Abstract
Pyrenophoric acid (P-Acid), P-Acid B, and P-Acid C are three phytotoxic sesquiterpenoids produced by the ascomycete seed pathogen Pyrenophora semeniperda, a fungus proposed as a mycoherbicide for biocontrol of cheatgrass, an extremely invasive weed. When tested in cheatgrass bioassays, these metabolites were able to delay seed germination, with P-Acid B being the most active compound. Here, we have investigated the cross-kingdom activity of P-Acid B and its mode of action, and found that it activates the abscisic acid (ABA) signaling pathway in order to inhibit seedling establishment. P-Acid B inhibits seedling establishment in wild-type Arabidopsis thaliana, while several mutants affected in the early perception as well as in downstream ABA signaling components were insensitive to the fungal compound. However, in spite of structural similarities between ABA and P-Acid B, the latter is not able to activate the PYR/PYL family of ABA receptors. Instead, we have found that P-Acid B uses the ABA biosynthesis pathway at the level of alcohol dehydrogenase ABA2 to reduce seedling establishment. We propose that the fungus P. semeniperda manipulates plant ABA biosynthesis as a strategy to reduce seed germination, increasing its ability to cause seed mortality and thereby increase its fitness through higher reproductive success.
Keywords: ABA2, ABA biosynthesis, abscisic acid, cross-kingdom activity, Pyrenophora semeniperda, pyrenophoric acid, pyrenophoric acid B, pyrenophoric acid C, PYR/PYL, seed germination
The fungus Pyrenophora semeniperda produces pyrenophoric acid B, a small molecule that exploits the plant ABA biosynthetic pathway to reduce seed germination, increasing its reproductive success.
Introduction
The fungus Pyrenophora semeniperda (Brittlebank and Adams) Shoemaker, a naturally occurring necrotrophic seed pathogen, has been proposed as a potential biocontrol agent against cheatgrass (Bromus tectorum) (Meyer et al., 2013). This invasive weed is dramatically altering the semi-arid shrubland ecosystems in the western USA, increasing wildfire frequency and intensity (Germino et al., 2016). Thus, the ability of P. semeniperda to produce toxins in vitro, which could potentially be useful in biological control, was investigated (Cimmino et al., 2015; Meyer et al., 2015). Pyrenophora semeniperda produces large amounts of cytochalasin B in solid cultures (Masi et al., 2014a), while spirocyclic γ-lactams, including spirostaphylotrichins A, C, D, R, V, and W, and triticone E, are produced when the fungus is grown in potato dextrose broth (Masi et al., 2014b). Furthermore, from the same fungal organic extract smaller quantities of cytochalasin A, cytochalasin F, and deoxaphomin were isolated together with a new phytotoxic sesquiterpenoid, named pyrenophoric acid (P-Acid) (Fig. 1A) (Masi et al., 2014a, c). Finally, these compounds were produced in B. tectorum seed culture together with another two phytotoxic sesquiterpenoids named pyrenophoric acids B and C (P-Acid B and P-Acid C, respectively) (Fig. 1A) (Masi et al., 2014d). P-Acid, P-Acid B, and P-Acid C were able to inhibit both root and coleoptile growth of B. tectorum (Masi et al., 2014c, d). In particular, P-Acid B was the most active sesquiterpenoid among the P-Acids studied (Masi et al., 2014d). It was able to reduce germination at 7 d after sowing and produced germination delay relative to the control treatment. P-Acid B was also more bioactive in reducing coleoptile and root growth than the other sesquiterpenoids, P-Acid and P-Acid C. Interestingly, the production of small molecules able to delay germination could benefit P. semeniperda, as its fitness is increased on slow-germinating or dormant seeds. This is because slow-germinating seeds cannot escape pathogen-caused mortality, thus increasing resources available for pathogen growth and reproduction (Cimmino et al., 2015; Meyer et al., 2015).
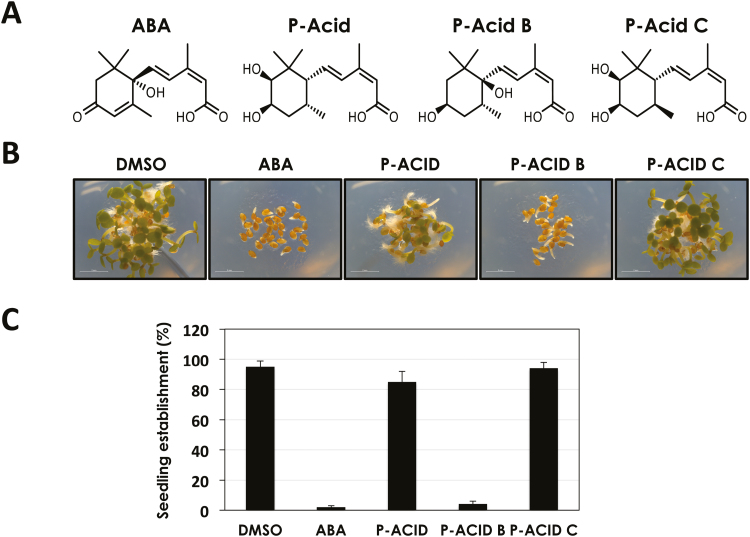
Fig. 1.
