Genetic metabolomics identifies a series of QTL-specific metabolic modules associated with quantitative resistance to clubroot, and highlights the possible roles of gluconasturtiin, citric acid, and two unknown compounds in partial resistance.
Keywords: Brassica napus, clubroot, dwarfing gene bzh, metabolomics, metabolic quantitative trait loci, oilseed rape, Plasmodiophora brassicae, quantitative resistance
Abstract
Plant disease resistance is often under quantitative genetic control. Thus, in a given interaction, plant cellular responses to infection are influenced by resistance or susceptibility alleles at different loci. In this study, a genetic linkage analysis was used to address the complexity of the metabolic responses of Brassica napus roots to infection by Plasmodiophora brassicae. Metabolome profiling and pathogen quantification in a segregating progeny allowed a comparative mapping of quantitative trait loci (QTLs) involved in resistance and in metabolic adjustments. Distinct metabolic modules were associated with each resistance QTL, suggesting the involvement of different underlying cellular mechanisms. This approach highlighted the possible role of gluconasturtiin and two unknown metabolites in the resistance conferred by two QTLs on chromosomes C03 and C09, respectively. Only two susceptibility biomarkers (glycine and glutathione) were simultaneously linked to the three main resistance QTLs, suggesting the central role of these compounds in the interaction. By contrast, several genotype-specific metabolic responses to infection were genetically unconnected to resistance or susceptibility. Likewise, variations of root sugar profiles, which might have influenced pathogen nutrition, were not found to be related to resistance QTLs. This work illustrates how genetic metabolomics can help to understand plant stress responses and their possible links with disease.
Introduction
Plant cellular responses to pathogens include constitutive and induced metabolic adjustments and the biosynthesis of a vast diversity of specialized small compounds (Dixon, 2001; Piasecka et al., 2015), whose toxicity can be challenged by tolerance mechanisms in pathogens (Osbourn et al., 1995; Enkerli et al., 1998, Coleman et al., 2011; Kettle et al., 2015; Pedras and Abdoli, 2017). The development of metabolomic approaches (Fiehn et al., 2000) led to renewed investigation of how pathogenic processes reprogram plant primary metabolism (Schultz et al., 2013; Rojas et al., 2014) and has provided opportunities to broaden our view about the roles of specialized metabolites possibly involved in plant defence (Heuberger, 2014; Tenenboim and Brotman, 2016). To take advantage of the large accessibility to metabolic phenotypes provided by these approaches, strategies are required to identify biochemical traits that are positively or negatively linked to pathogen and symptom development.
Metabolic profiling has often been used to compare pairs of plant genotypes showing contrasting resistance or susceptibility to a given pathogen strain, thus allowing the identification of candidate compounds putatively involved in plant defence or susceptibility (Ludwig-Müller et al., 1997; Lopez-Gresa, 2010; Sade et al., 2015; Yogendra et al., 2014). There can, however, be possible drawbacks to such comparative approaches, the first being that metabolic diversity among plant genotypes is controlled by a wealth of loci (Rowe et al., 2008; Chen et al., 2014), many of which might be unrelated to the resistance/susceptibility to a given pathogen strain. This problem can be solved by using near-isogenic lines, which allow the metabolic processes putatively connected to one given resistance locus to be specifically addressed (Gunnaiah, 2012). However, the metabolic comparison of resistant versus susceptible genotypes may be more complicated when examining quantitative resistances, controlled by the combined influence of sets of quantitative trait loci (QTLs). Quantitative resistance is widespread among plant–pathogen interactions (St Clair, 2010); most of the time, both in genotypes harbouring major resistance genetic factors and in apparently fully susceptible accessions, the level of symptoms is modulated by susceptibility or resistance alleles at low-effect QTLs, where each allele can provide a particular contribution to the complexity of metabolic responses (Corwin and Kliebenstein, 2017).
In this regard, the combination of metabolomics and quantitative genetics has been of great value in dissecting the roles of plant secondary metabolites in the interactions between plants and insects (Kliebenstein et al., 2002; Meihls et al., 2013). However, such approaches have been implemented only rarely in the field of plant–pathogen interactions (Denby et al., 2004; Koutouan et al., 2018). In the present study, we focused our efforts on deciphering the metabolic regulations triggered during the interaction between oilseed rape (Brassica napus) and the agent of clubroot, Plasmodiophora brassicae. This species is a eukaryotic (Rhizaria), telluric, and obligate biotroph, with an intracellular lifestyle and a host range mostly restricted to Brassicaceae species (Kageyama and Asano, 2009; Rolfe et al., 2016). After germination of P. brassicae spores in the soil, the infection starts with the primary infection of root hairs by primary plasmodia, and is followed by the colonization of hypertrophied root cortical cells by secondary plasmodes (secondary infection). This pathogenic process leads to the development of root galls in a few weeks, resulting in physiological disorders and subsequent substantial agronomic losses in Brassica crops worldwide. Genetic studies on the resistance of Brassicaceae species to different isolates of P. brassicae have highlighted major genetic resistance factors (Piao et al., 2009; Ueno et al., 2012; Hatakeyama et al., 2013; Kato et al., 2013; Chu et al., 2014; Rahman et al., 2014; Zhang et al., 2014; Yu et al., 2016; Zhang et al., 2016), and specific and broad-range QTLs (Manzanares-Dauleux et al., 2000a, 2003; Rocherieux et al. 2004; Jubault et al., 2008a; Chen et al., 2013; Tomita et al., 2013; Lee et al., 2016). As far as the information is accessible, genetic resistance factors appear to exert their control on pathogen development only after the completion of the primary phase (Kroll et al., 1983; Hatakeyama et al., 2013). One usual metabolic hallmark of susceptibility to clubroot infection is the accumulation of pathogen-synthesized trehalose, mostly during the later stages of infection (Keen and Williams, 1969; Broman et al., 2003; Gravot et al., 2011). In a previous study, we also pointed out that an accumulation of amino acids together with glutathione and S-methylcysteine during early secondary infection is correlated with susceptibility to clubroot in B. napus accessions (Wagner et al., 2012). However, the functional significance of these metabolic deviations remain unexplained.
The objective of the present study was to relate quantitative genetics to both metabolomics and pathogen-resistance traits, to get new insights into the metabolic modules associated with allelic variations at each QTL involved in the control of partial resistance. This work was focused on the progeny derived from two parental genotypes: Darmor-bzh, which displays a high level of partial resistance to the P. brassicae isolate eH, mainly controlled by the locus PbBn1, and Yudal, which shows an intermediate level of resistance to eH, controlled by several QTLs (Manzanares-Dauleux et al., 2003; Laperche et al., 2017; Aigu et al., 2018). We started the study with a histological and PCR-based evaluation of pathogen development over the duration of infection, to identify the earliest time point at which post-invasion partial resistance was distinguishable between the two parental genotypes. Then, at the earliest time point, metabolomic traits were investigated in roots of a segregating progeny derived from a Darmor-bzh × Yudal cross. Interlaced relationships between metabolites and the resistance-associated QTLs involved were then analysed using Cytoscape-based network representations.
Materials and methods
Plant and pathogen materials
Darmor-bzh is a winter dwarf line of B. napus, nearly isogenic to the genotype Darmor (Foisset et al., 1995), with an available genome sequence (Chalhoub et al., 2014). Yudal is a spring inbred line of B. napus selected from a Korean population. The segregating doubled haploid (DH) population used in this study was derived from an initial cross between the genotypes Darmor-bzh and Yudal (Foisset et al., 1996). The progeny, hereafter called the DY population, was first genotyped by Delourme et al. (2006). Based on these data, a subset of 130 DH lines was selected from this population, using the MapPop software (Brown and Vision, 2000) to optimize recombination events over the whole genome.
eH is a selection isolate of P. brassicae (Fähling et al., 2003). This isolate belongs to the most virulent pathotype, P1, according to Somé et al. (1996). This isolate was multiplied on the highly susceptible B. rapa spp. pekinensis cv. Granaat (genotype ECD5) and its pathogenicity profile was verified in parallel in each experiment, using the host differential set described by Somé et al. (1996).
Pathogenic assays
Most clubroot assays in this study (except the study of primary infection; see below) were performed using a previously reported procedure (Wagner et al., 2012). In brief, plants were sown in plates filled with a mixture of two-thirds compost and one-third vermiculite, then cultivated under controlled conditions (16 h light at 22 °C/8 h dark at 18 °C). Plantlets were inoculated 7 days after germination, using the eH isolate of P. brassicae (107 spores ml−1) according to Manzanares-Dauleux et al. (2000b), and non-inoculated plants were treated with distilled water (control treatment).
Evaluation of infection kinetics in genotypes Darmor-bzh and Yudal
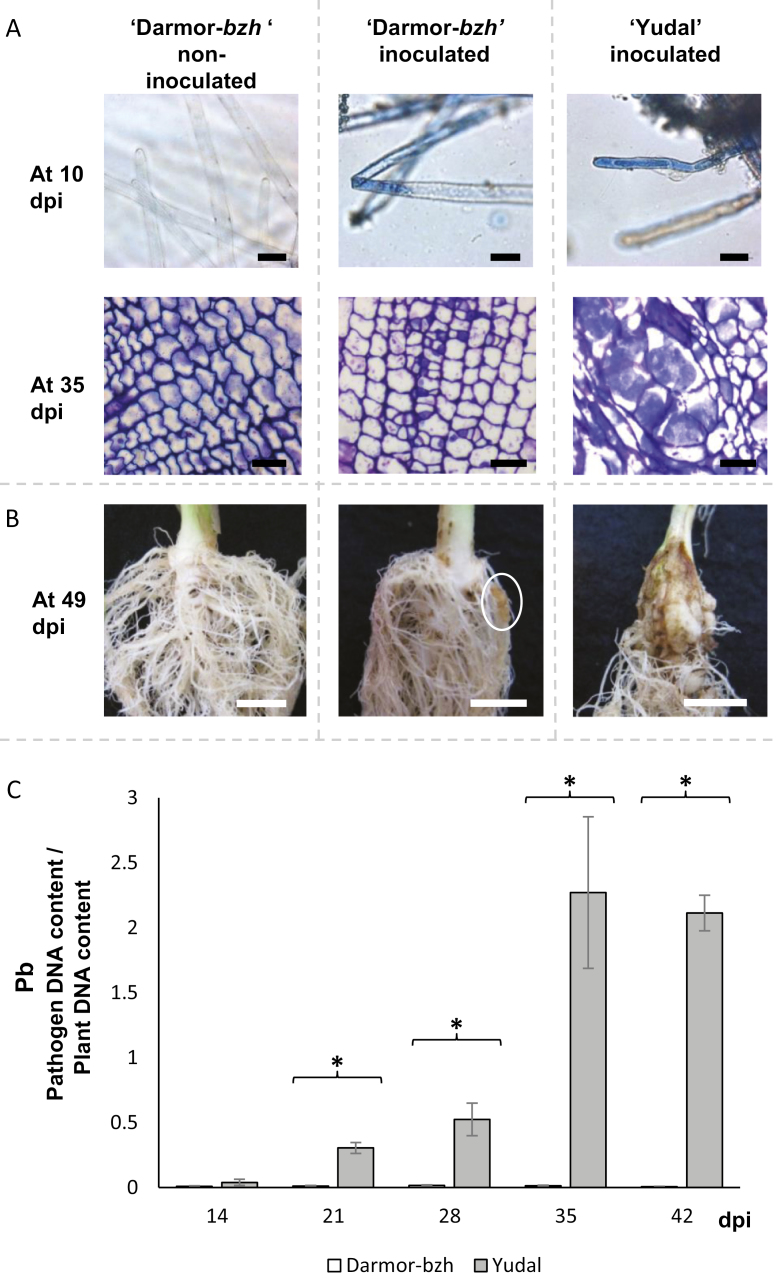
To study primary infection in root hairs (Fig. 1A), seedlings of genotypes Darmor-bzh and Yudal were germinated on moistened paper in petri dishes, and then cultivated hydroponically: the system consisted of a plastic pipette tip box and each plantlet was grown in one hole in the box. The root system of the plantlets was completely immersed in 400 ml of tap water (for non-inoculated plants) or 400 ml of an inoculum suspension at a concentration of 107 spores ml−1 (for inoculated plants). Plantlets were sampled at 10 days post-inoculation (dpi) and their roots were directly stained with 1% toluidine blue (Sigma Aldrich, St. Louis, MO, USA) for microscopy.
