Abstract
We have isolated a neurotrophin from the lamprey that permitted us to perform a phylogenetic analysis of the neurotrophin gene family that dates back more than 460 million years to the early vertebrate ancestors. The results show that the neurotrophin gene family was originally formed by two subsequent duplications. The duplication that formed nerve growth factor, neurotrophin-3, brain-derived neurotrophic factor, and neurotrophin-4/5 occurred after the split of lampreys but before the split of cartilaginous fish from the main vertebrate lineage. Compilation of chromosomal gene maps around the neurotrophins shows that they are located in paralogous regions, suggesting that the genes were formed at major duplication events possibly by complete genome doubling. Analysis of two isolated Trk receptor sequences shows similar results as for the lamprey neurotrophin. Multiple neurotrophin and Trk genes, including neurotrophin-6 and -7, have been found in bony fish, and we suggest that the extra genes were formed by an additional duplication in the bony fish lineage. Analysis of lamprey Trk mRNA expression in the adult brain shows that the genes are expressed in all regions analyzed so far. Together, the results suggest that the duplications of ancestral neurotrophin and Trk genes at an early vertebrate stage have permitted evolution to bring about differential neurotrophin and Trk expression, thereby allowing the formation of specific functions in selective neuronal populations.
Keywords: brain-derived neurotrophic factor, evolution, mRNA expression, gene duplication, molecular phylogeny, Lampetra fluviatilis, neurotrophin
The neurotrophins are structurally related neurotrophic proteins that regulate survival, growth, and morphological plasticity of neurons. The family includes nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), and neurotrophin-4/5 (NT-4/5) also known as just neurotrophin-4 or -5. All four genes have been found in amphibians, reptiles, and mammals (Hallböök et al., 1991). BDNF, NT-3, and NGF have also been found in fish. Two additional neurotrophins, NT-6 (Götz et al., 1994) and NT-7 (Nilsson et al., 1998; Lai et al., 1998), both related to NGF, have been found only in bony fish. The neurotrophins bind and activate the Trk receptors. TrkA is a receptor for NGF, NT-6, and -7, TrkB is a receptor for BDNF and NT-4/5, and TrkC is a receptor for NT-3 (Cordon-Cardo et al., 1991; Kaplan et al., 1991;Nebrada et al., 1991). These receptors mediate several of the neurotrophic functions of the neurotrophins.
The formation and early evolution of the neurotrophin and Trk receptor genes are poorly understood. The existence of several neurotrophins in fish implies origin from a common ancestor in the early vertebrate lineage (Hallböök et al., 1991; Götz and Schartl, 1994). It has been proposed that extensive gene duplications have occurred at the root of vertebrate evolution (Holland et al., 1994) and that these duplications may be the result of total or partial genome doubling (Ohno, 1970; Lundin, 1979, 1993). Doubling of the total genome (tetraploidization) would give new sets of gene copies in regions with preserved order, also called paralogous regions. Genes that have been formed as a result of the speciation process are orthologs.
We have isolated a neurotrophin from lamprey: Lampetra fluviatilis neurotrophin (Lf-NT). The lamprey diverged from the common vertebrate lineage some 460 million years ago (Halstead, 1993), and we have used molecular phylogenetic analysis to study whether the duplications leading to NGF, NT-3, BDNF, and NT-4/5 predated or postdated the divergence of lamprey from the lineage of jawed vertebrates. In addition to Lf-NT, we have isolated sequences encoding two lamprey Trk receptors: Lf-Trk1 and -Trk2. The Trk receptor sequences were analyzed using molecular phylogeny in relation to TrkA, -B, and -C, and the expression ofLf-Trk1 and -Trk2 mRNA in the adult lamprey was studied. The neurotrophins and Trk receptor genes in jawed vertebrates are expressed differentially (Lewin and Barde, 1996), and it is clear that the formation of multiple gene copies has permitted the evolution of differential expression and subsequent exclusive functions for the neurotrophins and their cognate Trk receptors. To test the hypothesis that the neurotrophin and Trk receptor genes were formed by duplications of larger genomic regions, we studied chromosome maps in the regions of the neurotrophin and Trk genes to see whether traces of original paralogous regions could be found. Based on the analysis of the neurotrophin and Trk receptor sequences, including the ones from lamprey, we used molecular phylogeny, genome organization, and mRNA expression and suggest a course of events for the formation and evolution of the extant neurotrophin and Trk receptor gene families.
MATERIALS AND METHODS
Isolation of lamprey neurotrophin. Approximately 1 gm of lamprey (L. fluviatilis) or hagfish (Myxine glutinosa) liver tissue was dissected, immediately frozen, and grained in liquid nitrogen. The fine powder was dissolved in lysis buffer (75 mm NaCl, 25 mm EDTA, pH 8), incubated with proteinase K (0.4 mg/ml) and 0.5% SDS for 4 hr at 37°C, and subsequently extracted with phenol/chloroform and precipitated with 2.5 m NH4Ac and 50% isopropanol. After mild centrifugation the DNA pellet was washed with 70% ethanol, dried, and dissolved in water to a concentration of 1 mg/ml. Degenerate oligonucleotides (Scandinavian Gene Synthesis AB, Nyköping, Sweden) representing all possible codons corresponding to the conserved neurotrophin regions with the amino acid sequences KQYF(F/Y)ET and WR(F/W)IRID were synthesized. PCR amplifications were run on 200–400 ng of purified genomic DNA using a PTC-100 thermal cycler (MJ Research, Watertown, MA). The program included two cycles with annealing temperatures of 43°C, two cycles at 48°C, and finally 30 cycles at 54°C. PCR fragments were ligated into a pBS KS+ cloning vector (Stratagene, La Jolla), which had been cleaved withEcoRV and incubated with dTTP and Taq-polymerase (Boehringer Mannheim, Mannheim, Germany) for 2 hr at 72°C to produce T-overhangs. The cloned PCR fragments were sequenced using an automated DNA sequencer based on the dye-terminator technique (ABI 373 stretch; Applied Biosystems, Foster City, CA). To isolate a DNA fragment encoding the entire sequence of the lamprey neurotrophin, 2 million clones of a L. fluviatilis lambda GEM-II genomic library, derived from lamprey blood, were screened using the PCR-derived fragment as a radioactively labeled probe. Positive clones were purified, subcloned, mapped, and sequenced according to standard molecular biology techniques.
Isolation of the lamprey Trk receptor fragments. Six micrograms of total RNA from lamprey spinal cord were used as a template for synthesis of first-strand cDNA (Pharmacia cDNA Synthesis Kit, Pharmacia, Piscataway, NJ). Six separate mixtures of 28-mer oligonucleotides representing combinations of all possible codons corresponding to the amino acid sequences NNG(N/D)Y and ENPQYF in the human TrkA sequence (Martin-Zanca et al., 1989) were synthesized. The 5′-oligonucleotide contained a XhoI restriction enzyme site, and the 3′-oligonucleotide contained an EcoRI restriction enzyme site to facilitate subcloning. Combinations of 5′- and 3′-oligonucleotides were used to prime the amplification of cDNA (50 ng) in the PCR by taq DNA polymerase (Boehringer Mannheim). PCR products of the expected sizes (∼450 bp) were restricted with XhoI and EcoRI, analyzed on a 2% agarose gel, and cloned into pBluescript KS+(Stratagene). The clones were sequenced and analyzed as described above.
Sequence and phylogeny analysis. Published sequences were retrieved from GenBank. Alignment and tree construction from sequences were performed using the program packages Wisconsin Package v. 9.1-UNIX from the Genetic Computer Group UW (Devereux et al., 1984) and the MUST package (Philippe, 1993) as well as PAUP v. 3.1.1. (see below). The generation of a phylogenetic tree is dependent on careful alignment of the sequences in a matrix that is used for the calculations, and nonhomologous characters should be excluded from the alignments. Three sequence matrices with aligned sequences were constructed: two with neurotrophin and one with Trk receptor amino acid sequences. The first neurotrophin matrix was constructed with the mature neurotrophin sequences and the other with sequences corresponding to amino acids 26–66 of the human NGF. This region was used for the second matrix because many of the fish sequences are partial. To construct the sequence matrices, homologous amino acid characters were used; therefore, insertions found in Lf-NT, -NT-4/5, -NT-6, and -NT-7 were removed. This was performed by first aligning all sequences and then manually removing the insertions on the basis of the alignment to construct the matrix. After this, the sequences were again aligned to ensure that the alignment in the matrix was correct. Conserved motifs including the six cysteines in the neurotrophin structure facilitated the alignment. The matrices used in this study can be achieved on request. Sequences for mature neurotrophin matrix are as follows: Lamprey (L. fluviatilis), Lf-NT GenBankAF071432; human, NGF (Ullrich et al., 1983), BDNF (Maisonpierre et al., 1991), NT-3 (Maisonpierre et al., 1990), NT-4/5 (also known as human NT-5) (Berkemeier et al., 1991); rat (Rattus norvegicus), NGF (Whittemore et al., 1988), BDNF (Maisonpierre et al., 1991), NT-3, (Ernfors et al., 1990), NT-4/5 (Ip et al., 1992) (also known as rat NT-4 or -5); chick (Gallus gallus), NGF (Ebendal et al., 1986), BDNF and NT-3 (Hallböök et al., 1993); African clawed toad (Xenopus laevis), NT-4 (Hallböök et al., 1991); platy fish (Xiphophorus maculata), NGF, BDNF, and NT-6 (Götz et al., 1994); carp (Cyprinus carpio), NT-7 (Lai et al., 1998); and zebra fish (Danio rerio), NT-7 (Nilsson et al., 1998). Additional sequences for the partial neurotrophin matrix are as follows: carp (C. carpio), NGF, BDNF, and NT-3 (Kullander et al., 1997); salmon (Salmo salar), NGF, BDNF, and NT-3 (Hallböök et al., 1991); ray (Raja clavata), BDNF and NT-3 (Hallböök et al., 1991); and hagfish (M. glutinosa), Mg-NT (GenBank AF071433).