Pyrenophoric acid B inhibits seed germination. (A) Structures of ABA, P-Acid, P-Acid B, and P-Acid C used in this work. (B) Pictures of Arabidopsis Col-0 seeds treated with DMSO, 1 μM ABA, and 100 μM of the different pyrenophoric acids, 3 d after being sown. (C) Quantification of seedling establishment 3 d after Col-0 seeds were sown on DMSO, 1 μM ABA, and 100 μM of the different pyrenophoric acids. Values represent the mean ±SD of 90 seeds used in three different experiments.
P-Acid, P-Acid B, and P-Acid C (Fig. 1A) are sesquiterpenoids closely related to abscisic acid (ABA) (Fig. 1A). ABA is a plant hormone involved in many physiological processes in plants including seed development and germination, root growth, and abscission, among others (Leung and Giraudat, 1998). It also has a central role in the adaptation to environmental stresses such as drought, high salinity, and low temperatures (Yamaguchi-Shinozaki and Shinozaki, 2006). However, ABA is not a plant-specific metabolite. It is produced by different organisms including fungi, suggesting that it is a ubiquitous and versatile small compound that can modulate a myriad of physiological functions (Hartung, 2010; Takezawa et al., 2011). In Arabidopsis thaliana (Arabidopsis), ABA biosynthesis begins with the conversion of zeaxanthin to all-trans-violaxanthin by a zeaxanthin epoxidase known as ABA1 (Marin et al., 1996; Audran et al., 1998). Next, in the rate-limiting step of ABA biosynthesis, 9-cis-epoxycarotenoid dioxygenase (NCED) enzymes break down C40 carotenoids to produce xanthoxin (Schwartz et al., 1997; Iuchi et al., 2000). Xanthoxin is further transformed into abscisic aldehyde by the Arabidopsis short-chain alcohol dehydrogenase ABA2 (González-Guzmán et al., 2002). Finally, abscisic aldehyde is used as substrate by the aldehyde oxidase AAO3, in a reaction that depends on the activity of the molybdenum cofactor sulfurase ABA3, to produce ABA (Schwartz et al., 1997; Seo et al., 2000; González-Guzmán et al., 2004). ABA is perceived by the PYR/PYL/RCAR family of ABA receptors, comprised of 14 members in the eudicot plants Arabidopsis and tomato, and 12 and 9 members in the monocot species rice and Brachypodium distachyon, respectively (Ma et al., 2009; Park et al., 2009; He et al., 2014; Pri-Tal et al., 2017). ABA-bound PYR/PYL/RCAR proteins interact with protein phosphatases type 2C (PP2Cs) and inhibit their activity, allowing the downstream phosphorylation cascades to activate the ABA response, including inhibition of seed germination and stomatal closure, among others.
Here we have studied the effect and the mode of action of the fungus-produced pyrenophoric acids on plant seed germination. Among P-Acids, P-Acid B was able to inhibit germination of Arabidopsis seeds. We have found that P-Acid B makes use of the ABA biosynthesis pathway at the level of ABA2 to activate ABA signaling and inhibit germination, increasing the probability of host seed mortality and thereby increasing pathogen fitness.
Materials and methods
Chemicals
Pyrenophoric acid and pyrenophoric acids B and C (Fig. 1A) were isolated from the solid B. tectorum (cheatgrass) seed culture of P. semeniperda (strain WRK10-22), as previously reported (Masi et al., 2014c, d)
Plant material and growth conditions
Arabidopsis thaliana seeds were surface sterilized with 30% bleach containing 0.02% Tween-20 for 10 min followed by rinsing with sterile water five times. Seeds were sown on solid or liquid Murashige and Skoog (MS) medium and stratified for 3 d at 4 °C. For seedling establishment experiments, seeds were sown on 24-well plates filled with 1 ml of MS medium+1% agar supplemented with the different chemicals dissolved in DMSO. Seeds were kept in a Sanyo incubator under long-day conditions at temperatures of 23 °C/21 °C (day/night) for 3 d. Pictures were taken with a Leica DMS1000 macroscope, and seedling establishment (seedlings with open and green cotyledons) was scored. For luciferase assays, seeds were incubated for 7 d under long-day conditions in liquid MS medium. The different mutants used in this work, 112458 (Gonzalez-Guzman et al., 2012), abi3-9 (Nambara et al., 2002), abi4-11 (Nambara et al., 2002), aba1-101 (Barrero et al., 2005), aba2-1 (Léon-Kloosterziel et al., 1996), nced3,5 (Frey et al., 2012), aba2-11 (González-Guzmán et al., 2002), aao3-2 (González-Guzmán et al., 2004), and aba3-1 (Léon-Kloosterziel et al., 1996), and the transgenic lines pMAPKKK18-LUC+ (Vaidya et al., 2017) and 35S-HAB1W385A (Dupeux et al., 2011) have been described previously. To generate the double nced2nced3 mutant, homozygous plants from the T-DNA lines SALK_090937C (nced2) and GK-129B08.01 (nced3) were crossed and the F2 selected on 100 mM NaCl plates. PCR-verified plants using the primers: FORNced2, ATGGTTTCTCTTCTTACAATGCCG; RVNced2, TTCCGGTTAACCATACCAATCTC; NewpROK2, GCCGATTTC GGAACCACCATC; FWNced3GABI, CCTAGTGTTCAGATCG CCGGA; RVNced3GABI, GAGATTCTCGTCAGACTCGTT; and GK-o8474, ATAATAACGCTGCGGACATCTACATTT were selfed and the F3 seed was used for the different experiments.
