CEPR2 interacts with some PYLs to promote their phosphorylation and degradation, whereas ABA inhibits this process. Thus, CEPR2 balances the growth regulation and stress response in Arabidopsis.
Keywords: Abscisic acid, CEPR2, degradation, phosphorylation, plasma membrane, PYR/PYLs
Abstract
Pyrabactin resistance 1 (PYR1)/PYR1-like (PYL) abscisic acid (ABA) receptors have been proved to be recruited in the plasma membrane (PM). In order to explain the roles of PYR/PYLs in the PM, PYL4 was used as bait to screen the PM-localized leucine-rich repeat receptor-like kinase family, and five members were found directly interacting with PYL4. Loss of function by T-DNA insertion in C-terminally encoded peptide receptor 2 (CEPR2) together with phloem intercalated with xylem (PXY) and PXY-Like 2 (PXL2) led to ABA hypersensitivity, while CEPR2 overexpression led to ABA insensitivity compared with the wild type, indicating the redundant and negative roles of CEPR2, PXY, and PXL2 in ABA signaling. The PYL4 proteins were strongly accumulated in cepr2/pxy/pxl2 compared with the wild type. Furthermore, higher phosphorylation levels accompanied by lower protein levels of PYL4 in CEPR2 overexpression lines were observed, indicating the requirement of phosphorylation of PYLs for degradation. Subsequently, MS and in vitro kinase assays demonstrated that CEPR2 phosphorylated PYL4 at Ser54, while this phosphorylation was diminished or even eliminated in the presence of ABA. Taken together, CEPR2 promotes the phosphorylation and degradation of PYLs in unstressed conditions, whereas ABA represses this process to initiate ABA response during times of stress.
Introduction
As sessile organisms, plants need a sophisticated regulatory network to survive unfavorable and changing environments. Abscisic acid (ABA) plays a critical role in plant stress response by the transcriptional induction of defense genes in different organs (Yoshida et al., 2014; Jones, 2016; Sah et al., 2016; Vishwakarma et al., 2017). In the presence of ABA, PYR1-like (PYL) receptors bind and inhibit protein phosphatases type 2C, such as ABA-insensitive 1 (ABI1) and ABI2, resulting in the release and activation of sucrose nonfermenting-1-related protein kinase 2s (SnRK2s) (Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009; Vlad et al., 2009; Cutler et al., 2010). Upon activation, SnRK2s phosphorylate and activate a group of transcription factors that mediate the expression of many ABA-responsive genes (Kang et al., 2002; Fujii et al., 2009; Yoshida et al., 2010). Among these, ABI3, ABI4, and ABI5 are essential in the control of seed germination and seedling establishment (Lopez-Molina et al., 2001; Finkelstein et al., 2002; Shu et al., 2013; Chen et al., 2014; Shu et al., 2016).
ABA receptors pyrabactin resistance 1 (PYR1)/PYLs have recently been reported to be targeted to the plasma membrane (PM) either through association with C2-domain ABA-related proteins (CARs) or via the E3 ubiquitin ligase ring finger of seed longevity 1 (RSL1) (Bueso et al., 2014; Rodriguez et al., 2014; Diaz et al., 2016). RSL1 can ubiquitinate PYLs for degradation by the 26S proteasome pathway. Partial loss of function of several members of the RSL1-like family leads to enhanced sensitivity to ABA-mediated inhibition of seed germination (Bueso et al., 2014); alternatively, FYVE1/FREE1, a component of the endosomal sorting complex, interacts with RSL1 and recruits PYL4 to endosomal compartments. Plants in which fyve1/free1 are knocked down exhibit impaired targeting of PYL4 for vacuolar degradation (Belda-Palazon et al., 2016). In contrast, different car triple mutants deficient in CAR1, CAR4, CAR5, and CAR9 genes show reduced sensitivity to ABA in seedling establishment (Rodriguez et al., 2014). However, the exact mechanisms of PYLs targeted to the PM through association with CARs or RSL1 are still unclear. Therefore, to better understand this process, new regulators involved in the CAR–PYL complex need to be identified.
Leucine-rich repeat receptor-like kinases (LRR-RLKs) represent the largest group of RLKs in the Arabidopsis genome (225 members), and they are grouped into 13 subfamilies on the basis of the organization of LRRs in their extracytoplasmic domain (Dievart and Clark, 2003; ten Hove et al., 2011). They play critical roles in many physiological and biological processes, including hormone perception, stress response, and plant development (Dievart and Clark, 2003; Torii, 2004; ten Hove et al., 2011). For example, brassinosteroid-insensitive 1 (BRI1) (Li and Chory, 1997; He et al., 2000) and BRI1-associated receptor kinase 1 (BAK1) bind to the plant hormones brassinosteroids (Li et al., 2002; Nam and Li, 2002), while flagellin-sensitive 2 (FLS2) and xanthomonas resistance 21 (XA21) mediate the sensing of flagellin and bacteria, respectively (Song et al., 1995; Gomez-Gomez and Boller, 2000). CEPR1 and CEPR2 recognize C-terminally encoded peptides (CEPs) to up-regulate nitrate transporter genes in roots (Tabata et al., 2014).
Moreover, receptor dead kinase 1 (RDK1) has been reported to interact with ABI1 in the PM and positively regulate ABA-mediated seed germination (Kumar et al., 2017). FERONIA (FER) interacts with ABI2 in the PM and facilitates signaling crosstalk between ABA and rapid alkalinization factor (RALF) peptide in Arabidopsis (Chen et al., 2016). Given the interaction of RDK1 with ABI1, and of FER1 with ABI2, we hypothesized that some LRR-RLKs may associate with PYR/PYLs in the PM. To test this directly, we conducted yeast two-hybrid experiments using the mating-based split ubiquitin system (MbSUS), a method for detecting interactions of PM proteins with PM or non-PM proteins. There are >600 membrane-bound RLK members in Arabidopsis. LRR-RLKs, as the well-researched subgroup of RLKs, play crucial roles in plant development, hormone signaling transduction, defense responses, and plant–environment interaction, and we focus on LRR-RLKs in this study (Yamaguchi et al., 2010; Sun et al., 2013; Yang et al., 2014; Dimitrov and Tax, 2018). Here we discovered an LRR-RLK CEPR2 in Arabidopsis, which interacted with and accelerated the phosphorylation and degradation of some of PYLs, at least PYL2 and PYL4. Thereby CEPR2 negatively regulated the sensitivity of Arabidopsis to ABA.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana (L.) Heynh. cv. ‘Columbia’ was used as the wild type (WT). The T-DNA insertion lines, SALK_014533C (cepr2), SALK_112416 (pxy), and SALK_095005C (pxl2), were obtained from the Arabidopsis Biological Resource Center (http://www.arabidopsis.org). Arabidopsis plants were grown as described previously (Chen et al., 2014), and the growth conditions of Arabidopsis in the greenhouse were 16 h light/8 h dark, 120 µmol m−2 s−1, 23 °C. The single, double, or triple mutants, cepr2, pxy, pxl2, cepr2/pxy, cepr2/pxl2, pxy/pxl2, and cepr2/pxy/pxl2, were analyzed by PCR using the primers listed in Supplementary Table S1 at JXB online. The transgenic plants containing 35S::CEPR2 or 35S::CEPR2-GFP were selected on 1/2 Murashige and Skoog (MS) medium (1% sucrose and 0.85% agar) supplemented with 50 mg l–1 kanamycin and confirmed by quantitative real-time PCR (qRT-PCR).
Root length measurements
Plants of different genotypes were grown under the same conditions in the greenhouse; the seeds were collected at the same time. For each comparison, seeds were planted on the same plate containing 1/2 MS medium with or without 1 µM ABA for 5.5 d. The root lengths of at least 20 seedlings were measured using a ruler and the mean was calculated. The experiment was performed with three biological repeats.
RNA extraction, RT–PCR, and qRT-PCR
Total RNAs from 7-day-old seedlings grown on 1/2 MS with or without 1 µM ABA were extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) or a Universal Plant Total RNA Extraction Kit (Spin-column)-I (BioTeke, Beijing, China) according to the manufacturer’s instructions. Reverse transcriptions were performed using PrimeScript reverse transcriptase with oligo(dT) primer using the Prime Script RT Enzyme MIX I (Takara, Osaka, Japan). qRT-PCR analysis was performed by ChamQ SYBR Color qPCR Master Mix (Q411, Vazyme, Nanjing, China) and Bio-Rad CFX96 (Bio-Rad, Hercules, CA, USA); UBC21 and UBQ10 were used as the internal controls. The cDNA used for reverse transcription–PCR (RT–PCR) was synthesized by a PrimeScript™ First-Strand cDNA Synthesis Kit (Takara). EF-1α was used as the internal control for RT–PCR. Primers are listed in Supplementary Table S1.
