Abstract
Tissue-specific gene transcription can be determined by the use of either positive-acting or negative-acting DNA regulatory elements. We have analyzed a promoter from the growth-associated protein 43 (GAP-43) gene and found that it uses both of these mechanisms to achieve its high degree of neuron-specific activity. Two novel transcription factor binding sites, designated Cx1 and Cx2, drive promoter activity in neurons from developing cerebral cortex but not in several other cell types. The promoter also contains an activator protein 1 (AP-1) site that contributes to activity in neurons. The AP-1 site can drive promoter activity in a wide range of non-neuronal cells that express little or no endogenous GAP-43, but only in the absence of a tissue-specific repressive element located downstream of the GAP-43 TATA box. These findings suggest that the GAP-43 repressive element plays an important role in allowing AP-1 signaling pathways to modulate activity of the GAP-43 gene in neurons, without also causing inappropriate activation by AP-1 transcription factors in other cell types.
Keywords: AP-1, Cx1, Cx2, GAP-43, gene, transcription, neuron, repressive element, NGFI-A, SNOG element, BIPPUR element, TATA box
The activation or repression of specific genes during the course of neuronal differentiation, and throughout adult life, often relies on signal-transducing pathways common to many neuronal and non-neuronal cell types. Yet activation of a particular signaling cascade can evoke a very different response in neurons than in non-neuronal cells. For example, a number of extracellular signaling events can activate AP-1 transcription factors (Sheng and Greenberg, 1990; Herschman, 1991), but the set of genes that are activated or repressed varies greatly depending on cell type and history. What mechanisms allow a specific gene to respond to a particular signaling pathway in one cell type, but not to the same signaling pathway in a different cellular context?
We have examined this issue using a neuron-specific promoter from the growth-associated protein 43 (GAP-43) gene. The GAP-43 gene, which codes for an axonal growth cone protein, is widely expressed in developing neurons during periods of axon elongation (Jacobson et al., 1986; Skene, 1989) and is also expressed in glial cells under some circumstances (Deloulme et al., 1993; Plantinga et al., 1993). Within neurons, GAP-43 expression declines as synaptic contacts are established but may be reactivated in response to axonal injury (Skene, 1992). Induction of GAP-43 after axotomy is correlated with activation of c-Jun, a common component of AP-1 transcription factors (Herdegen and Zimmermann, 1994; Schaden et al., 1994; Herdegen et al., 1997), and the GAP-43 gene contains a phylogenetically conserved AP-1 consensus sequence (Groen et al., 1995). Despite widespread expression of c-Jun and activation of AP-1-dependent pathways in many cell types, however, the GAP-43 gene is expressed only in the nervous system, and perhaps transiently in a few other cell types (Stocker et al., 1992; Heuss et al., 1995; Anchan et al., 1997).
Several potential mechanisms could contribute to cell type-specific activation of a commonly used response element such as an AP-1 site. First, the sequence of the response element itself might be recognized preferentially by some versions of the heterodimeric AP-1 transcription factor (Hai and Curran, 1991; Karin et al., 1997). Second, the effectiveness of AP-1 could be enhanced by cooperative binding with a second factor that is more restricted in its tissue distribution (Bassuk and Leiden, 1995). Thirdly, negative-acting and positive-acting transcription factors could compete for the same binding site (Igarashi et al., 1994). A fourth, more indirect mechanism is to use a tissue-selective repressive element to counter activity that would otherwise be driven by positive-acting elements recognized in many cell types (Chong et al., 1995; Schoenherr and Anderson, 1995a,b).
We show here that the GAP-43 promoter uses this fourth mechanism to produce neuron-specific activation through its AP-1 site. The conserved AP-1 element in the GAP-43 gene can serve as an effective target for activation in many different cell types, but the effect of this AP-1 mediated activation is counteracted in most cells by a separate repressive element that restricts promoter activity to neurons. The repressive element does not, however, account for all of the tissue specificity of the small GAP-43 promoter we have focused on. We also find that two novel protein binding sites located close to the AP-1 consensus element further contribute to neuron-specific gene expression by boosting promoter activity in some populations of neurons.
MATERIALS AND METHODS
DNA constructs. The wild-type 386 bp GAP-43 promoter/luciferase reporter gene construct pGL3A-386 has been described previously (Weber and Skene, 1997). The modified versions of the 386 bp promoter shown in Figure 1 were produced by PCR in whichHindIII restriction sites were added to the ends of the PCR primers. The E1b TATA box and transcription start site shown in Figure2, which includes bases −34 to +11 of the sequence published by Wu and coworkers (1987), was made by annealing complementary oligonucleotides and then cloning this fragment into the BglII andHindIII sites of the plasmid pGL3A-Basic (Weber and Skene, 1997) to obtain pGL3A-Viral TATA. For the additional constructs in Figure 2, BglII and BamHI sites were added to the ends of the AP-1/Cx region (the 90 bp sequence beginning 59 bp downstream of the GAP-43 TATA box) by PCR, and one or two copies of the AP-1/Cx region were cloned into the BglII site of pGL3A-TATA. The mutations described in the Figure 5 legend were introduced into partially complementary synthetic oligonucleotides that were extended by Klenow polymerase and then cloned into theXbaI and XmaI sites of the 386 bp GAP-43 promoter. The mutations used to disrupt the GAP-43 repressive element have already been described (Weber and Skene, 1997). Promoter constructs made by PCR or synthesized DNA were confirmed by DNA sequencing.
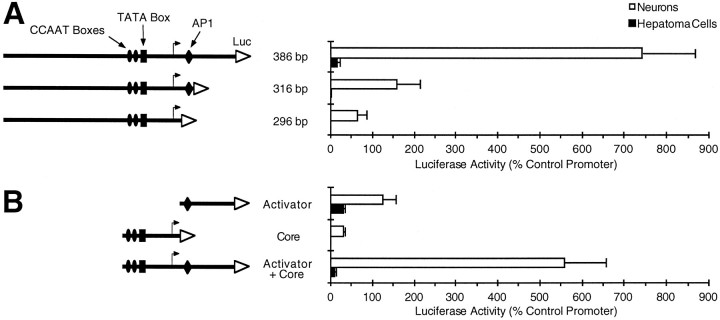
Fig. 1.
The activity of a neuron-specific 386 bp GAP-43 promoter depends on a synergy between a core promoter and a downstream activator region. The rat GAP-43 promoter constructs shown schematically were tested for the ability to drive the expression of a luciferase reporter gene (Luc) in primary neuronal cultures or hepatoma cells. The effect of deletions from the promoter’s 3′ end are shown inA, whereas a demonstration of the synergistic effect between the activator region and the core promoter is shown inB. Luciferase activity for each construct is normalized to the activity of a modified adenovirus promoter (see Materials and Methods). SEMs are based on at least three experiments. The neuronal cultures are dissociated cells from rat embryonic cerebral cortex treated with an antimitotic agent to kill the majority of non-neuronal cells. CCAAT, TATA, andAP-1 consensus sequences are labeled. The bent arrow designates the most 5′ transcription start site, which is located ∼45 bp downstream of the TATA box (Nedivi et al., 1992;Ortoft et al., 1993). The RNase protection assays performed by Ortoft and coworkers (1993) on human transcripts indicate that there are more dominant transcription start sites located ∼70 and 100 bp downstream of the TATA box (the transcription start sites for the rat promoter are very likely to be the same as for human, because the rat and human promoter sequences are highly conserved in this region). The 96 bp sequence that we refer to as the core promoter includes sequences from 6 bp upstream of the CCAAT boxes to 59 bp downstream of the TATA box. The 90 bp activator region includes the AP-1 consensus sequence.
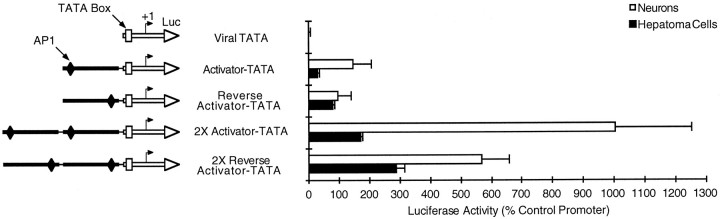
Fig. 2.
