Detergent-solubilized membrane extracts of mouse skeletal muscle were incubated with the pan-specific, anti-syntrophin antibody mAb 1351 coupled to Affi-Gel-10 agarose. After extensive washing with buffer containing 0.5 m NaCl and 1% Triton X-100, bound proteins were eluted with pH 11.5 buffer, neutralized, and resolved by SDS-PAGE. Immunoblotting of these syntrophin preparations with a polyclonal antibody to a region conserved in all known NaCh α subunits revealed a band of ∼250 kDa, consistent with the size of intact NaCh α subunit (Fig.1A, NaCh). Preabsorption of the NaCh antibody with the immunizing peptide eliminated this immunoreactivity (Fig. 1A,+pept). As expected, these preparations also contained full-length dystrophin (Kramarcy et al., 1994) and smaller bands, which likely correspond to shorter dystrophin isoforms (Ahn and Kunkel, 1993) (Fig. 1A, Dys). Eluates from control mouse IgG–agarose contained no detectable syntrophin, dystrophin, or NaCh. In the reciprocal experiment, immunoaffinity preparations of dystrophin contained both syntrophin and NaCh (Fig. 1B). Collectively, these data suggest that one or more syntrophins, dystrophin, and NaCh exist as a stable complex in detergent extracts of skeletal muscle and, presumably also, in situ. Similar results were also obtained with extracts of mouse cardiac muscle and with rat skeletal and cardiac muscle (data not shown).
Fig. 1.
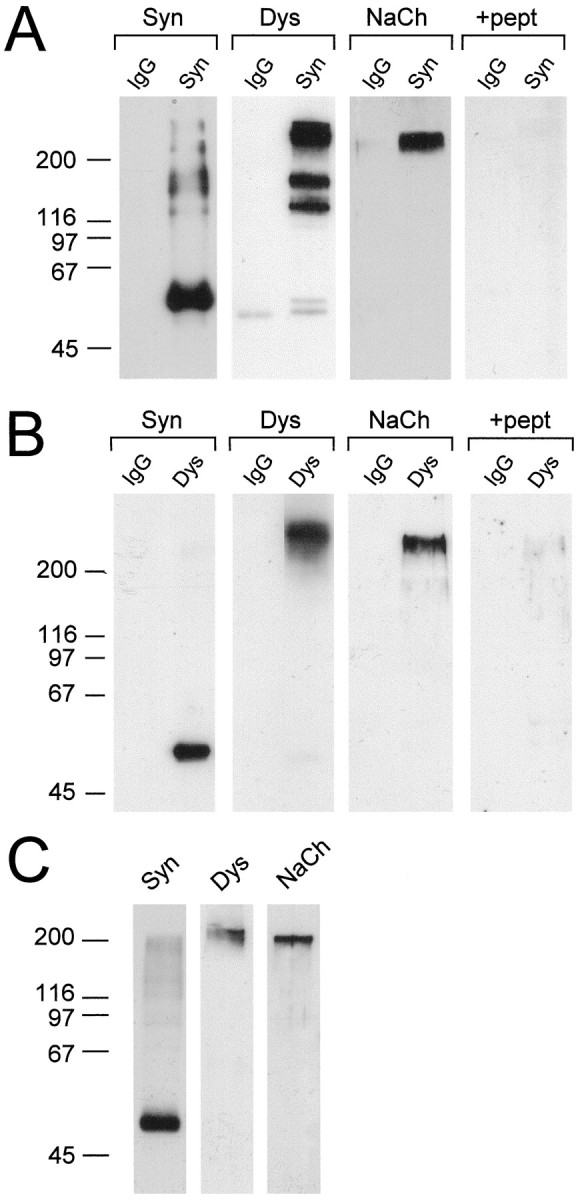
Syntrophin, dystrophin and NaChs form a stable complex in skeletal muscle and brain. Detergent-solubilized mouse skeletal muscle or brain membranes were incubated with Affi-Gel-10 agarose coupled to control mouse IgG (IgG), a pan-specific mAb to syntrophins (Syn), or an mAb to dystrophin (Dys). Immunoaffinity-purified complexes were analyzed by immunoblotting with mAbs specific for syntrophin and dystrophin or with a polyclonal antibody to NaChs. Immunoblots for NaCh were performed in the absence (NaCh), or presence (+pept) of excess immunizing peptide. A, Immunoaffinity-purified syntrophin preparations contain full-length dystrophin (and two shorter forms or proteolytic products of dystrophin) and NaCh. B, Immunoaffinity-purified dystrophin complexes contain both syntrophin and NaCh.C, Syntrophin preparations from brain contain dystrophin and NaCh. The positions of molecular mass markers (in kilodaltons) are shown at the left.
Because none of the known brain NaChs contain the consensus C-terminal sequence for binding to syntrophin PDZ domains, we also purified syntrophins from mouse brain as a control, in the expectation that these preparations would not contain NaChs. In fact, they also contained NaChs as well as dystrophin (Fig. 1C). These data suggest that syntrophins may also exist in a stable complex with brain NaChs, presumably through interactions that do not involve the PDZ domain.
The PDZ domain of syntrophin binds muscle, but not brain, NaChs
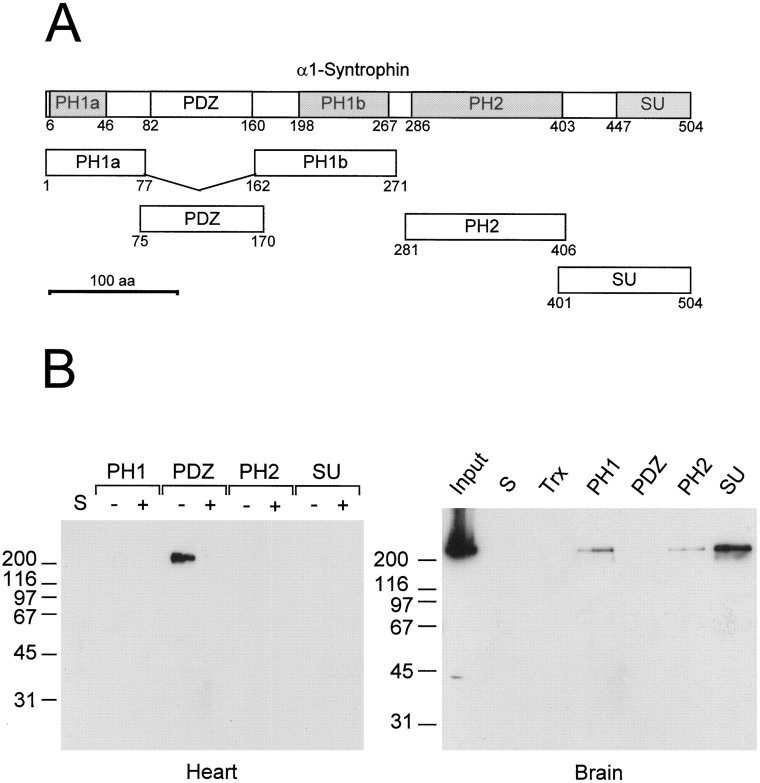
To determine which domains of syntrophin are necessary for binding to NaChs, we generated fusion proteins linking thioredoxin and a 15 amino acid epitope tag (S-Tag) to individual protein domains of α1-syntrophin (Fig.2A). The S-Tag permits recovery and detection of fusion proteins via its interaction with S-Protein. Approximately equal amounts of each fusion protein were incubated with Triton X-100 extracts of mouse cardiac muscle or brain and then isolated with S-Protein–agarose. On immunoblots of the eluates, bound fusion proteins were detected using HRP-conjugated S-Protein. Approximately equal amounts of fusion protein were captured in each case (data not shown). As expected, cardiac muscle NaCh was captured only by the PDZ domain fusion protein (Fig.2B, left). Amounts captured by other syntrophin domains were negligible. In several additional experiments these amounts were comparable to that captured by thioredoxin alone (data not shown). No NaCh was detected in eluates from S-Protein–agarose beads alone.