MbSUS, BiFC, and LCI assays
The MbSUS was performed as described in a previous study (Obrdlik et al., 2004). NubWT (wild-type) is the normal form of Nub without any mutant amino acids, which can interact with C-terminal ‘Cub’. Thus the group ‘Cub-ProteinA and NubWT’ is usually used as the positive control. NubG is a mutant form of Nub with weak binding affinity for Cub, and functional ubiquitin can only be reconstituted when NubG and Cub are in close proximity by fusion with proteins that interact. Thus, the group ‘Cub-ProteinA and NubG’ is usually used as the negative control and further used to identify whether the ProteinA has self-activating activity. The related ORFs were amplified from the first-strand cDNA with DNA polymerase by specific primers listed in Supplementary Table S1. For NubG fusions, pNXgate was cleaved with EcoRI/SmaI and transformed together with PCR products into the yeast THY.AP5 strain. Transformants were selected on synthetic dropout (SD) medium lacking tryptophan (W) and uracil (U). For CubPLV fusions, pMetYCgate was cleaved with PstI/HindIII and transformed together with PCR products into the THY.AP4 strain. Transformants were selected on SD medium lacking leucine (L). Several clones from each transformant were incubated on SD medium lacking leucine, tryptophan, and uracil (-WLU) at 30 °C for 3 d. For growth assays, diploid cells were replica-plated on SD medium -WLU and medium lacking leucine, tryptophan, uracil, adenine, and histidine (-WLUAH). Growth was monitored 3–6 d later.
For bimolecular fluorescence complementation (BiFC) assay, Nicotiana benthamiana plants were grown at a 16 h /8 h light/dark cycle at 26 °C for 30 d. The fourth, fifth, and sixth leaves were used for infiltration with the Agrobacterium tumefaciens GV3101. The A. tumefaciens suspensions adjusted to an OD600=1.0 in MMA medium (10 mM MgCl2, 50 mM MES, and 20 µM acetosyringone, pH 5.6) were kept at 28 °C in darkness for 3–5 h. Infiltrations were then conducted by gently pressing a 1 ml disposable syringe on the abaxial surface of fully expanded leaves with an approximate width of 3 cm at the middle region. Plants were subsequently regrown for another 36–60 h and imaged using an LSM51 confocal laser scanning microscope (Zeiss, Germany) at 488 nm.
For luciferase complementary imaging (LCI) assay, experiments were performed as previously described (Chen et al., 2008). Equal volumes of A. tumefaciens harboring pCAMBIA1300-nLUC and pCAMBIA1300-cLUC were mixed to a final concentration of OD600=1.0. Three different combinations of A. tumefaciens were infiltrated into three different positions in the same leaves of N. benthamiana and cultured for 60 h. Five minutes before detection, 0.2 mM luciferin (Promega, Madison, WI, USA) was uniformly infiltrated into the same positions as A. tumefaciens. Subsequently, luciferase activity was measured using a low-light cooled CCD imaging apparatus (Lumina II, Waltham, MA, USA).
Pull-down assay
To confirm the interaction between CEPR2 and PYLs, the fusion proteins β-glucuronidase (GST)–PYL2, GST–PYL4, or His-CEPR2KD (kinase domain, amino acids 642–977) proteins were expressed in the Escherichia coli Rosetta strain carrying a pGEX-4t-1-GST-PYL2, pGEX-4t-1-GST-PYL4, or pET30a-His-CEPR2KD-His construct. Transformant cells were cultured in 500 ml of Luria–Bertani medium at 37 °C to an OD at 600 nm of 1.0, at which time the protein expression was induced with 0.8 mM isopropyl-β-d-thiogalactopyranoside for 12 h at 16 °C. Then the bacterial cultures were separated by thoroughly centrifuging at a 6000 rpm for 5 min at 4 °C. A 5 ml aliquot of ddH2O was added to the centrifuge tube to re-suspend the pellet. The lysates were obtained by sonication using ultrasonic waves (JY92-II, Scientz Biotechnology Co., Ltd, Ningbo, China), with the parameters: operating power, 300 W; working time, 10 s; interval time, 5 s; cycles, 30. Lysates were clarified by centrifugation at 8000 rpm for 10 min at 4 °C. Then, His-CEPR2KD protein was purified with the His-Tagged Protein Purification Kit (CWBIO, Beijing, China), and GST–PYLs were purified with a Pierce Glutathione Spin Column (Thermo, Waltham, MA, USA). For pull-down assay, GST–PYLs (50 µg) and His-CEPR2KD (50 µg) were incubated 2 h at 4 °C with constant rocking in 1 ml of binding buffer (50 mM Tris–HCl, 150 mM NaCl, pH 8.0). Afterwards, GST proteins were purified with a Pierce Glutathione Spin Column, eluted, and analyzed with anti-His antibody (CWBIO, Beijing, China).
Protein degradation assay and half-life calculation
To detect whether CEPR2 accelerated the degradation of PYLs, cell-free assays were used to determine the degradation rate of PYL1, PYL2, and PYL4. Recombinant proteins GST–PYL1, GST–PYL2, or GST–PYL4 were expressed in the E. coli Rosetta strain and further purified with a Pierce Glutathione Spin Column (Thermo). Then 0.6 g samples of 7-day-old CEPR2 overexpression (CEPR2-OE), WT, and cepr2/pxy/pxl2 were ground in liquid nitrogen and mixed with 600 µl of non-denatured protein extract [Tris–HCl (pH 7.5), 25 mM; MgCl2, 10 mM; NaCl, 10 mM; DTT, 5 mM; phenylmethylsulfonyl fluoride (PMSF), 4 mM; ATP, 10 mM]. The crude extract was put on ice for 30 min and further centrifuged at 17 000 g for 15 min at 4 °C. The supernatant was centrifuged again, and the new supernatant was collected. Subsequently, 10 µl (~2 μg) of purified GST–PYL1, GST–PYL2, or GST–PYL4 were incubated with 240 µl of supernatant of CEPR2-OE, WT, and cepr2/pxy/pxl2 for 0, 10, 30, and 60 min at 22 °C. Then 40 µL reaction solutions from each time point were transferred to a new centrifuge tube, combined with 10 µl of 5× loading buffer, and boiled for 5 min. Anti-GST antibody was used to examine the protein level of PYLs. The spot densitometry was measured by Image J 1.36 (http://rsb.info.nih.gov/ij/). Data for all blots were combined using GraphPad Prism 5 to draw a single line, y=kt+b, and the half-life of PYL–LUC (luciferase) was calculated by the following equation: t=[y (0.5)–b]/k.
To further examine the degradation pathway of PYL4, the total proteins from 7-day-old seedlings of CEPR2-OE, WT, and cepr2/pxy/pxl2 grown on 1/2 MS were extracted and detected with anti-PYL4 antibody, kindly provided by Dr Xie (Yu et al., 2016). The protein synthesis of PYL4 was blocked by treatment with 100 μM cycloheximide (CHX) for 4 h, and then an inhibitor of the proteasome pathway (MG132, 100 μM) and an inhibitor of the vacuolar degradation pathway (E64d, 100 μM) were treated with seedlings of CEPR2-OE, WT, and cepr2/pxy/pxl2 for 4 h separately or together. PYL4 proteins were detected using an anti-PYL4 antibody, and Image J 1.36 was used to quantify the protein band.
Co-IP assay
To confirm the interactions of CEPR2 and PYLs, CARs, and RSL1. CEPR2ΔTM–GFP [transmembrane (TM) domain, amino acids 622–641, fused to green fluorescent protein] and PYL2-Myc, PYL4-Myc, CAR4-Myc, or RSL1-Myc were each expressed in N. benthamiana. Proteins were extracted and re-suspended in IP buffer [100 mM Tris–HCl, pH 7.4, 75 mM NaCl, 1 mM EDTA, 0.05% SDS, 0.1% Triton X-100, 10% glycerol, and protease inhibitor cocktail (Roche, Basel, Switzerland)]. Co-immunoprecipitation (Co-IP) was performed as previously described (Fiil et al., 2008). The protein extracts were then incubated with magnetic beads (Invitrogen Dynabeads™ Protein A; Carlsbad, CA, USA) and anti-GFP antibody (Sigma, Santa Clara, CA, USA; 1:300 dilution) at 4 °C for 5–6 h. The magnetic beads were recovered by centrifugation at 1000 g for 30 s and washing three times with pre-cooled Tris-buffered saline [TBS; 50 mM Tris–HCl (pH 7.5), 150 mM NaCl]. About 20 μl of sample buffer [50 mM Tris–HCl (pH 6.8), 2% SDS, 6% glycerol, 100 mM DTT, and 0.02% bromophenol blue] was added into the beads. After boiling for 5 min and centrifugation, the samples were loaded onto SDS–PAGE gels for western blot analysis and detected by anti-GFP (1:1000 dilution) and anti-Myc (Sigma1:300 dilution) antibodies.