The activator region is likely to contain transcription factor binding sites. The activator region that was defined in the previous figure was placed upstream of a TATA box and transcription start site that were borrowed from the adenovirus E1b promoter. As indicated in the schematics, the activator region was inserted as one or two copies in either the forward or reverse orientation. Promoter activity in neurons and hepatoma cells is reported in the graph (same as Fig. 1).
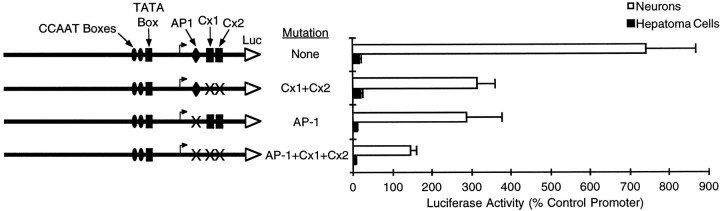
Fig. 5.
Transcription factor binding sites in the AP-1/Cx activator region are required for the majority of the activity of the 386 bp GAP-43 promoter. Promoter constructs with mutations in proposed transcription factor binding sites (AP-1, Cx1, Cx2) were tested for the ability to drive expression of a reporter gene in neurons and hepatoma cells (same as Fig. 1). The AP-1, Cx1, and Cx2 binding sites were altered by using mutations 1, 2, and 6, respectively, of Figure3.
Cell cultures and transfections. Primary cultures of dissociated rat cerebral cortex from embryonic day 18 were produced and cultured as described previously (Weber and Skene, 1997). Three days after plating, the cortical cultures were treated with 5 μm arabinose C for 24 hr to kill the majority of proliferating cells. HTC hepatoma cells (Thompson et al., 1966), RAT2 fibroblast-like cells (Topp, 1981), B1.1 Schwannoma cells (Anton et al., 1995), C6 glioma cells (Benda et al., 1968), PC12 chromaffin-like cells (Greene and Tischler, 1976), and CAD cells, a CNS catecholaminergic neuronal cell line (Qi et al., 1997), were all cultured and transfected with lipofectin reagent (Life Technologies, Gaithersburg, MD) as described previously (Weber and Skene, 1997). Cultures were harvested 2 d after transfection with 250 μl of Promega reporter lysis buffer (catalog #E397A; Promega, Madison, WI).
Luciferase and chloramphenicol acetyltransferase assays.Luciferase assays were conducted with a Turner luminometer (Promega) and either the Promega luciferase assay system (catalog #E1500) and 20 μl of cell lysate or 50 μl cell lysate, 180 μl of assay buffer (Brasier and Fortin, 1995), and injection of 100 μl of 2 mm luciferin. Promoter activity for each construct was determined using duplicate plates and a minimum of three independent experiments. The promoterless luciferase construct pGL3A-Basic was included in every experiment so that the minor luciferase signal driven by vector sequences could be subtracted from activity driven by promoter constructs. All promoter–luciferase constructs were cotransfected with a plasmid containing the Rous sarcoma virus promoter and the gene for chloramphenicol acetyltransferase (CAT) (Gorman et al., 1982). CAT enzyme activity, measured using tritiated acetate (Nordeen et al., 1987), was used to monitor transfection efficiency for each cell culture dish, and luciferase activity was normalized to the CAT activity. To compare promoter activities between cell types, luciferase expression for each promoter construct was normalized to a modified version of the adenovirus E1b promoter that has a deletion of a single G residue upstream of the TATA box (GGGGCGGGGC to GGGGCGGGC). This control promoter has a similar signal-to-noise ratio in neurons and hepatoma cells (8:1 and 9:1, respectively), but is approximately eightfold less active than the wild-type E1b promoter. Two other viral promoters were also tested with the luciferase reporter gene in both neurons and hepatoma cells. The herpes simplex virus thymidine kinase promoter had signal-to-noise ratio of 58:1 and 100:1 in neurons and hepatoma cells, respectively, and the Rous sarcoma virus promoter had signal-to-noise ratios of 2600:1 and 1800:1 in neurons and hepatoma cells, respectively. The observation that three different viral promoters expressed well in both cell types indicates that the strong preference for expression in neurons of the GAP-43 promoter is unlikely to be attributable to a poor transfection efficiency in hepatoma cells.
Protein extracts and electrophoretic mobility shift assays.Nuclear extracts from postnatal day 5 rat cerebral cortex or liver were prepared using the method of Gorski and coworkers (1986). The nuclear extracts were dialyzed against 25 mm HEPES, pH 7.9, 100 mm KCl, 0.1 mm EDTA, 10% glycerol, 0.1 mm phenylmethylsulfonal fluoride, and 1 mmdithiothreitol. Small scale whole-cell or nuclear extracts from cell cultures were made by the method of Dent and Latchman (1994). Total protein for all extracts was quantitated according to the Bradford method (Smith, 1987).
Electrophoretic mobility shift assays (EMSAs) were conducted using 5% polyacrylamide (29:1 ratio of acrylamide to bis-acrylamide) gels in low ionic strength buffer (Chodosh, 1988). Oligonucleotide probes were prepared and labeled with 32P as described previously (Weber and Skene, 1997). The unique identity of the rat repressive element probe, frog repressive element probe, rat AP-1 probe, and frog AP-1 probe of Figure 9 were confirmed by restriction digest withXbaI, Nci I, AluI, andHinfI restriction enzymes, respectively. Binding reactions were conducted for 60 min on ice using 20,000–50,000 cpm of radiolabeled probe (∼0.1–0.4 ng DNA), 2 μg poly(dI-dC), 15 mm HEPES, pH 7.9, 60 mm KCl, 12% glycerol, 1 mm EDTA, and 1 mm dithiothreitol in a reaction volume of 50 μl. The binding reactions used 15 ug of nuclear extracts from postnatal cerebral cortex, 10 ug of liver nuclear extract, or 35–60 μg of whole-cell extracts. Under our conditions, the strongest bands were achieved with 6 mm MgCl2 for AP-1 or Liv1, 1 mm MgCl2 for repressive element binding, and no MgCl2 for Cx1 and Cx2. Samples were loaded directly onto gels and run at 45 mA for ∼1 hr in a 4°C room. The gels were dried and then exposed to film overnight at −80°C with an intensifying screen.
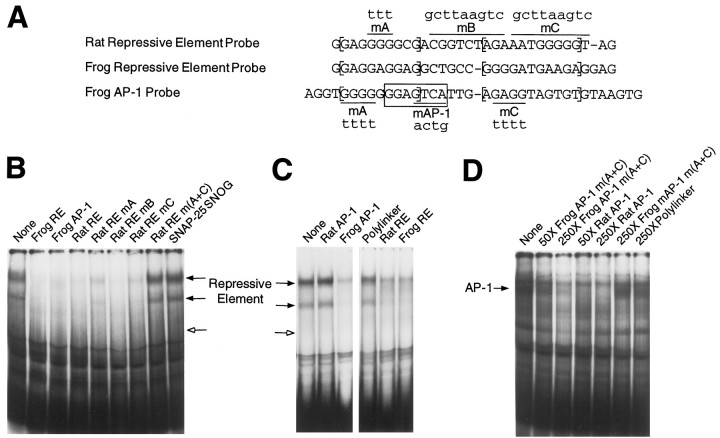
Fig. 9.