Fig. 2.
The PDZ domain of α1-syntrophin captures NaChs from muscle but not brain. A, Domain organization of α1-syntrophin and position of corresponding fusion protein constructs. α1-Syntrophin domains corresponding to those described byAdams et al. (1995) are shown drawn to scale; the first and last amino acids in each domain are numbered. Representedbelow are thioredoxin fusion proteins of individual syntrophin domains containing a 15 amino acid epitope tag (S-Tag) that binds to S-Protein. B, Affinity isolation of NaChs from mouse heart and brain with α1-syntrophin domain fusion proteins. The indicated α1-syntrophin fusion proteins (PH1,PDZ, PH2, and SU) were incubated with detergent-solubilized membranes of mouse heart or brain, with (+) or without (−) 10 μm SkM2 peptide, and S-Protein–agarose beads. Approximately equal amounts of each fusion protein were precipitated by the S-Protein-agarose beads, as determined by staining with Ponceau S and by blotting with S-Protein conjugated to HRP (data not shown). The PDZ domain of α1-syntrophin captured NaCh from heart but not from brain. Capture of NaCh from heart was inhibited completely by addition of SkM2 peptide. No NaCh was captured by thioredoxin alone (Trx) or by S-Protein-agarose beads alone (S).
Immunoblotting of the eluates from brain (Fig. 2B,right) revealed that the fusion protein containing the α1-syntrophin PDZ domain failed to capture NaChs, as expected. The other three fusion proteins did, however, capture NaChs. In several experiments the SU domain consistently bound more NaCh than either the PH1 or PH2 domains (Fig. 2B, right). Collectively, our results suggest that the syntrophin PDZ domain binds specifically to the C-terminal SXV motif of muscle NaChs, whereas other syntrophin domains contribute to the binding of brain NaChs.
The C terminus of muscle NaChs binds to the PDZ domain of syntrophins
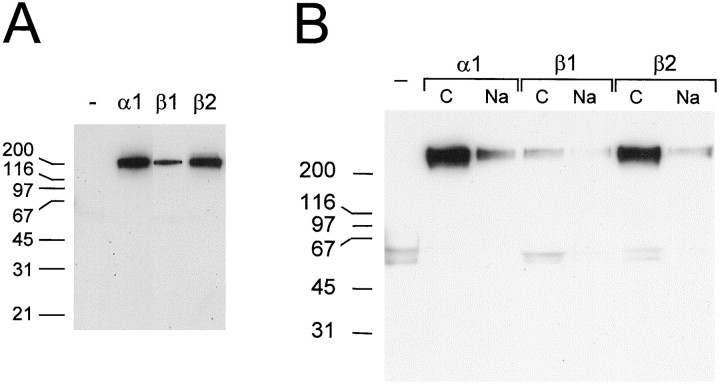
We next determined whether the C-terminal peptide binding activity of PDZ domains (Doyle et al., 1996) could account for interaction between SkM2 and syntrophin PDZ domains. Preincubation of syntrophin domain fusion protein with a peptide corresponding to the SkM2 NaCh C terminus completely blocked the ability of the PDZ domain to capture NaCh from cardiac extracts (Fig. 2B, left, lanes marked ±).
To determine whether the PDZ domains of all three syntrophins are capable of binding NaChs with a C-terminal SXV motif, PDZ domain fusion proteins were used to isolate NaChs from heart extracts. As shown in Figure 3A, all three captured NaChs from cardiac muscle, although β1-PDZ bound significantly less than either α1- or β2-PDZ. No NaCh was captured by S-Protein–agarose beads alone. In a separate experiment, syntrophin PDZ domain fusion proteins were preincubated with a non-SXV control peptide or a peptide corresponding to the SkM2 NaCh C terminus and then used to capture NaChs from cardiac muscle extracts. In the presence of control peptide (Fig. 3B, lanes C), all three syntrophin PDZ domains bound NaCh at levels comparable to those in the absence of peptide (Fig. 3A). The β1-PDZ domain, again, bound much less NaCh than the other two. As found previously for α1-PDZ (Fig. 2B, left), preincubation with the NaCh C-terminal peptide inhibited capture of NaCh by all three fusion proteins (Fig. 3B, lanes Na).
Fig. 3.
The C terminus is necessary for the interaction of the cardiac muscle NaCh with the PDZ domains of α1, β1, and β2-syntrophin. A, Affinity isolation of NaChs from heart with syntrophin PDZ domains. Detergent-solubilized mouse heart membranes were incubated with α1-, β1-, or β2-syntrophin PDZ domains and S-Protein–agarose. As in Figure 2, S-Protein–agarose beads pulled down approximately equal amounts of fusion protein (data not shown). α1- and β2-PDZ pulled down roughly equal amounts of NaCh, whereas β1-PDZ pulled down substantially less.B, The SkM2 NaCh C-terminal peptide inhibits binding to syntrophin PDZ domains. Syntrophin PDZ domain fusion proteins were added to cardiac muscle membrane extracts (as in A), except that PDZ fusion proteins were preincubated with 1 μm non-SXV control peptide (lanes C) or a peptide corresponding to the C-terminal 10 amino acids of the SkM2 NaCh (lanes Na). The SkM2 peptide almost completely blocks the interaction of cardiac NaChs with syntrophin PDZ domains. As inA, α1- and β2-PDZ pulled down roughly equal amounts of NaCh, whereas β1-PDZ pulled down substantially less.
