The high-yielding rice cultivar Takanari has fast photosynthetic induction owing to a high electron transport rate, stomatal conductance, and metabolic flux, leading to high daily carbon gain under fluctuating light.
Keywords: Chlorophyll fluorescence, metabolome, photosynthesis, photosynthetic induction, rice, stomatal conductance, sunfleck, transcriptome
Abstract
Leaves within crop canopies experience variable light over the course of a day, which greatly affects photosynthesis and crop productivity. Little is known about the mechanisms of the photosynthetic response to fluctuating light and their genetic control. Here, we examined gas exchange, metabolite levels, and chlorophyll fluorescence during the photosynthetic induction response in an Oryza sativa indica cultivar with high yield (Takanari) and a japonica cultivar with lower yield (Koshihikari). Takanari had a faster induction response to sudden increases in light intensity than Koshihikari, as demonstrated by faster increases in net CO2 assimilation rate, stomatal conductance, and electron transport rate. In a simulated light regime that mimicked a typical summer day, the faster induction response in Takanari increased daily CO2 assimilation by 10%. The faster response of Takanari was explained in part by its maintenance of a larger pool of Calvin–Benson cycle metabolites. Together, the rapid responses of electron transport rate, metabolic flux, and stomatal conductance in Takanari contributed to the greater daily carbon gain under fluctuating light typical of natural environments.
Introduction
To meet the increasing food demands of the growing human population, crop productivity needs to be increased (Ashraf and Akram, 2009). Photosynthesis is one important biochemical process supporting plant growth and grain yield, and improving photosynthesis is thus considered a major target for improving crop performance (Yamori et al., 2016b). In rice (Oryza sativa L.), the rate of steady-state leaf photosynthesis varies widely among cultivars (Kanemura et al., 2007). Quantitative trait loci (QTLs) and candidate genes associated with this variation have been reported (Adachi et al., 2011, 2013, 2017; Takai et al., 2013). In nature, however, steady-state conditions, especially of light intensity, are rarely observed, because of clouds, wind, and self-shading within the canopy (Slattery et al., 2018). In a soybean canopy, sunflecks contribute 40–90% of total daily photosynthetic photon flux density (PPFD) (Pearcy, 1990). However, understanding photosynthetic performance in dynamic light environments has received less attention than that under steady-state conditions, and for many crops, including rice, the patterns and mechanisms of dynamic light responses are unclear (Yamori, 2016; Vialet-Chabrand et al., 2017) and genotypic variation in these responses is little studied (Qu et al., 2016). Genotypic comparisons, in combination with molecular genomic techniques, can identify underlying genetic controls that could be exploited for crop improvement (Soleh et al., 2016; Yamori et al., 2016a; van Bezouw et al., 2019).
To make full use of sunflecks, plants need a rapid photosynthetic response to fluctuating light. However, upon sudden illumination, the photosynthetic rate in vivo typically shows a delayed response and requires several to tens of minutes to reach a new steady-state level (Scafaro et al., 2012; Yamori et al., 2012; Taylor and Long, 2017). Photosynthetic induction typically has three phases in response to an increase in PPFD (Pearcy, 1990). The initial phase occurs in the first 1–2 min and reflects an increase in electron transport. In particular, cyclic electron flow around PSI limits the speed of the photosynthetic response under fluctuating light (Yamori et al., 2016c).
The second phase reflects activation of Calvin–Benson cycle enzymes, including those that perform ribulose-1,5-bisphosphate (RuBP) carboxylation and RuBP regeneration (Sassenrath-Cole and Pearcy, 1992). Activation of Rubisco is a particularly limiting factor and responds slowly (5–10 min) after a step increase in PPFD (Pearcy, 1999; Yamori et al., 2012). Rubisco activation requires functional Rubisco activase, which is regulated by the ATP/ADP ratio and redox state in the chloroplast (Portis, 1995). Four enzymes of the Calvin–Benson cycle—phosphoribulokinase (PRK), glyceraldehyde 3-phosphate dehydrogenase (GAPDH), fructose 1,6-bisphosphatase (FBPase), and sedoheptulose 1,7-bisphosphatase (SBPase)—are activated by light via the ferredoxin/thioredoxin system (Buchanan, 1980). Activation of these enzymes occurs more rapidly than for Rubisco but can still take between 1 min and 10 min (Sassenrath-Cole and Pearcy, 1994). The Calvin–Benson cycle can also be limited by suboptimal levels of metabolites needed for enzyme regulation and activity. The regulation of metabolite pools by rapid increases in light intensity can lead to photosynthetic oscillations that disrupt efficient turnover of the Calvin–Benson cycle (Laisk et al., 1989; Zhu et al., 2013).
The third phase of photosynthetic induction reflects the speed of stomatal opening. Stomatal opening in response to light is mediated by signalling from phototropin to plasma membrane H+-ATPase of guard cells, which drives K+ uptake by activating inwardly rectifying K+ channels (Yamauchi et al., 2016). Stomatal opening is also affected by CO2 concentration, vapour pressure deficit, and temperature (Kaiser et al., 2017). Stomatal response to variable PPFD is slower by an order of magnitude than biochemical responses (Lawson et al., 2012), indicating that it is not always coupled to photosynthesis under dynamic conditions. Slow induction of stomatal conductance can limit the photosynthetic rate by 10–15% when leaves are transferred from low- to high-light conditions (McAusland et al., 2016). The relative importance of the three phases varies with the acclimation to light conditions (Pearcy, 1990). For example, when leaves receive saturating light after a brief period of shade following a long period of saturating light, Rubisco activity and stomatal conductance do not change considerably (Sassenrath-Cole and Pearcy, 1992).
Daily photosynthetic rate under fluctuating light conditions can be up to 35% lower than the optimal photosynthetic rate under constant light (Naumburg and Ellsworth, 2002). Understanding the genotypic differences in photosynthetic induction and underlying mechanisms would help rice breeding programmes aimed at increasing canopy biomass production. The rate of steady-state photosynthesis of the high-yielding indica cultivar Takanari is higher than those of most rice cultivars, including a popular Japanese commercial cultivar, Koshihikari (Taylaran et al., 2011). The higher rate in Takanari is due to its higher CO2 diffusion conductance and Rubisco content than in Koshihikari (Taylaran et al., 2011; Chen et al., 2014). Takanari also has higher chlorophyll and carotenoid content per leaf area, which may increase light-absorbing capacity (Chen et al., 2014).
In the present study, we analysed differences between Takanari and Koshihikari in the photosynthetic response to fluctuating light by simultaneously measuring chlorophyll fluorescence and gas exchange. We also characterized metabolite dynamics in the Calvin–Benson cycle, and the respiration and photorespiration pathways during photosynthetic induction. Finally, we analysed the dynamics of gene expression to understand the genetic and molecular mechanisms of the differences between the two cultivars in the photosynthetic response to fluctuating light.
Materials and methods
Plant materials and growth conditions
Seeds of O. sativa L. cv. Takanari and Koshihikari were sown in nursery boxes filled with artificial soil, and the seedlings were grown until the fourth to fifth leaf stage in a greenhouse. The seedlings were transplanted into 3 litre pots (one per pot) filled with a 1:1 (v/v) mixture of paddy soil (an alluvial clay loam) and upland soil (a diluvial volcanic ash). The pots were placed outside and the level of standing water was maintained at 2–4 cm above the soil. Fertilizer (0.5 g each of N as ammonium nitrate, P2O5, and K2O per pot) was applied at planting, and no additional fertilizer was applied. The pots were placed in the experimental field of Tokyo University of Agriculture and Technology in Fuchu, Japan (35°41'N, 139°29'E). The average daily temperature was 24.6 °C and the average maximum temperature was 29.0 °C during the growth period, both similar to the historical averages of the past 30 years (24.3 °C and 28.9 °C, respectively).
