Pyrenoid linker EPYC1 interacts with specific structures of the Rubisco small subunit. Modified plant Rubisco interacts with EPYC1 to form pyrenoid-like aggregates, a key feature of the algal CO2-concentrating mechanism.
Keywords: Arabidopsis thaliana, Chlamydomonas reinhardtii, chloroplast, CO2-concentrating mechanism, Nicotiana benthamiana, photosynthesis, pyrenoid
Abstract
Photosynthetic efficiencies in plants are restricted by the CO2-fixing enzyme Rubisco but could be enhanced by introducing a CO2-concentrating mechanism (CCM) from green algae, such as Chlamydomonas reinhardtii (hereafter Chlamydomonas). A key feature of the algal CCM is aggregation of Rubisco in the pyrenoid, a liquid-like organelle in the chloroplast. Here we have used a yeast two-hybrid system and higher plants to investigate the protein–protein interaction between Rubisco and essential pyrenoid component 1 (EPYC1), a linker protein required for Rubisco aggregation. We showed that EPYC1 interacts with the small subunit of Rubisco (SSU) from Chlamydomonas and that EPYC1 has at least five SSU interaction sites. Interaction is crucially dependent on the two surface-exposed α-helices of the Chlamydomonas SSU. EPYC1 could be localized to the chloroplast in higher plants and was not detrimental to growth when expressed stably in Arabidopsis with or without a Chlamydomonas SSU. Although EPYC1 interacted with Rubisco in planta, EPYC1 was a target for proteolytic degradation. Plants expressing EPYC1 did not show obvious evidence of Rubisco aggregation. Nevertheless, hybrid Arabidopsis Rubisco containing the Chlamydomonas SSU could phase separate into liquid droplets with purified EPYC1 in vitro, providing the first evidence of pyrenoid-like aggregation for Rubisco derived from a higher plant.
Introduction
Meeting the food demands of the rising global population is one of the most pressing challenges for plant science (Long et al., 2015; Simkin et al., 2019). The required increases are unlikely to be achieved entirely through traditional crop breeding approaches or agricultural land expansion, and agricultural sectors are increasingly looking to new genome engineering approaches for solutions (Borel et al., 2017; Waltz et al., 2017). Improving photosynthetic conversion efficiencies is a promising strategy to achieve this goal, and several recent examples in crops have demonstrated the capacity for photosynthetic enhancements in crop species using this approach (Kromdijk et al., 2016; Driever et al., 2017; Lopez-Calcagno et al., 2019). Much research is focused on overcoming the limitations of C3 photosynthesis, which is the most common form of CO2 assimilation in plants, including critical staple crops (e.g. rice, wheat, and soybean) (Simkin et al., 2019). In C3 plants, delivery of carbon dioxide (CO2) to chloroplasts occurs by passive diffusion, which limits photosynthetic efficiencies. A further limitation is imposed by the unfavourable catalytic properties of the primary carboxylase enzyme, Rubisco, located in the chloroplasts of leaf cells (Bathellier et al., 2018). Rubisco restricts CO2 assimilation in plants because it has a slow turnover rate and a relatively low affinity for CO2—it is only half-saturated with CO2 at concentrations inside C3 chloroplasts. To compensate, C3 plants invest up to 25% of leaf nitrogen into producing Rubisco (Parry et al., 2013). Rubisco also catalyses a competitive side reaction with oxygen (O2) that typically leads to substantial metabolic costs associated with a salvage process for the oxygenated product, called photorespiration (Busch et al., 2018). Photorespiration can result in a loss of productivity of up to 50% in C3 plants (South et al., 2018).
Several photosynthetic organisms, including cyanobacteria, algae, and a group of land plants called hornworts, have evolved biophysical CO2-concentrating mechanisms (CCMs) that actively increase the CO2 concentration around Rubisco. Researchers are currently working towards the introduction of these CCMs into C3 plants to increase the carboxylation efficiency of Rubisco and reduce photorespiration (Rae et al., 2017; Long et al., 2018). Engineering a biophysical CCM into higher plants is predicted to significantly enhance photosynthetic rates and productivity (Price et al., 2013; McGrath and Long, 2014; Yin and Struik, 2017). The eukaryotic algal CCM is composed of inorganic carbon (Ci) transporters at the plasma membrane and chloroplast envelope, which work together to deliver above ambient concentrations of CO2 to Rubisco within a chloroplastic microcompartment called the pyrenoid (Yamano et al., 2015; Mackinder et al., 2017; Zhan et al., 2018). Theoretical modelling approaches have demonstrated the requirement for a pyrenoid in algal systems (Badger et al., 1998) and shown that some form of microcompartment containing Rubisco would also be needed for a successful CCM in higher plant systems (Price et al., 2013).
The algal CCM is an attractive target for transfer as key components of the CCM localize appropriately in higher plants (Atkinson et al., 2016). Furthermore, the Rubisco small subunit (SSU; encoded by the rbcS nuclear gene family) of Chlamydomonas reinhardtii (hereafter Chlamydomonas) can complement severely SSU-deficient Arabidopsis mutants (Atkinson et al., 2017), with only small impacts on Rubisco function and assembly, and plant growth close to wild-type levels reported. In contrast, the replacement of tobacco Rubisco with cyanobacterial Rubisco produced poorer growing transplastomic plants, even when grown at greatly elevated CO2 concentrations, due to the low affinity of cyanobacterial Rubisco for CO2 and its low level of expression (Lin et al., 2014; Occhialini et al., 2016; Long et al., 2018). Thus, the challenges of introducing an algal-based CCM incorporating appropriate structural modifications that enable Rubisco aggregation might be readily testable without large perturbations to plant growth or a requirement for high CO2.
Recent work in Chlamydomonas has revealed new details about the structure and composition of the pyrenoid as a highly dynamic organelle with liquid-like properties rather than a static solid or crystalline structure (Engel et al., 2015; Freeman Rosenzweig et al., 2017; Mackinder et al., 2017). When the CCM is active, the pyrenoid matrix is composed primarily of Rubisco, Rubisco activase, and essential pyrenoid component 1 (EPYC1), a Rubisco linker protein important for Rubisco packaging in the pyrenoid (Mackinder et al., 2016). EPYC1 is an intrinsically disordered protein containing four repeated regions that are predicted to bind Rubisco via multiple low affinity interactions to promote phase transitions, which in turn allow dynamic internal rearrangement within the pyrenoid consistent with its liquid-like organization (Hyman et al., 2014; Freeman Rosenzweig et al., 2017; Yizhi et al., 2018). Pyrenoid formation is also dependent on the amino acid composition of the Rubisco SSU and, more specifically, on two surface-exposed α-helices, which differ markedly between Chlamydomonas and higher plants (Meyer et al., 2012). Recent work has shown that Rubisco from Chlamydomonas and EPYC1 are necessary and sufficient to induce the liquid–liquid phase separation observed in pyrenoids and that such interactions with EPYC1 are strongly influenced by the presence of the Chlamydomonas SSU (Wunder et al., 2018). Currently little is known regarding the potential interactive binding sites between EPYC1 and Rubisco.
Here we reveal new insights into the protein–protein interaction between EPYC1 and Rubisco through their expression in two heterologous systems: yeast and higher plants. When expressed in planta, EPYC1 appeared unstable. However, hybrid Rubisco complexes from Arabidopsis were able to phase separate with purified EPYC1 in vitro, thus recapitulating the liquid-like behaviour of the Chlamydomonas pyrenoid. We discuss the implications of these findings for ongoing efforts to introduce a functional algal CCM into a higher plant.
Materials and methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana, Col-0) seeds were sown on compost, stratified for 3 d at 4 °C, and grown at 20 °C, ambient CO2, 70% relative humidity, and 150 μmol photons m−2 s−1 in 12 h light, 12 h dark. For comparisons of different genotypes, plants were grown from seeds of the same age and storage history, harvested from plants grown in the same environmental conditions. Nicotiana benthamiana (L.) was grown at 20 °C with 150 μmol photons m−2 s−1 in 12 h light, 12 h dark.
Construct design and transformation
The coding sequence of EPYC1 was codon optimized for expression in higher plants using an online tool (www.idtdna.com/CodonOpt). All variants of EPYC1 were synthesized as Gblock fragments (IDT) and cloned directly into level 0 acceptor vectors (pAGM1299 and pICH41264) of the Plant MoClo system (Engler et al., 2014) or pB7WG2,0 vectors containing C- or N-terminal yellow fluorescent protein (YFP; Supplementary Table S1 at JXB online). To generate fusion proteins, gene expression constructs were assembled into binary level M acceptor vectors. Level M vectors were transformed into Agrobacterium tumefaciens (AGL1) for transient gene expression in N. benthamiana (Schöb et al., 1997) or stable insertion in Arabidopsis plants by floral dipping (Clough and Bent, 1998). Homozygous insertion lines were identified in the T3 generation using the pFAST-R selection cassette (Shimada et al., 2010).
DNA and leaf protein analyses
PCRs were performed as in McCormick and Kruger (2015) using gene-specific primers (Supplementary Table S2). Soluble protein was extracted from frozen leaf material of 21-d-old plants (sixth and seventh leaf) in 5× Bolt LDS sample buffer (ThermoFisher Scientific) with 200 mM DTT at 70 °C for 15 min. Extracts were centrifuged and the supernatants subjected to SDS–PAGE on a 4–12% (w/v) polyacrylamide gel and transferred to a nitrocellulose membrane. Membranes were probed with rabbit serum raised against wheat Rubisco at 1:10 000 dilution (Howe et al., 1982) or against EPYC1 at 1:2000 dilution (Mackinder et al., 2016), followed by horseradish peroxidase (HRP)-linked goat anti-rabbit IgG (Abcam) at 1:10 000 dilution, and visualized using Pierce ECL Western Blotting Substrate (Life Technologies).
