Plant phytochelatin synthases undergo evolutionarily rapid functional differentiation after duplication, allowing fast and precise adjustment of metal detoxification capacity by modulation of both transcription and enzymatic activity.
Keywords: Cadmium, divergence, gene duplication, giant reed, phytochelatin synthase, phytochelatins, subfunctionalization
Abstract
Phytochelatin synthases (PCSs) play pivotal roles in the detoxification of heavy metals and metalloids in plants; however, little information on the evolution of recently duplicated PCS genes in plant species is available. Here we characterize the evolution and functional differentiation of three PCS genes from the giant reed (Arundo donax L.), a biomass/bioenergy crop with remarkable resistance to cadmium and other heavy metals. Phylogenetic reconstruction with PCS genes from fully sequenced monocotyledonous genomes indicated that the three A. donax PCSs, namely AdPCS1-3, form a monophyletic clade. The AdPCS1-3 genes were expressed at low levels in many A. donax organs and displayed different levels of cadmium-responsive expression in roots. Overexpression of AdPCS1-3 in Arabidopsis thaliana and yeast reproduced the phenotype of functional PCS genes. Mass spectrometry analyses confirmed that AdPCS1-3 are all functional enzymes, but with significant differences in the amount of the phytochelatins synthesized. Moreover, heterogeneous evolutionary rates characterized the AdPCS1-3 genes, indicative of relaxed natural selection. These results highlight the elevated functional differentiation of A. donax PCS genes from both a transcriptional and an enzymatic point of view, providing evidence of the high evolvability of PCS genes and of plant responsiveness to heavy metal stress.
Introduction
Most transition elements, including the metalloid arsenic, are often collectively defined as ‘heavy metals’ (HMs), due to their high density (Clemens et al., 2002). Some of these elements (e.g. Cu, Mn, Ni, Zn, Fe) are essential nutrients for the majority of organisms, while others, such as Cd, Hg, Pb, and As, lack any known biological role and are toxic even at low concentrations (Tchounwou et al., 2012). HM pollution represents a threat to the environment as well to human populations (Järup, 2003; Thompson and Bannigan, 2008). Usually, HMs occur naturally in the environment as trace components of the earth’s crust (Fraústo da Silva and Williams 2001). Their presence as pollutants is therefore most often due to anthropogenic activities, such as mining, motorized transport, and industry (Nagajyoti et al., 2010; Tchounwou et al., 2012). The mechanisms underlying HM toxicity are still not completely understood. What is known is that excessive amounts of essential metals and traces of non-essential metals cause at least two major responses: (i) displacement of the correct cellular cofactors, for example, Cd is able to displace fundamental bivalent ions such as Zn2+, Fe2+, Cu2+, and Ca2+; and (ii) a cascade of aberrant reactions, with protein thiol groups binding the metal ions and producing reactive oxygen species (Rea et al., 2004). In the case of plants, toxic HM ions present in the rhizosphere are taken up into cells together with essential metal ions by relatively non-specific plasma membrane transporters (Luo et al., 2016). Among the HMs, possibly the most studied is Cd, given its high toxicity and widespread occurrence in soils worldwide (Tchounwou et al., 2012; Mahar et al., 2016). Uptake of Cd2+ from the rhizosphere is the result of ‘hitchhiking’ of this toxic ion on the plasma membrane transporters necessary for the uptake of essential metal ions, especially Mn and possibly Fe in rice (Redjala et al., 2009; Sasaki et al., 2012; Uraguchi and Fujiwara, 2013). Once in the cytoplasm, Cd has to be detoxified as soon as possible to prevent cellular damage. The major mechanism of intracellular Cd detoxification is based on phytochelatins (PCs), a family of cysteine-rich oligopeptides synthesized from glutathione (GSH) by the enzyme phytochelatin synthase (PCS), a γ-glutamylcysteine dipeptidyl transpeptidase (EC 2.3.2.15) (Grill et al., 1989; Vatamaniuk et al., 2004) belonging to clan CA of the papain-like cysteine proteases (Vivares et al., 2005; Romanyuk et al., 2006; Rea, 2012). Elevated HM concentrations in the cytoplasm and the presence of reduced GSH activate the PCS enzyme, readily starting the biosynthesis of PCs. PCs chelate the HM and the HM–PC complex is then transported into the vacuoles, where the HM can be detoxified (Song et al., 2014). PC biosynthesis carried out by PCSs represent the main mechanism for plants to detoxify the HMs present in their rhizosphere. PCS and PCS-like genes have been found in a wide range of organisms, from prokaryotes, such as cyanobacteria (Bhargava et al., 2005; Chaurasia et al., 2008), to eukaryotes, such as yeasts (Grill et al., 1985; Ha et al. 1999; Shine et al., 2015), plants, and animals (Clemens et al., 2001; Ray and Williams, 2011; Polak et al., 2014). PCSs are evolutionarily conserved in all land plants as well as in charophytes (Fontanini et al., 2018), their sister group, and recent evidence indicates that PC biosynthesis is a plesiomorphic character for plants (Degola et al., 2014; Petraglia et al., 2014). Most of our knowledge of PCS genes derives from Arabidopsis thaliana, whose genome encodes two different PCS genes, AtPCS1 (Vatamaniuk et al., 1999) and AtPCS2 (Cazalé and Clemens, 2001; Kühnlenz et al., 2014). AtPCS1 is the major PCS isoform, and loss-of-function mutations in the AtPCS1 gene render plants extremely sensitive to Cd stress. AtPCS2 is also active, but plays a minor role in Cd detoxification, as the phenotype of AtPCS2 mutants becomes visible only after AtPCS1 has been knocked out (Lee and Kang, 2005; Kühnlenz et al., 2014). More recently, two PCS genes from rice, OsPCS1 and OsPCS2, have been characterized in detail (Li et al., 2007; Das et al., 2017; Hayashi et al., 2017; Uraguchi et al., 2017; Yamazaki et al., 2018). The two isoforms have different specificities for Cd and As, and contribute differentially to detoxification of these HMs (Hayashi et al., 2017; Yamazaki et al., 2018).
Since the discovery of the PCS enzymes, the potential of exploiting them for bioengineering plant detoxification of HMs has attracted much attention. Many attempts to increase resistance to HMs in a number of plant species by overexpression of PCS genes from different sources resulted in a variety of experimental outcomes, ranging from decreased to enhanced resistance to HMs, with various types of relationship (positive, negative, or no relationship) to the accumulation of different HMs. As of 2015, Cd resistance was obtained in roughly one-quarter of the tested PCS-overexpressing lines, while in another quarter of the cases PCS overexpression caused hypersensitivity to HMs (Lee and Hwang, 2015). Early attempts to increase HM detoxification in Arabidopsis by overexpression of AtPCS1, for instance, unexpectedly resulted in higher sensitivity to Cd (Lee, 2003). Apparently, the overexpression of AtPCS1 might have caused as a side effect depletion of the intracellular pool of GSH, which, besides PCs, can play a relevant role in HM chelation (Jozefczak et al., 2012). Indeed, overexpression of both AtPCS and γ-glutamylcysteine synthetase, the enzyme catalyzing the first committed step of GSH biosynthesis, can overcome the problem, leading to enhanced HM resistance (Guo et al., 2008). By contrast, overexpression in Arabidopsis of PCS genes from other plant species directly resulted in higher Cd resistance (Guo et al., 2008; Liu et al., 2012; Fan et al., 2018), indicating that significant functional differences may exist among PCS enzymes. Despite these difficulties, the capacity to modulate HM detoxification through careful adjustments of PC biosynthesis is of extreme relevance for phytoremediation, a relatively new branch of science that uses plants and their associated microbes to reduce the concentration of HMs in soils and freshwaters (Krämer, 2005). Phytoremediation is now probably one of the most promising technologies at our disposal for the process of decontamination of water and soil from those harmful elements (Oyuela Leguizamo et al., 2017). Good candidate plant species for this technology should ideally be those with fast growth and remarkable biomass yield, as well as high ability to accumulate and inactivate HMs (Salt et al., 1998; Suresh and Ravishankar, 2004; Peuke and Rennenberg, 2005). Several hyperaccumulators (i.e. plant species that are able to accumulate HMs in their aerial parts at concentrations two to three orders of magnitude higher than normal plants; van der Ent et al., 2013) are highly tolerant to specific HMs, but they usually grow relatively slowly (Souza et al., 2013), thus limiting their overall effectiveness in phytoremediation. Recently, the giant reed (Arundo donax L.), a perennial rhizomatous grass of the Poaceae family, has been proposed as a potential phytoremediation species, as it is able to accumulate and tolerate high concentrations of HMs such as Ni, Cd, and As without showing major stress symptoms (Papazoglou et al., 2005, 2007; Papazoglou, 2007; Sabeen et al., 2013). The potential use of A. donax for phytoremediation has been confirmed for a variety of HM concentrations and conditions in soil (Guo and Miao, 2010; Mirza et al., 2011; Alshaal et al., 2013; Barbosa et al., 2015; Fernando et al., 2016; Atma et al., 2017), and especially for wastewaters and polluted aquatic environments (Mirza et al., 2010; Sagehashi et al., 2011; Kausar et al., 2012; Sabeen et al., 2013; Elhawat et al., 2014; Richveisová et al., 2014). Moreover, A. donax is a vigorous perennial plant that is capable of the high biomass yields necessary to rapidly remove HMs from soils. Although A. donax represents a promising non-crop plant for phytoremediation, to date no molecular characterization of the genes involved in HM detoxification has been carried out in this species. In this work we studied three recently diverged PCS genes of A. donax, and characterized them both in the species of provenance and in transgenic model organisms, in order to dissect their role in the molecular and physiological bases of resistance to Cd. Furthermore, we investigated whether functional diversification could be detected among the gene copies, to gain novel insights into the evolutionary trajectories and retention of recently duplicated PCS genes in plants.
Materials and methods
Plant materials, growth conditions, and stress treatment
Cohorts of Arundo donax L. cuttings (Sesto Fiorentino, Florence, Italy; 43°49′01.8ʺN, 11°11′57.0ʺE) and Arabidopsis thaliana L. Heynh. Col-0 wild-type and transgenic plants were used in this study. The procedures for stress treatments in A. donax were the same as those described previously (Fu et al., 2016; Li et al., 2017). Three biological replicates were used for all the treatments at every sampling time point. Sterilized seeds from AdPCS1-3 transgenic plants were sown on half-strength Murashige & Skoog solid medium supplemented with 1% sucrose, stratified at 4 °C for 3 days, and transferred to a growth chamber. After 3.5 days of growth, the young seedlings were transferred to square petri dishes containing the same medium as above (control), or supplemented with 150 µM CdSO4 (for HM treatment), and grown vertically for a further 10 days. Ten plants for Col-0 and two independent lines from each construct were grown in one plate, and at least 80 plants from each line and Col-0 were analyzed. All plants were grown in long-day conditions (16 h light/8 h dark) in a growth chamber at 23 °C with light intensity 100–120 µmol m–2 s–1 and 40% relative humidity.