Photosynthesis measurements
Gas exchange, chlorophyll fluorescence, and the redox state of reaction centre chlorophyll in PSI (P700) were measured simultaneously with a GFS-3000 gas exchange system and a Dual-PAM-100 fluorometer (both from Walz, Effeltrich, Germany) as described in Yamori et al. (2016c). Responses of several photosynthetic parameters to changes in light intensity were measured every 20 s at a CO2 concentration of 400 μmol mol−1. The quantum yield of PSI [Y(I)] and PSII [Y(II)] and non-photochemical quenching (NPQ) were analysed as previously described (Yamori et al., 2011). The electron transport rate (ETR) was calculated as ETR I (or ETR II)=0.5×0.84×Y(I) [or Y(II)], where 0.5 is the fraction of absorbed light reaching PSI or PSII, and 0.84 is the leaf absorptance.
The diurnal changes of the gas exchange rate were measured using a LI-6400XT portable photosynthesis system (LI-COR, Lincoln, NE, USA) to characterize a typical light environment in the canopy. PPFD and air temperature were recorded at 10 cm below the top of the canopy every 10 s from 06.00 h to 18.00 h using a DEFI-L photo quantum sensor (JFE, Japan) and a CS215 temperature sensor (Campbell Scientific, USA) at the experimental field of the Graduate School of Agriculture, Kyoto University (35°01'N, 135°47'E) on 1 July 2016. These ‘typical’ daily conditions for 12 h were replicated in the chamber of the LI-6400XT using an auto-measuring program. Plants in their pots were transferred to a dark room in the evening before the measurements. The sample leaf was enclosed within the leaf chamber at 05.55 h and was irradiated at 50 μmol photons m−2 s−1 for 5 min, and the auto-measuring program was initiated at 06.00 h. Net CO2 assimilation rate at a CO2 partial pressure of 400 μmol mol−1 (A400) and stomatal conductance (gs) were automatically recorded every 10 s for 12 h. Water use efficiency (WUE) was calculated as A400 divided by the transpiration rate. The total daily carbon gain was calculated as the integrated sum of A400 over the 12 h experiment.
Chlorophyll fluorescence in the paddy field
Plants were grown in a paddy field of the experimental farm of Kyoto University (34°44'N, 135°50'E). Diurnal changes of chlorophyll fluorescence parameters in the paddy field were measured with a Monitoring-PAM fluorometer (Waltz) as described in Ikeuchi et al. (2014) and Ishida et al. (2014). The leaf clips of the fluorometer were fixed on the south-facing upright leaves. The fluorescence parameters and PPFD of sunlight were recorded every 5 min from 05.00 h to 19.00 h on 19 August 2017. ETR II and NPQ were calculated as described above.
Metabolome analysis
Fully expanded leaves were covered with aluminium foil, and the plants were kept overnight in a dark cabinet (KG-50HLA; Koito Manufacturing, Tokyo, Japan). The next day, one leaf was quickly frozen in liquid nitrogen inside the dark cabinet. Other leaves were illuminated at 1100 μmol photons m−2 s−1 and frozen after 1, 5, 10, 30, or 60 min. Each sampled leaf was divided into two equal parts in liquid nitrogen, which were stored in two different centrifuge tubes at −80 °C. One half was used for metabolome analysis and the other half for transcriptome analysis.
Primary metabolites were quantified according to Miyagi et al. (2010) and Noguchi et al. (2018), with minor modifications. In brief, ~50 mg of frozen leaves was homogenized in 50% (v/v) methanol containing 50 μM 1,4-PIPES and 50 μM methionine sulfone. The homogenates were centrifuged at 4 °C at 22 000 g for 5 min and then at 14 000 g for 30 min in a 3 kDa cut-off filter (Millipore, Billerica, MA, USA), and the filtrate was used for analysis. Metabolites were quantified by capillary electrophoresis–triple quadrupole MS (CE–QQQ-MS; CE 7100, MS G6420A; Agilent Technologies, Santa Clara, CA, USA) in MRM mode. For MS stabilization, the sheath solutions [5 mM ammonium acetate in 50% (v/v) methanol for anions and 0.1% (v/v) formic acid in 50% methanol for cations] were applied to the capillary at 10 μl min−1 by an isocratic HPLC pump (Agilent 1200 series) equipped with a 1:100 splitter. Quantitative accuracy was determined with known concentrations of standard reference compounds in Agilent MassHunter software.
Kinetic modelling and simulation
The effect of PSI and PSII fluxes on the Calvin–Benson cycle was evaluated with a kinetic model for C3 plants (Pettersson and Ryde-Pettersson, 1988) with some modifications (see Supplementary Fig. S4 at JXB online for details). All rate equations of enzyme reactions with kinetic parameters are summarized in Supplementary Table S3. The ordinary differential equations and the initial concentrations of all metabolites are listed in Supplementary Table S4. The CO2 fixation rate in the Calvin–Benson cycle was simulated from the rates of ATP and NADPH synthesis in the photosystem using the kinetic model. To calculate the ordinary differential equations by a numerical method, the rates of ATP and NADPH synthesis at each time step were calculated from the fluxes of PSI and PSII. Values experimentally determined for Takanari or Koshihikari were used as inputs for the simulation (Fig. 1), and were interpolated by a smoothing spline technique in Curve Fitting Toolbox software (MATLAB R2018; MathWorks, Natick, MA, USA). The PSI and PSII fluxes were converted to linear and cyclic electron flows as:
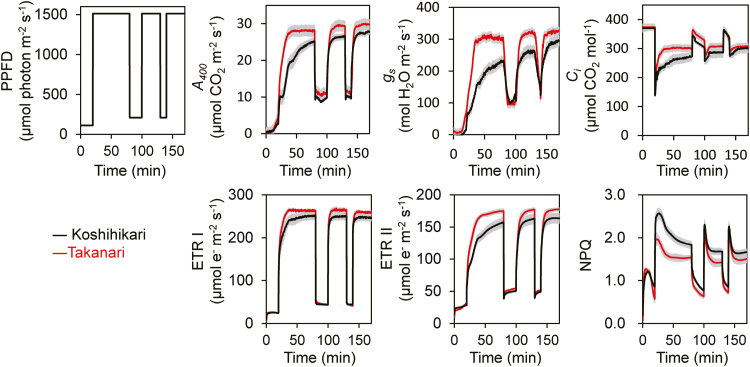
Fig. 1.