Growth analysis and photosynthetic measurements
Rosette growth rates were quantified using an in-house imaging system (Dobrescu et al., 2017). Maximum quantum yield of PSII (Fv/Fm) was measured using a Hansatech Handy PEA continuous excitation chlorophyll fluorimeter (Hansatech Instruments Ltd). (Maxwell and Johnson, 2000).
Co-immunoprecipitation (co-IP)
Rosettes of 35-day-old Arabidopsis plants expressing EPYC1 in a complemented Rubisco mutant background (S2Cr, 1AAtMOD, or 1AAt) were snap-frozen and ground in liquid N2. An equal volume of IP extraction buffer [100 mM HEPES (pH 7.5), 150 mM NaCl, 4 mM EDTA, 5 mM DTT, 0.4 mM phenylmethylsulfonyl fluoride (PMSF), 10% (v/v) glycerol, 0.1% (v/v) Triton X-100, and one Roche cOmplete EDTA-free protease inhibitor tablet per 10 ml] was added, samples were rotated at 4 °C for 15 min, centrifuged at 4 °C, and filtered through two layers of Miracloth (Merck). Each extract (2 ml) was pre-cleared by incubating with 50 µl of Protein A Dynabeads (ThermoFisher Scientific) pre-equilibrated in IP buffer for 1 h at 4 °C, before discarding the beads. Antibody-coated beads were generated by applying 3.5 µg of anti-EPYC1 antibody to 50 µl of Protein A Dynabeads, which were then rotated at 4 °C for 30 min. The antibody was cross-linked to the beads using Pierce BS3 cross-linking agent (Thermo Scientific). Each protein extract was incubated with the antibody-coated beads and rotated at 4 °C for 2 h. Unbound sample (flow-through) was discarded and the beads were washed four times with washing buffer [20 mM Tris–HCl (pH 8), 150 mM NaCl, 0.1% (w/v) SDS, 1% (v/v) Triton X-100, 2 mM EDTA]. Immunocomplexes were eluted by adding 50 µl of elution buffer (2× LDS sample buffer, 200 mM DTT) and heating for 15 min at 70 °C, before discarding beads and subjecting to SDS–PAGE and immunoblotting.
Immunogold labelling and electron microscopy
Leaf samples were taken from 21-day-old S2Cr and S2Cr_EPYC1 plants and fixed with with 4% (v/v) paraformaldehyde, 0.5% (v/v) glutaraldehyde, and 0.05 M sodium cacodylate (pH 7.2). Leaf strips (1 mm wide) were vacuum infiltrated with fixative three times for 15 min, then rotated overnight at 4 °C. Samples were rinsed three times with phosphate-buffered saline (PBS) then dehydrated sequentially by vacuum infiltrating with 50, 70, 80, and 90% ethanol (v/v) for 1 h each, then three times with 100%. Samples were infiltrated with increasing concentrations of LR White Resin [30, 50, and 70% (w/v)] mixed with ethanol for 1 h each, then with 100% resin three times. The resin was polymerized in capsules at 50 °C overnight. Sections (1 μm thick) were cut on a Leica Ultracut ultramicrotome, stained with Toluidine Blue, and viewed with a light microscope to select suitable areas for investigation. Ultrathin sections (60 nm thick) were cut from selected areas and mounted onto plastic-coated copper grids. Grids were blocked with 1% (w/v) BSA in TBSTT [Tris-buffered saline with 0.05% (v/v) Triton X-100 and 0.05% (v/v) Tween-20], incubated overnight with anti-Rubisco antibody in TBSTT at 1:250 dilution, and washed twice each with TBSTT and water. Incubation with 15 nm gold particle-conjugated goat anti-rabbit secondary antibody (Abcam) in TBSTT was carried out for 1 h at 1:200 dilution, before washing as before. Grids were stained in 2% (w/v) uranyl acetate then viewed in a JEOL JEM-1400 Plus transmission electron microscope. Images were collected on a GATAN OneView camera.
Confocal laser scanning microscopy
Leaves were imaged with a Leica TCS SP2 laser scanning confocal microscope (Leica) as in Atkinson et al. (2016).
Protein production, droplet sedimentation assay, and microscopy
Rubisco was purified from 25- to 30-day-old Arabidopsis rosettes using a combination of ammonium sulfate precipitation, ion-exchange chromatography, and gel filtration (Shivhare and Mueller-Cajar, 2017). Rubisco was purified from Chlamydomonas cells (CC-2677), and EPYC1 and EPYC1::GFP (green fluorescent protein) was produced in Escherichia coli and purified as described in Wunder et al. (2018). EPYC1–Rubisco droplets reconstituted at room temperature in 10 µl reactions for 5 min in buffer A [20 mM Tris–HCl (pH 8.0), and 50 mM NaCl] were separated at 4 °C from the bulk solution by centrifugation for 4 min at 21 100 g. Pellet (droplet) and supernatant (bulk solution) fractions were subjected to SDS–PAGE and Coomassie staining. For light and fluorescence microscopy, reaction solutions (5 µl) were imaged after 3–5 min with a Nikon Eclipse Ti Inverted Microscope using the settings for differential interference contrast and epifluorescence microscopy (using fluorescein isothiocyanate filter settings) with a ×100 oil-immersion objective focusing on the coverslip surface. The coverslips used were 22×22 mm (Superior Marienfeld, Germany) and fixed in one-well Chamlide CMS chamber for 22×22 coverslips (Live Cell Instrument, South Korea). ImageJ was used to pseudocolour all images.
Liquid chromatography–mass spectrometry
Cell lysate was prepared from Chlamydomonas cells according to Mackinder et al. (2016). Following membrane solubilization with 2% (w/v) digitonin, the clarified lysate was applied to 150 µl of Protein A Dynabeads that had been incubated with 20 µg of anti-EPYC1 antibody. The Dynabead–cell lysate was incubated for 1.5 h with rotation at 4 °C. The beads were then washed four times with IP buffer [50 mM HEPES, 50 mM KOAc, 2 mM Mg(OAc)2·4H2O, 1 mM CaCl2, 200 mM sorbitol, 1 mM NaF, 0.3 mM NA3VO4, Roche cOmplete EDTA-free protease inhibitor] containing 0.1% (w/v) digitonin. EPYC1 was eluted from the beads by incubating for 10 min in elution buffer [50 mM Tris–HCl, 0.2 M glycine (pH 2.6)], and the eluate was immediately neutralized with 1:10 (v/v) Tris–HCl (pH 8.5). A small amount of the eluate was run on an SDS–PAGE gel and stained with Coomassie (Supplementary Fig. S6A), and the remaining sample was used for LC-MS.
Intact protein LC-MS experiments were performed on a Synapt G2 Q-ToF instrument equipped with electrospray ionization (Waters Corp., Manchester, UK). LC separation was achieved using an Acquity UPLC equipped with a reverse phase C4 Aeris Widepore 50×2.1 mm HPLC column (Phenomenex, CA, USA), and a gradient of 5–95% acetonitrile (0.1% formic acid) over 10 min was employed. Data analysis was performed using MassLynx v4.1, and deconvolution was performed using MaxEnt.
Yeast two-hybrid (Y2H) assay
The two-hybrid plasmid vectors pGBKT7 and pGADT7 were used to detect interactions between proteins of interest. Genes were amplified using Q5 DNA polymerase (NEB) and cloned into each vector using the multiple cloning site, thus creating fusions with either the GAL4-DNA binding domain or activation domain, respectively (Supplementary Table S1). Competent yeast cells (Y2H Gold, Clontech) were prepared from a 50 ml culture grown in YPDA medium supplemented with kanamycin (50 µg ml−1). Cells were washed with ddH2O and a lithium acetate/TE solution [100 mM LiAc, 10 mM Tris-HCl (pH 7.5), 1 mM EDTA] before re-suspending in lithium acetate/TE solution. Cells were then co-transformed with binding and activation domain vectors by mixing 50 µl of competent cells with 1 µg of each plasmid vector and a polyethylene (PEG) solution [100 mM LiAc, 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 40% (v/v) PEG 4000]. Cells were incubated at 30 °C for 30 min, then subjected to a heat shock of 42 °C for 20 min. The cells were centrifuged, re-suspended in 500 µl of YPDA, and incubated at 30 °C for ~90 min, then centrifuged and washed in TE. The pellet was re-suspended in 200 µl of TE, spread onto SD-L-W (standard dextrose medium lacking leucine and tryptophan; Anachem), and grown for 3 d at 30 °C. Ten to fifteen of the resulting colonies were pooled per co-transformation and grown in a single culture for 24 h. The following day, 1 ml of culture was harvested, cell density (OD600) measured, centrifuged, and then diluted in TE to give a final OD600 of 0.5 or 0.1. Yeast cultures were then plated onto SD-L-W and SD-L-W-H (Anachem) containing different concentrations of the HIS3 inhibitor tri-aminotriazole (3-AT), and incubated for 3 d before assessing for the presence or absence of growth, as in van Nues and Beggs (2001). Protein extraction was carried out by re-suspending cells to an OD600 of 1 from an overnight liquid culture in a lysis buffer [50 mM Tris–HCl (pH 6), 4% (v/v) SDS, 8 M urea, 30% (v/v) glycerol, 0.1 M DTT, 0.005% (w/v) Bromophenol Blue], incubating at 65 °C for 30 min, and loading directly onto a 10% (w/v) Bis-Tris protein gel (Expedeon).
