Abstract
This article comments on:
Atkinson N, Velanis CN, Wunder T, Clarke DJ, Mueller-Cajar O, McCormick AJ. 2019. The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis-Chlamydomonas Rubisco through interactions with the algal Rubisco small subunit. Journal of Experimental Botany, 70, 5271–5285.
Keywords: carbon dioxide concentrating mechanism, carbon dioxide fixation, Chlamydomonas, EPYC1, pyrenoid, Rubisco, Yeast 2 hybrid
The chloroplast pyrenoid, an important component of the CO2 concentrating mechanism of algae, is a structure composed primarily of Rubisco. In Chlamydomonas, Rubisco in the pyrenoid is held together by the linker protein EPYC1. Atkinson et al., (2019) determined the regions of the Rubisco small subunit and EPYC1 that are important for the protein-protein interaction, thus making progress towards reconstruction of a pyrenoid in higher plants. Why is a protein soluble in one organism while its homologue in another species becomes part of a liquid-like cell structure? That is the question being addressed by Atkinson et al., (2019) in this issue of the Journal of Experimental Botany. It is even more striking when the protein is ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco), the most abundant soluble enzyme in plants and algae. In terrestrial plants, Rubisco behaves as a soluble protein found throughout the chloroplast stroma of leaf mesophyll cells. However, in most algae, Rubisco is found in a structure within the chloroplast called the pyrenoid.
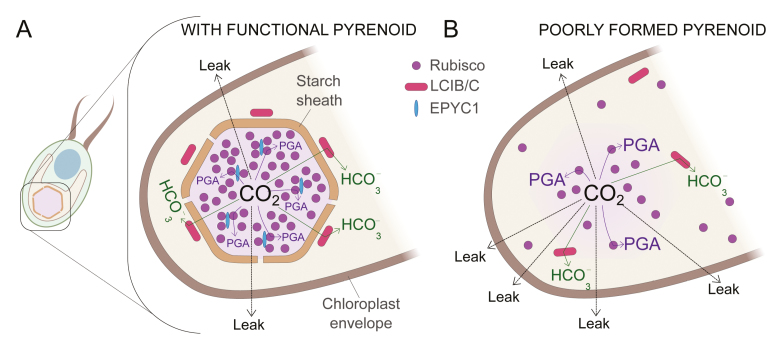
The physiological consequences of this packaging of Rubisco are profound. In general, algae with pyrenoids have a much higher affinity for inorganic carbon (Ci = CO2 + HCO3- + CO3-2) than terrestrial C3 plants. These algae are able to raise the CO2 concentration for Rubisco through the CO2 concentrating mechanism (CCM). Our current thinking is that the packaging of Rubisco is a requirement for the CCM (Mackinder, 2018; Moroney and Ynalvez, 2007; Spalding, 2008). However, since CO2 can readily cross cell membranes (Tolleter et al., 2017), how can a single-celled organism possibly concentrate CO2? Current CCM models have cells accumulating HCO3-, an ion that does not readily cross membranes, instead of CO2 directly. After taking up HCO3-, a key step in this process is the conversion of the accumulated HCO3- to CO2 by the action of the enzyme carbonic anhydrase (CA). This raises the CO2 concentration at Rubisco, which is located physically close to the CA. Then the Rubisco has a chance to fix the substrate CO2 before the CO2 diffuses away (Box 1A). If the pyrenoid is not correctly formed, the CO2 will inevitably leak out of the cell (Box 1B). Another proposed reason for the pyrenoid organization is that it facilitates the recapture of some of the CO2 as it leaks past Rubisco. In Chlamydomonas, the pyrenoid is surrounded by a starch sheath and the heteromeric protein LCIB/LCIC. The LCIB/C complex has been proposed to act as a CA, converting leaking CO2 to the less permeant HCO3-. Thus, CO2 generated by the CCM must pass through Rubisco and then a CA layer before it has a chance to leave the chloroplast (Box 1A).
Box 1.