To determine whether the C terminus alone of NaChs is sufficient for binding to syntrophins, biotinylated peptides corresponding to the C-terminal sequences of SkM1 (VRPGVKESLV), SkM2 (SPDRDRESIV), and the NMDA receptor 2B subunit (NR2B; EKLSSIESDV) were coupled to streptavidin–agarose and used to isolate syntrophins from cardiac muscle extracts. Figure 4 shows that the SkM1 and SkM2 peptides bound syntrophins well, with the SkM2 peptide binding more syntrophin than the SkM1 peptide. In this assay, the NR2B peptide, which also corresponds to the consensus sequence (Q/E)(S/T)XV-COOH for syntrophin PDZ binding, bound very small amounts of syntrophin. Streptavidin–agarose alone did not bind syntrophins detectably.
Fig. 4.
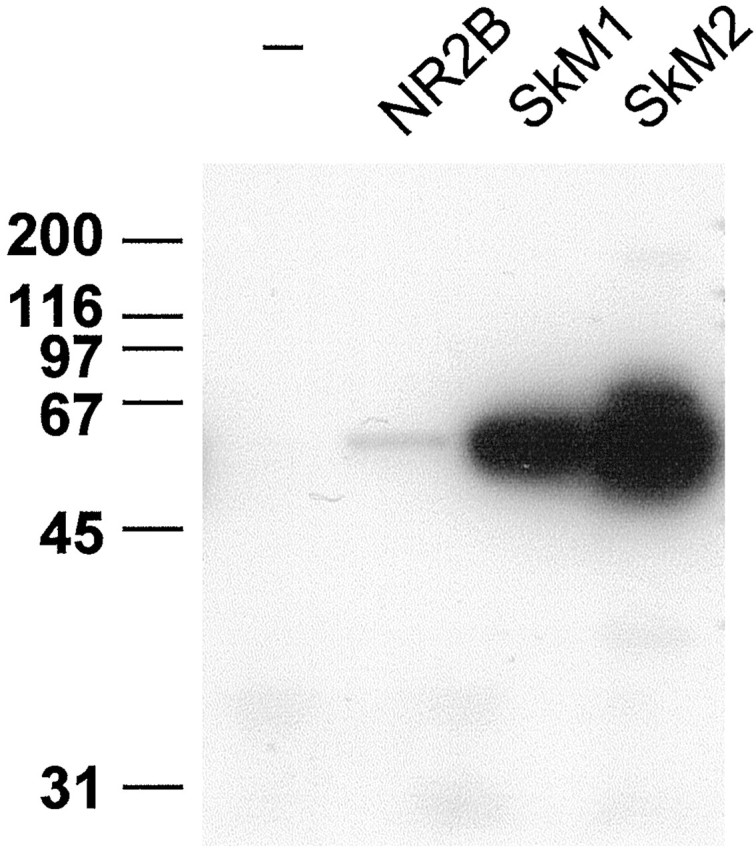
The C termini of muscle NaChs are sufficient for interaction with syntrophins. Approximately equal amounts of biotinylated peptides corresponding to the C-terminal 10 amino acids of the adult skeletal (SkM1) and cardiac (SkM2) muscle NaChs and the NMDA receptor 2B subunit (NR2B) were coupled to streptavidin–agarose beads. Detergent-solubilized heart membranes were incubated with peptide-conjugated beads or with beads alone (−). Bound proteins were immunoblotted for syntrophin. Both the SkM1 and SkM2 peptides bound substantial amounts of syntrophin, whereas NR2B bound much less.
The results described above do not exclude the possibility that an additional protein mediates the interaction between the NaCh C termini and syntrophin PDZ domains. To determine whether the interaction is direct, the biotinylated SkM1 and SkM2 peptides were coupled to streptavidin–HRP and used to overlay protein blots of α1-syntrophin domain fusion proteins (Fig.5A). A significant fraction of all three syntrophin PDZ domain fusion proteins consistently migrated in SDS-polyacrylamide gels as an ∼60 kDa band, approximately twice the size expected. The basis for this apparent dimerization is not known. Both peptides bound to the PDZ domain but not to the PH1, PH2, or SU domains. Notably, more SkM2 peptide bound to the α1-PDZ domain than did the SkM1 peptide. A peptide corresponding to the C terminus of Fas (NFRNEIQLSLV) (Sato et al., 1995) did not bind to any of the domains. The NaCh peptide sequences also appear to be specific for syntrophin PDZ domains. The SkM1 and SkM2 peptides, but not the Fas peptide, bound directly to the PDZ domains of all three syntrophins (Fig. 5B) but did not bind to the PDZ domain of nNOS. These data and the failure of brain NaChs to bind to syntrophin PDZ domains suggest that the C-terminal sequences of muscle NaCh α subunits are both necessary and sufficient for binding to syntrophin PDZ domains.
Fig. 5.

The C termini of muscle NaChs bind directly to syntrophin PDZ domains. A, Biotinylated peptides corresponding to the C-terminal 10 amino acids of the SkM1 and SkM2 NaChs or Fas bound to streptavidin–HRP were overlaid onto α1-syntrophin domain fusion proteins. The position and relative amount of each fusion protein was determined by blotting with S-Protein–HRP. The asterisk indicates the position of the PH1 fusion protein. Both the SkM1 and SkM2 peptides bind exclusively to the PDZ domain of α1-syntrophin, whereas the Fas peptide did not bind to any of the domains. B, SkM1, SkM2, and Fas C-terminal peptides were overlaid onto α1-, β1-, and β2-syntrophin PDZ domain fusion proteins and a GST fusion protein of nNOS (amino acids 1–150) containing the PDZ domain (NOS). The position and relative amount of each fusion protein were determined by blotting with S-Protein-HRP or with an mAb to GST (Anti-GST), followed by donkey anti-mouse-HRP (DAM–HRP). A significant fraction of all three syntrophin PDZ domain fusion proteins migrated as dimers. SkM1 and SkM2 peptides bound to the PDZ domains of all three syntrophins but not to the PDZ domain of nNOS. In contrast, the Fas peptide did not bind to any of the PDZ domains.