Fig. 1.
Time course of compatible interactions in the two genotypes of B. napus, Darmor-bzh and Yudal, inoculated with the eH isolate of P. brassicae. (A) Histopathological characterization of clubroot infection in the root hairs at 10 days post-infection (dpi) and from transverse sections of the roots at 35 dpi of inoculated and non-inoculated plants. Staining with 1% toluidine blue allows visualization of P. brassicae inside root hairs during the primary phase, and inside cortical cells during the secondary phase. Bars=25 μm. (B) Typical root symptoms at 49 dpi. Bars=1 cm. The white ellipse on the central picture indicates the presence of a small gall. (C) Dynamics of pathogen root invasion in Darmor-bzh and Yudal followed by quantitative PCR using DNA extracted from whole infected roots. The internal transcribed spacer region of P. brassicae (PbITS) was amplified and compared with the Cruciferin A gene of B. napus (BnCruA), and the ratio of pathogen to plant DNA (Pb) was calculated. Samples were analysed at 14, 21, 28, 35, and 42 dpi. Data are expressed as the mean ±SE of four replicates. Asterisks indicate significant differences in Pb values between Darmor-bzh and Yudal at the same sampling time (P<0.05, Wilcoxon-Mann-Whitney test). (This figure is available in colour at JXB online.)
Secondary infection kinetics was followed using a clubroot assay organized in a randomized complete block design with four replications. In each experiment, plants of Darmor-bzh and Yudal were inoculated with isolate eH and grown using previously reported procedures (Wagner et al., 2012). Root samples were collected at 14, 21, 28, 35, and 42 dpi. In each block, each sample consisted of a bulk of 27 plants, except for the first sampling point at 14 dpi, where 54 plants were collected to obtain enough plant material for further analyses. Six additional plants per block and per genotype were included in the experimental design to evaluate symptom severity at 49 dpi (Fig. 1B). At each sampling date, total roots were harvested and immediately frozen in liquid nitrogen and stored at –80 °C before further analyses.
Additional plants were included in this assay for histological evaluation of P. brassicae development during secondary infection. Roots of inoculated and non-inoculated plants of genotype Darmor-bzh were collected weekly from 14 to 35 dpi and immediately immersed in 2% glutaraldehyde in phosphate buffer (0.1 mM, pH 7.2) before subsequent histological investigations. Root samples previously fixed in glutaraldehyde were dehydrated according to Wagner et al. (2012) and embedded in Technovit 7100 resin (Heraeus Kulzer, Wehrheim, Germany) according to the supplier’s instructions. Thick sections (4 μm) were cut with a rotary microtome (Microm Microtech, Francheville, France) and stained with 1% toluidine blue O (Sigma Aldrich).
Clubroot assays with doubled haploid progeny
A clubroot assay was performed using isolate eH, the parental lines Darmor-bzh and Yudal, and 130 DH progeny lines. The experiment consisted of two independent biological replicates. For each biological replicate, parental lines were studied with 20 blocks of 18 inoculated plants and 10 blocks of 18 non-inoculated plants. DH lines were represented in each biological replicate by 18 inoculated plants. Samplings were performed at 21 dpi (corresponding to a time at which no clubroot symptoms are visible yet), during the first 3 hours after the beginning of the light phase. For each sample, 18 plants were washed, cut, pooled, frozen in liquid nitrogen, and then stored at –80 °C before subsequent extractions and molecular analyses. An extra 12 plants per genotype and per repetition were included in the experimental design to record symptoms at 49 dpi, using the scale described by Manzanares-Dauleux et al. (2000b). This allowed a disease index (DI) to be calculated for each genotype according to Buczacki et al. (1975).
DNA-based quantification of P. brassicae in infected tissues
Frozen root samples were ground to powder in liquid nitrogen using a mortar and pestle, and then freeze-dried. DNA was extracted using the DNeasy Animal Blood and Tissue Kit (Qiagen, Hilden, Germany). The DNA concentration was normalized using a Nanodrop ND-1000 Spectrophotometer (Labtech, Palaiseau, France). Pathogen DNA was quantified in planta using the primers PbITS6 and Pb4-1 designed by Sundelin et al. (2010) to specifically amplify the internal transcribed spacer (ITS) region of P. brassicae, and related to the amount of plant DNA using the Cruciferin A gene with the primers MDB510 and MDB511 (Wu et al., 2010). The melting curve of the PCR products was analysed at each time point to ensure amplification specificity. Real-time quantitative PCR was performed in a Light Cycler 480 (Roche Diagnostics GmbH, Mannheim, Germany) as described by Wagner et al. (2012). All DNA samples were analysed in triplicate in the same 384-well plate, and the experiment was repeated twice. Quantitative curves for the relative quantification of plant and pathogen DNA were established based on serial dilutions of DNA pools from mixed infected samples. Results were expressed as the ratio between the amount of pathogen DNA and plant DNA, hereafter referred to as Pb.
Metabolomic analyses
Targeted analyses of non-structural carbohydrates, polyols, organic acids, and amino acids were performed with procedures previously reported by Jubault et al. (2008b), Gravot et al. (2010), and Colinet et al. (2012). Non-targeted LC-MS analyses were performed as follows. Metabolites were extracted from 50 mg of dried plant material. A 1 ml volume of a methanol/water mixture (50/50, v/v) was added to the sample, acidified with 5% (v/v) trifluoroacetic acid, then filtered through a 0.45 µm filter. Samples were injected into a HPLC-LCQ Deca ion trap mass spectrometer (Thermo Fisher). Chromatogram files were analysed using the Bioconductor package XCMS (Smith et al., 2006). Peaks were detected using the centWave algorithm (Tautenhahn et al., 2008), using the following parameters: 700 ppm, peak width from 20 to 60 s, s/n threshold 5:1. Retention time correction was performed in three successive iterations of the functions ‘group’ (mzwd=0.25 and minfrac=0.2) and ‘retcor’ (decreasing the bw parameter from 80 to 40 and then 20). A total of 289 different mass signals (see also Supplementary Fig. S1 at JXB online) were obtained in all biological samples (246 HD lines and 59 parental lines). This raw dataset was then filtered as follows. First, the Bioconductor package CAMERA implemented in R (Kuhl et al., 2012) was used to identify 76 pc-groups, that is, sets of co-eluted mass signals with similar peak shapes. The mean number of mass tags in each pc-group was ~3.8 (range 1–33). In every single pc-group, the low-signal ion species that were highly correlated (Pearson’s correlation coefficient >0.75 among 301 samples) with another ion species of higher signal in the same pc-group were ignored in further analyses. This approach led to the selection of 186 ion species, that is, a mean of 2.5 non-correlated ion species for each pc-group of co-eluted ion species. These 186 ion species are hereafter referred to as analytes and named after their m/z (M) and retention time in seconds (T), for example, M365T432. Analyte peak areas were then normalized to take account of the mass of each sample (~50 mg). The structural identity of some of these analytes was investigated through additional tandem mass spectrometry (MS/MS), leading to the identification of 11 metabolites, including nine glucosinolates (neoglucobrassicin, gluconasturtiin, 4-methoxyglucobrassicin, glucobrassicin, 4-hydroxyglucobrassicin, glucoerucin, glucoraphenin, glucobrassicanapin, gluconapin), one phytoalexin (rutalexin), and one salicylate glucoside. High-resolution mass spectrometry of the fragments from M320 and 368 was investigated using a Shimadzu LCMS-IT-TOF instrument.
Data analysis
Mean-centred data from targeted and non-targeted analyses were pooled together in a single dataset, before subsequent statistical analyses. Principal component analysis (PCA) was performed on the data from the parental lines using the R package FactoMineR (Lê et al., 2008). Statistical analyses of phenotypic, molecular, and metabolic data from the DH lines were carried out using R software (R Core Team, 2016) and the package lme4 (Bates et al., 2015). A mixed linear model was defined as follows:
| (1) |
where Pijk is the mean value of the ith DH line sampled in the jth biological repetition, µ is the mean of all the data, Li is the DH effect, Rj is the repetition effect, and eijk is the residual. All effects were declared as random. Heritability (h2) and standard deviation were estimated from model (1), using the formula
| (2) |
where σ g2 is the genetic variance and σ e2 is the environmental variance. For each genotype and for each trait, adjusted means were considered according to model (1) and were used for QTL mining. Spearman correlation coefficients (α=0.05) were calculated between the metabolic contents of the DH lines and the Pb values.
