The expression of two cellulases, LcCEL2 and LcCEL8, is strongly associated with fruitlet abscission in litchi and is regulated by direct binding of an HD-Zip transcription factor to the gene promotors.
Keywords: Cell separation, cellulase, fruit abscission, HD-Zip, Litchi, chinensis, lychee
Abstract
Cellulases play important roles in the shedding of plant organs; however, little is yet known about the functions of cellulase genes during the process of organ abscission. Abnormal fruitlet abscission is a serious problem in the production of litchi (Litchi chinensis), an economically important fruit widely grown in South Asia. In this study, two abscission-accelerating treatments (carbohydrate stress and application of ethephon) were evaluated in litchi fruitlets. Cell wall degradation and cell separation were clearly observed in the abscission zones of treated fruitlets, consistent with enhanced cellulase activities and reduced cellulose contents. The expression of two cellulase genes (LcCEL2 and LcCEL8) was strongly associated with abscission. Floral organs of transgenic Arabidopsis overexpressing LcCEL2 or LcCEL8 showed remarkably precocious abscission. Electrophoretic mobility shift assays and transient expression experiments demonstrated that a novel homeodomain-leucine zipper transcription factor, LcHB2, could directly bind to and activate HD-binding cis-elements in the LcCEL2 and LcCEL8 promoters. Our results provide new information regarding the transcriptional regulation of the cellulase genes responsible for cell wall degradation and cell separation during plant organ shedding, and raise the possibility of future manipulation of litchi fruitlet abscission by modulation of the activities of these two cellulases.
Introduction
Litchi (Litchi chinensis) is an important tropical fruit crop that is currently cultivated in over 20 countries. Under normal conditions, litchi produces many inflorescences with 100–250 female flowers per inflorescence. However, more than 95% of the initial female flowers do not develop into mature fruit (Stern et al., 1995; Mitra et al., 2003). For normal-seeded litchi cultivars, three distinct waves of fruit abscission are observed (Yuan and Huang, 1988). The first occurs around the end of week 1 after full bloom with ~60% of fruitlet loss. The second wave occurs around the end of week 3 after full bloom, with about half of the remaining fruitlets being abscised. Soon after that, a small third wave begins. For aborted-seed cultivars, an additional pre-harvest abscission occurs 2–3 weeks before harvest and only ~60% of the remaining fruit survive (Yuan and Huang, 1988). In the period 2013–2015, the average yield of litchi in China was only 3.8 t ha–1 (Qi et al., 2016). The excessive abscission of flowers/fruitlets is one of the main factors responsible for the universally low productivity in litchi (Yuan and Huang, 1988; Mitra et al., 2003).
An essential event during abscission is the separation of specialized cells within the abscission zone (AZ), which is located at the base of the organ that will be shed (Patterson, 2001; Estornell et al., 2013). The breakdown of cell wall components in AZ cells is dependent upon the activities of cell wall-specific hydrolases (Roberts et al., 2002). Endo-(1,4)-β-D-glucanases (or cellulases, CELs) are responsible for cellulose degradation and are thought to be important for organ abscission (Sexton and Roberts, 1982). In bean (Phaseolus vulgaris), the structural integrity of the petiole AZ is lost during abscission owing to cellulase activity (Horton and Osborne, 1967), which is the result of de novo synthesis of a high-salt extractable cellulase named BEAN ABSCISSION CELLULASE (BAC) (Reid and Lewis, 1974; Tucker et al., 1991). In tomato (Solanum lycopersicum), three cellulase genes (SlCel1, SlCel2, and SlCel5) are found to be highly expressed in the pedicel AZs where cell separation occurs (Lashbrook et al., 1994; del Campillo and Bennett, 1996). Antisense suppression of SlCel1 mRNA accumulation reduces flower abscission by up to one third (Lashbrook et al., 1998), while knocking down the expression of SlCel2 results in greater force being required to remove fruits (Brummell et al., 1999). SlCel5 has been used as a cell wall-degradation marker gene for microarray analysis during ethylene-promoted tomato flower abscission (Wang et al., 2013). In Arabidopsis, there are 25 putative cellulase family members (Urbanowicz et al., 2007). A transcriptomic analysis using laser-capture microdissection has reported the induction of cellulase family members AtCel3 (At1g71380), AtCel5 (At1g22880), EGase10 (At1g75680), and EGase11 (At2g32990) in stamen AZs and their involvement in cell separation (Lashbrook and Cai, 2008). Recently, a newly discovered cellulase gene, AtCEL6 (At4g39010), has been shown to promote silique dehiscence by promoting cell disintegration in the separation layer (He et al., 2018). Overall, these results suggest that enhancement of the expression and activity of cellulases is important for cell separation during organ abscission.
To date, at least 10 different abscission-associated cellulase genes have been found in fruit crops, including ppEG1 and ppEG4 in peach (Prunus persica) (Trainotti et al., 1997, 2006), PaCel1 in avocado (Persea americana) (Tonutti et al., 1995), MdEG1 in apple (Malus×domestica) (Li and Yuan, 2008), and CitCEL3, CitCEL6 (CsCEL-a1), CitCEL10, CitCEL22, and CsCEL-b1 in citrus (Citrus sinensis) (Kazokas and Burns, 1998; Merelo et al., 2017). However, the functions of these cellulases during abscission as well as their underlying mechanisms are still largely unknown.
Promoter analysis of BAC gene during bean leaf abscission has shown that ethylene positively induces its expression, whereas auxin strongly suppresses expression (Koehler et al., 1996). However, because the core elements for ethylene and auxin responses have not been found in the BAC promoter (Koehler et al., 1996), these hormones probably affect BAC expression indirectly. Further analysis by Tucker et al. (2002) implicated three TGA-type basic leucine zipper (bZIP) transcription factors (TFs) in activating the BAC promoter. The Arabidopsis HD-Zip (homeodomain-leucine zipper) HDG11 can directly up-regulate the cellulase gene At2g32990 during the elongation of roots (Xu et al., 2014). Members of the HD-Zip gene family in higher plants encode TFs containing a DNA-binding homeodomain (HD) and an adjacent leucine-zipper domain (bZIP, also known as an LZ domain) (Ariel et al., 2007). The HD-Zip family comprises four subfamilies (HD-Zip I–IV), each of which plays specific roles in plant development (Ariel et al., 2007); however, whether they are involved in the shedding of plant organs has yet not been studied.
Carbohydrate deficiency and hormone effects are the two main factors considered to affect fruit drop in litchi (Yuan and Huang, 1988; Hieke et al., 2002). Comprehensive transcriptome profiling studies under carbohydrate stress and ethylene-induced abscission have been documented in our previous studies (Li et al., 2015a, 2015b) that identified many genes potentially involved in litchi fruitlet abscission. These studies revealed several genes with putative functions related to cell wall modification, as well as TFs such as HD-Zip genes. However, which genes are essential for abscission and how they are regulated is not yet known. In the present study, two cellulase genes (LcCEL2 and LcCEL8) and a novel HD-Zip TF (LcHB2) were identified as key genes associated with fruitlet abscission in litchi. Ectopic expression of LcCEL2 and LcCEL8 in Arabidopsis showed that these genes could induce precocious floral organ abscission. Further, LcHB2 was shown to directly bind the promoters and activate expression of LcCEL2 and LcCEL8. These results add to our understanding of the transcriptional regulatory mechanisms by which cellulase genes are involved in plant organ shedding.
Materials and methods
Plant materials and treatments
Several 12-year-old litchi trees (Litchi chinensis Sonn. cvs. ‘Zhumuru’ and ‘Kulin’) were randomly chosen in an orchard at the South China Agricultural University (Guangzhou, China) in 2015. Thirty fruitlet-bearing shoots of similar diameter (about 5–8 mm) growing in different directions were tagged on each tree. At 30 d after anthesis, half of the fruitlet-bearing shoots of ‘Zhumuru’ were treated by girdling (removing a 0.5-cm wide ring of bark and cambium from around the base of the branch) and by defoliation beyond the ring of girdling, hereafter referred to as the GPD treatment (‘girdling plus defoliation’; Supplementary Fig. S1A at JXB online). The remaining untreated shoots were used as the control. Half of the fruitlet-bearing shoots of ‘Kulin’ were dipped in a solution containing 250 mg l−1 ethephon and 0.05% Tween® 80 surfactant for 1 min (designated as the ETH treatment) and the remaining shoots were dipped in water and used as the control. Of the 15 treated shoots, three were used for monitoring the fruitlet abscission dynamics and the remainder were used for tissue sampling. We calculated the cumulative rate of fruitlet abscission according to our previous study (Kuang et al., 2012). Using a sharp razor blade, different regions of the fruitlet pedicels were sampled based around the fruitlet abscission zone (FAZ), which appears as a visible sunken ring of ~2 mm in length (Supplementary Fig. S1B). In addition to the FAZ, a region of the pedicel of ~2 mm in length between the FAZ and the fruitlet-bearing stem was cut and designated as the basal portion (BP). A region of similar length between the FAZ and the fruitlet was also excised and designated as the apical portion (AP). After separation, the FAZ, BP, and AP tissues were quickly frozen in liquid nitrogen and stored at –80 °C. Each tree was considered a biological replicate and three replicates were performed for each treatment.
Microscopic and histochemical observations
Samples of the FAZs (1 mm length) were fixed in 4% paraformaldehyde with 1% glutaraldehyde at 4 °C for 24 h. Samples were then dehydrated in an ethanol series and embedded in paraffin prior to cutting 10-μm sections. Sections were stained for morphological observation using 1% (w/v) Safranin O (Amresco, Solon, USA) and 1% (w/v) Fast Green FCF (Merck, Overijse, Belgium) (Zou et al., 2011). Insoluble polysaccharides were visualized by periodic acid–Schiff (PAS) staining (Feder and O’Brien, 1968), and cellulose was stained using 0.01% Calcofluor White (Sigma-Aldrich) and the carbohydrate-binding module CBM3a (PlantProbes, Leeds, UK).