Mammalian proteins bind to a potential frog GAP-43 repressive element and AP-1 site. A, Rat and frog repressive element probes for EMSAs are shown withbrackets enclosing an NGFI-A/EGR consensus sequence (Fig. 8, site A) and a purine-rich sequence that overlaps with site C of Figure 8. Sequences in the frog AP-1 probe that are similar to the bracketed regions of the repressive element probes are bracketed and discussed in Results. The AP-1 consensus sequence isboxed and should be read on the strand complementary to the sequence shown. Mutations in the EMSA probes are shown inlower case letters, and the wild-type sequences they replace are overlined or underlined.B, EMSAs with hepatoma whole-cell extracts and radiolabeled frog repressive element probe. Competitor probes were used at a 200-fold molar excess relative to the radiolabeled probe. Thehollow arrow marks the relative mobility of the SNOG element-specific band that binds to the rat repressive element, as we have shown previously (Weber and Skene, 1997), but an equivalent band was not obtained with the frog repressive element. The SNAP-25 SNOG competitor probe contains a high-affinity binding site for the SNOG element, but does not contain an NGFI-A/EGR consensus sequence.C, EMSAs with radiolabeled frog AP-1 probe and hepatoma whole-cell extracts. The sequence of the rat AP-1 competitor probe is given in Figure 3A. Note that repressive element-specific rather than AP-1-specific bands were obtained (see Results and Discussion). We verified the identity of each of the probes in question (see Materials and Methods) and conducted several repeat experiments to confirm these unexpected results. An independently synthesized batch of the frog AP-1 probe yielded the same results.D, Same as C except that the radiolabeled frog AP-1 probe contains mutations mA and mC and the magnesium concentration has been optimized to 6 mm rather than 1 mm (see Materials and Methods). When the EMSA inC was conducted at 6 mm magnesium rather than 1 mm, the repressive element binding was less intense, but we still could not detect any AP-1-specific bands (data not shown). Competitors were used at a 50-fold or 250-fold excess relative to the radiolabeled probe.
RESULTS
Identification of an activator region downstream of the GAP-43 TATA box
We have demonstrated previously that a 386 bp rat GAP-43 promoter has a strong preference for expression in neurons (Nedivi et al., 1992), primarily because of a repressive element that blocks promoter activity in non-neuronal cells (Weber and Skene, 1997). To identify and characterize additional cis-acting elements involved in the regulation of this promoter, we screened several subfragments of the 386 bp region for the ability to drive the expression of a luciferase reporter gene in transfected cell cultures (Fig.1). Deletion of 70 bp from the promoter’s 3′ end (to obtain the 316 bp construct of Fig.1A) results in a nearly fivefold loss of promoter activity in primary cultures of neurons from developing rat cerebral cortex. Deletion of an additional 20 bp, which includes an AP-1 consensus sequence (Eggen et al., 1994), produces a 296 bp promoter with <10% of the activity of the original 386 bp promoter. This dramatic loss of activity indicates that the 90 bp region spanned by these deletions contains one or more positive-acting elements required for maximal promoter activity in neurons.
On its own, this 90 bp putative activator region is able to drive only a low level of reporter gene expression (Fig. 1B). Strong promoter activity in neurons requires both the activator region and a core promoter, which contains CCAAT and TATA boxes that have been shown previously to be required for promoter activity (Weber and Skene, 1997). The synergistic interaction between the activator and the core promoter could be attributable to positive-acting transcription factors that bind to sequences in the activator region. However, the ability of the activator region to boost reporter gene expression also could be explained by the presence of multiple transcription start sites in this region (Ortoft et al., 1993) or by post-transcriptional effects, because the activator region is located downstream of the TATA box and the sense strand sequence of this region is transcribed into mRNA.
To investigate the mechanism by which the activator exerts its effects, we placed this region upstream of a heterologous, viral TATA box (Fig.2). In this context, a single copy of the activator region could elicit limited promoter activity, whereas two copies of the activator elicited much stronger promoter activity. The effects of the activator region were orientation dependent. When placed in the forward orientation, as either one or two copies, the activator region expressed five times more strongly in neurons than hepatoma cells. When placed in the reverse orientation, activity in hepatoma cells increased, whereas activity in neurons decreased (Fig. 2).
The activator region used in these constructs does not include the tissue-specific repressive element that accounts for the majority of the neuronal specificity of the 386 bp GAP-43 promoter (Weber and Skene, 1997). Therefore, the stronger activity in neurons, when the activator was tested in the forward orientation, suggests that the activator region may contain an additional element(s) that contributes to the tissue specificity of the GAP-43 promoter. The observation that two copies of the activator placed in reverse can still drive a substantial level of reporter gene expression indicates that the activator can stimulate transcription, because the ability of this region to act upstream of a TATA box, and in the reverse orientation, cannot be explained by post-transcriptional mechanisms mediated by the presence of activator sequences in the mRNA transcript.
Identification of protein binding sites in the activator region
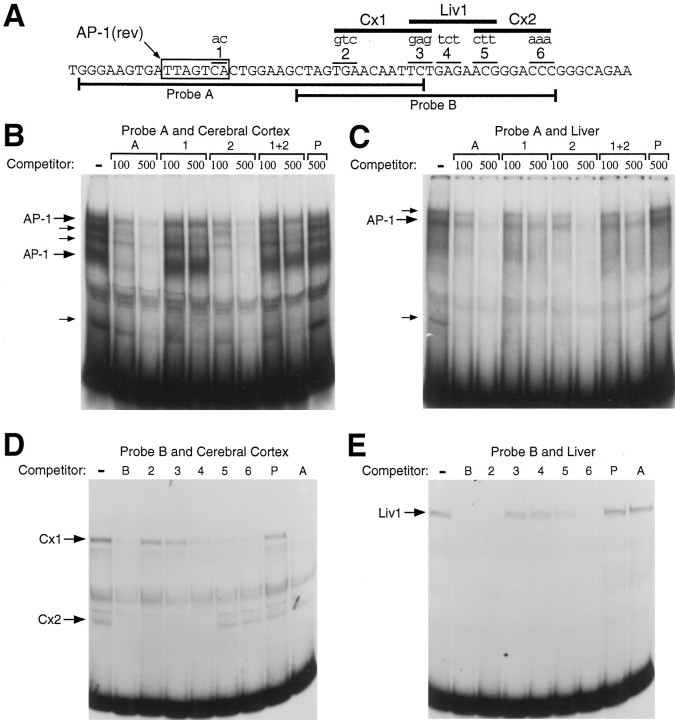
To identify potential transcription factor binding sites in the activator region, we tested small fragments of this sequence (Fig.3A) for the ability to bind proteins in EMSAs. Nuclear extract from postnatal rat cerebral cortex, where endogenous GAP-43 expression is high, contains several binding activities that recognize a small radiolabeled probe that includes the AP-1 consensus sequence (Fig. 3B). Competition experiments using unlabeled wild-type or mutated probes demonstrated that two of these binding activities are clearly dependent on the AP-1 sequence. Liver nuclear extracts contain a binding complex of exactly the same mobility as one of the AP-1 binding activities from cerebral cortex (Fig. 3C). Mutation of the AP-1 site slightly, but reproducibly, diminished the ability of an unlabeled probe to compete for the liver AP-1 binding activity. For both cortical and liver nuclear extracts, there were additional binding activities that do not appear to be dependent on the AP-1 sequence (Fig.3B,C).
Fig. 3.
Identification of protein binding sites in the AP-1/Cx activator region. A, The DNA sequences used to make probes for EMSAs are indicated by bars below the sequence for the first 60 bp of the 90 bp AP-1/Cx activator region. The AP-1 consensus sequence is boxed, and mutations introduced into the probes are shown in lower case letters. The sequence alteration used to mutate the AP-1 site ( TGACTAA to GTACTAA on the antisense strand) has been demonstrated previously to disrupt AP-1 binding and activity in another promoter (Lee et al., 1991). The location of novel putative transcription factor binding sites are indicated by barslabeled as Cx1, Liv1, and Cx2.B, EMSAs with radiolabeled probe A and nuclear extract from postnatal day 5 rat cerebral cortex. The first lane contains only the radiolabeled probe and nuclear extract, whereas the additional lanes include either a 100- or 500-fold molar excess of unlabeled wild-type probe A or probe A with mutations 1, 2, or the combination of mutations 1 and 2. P stands for a polylinker DNA with no similarity to the probe A sequence. If a mutation affects protein binding, then the competitor with that mutation should compete less effectively or not at all. AP-1 specific-bands are labeled withlarge arrows, and smaller arrows indicate additional sequence specific bands that do not appear to depend on the AP-1 sequence. C, EMSAs with probe A and liver nuclear extract. D, EMSAs with probe B and nuclear extract from postnatal cerebral cortex. Numbers designate probe B with mutations 2, 3, 4, 5, or 6. Competitor probes were used at a 500-fold molar excess. Cx1 refers to the binding site defined by mutations 2 and 3, whereas Cx2 refers to the binding site defined by mutations 5 and 6. E, EMSAs with probe B and liver nuclear extract. Competitors were used at a 50-fold molar excess in this case. Liv1 refers to the binding site defined by mutations 3, 4, and 5.