QTL analysis
The DY genetic map was obtained using an extended set of 356 DH lines (including the 130 DH lines previously described) and three sets of single nucleotide polymorphism (SNP) markers. The first two sets are those described by Delourme et al. (2013). The third set corresponds to a 60K Illumina Infinium array. In total, 29 319 markers were polymorphic and mapped. A reduced set of 3728 SNPs represented all unique positions. The 3728 SNPs were well spread over all linkage groups with on average one SNP every 0.56 cM. The biggest distance between two markers was present on chromosome C01, with 8.4 cM. The 19 linkage groups, named A01 to A10 for the A genome and C01 to C09 for the C genome, covered 2087.7 cM. QTL analyses were performed using the R/QTL package (Broman et al., 2003; Arends et al., 2010). Composite interval mapping was carried out for each of the 231 metabolites, as well as for Pb and the DI. The number of covariates was set according to a first run of simple interval mapping for the trait under investigation. Logarithm of the odds (LOD) thresholds were estimated using 1000 permutations (Churchill and Doerge, 1994), and QTL confidence intervals were defined according a LOD drop-off of one unit.
A meta-analysis was carried out using Biomercator V4.2 (Goffinet and Gerber, 2000, Arcade et al., 2004, Sosnowski et al., 2012). In this study, metaQTLs were defined as genetic regions where multiple QTLs were colocalized, and were obtained by integrating information from individual QTL detections (metabolites, DI, and Pb) using the Meta-analysis tool. The number of metaQTLs per linkage group was assessed according to the Veyrieras algorithm (Veyrieras et al., 2007) and the Bayesian Information Criterion. Each individual QTL contributed to one or several metaQTLs.
Cytoscape-based representation of QTL/trait networks
The full list of QTLs/phenotypic traits was treated as a directed network. In this network, nodes represent loci or traits, and edges represent statistically significant links between loci and phenotypic traits (from the QTL analysis described above). Edge widths were used to represent R2, that is, the percentage of variance explained by each locus for a given phenotypic trait. Edge colour was used to represent positive or negative allele effects on phenotypic traits. This network was represented graphically using Cytoscape software (Shannon et al., 2003).
Results and discussion
Partial resistance and metabolomic responses to clubroot infection in the Darmor-bzh and Yudal parental lines of B. napus
A first series of experiments was conducted to monitor pathogen development in the B. napus parental genotypes Darmor-bzh and Yudal. Primary plasmodia were observed in root hairs of both genotypes at 10 dpi (Fig. 1A). At 14 dpi, small secondary plasmodia were detected in the cortical cells of both Darmor-bzh and Yudal (data not shown). At 35 dpi, infected roots of Yudal displayed extensive hyperplasia and hypertrophied plant cells containing well-developed secondary plasmodia of P. brassicae (Fig. 1A). At the same sampling point in Darmor-bzh, secondary plasmodia were also observed, but they were much less developed than in Yudal, and the infected cells were of similar size or even smaller than uninfected cells in the non-inoculated plants. The DI at 49 dpi was 45 for Yudal; clubs were observed on the primary root system in the majority of plants (Fig. 1B) and most of the leaves were wilted. The DI for Darmor-bzh at the same time point was 28, and only a few small galls had developed on the secondary root system. Fully susceptible genotypes from the differential reference set (Somé et al., 1996) displayed DI values of 100. This confirmed that the Darmor-bzh and Yudal genotypes harbour partial resistance to eH.
The Pb value was similar in both genotypes at 14 dpi, the beginning of the secondary phase of infection (Fig. 1C). In Yudal, Pb increased by approximately two orders of magnitude between 14 and 35 dpi. By contrast, in the infected roots of Darmor-bzh, Pb remained consistently low throughout the whole time-course. Pb values of the two genotypes were significantly different at 21 dpi, after the beginning of the secondary infection in cortical cells, but before any cellular hypertrophy or club symptoms could be observed. The difference between the two genotypes remained significant at all time points from 21 dpi onwards. Our data thus indicate that Darmor-bzh displayed a higher level of partial resistance than Yudal, and that this resistance was triggered during the early phase of clubroot secondary infection (i.e. several days after the primary infection of root hairs). Such post-invasive resistance to clubroot is consistent with previous findings from several studies (Voorrips, 1992; Ueno et al., 2012; Lemarié et al., 2015; Xu et al., 2018). The time point 21 dpi was consequently chosen for analysing the genetic architecture of metabolic responses to clubroot infection.
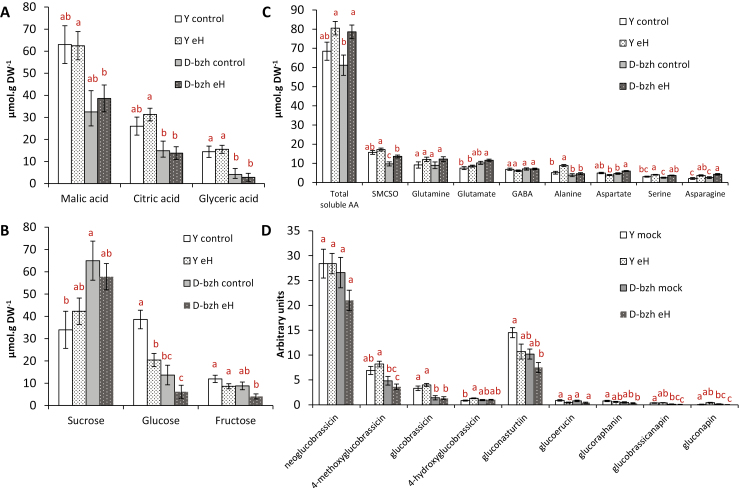
Next, root metabolic content was investigated in inoculated and non-inoculated roots of the parental Darmor-bzh and Yudal lines at 21 dpi, using a combination of GC-MS and LC-MS-based targeted and untargeted analyses (Supplementary Fig. S1 and Table S1). This resulted in a full dataset of 231 analytes, including 55 identified primary and secondary metabolites (Supplementary Fig. S1 and Table S1). The predominant primary metabolites in non-inoculated roots (Fig. 2) were malic acid, citric acid, and the soluble sugars sucrose and glucose (Fig. 2A, B), in accordance with previous studies (Wagner et al., 2012). The major amino acids were S-methylcysteine sulfoxide (SMCSO) (a typical non-proteinogenous sulfur amino acid from Brassica species), glutamine, and glutamate (Fig. 2C). The main glucosinolate-associated mass signals were related to indole glucosinolates with the most abundant compounds being 1-methoxy-3-indolyl glucosinolate (neoglucobrassicin) and 4-methoxy-3-indolyl glucosinolate (4-methoxyglucobrassicin), and to the aromatic glucosinolate gluconasturtiin (2-phenylethyl glucosinolate). In addition, a series of low-intensity mass signals were found associated with root aliphatic glucosinolate content (glucoerucin, glucoraphanin, glucobrassicanapin, and gluconapin) (Fig. 2D). Yudal differed from Darmor-bzh mainly in terms of its higher constitutive root content in total organic acids (P=0.031) and total glucosinolates (P=0.004), irrespective of inoculation with P. brassicae. Although total root sugars did not differ between the two non-inoculated genotypes, the ratio of sucrose to glucose was markedly higher in Darmor-bzh (5.2) than in Yudal (0.9). The major primary metabolic response to clubroot infection at 21 dpi was a slight increase of amino acid content in both genotypes, with a notable increase of alanine content in infected Yudal roots. This accumulation of alanine is consistent with the previously reported induction of fermentation processes and hypoxia responses during clubroot infection in a susceptible genotype of Arabidopsis thaliana (Gravot et al., 2016). In Yudal, inoculation also led to an important decrease in glucose content from 39 to 21 µmol g DW−1, and to a statistically significant (P=0.019) increase in trehalose content from 0.6 to 1 µmol g DW−1 (Supplementary Table S1), consistent with previous studies of this genotype (Wagner et al., 2012). The increasing accumulation of trehalose (presumably synthesized by P. brassicae) is usually observed during the secondary phase of clubroot infection, finally reaching 10–50 µmol g DW−1 in fully developed clubs (5–6 weeks post-inoculation in rapeseed) (Keen and Williams, 1969; Broman et al., 2003; Gravot et al., 2011; Wagner et al., 2012). In the present study, the slight increase in trehalose content in Yudal at 21 dpi is indicative of an already active secondary-phase development of P. brassicae.
Fig. 2.
Selected primary metabolite and glucosinolate contents in inoculated (with P. brassicae eH isolate) and non-inoculated (control) roots of the parental lines at 21 dpi. Y, Yudal, D-bzh, Darmor-bzh. (A) Organic acids. (B) Soluble carbohydrates. (C) Free amino acids. (D) Glucosinolates (arbitrary units, based on peak surface area and sample DW). For each condition, analyses were performed using 10 replicates (non-inoculated) or 20 replicates (inoculated with the eH isolate), each replicate consisting of at least 12 plants that were pooled for metabolite extractions. Error bars indicate SE. Different letters indicate significant differences in mean values (P<0.05, Tukey method), except for sucrose (α=0.062).
Concatenation of datasets from targeted and untargeted metabolic profiling, resulting in a full dataset of 231 analytes (Supplementary Table S1), broadened the view on possible metabolic contrasts between infected and non-infected roots of the two genotypes. PCA of this dataset (Supplementary Fig. S2) indicated that ‘genotype’ was the major factor influencing metabolic contrasts between samples (axis 1), followed by ‘inoculation’, which led to a clear metabolic shift in roots of Yudal (axis 2) and, to a lesser extent, in Darmor-bzh (axis 3).
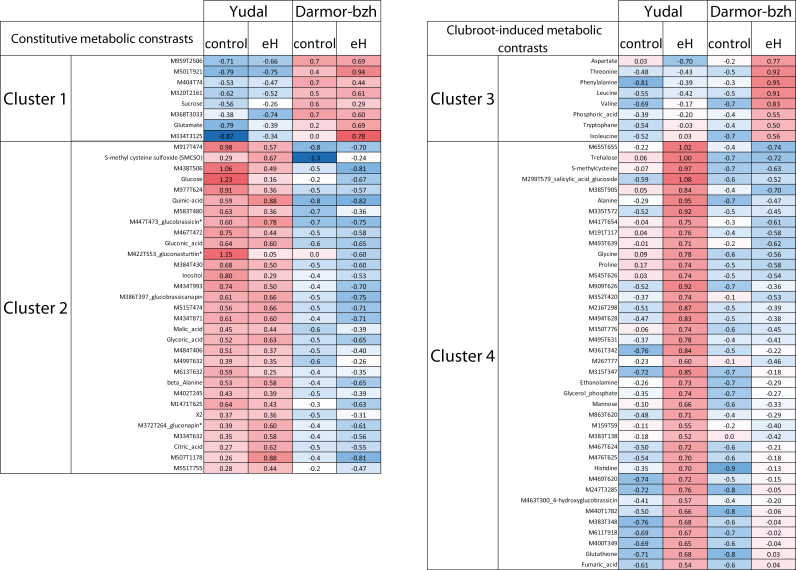
A further analysis of this extended metabolite dataset allowed us to identify four categories of analytes showing a contrast between the two genotypes in inoculated roots (Fig. 3): metabolites that are constitutively more accumulated in one of the two genotypes (cluster 1, Darmor-bzh; cluster 2, Yudal), and metabolites whose accumulation is triggered by P. brassicae inoculation in only one of the two genotypes (cluster 3, Darmor-bzh; cluster 4, Yudal). Cluster 4 included a large number of compounds that accumulated specifically in inoculated roots of the more susceptible accession, Yudal. This large set of metabolites included trehalose and S-methylcysteine, two analytes that we have previously reported as clubroot susceptibility biomarkers (Wagner et al., 2012); several additional compounds, such as a salicylic glucoside; and 27 unknown analytes, including M655T655 (with a mass signal ~10-fold higher in inoculated roots of Yudal compared with Darmor-bzh). Cluster 3 mostly included amino acids (threonine, phenylalanine, tryptophan, isoleucine, and valine) that were specifically triggered by P. brassicae inoculation in the more resistant accession Darmor-bzh.
