ZmPTF1 regulates drought tolerance in maize by promoting root development and ABA synthesis, by binding to the G-box in the promoter and activating the expression of NCEDs, CBF4, NAC081, and NAC30.
Keywords: Abscisic acid synthesis, drought stress, maize, root development, transcriptional regulation, ZmPTF1
Abstract
Drought stress is the most important environmental stress limiting maize production. ZmPTF1, a phosphate starvation-induced basic helix-loop-helix (bHLH) transcription factor, contributes to root development and low-phosphate tolerance in maize. Here, ZmPTF1 expression, drought tolerance, and the underlying mechanisms were studied by using maize ZmPTF1 overexpression lines and mutants. ZmPTF1 was found to be a positive regulator of root development, ABA synthesis, signalling pathways, and drought tolerance. ZmPTF1 was also found to bind to the G-box element within the promoter of 9-cis-epoxycarotenoid dioxygenase (NCED), C-repeat-binding factor (CBF4), ATAF2/NAC081, NAC30, and other transcription factors, and to act as a positive regulator of the expression of those genes. The dramatically upregulated NCEDs led to increased abscisic acid (ABA) synthesis and activation of the ABA signalling pathway. The up-regulated transcription factors hierarchically regulate the expression of genes involved in root development, stress responses, and modifications of transcriptional regulation. The improved root system, increased ABA content, and activated ABA-, CBF4-, ATAF2-, and NAC30-mediated stress responses increased the drought tolerance of the ZmPTF1 overexpression lines, while the mutants showed opposite trends. This study describes a useful gene for transgenic breeding and helps us understand the role of a bHLH protein in plant root development and stress responses.
Introduction
Drought stress is one of the most important environmental stresses worldwide, and it impacts agricultural productivity (Bray, 1987; Dai, 2013). Substantial reductions in maize productivity are observed under drought stress. For example, the USA suffered an agricultural drought in 2012, which caused a 12% decrease in maize production compared with production in 2011 (USDA, 2014). In the developing world, maize is grown by small-scale farmers in areas with minimal water input and management; therefore, the maize varieties they grow must have a good level of tolerance to drought stress.
The overall goal of genetic research to improve drought tolerance in crops is to develop plants that are capable of producing sufficient yields under drought conditions. In general, plants use three strategies to help mitigate the effects of drought stress: drought escape, drought avoidance, and drought tolerance. Drought-response genes can be grouped into those involved in signal transduction and those that are functional components (Jewell et al., 2010). Plant tolerance to drought is triggered by complex multicomponent signalling pathways. In the signal transduction network that connects the perceived stress signals with the expression of stress-responsive genes, transcription factors (TFs) play an essential role (Nakashima et al., 2014; Joshi et al., 2016).
Basic helix-loop-helix (bHLH) proteins are found throughout the three eukaryotic kingdoms and are defined by the bHLH signature domain, which consists of a basic region for DNA binding and the HLH region for dimerization (Ferre-D’Amare et al., 1993). According to their structural and biochemical properties, the known bHLH proteins from animals have been categorized into groups A to F (Atchley and Fitch, 1997). There are 225 members of the bHLH family of proteins in Arabidopsis, 211 members in rice, and 308 members in maize (Jin et al., 2014). The bHLH proteins in Arabidopsis are divided into 21 subfamilies (Toledo-Ortiz et al., 2003) and 15 separate clades (Buck and Atchley, 2003). Most of the plant bHLH proteins belong to what is Group B in animals (Atchley and Fitch, 1997; Li et al., 2006); these bHLH proteins bind to the G-box sequence CACGTG and are involved in plant morphogenesis and various abiotic and biotic stimulus responses.
Until now, studies of the possible roles of the bHLH TFs in plant adaptation to drought have focused on stomatal development, trichome development, root hair development, abscisic acid (ABA) sensitivity, and phytochrome interactions in terms of high-temperature-mediated plant structural adaptations (Castilhos et al., 2014). Three bHLH TFs, SPCH (SPEECHLESS), MUTE, and FAMA, control the basal pathway of stomatal development, and are influenced by SCRM (SCREAM)1 and SCRM2 (Kanaoka et al., 2008). GL3/AtbHLH001, EGL3/AtbHLH002, RHD6/AtbHLH083, bHLH54, bHLH66, bHLH69, and bHLH82 are required for trichrome morphogenesis and root hair development (Zhao et al., 2008; Karas et al., 2009; Bruex et al., 2012; Lin et al., 2015; Hwang et al., 2017). RD29, RD22, and MYC2 (OsbHLH148 in rice) have been implicated in the ABA signal transduction pathway (Abe et al., 2003; Msanne et al., 2011; Seo et al., 2011). In addition, AtAIB/AtbHLH17, AtbHLH92, and AtbHLH122 have been reported to be involved in the drought stress response (Li et al., 2007; Jiang et al., 2009; Babitha et al., 2013; Liu et al., 2014). However, in crop species, few bHLH TFs involved in stress responses have been reported, except for OsPTF1 and ZmPTF1, which provide tolerance to low-phosphate conditions in rice and maize (Yi et al., 2005; Li et al., 2011); OsbHLH148, which is involved in drought tolerance in rice (Seo et al., 2011); and TabHLH1, which is involved in osmotic and nutrient stress in wheat (Yang et al., 2016a, b).
In our previous work, we found that ZmPTF1 improved root development and increased the low-phosphate stress tolerance of maize (Li et al., 2011). Phosphorus (P) fertilizer applications can mitigate the negative impacts of drought on plant growth and metabolism, while P deficiency can exacerbate drought stress (dos Santos et al., 2006; Sardans and Peñuelas, 2012; Jin et al., 2015; Liu et al., 2015). Although the mechanism of plant adaptations to drought/osmotic stress or P deficiency has attracted much attention in recent decades, much less attention has been paid to evaluating the relationships and interactions between them. Because of the increased P-deficiency tolerance caused by the overexpression of ZmPTF1 in maize, we were interested in whether low-phosphate stress and drought stress share a common regulatory system, and were specifically interested in the role of ZmPTF1 in the drought stress response. In this study, the morphology, growth, and yield of maize under drought stress conditions were evaluated using ZmPTF1 transgenic lines and mutants. Similar to the P-deficiency tolerance observed previously, ZmPTF1 was found to be a positive regulator of root system development, ABA synthesis and signalling pathways, and drought stress tolerance. A downstream gene expression analysis of ZmPTF1 showed that ABA synthesis and ABA signalling were positively regulated by ZmPTF1 binding to the promoters of several key genes involved in ABA synthesis, such as the NCED genes in maize. The hierarchical regulation of gene expression by ZmPTF1 impacted root development and stress responses via the up-regulation of other TFs. In the ZmPTF1 overexpression lines, a dramatic up-regulation of the expression of these genes increased the ABA content and activated the core ABA signalling pathway. The present study reveals the function of ZmPTF1 in maize drought tolerance and provides information on a useful gene for maize transgenic breeding.
Materials and methods
Plant materials
The maize inbred line DH4866 [wild type (WT)] and its independent transgenic homozygous lines reported by Li et al. (2011) were used in this study (see Supplementary Fig. S1 at JXB online). For the drought stress assay and yield analysis in the field, T3 generation seedlings were used, and for the comparison of germination, the seeds of T4 generation plants were used. Mu mutants (ptf1-1, mu1046031; ptf1-2, mu1030095; ptf1-3, mu1040158) were kindly provided by the Maize Stock Centre and were backcrossed and self-pollinated for two generations. Uniform Mu is a special maize population developed specifically for genetics research by the introgression of active Mu transposons in the W22 inbred line background (McCarty et al., 2005).
Comparison of seed germination under different polyethylene glycol treatments
The maize seeds were surface sterilized and placed on damp filter paper in sterile culture flasks maintained at 28 °C in darkness. The filter papers were soaked in polyethylene glycol 8000 (PEG8000) solutions of different concentrations (0, 12%, 15%, and 18%). After 8 days of culture, the seedlings were subjected to a morphological analysis.
Plant culture, drought treatment, and morphological analysis
Hydroponic cultures and a root morphology analysis were performed as described by Li et al. (2018). Briefly, seeds were germinated on damp filter paper at 28 °C in darkness for 4 days, after which the seedlings were transferred to a nutrient solution and grown for 12 days. The roots were scanned with a scanner (Powerlook 1000, China) and analysed by using LA-S-type plant image analysis software (developed at Zhejiang University, China). The number of roots were counted, and the root lengths were measured. The roots and shoots were subsequently dried in an oven at 80 °C to a constant weight and then weighed. The plants were grown under a 32 °C/25 °C (day/night) temperature regimen at a photon flux density of 700 μmol m−2 s−1 with a 14 h/10 h light/dark cycle in a greenhouse with approximately 65% relative humidity.
A drought stress treatment was administered during the vegetative growth stage (V4 stage) and the reproductive growth stage (V10 stage) as described by Li et al. (2008). Maize seeds from different lines were sown in pots (25 cm dimeter×35 cm height) that contained homogeneous loam in May. An overexpression line and the WT line were separately sown on the left and the right sides of each pot. After germination, the seedlings were watered normally. For the plants that were treated at the reproductive stage, the seedlings were thinned to one plant per pot. When the plants reached the correct stage, half of them were subjected to drought stress by withholding watering, and the others were well watered and served as controls. In the drought stress treatment, the plants were analysed on the second [soil water content (SWC) ~14.5%] and fifth days of treatment, and on the second day (SWC ~22.4%) of recovery after watering for the vegetative stage experiment; the plants were analysed on day 0, on the third (SWC ~14.2%) and seventh days of treatment, and on the second day (SWC ~22.2%) of recovery after watering for the flowering stage experiment.