Results
EPYC1 interacts with the small subunit of Rubisco
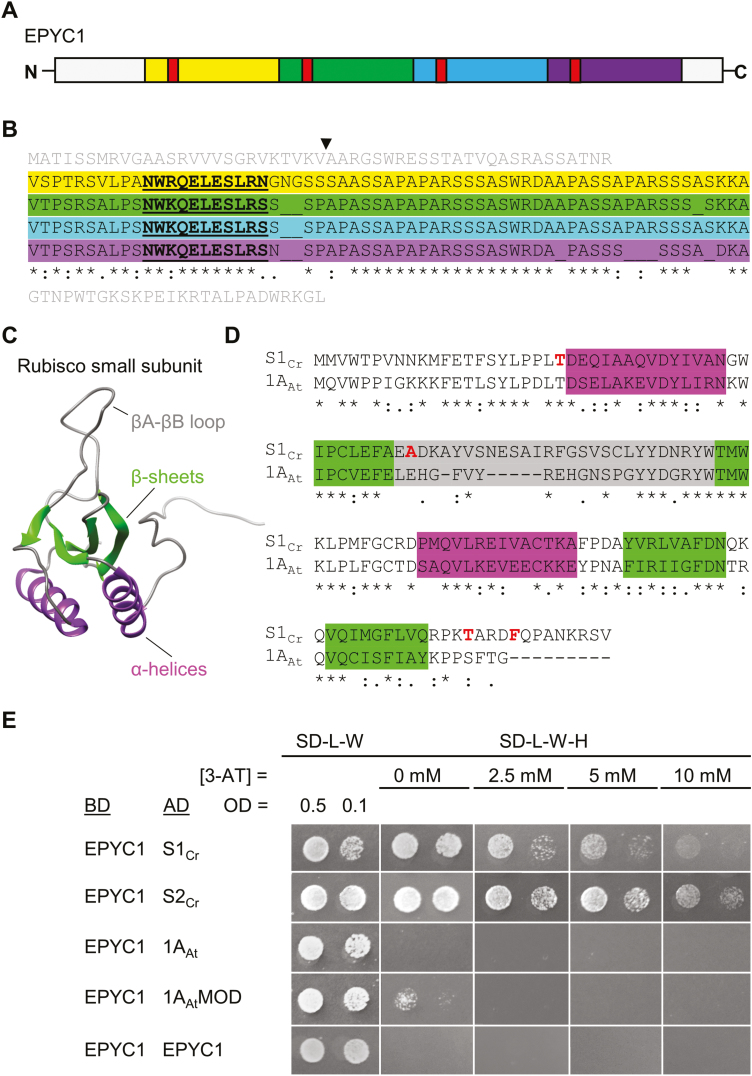
The two α-helices of the Chlamydomonas SSU have been proposed previously as potential binding sites for EPYC1 (Fig. 1A–D) (Meyer et al., 2012; Mackinder et al., 2016). We set out to test this hypothesis using a semi-quantitative Y2H approach. Chlamydomonas has two similar SSU homologues, S1Cr and S2Cr, which are the same size, have identical α-helices and β-sheets, but differ in amino acid sequence by four residues. EPYC1 showed a relatively strong protein–protein interaction with both S1Cr and S2Cr (i.e. growth at 10 mM 3-AT) (Fig. 1E). In contrast, EPYC1 did not interact with the 1A SSU from Arabidopsis (1AAt) but did interact weakly with a hybrid 1A SSU carrying the α-helices from Chlamydomonas (1AAtMOD) (Atkinson et al., 2017). The Y2H assay showed that EPYC1 does not interact with itself, other Chlamydomonas CCM components associated with the pyrenoid (i.e. LCIB, LCIC, and CAH3), or another intrinsically disordered protein found in the chloroplast stroma (AtCP12; Lopez-Calcagno et al., 2014) (Supplementary Fig. S1). These results indicate that EPYC1 is not prone to false-positive protein–protein interactions in Y2H assays.
Fig. 1.
EPYC1 interacts with the Rubisco small subunit. (A and B) EPYC1 consists of four near identical repeat regions (highlighted in yellow, green, blue, and purple), each containing a predicted α-helix (red, underlined). The putative cleavage site of the chloroplastic transit peptide between 26 (V) and 27 (A) is indicated. The N- and C- termini are shown in grey. (C) The predicted model of the small subunit of Rubisco (SSU; 1AAt shown) has similar features in algae and higher plants, including four β-sheet (green) and two α-helical (purple) regions. (D) Amino acid alignments of mature Arabidopsis SSU 1A (1AAt) and Chlamydomonas SSU 1 (S1Cr). Residues in red indicate the four amino acids that differ between the two Chlamydomonas SSUs, S1Cr and S2Cr (T/S, A/S, T/S, F/W). (E) Yeast two-hybrid interactions with EPYC1. Interaction strength is demonstrated by growth on increasing concentrations of the inhibitor 3-AT. Abbreviations: 3-AT, 3-amino-1,2,4-triazole; BD, binding domain; AD, activation domain; 1AAtMOD, modified 1AAt carrying the two α-helical regions from Chlamydomonas; SD-L-W, yeast synthetic minimal medium (SD medium) lacking leucine (L) and tryptophan (W); SD-L-W-H, SD medium lacking L, W, and histidine (H). See Supplementary Fig. S1 for Y2H additional controls.
The α-helices of the SSU are critical for interaction with EPYC1
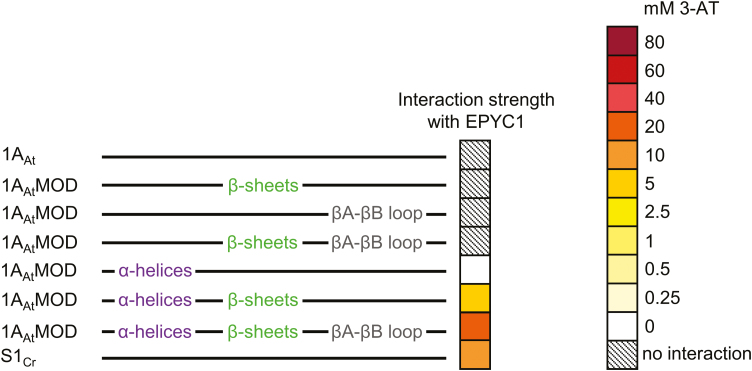
We next set out to identify the key domains on the Chlamydomonas SSU required for interaction with EPYC1. To isolate the structural components of the SSU, we generated a total of six different chimeric versions of 1AAt bearing residues from S1Cr associated with the three distinct β-sheets (βA, βC, and βD), the βA–βB loop, and the two α-helices (αA and αB) (Spreitzer, 2003) (Fig. 1C; Supplementary Fig. S2). As before, EPYC1 did not interact with 1AAt, and the addition of the β-sheets or the βA–βB loop from S1Cr, or both together, did not permit interaction (Fig. 2; Supplementary Fig. S3A). Interactions were only observed in the presence of the two α-helices from the Chlamydomonas SSU. The α-helices alone produced a minimal interaction (i.e. on 0 mM 3-AT), which was strengthened by the incorporation of the β-sheets and the βA–βB loop from S1Cr. Notably, the modified 1AAt variant with the α-helices, β-sheets, and βA–βB loop from Chlamydomonas (i.e. 79% sequence identity to S1Cr) showed a stronger interaction compared with S1Cr, indicating that SSUs can be engineered for increased affinity for EPYC1.
Fig. 2.
EPYC1 requires the α-helices of the Chlamydomonas SSU for interaction. Interaction is shown between EPYC1 and modified versions of 1AAt (top), in which different components (see Fig. 1C, D) have been switched for those from Chlamydomonas S1Cr (bottom). The heat map indicates interaction strength measured with yeast two-hybrid assays by the capacity for growth on increasing concentrations of 3-AT (mM). See Supplementary Fig. S2 for SSU sequences and Supplementary Fig. S3A for raw Y2H image data.
EPYC1 is a modular protein, and the predicted α-helices are important for interaction with the SSU
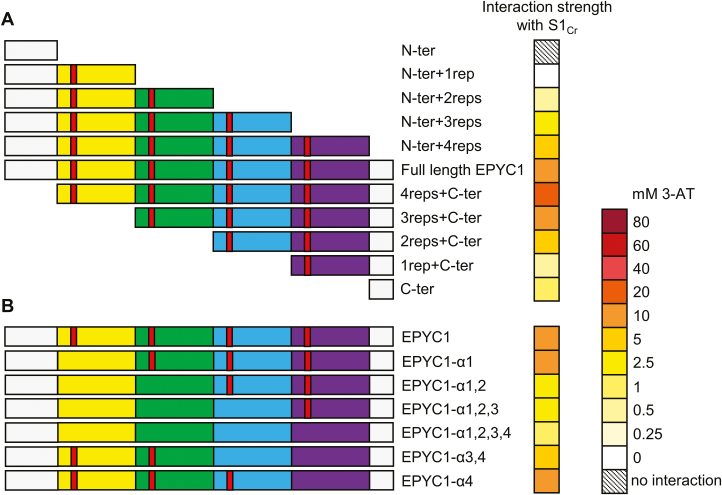
We initially generated a variety of truncated EPYC1 variants to characterize the key regions of EPYC1 required for interaction with the SSU. EPYC1 is a modular protein consisting of four highly similar repeat sequences flanked by shorter terminal regions. Thus, truncations were made to eliminate each region sequentially from either the N- or C-terminus direction (Fig. 3A; Supplementary Figs S3B, S4). Truncated EPYC1 variants were expressed well in yeast (Supplementary Fig. S5). In Y2H experiments, the EPYC1 N-terminus alone (N-ter) did not interact with S1Cr. Addition of the first EPYC1 repeat region was sufficient to detect interaction, and each subsequent repeat region correlated with growth at increased concentrations of 3-AT, indicating that EPYC1 is a modular protein and each repeat has an additive effect on SSU interaction. Addition of the C-terminal tail further increased the strength of the interaction. Interestingly, the C-terminus alone interacted with S1Cr, suggesting that SSU-binding sites are not limited to the repeat regions. Removal of the N-terminus also increased the interaction strength, which is consistent with the predicted role of the N-terminus as a chloroplastic transit peptide that would be cleaved during import into the chloroplast (Mackinder et al., 2016). Prediction tools ChloroP and PredAlgo suggest cleavage at residues 78 and 170, respectively (Emanuelsson et al., 2007). However, both predictions are unconvincing as they would result in cleavage within the repeat regions. To identify the potential cleavage site, we immunoprecipitated and analysed EPYC1 from Chlamydomonas using electrospray ionization-MS (ESI-MS). Intact protein ESI-MS analysis revealed several proteoforms of mature EPYC1 ranging from 29 622 Da to 30 621 Da (Supplementary Fig. S6). The molecular mass difference between proteoforms was 80 Da, suggesting variable phosphorylation states; an observation consistent with previous reports highlighting the highly phosphorylated nature of EPYC1 (Turkina et al., 2006; Wang et al., 2014). The highly post-translationally modified state of EPYC1 made determination of the precise molecular mass of the mature protein difficult. However, the smallest proteoform identified had a molecular mass of 29.6 kDa which, based on the theoretical mass of EPYC1, indicates a cleavage site between residues 26 (V) and 27 (A) (Fig. 1B).