The preparation of protoplasts and subcellular localization analysis
To confirm the PM localization of CEPR2, 3-week-old transgenic plants containing 35S::CEPR2-GFP were used to prepare the protoplasts. After removing the lower epidermis with sellotape, the leaves were incubated with dissociation solution (1% cellulase R10, 0.25% macerozyme R10, and 400 mM mannitol) in a rotator at 40 rpm at 23 °C for 3–5 h, and then centrifuged at 100 g for 3 min. The protoplasts were re-suspended with W5 solution (150 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, 2 mM MES) and imaged by LSCM51 (Zeiss, Germany) at 488 nm.
In vitro kinase assay
In order to test whether CEPR2 phosphorylates PYLs in vitro, PYL2–GST and PYL4–GST (50 µg each) were each incubated with CEPR2KD-His (0.5 µg) with or without 10 µM ABA in 50 µl of reaction buffer (25 mM HEPES, pH 7.2, 1 mM DTT, 50 mM NaCl, 2 mM EGTA, 5 mM MgSO4, 50 µM ATP). The reaction mixtures were incubated at 30 °C for 0–120 min and terminated by adding 5× loading buffer; 0 min without reaction was used as the negative control. Proteins were then fractionated by SDS–PAGE and Mn2+-phos-tag-PAGE (50 µM phos-tag and 100 µM Mn2+). Calf intestinal alkaline phosphatase (CIAP; Promega), at 0.01 U µl–1, was added in the reaction buffer after incubating for 30 min, and incubated for another 30 min at 37 °C to remove the phosphoryl group(s) of PYLs. To further identify the phosphorylation site(s) of PYL4, the protein sample incubated in a kinase buffer containing CEPR2KD for 1 h was separated by SDS–PAGE, and the target protein was subsequently cut and identified by ‘phosphorylation MS’ (phosphorylation site identification by MS).
The degradation of PYL4 in a cell-free system
Seven-day-old seedlings of OE#9 grown on 1/2 MS medium were harvested, and then ground into power in liquid nitrogen. The total proteins were extracted in buffer [5 mM DTT, 10 mM NaCl, 25 mM Tris-HCl (pH 7.5), 10 mM ATP, 4 mM PMSF, and 10 mM MgCl2], and the supernatants were collected after two centrifugations (12 000 rpm, 10 min, 4 °C). Then, the purified PYL4Normal–GST (1 μg), PYL4S86AS88A–GST (1 μg), or PYL4S54A–GST (1 μg) was incubated in total proteins (0.5 mg, 100 μl) at 22 °C for 0, 15, 30, 45, or 60 min. The reactions were terminated by adding 5× loading buffer, and the protein abundance of PYL4–GST was detected by anti-GST antibody.
Statistical analysis
All experiments were performed at least three times. Error bars in each graph indicate the mean values ±SE of replicates. Statistically significant differences between measurements were determined using Student’s t-test (*P<0.05; **P<0.01; ***P<0.001) or one-way ANOVA (P<0.05; LSD and Duncan test) in Statistical Product and Service Solutions Statistics software (SPSS 24; IBM, Armonk, NY, USA).
Results
Identification of LRR-RLKs as interacting partners of PYR/PYLs
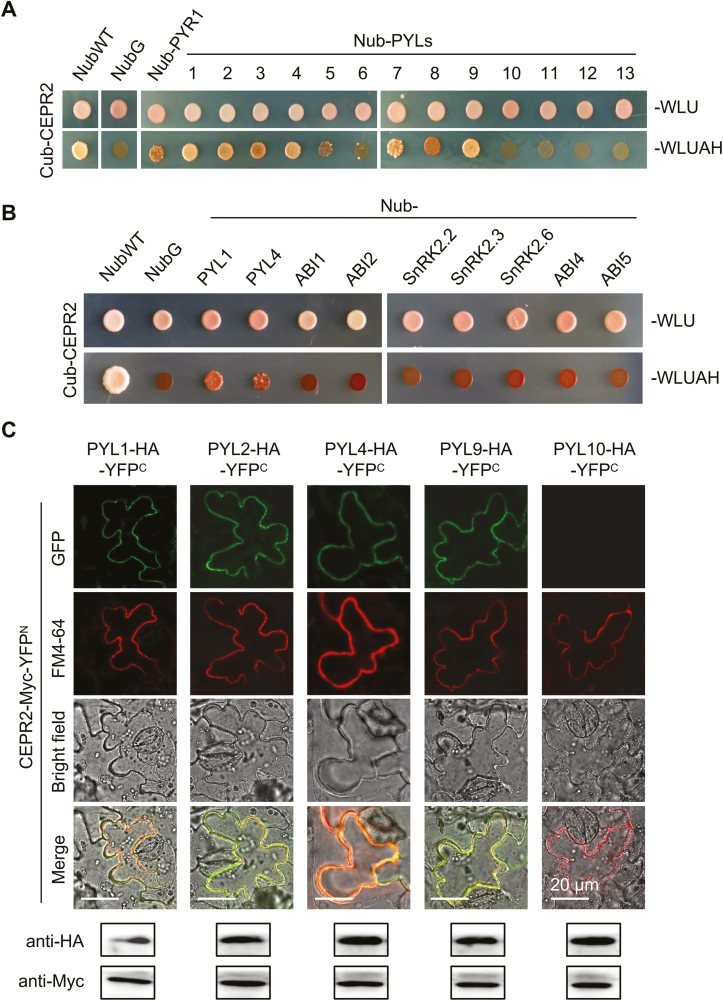
In order to isolate the LRR-RLKs that interact with PYR/PYLs, PYL4 was used as the bait protein for the MbSUS experiment. In the initial screening process, five members were identified to interact with PYL4, namely CEPR2, PXL1, RGI2, GSO1, and an LRR-RLK with unknown function. Subsequent screening is ongoing and the relevant data are not shown. Here, CEPR2 (At1g72180), one of the receptors of C-terminally encoded peptides (CEPs), is selected to investigate the interaction mechanism of CEPR2 and PYL4, and the roles that CEPR2 and PYL4 play in the ABA signaling pathway were further analyzed. We then examined the interactions between CEPR2 and all PYR/PYLs members, and found that 9 of 14 PYR/PYLs members interacted with CEPR2 in MbSUS assays (Fig. 1A; Supplementary Fig. S1). Among them, PYL1, PYL2, PYL3, PYL4, PYL7, and PYL9 interacted strongly, while PYR1, PYL5, and PYL8 interacted weakly with CEPR2 (Fig. 1A; Supplementary Fig. S1). Interestingly, CEPR2 did not interact with other components in the ABA signaling pathway (Fig. 1B), indicating the relatively specific interaction between CEPR2 and some of the ABA receptors. PYL1, PYL2, PYL4, PYL9, and PYL10 were then selected for BiFC experiments to confirm the interactions. The corresponding constructs were co-delivered into tobacco epidermal leaf cells, and western blot assays showed that CEPR2 and all PYLs could be expressed in tobacco epidermal cells. However, except for CEPR2–PYL10, co-localization of reconstituted yellow fluorescent proteins (YFPs) and the PM marker FM4-64 occurred on the PM (Fig. 1C), indicating that CEPR2 could interact with PYL1, PYL2, PYL4, and PYL9, but not PYL10. Although CEPR2 has previously been reported as a receptor of CEPs, its PM localization still lacks direct evidence. Here, the fluorescent signal of 35S::CEPR2-GFP appeared only in the PM, which confirmed the PM localization of CEPR2 (Supplementary Fig. S2).
Fig. 1.