Fig. 3.
Constitutive and clubroot-induced root metabolomic differences between inoculated (control) and non-inoculated (with P. brassicae eH) roots of Yudal and Darmor-bzh. Cluster 1: metabolites constitutively accumulated at a higher level in Darmor-bzh than in Yudal; cluster 2: metabolites constitutively accumulated at a higher level in Yudal than in Darmor-bzh; cluster 3: metabolites accumulated at a higher level in inoculated roots of Darmor-bzh compared with non-inoculated roots and compared with inoculated roots of Yudal; cluster 4: metabolites accumulated at a higher level in inoculated roots of Yudal compared with non-inoculated roots and compared with inoculated roots of Darmor-bzh. Metabolomic data are presented as mean-centred values from 10 replicates (non-inoculated) or 20 replicates (inoculated with the eH isolate), each replicate sample consisting of pooled roots from 12 plants. Red indicates higher levels of accumulation and blue indicates lower levels. XCMS-generated mass-tags were named based on their m/z (M) and retention time (T). Statistically significant differences (P<0.05) were inferred from Student’s t-test.
The biological role of the above-described metabolic features at 21 dpi in the expression of clubroot resistance (or susceptibility) in Darmor-bzh and Yudal is difficult to decipher. Indeed, any compound within these four clusters can a priori promote or impede the infection process; for instance, glucobrassicin and other indole glucosinolates may support the biosynthesis of auxin, thus possibly favouring gall development (Ludwig-Müller, 2009), or alternatively might support the synthesis of defensive phytoalexins (Clay et al., 2009; Klein et al., 2017). Moreover, Darmor-bzh and Yudal displayed compatible interactions with isolate eH, and accordingly it is expected that both genotypes would show some metabolic responses associated with susceptibility. Conversely, previous studies (Laperche et al., 2017; Aigu et al., 2018) have highlighted that both Darmor-bzh and Yudal harbour resistance alleles at different QTLs, and consequently both genotypes were expected to exhibit metabolic features associated with resistance (the mechanisms of which could be either similar or different in the two accessions). To extend the understanding of the relationship between partial resistance and metabolic traits, it was necessary to compare the mapping of QTLs associated with the control of disease traits and metabolic profiles in the two genotypes. To this end, a genetic analysis of Pb, DI, and metabolite patterns was conducted on a 130 DH progeny derived from the cross Darmor-bzh × Yudal.
QTLs controlling pathogen infection and the root metabolome
PCR-based quantification of pathogen relative growth (expressed here as Pb) was performed at 21 dpi in the progeny, where this variable showed a continuous pattern of distribution (Fig. 4A), suggesting polygenic control of pathogen growth. The distribution of DI at 49 dpi in the progeny showed a continuous but near bimodal shape, suggesting the involvement of a major-gene resistance in the control of the club phenotype (Fig. 4B). The values of DI were plotted against the values of Pb for each DH line (Fig. 4C), revealing that DI and Pb were not linearly correlated. This may reflect a saturation in the evaluation of symptoms with the DI scale. The DI takes account of the proportion of galls relative to asymptomatic roots, but it does not take account of the size of fully developed clubs, and thus does not distinguish nuances between highly susceptible genotypes. This argument may, however, not explain everything, as a low Pb value at 21 dpi in a given recombinant line did not allow the prediction of the final DI at 49 dpi, which may also suggest the existence of genetic factors exerting their effects after 21 dpi.
Fig. 4.
Quantitative analysis of resistance to infection with the P. brassicae eH isolate in the Darmor-bzh × Yudal DH progeny. (A) Frequency distribution of the pathogen–plant genomic DNA ratio (Pb) in the Darmor-bzh × Yudal DH population. The arrows indicate the mean values of the parental lines (D, Darmor-bzh; Y, Yudal). (B) Frequency distribution of the mean disease index (DI) in the Darmor-bzh × Yudal DH population. The arrows indicate the mean values of the parental lines. (C) DI values at 49 dpi plotted against Pb values at 21 dpi for each DH line.
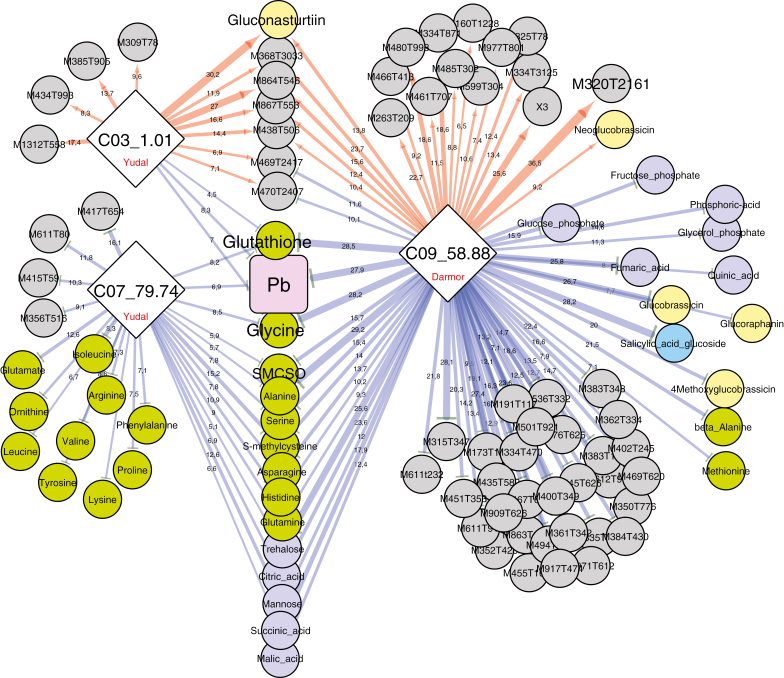
Genetic analysis by composite interval mapping of Pb led to the identification (Supplementary Table S2) of one moderate-effect QTL (R2=27.9%) on chromosome C09 (resistance allele from Darmor-bzh) and two weak-effect QTLs on chromosomes C03 and C07 (resistance allele from Yudal). DI was found to be under the control of one major QTL (R2=78.8%) on C09, consistent with the phenotype distribution curve (Fig. 4B). The moderate-effect QTL C09_Pb and the major-effect QTL C09_DI, both detected at 59 cM, colocalized with the major resistance locus, first described as Pb-Bn2 by Manzanares-Dauleux et al. (2003) and further reported by Laperche et al. (2017) and Aigu et al. (2018). Four additional minor QTLs were found to be involved in the control of DI; these were located on chromosomes A01, A10, C02, and C03. The latter two QTLs, on C02 (position=130.8 cM) and C03 (position=35.2 cM), colocalized with QTLs involved in resistance to isolate eH reported by Laperche et al. (2017) and Aigu et al. (2018). In those previous studies, the genetic effect of these QTLs was found to be enhanced by decreasing the nitrogen supply (Aigu et al. 2018). Plants were cultivated under a non-limited nitrogen supply in the present study, and consequently the two QTLs on chromosomes C02 and C03 had only a low effect on resistance. Finally, the genetic architectures of Pb and DI were overlapping only for the QTL on C09 (Pb-Bn2). Other loci controlling Pb (C03, position=2.5 cM, and C07, position=67.7 cM) were also found to be involved in delaying the progression of P. brassicae infection during the early phase, without affecting the final development of clubroot symptoms. By contrast, for example, the resistance QTL on C02 appeared to control final symptom development but without any effect on the early progression of P. brassicae infection (Supplementary Table S2). These results can be compared with other reports of age-related or ontogenetically conditional resistance loci in quantitative resistance to pathogens (examples are given in Corwin and Kliebenstein, 2017). The fact that Pb and DI do not measure equivalent traits (the first is indicative of pathogen development, whereas the second is indicative of infection-triggered cellular proliferation) may also explain why the genetic architecture of club development overlaps only partially with that of P. brassicae multiplication. Similar phenomena have been previously reported for clubroot resistance in B. napus by Aigu et al. (2018).
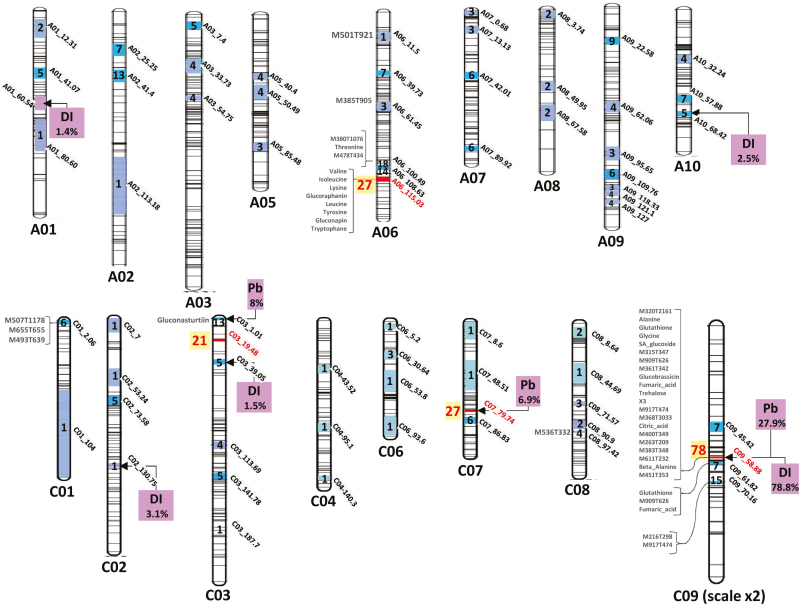
Targeted and untargeted metabolite profiling was therefore performed on inoculated root samples of the 130 DH lines at 21 dpi together with the parental lines (detailed results are presented in Supplementary Table S1). QTL analysis was performed for the 231 analytes. No QTL was detected for 55 analytes (including sucrose; the other 54 analytes were unidentified compounds). Nevertheless, 374 metabolite QTLs (mQTLs) were detected (details are given in Supplementary Table S2). One to seven QTLs (most often one or two QTLs) were detected for each analyte. The mean R2 per QTL was 12% (range 1.2–66.4%). To simplify further analysis of colocalizations, both mQTLs and resistance QTLs (controlling either Pb or DI) were processed by a QTL meta-analysis, allowing their aggregation on a set of 70 consensus genetic intervals (metaQTLs) (Fig. 5, Supplementary Table S3, Supplementary Figs S3 and S4). When necessary, some mQTLs were simultaneously attributed to two adjacent metaQTLs (details of weighting factors are given in Supplementary Table S3). Such cases were taken into account in further analysis when weighting factors exceeded 20%.