How the pyrenoid reduces CO2 leakage in Chlamydomonas reinhardtii.A) In a functional pyrenoid, a starch sheath forms around the pyrenoid, which contains most of the Rubisco (indicated with purple). Rubisco interacts with EPYC1 (shown in blue) and that interaction aids in the formation of the pyrenoid. In this figure HCO3- uptake and its subsequent conversion to CO2 with the help of carbonic anhydrase has not been shown for the sake of simplicity. The Rubisco product glycerate-3-phosphate (PGA), shown by purple arrows, forms when CO2 concentrates in the pyrenoid. CO2 that leaks past Rubisco is sometimes recaptured and converted to HCO3- (indicated by green arrows) by LCIB/C (red), a stromal carbonic anhydrase, or exits the chloroplast entirely (dotted arrows). B) Absence of EPYC1 prevents the formation of a pyrenoid and accumulated HCO3- once converted to CO2 easily leaks out and is not recaptured.
In Chlamydomonas, the protein sequence of the Rubisco small subunit (SSU) and the protein EPYC1 (essential pyrenoid component 1) are important to the formation of the pyrenoid (Mackinder et al., 2016; Meyer et al., 2012). The first evidence came from the work of Meyer et al., (2012), who showed that Chlamydomonas cells expressing the Arabidopsis SSU instead of the Chlamydomonas SSU failed to form a pyrenoid and failed to grow in ambient CO2. However, it was interesting that the Rubisco in these cells, consisting of the Chlamydomonas large subunit (LSU) and Arabidopsis SSU, was still enzymatically active. Meyer et al., (2012) also identified regions in the Rubisco SSU necessary for pyrenoid formation. Replacing specific Chlamydomonas SSU α-helices with the corresponding sequence from plant Rubisco SSU resulted in cells without pyrenoids and with defective CCMs. In 2016, Mackinder et al. found that EPYC1 was also required for pyrenoid formation. EPYC1 is a linker protein which binds to Rubisco in Chlamydomonas and facilitates the liquid-like formation (Küken et al., 2018; Rosenzweig et al., 2017). It is not present in terrestrial plants. Loss of EPYC1 results in Chlamydomonas cells being unable to form a normal pyrenoid and develop a functional CCM (Mackinder et al., 2016).
The work described by Atkinson et al., (2019) greatly extends these studies by investigating which regions of each protein are required for SSU-EPYC1 binding. Chlamydomonas has two genes encoding the SSUs, designated S1Cr and S2Cr. Atkinson et al. (2019) used quantitative yeast two-hybrid (Y2H) experiments to show that EPYC1 strongly interacts with both Chlamydomonas SSU homologs but not with the Arabidopsis SSU (1AAt). They then systematically replaced parts of the Chlamydomonas SSU with the corresponding regions of the Arabidopsis SSU. They found that multiple parts of the Chlamydomonas SSU contributed to the SSU-EPYC1 interactions. Substituting in the two α-helices from the Chlamydomonas SSU into the Arabidopsis SSU was essential for interaction, while adding in the β sheets or the βA-βB loop region greatly increased SSU-EPYC1 binding. Atkinson et al., (2019) repeated these Y2H experiments but with modifications of EPYC1. EPYC1 has four repeat regions with short terminus regions. They found that each of the repeated regions and the C terminus contributed to the binding of EPYC1 to the SSU. Thus, large portions of each protein were important to the strength of the protein-protein interaction. They also found that a mixture of EPYC1 and the Chlamydomonas SSU could phase separate and form liquid droplets in vitro, indicating that a large number of components may not be needed to form a pyrenoid-like structure.
The question arises: can researchers reconstruct a pyrenoid in higher plants? Photosynthesis modelling suggests that introducing algal CCM bicarbonate transporters into C3 plants and packaging Rubisco could lead to a significant increase in photosynthetic efficiency (Furbank et al., 2013; McGrath and Long, 2014; Zhu et al., 2010). Atkinson et al. (2019) took a significant step towards building a pyrenoid by expressing EPYC1 in Arabidopsis wild type plants as well as Arabidopsis plants expressing the Chlamydomonas SSU. However, no Rubisco aggregation was seen in either; instead, an even distribution of Rubisco was seen throughout the chloroplast. It was encouraging that they were able to express EPYC1 in plants, although they postulate that the amount of EPYC1 present was still too low to expect liquid phase separation to occur, which is due to EPYC1-Rubisco interactions. An in vitro assay developed by Wunder et al., (2018) showed that a critical EPYC1:Rubisco ratio is required before phase separation occurs; the EPYC1 producing plants in this paper may not have enough of the linker protein.