Sequence homology searches for novel PCS genes in A. donax
PCS homologs were mined from the A. donax transcriptome (Sablok et al., 2014) using the BLAST+suite (Camacho et al., 2009). Oryza sativa PCS2 homolog (LOC_Os06g01260) was used as query to perform a BLASTn search, due to its close evolutionary distance from Arundo. Transcripts were aligned using the ClustalW algorithm with the BioEdit7 suite (Hall, 1999) and manually checked for the presence of the typical triad of PCS catalytic residues (based on A. thaliana PCS sequence: Cys56, His162, Asp180). Three complete coding DNA sequences (CDSs) were identified and classified as putative A. donax PCS1, PCS2, and PCS3 (henceforth called AdPCS1, AdPCS2, and AdPCS3).
Genomic DNA extraction and cloning
Genomic DNAs from Arundo accessions/species growing in a growth chamber were isolated using the hexadecyltrimethylammonium bromide method (Doyle and Doyle, 1987). The last intron of AdPCS1, AdPCS2, and AdPCS3 was amplified using Phusion High Fidelity DNA Polymerase (Thermo Scientific) with the primers listed in Supplementary Table S1 at JXB online. After A-tailing with Taq polymerase (Sigma), amplicons were cloned into the pGEM-T vector and at least three clones were bidirectionally sequenced for each amplicon.
Total RNA extraction and real-time PCR analyses
Total RNA extraction and cDNA synthesis were performed as previously reported (Fu et al., 2016). Semi-quantitative RT–PCR was carried out for different organs using the A. donax glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene as a reference (Li et al., 2017) and AtAct2 for transgenic analysis. For stress treatments, qRT–PCR was conducted with Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen) using a putative A. donax actin as a reference gene (Fu et al., 2016) in a Bio-Rad C1000 Thermal Cycler detection system. All reactions for qRT–PCR analyses were performed in triplicate and the 2−ΔΔCT method was used to calculate fold changes. Primers used for PCR are listed in Supplementary Table S1.
Phylogenetic reconstruction
A BLASTp search was conducted using AtPCS1 as query against the proteome of all Poaceae and Brassicaceae species present in the Phytozome v.12 database (Goodstein et al., 2012). The resulting homologs were manually checked for the presence of the canonical catalytic triad (Cys56, His162, Asp180) as well as for N- and C-terminal domain integrity. The Phragmites australis PCS sequence was additionally retrieved from the nr GenBank database (Zhao et al., 2014). Sequences were then aligned using the MAFFT program (algorithm E-INS-i, dedicated to proteins with two or more conserved domains; Katoh and Standley, 2013) and processed using GBlocks (Talavera and Castresana, 2007) with the following parameters: minimum number of sequences for a conserved position: 13; minimum number of sequences for a flanking position: 13; maximum number of contiguous non-conserved positions: 8; minimum length of a block: 5; allowed gap positions: with half; use similarity matrices: yes. Maximum likelihood (ML) phylogenetic reconstruction was performed with the PhyML 3.0 web server (Guindon and Gascuel, 2003; Guindon et al., 2010). The best-fitting evolutionary model for Poaceae ML reconstruction was evaluated using the Smart Model Selection method (Lefort et al., 2017) and the ProtTest 2.4 Server (Abascal et al., 2005). Statistical support was calculated based on the Shimodaira–Hasegawa-like Approximate Likelihood-Ratio test (SH-aLRT) for branches (Anisimova and Gascuel, 2006). Branches with SH-aLRT<0.05 were collapsed in the representation of the final tree. Bayesian inference (BI) phylogenetic reconstruction was performed for both alignments with MrBayes v3.2 (Ronquist et al., 2012), using two independent runs, each with one cold and three heated chains over 5 000 000 generations. The program was allowed to average over the first 10 amino acid rate matrix models by specifying the setting: aamodelpr=mixed. Trees were sampled every 1000 generations and posterior probabilities of splits were obtained from the 50% majority rule consensus of the sampled trees, discarding the first 25% as burn-in. Poaceae ML and BI trees were combined using the program TreeGraph 2 (Stöver and Müller, 2010). The phylogenetic reconstruction of the last intron of AdPCS1-3 from different Arundo species/accessions was carried out by ML as described above.
Analyses of molecular evolution
Different programs of the HyPhy package (Kosakovsky Pond et al., 2005) were used for molecular evolution analyses of the codon-aligned AdPCS CDS (502 codons) using the HKY85 model. GARD (Kosakovsky Pond et al., 2006) was used to infer putative recombination sites with no site-to-site rate variation and two rate classes. The BUSTED program (Murrell et al., 2015) was used for testing gene-wide episodic diversifying selection in all branches of the AdPCS1-3 phylogeny inferred with the neighbor-joining algorithm. In addition, a total of three branches were formally tested for diversifying selection using the aBSREL program (Smith et al., 2015). Significance was assessed using the likelihood ratio test at a threshold of P≤0.05, after correcting for multiple testing. Analysis of selective pressures acting on the single alignment sites was carried out with the FUBAR program (Murrell et al., 2013) using a posterior probability of 0.9 as cutoff. Relaxation/intensification of selection on specific branches of the tree was assessed with the RELAX program (Wertheim et al., 2015).
Molecular dating
Patristic distances (number of substitutions per site) among sequences of the last intron of AdPCS1-3 were calculated with T-REX software (Boc et al., 2012). Representative sequences for each gene were used to estimate divergence time (T) using the formula T=K/2r, where r is the absolute rate of substitutions per site per year and K is the estimated numbers of substitutions per site between homologous sequences. The value of r used (9E-09) corresponds to the rate for the third intron of the Adh gene (Zhu and Ge, 2005), corrected for lineage-specific evolution of Poaceae (Wang et al., 2015).
Yeast complementation assay
Full-length AdPCS1, AdPCS2, and AdPCS3 CDSs were amplified with primers listed in Supplementary Table S1, cloned into pENTR/D-Topo, recombined into the pYES-DEST52 vector (Invitrogen™), and transformed into Cd-sensitive Saccharomyces cerevisiae strain YK44 (ura3-52 his3-200, ∆ZRCDCot1, mating type α) using the lithium acetate method (Gietz and Schiestl, 2007). The culture for each transformant and spotting on YPGAL solid medium supplemented with or without 100 µM CdSO4 were as previously reported (Zhao et al., 2014). All experiments were independently repeated four times.
Expression of recombinant protein and PCS activity assay
The coding sequences of full-length AdPCS1, AdPCS2, and AdPCS3 proteins were amplified from cDNA using primers listed in Supplementary Table S1 and cloned into the expression vector pET28b in-frame with an N-terminal 6xHis-tag. The expression plasmids were transformed into Escherichia coli Rosetta (DE3) cells, which were induced with 0.5 mM isopropyl ß-D-thiogalactopyranoside and cultured overnight at room temperature. The cells were collected by centrifugation and the soluble fraction of recombinant protein was purified as previously described (Pilati et al., 2014), quantified with the Quant-iT Protein Assay Kit (Thermo Fisher Scientific), and verified on 10% SDS-PAGE. The PCS activity assay was performed in 100 µl reaction buffer (200 mM HEPES-NaOH pH 8.0, 10 mM β-mercaptoethanol, 12.5 mM GSH, and 100 μM CdSO4) containing recombinant protein (500 ng ml−1) at 35 °C for 60 min, then stopped by adding 25 µl 10% (v/v) trifluoroacetic acid and immediately analyzed for production of PCs with high-performance liquid chromatography–electrospray ionization tandem mass spectrometry (HPLC-ESI-MS-MS; Bellini et al., 2019).
Mass spectrometry analyses of production of PCs by AdPCS1-3 in yeast and in vitro
Single colonies from each yeast transformant line were precultured overnight in YSD-U selective medium. A 500 μl aliquot (OD600=0.5) of each line was inoculated into YPGAL containing 100 μM CdSO4 and 1 mM GSH, and cultured for 24 h. Afterwards, 2 ml of each culture (OD600=2) was pelleted, washed once with sterile water, and resuspended with 200 μl 5% (w/v) sulfosalicylic acid. Cells were ground using glass beads with a tissue lyser, the recovered supernatant was rapidly frozen in liquid nitrogen, and a 10 μl aliquot was used for mass spectrometry (MS) analysis as previously described (Petraglia et al., 2014). For recombinant AdPCS1-3 enzymes, thiol-peptides were characterized and quantified by HPLC-ESI-MS-MS using an Agilent 1290 Infinity UHPLC (Santa Clara, CA, USA) with a thermostated autosampler, a binary pump, and a column oven, coupled to an AB Sciex API 4000 triple quadrupole mass spectrometer (Concord, ON, Canada) equipped with a Turbo-V ion spray source (Concord, ON, Canada). Chromatographic separation was performed by using a reverse-phase Waters (Milford, MA, USA) X-Select® HSS T3 2.5 µm XP-C18, 100×3 mm HPLC column, protected by a guard cartridge. Separation was achieved using a gradient solvent system (solvent A, acetonitrile with 0.1% v/v formic acid; solvent B, water with 0.1% v/v FA) as follows (Bellini et al., 2019): solvent A was set at 2% for 5 min, raised with a linear gradient to 44% in 4.5 min, and then raised with a linear gradient to 95% in 1 min. Solvent A was maintained at 95% for 1 min before column re-equilibration (2.5 min). The flow rate and column oven temperature were set to 300 μl min−1 and 30 °C, respectively. The identification and quantification of thiol-peptides (GSH and PCn) was performed by tandem mass spectrometry (MS/MS) using certified standards (GSH, PC2-4; AnaSpec Inc., Fremont, CA, USA) to build external calibration curves and certified glycine-13C2,15N-labelled GSH (Sigma-Aldrich, Saint Louis, MO, USA) and glycine-13C2,15N-labelled PC2 (AnaSpec Inc., Fremont, CA, USA) as internal standards. The calibration curves were built by plotting the peak area ratio (analyte/internal standard) against the concentration ratio (analyte/internal standard). For accurate quantification of thiol-peptides using the present HPLC-ESI-MS-MS method, each sample was run at dilutions of 100, 10−1, and 10−2. System control and data acquisition and processing were carried out by using AB Sciex Analyst® version 1.6.3 software.