The Trk matrix was constructed using published TrkA, -B, and -C sequences from representative species, including the lamprey. To construct a Trk matrix with homologous characters, a 17 amino acid insertion in the analyzed region of the TrkB sequences compared with TrkA and -C was removed. The procedure was similar to that of the construction of the neurotrophin matrices. Trk-receptor sequences are from human TrkA (Martin-Zanca et al., 1989), TrkB (Shelton et al., 1995), and TrkC (Mcgregor et al., 1994); rat TrkA (Meakin et al., 1992), TrkB (Middlemas et al., 1991), and TrkC (Merliot et al., 1992); chick TrkA (Schröpel et al., 1995), TrkB (Vinh et al., 1994), and TrkC (Williams et al., 1993); zebra fish TrkB1, TrkB2, TrkC1, and TrkC2 (Martin et al., 1995); lamprey Lf-Trk1 (GenBank AF071435) and Lf-Trk2 (GenBank AF071434).
For the tree constructions (all three consensus trees), the maximum parsimony method was used, which is a character-state method that treats each site as an independent character. Sequences were imported to PAUP v. 3.1.1 (Swofford, 1993) for an unbiased parsimonious analysis. The neurotrophin and Trk matrices were analyzed using unweighted characters and with the exclusion of uninformative characters. The phylogenetic trees were generated by the branch-and-bound algorithm. This algorithm is guaranteed to find the shortest parsimonious tree (Swofford and Olsen, 1990; Swofford, 1993). Trees were generated without a specified outgroup but were oriented according to accepted views of the relationships between vertebrate species, with lamprey or hagfish at the base. Support for individual clades was evaluated by bootstrapping (Felsenstein, 1985), where 500 replicates were analyzed using the branch-and-bound algorithm. Consensus trees were constructed, and node values indicate how often a particular branch is found among the analyzed trees. A node present in <50% of the trees was regarded as unsupported and collapsed into a polytomy (more than two branches from a node). Each of the three consensus trees had the same topology as one or several of the shortest trees among the bootstrap replicates. The consistency index (CI) for the shortest trees is indicated in the figure legend and is a measure of the amount of homoplasy exhibited by the set of characters (amino acids), defined as the maximum conceivable number of state changes divided by the actual, calculated number of state changes (Kluge and Farris, 1969). Homoplasy is defined as a collection of phenomena that leads to similarities in character states for reasons other than inheritance from a common ancestor. These include convergence, parallelism, and reversal.
Estimation of time periods. All dates are given as million years before present (MYBP) and have been obtained from the fossil record (Benton, 1993), where the earliest representatives of two lineages establish a minimum time of divergence of these lineages. A recent revised timescale based on molecular data (Kumar and Hedges, 1998) agrees with most early and late fossil-based times but indicates major gaps in the Mesozoic fossil record (70–250 MYBP).
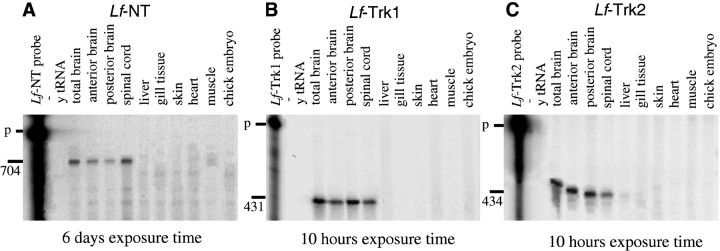
RNA preparation and RNase protection assay. Total RNA was isolated from homogenized lamprey tissue and control tissue as indicated in Figure 4. RNA was prepared as described previously (Karlsson and Hallböök, 1998). Briefly, the tissues were homogenized with extraction buffer containing 4 mguanidinium isothiocyanate and 2% β-mercaptoethanol. Total RNA was then isolated and purified by acid phenol–chloroform extraction and precipitated with ethanol. Concentration was determined spectrophotometrically. Few genes have been isolated from river lamprey, and no complementary reference probe such as glyceraldehyde-3-phosphate dehydrogenase was available; therefore the integrity of the RNA was carefully checked on an agarose gel before it was submitted for RNase protection assay (RPA). The cRNA probe for Lf-NT was prepared from a 704 bpSmaI/BamHI fragment cloned in pBS-KS, which contains most of the mature neurotrophin sequence. The probes forLf-Trk1 and -Trk2 were synthesized from respective PCR fragments. The RPA was performed using the RPAII ribonuclease protection assay kit (Ambion, Austin, TX) according to the manufacturer’s recommendations. cRNA probes were labeled with32P-UTP (Amersham, Buckinghamshire, UK) by in vitro transcription and hybridized to 10 μg of total RNA. After hybridization the samples were treated with ribonuclease, and the protected cRNA fragments were analyzed on a 5% denaturing polyacrylamide gel (Karlsson and Hallböök, 1998). The mRNA levels were visualized using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA) (see Fig. 5).
Fig. 4.
Expression of Lf-NT and Trk receptor mRNA in adult lamprey. RNase protection assay was used to analyze the levels of (A) Lf-NT, (B) Lf-Trk1, and (C) Lf-Trk2 mRNA in total RNA prepared from adult lamprey. Total RNA (15 μg) prepared from the indicated tissue was analyzed, and protected RNA fragments were visualized on 5% denaturing polyacrylamide gels that were exposed to PhosphorImager screens and subsequently scanned by a PhosphorImager. Note that exposure times for the Lf-NT gel (A) was 6 d and for the Trk gels (B, C) overnight (10 hr), indicating lower levels ofLf-NT mRNA. Yeast tRNA (y tRNA) and RNA from total 4-d-old chicken embryos (chick embryo) were used as negative controls. The protectedLf-NT fragment is 704 bases long, theLf-Trk1 fragment is 431 bases, and theLf-Trk2 is 434 bases. p, Undigested RPA probe.
Fig. 5.
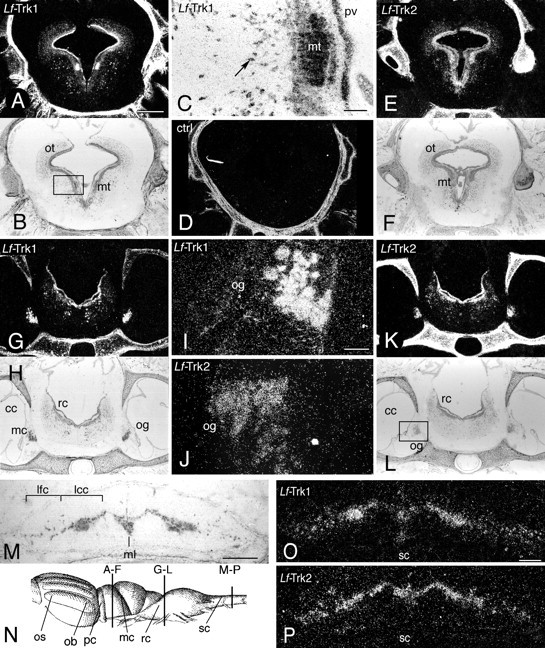
Expression of Lf-Trk1 and -Trk2 mRNA in adult lamprey nervous system as revealed by in situ hybridization analysis. In situhybridization was used to study the localization ofLf-Trk1 and -Trk2 mRNA in the nervous system of adult lamprey. Labeling for Lf-Trk1 and -Trk2 was found throughout the brain, and three different levels are depicted in the panel: the level of the optic tectum (A–F), brain stem at the level of the otic ganglia and inner ear (G–L), and rostral spinal cord (M–P). Dark-field and corresponding bright-field micrographs of transverse sections are shown to facilitate orientation.A, B, Intense labeling for Lf-Trk1 mRNA is shown in the optic tectum over the periventricular zone, over scattered cells in the central zone, and over cells in the mesencephalic tegmentum. C, High power magnification of cells in the mesencephalic tegmentum in the region indicated by abox in B. The arrow is pointing at one of the strongly labeled scattered cells in the central zone. D, A control shows that coldLf-Trk1 oligonucleotide in 100 times excess can out-compete the signal from the 35S-labeled oligonucleotide probe. Note that the lamprey vertebral structures scatter light when viewed by dark-field microscopy. E, F, Intense labeling for Lf-Trk2 mRNA in the optic tectum over the periventricular zone and over scattered cells in the central zone.G–L, Labeling for Lf-Trk1 and -Trk2 mRNA in the brain stem and otic ganglia. I, J, High-power magnification of cells in the otic ganglia as indicated by thebox in L. M, Bright-field micrograph of the spinal cord. N, Schematic diagram of the lamprey brain showing the levels for the depicted panels.O, P, Labeling for Lf-Trk1 and -Trk2 mRNA, respectively, in the spinal cord. cc, Ciliary chamber of the inner ear; lcc, lateral cell column;lfc, lateral fiber column; mc, macula communis; mc, mesencephalon; mt, mesencephalic tegmentum; og, otic ganglia;ob, olfactory bulb; os, olfactory sac;ot, optic tectum; pc, prosencephalon;pv, periventricular zone; rc, rhombencephalon; sc, spinal cord. Scale bars (shown inA for A, B, D–H, K, L), 680 μm; (shown in C for C, I, J), 100 μm; (shown in M for M, O, P), 200 μm.