The field experiment was carried out in accordance with the same protocol, and in the same field, as those of Li et al. (2008). The experiment was carried out in an experimental field under a rain shelter that was rolled up on sunny days in Jinan (117°29′ E, 36°54′ N). The trial plots were arranged in a randomized complete block design with four replications. Forty seeds of each homozygous transgenic or WT line were sown in a double row in each plot in May. Each plot was 2.5 m in length, with a width of 0.6 m between rows, and 25 cm between the plants in each row. The plants were thinned at the three-leaf stage to ensure the desired experimental density (66 700 plants/ha). At the V9 stage, the plants were subjected to drought stress for 6 weeks by maintaining the SWC at a depth of 40 cm at between 15% and 17% during the treatment. The plants grown under normal conditions were designated as the controls. After 6 weeks of drought stress treatment, the plants were well watered until harvest for yield calculations.
Real-time RT–PCR of the candidate genes, identification of differentially expressed genes, and data analysis
Real-time reverse transcription–PCR (RT–PCR) of the candidate genes, identification of differentially expressed genes (DEGs), and data analysis were performed as described by Li et al. (2018). Three biological replicates were used. The primer sequences used in this study are shown in Supplementary Table S1. For the DEGs, the roots from the ZmPTF1 transgenic (L+4) and WT lines cultured in nutrient solutions for 12 days were used for transcriptome analysis. A total of 0.5 g of roots (collected from 25 plants) was used for one RNA library. The total RNA was extracted as described for molecular cloning (Chomczynski and Sacchi, 1987). Tag preparation, DNA purification, and Illumina sequencing were performed by BGI Tech Solutions Co., Ltd (Shenzhen, China), according to standard procedures. The bioinformatics analysis for digital gene expression profiling was performed according to the bioinformatics analysis procedure of BGI Tech. The criteria of false discovery rate ≤0.001 and the absolute value of log2 ratio ≥1 were used as thresholds to judge the significance of differences in gene expression (data deposited in Dryad Digital Repository: https://doi.org/10.5061/dryad.7nr377v).
Sequence analysis
Clustal W2 (https://www.ebi.ac.uk/Tools/msa/clustalw2/) and MEGA 5 (Tamura et al., 2011) were used for sequence analysis of the bHLH TFs. PlantCARE (Lescot et al., 2002) was used for cis-element identification in the promoter.
ABA determination by using UPLC-MS/MS
Plants cultured in a normal nutrient solution for 11 days and then transplanted into nutrient solutions with or without 15% PEG8000 for another 24 h were used for ABA quantification. Roots and leaves were collected and flash frozen in liquid nitrogen. The extraction and determination of ABA were performed as described by Fu et al. (2012). Five independent biological replicates were used, and each biological replicate was collected from five plants.
Yeast-one-hybrid analysis and electrophoretic mobility shift assays
Promoter fragments were amplified and inserted into a pLacZi vector, and the coding region of ZmPTF1 was inserted into pGADT7 (Supplementary Table S1). These vectors were used as the bait and prey in a yeast one-hybrid analysis. The yeast strain AH109 was used as the host strain, and the experiment was performed according to the Yeast Protocols Handbook (Clontech). An ONPG β-galactosidase assay and X-gal staining were carried out as described in the Yeast Protocols Handbook (Clontech). At least three biological replicates were used for the assay. For the electrophoretic mobility shift assays (EMSAs), ZmPTF1 was inserted into pET30a for expression. Probes were generated using a DIG Gel Shift Kit (Roche, China). The sequences of the probes and the mutated probes used in this assay are shown in Supplementary Table S2. The mutated probes were artificially synthesized. All experiments were repeated three times.
Results
Expression pattern and sequence analysis of ZmPTF1
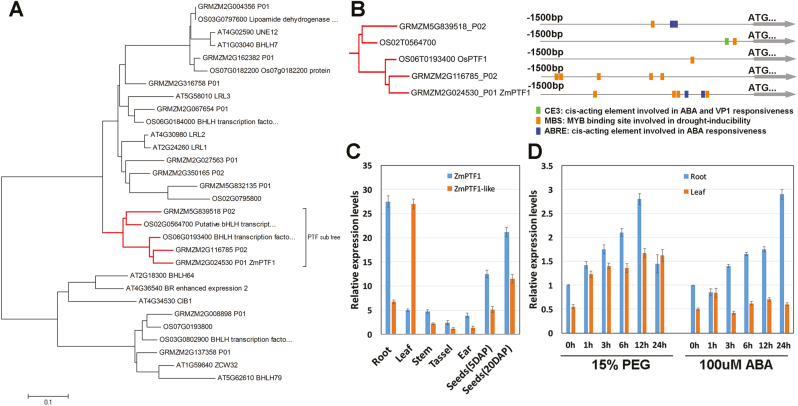
The sequence alignment showed that ZmPTF1 (GRMZM2G024530) and ZmPTF1-like (GRMZM2G116785) were the corresponding genes of OsPTF1, and the members of this PTF subgroup were highly similar to LRL (LJRHL1-LIKE)1, LRL2, and LRL3 in Arabidopsis (Fig. 1A) (Karas et al., 2009). As shown in Fig. 1B, the ZmPTF1 gene has four MBSs (MYB binding sites involved in drought inducibility) and two ABREs (cis-acting elements involved in ABA responsiveness), whereas OsPTF1 has only one MBS in its promoter region. The expression level of ZmPTF1 was relatively high in the roots and seeds, and that of ZmPTF1-like was higher in the leaves than in the other plant organs (Fig. 1C). The expression of ZmPTF1 was induced not only by low-phosphate stress (Li et al., 2011) but also by the PEG and ABA treatments, especially in the roots (Fig. 1D). In response to treatment with 15% PEG or 100 µM ABA, ZmPTF1 expression increased 2- to 3-fold at 12 h and 24 h in the roots, while in the leaves it was induced by PEG treatment but not ABA treatment. The expression pattern and promoter element analysis indicated that ZmPTF1 may participate in the drought stress response.
Fig. 1.
Sequence analysis and expression analysis of ZmPTF1. (A) Phylogenetic tree of the deduced amino acid sequences of the bHLH transcription factors in Arabidopsis, rice, and maize. (B) Promoter analysis of ZmPTF1 and its homologues showed a distribution of drought-related cis-elements in the promoters. (C) Expression analysis of ZmPTF1 and ZmPTF1-like in different organs and stages of the maize inbred line DH4866. The roots, leaves, and stems were collected from three-leaf-stage maize plants, the tassels and ears were collected from plants at the V9 stage, and the seeds were collected from plants after pollination. All of the DH4866 plants used were grown under normal conditions. (D) Expression analysis of ZmPTF1 in plants at the three-leaf stage subjected to 15% PEG8000 or 100 µM ABA treatments via a hydroponic culture system. The transcript levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001), with maize Actin1 (NM_001155179.1) as an internal control. Three biological replicates were used for the experiment. (This figure is available in colour at JXB online.)
Overexpression of ZmPTF1 improved maize root growth by promoting lateral root development
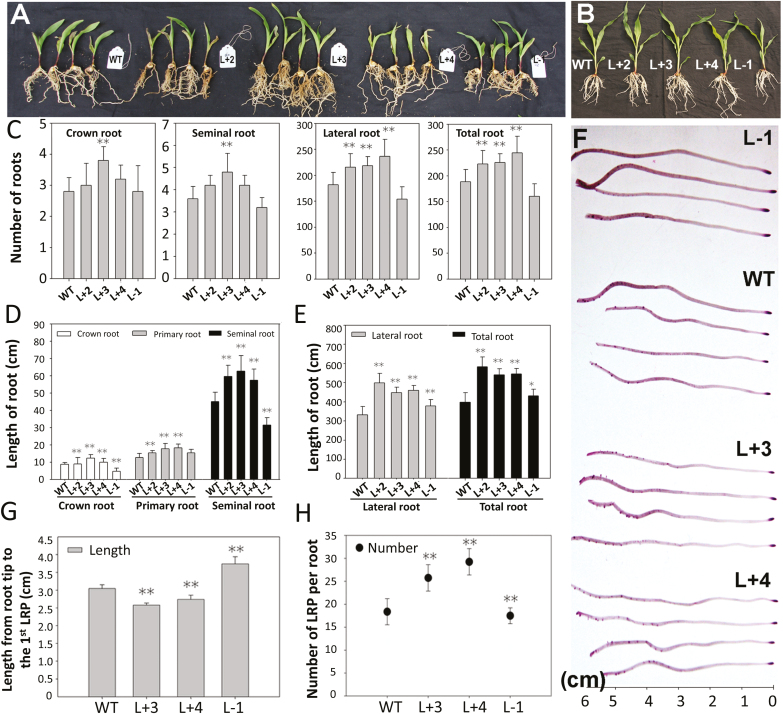
The growth and development of maize plants were observed throughout their entire life cycle. As shown in Fig. 2 and Supplementary Fig. S2A, B, the overexpression lines had a more developed root system than the WT and antisense lines at the V4 stage when cultured in either nutrient solution, sand, or soil. At the reproductive stage, the brace root volume and total root volume of the overexpression lines were greater than those of the WT line (Supplementary Fig. S2C–H).
Fig. 2.
Overexpression of ZmPTF1 promotes root system development. (A, B) Morphology of the seedlings of the ZmPTF1 overexpression and antisense lines and the wild-type (WT) line grown in sand (A) or nutrient solutions (B). (C–E) Root number and root length of the different lines of the plants grown in nutrient solutions. (F) Distribution of the lateral root primordia in seedlings at the two-leaf stage from the ZmPTF1 overexpression and antisense lines and the WT. (G) Distance from the first lateral primordium to the root tip. (H) Number of lateral root primordia per seminal root. L+2, L+3, and L+4 are the ZmPTF1 overexpression lines, L-1 is the antisense line, and WT is the control, DH4866. Values are means ±SD; six biological replicates were used for the experiment. Asterisks indicate significant differences between the transgenic and WT lines according to t-tests: *P<0.05, **P<0.01. (This figure is available in colour at JXB online.)
