Abstract
The transport of mRNAs into developing dendrites and axons may be a basic mechanism to localize cytoskeletal proteins to growth cones and influence microfilament organization. Using isoform-specific antibodies and probes for in situ hybridization, we observed distinct localization patterns for β- and γ-actin within cultured cerebrocortical neurons. β-Actin protein was highly enriched within growth cones and filopodia, in contrast to γ-actin protein, which was distributed uniformly throughout the cell. β-Actin protein also was shown to be peripherally localized after transfection of β-actin cDNA bearing an epitope tag. β-Actin mRNAs were localized more frequently to neuronal processes and growth cones, unlike γ-actin mRNAs, which were restricted to the cell body. The rapid localization of β-actin mRNA, but not γ-actin mRNA, into processes and growth cones could be induced by dibutyryl cAMP treatment. Using high-resolution in situ hybridization and image-processing methods, we showed that the distribution of β-actin mRNA within growth cones was statistically nonrandom and demonstrated an association with microtubules. β-Actin mRNAs were detected within minor neurites, axonal processes, and growth cones in the form of spatially distinct granules that colocalized with translational components. Ultrastructural analysis revealed polyribosomes within growth cones that colocalized with cytoskeletal filaments. The transport of β-actin mRNA into developing neurites may be a sequence-specific mechanism to synthesize cytoskeletal proteins directly within processes and growth cones and would provide an additional means to deliver cytoskeletal proteins over long distances.
Keywords: mRNA localization, actin isoforms, cytoskeleton, growth cones, axonal transport, in situhybridization
Elongating axons and dendrites terminate in growth cones, which are specialized motile structures that respond to extracellular cues and control neurite outgrowth. The cytoskeletal organization of the growth cone is unique from the perikarya both in its protein composition and organization of cytoskeletal filaments. Lamellipodia and filopodial protrusions of the growth cone contain bundles of actin filaments oriented with their barbed ends at the plasma membrane (Gordon-Weeks, 1987). Several actin binding proteins have been localized within growth cones, such as actin depolymerization factor (Bamburg and Bray, 1987), filamen, α-actinin (Letourneau and Shattuck, 1989), and myosins (Bridgman and Dailey, 1989; Wang et al., 1996; Evans et al., 1997). A major challenge of neuronal cell biology is to identify how the protein composition of the growth cone differs from the perikarya and to identify mechanisms involved in this sorting and assembly.
One mechanism to provide growth cones with a distinct cytoskeletal composition from the perikarya is to actively transport specific proteins into processes and growth cones after their synthesis within the cell body. For example, it generally has been assumed that actin and tubulin are synthesized in the cell body and transported by polymer sliding (Lasek, 1986) or monomer and/or oligomer transport (Nixon, 1987; Okabe and Hirokawa, 1990; Sabry et al., 1995; Takeda et al., 1995). The transport of mRNAs may provide an additional mechanism for the localization of newly synthesized cytoskeletal proteins and could promote their enrichment within a peripheral compartment via local synthesis. The availability of mRNAs within processes and growth cones could allow the neuron to circumvent the need to transport newly synthesized proteins from the cell body, thus providing the growth cone with autonomous control of its own structure. Contemporary models for cytoskeletal transport have not considered this alternative mechanism (Takeda et al., 1995; Tanaka and Sabry, 1995).
Ultrastructural analysis has demonstrated the presence of polyribosomes in growth cones of developing hippocampal neurons (Deitch and Banker, 1993). Isolated growth cones have been shown to incorporate radiolabeled amino acids into proteins (Davis et al., 1992). A heterogeneous population of mRNAs was shown to exist in dendritic growth cones, which included microtubule-associated protein, MAP2, and internexin (Crino and Eberwine, 1997). We have developed digital imaging methods for in situ hybridization as a high-resolution approach to reveal whether specific mRNAs are localized to growth cones of developing neurons in culture. β-Actin mRNA previously has been localized to the peripheral cytoplasm of non-neuronal cells (Cheng and Bjerknes, 1989; Sundell and Singer, 1991;Kislauskis et al., 1993, 1994). Actin isoforms are sorted within the cytoplasm, and β-actin may have a specific role in regions of motile cytoplasm (Herman and D’Amore, 1985; Otey et al., 1986; Shuster and Herman, 1995; Von Arx et al., 1995; Yao and Forte, 1995). We demonstrated that sequence-specific isoform localization patterns exist in neurons at both the mRNA and protein levels. The β-actin isoform was found to be highly enriched within growth cones. β-Actin mRNAs also were observed within growth cones, and their localization into processes and growth cones was a sequence-specific pattern and correlated spatially at high resolution with the presence of translational components and the microtubular cytoskeleton.
MATERIALS AND METHODS
Cell culture. The method of neuronal culture has been described in detail (Goslin and Banker, 1991) and modified for use with cortical neurons in our laboratory (Kosik and Finch, 1987). Cerebral cortex was dissected from embryonic day 19 rats and digested with 0.25% trypsin in HBSS. Tissue was washed twice in HBSS, placed in minimal essential media (MEM) with 10% fetal bovine serum, and mechanically dissociated by pipetting. Neurons were plated at low density (3000 cells/cm2) on poly-l-lysine-coated coverslips (0.1 mg/ml, overnight) and cultured for 4 d. After neurons had attached to the substrate (4 hr), coverslips were inverted onto a monolayer of astrocytes. The coculture of neurons with glia using this sandwich technique has been observed previously to promote neuronal development (Goslin and Banker, 1991). Astrocytes were prepared from postnatal day 1 rat cortex by culturing dissociated cortex in MEM with 10% horse serum on untreated tissue culture plates. Under these conditions neurons will not attach to the substrate, and the major cell type cultured is GFAP-positive Type-1 astrocytes. The coculture was maintained in glutamate-free MEM with N2 supplements, which included transferrin (100 μg/ml), insulin (5 μg/ml), progesterone (20 nm), putrescine (100 μm), and selenium dioxide (30 nm). In addition, extra glucose (600 mg/l), sodium pyruvate (1 mm), and ovalbumin (0.1%) were used.
To induce β-actin mRNA transport, we cultured cells for 4 d as described above and then transferred them to MEM for 3 hr. Dibutyryl cAMP (db-cAMP) was added to the medium at concentrations of 5, 10, 25, and 100 μg/ml for periods of 10, 20, 30, 45, 60, and 120 min. Then neurons were fixed in paraformaldehyde (4% in PBS with 5 mm MgCl2) for 15 min at room temperature.
Construction of hemagglutinin (HA)–actin cDNA and transfection.Full-length β-actin cDNA containing all coding and untranslated sequences was subcloned into a Rous sarcoma virus (RSV) vector for transfection studies (Kislauskis et al., 1993, 1994). Advantages of this vector are the powerful and promiscuous transcriptional activity of the Rous sarcoma virus long-terminal repeat and SV40 processing signals (small T intron and polyadenylation signal), which allow efficient expression of the reporter in eukaryotic cells. Two sequences of the HA epitope tag (Wilson et al., 1984) were subcloned into the β-actin cDNA directly upstream of the 3′-untranslated region (UTR). The tag encodes for nine amino acids and was subcloned into full-length actin cDNA by using PCR to introduce restriction sites such that the tag was inserted close to the translation termination codon. Sequence analysis confirmed that the HA tag was in the proper reading frame with β-actin.
To establish a working transfection protocol, we evaluated several published methods for transfer efficiency, toxicity, and effects on endogenous mRNA and protein localization. The best results were obtained with a modification of the recently developed lipofection method withN-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate (DOTAP) (Boehringer Mannheim, Indianapolis, IN), developed for use with primary neuronal cultures (Kaech et al., 1996;Marsden et al., 1996). Briefly, 2 μg of DNA (in 50 μl of 0.15m NaCl and 20 mm HEPES, pH 7.4) and 5 μl of DOTAP (in 50 μl of NaCl/HEPES) were mixed together at room temperature. Recently trypsinized cells (1 × 106, 200 μl of MEM) were mixed with the DOTAP/DNA suspension at 37°C for 1 hr. Then the transfected cells were plated on poly-l-lysine-coated coverslips in the presence of 10% FBS for 2 hr and transferred to N2 media, as described. The cells were cultured for 4 d and then fixed. A transfection efficiency of ∼1–3% was observed, using a monoclonal antibody to HA (Boehringer Mannheim). This approach was nontoxic to the neurons because transfected cells had identical morphology, localization of microfilaments, microtubules, β-actin protein, and mRNA.
Probe preparation. Amino group modified oligonucleotides were made on a DNA synthesizer, having modification at five positions within the sequence. Ten oligonucleotide sequences (50 bases each) complementary to β-actin or γ-actin 3′-untranslated sequences (Nudel et al., 1983; Brown et al., 1990) were chemically labeled with digoxigenin or biotin succinimide ester (Boehringer Mannheim). To ensure isoform specificity, we selected probes from unique regions within the 3′-UTR. Oligonucleotide probes also were made to adult rat 18S ribosomal RNA. Oligonucleotide probes complementary to β-galactosidase mRNA were used as a control. Oligo-dT [50 nucleotides (nt)] was labeled with biotin or digoxigenin to probe poly(A+) mRNA, as described previously (Bassell et al., 1994). Probes were purified by using a 20 ml G-50 column, and the collected fractions were blotted onto nitrocellulose and detected with an antidigoxigenin or streptavidin alkaline phosphatase conjugates (Boehringer Mannheim). Positive fractions were lyophilized, combined, and resuspended in water.