Responses of photosynthetic parameters to changes in light intensity. PPFD, photosynthetic photon flux density; A400, net CO2 assimilation rate at a CO2 partial pressure of 400 μmol mol−1; gs, stomatal conductance; Ci, intercellular CO2 concentration; ETR I, photosynthetic electron transport around PSI; ETR II, photosynthetic electron transport around PSII; NPQ, non-photochemical quenching. These parameters were measured simultaneously with the GFS-300 and Dual-PAM-100 measuring system under a CO2 concentration of 400 μmol mol−1 and 25 °C. Values are means ±SE (n=4). (This figure is available in colour at JXB online.)
where v is flux rate. Three protons are transported to the thylakoid lumen per electron in the linear electron flow and two protons in the cyclic electron flow. Since three molecules of ATP are produced per 14 protons by ATP synthase, 9/14 ATP in the linear electron flow and 3/7 ATP in the cyclic electron flow are produced in photosynthesis. Similarly, we assumed that 1/2 NADPH is produced by NADP reductase per electron in the linear electron flow. The generation of ATP and NADPH was calculated as:
Since the model of Pettersson and Ryde-Pettersson (1988) was developed for a different plant species, the Vmax and rate constants were determined by fitting our experimental data, except that the Vmax values of reactions r4, r5, r7, r8, and r10–r12 were fixed at 105 μM min−1, because these were equilibrium reactions. First, the Vmax (r1, r6, and r9) and rate constants (r2, r3, and r13–r16) were optimized by the fmincon function of MATLAB R2018 to minimize the residual sum of squares of CO2 fixation rate between the measured and simulated data for Koshihikari (Fig. 1). All Vmax values and rate constants for the kinetic model are listed in Supplementary Table S3. Then, using the optimized Vmax and rate constants, the metabolism of the Calvin–Benson cycle in Takanari was simulated using the PSI and PSII fluxes of Takanari as inputs. The ode15s solver was used to solve the ordinary differential equations.
Transcriptome analysis
Frozen leaf samples were homogenized in a TissueLyser II apparatus (Qiagen, Hilden, Germany), and total RNA was extracted with a Maxwell 16 LEV Plant RNA kit in a Maxwell 16 automated purification system (both Promega, Madison, WI, USA). RNA concentrations were measured with a Quant-iT RNA Assay kit (Thermo Fisher Scientific, Hampton, NH, USA). An automated liquid handling system (Freedom EVO 150; TECAN, Mannedorf, Switzerland) and a thermal cycler (ODTC 384; Inheco, Munich, Germany) were utilized for RNA sequencing (RNA-Seq) library preparation. Selective depression of abundant transcripts such as rRNAs was conducted as described in Nagano et al. (2015). Then, the RNA-Seq library was prepared as described in Ishikawa et al. (2017). Single-end 50 bp reads were sequenced on a Hiseq 2500 sequencer (Illumina, Hayward, CA, USA) by Macrogen Co. All RNA-Seq data are deposited in the Sequence Read Archive (SRA) under accession number PRJNA511617.
Quality control and mapping of RNA-Seq data were conducted as described in Ishikawa et al. (2017), with reference sequences of IRGSP-1.0_transcript (Kawahara et al., 2013). All transcriptome analysis was conducted in R v. 3.4.2 software (R Core Team, 2017) according to the script at https://github.com/naganolab/Rice_light_induction_RNA-Seq.git. In brief, differentially expressed genes (DEGs) were analysed with the R packages TCC (Sun et al., 2013) and edgeR (Robinson et al., 2010). Principal component analysis (PCA) was conducted with the prcom function. Genes were considered to be related to photosynthesis if they were annotated as ‘Photosynthesis’ (map00195), ‘Photosynthesis—antenna proteins’ (map00196), ‘Carbon fixation in photosynthetic organisms’ (map00710), or ‘Photorespiration’ (M00532) in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2017). The database of Gene Ontology (GO) terms in RAP-DB (Kawahara et al., 2013) was used, and GO enrichment was analysed with the function fisher.test.
Results
Response of photosynthesis to fluctuating light
At steady-state conditions (1500 μmol photons m−2 s−1), the CO2 response of the CO2 assimilation rate, ETR I, and ETR II tended to be greater in Takanari than in Koshihikari (Supplementary Fig. S1; Supplementary Table S1), as previously reported (Takai et al., 2013). Under fluctuating light (100 μmol photons m−2 s−1 for 20 min, then 1500 μmol photons m−2 s−1 for 60 min), the induction of A400 was much faster in Takanari than in Koshihikari (Fig. 1). The A400 relative to the maximum value also showed a faster response in Takanari than in Koshihikari (Supplementary Fig. S2). At the second and third irradiations with high light after 20 min and 10 min exposure to low light, respectively, A400 also reached a higher value in Takanari than in Koshihikari. The increases in gs and ETR II upon irradiation were much faster in Takanari than in Koshihikari, and the increase in ETR I was larger in Takanari (Fig. 1, Supplementary Fig. S2). The intercellular CO2 concentration (Ci), especially in the high-light phases, was also higher in Takanari than in Koshihikari. NPQ was consistently lower in Takanari than in Koshihikari and reached steady state more quickly in Takanari (Fig. 1). These results indicate that during photosynthetic induction, Takanari was able to use more light energy to drive electron transport to generate ATP and NADPH than Koshihikari without dissipating much excess energy.
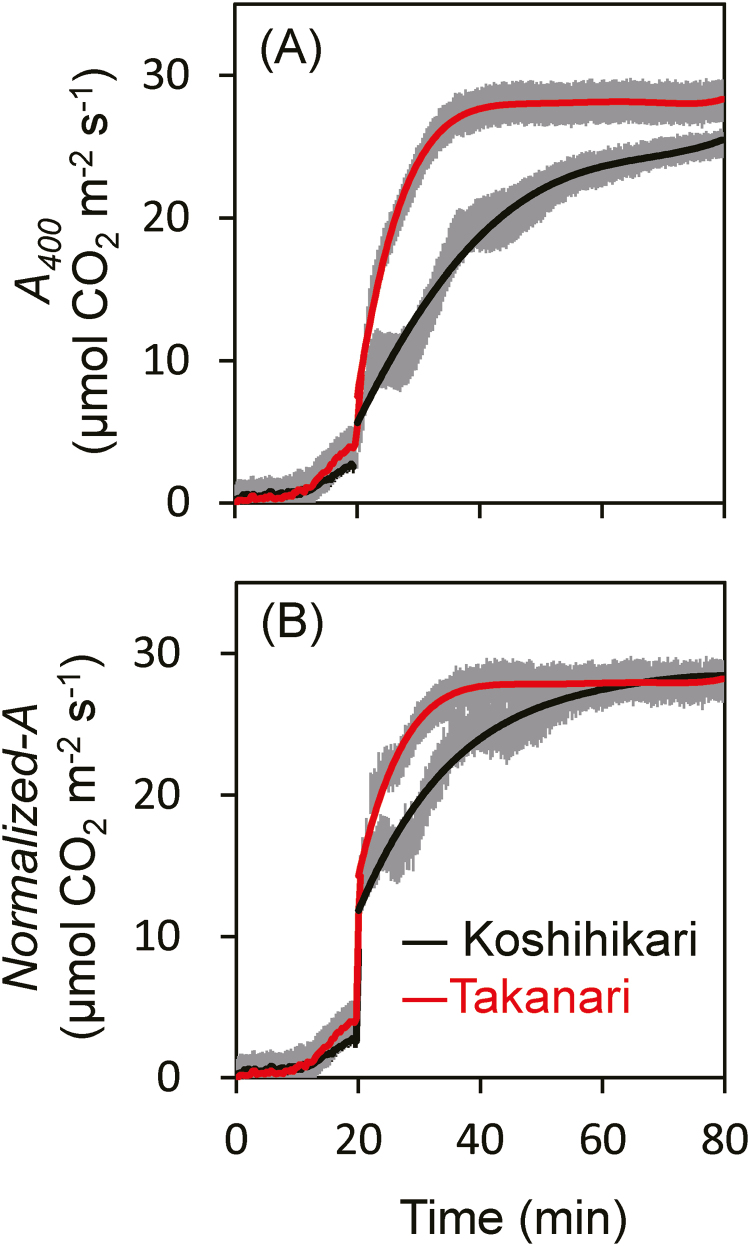
The rate of the first photosynthetic induction was estimated from the time required to reach 50% (t50) and 80% (t80) of full photosynthetic induction (Supplementary Table S2). The values of t50 for ETR I and ETR II were similar between Koshihikari and Takanari, but those of t50 for A400 and gs were smaller in Takanari. Those of t80 for ETR I were similar between Koshihikari and Takanari, but those of t80 for ETR II, A400, and gs were smaller in Takanari, showing that photosynthetic induction was significantly faster in Takanari than in Koshihikari. To remove the effects of the differences in Ci between Koshihikari and Takanari, the photosynthetic rates were normalized to a Ci of 300 μmol mol−1 (Woodrow and Mott, 1989; Yamori et al., 2012). Photosynthetic induction differed considerably between Koshihikari and Takanari at 400 μmol mol−1 CO2, but the difference was smaller under an assumption of Ci=300 μmol mol−1 (Fig. 2).