CEPR2 physically interacts with PYR/PYLs. (A) The MbSUS (mating-based split ubiquitin system) assay was performed to determine the interactions of CEPR2 with all members of PYLs. Images were taken after culturing at 30 °C for 4 d. ‘Cub-CEPR2 and NubWT’ were used as positive control, and ‘Cub-CEPR2 and NubG’ were used as negative control. (B) MbSUS assay showing the interactions of CEPR2 with other ABA signaling components. (C) The BiFC assay was performed in N. benthamiana to further confirm the interactions between CEPR2 and PYLs. Western blot assays were performed to verify whether CEPR2 and PYLs could be expressed in N. benthamiana. Anti-Myc antibody was used to examine the expression level of CEPR2, and anti-HA was used to examine the expression level of PYLs. PYL10, which does not interact with CEPR2 in the MbSUS assay, was used as a negative control. FM4-64 was used as a PM dye. FM4-64, N-(3-triethylammomiumpropyl) 4-(p-diethylaminophenylhexa-trienyl); PM, plasma membrane; BiFC, bimolecular fluorescence complementation. (This figure is available in color at JXB online.)
CEPR2 negatively and redundantly regulates ABA signaling with PXY and PXL2 in Arabidopsis
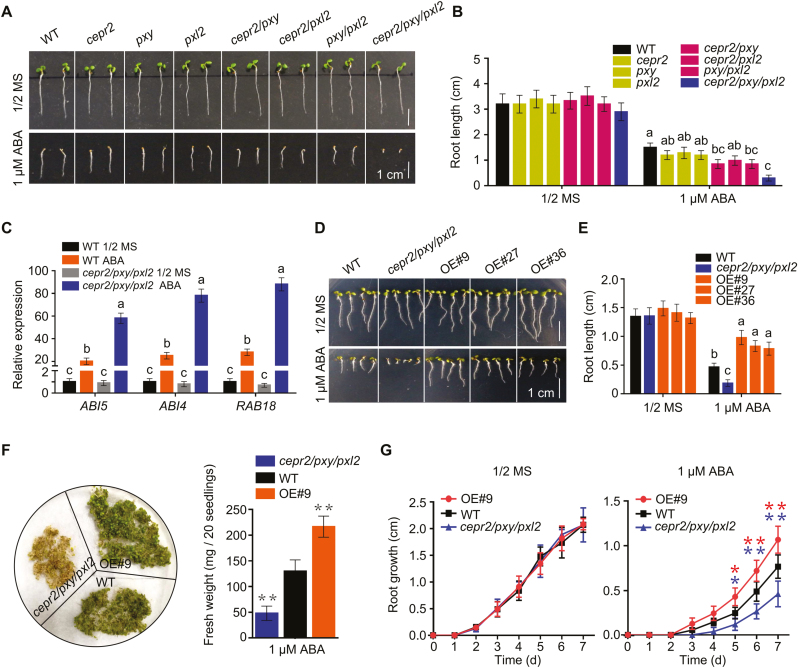
To further determine the roles of CEPR2 in ABA signaling, a T-DNA insertion mutant of CEPR2 was obtained (Supplementary Fig. S3A, B). The cepr2 single mutant showed no difference in response to ABA compared with the WT (Fig. 2A, B), suggesting the functional redundancy of CEPR2 with other LRR-RLKs. Hence, two members, phloem intercalated with xylem (PXY) and PXY-Like 2 (PXL2), that had a closer relationship with CEPR2 were selected and used for further investigation (Supplementary Fig. S3A, B). T-DNA knockout mutants of PXY and PXL2 were identified. Then, different double and triple mutants were constructed, and the cepr2/pxy/pxl2 triple mutant showed shorter root length, lower fresh weight, and slower root growth than the WT in the presence of ABA (Fig. 2A, B, F, G). Moreover, ABI4, ABI5, and RAB18 were dramatically up-regulated in cepr2/pxy/pxl2 mutants in the presence of ABA. Taken together, these results demonstrated the negative and redundant roles of CEPR2, PXY, and PXL2 in ABA signaling.
Fig. 2.
CEPR2 negatively and redundantly regulates ABA signaling with PXY and PXL2. (A) Seedling phenotypes of WT, single, double, or triple mutants of cepr2, pxy, and pxl2 grown on 1/2 MS or 1 µM ABA for 7 d. (B) The primary root length of seedlings shown in (A). Error bars indicate the SE (n=6). P<0.05, one-way ANOVA. Statistical differences are indicated by lower case letters above the columns, and different letters represent different significance. (C) The expression levels of ABA signaling marker genes in WT and cepr2/pxy/pxl2 lines grown on 1/2 MS or 1 µM ABA for 7 d were analyzed by qRT-PCR. Error bars indicate the SE (n=3). P<0.01, one-way ANOVA. (D) Seedling phenotypes of WT, cepr2/pxy/pxl2, and three CEPR2-overexpressing lines grown on 1/2 MS or 1 µM ABA for 7 d. (E) The primary root length of seedlings shown in (D). Error bars indicate the SE (n=3). P<0.01, one-way ANOVA. (F) The phenotype and fresh weight of WT, OE#9, and cepr2/pxy/pxl2 lines grown on 1/2 MS with 1 µM ABA for 7 d. Error bars indicate the SE (n=5). Asterisks indicate significant differences compared with the WT treated with ABA. **P<0.01, Student’s t-test. (G) The primary root growth of OE#9, WT, and cepr2/pxy/pxl2 lines grown on 1/2 MS or 1 µM ABA. Error bars indicate the SE (n=3). Asterisks indicate significant differences compared with the WT treated with ABA. *P<0.05, **P<0.01, Student’s t-test. (This figure is available in color at JXB online.)
At the same time, three CEPR2-OE lines were generated, and the expression levels were detected by RT–PCR and qRT-PCR (Supplementary Fig. S3C, D). In the presence of ABA, the OE lines showed longer root length, higher fresh weight, and faster root growth than the WT (Fig. 2D–G), which further indicated the negative roles of CEPR2 in ABA signaling.
CEPR2 promotes the degradation of PYLs
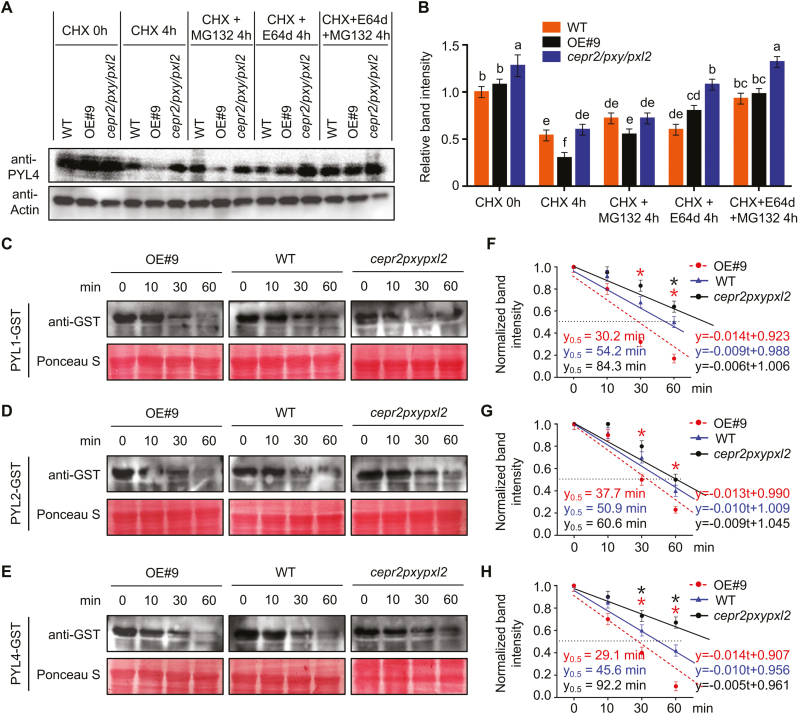
To determine the effects of CEPR2 on PYLs, we examined the transcription level of PYL4 in different seedlings. However, no statistically significant differences were found among OE#9, WT, and cepr2/pxy/pxl2 seedlings grown on 1/2 MS or 1 μM ABA medium (Supplementary Fig. S4). Then the protein levels of PYL4 were detected in OE#9, WT, and cepr2/pxy/pxl2 lines by anti-PYL4, and the antibody provided by Dr Xie was specific to PYL4 protein (Supplementary Fig. S5). We treated 7-day-old seedlings of OE#9, WT, and cepr2/pxy/pxl2 with 100 μM CHX to block protein synthesis and to investigate the half-life of PYL4. We found that PYL4 protein levels diminished after CHX treatment, whereas the protein content of PYL4 in OE#9 decreased faster than that in the WT and cepr2/pxy/pxl2, suggesting that CEPR2 can promote the degradation of PYL4 (Fig. 3A). To further investigate the degradation pathway(s) of PYL4, OE#9, WT, and cepr2/pxy/pxl2 seedlings were treated with 100 μM MG132 (an inhibitor of the 26S proteasome pathway) and 100 μM E64d (an inhibitor of vacuolar hydrolases) separately or together. As a result, the degradation of PYL4 slowed after MG132 or E64d treatment compared with the samples with CHX treatment. Moreover, there were no statistically significant differences between MG132+E64d and control (Fig. 3A, B). These results indicated that CEPR2 promoted the degradation of PYL4 through both the 26S proteasome and vacuolar degradation pathways.