Hybridization. Cells were washed in PBS containing 5 mm MgCl2 and then equilibrated in 40% formamide (Sigma), 1× SSC, and 10 mm sodium phosphate, pH 7.0, at room temperature for not more than 10 min. Probe mixture (15 ng) was dried down with Escherichia coli tRNA (10 μg) and sonicated salmon sperm DNA (10 μg) and then suspended in 10 μl of 80% formamide containing 20 mm sodium phosphate, pH 7.0. Probes were mixed with 10 μl of hybridization buffer (20% dextran sulfate, 2× SSC, 0.4% BSA, and 20 mm sodium phosphate, pH 7.0). Coverslips were placed cell-side-down on Parafilm containing 20 μl of probe mixture and hybridized for 3 hr at 37°C. After hybridization, coverslips were washed for 20 min in 40% formamide/1× SSC at 37°C and then were given three 10 min washes in 1× SSC on a rotary shaker at room temperature.
The specificity of actin mRNA probes was demonstrated with both positive and negative controls. The peripheral localization of β-actin mRNA in lamellae of fibroblast-like cells present in the cortical culture (data not shown) was similar to previous studies in fibroblasts from chicken embryos (Kislauskis et al., 1993, 1994). No signal was obtained when actin oligonucleotide probes were omitted from the hybridization or when digoxigenin- or biotin-labeled oligonucleotide probes to β-galactosidase mRNA were used (data not shown). As an alternative negative control, the hybridization signal with labeled actin probes was eliminated by competition with an excess amount of unlabeled actin probe (data not shown).
Immunofluorescence. Digoxigenin-labeled probes were detected by using a monoclonal antibody to digoxigenin conjugated to Cy3 (Jackson ImmunoResearch, West Grove, PA) and viewed with a Cy3 filter (Chroma Technology, Brattleboro, VT). Biotin-labeled probes were detected by using streptavidin–Cy5 and viewed with a Cy5 filter (Chroma Technology). Rabbit polyclonal antibodies to EF1α (Sanders et al., 1996) and 60S ribosomal proteins (Horne and Hesketh, 1990) were detected with donkey antibody to rabbit IgG conjugated to Cy5 (Jackson ImmunoResearch). F-actin was detected by using tetramethylrhodamine isothiocyanate (TRITC)-labeled phalloidin (Molecular Probes, Eugene, OR). The β-actin isoform was detected by using an affinity-selected anti-β-actin IgG that was depleted of all other isoactin cross-reactivity by selective absorption, as described in Hoock et al., 1991. β-Actin also was detected by using a monoclonal antibody (Sigma). γ-Actin was detected by using a polyclonal antibody from rabbit specific for the N terminus of γ-actin (obtained from J. C. Bulinski, Columbia University, New York, NY). Microtubules were detected by using a monoclonal antibody to tubulin (Amersham, Arlington Heights, IL) or a rabbit polyclonal antibody (Accurate Antibodies, Westbury, NY). All secondary antibodies were affinity-purified donkey antibodies to mouse or rabbit IgG conjugated to fluorochrome (Jackson ImmunoResearch). Fluorochrome separation for Cy3 and FITC was achieved via a FITC filter equipped with an emission barrier between 520 and 560 nm. Antibody incubations were for 1 hr at 37°C in Tris-buffered saline (TBS) with 1% BSA and 0.1% Triton X-100 and were followed by several washes in buffer on a rotary shaker. Immunofluorescence was viewed with an Olympus-BX60 microscope equipped with a 60× Plan-Neofluar objective and Nomarski (differential interference contrast; DIC) optics.
Digital imaging microscopy—two-dimensional. Cells were viewed as above, using a 100 W mercury arc lamp, and filtered with HiQ bandpass filters (Chroma Technology). The images were captured with a cooled CCD camera (Photometrics, Tucson, AZ) with a 35 mm shutter and processed by Metamorph 2.0 (Universal Imaging, Media, PA) running on a 486DX2 processor (Intel). For analysis of hybridization intensity within processes, 50 neurons were randomly selected under the fluorescein filter, which displayed immunostaining for tubulin. After each selection the cartridge was shifted to the rhodamine filter to visualize actin mRNA. Images were captured under both fluorescein and rhodamine filters.
Image processing by Metamorph was used similarly to evaluate colocalization of actin mRNA and EF1α or 60S ribosomal proteins. In these experiments, actin mRNA was detected with TRITC as described above, and rabbit polyclonal antibodies to EF1α (Sanders et al., 1996) or the 60S ribosomal subunit (Horne and Hesketh, 1990) were detected in fluorescein or Cy5.
Digital imaging microscopy—three dimensional. Cells were viewed with a Nikon Diaphot 300 microscope and 60×/1.4 objective equipped for epifluorescence and modified to obtain images along thez-axis (Fay et al., 1989; Carrington et al., 1995). Images at each focal plane (100 or 250 nm steps) were acquired with a thermoelectrically cooled CCD camera (model 220, Photometrics). This resulted in acquisition of an image set of the cell in three dimensions (composed of 20 optical sections). Pixels had an x andy dimension of 100 nm. Changes in position of the focus (to control z-axis positioning) were effected by a computer-controlled stepper motor (Ludl), using feedback from an eddy current sensor. We then switched to a computer-controlled piezoelectric positioning device from Scanalytics, which provided highly accurate movements of the objective (10 nm). The data presented in Figures 6 and7 used this technology. The duration of exposure of the specimen to the excitation source was controlled by a computerized shutter. Image acquisition software was written by the Biomedical Imaging Group for use with this workstation and has been described previously (Fay et al., 1989; Carrington et al., 1995).
Fig. 6.
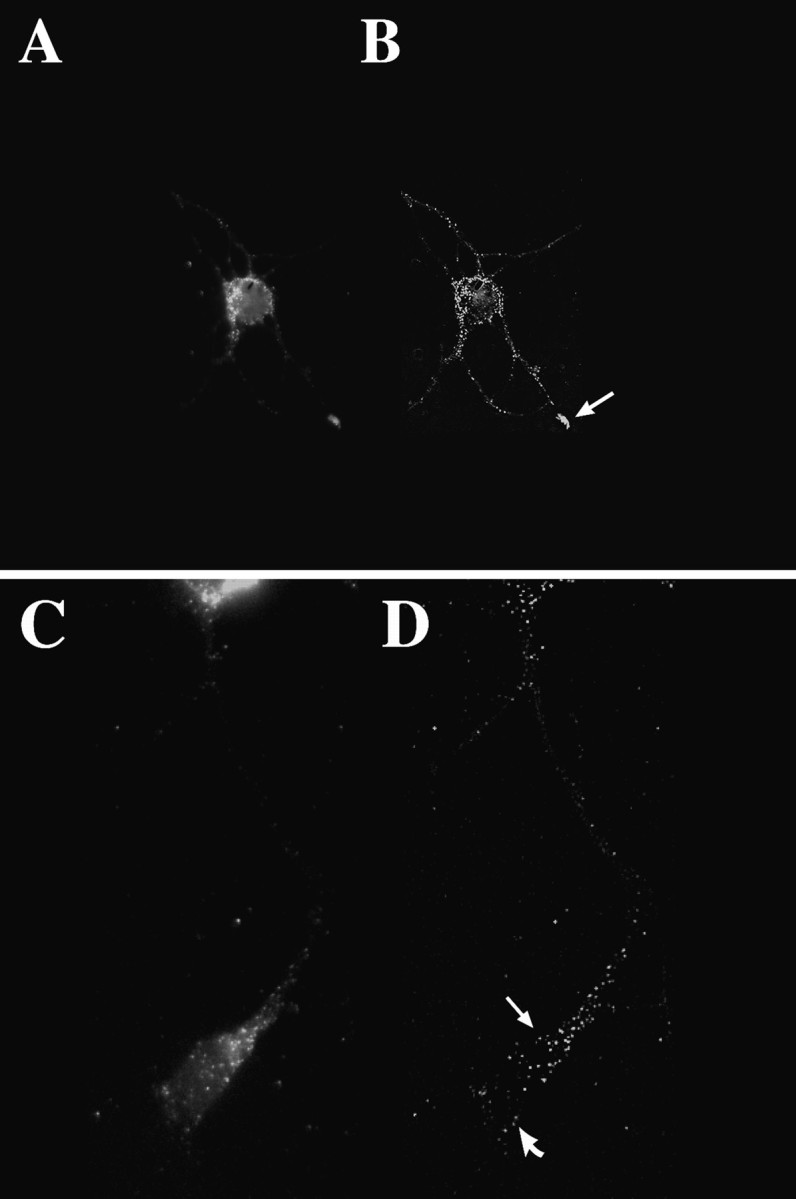
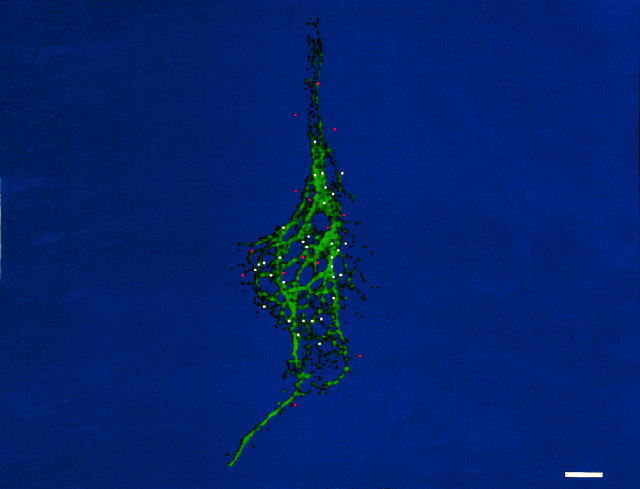
Visualization of β-actin mRNA by three-dimensional digital imaging microscopy. To visualize actin mRNA in cortical neurons (4 d in culture) with higher resolution than conventional epifluorescence microscopy, we took a series of optical sections (100 or 250 nm) from each cell and further processed them by using deconvolution algorithms and applying a point spread function (Fay et al., 1989). A, Localization of β-actin mRNA within a single optical section (250 nm) of a cell body and minor processes (unprocessed image). The fluorescence intensity within the neurite shaft was low. A concentration of β-actin mRNA was observed within the growth cone. B, The same image after restoration. A punctate distribution was observed throughout most of the processes. Note the concentration of β-actin mRNA within one of the growth cones (arrow). C,D) Localization of β-actin mRNA within an axon and its growth cone. The cell body is at the top of the image, and the axon extends downward, terminating in an elaborate growth cone. Note the concentration of β-actin mRNA granules within the central domain (arrow) and few granules within peripheral regions (curved arrow). Scale bar, 10 μm.