Fig. 3.
EPYC1 repeat regions contribute to interaction with the Rubisco small subunit. (A) Decreasing the number of EPYC1 repeat regions reduced the strength of interaction with S1Cr. (B) The predicted α-helical region in each repeat (red) is important for interaction with S1Cr. These were eliminated by matutation to seven alanines in each of the different EPYC1 variants. The heat map indicates interaction strength measured with yeast two-hybrid assays by the capacity for growth on increasing concentrations of 3-AT (mM). See Supplementary Fig. S3B and C for raw Y2H image data.
We hypothesized that the interaction between EPYC1 and the SSU may be mediated through a predicted α-helix conserved in each of the four repeats, which together would allow EPYC1 to bind at least four Rubisco complexes (Mackinder et al., 2016; Freeman Rosenzweig et al., 2017). The relative contribution of each of the four domains was analysed by eliminating the predicted α-helical structure through mutation of the residues ‘RQELESL’ in the first repeat and ‘KQELESL’ in the subsequent three repeats to seven alanines. Mutation of a single helix did not have an impact on interaction strength. However, sequentially weaker interactions with S1Cr were observed with increasing mutations of the α-helical regions (Fig. 3B; Supplementary Figs S3C, S4). If all four α-helices were mutated, the interaction was not eradicated completely. The latter finding supported our evidence for an additional SSU-binding site(s) on the C-terminus, as in the absence of all α-helices the interaction strength was reduced to the same extent as the C-terminus alone (Fig. 3A). Overall, our data suggest that EPYC1 has at least five SSU interaction sites, located in each of its four repeat regions and the C-terminus, respectively.
EPYC1 can be modified to improve interaction strength with the SSU
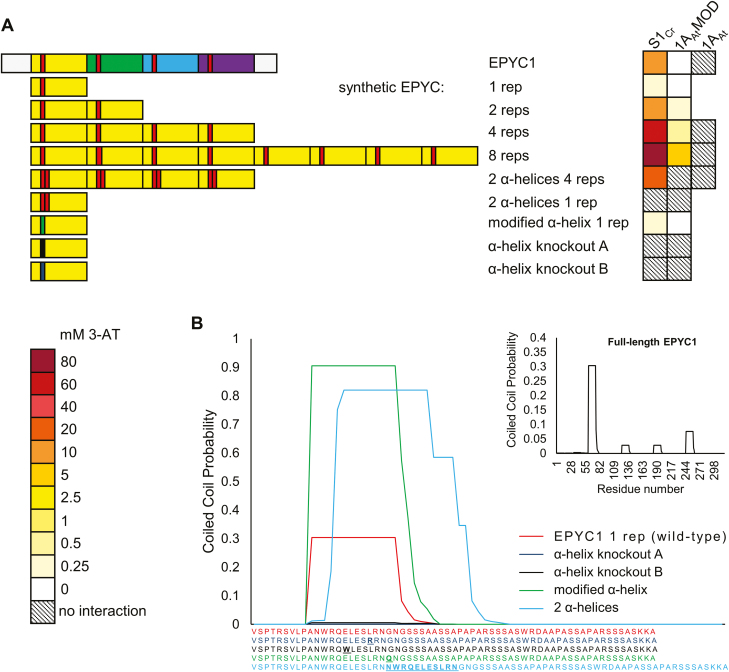
Analysis of EPYC1 with PCOILS (https://toolkit.tuebingen.mpg.de/#/tools/pcoils) suggested that the putative α-helices of EPYC1 might behave like coiled-coil domains, with the first repeat showing the highest predicted value (Fig. 4) (Gruber et al., 2006; Zimmermann et al., 2018). Thus, we hypothesized that the first repeat region could be a useful target scaffold to engineer a synthetic EPYC1 with increased affinity for SSU interaction. We constructed four synthetic EPYC1 variants containing one, two, four, or eight copies of the first repeat in tandem (Fig. 4A; Supplementary Fig. S3D). Four copies of the first repeat (4 rep) showed a stronger interaction strength with S1Cr and 1AAtMOD compared with native mature EPYC1. The strongest interaction was observed for the variant with eight repeats (8 rep), which grew on the maximum 3-AT concentrations tested (80 mM).
Fig. 4.
EPYC1 can be modified to increase the interaction strength with SSUs. (A) Synthetic variants of EPYC1 are based on the first repeat regions (yellow) and the predicted α-helix (red). The heat map indicates interaction strength measured with yeast two-hybrid assays by the capacity for growth on increasing concentrations of 3-AT (mM). See Supplementary Fig. S3D for raw Y2H image data. (B) Predicted coiled-coil domain strengths for synthetic variants of the first repeat region of EPYC1 using the PCOILS bioinformatic tool. Matching colour-coded amino acid sequences are shown below, with residues that differ from the wild-type sequence shown in bold. The inlay shows the prediction for full-length EPYC1.
Using the single copy variant (1 rep), we compared modifications of the α-helix region for interaction strength based on predictions from the PCOILS tool. Duplication of the α-helical region (SVLPANWRQELESLRNNWRQELESLRNGNGSS) or a G–Q substitution near the α-helix (NWRQELESLRNQ) predicted an increased probability of coiled-coil behaviour (Fig. 4B). In contrast to the predictions by PCOILS, the former modification eradicated the interaction, while the latter did not change the interaction strength compared with the native 1 rep variant. Finally, we attempted to knock out the interaction with either an L–R or E–W substitution within the α-helix (NWRQELESRRN or NWRQWLESLRN). Both substitutions eradicated the interaction. Our results suggest that EPYC1 α-helices do not behave like traditional coiled-coil domains, but that even single point mutations within the α-helix can affect interaction. These results support those presented in Fig. 2.
EPYC1 requires a native chloroplastic transit peptide for appropriate localization in plants
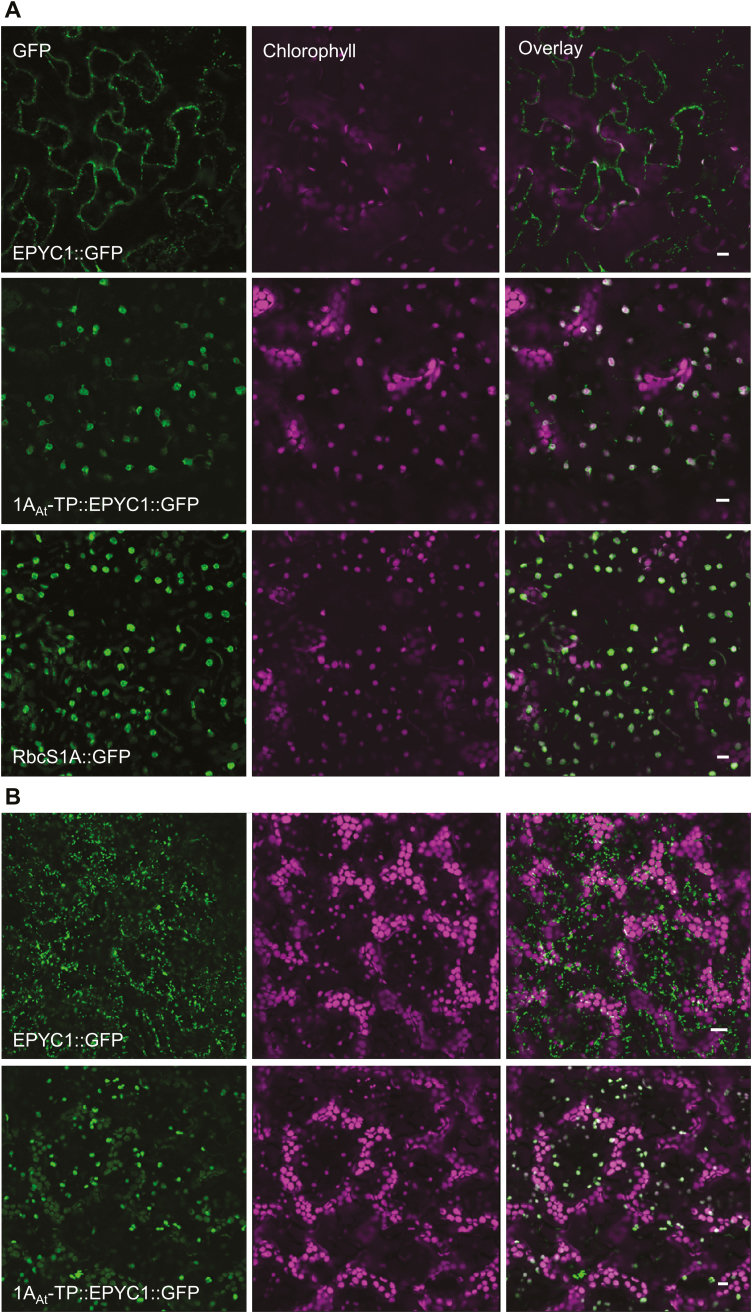
EPYC1 was codon optimized for nuclear expression in higher plants (Supplementary Fig. S7), and binary expression vectors were constructed whereby EPYC1 was C-terminally fused to GFP and expressed under the control of the 35S constitutive promoter. The localization of EPYC1::GFP was then visualized in agro-infiltrated N. benthamiana leaves and in stably transformed Arabidopsis plants. Unlike other chloroplast CCM components expressed in plants thus far (Atkinson et al., 2016), EPYC1 was not able to localize to the chloroplast in either N. benthamiana or Arabidopsis, with fluorescent signals absent from the chloroplast (Fig. 5). The 1AAt chloroplastic transit peptide (1AAt-TP) was therefore added to the N-terminus of the full-length EPYC1::GFP. Preservation of the full native EPYC1 leader sequence was based on a conservative strategy to avoid removing any potential interaction sites. Furthermore, our Y2H results indicated that the presence of the N-terminus did not strongly affect interaction with the Rubisco SSU (Fig. 3). Fusion to 1AAt-TP resulted in re-localization of EPYC1::GFP to the chloroplast stroma in both N. benthamiana and Arabidopsis.