Luciferase imaging
pMAPKKK18-LUC + seedlings were grown in 24-well plates filled with 1 ml of liquid MS medium for 7 d. MS medium was changed to MS medium supplemented with 100 µM d-luciferin, potassium salt (GoldBio), and the different treatments: 25 µM ABA, 200 µM P-Acid B, or DMSO as control, and incubated for 6 h. Luminescence was recorded with a LAS-3000 imager (Fujifilm) equipped with a CCD camera using 2 min exposures. Eight-bit images were transformed in rainbow false color and quantified using Fiji (Schindelin et al., 2012).
Protein purification
The different expression clones were described previously (Antoni et al., 2012; Castillo et al., 2015). Protein induction was performed by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to 250 ml cultures of BL21(DE3) cells at OD=0.6–0.8. Cells were grown overnight at 16 °C and collected by centrifugation. Cells were lysed in 50 mM Tris–HCl (pH 7.6), 250 mM KCl, 10 mM 2-mercaptoethanol, 10 mM imidazole, 0.1% Tween-20, 10% glycerol, and freeze/thawed at –80 °C twice. After sonication, the protein extract was cleared by centrifugation and applied into 1 ml of Ni-NTA agarose. Ni-NTA beads were washed three times with 10 column volumes of 50 mM Tris–HCl (pH 7.6), 250 mM KCl, 10 mM 2-mercaptoethanol, 20% glycerol, 0.1% Tween-20 buffer supplemented with 30 mM imidazole. Proteins were eluted from the Ni-NTA beads with the same buffer containing 250 mM imidazole.
PP2C enzymatic assays
For PP2C assays, 1 µM dNHAB1 and 2 µM PYR/PYL were used (Santiago et al., 2009). Proteins were incubated with the different chemicals (DMSO, ABA, or P-Acid B) for 10 min in a volume of 50 µl containing 10 mM MnCl2. Reactions were initiated by the addition of 50 µl of 50 mM Tris pH 8.0, 50 mM p-nitrophenyl phosphate (pNPP). Dephosphorylation of pNPP was followed by monitoring the absorbance at 405 nm during 20 min. The activity of dNHAB1 in the absence of any receptor was set to 100%.
Results
Effect of pyrenophoric acids on seed germination
P-Acid, P-Acid B, and P-Acid C (Fig. 1A) were purified from an organic extract of P. semeniperda (strain WRK10-22) grown in solid B. tectorum (cheatgrass) seed culture following the procedure described previously (Masi et al., 2014c, d) and identified by comparing the physical and spectroscopic properties with those previously reported (Masi et al., 2014c, d). To assess the effects of P-Acids in planta, Arabidopsis wild-type Col-0 seeds were germinated in the presence of 100 µM of each P-Acid, 1 µM ABA, or DMSO as a control. P-Acid and P-Acid C did not have any effect on seed germination, while P-Acid B severely delayed germination and markedly inhibited seedling establishment (Fig. 1B). At day 3 after sowing, 100% of Col-0 seedlings established under control (DMSO), P-Acid, or P-Acid C conditions. However, the P-Acid B treatment reduced seedling establishment to 10%, similar to a treatment with 1 µM ABA (Fig. 1C). Therefore, it seems that among P-Acids, P-Acid B, produced by the fungus P. semeniperda, has cross-kingdom activity, as it is able to inhibit plant seed germination.
Pyrenophoric acid B activates the ABA signaling pathway
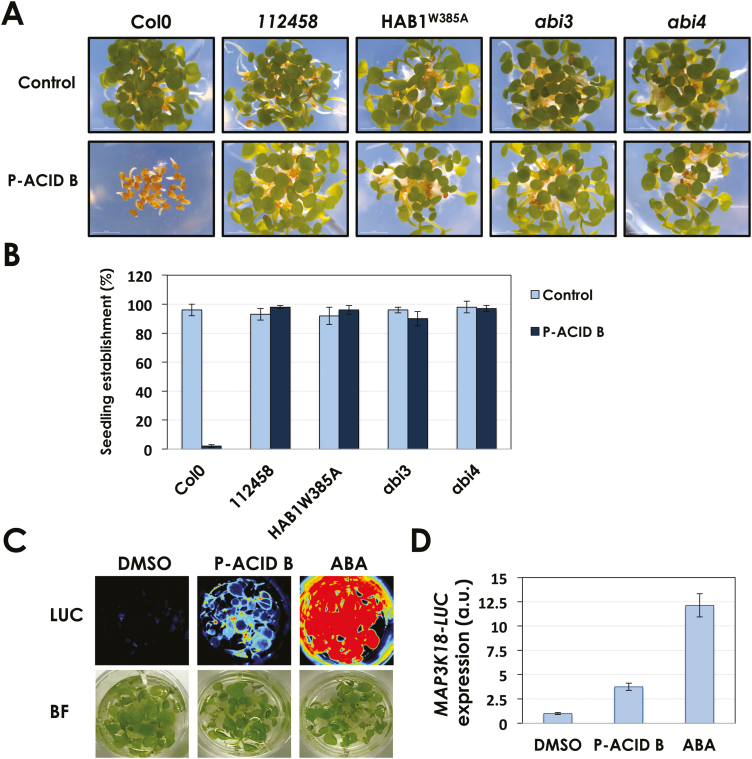