Measurement of cellulase activity and cellulose content
Cellulose contents were determined using the anthrone method (Viles and Silverman, 1949). Cellulase activities were measured using a tissue-blotting and gel-diffusion method (Bourgault and Bewley, 2002; Yang et al., 2015) for litchi FAZs and by the DNS (3,5-dinitrosalicylic acid) method for Arabidopsis leaves (Wang et al., 1998), respectively.
Sequence analysis and quantitative real-time PCR (qRT-PCR)
Protein sequences of all candidate genes were retrieved from the litchi genome database (http://litchidb.genomics.cn/page/species/index.jsp), the Arabidopsis genome database (https://www.arabidopsis.org/index.jsp), and the plant genomics resource (Phytozome version 12.1; https://phytozome.jgi.doe.gov/pz/portal.html). Multiple alignments were performed using the ClustalW 1.83 (Thompson et al., 1994) and GeneDoc software (Nicholas, 1997). Phylogenetic trees were constructed using neighbor-joining analysis in MEGA 7 (Tamura et al., 2011) with the Poisson correction model, pairwise deletion method, and bootstrapping with 1000 replicates. Total RNA was isolated using a Column Plant RNAout 2.0 Kit (Tiandz, Beijing, China). qRT-PCR was performed using GoTaq® qPCR Master Mix (Promega) on a CFX96 Real-Time PCR System (Bio-Rad). Gene expression levels were normalized using LcEF-1α and LcGAPDH as the internal reference genes for litchi (Zhong et al., 2011) and AtUBQ for Arabidopsis (Ying et al., 2016). Three biological replicates were performed. All primers used in this study are listed in Supplementary Table S1.
Subcellular localization analysis
Coding sequences of LcCEL2 (1863 bp), LcCEL8 (1491 bp), and LcHB2 (684 bp), each lacking a stop codon, were subcloned into the pEAQ-HT-GFP vector (Ye et al., 2016) and fused in-frame with the green fluorescent protein (GFP) sequence under the control of the CaMV 35S promoter using a ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). Agrobacterium tumefaciens strain GV3101 harboring LcCEL2-GFP, LcCEL8-GFP, LcHB2-GFP, or the positive control were individually infiltrated into the abaxial side of leaves of tobacco (Nicotiana benthamiana) or inoculated into plasmolytic epidermal cells of onion (Allium cepa). GFP fluorescence signals were visualized using an Axioskop 2 Plus fluorescence microscope (Zeiss). All assays were performed with at least three replications.
Generation of transgenic plants and BCECF fluorescence analyses
Coding sequences of LcCEL2 and LcCEL8, each lacking a stop codon, were subcloned under the control of the CaMV 35S promoter into the pCAMBIA1302 vector, and then these constructs were individually transformed into Arabidopsis Columbia-0 (Col-0) ecotype plants using the floral dip method (Clough and Bent, 1998). Further phenotypic analysis and BCECF [2',7'-Bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein] fluorescence assays were conducted as described by Ying et al. (2016). Flower position numbers were counted from the first flower with visible white petals at the top of the inflorescence.
Histochemical GUS assays
Genomic DNA was extracted from litchi leaves following the CTAB protocol (Puchooa, 2004). The promoter fragments of LcCEL2 (2215 bp), LcCEL8 (2192 bp), and LcHB2 (2048 bp) were subcloned into the pCAMBIA1391 vector upstream of the GUS (β- glucuronidase)-coding region (Supplementary Fig. S2A). These constructs were then individually transformed into Arabidopsis Col-0 plants. Inflorescences, siliques, and cauline leaves were incubated in GUS staining buffer (Waryong, Beijing, China) for 2–4 h at 37 °C in the dark. The samples were then decolorized in 100% ethanol, cleared in transparent solution (30 g Chloral hydrate, 10 ml glycerol, 30 ml water) overnight, and visualized using a Zeiss SV11 stereoscope.
Dual-luciferase reporter assays
Diagrams of the effector and reporter vectors used for dual-luciferase reporter assays are shown in Supplementary Fig. S2B, C. The effector and reporter plasmid constructs were co-transformed into tobacco leaves using Agrobacterium infiltration. After 2–3 d, activities of the LUC and REN luciferases were measured using the Dual-Luciferase® Reporter Assay System (Promega) on a Luminoskan Ascent Microplate Luminometer (ThermoFisher Scientific). The results were calculated as the ratio of fluorescence of LUC to that of REN. At least six biological replications were performed for each combination of effector and reporter.
Electrophoretic mobility shift assays (EMSA)
LcHB2 was cloned into the pET-28a(+) vector and expressed in E. coli BL21 (DE3) cells. Recombinant proteins were purified and used for EMSAs along with biotin-labeled fragments (~50 bp) of the LcCEL2 and LcCEL8 promoters. The same fragment, but unlabeled, was used as a competitor, while a probe within the mutant HD-binding cis-elements (interchanging A with G or T with C) was used as a mutant competitor in the assay. EMSAs were performed using a LightShift™ Chemiluminescent EMSA Kit (ThermoFisher Scientific). After cross-linking, the membrane was detected by the chemiluminescence method on a ChemiDoc™ MP Imaging System (Bio-Rad).
Accession numbers
Gene accession numbers are listed in Supplementary Tables S2–S4.
Results
Effects of carbohydrate stress and ethephon treatments on litchi fruitlet abscission
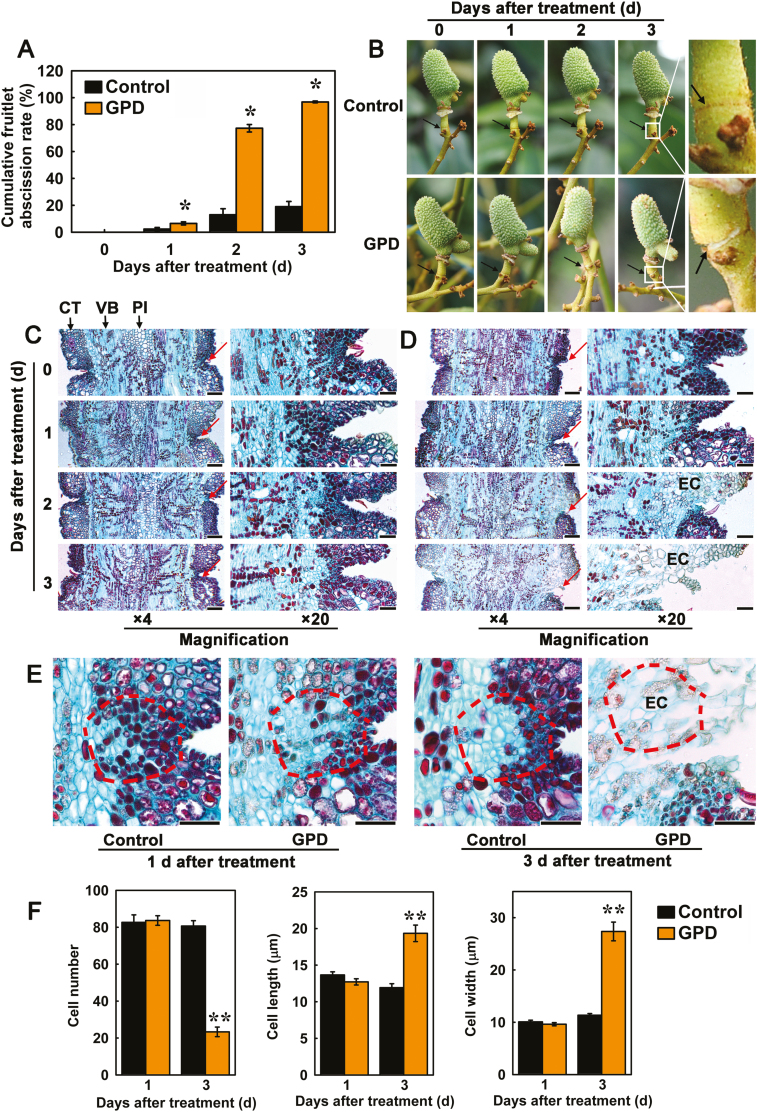
To determine the effect of girdling plus defoliation (GPD), which leads to carbohydrate stress, and application of ethephon (ETH) on fruitlet abscission, the rate of fruitlet drop under each treatment was recorded each day for 5 d after the treatments. Compared to the control, GPD dramatically accelerated fruitlet drop, with a rate of 77.27% at day 2 and up to 96.84% at day 3 (Fig. 1A). At day 3, a clear white ring (the abscission ring) could be seen to have formed across the FAZ, which was caused by fracturing of the epidermis (Fig. 1B). Although the fruitlets remain attached to the branch, they could easily be detached with only a light external force. Under the ETH treatment, the rate of abscission gradually increased and reached a maximum of 92.1% at day 5, when visible abscission rings were again observed (Supplementary Fig. S3A, B). In contrast, fruitlet drop was much slower in the controls, with a cumulative abscission rate of only 18.99% at day 3 in the control for GPD and of 14.5% at day 5 in the control for the ETH treatment.
Fig. 1.
Effects of carbohydrate stress treatment (girdling plus defoliation, GPD) on the cumulative fruitlet abscission rate, phenotypic performance, and cell separation in the fruitlet abscission zone (FAZ) of litchi. (A) Cumulative fruitlet abscission rate. Data are means (±SE) from three replicates. Significant differences between GPD and control branches were determined using Student’s t-test: *P<0.05. (B) Phenotypic characteristics of FAZs during the fruitlet abscission process. The images on the right show magnifications of the FAZs at 3 d after GPD treatment. Arrows indicate the location of the abscission layers. (C, D) Longitudinal sections of the FAZs from fruitlets from control (C) and GPD-treated (D) branches stained with Safranin O and Fast Green at 4× and 20× magnification, respectively. Arrows indicate the location of the abscission layers. CT, cortex; EC, expanding cells; PI, pith; VB, vascular bundle. Scale bars are 200 μm at 4× magnification and 50 μm at 20× magnification. (E) Longitudinal sections of FAZs from control and GPD-treated branches at 1 d and 3 d after treatment. Scale bars are 50 μm. (F) Cell number, cell length, and cell width of the FAZs. A vision field of ~10 000 μm2 (shown by the dashed lines in E) were used to count cell numbers. Up to 90 cells were used to measure the length and width. Significant differences between GPD and control branches were determined using Student’s t-test: **P<0.01, for at least three longitudinal sections.