Fig. 2.
Response of normalized CO2 assimilation rate after an increase in light intensity. (A) Leaves exposed to 100 μmol photons m−2 s−1 at a CO2 partial pressure of 400 μmol mol−1 for 20 min were illuminated with 1500 μmol photons m−2 s−1 (the data are obtained from Fig. 1). (B) To remove the effects of changes in intercellular CO2 concentration (Ci), the photosynthesis rates were normalized to a Ci of 300 μmol mol−1. Values are means ±SE (n=4). (This figure is available in colour at JXB online.)
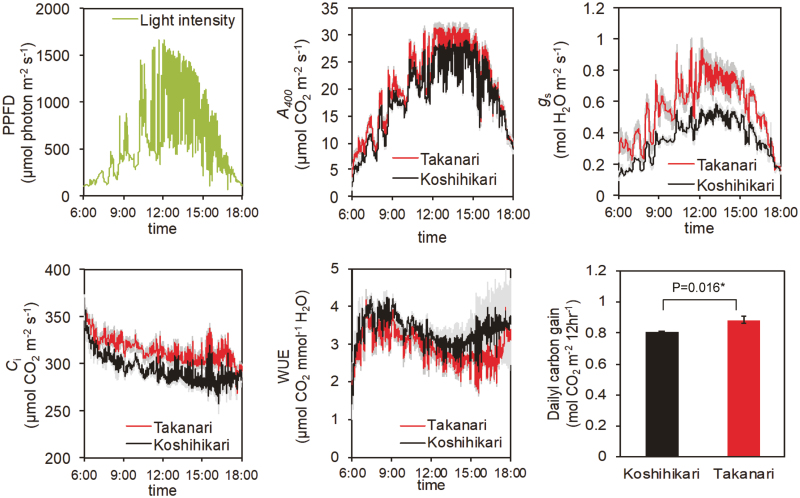
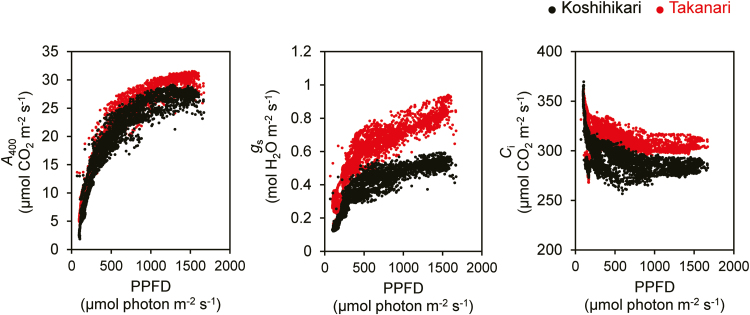
The auto-measuring program allowed repeated gas exchange measurements under light intensity and temperature conditions mimicking those in the field. A400 showed typical diurnal changes that followed the PPFD pattern, reaching a maximum at around noon and decreasing in the late afternoon (after 15.00 h; Fig. 3). Takanari had a greater A400 during most of the day than Koshihikari, except for late afternoon, when A400 was similar between cultivars. gs showed a similar pattern but with a much larger difference between cultivars. Ci was always higher and WUE was always lower in Takanari. The daily carbon gain (the cumulative value of A400 in a day) was ~10% higher in Takanari than in Koshihikari. When the photosynthetic parameters were considered as a function of PPFD, the cultivar differences in A400, gs, and Ci were apparent above 500 μmol photons m−2 s−1 (Fig. 4). The greater ETR II and lower NPQ in Takanari over the course of a day in the paddy field (Supplementary Fig. S3) suggests that the daily carbon gain would be greater in Takanari under natural field conditions.
Fig. 3.
Diurnal patterns of photosynthesis and daily carbon gain. PPFD, photosynthetic photon flux density; A400, net CO2 assimilation rate at a CO2 partial pressure of 400 μmol mol−1; gs, stomatal conductance; Ci, intercellular CO2 concentration; WUE, water use efficiency. PPFD data were obtained at the top of the rice canopy on 1 July 2016. Gas exchange measurements were conducted with LI-6400 under a CO2 concentration of 400 μmol mol−1. Values are means ±SE (n=4). (This figure is available in colour at JXB online.)
Fig. 4.
Relationships between gas exchange parameters and light intensity. PPFD, photosynthetic photon flux density; A400, net CO2 assimilation rate at a CO2 partial pressure of 400 μmol mol−1; gs, stomatal conductance; Ci, intercellular CO2 concentration. These relationships were obtained from Fig. 2. (This figure is available in colour at JXB online.)
Response of metabolites to light
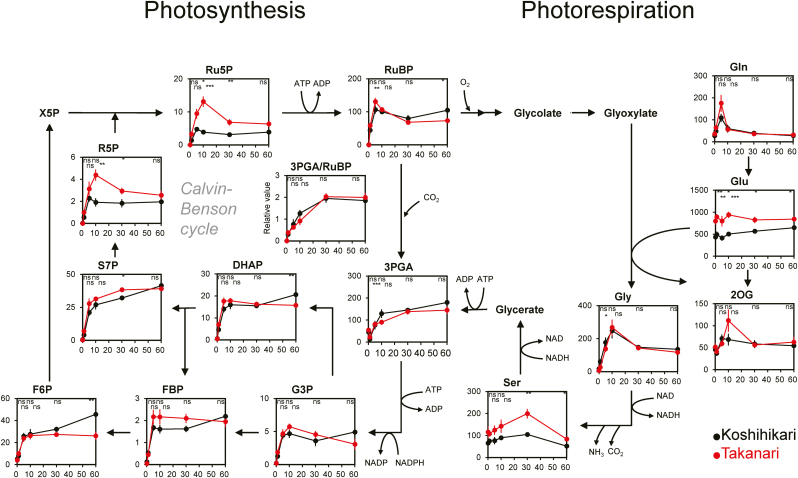
Most metabolites in the Calvin–Benson cycle showed similar responses during photosynthetic induction in both cultivars: their contents rapidly increased (within 5 min) upon irradiation, peaked at 10 min, and reached a steady state in 30 min (Fig. 5). The 3-phosphoglycerate (3PGA)/RuBP ratio increased after irradiation and plateaued within 30 min, with no cultivar differences. However, the contents of ribose 5-phosphate (R5P) and ribulose 5-phosphate (Ru5P) at 10 min and 30 min upon irradiation were significantly greater in Takanari than in Koshihikari.