In order to gain further insight into the mode of action of P-Acid B, we used several mutants affected in the early and late steps of ABA perception and signaling. We used the 112458 sextuple mutant impaired in PYR1, PYL1, PYL2, PYL4, PYL5, and PYL8 ABA receptors (Gonzalez-Guzman et al., 2012); 35S:HAB1W385A, a transgenic line expressing an engineered mutant form of the PP2C HAB1 unable to interact with ABA receptors (Dupeux et al., 2011); and abi3 and abi4 that carry mutations in two transcription factors central to the activation of ABA signaling (Nambara et al., 2002). All mutants tested were insensitive to P-Acid B application (Fig. 2A). While only 10% of Col-0 seedlings established in the presence of 100 µM P-Acid B, all the mutants reached nearly 100% seedling establishment 3 d after the treatment (Fig. 2B). These results indicate that the pathogen-produced P-Acid B uses the plant ABA signaling pathway to inhibit seed germination. To elucidate if P-Acid B could activate ABA-dependent gene expression, we used an ABA reporter line that carries the ABA-inducible promoter pMAPKKK18 (Okamoto et al., 2013) fused to the firefly luciferase enzyme. This reporter line is highly activated by ABA (Fig. 2C, D) (Vaidya et al., 2017). P-Acid B was able to activate the reporter line since pMAPKKK18-LUC+ seedlings treated with 200 µM P-Acid B during 6 h showed an ~4.5-fold increase on LUC activity due to the activation of the ABA-responsive promoter pMAPKKK18 (Fig. 2D). In conclusion, P-Acid B is able to activate ABA signaling, including ABA-dependent gene expression, to inhibit seed germination.
Fig. 2.
Pyrenophoric acid B (P-Acid B) activates the ABA signaling pathway to inhibit seed germination. (A) Pictures of Arabidopsis seeds treated with 100 μM P-Acid B or DMSO as control 3 d after being sown. Col-0 seed was used along with mutants in the ABA signaling pathway. (B) Quantification of seedling establishment 3 d after seeds were sown on DMSO or 100 μM P-Acid B. Values represent the mean ±SD of 90 seeds used in three different experiments. (C) Luminescence detection (LUC) of seedlings of the ABA reporter line pMAPKKK18-LUC+ treated with DMSO as control, 200 μM P-Acid B, or 25 μM ABA for 6 h. The image of the seedlings under bright light (BF) is also included. (D) Quantification of the pMAPKKK18-LUC+ expression shown in (C).
Pyrenophoric acid B does not activate ABA receptors
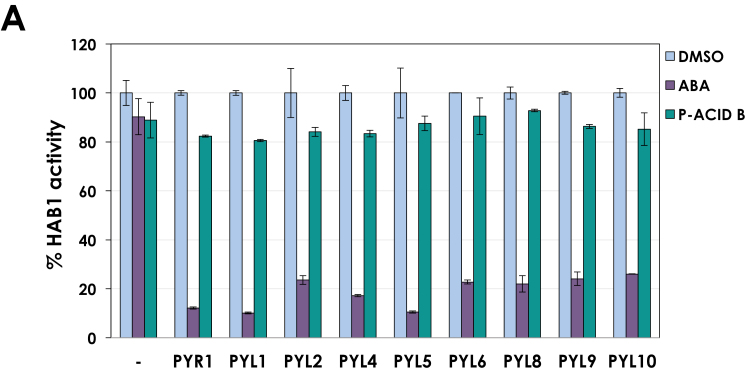
P-Acid B inhibits seed germination, and the sextuple mutant 112458 is insensitive to the application of P-Acid B (Fig. 2A, B). In addition, P-Acids are sesquiterpenoids with structural similarities to ABA (Fig. 1A). We reasoned that P-Acid B could activate the ABA signaling pathway by directly binding to ABA receptors. To investigate this possibility, we used PP2C phosphatase assays with different ABA receptors and the PP2C phosphatase HAB1. The PYR/PYL/RCAR family of ABA receptors binds ABA within a hydrophobic cavity that defines the ABA-binding pocket (Melcher et al., 2009; Santiago et al., 2009). After ABA binding, allosteric changes induced in ABA receptors facilitate their interaction with PP2Cs, inhibiting phosphatase activity. Therefore, the binding of small molecules to ABA receptors can be followed by in vitro PP2C phosphatase assays as reported elsewhere (Park et al., 2009; Okamoto et al., 2013; Vaidya et al., 2017). We performed in vitro PP2C phosphatase assays using recombinant PYR1, PYL1, PYL2, PYL4, PYL5, PYL6, PYL8, and PYL10. As expected, ABA bound to all receptors tested, producing the concomitant inhibition of HAB1 phosphatase activity by 80–90%. However, P-Acid B was not able to inhibit HAB1 in the presence of any of the receptors tested (Fig. 3), strongly suggesting that despite structural and functional similarities to ABA, P-Acid B does not activate ABA receptors.
Fig. 3.