Cell disruption and separation initiate in the FAZs during abscission
Microscopic observations revealed clear evidence of cell breakdown in the FAZ during litchi fruitlet abscission. The FAZ usually included 7–10 layers of small, dense, oblong cells (Fig. 1C–E; Supplementary Fig. S3C–E). At 2 d following GPD treatment, cortical cells in the FAZ became enlarged and less Safranin O/Fast Green staining was absorbed. Separation of cells in the epidermis and cortex then occurred at the abscission site (Fig. 1D). Soon afterwards, the FAZ cells expanded further and disintegrated, leading to the formation of an intercellular space. As a result, a fracture appeared in the FAZ and further progressed toward the vascular bundle and pith, indicating that cell separation occurred at the FAZ after GPD treatment (Fig. 1D, E). In the FAZ from ETH-treated branches, enlarged and distorted cells appeared on day 3, and the separation layer was visible on day 5 (Supplementary Fig. S3D, E). In contrast, the FAZ of the controls showed no cell expansion and separation at day 3 after GPD and day 5 after ETH (Fig. 1C, E; Supplementary Fig. S3C, E). Both treatments significantly decreased the number of FAZ cells (only 29% of the control at day 3 after GPD and 49% of the control at day 5 after ETH treatment), and the cells were longer and wider than those in the controls (Fig. 1F; Supplementary Fig. S3F). All the results indicated that the enlargement of FAZ cells was involved in the processes of cell disruption and separation during fruitlet abscission.
Cellulose degradation occurs upon disruption of FAZ cells during abscission
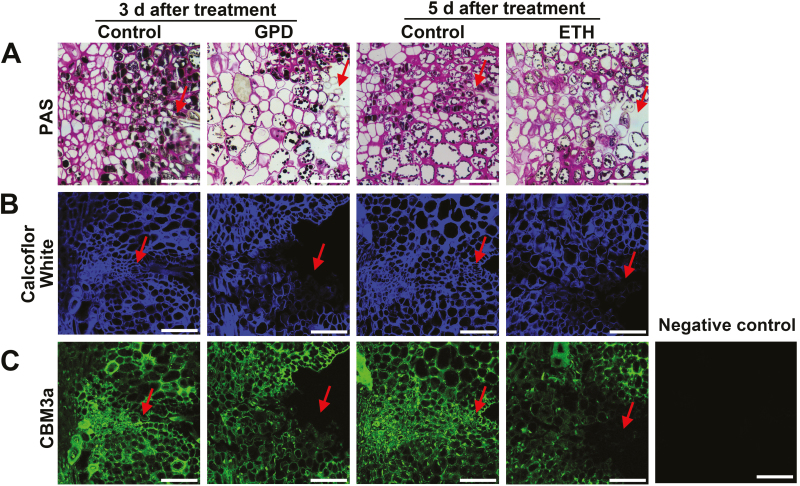
PAS staining was used to characterize the anatomical locations of insoluble polysaccharides in the FAZs after the GPD and ETH treatments. The walls of the separation layer cells in the FAZs showed a decreased affinity for PAS compared to those of the controls, indicating lower contents of insoluble cell wall polysaccharides in the FAZ cells (Fig. 2A). Calcofluor White staining and CBM3a labeling were further used to investigate the changes in cellulose contents. In control the FAZs, Calcofluor White staining of the walls of cells in the separation layer remained high (Fig. 2B), and no clear differences in CBM3a labeling were detected throughout the experiment (Fig. 2C). In contrast, at later stages following the GPD and ETH treatments, remarkably low intensities of Calcofluor White and CBM3a labeling signals were observed in FAZ cells, specifically where cell separation occurred (Fig. 2B, C).
Fig. 2.
Localization of polysaccharides and cellulose epitopes in fruitlet abscission zone (FAZ) cells during abscission in litchi. (A) Periodic acid–Schiff reactive (PAS) staining for insoluble carbohydrates in longitudinal sections of the FAZs. (B) Calcofluor White staining for cellulose in longitudinal sections of the FAZs. Calcofluor White is a special fluorescent stain that binds strongly to cellulose (Hughes and McCully, 1975). (C) Staining with antibody CBM3a in longitudinal sections of the FAZs. CBM3a is a CBM (carbohydrate-binding module) probe for crystalline cellulose (Blake et al., 2006). The micrographs show the FAZs from fruitlets of control branches and branches at 3 d after girdling plus defoliation (GPD) or 5 d after treatment with ethephon (ETH). Arrows indicate the location of the abscission layer. Scale bars are 50 μm.
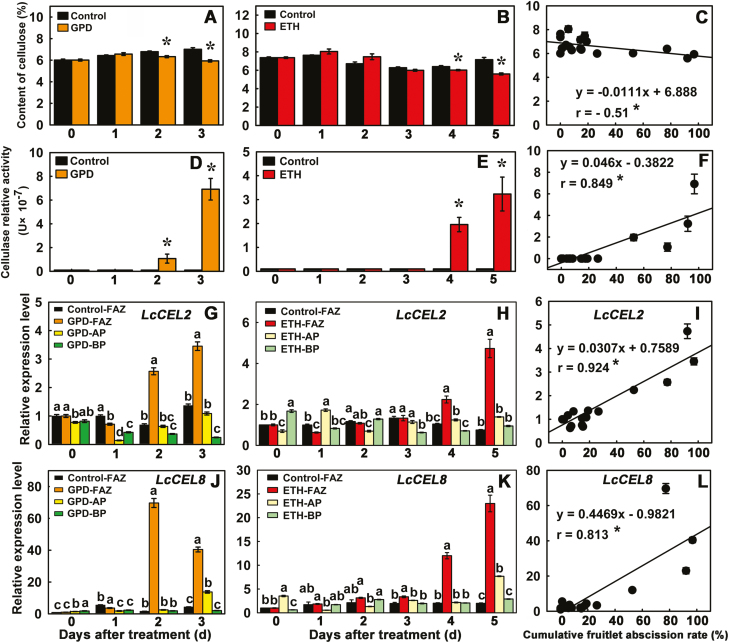
Cellulose concentrations in the FAZs at day 3 after the GPD treatment and at day 5 after the ETH treatment were much less than those of the controls (Fig. 3A, B). Further, the concentration of cellulose was significantly negatively correlated with fruitlet abscission rates (r=−0.51) (Fig. 3C). Compared with the control, cellulase activities in the FAZs were significantly higher at day 2 after the GPD treatment and at day 4 after the ETH treatment, and reached their highest levels at day 3 (~7.2-fold) after GPD and at day 5 (~3.2-fold) after ETH treatment (Fig. 3D, E). Cellulase activities were significantly and positively correlated with fruitlet abscission rates (r=0.849) (Fig. 3F). In contrast, control FAZs exhibited extremely low or non-detectable cellulase activity. Collectively, these results indicated that the increase in cellulase activity in the FAZ cells caused loss of cellulose content in cell walls, and directly resulted in the enhancement of fruitlet abscission.
Fig. 3.
Effects of two abscission-accelerating treatments on cellulose content (A-C), cellulase activity (D-F), and gene expression of LcCEL2 (G–I) and LcCEL8 (J–L) during fruitlet abscission in litchi. (A, D, G, J) Samples collected after girdling plus defoliation (GPD). (B, E, H, K) Samples collected after treatment with ethephon (ETH). (C, F, I, L) Correlations between the cumulative fruitlet abscission rate and (C) cellulose content, (F) cellulase activity, and expression of (I) LcCEL2 and (L) LcCEL8 in the fruitlet abscission zone. Data are means (±SE) from three replicates. Significant differences between treated and control branches in (A, B, D, E) were determined using Student’s t-test: **P<0.01. Different letters in (G, H, J, K) indicate significant differences as determined using Duncan’s multiple range test (P<0.05). The correlation coefficients (r) are significant at *P<0.05 (C, F, I, L). FAZ, fruitlet abscission zone; AP, apical portion of pedicel; BP, basal portion of pedicel (see Methods and Supplementary Fig. S1B).
Two cellulase genes are associated with fruitlet abscission
A total of 20 cellulase genes, designated as LcCEL1–LcCEL20, were identified in the litchi genome (Supplementary Table S2). The expression of three putative target genes (LcCEL2, LcCEL8, and LcCEL9) was found to be significantly higher during fruitlet abscission according to our previous RNA-Seq transcriptome analysis (Supplementary Fig. S4, Supplementary Table S5; Li et al., 2015b). To further confirm the expression of these genes, we performed qRT-PCR analysis and found that the expression of LcCEL2 and LcCEL8 was induced in FAZ tissues during the abscission process. Compared with the control, the expression of LcCEL2 in FAZs after GPD treatment significantly increased from day 2 and reached its highest level at day 3 (~3.5-fold) (Fig. 3G). Expression of LcCEL8 in FAZs increased more dramatically, by ~70-fold at day 2 and ~40-fold at day 3 after the GPD treatment (Fig. 3J). During ETH-induced abscission, both LcCEL8 and LcCEL2 expression in FAZs strongly increased from day 4 onward and reached their highest levels (~23-fold and ~4.7-fold, respectively) at day 5 (Fig. 3H, K). Thus, LcCEL2 and LcCEL8 expression in FAZs was significantly correlated with fruitlet abscission rates (r=0.924 and 0.813, respectively) (Fig. 3I, L). Moreover, when the expressions of LcCEL8 and LcCEL2 were examined in the two pedicel regions flanking the FAZs, the apical portion (AP) at the fruitlet side and the basal portion (BP) at the proximal pedicel side (Supplementary Fig. S1B), LcCEL2 and LcCEL8 transcripts accumulated exclusively in the FAZ under both GPD and ETH treatments (Fig. 3G, H, J, K). Although transcriptomic data from abscising fruitlets showed higher RPKM values for LcCEL9 under GPD and ETH treatments than in the control group (Supplementary Fig. S4, Supplementary Table S5), the accumulation of LcCEL9 transcripts could not be detected by qRT-PCR. We therefore chose LcCEL2 and LcCEL8 for further analysis as these two genes could be strongly associated with fruitlet abscission.