Fig. 3.
CEPR2 accelerates the degradation of PYLs. (A) The PYL4 protein levels in 7-day-old seedlings of OE#9, WT, and cepr2/pxy/pxl2 grown on 1/2 MS with or without CHX, CHX+MG132, CHX+E64d, or CHX+MG132+E64d for 4 h were detected by anti-PYL4 antibody. CHX, cycloheximide; MG132, an inhibitor of the 26S proteasome degradation pathway; E64d, an inhibitor of the vacuolar degradation pathway. (B) The PYL4 protein levels shown in (A) were quantified by Image J. Error bars indicate the SE (n=3). P<0.05, one-way ANOVA. (C–E) Cell-free assays showing the degradation rate of PYL1–GST, PYL2–GST, and PYL4–GST incubated with the supernatant of CEPR2-OE#9, WT, or cepr2pxypxl2. The degradation rate of GST–PYLs was detected by anti-GST antibody. Ponceau staining of Rubisco indicates equal loading. (F–H) The regression line represents the degradation rate based on the intensity of the band quantified by ImageJ, and y=0.5 reflects the half-life of PYL–GST proteins. Error bars indicate the SE (n=3). Asterisks indicate significant differences compared with the WT. *P<0.05, Student’s t-test. (This figure is available in color at JXB online.)
Due to the lack of antibodies against PYL1 and PYL2, either PYL1, PYL2, or PYL4 was translationally fused to GST to further confirm the degradation effects of CEPR2 on PYLs. Recombinant PYL1, PYL2, and PYL4 were purified and incubated with supernatant of CEPR2-OE#9, WT, and cepr2pxypxl2 for cell-free assays. Then we used anti-GST antibody to examine the PYL–GST fusions at different time points. PYL1–GST, PYL2–GST, and PYL4–GST were degraded more rapidly in OE#9 than in the WT, whereas the degradation of PYL–GST fusions was slower in cepr2pxypxl2 (Fig. 3C–E). The protein level and half-life of PYL–GST fusions were then measured according to the method of a previous study (Gilkerson et al., 2015). As shown in Fig. 3F–H, PYL–GST proteins were all reduced over time in OE#9, WT, and cepr2pxypxl2, while the decrease of PYL–GST proteins was much faster and their half-lives were much shorter in OE#9. Taken together, these results illustrated the accelerated degradation of PYL1, PYL2, and PYL4 by CEPR2.
The kinase domain of CEPR2 is required for the interaction with PYLs
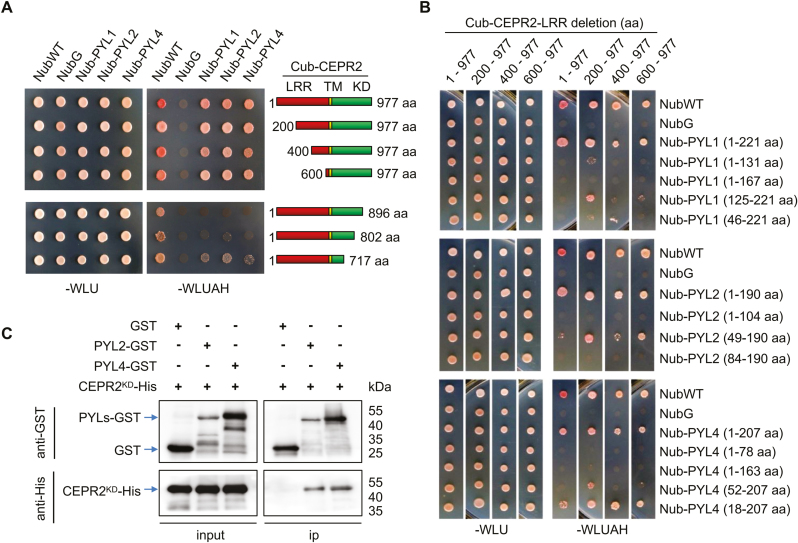
We further sought to identify the core components of CEPR2 involved in interaction with PYLs. As shown in Fig. 4A and Supplementary Fig. S6A, in comparison with full-length CEPR2, CEPR2 with the LRR domain deleted also interacted with PYLs. Then, CEPR2 was truncated by the KD domain. As shown in Fig. 4A and Supplementary Fig. S6B, when CEPR2 was truncated by 81 amino acids (KD, amino acids 1–896) and 175 amino acids (KD, amino acids 1–802), the interactions were diminished, indicating the important role of these regions in the interaction of CEPR2 with PYLs. Thus, the interaction required the kinase domain of CEPR2.
Fig. 4.
Identification of the interaction domain of CEPR2 and PYLs. (A) The interaction of full-length or truncated CEPR2 from the LRR domain or KD domain with PYLs in MbSUS assay. LRR, leucine-rich repeat receptor-like domain; TM, transmembrane domain; KD, kinase domain. (B) The interaction of full-length or truncated PYLs with full-length or truncated CEPR2 from the LRR domain in MbSUS assay. (C) The interaction between PYLs and CEPR2KD was verified by pull-down assay using purified GST, PYL2–GST, PYL4–GST, and CEPR2KD-His expressed in Escherichia coli. CEPR2KD-His proteins were incubated with immobilized PYL2–GST, PYL4–GST, or GST, and the proteins immunoprecipitated with GST beads were detected using anti-His antibody. PYL2–GST and PYL4–GST were detected with anti-GST antibody, and CEPR2KD-His proteins were detected with anti-His antibody. (This figure is available in color at JXB online.)
Similarly, different numbers of amino acids were deleted from the C- and N-terminus of PYL1, PYL2, and PYL4. Upon deletion of amino acids from the C- or N-terminus of PYLs, their interaction with CEPR2 diminished or disappeared (Fig. 4B; Supplementary Fig. S6C–E), which indicated that the full length of PYLs is required for interaction with CEPR2. Then, we purified recombinant GST-tagged PYL2 (PYL2–GST), PYL4 (PYL4–GST), and His-labeled kinase domain of CEPR2 (CEPR2KD-His) to perform a GST pull-down assay. The ability of GST-tagged protein to pull down the other protein would indicate the existence of an interaction between the two proteins. Our results suggested that CEPR2KD could be pulled down by PYL2–GST and PYL4–GST (Fig. 4C), also demonstrating that the full length of PYLs could directly interact with CEPR2KD.
CEPR2 phosphorylates PYLs in the absence of ABA
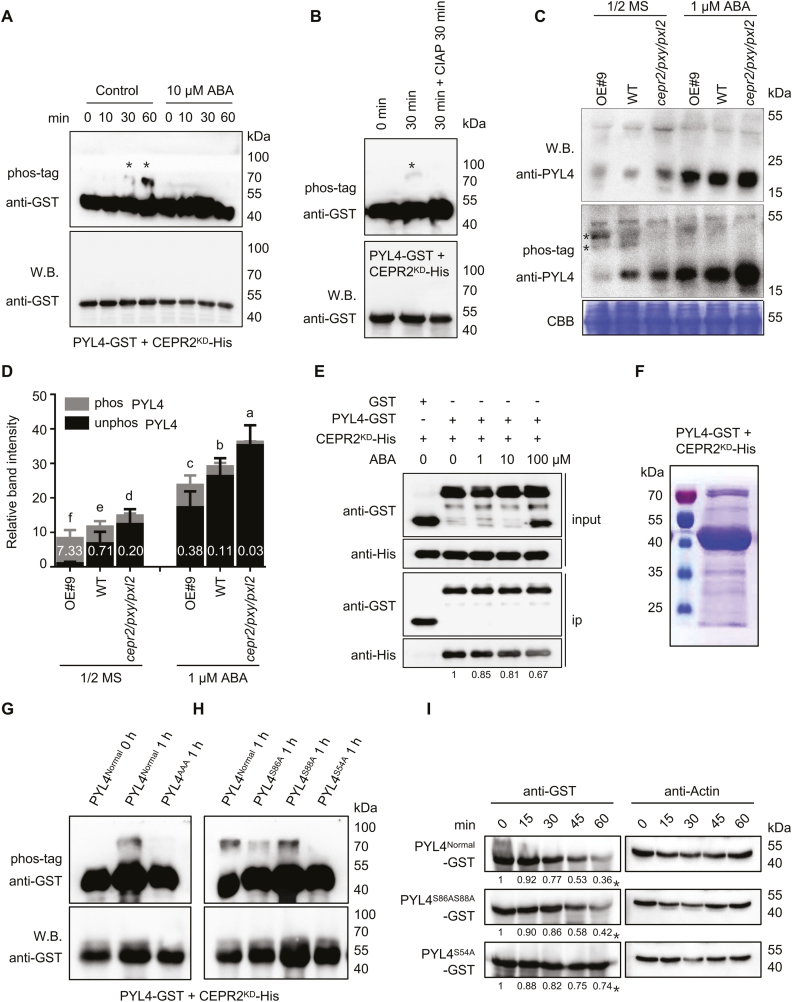
It is reasonable to hypothesize that CEPR2 may act as a protein kinase to phosphorylate PYLs. To verify this hypothesis, an in vitro kinase assay was performed; the results showed that the phosphorylated PYL4 was detected at 30 min when PYL4 was incubated with CEPR2KD in the kinase buffer (Fig. 5A). Moreover, CIAP successfully removed the phosphoryl group of PYL4, indicating that PYL4 is a direct substrate of CEPR2 (Fig. 5B). However, the phosphorylated PYL2 was detected at 120 min, and this phosphorylation can also be inhibited by CIAP (Supplementary Fig. S7), suggesting the low phosphorylation ability of CEPR2 towards PYL2. Surprisingly, in the presence of ABA, the phosphorylated PYL4 and PYL2 were no longer detected (Fig. 5A; Supplementary Fig. S7A), which indicated that ABA inhibited this phosphorylation in vitro.