Growth kinetic analyses of yeast transformed with AdPCS1-3
Single colonies from each transformant were grown overnight in YSD-U medium and each aliquot with OD600=0.5 was inoculated for further growth in YPGAL medium. During the following 48 h culture at 30 °C, 1 ml aliquots were taken at fixed time intervals (0, 4, 8, 12, 16, 20, 24, 36, and 48 h) and their OD600 measured. All experiments were done in quadruplicate.
Overexpression of AdPCS1-3 in Arabidopsis thaliana
The same pENTR clones mentioned above for the yeast analyses were individually recombined into the pK7WG2 vector by the GATEWAY LR (attL and attR recombination sites) reaction and transformed into Agrobacterium tumefaciens strain GV3101-pMP90, then transformed into A. thaliana by the floral dip method (Clough and Bent, 1998). T4 homozygote seeds from two independent single-copy lines for each construct were used for all downstream analyses.
Statistical analyses
Unless otherwise stated, for each statistical test a threshold of P<0.05 was applied to determine statistical significance. The data were analyzed using the PAST3 statistical package (Hammer et al., 2001). For statistical analysis of differences in total PC production among yeast strains overexpressing the three AdPCS enzymes, the sum of PC2 to PC5 products was used. In the case of recombinant enzymes, the sum of PC2 to PC4 products was used. One-way ANOVA with Tukey’s pairwise correction was applied to the net PC2–PC5 or PC2–PC4 quantities to test for the statistical significance of mean differences.
Results
The A. donax genome encodes at least three different PCS gene copies
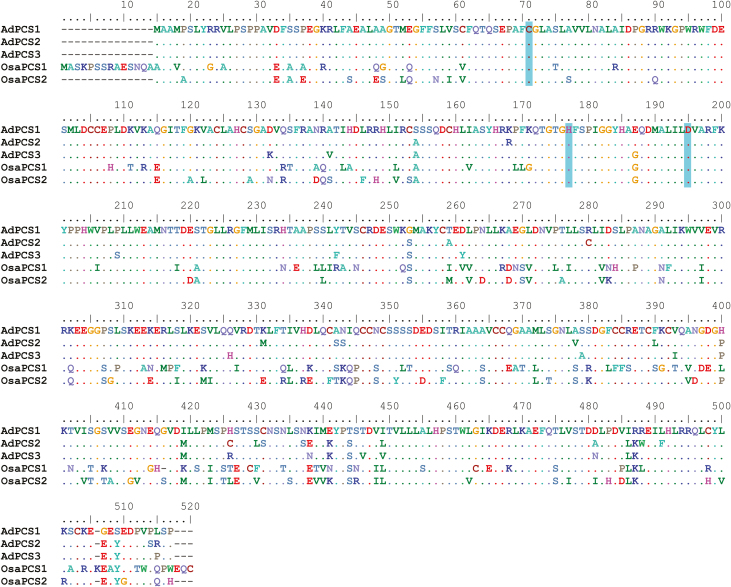
All three putative AdPCS full-length CDSs isolated contained a single open reading frame, encoding a protein of 502 residues and a predicted molecular weight of ~55 kDa. Once translated, the three proteins showed an average identity with A. thaliana PCS1 (NCBI accession number NP_199220) of 55% and an average similarity of 70%. This level of divergence was higher than those usually observed among alleles of single genetic loci in plants (e.g. Brown et al., 2004; Tatarinova et al., 2016; Zhao et al., 2017). Phylogenetic analyses of the last intron of AdPCS1-3 isolated from four different Arundo species clearly showed that divergence among sequences predated the evolutionary split among species and ruled out recombination (Supplementary Fig. S1). It is thus almost certain that the isolated CDSs are not alleles of a single gene, but different genes encoded in the A. donax genome. In line with previous reports (Matsumoto et al., 2004), the N-terminal of the AdPCS1-3 proteins was more conserved than the C-terminal; this was also the case for the rice paralogs OsPCS1 (Os05g0415200) and OsPCS2 (Os06g0102300) (Fig. 1). AdPCS1-3 proteins are significantly less divergent than other duplicated PCSs from previous studies (Supplementary Table S2), displaying only 25–29 substitutions. All the canonical features of PCSs were present, namely the catalytic triad Cys56, His162, and Asp180, and the lengths of both N- and C-terminal domains were comparable to those of previously validated PCSs (Fig. 1). In addition, in the Brassicaceae, we detected a PCS duplication present in the genomes of all species that have been fully sequenced to date (Supplementary Fig. S2), suggesting that retention of PCS duplicates for long evolutionary time spans is a relatively common phenomenon in angiosperms.
Fig. 1.
Multiple sequence alignment of A. donax and O. sativa PCS proteins. Dashes indicate gaps; dots indicate residues identical to those in the sequence of AdPCS1. The amino acids of the catalytic triad (Cys56, His162, and Asp180 in A. thaliana, corresponding to positions 57, 163, and 181 in the AdPCS1-3 proteins) are highlighted. (This figure is available in colour at JXB online.)
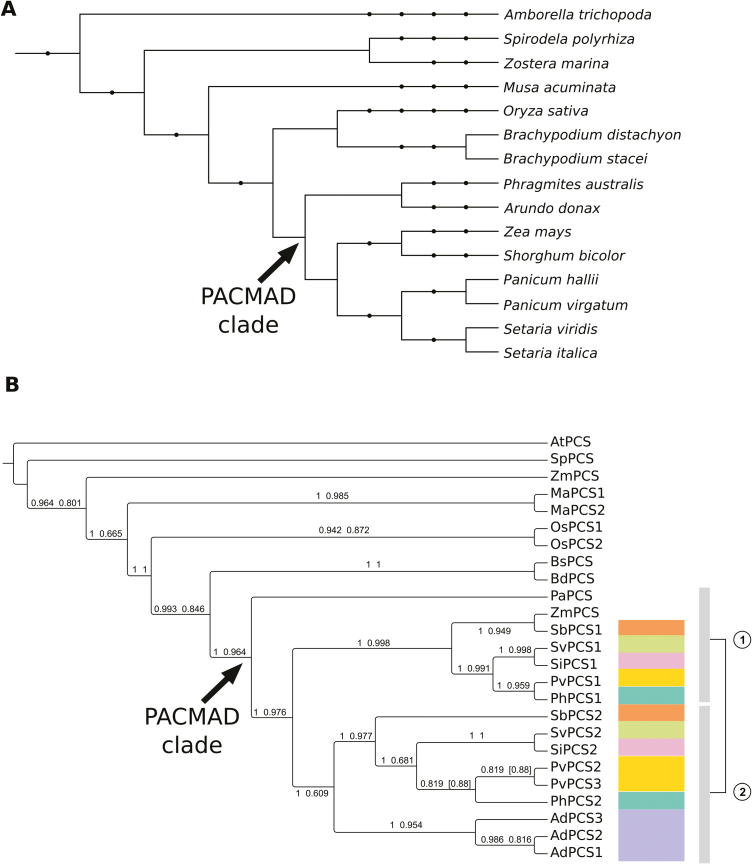
Two different statistical methods suggested JTT+G+I (Jones et al, 1992) as the best-fitting evolutionary model for the alignment used for the phylogenetic reconstruction of monocots and Amborella trichopoda PCS proteins shown in Fig. 2. Accession numbers for all sequences used are reported in Supplementary Table S3. Using A. trichopoda PCS as an outgroup to polarize the tree, P. australis PCS was basal to the Panicoideae clade. With the exception of Zea mays, which has only one PCS gene (which could be the result of mis-annotation), all other Panicoideae species had at least two PCS genes, always present in two distinct clades, clearly indicating an early duplication event predating the Panicoideae radiation (Fig. 2). A further species-specific duplication was evident in Panicum virgatum. The A. donax sequences characterized in this study grouped together at the base of only one of the two Panicoideae clades, indicating that the three gene copies originated after the split of A. donax from the other members of the PACMAD clade. The two PCS copies present in O. sativa (Yamazaki et al., 2018) clustered together, indicating a lineage-specific duplication independent from the major duplication event in the Panicoideae.
Fig. 2.
Phylogenetic reconstruction of monocotyledonous PCS proteins. (A) Cladogram representing the taxonomy of the species used for phylogenetic reconstruction. Internal nodes indicate high-order taxonomic groups corresponding to NCBI taxonomy. The PACMAD clade is indicated with an arrow. The taxonomic node at the root of the tree is Spermatophyta. (B) Cladogram of the relationships among PCS protein sequences from fully sequenced plant genomes of monocotyledonous species (plus A. trichopoda, the most basal angiosperm species known to date, used here to root the tree) from the Phytozome 12 database. The PCS protein of Phragmites australis, the closest relative of A. donax for which a PCS gene has been functionally characterized, is also included. Phylogenetic reconstruction was carried out with both the BI (MrBayes) and ML (PhyML) methods. The tree reports the BI topology and it is rooted in correspondence of A. trichopoda. Numbers above the branches are Bayesian posterior probabilities (first value) and approximate likelihood ratio test bootstrap supports (second value). Values in square brackets indicate topological differences between methods. The duplication in the PACMAD clade (arrow) is represented by the two vertical bars to the right of the cladogram, each corresponding to a different clade of the duplication. Species with duplicated PCSs in the PACMAD clade are color-coded to simplify identification of duplicates in the tree. The analyzed organisms and the relative abbreviations used in the tree are reported in Supplementary Table S3. (This figure is available in colour at JXB online.)