Syntrophin PDZ domains bind preferentially to SkM2 NaCh C-terminal peptides
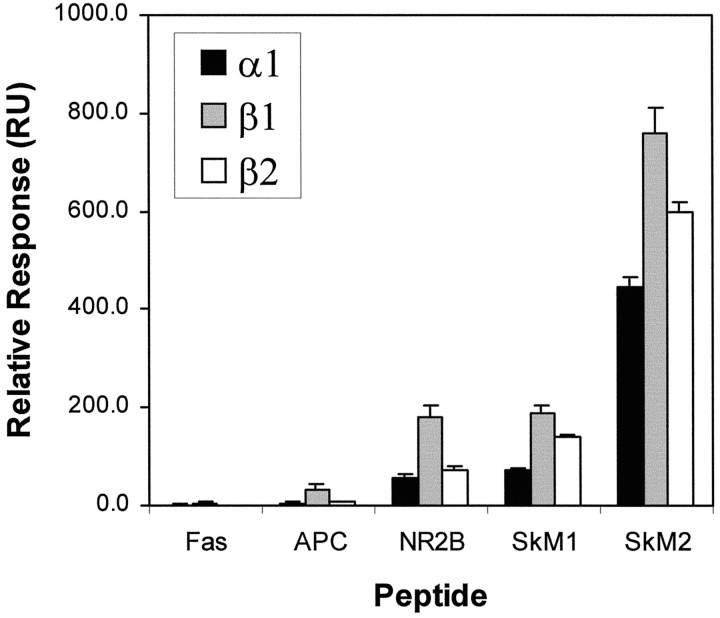
Relative strengths of the interactions between C-terminal S/TXV sequences and syntrophin PDZ domains were measured using surface plasmon resonance. The three syntrophin PDZ domains were tested against biotinylated C-terminal peptides from the following proteins: SkM1, SkM2, NR2B, Fas, and APC (HSGSYLVTSV). As shown in Figure6, all three syntrophin PDZs bound best to the SkM2 NaCh peptide. Each also bound the SkM1 NaCh peptide and, interestingly, the NR2B peptide, albeit to a lesser extent than to the SkM2 peptide. In contrast, binding to both Fas and APC peptides was minimal, suggesting that amino acids upstream of S/TXV may also influence binding of these C-terminal peptides to syntrophin PDZ domains.
Fig. 6.
Syntrophin PDZ domains bind to specific C-terminal SXV peptides. The ability of syntrophin PDZ domain fusion proteins to bind to SkM1, SkM2, NR2B, Fas, and APC peptides was quantitated by surface plasmon resonance. Data for the binding of 500 nmα1-PDZ (black bars), β1-PDZ (gray bars), and β2-PDZ (open bars) are taken from a single time point in the steady-state part of the binding reaction and are the average of two separate determinations. The relative response (resonance units, RU) corresponds to the amount of fusion protein bound and was normalized to the amount of peptide immobilized in each flow cell. Error bars represent SEM.
To address further the role of sequences upstream of S/TXV, we used pull-down assays to determine whether syntrophin PDZ domain fusion proteins could interact with three members of the shakerfamily of K+ channels: Kv1.4 (VETDV), Kv1.1 (LLTDV), and Kv1.2 (MLTDV). Kv1.4 has been shown previously to bind to the second PDZ domain of PSD-95 (Kim et al., 1995). We found that all three syntrophin PDZ domains captured substantial amounts of Kv1.4 from detergent extracts of mouse brain membranes but very little of either Kv1.1 or Kv1.2, as detected by immunoblotting (data not shown). These results suggest that an E at the −3 position from the C terminus of the S/TXV peptide is important for strong syntrophin PDZ domain interaction. These results also show that syntrophin PDZ domains can interact with sequences with either S or T at the −2 position. A summary of the syntrophin PDZ domain sequence preferences, based on these findings, is presented in Figure7.
Fig. 7.
Summary of the binding of syntrophins to C-terminal SXV sequences. Residues in common with those in SkM1 or SkM2 are shaded. The consensus sequence for binding to syntrophin PDZ domains is shown below theline, with residues important for strongest binding to syntrophin PDZ domains in bold. The methods used to conclude whether a given sequence binds to syntrophin PDZ domains are indicated at the right. a, Immunoaffinity copurification; b, affinity isolation with PDZ fusion proteins; c, peptide overlay assays; d, surface plasmon resonance; e, peptide affinity chromatography. RBSC, Rat brain sodium channel.
K+ channels bind to syntrophin PDZ domains but do not copurify with syntrophins
Because syntrophin PDZs can interact with shakerK+ channels (especially Kv1.4; see above), we asked whether these proteins are present in syntrophin preparations. In contrast to our results with NaChs, we were unable to detect K+ channels in syntrophin preparations from brain, even though we could readily detect them in the membrane extracts (data not shown). Thus, if K+ channels exist in complexes with syntrophins in situ, the complexes may be of such low affinity that they dissociate after detergent solubilization. It is noteworthy that although syntrophin PDZ–K+ channel complexes did not form in the extracts under our conditions of immunopurification, they could form under pulldown conditions (i.e., a large excess of PDZ fusion protein added to the extracts). From this, we conclude that the presence of NaChs in syntrophin preparations reflects specific complexes that exist in situ, before detergent solubilization.
The C-terminal peptide of SkM2 inhibits binding of the α1-syntrophin PDZ domain to nNOS
The PDZ domain of α1-syntrophin and the second PDZ domain of PSD-95 bind directly to nNOS fusion proteins containing a PDZ domain, probably by forming PDZ–PDZ heterodimers (Brenman et al., 1996a). Because the PDZ domain of nNOS is internal to the protein, the interaction cannot be via C-terminal binding. The interaction of nNOS and PSD-95 is inhibited by the C-terminal peptide of NR2B (Brenman et al., 1996a), which binds strongly to the second PDZ domain of PSD-95 (Kornau et al., 1995). This suggests that the two binding reactions involve overlapping sites or that there is strong allosteric coupling between sites. To determine whether syntrophin PDZ domains exhibit this property, we tested NaCh C-terminal peptides for inhibition of the binding of the α1-syntrophin PDZ domain to an nNOS PDZ-containing fusion protein (GST–nNOS 1–299) (Brenman et al., 1996a). First, we tested whether an NaCh C-terminal peptide could inhibit the binding of nNOS fusion protein to α1-PDZ in overlay assays. As shown in Figure8A, SkM1 peptide (1 μm) completely inhibited the binding of nNOS to α1-PDZ. In contrast, addition of the same concentration of either non-SXV peptides or NR2B had no effect. We also tested the ability of the SkM1 peptide to inhibit α1-PDZ binding to nNOS using surface plasmon resonance. GST-nNOS (1–299) fusion protein was immobilized on a sensor chip, and α1-PDZ was injected onto the surface of the chip. In the absence of added peptide, injection of α1-PDZ resulted in a large increase in the relative response, indicative of α1-PDZ binding to the nNOS fusion protein (Fig. 8B, top trace). Addition of 1 μm peptide decreased the observed response to approximately half of the control response (Fig.8B, middle trace). The response was further reduced when 10 μm peptide was added (Fig.8B, bottom trace). These data demonstrate that NaChs and nNOS bind to the same or overlapping sites on the PDZ domain of α1-syntrophin. Thus, it appears unlikely that nNOS and NaChs bind simultaneously to syntrophin PDZ domains.
Fig. 8.