Fig. 7.
Colocalization of β-actin mRNA with microtubules. β-Actin mRNA was detected with rhodamine, and tubulin protein was detected with fluorescein; then the two processed images were superimposed. Pixels that contained both fluorochromes appearedwhite in optical sections, whereas red pixels denote probe that is not within the same pixel as anti-tubulin (green pixels). The majority of β-actin mRNA granules colocalized with microtubules (white pixels). Scale bar, 5 μm.
To visualize the punctate distribution of β-actin mRNA in neurons at a high resolution, we deconvolved the optical sections, using a point spread function derived from a latex bead (0.19 μm in diameter with attached fluorochromes). A point spread function was obtained with the 60× objective, which was the only objective used in this analysis. This process removes the out-of-focus light for each image and reassigns photons to their original point sources, resulting in higher sensitivity and resolution. This process is based on the regularization theory and is formulated to obtain an estimate of the molecular distribution inside a cell, which represents a balance between finding the estimate that represents a least-squares best fit to the data and an estimate that is smoothest, as defined by the L2norm. This algorithm has been described in considerable detail (Fay et al., 1989; Carrington et al., 1995). The estimate of molecular distribution was found by using this theory in an interactive manner; typically, satisfactory restorations took 50 iterations (further iterations produced no detectable improvement in the clarity within the image). For double-label experiments, actin mRNA was detected with Cy3, and tubulin protein was detected with fluorescein. Then the two images from the same section were superimposed. Both three-dimensional image sets were registered alongx–y–z-axes by using beads present in the mounting medium with fluorescein and rhodamine attached on the same beads, which were used as fiduciary markers to ensure a precise correspondence of pixel coordinates between the RNA and protein images.
Previous estimates of detection efficiency of nonisotopic methods used in our lab indicate that ∼60% of the actin mRNAs are being detected (Singer et al., 1989). The hybridization of multiple oligonucleotide probes to a target mRNA molecule, detected with fluorochrome-conjugated antibodies, gives a very high density of fluorescent signal that can be distinguished from nonspecific background by digital microscopy (Taneja et al., 1992). Using this method, we detected 200–400 signals (granules) within growth cones. Because the fluorescent signal of each granule is considerably above noise levels, mRNAs that are less abundant, e.g., <100 granules, also should be amenable to this technology.
Electron microscopy. Cells were washed briefly in HBSS and transferred to glutaraldehyde (2.5% in 0.1 m cacodylate, pH 7.5) for fixation at room temperature for 15 min. Samples were post-fixed with 1% osmium tetroxide, followed by uranyl acetate, dehydrated through a graded series of ethanol, and embedded in LX112 resin (Ladd Research, Burlington, VT). Ultrathin sections were cut parallel to the monolayer on a Reichert Ultracut E, with uranyl acetate, followed by Reynold’s lead citrate. Neurons were identified at low magnification with a transmission electron microscope (JEOL 1200EX) at 80 kV.
RESULTS
Enrichment of the β-actin isoform within growth cones
The use of primary neuronal cultures to study mRNA and protein localization permits direct visualization of individual cells and their complete sets of processes. Shortly after the neurons have attached to the substrate, they extend actin-rich lamellae. These lamellipodia consolidate to form a relatively symmetric array of minor neurites. Within the first 24 hr one of the minor neurites becomes significantly longer than the others and begins to assume the morphology of an axon. Within the next few days the other neurites develop the tapering and branching characteristics of dendrites. This type of culture system has been used previously to study the localization and sorting of mRNAs and cytoskeletal proteins to dendrites and/or axons and has demonstrated similar distributions to those observed in vivo (Matus et al., 1981; Kosik and Finch, 1987; Garner et al., 1988; Kleiman et al., 1990). We have described the use of cerebrocortical cultures to study the segregation of most poly(A+) mRNA to the somatodendritic compartment (Bassell et al., 1994).
To visualize β-actin and γ-actin proteins within neurons, we used isoform-specific polyclonal antibodies (Otey et al., 1986; Hoock et al., 1991) for immunofluorescence localization. In the somatodendritic compartment, β-actin labeling was highly enriched in the distal tips of minor neurites and growth cones, but only weak staining was observed within the cell body and proximal segments (Fig.1C). Within axonal growth cones, β-actin protein was highly enriched within the peripheral margin of the growth cone and a subset of filopodia (Fig.1G). In contrast to β-actin, γ-actin was distributed throughout the cell body, all processes, and filopodia (Fig.1A,E). The distribution for γ-actin resembled the distribution of phalloidin labeling throughout the cell.
Fig. 1.
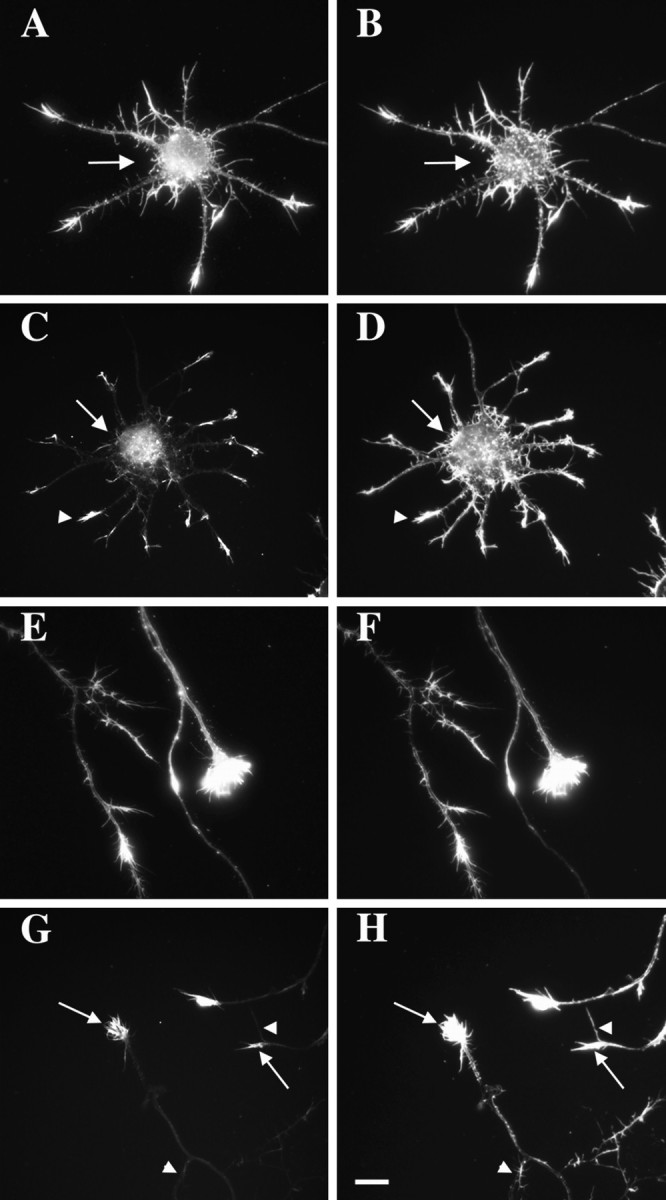
Localization of actin isoforms within cultured neurons. Cortical neurons cultured for 4 d were double-labeled with an isoform-specific antibody to β-actin or γ-actin (left column) and phalloidin–TRITC (right column). A, B, γ-Actin was distributed throughout the cell body (arrow) and neurites and resembled phalloidin staining. C,D, Localization of β-actin at tips of minor neurites (arrowhead). Low levels of β-actin were observed in the cell body (arrow). Phalloidin labeled actin filaments throughout the cell body (arrow) and minor neurites (arrowhead). E,F, γ-Actin was distributed throughout the axon and growth cone, as was phalloidin staining. G,H, β-Actin was enriched within axonal growth cones (arrows). Only weak labeling was observed in the axon shaft. Not all filopodia were labeled (arrowheads) despite the presence of F-actin (phalloidin). Scale bar, 8.5 μm.
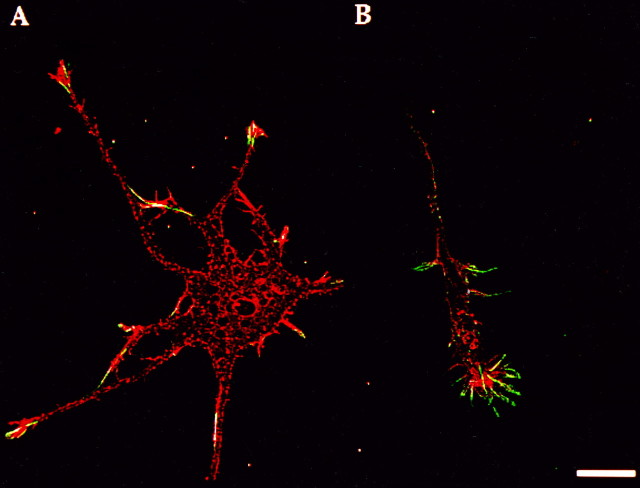
The distribution of β-actin within processes and growth cones also was studied in optical sections acquired with a cooled CCD camera. This approach permitted comparison of signal intensity differences between the cell body, the thickest region of the cell, and the comparatively thinner neuronal processes. β-Actin was not apparent within the cell body, and signal was confined solely to distal neurites and growth cones in optical sections (100 nm) (Fig.2A). In axonal growth cones (Fig. 2B) β-actin was present in the peripheral region, whereas only a weak signal was observed in the central region of the growth cone. β-Actin also was localized to filopodia that projected from other segments of the axon but was virtually absent in the axonal shaft itself (Fig.2B). β-Actin labeling overlapped in part with phalloidin staining, as indicated by the appearance of white/yellow pixels, which suggests that both labels occupy the same pixel element (100 nm each side). β-Actin labeling at the extreme ends of filopodia or distal neurites did not colocalize with phalloidin (green pixels), perhaps because of the presence of short β-actin oligomers associated with the membrane that are not labeled by phalloidin (Shuster and Herman, unpublished data) or a localized pool of unpolymerized actin (Cao et al., 1993).