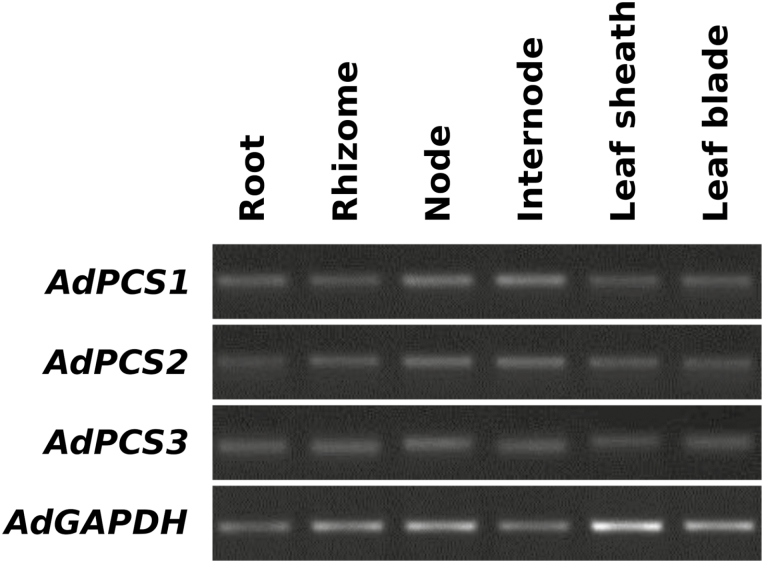
Expression of the AdPCS 1-3 genes is responsive to HM stress
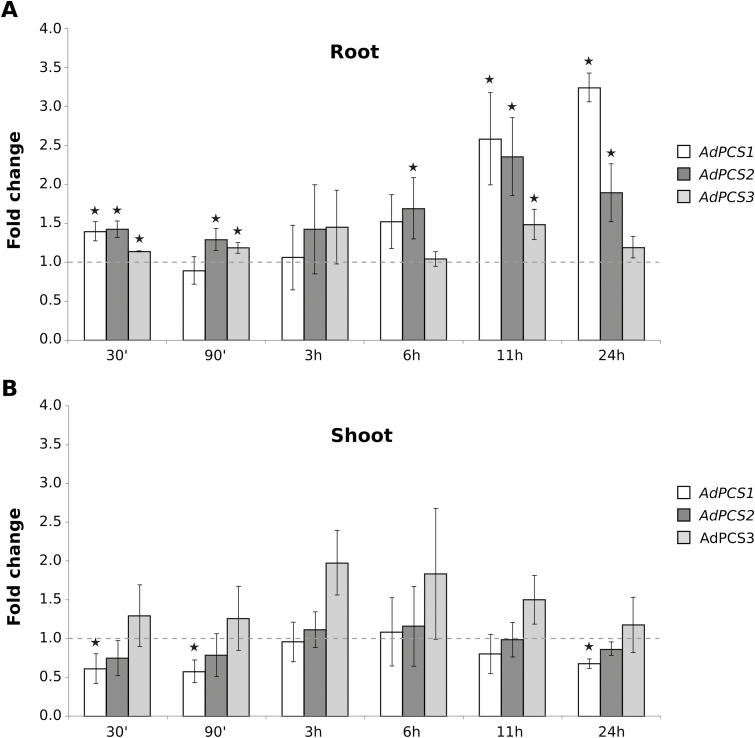
AdPCS1-3 transcript expression levels were analyzed in both physiological and stressed conditions. In normal conditions, AdPCS1-3 were expressed in all organs analyzed, as indicated by semi-quantitative RT-PCR analyses (Fig. 3). In response to treatment with a concentration of 500 μM CdSO4, the three transcripts reacted independently and in a tissue-specific manner. In the shoot, expression of the three genes did not change in response to CdSO4, with the exception of a very modest, although statistically significant, decrease of AdPCS1 expression in the first 90 min of stress application (Fig. 4). By contrast, in root, all three genes showed significant increases of expression at various time points, with a general trend towards up-regulation, especially after 6 h after stress onset. AdPCS1 was the most reactive gene in root, with its expression level reaching 3-fold higher than in the absence of stress. Notably, AdPCS1 transcriptional up-regulation was most prominent in root between 11 and 24 h after stress application, whereas in shoot the gene was either unresponsive or mildly down-regulated by the presence of Cd2+ at these time points (Fig. 4). AdPCS3 responded so little to CdSO4 treatment in absolute terms (maximal fold change 1.5) that its expression can be considered practically not inducible by HM.
Fig. 3.
Expression pattern of AdPCS1-3 in different organs/tissues of A. donax as demonstrated by semi-quantitative RT–PCR. Expression levels of AdPCS1-3 transcripts were measured in physiological, unstressed conditions. The AdGAPDH gene was used as a normalization reference, using 26 PCR cycles, while 33 cycles were used for AdPCS1-3.
Fig. 4.
Responsivity of AdPCS1, AdPCS2, and AdPCS3 to Cd stress. Time-course variation by real-time PCR of AdPCS1-3 expression in A. donax roots (A) and shoots (B) in response to HM stress resulting from treatment of the root system with 500 µM CdSO4. The Y-axis reports variations in transcript level compared with the non-stressed condition (0 h, indicated by the continuous line at Y=1). Bars indicate SD (n=3 biological replicates); asterisks indicate statistically significant differences from untreated controls (P<0.05, t-test corrected with false discovery rate).
Overexpression of AdPCS genes in A. thaliana enhances Cd sensitivity
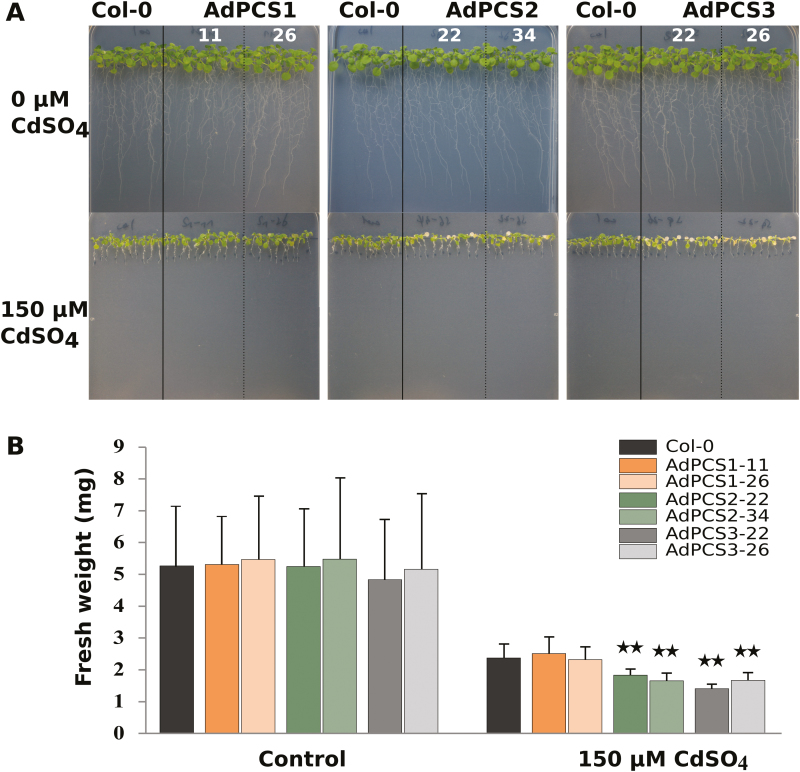
Expression of AdPCS1-3 was confirmed by semi-quantitative RT–PCR in two A. thaliana transgenic lines per construct representing independent transformation events (Supplementary Fig. S3). No differences in growth could be detected in the absence of Cd among A. thaliana Col-0 untransformed controls and transgenic lines transformed with AdPCS1-3 CDSs. Independent lines overexpressing either the AdPCS2 or AdPCS3 CDS exhibited a significant reduction in growth (measured as the fresh weight of the aerial part) compared with Col-0 when treated with 150 µM CdSO4 in the growth medium (Fig. 5A, B). In addition, chlorosis of transgenic plants overexpressing AdPCS2 or AdPCS3 was clearly visible (Fig. 5B). By contrast, the transgenic plants overexpressing AdPCS1 closely resembled the Col-0 ecotype and did not display any discernible phenotype in terms of fresh weight or pigmentation.
Fig. 5.
Phenotype of A. thaliana plants overexpressing AdPCS1-3 in the presence of Cd. (A) Phenotype of two independent transgenic lines per construct and the control untransformed line (Col-0) grown on vertical plates for 10 days without (top) or with (bottom) the addition of 150 µM CdSO4. (B) Fresh weight of the plants shown in A. Bars indicate the SD of n=3 biological replicates; asterisks indicate statistically significant differences from the untreated controls (P<0.05, t-test corrected with false discovery rate). (This figure is available in colour at JXB online.)
Expression of AdPCS1-3 in yeast confers Cd resistance
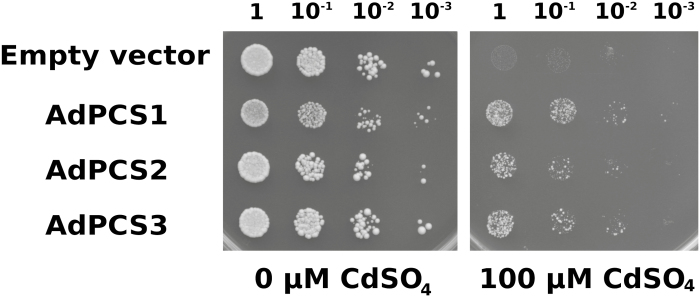
AdPCS1-3 CDSs subcloned in the pYES-DEST52 vector were transformed into the S. cerevisiae YK44 highly Cd sensitive strain, previously used for the same purpose by Zhao et al. (2014). We observed that the growth of AdPCS1 transformants was delayed compared with AdPCS2-3 transformants. To further investigate the basis of this difference, a growth analysis was conducted on YPGAL media without any HM stress (Supplementary Fig. S4). The results clearly showed that the AdPCS1 transformant had a longer lag phase, resulting in slower initial growth. This difference was rescued after 24 h, with the growth kinetics of the AdPCS1 transformant becoming normal, indicating that the difference resulted from decreased translational efficiency in the yeast heterologous system. Comparison of OD-normalized cultures showed that AdPCS1-3 transformants acquired equivalent levels of Cd resistance in the presence of 100 µM CdSO4 (Fig. 6). This result indicates that the three different CDSs code for functional PCSs and that A. donax possesses at least three different genes for PCS enzymes.
Fig. 6.
Growth of yeast cells expressing AdPCS1-3 CDSs. The four yeast strains, transformed with AdPCS1-3 CDSs or the empty vector, were plated in serial dilutions (dilution factors are indicated above the image) in either the absence or presence of 100 µM CdSO4. Four independent replicates were performed; the picture shows a representative example of one replicate.
AdPCS1-3 produce phytochelatins in yeast and in vitro
To prove that Cd resistance was due to PC synthesis, MS analyses were carried out. As expected, all three PCS enzymes produced detectable amounts of PCs in yeast (Supplementary Fig. S5). There was significant heterogeneity in the mean quantities of the major PC polymerization forms (PC2 to PC5) produced by the strains overexpressing different AdPCS genes (one-way ANOVA, F2, 6=5.149, P=0.042). We attempted, without success, to normalize the amounts of enzyme in the AdPCS1-3 transformants by western blotting using an antibody raised against Arabidopsis PCS1 (data not shown). We thus could not rule out the possibility that the cause of the differences in total PCs among yeast transformants could lie in differences in the translational efficiency of the AdPCS transgenes in the yeast heterologous system (see above).