Pyrenophoric acid B (P-Acid B) does not activate ABA receptors. PP2C phosphatase assay using HAB1 (1 μM) and different ABA receptors (2 μM) in the presence of DMSO as control, 10 μM ABA, or 100 μM P-Acid B. HAB1 phosphatase activity was set to 100% in the absence of receptor, and pNPP was used as a chromogenic substrate. Values represent means ±SD (n=3).
P-Acid B exploits the plant ABA biosynthetic pathway to inhibit seed germination
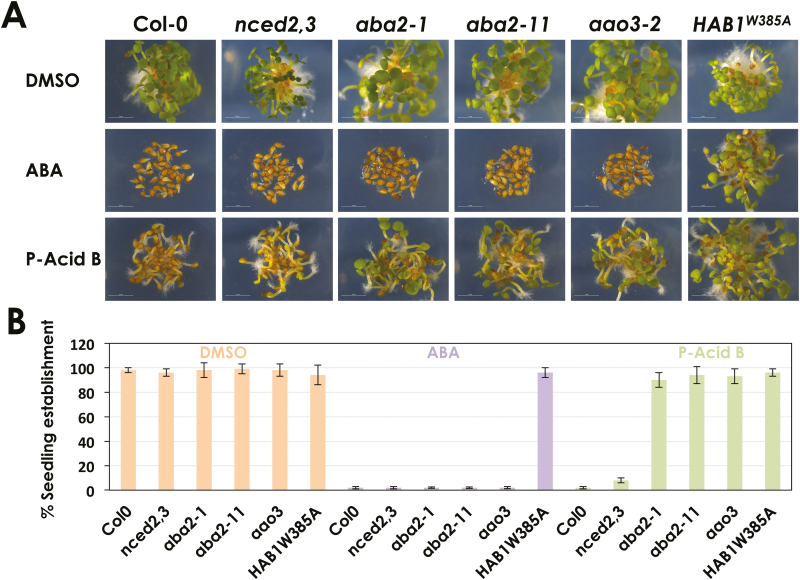
P-Acid B inhibits seed germination, and ABA-insensitive mutants are also insensitive to P-Acid B, suggesting that P-Acid B needs an intact ABA signaling pathway to inhibit seed germination. To better understand P-Acid B activity, we analyzed the effect of P-Acid B treatment on the germination of mutants affected in different steps of the ABA biosynthetic pathway. We used aba1-101, compromised in the early steps of ABA biosynthesis (Barrero et al., 2005), the double mutants nced2nced3 (nced2,3) and nced3,5, impaired in the production of xanthoxin, aba2-1 and aba2-11, loss-of-function alleles of ABA2 that cannot convert xanthoxin into abscisic aldehyde (Cheng et al., 2002; González-Guzmán et al., 2002), and lastly aao3-2 and aba3-1, blocked in the last step of ABA biosynthesis where abscisic aldehyde is oxidized to ABA (Schwartz et al., 1997; González-Guzmán et al., 2004). As expected, while all the genotypes germinated and established 3 d after sowing in control (DMSO) conditions, 1 µM ABA treatment inhibited germination and seedling establishment in the wild-type Col-0 and also in all the ABA-deficient mutants tested (Fig. 4A, B; Supplementary Fig. S1 at JXB online). As in previous experiments, P-Acid B delayed germination and severely affected seedling establishment of Col-0 seeds. Interestingly, aba2-1, aba2-11, aao3-2, and aba3-1 mutants were resistant to P-Acid B treatment, indicating that ABA biosynthesis from ABA2 onwards is required for P-Acid B bioactivity on seed germination. However, nced2,3 which is impaired in the upstream step of ABA biosynthesis that precedes ABA2 activity, was sensitive to P-Acid B, with a clear reduction in seedling establishment in response to this treatment (Fig. 4A, B). This result was confirmed using a different NCED double mutant, nced3,5, and also the mutant aba1-101, impaired in the first step of ABA biosynthesis (Supplementary Fig. S1). Thus, while P-Acid B reduced seedling establishment of Col-0, aba1-101, nced2,3, and nced3,5 seeds to <10%, the compound was unable to produce any effect on the other ABA biosynthetic mutants aba2-1, aba2-11, aao3-2, and aba3-1 (Fig. 4A, B; Supplementary Fig. S1). Therefore, P-Acid B can complement the ABA biosynthetic defect of aba1-101, nced2,3, and nced3,5 but requires downstream enzymatic steps. These results suggest that P-Acid B requires ABA2 activity to inhibit seed germination and establishment. Actually, since P-Acid B is structurally related to xanthoxin and ABA2 is able to convert xanthoxin-related compounds into abscisic aldehyde (González-Guzmán et al., 2002; Nambara and Marion-Poll, 2005), we hypothesized that P-Acid B could be used by ABA2 as a substrate to produce ABA or an ABA-mimic, thus explaining the activation of the ABA signaling pathway by P-Acid B and its negative effect on seed germination.
Fig. 4.
Pyrenophoric acid B (P-Acid B) uses the ABA biosynthesis pathway to inhibit seed germination. (A) Pictures of Arabidopsis seeds treated with 1 μM ABA, 100 μM P-Acid B, or DMSO as control, 3 d after being sown. Col-0 was used along with mutants in the ABA biosynthesis pathway. (B) Quantification of seedling establishment 3 d after seeds were sown on DMSO, 1 μM ABA, or 100 μM P-Acid B. Values represent means ±SD of 90 seed used in three different experiments.