Fig. 2.
Localization of β-actin protein visualized in optical sections via image processing. Cortical neurons cultured for 4 d were double-labeled with phalloidin (rhodamine) and an isoform-specific antibody to β-actin (fluorescein). Images were superimposed after restoration and then registered (see Materials and Methods). Shown here is a single optical section (250 nm) from thez-series. A, Phalloidin labeled actin filaments throughout the cell body and neurites, whereas the β-actin isoform was concentrated within the distal tips of minor processes. The overlap is indicated by the presence of white pixels. B, In distal axons, phalloidin labeling is distributed throughout the neurite and growth cone, whereas the β-actin isoform is localized to the peripheral margin; note the apparent fibrillar distribution within filopodia. Scale bar, 10 μm.
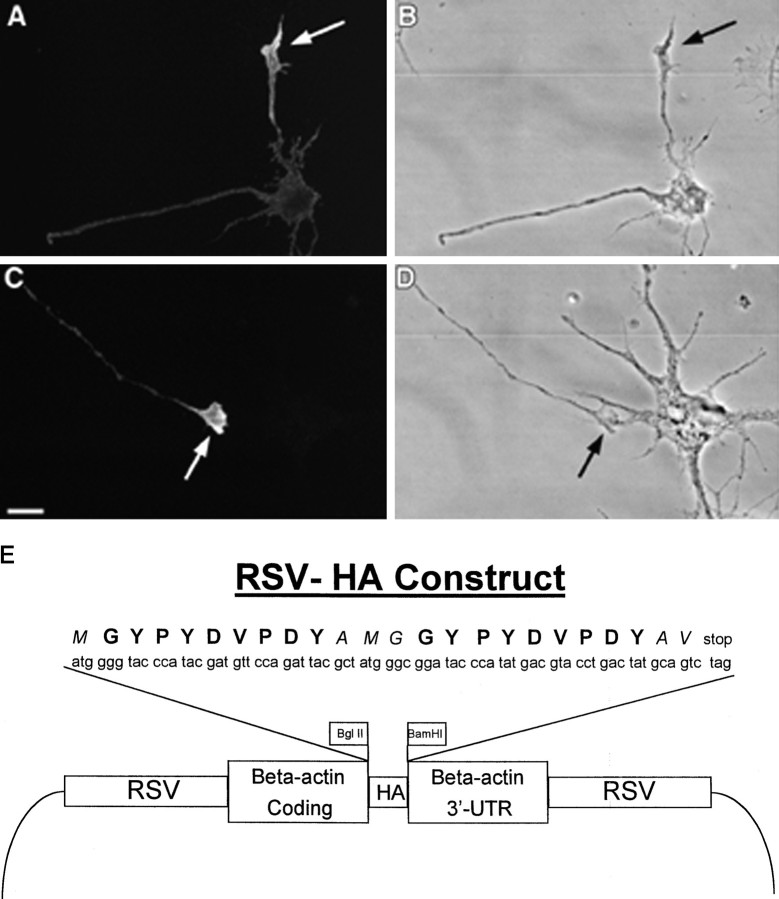
The isoactin-specific antibody used to localize β-actin recognizes the N terminus, which differs from γ-actin in six amino acids (Hoock et al., 1991). It is possible that the observed binding of the β-actin-specific antibody may not reflect the actual distribution of β-actin within the cell. Cytoplasmic actins are modified post-translationally and interact with distinct actin binding proteins (Herman, 1993; Shuster and Herman, 1995). β-Actin within growth cones could be in a modified form that is favorably recognized by the antibody. In consideration of these issues, β-actin cDNA was epitope-tagged at the C terminus with two HA sequences and transfected into cortical neurons. The antibody used to detect the chimeric β-actin recognizes the nine amino acid HA sequence. In transfected cells, HA-actin was highly localized to growth cones of both minor processes and axons and was virtually undetectable within the cell body (Fig. 3A,C).
Fig. 3.
Localization of HA–actin protein. Cortical neurons were transfected with an RSV vector containing β-actin bearing an HA epitope tag. A, Detection of HA–actin within the growth cone of a minor process (arrow).B, Differential interference contrast (DIC) optics.C, Detection of HA–actin within an axonal growth cone (arrow) near another neuron. D, DIC optics. E, Schematic drawing showing the location of the HA sequences between the coding region and the 3′-UTR. Scale bar, 10 μm.
β-Actin mRNA isoforms are preferentially localized to processes and growth cones
The ability to detect two mRNA species by using different probes allowed for direct visualization of mRNA sorting within the same cell. Considerable methodological development was required to ensure that both probes were hybridized and detected with the same efficiency. Several monoclonal and polyclonal antibodies to digoxigenin, antibodies to biotin, and avidin reagents, along with several fluorochrome-conjugated antibodies, were screened for their ability to detect poly(A+) mRNA in samples hybridized simultaneously with biotin-labeled oligo-dT (50 nt) and digoxigenin-labeled oligo-dT (50 nt) probes. This approach enabled the detection of poly(A+) mRNA by double-label fluorescence, e.g., biotin probe detected in Cy2 and digoxigenin probe detected in Cy3. The presence of several discrete foci of poly(A+) RNA in the nucleus, as well as discrete RNA granules in the cytoplasm, allowed visual assessment of whether the two detection methods resulted in morphologically similar fluorescent signals within cells (data not shown). The selected method detected digoxigenin by using monoclonal antibody conjugated to Cy3 (Jackson ImmunoResearch) and biotin by using streptavidin conjugated to Cy5 (Jackson ImmunoResearch). Another feature of the double-label in situ hybridization methodology is that the probes were chemically labeled by coupling hapten to modified amino groups in the probe (see Materials and Methods). Each probe used was exactly 50 nt in length and contained exactly five haptens. This approach limited the variability in detection efficiency, which can result from end labeling by using terminal transferase.
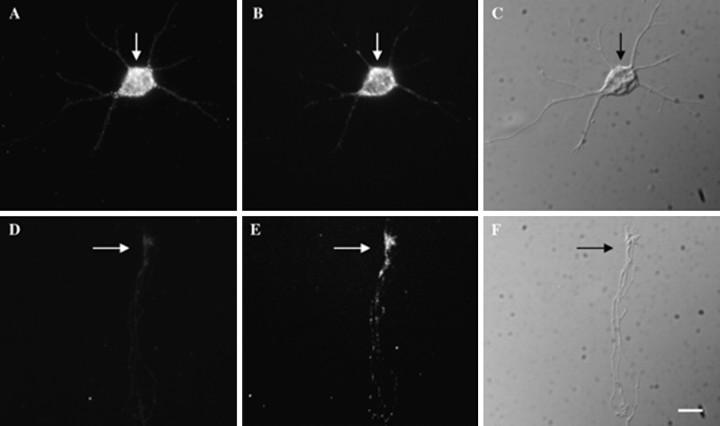
To investigate the distribution of actin mRNA isoforms within neuronal processes and/or growth cones, we labeled oligonucleotide probes to isoform-specific 3′-UTR sequences with either biotin- or digoxigenin-modified nucleotides, as described above. In addition, β- and γ-actin probes were of the same length and guanine and cytosine (GC) content. Via the double-labeling approach β- and γ-actin mRNAs exhibited similarities as well as differences in their intracellular distribution. Hybridization to both β- and γ-actin mRNA was highly punctate, suggesting the presence of RNA granules similar to that observed in other cell types (for review, see Bassell and Singer, 1997). Both mRNAs were present within the cell body (Fig.4A,B). The hybridization signal within the cell body was similar between the β- and γ-actin probes. This suggests that β- and γ-actin mRNA are of comparable abundance within the cell body. However, β-actin mRNAs frequently were localized to the cell body and processes, whereas γ-actin mRNAs were confined to the cell body (Fig.4D,E). It is unlikely that these different localization patterns could be attributable to differences in the tightness of binding of probes. As discussed above, the probes were of identical length, GC content, and hapten incorporation. This pattern also was observed with a different set of β and γ probes complementary to a more distal part of the 3′-UTR (data not shown) or when the detection scheme was reversed, e.g., β-actin probe labeled with biotin. We conclude that the presence of β, but not γ, within distal processes and growth cones reflects sorting of specific mRNA sequences within the cell.
Fig. 4.
Intraneuronal distribution of β-actin and γ-actin mRNA. Cortical neurons were cultured for 4 d, at which time most neurons have distinguishable axonal and dendritic processes.A, Hybridization of biotinated probes to γ-actin mRNA within the cell body (arrow). B, Hybridization of digoxigenin-labeled probes to β-actin mRNA within the cell body (arrow). C, Differential interference contrast (DIC) microscopy of the cell body (arrow), minor neurites, and a single axon. The axon is considerably longer than the minor neurites and cannot be photographed in entirety at this magnification. Shown here is the initial segment.D, Absence of γ-actin mRNAs from the axonal growth cone (arrow). E, Localization of β-actin mRNA within the axonal growth cone (arrow).F, DIC image of axonal growth cone (arrow) from this axon. The axon in this cell is ∼150 μm in length. Scale bar, 10 μm.