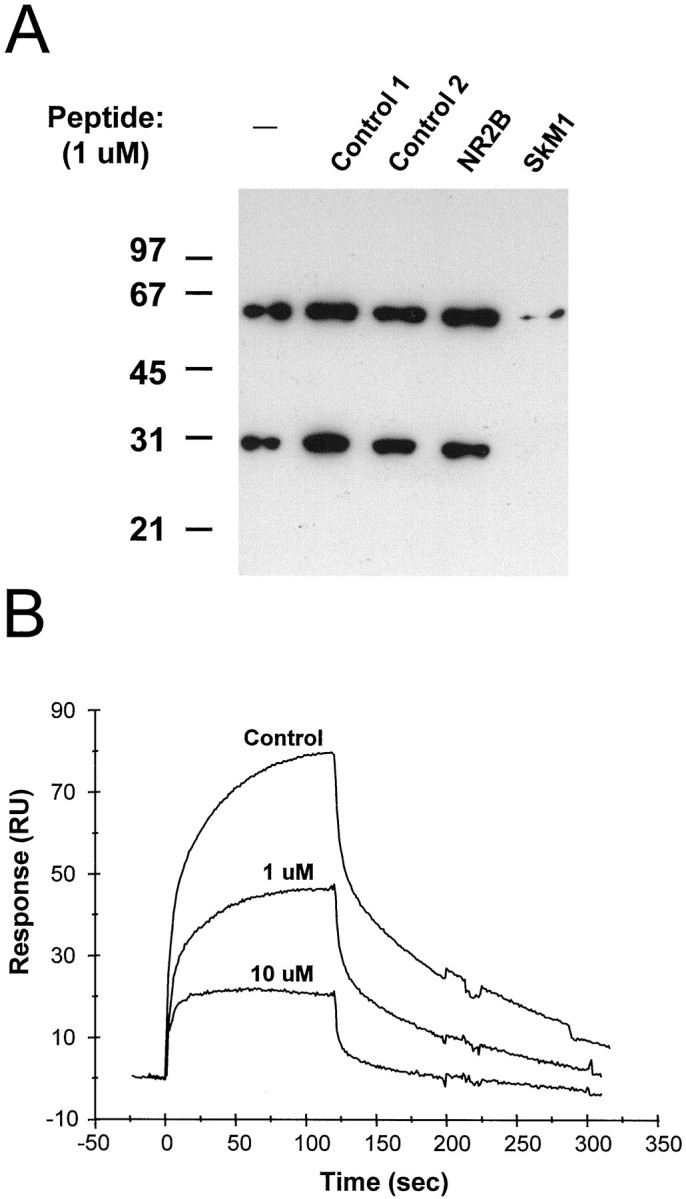
Competition of nNOS binding to α1-syntrophin PDZ domain by the SkM1 NaCh C-terminal peptide. A, Purified α1-syntrophin PDZ domain fusion protein was resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Individuallanes were incubated with a GST–nNOS (1–299) fusion protein either alone (−) or in the presence of 1 μmcontrol non-SXV peptides (Control 1, 2), NR2B C-terminal peptide (NR2B), or SkM1 C-terminal peptide (SkM1). nNOS bound to both the monomer and dimer of α1-PDZ. Binding was almost completely blocked by the SkM1 peptide.B, Surface plasmon resonance. GST–nNOS (1–299) fusion protein was injected onto the surface of a sensor chip coated with an mAb to GST, followed by injection of purified α1-syntrophin PDZ domain fusion protein in the absence (top trace) or presence of the indicated concentrations of SkM1 C-terminal peptide (middle and bottom traces). Binding of α1-PDZ to GST-nNOS was measured in real time as an increase in resonance units (RU). One micromolar SkM1 peptide reduced the binding by approximately half, and 10 μmpeptide almost completely inhibited the binding of α1-PDZ to nNOS (the remaining response is attributable primarily to bulk changes in refractive index).
DISCUSSION
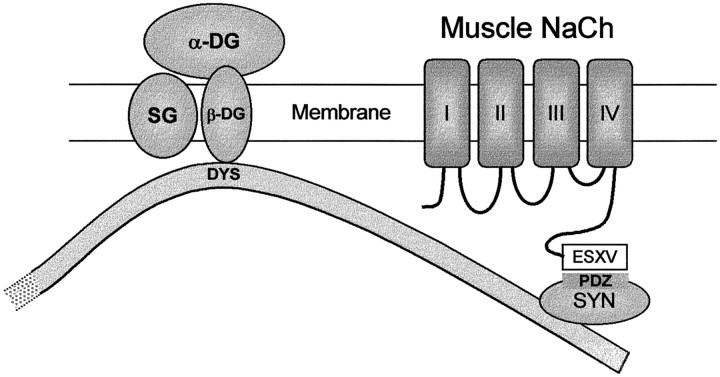
We have shown that syntrophins bind to adult skeletal (SkM1) and cardiac (SkM2) muscle NaChs. This interaction is mediated by direct binding of the PDZ domains of syntrophins to the (S/T)XV C terminus of these NaChs and may link NaChs to the extracellular matrix and the cortical actin network. Formation of this complex (Fig.9) may be relevant to the proper localization and physiological function of NaChs in muscle.
Fig. 9.
Model of NaCh interactions with the DAPC in muscle. Syntrophin (SYN) is shown bound to the C-terminal domain of dystrophin (DYS). Dystrophin associates directly with β-dystroglycan (β-DG), which in turn binds α-dystroglycan (α-DG). The PDZ domain of syntrophin (rectangle) is bound to the C-terminal SXV sequence of a NaCh. SG, Sarcoglycan complex.
The C termini of the SkM1 and SkM2 NaChs are sufficient to bind detergent-solubilized syntrophins from muscle, to inhibit the binding of NaChs to syntrophin PDZ domain fusion proteins, and to bind directly to syntrophin PDZ domains. To a lesser extent, syntrophin PDZ domains also interact with Kv1.4 channels and the NMDA receptor 2B subunit. Binding to Fas or APC C-terminal peptides was negligible. Thus, syntrophin PDZ domains exhibit a high degree of specificity for C-terminal peptides (see Fig. 7).