Fig. 5.
Expression of GFP-fused EPYC1 with and without the 1AAt chloroplastic transit peptide (1AAt-TP). (A) The constructs were expressed transiently in Nicotiana benthamiana, alongside a GFP-fused Arabidopsis 1A small subunit of Rubisco (RbcS1A::GFP). (B) Stable expression in Arabidopsis. Green and purple signals are GFP and chlorophyll autofluoresence, respectively. Overlapping signals are white. Scale bar=10 µm.
EPYC1 is unstable in higher plants but does not affect plant growth performance
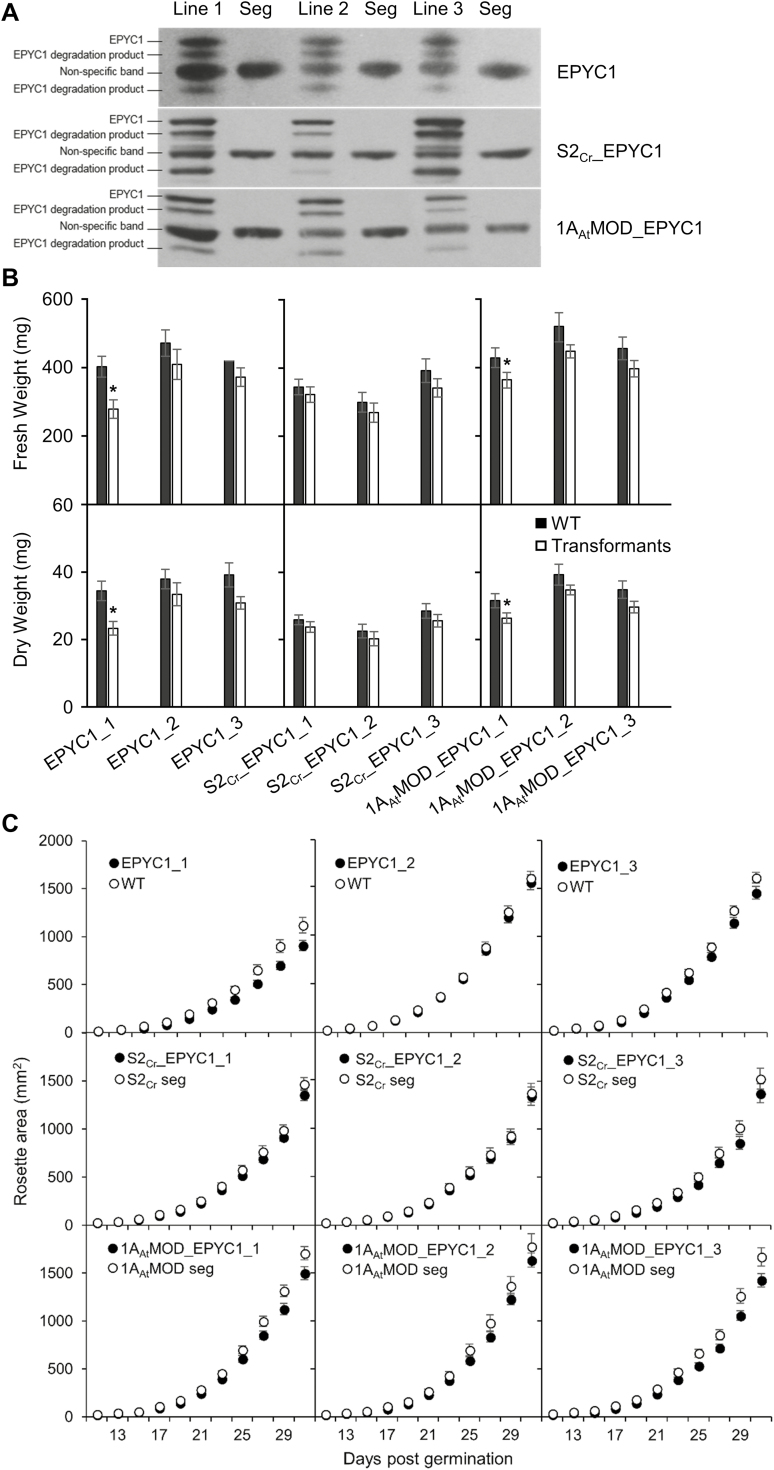
Wild-type Arabidopsis plants and two Rubisco small subunit (1a3b) mutant lines complemented with S2Cr or 1AAtMOD, previously made by Atkinson et al. (2017) (Supplementary Fig. S2), were transformed with 1AAt-TP::EPYC1 (lacking a GFP tag) (see Supplementary Fig. S7 for the plasmid map). We selected three homozygous T3 lines from each background for further analyses (EPYC1_1-3, S2Cr_EPYC1_1-3, and 1AAtMOD_EPYC1_1-3). Immunoblots against 1AAt-TP::EPYC1 in Arabidopsis produced a dominant band of ~34 kDa [slightly smaller than the mature native Chlamydomonas isoform (35 kDa)], suggesting cleavage of both 1AAt-TP and a portion of the N-terminal region of EPYC1 (the antibody serum targets the C-terminus of EPYC1) (Emanuelsson et al., 2007) (Fig. 6A; Supplementary Fig. S8A). Densitometry analysis showed that protein levels of EPYC1 in our highest expressing Arabidopsis lines were ~14 times lower than those in Chlamydomonas in relation to the Rubisco LSU (Supplementary Fig. S8B). Based on the reported ratio of ~1:6 for EPYC1 to Rubisco LSU in Chlamydomonas under low CO2 (Mackinder et al., 2016), the stoichiometry of EPYC1 to the Arabidopsis LSU in our transgenic line could therefore be estimated as 1:84. This ratio is also lower than the observed occurrence of between one and four EPYC1 peptides per Rubisco (i.e. eight LSUs) in phase-separated material in the in vitro reconstituted pyrenoidal system (Wunder et al., 2018). In addition to a non-specific band at 29 kDa, several smaller bands were also evident for EPYC1 in Arabidopsis (Fig. 6A). Additional bands were not observed for EPYC1 extracted from Chlamydomonas or yeast (Supplementary Fig. S8A), suggesting that EPYC1 may be targeted by plant proteases.
Fig. 6.
Growth of Arabidopsis plant lines expressing EPYC1 fused with the 1AAt-TP in either the wild-type (WT), S2Cr, or the 1AAtMOD background. (A) Immunoblots show the relative EPYC1 expression levels in three independently transformed lines (T3) per background, compared with their corresponding segregants (seg) lacking EPYC1. (B) Plants were harvested at 31 d, and the fresh (FW) and dry (DW) weights were measured. (C) Rosette growth of the nine transformed lines. Values are the means ±SE of measurements made on 12 (FW, DW) or 16 (growth assays) rosettes. Asterisks indicate significant difference in FW or DW between transformed lines and segregants (P<0.05) as determined by Student’s paired sample t-tests.
Growth analyses showed a slightly reduced growth phenotype (i.e. area, FW, and DW) for some plants expressing 1AAt-TP::EPYC1 compared with their corresponding segregants, but the observed decrease was not consistently significant (Fig. 6B, C). Similarly, the photosynthetic efficiency of plants expressing 1AAt-TP::EPYC1 (measured by dark-adapted leaf fluorescence; Fv/Fm) was unaffected (Supplementary Table S3). Therefore, constitutive expression of EPYC1 in the chloroplast did not appear to impact plant growth under the conditions tested.