To identify additional protein binding sites in the activator region, we tested a second small radiolabeled probe corresponding to a portion of the activator region downstream of the AP-1 consensus sequence (Fig.3A, Probe B). This probe is recognized by two distinct protein complexes present in nuclear extracts from postnatal cerebral cortex. We refer to these binding activities as Cx1 and Cx2, because they were detected in nuclear extracts from cerebral cortex (Fig. 3D) but not from liver (Fig. 3E). Competition with unlabeled oligonucleotides containing a series of small (3 bp) mutations showed that Cx1 and Cx2 recognize distinct sequences within Probe B (Fig. 3D). EMSAs with liver extracts revealed an additional protein complex not detected in cortical extracts. This third binding activity, which we name Liv1, recognizes a DNA sequence that overlaps the Cx1 and Cx2 binding sites. The striking difference in protein binding patterns between cortical and liver nuclear extracts suggests that the region immediately downstream of the AP-1 site could contribute to the neuron-specific activity of the GAP-43 promoter.
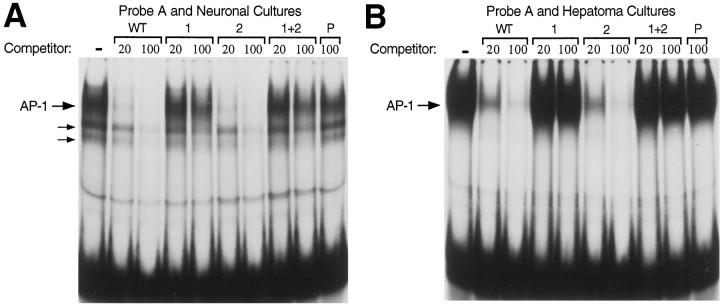
Because our functional promoter assays are done by transient transfection of cell cultures, we repeated the EMSA experiments using protein extracts from cells cultured under the same conditions as for our promoter assays. Very strong AP-1-specific binding of the same electrophoretic mobility was detected in both the neuronal and hepatoma cell cultures (Fig. 4). When the probe for sequences downstream of the AP-1 site was used with neuronal extracts, Cx1 binding was detected, but not Cx2 (data not shown). The inability to detect Cx2 could be because of either its absence in the neuronal cultures or perhaps some difference in the procedures for preparing protein extracts from cell cultures rather than cerebral cortex. The Liv1-specific binding detected with liver extracts was also detected with hepatoma protein extracts (data not shown).
Fig. 4.
AP-1-specific binding in protein extracts from neuronal and hepatoma cultures. A, EMSAs with probe A of Figure 3 and whole-cell extract from cultures of embryonic rat cerebral cortex. B, Same as A except with whole-cell extracts from hepatoma cell cultures. Nuclear extracts from the neuronal and hepatoma cell cultures gave similar results (data not shown).
Taken together, the EMSA experiments indicate that the GAP-43 gene’s AP-1 site can be recognized in both neuronal and non-neuronal cells, but that protein binding sites downstream of the AP-1 site may be used quite differently in neurons and non-neuronal cells. It should also be noted, however, that there might be some tissue-specific differences in what versions of the AP-1 transcription factor preferentially bind the AP-1 site and are best able to interact with other transcription factors involved in the regulation of the GAP-43 gene.
Identification of positive-acting elements in the activator region
To determine whether the protein binding sites identified by EMSAs are important for the activity of the 386 bp GAP-43 promoter in neurons, we modified the promoter with the same small mutations that disrupted protein binding to the AP-1, Cx1, and Cx2 sites (Fig.5). Combined mutation of the Cx1 and Cx2 sites reduced promoter activity in transfected neurons by more than twofold. Mutation of the AP-1 consensus sequence resulted in a similar loss of activity. A combination of mutations in the AP-1, Cx1, and Cx2 sites abolished the majority of the GAP-43 promoter’s activity in neurons (Fig. 5). However, the remaining neuron-specific activity is still higher than when this entire region was deleted (Fig.1A, 296 bp), indicating that there may still be additional cis-acting elements in this activator region.
Because the AP-1 and Cx sites are located downstream of the TATA box, the loss of activity caused by mutations in these sites could result from post-transcriptional effects attributable to an altered mRNA sequence or perturbation of transcription start sites. However, the observation that the same small mutations that reduce promoter activity also cause a loss of protein binding is more consistent with the proposal that this region contains a cluster of positive-acting transcription factor binding sites. We have designated this region, which is required for the activity of the 386 bp promoter in neurons, as the AP-1/Cx region.
Evaluation of the tissue specificity of the AP-1/Cx activator region
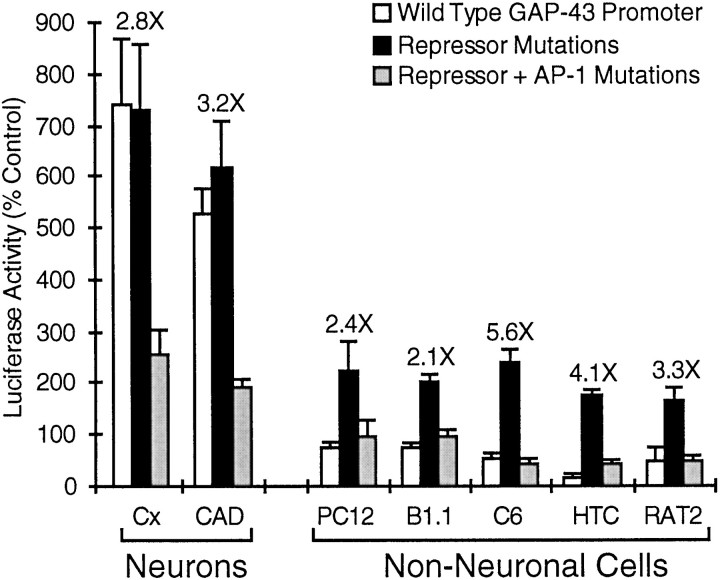
Mutational analysis demonstrates that the AP-1 site activates the 386 bp GAP-43 promoter in neurons but not in hepatoma cells (Fig. 5), yet EMSA assays detected very robust AP-1 binding activity in hepatoma cell extracts (Fig. 4B), suggesting that some additional factor(s) prevents or counteracts the actions of AP-1 on the GAP-43 promoter. We have shown previously that the majority of the tissue specificity of the 386 bp GAP-43 promoter can be accounted for by a repressive element located between the TATA box and the AP-1/Cx activator region (Weber and Skene, 1997). Mutations in this repressive element result in a 3-fold to 10-fold increase in promoter activity in various non-neuronal cells. Figure 6shows that this activation of the 386 bp promoter in non-neuronal cells is dependent on the AP-1 site, indicating that the AP-1 motif in the GAP-43 promoter can be recognized by positive-acting AP-1 factors in many different cell types. The five non-neuronal cell lines used (naive PC12 cells, a chromaffin cell-derived line; B1.1, Schwannoma cells; C6, glioma cells; HTC, hepatoma cells; RAT2, fibroblast-like cell line) were chosen for their lack of endogenous GAP-43 expression. The three neural but non-neuronal cell lines (PC12, B1.1, C6) may express low levels of endogenous GAP-43, but under our culture conditions we detected GAP-43 protein only in the primary cortical neuronal cultures and the CAD neuronal cell line (Weber and Skene, 1997).
Fig. 6.
Evaluation of the activity of the AP-1/Cx activator region in a wide range of cell types. We have shown previously that mutation of a repressive element located downstream of the TATA box of the 386 bp GAP-43 promoter results in an increase in promoter activity in non-neuronal cells (Weber and Skene, 1997). Here we compare the activity of the wild-type 386 bp GAP-43 promoter, the GAP-43 promoter with mutations in the previously characterized repressive element, and the GAP-43 promoter with mutations in both the repressive element and the AP-1 site (same AP-1 mutation as Fig. 3). These promoter–reporter gene constructs were tested for activity in primary neuronal cultures (Cx for neuronal cultures from rat embryonic cerebral cortex), a neuronal cell line with high levels of endogenous GAP-43 (CAD cells), and five non-neuronal cell lines: PC12, B1.1, C6, HTC, and RAT2 (discussed in Results). Note that in each of the non-neuronal cell types, mutation of the AP-1 site eliminates most or all of the activity that had been achieved by disruption of the repressive element.