The number of roots, root length, and number of lateral root primordia of the maize plants cultured in nutrient solutions were evaluated. In the overexpression lines, a significant increase in both the number of lateral roots and the length of the seminal and lateral roots was observed (Fig. 2C–E). The data showed that the improved root system of the ZmPTF1 overexpression lines was mainly caused by the number and length of the roots, especially the lateral roots. The analysis of the lateral root primordia, determined by Feulgen staining, showed that the emergence of lateral root primordia occurred earlier in the overexpression lines than in the WT line, meaning that the distance from the first lateral root primordium to the root tip was shorter in the overexpression lines than in the WT (Fig. 2F–H). The number of lateral root primordia in the overexpression lines was 141–161% greater than that in the WT, and the number of lateral roots was 122–138% greater than that in the WT. In addition, more than one internode from which adventitious roots (brace roots in maize) emerged was observed in the mature plants in the field (Supplementary Fig. S2C–H). With the overexpression of ZmPTF1, the numbers of lateral roots, brace roots, and seminal roots increased, which promoted plant growth.
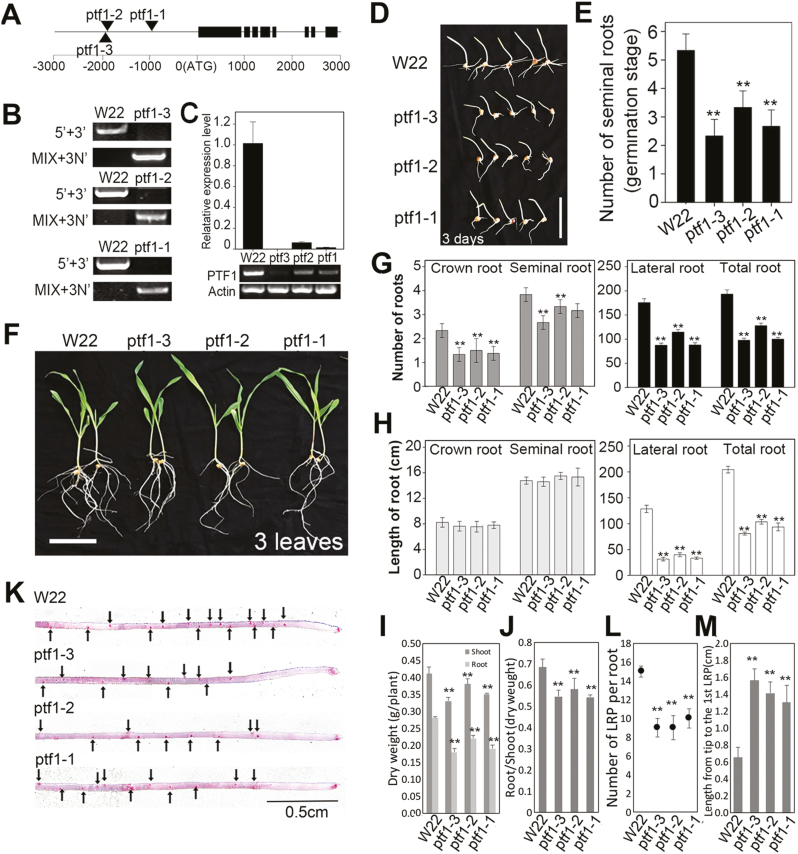
Knockdown of ZmPTF1 slowed root system development by reducing the number and growth of lateral roots
Since overexpression of ZmPTF1 promoted root development and increased the number and growth of lateral roots, we wondered whether the mutant of ZmPTF1 would be affected in root development. The root morphology of three ZmPTF1 Mu mutants was used to test the correlation between ZmPTF1 expression levels and root development. As shown in Fig. 3, all three mutations occurred upstream of ZmPTF1 (Fig. 3A, B) and significantly reduced the expression of ZmPTF1, especially ptf1-3 (Fig. 3C). When seeds were germinated on filter paper for 8 days (Fig. 3D, E), the root growth and development of the mutants were significantly reduced. When the seeds were grown in a nutrient solution (Fig. 3F–J), a dramatically reduced number and length of lateral roots were observed in the mutants, and a slight reduction in both seminal root and crown root development was observed, in contrast to that of the WT line W22. A 50.3–76.7% reduction in lateral root number and a 36.6–51.2% reduction in lateral root length compared with the WT were observed; the marked reduction in lateral root development led to an undeveloped root system with reduced biomass, number of total roots, and root surface area and volume. As shown in Fig. 3K–M, the numbers of lateral root primordia of the mutants were significantly reduced, and the length from the first lateral root to the root tip was longer in the mutants than in W22.
Fig. 3.
ZmPTF1 Mu mutants showed poor root system development. (A) Genetic map of the ZmPTF1 Mu mutants from the maize stock centre. The triangles represent the insertions. (B) Molecular identification of the ZmPTF1 Mu mutants (ptf1-1, mu1046031; ptf1-2, mu1030095; ptf1-3, mu1040158) with specific combinations of primers. (C) Relative ZmPTF1 transcript levels in the roots of Mu mutants ptf1-1, ptf1-2, and ptf1-3 and the wild-type (WT) control W22 determined by real-time RT–PCR (upper panel) and RT–PCR (lower panel). (D, E) Seedlings of the ZmPTF1 Mu mutants and WT during the germination period. (F) Seedlings of the ZmPTF1 Mu mutants and WT cultured in nutrient solutions. (G–J) Root number (G), root length (H), biomass (I) and the root/shoot ratio (J) of the ZmPTF1 Mu mutants and WT cultured in nutrient solutions. (K) Distribution of the lateral root primordia and distance from the root tip to the first lateral root primordium of seedlings of the ZmPTF1 Mu mutants and WT. (L, M) Number of lateral root primordia per seminal root (L) and distance from the first lateral primordium to the root tip (M). Values are means ±SD; three biological replicates were used for the gene expression analysis and six biological replicates were used for the morphological analysis. Asterisks indicate significant differences between the mutant and WT lines according to t-tests: **P<0.01. (This figure is available in colour at JXB online.)
Overexpression of ZmPTF1 enhanced osmotic/drought stress tolerance and yields under drought stress
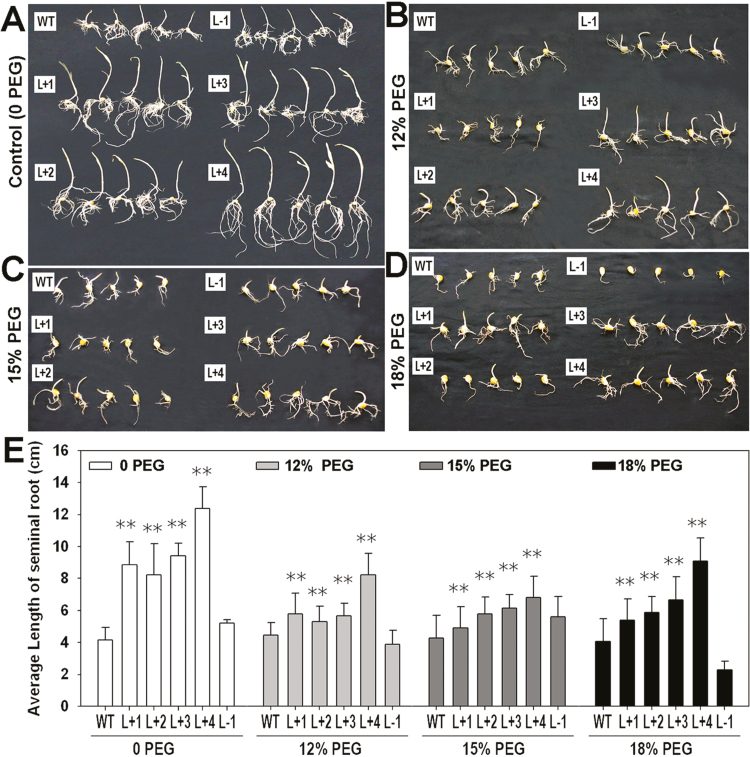
The osmotic/drought stress tolerance of the ZmPTF1 transgenic plants was examined at the seed germination stage, at the V4 stage, and at the V10 stage. When seeds were germinated on filter paper, compared with the WT and L-1 lines, the overexpression lines had longer shoots and a more robust root system. The root length of the overexpression lines was 190–293% greater than that of the WT line (Fig. 4A, E). When the seeds were germinated in PEG solutions, the germination rates of all lines decreased with increasing PEG concentrations. However, the overexpression lines maintained a relatively higher germination rate and level of growth on filter paper soaked in the PEG solutions than the other lines (Fig. 4B–E), and the WT and antisense lines showed limited germination in the 18% PEG solution. The seeds from the overexpression lines were able to germinate in the 18% PEG solution, which was shown when the coleoptiles and radicles penetrated the seed coat and continued growing.
Fig. 4.
Overexpression of ZmPTF1 enhances tolerance to osmotic stress during the germination stage. (A–D) Seeds of the ZmPTF1 overexpression and antisense lines and the wild type (WT) germinated on filter paper soaked in different solutions of PEG8000: (A) water without PEG8000; (B) 12% PEG8000; (C) 15% PEG8000; (D) 18% PEG8000. (E) Average length of the seminal roots of the seedlings in A–D. The lines used are those described in Fig. 2. Values are the means ±SD; six biological replicates were used for the experiment. Asterisks indicate significant differences between the transgenic and WT lines according to t-tests : **P<0.01. (This figure is available in colour at JXB online.)