Ectopic expression of LcCEL2 and LcCEL8 activate abscission of floral organs in Arabidopsis
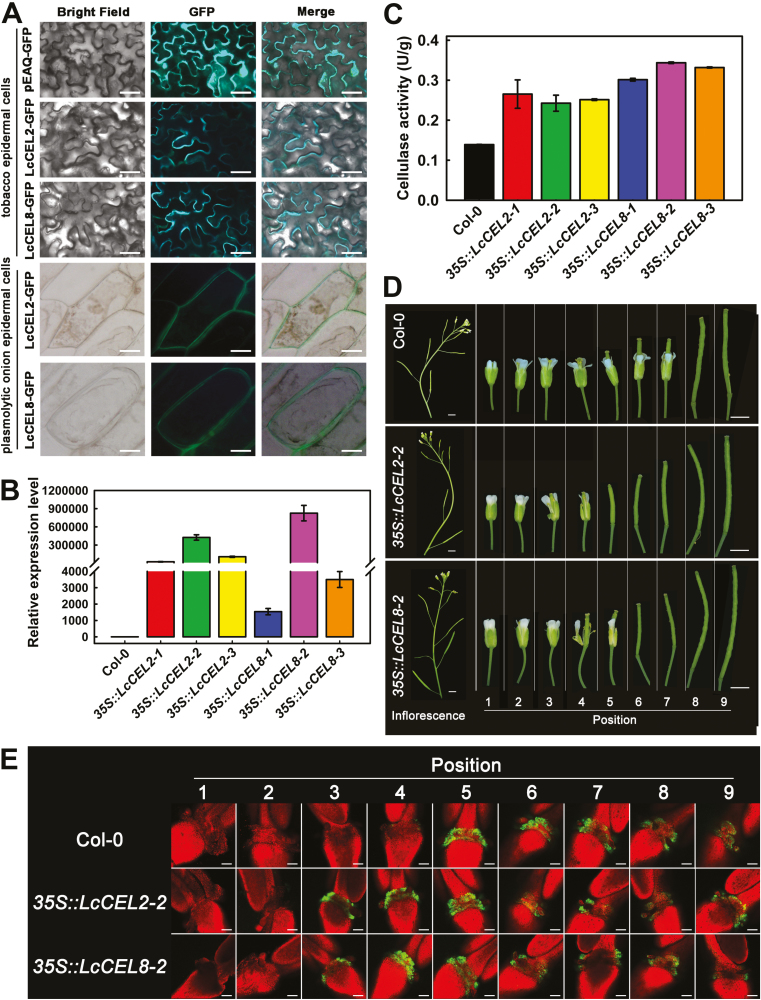
To further investigate the functions of LcCEL2 and LcCEL8, the in vivo subcellular localizations of the LcCEL2 (621 aa) and LcCEL8 (497 aa) proteins were examined. We found that LcCEL2-GFP and LcCEL8-GFP were exclusively localized to cell walls when transiently expressed in tobacco leaves and plasmolytic onion epidermal cells (Fig. 4A). This suggested that LcCEL2 and LcCEL8 are cell wall-localized proteins, which agrees with their potential functions in cell wall degradation. We then transgenically expressed LcCEL2 and LcCEL8 under the control of the CaMV 35S promoter in Arabidopsis. Homozygous 35S::LcCEL2 and 35S::LcCEL8 transgenic lines were obtained for further experiments. Both cellulase activities and expression of LcCEL2 and LcCEL8 in the transgenic lines were significantly higher than those of the corresponding wild-type (Col-0) (Fig. 4B, C). The phenotypes of the flowers or siliques at specific flower positions on Col-0, 35S::LcCEL8, and 35S::LcCEL2 transgenic plants were examined. Both 35S::LcCEL2 and 35S::LcCEL8 plants dropped their flowers first at position 5 or 6 (Fig. 4D; Supplementary Fig. 5A, B). In contrast, Col-0 plants first abscised their flowers at position 8 (Fig. 4D). This suggested that transgenic expression of LcCEL2 and LcCEL8 in Arabidopsis could induce precocious abscission of floral organs.
Fig. 4.
Functional analysis of litchi LcCEL2 and LcCEL8. (A) Subcellular localization of LcCEL2-GFP and LcCEL8-GFP fusion proteins in tobacco leaves and plasmolytic onion epidermal cells. (B) Expression of LcCEL2 and LcCEL8 in leaves of different transgenic Arabidopsis plants. The 35S::LcCEL2-1, 35S::LcCEL2-2, and 35S::LcCEL2-3 lines ectopically expressed LcCEL2 under the control of the CaMV 35S promoter in wild-type plants (Col-0). The 35S::LcCEL8-1, 35S::LcCEL8-2, and 35S::LcCEL8-3 lines ectopically expressed LcCEL8 under the control of the CaMV 35S promoter in Col-0. AtUBQ was used as an internal control for qRT-PCR analysis. The y-axis represents the fold-change in the expression levels relative to Col-0. Data are means (±SE) from three replicates. (C) Cellulase activity in leaves of different transgenic Arabidopsis plants. Data are means (±SE) from three replicates. (D) Phenotypes of floral organ abscission in transgenic Arabidopsis lines. Position numbers were counted from the first flower with visible white petals at the top of the inflorescence. (E) BCECF fluorescence micrographs of floral organ abscission zones in transgenic Arabidopsis lines. The images represent BCECF fluorescence merged with chlorophyll autofluorescence. An increase in pH is indicated by green fluorescence; chlorophyll autofluorescence is in red. Representative images are shown from 3–4 replicates in total. Scale bars are 25 μm (A), 3 mm (D), and 100 μm (E).
Sundaresan et al. (2014) demonstrated that cytosolic pH increases in AZ cells concomitant with organ abscission. We also observed an early increase in the cytosolic pH of AZ cells of the floral organs using the pH-sensitive indicator BCECF. Green fluorescence from BCECF was normally observed in the AZ of flowers beginning at position 5 in the wild-type plants (Fig. 4E). In contrast, the lines expressing 35S::LcCEL2 and 35S::LcCEL8 showed much earlier fluorescence signals that could be detected first in flowers at position 3 (Fig. 4E; Supplementary Fig. 5C), consistent with the precocious abscission phenotypes of these transgenic lines.
Next, the 5´ sequences upstream of the transcription start sites of LcCEL2 and LcCEL8 genes were amplified from the genome of litchi. The sizes of fragments amplified from these genomic regions were 2215 bp and 2192 bp for LcCEL2 and LcCEL8, respectively (Supplementary Dataset S1). We then transgenically expressed a GUS reporter gene fused downstream of the native LcCEL2 and LcCEL8 promoters to assess the strength and specificity of their expression in Arabidopsis. Both LcCEL2 and LcCEL8 were successfully expressed in Arabidopsis floral AZs during organ abscission (Fig. 5). GUS signals in proCEL2::GUS plants were strongly detected in AZs of flowers from position 5 to 9, and were also apparent in stigma tissues, anther filaments, and petals, and in the dehiscence and seed-shedding zones of the siliques (turning yellow), encompassing the replum, the valve margins, and the funiculi (Fig. 5A, C, D). GUS signals in proCEL8::GUS plants were exclusively in the abscission regions, such as the flower AZs from position 3 onward, the dehiscence and seed-shedding zones of the siliques, and the base of cauline leaves (Fig. 5B, E–G). Collectively, these data demonstrated that LcCEL2 and LcCEL8 probably have important functions during the abscission of plant organs.
Fig. 5.
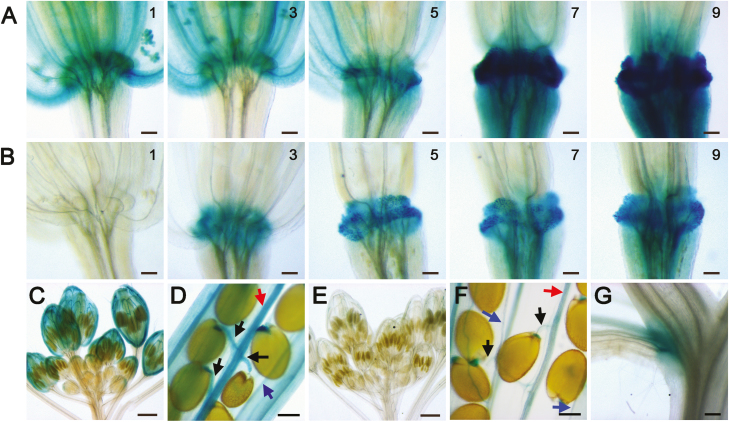
GUS expression driven by the litchi LcCEL2 and LcCEL8 promoters in transgenic Arabidopsis. (A) GUS expression in the abscission zone at positions 1, 3, 5, 7, and 9 along the inflorescence for proLcCEL2::GUS. (B) GUS expression in the abscission zone at positions 1, 3, 5, 7, and 9 along the inflorescence for proLcCEL8::GUS. Position numbers were counted from the first flower with visible white petals at the top of the inflorescence. (C, D) GUS expression in (C) floral buds and (D) siliques (turning yellow) for proLcCEL2::GUS. (E–G) GUS expression in (E) floral buds, (F) silique (turning yellow), and (G) cauline leaves for proLcCEL8::GUS. Arrows indicate funiculi (black), replums (red), and valve margins (blue). Scale bars are 100 μm (A, B, D, F), 300 μm (G), and 500 μm (C, E).