Fig. 5.
Changes of metabolites for photosynthesis and photorespiration during photosynthetic induction. Vertical and horizontal axes represent metabolite content (μmol m−2) and times after irradiation (min), respectively. Asterisks indicate significance at *P<0.05, **P<0.01, and ***P<0.001 between cultivars (t-test). ns, no significant difference. Values are means ±SE (n=6). Abbreviations: X5P, xylulose 5-phosphate; Ru5P, ribulose 5-phosphate; RuBP, ribulose 1,5-bisphosphate; 3PGA, 3-phosphoglycerate; G3P, glyceraldehyde 3-phosphate; DHAP, dihydroxyacetone phosphate; FBP, fructose 1,6-bisphosphate; F6P, fructose 6-phosphate; S7P, sedoheptulose 7-phosphate; R5P, ribose 5-phosphate. (This figure is available in colour at JXB online.)
Simulations using the adjusted model of Pettersson and Ryde-Pettersson (1988) showed that the RuBP conversion to 3PGA by Rubisco (r1) upon irradiation was more rapid in Takanari than in Koshihikari (Supplementary Fig. S4), indicating that the photosynthetic metabolic flow in the Calvin–Benson cycle during photosynthetic induction was faster in Takanari, as was the CO2 assimilation rate (Fig. 1).
In the photorespiratory pathway, the content of Gly increased rapidly upon irradiation in both Takanari and Koshihikari, similar to that of RuBP (Fig. 5). The contents of Glu and Ser upon irradiation were higher in Takanari.
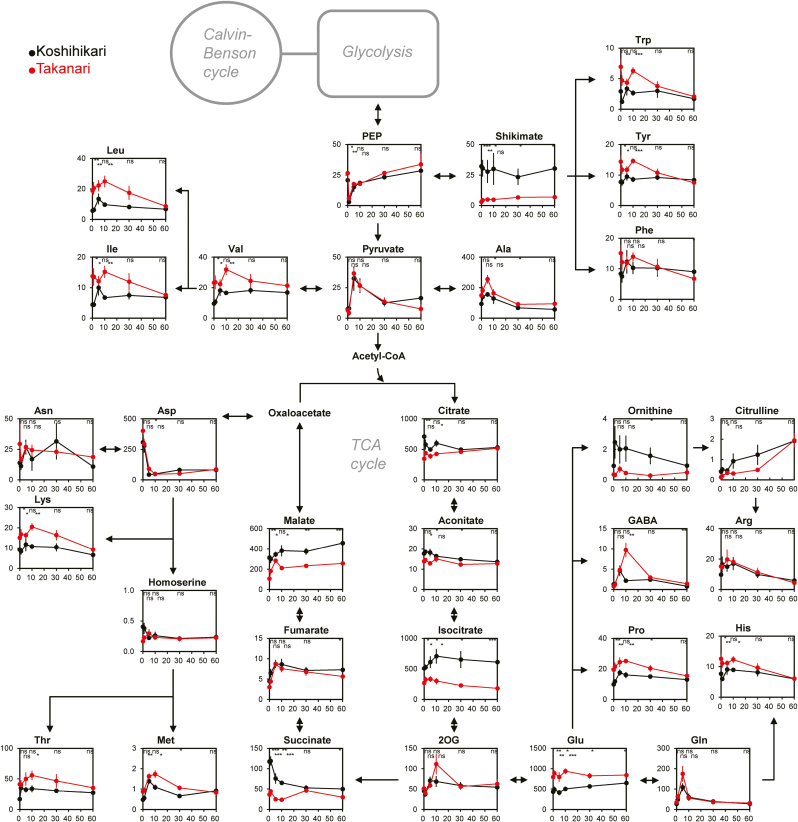
Most of the other metabolites were less affected by irradiation (Fig. 6). At all time points after irradiation, the contents of Leu, Ile, Val, Trp, Tyr, Pro, His, Glu, Lys, and Met were greater in Takanari. In contrast, the contents of several metabolites in the tricarboxylic acid (TCA) cycle (citrate, isocitrate, succinate, and malate) and shikimate were higher in Koshihikari.
Fig. 6.
Changes of metabolites for the TCA cycle and amino acid synthesis during photosynthetic induction. Vertical and horizontal axes represent metabolite content (μmol m−2) and times after illumination (min), respectively. Asterisks indicate significance at *P<0.05, **P<0.01, and ***P<0.001 between cultivars (t-test). ns, no significant difference. Values are means ±SE (n=6). PEP, phosphoenolpyruvate; 2OG, 2-oxoglutarate;. (This figure is available in colour at JXB online.)
Response of gene expression to light
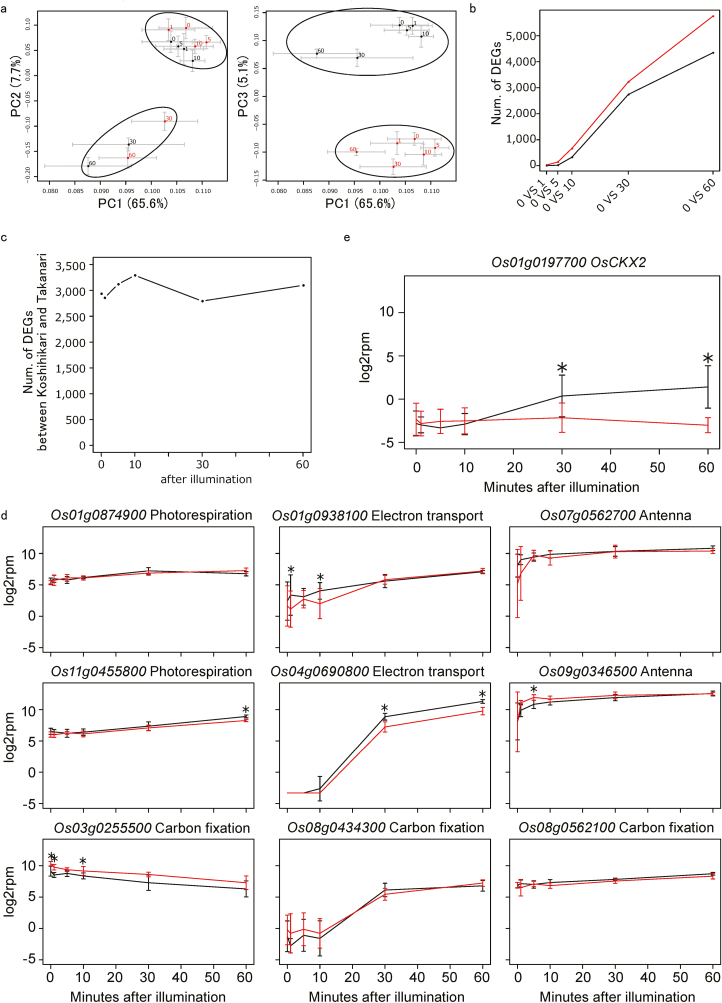
In PCA of RNA-Seq data obtained during photosynthetic induction, PC1 and PC2 showed two distinct clusters regardless of cultivar. One cluster consisted of samples at 0, 1, 5, and 10 min after irradiation, and the other consisted of samples at 30 min and 60 min (Fig. 7a). PC3 represented the difference between Koshihikari and Takanari. The number of DEGs between before (0 min) and after irradiation (1, 5, 10, 30, and 60 min) increased with time and was greater in Takanari at all time points (Fig. 7b), whereas the number of DEGs at each time point was similar between the cultivars (Fig. 7c).