Discussion
The fungus P. semeniperda is a plant pathogen specialized in infecting seed tissues. Due to its ability to infect cheatgrass, one of the most invasive plant species in the USA, lately it has been considered as a biocontrol agent for invasive grass species. For this purpose, P. semeniperda produces a series of toxins that kill the host cells, including the mycotoxin cytochalasin B, which impedes the correct polymerization of actin filaments, leading to cell death (Scherlach et al., 2010). However, cytochalasin B is not the only low molecular weight compound produced by P. semeniperda that has a role in infection. Recently, a series of metabolites was isolated and identified from P. semeniperda cultures and, among them, P-Acid, P-Acid B, and P-Acid C were found (Masi et al., 2014c, d). P-Acids are bioactive compounds able to inhibit seed germination and coleoptile growth, with P-Acid B the most active of these compounds (Masi et al., 2014d). In this work, we have investigated the molecular mechanism underlying the mode of action of P-Acids. We have found that P-Acid B can inhibit A. thaliana seed germination through the ABA signaling pathway. P-Acid B needs an intact ABA signaling pathway to inhibit seed germination since mutants affected in the first steps of ABA perception as well as mutants impaired in central transcription factors essential for the activation of ABA responses were insensitive to P-Acid B application.
P-Acids are ABA-related compounds with structural similarities to the plant hormone. It has been previously reported that ABA receptors can accommodate ABA-like molecules within their hydrophobic pocket (Kepka et al., 2011; Benson et al., 2015). For example, the ABA degradation by-product, phaseic acid (PA), is able to bind to certain ABA receptors (Weng et al., 2016). PA has submicromolar activity towards the ABA receptors PYL3, PYL5, and PYL6, and a role for PA on water stress tolerance has been proposed (Lozano-Juste and Cutler, 2016; Rodriguez, 2016; Weng et al., 2016). Other hormone receptors can also accommodate hormone analogs or different forms of the hormone. GID1, the gibberellic-acid receptor, binds different gibberellins (Murase et al., 2008; Shimada et al., 2008), and the jasmonic acid (JA) receptor COI1 is activated not only by its plant derived ligand, JA-Ile, but also by the JA analog coronatin, produced by biotrophic bacteria (Sheard et al., 2010). Therefore, we reasoned that the ABA-related sequiterpenoid P-Acid B could directly bind to ABA receptors. However, we could not find activation of any of the ABA receptors studied, indicating that P-Acid B, despite its structural similarities to ABA, does not activate ABA receptors directly, suggesting a different mechanism of action. An alternative hypothesis for P-Acid B bioactivity is acting as a precursor of ABA, feeding the biosynthetic pathway and promoting ABA accumulation. This hypothesis proved to be correct since aba3-1, aao3-2, aba2-1, and aba2-11 mutants affected in ABA biosynthesis could establish in the presence of P-Acid B while Col-0 plants could not. Therefore, P-Acid B may require de novo synthesis of ABA to inhibit seedling establishment. In order to pinpoint the entrance of P-Acid B into the ABA biosynthetic pathway, we have compared sensitivity to P-Acid B of mutants impaired in different steps of ABA biosynthesis. Thus, when we analyzed the effect of P-Acid B on the aba1-101, nced2,3, and nced3,5 mutants, we found that these strains are not resistant to P-Acid B, while aba2, aao3, and aba3 single mutants could germinate and establish in the presence of P-Acid B. These data suggest that P-Acid B enters the ABA biosynthetic pathway downstream of NCED and requires ABA2, AAO3, and ABA3 for bioactivity. AAO3 encodes an aldehyde oxidase and might restore the carboxylic group of P-Acid B if it was reduced to aldehyde by the reductive ability of plant cells for exogenous molecules. Indeed, the reduction of non-activated carboxylic groups in different substrates has been described for at least nine species of cultured plant cells (Villa and Molinari, 2008). ABA2 is a member of the short-chain alcohol dehydrogenase protein family crucial for ABA biosynthesis (Cheng et al., 2002; González-Guzmán et al., 2002). It catalyzes the conversion of xanthoxin into abscisic aldehyde that is used by the AAO3 protein to produce ABA. Accordingly, the levels of ABA in aba2 mutants are severely reduced (Léon-Kloosterziel et al., 1996; González-Guzmán et al., 2002), indicating that this is a key step in the basal production of ABA in plants. The conversion of xanthoxin into abscisic aldehyde by ABA2 involves not one but several catalytic steps. ABA2 is able to oxidize the 4'-hydroxyl group of xanthoxin into a 4'-keto group but it also isomerizes the 1',2'-epoxide into a 1'-hydroxy-2'-ene group (González-Guzmán et al., 2002). Due to structural similarities between xanthoxin and P-Acid B, we hypothesized that ABA2 could oxidize the 4'-hydroxyl group and dehydrogenate C-2' and C-3' of P-Acid B, generating ABA. Actually, the enzyme ABA2 can use certain alcohol substrates different from xanthoxin, being able to oxidize 3,5,5′-trimethylcyclohexanol alcohol, although with higher Km values (González-Guzmán et al., 2002). Taken together, we propose P-Acid B as a fungal compound with cross-kingdom activity that might be able to use the plant enzyme ABA2 to induce the synthesis of ABA and inhibit seed germination. A body of evidence indicates that the fungus P. semeniperda can benefit from reducing seed germination and seedling establishment through P-Acid B production. P. semeniperda reproductive success is often associated with slow germinating or dormant seeds (Finch-Boekweg et al., 2016), while fast germinating seeds usually tolerate P. semeniperda infection and develop into normal plants (Finch-Boekweg et al., 2016). Additionally, the distribution of P. semeniperda populations is correlated with environmental conditions that reduce seed germination rates and, therefore, P. semeniperda is found more frequently in dry scenarios rather than humid ones. Dry conditions reduce seed germination and induce post-germination growth arrest (Beckstead et al., 2007, 2016; Meyer et al., 2015).