The amount of hybridization signal for β-actin varied among growth cones, and the morphology of the growth cone correlated with the extent of actin mRNA signal. A weak hybridization signal was observed in spindly growth cones that lacked a lamellar morphology (data not shown). In contrast, growth cones with a flattened lamellar morphology and defined central and peripheral regions had a strong signal. Large axonal growth cones with well spread lamella contained the strongest hybridization signal, showing even greater intensity than the cell body. Actin mRNAs frequently were detected in axonal growth cones after neurites had differentiated into axonal and dendritic processes (Fig.4E). Actin mRNA in distal axons and growth cones was often >100 μm from the cell body (Fig. 4E). Lamellar growth cones of minor neurites, which had not differentiated into dendrites, also were observed to contain actin mRNA. In contrast, actin mRNAs were not detectable within axons or dendrites beyond the proximal segment after several additional days in culture, when very few growth cones were present (data not shown; also see Kaech et al., 1997). This suggests a correlation between β-actin mRNA localization and growth cone-directed neurite outgrowth.
We observed that β-actin mRNA levels within processes and growth cones could be reduced over a few hours by changing the media from N2 supplements to MEM (Fig. 5A). This brief deprivation of N2 supplements did not result in adverse effects on neuronal morphology or cytoskeletal integrity, as judged by DIC microscopy or immunofluorescence staining for actin and tubulin. The objective was to have the cells in a quiescent state and sensitive to signal transduction mechanisms, which can promote localization of β-actin mRNA. Treatment of these N2-deprived cells with db-cAMP, a membrane-permeable analog of cAMP, resulted in a dramatic efflux of β-actin mRNA into processes and growth cones. An increase of β-actin mRNA within the proximal segment of processes could be observed after 15 min in db-cAMP (Fig. 5C). An increase of β-actin mRNA within growth cones was observed as early as 30 min. After 1 hr, axonal growth cones contained levels of β-actin mRNA that exceeded their level in cells grown in N2-supplemented media (Fig.5E). These responses were elicited by >85% of the cells. An identical response was observed with forskolin treatment (data not shown). The rapid transport of β-actin mRNAs within processes was still observed by previous treatment of the cells with actinomycin D, suggesting that preexisting β-actin mRNA can be recruited into processes (data not shown). β-Actin mRNAs transported into processes may have originated from the nucleus and/or cell body. γ-Actin mRNAs were not observed to be transported into processes during db-cAMP treatment, which provides further evidence for the hypothesis that the localization of mRNAs from the cell body into processes is a sequence-specific pattern unlikely due to leakage of mRNAs from the cell body (Fig. 5G).
Fig. 5.
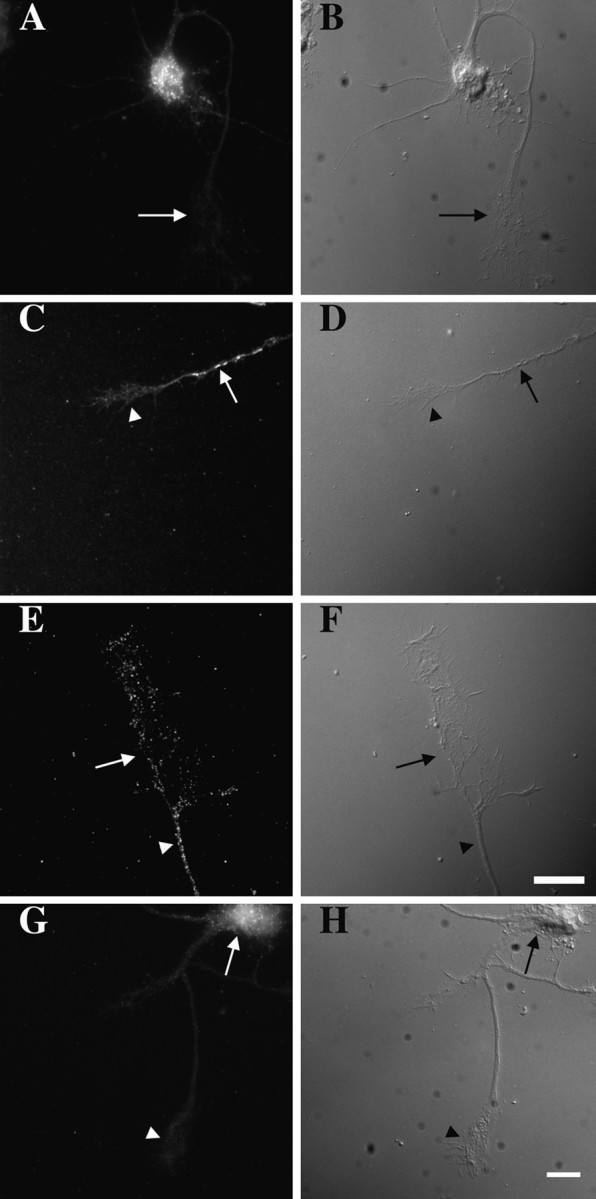
Transport of β-actin mRNA into processes after treatment with db-cAMP. Cortical neurons cultured for 4 d in N2 supplements were transferred to MEM for 3 hr. Cells were fixed and hybridized with digoxigenin-labeled probes specific to β-actin mRNA. Probes were detected by using fluorochrome-conjugated antibodies, and images were acquired with a cooled CCD camera (see Materials and Methods). A, β-Actin mRNA was detected in the cell body, but the signal no longer was observed in growth cones (arrow). C, After 15 min in db-cAMP, β-actin mRNA granules were observed in processes (arrow) but were not yet detectable within growth cones (arrowhead). Shown here is a signal within an axonal process. E, After 1 hr, β-actin mRNA granules were observed within growth cones. Shown here is a hybridization signal within the distal axon (arrowhead) and growth cone (arrow). G, γ-Actin mRNA was confined to the cell body (arrow). No signal was observed within the axonal growth cone (arrowhead). Shown here is a cell after 1 hr of treatment with db-cAMP. B, D, F, H, DIC optics. Scale bar, 10 μm.
The distribution of β-actin mRNA within processes is in granules that correlate spatially with microtubules
To visualize the punctate distribution of β-actin mRNA in neurons at a high resolution and sensitivity, we used digital imaging microscopy (DIM) to capture a series of optical sections and restore them by the reassignment of photons to their original point sources (Fay et al., 1989; Carrington et al., 1995). This approach revealed the punctate or granular nature of actin mRNA. These granules were present in the cell body and extended into distal neurites and growth cones. Also, these granules were present in the cell body and extended into distal tips of minor neurites (Fig.6A,B). The density of β-actin mRNA granules within distal tips and growth cones was of comparable visual intensity to the cell body, and there was no evidence that the signal decreased in a proximodistal gradient. On the contrary, the signal within growth cones often exceeded signal intensity within the cell body. Within larger axonal growth cones, β-actin mRNA granules were concentrated within the central region (Fig.6D, arrow), although a few granules also were observed in the peripheral margin (Fig. 6D, curved arrow). The size and quantity of actin RNA granules within growth cones were estimated by DIM analysis. The majority of granules (52%) occupied a volume of less than three voxels (100 nm on a side). Large axonal growth cones contained between 300 and 500 actin RNA granules (Fig. 6D, showing one optical section).
β-Actin mRNA granules within growth cones colocalized with tubulin protein, as suggested by the presence of white pixels after FITC (tubulin) and TRITC (mRNA) images were superimposed (Fig.7). Because both β-actin mRNA and microtubules are heavily concentrated within the central region of the growth cone, it is possible to conclude inappropriately that there is a nonrandom association. One could argue that the mere density of both signals in this region could result fortuitously in a coincidence between fluorochromes. We therefore used image processing to measure the distance between each granule and the microtubule and performed a statistical analysis that compared the observed signals with randomized signals. After image acquisition and restoration, the images were thresholded to separate the signal from the background. The mRNA granules were converted into individual voxels by thresholding (defining minimum pixel intensity) the mRNA image and then replacing each RNA granule by one voxel at its brightest location. This reduced the size of each actin RNA granule to 100 nm on a side (Fig. 7). The microtubule data also were thresholded to remove background fluorescence, resulting in the appearance of more distinct filament-like structures. We then calculated the distance from each actin mRNA voxel to the nearest voxel in the microtubule image. A histogram was produced that described the shortest measured distance between an mRNA granule and the nearest microtubule (Fig.8). The majority of actin mRNA granules (77%) occupied the same or adjacent voxel as tubulin (Figs. 7, 8). Actin mRNA granules were observed less frequently at distances further away from microtubules. Granules that were located within the peripheral margin of the growth cone were frequently not within the same pixel as tubulin (red granules). The quantitative colocalization analysis suggested that a majority of granules that occupied the same voxel or adjacent voxel as tubulin were <200 nm from the microtubule.
Fig. 8.
Distance between β-actin mRNA granules and microtubules, as compared with randomized signals. The distance of β-actin mRNA (brightest voxels) to the nearest tubulin voxel was compared with a randomized distribution. This analysis was performed on a three-dimensional data set from 100 nm optical sections. The mean and SD of the random distribution are shown. The observed distribution of β-actin mRNA is significantly closer to the microtubules than a random distribution.