In situ hybridization analysis. Lf-NT,Lf-Trk1, and -Trk2 mRNA expression were studied by in situ hybridization analysis of 10 μm cryostat transverse sections of adult lamprey. The sections were cut through the complete head and were collected on poly-l-lysine-coated glass slides. Synthetic oligonucleotides (Scandinavian Gene Synthesis AB) complementary to the mRNA were used as probes. The Lf-NT probe is antisense to a 47-nucleotide-long sequence corresponding to amino acids 57–72 in the Lf-NT protein. Lf-Trk1 and -Trk2 probes are antisense to the regions corresponding to amino acids 408–424 of the rat TrkB sequence, which is in the extracellular region of the receptors. The probes will therefore recognize all types of receptor mRNAs encoding both putative truncated and tyrosine kinase domain-containing Trk receptors. Specificity controls were performed by the addition of unlabeled oligonucleotides at >100 times excess together with the labeled probes to outcompete the specific signals (see Fig. 6D). An unrelated probe was also used as a control for unspecific labeling (data not shown).
Fig. 6.
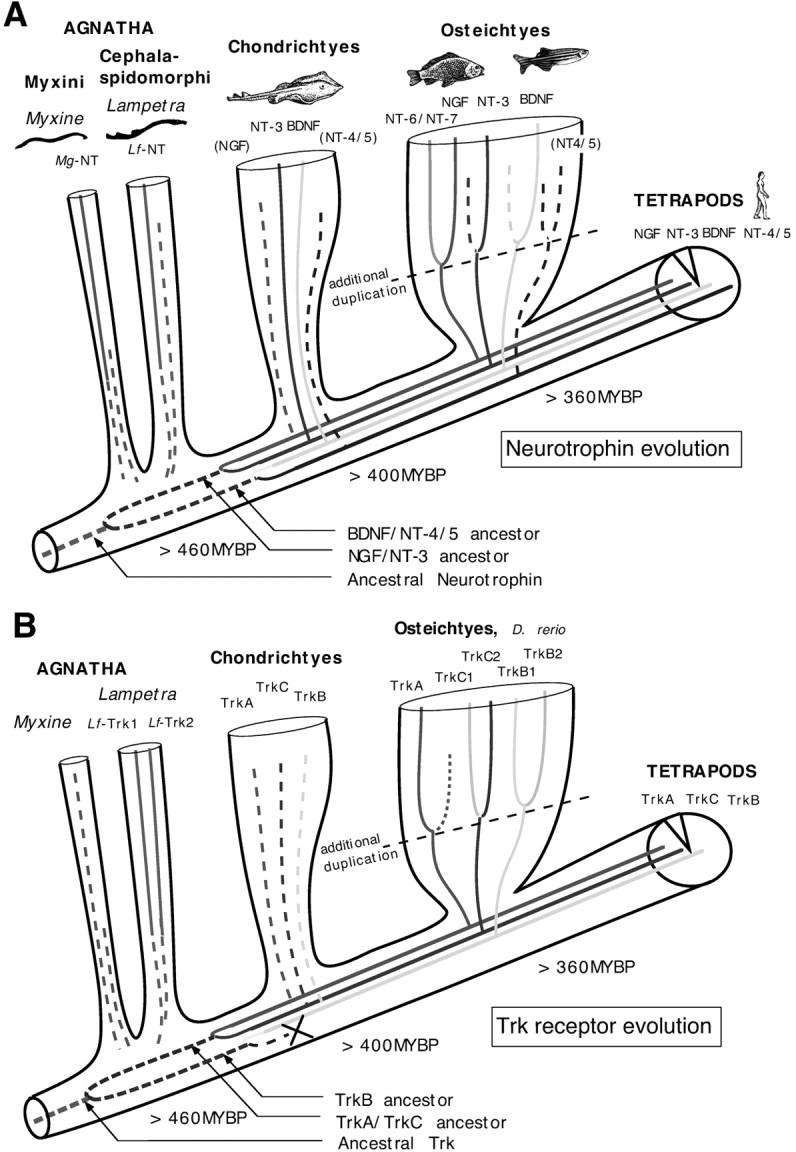
Schematic illustration of the phylogenetic relationship between jawless fish, jawed fish, the common vertebrate lineage, and the suggested gene duplications leading to the extant neurotrophin and Trk receptor gene homologs. The diagram illustrates the divergence of branches representing jawless fish (Agnatha), cartilage fish (Chondrichtyes), and bony fish (Osteichtyes) from the tetrapod lineage. The neurotrophin (A) and Trk receptor (B) gene families are illustrated within the branches, with suggested duplications shown as line forks. Two subsequent duplication events are suggested in the early chordate/vertebrate lineage. Our results strongly suggest that the second duplication occurred after the divergence of lampreys (Cephalaspidomorphi) but before the divergence of cartilaginous fish and gave rise to the extant neurotrophin and Trk gene families. We suggest that an additional duplication occurred in the bony fish lineage giving rise to the hitherto found NGF, TrkB, and TrkC paralogs in bony fish. It is not clear whether all bony fishes were affected or whether the duplication occurred exclusively in the branch including carp, zebra, and platy fish (Euteleostei). The dates of the divergence of ancestors to extant jawless and ray-finned fish from the common vertebrate lineage are based on fossil records (Benton, 1993).MYBP, Million years before present.
The oligonucleotide probes (50 ng) were labeled at the 3′ end with35S-ATP (Amersham) using terminal deoxynucleotidyl transferase (Promega, Madison, WI) and were purified on Nensorb 20 columns before use (DuPont NEN, Wilmington, DE). In situhybridization was performed as described previously (Hallböök et al., 1990). 35S-labeled oligonucleotide probe (106 cpm/slide) was diluted in hybridization solution (50% formamide, 4× SSC, 0.1× Denhardt’s solution, 0.1% sarcosyl, 0.2 m phosphate buffer, pH 7.0, 10% dextrane sulfate, 0.275 mg/ml yeast tRNA, 0.5 mg/ml salmon sperm DNA, and 0.2 m dithiothreitol) and hybridized to the sections in a humidified chamber at 43°C for 14–16 hr. After four 30 min washes in 1× SSC at 55°C, a short rinse in cold water, and dehydration in ethanol, the sections were dipped in photoemulsion (NTB2 Kodak Photo Emulsion) and exposed for 4–6 weeks at 4°C. The sections were developed in D19 developer (Kodak), counterstained with cresyl violet, and viewed in a Zeiss Axiophot photomicroscope. Black and white photo negatives were scanned using a Nikon Super CoolScan LS-1000, and Figure 5 was composed using Adobe Photoshop v.3.0.4. Manipulations and minor contrast and shade adjustments (<5%) were performed to equalize the different panels.
RESULTS
The neurotrophin protein cores have stretches of amino acids that are conserved from fish to mammals. These conserved sequences are suitable as templates for designing degenerate oligonucleotides for PCR amplification of neurotrophin DNA fragments from genomic DNA.