Fig. 5.
Map of QTLs involved in the control of clubroot resistance traits (Pb and DI) and metabolite QTLs (mQTLs) involved in the control of root metabolite contents at 21 dpi. Inside the chromosomes, the confidence intervals (following metaQTL analysis) and the number of overlapping mQTLs (light blue <5, blue <20, red >20) are indicated. At the left side of the chromosomes, mQTLs associated with R2 >20% are indicated. At the right side of the chromosomes, purple boxes indicate QTLs controlling the relative content of pathogen DNA at 21 dpi (Pb; see the Materials and methods for details), and QTLs controlling the clubroot disease index (DI) at 49 dpi. Additional details of individual QTLs are given in the Supplementary data.
Among the 70 metaQTLs identified, 18 contained only one QTL. This included the metaQTL A01_60.54, which exerted a small effect on DI (R2=1.4%) but was not associated with any mQTL, and the QTL A06_11.5, which exerted strong control (R2=40%) on the content of the unidentified root compound M501T921.
By contrast, four genomic regions were involved in the control of more than 20 mQTLs (Fig. 5). The metaQTL C09_58.88 was a major ‘hot-spot’ containing 78 mQTLs, including 35 mQTLs with R2>15%. This metalocus colocalized with a major resistance QTL controlling DI and Pb (R2=78.8% and 27.9%, respectively). The metaQTL C07_79.74 was associated with 27 mQTLs, and colocalized with a resistance QTL contributing to the control of Pb (resistance allele from Yudal, R2=6.9%). This locus included the major mQTL controlling gluconasturtiin (R2=30.2%), with the Yudal allele being associated with a higher gluconasturtiin content.
Three adjacent metaQTLs (A06_100.49, A06_108.63, and A06_115.03) were found on chromosome A06 along a 15 cM genomic region. Altogether, the metaQTLs from this region were involved in the control of 59 metabolites. No clubroot resistance QTL was detected in this region in our study. A06_115.03 corresponded to the bzh genomic region introduced into the oilseed rape genotype Darmor-bzh from a mutant line obtained by chemical mutagenesis of the genotype Primor (Foisset et al., 1995). Some of the metabolic features observed in the present study were likely caused by this dwarfism allele (bzh), which corresponds to the DELLA-encoding gene BnaA06g34810D (genetic position 114.3 cM, physical position 23008797-23010993), which is involved in the repression of gibberellin responses (Zhao et al., 2017). The metaQTL C03_19.48 contained 21 mQTLs. The confidence intervals calculated from the meta-analysis suggested that this metabolic hot-spot did not include the two resistance QTLs detected nearby on the same chromosome: one resistance QTL exerting a moderate effect on Pb (R2=8.9%) and one resistance QTL exerting a low effect on DI (R2=1.5%) (Fig. 5).
The fact that two out of the four hot-spot metabolic metaQTLs were genetically unrelated to partial resistance can be viewed as supporting the hypothesis that not every single metabolic contrast observed between Yudal and Darmor-bzh was necessarily connected to the fate (i.e. resistance or susceptibility) of the interaction with P. brassicae. Furthermore, from this dataset it also appeared that several compounds linked to resistance-related metaQTLs were also under the genetic control of other resistance-unrelated metaQTLs, thus questioning the putative functional link of those compounds with resistance. A network-based approach was thus necessary to shed more light on the complexity of locus/metabolite/resistance relationships.
Network-based analysis of links between metabolite fingerprints and resistance QTLs
Similarities and differences between metabolic patterns controlled by each of the four hot-spots were compared by a Cytoscape-drawn directed network (represented in Supplementary Fig. S5). This representation highlighted not only how each hot-spot or metaQTL displayed its own pattern of metabolites, but also allowed an effective visualization of metabolites that are under the simultaneous control of two or even three of the four metaQTLs.
A first example is a series of amino acids (represented in green in the lower left of Supplementary Fig. S5), which were controlled by several mQTLs clustered under the metaQTL C07_79.74 (which is also associated with clubroot resistance) and other mQTLs clustered under the metaQTL A06_115.03 associated with the mutation bzh. Then, the rise of the amino acid contents could be independently triggered by two independent processes: an enhanced root metabolic sink caused by P. brassicae when a susceptible allele is present at C07_79.74 (Keen and Williams, 1969; Li et al., 2018), or an influence of the dwarfism gene bzh on shoot-to-root metabolite allocation (in line with Elias et al. 2012).
Fig. 6 illustrates the similarities and differences between metabolic patterns observed at 21 dpi associated with each of the three QTLs controlling pathogen development at 21 dpi (expressed as Pb). The amounts of glutathione and glycine are lower (indicated by blue arrows in Fig. 6) in the presence of alleles that decrease Pb at the three metaQTLs C03_1.01, C07_79.74, and C09_58.88. The apparently pivotal significance of these two nitrogen-containing compounds is consistent with previous results highlighting these compounds as susceptibility biomarkers (Wagner et al., 2012). In this previous study, Wagner et al. (2012) also identified additional susceptibility-related metabolites, including S-methylcysteine, trehalose, and alanine, which in the present study have been found to be regulated by the QTLs C07_79.74 and C09_58.88. As already mentioned, a large number of specific metabolic features were found to be associated with the metaQTL C09_58.88 (comprising one major-effect resistance QTL and 78 mQTLs). For a large number of these compounds, their increased accumulation was controlled by the resistance allele (indicated by red arrows in Fig. 6), suggesting that some of these unidentified compounds might be good candidates for causal factors involved in defence (detailed below). The metaQTL C03_1.01 included a major effect mQTL (R2=30.2%) controlling gluconasturtiin content, and a set of 13 mQTLs. The increased accumulation of gluconasturtiin was controlled by the resistance allele, which may suggest a positive role exerted by this glucosinolate on partial resistance. The resistance allele at QTL C07_79.74 was clearly involved in repressing the content of a series of amino acids, some of which were also repressed by C09_58.88, whereas others were specifically controlled by QTL C07_79.74.
Fig. 6.
Sub-network representing mQTLs connected to the three QTLs involved in the control of the pathogen–plant genomic DNA ratio (Pb) measured at 21 dpi. Nodes represent QTLs controlling the Pb trait (diamonds) and metabolites (circles), and edges represent the partial quantitative effect of metaQTLs on traits. The biochemical category of the metabolites is indicated by the colours of the circles: green, amino acids; pale yellow, glucosinolates; dark purple-blue, sugars, polyols and organic acids; bright blue, hormone-related compounds; grey, non-identified compounds. The width of the edges indicates the quantitative effect of each QTL (R2) for a given QTL. Red and blue edges indicate positive and negative effects, respectively, on trait values (metabolite or Pb), controlled by Darmor or Yudal alleles at each QTL (as indicated in the diamonds).
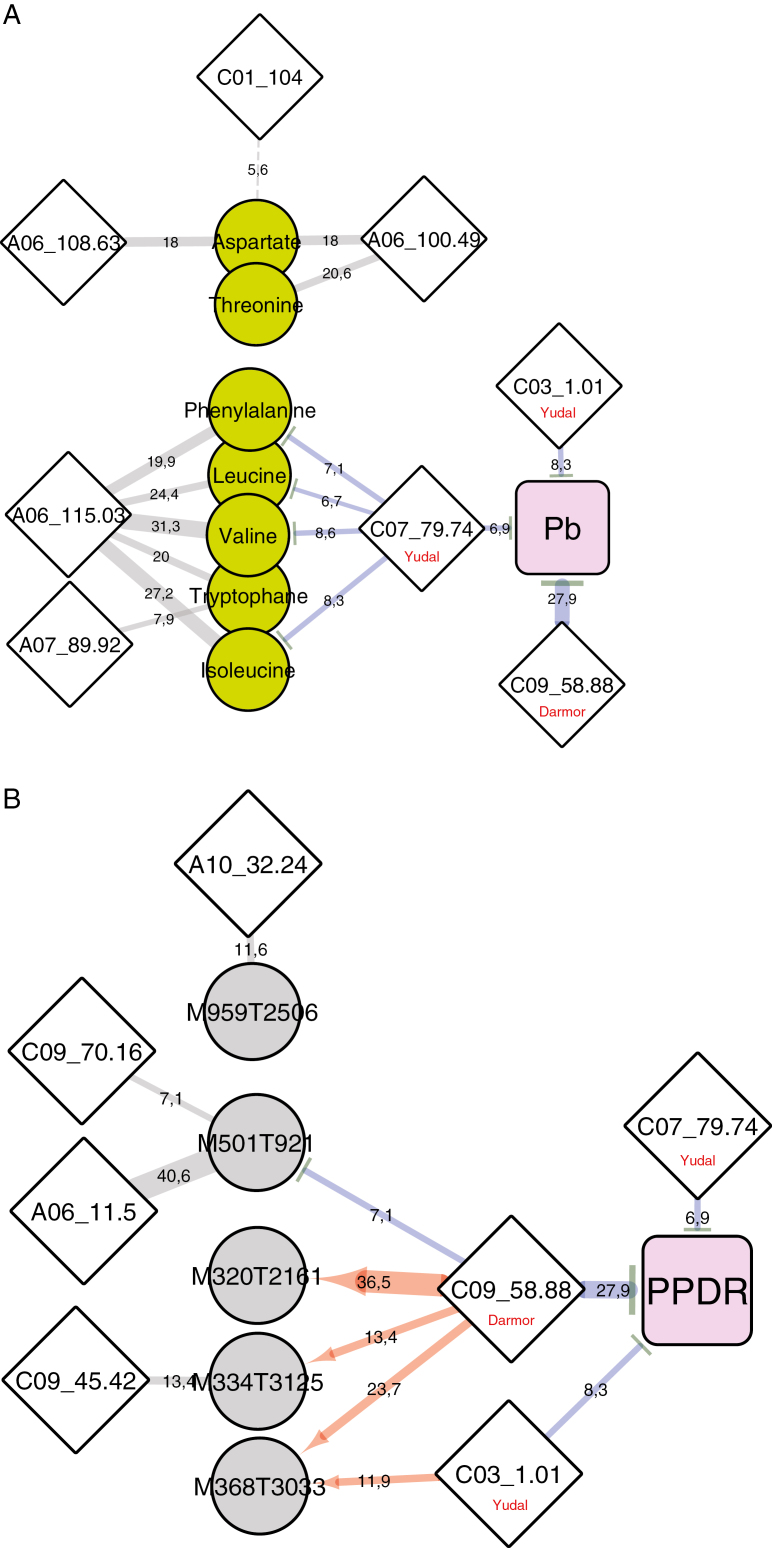
A similar network connecting mQTLs at 21 dpi to QTL involved in the control of DI at 49 dpi (Supplementary Fig. S6), indicated that at 21 dpi only the major QTL, Pb-Bn2 (C09_58.88), was associated with a rich metabolic network. By contrast, minor QTLs involved in the control of DI displayed only faint metabolic fingerprints. This result was consistent with the fact that those minor QTLs did not colocalize with any QTLs controlling pathogen development (i.e. Pb) at 21 dpi. Together, these results indicate that for the QTLs with a low effect on DI, underlying resistance processes are set up after 21 dpi.