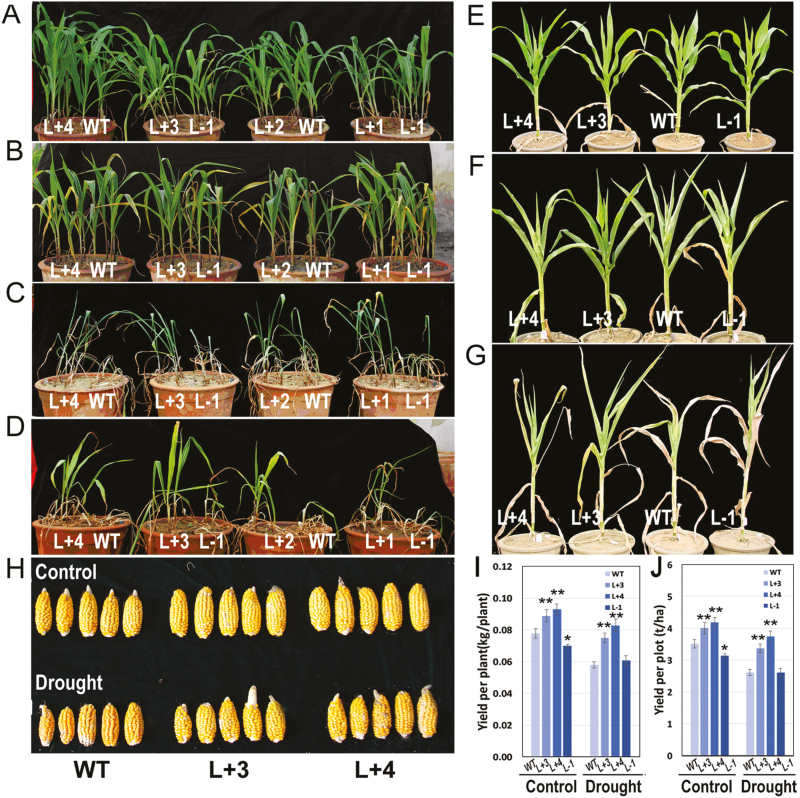
At the seedling stage (Fig. 5A), all plants showed leaf wilting caused by a 2-day water shortage (Fig. 5B) and exhibited severe wilting after another 3 days (Fig. 5C) of withholding watering. The WT and L-1 lines showed severe dehydration and died, whereas the overexpression lines maintained relatively better growth. After they were rewatered (Fig. 5D), the overexpression lines recovered to normal conditions, whereas few WT and antisense plants survived. At the flowering stage (Fig. 5E–G, Supplementary Fig. S3), the field-grown plants overexpressing ZmPTF1 demonstrated improved drought tolerance, producing a greater yield per plant and yield per plot than the WT line; this was especially the case for the L+4 line, which produced a kernel yield 147% greater than the WT (Fig. 5H–I).
Fig. 5.
Overexpression of ZmPTF1 increases tolerance to drought stress during the seedling and flowering stages. (A–D) Seedlings of the ZmPTF1 overexpression and antisense lines and the wild type (WT) without watering for (A) 0, (B) 2, and (D) 5 days, and (D) after rewatering for 2 days. (E–G) Plants of the ZmPTF1 overexpression and antisense lines and the WT without watering for (E) 0, (F) 3, and (G) 7 days during the flowering phase. (H–J) Ears (H) and yields (I, J) of the ZmPTF1 overexpression and antisense lines and the WT under normal (control) and drought stress conditions in the field. The lines used are those described in Fig. 2. Values are means ±SD; six biological replicates were used for the experiment. Asterisks indicate significant differences between the transgenic and WT lines according to t-tests: *P<0.05, **P<0.01. (This figure is available in colour at JXB online.0029
Overexpression of ZmPTF1 enhanced root development and stress responses, and modified transcriptional regulation
To elucidate the regulatory network of ZmPTF1, the DEGs in the roots of the ZmPTF1 overexpression line L+4 and the WT cultured in nutrient solutions for 12 days were analysed (data deposited in Dryad Digital Repository: https://doi.org/10.5061/dryad.7nr377v). The data showed that 761 genes were differentially expressed between L+4 and the WT, with 532 DEGs up-regulated and 229 down-regulated in the roots of L+4 compared with the WT (Supplementary Table S3–S8); 35 of them were validated via real-time RT–PCR. The DEGs were enriched in the following three Gene Ontology (GO) categories: response to stimuli, growth and development, and transcriptional regulation (Fig. S4, Supplementary Table S3). Interestingly, 88 up-regulated genes functioned in at least two biological processes and were significantly different from the down-regulated genes. For example, 15 genes were involved in all three of the aforementioned GO categories, 59 genes were involved in the response to stimuli and transcriptional regulation, 26 genes were involved in growth and development and transcriptional regulation, and 33 genes were involved in the response to stimuli and growth and development. GO analysis indicated that ZmPTF1 works as a positive regulator of stress responses as well as growth and development. ZmPTF1 might play an important role in the activation of certain key regulatory networks in these biological processes (Supplementary Table S3).
As shown in Supplementary Table S4, the phytohormone-mediated morphology, especially in relation to auxin, was significantly affected by ZmPTF1 overexpression. The expression of orthologues of IAA3 and ARF6 and four SAUR genes was significantly altered, with SHY2/IAA3 (GRMZM2G115357, 5.84-fold, L+4/WT), SAUR32 (GRMZM2G414727, 3.91-fold), SAUR55 (GRMZM2G430052, 2.26-fold), and SAUR71 (GRMZM2G146108, 364-fold) significantly up-regulated, and ARF6 (GRMZM2G081158, 0.44-fold) and SAUR53 (GRMZM2G442000, 0.37-fold) down-regulated. In addition to the auxin signalling genes, three BRH1 genes (GRMZM2G044537, GRMZM2G071277, and GRMZM2G318408) and two GID-like genes (GRMZM2G173630 and GRMZM2G440543) were up-regulated by ZmPTF1. The NAC TFs, which were named after the abnormal morphology of the mutants NAM, ATAF, and CUC, were significantly induced by ZmPTF1. As summarized in Supplementary Table S4, seven NAC genes were induced by ZmPTF1, including the orthologues of NAC1 (GRMZM2G063522, 2.40-fold), NAC30 (AC212859.3_FG008, 257-fold), NAC047 (GRMZM2G011598, 2.52-fold), and ATAF2/NAC081 (GRMZM2G127379, 3.97-fold; GRMZM2G068973, 4.85-fold; GRMZM2G162739, 3.89-fold; and GRMZM2G3470434, 20-fold).
Genes involved in the response to stimuli were active in the ZmPTF1 overexpression line, including ABA synthesis and signalling genes as well as AP2/DREBP, WRKY, NAC, and bHLH TFs (Supplementary Table S5 and 6). For ABA synthesis, two NCED9 homologues were dramatically induced, and the orthologues of PYL/RCAR, ABO3, ABFs, CBF4, and ERF1 involved in the ABA core signalling pathway and ABA response were also active. In addition to the ABA-dependent pathway, TFs involved in the ABA-independent stress response pathways were also active. As shown in Supplementary Table S6, 18 AP2/DREBP, 12 WRKY, 11 NAC, 6 MYB, and 6 bHLH TFs, as well as some other key genes involved in drought and phosphate responses, such as those that encode phosphate and potassium transporters, the oxidative stress 3 protein, peroxidase, and molecular chaperones, were induced in the ZmPTF1 overexpression lines. Thus, a hierarchical regulation of TFs by ZmPTF1 was found in response to environmental stress.
ZmPTF1 binds to the G-box element of maize NCED, CBF4, ATAF2, and NAC30, and acts as a positive regulator of the expression of these genes
In rice, Yi et al. (2005) showed that OsPTF1 is able to bind to the G-box. ZmPTF1 has conserved residues in its bHLH domain for the recognition of the G-box, similar to OsPTF1. The DEGs were analysed for the presence of a G-box element in their promoter regions. As described above, the genes involved in the three GO categories had a high G-box frequency in their promoter region (Supplementary Fig. S4). Among the 175 up-regulated genes that respond to stimuli, 115 (65.71%) had a G-box element, and 61 (34.86%) had more than two G-box elements. Among the 70 up-regulated genes involved in growth and development, 53 (75.71%) had a G-box, and 28 (40%) had more than two G-boxes. Regarding transcriptional regulation, among the 96 up-regulated genes, 68 (70.83%) had a G-box and 30 (31.25%) had more than two G-boxes. These values are significantly higher than the average level and the levels in the down-regulated genes in these three GO categories. Interestingly, all 15 up-regulated genes involved in the response to stimuli, growth and development, and transcriptional regulation harboured a G-box element in their promoter, and eight of them (53.33%) had more than two G-box elements.