Then the distance between actin mRNA granules and microtubules within the growth cone was compared with the distance between randomly distributed signals and microtubules (Fig. 8). The total three-dimensional data set of actin mRNA signals was randomized within the same volume occupied by actin mRNA (including central and peripheral regions of growth cones). Ninety-nine such random sets were created and analyzed. This produced 99 histograms, which yielded a measure of the statistical variability inherent in the histogram measurements. The difference of each histogram from an average reference histogram was calculated by computing the sum of the squares of the differences between the two curves. The actual data histogram had a larger difference than did the histograms from randomly distributed signals. Of these randomly distributed signals 24% was localized within the same or adjacent voxel as tubulin, compared with 77% for β-actin RNA. This statistical analysis demonstrated, with 99% confidence, that β-actin mRNA granules were significantly closer to microtubules in growth cones than randomly positioned signals (Fig.8). Therefore, despite the high density of microtubules within the growth cone, randomly distributed signals frequently were found in voxels that did not contain anti-tubulin and were at least 200 nm away from the microtubule. This method of statistical analysis was applied to three cells imaged in three-dimensional format for actin mRNA and tubulin protein. A similar type of nearest neighbor analysis and comparison to randomized signals was done previously to show that the majority of poly(A+) mRNA colocalized with microfilaments in fibroblast cells (Taneja et al., 1992).
Various controls were taken to avoid overestimation of the cell volume during randomization. The total actin RNA three-dimensional data set that was obtained from 20 sections was randomized within the central 10 sections. This reduced the cell volume by 50%, which would ensure that randomized signals would be further restricted to the region with the highest microtubule content, yielding a conservative statistical analysis (Fig. 8). As another control for variations in cell volume, the statistical analysis also was performed with cell border tracings, which were decreased by four pixels. Even when the RNA signals were randomized only within the central region of the growth cone (not permitted to enter the microtubule-deficient peripheral region), the distribution of randomly distributed RNA still did not colocalize with microtubules under these volume restrictions.
We also analyzed RNA and tubulin colocalization over a wide range of threshold values. The lowest threshold value was chosen subjectively such that any lower threshold would include too much background, connecting objects when they should not be connected. The highest threshold was one such that objects started to disappear from the image. The data pair discussed in this paper were examined at 49 different threshold combinations. All threshold combinations showed (to a 99% confidence level) that the mRNA distribution was not random with respect to the distribution of microtubules. Similar statistical analyses have been applied to other biological systems (Taneja et al., 1992).
Spatial correlation of β-actin mRNA with protein synthesis components in growth cones
Cortical neurons cultured for 4 d on coverslips were fixed in glutaraldehyde and processed for electron microscopy (see Materials and Methods). Analysis of thin sections revealed the presence of polyribosomes within growth cones (Fig.9). It was frequently possible to identify a particular neurite as an axon on the basis of its long and thin morphology. Polyribosomes were more prominent just at the end of the axon or within the growth cone palm (Fig. 9) and rarely were observed within the peripheral region or filopodia. Polyribosomes at the end of the process frequently were juxtaposed to microtubules, as were mitochondria. Polyribosomes within the growth cone palm frequently were observed adjacent to microtubules and only rarely were associated with other cytoskeletal structures at the base of filopodia, possibly microfilaments. It was not possible to visualize polyribosomes afterin situ hybridization and immunogold labeling because of the loss of ultrastructural morphology resulting from formamide treatment. However, it was possible to colocalize β-actin mRNA with components of the polyribosomal complex via digital imaging microscopy. β-Actin mRNA granules colocalized with the punctate labeling of EF1α and ribosomal proteins (Fig. 10), suggesting that RNA granules are associated with translational components. The distribution, spacing, and morphology of groups of granules mirrored the pattern of EF1α (Fig. 10A,B, arrows) or 60S ribosomal proteins (Fig. 10C,D, arrows), suggesting a direct correlation. Some of the punctate label for ribosomal proteins and elongation factor did not colocalize with actin RNA, perhaps suggestive of polyribosomes that encode other mRNA species (Fig. 10, arrowheads).
Fig. 9.
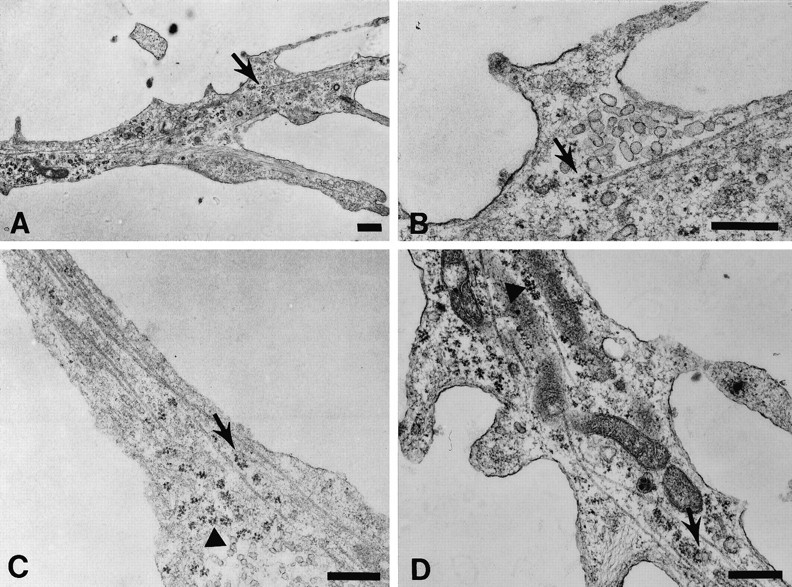
Ultrastructural visualization of polyribosomes within growth cones. Cortical neurons were grown on coverslips and processed for thin sectioning parallel to the monolayer, as described in Materials and Methods. A, An axonal growth cone viewed at low magnification. Polyribosomes are observed in the distal segment of the axon and central region of the growth cone. An area of interest (arrow) adjacent to filopodia is photographed at higher magnification in B to visualize the proximity of a polyribosome to a microtubule (arrow).C, Central region of a growth cone from another neuron that contains polyribosomes in clusters (arrowhead) and between microtubules (arrow). D, A large axonal growth cone from a third cell with polyribosomes near microtubules (arrowhead) and in clusters between microtubules (arrow). Note other membranous organelles and mitochondria. Scale bar, 0.5 μm.
Fig. 10.
Colocalization of β-actin mRNA and translational components. Shown are double labeling for β-actin mRNA (rhodamine) and EF1α or 60S ribosomes (fluorescein).A, EF1α and (B) β-actin mRNA in the distal field of an axonal process and its terminal branches. Punctate fluorescence for EF1α colocalized with β-actin mRNA granules (arrows). Image processing indicated that β-actin mRNA and EF1α occupied the same pixel coordinates. Note the identical spacing between the punctate distribution patterns in both rhodamine (β-actin mRNA) and fluorescein (EF1α). A granule containing EF1α does not colocalize with β-actin mRNA (arrowheads). C, D, Ribosomal subunit (60S) and actin mRNA in an axonal growth cone.C, Cluster of four granules that contain 60S protein (arrow) and (D) actin mRNA (arrow). A granule containing the 60S subunit does not contain actin mRNA (arrowheads). Scale bar, 5 μm.
DISCUSSION
Actin mRNA and protein isoform localization
The β-actin isoform was shown to be highly enriched within growth cones and filopodia of developing dendritic and axonal processes. These results were obtained by using both an isoform-specific antibody to β-actin and transfection of epitope-tagged β-actin. The localization of γ-actin, in contrast, was not preferentially sorted away from the perikarya and was distributed throughout the neuron. These results suggest that growth cones may be enriched for microfilaments composed of the β-isoform and imply that a sorting mechanism is necessary to achieve this spatial segregation. Both protein and mRNA transport mechanisms may function to localize newly synthesized β-actin to distal neurites and growth cones. One mechanism could be transport of actin monomers or small oligomers after their synthesis in the cell body (Okabe and Hirokawa, 1990). Because β-actin mRNA within the neuron was observed in the cell body as well as in the processes, a population of β-actin protein could be transported into the neurite post-translationally. One then would have to assert that the inability to detect β-actin protein in the cell body or proximal neurite occurred because the transported form of actin was inaccessible to both types of antibodies used (β-actin, HA tag). In this model the localization of β-actin mRNA into processes would provide a second mechanism for cytoskeletal delivery. An alternative model is that β-actin mRNAs are not translated until they leave the cell body. They could be translated during transport or, perhaps, not until they reach the growth cone, permitting the enrichment of the β-actin isoform within the peripheral margin of growth cones. An example of this type of mechanism has been demonstrated in Drosophila oocytes, wherenos mRNA is translated when it is localized at the posterior pole and not from unlocalized RNA distributed throughout the oocyte (Gavis and Lehman, 1992). Sequences within the 3′-UTR have been implicated in localization-dependent translation (Gavis and Lehman, 1992).
Future work will clarify how protein transport and mRNA transport mechanisms contribute to the sorting of β-actin protein to growth cones; at the present time it is unclear as to their individual contributions. In this report we present the first evidence for the presence of β-actin mRNAs within developing dendritic and axonal growth cones. Growth cones were shown to contain polyribosomes, providing morphological evidence that protein synthesis does occur. Under digital imaging microscopy, β-actin mRNA within growth cones colocalized with components of the polyribosomal complex, suggesting the local synthesis of β-actin protein. The observed localization of β-actin mRNA into growth cones could not be attributed to diffusion or the presence of a “leaky barrier” of mRNA from the cell body for the following reasons. First, β-actin mRNAs demonstrated a statistically nonrandom association for microtubules within growth cones. Second, the localization of actin mRNA into growth cones was a sequence-specific pattern. β-Actin mRNA was observed in growth cones, whereas γ-actin mRNA was restricted to the cell body. Furthermore, γ-actin mRNA transport was not affected by db-cAMP as was β-actin, further suggestive of sequence specificity in the RNA localization signal. Sequence differences in the untranslated regions between γ- and β-actin mRNAs could be involved in mediating this response. The 3′-UTR has been shown to confer differences between β- and α-cardiac actin mRNAs in developing muscle cultures (Kislauskis et al., 1993).