Lamprey neurotrophin (Lf-NT)
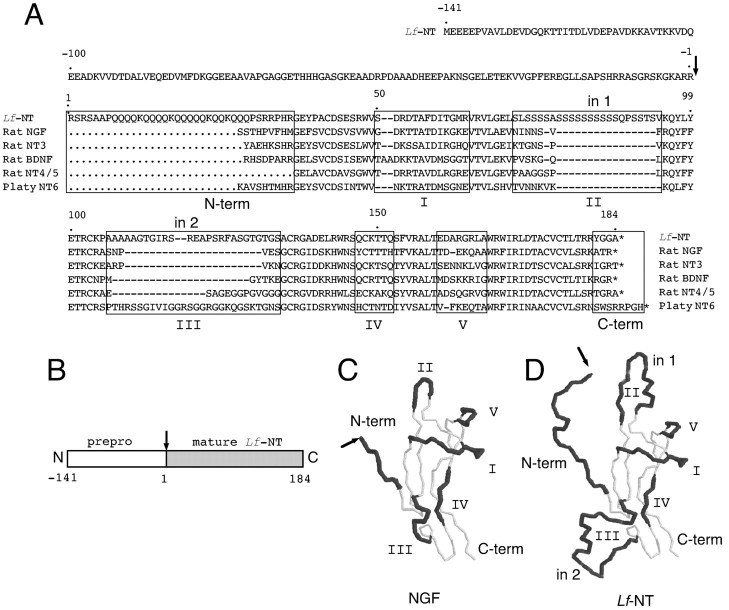
Approximately 150 different PCR fragments amplified from lamprey genomic DNA were cloned, and one 228 bp fragment contained a sequence that showed similarity to the conserved regions of the neurotrophins. The fragment was used as a probe to screen a genomic lamprey DNA library. One isolated phage clone (clone 12) containing a 9 kb fragment hybridized with the lamprey PCR fragment. The fragment was mapped and subcloned, and the region around the hybridizing part was sequenced. An open reading frame encoding 324 amino acid residues matching the PCRLf-NT neurotrophin sequence was found in one exon. The typical neurotrophin structure with six conserved cysteine residues forming the characteristic cysteine knot structure and a pro-region with a proteolytic cleavage sites in front of the mature sequence was found in Lf-NT (Fig.1A,B). Cleavage would give a 184 amino acid mature protein. When compared with mammalian neurotrophins, Lf-NT was shown to contain two amino acid insertions, both of which are situated in variable regions (Ibáñez et al., 1990). One insertion consisting of 16 amino acids, mostly serines, was found in the variable region II, where no insertion has previously been found. The other insertion of 20 amino acids was found in variable region III, where insertions have been found in NT-6, NT-7, and the mammalian versions of NT-4/5 (Fig.1A). Both insertions are in the loop regions, which can tolerate extra residues without disturbing the general neurotrophin structure. The structure allows the insertions to bulge out from the core (Fig. 1C,D). Lf-NT has a long glutamine- and lysine-rich amino terminus extending 36 amino acids beyond the conserved neurotrophin motif. The amino terminus of the neurotrophins is one of the variable regions and has been shown to be important for the specificity and activation of TrkA by NGF. The variable regions I, IV, and V in Lf-NT appear normal in comparison with other neurotrophins. The relation of Lf-NT to the other known neurotrophins was studied using phylogenetic analysis.
Fig. 1.
Lf-NT amino acid sequence.A, Prepro-amino acid sequence of Lf-NT. The mature protein sequence is compared with NGF, NT-3, BDNF, and NT-4/5 from rat as well as platy fish NT-6. Regions that are variable between neurotrophins (variable regions I–V) areboxed and marked by roman numerals. The two insertions in variable regions II andIII are indicated (in 1, in 2).B, The complete coding region is located in one 3′-exon and includes signal sequence, a pro-region, and the mature neurotrophin indicated by the shaded box. The putative proteolytic cleavage site is marked by an arrow. C, Three-dimensional structure model of the peptide backbone of a NGF protomer with the variable regions indicated by a shaded backbone and roman numerals.D, A model of Lf-NT where the two insertions in Lf-NT are introduced into the NGF backbone to illustrate their positions. N-term, Amino terminus;C-term, C terminus.
Phylogenetic analysis
Neurotrophin phylogeny
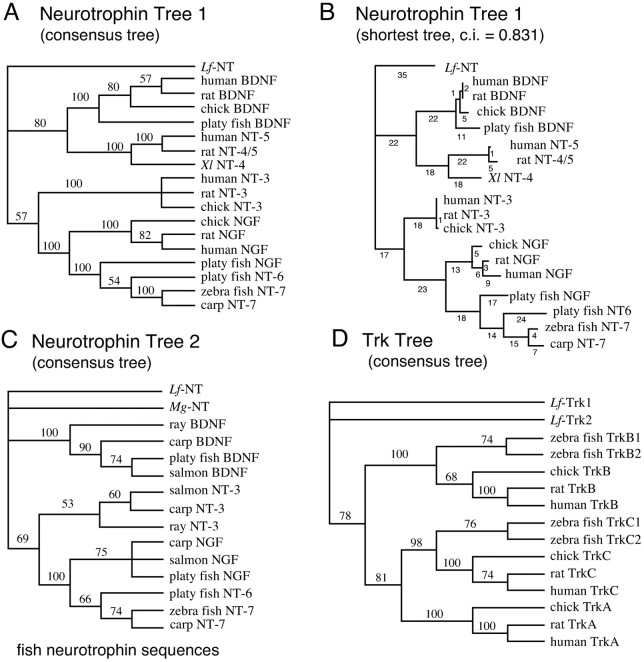
Two different trees were generated. One tree was based on a matrix with the mature neurotrophin amino acid sequences including theLf-NT, human, rat, and chicken orthologs of NGF, BDNF, and NT-3 as well as the human, rat, and Xenopus laevis NT-4/5. The matrix also contained NGF and NT-6 from platy fish and NT-7 from carp and zebra fish. The second tree was based on a matrix with neurotrophin sequences from species that diverged early from the vertebrate lineage: jawless, cartilaginous, and bony fish. Many of the fish neurotrophin sequences are partial, so this matrix includes 43 amino acids from the mature neurotrophins. We have shown previously that trees constructed from a similarly sized matrix can resolve phylogenetic relationships between mammalian neurotrophins (Kullander et al., 1997). The representation of jawless fish in the analysis was increased by isolation of a neurotrophin fragment from hagfish (M. glutinosa) genomic DNA using the degenerate primer PCR strategy. Hagfish is thought to have diverged very early from the common vertebrate lineage, similar to the lamprey. We were able to find one sequence encoding a 43 amino acid neurotrophin fragment. Several independent clones of the fragment were sequenced, and the sequence was given the name M. glutinosa neurotrophin (Mg-NT).Mg-NT has the characteristic features of a neurotrophin, but does not show a significantly higher degree of identity to any of the other neurotrophins, including Lf-NT. The fragment spans the variable region III and does not carry any insertion.
Bootstrap resampling was used to assess the reliability of each node in the trees, and the bootstrap percentages are indicated on the branches at the nodes in Figure 2. The consensus phylogenetic trees (Fig. 2A,C) show several significant features.
Fig. 2.
Phylogenetic trees generated with neurotrophin and Trk receptor amino acid sequences. A,Neurotrophin Tree 1, Consensus tree based on a matrix with the mature neurotrophin amino acid sequences from representative species including human, rat, chicken, and lamprey. B, One of the shortest trees from the search giving the consensus tree inA. Branch lengths are indicated below branches.C, Neurotrophin Tree 2, Consensus tree based on a matrix with fish neurotrophin amino acid sequences.D, Trk Tree, Consensus tree based on a matrix with Trk receptor sequences including Lf-Trk1 and -Trk2. The depicted consensus trees are bootstrap consensus trees and were generated using PAUP v.3.1.1 from 500 unrooted branch-and-bound replicates of the most parsimonious trees, with sampling excluding uninformative characters. Bootstrap percentages are indicated on branches in the tree and indicate how often a particular branch was found in the bootstrap analysis. If a branch is found in <50% of the trees it is collapsed into a polytomy. The consensus trees show the relations between the taxa, and branch lengths in those trees do not reflect distances between taxa. The shortest trees found when analyzing the matrices had high consistency index (CI) values indicating a low degree of homoplasy (CI tree 1 = 0.831, tree 2 = 0.829, and Trk tree = 0.844).
(1) Tree 1: NGF and NT-3 as well as BDNF and NT-4/5, respectively, clearly were formed by duplications of common precursor genes. The bootstrap value (BV) for the nodes joining NGF and NT-3 (BV 57%) as well as BDNF and NT-4/5 (BV 80%) are high enough for us to conclude that the topology is correct (Fig. 2A).
(2) Tree 1 and 2: The same topology is seen in both trees. In tree 2, it is clear that ray NT-3 and BDNF, which are representatives of early jawed vertebrates, join the respective gene groups. This shows that the gene duplication giving rise to NGF and NT-3 as well as BDNF and NT-4/5 must have occurred before the split of cartilaginous fish from the common vertebrate lineage (Fig. 2A,C).
(3) Tree 1 and 2: Lf-NT clearly diverged from a common neurotrophin precursor before the split into NGF and NT-3 as well as BDNF and NT-4/5. The nodes joining these neurotrophins excludeLf-NT (tree 1), and in tree 2, they exclude bothLf-NT and Mg-NT. This shows that the gene duplication giving rise to NGF and NT-3 as well as BDNF and NT-4/5 must have occurred after the divergence of jawless fish (Cephalaspidomorphi) from the common vertebrate lineage.
(4) Tree 1 and 2: Lf-NT (Fig. 2, trees 1 and 2) andMg-NT (Fig. 2C, tree 2) form a polytomy with the branches to NGF/NT-3 and BDNF/NT-4/5 groups, indicating that the resolution in the tree is not sufficient to resolve the associations between Lf-NT and precursors of NGF/NT-3 or BDNF/NT-4/5.
(5) Tree 1 and 2: The topology suggests that NT-6 and NT-7 are the results of a duplication of an ancestral fish NGF gene. Both genes form a monophyletic group with fish NGF (BV 100%), excluding tetrapod NGF sequences, suggesting that the gene duplication giving rise to fish NGF, NT-6, and NT-7 occurred after the divergence of bony fish from the common vertebrate lineage. Furthermore, in both neurotrophin tree 1 and 2 the platy fish NT-6 and carp and zebra fish NT-7 sequences group and exclude the NGF sequences (Fig. 2A–C).