LcHB2 activates the expression of LcCEL2 and LcCEL8 by directly binding to their promoters
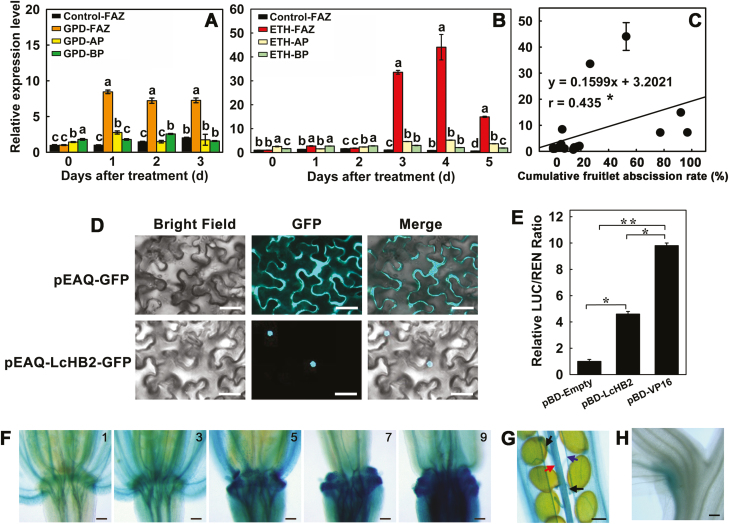
To characterize potential regulators of LcCEL2 and LcCEL8, their promoter sequences were further analysed for core cis-elements and other motifs. We found that the promoters of both genes contained two HD-binding cis-elements (Supplementary Dataset S1). The LcCEL2 promoter had two HD-binding cis-elements: AAATTAAA at position –95 to –46 relative to the start of transcription, and AAATTAGT at position –725 to –676 relative to the start of transcription. Interestingly, the LcCEL8 gene promoter contained binding sites for HD-Zip TFs at position –1589 to –1540 (TAAATGCA) relative to the transcription start site and at position –2076 to –2027 (AAATTAGT) relative to the transcription start site. The promoter regions of both LcCEL2 and LcCEL8 contained the AAATTAGT binding motif. We hypothesized that LcCEL2 and LcCEL8 might be directly regulated by HD-Zip TFs in litchi. In our previous RNA-Seq analyses, we identified an HD-ZIP unigene (L10059562) with significantly increased expression after ethylene induction of fruitlet abscission (Li et al., 2015b). In the present study, we cloned the full-length cDNA and designated this gene as LcHB2. Phylogenetic analysis showed that LcHB2 clustered with ATHB7, ATHB12, and CsHB18 in the HD-Zip I subfamily (Supplementary Fig. S6). LcHB2 was expressed exclusively and strongly in the FAZs, with expression highest at day 1 after GPD (~8.5-fold) (Fig. 6A) and at day 4 after ETH treatment (~44-fold) (Fig. 6B). The expression of LcHB2 in the FAZs was significantly positively correlated with the cumulative fruitlet abscission rate (Fig. 6C). In vivo subcellular localization analysis revealed that the LcHB2 protein was located in the nucleus (Fig. 6D). Furthermore, dual-luciferase assays showed that the LcHB2 protein acted as a transcriptional activator in vivo (Fig. 6E). These results demonstrated that LcHB2 was a genuine TF that could activate the expression of downstream genes. In addition, LcHB2 promoter activity was examined using a promoter::GUS transgene (proLcHB2::GUS). The GUS signals were strongly detected in the abscission regions, such as the flower AZs from position 5 onward, the dehiscence zones of the siliques (replum and valve margins), the seed-shedding zone (funiculi), and the base of cauline leaves (Fig. 6F–H). The GUS signal also appeared in anther filaments and petals (Fig. 6F).
Fig. 6.
Expression patterns, subcellular localization, and the transcriptional activation function of LcHB2 protein in litchi. (A, B) Effects of girdling plus defoliation (GPD) and treatment with ethephon (ETH) on the expression of LcHB2 in the fruitlet abscission zones (FAZs) and two pedicel regions flanking the FAZs, the apical portion (AP) at the fruitlet side above the FAZ and the basal portion (BP) below the FAZ. (C) Correlation between the cumulative fruitlet abscission rate and the expression level of LcHB2 in the FAZ. Data in (A–C) are means (±SE) from three replicates. Different letters indicate significant differences as determined using Duncan’s multiple range test (P<0.05). The correlation coefficient (r) is significant at *P<0.05. (D) Nuclear localization of LcHB2-GFP in tobacco leaves. Scale bars are 25 μm. (E) Transcriptional activation function of LcHB2 in tobacco leaves, as indicated by the ratio of LUC to REN expression. Data are means (±SE) from six replicates. Significant differences between means were determined using Student’s t-test: * P<0.05, ** P<0.01. (F–H) GUS expression driven by the LcHB2 promoter in transgenic Arabidopsis. (F) GUS expression in the abscission zone at positions 1, 3, 5, 7, and 9 along the inflorescence for proLcHB2::GUS. Position numbers were counted from the first flower with visible white petals at the top of the inflorescence. (G, H) GUS expression in (G) the siliques (turning yellow) and (H) cauline leaves for proLcHB2::GUS. Arrows indicate funiculi (black), replums (red), and valve margins (blue). Scale bars are 100 μm (F, G), and 300 μm (H).
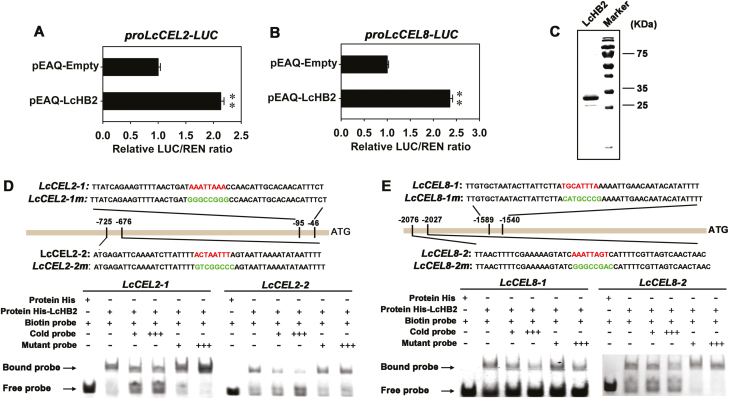
To examine the function of LcHB2 in the regulation of expression of LcCEL2 and LcCEL8, we performed transient expression assays using a dual-luciferase system. Compared with the empty-vector control, the ratio of LUC/REN expression was significantly increased when the pEAQ-LcHB2 effector construct was co-transfected with either the proLcCEL2-LUC or the proLcCEL8-LUC reporter construct (Fig. 7A, B), confirming that LcHB2 is a transcriptional activator of LcCEL2 and LcCEL8. EMSAs were performed to test whether LcHB2 could bind to the HD-binding cis-elements in the LcCEL2 and LcCEL8 promoters. Four DNA fragments (~50 bp) containing HD-binding cis-elements in the promoter regions of LcCEL2 and LcCEL8 were synthesized and labeled with biotin. The recombinant histidine-tagged (His)-LcHB2 fusion protein was expressed in E. coli BL21 (DE3) cells and purified (Fig. 7C). We found that recombinant LcHB2 protein could bind strongly to the LcCEL2 and LcCEL8 promoter fragments, and that this binding could be abolished by high amounts of competitive unlabeled sequences that were otherwise identical, but not by promoter sequences containing mutant binding sites (Fig. 7D, E). Mobility shifts were not observed if the LcCEL2 and LcCEL8 promoter fragments were incubated with poly-His alone (Fig. 7D, E). Thus, LcCEL2 and LcCEL8 are direct targets of LcHB2, which activates the expression of these two genes by directly binding to their promoters.
Fig. 7.
Binding of LcHB2 to LcCEL2 and LcCEL8 promoters and activation of their activities. LcHB2 activates the expression of LcCEL2 (A) and LcCEL8 (B) in vivo as indicated by transient dual-luciferase reporter assays in tobacco leaves. The ratio of LUC/REN expression of the empty vector (pEAQ) plus promoter was used as a calibrator (set to 1). Data are means (±SE) from six replicates. Significant differences between means were determined using Student’s t-test: ** P<0.01. (C) SDS-PAGE gel stained with Coomassie Brilliant Blue demonstrating affinity purification of the recombinant LcHB2 protein. (D, E) Electrophoretic mobility shift assays (EMSAs) showing the association of LcHB2 with the LcCEL2 and LcCEL8 promoters in vitro. Sequences of both the wild-type and mutant probes are shown at the top. The core HD-binding cis-elements are indicated in red and the mutant elements are indicated in green. Shifted bands, suggesting the formation of DNA–protein complexes, are indicated by arrows. ‘–’ and ‘+’ represent an absence or presence, respectively. ‘+++’ indicates increasing amounts of unlabeled or mutated probes introduced for competition. A non-biotin-labeled probe was added as an unlabeled competitor.
Discussion
The fruitlet abscission zone (FAZ) in litchi where fruitlets detach from maternal plants is located at the pedicel–fruitlet junction (Supplementary Fig. S1B) and consists of 7–10 layers of small cells with dense cytoplasm that are histologically distinct from their surrounding cells (Fig. 1). We explored the changes in the microscopic structure of FAZs during fruitlet abscission under two abscission-accelerating treatments, namely carbohydrate deficiency (caused by girdling plus defoliation, GPD) and application of ethephon (ETH). GPD completely blocks the carbohydrate transport to fruits and leads to a serious carbohydrate stress for fruit development (Obeso, 1998; Gómez-Cadenas et al., 2000), while ethylene is known to accelerate abscission of plant organs (Taylor and Whitelaw, 2001). Cell separation in FAZs was clearly observed following these two treatments in our study. The disruption of cells initiated in the stem cortex and epidermis and spread toward the vascular bundle and pith regions in the FAZ. Significant increases in the sizes of FAZ cells were also observed (Fig. 1; Supplementary Fig. S3). Similar enlargement in abscission zone cells has previously been observed in flower AZs of soybean (Glycine max; Oberholster et al., 1991) and tomato (Tabuchi et al., 2001), in floral AZs of Arabidopsis (Shi et al., 2011), in leaf AZs of olive (Olea europaea; Kitsaki et al., 1999), and in fruit AZs of apple (Pandita and Jindal, 1991) and citrus (cv. ‘Shamouti’, Huberman et al., 1988; cv. ‘Satsuma Mandarin’ and ‘Kiyomi’, Li et al., 2017). Enlargement of separation zone cells is the result of tension across intact walls that is released during separation (Sexton and Roberts, 1982), and it may be the earliest conspicuous structural change that is characteristic of organ shedding. The enlargement of cells may generate the tension needed for final rupture of the FAZ in litchi.