These results can be rationalized on the basis of the crystal structure of a PDZ domain complexed with its cognate ligand (Doyle et al., 1996). In the third PDZ domain of PSD-95, a loop formed by the conserved GLGF sequence forms a hydrophobic pocket which binds the C-terminal Val (0 position) of the peptide. Side chains of residues at positions −2 and −3 make contacts with side chains in the peptide-binding pocket of the PDZ domain, whereas the side chain of the residue at position −1 points away from the binding pocket and therefore is not predicted to influence binding. Our studies clearly demonstrate that C-terminal SXV sequences are important for binding syntrophin PDZ domains (Fig. 7). For example, brain NaChs do not bind to syntrophin PDZ domains but have C-terminal sequences (especially RBSC II and RBSC VI) that are highly similar to muscle NaChs, except that they lack a Val at the 0 position. The residue at the −3 position also appears to be essential for binding to syntrophin PDZ domains, because all of the peptides that bound had a Glu at this position. For example, the last three amino acids of Fas are identical to SkM1, yet the Fas peptide, which has a Gln at the −3 position, did not bind to the PDZ domain of any of the syntrophins. Finally, the residue at position −4 may be important for binding of peptides to syntrophin PDZ domains. The SkM2 NaCh peptide, which has an Arg at −4, bound approximately fourfold better (as measured by the relative response; Fig. 6) to syntrophin PDZs than either the SkM1 or NR2B peptides, which have a Lys or Ile, respectively, at this position. It has been suggested that a Lys at position −4 can bind a Glu in the loop between the βB and βC strands of the PDZ domain (Songyang et al., 1997), a residue conserved in the syntrophin PDZ domain. Perhaps then, an Arg at the −4 position confers stronger binding than a corresponding Lys. Interestingly, syntrophins contain GLGI in place of the well conserved GLGF sequence. Our results demonstrate that PDZ domains containing GLGI can also bind C-terminal peptides with an SXV consensus sequence.
A surprising result of our studies was the finding that brain NaChs, which lack the SXV C-terminal consensus sequence, also copurify with syntrophins. As expected, the PDZ domain of α1-syntrophin did not bind any brain NaChs but did bind to the SXV-containing muscle NaChs. We did find, however, that NaChs from brain were captured by the SU and PH1 domains, and to a lesser extent by the PH2 domain, of α1-syntrophin. The SU domain consistently captured more NaCh than either the PH1 or PH2 domains. Collectively, our results suggest that other domains in syntrophin can contribute to binding brain NaChs.
If the interaction of the PH and SU domains of syntrophin with brain NaChs is direct, it may occur via binding to other intracellular domains of NaChs. Alternatively, other proteins associated with NaChs may mediate this interaction. The two unrelated auxiliary β subunits of NaChs, designated β1 and β2, are possibilities (Isom et al., 1992, 1995). Another possibility is ankyrin, which copurifies with NaChs from brain (Srinivasan et al., 1988). In skeletal muscle, ankyrin is colocalized with NaChs in the troughs of the postjunctional folds (Flucher and Daniels, 1989); in brain, ankyrin is colocalized with NaChs at the nodes of Ranvier on myelinated axons (Kordeli et al., 1990). In any case, our results suggest that NaChs in brain, like those in muscle, may be associated with the dystrophin or related proteins.
What is the functional significance of the association of NaChs with syntrophin and dystrophin? The ability of syntrophin to link NaChs to dystrophin and the DAPC is reminiscent of the AChR-associated protein rapsyn. Cotransfection experiments have demonstrated that rapsyn can cluster AChRs (Froehner et al., 1990; Phillips et al., 1991; Apel et al., 1995) and α- and β-dystroglycan (Apel et al., 1995, 1997), extracellular and transmembrane proteins, respectively, of the DAPC (Ervasti and Campbell, 1991). Thus, rapsyn may function as a link between AChRs and the DAPC. Syntrophins may play an analogous role in the anchoring or clustering of NaChs. Because dystrophin binds actin (Hemmings et al., 1992), and α-dystroglycan binds both laminin (Ibraghimov-Beskrovnaya et al., 1992; Ervasti and Campbell, 1993; Gee et al., 1993) and agrin (Bowe et al., 1994; Campanelli et al., 1994;Gee et al., 1994; Sugiyama et al., 1994), the interaction with syntrophins may be sufficient to link NaChs to the cortical actin network and the extracellular matrix. Consistent with this idea, CHO cells expressing a neural agrin isoform can cluster NaChs at points of contact with muscle cells (Sharp and Caldwell, 1996).
Finally, syntrophins may modulate the function of NaChs. Evidence supporting such a notion is the recent demonstration that the interaction of the SXV-containing C terminus of the K+ channel α subunit Kv1.1 with the cytoskeleton regulates the extent of inactivation conferred by the β subunit (Jing et al., 1997). Such functional changes may be especially important when considering myopathies such as DMD. For example, patients with DMD suffer from conduction disturbances and heart block, suggesting an important role for dystrophin in the cardiac conduction system (Bies et al., 1992). The finding that syntrophin links NaChs to dystrophin provides a plausible explanation for these defects; a reduction in sarcolemmal syntrophin may lead to aberrant NaCh distribution or function in cardiac cells. Our results, taken together, emphasize the importance of syntrophins as multidomain scaffolds that link NaChs to the DAPC and, hence, to the actin cytoskeleton and the extracellular matrix.
Footnotes
This work was supported by grants from the National Institutes of Health (S.R.L., J.H.C., R.S., and S.C.F.) and the Muscular Dystrophy Association (R.S. and S.C.F.). S.H.G. is supported by a Human Frontier Science Program Organization Postdoctoral Fellowship. We thank Dr. Brian Kay and Stacy Sekely (University of North Carolina at Chapel Hill) for providing APC and Fas peptides, Karen Christopherson, Jay Brenman, and Dr. David Bredt (University of California at San Francisco) for nNOS PDZ domain fusion proteins, and Dr. James Trimmer (State University of New York at Stony Brook) for antibodies to K+ channel subunits. We especially thank Dr. Christian R. Lombardo of the University of North Carolina Macromolecular Interactions Facility for help with the collection and analysis of surface plasmon resonance data and Donna Krzemien for technical assistance. Finally, we thank the reviewers for their comments and suggestions.