Carp and zebra fish are closely related and belong to the same family (Family Cyprinidae), whereas platy fish (F.Poeciliidae) are distantly related. The divergence between the two branches occurred very early in the bony fish tree, and the branch point is at sub-cohort level (Euteleostei). Carp and zebra fish belong to the superorder Ostariophysi, and the platy fish belongs to Acanthopterygii. Fossils separating the two superorders date back to 160 MYBP (Patterson, 1993). Fossils separating carp and zebra fish ancestors (F. Cyprinidae) go back 40 MYBP. Phylogenetically, the difference between zebra fish and platy fish is much greater than between carp and zebra fish. The mutation rate for NT-7 has produced 11 amino acid changes in homologous amino acid positions between carp and zebra fish during 40 million years. With this mutation rate, 43 changes would have accumulated during 160 million years. This number is close to the number of changes found between platy fish NT-7 and zebra fish NT-6 sequences, which have 37 differences in homologous positions. Together this suggests that NT-6 and NT-7 relate to each other in a way that is similar to how the species they were isolated from relate to each other and that NT-6 and NT-7 may be the result of the speciation process.
Trk receptor phylogeny
To generate a tree based on Trk receptor genes that is comparable to the neurotrophin trees, we isolated DNA fragments encoding Trk receptors from lamprey. Two fragments of ∼400 bp, with sequence similarity to the Trk receptors, were identified using degenerate primer PCR amplification with cDNA from adult lamprey brain as template. The Trk receptors encoded by the two fragments (132 and 133 amino acids), which span the transmembrane and intracellular regions, were named L. fluviatilis Trk1 and -2 (Lf-Trk1, -Trk2). The sequences of the two fragments were different from each other and from TrkA, -B, and -C. The degree of identity between the Lf-Trks and other vertebrate Trks is in the same range as shown by comparison with the rat Trk sequences.Lf-Trk1 and -Trk2 show a somewhat higher degree of identity to each other and to TrkC (Table 1). A careful comparison of the relationship between the sequences was performed using molecular phylogenetic analysis.
Table 1.
Similarity of Lf-Trk1, Lf-Trk2, TrkA, -B, and -C amino acid sequences
| TrkC | TrkB | TrkA | Lf-Trk2 | |
|---|---|---|---|---|
| Lf-Trk1 | 56.6 | 48.5 | 38.5 | 60.3 |
| Lf-Trk2 | 53.5 | 50.3 | 37.8 | |
| TrkA | 40.3 | 37.3 | ||
| TrkB | 49.6 |
The amino acid sequences of Lf-Trk1 andLf-Trk2 were compared with those of rat TrkA, -B, and -C. The degree of identity was calculated using the GAP program in the GCG software package and is shown as percent amino acid identity in the table.
A tree was generated using a matrix with Trk receptor sequences including the Lf-Trk1 and -Trk2, human, rat, and chicken orthologs of TrkA, -B, and -C, as well as two TrkB and two TrkC sequences found in zebra fish (TrkB1, -2 and TrkC1, -2). The tree shows several significant features (Fig. 2D).
(1) The similarity to the neurotrophin tree topology is striking, and the tree suggests that the Trk genes were formed by two duplications of an ancestral Trk gene. Recently a tyrosine kinase receptor with similarity to the Trk receptors was isolated from the snailLymnea stagnalis, which may be related and may represent this ancestral Trk gene (Kesteren et al., 1998).
(2) TrkA and TrkC were formed by a duplication of an ancestral precursor gene. The bootstrap values for the nodes joining TrkA with TrkC and excluding TrkB are high enough (81%) for us to conclude that this topology is correct. Most likely the TrkB precursor was duplicated into TrkB and a fourth Trk that was lost during the course of evolution. The duplications occurred before the split of bony fish from the common vertebrate lineage (Fig. 2D).
(3) The tree suggests that Lf-Trk1 and -Trk2 diverged from the mammalian, bird, and fish Trk genes, before the split of Trk precursor into TrkA, -B, and -C. The nodes joining the different Trks, as well as the node joining the precursors to TrkA/C and TrkB, exclude the Lf-Trk receptors.
(4) TrkB1 and -2 as well as TrkC1 and -2 found in zebra fish were formed by duplications of the fish TrkB and TrkC genes, after the split of the zebra fish ancestor from the common lineage of vertebrates.
Neurotrophin and Trk genes in paralogous regions
We compared the chromosome maps in the regions of human and mouse neurotrophin and Trk receptor genes and found that several paralogous genes are located in the vicinity of the neurotrophin and Trk receptor genes, respectively (Fig.3A,B). The paralogous conservation is most obvious around the neurotrophin genes. In the human genome, neurotrophin genes are on chromosomes 1, 11, 12, and 19, and in the mouse on chromosomes 3, 2, 6, and 7. In particular, genes in the potassium channel (KCN), ras kinase (RAS), and thyroid-stimulating hormone β polypeptide families (TSHB, FSHB, and LHB) are all linked to the region around the neurotrophins. Several other gene families can be found in the region on two or more of the chromosomes (Fig.3A).
Fig. 3.
Gene maps with paralogous genes in the regions of neurotrophin and Trk receptor genes on human chromosomes and their orthologous regions in the mouse genome. A, Paralogous regions around the neurotrophin genes on human chromosomes (HSA) 1, 11, 12, and 19 and their orthologous regions on the mouse chromosomes (MMU). B, Paralogous regions around the Trk receptor genes on human chromosomes 1, 9, (5), and 15 and their orthologous regions in the mouse genome. Different members of each gene family are positioned on the same row in the diagram and are named according to the Genome Database nomenclature (http://www.gene.ucl.ac.uk/nomenclature/). The position of each paralogous gene in the diagram represents its corresponding relative location on the chromosome. Locally duplicated genes are placed under each other. Genes marked with * have a position that deviates from the exact corresponding location but is in the correct region; ** indicates that the location of the gene is on the correct chromosome but is not determined; and *** indicates that chromosomal localization is not determined. The human chromosomes and their orthologous mouse regions are double underlined, and the neurotrophin and Trk paralogs are boxed. AGC, Aggrecans;ALDH, aldehyde dehydrogenases; ANX, annexins; ARH, Aplysia ras-related homologs; ATP, Na+/K+ transporting ATPases;CAP, calpains; CDKN, cyclin-dependent kinase inhibitors; CRABP, cellular retinoic acid-binding proteins; CSPG, versicans; CTS, cathepsins; FDPSL, farnesyl diphosphate synthase-like proteins; FSHB, follicle-stimulating hormone β;GNA, guanine nucleotide-binding proteins;GYS, glycogen synthases; INSR, insulin receptor; IGF1R, insulin-like growth factor 1 receptor;KCN, potassium voltage-gated channels;LDH, lactate dehydrogenases; LHB, luteinizing hormone β; MAP, microtuble-associated proteins; MEF, MADS box transcription factor enhancer factors; PACE, paired basic amino acid cleaving enzymes; PCSK, subtilisins; PK, pyruvate kinases; PTH, parathyroid hormone; RAS, viral kinase oncogene homologs; SIAT, sialyltransferases; SYT, synaptotagmins;TCF, transcription factors II; TFCOUP(ERBAL), transcription factor chicken ovalbumin upstream promoter; THBS, thrombospondins; TLE, transducin-like enhancers of split; TNNT, troponins;TPM, tropomyosins; TSHB, thyroid-stimulating hormone β.
TrkA, -B, and -C are located on chromosomes 1q, 9q, and 15q in man, and 3, 13, and 7 in mouse. There are several paralogous genes in two or more of the chromosomes (Fig. 3B), but the regions around the Trk genes are less predictable than the ones around the neurotrophin genes. The cysteine proteases in the cathepsin family and the tropomyosin genes are Trk-linked. According to the hypothesis that the Trk genes were formed after two subsequent duplications, four genes existed originally, one of which most likely has been silenced during evolution. The compilation of the gene maps shows that several translocations and inversions have occurred. TrkB is located on HSA 9, but this paralogous region is split on HSA 5 and HSA 9, indicating a translocation of part of this region. A similar situation is found on MMU7 and MMU2 in the vicinity of the BDNF gene and in MMU7 and MMU9 in the mouse TrkC region (Fig. 3B).
Lf-NT, Lf-Trk1, and Lf-Trk2 expression in lamprey
Lf-NT, Lf-Trk1, and Lf-2 mRNA expression were analyzed in adult lamprey using RPA. In general, theLf-NT mRNA level was low, as shown by the long exposure time required to visualize bands, and among the analyzed tissues the highest levels were found in the nervous system in RNA prepared from total brain, diencephalon including the optic tectum, hindbrain, and spinal cord. Very low levels could be detected in heart and muscle, and levels were below the detection limit in gill tissue, liver, and skin (Fig.4A).
Lampreys have a laminar brain where the majority of the neuron cell bodies are located in the periventricular zone, with few scattered neurons within the central and superficial zones (Butler and Hodos, 1996). In situ hybridization using a probe toLf-NT mRNA showed a very weak labeling above background levels over the periventricular gray. The labeling could be out-competed with an excess of cold oligonucleotides, but the just slightly higher density of silver grains did not allow closer regional or cellular resolution (data not shown).