In summary, we propose that the fungus P. semeniperda produces pyrenophoric acid B to inhibit seed germination through the activation of plant ABA biosynthesis, in order to increase its fitness by increasing the probability of pathogen-caused seed mortality, thereby increasing resources available for pathogen reproduction.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Pyrenophoric acid B (P-Acid B) requires ABA2 and onwards to inhibit seedling establishment.
Acknowledgements
This research was funded in part by Grant JFSP-11-S-206 to SM from the Joint Fire Sciences Program of the US Departments of Agriculture and Interior and, in part, to MM, by Programme STAR 2017 financially supported by UniNA and Compagnia di San Paolo. JL-J is funded by a Marie-Sklodowska Curie Reintegration Grant H2020-MSCA-707477. Work in the PLR lab is supported by RTC-2017-6019-2 and BIO2017-82503-R grants from Ministerio de Ciencia, Innovación y Universidades. MAF is a recipient of a FPU fellowship from MECD. AE is associated to the Istituto di Chimica Biomolecolare del CNR, Pozzuoli, Italy. We would like to thank Ebe Merilo (University of Tartu) for sharing nced3,5 and aba3-1 mutant seeds.
References
- Antoni R, Gonzalez-Guzman M, Rodriguez L, Rodrigues A, Pizzio GA, Rodriguez PL. 2012. Selective inhibition of clade A phosphatases type 2C by PYR/PYL/RCAR abscisic acid receptors. Plant Physiology 158, 970–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audran C, Borel C, Frey A, Sotta B, Meyer C, Simonneau T, Marion-Poll A. 1998. Expression studies of the zeaxanthin epoxidase gene in Nicotiana plumbaginifolia. Plant Physiology 118, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Piqueras P, González-Guzmán M, Serrano R, Rodríguez PL, Ponce MR, Micol JL. 2005. A mutational analysis of the ABA1 gene of Arabidopsis thaliana highlights the involvement of ABA in vegetative development. Journal of Experimental Botany 56, 2071–2083. [DOI] [PubMed] [Google Scholar]
- Beckstead J, Meyer SE, Ishizuka TS, McEvoy KM, Coleman CE. 2016. Lack of host specialization on winter annual grasses in the fungal seed bank pathogen Pyrenophora semeniperda. PLoS One 11, e0151058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead J, Meyer SE, Molder CJ, Smith C. 2007. Germination rate determines pathogen-caused seed mortality. Annals of Botany 99, 907–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson CL, Kepka M, Wunschel C, Rajagopalan N, Nelson KM, Christmann A, Abrams SR, Grill E, Loewen MC. 2015. Abscisic acid analogs as chemical probes for dissection of abscisic acid responses in Arabidopsis thaliana. Phytochemistry 113, 96–107. [DOI] [PubMed] [Google Scholar]
- Castillo MC, Lozano-Juste J, González-Guzmán M, Rodriguez L, Rodriguez PL, León J. 2015. Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Science Signaling 8, ra89. [DOI] [PubMed] [Google Scholar]
- Cheng WH, Endo A, Zhou L, et al. 2002. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. The Plant Cell 14, 2723–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino A, Masi M, Evidente M, Superchi S, Evidente A. 2015. Fungal phytotoxins with potential herbicidal activity: chemical and biological characterization. Natural Product Reports 32, 1629–1653. [DOI] [PubMed] [Google Scholar]
- Dupeux F, Antoni R, Betz K, et al. 2011. Modulation of abscisic acid signaling in vivo by an engineered receptor-insensitive protein phosphatase type 2C allele. Plant Physiology 156, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch-Boekweg H, Gardner JS, Allen PS, Geary B. 2016. Postdispersal infection and disease development of Pyrenophora semeniperda in Bromus tectorum seeds. Phytopathology 106, 236–243. [DOI] [PubMed] [Google Scholar]
- Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, Berger A, Sechet J, To A, North HM, Marion-Poll A. 2012. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. The Plant Journal 70, 501–512. [DOI] [PubMed] [Google Scholar]
- Germino MJ, Chambers JC, Brown CS, eds.2016. Exotic brome-grasses in arid and semiarid ecosystems of the Western US: causes, consequences, and management implications. Cham: Springer International Publishing. [Google Scholar]
- González-Guzmán M, Abia D, Salinas J, Serrano R, Rodríguez PL. 2004. Two new alleles of the abscisic aldehyde oxidase 3 gene reveal its role in abscisic acid biosynthesis in seeds. Plant Physiology 135, 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL. 2002. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. The Plant Cell 14, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guzman M, Pizzio GA, Antoni R, et al. 2012. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. The Plant Cell 24, 2483–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung W. 2010. The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Functional Plant Biology 37, 806–812. [Google Scholar]