Fig. 7.
Transciptomic changes during photosynthetic induction. (a) Principal component analysis for the time series RNA-Seq of Koshihikari and Takanari after illumination. (b) The numbers of differentially expressed genes (DEGs) in Koshihikari and Takanari between before (0 min) and after (1, 5, 10, 30, and 60 min) irradiation. False discovery rate=0.05. (c) The numbers of DEGs between Koshihikari and Takanari at each time point after irradiation. (d) Expression of genes relating to photosynthesis during photosynthetic induction. (e) Expression of OsCKX2 during photosynthetic induction. Values are means ±SE (n=6). *indicates that the gene was differentially expressed between Koshihikari and Takanari (adjusted P-value <0.05). (This figure is available in colour at JXB online.)
Among 193 genes related to photosynthesis in the KEGG database (Kanehisa et al., 2017), expression of nine genes changed significantly after irradiation (Fig. 7d); of these, the expression of eight genes increased. For example, the expression of Os04g0690800, which encodes PSII subunit S (PsbS), involved in photoprotection, was low until 10 min after irradiation and increased greatly at 30 min, especially in Koshihikari. The expression of Os03g0255500, which encodes phosphoenolpyruvate carboxykinase (PEPCK), which catalyses the conversion of oxaloacetate to phosphoenolpyruvate with release of CO2, decreased after irradiation. Although differences between the cultivars were significant at several time points, the overall trend of each gene was similar between them. In GO enrichment analysis of the DEGs, ‘cytokinin metabolic process’ (GO:0009690, adjusted P=0.017) and ‘cytokinin dehydrogenase activity’ (GO:0019139, adjusted P=0.017) were enriched only in Koshihikari at 30 min after irradiation. These GO terms are assigned to cytokinin oxidase/dehydrogenase (CKX) genes. Among all 11 CKX genes, we found the largest difference between Koshihikari and Takanari in Os01g0197700 (OsCKX2) (Fig. 7e), whose expression level at 30 min upon irradiation increased in Koshihikari but remained low in Takanari.
Discussion
Our metabolome analysis and simultaneous measurements of gas exchange and chlorophyll fluorescence clearly indicated a faster photosynthetic induction and response to fluctuating light in Takanari than in Koshihikari (Figs 1–4; Supplementary Figs S2–S4). The faster photosynthetic induction and greater CO2 assimilation rate at steady state contributed to the higher daily carbon gain in Takanari than in Koshihikari (Fig. 3). Our results suggest that the improvement of photosynthesis under fluctuating light offers an important way to enhance rice productivity.
Upon irradiation, electron transport in the chloroplast thylakoid membrane is activated to generate NADPH and ATP, accelerating the Calvin–Benson cycle reactions to increase RuBP levels (Yamori, 2016; Yamori et al., 2016b). The responses of both ETR I and ETR II upon irradiation were faster in Takanari than in Koshihikari, leading to the higher photosynthetic induction (Fig. 1; Supplementary Fig. S2). In the field environment, particularly when the leaves were exposed to high-intensity sunlight, ETR II was higher and NPQ response was lower in Takanari (Supplementary Fig. S3). As a consequence, Takanari directed more photon energy to NADPH and ATP production and less to heat loss through photoquenching.
Takanari had a faster increase in R5P and Ru5P in the Calvin–Benson cycle within 10 min upon irradiation than Koshihikari (Fig. 5). Their up-regulation can provide abundant RuBP to support CO2 fixation by Rubisco, leading to the similar 3PGA/RuBP ratio and higher CO2 fixation rate (Fig. 5; Supplementary Fig. S4). As mentioned in the Introduction, the activity of enzymes such as PRK, GAPDH, FBPase, and SBPase in the Calvin–Benson cycle is regulated by the ferredoxin/thioredoxin system (Buchanan, 1980). Rubisco activase activity is regulated by the ATP/ADP ratio and redox state in the chloroplast (Portis, 1995). The higher electron transport in the thylakoid membrane in Takanari probably leads to a faster increase in enzymatic activities, accelerating Calvin–Benson cycle reactions; this possibility should be tested by direct measurement of enzymatic activities during photosynthetic induction.
Stomatal conductance considerably affects photosynthetic induction (Lawson et al., 2012). Takanari had a greater response of gs upon irradiation than Koshihikari, which reflected the smaller t80 for gs (Figs 1, 2; Supplementary Table S2), and always had greater gs under fluctuating light over the course of a day (Fig. 3). The greater response of gs to light could boost rapid CO2 uptake from the air into leaves, accelerating photosynthetic induction in Takanari. The quick stomatal opening should increase the risk of water deficit in paddy rice. The water deficit is also suggested by the lower WUE in Takanari (Fig. 3). However, since Takanari has higher hydraulic conductance from roots to leaves owing to its higher root surface area than in Koshihikari (Taylaran et al., 2011), the higher water supply to leaves could support rapid stomatal opening in Takanari.
Which trait most affects the difference in photosynthetic induction between cultivars? After a sudden increase in light intensity, the Ci values dropped sharply, and the recovery was faster in Takanari than in Koshihikari (Fig. 1). In the 12 h experiment, Ci was also constitutively higher in Takanari (Fig. 3). The higher Ci can simply increase A in plants. In addition, the higher atmospheric CO2 concentration increases the apparent rate of Rubisco activity and decreases NPQ during photosynthetic induction (Kaiser et al., 2017). Because of these reasons, the higher Ci in Takanari upon irradiation may accelerate photosynthetic induction relative to Koshihikari. Elimination of the Ci difference between the cultivars decreased the A difference (Fig. 2), suggesting that the much higher gs in Takanari is a primary factor in the rapid increase in A during photosynthetic induction.
Transcriptomic responses to light continued even at 60 min after irradiation (Fig. 7a), whereas most metabolomic responses to light occurred within 10 min and the cultivar difference in photosynthetic induction was most prominent in the first 30 min (Figs 1, 5), suggesting that gene expression in response to a sudden increase in light intensity does not directly affect photosynthetic induction. Exposure to fluctuating light for 3 d alters the expression profiles of genes involved in photoprotection, photosynthesis, and photorespiration, as well as pigment, prenylquinone, and vitamin metabolism, in Arabidopsis (Schneider et al., 2019). Acclimatization to fluctuating light rather than short-term responses of gene expression patterns in rice would be of interest in future research.
OsCKX2 encodes cytokinin oxidase/dehydrogenase, which mediates cytokinin degradation; we found a significant difference in OsCKX2 expression between Koshihikari and Takanari. Ashikari et al. (2005) reported that reduced expression of OsCKX2 in the panicles of the indica cultivar Habataki, a sister cultivar of Takanari, relative to that in Koshihikari causes cytokinin accumulation in inflorescence meristems, increasing the number of reproductive organs and grain yield. Thus, Takanari leaves might accumulate higher cytokinin levels than Koshihikari leaves. Since cytokinin suppresses stomatal closure triggered by abscisic acid (Stoll et al., 2000; Tanaka et al., 2006), the higher cytokinin level in Takanari leaves may accelerate stomatal opening during photosynthetic induction, although the effect of cytokinin on stomatal opening is still under debate (Daszkowska-Golec and Szarejko, 2013).