During swelling and expansion, FAZ cells lose their anisotropic properties, suggesting that great changes are taking place in the cell wall architecture. High cellulase activity appears to be characteristic of developmental processes during which cell wall architecture is disrupted, including organ abscission (Sexton et al., 1980; Brummell et al., 1999) and pod dehiscence (Chauvaux et al., 1997). Thus, it was not surprising to find cellulase activity associated with litchi fruitlet abscission (Figs 3, 4), because increased activity seems to be a feature of separating AZs (Sexton et al., 1980; Trainotti et al., 1998a, 1998b; Brummell et al., 1999). We have assumed that the natural substrate for the cellulase that we measured is cellulose, the predominant insoluble cell wall polysaccharide that is composed of D-glucose moieties joined by β(1→4) linkages into a linear molecule (O’Brien et al., 1964). In our present study, the strong loss in intensity of PAS, Calcofluor White, and CBM3a staining in walls of abscising FAZ cells that we observed after GPD or ETH treatment suggested that the polysaccharide constituents, or to be more exact, cellulose, decreased exclusively at the onset of cell separation (Fig. 2). We also found increases in cellulase activities and corresponding decreases in cellulose contents in abscising FAZs (Fig. 3A, B), confirming that cellulases are involved in degrading cellulose during cell separation of in the abscission zone of litchi fruitlets.
The accumulation of a cellulase-encoding mRNA induced by ethylene treatment was first reported in abscising bean leaves (Tucker et al., 1988). Since then, several studies have demonstrated that cellulase activity and gene expression are correlated with events in AZs during shedding of plant organs (in avocado, Tonutti et al., 1995; in tomato, del Campillo and Bennett, 1996; in pepper, Capsicum annuum, Trainotti et al., 1997; and in citrus, Kazokas and Burns, 1998). Cellulases are encoded by a multigene family, as confirmed in our study that identified 20 cellulase genes in the litchi genome. Among these, the abscission-specific expression of LcCEL2 and LcCEL8 were followed by enhanced cellulase activities and subsequently by decreased cellulose contents in FAZ cells (Fig. 3). Although many abscission-related cellulase genes have also been identified in the FAZ tissues of several other fruit crop species (Trainotti et al., 1997; Li et al., 2010; Merelo et al., 2017), our understanding of the functions of these cellulases during fruit abscission is limited. Here, overexpression of LcCEL2 and LcCEL8 in Arabidopsis was able to accelerate floral organ abscission, and analysis of GUS-promoter fusion lines further showed that these genes were expressed in cells in the AZ during floral organ abscission and silique dehiscence (Figs 4D, E, 5; Supplementary Fig. S5), suggesting that LcCEL2 and LcCEL8 are functionally involved in abscission.
Interestingly, the LcCEL2 and LcCEL8 proteins were not highly conserved with each other and showed only 49% similarity. Compared to cellulases in other plants (Supplementary Fig. S7), LcCEL8 had higher similarity to abscission-related cellulases in citrus (CitCEL6; Merelo et al., 2017), peach (PpEG1; Trainotti et al., 1997), tomato (SlCel5; Kalaitzis et al., 1999), and pepper (cCel2; Trainotti et al., 1998b), while LcCEL2 was more closely related to cotton (Gossypium hirsutum) GhCel1 (Mishra et al., 2008) and peach PpEG4 (Trainotti et al., 2006). These data might not seem surprising if the comparisons refer only to the structure of the polypeptides rather than to their functions in vivo. Still, proteins from the subfamilies to which LcCEL2 and LcCEL8 belong seem more likely to be involved in organ abscission than do members of other cellulase families. Notably, the LcCEL2 and LcCEL8 promoters could drive GUS expression not only in the floral AZ but also in the dehiscence zones of the siliques in Arabidopsis, encompassing the replum, the valve margins, and the funiculi. (Fig. 5), indicating that LcCEL2 and LcCEL8 might also affect silique dehiscence in transgenic Arabidopsis. In addition, the GUS signal in proLcCEL2::GUS plants was present in the stigmatic tissues, anther filaments, and petals, suggesting that LcCEL2 might also affect floral organ development.
HD-Zip TFs are a large class of plant TFs that are widely involved in the regulation of different growth and developmental processes. Although no study has directly confirmed whether HD-Zip TFs control plant organ shedding, high-throughput gene expression analyses have shown that HD-Zip genes encode differentially expressed TFs that might have central roles in the shedding of organs (e.g. in tomato flowers, Meir et al., 2010; apple fruitlets, Zhu et al., 2011; Heo et al., 2016; olive fruit, Gil-Amado and Gomez-Jimenez, 2013; soybean leaves, Kim et al., 2016; and rose petals, Gao et al., 2016). Indeed, transcripts of a putative HD-Zip gene (LcHB2) did show significantly increased abundance during the abscission of litchi fruitlets upon carbohydrate stress or application of ethylene (Fig. 6). LcHB2 belongs to one of the four subfamilies (I–IV) of HD-Zip TFs, which show binding preferences for variant HD-binding motifs (Sessa et al., 1993, 1998; Palena et al., 2001; Lin et al., 2008; Tominaga-Wada et al., 2009; Xu et al., 2014). The Arabidopsis cellulase gene At2g32990 contains three such HD-binding motifs in its promoter that allow its transcription to be directly regulated by an HD-Zip IV TF (HDG11) to promote root elongation and enhance drought tolerance (Xu et al., 2014). It is noteworthy that At2g32990 has also been reported to be involved in cell wall remodeling in Arabidopsis stamen AZs (Lashbrook and Cai, 2008).
In our study, LcCEL2 and LcCEL8 associated with fruitlet abscission in litchi also contained two HD-binding cis-elements in their promoters. Further experiments confirmed that LcHB2 could directly bind with promoter segments from LcCEL2 and LcCEL8 that contained an HD-binding cis-element and hence serve as a transcriptional activator for these genes (Fig. 7). Notably, accumulation of LcHB2 transcripts occurred earlier than LcCEL2 and LcCEL8 transcripts, which further supports the hypothesized role of LcHB2 as an upstream regulator of these two cellulase genes. HD-Zip TFs participate in responses to environmental cues including abiotic stress and stresses mediated by ABA (Johannesson et al., 2003; Olsson et al., 2004; Ariel et al., 2007, 2010; Romani et al., 2016), in leaf or floral development (Kim et al., 2007; Lin et al., 2008), root elongation (Miao et al., 2018), flower senescence (Chang et al., 2014; Lü et al., 2014), and fruit ripening (Lin et al., 2008; Jiang et al., 2017). Here, we demonstrated that an HD-Zip TF is also involved in organ shedding through regulation of the expression of cellulase genes. Interestingly, the regulation of abscission- or dehiscence-related cellulase genes by HD-Zip TFs is probably conserved among plant species, as at least one HD-binding cis-element was found in the promoter regions of 17 cellulase-encoding genes from six plant species (Supplementary Fig. S8, Supplementary Dataset S1).
Taken our results together, we propose that LcHB2 may act as a positive regulator of fruitlet abscission through directly activating LcCEL2 and LcCEL8 in litchi. When fruitlets sense abscission signals such as carbohydrate deficiency or ethylene stimuli, the expression levels of LcHB2 are up-regulated. LcCEL2 and LcCEL8 expressed specifically i the FAZ are induced by LcHB2 via direct binding to their promoters, and cellulase activities are increased. Cellulose contents are therefore reduced, and ultimately fruitlets abscise due to cell wall degradation and cell separation in the FAZ.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Illustration of girdling and the pedicel samples examined in this study.
Fig. S2. Constructs used in this study.
Fig. S3. Effects of ethephon treatment on cumulative fruitlet abscission rate, phenotypic performance, and cell separation in the FAZ.
Fig. S4. Expression profiles of 20 cellulase genes from litchi after GPD and ETH treatments.
Fig. S5. Overexpression of LcCEL2 and LcCEL8 causes earlier floral organ abscission in Arabidopsis.
Fig. S6. Multiple sequence alignments and phylogenetic analysis of LcHB2 with other plant HD-Zip proteins.
Fig. S7. Multiple sequence alignments and phylogenetic analysis of LcCEL2 and LcCEL8.
Fig. S8. Diagram of 17 promoters of abscission-related cellulase genes from various plant species.
Table S1. List of primers used in this study.
Table S2. Characteristics of cellulase and HD-Zip proteins from litchi.
Table S3. Presence of cellulase genes identified in this study in other plant species.
Table S4. Presence of HD-Zip genes identified in this study in other plant species.
Table S5. RPKM values of 20 cellulase genes of litchi identified in the transcriptomes of plants in the GPD and ETH treatments.
Dataset S1. Nucleotide sequences of the promoters of LcHB2 and 19 cellulase genes.
Author contributions
CL, JL, HW, and MZ designed the study; CL, XM, ZW, PY, and MP performed the experiments; MZ and XN provided critical technical assistance; CL and JL wrote the manuscript with contributions from all the authors; CL, JL, MZ, HW, and RX supervised the project, interpreted the data, and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by funds from the Natural Science Foundation of China (Grant Nos 31772269, 31600152, and 31070159), the Postdoctoral Science Foundation of China (Grant No. 2015M572329), the Innovation Team Project of the Department of Education of Guangdong Province (2016KCXTD011), the Guangzhou Science and Technology Key Project (201804020063), and the Outstanding Talent Program of the Ministry of Agriculture, and the China Agricultural Research System (CARS-33-11). We would like to thank Dr Chunlin Shi (Department of Biosciences, University of Oslo) for critically reading and assisting with drafting this manuscript, and Mary Ann Cushman (Alum Canyon Editing, LLC) for her helpful and critical English language editing.