In each of the non-neuronal cell types tested, mutation of the AP-1 site, in the context of a GAP-43 promoter in which the repressive element had already been eliminated, resulted in a loss of activity nearly equal in magnitude to the activity that had been gained by mutation of the repressive element. This comparison indicates that the AP-1 element is capable of driving GAP-43 promoter activity in non-neuronal cells but is normally prevented from doing so by the tissue-specific GAP-43 repressive element.
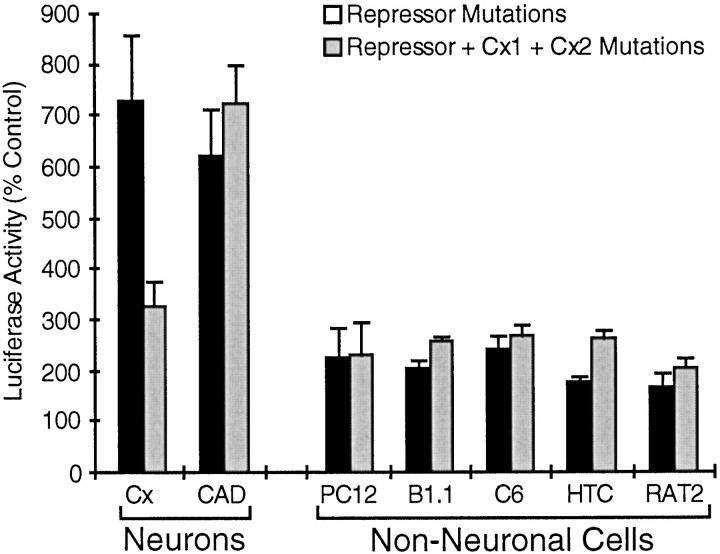
We used the same cell culture systems to evaluate the functional contribution of the protein binding sites located immediately downstream of the AP-1 site. A mutation in the Liv1 binding site (Fig.3, mutation 4) appeared to have no effect in our assay systems, even when combined with the repressive element mutations (data not shown). Mutation of the Cx1 and Cx2 sites in the context of the GAP-43 promoter already had the repressive element mutations resulted in an approximately twofold loss of activity in primary neuronal cultures from rat embryonic cerebral cortex (Cx) (Fig. 7). However, these mutations did not result in a loss of activity in a neuronal cell line (CAD cells, a CNS catecholaminergic cell line) (Qi et al., 1997) or in any of the five non-neuronal cell lines tested. Thus, in contrast to the AP-1 element, the Cx1 and/or Cx2 site(s) do not appear to be activated in non-neuronal cells and so do not depend on the GAP-43 repressive element to restrict their activity to neurons. The contribution of the Cx elements to neuron-specific promoter activity appears to differ among different populations of neurons, which may reflect differences in subtypes of neurons or other differences between primary cortical cultures and immortalized CAD cells.
Fig. 7.
The Cx1 and Cx2 sites contribute to neuron-specific expression of the GAP-43 promoter. The 386 bp GAP-43 promoter with mutations in the repressive element is compared with the same promoter with additional mutations in the Cx1 and Cx2 sites (same methods as in Fig. 6).
Comparison of mammalian and amphibian promoter sequences
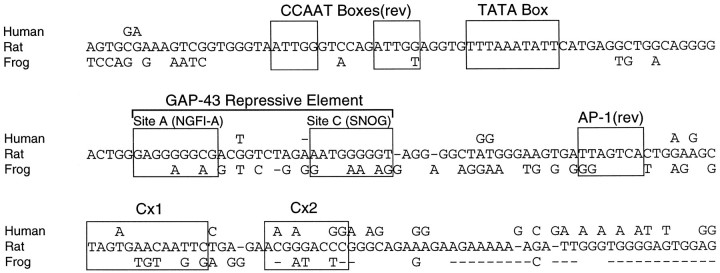
The 386 bp GAP-43 promoter from rat is preferentially expressed in the developing nervous system of transgenic zebrafish, indicating that one or more of the cis-acting elements that regulate this promoter must be highly conserved among vertebrates (Reinhard et al., 1994). To identify phylogenetically conserved cis-regulatory elements in this promoter, we compared sequences from the corresponding regions of the human, rat, and frog GAP-43 genes (Fig.8). In the region encompassing the CCAAT and TATA boxes, there is a 78 bp stretch of human sequence that is absolutely identical to the rat sequence, and the equivalent amphibian sequence is >80% identical. This high degree of sequence conservation suggests that the functional role of these elements is likely to be critical for proper regulation of the GAP-43 gene.
Fig. 8.
Comparison of mammalian and amphibian GAP-43 sequences. Human (Ortoft et al., 1993), rat (GenBank accession numberM88356; Nedivi et al., 1992), and Xenopus (GenBank accession number Y09834; submitted by L. H. Schrama, Rudolf Magnus Institute for Neuroscience, Utrecht, The Netherlands) GAP-43 sequences surrounding and downstream of the GAP-43 TATA box are shown. The human and frog sequences are reported only where they deviate from the rat sequence in our alignment. A dash indicates a space inserted to obtain the best alignment. Conservation of the boxed elements is discussed in Results, and consensus sequences that should be read on the complementary strand are marked as reverse (rev).
The GAP-43 AP-1 site also appears to be phylogenetically conserved. In the alignment shown in Figure 8, the human and rat genes have the same AP-1 sequence, but the corresponding frog sequence deviates by 2 bp. However, this frog sequence (TGACTCC as read on the antisense strand) has been identified as an AP-1 site in the promoter of the rat JE gene (Timmers et al., 1990) and its homolog the human monocyte chemotactic protein 1 (MCP-1) gene (Shyy et al., 1995). These sequence comparisons suggest that the frog GAP-43 gene is likely to have an AP-1 binding site in the same location, relative to the TATA box, as the rat and human genes.
Downstream of the AP-1 site, the human and frog sequences have no obvious similarities to the rat Cx2 binding site defined in Figure 3. The Cx1 binding site, however, is likely to be present in the human sequence, because the rat and human sequences have 12 identical bp within a 13 bp region spanning the Cx1 site. It is less obvious whether there is a Cx1 site in the frog sequence. At the corresponding location in the frog, only 8 of the 13 bp are identical to the rat sequence, and we have not determined whether this similarity is sufficient to allow binding of either the mammalian Cx1 factor or any corresponding protein from frog.
In the region of the GAP-43 repressive element, the human sequence is nearly identical to that of the rat, whereas the frog sequence deviates enough to suggest that only one of the two factors that bind to the rat sequence is likely to also bind to the frog sequence. We found previously that this region of the rat GAP-43 gene is recognized by at least two distinct protein factors. One of these factors recognizes a single site of ∼9 bp and also binds to sequences found downstream of the 25 kDa synaptosome-associated protein (SNAP-25) and neuronal nitric oxide synthase (nNOS) gene’s TATA boxes, allowing us to derive the SNOG consensus sequence (A/G)ATG(A/G)GGG(C/T) (Weber and Skene, 1997). The rat and human promoters both contain this sequence (Fig. 8, siteC), but the frog sequence varies enough from the consensus that it is unclear whether the SNOG element is conserved between amphibians and mammals.
A second factor that recognizes the rat GAP-43 repressive element binds to a much larger site that includes sequences in both site A and site C of Figure 8 (Weber and Skene, 1997). Site A corresponds to a consensus binding sequence for members of the NGFI-A/EGR family of transcription factors. The equivalent frog sequence varies from the 9 bp rat site A sequence by an adenosine instead of a guanosine in the fifth position and an adenosine instead of a cytidine in the eighth position. Intriguingly, a study using recombinant members of the NGFI-A/EGR family of transcription factors to select random DNA sequences has demonstrated that there is a considerable degree of variability in the DNA sequences to which these proteins bind (Swirnoff and Milbrandt, 1995), and both the rat and frog site A fit the experimentally derived NGFI-A/EGR consensus sequence. Although the originally described binding sequence for this family was GCGGGGGCG, the selection experiments showed that some members of the family would choose an adenosine in the second or eighth position ∼5–10% of the time and an adenosine in the fifth position anywhere from 24 to 31% of the time.