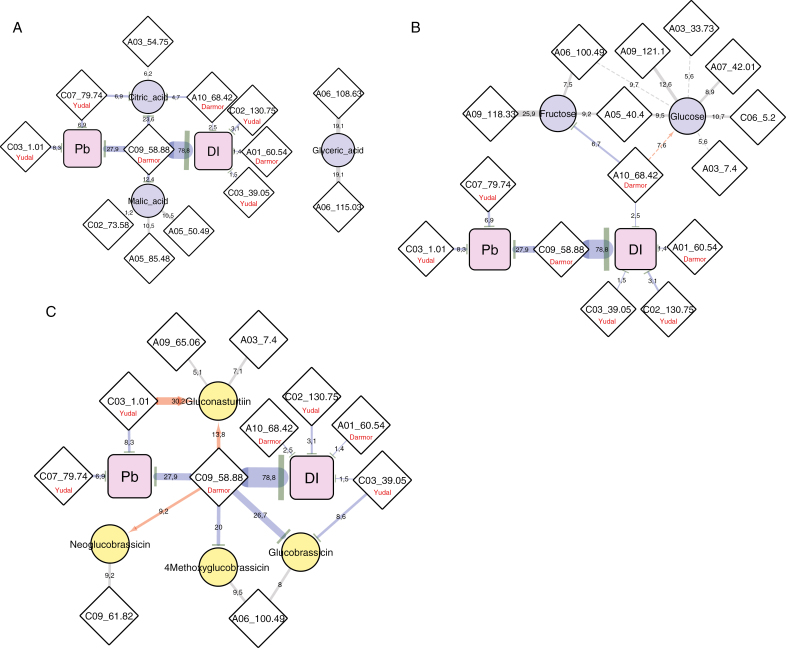
In the first part of this study, comparative targeted analysis from parental lines had highlighted constitutively higher contents of organic acids, glucose, and glucosinolates in the accession Yudal compared with Darmor-bzh (Fig. 2), raising the question of the role of these compounds during P. brassicae infection. Citric acid, which accumulated at constitutively higher levels in the roots of Yudal, was found to be under the control of four QTLs, three of which were involved in the control of clubroot resistance traits (Fig. 7A). For each of these three QTLs, the resistance allele was associated with a decrease of citric acid content, thus suggesting that constitutively high citric acid levels might contribute to susceptibility. Indeed, with levels of more than 20 µmol g DW−1 in roots, this compound might be a primary source of ready-to-use organic carbon for P. brassicae. By contrast, glucose content was under the control of seven QTLs (Fig. 7B), of which only one (R2=7.6%) aggregated with the metaQTL A10_68.42, which colocalized with a QTL involved in the control of DI. At this locus, the resistance allele was from Darmor-bzh and was associated with an increase of glucose content, which is inconsistent with the initial hypothesis that higher glucose contents in Yudal may favour pathogen development.
Fig. 7.
Sub-networks representing interconnected genetic architectures of clubroot resistance traits with (A) organic acids, (B) non-structural carbohydrates, and (C) glucosinolates. Nodes represent metabolites (circles), resistance traits (squares), or loci defined from the QTL meta-analysis (diamonds). The width of the edges indicates the quantitative effect of each QTL (R2) for a given QTL. When metaQTLs are associated with the control of Pb or DI, red and blue directed edges indicate positive or negative effects, respectively, on trait values (metabolite, Pb, or DI), exerted by Darmor or Yudal alleles at the QTL (as indicated in the diamonds).
The constitutively high level of gluconasturtiin in Yudal was mostly controlled by the Yudal allele at the metaQTL C03_1.01, which also contributed to the reduction of Pb at 21 dpi (Fig. 7C). To a minor extent, the Darmor resistance allele at the metaQTL C09_58.88 also contributed to increase the content of gluconasturtiin. This genetic architecture thus suggests that high root gluconasturtiin contents might contribute to partial resistance. By contrast, neoglucobrassicin (the most abundant indole glucosinolate in roots) was under the control of only two QTLs with low R2, which did not clearly support a role for this molecule in clubroot resistance or susceptibility. In addition, low contents of the indole glucosinolates glucobrassicin and 4-methoxyglucobrassicin were mostly controlled by the resistance alleles (Supplementary Fig. S6C) at two loci, C09_58.88 (resistance allele provided by Darmor-bzh) and, to a minor extent, C03_39.05 (resistance allele provided by Yudal). This genetic architecture suggests that low levels of these indole glucosinolates could be associated with the reduction of clubroot development. This conclusion would be consistent with the possible role of clubroot-induced nitrilase in converting indole-3-acetonitrile (produced by the myrosinase-mediated degradation of glucobrassicin) into indole-3-acetic acid (Ludwig-Müller et al., 1999; Ludwig-Müller, 2009). The importance of this biosynthetic pathway is, however, still a matter of debate (Malka and Chen, 2017).
From the comparative non-targeted metabolite analysis between parental lines (Fig. 3), a number of amino acids were identified (those in cluster 3) that were induced by clubroot infection only in the clubroot-resistant genotype Darmor-bzh. The QTL/metabolite sub-network represented in Fig. 8A indicates that the variations of these metabolites were mostly controlled by QTLs that are not associated with the control of clubroot resistance traits (mostly the bzh metaQTL A06_115.03). To a minor extent, the contents of phenylalanine, leucine, valine, tryptophan, and isoleucine were partly controlled by the QTL C07_79.74. However, at this locus, the resistance allele from Yudal was involved in repressing metabolite content. Thus, if the accumulation of these amino acids specifically in inoculated roots of Darmor-bzh (Fig. 3) had something to do with pathogen development, this would be linked to a susceptibility response. Few compounds that were constitutively accumulated in Darmor-bzh were also initially highlighted, most of them being unidentified (Fig. 3, cluster 1). The genetic control network (Fig. 8B) of M320T2161 and M368T3033, which were associated with remarkably high R2 values (36.5% and 23.7%, respectively), indicated that the constitutively high levels of those two compounds in Darmor-bzh was related to the presence of the Darmor-bzh resistance allele at the metaQTL C09_58.88. This ‘coincidence’ consistently suggested a possible role for these two unidentified molecules as candidate key players in the partial resistance driven by C09_58.88. Additional high-resolution MS/MS approaches allowed more precise m/z measurements of parental ions, and further investigation of their corresponding ion fragments (Supplementary Fig. S7). These fingerprints did not, however, match any referenced molecule available in public databases. Additional semiprep-scale purification of these compounds would be required for further investigations of their chemical structures by nuclear magnetic resonance spectroscopy.
Fig. 8.
Sub-networks representing interconnected genetic architectures of the pathogen–plant genomic DNA ratio (Pb) at 21 dpi with clusters of compounds highlighted in Fig. 3. (A) Metabolites accumulated during early clubroot infection only in Darmor-bzh and not in Yudal. (B) Metabolites accumulated at constitutive higher levels in the roots of Darmor-bzh (compared with Yudal). The width of the edges is indicative of the quantitative effect of each QTL (R2) for a given QTL. Red and blue edges indicate positive and negative effects, respectively, on trait values (metabolite or Pb), controlled by the Darmor allele at the metaQTL C09_58.88, or the Yudal allele at the metaQTLs C07_79.74 and C03_1.01.
Conclusions
Allelic variations affecting plant metabolism can potentially affect the plant/pathogen trophic relationship, regulate plant defence responses, or modulate the negative consequences of pathogen development. However, identifying connections between given metabolic features and disease resistance can be a difficult task when dealing with quantitative resistances controlled by series of resistance QTLs. In this work, metabolomic analysis of the B. napus genotypes Darmor-bzh and Yudal, which express contrasting degrees of clubroot resistance/susceptibility, highlighted differentiated constitutive metabolic features in non-inoculated plants, and dissimilar inducible responses to infection. A detailed analysis of metabolic and pathogen responses in the progeny of those two accessions allowed comparison of the QTL architectures governing clubroot resistance traits and metabolomic profiles.
Conclusions from the present work should be nuanced by possible inaccuracies in the estimation of QTL colocalizations. In addition, some QTLs may have been missed in the analysis, due to the limited number of recombinant lines in this study (130 DH lines). However, from a broad perspective, our approach allowed us to build an interesting contrasted map of metabolic features associated with different metaQTLs. To the best of our knowledge, this work is the first report of network (Cytoscape)-based representation of multi-trait QTL interactions for the investigation of plant responses to a pathogen. As shown here, this approach can help to test hypotheses, and to get a clear view of the complexity of genetic networks influencing metabolic responses and disease resistance. Our main conclusions are that (i) contrasting metabolic features are associated with metaQTLs controlling pathogen development, suggesting that those resistance loci may mobilize different physiological responses; (ii) a significant part of the genotype-specific metabolic responses to infection is not genetically connected to either resistance or susceptibility; and (iii) several constitutively accumulated metabolites may be linked to partial resistance.
From the seminal works of Keen and Williams (1969) to the recent study of Walerowski et al. (2018), it has been clearly shown that modulation of the carbohydrate economy plays a major role in the development of clubroot. It would thus have been conceivable that allelic variations affecting the amount of soluble carbohydrates could affect the course of infection. However, despite differences in the accumulation of glucose, fructose, and sucrose between the roots of Yudal and Darmor-bzh, these accumulations were found not to be related to the genetic factors controlling partial resistance. By contrast, our work underlined that partial resistance to clubroot is genetically related to the control of constitutive accumulation of citric acid (which may represent a consistent source of organic carbon for P. brassicae), gluconasturtiin, and the two unknown analytes M320T2161 and M368T3033.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Full dataset of metabolomic analyses of the parental lines and in the Darmor-bzh × Yudal progeny.