Correspondence should be addressed to Dr. Stanley C. Froehner or Dr. Robert Sealock, Department of Physiology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599-7545
REFERENCES
- 1.Adams ME, Butler MH, Dwyer TM, Peters MF, Murnane AA, Froehner SC. Two forms of mouse syntrophin, a 58 kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron. 1993;11:531–540. doi: 10.1016/0896-6273(93)90157-m. [DOI] [PubMed] [Google Scholar]
- 2.Adams ME, Dwyer TM, Dowler LL, White RA, Froehner SC. Mouse α1- and β2-syntrophin gene structure, chromosome localization, and homology with a discs large domain. J Biol Chem. 1995;270:25859–25865. doi: 10.1074/jbc.270.43.25859. [DOI] [PubMed] [Google Scholar]
- 3.Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 4.Ahn AH, Kunkel LM. Syntrophin binds to an alternatively spliced exon of dystrophin. J Cell Biol. 1995;128:363–371. doi: 10.1083/jcb.128.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn AH, Yoshida M, Anderson MS, Feener CA, Selig S, Hagiwara Y, Ozawa E, Kunkel LM. Cloning of human basic A1, a distinct 59 kDa dystrophin-associated protein encoded on chromosome 8q23–24. Proc Natl Acad Sci USA. 1994;91:4446–4450. doi: 10.1073/pnas.91.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahn AH, Freener CA, Gussoni E, Yoshida M, Ozawa E, Kunkel LM. The three human syntrophin genes are expressed in diverse tissues, have distinct chromosomal locations, and each bind to dystrophin and its relatives. J Biol Chem. 1996;271:2724–2730. doi: 10.1074/jbc.271.5.2724. [DOI] [PubMed] [Google Scholar]
- 7.Angelides KJ. Fluorescently labelled Na+ channels are localized and immobilized to synapses of innervated muscle fibres. Nature. 1986;321:63–66. doi: 10.1038/321063a0. [DOI] [PubMed] [Google Scholar]
- 8.Apel ED, Roberds SL, Campbell KP, Merlie JP. Rapsyn may function as a link between the acetylcholine receptor and the agrin-binding dystrophin-associated glycoprotein complex. Neuron. 1995;15:115–126. doi: 10.1016/0896-6273(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 9.Apel ED, Glass DJ, Moscoso LM, Yancopoulos GD, Sanes JR. Rapsyn is required for MuSK signaling and recruits synaptic components to a MuSK-containing scaffold. Neuron. 1997;18:623–635. doi: 10.1016/s0896-6273(00)80303-7. [DOI] [PubMed] [Google Scholar]
- 10.Beam KG, Caldwell JH, Campbell DT. Na channels in skeletal muscle concentrated near the neuromuscular junction. Nature. 1985;313:588–590. doi: 10.1038/313588a0. [DOI] [PubMed] [Google Scholar]
- 11.Bekele-Arcuri Z, Matos MF, Manganas L, Strassle BW, Monaghan MM, Rhodes KJ, Trimmer JS. Generation and characterization of subtype-specific monoclonal antibodies to K+ channel α- and β-subunit polypeptides. Neuropharmacology. 1996;35:851–865. doi: 10.1016/0028-3908(96)00128-1. [DOI] [PubMed] [Google Scholar]
- 12.Betz WJ, Caldwell JH, Kinnamon SC. Increased sodium conductance in the synaptic region of rat skeletal muscle fibres. J Physiol (Lond) 1984;352:189–202. doi: 10.1113/jphysiol.1984.sp015286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bies RD, Friedman D, Roberts R, Perryman MB, Caskey CT. Expression and localization of dystrophin in human cardiac Purkinje fibers. Circulation. 1992;86:147–153. doi: 10.1161/01.cir.86.1.147. [DOI] [PubMed] [Google Scholar]
- 14.Bowe MA, Fallon JR. The role of agrin in synapse formation. Annu Rev Neurosci. 1995;18:443–462. doi: 10.1146/annurev.ne.18.030195.002303. [DOI] [PubMed] [Google Scholar]
- 15.Bowe MA, Deyst KA, Leszyk JD, Fallon JR. Identification and purification of an agrin receptor from torpedo postsynaptic membranes: a heteromeric complex related to the dystroglycans. Neuron. 1994;12:1173–1180. doi: 10.1016/0896-6273(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 16.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu ZQ, Huang F, Xia HH, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and α1-syntrophin mediated by PDZ domains. Cell. 1996a;84:757–767. doi: 10.1016/s0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 18.Brenman JE, Christopherson KS, Craven SE, McGee AW, Bredt DS. Cloning and characterization of postsynaptic density 93, a nitric oxide synthase interacting protein. J Neurosci. 1996b;16:7407–7415. doi: 10.1523/JNEUROSCI.16-23-07407.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler MH, Douville K, Murnane AA, Kramarcy NR, Cohen JB, Sealock R, Froehner SC. Association of the Mr 58,000 postsynaptic protein of electric tissue with Torpedo dystrophin and the Mr 87,000 postsynaptic protein. J Biol Chem. 1992;267:6213–6218. [PubMed] [Google Scholar]
- 20.Caldwell JH, Campbell DT, Beam KG. Na channel distribution in vertebrate skeletal muscle. J Gen Physiol. 1986;87:907–932. doi: 10.1085/jgp.87.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campanelli JT, Roberds SL, Campbell KP, Scheller RH. A role for dystrophin-associated glycoproteins and utrophin in agrin-induced AChR clustering. Cell. 1994;77:663–674. doi: 10.1016/0092-8674(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 22.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 23.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 24.Dugandzija-Novakovic S, Koszowski AG, Levinson SR, Shrager P. Clustering of Na+ channels and node of Ranvier formation in remyelinating axons. J Neurosci. 1995;15:492–503. doi: 10.1523/JNEUROSCI.15-01-00492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dwyer TM, Froehner SC. Direct binding of Torpedo syntrophin to dystrophin and the 87 kDa dystrophin homologue. FEBS Lett. 1995;375:91–94. doi: 10.1016/0014-5793(95)01176-f. [DOI] [PubMed] [Google Scholar]
- 26.Ervasti JM, Campbell KP. Membrane organization of the dystrophin-glycoprotein complex. Cell. 1991;66:1121–1131. doi: 10.1016/0092-8674(91)90035-w. [DOI] [PubMed] [Google Scholar]
- 27.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fertuck HC, Salpeter MM. Localization of acetylcholine receptor by 125I-labeled α-bungarotoxin binding at mouse motor endplates. Proc Natl Acad Sci USA. 1974;71:1376–1378. doi: 10.1073/pnas.71.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher RJ, Fivash M. Surface plasmon resonance based methods for measuring the kinetics and binding affinities of biomolecular interactions. Curr Opin Biotechnol. 1994;5:389–395. doi: 10.1016/0958-1669(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 30.Flucher BE, Daniels MP. Distribution of Na+ channels and ankyrin in neuromuscular junctions is complementary to that of acetylcholine receptors and the 43 kd protein. Neuron. 1989;3:163–175. doi: 10.1016/0896-6273(89)90029-9. [DOI] [PubMed] [Google Scholar]
- 31.Froehner SC, Murnane AA, Tobler M, Peng HB, Sealock R. A postsynaptic Mr 58,000 (58K) protein concentrated at acetylcholine receptor-rich sites in Torpedo electroplaques and skeletal muscle. J Cell Biol. 1987;104:1633–1646. doi: 10.1083/jcb.104.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Froehner SC, Luetje CW, Scotland PB, Patrick J. The postsynaptic 43K protein clusters muscle nicotinic acetylcholine receptors in Xenopus oocytes. Neuron. 1990;5:403–410. doi: 10.1016/0896-6273(90)90079-u. [DOI] [PubMed] [Google Scholar]