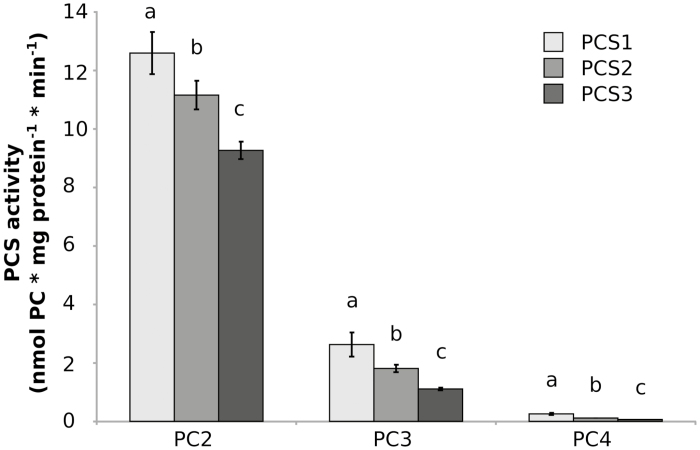
Recombinant AdPCS1-3 proteins purified from E. coli (Supplementary Fig. S6) were further assayed in vitro to compare their enzymatic activity in the absence of confounding effects due to in vivo differences in expression. The specific activities of AdPCS1-3 enzymes for the major PC polymerization forms (PC2 to PC4, either as gross total or individually) were significantly heterogeneous (one-way ANOVA; for total PCs: F2, 12=98.35, P=3.62E-05; for PC2: F2, 12=49.2, P=1.65E-06; for PC3: F2, 12=46.6, P=2.34E-03; for PC4: F2, 12=98.39, P=3.61E-05). Significant differences were detected among the amounts of both total PCs and single polymerization forms produced in all pairwise comparisons among enzymes (Tukey’s post-hoc test; total PCs: P<0.00024; PC2: P<0.00304; PC3: P<0.00234; PC4: P<0.01109 for all comparisons; Fig. 7). The total PCs produced ranged (mean ±SD) from 10.43±0.29 to 15.47±0.73 nmol mg protein−1 min−1. The rank in terms of total production was AdPCS1>AdPCS2>AdPCS3 (Fig. 7). Only minor differences in the spectrum of PCs produced (as referred to PC1 for each enzyme) were detected.
Fig. 7.
Spectra of phytochelatins (PCs) produced by recombinant AdPCS1-3 proteins in vitro. Average amounts of PCs (PC2 to PC4, either individually or as gross total) produced by AdPCS1-3 enzymes purified from E. coli. Five independent replicates were performed. For each construct, bars marked with the same letter do not significantly differ from each other (Tukey–Kramer test, P>0.05). Bars represent the SD.
AdPCS1-3 genes evolve at different evolutionary rates
To shed additional light on the functional differences observed among the AdPCS1-3 genes and reconstruct their evolutionary history, we carried out a series of analyses on the patterns of molecular evolution of their sequences. The three genes did not show significant evidence of either gene-wide (likelihood ratio test, P =0.859) or branch-specific (likelihood ratio test, P=1.00 for AdPCS1, 1.00 for AdPCS2, 0.15 for AdPCS3) episodic diversifying selection.
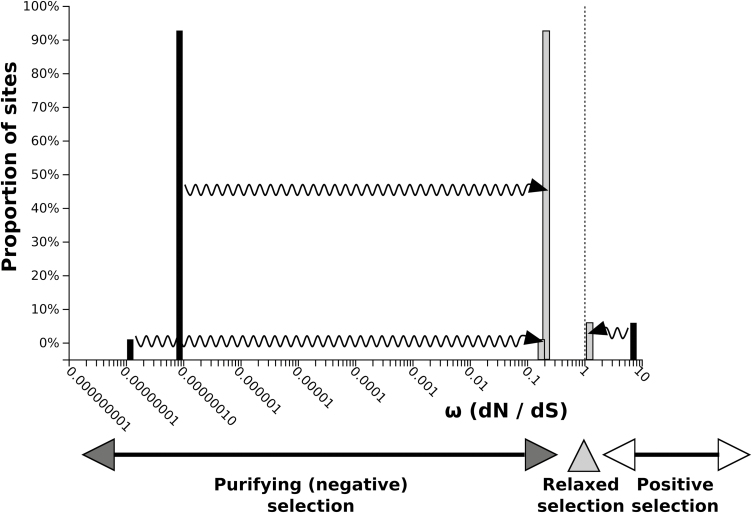
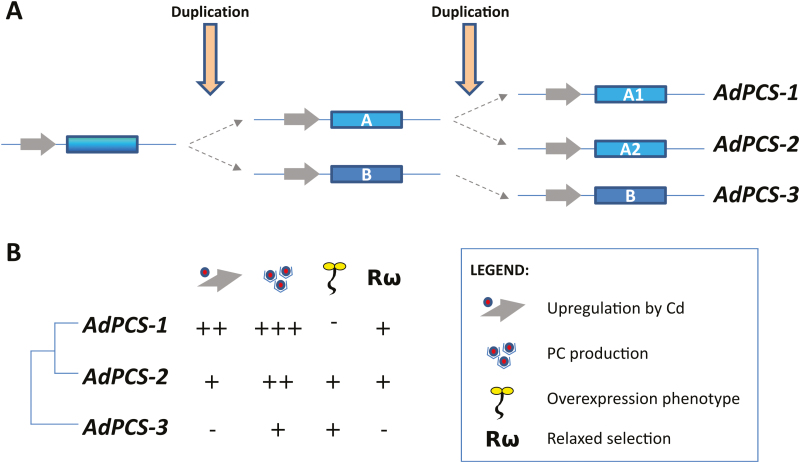
Therefore, there is no evidence that any sites have experienced diversifying selection along the test branch(es). A putative break-point, indicative of either rate variation or topological incongruence due to recombination, was identified (position 296 of the alignment; corrected Akaike Information Criterion score of the best fitting GARD model: 5362.96; AICc score of null model: 5365.01). The Kishino–Hasegawa topological incongruence test resulted negative (Shimodaira–Hasegawa test applied to the left partition, P=0.997; applied to the right partition, P=1.000), suggesting that the different domains of AdPCS genes underwent evolutionary rate variation rather than recombination. Two additional findings support evolutionary rate heterogeneity among AdPCS sequences. First, episodic positive/diversifying selection was detected at four sites, all of them located at the C-terminus of the proteins (Supplementary Table S4), while the 29 sites under episodic negative/purifying selection were distributed throughout the alignment. Second, the sequences of AdPCS1 and AdPCS2 displayed statistically significant relaxation (K=0.10, P=0.030, likelihood ratio=4.72) compared with AdPCS3 (Fig. 8). Given such evolutionary rate variation, CDS could not be reliably used for estimation of duplication time. Theoretically more reliable estimations of divergence times from nucleotide divergence of the AdPCS1-3 last introns varied depending on the pairs of genes used (Supplementary Tables S5 and S6), providing an approximate time range for AdPCS1-3 duplications. These and the results described above (i.e. phylogeny, transcriptional responsiveness to Cd) allowed reconstruction of the plausible events leading to evolution of the three homeologous PCS genes characterized in this study. According to the evolutionary model shown in Fig. 9A, we propose that two rounds of polyploidization happened in rapid succession, in a time range between ~8.5±0.1 and ~11.3±0.1 million years ago. The first duplication generated PCS ancestral copies A and B. While PCS B evolved by mutation/drift into present-day AdPCS3, the second duplication of PCS A gave rise to AdPCS1 and AdPCS2. During the whole process, functional divergence among the resulting copies involved the changes in gene transcription, enzyme activity, and evolutionary rates that characterize the present-day AdPCS1-3 genes (Fig. 9B).
Fig. 8.
Relaxation of selective pressure acting on AdPCS1 and AdPCS2. The distribution of ω (omega ratio of non-synonymous to synonymous substitution rates) across alignment sites is shown by the black bars for the null model in which the same ω distribution is assumed for the AdPCS3 (reference) and AdPCS1-2 (test) branches. The grey bars represent the ω distribution for the alternative model in which different ω distributions are assumed for the reference and test branches. The arrows indicate the direction of ω classes variation from the null model to the alternative model distribution. The shift observed towards ω values closer to neutrality (ω=1, vertical dotted line) indicates relaxation of the selective pressure acting on AdPCS1 and AdPCS2 compared with AdPCS3.
Fig. 9.
Evolutionary model and summary of functional diversification among AdPCS1-3. (A) Scheme of the two rounds of duplication giving rise to AdPCS1-3. (B) Functional features of AdPCS1-3 with respect to transcriptional up-regulation by Cd, total amount of PCs produced, and the presence of reduced growth and a chlorotic phenotype when overexpressed in A. thaliana, and relaxation of the evolutionary constraints of purifying selection acting on the single genes. – indicates (near) absence and + indicates presence, with more + indicating more pronounced features. (This figure is available in colour at JXB online.)