β-actin mRNAs have been shown previously to be localized to the leading edge of myoblasts, whereas cardiac and γ-actin mRNAs are perinuclear (Hill and Gunning, 1993; Kislauskis et al., 1993). The enrichment of β-actin at the cell periphery is consistent with a specific function for this isoform in regions of motile cytoplasm (Hoock et al., 1991; Hill and Gunning, 1993). β-Actin may be the predominant isoform at submembranous sites, where it binds ezrin, and β-actin polymerization dynamics could be sensitive to signaling events across the membrane (Shuster and Herman, 1995). In neurons, immunohistochemical localization of β- and γ-actin in tissue sections of developing rat brain has shown that β-actin protein is specific to growing axons and is deplete in mature neurons (Weinberger et al., 1996). Actin and tubulin have been found in a cDNA library from squid axoplasm (Kaplan et al., 1992) and more recently in biochemical fractions of rat sympathetic axons (Olink-Coux and Hollenbeck, 1996). The axonal synthesis of actin was demonstrated by analysis of proteins synthesized after radiolabeled amino acid incubation (Koenig and Adams, 1982; Koenig, 1989, 1991). Here we report on the intracellular localization of mRNA and protein isoforms within processes and growth cones of differentiating mammalian neurons growing in culture.
RNA granules and microtubules
The distribution of β-actin mRNA within processes and growth cones was highly granular. In situ hybridization studies have revealed similar nonhomogeneous patterns for a variety of mRNAs, including “punctate” actin mRNA (Sundell and Singer, 1990), “island-like structures” of Xlsirt RNA (Kloc et al., 1993), formation of bicoid RNA “particles” (Ferrandon et al., 1994), and “granules” of myelin basic protein mRNA (Ainger et al., 1993). RNA granules in oligodendrocytes and neurons colocalized with translational components, suggesting the presence of a supramolecular complex that could contain many mRNA molecules (Barbarese et al., 1995;Knowles et al., 1996).
In neurons, microtubules play a dominant role in the maintenance of poly(A+) mRNA within neuronal processes (Bassell et al., 1994) and in the transport of RNA granules containing poly(A+) mRNA (Knowles et al., 1996). These results obtained in neuronal cells are in contrast to the predominant involvement of microfilaments in both the transport and anchoring phases of β-actin mRNA from the nucleus to the fibroblast lamellae (Sundell and Singer, 1991), which have structural and functional similarities to neuronal growth cones. In this study we show an association of β-actin mRNA with microtubules within growth cones. Although we cannot discriminate between those mRNAs that are being transported versus those that have been anchored, it is likely that microtubules are involved in some component of β-actin mRNA localization in neurons. It is possible that β-actin mRNA granules can interact with both microfilaments and microtubules and that the preferential usage of a particular filament system is characteristic of the specific cell type. This mechanism could involvecis-acting elements within the actin mRNA 3′-UTR that interact with a set of cell type-specific RNA binding proteins and/or motor proteins that promote preferential usage on one filament system (Bassell and Singer, 1997).
Implications of localized β-actin synthesis
Actin protein within growth cones is highly dynamic, and there is rapid exchange between monomer and F-actin (Okabe and Hirokawa, 1990,1991; Sanders and Wang, 1991). Actin polymerization within growth cones proceeds by the addition of monomer at the barbed end adjacent to the plasma membrane, which is followed by a retrograde flow of actin and its disassembly at the rear (Small et al., 1978; Forscher and Smith, 1988; Okabe and Hirokawa, 1991). Because these filaments are relatively short, the amount of time a monomer spends in this cycle could be <1 hr, inferred from measurements that use fluorescence recovery after photobleaching (Okabe and Hirokawa, 1991). Once actin filaments are disassembled in the central region of the growth cone, monomers could diffuse away, become recycled, or be incorporated into the cortical cytoskeleton of the axonal shaft (Okabe and Hirokawa, 1991; Sanders and Wang, 1991). This dynamic cycle may necessitate a local mechanism to replenish the G-actin pool.
Quantitative imaging analysis with deconvolution of optical sections estimated that there were between 300 and 500 actin RNA granules within a large axonal growth cone. Because the granules may contain more than a single actin mRNA, it is possible that the number of actin mRNA molecules is considerably higher. If ribosomes are spaced apart by 15 nucleotides and move at a rate of five amino acids per second (Darnell, 1995), we estimate that one actin monomer could be synthesized per second per mRNA (Kislauskis et al., 1997). Therefore, 30,000 actin monomers could be synthesized locally per minute (assuming one mRNA per granule). If one assumes that the growth cone may contain 107 molecules (actin concentration of 50 μm; growth cone volume of 1 pl), local synthesis could contribute 10% of the total actin content in 1 hr. This number would be higher if granules contain more than one mRNA molecule (Barbarese et al., 1995; Knowles et al., 1996). Therefore, it is reasonable to expect that maximal rates of actin polymerization during stimulated process outgrowth could depend on local synthesis. This hypothesis has been tested in fibroblasts, where cells having localized β-actin mRNA were more motile, and delocalization of β-actin mRNA from lamellae could reduce motility (Kislauskis et al., 1997). The active transport of β-actin mRNA into growth cones, perhaps in response to physiological signals, may play a role in the localization of specific cytoskeletal proteins to growth cones during process outgrowth. This mechanism may be as essential to neuronal asymmetry as the post-translational transport of cytoskeletal complexes from the perikarya.
Footnotes
This work was supported by a Basil O’Connor award, March of Dimes Foundation, to G.J.B., French Foundation for Alzheimer’s Research, to G.J.B., and American Health Assistance Foundation, to G.J.B. and K.S.K. We thank Frank Macaluso of the Analytical Imaging Facility, Albert Einstein College of Medicine, for sample processing and thin sectioning, Jeanette Bulinski for providing antibody to γ-actin, Wim Moller for providing antibody to EF1α, and John Hesketh for providing antibody to ribosomal protein. We thank Mary Weitzman for literature searches.
Correspondence should be addressed to Dr. Gary Bassell, Department of Anatomy and Structural Biology, Albert Einstein College of Medicine, 1300 Morris Park Avenue, Bronx, NY 10461.
REFERENCES
- 1.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous MBP mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamburg JR, Bray D. Distribution and cellular localization of actin depolymerization factor. J Cell Biol. 1987;105:2817–2825. doi: 10.1083/jcb.105.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbarese E, Koppel DE, Deutscher MP, Smith CL, Ainger K, Morgan F, Carson JH. Protein translation components are colocalized in granules in oligodendrocytes. J Cell Sci. 1995;108:2781–2790. doi: 10.1242/jcs.108.8.2781. [DOI] [PubMed] [Google Scholar]
- 4.Bassell GJ, Singer RH. mRNA and cytoskeletal filaments. Curr Opin Cell Biol. 1997;9:109–115. doi: 10.1016/s0955-0674(97)80159-7. [DOI] [PubMed] [Google Scholar]
- 5.Bassell GJ, Singer RH, Kosik KS. Association of poly(A+) mRNA with microtubules in cultured neurons. Neuron. 1994;12:571–582. doi: 10.1016/0896-6273(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 6.Bridgman PC, Dailey ME. The organization of myosin and actin in rapid frozen nerve growth cones. J Cell Biol. 1989;108:95–109. doi: 10.1083/jcb.108.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown CW, McHugh KM, Lessard JL. A cDNA sequence encoding cytoskeletal gamma actin from rat. Nucleic Acids Res. 1990;18:5312–5317. doi: 10.1093/nar/18.17.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao LG, Fishkind DJ, Wang Y. Localization and dynamics of nonfilamentous actin in cultured cells. J Cell Biol. 1993;123:173–181. doi: 10.1083/jcb.123.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrington WA, Lynch RM, Moore ED, Isenberg G, Fogarty KE, Fay FS. Superresolution three-dimensional images of fluorescence in cells. Science. 1995;268:1483–1486. doi: 10.1126/science.7770772. [DOI] [PubMed] [Google Scholar]
- 10.Cheng H, Bjerknes M. Asymmetric distribution of actin mRNA and cytoskeletal pattern generation in polarized epithelial cells. J Mol Biol. 1989;210:541–549. doi: 10.1016/0022-2836(89)90130-7. [DOI] [PubMed] [Google Scholar]
- 11.Crino PB, Eberwine J. Molecular characterization of the dendritic growth cone. Neuron. 1997;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 12.Darnell JE, Lodish H, Baltimore D, Berk A, Zipursky SL, Matsudaira P. Molecular cell biology (Greeman WH, ed). Scientific American; New York: 1995. [Google Scholar]
- 13.Davis L, Ping D, DeWitt M, Kater SB. Protein synthesis within neuronal growth cones. J Neurosci. 1992;12:4867–4877. doi: 10.1523/JNEUROSCI.12-12-04867.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deitch JS, Banker GA. An electron microscopic analysis of hippocampal neurons developing in culture: early stages in the emergence of polarity. J Neurosci. 1993;13:4301–4315. doi: 10.1523/JNEUROSCI.13-10-04301.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans LL, Hammer J, Bridgman PC. Subcellular localization of myosin V in nerve growth cones and outgrowth from dilute lethal neurons. J Cell Sci. 1997;11:430–449. doi: 10.1242/jcs.110.4.439. [DOI] [PubMed] [Google Scholar]