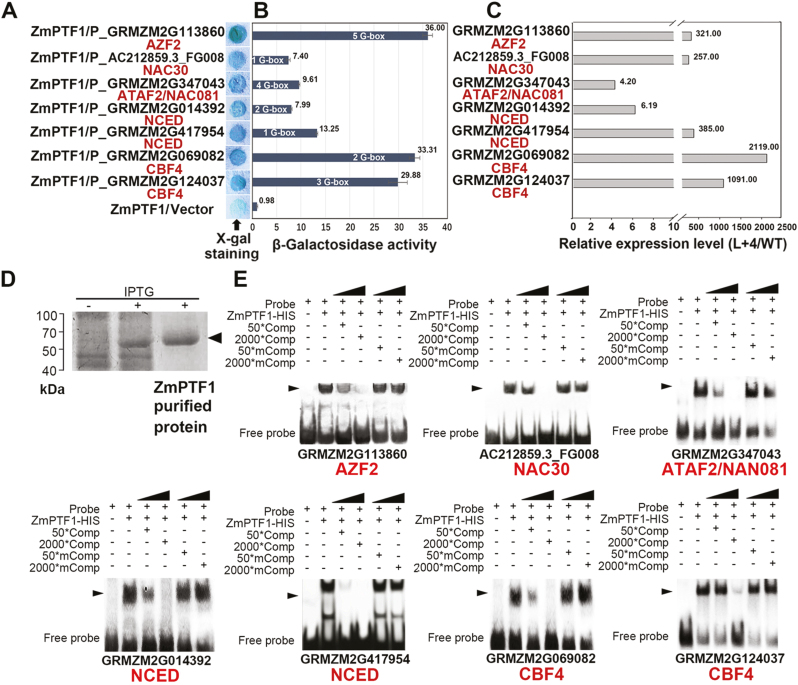
To investigate whether ZmPTF1 directly regulates the candidate genes, the binding affinity between ZmPTF1 and the promoters of seven candidate genes was assessed by employing a yeast one-hybrid assay, and was confirmed by EMSA (Fig. 6). These seven genes are named according to their orthologues in Arabidopsis: AZF2 (GRMZM2G113860), NAC30 (AC212859.3_FG008), ATAF2/NAC081 (GRMZM2G347043), NCED9 (GRMZM2G014392 and GRMZM2G417954), and CBF4 (GRMZM2G069082 and GRMZM2G124037). As shown in Fig. 6A, ZmPTF1 bound to the promoter and drove the expression of the reporter genes, albeit to different extents, when the G-box was in the promoter region. The colour intensity that developed in an ONPG β-galactosidase transactivation assay showed that the ability of ZmPTF1 to bind to these promoters ranged from 7.40 (NAC30) to 36.00 (AZF2). The binding affinity between ZmPTF1 and the promoters of the two CBF4 genes and the two NCED9 genes was high, with values of 33.31 and 29.88, and 7.99 and 13.25, respectively (Fig. 6B). These expression levels corresponded to the expression levels of the candidate genes in the overexpression line L+4, in which they were dramatically up-regulated by 4.20- to 2119-fold relative to the WT (Fig. 6C). The results showed that ZmPTF1 bound to the promoter of the maize genes NCED, CBF4, ATAF2, and NAC30 and acted as a positive regulator of the expression of these genes. The EMSA assay confirmed that the binding of ZmPTF1 to these promoters was specific and stable (Fig. 6E).
Fig. 6.
ZmPTF1 binds to the G-box in the promoter region of the candidate target genes and activates their expression. (A, B) Yeast one-hybrid analysis of ZmPTF1 and the seven candidate target genes using X-gal staining (A) and β-galactosidase activity and G-box number (B). (C) Overexpression of ZmPTF1 activated the expression of seven candidate genes from the DEG analysis. (D) ZmPTF1 protein expressed in E. coli. (E) Electrophoretic mobility shift assay for the specific binding of ZmPTF1 and the promoter motifs of the candidate target genes. The probes and mutant probes were generated using a DIG Gel Shift Kit (Roche). Values are means ±SD; at least three replicates were performed for the experiment. (This figure is available in colour at JXB online.)
ZmPTF1 affects ABA biosynthesis by regulating NCED family genes
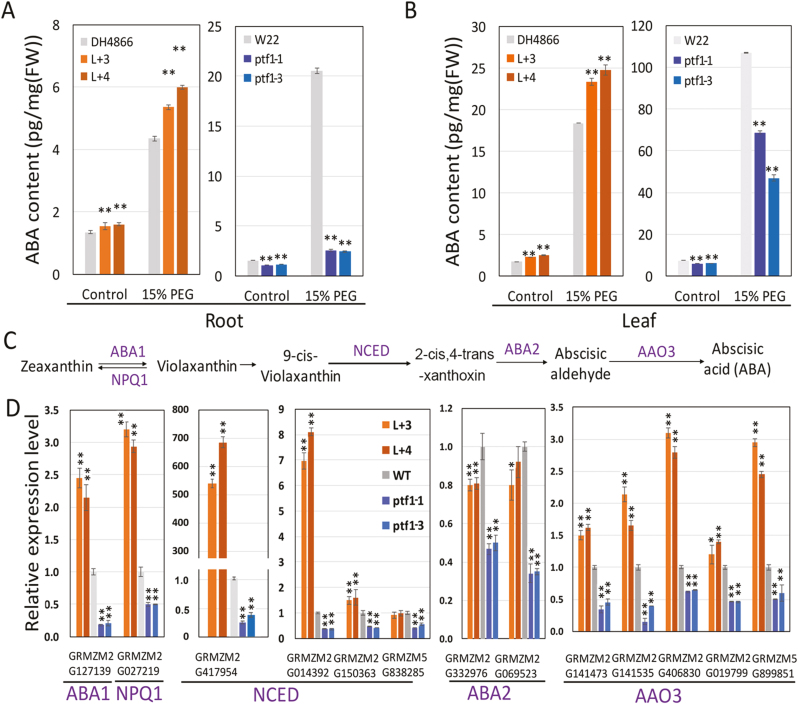
The ABA contents in the roots and leaves of the ZmPTF1 overexpression and WT (DH4866) lines, and in the Mu insertion mutant and WT (W22) lines, were determined. As shown in Fig. 7A and B, the ABA contents of the overexpression line were 20% and 25% higher in the roots and leaves, respectively, than those of the WT, while the ABA contents were lower in the mutants than in the WT under normal culture conditions. When the plants were subjected to treatment with 15% PEG, the ABA content in the plants increased dramatically, by 11-fold in the leaves and 3-fold in the roots compared with the control, and the ABA contents in the overexpression lines were higher than those in the WT. When ZmPTF1 was knocked down, the drought-induced increase in ABA was dramatically weakened, especially in the roots. This finding indicated that increasing the expression level of ZmPTF1 enhanced ABA accumulation in both the roots and leaves, while the knockdown of ZmPTF1 reduced ABA accumulation, especially when the plants were subjected to drought stress treatment.
Fig. 7.
ABA content and synthesis were greatly affected by ZmPTF1. (A, B) ABA contents in the roots (A) and leaves (B) of the ZmPTF1 overexpression lines, wild-type (WT) DH4866 plants, ZmPTF1 Mu insertion lines, and WT W22 plants under normal conditions and under treatment with 15% PEG8000. Plants were cultured in a normal nutrient solution for 11 days and then transplanted into nutrient solutions with and without 15% PEG8000 for another 24 h. The leaves and roots were used for ABA content analysis. (C) The ABA biosynthesis pathway and the enzymes that catalyse each step. (D) Expression levels of the key genes involved in ABA biosynthesis in the roots of the ZmPTF1 overexpression lines, WT DH4866 plants, ZmPTF1 Mu insertion lines, and WT W22 plants under normal conditions, determined using real-time RT–PCR. The roots from plants cultured in a normal nutrient solution were used for gene expression analysis. The levels of the gene transcripts were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001), with maize Actin1 (NM_001155179.1) as an internal control, and the levels of WT (DH4866 and W22) expression were set as 1-fold. L+3 and L+4 are ZmPTF1 overexpression lines in the DH4866 background, and ptf1-1 and ptf1-3 are the ZmPTF1 Mu insertion lines in the W22 background. Values are means ±SD; five independent biological replicates were used for ABA determination, and three biological replicates were used for real-time RT–PCR. Asterisks indicate significant differences between the transgenic and WT lines according to t-tests: **P<0.01. (This figure is available in colour at JXB online.)
In plants, ABA can be synthesized directly through the mevalonic acid pathway. The expression levels of the genes coding for the enzymes that catalyse the conversion of zeaxanthin to ABA in the ZmPTF1 overexpression lines, mutants, and WT lines were compared. These genes were homologues of ABA1, NPQ1, NCED, ABA2, and AAO3 (Fig. 7C). The results showed that the main step regulated by ZmPTF1 was the step controlled by the expression of members of the NCED gene family. In this subgroup, maize has four members that are similar to Arabidopsis NCED2, 3, 5, and 9. Among these four genes, two (GRMZM2G150363 and GRMZM2G014392) were dramatically up-regulated in the ZmPTF1 overexpression lines. The cleavage of 9-cis-epoxycarotenoids, which is catalysed by NCED, has been reported to be the rate-limiting step of ABA synthesis (Schwartz et al., 1997; Burbidge et al., 1999). The promoters of the four NCED genes in maize were analysed, and two G-box elements were found in the promoter region of GRMZM2G014392 and GRMZM5G150363. As shown in Figs 6, 7D, and Supplementary Fig. S5, ZmPTF1 can bind to the promoter of NCED9 genes. The expression of the two NCEDs with G-box elements was highly regulated by ZmPTF1, while that of the other two NCEDs was not. This finding indicated that ZmPTF1 acted as a positive regulator of ABA synthesis by directly binding to the promoter region of NCEDs, and the increased expression of NCEDs enhanced the ABA content by increasing ABA synthesis, which contributes to the drought stress tolerance of maize. Beside the NCEDs, most of the genes involved in ABA synthesis were expressed at a higher level in the ZmPTF1 overexpression lines except for ABA2, which had an expression level approximately 0.8–0.91-fold that in the WT. In Arabidopsis, aba2 mutants are insensitive to sucrose and glucose (Lin et al., 2007). ZmPTF1 overexpression led to lower levels of glucose and sucrose in the leaves, and higher levels in the roots, compared with WT (Li et al., 2011). Plants might have a fine-tuning regulation in maintaining ABA levels in response to stresses through regulation of primary metabolic changes, maybe through ABA2.