Discussion
Given the reported high resistance of A. donax to Cd and other HMs (e.g. Papazoglou et al., 2007; Mirza et al., 2011) and its relevance as a biomass/bioenergy crop (Angelini et al., 2009), this species has been repeatedly suggested as a strong candidate for phytoremediation (Fernando et al., 2016). To date, however, the molecular bases for the high resistance of A. donax to HMs has not been functionally investigated in detail. The fundamental role of the PCS genes in HM detoxification is well established in a number of higher plants (Clemens, 2006; Yadav, 2010); thus, the characterization of the transcriptional and enzymatic activity of the three putative PCS genes carried out in this study constitute an important step towards the dissection of the mechanisms underlying the resistance of A. donax to HMs. The results obtained demonstrate that all the AdPCS isoforms characterized are fully functional, as they are transcribed in various A. donax organs (Fig. 3), can confer enhanced resistance to Cd in yeast (Fig. 6), and they all synthesize PCs (Fig. 7, Supplementary Fig. S5). However, they also display marked functional differences in the amount and, to a lower extent, levels of polymerization of PCs they produce, as well as in their transcriptional regulation upon exposure to HM stress (Figs 4 and 7). In addition, PCS paralogs previously characterized in other species (Lotus japonicus, O. sativa, A. thaliana, and Morus notabilis) were found to display functional differentiation in terms of the amount of PCs produced and the specificity of metal-mediated activation, as well as differential regulation of transcription in response to HMs (Lee and Kang, 2005; Blum et al., 2007; Ramos et al., 2008; Moudouma et al., 2012; Kühnlenz et al., 2014; Fan et al., 2018; Yamazaki et al., 2018). In particular, the recent work carried out in O. sativa var. japonica clearly demonstrates that OsPCS1 and OsPCS2 have specialized roles in Cd and As resistance. OsPCS2 is the major isozyme for biosynthesis of PCs and is the most responsive to Cd (Hayashi et al., 2017; Yamazaki et al., 2018), while OsPCS1 plays a major role in allowing the accumulation of As in rice grains but is also important in resistance to both As and Cd (Li et al., 2007; Hayashi et al., 2017; Uraguchi et al., 2017). This high level of functional divergence parallels the medium to high sequence divergence of PCS paralogs from these species (52–54% identity between LjPCS1 and LjPCS2/LjPCS3; 72% identity between OsPCS1 and OsPCS2; 84% identity between AtPCS1 and AtPCS2) compared with that among AdPCS1-3 (94–95%), indicating that they originated from more ancient duplication events than AdPCS1-3. For instance, two PCS paralogs were found in the genomes of all Brassicaeae analyzed in our study (Supplementary Fig. S1), and functional AtPCS1 and AtPCS2 orthologs have been characterized in Arabidopsis halleri and Noccaea caerulescens (formerly named Thlaspi caerulescens; Meyer et al., 2011). The AtPCS1 and AtPCS2 paralogs thus originated from a duplication predating the divergence between the Arabidopsis and Noccaea genera, which has been estimated to have taken place in the Oligocene about 27 million years ago (Huang et al., 2016). Unfortunately, the lack of a reference genome makes it difficult to infer the timing and mechanism of the AdPCS1-3 duplication. The best estimates we could obtain place both duplication events at roughly 9–12 million years ago, corroborating the notion that the AdPCS1-3 duplications are more recent than those of previously characterized PCS genes. The increased relaxation of selective pressure, closer phylogenetic relationship of AdPCS1 and AdPCS2 compared with AdPCS3, and the overall low sequence divergence among AdPCS1-3 proteins together suggest that two rounds of duplication took place in close succession: the first duplication gave rise to two ancestral PCS copies (PCS A and B in Fig. 9), followed relatively soon after by a second duplication responsible for the evolution of AdPCS1 and AdPCS2 from the ancestral PCS A copy. Another example of such an evolutionary pattern has been reported for LjPCS1-3, but in this case the second duplications happened much later than the first (Ramos et al., 2007). Given the transcriptional responsiveness of PCS genes to HMs in other species (e.g. Moudouma et al., 2012; Yamazaki et al., 2018), the ancestral PCS existing before both duplications may have been up-regulated by Cd stress, like the present-day AdPCS1 and AdPCS2 genes; thus, we propose that the PCS B ancestral copy underwent subfunctionalization of regulatory promoter elements after divergence. This scenario is corroborated by the fact that stress-responsive genes tend to be over-represented among paralogous pairs with different expression levels and reduced conservation of cis-acting regulatory elements (Hoffmann and Palmgren, 2016). Interestingly, transcriptional differences, with one copy dominant over the other, seem to be a relatively common feature of duplicated PCS genes across different plant families and originating from independent duplication events (e.g. Lee and Kang, 2005; Fan et al., 2018; Yamazaki et al., 2018), suggesting that transcriptional divergence may be a relevant mechanism for long-term retention of multiple PCSs. In addition to non-coding regulatory regions, subfunctionalization is known to be a common outcome for protein-coding sequences of retained duplicates (Hoffmann and Palmgren, 2016). Even in the absence of pseudogenization, duplicated genes can undergo functional decay for millions of years simply due to genetic drift (Panchy et al., 2016). The presence of several sites under purifying selection and some under intensified/positive selection (Supplementary Table S4), however, indicates that this is not the case for the AdPCS1-3 genes, further suggesting that loss of any of the three gene copies, including AdPCS1, would have a detrimental effect on plant fitness (Panchy et al., 2016). It is noteworthy that the relatively large functional variation among AdPCS isoforms required a limited number of amino acid substitutions (<30) to take place (Supplementary Table S2). The fact that the majority of the substitutions took place in the C-terminal domain of the protein suggests that regulation rather than core catalytic activity is affected, in agreement with previous reports demonstrating that this fast-evolving domain is responsible for metal sensing and enzyme stability (Cobbett and Goldsbrough, 2002; Ruotolo et al., 2004).
Taken together, these results strongly suggest that AdPCS1, ADPCS2, and AdPCS3 most likely contribute to Cd detoxification in A. donax. The presence of multiple PCS copies seems to be advantageous as on the one hand redundancy provides higher overall levels of PC biosynthesis, while on the other hand functional specialization provides increased flexibility in HM resistance (Panchy et al., 2016). According to the ubiquitous and conserved level of constitutive expression of the three isozymes in all A. donax organs/tissues but the lower PC biosynthetic activity of AdPCS3, it seems that AdPCS1 and AdPCS2 play a major role in basal detoxification of Cd and possibly other HMs. Transcriptional inducibility by Cd further suggests that AdPCS1 and AdPCS2 provide the most relevant contribution to HM detoxification in roots at high metal concentrations (Fig. 4). A. donax accumulates the majority of absorbed Cd in the roots and rhizomes (Sagehashi et al., 2011; Yu et al., 2018), implying that below-ground organs act as the main centers of bioaccumulation (Bonanno, 2012). AdPCS1 and AdPCS2 thus likely contribute to avoidance of metal toxicity by preventing Cd sequestration in non-photosynthetic tissues. While the reduced growth observed for AdPCS2- and AdPCS3-overexpressing plants in Arabidopsis is in line with former reports (Lee, 2003), the reason why overexpression of AdPCS1 does not cause any visible phenotype under the conditions tested is currently not clear. Although the transcriptional levels of the AdPCS1 transgene were similar to those of AdPCS2 and AdPCS3, the amount of AdPCS1 protein expression in this heterologous system may be limited by translational efficiency, analogous to what was detected in yeast. Further studies will be needed to investigate this point.
More generally, the AdPCS1-3 triplication is, to the best of our knowledge, by far the most recent set of paralogs functionally characterized in plants until now. The results obtained therefore provide novel insights into the mechanisms of functional diversification that, ultimately, are responsible for long-term retention of PCS duplicates in plants. In fact, they demonstrate that functional diversification happens relatively early after duplication and entails a small number of amino acid changes in the C-terminal domain of PCS, suggesting a mechanism underlying the flexibility of species with multiple PCSs to adapt to HM stress.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in this study.
Table S2. Amino acid divergence among PCS duplicates from different species.
Table S3. List of species and PCS accession numbers used for phylogenetic reconstruction.
Table S4. Codons of AdPCS1-3 under episodic positive/diversifying selection or episodic negative/purifying selection as detected by FUBAR software.
Table S5. Summary of sequences of the AdPCS1-3 last intron from different Arundo species/accessions.
Table S6. Estimated divergence times among AdPCS genes.
Fig. S1. Maximum likelihood phylogenetic tree of the AdPCS1-3 last intron from four Arundo species.
Fig. S2. Bayesian inference phylogenetic tree of PCS proteins present in Brassicaceae with fully sequenced genomes.
Fig. S3. Semi-quantitative RT–PCR of AdPCS1-3 transcription levels in two A. thaliana transgenic lines.
Fig. S4. Liquid growth kinetics of yeast strains overexpressing AdPCS1-3 CDS in non-stressed conditions.
Fig. S5. Total production of PCs (PC2-5) in yeast lines overexpressing AdPCS1, AdPCS2, or AdPCS3 CDS.
Fig. S6. SDS-PAGE of recombinant AdPCS1-3 enzymes purified from E. coli.
Fig. S7. Summary and sequence alignment of the AdPCS1-3 last intron from different Arundo species.
Acknowledgements
This work has been funded by the Autonomous Province of Trento through core funding of the Ecogenomics group of Fondazione E. Mach. LS was the recipient of a PhD fellowship from the University of Parma. This work was partly supported by MIUR-PRIN 2015 funds (prot. 20158HTL58, PI Prof. Luigi Sanità di Toppi) and by the Evolutionary Biology and Ecology PhD course of the University of Parma. The authors thank Enrico Barbaro (Fondazione E. Mach) for technical support in sampling of A. donax material, Andrea Faccini (Centro Interdipartimentale Misure ‘G. Casnati’, University of Parma) for assistance in mass spectrometry analyses, Stefania Pilati for help in recombinant protein purification, Massimo Pindo for technical support in Sanger sequencing, and Dietrich H. Nies for the kind gift of the YK44 yeast strain.
Glossary
Abbreviations
- BI
Bayesian inference
- CDS
coding DNA sequence
- GSH
glutathione
- HM
heavy metal
- ML
maximum likelihood
- PC
phytochelatin
- PCS
phytochelatin synthase
Conflict of interest
The authors declare no conflict of interest.