Table S2. Detailed results from the QTL analysis.
Table S3. Detailed results from the metaQTL analysis.
Table S4. Formatted network data for Cytoscape analysis.
Table S5. Node attributes for Cytoscape analysis.
Fig. S1. Overview of the metabolomic workflow.
Fig. S2. PCA representation of metabolomic profiles.
Fig. S3. Detailed graphic view of mQTL colocalizations on the genome A.
Fig. S4. Detailed graphic view of mQTL colocalizations on the genome C.
Fig. S5. Sub-network representing mQTLs connected to the four genomic ‘hot-spots’ (>20 colocalized mQTLs).
Fig. S6. Sub-network representing mQTLs connected to the five QTL involved in the control of disease index.
Fig. S7. MS/MS high-resolution spectra (negative mode) of compounds corresponding to the two mass tags M320T2161 and M368T3033.
Acknowledgements
The authors wish to acknowledge P2M2 and ThalassOMICS, two CORSAIRE metabolomics analytical facilities from the BIOGENOUEST core facilities network (http://www.biogenouest.org/). Thanks are also due to the CRB BrACySol for providing seeds, and colleagues of the IGEPP platform ‘Serres-Installations experimentales’.
Glossary
Abbreviations:
- DH
doubled haploid
- DI
disease index
- dpi
days post-inoculation
- Pb
pathogen–plant genomic DNA ratio
- GABA
gamma-aminobutyric acid
- PCA
principal component analysis
- QTL
quantitative trait locus
- SMCSO
S-methylcysteine sulfoxide
- SNP
single nucleotide polymorphism
References
- Aigu Y, Laperche A, Mendes J, Lariagon C, Guichard S, Gravot A, Manzanares‐Dauleux MJ. 2018. Nitrogen supply exerts a major/minor switch between two QTLs controlling Plasmodiophora brassicae spore content in rapeseed. Plant Pathology 67, 1574–1581. [Google Scholar]
- Arcade A, Labourdette A, Falque M, Mangin B, Chardon F, Charcosset A, Joets J. 2004. BioMercator: integrating genetic maps and QTL towards discovery of candidate genes. Bioinformatics 20, 2324–2326. [DOI] [PubMed] [Google Scholar]
- Arends D, Prins P, Jansen RC, Broman KW. 2010. R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26, 2990–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67, 1–48. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. [DOI] [PubMed] [Google Scholar]
- Brown D, Vision T. 2000. MapPop 1.0: software for selective mapping and bin mapping. http://www.bio.unc.edu/faculty/vision/lab/mappop/
- Buczacki ST, Toxopeus H, Mattusch P, Johnston TD, Dixon GR, Hobolth LA. 1975. Study of physiologic specialization in Plasmodiophora brassicae: proposals for attempted rationalization through an international approach. Transactions of the British Mycological Society 65, 295–303. [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, et al. 2014. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chen J, Jing J, Zhan Z, Zhang T, Zhang C, Piao Z. 2013. Identification of novel QTLs for isolate-specific partial resistance to Plasmodiophora brassicae in Brassica rapa. PLoS One 8, e85307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, et al. 2014. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nature Genetics 46, 714–721. [DOI] [PubMed] [Google Scholar]
- Chu M, Song T, Falk KC, et al. 2014. Fine mapping of Rcr1 and analyses of its effect on transcriptome patterns during infection by Plasmodiophora brassicae. BMC Genomics 15, 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. 2009. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JJ, Wasmann CC, Usami T, White GJ, Temporini ED, McCluskey K, VanEtten HD. 2011. Characterization of the gene encoding pisatin demethylase (FoPDA1) in Fusarium oxysporum. Molecular Plant-Microbe Interactions 24, 1482–1491. [DOI] [PubMed] [Google Scholar]
- Colinet H, Larvor V, Laparie M, Renault D. 2012. Exploring the plastic response to cold acclimation through metabolomics. Functional Ecology 26, 711–722. [Google Scholar]
- Corwin JA, Kliebenstein DJ. 2017. Quantitative resistance: more than just perception of a pathogen. The Plant Cell 29, 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delourme R, Falentin C, Fomeju BF, et al. 2013. High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics 14, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delourme R, Falentin C, Huteau V, et al. 2006. Genetic control of oil content in oilseed rape (Brassica napus L.). Theoretical and Applied Genetics 113, 1331–1345. [DOI] [PubMed] [Google Scholar]
- Denby KJ, Kumar P, Kliebenstein DJ. 2004. Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. The Plant Journal 38, 473–486. [DOI] [PubMed] [Google Scholar]
- Develey‐Rivière M, Galiana E. 2007. Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytologist 175, 405–416. [DOI] [PubMed] [Google Scholar]
- Dixon RA. 2001. Natural products and plant disease resistance. Nature 411, 843–847. [DOI] [PubMed] [Google Scholar]
- Elias AA, Busov VB, Kosola KR, et al. 2012. Green revolution trees: semidwarfism transgenes modify gibberellins, promote root growth, enhance morphological diversity, and reduce competitiveness in hybrid poplar. Plant Physiology 160, 1130–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkerli J, Bhatt G, Covert SF. 1998. Maackiain detoxification contributes to the virulence of Nectria haematococca MP VI on chickpea. Molecular Plant-Microbe Interactions 11, 317–326. [Google Scholar]
- Fähling M, Graf H, Siemens J. 2003. Pathotype separation of Plasmodiophora brassicae by the host plant. Journal of Phytopathology 151, 425–430. [Google Scholar]
- Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. 2000. Metabolite profiling for plant functional genomics. Nature Biotechnology 18, 1157–1161. [DOI] [PubMed] [Google Scholar]
- Figueiredo A, Fortes AM, Ferreira S, Sebastiana M, Choi YH, Sousa L, Acioli-Santos B, Pessoa F, Verpoorte R, Pais MS. 2008. Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. Journal of Experimental Botany 59, 3371–3381. [DOI] [PubMed] [Google Scholar]
- Foisset N, Delourme R, Barret P, Hubert N, Landry BS, Renard M. 1996. Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled-haploid progeny. Theoretical and Applied Genetics 93, 1017–1025. [DOI] [PubMed] [Google Scholar]
- Foisset N, Delourme R, Barret P, Renard M. 1995. Molecular tagging of the dwarf BREIZH (Bzh) gene in Brassica napus. Theoretical and Applied Genetics 91, 756–761. [DOI] [PubMed] [Google Scholar]
- Forsyth A, Mansfield JW, Grabov N, de Torres M, Sinapidou E, Grant MR. 2010. Genetic dissection of basal resistance to Pseudomonas syringae pv. phaseolicola in accessions of Arabidopsis. Molecular Plant-Microbe Interactions 23, 1545–1552. [DOI] [PubMed] [Google Scholar]
- Gilmour AR, Gogel BJ, Cullis BR, Thompson R. 2009. ASRemlUser guide release 3.0. Hemel Hempstead: VSN International Ltd; https://www.vsni.co.uk. [Google Scholar]
- Goffinet B, Gerber S. 2000. Quantitative trait loci: a meta-analysis. Genetics 155, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravot A, Dittami SM, Rousvoal S, Lugan R, Eggert A, Collén J, Boyen C, Bouchereau A, Tonon T. 2010. Diurnal oscillations of metabolite abundances and gene analysis provide new insights into central metabolic processes of the brown alga Ectocarpus siliculosus. New Phytologist 188, 98–110. [DOI] [PubMed] [Google Scholar]
- Gravot A, Grillet L, Wagner G, Jubault M, Lariagon C, Baron C, Deleu C, Delourme R, Bouchereau A, Manzanares-Dauleux MJ. 2011. Genetic and physiological analysis of the relationship between partial resistance to clubroot and tolerance to trehalose in Arabidopsis thaliana. New Phytologist 191, 1083–1094. [DOI] [PubMed] [Google Scholar]
- Gravot A, Richard G, Lime T, et al. 2016. Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot. BMC Plant Biology 16, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnaiah R, Kushalappa AC, Duggavathi R, Fox S, Somers DJ. 2012. Integrated metabolo-proteomic approach to decipher the mechanisms by which wheat QTL (Fhb1) contributes to resistance against Fusarium graminearum. PLoS One 7, e40695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K, Suwabe K, Tomita RN, Kato T, Nunome T, Fukuoka H, Matsumoto S. 2013. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One 8, e54745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuberger AL, Robison FM, Lyons SM, Broeckling CD, Prenni JE. 2014. Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Frontiers in Plant Science 5, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubault M, Hamon C, Gravot A, Lariagon C, Delourme R, Bouchereau A, Manzanares-Dauleux MJ. 2008a. Differential regulation of root arginine catabolism and polyamine metabolism in clubroot-susceptible and partially resistant Arabidopsis genotypes. Plant Physiology 146, 2008–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubault M, Lariagon C, Simon M, Delourme R, Manzanares-Dauleux MJ. 2008b. Identification of quantitative trait loci controlling partial clubroot resistance in new mapping populations of Arabidopsis thaliana. Theoretical and Applied Genetics 117, 191–202. [DOI] [PubMed] [Google Scholar]
- Kageyama K, Asano T. 2009. Life cycle of Plasmodiophora brassicae. Journal of Plant Growth Regulation 28, 203–211. [Google Scholar]
- Kato T, Hatakeyama K, Fukino N, Matsumoto S. 2013. Fine mapping of the clubroot resistance gene CRb and development of a useful selectable marker in Brassica rapa. Breeding Science 63, 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen NT, Williams PH. 1969. Translocation of sugars into infected cabbage tissues during clubroot development. Plant Physiology 44, 748–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettle AJ, Batley J, Benfield AH, Manners JM, Kazan K, Gardiner DM. 2015. Degradation of the benzoxazolinone class of phytoalexins is important for virulence of Fusarium pseudograminearum towards wheat. Molecular Plant Pathology 16, 946–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ. 2009. Genetical metabolomics: closing in on phenotypes. Current Opinion in Plant Biology 12, 223–230. [DOI] [PubMed] [Google Scholar]
- Klein AP, Sattely ES. 2017. Biosynthesis of cabbage phytoalexins from indole glucosinolate. Proceedings of the National Academy of Sciences, USA 114, 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T. 2002. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 161, 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutouan C, Clerc VL, Baltenweck R, et al. 2018. Link between carrot leaf secondary metabolites and resistance to Alternaria dauci. Scientific Reports 8, 13746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll TK, Lacy GH, Moore LD. 1983. A quantitative description of the colonization of susceptible and resistant radish plants by Plasmodiophora brassicae. Journal of Phytopathology 108, 97–105. [Google Scholar]