Using RPA, high levels of Lf-Trk1 and -Trk2 mRNA were found in brain and spinal cord but could not be detected in the other tissues (Fig. 4B,C). In situ hybridization using probes to Lf-Trk1 and -Trk2 mRNA confirmed the RPA and showed labeling in brain over cells in the periventricular zone and over scattered cells in the central and peripheral zones. The patterns by the two probes were both overlapping and specific and were found in all regions of the brain: olfactory bulbs, forebrain, midbrain including the optic tectum and hindbrain, and spinal cord. The labeling with both probes could be out-competed with an excess of cold oligonucleotides (Fig. 5D). When the two patterns were compared, most regions seemed to be labeled by either or both probes. Figure 5 shows three levels of the brain as indicated in the schematic diagram of the lamprey brain shown in Figure5N: mesencephalon in the level of the optic tectum (Fig.5A–F), medulla in the level of the otic foramen (Fig. 5G–L), and the rostral spinal cord (Fig.5M–P). The Lf-Trk1 pattern was conspicuous in the mesencephalon, with strong labeling over the innermost layer of the periventricular gray and over the mesencephalic tegmentum (Fig.5A–C). Cells in the mesencephalic tegmentum have been suggested to comprise the midbrain locomotor region, which is a command generator for sequential movements in the lamprey (Butler and Hodos, 1996). Some of the individually scattered cells in the central zone of the midbrain were strongly labeled with either of the Lf-Trk probes (Fig. 5C,E). Sections through the medulla showed labeling for Lf-Trk1 and -Trk2 mRNA over the region of the motor column (Fig. 5K,L) with labeling over large cells, which were tentatively identified as Müller cells. Strong labeling for Lf-Trk1 and -Trk2 mRNA was found in the spinal cord. Lf-Trk2 labeling was uniform in medial and lateral portions of the cord over both small and large cells around the central canal, in the lateral cell column, and over scattered small cells in the lateral fiber column (Fig. 5O). Lf-Trk1 labeling was stronger over the lateral cell column (Fig.5P). The organization of the lamprey brain and spinal cord is not as strict as the mammalian, and the position of a particular cell type varies. To identify a neuronal population, tracing must be performed, and tracing studies in combination with in situ hybridization histochemistry will allow the identification of populations of neurons expressing Lf-Trk1 and -Trk2 receptors.
Lf-NT, Lf-Trk1, and Lf-Trk2 are expressed in the otic system
In situ hybridization analysis using theLf-NT probe on sections through the ciliary chamber of the lamprey inner ear revealed weak Lf-NT mRNA labeling over epithelium in the macula communis of the lamprey inner ear. The labeling could be out-competed with cold oligonucleotides (data not shown). The in situ hybridization analysis has a limited sensitivity, and only cells with a relatively high level ofLf-NT mRNA expression will be labeled. Using the probes forLf-Trk1 and -Trk2, strong labeling was found over cells in a ganglionic structure between the macula communis and the otic foramen (Fig. 5G–L), which most likely represent neurons in the otic ganglion. The cranial ganglia are closely positioned and absolute identification of neurons in the different ganglia mediating electroreceptive, gustatory, and lateral line sensory information would require retrograde filling from the respective nerves.
DISCUSSION
In this report we show results that suggest that the neurotrophin and Trk gene families were originally formed by duplications of ancestral neurotrophin and Trk receptor genes in the early vertebrate lineage. The results suggest that duplications occurred by means of genome doubling (tetraploidization) or of duplication of larger parts of the ancestral genome, including both ancestral neurotrophin and Trk receptor genes.
The isolation of a neurotrophin from lamprey permitted us to perform a phylogenetic analysis of the neurotrophin gene family that dates back at least 460 million years (Halstead, 1993) to the early vertebrate lineage. Our data suggest that two subsequent duplications of a putative ancestor gave rise to NGF, NT-3, BDNF, and NT-4/5, and the phylogenetic tree topologies (Fig. 2) show that the second of the duplications occurred after the divergence ofCephalaspidomorphi (460 MYBP) (Fig.6), which includes lampreys, and before the divergence of cartilaginous fish (400 MYBP) from the common vertebrate lineage. Thus neurotrophins in ancestors of extant jawless fish represent precursors to the higher vertebrate neurotrophins. This conclusion is supported by the addition of the neurotrophin sequence from hagfish, which is thought to have diverged even earlier from the vertebrate lineage. The isolation of Lf-Trk1 and -Trk2 and subsequent phylogenetic analysis of the Trk sequences generated a tree with a topology strikingly similar to that of the neurotrophin trees. This suggests that both neurotrophin and Trk gene families were formed in a similar way and during the same time period. On the basis of these results we suggest that the two gene families were formed by the same duplication events and that this has allowed the neurotrophin and Trk receptors to co-evolve. Figure 6A,B shows an illustration of the suggested course of events underlying the formation of the gene families.
Our data suggest that at least one duplication event occurred after the split of lamprey from the common vertebrate lineage. For obvious reasons this is the second of the two duplications and if the first duplication event predated the divergence of jawless fish,Lf-NT and Mg-NT should ideally associate with either NGF/NT-3 or BDNF/NT-4/5. In the neurotrophin trees,Lf-NT and Mg-NT form a polytomy with the branches to NGF/NT-3 and BDNF/NT-4/5 groups, indicating that the resolution is not sufficient to resolve the associations between Lf-NT and precursors to NGF/NT-3 or BDNF/NT-4/5. Interestingly, neitherLf-Trk1 nor -Trk2 are joined to either imminent TrkA/TrkC and TrkB precursors but rather join a hypothetical ancestor. The separation is supported by a relatively high BV of 78% for the node. This would suggest that both duplications could have occurred after the split of lamprey, but taken together with other results this is less likely. (1) Lf-Trk1 is slightly more similar to TrkC than to TrkB (Table 1). (2) It is known that taxa with long branch lengths have a tendency to associate (Eernisse and Kluge, 1993) and when the Trk tree is constructed with Lf-Trk1 and not Lf-Trk2, a polytomous branch is formed with Lf-Trk1 and the TrkA/C precursor (data not shown). This would suggest that Lf-Trk1 and -Trk2 are orthologs to the TrkA/C and TrkB ancestors, respectively. Alternatively, Lf-Trk1 and -Trk2 were formed by an independent duplication in the lamprey lineage. (3) Analysis of the gene for DNA-binding chromosomal high mobility group proteins (Sharman et al., 1997), members in the complement family (Nonaka and Takahashi, 1992; Hughes, 1994), and the lactate dehydrogenase (Stock and Whitt, 1992) support one duplication event after the split of lampreys. Furthermore, indirect evidence for one duplication after the split of lampreys is that the second duplication event has been assigned the role to have permitted the evolution of jaws and fins, whereas a previous duplication event has tentatively been assigned to have permitted the evolution of epidermal placodes, the neural crest, and neural crest derivatives as well as paired sensory organs (Holland et al., 1994; Holland and Garcia-Fernandez, 1996). These structures evolved before the split of lampreys, and consequently the first duplication event should have occurred before this split.
Two subsequent major duplication events produced four copies of each gene in the duplicated regions. The phylogenetic analyses suggest that two subsequent duplications gave rise to NGF, NT-3, BDNF, and NT-4/5. Similarly, two duplications gave rise to TrkA, TrkC, TrkB, and the extinct TrkB sister gene. Silencing of paralogous genes is common (Li, 1982), and examples can be found in the four vertebrate Hox clusters. The compilation of paralogous genes in the regions of neurotrophin and Trk receptor genes from the human and mouse chromosome maps (Fig. 3) supports the hypothesis that the duplication events leading to the formation of the gene families involved duplication of large regions of the genome, likely via tetraploidization. The fact that NT-4/5 is located in a region with paralogy to the other neurotrophins and acts via TrkB suggests that the TrkB sister gene was silenced shortly after the second duplication. The location of the Trk-linked paralogous genes (MEF, INSR, and TPM) on HAS 19 p13 tentatively suggests that the TrkB sister gene once was located in this region. So far, the prime examples of chromosomal regions likely to have been conserved after the proposed genome doublings are found in human chromosomes 2, 7, 12, and 17 and their orthologous counterpart in the mouse, with the presence of Hox clusters on all four chromosomes associated with collagen genes, clusters of integrin genes (Lundin, 1993; Wang et al., 1995), and genes for intermediate filaments. Another example comprises a region around the PBX and Notch genes in the human genome with regional paralogies in chromosomes 1, 6, and 9 and a small region of chromosome 19 (Katsanis et al., 1996). Obviously, the paralogies are just traces left after extensive chromosomal rearrangements. Translocations, insertions, and inversions have changed to a great extent the structure of chromosomal regions. However, random nonordered duplications would rarely produce paralogous regions of the kind we find here. Single gene duplications most often are produced by tandem duplications. The human NT-5 (NT-4/5) and its tandem duplicated pseudogenes is one such example (Berkemeier et al., 1991).
The topology of the neurotrophin trees, joining NT-6 and NT-7 sequences on a separate branch in the NGF group and the fact that NT-6 and NT-7 sequences relate to each other in the same way as the fish species they were isolated from (Fig. 3), suggests that platy fish NT-6 and carp and zebra fish NT-7 are orthologs rather than separate genes. We suggest that the genes were formed by a duplication of an ancestral fish NGF gene. Furthermore, the existence of two zebra fish TrkB (B1 and B2) and TrkC (C1 and C2) genes suggests that an additional tetraploidization may have occurred in the fish lineage before the split of the fish superorders Ostariophysi and Acanthopterygii. This is indeed supported by recent findings of multiple extra hox gene clusters (Prince et al., 1998) and Notch genes (Westin and Lardelli, 1997) in zebra fish.