References
- Abascal F, Zardoya R, Posada D. 2005. ProtTest : selection of best-fit models of protein evolution. Bioinformatics 21, 2104–2105. [DOI] [PubMed] [Google Scholar]
- Alshaal T, Domokos-Szabolcsy É, Márton L, Czakó M, Kátai J, Balogh P, Elhawat N, El-Ramady H, Fári M. 2013. Phytoremediation of bauxite-derived red mud by giant reed. Environmental Chemistry Letters 11, 295–302. [Google Scholar]
- Angelini LG, Ceccarini L, Nassi o Di Nasso N, Bonari E. 2009. Comparison of Arundo donax L. and Miscanthus x giganteus in a long-term field experiment in Central Italy: analysis of productive characteristics and energy balance. Biomass and Bioenergy 33, 635–643. [Google Scholar]
- Anisimova M, Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Systematic Biology 55, 539–552. [DOI] [PubMed] [Google Scholar]
- Atma W, Larouci M, Meddah B, Benabdeli K, Sonnet P. 2017. Evaluation of the phytoremediation potential of Arundo donax L. for nickel-contaminated soil. International Journal of Phytoremediation 19, 377–386. [DOI] [PubMed] [Google Scholar]
- Barbosa B, Boléo S, Sidella S, Costa J, Duarte MP, Mendes B, Cosentino SL, Fernando AL. 2015. Phytoremediation of heavy metal-contaminated soils using the perennial energy crops Miscanthus spp. and Arundo donax L. Bioenergy Research 8, 1500–1511. [Google Scholar]
- Bellini E, Borsò M, Betti C, Bruno L, Andreucci A, Ruffini Castiglione M, Saba A, Sanità di Toppi L. 2019. Characterization and quantification of thiol-peptides in Arabidopsis thaliana using combined dilution and high sensitivity HPLC-ESI-MS-MS. Phytochemistry 164, 215–222. [DOI] [PubMed] [Google Scholar]
- Bhargava P, Srivastava AK, Urmil S, Rai LC. 2005. Phytochelatin plays a role in UV-B tolerance in N2-fixing cyanobacterium Anabaena doliolum. Journal of Plant Physiology 162, 1220–1225. [DOI] [PubMed] [Google Scholar]
- Blum R, Beck A, Korte A, Stengel A, Letzel T, Lendzian K, Grill E. 2007. Function of phytochelatin synthase in catabolism of glutathione-conjugates. The Plant Journal 49, 740–749. [DOI] [PubMed] [Google Scholar]
- Boc A, Diallo AB, Makarenkov V. 2012. T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Research 40, W573–W579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonanno G. 2012. Arundo donax as a potential biomonitor of trace element contamination in water and sediment. Ecotoxicology and Environmental Safety 80, 20–27. [DOI] [PubMed] [Google Scholar]
- Brown GR, Gill GP, Kuntz RJ, Langley CH, Neale DB. 2004. Nucleotide diversity and linkage disequilibrium in loblolly pine. Proceedings of the National Academy of Sciences, USA 101, 15255–15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalé AC, Clemens S. 2001. Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS Letters 507, 215–219. [DOI] [PubMed] [Google Scholar]
- Chaurasia N, Mishra Y, Rai LC. 2008. Cloning expression and analysis of phytochelatin synthase (pcs) gene from Anabaena sp. PCC 7120 offering multiple stress tolerance in Escherichia coli. Biochemical and Biophysical Research Communications 376, 225–230. [DOI] [PubMed] [Google Scholar]
- Clemens S. 2006. Evolution and function of phytochelatin synthases. Journal of Plant Physiology 163, 319–332. [DOI] [PubMed] [Google Scholar]
- Clemens S, Palmgren MG, Krämer U. 2002. A long way ahead: understanding and engineering plant metal accumulation. Trends in Plant Science 7, 309–315. [DOI] [PubMed] [Google Scholar]
- Clemens S, Schroeder JI, Degenkolb T. 2001. Caenorhabditis elegans expresses a functional phytochelatin synthase. European Journal of Biochemistry 268, 3640–3643. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cobbett C, Goldsbrough P. 2002. Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annual Review of Plant Biology 53, 159–182. [DOI] [PubMed] [Google Scholar]
- Das N, Bhattacharya S, Bhattacharyya S, Maiti MK. 2017. Identification of alternatively spliced transcripts of rice phytochelatin synthase 2 gene OsPCS2 involved in mitigation of cadmium and arsenic stresses. Plant Molecular Biology 94, 167–183. [DOI] [PubMed] [Google Scholar]
- Degola F, De Benedictis M, Petraglia A, Massimi A, Fattorini L, Sorbo S, Basile A, Sanità di Toppi L. 2014. A Cd/Fe/Zn-responsive phytochelatin synthase is constitutively present in the ancient liverwort Lunularia cruciata (L.) Dumort. Plant & Cell Physiology 55, 1884–1891. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11–15. [Google Scholar]
- Elhawat N, Alshaal T, Domokos-Szabolcsy É, et al. 2014. Phytoaccumulation potentials of two biotechnologically propagated ecotypes of Arundo donax in copper-contaminated synthetic wastewater. Environmental Science and Pollution Research International 21, 7773–7780. [DOI] [PubMed] [Google Scholar]
- Fan W, Guo Q, Liu C, Liu X, Zhang M, Long D, Xiang Z, Zhao A. 2018. Two mulberry phytochelatin synthase genes confer zinc/cadmium tolerance and accumulation in transgenic Arabidopsis and tobacco. Gene 645, 95–104. [DOI] [PubMed] [Google Scholar]
- Fernando AL, Barbosa B, Costa J, Papazoglou EG. 2016. Giant reed (Arundo donax L.): A multipurpose crop bridging phytoremediation with sustainable bioeconomy. In: Prasad MNV, ed. Bioremediation and bioeconomy. Amsterdam: Elsevier, 77–95. [Google Scholar]
- Fontanini D, Andreucci A, Ruffini Castiglione M, et al. 2018. The phytochelatin synthase from Nitella mucronata (Charophyta) plays a role in the homeostatic control of iron(II)/(III). Plant Physiology and Biochemistry 127, 88–96. [DOI] [PubMed] [Google Scholar]
- Fraústo da Silva JJR, Williams RJP. 2001. The biological chemistry of the elements: the inorganic chemistry of life. Oxford: Oxford University Press. [Google Scholar]
- Fu Y, Poli M, Sablok G, Wang B, Liang Y, La Porta N, Velikova V, Loreto F, Li M, Varotto C. 2016. Dissection of early transcriptional responses to water stress in Arundo donax L. by unigene-based RNA-seq. Biotechnology for Biofuels 9, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2, 31–34. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Löffler S, Winnacker EL, Zenk MH. 1989. Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific gamma-glutamylcysteine dipeptidyl transpeptidase (phytochelatin synthase). Proceedings of the National Academy of Sciences, USA 86, 6838–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E, Winnacker EL, Zenk MH. 1985. Phytochelatins: the principal heavy-metal complexing peptides of higher plants. Science 230, 674–676. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Guo J, Dai X, Xu W, Ma M. 2008. Overexpressing GSH1 and AsPCS1 simultaneously increases the tolerance and accumulation of cadmium and arsenic in Arabidopsis thaliana. Chemosphere 72, 1020–1026. [DOI] [PubMed] [Google Scholar]
- Guo ZH, Miao XF. 2010. Growth changes and tissues anatomical characteristics of giant reed (Arundo donax L.) in soil contaminated with arsenic, cadmium and lead. Journal of Central South University of Technology 17, 770–777. [Google Scholar]
- Ha SB, Smith AP, Howden R, Dietrich WM, Bugg S, O’Connell MJ, Goldsbrough PB, Cobbett CS. 1999. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. The Plant Cell 11, 1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41, 95–98. [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4, 1–9. [Google Scholar]
- Hayashi S, Kuramata M, Abe T, Takagi H, Ozawa K, Ishikawa S. 2017. Phytochelatin synthase OsPCS1 plays a crucial role in reducing arsenic levels in rice grains. The Plant Journal 91, 840–848. [DOI] [PubMed] [Google Scholar]
- Hoffmann RD, Palmgren M. 2016. Purifying selection acts on coding and non-coding sequences of paralogous genes in Arabidopsis thaliana. BMC Genomics 17, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Sun R, Hu Y, et al. 2016. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Molecular Biology and Evolution 33, 394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järup L. 2003. Hazards of heavy metal contamination. British Medical Bulletin 68, 167–182. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Jozefczak M, Remans T, Vangronsveld J, Cuypers A. 2012. Glutathione is a key player in metal-induced oxidative stress defenses. International Journal of Molecular Sciences 13, 3145–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausar S, Mahmood Q, Raja IA, Khan A, Sultan S, Gilani MA, Shujaat S. 2012. Potential of Arundo donax to treat chromium contamination. Ecological Egineering 42, 256–259. [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22, 3096–3098. [DOI] [PubMed] [Google Scholar]
- Krämer U. 2005. Phytoremediation: novel approaches to cleaning up polluted soils. Current Opinion in Biotechnology 16, 133–141. [DOI] [PubMed] [Google Scholar]
- Kühnlenz T, Schmidt H, Uraguchi S, Clemens S. 2014. Arabidopsis thaliana phytochelatin synthase 2 is constitutively active in vivo and can rescue the growth defect of the PCS1-deficient cad1-3 mutant on Cd-contaminated soil. Journal of Experimental Botany 65, 4241–4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. 2003. Overexpression of Arabidopsis phytochelatin synthase paradoxically leads to hypersensitivity to cadmium stress. Plant Physiology 131, 656–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DB, Hwang S. 2015. Tobacco phytochelatin synthase (NtPCS1) plays important roles in cadmium and arsenic tolerance and in early plant development in tobacco. Plant Biotechnology Reports 9, 107–114. [Google Scholar]
- Lee S, Kang BS. 2005. Expression of Arabidopsis phytochelatin synthase 2 is too low to complement an AtPCS1-defective Cad1-3 mutant. Molecules and Cells 19, 81–87. [PubMed] [Google Scholar]
- Lefort V, Longueville JE, Gascuel O. 2017. SMS: smart model selection in PhyML. Molecular Biology and Evolution 34, 2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guo J, Xu W, Ma M. 2007. RNA interference-mediated silencing of phytochelatin synthase gene reduce cadmium accumulation in rice seeds. Journal of Integrative Plant Biology 49, 1032–1037. [Google Scholar]
- Li M, Xu J, Algarra Alarcon A, Carlin S, Barbaro E, Cappellin L, Velikova V, Vrhovsek U, Loreto F, Varotto C. 2017. In planta recapitulation of isoprene synthase evolution from ocimene synthases. Molecular Biology and Evolution 34, 2583–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Gu C, Chen F, Yang D, Wu K, Chen S, Jiang J, Zhang Z. 2012. Heterologous expression of a Nelumbo nucifera phytochelatin synthase gene enhances cadmium tolerance in Arabidopsis thaliana. Applied Biochemistry and Biotechnology 166, 722–734. [DOI] [PubMed] [Google Scholar]
- Luo ZB, He J, Polle A, Rennenberg H. 2016. Heavy metal accumulation and signal transduction in herbaceous and woody plants: paving the way for enhancing phytoremediation efficiency. Biotechnology Advances 34, 1131–1148. [DOI] [PubMed] [Google Scholar]
- Mahar A, Wang P, Ali A, Awasthi MK, Lahori AH, Wang Q, Li R, Zhang Z. 2016. Challenges and opportunities in the phytoremediation of heavy metals contaminated soils: a review. Ecotoxicology and Environmental Safety 126, 111–121. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Shiraki K, Tsuji N, Hirata K, Miyamoto K, Takagi M. 2004. Functional analysis of phytochelatin synthase from Arabidopsis thaliana and its expression in Escherichia coli and Saccharomyces cerevisiae. Science and Technology of Advanced Materials 5, 377–381. [Google Scholar]
- Meyer CL, Peisker D, Courbot M, Craciun AR, Cazalé AC, Desgain D, Schat H, Clemens S, Verbruggen N. 2011. Isolation and characterization of Arabidopsis halleri and Thlaspi caerulescens phytochelatin synthases. Planta 234, 83–95. [DOI] [PubMed] [Google Scholar]