The ABA-dependent stress response is active in ZmPTF1 overexpression lines
With respect to plant responses to abiotic stress, the ABA-dependent stress response has been well elucidated. Based on the GO analysis, the response to the ABA stimulus was largely enhanced in the ZmPTF1 overexpression lines (Supplementary Tables S3 and S5). With respect to the previously described 15 genes involved in the response to stimuli, growth and development, and transcriptional regulation, four were involved in the ABA signalling pathway, and ABF homologues were identified (Supplementary Table S5). Three PYR/PYL ABA receptor genes were differentially expressed (GRMZM2G377904, 4.67-fold; GRMZM2G050512, 3.58-fold; and GRMZM2G446858, 0.32-fold). In addition, an ABF gene (GRMZM2G033413) was up-regulated 2.19-fold, and the ABO3/WRKY63 gene (GRMZM2G158328) was up-regulated 663-fold. Furthermore, both the WRKY TF and the MEKK-MPK system were active, which lead to the activation of genes with an ABRE cis-element. Two CBF4 genes (GRMZM2G124037 and GRMZM2G069082) that are in the ABA signalling pathway were dramatically up-regulated, by 1091-fold and 2119-fold, respectively, in the overexpression lines compared with the WT.
Discussion
ZmPTF1 overexpression enhances drought tolerance by increasing ABA accumulation and activating ABA signalling
ZmPTF1 overexpression led to an accumulation of ABA in both the roots and leaves under normal conditions (Fig. 7), which suggests that the biosynthesis of ABA may be greater in the overexpression lines than in the WT. Treatment with PEG solution induced an increase in the ABA contents in the roots and more pronouncedly in the leaves. A critical function of ABA is its mediation of cellular responses to environmental stresses, especially drought stress (Yamaguchi-Shinozaki and Shinozaki, 2006; Qin et al., 2011; Finkelstein, 2013; Sah et al., 2016). A number of genes involved in ABA biosynthesis have been identified in higher plants (Finkelstein, 2013; Endo et al., 2014), and the cleavage of 9-cis-epoxycarotenoids catalysed by NCED has been shown to be the rate-limiting step in the biosynthetic pathway (Qin and Zeevaart, 1999). The first NCED gene (VP14) was cloned from maize and shown to cleave violaxanthin or neoxanthin to form xanthoxin (Tan et al., 1997). NCED genes belong to a multigene family, and nine NCEDs have been identified in Arabidopsis. Functional analyses have indicated that five of them (AtNCED2, 3, 5, 6, and 9) are most likely involved in ABA biosynthesis (Lefebvre et al., 2006; Frey et al., 2012). In the ZmPTF1 overexpression lines, the expression of NCED genes was dramatically up-regulated compared with that of ABA1, NPQ1, and AAO3 (and the expression of ABA2 was slightly down-regulated); this finding is consistent with the accumulation of ABA. bHLH TFs have been implicated in the ABA signal transduction pathway (Abe et al., 2003; Msanne et al., 2011; Kazan and Manners, 2013), and bHLH122 may bind to G-box/E-box cis-elements in the CYP707A3 promoter and repress the expression of the promoter, thereby leading to increased cellular ABA levels (Liu et al., 2014). In contrast, overexpression of ZmPTF1 increases the expression of NCED gene family members by binding to the G-box elements in their promoters, which suggests that ZmPTF1 is an upstream regulator of ABA synthesis. The up-regulated expression of the NCED genes led to a relatively higher concentration of ABA in the transgenic lines than in the WT lines.
ABA regulates the stress response via the ‘core signalling pathway’ (Cutler et al., 2010), which includes the PYR/PYL/RCAR receptor, PP2C proteins, SnRK2 family members, AREB/ABF TFs, and the ABA-activated signalling pathway. In this study, the DEG analysis revealed that the ABA core signalling pathway and the ABA-activated signalling pathway were active, which included genes of the up-regulated PYR/PYL/RCAR family of proteins; an ABO3-like WRKY TF; an ABF4-like bZIP TF; ERD1; and a number of bHLH, WRKY, NAC, and ERF/AP2 TFs (Supplementary Table S5). ZmPTF1 contributes to drought stress tolerance by increasing ABA accumulation and by activating ABA signalling.
ZmPTF1 is involved in the development of the root system of maize and contributes to maize yields
bHLH TFs constitute one of the largest families of TFs and play important roles in development and the stress response (Castilhos et al., 2014). ZmPTF1 and ZmPTF1-Like are orthologues of OsPTF1, which is a monocotyledon-specific protein with a high similarity to Arabidopsis LRL1, 2, and 3, which are required for root hair development (Zhao et al., 2008; Karas et al., 2009; Bruex et al., 2012). In this study, ZmPTF1 played roles in root development primarily via the regulation of root number. When ZmPTF1 was knocked down, the lateral root primordia were significantly reduced in number, and the length from the first lateral root to the root tip was longer than that in the WT. The dramatic reduction in lateral root number led to a smaller root system in the mutant plants. Overexpression of ZmPTF1 improved maize root growth by promoting lateral root development, and ZmPTF1 was found to act as a positive regulatory factor of root development.
Regarding the genes downstream of ZmPTF1, eight genes involved in the auxin signalling pathway were differentially expressed, including four SAUR genes, SHY2/IAA3, an ARF6 homologue, and two auxin-responsive family genes (Supplementary Table S4). SHY2/IAA3 was reported to affect auxin-dependent root growth, lateral root formation, and the timing of gravitropism (Tian et al., 2002; Chaabouni et al., 2009; Goh et al., 2012; Lavenus et al., 2013), and ARF6 was shown to act redundantly with ARF8 to control stamen elongation and flower maturation (Nagpal et al., 2005). SAUR proteins have been proposed to modulate auxin transport and cell expansion by an unknown mechanism (Ren and Gray, 2015). NAC TFs are important in the morphological development process, such as the involvement of NAC1 in shoot apical meristem and lateral root formation (Xie et al., 2000; Wang et al., 2006; Li et al., 2012; Chen et al., 2016); the involvement of ATAF2 in the regulation of auxin and brassinosteroid synthesis via the regulation of NIT2 (Huh et al., 2012), BAS1 (CYP734A1), and SOB7 (CYP72C1) (Peng et al., 2015); and the involvement of NAC30/VND7 in the stress response, xylem formation, and lateral organ development (Yamaguchi et al., 2011; Reusche et al., 2012). Six NAC genes were induced by ZmPTF1, including the orthologues of NAC1, NAC30, NAC047, and ATAF2/NAC081. Hormone- and NAC-mediated processes may contribute to root development, and they altered stress responses in the ZmPTF1 overexpression lines. In addition to auxin, gibberellins and brassinosteroids mediate these processes. SLEEPY1 (SLY1), an F-box gene (Hauvermale et al., 2014), and three BRI genes were affected by the overexpression of ZmPTF1.
Certain root parameters, including small fine-root diameters, long specific root lengths, and considerable lateral root density, are associated with maintaining plant productivity under drought conditions (Lynch, 2013; Zhan et al., 2015). In maize, the seminal roots are of the greatest importance during the early growth of the seedling and determine the depth of the root system. The lateral roots are the most active in water and nutrient uptake, while adventitious roots are the most important organs for adapting to the environment. For example, brace roots can keep maize plants from falling over (Hochholdinger, 2009). There are several reasons why the overexpression of ZmPTF1 enhanced maize root development and yields. First, the lateral root number and length increased, which resulted in a larger volume and surface area of the root system, thereby increasing the absorption of water and nutrients. Second, the growth and elongation of the seminal roots and primary roots were promoted under both optimal and osmotic stress conditions, which reduced the inhibition of plant root growth during drought/osmotic stress and was beneficial to plant tolerance to drought stress environments. Third, the brace roots of the overexpression lines were more robust than those of the WT, which led to an increase in lodging tolerance (anchor strength is mainly contributed by adventitious roots). Thus, the improved root systems of the ZmPTF1 overexpression lines contributed to these lines’ higher yields and drought stress tolerance.
Hierarchical regulation by ZmPTF1 plays multiple roles in the stress response and root development
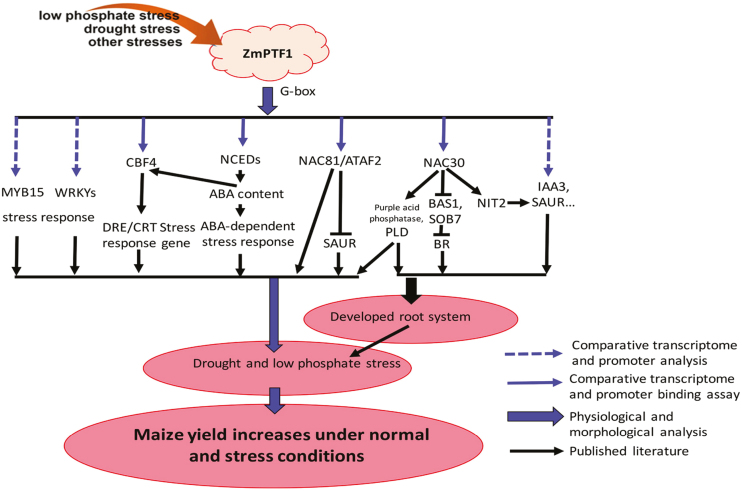
ZmPTF1 overexpression leads to the activation of the ABA-dependent stress response pathway as well as the ABA-independent stress response pathway. Based on the analysis of the transcriptome and promoter motifs of the DEGs, a schematic of the primary role of ZmPTF1 in the crosstalk network during abiotic stress and root development is shown in Fig. 8, in which ZmPTF1 binds to the G-box element as a heterodimer and/or a homodimer and regulates the expression of downstream genes such as NCEDs.
Fig. 8.