Binding by the factor that recognizes the NGFI-A/EGR-like sequence (site A) also requires a second sequence (site C) that overlaps the SNOG element (Weber and Skene, 1997). Similarity between the rat and frog sequences in the general region of site C is less obvious, but both the rat and frog sequences have a conserved thymidine that is surrounded by several purines on either side. Taking into account that very large binding sites can accommodate a substantial amount of variability in the sequences to which they will bind (Schoenherr et al., 1996), the sequence similarities in site A and the general region of site C suggest that the frog gene is likely to have a binding site for at least one of the factors that recognizes the rat GAP-43 repressive element and that this binding site is located the same distance downstream of the TATA box in human, rat, and frog genes.
Binding of mammalian proteins to frog GAP-43 promoter sequences
To determine whether any of the mammalian proteins that bind the rat GAP-43 repressive element can actually bind to similar sequences in the frog gene, the rat and frog repressive element probes shown in Figure 9A were used as competitors in EMSAs. The sequence-specific bands obtained with hepatoma extracts and the frog repressive element (Fig. 9B) had the same mobilities as the complexes we observed previously with the rat GAP-43 repressive element (Weber and Skene, 1997), except that we did not detect a band of the same mobility as the complex that bound to only the SNOG consensus. Unlabeled rat and frog repressive elements competed about equally well for the radiolabeled probe. Moreover, a combination of mutations in both site A and site C of the rat repressive element were required to fully disrupt its ability to compete for the frog repressive element.
Quite surprisingly, a DNA probe spanning the frog AP-1 site was a highly effective competitor for the radiolabeled frog (Fig.9B) or rat (data not shown) repressive element. Moreover, the radiolabeled frog AP-1 probe produced bands of the same mobility as the repressive element probe (Fig. 9C). The rat AP-1 probe does not compete for these bands, but the frog and rat repressive element probes do (Fig. 9C).
Careful comparison of the frog AP-1 probe sequence with the frog and rat repressive elements provides an explanation for this unexpected binding pattern (Fig. 9A). Our mutational analysis indicated that the GAP-43 repressive element comprises two parts: site A, which fits the NGFI-A/EGR consensus sequence, and a highly purine-rich site C. The rat and frog site A define a consensus sequence of GAGG(A/G)GGG(A/C)G. Overlapping the frog AP-1 site is a sequence that varies from this site A consensus by only one base. Downstream of site A, the rat and frog repressive elements share a conserved thymidine flanked by five purines on either side. The equivalent sequences in the vicinity of the frog AP-1 site also have a thymidine in the middle of a purine-rich sequence. These sequence comparisons, along with our EMSA results, indicate that the frog GAP-43 gene contains two complete copies of one of the protein binding sites found in the rat GAP-43 repressive element.
Although the frog AP-1 probe clearly bound one of the factors that recognizes the GAP-43 repressive element, it showed no binding mediated by the AP-1 consensus (Fig. 9C). The apparent lack of AP-1-specific binding could be attributable to interference by the factor that binds to the repressive element-like sequence that overlaps the frog AP-1 site. To eliminate this potential interference, we made a frog AP-1 probe in which the sequences surrounding the AP-1 site were disrupted (Fig. 9A, mutations A and C). With this new probe, we detected a band of the same mobility as had been obtained with the rat AP-1 probe in Figure 4B (Fig. 9D). The frog AP-1 sequence, which varies by 2 bp from the equivalent rat sequence, is bound only weakly by the mammalian proteins that had produced very robust binding to the rat GAP-43 AP-1 site. However, this binding is AP-1 specific, because the rat AP-1 probe was an extremely effective competitor, and mutation of the frog AP-1 site eliminated its ability to compete.
These binding assays indicate that mammalian proteins can recognize at least some of the frog GAP-43 sequences. The frog gene contains two complete copies of the binding site for one of the factors that recognizes the rat GAP-43 repressive element, and the second copy can interfere with binding to the AP-1 site that it overlaps.
DISCUSSION
Neuron-specific gene transcription can be accomplished by a combination of positive and negative methods
Tissue-specific gene transcription can be achieved by two basic methods: (1) the use of positive-acting DNA elements that are recognized by transcription factors present in certain cell types, but not others, and (2) the use of negative-acting DNA elements to prevent transcription, in inappropriate cell types, that would otherwise be driven by widely recognized positive-acting DNA elements. As an example of the first method, binding sites for Pit-1, a transcription factor that is restricted to cells of the pituitary gland in adults, play a key role in determining the pituitary-specific expression of the growth hormone and prolactin genes (Lefevre et al., 1987; Nelson et al., 1988;Ingraham et al., 1990). A good example of the second method is the repressive element 1/neuron-restrictive silencer element (RE1/NRSE), which binds a factor, the RE1-silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF), that prevents transcription from the type II sodium channel and SCG10 promoters in non-neural cells (Kraner et al., 1992; Mori et al., 1992; Chong et al., 1995; Schoenherr and Anderson, 1995a,b).
The 386 bp GAP-43 promoter we have studied here uses both methods for achieving neuron-specific expression. Two novel transcription factor binding sites, Cx1 and Cx2, boost promoter activity in neuronal cultures from embryonic rat cerebral cortex, but not in any of several other cell types that we tested. The majority of the tissue specificity of this promoter, however, is conferred by a repressive element located downstream of the GAP-43 TATA box (Weber and Skene, 1997). We have now shown that the GAP-43 repressive element, which is unrelated to the RE1/NRSF element, is sufficient to block the activity in non-neuronal cells that would otherwise be driven by a highly promiscuous AP-1 element. This strategy allows the GAP-43 gene to take advantage of a signal transduction pathway(s) present in a wide range of tissue types yet still remain highly tissue specific.
Regulation of GAP-43 transcription by AP-1
AP-1 transcription factors are found in most cell types and are activated in response to a fairly wide range of extracellular stimuli (Sheng and Greenberg, 1990; Herschman, 1991). The response of individual genes to AP-1 activation depends on cell type and history (Morgan and Curran, 1995), suggesting that transcription factors that can cooperate or interfere with AP-1 driven activity are likely to be important determinants of promoter specificity. The tissue-specific GAP-43 repressive element should allow signaling events associated with neuronal differentiation to use the AP-1 signaling pathway(s) without the undesired side effect of activating GAP-43 transcription outside of the nervous system. The repressive element may also play a role in preventing AP-1 from causing overexpression of the GAP-43 gene in glial cells. Primary cultures of Schwann cells that express endogenous GAP-43 fail to express a small GAP-43 promoter that includes the AP-1 site (Plantinga et al., 1994), despite the fact that cultured Schwann cells express high levels of c-Jun (De Felipe and Hunt, 1994). Instead, transcriptional activity is driven by sequences immediately adjacent to the GAP-43 protein coding region (a proposed 230 bp TATA-less promoter).
Correlative evidence suggests that AP-1 transcription factors may contribute to activation of the GAP-43 gene in neurons during axon outgrowth. One of the components of AP-1, c-Jun, is highly elevated during axon regeneration by dorsal root ganglion neurons (Herdegen and Zimmermann, 1994; Schaden et al., 1994). Moreover, bothc-Jun (Herdegen and Zimmermann, 1994) and GAP-43 (Van der Zee et al., 1989; Schreyer and Skene, 1991) will remain elevated for months if the regenerating axons are prevented from reinnervating their target tissue. c-Jun and GAP-43 are also elevated in adult retinal ganglion cells under conditions conducive to axon regeneration (Herdegen and Zimmermann, 1994; Schaden et al., 1994).