- 16.Fay FS, Carrington W, Fogarty KE. Three-dimensional molecular distribution in single cells analyzed using the digital imaging microscope. J Microsc. 1989;153:133–149. [PubMed] [Google Scholar]
- 17.Ferrandon D, Elphick L, Nusslein-Volhard C, St. Johnson D. Staufen protein associates with the 3’ UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 18.Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garner CC, Tucker RP, Matus A. Selective localization of mRNA for MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 20.Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71:301–313. doi: 10.1016/0092-8674(92)90358-j. [DOI] [PubMed] [Google Scholar]
- 21.Gordon-Weeks PR. The cytoskeletons of isolated neuronal growth cones. Neuroscience. 1987;21:977–989. doi: 10.1016/0306-4522(87)90052-2. [DOI] [PubMed] [Google Scholar]
- 22.Goslin K, Banker G. Culturing nerve cells. MIT; London: 1991. [Google Scholar]
- 23.Herman IM. Actin isoform diversity and function. Curr Opin Cell Biol. 1993;5:48–56. doi: 10.1016/s0955-0674(05)80007-9. [DOI] [PubMed] [Google Scholar]
- 24.Herman IM, D’Amore PA. Microvascular pericytes contain muscle and nonmuscle actins. J Cell Biol. 1985;101:43–52. doi: 10.1083/jcb.101.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill MA, Gunning P. Beta and gamma actin mRNAs are differentially located within myoblasts. J Cell Biol. 1993;122:825–832. doi: 10.1083/jcb.122.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoock TC, Newcomb PM, Herman IM. Beta actin and its mRNA are localized at the plasma membrane and the regions of moving cytoplasm during the cellular response to injury. J Cell Biol. 1991;112:653–664. doi: 10.1083/jcb.112.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horne S, Hesketh J. Immunological localization of ribosomes in striated rat muscle. Biochem J. 1990;268:231–236. doi: 10.1042/bj2680231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaech S, Kim JB, Cariola M, Ralston E. Improved lipid mediated gene transfer into primary cultures of hippocampal neurons. Mol Brain Res. 1996;233:66–69. doi: 10.1016/0169-328x(95)00238-n. [DOI] [PubMed] [Google Scholar]
- 29.Kaech S, Fischer M, Doll T, Matus A. Isoform specificity in the relationship of actin to dendritic spines. J Neurosci. 1997;17:9565–9572. doi: 10.1523/JNEUROSCI.17-24-09565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaplan B, Gioio AE, Perrone-Capano C, Crispino M, Giuditta A. β-Actin and β-tubulin are components of a heterogeneous mRNA population present in the giant squid axon. Mol Cell Neurosci. 1992;3:133–144. doi: 10.1016/1044-7431(92)90017-v. [DOI] [PubMed] [Google Scholar]
- 31.Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3′-untranslated sequences sort α-cardiac and β-cytoplasmic actin messenger RNAs to different cytoplasmic compartment. J Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kislauskis EH, Zhu X, Singer RH. β-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kleiman R, Banker G, Steward O. Differential subcellular localization of particular mRNAs in hippocampal neurons in culture. Neuron. 1990;5:821–830. doi: 10.1016/0896-6273(90)90341-c. [DOI] [PubMed] [Google Scholar]
- 35.Kloc M, Spohr G, Etkin LD. Translocation of repetitive RNA sequences with the germ plasm in Xenopus oocyte. Science. 1993;262:1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- 36.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16:7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koenig E. Cycloheximide-sensitive methionine labeling of proteins in goldfish retinal ganglion cell axons in vitro. Brain Res. 1989;481:119–123. doi: 10.1016/0006-8993(89)90491-5. [DOI] [PubMed] [Google Scholar]
- 38.Koenig E. Evaluation of local synthesis of axonal proteins in the goldfish Mauthner cell axon and axons of dorsal and ventral roots of the rat in vitro. Mol Cell Neurosci. 1991;2:384–394. doi: 10.1016/1044-7431(91)90025-j. [DOI] [PubMed] [Google Scholar]
- 39.Koenig E, Adams P. Local protein synthesizing activity in axonal fields regenerating in vitro. J Neurochem. 1982;39:386–400. doi: 10.1111/j.1471-4159.1982.tb03960.x. [DOI] [PubMed] [Google Scholar]
- 40.Kosik KS, Finch EA. MAP2 and tau segregate into dendritic and axonal domains after the elaboration of morphologically distinct neurites: an immunocytochemical study of cultured rat cerebrum. J Neurosci. 1987;7:3142–3153. doi: 10.1523/JNEUROSCI.07-10-03142.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lasek RJ. Polymer sliding in axons. J Cell Sci. 1986;5:161–179. doi: 10.1242/jcs.1986.supplement_5.10. [DOI] [PubMed] [Google Scholar]
- 42.Letourneau PC, Shattuck TA. Distribution of actin-associated proteins in growth cones. Development. 1989;105:509–519. doi: 10.1242/dev.105.3.505. [DOI] [PubMed] [Google Scholar]
- 43.Marsden K, Doll T, Ferralli J, Botteri F, Matus A. Transgenic expression of embryonic MAP2 in adult mouse brain: implications for neuronal polarization. J Neurosci. 1996;16:3265–3273. doi: 10.1523/JNEUROSCI.16-10-03265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matus A, Bernhardt R, Hugh-Jones T. High molecular weight MAPs are preferentially associated with dendritic microtubules in brain. Proc Natl Acad Sci USA. 1981;78:3010–3014. doi: 10.1073/pnas.78.5.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nixon RA. The axonal transport of cytoskeletal proteins: a reappraisal. In: Bisby MA, Smith RS, editors. Axonal transport. Liss; New York: 1987. pp. 175–200. [Google Scholar]
- 46.Nudel U, Zakut R, Shani M, Neuman S, Levy Z, Yaffe D. The nucleotide sequence of rat cytoplasmic beta actin gene. Nucleic Acids Res. 1983;11:1759–1771. doi: 10.1093/nar/11.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okabe S, Hirokawa N. Turnover of fluorescently labeled tubulin and actin in the axon. Nature. 1990;343:479–482. doi: 10.1038/343479a0. [DOI] [PubMed] [Google Scholar]
- 48.Okabe S, Hirokawa N. Actin dynamics in growth cones. J Neurosci. 1991;11:1918–1929. doi: 10.1523/JNEUROSCI.11-07-01918.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons. J Neurosci. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otey CA, Lessard JL, Bulinski JC. Immunolocalization of the gamma isoform of nonmuscle actin. J Cell Biol. 1986;102:1732–1737. doi: 10.1083/jcb.102.5.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sabry J, O’Connor TP, Kirschner MW. Axonal transport of tubulin in Ti1 pioneer neurons. Neuron. 1995;14:1247–1256. doi: 10.1016/0896-6273(95)90271-6. [DOI] [PubMed] [Google Scholar]
- 52.Sanders MC, Wang Y-L. Assembly of actin-containing cortex occurs at distal regions of growing neurites in PC12 cells. J Cell Sci. 1991;100:771–780. doi: 10.1242/jcs.100.4.771. [DOI] [PubMed] [Google Scholar]
- 53.Sanders J, Brandsma M, Janssen GM, Dijk J, Moller W. Immunofluorescence studies of human fibroblasts demonstrate the presence of the complex of EF1α in the rough endoplasmic reticulum. J Cell Sci. 1996;109:1113–1117. doi: 10.1242/jcs.109.5.1113. [DOI] [PubMed] [Google Scholar]
- 54.Shuster CB, Herman IM. Indirect association of ezrin with F-actin: isoform specificity and calcium sensitivity. J Cell Biol. 1995;128:837–848. doi: 10.1083/jcb.128.5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singer RH, Langevin GL, Lawrence JB. Ultrastructural visualization of cytoskeletal mRNAs and their associated proteins using double label in situ hybridization. J Cell Biol. 1989;108:2343–2353. doi: 10.1083/jcb.108.6.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Small JV, Isenberg G, Celis JE. Polarity of actin at the leading edge of cultured cells. Nature. 1978;272:638–639. doi: 10.1038/272638a0. [DOI] [PubMed] [Google Scholar]
- 57.Sundell CL, Singer RH. Actin mRNA localizes in the absence of protein synthesis. J Cell Biol. 1990;111:2397–2403. doi: 10.1083/jcb.111.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin mRNAs. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- 59.Takeda S, Hirokawa N. Tubulin dynamics in neuronal axons. Neuron. 1995;14:1257–1264. doi: 10.1016/0896-6273(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka E, Sabry J. Cytoskeletal rearrangements during growth cone guidance. Cell. 1995;83:171–176. doi: 10.1016/0092-8674(95)90158-2. [DOI] [PubMed] [Google Scholar]
- 61.Taneja KL, Lifshitz M, Fay FS, Singer RH. Poly(A+) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J Cell Biol. 1992;119:1245–1260. doi: 10.1083/jcb.119.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Von Arx P, Bantle S, Soldati T, Perriard J. Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J Cell Biol. 1995;131:1759–1773. doi: 10.1083/jcb.131.6.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang FS, Wolenski JS, Cheney RE, Mooseker MS, Jay DG. Function of myosin V in filopodial extension of neuronal growth cones. Science. 1996;273:660–662. doi: 10.1126/science.273.5275.660. [DOI] [PubMed] [Google Scholar]
- 64.Weinberger R, Schevzov G, Jeffrey P, Gordon K, Hill M, Gunning P. The molecular composition of neuronal microfilaments is spatially and temporally regulated. J Neurosci. 1996;16:238–252. doi: 10.1523/JNEUROSCI.16-01-00238.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly MR, Lerner RA. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- 66.Yao X, Forte JG. Polarized distribution of actin isoforms in gastric parietal cells. Mol Biol Cell. 1995;6:541–557. doi: 10.1091/mbc.6.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]