Lf-NT mRNA is expressed in the lamprey sensory epithelium of the inner ear. In higher vertebrates, BDNF and NT-3 are expressed in the inner ear (Pirvola et al., 1992; Fritzsch et al., 1997) and support sensory innervation from the cochleo vestibular (otic) ganglia. TrkB as well as TrkC are expressed in the otic ganglia, and accordingly we have found Lf-Trk receptor expression in lamprey otic ganglia (Fig. 6I,J). Lf-Trk1 and -Trk2 mRNA is expressed in all regions, and Lf-NT is expressed at low levels in the adult lamprey CNS. Hence, Lf-Trk receptor expression is found over cells of the reticular formation, which is considered to be an evolutionarily old part of the vertebrate brain (Cruce and Newman, 1984). Our results show that Lf-NT,Lf-Trk1, and -Trk2 represent precursors to the higher vertebrate neurotrophin and Trk receptor genes. The similarity betweenLf-NT and BDNF/NT-3 expression suggests that BDNF and NT-3 have a more archetypal expression pattern or function compared with NGF. This is in agreement with the higher degree of conservation of the BDNF and NT-3 sequences compared with NGF during higher vertebrate evolution (Hallböök et al., 1991; Götz and Schartl, 1994) and the fact that lampreys do not have the cell types that are associated with NGF/TrkA expression. Jawless fish do not have any sympathetic ganglia, and the nociceptive sensory system is poorly developed (Butler and Hodos, 1996). The forebrain of a lamprey is small and has no cortex, hippocampal formation, or cholinergic basal forebrain nuclei, which in the mammalian brain contain neurons that express NGF or TrkA.
On the basis of the expression of the Lf-NT,Lf-Trk1, and Lf-Trk2 receptors, phylogenetic analyses, and genome organization, we propose that ancestral neurotrophin/Trk receptor archetype genes were expressed in all regions of the ancestral nervous system and that the duplications of these ancestor genes at an early vertebrate stage have permitted the evolution of differential neurotrophin and Trk receptor expression, thereby allowing specific functions in selective neuronal populations to form.
Footnotes
This work was supported by the Swedish Medical Research Council (B95–13R-11098 to K.K.; K-98–12X12187 to F.H.) and the Swedish Natural Research Council (B-BU 08904–305 to F.H.; BMH4 CT96 0976 to E.U). We thank Natalie von der Lehr for help with isolation of the lamprey Trks, and Abdel ElAmira, Lennart Brodin, Bernd Fritzsch, Sten Grillner, and Dan Larhammar for lamprey tissue, the genomic library, and valuable discussions. We also thank Nancy Ip and Carlos Ibáñez for providing the NT-7 sequences before publishing. We thank anonymous reviewers for their constructive comments.
Correspondence should be addressed to Dr. Hallböök, Department of Neuroscience, Box 587, BMC, Uppsala University, S-751 23 Uppsala, Sweden.
Dr. Kullander’s present address: EMBL, Meyerhofstrasse 1, 69117 Heidelberg, Germany.
REFERENCES
- 1.Benton MJ. The fossil record 2. Chapham and Hall; London: 1993. [Google Scholar]
- 2.Berkemeier LR, Winslow JW, Kaplan DR, Nikolics K, Goeddel DV, Rosenthal A. Neurotrophin-5: a novel neurotrophic factor that activates trk and trkB. Neuron. 1991;7:857–866. doi: 10.1016/0896-6273(91)90287-a. [DOI] [PubMed] [Google Scholar]
- 3.Butler AB, Hodos W. Comparative vertebrate neuroanatomy; evolution and adaptation. Wiley; New York: 1996. [Google Scholar]
- 4.Cordon-Cardo C, Tapley P, Jing SQ, Nanduri V, O’Rourke E, Lamballe F, Kovary K, Klein R, Jones KR, Reichardt LF, Barbacid M. The trk tyrosine protein kinase mediates the mitogenic properties of nerve growth factor and neurotrophin-3. Cell. 1991;66:173–183. doi: 10.1016/0092-8674(91)90149-s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruce WLR, Newman DB. Evolution of motor systems: the reticulospinal pathways. Am Zool. 1984;24:733–753. [Google Scholar]
- 6.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acid Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebendal T, Larhammar D, Persson H. Structure and expression of the chicken β nerve growth factor. EMBO J. 1986;5:1483–1487. doi: 10.1002/j.1460-2075.1986.tb04386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eernisse DJ, Kluge AG. Taxonomic congruence versus total evidence, and amniote phylogeny and amniote phylogeny inferred from fossils, molecules and morphology. Mol Biol Evol. 1993;10:1170–1195. doi: 10.1093/oxfordjournals.molbev.a040071. [DOI] [PubMed] [Google Scholar]
- 9.Ernfors P, Ibáñez CF, Ebendal T, Olson L, Persson H. Molecular cloning and neurotrophic activities of a protein with structural similarities to β-nerve growth factor: developmental and topographical expression in the brain. Proc Natl Acad Sci USA. 1990;87:5454–5458. doi: 10.1073/pnas.87.14.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein J. Confidence limits on phylogenies: an approach using bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Fritzsch B, Silossantiago I, Bianchi LM, Fariñas I. The role of neurotrophic factors in regulating the development of inner ear innervation. Trends Neurosci. 1997;20:159–164. doi: 10.1016/s0166-2236(96)01007-7. [DOI] [PubMed] [Google Scholar]
- 12.Götz R, Schartl M. The conservation of neurotrophic factors during vertebrate evolution. Comp Biochem Physiol. 1994;108:1–10. doi: 10.1016/1367-8280(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 13.Götz R, Koster R, Winkler C, Raulf F, Lottspeich F, Schartl M, Thoenen H. Neurotrophin-6 is a new member of the nerve growth factor family. Nature. 1994;372:266–269. doi: 10.1038/372266a0. [DOI] [PubMed] [Google Scholar]
- 14.Hallböök F, Ayer-Lelièvre C, Ebendal T, Persson H. Expression of nerve growth factor receptor mRNA during early development of the chicken embryo: emphasis on cranial ganglia. Development. 1990;108:693–704. doi: 10.1242/dev.108.4.693. [DOI] [PubMed] [Google Scholar]
- 15.Hallböök F, Ibàñez C, Persson H. Evolutionary studies of the nerve growth factor family reveal a novel member abundantly expressed in Xenopus ovary. Neuron. 1991;6:845–858. doi: 10.1016/0896-6273(91)90180-8. [DOI] [PubMed] [Google Scholar]
- 16.Hallböök F, Ibañez CF, Ebendal T, Persson H. Cellular localization of brain-derived neurotrophic factor and neurotrophin-3 mRNA in the early chicken embryo. Eur J Neurosci. 1993;5:1–14. doi: 10.1111/j.1460-9568.1993.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 17.Halstead LB. Agnatha. In: Benton MJ, editor. The fossil record 2. Chapman and Hall; London: 1993. pp. 573–581. [Google Scholar]
- 18.Holland PW, Garcia-Fernandez J. Hox genes and chordate evolution. Dev Biol. 1996;173:382–395. doi: 10.1006/dbio.1996.0034. [DOI] [PubMed] [Google Scholar]
- 19.Holland PW, Garcia FJ, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Development [Suppl] 1994;1994:125–133. [PubMed] [Google Scholar]
- 20.Hughes AL. Phylogeny of the C3/C4/C5 complement-component gene family indicate that C5 diverged first. Mol Biol Evol. 1994;11:417–425. doi: 10.1093/oxfordjournals.molbev.a040123. [DOI] [PubMed] [Google Scholar]
- 21.Ibáñez CF, Hallböök F, Ebendal T, Persson H. Structure-function studies of nerve growth factor. EMBO J. 1990;9:1477–1483. doi: 10.1002/j.1460-2075.1990.tb08265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ip NY, Ibanez CF, Nye SH, Mcclain J, Jones PF, Gies DR, Belluscio L, Lebeau MM, Espinosa R, Squinto SP, Persson H, Yancopoulos GD. Mammalian neurotrophin-4: structure, chromosomal localization, tissue distribution, and receptor specificity. Proc Natl Acad Sci USA. 1992;89:3060–3064. doi: 10.1073/pnas.89.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan DR, Hempsted BL, Martin-Zanca D, Chao MV, Parada LF. The trk proto-oncogene product: a signal transducing receptor for nerve growth factor. Science. 1991;252:554–558. doi: 10.1126/science.1850549. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson M, Hallböök F. Kainic acid, tertrodotoxin and light modulate BDNF expression in retinal ganglion cells and their tectal target. Neuroscience. 1998;83:137–150. doi: 10.1016/s0306-4522(97)00340-0. [DOI] [PubMed] [Google Scholar]