If c-Jun is a positive regulator of the GAP-43 gene, it may need to work in cooperation with other transcription factors, including other members of the AP-1 family. Differences in the composition of AP-1 can determine its binding affinity and which intracellular signaling pathways it responds to (Hai and Curran, 1991; Kerppola and Curran, 1994; Gass and Herdegen, 1995). Identification of the GAP-43 AP-1 site as a functional regulatory element opens up a new avenue for determining what version(s) of AP-1 is involved in regulation of the GAP-43 gene.
Regulation of GAP-43 transcription by Cx1 and Cx2
The Cx1 and/or Cx2 elements are important for activity in primary neuronal cultures from embryonic rat cerebral cortex but are not used in any of the non-neuronal cells we have tested. Under our culture conditions, the Cx1 and Cx2 sites do not appear to be used in CAD cells, although this murine CNS catecholaminergic neuronal cell line has high levels of endogenous GAP-43 (Qi et al., 1997; Weber and Skene, 1997) and expresses the 386 bp GAP-43 promoter construct nearly as well as the primary neuronal cultures. This differential use of the Cx1 and/or Cx2 elements could reflect differences in neuronal subtype. Alternatively, these elements might be used only at certain stages of neuronal differentiation or in response to specific signaling events that differ between our two neuronal culture systems.
Phylogenetic conservation of cis-acting elements proximal to the GAP-43 TATA box
The regulation of the endogenous GAP-43 gene involves many additional regulatory elements outside of the small promoter region on which we have focused here (Nedivi et al., 1992; Ortoft et al., 1993;Perrone-Bizzozero et al., 1993; Eggen et al., 1994; Reinhard et al., 1994; Vanselow et al., 1994; Weber and Skene, 1997). However, the high degree of phylogenetic conservation (Groen et al., 1995) in this small region indicates that many of the elements we have characterized are likely to play a critical role in the regulation of the GAP-43 gene and perhaps in other genes involved in neuronal differentiation and axon outgrowth.
The neuron-specific activity of the 386 bp rat GAP-43 promoter is determined by a combinatorial code involving both positive (Cx1 and Cx2) and negative (the repressive element) tissue-specific elements and an AP-1 site that can be recognized in a wide range of cell types. Although we have not yet evaluated the novel Cx1 and Cx2 sites separately, phylogenetic comparisons suggest that the Cx1 site is likely to be the more important regulatory element. The rat Cx2 sequence is not present in the human or frog promoters, whereas the Cx1 site is conserved between rat and human promoters and has at least some similarities between the rat and frog sequences.
The rat GAP-43 repressive element has binding sites for two different factors. Binding by one of these factors depends only on the 9 bp SNOG consensus sequence (G/A)ATG(G/A)GGG (C/T), which is also found in the SNAP-25 and neuronal NOS genes (Weber and Skene, 1997). The second factor that binds to the GAP-43 repressive element recognizes a bipartite purine-rich (BIPPUR) element. The first part of this element has a striking similarity to the consensus sequence for the NGFI-A/EGR family of transcription factors (Swirnoff and Milbrandt, 1995). The second part is a purine-rich sequence that overlaps the SNOG element in the rat and human GAP-43 genes.
Although the Xenopus GAP-43 gene does not contain a SNOG element, it does contain two complete copies of the BIPPUR element. The second copy appears to be able to interfere with binding to the frog AP-1 site. The ability of mammalian proteins to recognize these amphibian BIPPUR elements strongly suggests that this element plays an important phylogenetically conserved role in the regulation of the GAP-43 gene.
We have not established a causal relationship between protein binding and inhibition of transcription by the GAP-43 repressive element. However, the factor that binds to the BIPPUR element is a good candidate for a repressor, because its binding is well correlated with transcriptional repression in non-neuronal cells (Weber and Skene, 1997). The functional role of the SNOG element is currently unclear. However, identification of the BIPPUR and SNOG elements as distinct components of the mammalian GAP-43 repressive element should facilitate the identification of these elements in other neuronal genes and lead to a clarification of the regulatory roles of the factors that bind these elements.
Footnotes
This work was supported by National Institutes of Health Grant EY07397. The CAD cells were kindly provided by Dona Chikaraishi and Yanping Qi, and the B1.1 Schwannoma cells were kindly provided by Bill Matthew.
Correspondence should be addressed to Pate Skene, Box 3209, Duke University Medical Center, Durham, NC 27710.
Dr. Weber’s present address: Beth Israel Deaconess Medical Center, 330 Brookline Avenue, RW663, Boston, MA 02215.
REFERENCES
- 1.Anchan RM, Drake DP, Haines CF, Gerwe EA, LaMantia AS. Disruption of local retinoid-mediated gene expression accompanies abnormal development in the mammalian olfactory pathway. J Comp Neurol. 1997;379:171–184. [PubMed] [Google Scholar]
- 2.Anton ES, Hadjiargyrou M, Patterson PH, Matthew WD. CD9 plays a role in Schwann cell migration in vitro. J Neurosci. 1995;15:584–595. doi: 10.1523/JNEUROSCI.15-01-00584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassuk AG, Leiden JM. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 4.Benda P, Lightbody J, Sato G, Levine L, Sweet W. Differentiated rat glial cell strain in tissue culture. Science. 1968;161:370–371. doi: 10.1126/science.161.3839.370. [DOI] [PubMed] [Google Scholar]
- 5.Brasier AR, Fortin JJ. Nonisotopic assays for reporter gene activity. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; New York: 1995. pp. 9.7.12–9.7.14. [DOI] [PubMed] [Google Scholar]
- 6.Chodosh LA. Mobility shift DNA-binding assay using gel electrophoresis. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; New York: 1988. pp. 12.2.1–12.2.10. [DOI] [PubMed] [Google Scholar]
- 7.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 8.De Felipe C, Hunt SP. The differential control of c-Jun expression in regenerating sensory neurons and their associated glial cells. J Neurosci. 1994;14:2911–2923. doi: 10.1523/JNEUROSCI.14-05-02911.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deloulme JC, Laeng P, Janet T, Sensenbrenner M, Baudier J. Expression of neuromodulin (GAP-43) and its regulation by basic fibroblast growth factor during the differentiation of O-2A progenitor cells. J Neurosci Res. 1993;36:147–162. doi: 10.1002/jnr.490360205. [DOI] [PubMed] [Google Scholar]
- 10.Dent CL, Latchman DS. The DNA mobility shift assay. In: Latchman DS, editor. Transcription factors. Oxford UP; Oxford: 1994. pp. 1–26. [Google Scholar]
- 11.Eggen BJL, Nielander HB, Rensen-de Leeuw MG, Schotman P, Gispen WH, Schrama LH. Identification of two promoter regions in the rat B-50/GAP-43 gene. Mol Brain Res. 1994;23:221–234. doi: 10.1016/0169-328x(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 12.Gass P, Herdegen T. Neuronal expression of AP-1 proteins in excitotoxic-neurodegenerative disorders and following nerve fiber lesions. Prog Neurobiol. 1995;47:257–290. [PubMed] [Google Scholar]
- 13.Gorman CM, Merlino GT, Willingham MC, Pastan I, Howard BH. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci USA. 1982;79:6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 15.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groen PC, Eggen BJL, Gispen WH, Schotman P, Schrama LH. Cloning and promoter analysis of the human B-50/GAP-43 gene. J Mol Neurosci. 1995;6:109–119. doi: 10.1007/BF02736770. [DOI] [PubMed] [Google Scholar]
- 17.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herdegen T, Zimmermann M. Expression of c-Jun and JunD transcription factors represent specific changes in neuronal gene expression following axotomy. Prog Brain Res. 1994;103:153–171. doi: 10.1016/s0079-6123(08)61135-8. [DOI] [PubMed] [Google Scholar]