Fig. 5.
CEPR2 phosphorylates PYL4 in vitro and in vivo. (A) In vitro kinase assays showed that PYL4 was phosphorylated by CEPR2KD, while the phosphorylated PYL4 disappeared under ABA treatment. Western blot assay was used to detect the loading control and unphosphorylated PYL4. Phos-tag (50 µM) was used to isolate the phosphorylated forms of PYL4 in this kinase assay. The position of phosphorylated PYL4 is indicated by asterisks. (B) CIAP successfully removed the phosphoryl group of PYL4 in this kinase assay. CIAP, calf intestinal alkaline phosphatase, which is used to remove phosphate group(s) of proteins via dephosphorylation. (C) The phosphorylation levels of PYL4 in OE#9, WT, and cepr2/pxy/pxl2 seedlings grown on 1/2 MS or 1 µM ABA for 7 d were detected by anti-PYL4 antibody. Asterisks represent two different forms of phosphorylated PYL4, and the band above the arrows is a non-specific band. (D) The PYL4 protein levels shown in (C) were quantified by Image J. The phos PYL4/unphos PYL4 ratios were labeled in the columns in white. Error bars indicate the SE (n=3). P<0.05, one-way ANOVA. Statistical differences are indicated by lower case letters above the columns, and different letters represent different significance. (E) The pull-down assay showing the interaction of CEPR2KD-His and PYL4–GST with or without different concentrations of ABA. CEPR2KD-His was incubated with immobilized GST or PYL4–GST, and the proteins immunoprecipitated with GST beads were detected using anti-His antibody. (F) Phosphorylated PYL4 incubated in kinase buffer for 1 h was isolated by SDS–PAGE for phosphorylation site identification by MS. (G) All three serines identified by MS were artificially mutated into alanine (PYL4AAA), and the PYL4AAA protein completely lacked the ability to be phosphorylated by CEPR2KD in the in vitro kinase experiment. (H) Ser54, Ser86, and Ser88 of PYL4 were each artificially mutated into alanine, and only PYL4S54A completely lacked the ability to be phosphorylated by CEPR2KD in the in vitro kinase experiment. (I) A cell-free system was used to determine the degradation rate of PYL4 proteins in OE#9 lines. The protein levels of PYL4Normal–GST, PYL4S86AS88A–GST, and PYL4S54A–GST were detected by anti-GST antibody. (This figure is available in color at JXB online.)
The phosphorylation levels of PYL4 were determined in OE#9, WT, and cepr2/pxy/pxl2 seedlings grown on 1/2 MS or 1 μM ABA. As shown in Fig. 5C, in spite of ABA treatment, the phosphorylated signals of PYL4 were stronger in OE#9 but weaker in cepr2/pxy/pxl2 than in the WT, indicating that CEPR2 promoted the phosphorylation of PYL4. In contrast, the unphosphorylated levels of PYL4 were obviously lower in OE#9 but higher in cepr2/pxy/pxl2 mutants (Fig. 5C, D). Moreover, the total protein levels of PYL4 were significantly lower in OE#9 but higher in cepr2/pxy/pxl2 mutants (Fig. 5D). Taken together, these results indicated that the higher phosphorylation levels were accompanied by the lower protein levels of PYL4. At the same time, phosphorylated PYL4 was significantly decreased in OE#9 in the presence of ABA, indicating that the phosphorylation was also inhibited by ABA in vivo (Fig. 5C).
Previous studies have shown that ABA can induce the ‘gate’ closure of PYLs, which greatly changed their three-dimensional structure (Melcher et al., 2009; Nishimura et al., 2009). In order to answer the question of whether the interaction of CEPR2 and PYL4 was inhibited by ABA, a pull-down assay was performed with or without different concentrations of ABA. CEPR2KD-His was incubated with immobilized GST or PYL4–GST for 1 h, and the proteins immunoprecipitated with GST beads were detected using anti-His antibody. As shown in Fig. 5E, the interaction between PYL4 and CEPR2 under different ABA conditions was much weaker than that without ABA, indicating that ABA-bound PYL4 interacted less with CEPR2.
CEPR2 phosphorylates PYL4 at Ser54
Finally, an in vitro kinase assay was performed to identify the site(s) of phosphorylation of PYL4 by CEPR2. The protein samples incubated in kinase buffer for 1 h were separated by SDS–PAGE to isolate the phosphorylated PYL4 (Fig. 5F). Subsequently, the target proteins were excised from the gel and analyzed by phosphorylation MS, and three putative phosphorylation sites, Ser54, Ser86, and Ser88, were identified (Supplementary Fig. S8). Then, the in vitro kinase assay was performed again to identify the exact phosphorylation site(s) of PYL4. First, the phosphorylated PYL4 disappeared when three serine residues were all artificially mutated into alanine residues (Fig. 5G), which indicated that at least one serine residue was the site of phosphorylation of PYL4 by CEPR2. Secondly, Ser54, Ser86, and Ser88 were each artificially mutated into alanine residues, and only PYL4S54A completely abolished the ability to be phosphorylated by CEPR2KD (Fig. 5H). Taken together, CEPR2 phosphorylates PYL4 at the crucial target site Ser54.
In order to test whether this phosphorylation of PYL4 is a prerequisite for its degradation, we conducted the cell-free experiment in the OE#9 line. The protein levels of PYL4Normal–GST, PYL4S86AS88A–GST, and PYL4S54A–GST were detected by anti-GST antibody. As shown in Fig. 5I, there was no significant difference in the protein degradation rate between PYL4Normal–GST and PYL4S86AS88A–GST, while the protein degradation rate of PYL4S54A–GST was much lower than that of PYL4Normal–GST, indicating that the phosphorylation of PYL4 at Ser54 was the cause of its degradation.