Glossary
Abbreviations:
- AP
apical portion
- AZ
abscission zone
- BAC
BEAN ABSCISSION CELLULASE
- BCECF
2′,7′-Bis(2-carboxyethyl)-5-(and-6)-carboxyfluorescein
- BP
basal portion
- bZIP
basic leucine zipper
- CBD
cellulose-binding domain
- CBM
carbohydrate-binding module
- CEL
cellulase
- ETH
ethephon
- FAZ
fruit abscission zone
- GFP
green fluorescent protein
- GPD
girdling plus defoliation
- GUS
β-glucuronidase
- HD
homeodomain
- HD-Zip
homeodomain-leucine zipper
- LZ
leucine-zipper
- PAS
periodic acid-Schiff stain
- pI
isoelectric point
- qRT-PCR
quantitative real-time PCR
- TF
transcription factor
References
- Ariel F, Diet A, Verdenaud M, Gruber V, Frugier F, Chan R, Crespi M. 2010. Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. The Plant Cell 22, 2171–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel FD, Manavella PA, Dezar CA, Chan RL. 2007. The true story of the HD-Zip family. Trends in Plant Science 12, 419–426. [DOI] [PubMed] [Google Scholar]
- Blake AW, McCartney L, Flint JE, Bolam DN, Boraston AB, Gilbert HJ, Knox JP. 2006. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding modules in prokaryotic enzymes. The Journal of Biological Chemistry 281, 29321–29329. [DOI] [PubMed] [Google Scholar]
- Bourgault R, Bewley JD. 2002. Gel diffusion assays for endo-beta-mannanase and pectin methylesterase can underestimate enzyme activity due to proteolytic degradation: a remedy. Analytical Biochemistry 300, 87–93. [DOI] [PubMed] [Google Scholar]
- Brummell DA, Hall BD, Bennett AB. 1999. Antisense suppression of tomato endo-1,4-beta-glucanase Cel2 mRNA accumulation increases the force required to break fruit abscission zones but does not affect fruit softening. Plant Molecular Biology 40, 615–622. [DOI] [PubMed] [Google Scholar]
- Chang X, Donnelly L, Sun D, Rao J, Reid MS, Jiang CZ. 2014. A Petunia homeodomain-leucine zipper protein, PhHD-Zip, plays an important role in flower senescence. PLoS ONE 9, e88320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvaux N, Child R, John K, Ulvskov P, Borkhardt B, Prinsen E, Van Onckelen HA. 1997. The role of auxin and cell separation in the dehiscence zone of oilseed rape pods. Journal of Experimental Botany 48, 1423–1429. [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip, a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- del Campillo E, Bennett AB. 1996. Pedicel breakstrength and cellulase gene expression during tomato flower abscission. Plant Physiology 111, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estornell LH, Agusti J, Merelo P, Talon M, Tadeo FR. 2013. Elucidating mechanisms underlying organ abscission. Plant Science 199, 48–60. [DOI] [PubMed] [Google Scholar]
- Feder NED, O’Brien TP. 1968. Plant microtechnique, some principles and new methods. American Journal of Botany 55, 123–142. [Google Scholar]
- Gao Y, Liu C, Li X, Xu H, Liang Y, Ma N, Fei Z, Gao J, Jiang CZ, Ma C. 2016. Transcriptome profiling of petal abscission zone and functional analysis of an Aux/IAA family gene RhIAA16 involved in petal shedding in rose. Frontiers in Plant Science 7, 1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilamado JA, Gomez-Jimenez MC. 2013. Transcriptome analysis of mature fruit abscission control in olive. Plant & Cell Physiology 54, 244–269. [DOI] [PubMed] [Google Scholar]
- Gómez-Cadenas A, Mehouachi J, Tadeo FR, Primo-Millo E, Talon M. 2000. Hormonal regulation of fruitlet abscission induced by carbohydrate shortage in citrus. Planta 210, 636–643. [DOI] [PubMed] [Google Scholar]
- He H, Bai M, Tong P, Hu Y, Yang M, Wu H. 2018. CELLULASE6 and MANNANASE7 affect cell differentiation and silique dehiscence. Plant Physiology 176, 2186–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo S, Hwang JH, Jun JH, Lee HJ. 2016. Abscission-related genes revealed by RNA-Seq analysis using self-abscising apple (Malus×domestica). Journal of Pomology & Horticultural Science 91, 271–278. [Google Scholar]
- Hieke S, Menzel CM, Doogan VJ, Ludders P. 2002. The relationship between yield and assimilate supply in lychee (Litchi chinensis Sonn.). Journal of Horticultural Science & Biotechnology 77, 326–332. [Google Scholar]
- Horton RF, Osborne DJ. 1967. Senescence, abscission and cellulase activity in Phaseolus vulgaris. Nature 214, 1086–1088. [Google Scholar]
- Huberman ME, Zamski E, Goren R. 1988. Anatomical changes induced by ethylene in the abscission zone of citrus leaf and fruit explants. Israel Journal of Plant Sciences 37, 107–121. [Google Scholar]
- Hughes J, McCully ME. 1975. The use of an optical brightener in the study of plant structure. Stain Technology 50, 319–329. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Liu C, Yan D, et al. 2017. MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. Journal of Experimental Botany 68, 1055–1069. [DOI] [PubMed] [Google Scholar]
- Johannesson H, Wang Y, Hanson J, Engström P. 2003. The Arabidopsis thaliana homeobox gene ATweHB5 is a potential regulator of abscisic acid responsiveness in developing seedlings. Plant Molecular Biology 51, 719–729. [DOI] [PubMed] [Google Scholar]
- Kalaitzis P, Hong SB, Solomos T, Tucker ML. 1999. Molecular characterization of a tomato endo-beta-1,4-glucanase gene expressed in mature pistils, abscission zones and fruit. Plant & Cell Physiology 40, 905–908. [DOI] [PubMed] [Google Scholar]
- Kazokas WC, Burns JK. 1998. Cellulase activity and gene expression in citrus fruit abscission zones during and after ethylene treatment. Journal of the American Society for Horticultural Science 123, 781–786. [Google Scholar]
- Kim J, Yang J, Yang R, Sicher RC, Chang C, Tucker ML. 2016. Transcriptome analysis of soybean leaf abscission identifies transcriptional regulators of organ polarity and cell fate. Frontiers in Plant Science 7, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Son O, Kim MR, Nam KH, Kim GT, Lee MS, Choi SY, Cheon CI. 2007. ATHB23, an Arabidopsis class I homeodomain-leucine zipper gene, is expressed in the adaxial region of young leaves. Plant Cell Reports 26, 1179–1185. [DOI] [PubMed] [Google Scholar]
- Kitsaki CK, Drossopoulos JB, Aivalakis G, Anastasiadou F, Delis C. 1999. In vitro studies of ABA and ethephon induced abscission in olive organs. Journal of Pomology & Horticultural Science 74, 19–25. [Google Scholar]
- Koehler SM, Matters GL, Nath P, Kemmerer EC, Tucker ML. 1996. The gene promoter for a bean abscission cellulase is ethylene-induced in transgenic tomato and shows high sequence conservation with a soybean abscission cellulase. Plant Molecular Biology 31, 595–606. [DOI] [PubMed] [Google Scholar]
- Kuang JF, Wu JY, Zhong HY, Li CQ, Chen JY, Lu WJ, Li JG. 2012. Carbohydrate stress affecting fruitlet abscission and expression of genes related to auxin signal transduction pathway in litchi. International Journal of Molecular Sciences 13, 16084–16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Cai S. 2008. Cell wall remodeling in Arabidopsis stamen abscission zones: temporal aspects of control inferred from transcriptional profiling. Plant Signaling & Behavior 3, 733–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Giovannoni JJ, Hall BD, Fischer RL, Bennett AB. 1998. Transgenic analysis of tomato endo-β-1,4-glucanase gene function role of cel1 in floral abscission. The Plant Journal 13, 303–310. [Google Scholar]
- Lashbrook CC, Gonzalez-Bosch C, Bennett AB. 1994. Two divergent endo-beta-1,4-glucanase genes exhibit overlapping expression in ripening fruit and abscising flowers. The Plant Cell 6, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang Y, Huang X, Li J, Wang H, Li J. 2015a. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Frontiers in Plant Science 6, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wang Y, Ying P, Ma W, Li J. 2015b. Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Frontiers in Plant Science 6, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JG, Yuan RC. 2008. NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘Delicious’ apples. Journal of Plant Growth Regulation 27, 283–295. [Google Scholar]
- Li JG, Zhu H, Yuan RC. 2010. Profiling the expression of genes related to ethylene biosynthesis, ethylene perception, and cell wall degradation during fruit abscission and fruit ripening in apple. Journal of the American Society for Horticultural Science 135, 391–401. [Google Scholar]
- Li X, Kitajima A, Kataoka K, Takisawa R, Nakazaki T. 2017. Anatomical observations of the citrus fruit abscission zone and morphological changes of the cells during secondary physiological fruit drop. Journal of the Japanese Society for Horticulture Science 86, 447–455. [Google Scholar]
- Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. 2008. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. The Plant Journal 55, 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü P, Zhang C, Liu J, Liu X, Jiang G, Jiang X, Khan MA, Wang L, Hong B, Gao J. 2014. RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose (Rosa hybrida) petal senescence. The Plant Journal 78, 578–590. [DOI] [PubMed] [Google Scholar]