- 19.Herdegen T, Skene P, Bahr M. The c-Jun transcription factor: bipotential mediator of neuronal death, survival and regeneration. Trends Neurosci. 1997;20:227–231. doi: 10.1016/s0166-2236(96)01000-4. [DOI] [PubMed] [Google Scholar]
- 20.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 21.Heuss D, Engelhardt A, Gobel H, Neundorfer B. Light-microscopic study of phosphoprotein B-50 in myopathies. Virchows Arch. 1995;426:69–76. doi: 10.1007/BF00194700. [DOI] [PubMed] [Google Scholar]
- 22.Igarashi K, Kataoka K, Itoh K, Hayashi N, Nishizawa M, Yamamoto M. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature. 1994;367:568–572. doi: 10.1038/367568a0. [DOI] [PubMed] [Google Scholar]
- 23.Ingraham HA, Albert VR, Chen RP, Crenshaw EB, Elsholtz HP, He X, Kapiloff MS, Mangalam HJ, Swanson LW, Treacy MN, Rosenfeld MG. A family of POU-domain and Pit-1 tissue-specific transcription factors in pituitary and neuroendocrine development. Annu Rev Physiol. 1990;52:773–791. doi: 10.1146/annurev.ph.52.030190.004013. [DOI] [PubMed] [Google Scholar]
- 24.Jacobson RD, Virag I, Skene JHP. A protein associated with axon growth, GAP-43, is widely distributed and developmentally regulated in rat CNS. J Neurosci. 1986;6:1843–1855. doi: 10.1523/JNEUROSCI.06-06-01843.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 26.Kerppola TK, Curran T. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene. 1994;9:675–684. [PubMed] [Google Scholar]
- 27.Kraner SD, Chong JA, Tsay HJ, Mandel G. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 28.Lee ME, Dhadly MS, Temizer DH, Clifford JA, Yoshizumi M, Quertermous T. Regulation of endothelin-1 gene expression by Fos and Jun. J Biol Chem. 1991;266:19034–19039. [PubMed] [Google Scholar]
- 29.Lefevre C, Imagawa M, Dana S, Grindlay J, Bodner M, Karin M. Tissue-specific expression of the human growth hormone gene is conferred in part by the binding of a specific trans-acting factor. EMBO J. 1987;6:971–981. doi: 10.1002/j.1460-2075.1987.tb04847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan JI, Curran T. Immediate-early genes: ten years on. Trends Neurosci. 1995;18:66–67. [PubMed] [Google Scholar]
- 31.Mori N, Schoenherr C, Vandenbergh DJ, Anderson DJ. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 32.Nedivi E, Basi GS, Virag-Akey I, Skene JH. A neural-specific GAP-43 core promoter located between unusual DNA elements that interact to regulate its activity. J Neurosci. 1992;12:691–704. doi: 10.1523/JNEUROSCI.12-03-00691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson C, Albert VR, Elsholtz HP, Lu LI, Rosenfeld MG. Activation of cell-specific expression of rat growth hormone and prolactin genes by a common transcription factor. Science. 1988;239:1400–1405. doi: 10.1126/science.2831625. [DOI] [PubMed] [Google Scholar]
- 34.Nordeen SK, Green PP, Fowlkes DM. A rapid, sensitive, and inexpensive assay for chloramphenicol acetyltransferase. DNA. 1987;6:173–178. doi: 10.1089/dna.1987.6.173. [DOI] [PubMed] [Google Scholar]
- 35.Ortoft E, Pahlman S, Andersson G, Parrow V, Betsholtz C, Hammerling U. Human GAP-43 gene expression: multiple start sites for initiation of transcription in differentiating human neuroblastoma cells. Mol Cell Neurosci. 1993;4:549–561. doi: 10.1006/mcne.1993.1068. [DOI] [PubMed] [Google Scholar]
- 36.Perrone-Bizzozero NI, Cansino VV, Kohn DT. Post-transcriptional regulation of GAP-43 gene expression in PC12 cells through protein kinase C-dependent stabilization of the mRNA. J Cell Biol. 1993;120:1263–1270. doi: 10.1083/jcb.120.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plantinga LC, Verhaagen J, Gispen WH. The expression of B-50/GAP-43 in Schwann cells. Ann NY Acad Sci. 1993;679:412–417. doi: 10.1111/j.1749-6632.1993.tb18331.x. [DOI] [PubMed] [Google Scholar]
- 38.Plantinga LC, Schrama LH, Eggen BJL, Gispen WH, Verhaagen J, Lemke G. B-50/GAP-43 mRNA expression in cultured primary Schwann cells is regulated by cyclic AMP. NeuroReport. 1994;5:2465–2468. doi: 10.1097/00001756-199412000-00015. [DOI] [PubMed] [Google Scholar]
- 39.Qi Y, Wang JKT, McMillian M, Chikaraishi DM. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinhard E, Nedivi E, Wegner J, Skene JH, Westerfield M. Neural selective activation and temporal regulation of a mammalian GAP-43 promoter in zebrafish. Development. 1994;120:1767–1775. doi: 10.1242/dev.120.7.1767. [DOI] [PubMed] [Google Scholar]
- 41.Schaden H, Stuermer CA, Bahr M. GAP-43 immunoreactivity and axon regeneration in retinal ganglion cells of the rat. J Neurobiol. 1994;25:1570–1578. doi: 10.1002/neu.480251209. [DOI] [PubMed] [Google Scholar]
- 42.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995a;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 43.Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Curr Opin Neurobiol. 1995b;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 44.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreyer DJ, Skene JHP. Fate of GAP-43 in ascending spinal axons of DRG neurons after peripheral nerve injury: delayed accumulation and correlation with regenerative potential. J Neurosci. 1991;11:3738–3751. doi: 10.1523/JNEUROSCI.11-12-03738.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 47.Shyy JY-J, Lin M-C, Han J, Lu Y, Petrime M, Chien S. The cis-acting phorbol ester “12-O-tetradecanoylphorbol 13-acetate”-responsive element is involved in shear stress-induced monocyte chemotactic protein 1 gene expression. Proc Natl Acad Sci USA. 1995;92:8069–8073. doi: 10.1073/pnas.92.17.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skene JHP. Axonal growth-associated proteins. Annu Rev Neurosci. 1989;12:127–156. doi: 10.1146/annurev.ne.12.030189.001015. [DOI] [PubMed] [Google Scholar]
- 49.Skene JHP. Retrograde pathways controlling expression of a major growth cone component in the adult CNS. In: Letourneau PC, Kater SB, Macagno ER, editors. The nerve growth cone. Raven; New York: 1992. pp. 463–465. [Google Scholar]
- 50.Smith JA. Quantitation of proteins. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; New York: 1987. p. 10.1.1. [Google Scholar]
- 51.Stocker KM, Baizer L, Ciment G. Transient expression of GAP-43 in non-neuronal cells of the embryonic chicken limb. Dev Biol. 1992;149:406–414. doi: 10.1016/0012-1606(92)90295-r. [DOI] [PubMed] [Google Scholar]
- 52.Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson EB, Tomkins GM, Curran JF. Induction of tyrosine alpha-ketoglutarate transaminase by steroid hormones in a newly established tissue culture cell line. Proc Natl Acad Sci USA. 1966;56:296–303. doi: 10.1073/pnas.56.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timmers HTM, Pronk JG, Bos JL, van der Eb AJ. Analysis of the rat JE gene promoter identifies an AP-1 binding site essential for basal expression but not for TPA induction. Nucleic Acids Res. 1990;18:23–34. doi: 10.1093/nar/18.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Topp WC. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981;113:408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- 56.Van der Zee CE, Nielander HB, Vos JP, Lopes da Silva S, Verhaagen J, Oestreicher AB, Schrama LH, Schotman P, Gispen WH. Expression of growth-associated protein B-50 (GAP43) in dorsal root ganglia and sciatic nerve during regenerative sprouting. J Neurosci. 1989;9:3505–3512. doi: 10.1523/JNEUROSCI.09-10-03505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanselow J, Grabczyk E, Ping J, Baetscher M, Teng S, Fishman MC. GAP-43 transgenic mice: dispersed genomic sequences confer a GAP-43-like expression pattern during development and regeneration. J Neurosci. 1994;14:499–510. doi: 10.1523/JNEUROSCI.14-02-00499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weber JRM, Skene JHP. Identification of a novel repressive element that contributes to neuron-specific gene expression. J Neurosci. 1997;17:7583–7593. doi: 10.1523/JNEUROSCI.17-20-07583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu L, Rosser DS, Schmidt MC, Berk A. A TATA box implicated in E1A transcriptional activation of a simple adenovirus 2 promoter. Nature. 1987;326:512–515. doi: 10.1038/326512a0. [DOI] [PubMed] [Google Scholar]