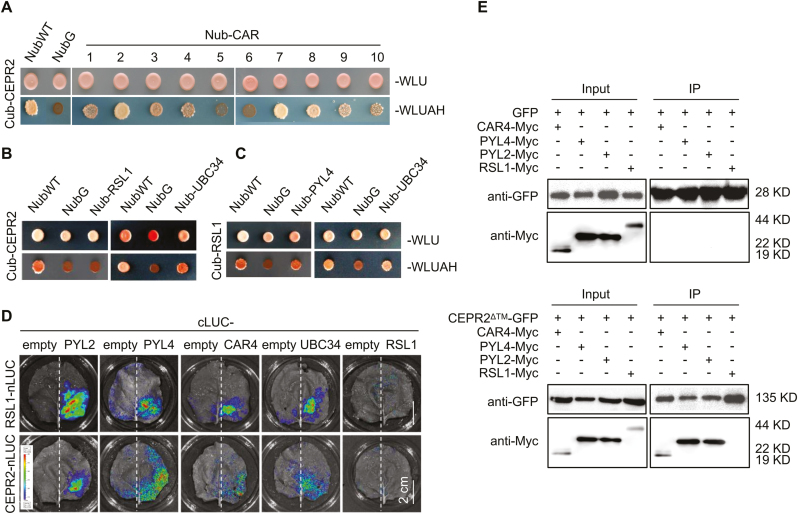
CEPR2 interacts with CARs and UBC34
It is known that CARs and RSL1 interact with PYLs and recruit them to the PM, but the mechanism is still unclear (Rodriguez et al., 2014; Diaz et al., 2016). To determine whether CARs and RSL1 are essential for the interaction between PM-localized CEPR2 and cytoplasm-localized PYLs, the interactions between CEPR2 and CARs were verified by MbSUS, LCI, and Co-IP assays (Fig. 6A, D, E). Encouragingly, CEPR2 did interact with several members of CARs (Fig. 6A). However, no interaction between CEPR2 and RSL1 was found in yeast or N. benthamiana (Fig. 6B, D, E). To further determine how CEPR2 accelerates the degradation of PYLs, we analyzed all the predicted proteins obtained from http://bar.utoronto.ca/interactions/cgi-bin/arabidopsis_interactions_viewer.cgi?input=AT1G72180&qbar=yes, which indicated that UBC34, a ubiquitin E2 conjugating enzyme, showed strong interaction with CEPR2. Also the interactions of CEPR2–UBC34 and UBC34–RSL1 were confirmed via MbSUS and LCI assays (Fig. 6B–D). Taken together, the complex of CEPR2–CARs–PYLs might be involved in the movement of PYLs from the cytoplasm to the PM, and the CEPR2–UBC34–RSL1 complex might be involved in the degradation of PYLs in the PM.
Fig. 6.
Identification of interactions between different proteins, including CEPR2, CARs, UBC34, and RSL1. (A–C) The interactions of CEPR2 and CARs (A), CEPR2 and UBC34 (B), and RSL1 and UBC34 (C) were detected in MbSUS assays. (D) LCI assays detected the interactions of CEPR2 or RSL1 with CAR4, UBC34, or RSL1 in N. benthamiana. LCI, firefly luciferase complementation imaging. (E) Co-IP analysis of PYL2-Myc, PYL4-Myc, CAR4-Myc, or RSL1-Myc with GFP or CEPR2ΔTM–GFP. The proteins immunoprecipitated by CEPR2ΔTM–GFP were detected using anti-Myc antibody. (This figure is available in color at JXB online.)
Discussion
CEPR2 physically interacts with ABA receptors in the PM
PYR/PYLs play important roles in ABA signaling and have been shown to be recruited to the PM by CARs. However, the mechanism remains unclear. The PM-localized LRR-RLKs have been shown to play critical roles in the transduction of various environmental and developmental signals (Dievart and Clark, 2003). Recently, RDK1 was reported to interact with ABI1 in the PM and positively regulate ABA-mediated seed germination (Kumar et al., 2017); FER interacts with ABI2 in the PM to regulate the ABA-mediated stress response (Chen et al., 2016). In this study, the interactions of CEPR2–PYLs and CEPR2–CARs in the PM were identified (Figs 1, 6). These results indicated that the ABA receptors recruited to the PM by CARs might be regulated by CEPR2.
CEPR2 phosphorylates PYL4 at Ser54 in the PM for degradation
Recently, EL1-like (AEL) kinase was reported to phosphorylate most of the PYLs in the nucleus (Chen et al., 2018). Moreover, cytosolic TOR kinase was also discovered to phosphorylate PYLs, but the phosphorylated forms of PYLs can still be detected in tor mutants, so the fascinating question is how many kinases can phosphorylate PYLs in Arabidopsis (Wang et al., 2018). Therefore, CEPR2 is the first reported PM-localized kinase that interacts with ABA receptors. The interaction between CEPR2 and PYLs requires the kinase domain of CEPR2, indicating the phosphorylated regulation of PYLs by CEPR2 (Fig. 4). Further phosphorylation experiments of the effects of CEPR2 on PYLs in vivo and in vitro proved the phosphorylated regulation of PYLs by CEPR2 (Fig. 5). The phosphorylation MS and in vitro kinase assay proved the crucial role of Ser54 in the phosphorylation of PLY4 by CEPR2 (Fig. 5F–H). Interestingly, this site of phosphorylation of PYL4 by CEPR2 was not the same site as those for TOR or AEL, which indicated that PYLs were strictly regulated by phosphorylation in Arabidopsis.
Studies have shown that specific site phosphorylation of substrates is the marker for degradation (Filipcik et al., 2017). We found that the presence of higher levels of CEPR2 induced the phosphorylation of PYL4 and reduced its protein levels in OE#9 (Fig. 5C), indicating the requirement of phosphorylation of PYLs by CEPR2 for their protein degradation. RSL1, a PM-anchored Ring-type E3 ligase, performs the attachment of ubiquitin to PYR1 and PYL4 to promote their degradation via the 26S proteasome protein degradation pathway (Bueso et al., 2014; Belda-Palazon et al., 2016). FYVE1/FREE1, a component of the endosomal sorting complex, interacts with RSL1 and recruits PYL4 to endosomal compartments for vacuolar degradation (Belda-Palazon et al., 2016). The interference function of RSL1 led to increased sensitivity to ABA compared with the WT (Bueso et al., 2014), which is a similar phenotype to that of the cepr2/pxy/pxl2 mutant, suggesting similar effects of RSL1 and CEPR2 on PYLs. Therefore, the accelerated degradation of PYLs by CEPR2 might be associated with RSL1. However, our data showed that CEPR2 did not interact with RSL1 (Fig. 6B, D, E). Thus, other intermediates that interact with CEPR2 might function in the degradation complex of PYLs. UBC34, a ubiquitin E2 conjugating enzyme, was verified to interact with CEPR2 and RSL1 in MbSUS and LCI assays (Fig. 6B–D), suggesting that CEPR2 might accelerate the degradation of PYLs by recruiting UBC34 and then RSL1.
The balance between PYL phosphorylation and stability
AEL can phosphorylate PYR1 at Ser152 and PYL1 at Ser182, causing a decrease in their protein stability (Chen et al., 2018). However, cytosolic ABA receptor kinase 1 (CARK1) phosphorylates PYR1 and PYL8 at T78 and at T77 respectively, which can enhance the protein stability of PYR1 and PYL8 (Zhang et al., 2018). It is of interest to find that the phosphorylation of PYR1 at different sites determines the different protein stability. Thus, we can make an assumption that the phosphorylation of PYL8 at T77 can enhance its protein stability, and further study should be performed to identify whether phosphorylation at some certain site can reduce its protein stability. Similarly, phosphorylation at S54 promotes degradation of PYL4, and the site that can enhance protein stability awaits further study. It would be intriguing to investigate the regulation mechanism in optimizing the balance between different phosphorylation sites and protein stability.
ABA inhibits the phosphorylation of PYLs by CEPR2 to activate ABA signaling during times of stress
CAR proteins anchor and function in a cluster in generating strong positive membrane curvature in a Ca2+-dependent manner (Diaz et al., 2016). This structure acts as a signal platform and participates in the recruitment of PYLs to the PM. Moreover, our data showed that both CEPR2 and RSL1 could interact with CAR4 (Fig. 6). Thus, the platform formed by CAR clustering may recruit PYLs for phosphorylation by CEPR2 and then ubiquitination by RSL1. However, the exact complex between CEPR2, CARs, UBC34, and RSL1 for PYL function under different conditions needs further investigations.
ABA inhibited the phosphorylation and degradation of PYL4 in vitro and in vivo (Fig. 5A, C, D). In addition, the intensity of interaction between CEPR2 and PYL4 was decreased in the presence of ABA (Fig. 5E). These results suggested that the ABA-bound PYL4 successfully inhibits the phosphorylation by CEPR2 during times of stress. Taken together, although many investigations are needed in the future, our results here indicated that the phosphorylation of PYL2/PYL4 by CEPR2 might promote their ubiquitination for degradation, resulting in repressed ABA signaling. However, ABA-bound PYLs successfully prevent this process and activate ABA signaling during times of stress. In short, plants utilize this phospho-regulatory mechanism to optimize the balance of growth and stress responses (Supplementary Fig. S9).