Advanced mapping populations for genetic analysis, such as backcrossed inbred lines and chromosome segment substitution lines derived from a cross between Koshihikari and Takanari, have been developed (Takai et al., 2014, Fukuda et al., 2018). The use of these mapping populations could contribute to elucidation of the genetic and molecular mechanisms underlying the differences in photosynthetic response to fluctuating light between the two cultivars.
In conclusion, the present study clearly showed that the high-yielding indica cultivar Takanari has higher photosynthetic induction than the commercial japonica cultivar Koshihikari, resulting in a higher daily carbon gain under fluctuating light resembling natural light. The higher photosynthetic induction is related to a greater electron transport rate and larger metabolic flow of the Calvin–Benson cycle. These higher values in Takanari would be caused primarily by its higher stomatal conductance. In the future, the genetic factors related to each physiological trait should be examined in a population developed from a Koshihikari/Takanari cross. Such studies will contribute to increasing the photosynthetic capacity under fluctuating light, as occurs naturally in the field.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Differences in steady-state photosynthetic characteristics between Koshihikari and Takanari.
Table S2. Time to reach 50% (t50) or 80% (t80) of the final steady-state photosynthesis after an increase in irradiance.
Table S3. Rate equations for the kinetic model of the Calvin–Benson cycle.
Table S4. Differential equations and initial concentrations.
Fig. S1. CO2 response curves of gas exchange and chlorophyll fluorescence parameters.
Fig. S2. Relative changes of photosynthetic inductions.
Fig. S3. Diurnal patterns of chlorophyll fluorescence parameters in the paddy field.
Fig. S4. Flux analysis of the Calvin–Benson cycle reaction.
Fig. S5. Expression levels of CKX genes during photosynthetic induction.
Author contributions
SA and WY designed the experiments; WY, YT, SK, and SO analysed gas exchange; SA and WY performed the sampling and extraction for metabolome and transcriptome analysis; AM and MK-Y conducted the metabolome analysis; MK, AT, and AN conducted transcriptome analysis; YT and HS conducted flux analysis; SA and WY prepared the manuscript; and AM, YT, and AN provided critical feedback on the drafts.
Acknowledgements
This work was supported in part by the Japan Science and Technology Agency, PRESTO (grant nos JPMJPR13B1, JPMJPR16Q5, JPMJPR12B6, and JPMJPR13BB to SA, YT, AN, and WY), by JSPS KAKENHI (grant nos 16H06552, 18H02185, and 18KK0170 to WY, and 16K18643 to SA), and by the Japan Science and Technology Agency, CREST grant no. JPMJCR15O2 (to SA and AN). We are grateful to Ms Y. Yamashita for assistance with metabolite and RNA extraction.
References
- Adachi S, Nakae T, Uchida M, et al. 2013. The mesophyll anatomy enhancing CO2 diffusion is a key trait for improving rice photosynthesis. Journal of Experimental Botany 64, 1061–1072. [DOI] [PubMed] [Google Scholar]
- Adachi S, Nito N, Kondo M, Yamamoto T, Arai-Sanoh Y, Ando T, Ookawa T, Yano M, Hirasawa T. 2011. Identification of chromosomal regions controlling the leaf photosynthetic rate in rice by using a progeny from japonica and high-yielding indica varieties. Plant Production Science 14, 118–127. [Google Scholar]
- Adachi S, Yoshikawa K, Yamanouchi U, Tanabata T, Sun J, Ookawa T, Yamamoto T, Sage RF, Hirasawa T, Yonemaru J. 2017. Fine mapping of carbon assimilation rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Frontiers in Plant Science 8, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M, Sakakibara H, Lin S, Yamamoto T, Takashi T, Nishimura A, Angeles ER, Qian Q, Kitano H, Matsuoka M. 2005. Cytokinin oxidase regulates rice grain production. Science 309, 741–745. [DOI] [PubMed] [Google Scholar]
- Ashraf M, Akram NA. 2009. Improving salinity tolerance of plants through conventional breeding and genetic engineering: an analytical comparison. Biotechnology Advances 27, 744–752. [DOI] [PubMed] [Google Scholar]
- Buchanan BB. 1980. Role of light in the regulation of chloroplast enzymes. Annual Review of Plant Physiology 31, 341–374. [Google Scholar]
- Chen CP, Sakai H, Tokida T, Usui Y, Nakamura H, Hasegawa T. 2014. Do the rich always become richer? Characterizing the leaf physiological response of the high-yielding rice cultivar Takanari to free-air CO2 enrichment. Plant & Cell Physiology 55, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszkowska-Golec A, Szarejko I. 2013. Open or close the gate—stomata action under the control of phytohormones in drought stress conditions. Frontiers in Plant Science 4, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Kondo K, Ikka T, Takai T, Tanabata T, Yamamoto T. 2018. A novel QTL associated with rice canopy temperature difference affects stomatal conductance and leaf photosynthesis. Breeding Science 68, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Uebayashi N, Sato F, Endo T. 2014. Physiological functions of PsbS-dependent and PsbS-independent NPQ under naturally fluctuating light conditions. Plant & Cell Physiology 55, 1286–1295. [DOI] [PubMed] [Google Scholar]
- Ishida S, Uebayashi N, Tazoe Y, Ikeuchi M, Homma K, Sato F, Endo T. 2014. Diurnal and developmental changes in energy allocation of absorbed light at PSII in field-grown rice. Plant & Cell Physiology 55, 171–182. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Kashima M, Nagano AJ, Ishikawa-Fujiwara T, Kamei Y, Todo T, Mori K. 2017. Unfolded protein response transducer IRE1-mediated signaling independent of XBP1 mRNA splicing is not required for growth and development of medaka fish. eLife 6, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser E, Kromdijk J, Harbinson J, Heuvelink E, Marcelis LF. 2017. Photosynthetic induction and its diffusional, carboxylation and electron transport processes as affected by CO2 partial pressure, temperature, air humidity and blue irradiance. Annals of Botany 119, 191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. 2017. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research 45, D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemura T, Homma K, Ohsumi A, Shiraiwa T, Horie T. 2007. Evaluation of genotypic variation in leaf photosynthetic rate and its associated factors by using rice diversity research set of germplasm. Photosynthesis Research 94, 23–30. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, et al. 2013. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laisk A, Eichelmann H, Oja V, Eatherall A, Walker DA. 1989. A mathematical model of the carbon metabolism in photosynthesis. Difficulties in explaining oscillations by fructose 2,6-bisphosphate regulation. Proceedings of the Royal Society B: Biological Sciences 237, 389–415. [Google Scholar]
- Lawson T, Kramer DM, Raines CA. 2012. Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Current Opinion in Biotechnology 23, 215–220. [DOI] [PubMed] [Google Scholar]
- McAusland L, Vialet-Chabrand S, Davey P, Baker NR, Brendel O, Lawson T. 2016. Effects of kinetics of light-induced stomatal responses on photosynthesis and water-use efficiency. New Phytologist 211, 1209–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi A, Takahashi H, Takahara K, Hirabayashi T, Nishimura Y, Tezuka T, Kawai-Yamada M, Uchimiya H. 2010. Principal component and hierarchical clustering analysis of metabolites in destructive weeds; polygonaceous plants. Metabolomics 6, 146–155. [Google Scholar]