- He Y, Hao Q, Li W, Yan C, Yan N, Yin P. 2014. Identification and characterization of ABA receptors in Oryza sativa. PLoS One 9, e95246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. 2000. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiology 123, 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepka M, Benson CL, Gonugunta VK, Nelson KM, Christmann A, Grill E, Abrams SR. 2011. Action of natural abscisic acid precursors and catabolites on abscisic acid receptor complexes. Plant Physiology 157, 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M. 1996. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. The Plant Journal 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. 1998. Abscisic acid signal transduction. Annual Review of Plant Physiology and Plant Molecular Biology 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Lozano-Juste J, Cutler SR. 2016. Hormone signalling: ABA has a breakdown. Nature Plants 2, 16137. [DOI] [PubMed] [Google Scholar]
- Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. 2009. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A. 1996. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. The EMBO Journal 15, 2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Masi M, Evidente A, Meyer S, Nicholson J, Muñoz A. 2014a Effect of strain and cultural conditions on the production of cytochalasin B by the potential mycoherbicide Pyrenophora semeniperda (Pleosporaceae, Pleosporales). Biocontrol Science and Technology 24, 53–64. [Google Scholar]
- Masi M, Meyer S, Cimmino A, Andolfi A, Evidente A. 2014c Pyrenophoric acid, a phytotoxic sesquiterpenoid penta-2,4-dienoic acid produced by a potential mycoherbicide, Pyrenophora semeniperda. Journal of Natural Products 77, 925–930. [DOI] [PubMed] [Google Scholar]
- Masi M, Meyer S, Cimmino A, Clement S, Black B, Evidente A. 2014d Pyrenophoric acids B and C, two new phytotoxic sesquiterpenoids produced by Pyrenophora semeniperda. Journal of Agricultural and Food Chemistry 62, 10304–10311. [DOI] [PubMed] [Google Scholar]
- Masi M, Meyer S, Clement S, Andolfi A, Cimmino A, Evidente A. 2014b Spirostaphylotrichin W, a spirocyclic γ-lactam isolated from liquid culture of Pyrenophora semeniperda, a potential mycoherbicide for cheatgrass (Bromus tectorum) biocontrol. Tetrahedron, 70, 1497–1501. [Google Scholar]
- Melcher K, Ng LM, Zhou XE, et al. 2009. A gate–latch–lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer SE, Clement S, Beckstead J. 2013. Annual brome control using a native fungal seed pathoge n. https://www.fs.usda.gov/treesearch/pubs/43772 [Google Scholar]
- Meyer SE, Masi M, Clement S, Davis TL, Beckstead J. 2015. Mycelial growth rate and toxin production in the seed pathogen Pyrenophora semeniperda: resource trade-offs and temporally varying selection. Plant Pathology 64, 1450–1460. [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. 2008. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P. 2002. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana. Genetics 161, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M, Peterson FC, Defries A, Park S-Y, Endo A, Nambara E, Volkman BF, Cutler SR. 2013. Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proceedings of the National Academy of Sciences, USA 110, 12132–12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, et al. 2009. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pri-Tal O, Shaar-Moshe L, Wiseglass G, Peleg Z, Mosquna A. 2017. Non-redundant functions of the dimeric ABA receptor BdPYL1 in the grass Brachypodium. The Plant Journal 92, 774–786. [DOI] [PubMed] [Google Scholar]
- Rodriguez PL. 2016. Abscisic acid catabolism generates phaseic acid, a molecule able to activate a subset of ABA receptors. Molecular Plant 9, 1448–1450. [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. 2009. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. [DOI] [PubMed] [Google Scholar]
- Scherlach K, Boettger D, Remme N, Hertweck C. 2010. The chemistry and biology of cytochalasans. Natural Product Reports 27, 869–886. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. 1997. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Seo M, Peeters AJM, Koiwai H, Oritani T, Marion-Poll A, Zeevaart JAD, Koornneef M, Kamiya Y, Koshiba T. 2000. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proceedings of the National Academy of Sciences, USA 97, 12908–12913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. 2010. Jasmonate perception by inositol-phosphate-potentiated COI1–JAZ co-receptor. Nature 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. 2008. Structural basis for gibberellin recognition by its receptor GID1. Nature 456, 520–523. [DOI] [PubMed] [Google Scholar]
- Takezawa D, Komatsu K, Sakata Y. 2011. ABA in bryophytes: how a universal growth regulator in life became a plant hormone? Journal of Plant Research 124, 437–453. [DOI] [PubMed] [Google Scholar]
- Vaidya AS, Peterson FC, Yarmolinsky D, et al. 2017. A rationally designed agonist defines subfamily IIIA abscisic acid receptors as critical targets for manipulating transpiration. ACS Chemical Biology 12, 2842–2848. [DOI] [PubMed] [Google Scholar]
- Villa R, Molinari F. 2008. Reduction of carbonylic and carboxylic groups by plant cell cultures. Journal of Natural Products 71, 693–696. [DOI] [PubMed] [Google Scholar]
- Weng JK, Ye M, Li B, Noel JP. 2016. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 166, 881–893. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57, 781–803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.