- Meir S, Philosoph-Hadas S, Sundaresan S, Selvaraj KS, Burd S, Ophir R, Kochanek B, Reid MS, Jiang CZ, Lers A. 2010. Microarray analysis of the abscission-related transcriptome in the tomato flower abscission zone in response to auxin depletion. Plant Physiology 154, 1929–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merelo P, Agustí J, Arbona V, et al. 2017. Cell wall remodeling in abscission zone cells during ethylene-promoted fruit abscission in citrus. Frontiers in Plant Science 8, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao ZQ, Zhao PX, Mao JL, Yu LH, Yuan Y, Tang H, Liu ZB, Xiang CB. 2018. HOMEOBOX PROTEIN52 mediates the crosstalk between ethylene and auxin signaling during primary root elongation by modulating auxin transport-related gene expression. The Plant Cell 30, 2761–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Khare S, Trivedi PK, Nath P. 2008. Effect of ethylene, 1-MCP, ABA and IAA on break strength, cellulase and polygalacturonase activities during cotton leaf abscission. South African Journal of Botany 74, 282–287. [Google Scholar]
- Mitra SK, Pereira LS, Pathak PK, Majumdar D. 2003. Fruit abscission pattern of lychee cultivars. Acta Horticulturae 665, 215–218. [Google Scholar]
- Nicholas KB. 1997. GeneDoc, analysis and visualization of genetic variation. EMBNEW. NEWS 4, 14. [Google Scholar]
- O’Brien TP, Feder N, McCully ME. 1964. Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma 59, 368–373. [Google Scholar]
- Oberholster SD, Peterson CM, Dute RR. 1991. Pedicel abscission of soybean, cytological and ultrastructural changes. Canadian Journal of Botany 69, 2177–2186. [Google Scholar]
- Obeso JR. 1998. Effects of defoliation and girdling on fruit production in Ilex aquifolium. Functional Ecology 12, 486–491. [Google Scholar]
- Olsson AS, Engström P, Söderman E. 2004. The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Molecular Biology 55, 663–677. [DOI] [PubMed] [Google Scholar]
- Palena CM, Tron AE, Bertoncini CW, Gonzalez DH, Chan RL. 2001. Positively charged residues at the N-terminal arm of the homeodomain are required for efficient DNA binding by homeodomain-leucine zipper proteins. Journal of Molecular Biology 308, 39–47. [DOI] [PubMed] [Google Scholar]
- Pandita VK, Jindal KK. 1991. Enzymatic and anatomical changes in abscission zone cells of apple fruits induced by ethephon. Biologia Plantarum 33, 20–25. [Google Scholar]
- Patterson SE. 2001. Cutting loose. abscission and dehiscence in Arabidopsis. Plant Physiology 126, 494–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchooa D. 2004. A simple, rapid and efficient method for the extraction of genomic DNA from lychee (Litchi chinensis Sonn.). African Journal of Biotechnology 3, 253–255. [Google Scholar]
- Qi WE, Chen HB, Li WW, Zhang HJ. 2016. Development situation, trend and suggestions of Chinese litchi industry. Guangdong Agricultural Sciences 43, 173–179. [Google Scholar]
- Reid PD, Lewis LN. 1974. Cellulase and abscission in the red kidney bean (Phaseolus vulgaris). Plant Physiology 53, 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JA, Elliott KA, Gonzalez-Carranza ZH. 2002. Abscission, dehiscence, and other cell separation processes. Annual Review of Plant Biology 53, 131–158. [DOI] [PubMed] [Google Scholar]
- Romani F, Ribone PA, Capella M, Miguel VN, Chan RL. 2016. A matter of quantity: common features in the drought response of transgenic plants overexpressing HD-Zip I transcription factors. Plant Science 251, 139–154. [DOI] [PubMed] [Google Scholar]
- Sessa G, Morelli G, Ruberti I. 1993. The ATHB-1 and -2 HD-ZIP domains homodimerize forming complexes of different DNA binding specificities. The EMBO Journal 12, 3507–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Steindler C, Morelli G, Ruberti I. 1998. The Arabidopsis ATHB-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Molecular Biology 38, 609–622. [DOI] [PubMed] [Google Scholar]
- Sexton R, Durbin ML, Lewis LN, Thomson WW. 1980. Use of cellulase antibodies to study leaf abscission. Nature 283, 873–874. [Google Scholar]
- Sexton R, Roberts JA. 1982. Cell biology of abscission. Annual Review of Plant Biology 33, 133–162. [Google Scholar]
- Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA. 2011. Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. The Plant Cell 23, 2553–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern RA, Kigel J, Tomer E, Gazit S. 1995. ‘Mauritius’ lychee fruit development and reduced abscission after treatment with the auxin 2,4,5-TP. Journal of the American Society for Horticultural Science 120, 65–70. [Google Scholar]
- Sundaresan S, Philosoph-Hadas S, Riov J, Belausov E, Kochanek B, Tucker ML, Meir S. 2014. Abscission of flowers and floral organs is closely associated with alkalization of the cytosol in abscission zone cells. Journal of Experimental Botany 66, 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T, Ito S, Arai N. 2001. Anatomical studies of the abscission process in the tomato pedicels at flowering stage. Journal of the Japanese Society for Horticultural Science 70, 63–65. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JE, Whitelaw CA. 2001. Signals in abscission. New Phytologist 151, 323–340. [Google Scholar]
- Thompson DS, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. 2009. The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. The Plant Journal 60, 564–574. [DOI] [PubMed] [Google Scholar]
- Tonutti P, Cass LG, Christoffersen RE. 1995. The expression of cellulase gene family members during induced avocado fruit abscission and ripening. Plant, Cell & Environment 18, 709–713. [Google Scholar]
- Trainotti L, Ferrarese L, Casadoro G. 1998a. Characterization of cCel3, a member of the pepper endo-beta-1,4-glucanase multigene family. Hereditas 128, 121–126. [DOI] [PubMed] [Google Scholar]
- Trainotti L, Ferrarese L, Poznanski E, Vecchia FD. 1998b. Endo-β-1,4-glucanase activity is involved in the abscission of pepper flowers. Journal of Plant Physiology 152, 70–77. [Google Scholar]
- Trainotti L, Pavanello A, Zanin D. 2006. PpEG4 is a peach endo-beta-1,4-glucanase gene whose expression in climacteric peaches does not follow a climacteric pattern. Journal of Experimental Botany 57, 589–598. [DOI] [PubMed] [Google Scholar]
- Trainotti L, Spolaore S, Ferrarese L, Casadoro G. 1997. Characterization of ppEG1, a member of a multigene family which encodes endo-beta-1,4-glucanase in peach. Plant Molecular Biology 34, 791–802. [DOI] [PubMed] [Google Scholar]
- Tucker ML, Baird SL, Sexton R. 1991. Bean leaf abscission: tissue-specific accumulation of a cellulase mRNA. Planta 186, 52–57. [DOI] [PubMed] [Google Scholar]
- Tucker ML, Sexton R, Del Campillo E, Lewis LN. 1988. Bean abscission cellulase: characterization of a cDNA clone and regulation of gene expression by ethylene and auxin. Plant Physiology 88, 1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker ML, Whitelaw CA, Lyssenko NN, Nath P. 2002. Functional analysis of regulatory elements in the gene promoter for an abscission-specific cellulase from bean and isolation, expression, and binding affinity of three TGA-type basic leucine zipper transcription factors. Plant Physiology 130, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowicz BR, Bennett AB, Del Campillo E, et al. 2007. Structural organization and a standardized nomenclature for plant endo-1,4-beta-glucanases (cellulases) of glycosyl hydrolase family 9. Plant Physiology 144, 1693–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viles FJ Jr, Silverman L. 1949. Determination of starch and cellulose with anthrone. Analytical Chemistry 21, 950–953. [Google Scholar]
- Wang L, Liu GS, Wang LS, Zhang ZH, Hou JH, Guo HM. 1998. The optimal conditions for cellulase activity measurement with DNS method. Journal of Henan Normal University 26, 66–69. [Google Scholar]
- Wang X, Liu D, Li A, Sun X, Zhang R, Wu L, Liang Y, Mao L. 2013. Transcriptome analysis of tomato flower pedicel tissues reveals abscission zone-specific modulation of key meristem activity genes. PLoS ONE 8, e55238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Cai XT, Wang Y, Xing L, Chen Q, Xiang CB. 2014. HDG11 upregulates cell-wall-loosening protein genes to promote root elongation in Arabidopsis. Journal of Experimental Botany 65, 4285–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhong X, Fan Y, Wang H, Li J, Huang X. 2015. Burst of reactive oxygen species in pedicel-mediated fruit abscission after carbohydrate supply was cut off in longan (Dimocarpus longan). Frontiers in Plant Science 6, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye YJ, Xiao YY, Han YC, Shan W, Fan ZQ, Xu QG, Kuang JF, Lu WJ, Lakshmanan P, Chen JY. 2016. Banana fruit VQ motif-containing protein5 represses cold-responsive transcription factor MaWRKY26 involved in the regulation of JA biosynthetic genes. Scientific Reports 6, 23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying P, Li C, Liu X, Xia R, Zhao M, Li J. 2016. Identification and molecular characterization of an IDA-like gene from litchi, LcIDL1, whose ectopic expression promotes floral organ abscission in Arabidopsis. Scientific Reports 6, 37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan RC, Huang HB. 1988. Litchi fruit abscission, its patterns, effect of shading and relation to endogenous abscisic acid. Scientia Horticulturae 36, 281–292. [Google Scholar]
- Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, Chen JY, Lu WJ, Li JG. 2011. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Reports 30, 641–653. [DOI] [PubMed] [Google Scholar]
- Zhu H, Dardick CD, Beers EP, Callanhan AM, Xia R, Yuan R. 2011. Transcriptomics of shading-induced and NAA-induced abscission in apple (Malus domestica) reveals a shared pathway involving reduced photosynthesis, alterations in carbohydrate transport and signaling and hormone crosstalk. BMC Plant Biology 11, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou LP, Sun XH, Zhang ZG, Liu P, Wu JX, Tian CJ, Qiu JL, Lu TG. 2011. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiology 156, 1589–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.