CEPR2 may inhibit PYL4 activity by phosphorylation and nitration
Nitric oxide (NO) as important signaling molecule plays important roles in plants. The excessive accumulation of NO in Arabidopsis leads to the nitration of PYL4 at a tyrosine residue, and reduces its activity through ubiquitin degradation (Castillo et al., 2015). CEPR2 can increase the absorption of NO3− by up-regulating NRT1.1, NRT2.1, and NRT3.1 (Tabata et al., 2014). As a precursor of NO, NO3− may promote PYL4 degradation by nitration modification, which is consistent with the CEPR2-mediated inactivation of PYL4 by phosphorylation in this study. Therefore, CEPR2 may inhibit PYL4 activity by phosphorylation and nitration modification. This awaits further research.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. CEPR2 interacts with PYR/PYLs in yeast.
Fig. S2. CEPR2 is localized in the plasma membrane.
Fig. S3. Identification of different mutants and CEPR2-OE lines.
Fig. S4. CEPR2 does not regulate the transcript levels of PYL4.
Fig. S5. Anti-PYL4 antibody can specifically recognize PYL4.
Fig. S6. Identification of the interaction domain of CEPR2 and PYLs by LCI.
Fig. S7. CEPR2 phosphorylates PYL2 in vitro.
Fig. S8. Identification of the phosphorylation sites of PYL4 by phosphorylation MS.
Fig. S9. CEPR2-mediated phosphorylation optimizes the balance of growth regulation and stress response in Arabidopsis.
Table S1. Primers used in this study.
Author contributions
CW and CZ conceived the original screening and research plans; ZY, DZ, YX, SJ, and LZ performed experiments; CW and ZY analyzed the data, produced the figures, and wrote the article; SZ, GY, JH, and KY provided some important suggestions; CW and CZ supervised and complemented the writing. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Dr Qi Xie (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for kindly providing PYL4 antibody; Dr Yan Zhang (College of Life Sciences, Shandong Agricultural University) for the MbSUS plasmids; and Dr. Gang Li (College of Life Sciences, Shandong Agricultural University) for the LCI plasmids. The authors also thank the biomass spectrometry laboratory of the China Agricultural University for mass spectrometry sequencing and data analysis. This work was supported by the Major Program of Shandong Province Natural Science Foundation (ZR2018ZB0212) and the National Natural Science Foundation of China (grant nos 31570271 and 31471425). The authors declare that they have no conflict of interest.
References
- Belda-Palazon B, Rodriguez L, Fernandez MA, et al. 2016. FYVE1/FREE1 interacts with the PYL4 ABA receptor and mediates its delivery to the vacuolar degradation pathway. The Plant Cell 28, 2291–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueso E, Rodriguez L, Lorenzo-Orts L, Gonzalez-Guzman M, Sayas E, Muñoz-Bertomeu J, Ibañez C, Serrano R, Rodriguez PL. 2014. The single-subunit RING-type E3 ubiquitin ligase RSL1 targets PYL4 and PYR1 ABA receptors in plasma membrane to modulate abscisic acid signaling. The Plant Journal 80, 1057–1071. [DOI] [PubMed] [Google Scholar]
- Castillo MC, Lozano-Juste J, González-Guzmán M, Rodriguez L, Rodriguez PL, León J. 2015. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Science Signaling 8, ra89. [DOI] [PubMed] [Google Scholar]
- Chen C, Wu C, Miao J, Lei Y, Zhao D, Sun D, Yang G, Huang J, Zheng C. 2014. Arabidopsis SAG protein containing the MDN1 domain participates in seed germination and seedling development by negatively regulating ABI3 and ABI5. Journal of Experimental Botany 65, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. 2008. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiology 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Qu L, Xu ZH, Zhu JK, Xue HW. 2018. EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Molecular Plant 11, 706–719. [DOI] [PubMed] [Google Scholar]
- Chen J, Yu F, Liu Y, et al. 2016. FERONIA interacts with ABI2-type phosphatases to facilitate signaling cross-talk between abscisic acid and RALF peptide in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, E5519–E5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Diaz M, Sanchez-Barrena MJ, Gonzalez-Rubio JM, et al. 2016. Calcium-dependent oligomerization of CAR proteins at cell membrane modulates ABA signaling. Proceedings of the National Academy of Sciences, USA 113, E396–E405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diévart A, Clark SE. 2003. Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Current Opinion in Plant Biology 6, 507–516. [DOI] [PubMed] [Google Scholar]
- Dimitrov I, Tax FE. 2018. Lateral root growth in Arabidopsis is controlled by short and long distance signaling through the LRR RLKs XIP1/CEPR1 and CEPR2. Plant Signaling & Behavior 13, e1489667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiil BK, Qiu JL, Petersen K, Petersen M, Mundy J. 2008. Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protocols 2008, pdb.prot5049. [DOI] [PubMed] [Google Scholar]
- Filipčík P, Curry JR, Mace PD. 2017. When worlds collide—mechanisms at the interface between phosphorylation and ubiquitination. Journal of Molecular Biology 429, 1097–1113. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD. 2002. Abscisic acid signaling in seeds and seedlings. The Plant Cell 14(Suppl), S15–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. 2009. In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson J, Kelley DR, Tam R, Estelle M, Callis J. 2015. Lysine residues are not required for proteasome-mediated proteolysis of the auxin/indole acidic acid protein IAA1. Plant Physiology 168, 708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. 2000. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- He Z, Wang ZY, Li J, Zhu Q, Lamb C, Ronald P, Chory J. 2000. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- Jones AM. 2016. A new look at stress: abscisic acid patterns and dynamics at high-resolution. New Phytologist 210, 38–44. [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. 2002. Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. The Plant Cell 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D, Kumar R, Baek D, Hyun TK, Chung WS, Yun DJ, Kim JY. 2017. Arabidopsis thaliana RECEPTOR DEAD KINASE1 functions as a positive regulator in plant responses to ABA. Molecular Plant 10, 223–243. [DOI] [PubMed] [Google Scholar]
- Li J, Chory J. 1997. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences, USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. 2009. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. 2009. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326, 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, El-Bakkoury M, Hamacher T, et al. 2004. K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proceedings of the National Academy of Sciences, USA 101, 12242–12247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez L, Gonzalez-Guzman M, Diaz M, et al. 2014. C2-domain abscisic acid-related proteins mediate the interaction of PYR/PYL/RCAR abscisic acid receptors with the plasma membrane and regulate abscisic acid sensitivity in Arabidopsis. The Plant Cell 26, 4802–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah SK, Reddy KR, Li J. 2016. Abscisic acid and abiotic stress tolerance in crop plants. Frontiers in Plant Science 7, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Chen Q, Wu Y, et al. 2016. ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. The Plant Journal 85, 348–361. [DOI] [PubMed] [Google Scholar]
- Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q. 2013. ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in arabidopsis. PLoS Genetics 9, e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, et al. 1995. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J. 2013. Structural basis for flg22-induced activation of the Arabidopsis FLS2–BAK1 immune complex. Science 342, 624–628. [DOI] [PubMed] [Google Scholar]
- Tabata R, Sumida K, Yoshii T, Ohyama K, Shinohara H, Matsubayashi Y. 2014. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science 346, 343–346. [DOI] [PubMed] [Google Scholar]
- ten Hove CA, Bochdanovits Z, Jansweijer VM, Koning FG, Berke L, Sanchez-Perez GF, Scheres B, Heidstra R. 2011. Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Molecular Biology 76, 69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii KU. 2004. Leucine-rich repeat receptor kinases in plants: structure, function, and signal transduction pathways. International Review of Cytology 234, 1–46. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. 2009. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma K, Upadhyay N, Kumar N, et al. 2017. Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Frontiers in Plant Science 8, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad F, Rubio S, Rodrigues A, Sirichandra C, Belin C, Robert N, Leung J, Rodriguez PL, Laurière C, Merlot S. 2009. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. The Plant Cell 21, 3170–3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao Y, Li Z, et al. 2018. Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Molecular Cell 69, 100–112.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Huffaker A, Bryan AC, Tax FE, Ryan CA. 2010. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. The Plant Cell 22, 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wu K, Gao P, Liu X, Li G, Wu Z. 2014. GsLRPK, a novel cold-activated leucine-rich repeat receptor-like protein kinase from Glycine soja, is a positive regulator to cold stress tolerance. Plant Science 215–216, 19–28. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. 2010. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. The Plant Journal 61, 672–685. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Mogami J, Yamaguchi-Shinozaki K. 2014. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Current Opinion in Plant Biology 21, 133–139. [DOI] [PubMed] [Google Scholar]
- Yu F, Lou L, Tian M, et al. 2016. ESCRT-I component VPS23A affects ABA signaling by recognizing ABA receptors for endosomal degradation. Molecular Plant 9, 1570–1582. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li X, Li D, et al. 2018. CARK1 mediates ABA signaling by phosphorylation of ABA receptors. Cell Discovery 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.