- 25.Katsanis N, Fitzgibbon J, Fisher EMC. Paralogy mapping: identification of a region in the human MHC triplicated onto human chromosomes 1 and 9 allows the prediction and isolation of novel PBX and NOTCH loci. Genomics. 1996;35:101–108. doi: 10.1006/geno.1996.0328. [DOI] [PubMed] [Google Scholar]
- 26.Kesteren RE, Fainzilber M, Hauser G, van Minnen J, Vreugdenhill E, Smit AB, Ibáñez CF, Garaerts PM, Bulloch AGM. Early evolutionary origin of the neurotrophin receptor family. EMBO J. 1998;17:2534–2542. doi: 10.1093/emboj/17.9.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kluge AG, Farris JS. Quantitative phylogenetics and the evolution of anurans. Syst Zool. 1969;18:1–32. [Google Scholar]
- 28.Kullander K, Carlson B, Hallböök F. Molecular phylogeny and evolution of the neurotrophins from monotremes and marsupials. J Mol Evol. 1997;45:311–321. doi: 10.1007/pl00006235. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- 30.Lai KO, Fu WY, Ip FCF, Ip NY. Cloning and expression of a novel neurotrophin, NT-7 from carp. Mol Cell Neurosci. 1998;11:64–76. doi: 10.1006/mcne.1998.0666. [DOI] [PubMed] [Google Scholar]
- 31.Lewin GR, Barde YA. Physiology of the neurotrophins. Annu Rev Neurosci. 1996;19:289–317. doi: 10.1146/annurev.ne.19.030196.001445. [DOI] [PubMed] [Google Scholar]
- 32.Li WH. Evolutionary changes of duplicated genes. Curr Top Biol Med Res. 1982;6:55–92. [PubMed] [Google Scholar]
- 33.Lundin LG. Evolutionary conservation of large chromosomal segments reflected in mammalian gene maps. Clin Genet. 1979;16:72–81. doi: 10.1111/j.1399-0004.1979.tb00855.x. [DOI] [PubMed] [Google Scholar]
- 34.Lundin LG. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics. 1993;16:1–19. doi: 10.1006/geno.1993.1133. [DOI] [PubMed] [Google Scholar]
- 35.Maisonpierre PC, Belluscio L, Squinto S, Ip NY, Furth ME, Lindsay RM, Yancopoulos GD. Neurotrophin-3: a neurotrophic factor related to NGF and BDNF. Science. 1990;247:1446–1451. doi: 10.1126/science.247.4949.1446. [DOI] [PubMed] [Google Scholar]
- 36.Maisonpierre PC, Le BM, Espinosa Rd, Ip NY, Belluscio L, de la Monte S, Squinto S, Furth ME, Yancopoulos GD. Human and rat brain-derived neurotrophic factor and neurotrophin-3: gene structures, distributions, and chromosomal localizations. Genomics. 1991;10:558–568. doi: 10.1016/0888-7543(91)90436-i. [DOI] [PubMed] [Google Scholar]
- 37.Martin SC, Marazzi G, Sandell JH, Heinrich G. Five Trk receptors in the zebrafish. Dev Biol. 1995;169:745–758. doi: 10.1006/dbio.1995.1184. [DOI] [PubMed] [Google Scholar]
- 38.Martin-Zanca D, Oskam R, Mitra G, Copeland T, Barbacid M. Molecular and biochemical characterization of the human trk proto-oncogene. Mol Cell Biol. 1989;9:24–33. doi: 10.1128/mcb.9.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mcgregor LM, Baylin SB, Griffin CA, Hawkins AL, Nelkin BD. Molecular cloning of the cDNA for human TrkC (NTRK3), chromosomal assignment, and evidence for a splice variant. Genomics. 1994;22:267–272. doi: 10.1006/geno.1994.1383. [DOI] [PubMed] [Google Scholar]
- 40.Meakin SO, Suter U, Drinkwater CC, Welcher AA, Shooter EM. The rat trk protooncogene product exhibits properties characteristic of the slow nerve growth factor receptor. Proc Natl Acad Sci USA. 1992;89:2374–2378. doi: 10.1073/pnas.89.6.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merliot J-P, Ernfors P, Jaber M, Persson H. Molecular cloning of rat trkC and identification of cells expressing mRNAs for members of the trk family in the rat central nervous system. Neuroscience. 1992;51:513–532. doi: 10.1016/0306-4522(92)90292-a. [DOI] [PubMed] [Google Scholar]
- 42.Middlemas DS, Lindberg RA, Hunter T. trkB, a neural receptor protein kinase: evidence for a full length and two truncated receptors. Mol Cell Biol. 1991;11:143–153. doi: 10.1128/mcb.11.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nebrada AR, Kaplan DR, Martin-Zanca D, Parada LF, Santos E. Induction by NGF of meiotic maturation of Xenopus oocytes expressing the trk proto-oncogene product. Science. 1991;252:568–560. doi: 10.1126/science.1850550. [DOI] [PubMed] [Google Scholar]
- 44.Nilsson A-S, Fainzilber M, Falck P, Ibáñez CF. Neurotrophin-7: a novel member of the neurotrophin family from the zebra fish. FEBS Lett. 1998;424:285–290. doi: 10.1016/s0014-5793(98)00192-6. [DOI] [PubMed] [Google Scholar]
- 45.Nonaka M, Takahashi M. Complete cDNA sequence of the third component of complement of lamprey. J Immunol. 1992;148:3290–3295. [PubMed] [Google Scholar]
- 46.Ohno S. Evolution by gene duplication. Springer; New York: 1970. [Google Scholar]
- 47.Patterson C. Osteichtyes: teleostei. In: Benton MJ, editor. The fossil record 2. Chapman and Hall; London: 1993. pp. 621–656. [Google Scholar]
- 48.Philippe H. MUST, a computer package of management utilities sequences and trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pirvola U, Ylikoski J, Palgi J, Lehtonen E, Arumae U, Saarma M. Brain-derived neurotrophic factor and neurotrophin 3 mRNAs in the peripheral target fields of developing inner ear ganglia. Proc Natl Acad Sci USA. 1992;89:9915–9919. doi: 10.1073/pnas.89.20.9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prince VE, Joly J, Ekker M, Ho R. Zebrafish hox genes: genome organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- 51.Schröpel A, von Schank D, Dechant G, Barde Y-A. Early expression of the nerve growth factor receptor ctrkA in chick sympathetic and sensory ganglia. Mol Cell Neurosci. 1995;6:544–557. doi: 10.1006/mcne.1995.0006. [DOI] [PubMed] [Google Scholar]
- 52.Sharman AC, Hay SA, Holland PW. Cloning and analysis of an HMG gene from the lamprey Lampetra fluviatilis: gene duplication in vertebrate evolution. Gene. 1997;184:99–105. doi: 10.1016/s0378-1119(96)00580-x. [DOI] [PubMed] [Google Scholar]
- 53.Shelton DL, Sutherland J, Gripp J, Camerato T, Armanini MP, Phillips HS, Carroll K, Spencer SD, Levinson AD. Human trks: molecular cloning, tissue distribution, and expression of extracellular domain immunoadhesins. J Neurosci. 1995;15:477–491. doi: 10.1523/JNEUROSCI.15-01-00477.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stock DW, Whitt GS. Evolutionary implications of the cDNA sequence of the single lactate dehydrogenase of a lamprey. Proc Natl Acad Sci USA. 1992;89:1799–1803. doi: 10.1073/pnas.89.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swofford DL. Phylogenetic analysis using parsimony. Illinois Natural History Survey; Champaign, IL: 1993. [Google Scholar]
- 56.Swofford DL, Olsen GJ. Phylogeny reconstruction. In: Hillis DM, Moritz C, editors. Molecular systematics. Sinauer; Sunderland, MA: 1990. pp. 411–501. [Google Scholar]
- 57.Ullrich A, Gray A, Berman C, Dull TJ. Human β-nerve growth factor gene highly homologous to that of mouse. Nature. 1983;303:821–825. doi: 10.1038/303821a0. [DOI] [PubMed] [Google Scholar]
- 58.Vinh NQ, Erdmann KS, Heumann R. Cloning and sequence analysis of a cDNA encoding a novel truncated form of the chicken TrkB receptor. Gene. 1994;149:383–384. doi: 10.1016/0378-1119(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 59.Wang W, Wu W, Desai T, Ward D, Kaufman S. Localization of the alpha 7 integrin gene (ITGA7) on human chromosome 12q13: clustering of integrin and Hox genes implies parallel evolution of these gene families. Genomics. 1995;26:568–570. doi: 10.1016/0888-7543(95)80176-m. [DOI] [PubMed] [Google Scholar]
- 60.Westin J, Lardelli M. Three novel Notch genes in zebrafish: implications for vertebrate notch gene evolution. Dev Genes Evol. 1997;207:51–67. doi: 10.1007/s004270050091. [DOI] [PubMed] [Google Scholar]
- 61.Whittemore SR, Friedman PL, Larhammar D, Persson H, Gonzalez CM, Holets V. Rat beta-nerve growth factor sequence and site of synthesis in adult hippocampus. J Neurosci Res. 1988;20:403–410. doi: 10.1002/jnr.490200402. [DOI] [PubMed] [Google Scholar]
- 62.Williams R, Bäckström A, Ebendal T, Hallböök F. Molecular cloning and cellular localization of trkC in the chicken embryo. Dev Brain Res. 1993;75:235–252. doi: 10.1016/0165-3806(93)90028-9. [DOI] [PubMed] [Google Scholar]