Schematic of the possible role of ZmPTF1 in the crosstalk network of the abiotic stress response and root system development. This is a suggested working model of ZmPTF1 based on the morphological, physiological, promoter binding, and downstream gene expression analyses in this study and the literature. The expression of ZmPTF1 is induced by abiotic stresses such as drought and low-phosphate stress. ZmPTF1 could then bind to the G-box cis-element in the promoter of NCEDs to increase the synthesis of ABA and activate the ABA signalling pathway. Based on the promoter binding assay, ZmPTF1 could bind to the G-box cis-element in the promoter of CBF4, ATAF2/NAC81, NAC30, IAA3, WRKY, and MYB15 and dramatically increase the expression of these transcription factors. The stress response processes mediated by these genes were more active in the ZmPTF1 overexpression line compared with the WT. These transcription factors were shown to be positive regulators of the stress response and/or root growth and development. ZmPTF1 contributes to drought tolerance mainly by promoting root system development and activating the stress response pathways. (This figure is available in colour at JXB online.)
As summarized in Fig. 8 and Supplementary Table S6, the AP/DREB, WRKY, NAC, and bHLH TFs were significantly affected by ZmPTF1. These TFs have been reported to be regulators of the response to environmental stress and development. Orthologues of RAP2.3/ERF72, RAP2.4, RAP2.5/ATERF4, and ATERF7 were regulated by ZmPTF1. These ERF TFs could act as regulators or downstream members of the ethylene, ABA, or jasmonic acid signalling pathways (Yang et al., 2005; Lingam et al., 2011; Papdi et al., 2015; Gasch et al., 2016). CBF4 is the only known CBF gene that is definitively involved in the ABA-dependent signalling pathways, cold acclimation, and drought adaptation (Haake et al., 2002). In the ZmPTF1 overexpression lines, the DREB/CBF genes that function in the abiotic stress response were active, and the dramatic up-regulation of ERFs and CBF4 activated the ethylene-, ABA-, and CBF4-mediated stress response pathways. Another kind of TF that was significantly induced by ZmPTF1 was the WRKY TFs; these TFs included orthologues of AtWRKY11, 33, 40, 41, 46, 51, 56, 57, 63, and 72. WRKY TFs were identified to have important roles in plant tolerance to both biotic and abiotic stresses (Zheng et al., 2006; Jiang and Deyholos, 2009; Lai et al., 2011). The overexpression of ZmPTF1 could lead to increased expression levels of MYB15 and AZF2 in maize. Both of these genes were reported to be involved in the regulation of genes involved in osmotic stress and ABA-mediated responses (Sakamoto et al., 2004; Kodaira et al., 2011).
The improved root system, increased ABA content, activated ABA and CBF4 signalling, and ATAF2- and NAC30-mediated stress responses increased the drought stress tolerance of the ZmPTF1 overexpression lines. It is concluded that ZmPTF1 functions as a TF that is involved in the gene response to stimuli, growth and development, and transcriptional regulation, and promotes ABA synthesis. All of these effects led to improved maize root system development and an increased ability to respond to stress.
Overexpression of ZmPTF1 is a valuable strategy to enhance the drought tolerance and yield potential of maize
Crop improvement represents a traditional method for increasing yields and enhancing stress tolerance. Ideally, drought tolerance should be achieved without yield penalties. The performance of maize hybrids during the past 70 years has demonstrated that yield potential and stress tolerance are associated traits (Lopes et al., 2011; Claeys and Inze, 2013). When molecular breeding techniques that involve the transfer of one or more coding DNA regions/CRISPR gRNAs/RNAi constructs into an elite cultivar are adopted, they can be useful for improving crop tolerance to environmental stresses and increasing yields. Many successful applications of introducing useful genes into crop species have resulted in improved environmental stress tolerance (Anami et al., 2009; Jewell et al., 2010); however, few transgenic plants have produced higher yields and exhibited increased environmental stress tolerance under normal and stress conditions. The initial attempts to develop transgenic plants with abiotic stress tolerance mainly focused on the genes responsible for the modification of a single component, such as water channel proteins, transporters, key enzymes involved in osmolyte biosynthesis, and detoxification enzymes, which would provide increased tolerance to salt or drought stress. However, this approach does not consider the many genes that are simultaneously involved in abiotic stress tolerance, or the lack of sustainability of single-gene tolerance. Regulatory proteins have been analysed because they trigger cascades of genes that act together to enhance tolerance towards multiple stresses. However, the altered expression of downstream genes in all organs and at all development stages can cause abnormalities in those plants grown under normal conditions. In this study, ZmPTF1 overexpression in the inbred line DH4866 improved drought stress tolerance without affecting the yield potential. The yield increased under both normal and drought stress conditions, meaning that high yield and high drought tolerance were achieved concurrently. ZmPTF1 could be used to improve the viability of agricultural crop species grown in soils with sufficient or insufficient amounts of water and nutrients.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Real-time RT–PCR analysis of ZmPTF1 expression in the roots of the maize transgenic lines used in this paper.
Fig. S2. Biomass and morphological analysis of the ZmPTF1 overexpression and antisense lines and the WT plants.
Fig. S3. Effects of drought stress on the physiological parameters of the ZmPTF1 overexpression and antisense lines and the WT plants.
Fig. S4. Overexpression of ZmPTF1 activated stress responses and modified transcriptional regulation.
Fig. S5. Expression levels of the key genes involved in ABA biosynthesis in the roots of different lines under normal and 15% PEG8000 treatment conditions.
Table S1. Primers used in this study.
Table S2. Sequences of probes and mutated probes used in the electrophoretic mobility shift assay.
Table S3. GO enrichment analysis between the ZmPTF1 overexpression line (L+4) and WT.
Table S4. Differentially expressed key genes involved in plant growth and development between the ZmPTF1 overexpression line (L+4) and WT.
Table S5. Differentially expressed key genes involved in the ABA metabolic and signalling pathway between the ZmPTF1 overexpression line (L+4) and WT.
Table S6. Differentially expressed key genes between the ZmPTF1 overexpression line (L+4) and WT.
Table S7. All the differentially expressed genes between the ZmPTF1 overexpression line (L+4) and WT.
Table S8. Results of real-time RT–PCR to validate the results of the RNA-seq analysis in the ZmPTF1 overexpression lines L+3 and L+4, and WT.
Acknowledgements
We thank Jinfang Chu (IGDB, Chinese Academy of Sciences) for ABA determination. This work was supported by the China National Natural Science Foundation (31571674) and National Major Projects for Genetically Modified Organisms Breeding in China (2016ZX08003004-003).
Data deposition
The RNA-seq data are available at Dryad Digital Repository. https://doi.org/10.5061/dryad.7nr377v
Author contributions
ZL and JZ designed the experiments. ZL, CL, YZ, BW, and QR conducted the experiments and analysed the results. JZ supervised the experiments. ZL wrote the paper and JZ revised the paper.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anami S, De Block M, Machuka J, Van Lijsebettens M. 2009. Molecular improvement of tropical maize for drought stress tolerance in sub-Saharan Africa. Critical Reviews in Plant Sciences 28, 16–35. [Google Scholar]
- Atchley WR, Fitch WM. 1997. A natural classification of the basic helix-loop-helix class of transcription factors. Proceedings of the National Academy of Sciences, USA 94, 5172–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitha KC, Ramu SV, Pruthvi V, Mahesh P, Nataraja KN, Udayakumar M. 2013. Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Research 22, 327–341. [DOI] [PubMed] [Google Scholar]
- Bray EA. 1987. Plant responses to water deficit. Trends in Plant Science 2, 48–54. [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, et al. 2012. A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genetics 8, e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck MJ, Atchley WR. 2003. Phylogenetic analysis of plant basic helix-loop-helix proteins. Journal of Molecular Evolution 56, 742–750. [DOI] [PubMed] [Google Scholar]
- Burbidge A, Grieve TM, Jackson A, Thompson A, McCarty DR, Taylor IB. 1999. Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14. The Plant Journal 17, 427–431. [DOI] [PubMed] [Google Scholar]
- Castilhos G, Lazzarotto F, Spagnolo-Fonini L, Bodanese-Zanettini MH, Margis-Pinheiro M. 2014. Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Science 223, 1–7. [DOI] [PubMed] [Google Scholar]
- Chaabouni S, Jones B, Delalande C, Wang H, Li Z, Mila I, Frasse P, Latché A, Pech JC, Bouzayen M. 2009. Sl-IAA3, a tomato Aux/IAA at the crossroads of auxin and ethylene signalling involved in differential growth. Journal of Experimental Botany 60, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cheng J, Chen L, Zhang G, Huang H, Zhang Y, Xu L. 2016. Auxin-independent NAC pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiology 170, 2136–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Analytical Biochemistry 162, 156–159. [DOI] [PubMed] [Google Scholar]
- Claeys H, Inzé D. 2013. The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiology 162, 1768–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Dai A. 2013. Increasing drought under global warming in observations and models. Nature Climate Change 3, 52–58. [Google Scholar]
- dos Santos MG, Ribeiro RV, de Oliveira RF, Machado EC, Pimentel C. 2006. The role of inorganic phosphate on photosynthesis recovery of common bean after a mild water deficit. Plant Science 170, 659–664. [Google Scholar]
- Endo A, Okamoto M, Koshiba T. 2014. ABA biosynthetic and catabolic pathways. In: Zhang D-P, ed. Abscisic acid: metabolism, transport and signaling. Dordrecht: Springer, 21–45. [Google Scholar]
- Ferré-D’Amaré AR, Prendergast GC, Ziff EB, Burley SK. 1993. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38–45. [DOI] [PubMed] [Google Scholar]
- Finkelstein R. 2013. Abscisic acid synthesis and response. The Arabidopsis Book 11, e0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey A, Effroy D, Lefebvre V, Seo M, Perreau F, Berger A, Sechet J, To A, North HM, Marion-Poll A. 2012. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. The Plant Journal 70, 501–512. [DOI] [PubMed] [Google Scholar]
- Fu J, Chu J, Sun X, Wang J, Yan C. 2012. Simple, rapid, and simultaneous assay of multiple carboxyl containing phytohormones in wounded tomatoes by UPLC-MS/MS using single SPE purification and isotope dilution. Analytical Sciences 28, 1081–1087. [DOI] [PubMed] [Google Scholar]