- 33.Gautam M, Noakes PG, Mudd J, Nichol M, Chu GC, Sanes JR, Merlie JP. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature. 1995;377:232–236. doi: 10.1038/377232a0. [DOI] [PubMed] [Google Scholar]
- 34.Gee SH, Blacher RW, Douville PJ, Provost PR, Yurchenco PD, Carbonetto S. Laminin-binding protein 120 from brain is closely related to the dystrophin-associated glycoprotein, dystroglycan, and binds with high affinity to the major heparin binding domain of laminin. J Biol Chem. 1993;268:14972–14980. [PubMed] [Google Scholar]
- 35.Gee SH, Montanaro F, Lindenbaum MH, Carbonetto S. Dystroglycan-α, a dystrophin-associated glycoprotein, is a functional agrin receptor. Cell. 1994;77:675–686. doi: 10.1016/0092-8674(94)90052-3. [DOI] [PubMed] [Google Scholar]
- 36.Haimovich B, Schotland DL, Fieles WE, Barchi RL. Localization of sodium channel subtypes in adult rat skeletal muscle using channel-specific monoclonal antibodies. J Neurosci. 1987;7:2957–2966. doi: 10.1523/JNEUROSCI.07-09-02957.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hemmings L, Kuhlman PA, Critchley DR. Analysis of the actin-binding domain of α-actinin by mutagenesis and demonstration that dystrophin contains a functionally homologous domain. J Cell Biol. 1992;116:1369–1380. doi: 10.1083/jcb.116.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ibraghimov-Beskrovnaya O, Ervasti JM, Leveille CJ, Slaughter CA, Sernett SW, Campbell KP. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 39.Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA. Primary structure and functional expression of the β1 subunit of the rat brain sodium channel. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 40.Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BF, Scheuer T, Catterall WA. Structure and function of the β2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 41.Jing J, Peretz T, Singer-Lahat D, Chikvashvili D, Thornhill WB, Lotan I. Inactivation of a voltage dependent K+ channel by β subunit: modulation by a phosphorylation-dependent interaction between the distal C terminus of α subunit and cytoskeleton. J Biol Chem. 1997;272:14021–14024. doi: 10.1074/jbc.272.22.14021. [DOI] [PubMed] [Google Scholar]
- 42.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 43.Kim E, Cho KO, Rothschild A, Sheng M. Heteromultimerization and NMDA receptor-clustering activity of Chapsyn-110, a member of the PSD-95 family of proteins. Neuron. 1996;17:103–113. doi: 10.1016/s0896-6273(00)80284-6. [DOI] [PubMed] [Google Scholar]
- 44.Kordeli E, Davis J, Trapp B, Bennett V. An isoform of ankyrin is localized at nodes of Ranvier in myelinated axons of central and peripheral nerves. J Cell Biol. 1990;110:1341–1352. doi: 10.1083/jcb.110.4.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 46.Kramarcy NR, Vidal A, Froehner SC, Sealock R. Association of utrophin and multiple dystrophin short forms with the mammalian M(r) 58,000 dystrophin-associated protein (syntrophin). J Biol Chem. 1994;269:2870–2876. [PubMed] [Google Scholar]
- 47.Lupa MT, Caldwell JH. Sodium channels aggregate at former synaptic sites in innervated and denervated regenerating muscles. J Cell Biol. 1994;124:139–147. doi: 10.1083/jcb.124.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McMahan UJ. The agrin hypothesis. Cold Spring Harb Symp Quant Biol. 1990;55:407–418. doi: 10.1101/sqb.1990.055.01.041. [DOI] [PubMed] [Google Scholar]
- 49.Morelock MM, Ingraham RH, Betageri R, Jakes S. Determination of receptor-ligand kinetic and equilibrium binding constants using surface plasmon resonance: application to the lck SH2 domain and phosphotyrosyl peptides. J Med Chem. 1995;38:1309–1318. doi: 10.1021/jm00008a009. [DOI] [PubMed] [Google Scholar]
- 50.Myszka DG, Arulanantham PR, Sana T, Wu Z, Morton TA, Ciardelli TL. Kinetic analysis of ligand binding to interleukin-2 receptor complexes created on an optical biosensor surface. Protein Sci. 1996;5:2468–2478. doi: 10.1002/pro.5560051209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8:50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 52.O’Shannessy DJ. Determination of kinetic rate and equilibrium binding constants for macromolecular interactions: a critique of the surface plasmon resonance literature. Curr Opin Biotechnol. 1994;5:65–71. doi: 10.1016/s0958-1669(05)80072-2. [DOI] [PubMed] [Google Scholar]
- 53.Peters MF, Kramarcy NR, Sealock R, Froehner SC. β2-Syntrophin: localization at the neuromuscular junction in skeletal muscle. NeuroReport. 1994;5:1577–1580. [PubMed] [Google Scholar]
- 54.Peters MF, Adams ME, Froehner SC. Differential association of syntrophin pairs with the dystrophin complex. J Cell Biol. 1997;138:81–93. doi: 10.1083/jcb.138.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phillips WD, Kopta C, Blount P, Gardner PD, Steinbach JH, Merlie JP. ACh receptor-rich membrane domains organized in fibroblasts by recombinant 43-kilodalton protein. Science. 1991;251:568–570. doi: 10.1126/science.1703661. [DOI] [PubMed] [Google Scholar]
- 56.Sato T, Irie S, Kitada S, Reed JC. FAP-1: a protein tyrosine phosphatase that associates with Fas. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 57.Sealock R, Butler MH, Kramarcy NR, Gao KX, Murnane AA, Douville K, Froehner SC. Localization of dystrophin relative to acetylcholine receptor domains in electric tissue and adult and cultured skeletal muscle. J Cell Biol. 1991;113:1133–1144. doi: 10.1083/jcb.113.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharp AA, Caldwell JH. Aggregation of sodium channels induced by a postnatally upregulated isoform of agrin. J Neurosci. 1996;16:6775–6783. doi: 10.1523/JNEUROSCI.16-21-06775.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Songyang Z, Fanning AS, Fu C, Xu J, Marfatia SM, Chishti AH, Crompton A, Chan AC, Anderson JM, Cantley LC. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 60.Srinivasan Y, Elmer L, Davis J, Bennett V, Angelides K. Ankyrin and spectrin associate with voltage-dependent sodium channels in brain. Nature. 1988;333:177–180. doi: 10.1038/333177a0. [DOI] [PubMed] [Google Scholar]
- 61.Sugiyama J, Bowen DC, Hall ZW. Dystroglycan binds nerve and muscle agrin. Neuron. 1994;13:103–115. doi: 10.1016/0896-6273(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 62.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang B, Ibraghimov-Beskrovnaya O, Moomaw CR, Slaughter CA, Campbell KP. Heterogeneity of the 59 kDa dystrophin-associated protein revealed by cDNA cloning and expression. J Biol Chem. 1994;269:6040–6044. [PubMed] [Google Scholar]
- 64.Yang B, Jung D, Rafael JA, Chamberlain JS, Campbell KP. Identification of α-syntrophin binding to syntrophin triplet, dystrophin, and utrophin. J Biol Chem. 1995;270:4975–4978. doi: 10.1074/jbc.270.10.4975. [DOI] [PubMed] [Google Scholar]