EPYC1 interacts with Rubisco in wild-type and complemented Arabidopsis SSU mutants
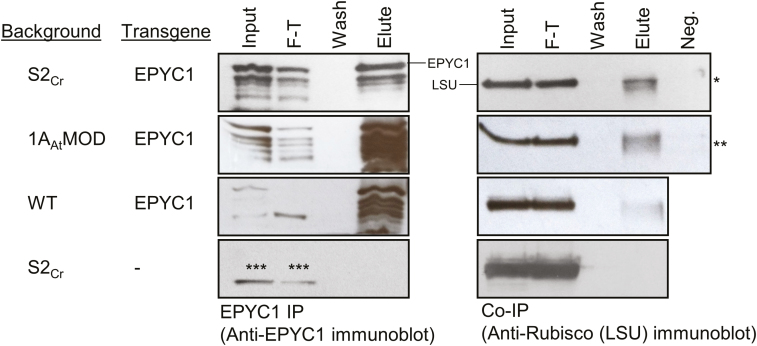
Having shown that specific SSUs can interact with EPYC1 in a Y2H system, we then investigated whether the interactions with Rubisco would occur in planta in two complemented 1a3b mutants and wild-type plant lines expressing EPYC1 (S2Cr_EPYC1_1, 1AAtMOD_EPYC1_1, and EPYC1_1, respectively). We immunoprecipitated EPYC1 from each of these lines using anti-EPYC1 antibody attached to Protein A-coated beads, and analysed the elutes by immunoblot using antibodies against EPYC1 or Rubisco (Fig. 7). Unexpectedly, the LSU was detected in the elutes of S2Cr_EPYC1 and 1AAtMOD_EPYC1 lines, as well as the wild-type expressing EPYC1. To ensure that the observed co-IP was not a result of Rubisco promiscuity or non-specific binding onto the beads or antibodies, several negative controls were included. Rubisco was not detected in the eluate of pull-downs with anti-HA-coated beads or beads with no antibody, or in the eluate from S2Cr plants not transformed with EPYC1. Therefore, these results indicated that EPYC1 was able to interact with Rubisco in transformed plant lines in the absence of a Chlamydomonas or Chlamydomonas-like SSU. It was not possible to fully quantify the relative strength of the interactions due to the inherent variation in EPYC1 expression levels between the three lines tested. Nevertheless, the levels of EPYC1 eluted in the EPYC1 IP assays were similar, while the greater amounts of Rubisco eluted in the 1AAtMOD_EPYC1 and S2Cr_EPYC1 co-IP assays could suggest a stronger interaction with EPYC1 in those lines than in the wild-type background.
Fig. 7.
Co-immunoprecipitation of Rubisco with EPYC1. EPYC1 interacts with Rubisco in transgenic S2Cr, 1AAtMOD, and wild-type (WT) lines expressing EPYC1 fused with the 1AAt-TP. Co-immunoprecipitation was performed using Protein-A coated beads that had been cross-linked to anti-EPYC1 antibody. The input, flow-through (F-T), fourth wash, and boiling elute were run on an SDS–PAGE gel, transferred to a nitrocellulose membrane, and probed with either anti-Rubisco or anti-EPYC1 antibody. Negative controls (Neg.) were carried out by replacing the anti-EPYC1 antibody on the Protein A beads with either anti-HA antibody (*) or no antibody (**) and proceeding with IP as before (only the eluted sample is shown). Asterisks (***) indicate a non-specific band observed with the anti-EPYC1 antibody in all samples including the control line not expressing EPYC1 (S2Cr).
Bimolecular fluorescence complementation (BiFC) analysis was carried out to provide additional information about the EPYC1–Rubisco interaction in vivo. Three SSUs (1AAt, S2Cr, and 1AAtMOD) and EPYC1, each fused at the C-terminus to either YFPN or YFPC, were transiently co-expressed in N. benthamiana (Walter et al., 2004). Consistent with immunoprecipitation results (Fig. 7), a BiFC signal for reconstituted YFP fluorescence was observed with plants co-expressing EPYC1 and each of the three SSUs, regardless of which protein was fused to YFPN and which to YFPC (Fig. S9). A negative control, AtCP12::YFPC, unexpectedly produced a BiFC signal with 1AAt::YFPN. However this interaction may be artefactual because there was no interaction between 1AAt::YFPC and AtCP12::YFPN.
Immunogold labelling revealed no aggregation of Rubisco in plants expressing EPYC1
To investigate the effect of EPYC1 on Rubisco aggregation in planta, we examined the localization of Rubisco in the chloroplast of S2Cr complemented 1a3b mutants expressing the highest levels of EPYC1 (S2Cr_EPYC1_1). Immunogold labelling of Rubisco revealed an even distribution of gold particles throughout the chloroplast when visualized by TEM similar to the S2Cr control not expressing EPYC1 (Supplementary Fig. S10), indicating that co-expression of EPYC1 and the Chlamydomonas SSU did not induce detectable rigid aggregations of Rubisco in these transformants.
EPYC1 phases separate with Rubisco from Arabidopsis SSU mutants complemented with S2Cr
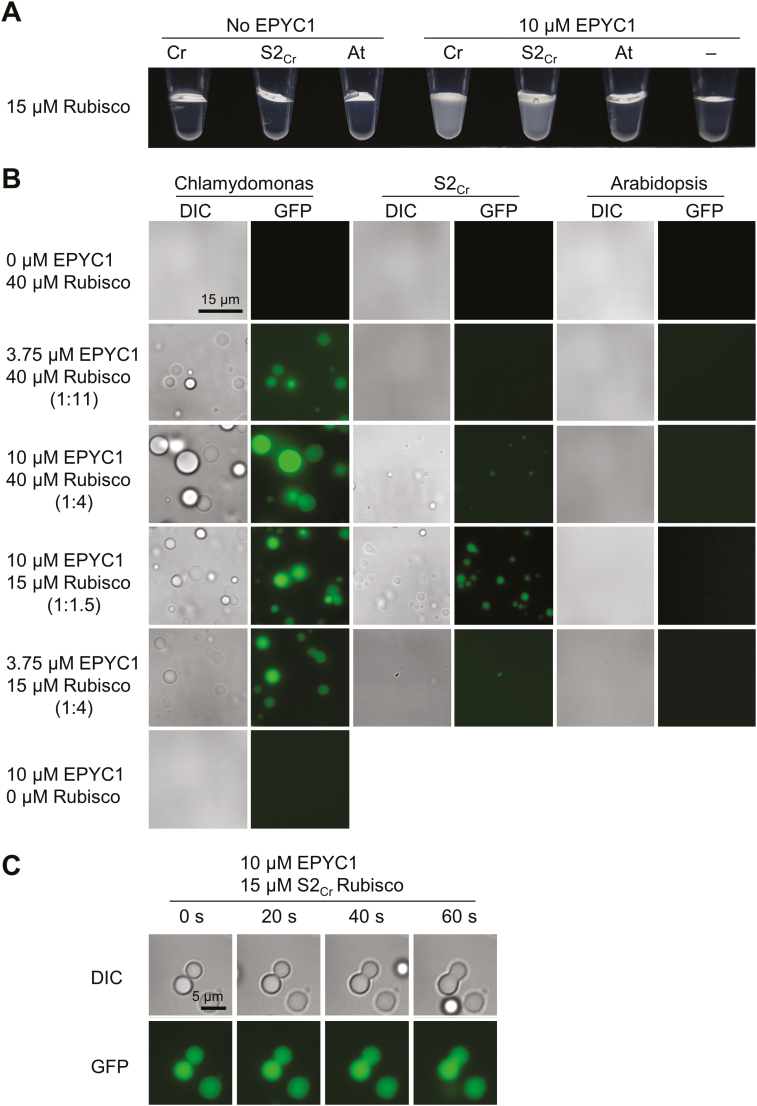
Current models of pyrenoid formation are based on specific weak multivalent interactions that promote liquid-like phase separation (Hyman et al., 2014; Freeman Rosenzweig et al., 2017). To observe if such interactions could occur with hybrid plant-derived Rubisco, we next examined if Rubisco from Arabidopsis 1a3b mutants complemented with S2Cr was able to facilitate liquid–liquid phase separation with EPYC1 using an in vitro assay developed by Wunder et al. (2018). Rubisco was extracted from Chlamydomonas, Arabidopsis wild-type plants, and S2Cr lines. The hybrid Rubisco complexes in S2Cr lines consisted of the Arabidopsis LSU and a mixed population of Arabidopsis SSUs and S2Cr (~1:1) (Atkinson et al., 2017). Similarly to Chlamydomonas Rubisco, hybrid plant Rubisco was able to demix with EPYC1 and formed liquid-like droplets of comparable size, albeit at slightly higher EPYC1:Rubisco ratios (Fig. 8A, B). In contrast, wild-type Arabidopsis Rubisco did not phase separate under similar conditions, indicating that the presence of S2Cr was critical for aggregation. In solutions containing Chlamydomonas or hybrid plant Rubisco, the droplets fused into a large homogeneous droplet (coalescence), supporting their liquid nature (Fig. 8C) (Hyman et al., 2014). Analysis by SDS–PAGE confirmed that both EPYC1 and Rubisco had entered the droplets (Supplementary Fig. S11).
Fig. 8.
Hybrid Arabidopsis Rubisco carrying a Chlamydomonas small subunit is able to phase separate and form liquid droplets with EPYC1. (A) Addition of EPYC1 to Rubisco results in turbidity in Chlamydomonas (Cr) and hybrid (S2Cr), but not Arabidopsis (At) Rubisco (shown at ~3 min after mixing at room temperature). (B) The turbidity is caused by the formation of spherical droplets. Fluoresence in samples containing EPYC1 is due to the inclusion of EPYC1::GFP (0.25 µM). (C) Droplets from S2Cr Rubisco and EPYC1 fuse by coalescence. See Supplementary Fig. S11 for droplet sedimentation analysis.