These results indicate that with a higher EPYC1: Rubisco expression, a packaging of Rubisco might be engineered in a C3 plant, as EPYC1 doesn’t appear to need additional proteins to bind to Rubisco in vitro. However, in vivo, other components might be required (Ma et al., 2011). Mackinder et al., (2017) showed that in Chlamydomonas, EPYC1 interacts with a protein kinase and 14-3-3 proteins, suggesting a role of phosphorylation in Rubisco-EPYC1 assembly. This finding fits with earlier studies showing that EPYC1 is a phosphoprotein (Turkina et al., 2006). The Atkinson et al., (2019) paper highlights the residues needed for EPYC1-Rubisco interaction in order to obtain a liquid-like pyrenoid. Thus, both EPYC1 and modified Rubiscos have been successfully expressed in heterologous systems of yeast and Arabidopsis by Atkinson et al., (2019) to show that the strength of the EPYC1-Rubisco interaction can be manipulated. This is a big step towards the end goal of organizing Rubisco in C3 plants into a pyrenoid-like structure. Rubisco organized in this fashion should be more efficient at fixing CO2 if a functional CCM is introduced into C3 plants. Rubisco in a pyrenoid-like structure should better capture CO2 generated by a CCM thus preventing CO2 leakage (Box 1A).
References
- Atkinson N, Velanis CN, Wunder T, Clarke DJ, Mueller-Cajar O, McCormick AJ. 2019. The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis-Chlamydomonas Rubisco through interactions with the algal Rubisco small subunit. Journal of Experimental Botany 70, 5283–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT, Caemmerer S, Price GD. 2013. CO2-concentrating mechanisms in crop plants to increase yield. Applying photosynthesis research to improvement of food crops. ACIAR Proceedings 140, 130–137. [Google Scholar]
- Küken A, Sommer F, Yaneva-Roder L, Mackinder LC, Höhne M, Geimer S, Jonikas MC, Schroda M, Stitt M, Nikoloski Z. 2018. Effects of microcompartmentation on flux distribution and metabolic pools in Chlamydomonas reinhardtii chloroplasts. eLife 7, e37960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Pollock SV, Xiao Y, Cunnusamy K, Moroney JV. 2011. Identification of a novel gene, CIA6, required for normal pyrenoid formation in Chlamydomonas reinhardtii. Plant Physiology 156, 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM, Chen C, Leib RD, et al. . 2017. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LC, Meyer MT, Mettler-Altmann T, et al. . 2016. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proceedings of the National Academy of Sciences, USA 113, 5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM. 2018. The Chlamydomonas CO2 -concentrating mechanism and its potential for engineering photosynthesis in plants. New Phytologist 217, 54–61. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Long SP. 2014. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiology 164, 2247–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MT, Genkov T, Skepper JN, Jouhet J, Mitchell MC, Spreitzer RJ, Griffiths H. 2012. Rubisco small-subunit α-helices control pyrenoid formation in Chlamydomonas. Proceedings of the National Academy of Sciences 109, 19474–19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Ynalvez RA. 2007. Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryotic Cell 6, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig ES, Xu B, Kuhn Cuellar L, et al. . 2017. The Eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH. 2008. Microalgal carbon-dioxide-concentrating mechanisms: Chlamydomonas inorganic carbon transporters. Journal of Experimental Botany 59, 1463–1473. [DOI] [PubMed] [Google Scholar]
- Tolleter D, Chochois V, Poiré R, Price GD, Badger MR. 2017. Measuring CO2 and HCO3- permeabilities of isolated chloroplasts using a MIMS-18O approach. Journal of Experimental Botany 68, 3915–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkina MV, Blanco-Rivero A, Vainonen JP, Vener AV, Villarejo A. 2006. CO2 limitation induces specific redox-dependent protein phosphorylation in Chlamydomonas reinhardtii. Proteomics 6, 2693–2704. [DOI] [PubMed] [Google Scholar]
- Wunder T, Le Hung SC, Mueller-Cajar O. 2018. Reconstitution of the liquid liquid phase separation underlying the microalgal Rubisco supercharger. Biophysical Journal 114, 61a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Long SP, Ort DR. 2010. Improving photosynthetic efficiency for greater yield. Annual Review of Plant Biology 61, 235–261. [DOI] [PubMed] [Google Scholar]