- Kuhl C, Tautenhahn R, Böttcher C, Larson TR, Neumann S. 2012. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Analytical Chemistry 84, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laperche A, Aigu Y, Jubault M, Ollier M, Guichard S, Glory P, Strelkov SE, Gravot A, Manzanares-Dauleux MJ. 2017. Clubroot resistance QTL are modulated by nitrogen input in Brassica napus. Theoretical and Applied Genetics 130, 669–684. [DOI] [PubMed] [Google Scholar]
- Lê S, Josse J, Husson F. 2008. FactoMineR: an R package for multivariate analysis. Journal of Statistical Software 25, 1–18. [Google Scholar]
- Lee J, Izzah NK, Choi BS, et al. 2016. Genotyping-by-sequencing map permits identification of clubroot resistance QTLs and revision of the reference genome assembly in cabbage (Brassica oleracea L.). DNA Research 23, 29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemarié S, Robert-Seilaniantz A, Lariagon C, et al. 2015. Camalexin contributes to the partial resistance of Arabidopsis thaliana to the biotrophic soilborne protist Plasmodiophora brassicae. Frontiers in Plant Science 6, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li X, Xuan Y, Jiang J, Wei Y, Piao Z. 2018. Genome wide identification and expression profiling of SWEET genes family reveals its role during Plasmodiophora brassicae-induced formation of clubroot in Brassica rapa. Frontiers in Plant Science 9, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente F, Alonso-Blanco C, Sánchez-Rodriguez C, Jorda L, Molina A. 2005. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. The Plant Journal 43, 165–180. [DOI] [PubMed] [Google Scholar]
- López-Gresa MP, Maltese F, Bellés JM, Conejero V, Kim HK, Choi YH, Verpoorte R. 2010. Metabolic response of tomato leaves upon different plant–pathogen interactions. Phytochemical Analysis 21, 89–94. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J. 2009. Glucosinolates and the clubroot disease: defense compounds or auxin precursors? Phytochemistry Reviews 8, 135–148. [Google Scholar]
- Ludwig-Müller J, Pieper K, Ruppel M, Cohen JD, Epstein E, Kiddle G, Bennett R. 1999. Indole glucosinolate and auxin biosynthesis in Arabidopsis thaliana (L.) Heynh. glucosinolate mutants and the development of clubroot disease. Planta 208, 409–419. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J, Schubert B, Pieper K, Ihmig S, Hilgenberg W. 1997. Glucosinolate content in susceptible and resistant Chinese cabbage varieties during development of clubroot disease. Phytochemistry 44, 407–414. [Google Scholar]
- Malka SK, Cheng Y. 2017. Possible interactions between the biosynthetic pathways of indole glucosinolate and auxin. Frontiers in Plant Science 8, 2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares-Dauleux M, Delourme R, Baron F, Thomas G. 2000a. Mapping of one major gene and of QTLs involved in resistance to clubroot in Brassica napus. Theoretical and Applied Genetics 101, 885–891. [Google Scholar]
- Manzanares-Dauleux MJ, Delourme R, Glory P, Giboulot A, Thomas G. 2003. Mapping QTLs and major resistance genes to clubroot (Plasmodiophora brassicae) in Brassica napus. In: 13th Crucifer Genetics Workshop UC Davis, California, 23–26. [Google Scholar]
- Manzanares-Dauleux MJ, Divaret I, Baron F, Thomas G. 2000b. Evaluation of French Brassica oleracea landraces for resistance to Plasmodiophora brassicae. Euphytica 113, 221. [Google Scholar]
- Meihls LN, Handrick V, Glauser G, et al. 2013. Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. The Plant Cell 25, 2341–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muangprom A, Thomas SG, Sun TP, Osborn TC. 2005. A novel dwarfing mutation in a green revolution gene from Brassica rapa. Plant Physiology 137, 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn A, Bowyer P, Lunness P, Clarke B, Daniels M. 1995. Fungal pathogens of oat roots and tomato leaves employ closely related enzymes to detoxify different host plant saponins. Molecular Plant-Microbe Interactions 8, 971–978. [DOI] [PubMed] [Google Scholar]
- Pedras MS, Abdoli A. 2017. Biotransformation of rutabaga phytoalexins by the fungus Alternaria brassicicola: unveiling the first hybrid metabolite derived from a phytoalexin and a fungal polyketide. Bioorganic & Medicinal Chemistry 25, 557–567. [DOI] [PubMed] [Google Scholar]
- Piao ZY, Ramchiary N, Lim YP. 2009. Genetics of clubroot resistance in Brassica species. Journal of Plant Growth Regulation 28, 252–264. [Google Scholar]
- Piasecka A, Jedrzejczak-Rey N, Bednarek P. 2015. Secondary metabolites in plant innate immunity: conserved function of divergent chemicals. New Phytologist 206, 948–964. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; https://www.R-project.org/. [Google Scholar]
- Rahman H, Peng G, Yu F, Falk KC, Kulkarni M, Selvaraj G. 2014. Genetics and breeding for clubroot resistance in Canadian spring canola (Brassica napus L.). Canadian Journal of Plant Pathology 36(Suppl. 1), 122–134. [Google Scholar]
- Rocherieux J, Glory P, Giboulot A, Boury S, Barbeyron G, Thomas G, Manzanares-Dauleux MJ. 2004. Isolate-specific and broad-spectrum QTLs are involved in the control of clubroot in Brassica oleracea. Theoretical and Applied Genetics 108, 1555–1563. [DOI] [PubMed] [Google Scholar]
- Rojas CM, Senthil-Kumar M, Tzin V, Mysore KS. 2014. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Frontiers in Plant Science 5, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe SA, Strelkov SE, Links MG, et al. 2016. The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host Brassica spp. BMC Genomics 17, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. 2008. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. The Plant Cell 20, 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade D, Shriki O, Cuadros-Inostroza A, Tohge T, Semel Y, Haviv Y, Willmitzer L, Fernie AR, Czosnek H, Brotman Y. 2015. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 11, 81–97. [Google Scholar]
- Schultz JC, Appel HM, Ferrieri AP, Arnold TM. 2013. Flexible resource allocation during plant defense responses. Frontiers in Plant Science 4, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical Chemistry 78, 779–787. [DOI] [PubMed] [Google Scholar]
- Somé A, Manzanares MJ, Laurens F, Baron F, Thomas G, Rouxel F. 1996. Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single‐spore isolates. Plant Pathology 45, 432–439. [Google Scholar]
- Sosnowski O, Charcosset A, Joets J. 2012. BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28, 2082–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Clair DA. 2010. Quantitative disease resistance and quantitative resistance loci in breeding. Annual Review of Phytopathology 48, 247–268. [DOI] [PubMed] [Google Scholar]
- Sundelin T, Christensen CB, Larsen J, Møller K, Lübeck M, Bødker L, Jensen B. 2010. In planta quantification of Plasmodiophora brassicae using signature fatty acids and real-time PCR. Plant Disease 94, 432–438. [DOI] [PubMed] [Google Scholar]
- Tautenhahn R, Böttcher C, Neumann S. 2008. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics 9, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenboim H, Brotman Y. 2016. Omic relief for the biotically stressed: metabolomics of plant biotic interactions. Trends in Plant Science 21, 781–791. [DOI] [PubMed] [Google Scholar]
- Tomita H, Shimizu M, Asad-ud Doullah M, Fujimoto R, Okazaki K. 2013. Accumulation of quantitative trait loci conferring broad-spectrum clubroot resistance in Brassica oleracea. Molecular Breeding 32, 889–900. [Google Scholar]
- Ueno H, Matsumoto E, Aruga D, Kitagawa S, Matsumura H, Hayashida N. 2012. Molecular characterization of the CRa gene conferring clubroot resistance in Brassica rapa. Plant Molecular Biology 80, 621–629. [DOI] [PubMed] [Google Scholar]
- Veyrieras JB, Goffinet B, Charcosset A. 2007. MetaQTL: a package of new computational methods for the meta-analysis of QTL mapping experiments. BMC Bioinformatics 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. 1992. Root hair infection by Plasmodiophora brassicae in clubroot resistant and susceptible genotypes of Brassica oleracea, B. rapa and Brassica napus. Netherlands Journal of Plant Pathology 98, 361–368. [Google Scholar]
- Wagner G, Charton S, Lariagon C, et al. 2012. Metabotyping: a new approach to investigate rapeseed (Brassica napus L.) genetic diversity in the metabolic response to clubroot infection. Molecular Plant-Microbe Interactions 25, 1478–1491. [DOI] [PubMed] [Google Scholar]
- Walerowski P, Gündel A, Yahaya N, Truman W, Sobczak M, Olszak M, Rolfe S, Borisjuk L, Malinowski R. 2018. Clubroot disease stimulates early steps of phloem differentiation and recruits SWEET sucrose transporters within developing galls. The Plant Cell 30, 3058–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Zhang L, Wu Y, Cao Y, Lu C. 2010. Comparison of five endogenous reference genes for specific PCR detection and quantification of Brassica napus. Journal of Agricultural and Food Chemistry 58, 2812–2817. [DOI] [PubMed] [Google Scholar]
- Xu L, Yang H, Ren L, Chen W, Liu L, Liu F, Zeng L, Yan R, Chen K, Fang X. 2018. Jasmonic acid-mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Frontiers in Plant Science 9, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogendra KN, Kushalappa AC, Sarmiento F, Rodriguez E, Mosquera T. 2014. Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Functional Plant Biology 42, 284–298. [DOI] [PubMed] [Google Scholar]
- Yu F, Zhang X, Huang Z, et al. 2016. Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot resistance gene Rcr1 through bulked segregant RNA sequencing. PLoS One 11, e0153218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Feng J, Hwang SF, Strelkov SE, Falak I, Huang X, Sun R. 2016. Mapping of clubroot (Plasmodiophora brassicae) resistance in canola (Brassica napus). Plant Pathology 65, 435–440. [Google Scholar]
- Zhang T, Zhao Z, Zhang CY, Pang WX, Choi SR, Lim YP, Piao ZY. 2014. Fine genetic and physical mapping of the CRb gene conferring resistance to clubroot disease in Brassica rapa. Molecular Breeding 34, 1173–1183. [Google Scholar]
- Zhao B, Haitao L, Juanjuan L, Wang B, Dai C, Wang J, Liu K. 2017. Brassica napus DS-3, encoding a DELLA protein, negatively regulates stem elongation through gibberellin signaling pathway. Theoretical and Applied Genetics 130, 727–741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.