- Gasch P, Fundinger M, Müller JT, Lee T, Bailey-Serres J, Mustroph A. 2016. Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. The Plant Cell 28, 160–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. 2012. Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 367, 1461–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. 2002. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiology 130, 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM. 2014. The roles of the GA receptors GID1a, GID1b, and GID1c in sly1-independent GA signaling. Plant Signaling & Behavior 9, e28030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F. 2009. The maize root system: morphology, anatomy, and genetics. In: Bennetzen JL, Hake SC, eds. Handbook of maize: its biology. New York: Springer, 145–160. [Google Scholar]
- Huh SU, Lee SB, Kim HH, Paek KH. 2012. ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Molecules and Cells 34, 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Choi HS, Cho HM, Cho HT. 2017. Tracheophytes contain conserved orthologs of a basic helix-loop-helix transcription factor that modulate ROOT HAIR SPECIFIC genes. The Plant Cell 29, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell MC, Campbell BC, Godwin ID. 2010. Transgenic plants for abiotic stress resistance. In: Kole C, Michler CH, Abbott AG, Hall TC, eds. Transgenic crop plants. Heidelberg: Springer, 67–132. [Google Scholar]
- Jiang Y, Deyholos MK. 2009. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology 69, 91–105. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Deyholos MK. 2009. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Molecular Genetics and Genomics 282, 503–516. [DOI] [PubMed] [Google Scholar]
- Jin J, Lauricella D, Armstrong R, Sale P, Tang C. 2015. Phosphorus application and elevated CO2 enhance drought tolerance in field pea grown in a phosphorus-deficient vertisol. Annals of Botany 116, 975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Zhang H, Kong L, Gao G, Luo J. 2014. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Research 42, D1182–D1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R, Wani SH, Singh B, Bohra A, Dar ZA, Lone AA, Pareek A, Singla-Pareek SL. 2016. Transcription factors and plants response to drought stress: current understanding and future directions. Frontiers in Plant Science 7, 1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaoka MM, Pillitteri LJ, Fujii H, Yoshida Y, Bogenschutz NL, Takabayashi J, Zhu JK, Torii KU. 2008. SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to Arabidopsis stomatal differentiation. The Plant Cell 20, 1775–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K. 2009. Conservation of Lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiology 151, 1175–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2013. MYC2: the master in action. Molecular Plant 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Kodaira KS, Qin F, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. 2011. Arabidopsis Cys2/His2 zinc-finger proteins AZF1 and AZF2 negatively regulate abscisic acid-repressive and auxin-inducible genes under abiotic stress conditions. Plant Physiology 157, 742–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Li Y, Wang F, Cheng Y, Fan B, Yu JQ, Chen Z. 2011. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. The Plant Cell 23, 3824–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L. 2013. Lateral root development in Arabidopsis: fifty shades of auxin. Trends in Plant Science 18, 450–458. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A. 2006. Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. The Plant Journal 45, 309–319. [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S. 2002. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wei A, Song C, Li N, Zhang J. 2008. Heterologous expression of the TsVP gene improves the drought resistance of maize. Plant Biotechnology Journal 6, 146–159. [DOI] [PubMed] [Google Scholar]
- Li H, Sun J, Xu Y, Jiang H, Wu X, Li C. 2007. The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Molecular Biology 65, 655–665. [DOI] [PubMed] [Google Scholar]
- Li J, Guo G, Guo W, Guo G, Tong D, Ni Z, Sun Q, Yao Y. 2012. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.). BMC Plant Biology 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Duan X, Jiang H, et al. 2006. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiology 141, 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gao Q, Liu Y, He C, Zhang X, Zhang J. 2011. Overexpression of transcription factor ZmPTF1 improves low phosphate tolerance of maize by regulating carbon metabolism and root growth. Planta 233, 1129–1143. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu C, Zhang Y, Wang B, Ran Q, Zhang J. 2018. Data from: The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and ABA synthesis. Dryad Digital Repository. 10.5061/dryad.7nr377v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang X, Zhao Y, Li Y, Zhang G, Peng Z, Zhang J. 2018. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnology Journal 16, 86–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. 2007. Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiology 143, 745–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Ohashi Y, Kato M, Tsuge T, Gu H, Qu LJ, Aoyama T. 2015. GLABRA2 directly suppresses basic helix-loop-helix transcription factor genes with diverse functions in root hair development. The Plant Cell 27, 2894–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P. 2011. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. The Plant Cell 23, 1815–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Wang Y, Pan K, Jin Y, Li W, Zhang L. 2015. Effects of phosphorus application on photosynthetic carbon and nitrogen metabolism, water use efficiency and growth of dwarf bamboo (Fargesia rufa) subjected to water deficit. Plant Physiology and Biochemistry 96, 20–28. [DOI] [PubMed] [Google Scholar]
- Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, Li WX. 2014. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytologist 201, 1192–1204. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25,402–408. [DOI] [PubMed] [Google Scholar]
- Lopes MS, Araus JL, van Heerden PD, Foyer CH. 2011. Enhancing drought tolerance in C4 crops. Journal of Experimental Botany 62, 3135–3153. [DOI] [PubMed] [Google Scholar]
- Lynch JP. 2013. Steep, cheap and deep: an ideotype to optimize water and N acquisition by maize root systems. Annals of Botany 112, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, et al. 2005. Steady-state transposon mutagenesis in inbred maize. The Plant Journal 44, 52–61. [DOI] [PubMed] [Google Scholar]
- Msanne J, Lin J, Stone JM, Awada T. 2011. Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107. [DOI] [PubMed] [Google Scholar]
- Nagpal P, Ellis CM, Weber H, et al. 2005. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K. 2014. The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Frontiers in Plant Science 5, 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C, Pérez-Salamó I, Joseph MP, Giuntoli B, Bögre L, Koncz C, Szabados L. 2015. The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. The Plant Journal 82, 772–784. [DOI] [PubMed] [Google Scholar]
- Peng H, Zhao J, Neff MM. 2015. ATAF2 integrates Arabidopsis brassinosteroid inactivation and seedling photomorphogenesis. Development 142, 4129–4138. [DOI] [PubMed] [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K. 2011. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant & Cell Physiology 52, 1569–1582. [DOI] [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD. 1999. The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proceedings of the National Academy of Sciences, USA 96, 15354–15361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Gray WM. 2015. SAUR proteins as effectors of hormonal and environmental signals in plant growth. Molecular Plant 8, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche M, Thole K, Janz D, Truskina J, Rindfleisch S, Drübert C, Polle A, Lipka V, Teichmann T. 2012. Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. The Plant Cell 24, 3823–3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah SK, Reddy KR, Li J. 2016. Abscisic acid and abiotic stress tolerance in crop plants. Frontiers in Plant Science 7, 571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. 2004. Arabidopsis Cys2/His2-type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiology 136, 2734–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardans J, Peñuelas J. 2012. The role of plants in the effects of global change on nutrient availability and stoichiometry in the plant-soil system. Plant Physiology 160, 1741–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz SH, Tan BC, Gage DA, Zeevaart JA, McCarty DR. 1997. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Seo JS, Joo J, Kim MJ, Kim YK, Nahm BH, Song SI, Cheong JJ, Lee JS, Kim JK, Choi YD. 2011. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. The Plant Journal 65, 907–921. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Schwartz SH, Zeevaart JAD, McCarty DR. 1997. Genetic control of abscisic acid biosynthesis in maize. Proceedings of the National Academy of Sciences, USA 94, 12235–12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Uhlir NJ, Reed JW. 2002. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. The Plant Cell 14, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. The Plant Cell 15, 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA 2014. Crop Production 2014 Summary. Washington DC: National Agricultural Statistics Service, USDA; https://www.census.gov/history/pdf/cropan15.pdf [Google Scholar]
- Wang Y, Duan L, Lu M, Li Z, Wang M, Zhai Z. 2006. Expression of NAC1 up-stream regulatory region and its relationship to the lateral root initiation induced by gibberellins and auxins. Science in China. Series C, Life Sciences 49, 429–435. [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH. 2000. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes & Development 14, 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T. 2011. VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. The Plant Journal 66, 579–590. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. 2006. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology 57, 781–803. [DOI] [PubMed] [Google Scholar]
- Yang T, Hao L, Yao S, Zhao Y, Lu W, Xiao K. 2016a. TabHLH1, a bHLH-type transcription factor gene in wheat, improves plant tolerance to Pi and N deprivation via regulation of nutrient transporter gene transcription and ROS homeostasis. Plant Physiology and Biochemistry 104, 99–113. [DOI] [PubMed] [Google Scholar]
- Yang T, Yao S, Hao L, Zhao Y, Lu W, Xiao K. 2016b. Wheat bHLH-type transcription factor gene TabHLH1 is crucial in mediating osmotic stresses tolerance through modulating largely the ABA-associated pathway. Plant Cell Reports 35, 2309–2323. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. 2005. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology 58, 585–596. [DOI] [PubMed] [Google Scholar]
- Yi K, Wu Z, Zhou J, Du L, Guo L, Wu Y, Wu P. 2005. OsPTF1, a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiology 138, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan A, Schneider H, Lynch JP. 2015. Reduced lateral root branching density improves drought tolerance in maize. Plant Physiology 168, 1603–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. 2008. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135, 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Qamar SA, Chen Z, Mengiste T. 2006. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. The Plant Journal 48, 592–605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.