- Nagano AJ, Honjo MN, Mihara M, Sato M, Kudoh H. 2015. Detection of plant viruses in natural environments by using RNA-Seq. Methods in Molecular Biology 1236, 89–98. [DOI] [PubMed] [Google Scholar]
- Naumburg E, Ellsworth DS. 2002. Short-term light and leaf photosynthetic dynamics affect estimates of daily understory photosynthesis in four tree species. Tree Physiology 22, 393–401. [DOI] [PubMed] [Google Scholar]
- Noguchi K, Tsunoda T, Miyagi A, et al. 2018. Effects of elevated atmospheric CO2 on respiratory rates in mature leaves of two rice cultivars grown at a Free-Air CO2 enrichment site and analyses of the underlying mechanisms. Plant & Cell Physiology 59, 637–649. [DOI] [PubMed] [Google Scholar]
- Pearcy RW. 1990. Sunflecks and photosynthesis in plant canopies. Annual Review of Plant Biology 41, 421–453. [Google Scholar]
- Pearcy RW. 1999. Responses of plants to heterogeneous light environments. In: Pugnaire F, Valladares F, eds. Handbook of functional plant ecology. New York: Marcel Dekker, 269–314. [Google Scholar]
- Pettersson G, Ryde-Pettersson U. 1988. A mathematical model of the Calvin photosynthesis cycle. European Journal of Biochemistry 175, 661–672. [DOI] [PubMed] [Google Scholar]
- Portis AR., Jr 1995. The regulation of Rubisco by Rubisco activase. Journal of Experimental Botany 46, 1285–1291. [DOI] [PubMed] [Google Scholar]
- Qu M, Hamdani S, Li W, Wang S, Tang J, Chen Z, Song Q, Li M, Zhao H, Chang T. 2016. Rapid stomatal response to fluctuating light: an under-explored mechanism to improve drought tolerance in rice. Functional Plant Biology 43, 727–738. [DOI] [PubMed] [Google Scholar]
- R Core Team 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; https://www.r-project.org/ [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassenrath-Cole GF, Pearcy RW. 1992. The role of ribulose-1,5-bisphosphate regeneration in the induction requirement of photosynthetic CO2 exchange under transient light conditions. Plant Physiology 99, 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassenrath-Cole GF, Pearcy RW. 1994. Regulation of photosynthetic induction state by the magnitude and duration of low light exposure. Plant Physiology 105, 1115–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafaro AP, Yamori W, Carmo-Silva AE, Salvucci ME, von Caemmerer S, Atwell BJ. 2012. Rubisco activity is associated with photosynthetic thermotolerance in a wild rice (Oryza meridionalis). Physiologia Plantarum 146, 99–109. [DOI] [PubMed] [Google Scholar]
- Schneider T, Bolger A, Zeier J, Preiskowski S, Benes V, Trenkamp S, Usadel B, Farré EM, Matsubara S. 2019. Fluctuating light interacts with time of day and leaf development stage to reprogram gene expression. Plant Physiology 179, 1632–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery RA, Walker BJ, Weber APM, Ort DR. 2018. The impacts of fluctuating light on crop performance. Plant Physiology 176, 990–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleh MA, Tanaka Y, Nomoto Y, Iwahashi Y, Nakashima K, Fukuda Y, Long SP, Shiraiwa T. 2016. Factors underlying genotypic differences in the induction of photosynthesis in soybean [Glycine max (L.) Merr]. Plant, Cell & Environment 39, 685–693. [DOI] [PubMed] [Google Scholar]
- Stoll M, Loveys B, Dry P. 2000. Hormonal changes induced by partial rootzone drying of irrigated grapevine. Journal of Experimental Botany 51, 1627–1634. [DOI] [PubMed] [Google Scholar]
- Sun J, Nishiyama T, Shimizu K, Kadota K. 2013. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T, Adachi S, Taguchi-Shiobara F, et al. 2013. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Scientific Reports 3, 2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai T, Ikka T, Kondo K, et al. 2014. Genetic mechanisms underlying yield potential in the rice high-yielding cultivar Takanari, based on reciprocal chromosome segment substitution lines. BMC Plant Biology 14, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sano T, Tamaoki M, Nakajima N, Kondo N, Hasezawa S. 2006. Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. Journal of Experimental Botany 57, 2259–2266. [DOI] [PubMed] [Google Scholar]
- Taylaran RD, Adachi S, Ookawa T, Usuda H, Hirasawa T. 2011. Hydraulic conductance as well as nitrogen accumulation plays a role in the higher rate of leaf photosynthesis of the most productive variety of rice in Japan. Journal of Experimental Botany 62, 4067–4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SH, Long SP. 2017. Slow induction of photosynthesis on shade to sun transitions in wheat may cost at least 21% of productivity. Philosophical Transactions of the Royal Society B: Biological Sciences 372, 20160543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bezouw RFHM, Keurentjes JJB, Harbinson J, Aarts MGM. 2019. Converging phenomics and genomics to study natural variation in plant photosynthetic efficiency. The Plant Journal 97, 112–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialet-Chabrand S, Matthews JS, Simkin AJ, Raines CA, Lawson T. 2017. Importance of fluctuations in light on plant photosynthetic acclimation. Plant Physiology 173, 2163–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodrow IE, Mott KA. 1989. Rate limitation of non-steady-state photosynthesis by ribulose-1,5-bisphosphate carboxylase in spinach. Australian Journal of Plant Physiology 16, 487–500. [Google Scholar]
- Yamauchi S, Takemiya A, Sakamoto T, Kurata T, Tsutsumi T, Kinoshita T, Shimazaki K. 2016. The plasma membrane H+-ATPase AHA1 plays a major role in stomatal opening in response to blue light. Plant Physiology 171, 2731–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W. 2016. Photosynthetic response to fluctuating environments and photoprotective strategies under abiotic stress. Journal of Plant Research 129, 379–395. [DOI] [PubMed] [Google Scholar]
- Yamori W, Irving LJ, Adachi A, Busch FA. 2016a Strategies for optimizing photosynthesis with biotechnology to improve crop yield. In: Pessarakli M, ed. Handbook of Photosynthesis, 3rd edn. Boca Raton, FL: CRC Press, 741–759. [Google Scholar]
- Yamori W, Kondo E, Sugiura D, Terashima I, Suzuki Y, Makino A. 2016b Enhanced leaf photosynthesis as a target to increase grain yield: insights from transgenic rice lines with variable Rieske FeS protein content in the cytochrome b6/f complex. Plant, Cell & Environment 39, 80–87. [DOI] [PubMed] [Google Scholar]
- Yamori W, Makino A, Shikanai T. 2016c A physiological role of cyclic electron transport around photosystem I in sustaining photosynthesis under fluctuating light in rice. Scientific Reports 6, 20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamori W, Masumoto C, Fukayama H, Makino A. 2012. Rubisco activase is a key regulator of non-steady-state photosynthesis at any leaf temperature and, to a lesser extent, of steady-state photosynthesis at high temperature. The Plant Journal 71, 871–880. [DOI] [PubMed] [Google Scholar]
- Yamori W, Takahashi S, Makino A, Price GD, Badger MR, von Caemmerer S. 2011. The roles of ATP synthase and the cytochrome b6/f complexes in limiting chloroplast electron transport and determining photosynthetic capacity. Plant Physiology 155, 956–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Wang Y, Ort DR, Long SP. 2013. e-Photosynthesis: a comprehensive dynamic mechanistic model of C3 photosynthesis: from light capture to sucrose synthesis. Plant, Cell & Environment 36, 1711–1727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.