Discussion
EPYC1 is required for pyrenoid aggregation and functionality of the CCM in Chlamydomonas and will probably be a crucial component for introducing an algal-based CCM into higher plants. Previously, EPYC1 has been shown to interact with Rubisco, but the site(s) of the interaction was unclear. Mackinder et al. (2016) hypothesized that interaction could be mediated through the α-helices of the SSU, as previous work had shown that exchanging the α-helices of the Chlamydomonas SSU with those from spinach inhibited pyrenoid formation (Meyer et al., 2012). In addition, Wunder et al. (2018) have demonstrated that the Chlamydomonas SSU is important for the liquid-like aggregation typical of an algal pyrenoid. Here we have characterized this interaction by expressing EPYC1 and different SSUs together in yeast and higher plants. We have shown that the α-helical regions of the Chlamydomonas SSU are necessary and sufficient for interaction with EPYC1, and that introduction of the α-helices into an Arabidopsis SSU enabled interaction with EPYC1. Further inclusion of additional features from the Chlamydomonas SSU strengthened the interaction in an additive fashion, suggesting that the conformation of the SSU could have evolved, in part, to present the α-helical region to EPYC1. Although a stronger interaction may not be optimal for pyrenoid phase transitions in Chlamydomonas (Freeman Rosenzweig et al., 2017), our results showed that the strength of the EPYC1–SSU interaction could be further increased by engineering the SSU alone. This may be required for optimizing Rubisco aggregation in higher plants while retaining features of the higher plant SSU (Atkinson et al., 2017).
We established key regions of EPYC1 that affect binding strength with the SSU. Previously Mackinder et al. (2016) speculated that the Rubisco-binding sites in EPYC1 were located within the four repeats. Our data supported this hypothesis and identified the putative α-helices in each repeat as key sites for SSU interaction. Our MS data suggested that the N-terminus comprised a chloroplastic transit peptide (~26 residues in length) that is cleaved during import and maturation in Chlamydomonas (Emanuelsson et al., 2007). We also showed that the C-terminal region of EPYC1 probably contains an additional SSU interaction site(s) as this region could interact with the SSU independently as well as increase the strength of interaction when present with the four repeats. In support of this hypothesis, Wunder et al. (2018) demonstrated that a single EPYC1 repeat fragment carrying the C-terminal region was sufficient to demix with Rubisco (i.e. at least two interaction sites are required to aggregate Rubisco). Based on our results, we cannot rule out the existence of additional SSU-binding sites in EPYC1. Thus, models in which EPYC1 has precisely four binding sites for Rubisco are probably too simplistic (Mackinder et al., 2016). Freeman Rosenzweig et al. (2017) proposed a ‘magic number’ hypothesis, whereby the stoichiometry between the number of available EPYC1-binding sites and the surrounding Rubisco complexes defined the aggregation state of Rubisco. In their model, an odd number of binding sites was preferable for aggregation. Additional binding sites in EPYC1, for example in the C-terminal region, may help to explain such an observation. Furthermore, our data suggested that the interaction strength of EPYC1 can be modified from its native state, for example by duplicating the first repeat region. This may provide additional options for optimizing Rubisco aggregation in heterologous environments, such as higher plant chloroplasts.
Previous work has shown that most CCM components from Chlamydomonas localized to the same subcellular locations when expressed in plants (Atkinson et al., 2016). Thus, we were surprised that a native chloroplastic transit peptide was required for expression of EPYC1 in higher plant chloroplasts. One explanation is that EPYC1 may be mistargeted to the mitochondrion, as chloroplast transit peptides from Chlamydomonas are known to resemble mitochondrial transit peptides in higher plants (Franzén et al., 1990). Alternatively, the N-terminus might undergo proteolytic degradation when expressed in plants, thus inhibiting chloroplastic import (Doran, 2006). Even when redirected to the chloroplast with a native transit peptide, the presence on immunoblots of several smaller bands suggested that EPYC1 may be partially targeted by proteases in the chloroplast (Fig. 6; Suppleentary Fig. S8) (Nishimura et al., 2017). For future work, it may be desirable to identify and modify motifs in EPYC1 recognized by native proteases to enhance EPYC1 stability, and in addition reduce the length of the native N-terminus by eliminating the sequence upstream of the putative transit peptide cleavage site (Fig. 1B).
Previous work has shown that EPYC1 can phase separate with heterologous Rubiscos of cyano- and chemoautotrophic bacterial origin (Wunder et al., 2018). Even though droplet formation was less efficient for those cases, it suggests a certain promiscuity of the EPYC1–Rubisco interaction. Likewise, a residual compatibility with plant Rubisco might account for our observations made with EPYC1 expressed in planta (Fig. 7: Supplementary Fig. S9). EPYC1 may interact differently with free SSUs—as in yeast—and SSUs assembled into the octomeric (L8S8) Rubisco complex. Whilst no interactions with EPYC1 were evident for 1AAt in yeast (Fig. 1), and similarly no phase separation was observed in vitro with wild-type Arabidopsis Rubisco and EPYC1 (Fig. 8), the high abundance of Rubisco in plant chloroplasts may increase the likelihood of potential interactions between the positively charged EPYC1 peptide and any negatively charged moieties on the LSU and/or SSU (Wunder et al., 2018). Further work will be required to distinguish whether the interactions observed in planta were indicative of true liquid–liquid phase separation or symptomatic of residual interactions due to the abundance of Rubisco (Hyman et al., 2014)—the average concentration of Rubisco in plant chloroplasts is estimated at ~500 µM (Jensen and Bahr, 1977; Harris and Königer, 1997), which is similar to that of the pyrenoid in the Chlamydomonas chloroplast (~628 µM Rubisco) (Freeman Rosenzweig et al., 2017).
Previously, Rubisco aggregates were observed in transgenic tobacco plants expressing cyanobacterial Rubisco and the β-carboxysomal assembly protein CcmM35 (Lin et al., 2014). However, CcmM35 cross-links Rubisco into a highly organized and rigid paracrystalline array in β-carboxysomes (Faulkner et al., 2017), which differs significantly from the dynamic and more disordered phase separation observed during the formation of pyrenoidal Rubisco aggregates mediated by EPYC1 (Engel et al., 2015; Freeman Rosenzweig et al., 2017; Wunder et al., 2018). Promisingly, the observed interactions between EPYC1 and SSUs in yeast and recombinant EPYC1 with hybrid plant Rubisco suggest that EPYC1 does not require additional modifications or chaperones for binding. However, the low EPYC1:Rubisco protein ratio in our transgenic Arabidopsis plants, which was probably compounded by proteolytic degradation of EPYC1, was insufficient to enable detectable phase separation (Supplementary Fig. S8). Plants typically produce 4- to 5-fold more Rubisco than Chlamydomonas (30% versus 6.6% total protein per cell, respectively) (Parry et al., 2013; Hammel et al., 2018). Modification of EPYC1:Rubisco in the transgenic plants to give a more favourable ratio (i.e. through increased expression of EPYC1, reduction in overall Rubisco, and/or increased abundance of a pyrenoid-compatible SSU) may help to further protect EPYC1 from degradation by assembling EPYC1 with Rubisco into a pyrenoid-like structure, which might shield EPYC1 from proteases in the chloroplast. Reducing nitrogen investment in Rubisco while increasing Rubisco operating efficiency, as seen in Chlamydomonas and other photosynthetic organisms with CCMs (Rae et al., 2017), is in line with our overarching goal. Such modifications could be achieved using CRISPR (clustered regularly interspaced short palindromic repeats) to target the native Arabidopsis SSU family (Pottier et al., 2018; Khumsupan et al., 2019). Furthermore, our data show that synthetic variants of EPYC1 could be developed to better enable aggregation of Rubisco in higher plants (Fig. 7). Recently developed tools for expressing plant Rubisco in E. coli offer the opportunity to more rapidly fine-tune this relationship for expression in plants going forward (Aigner et al., 2017), and further explore any potential interactions between EPYC1 and other regions on the Rubisco complex. Overall, this study has demonstrated the merits of testing protein–protein interactions in different heterologous systems to improve our understanding of the algal CCM and progress current efforts to introduce a functional algal-based CCM into a higher plant.
Supplementary data
Supplementary data are available at JXB online.
Table S1. List of vectors used in this study.
Table S2. List of primers used in this study.
Table S3. Maximum quantum yield of PSII (Fv/Fm) measurements for EPYC1-expressing plants from three backgrounds.
Fig. S1. Yeast two-hybrid interaction controls.
Fig. S2. Modified variants of the Arabidopsis 1A small subunit of Rubisco.
Fig. S3. Semi-quantitative yeast two-hybrid interaction assays.
Fig. S4. Modified variants of EPYC1.
Fig. S5. Immunoblot of EPYC1 and N-terminus truncated variants in yeast.
Fig. S6. Immunoprecipitation and intact protein MS of mature EPYC1 from Chlamydomonas reinhardtii.
Fig. S7. Sequence map of the binary vector carrying 1AAt-TP::EPYC1.
Fig. S8. Immunoblot analysis of EPYC1 and Rubisco produced and extracted from Arabidopsis, Chlamydomonas, and yeast.
Fig. S9. Bimolecular fluorescence complementation assays.
Fig. S10. Immunogold labelling of Rubisco in Arabidopsis plants expressing EPYC1.
Fig. S11. Droplet sedimentation analysis by SDS–PAGE for three different Rubiscos with EPYC1.
Acknowledgements
This work was funded by the UK BBSRC (BB/M006468/1) and Leverhulme Trust (RPG-2017-402). TEM was carried out with the support of the Wellcome Trust Multi User Equipment Grant (WT104915MA). We thank Sinead Collins and Rasmus Lindberg (University of Edinburgh) for supplying Chlamydomonas reinhardtii cultures.