- Mirza N, Mahmood Q, Pervez A, Ahmad R, Farooq R, Shah MM, Azim MR. 2010. Phytoremediation potential of Arundo donax in arsenic-contaminated synthetic wastewater. Bioresource Technology 101, 5815–5819. [DOI] [PubMed] [Google Scholar]
- Mirza N, Pervez A, Mahmood Q, Shah MM, Shafqat MN. 2011. Ecological restoration of arsenic contaminated soil by Arundo donax L. Ecological Engineering 37, 1949–1956. [Google Scholar]
- Moudouma CFM, Gloaguen V, Riou C, Forestier L, Saladin G. 2012. High concentration of cadmium induces AtPCS2 gene expression in Arabidopsis thaliana (L.) Heynh ecotype Wassilewskija seedlings. Acta Physiologiae Plantarum 34, 1083–1091. [Google Scholar]
- Murrell B, Moola S, Mabona A, Weighill T, Sheward D, Kosakovsky Pond SL, Scheffler K. 2013. FUBAR: a Fast, Unconstrained Bayesian AppRoximation for inferring selection. Molecular Biology and Evolution 30, 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell B, Weaver S, Smith MD, et al. 2015. Gene-wide identification of episodic selection. Molecular Biology and Evolution 32, 1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagajyoti PC, Lee KD, Sreekanth TVM. 2010. Heavy metals, occurrence and toxicity for plants: a review. Environmental Chemistry Letters 8, 199–216. [Google Scholar]
- Oyuela Leguizamo MA, Fernández Gómez WD, Sarmiento MCG. 2017. Native herbaceous plant species with potential use in phytoremediation of heavy metals, spotlight on wetlands – a review. Chemosphere 168, 1230–1247. [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH. 2016. Evolution of gene duplication in plants. Plant Physiology 171, 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazoglou EG. 2007. Arundo donax L. stress tolerance under irrigation with heavy metal aqueous solutions. Desalination 211, 304–313. [Google Scholar]
- Papazoglou EG, Karantounias GA, Vemmos SN, Bouranis DL. 2005. Photosynthesis and growth responses of giant reed (Arundo donax L.) to the heavy metals Cd and Ni. Environment International 31, 243–249. [DOI] [PubMed] [Google Scholar]
- Papazoglou EG, Serelis KG, Bouranis DL. 2007. Impact of high cadmium and nickel soil concentration on selected physiological parameters of Arundo donax L. European Journal of Soil Biology 43, 207–215. [Google Scholar]
- Petraglia A, De Benedictis M, Degola F, Pastore G, Calcagno M, Ruotolo R, Mengoni A, Sanità di Toppi L. 2014. The capability to synthesize phytochelatins and the presence of constitutive and functional phytochelatin synthases are ancestral (plesiomorphic) characters for basal land plants. Journal of Experimental Botany 65, 1153–1163. [DOI] [PubMed] [Google Scholar]
- Peuke AD, Rennenberg H. 2005. Phytoremediation. EMBO Reports 6, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati S, Brazzale D, Guella G, Milli A, Ruberti C, Biasioli F, Zottini M, Moser C. 2014. The onset of grapevine berry ripening is characterized by ROS accumulation and lipoxygenase-mediated membrane peroxidation in the skin. BMC Plant Biology 14, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak N, Read DS, Jurkschat K, Matzke M, Kelly FJ, Spurgeon DJ, Stürzenbaum SR. 2014. Metalloproteins and phytochelatin synthase may confer protection against zinc oxide nanoparticle induced toxicity in Caenorhabditis elegans. Comparative Biochemistry and Physiology. Part C: Toxicology & Pharmacology 160, 75–85. [DOI] [PubMed] [Google Scholar]
- Ramos J, Clemente MR, Naya L, Loscos J, Pérez-Rontomé C, Sato S, Tabata S, Becana M. 2007. Phytochelatin synthases of the model legume Lotus japonicus. A small multigene family with differential response to cadmium and alternatively spliced variants. Plant Physiology 143, 1110–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J, Naya L, Gay M, Abián J, Becana M. 2008. Functional characterization of an unusual phytochelatin synthase, LjPCS3, of Lotus japonicus. Plant Physiology 148, 536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Williams DL. 2011. Characterization of the phytochelatin synthase of Schistosoma mansoni. PLoS Neglected Tropical Diseases 5, e1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea PA. 2012. Phytochelatin synthase: of a protease a peptide polymerase made. Physiologia Plantarum 145, 154–164. [DOI] [PubMed] [Google Scholar]
- Rea PA, Vatamaniuk OK, Rigden DJ. 2004. Weeds, worms, and more. Papain’s long-lost cousin, phytochelatin synthase. Plant Physiology 136, 2463–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redjala T, Sterckeman T, Morel JL. 2009. Cadmium uptake by roots: contribution of apoplast and of high- and low-affinity membrane transport systems. Environmental and Experimental Botany 67, 235–242. [Google Scholar]
- Richveisová BM, Dürešová Z, Horník M, Augustín J, Pipíška M. 2014. Distribution of zinc and cadmium in tissues of giant reed (Arundo donax L.): sequential extraction - radiometric study. Nova Biotechnologica et Chimica 13, 38–47. [Google Scholar]
- Romanyuk ND, Rigden DJ, Vatamaniuk OK, Lang A, Cahoon RE, Jez JM, Rea PA. 2006. Mutagenic definition of a papain-like catalytic triad, sufficiency of the N-terminal domain for single-site core catalytic enzyme acylation, and C-terminal domain for augmentative metal activation of a eukaryotic phytochelatin synthase. Plant Physiology 141, 858–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruotolo R, Peracchi A, Bolchi A, Infusini G, Amoresano A, Ottonello S. 2004. Domain organization of phytochelatin synthase: functional properties of truncated enzyme species identified by limited proteolysis. Journal of Biological Chemistry 279, 14686–14693. [DOI] [PubMed] [Google Scholar]
- Sabeen M, Mahmood Q, Irshad M, Fareed I, Khan A, Ullah F, Hussain J, Hayat Y, Tabassum S. 2013. Cadmium phytoremediation by Arundo donax L. from contaminated soil and water. BioMed Research International 2013, 324830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablok G, Fu Y, Bobbio V, et al. 2014. Fuelling genetic and metabolic exploration of C3 bioenergy crops through the first reference transcriptome of Arundo donax L. Plant Biotechnology Journal 12, 554–567. [Google Scholar]
- Sagehashi M, Liu C, Fujii T, Fujita H, Sakai Y, Hu H-Y, Sakoda A. 2011. Cadmium removal by the hydroponic culture of giant reed (Arundo donax) and its concentration in the plant. Journal of Water and Environment Technology 9, 121–127. [Google Scholar]
- Salt DE, Smith RD, Raskin I. 1998. Phytoremediation. Annual Review of Plant Physiology and Plant Molecular Biology 49, 643–668. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF. 2012. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. The Plant Cell 24, 2155–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine AM, Shakya VPS, Idnurm A. 2015. Phytochelatin synthase is required for tolerating metal toxicity in a basidiomycete yeast and is a conserved factor involved in metal homeostasis in fungi. Fungal Biology and Biotechnology 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Molecular Biology and Evolution 32, 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WY, Mendoza-Cózatl DG, Lee Y, Schroeder JI, Ahn SN, Lee HS, Wicker T, Martinoia E. 2014. Phytochelatin–metal(loid) transport into vacuoles shows different substrate preferences in barley and Arabidopsis. Plant, Cell & Environment 37, 1192–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza LA, Piotto FA, Nogueirol RC, Azevedo RA. 2013. Use of non-hyperaccumulator plant species for the phytoextraction of heavy metals using chelating agents. Scientia Agricola 70, 290–295. [Google Scholar]
- Stöver BC, Müller KF. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh B, Ravishankar GA. 2004. Phytoremediation—a novel and promising approach for environmental clean-up. Critical Reviews in Biotechnology 24, 97–124. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology 56, 564–577. [DOI] [PubMed] [Google Scholar]
- Tatarinova TV, Chekalin E, Nikolsky Y, Bruskin S, Chebotarov D, McNally KL, Alexandrov N. 2016. Nucleotide diversity analysis highlights functionally important genomic regions. Scientific Reports 6, 35730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. 2012. Heavy metals toxicity and the environment. Molecular, Clinical and Environmental Toxicology 101, 133–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J, Bannigan J. 2008. Cadmium: toxic effects on the reproductive system and the embryo. Reproductive Toxicology 25, 304–315. [DOI] [PubMed] [Google Scholar]
- Uraguchi S, Fujiwara T. 2013. Rice breaks ground for cadmium-free cereals. Current Opinion in Plant Biology 16, 328–334. [DOI] [PubMed] [Google Scholar]
- Uraguchi S, Tanaka N, Hofmann C, et al. 2017. Phytochelatin synthase has contrasting effects on cadmium and arsenic accumulation in rice grains. Plant & Cell Physiology 58, 1730–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ent A, Baker AJM, Reeves RD, Pollard AJ, Schat H. 2013. Hyperaccumulators of metal and metalloid trace elements: facts and fiction. Plant and Soil 362, 319–334. [Google Scholar]
- Vatamaniuk OK, Mari S, Lang A, Chalasani S, Demkiv LO, Rea PA. 2004. Phytochelatin synthase, a dipeptidyltransferase that undergoes multisite acylation with γ-glutamylcysteine during catalysis: stoichiometric and site-directed mutagenic analysis of Arabidopsis thaliana PCS1-catalyzed phytochelatin synthesis. Journal of Biological Chemistry 279, 22449–22460. [DOI] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lu YP, Rea PA. 1999. AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proceedings of the National Academy of Sciences, USA 96, 7110–7115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivares D, Arnoux P, Pignol D. 2005. A papain-like enzyme at work: native and acyl–enzyme intermediate structures in phytochelatin synthesis. Proceedings of the National Academy of Sciences, USA 102, 18848–18853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang J, Jin D, Guo H, Lee TH, Liu T, Paterson AH. 2015. Genome alignment spanning major Poaceae lineages reveals heterogeneous evolutionary rates and alters inferred dates for key evolutionary events. Molecular Plant 8, 885–898. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Molecular Biology and Evolution 32, 820–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SK. 2010. Heavy metals toxicity in plants: an overview on the role of glutathione and phytochelatins in heavy metal stress tolerance of plants. South African Journal of Botany 76, 167–179. [Google Scholar]
- Yamazaki S, Ueda Y, Mukai A, Ochiai K, Matoh T. 2018. Rice phytochelatin synthases OsPCS1 and OsPCS2 make different contributions to cadmium and arsenic tolerance. Plant Direct 2, e00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Sheng L, Zhang C, Deng H. 2018. Physiological response of Arundo donax to cadmium stress by Fourier transform infrared spectroscopy. Spectrochimica Acta. Part A: Molecular and Biomolecular Spectroscopy 198, 88–91. [DOI] [PubMed] [Google Scholar]
- Zhao C, Xu J, Li Q, Li S, Wang P, Xiang F. 2014. Cloning and characterization of a Phragmites australis phytochelatin synthase (PaPCS) and achieving Cd tolerance in tall fescue. PLoS ONE 9, e103771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Zhang J, Qian C, Zhou Q, Zhao X, Chen G, Ma XF. 2017. SNP discovery and genetic variation of candidate genes relevant to heat tolerance and agronomic traits in natural populations of sand rice (Agriophyllum squarrosum). Frontiers in Plant Science 8, 536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ge S. 2005. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytologist 167, 249–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.