References
- Aigner H, Wilson RH, Bracher A, Calisse L, Bhat JY, Hartl FU, Hayer-Hartl M. 2017. Plant RuBisCo assembly in E. coli with five chloroplast chaperones including BSD2. Science 358, 1272–1278. [DOI] [PubMed] [Google Scholar]
- Atkinson N, Feike D, Mackinder LC, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ. 2016. Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components. Plant Biotechnology Journal 14, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N, Leitão N, Orr DJ, Meyer MT, Carmo-Silva E, Griffiths H, Smith AM, McCormick AJ. 2017. Rubisco small subunits from the unicellular green alga Chlamydomonas complement Rubisco-deficient mutants of Arabidopsis. New Phytologist 214, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Andrews TJ, Whitney SM, Ludwig M, Yellowlees DC, Leggat W, Price GD. 1998. The diversity and coevolution of Rubisco, plastids, pyrenoids, and chloroplast-based CO2-concentrating mechanisms in algae. Canadian Journal of Botany 76, 1052–1071. [Google Scholar]
- Bathellier C, Tcherkez G, Lorimer GH, Farquhar GD. 2018. Rubisco is not really so bad. Plant, Cell & Environment 41, 705–716. [DOI] [PubMed] [Google Scholar]
- Borel B. 2017. CRISPR, microbes and more are joining the war against crop killers. Nature 543, 302–304. [DOI] [PubMed] [Google Scholar]
- Busch FA, Sage RF, Farquhar GD. 2018. Plants increase CO2 uptake by assimilating nitrogen via the photorespiratory pathway. Nature Plants 4, 46–54. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dobrescu A, Scorza LCT, Tsaftaris SA, McCormick AJ. 2017. A ‘Do-It-Yourself’ phenotyping system: measuring growth and morphology throughout the diel cycle in rosette shaped plants. Plant Methods 13, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doran PM. 2006. Foreign protein degradation and instability in plants and plant tissue cultures. Trends in Biotechnology 24, 426–432. [DOI] [PubMed] [Google Scholar]
- Driever SM, Simkin AJ, Alotaibi S, Fisk SJ, Madgwick PJ, Sparks CA, Jones HD, Lawson T, Parry MAJ, Raines CA. 2017. Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philosophical Transactions of the Royal Society B: Biological Sciences 372, 20160384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nature Protocols 2, 953–971. [DOI] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W. 2015. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4, e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JD, Patron NJ, Marillonnet S. 2014. A golden gate modular cloning toolbox for plants. ACS Synthetic Biology 3, 839–843. [DOI] [PubMed] [Google Scholar]
- Faulkner M, Rodriguez-Ramos J, Dykes GF, Owen SV, Casella S, Simpson DM, Beynon RJ, Liu LN. 2017. Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes. Nanoscale 9, 10662–10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzén LG, Rochaix JD, von Heijne G. 1990. Chloroplast transit peptides from the green alga Chlamydomonas reinhardtii share features with both mitochondrial and higher plant chloroplast presequences. FEBS Letters 260, 165–168. [DOI] [PubMed] [Google Scholar]
- Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, et al. 2017. The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber M, Söding J, Lupas AN. 2006. Comparative analysis of coiled-coil prediction methods. Journal of Structural Biology 155, 140–145. [DOI] [PubMed] [Google Scholar]
- Hammel A, Zimmer D, Sommer F, Mühlhaus T, Schroda M. 2018. Absolute quantification of major photosynthetic protein complexes in Chlamydomonas reinhardtii using quantification concatamers (QconCATs). Frontiers in Plant Science 9, 1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris GC, Königer M. 1997. The ‘high’ concentrations of enzymes within the chloroplast. Photosynthesis Research 54, 5–23. [Google Scholar]
- Howe CJ, Auffret AD, Doherty A, Bowman CM, Dyer TA, Gray JC. 1982. Location and nucleotide sequence of the gene for the proton-translocating subunit of wheat chloroplast ATP synthase. Proceedings of the National Academy of Sciences, USA 79, 6903–6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, Weber CA, Jülicher F. 2014. Liquid–liquid phase separation in biology. Annual Review of Cell and Developmental Biology 30, 39–58. [DOI] [PubMed] [Google Scholar]
- Jensen RG, Bahr JT. 1977. Ribulose 1,5-bisphosphate carboxylase-oxygenase. Annual Review of Plant Physiology, 28, 379–400. [Google Scholar]
- Khumsupan P, Donovan S, McCormick AJ. 2019. CRISPR/Cas in Arabidopsis: overcoming challenges to accelerate improvements in crop photosynthetic efficiencies. Physiologia Plantarum 166, 428–437. [DOI] [PubMed] [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP. 2016. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354, 857–861. [DOI] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MA, Hanson MR. 2014. A faster Rubisco with potential to increase photosynthesis in crops. Nature 513, 547–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Hee WY, Sharwood RE, et al. 2018. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nature Communications 9, 3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Marshall-Colon A, Zhu XG. 2015. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161, 56–66. [DOI] [PubMed] [Google Scholar]
- López-Calcagno PE, Fisk S, Brown KL, Bull SE, South PF, Raines CA. 2019. Overexpressing the H-protein of the glycine cleavage system increases biomass yield in glasshouse and field-grown transgenic tobacco plants. Plant Biotechnology Journal 17, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Calcagno PE, Howard TP, Raines CA. 2014. The CP12 protein family: a thioredoxin-mediated metabolic switch? Frontiers in Plant Science 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, Jonikas MC. 2016. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proceedings of the National Academy of Sciences, USA 113, 5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, Jonikas MC. 2017. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. 2000. Chlorophyll fluorescence—a practical guide. Journal of Experimental Botany 51, 659–668. [DOI] [PubMed] [Google Scholar]
- McCormick AJ, Kruger NJ. 2015. Lack of fructose 2,6-bisphosphate compromises photosynthesis and growth in Arabidopsis in fluctuating environments. The Plant Journal 81, 670–683. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Long SP. 2014. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiology 164, 2247–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MT, Genkov T, Skepper JN, Jouhet J, Mitchell MC, Spreitzer RJ, Griffiths H. 2012. Rubisco small-subunit α-helices control pyrenoid formation in Chlamydomonas. Proceedings of the National Academy of Sciences, USA 109, 19474–19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Kato Y, Sakamoto W. 2017. Essentials of proteolytic machineries in chloroplasts. Molecular Plant 10, 4–19. [DOI] [PubMed] [Google Scholar]
- Occhialini A, Lin MT, Andralojc PJ, Hanson MR, Parry MA. 2016. Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2. The Plant Journal 85, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MA, Andralojc PJ, Scales JC, Salvucci ME, Carmo-Silva AE, Alonso H, Whitney SM. 2013. Rubisco activity and regulation as targets for crop improvement. Journal of Experimental Botany 64, 717–730. [DOI] [PubMed] [Google Scholar]
- Pottier M, Gilis D, Boutry M. 2018. The hidden face of rubisco. Trends in Plant Science 23, 382–392. [DOI] [PubMed] [Google Scholar]
- Price GD, Pengelly JJ, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR. 2013. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. Journal of Experimental Botany 64, 753–768. [DOI] [PubMed] [Google Scholar]
- Rae BD, Long BM, Förster B, Nguyen ND, Velanis CN, Atkinson N, Hee WY, Mukherjee B, Price GD, McCormick AJ. 2017. Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants. Journal of Experimental Botany 68, 3717–3737. [DOI] [PubMed] [Google Scholar]
- Schöb H, Kunz C, Meins F Jr. 1997. Silencing of transgenes introduced into leaves by agroinfiltration: a simple, rapid method for investigating sequence requirements for gene silencing. Molecular & General Genetics 256, 581–585. [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I. 2010. A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. The Plant Journal 61, 519–528. [DOI] [PubMed] [Google Scholar]
- Shivhare D, Mueller-Cajar O. 2017. In vitro characterization of thermostable CAM Rubisco activase reveals a Rubisco interacting surface loop. Plant Physiology 174, 1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Lopez-Calcagno PE, Raines CA. 2019. Feeding the world: improving photosynthetic efficiency for sustainable crop production. Journal of Experimental Botany 70, 1119–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Lopez-Calcagno PE, Raines CA, Ort DR. 2018. Optimizing photorespiration for improved crop productivity. Journal of Integrative Plant Biology 60, 1217–1230. [DOI] [PubMed] [Google Scholar]
- Spreitzer RJ. 2003. Role of the small subunit in ribulose-1,5-bisphosphate carboxylase/oxygenase. Archives of Biochemistry and Biophysics 414, 141–149. [DOI] [PubMed] [Google Scholar]
- Turkina MV, Blanco-Rivero A, Vainonen JP, Vener AV, Villarejo A. 2006. CO2 limitation induces specific redox-dependent protein phosphorylation in Chlamydomonas reinhardtii. Proteomics 6, 2693–2704. [DOI] [PubMed] [Google Scholar]
- van Nues RW, Beggs JD. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157, 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Waltz E. 2017. Digital farming attracts cash to agtech startups. Nature Biotechnology 35, 397–398. [DOI] [PubMed] [Google Scholar]
- Wang H, Gau B, Slade WO, Juergens M, Li P, Hicks LM. 2014. The global phosphoproteome of Chlamydomonas reinhardtii reveals complex organellar phosphorylation in the flagella and thylakoid membrane. Molecular & Cellular Proteomics 13, 2337–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder T, Cheng SLH, Lai SK, Li HY, Mueller-Cajar O. 2018. The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nature Communications 9, 5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Sato E, Iguchi H, Fukuda Y, Fukuzawa H. 2015. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 112, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Struik PC. 2017. Can increased leaf photosynthesis be converted into higher crop mass production? A simulation study for rice using the crop model GECROS. Journal of Experimental Botany 68, 2345–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhi Z, Hélène L, Antoine S, Régine L, Brigitte G. 2018. Exploring intrinsically disordered proteins in Chlamydomonas reinhardtii. Scientific Reports 8, 6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Marchand CH, Maes A, et al. 2018. Pyrenoid functions revealed by proteomics in Chlamydomonas reinhardtii. PLoS One 13, e0185039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann L, Stephens A, Nam SZ, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. Journal of Molecular Biology 430, 2237–2243. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.