Abstract
Maintaining mitochondrial proteome integrity is especially important under stress conditions to ensure a continued ATP supply for protection and adaptation responses in plants. Deg/HtrA proteases are important factors in the cellular protein quality control system, but little is known about their function in mitochondria. Here we analyzed the expression pattern and physiological function of Arabidopsis thaliana DEG10, which has homologs in all photosynthetic eukaryotes. Both expression of DEG10:GFP fusion proteins and immunoblotting after cell fractionation showed an unambiguous subcellular localization exclusively in mitochondria. DEG10 promoter:GUS fusion constructs showed that DEG10 is expressed in trichomes but also in the vascular tissue of roots and aboveground organs. DEG10 loss-of-function mutants were impaired in root elongation, especially at elevated temperature. Quantitative proteome analysis revealed concomitant changes in the abundance of mitochondrial respiratory chain components and assembly factors, which partially appeared to depend on altered mitochondrial retrograde signaling. Under field conditions, lack of DEG10 caused a decrease in seed production. Taken together, our findings demonstrate that DEG10 affects mitochondrial proteostasis, is required for optimal root development and seed set under challenging environmental conditions, and thus contributes to stress tolerance of plants.
Keywords: Arabidopsis, Deg proteases, mitochondria, proteome, root, seed yield, temperature stress
The protease DEG10 is located in mitochondria and contributes to mitochondrial proteostasis. Mutants lacking DEG10 showed impaired root development under temperature stress and reduced seed production in the field.
Introduction
Protein turnover is an important factor for cellular homeostasis and metabolic flexibility in plants, enabling them to survive, grow, and reproduce under constantly changing and often challenging environmental conditions. Proteins are ubiquitously damaged by highly reactive chemicals generated as byproducts of normal cellular metabolism or resulting from impaired cellular functions under stress. To maintain proteostasis and prevent the formation of cytotoxic protein aggregates, an elaborate system for protein quality control monitors proper protein function and triggers chaperone-mediated repair or proteolytic removal of damaged proteins (Seki et al., 2002; Sakurai et al., 2005).
Respiratory ATP production is the main function of mitochondria, but numerous other metabolic pathways and biological processes, such as regulation of cell proliferation and differentiation, also depend on mitochondrial functions (Todd and Gifford, 2002). Metabolism and ATP production in plant mitochondria depend on the availability of photoassimilates and their efficient translocation to sink tissues, which are often impaired under stressful environmental conditions (Fox and Weisberg, 2011). Mitochondrial respiration is especially important in photosynthetically inactive cells, but it can generate reactive oxygen species (ROS) as byproducts that result in protein damage (Hothorn et al., 2008; Bates et al., 2014; Pinheiro et al., 2015). Many types of stress increase ROS production and thereby increase the need for repair or degradation of oxidatively damaged proteins. The importance of proteolysis and protein turnover for plant mitochondrial function was highlighted previously by deletion of the ATP-dependent mitochondrial FtsH4 protease, which resulted in elevated ROS levels and altered organelle and leaf morphology under short-day conditions (Gibala et al., 2009).
The ATP-independent Deg/HtrA serine endoproteases (for degradation of periplasmic proteins and high temperature requirement A, respectively), hereafter Deg proteases, represent an important group of protein quality control factors with dual functions as chaperones and proteases in prokaryotes and eukaryotic organelles (Clausen et al., 2011; Schuhmann and Adamska, 2012). The 16 Deg proteases encoded in the genome of the model plant Arabidopsis thaliana are targeted to various subcellular compartments, including chloroplasts, mitochondria, the nucleus, and peroxisomes (Schuhmann and Adamska, 2012; Tanz et al., 2014). In plastids, Deg proteases participate in protein quality control of the photosynthetic machinery and the selective replacement of photodamaged proteins, including core subunits of the photosynthetic electron transport chain (Schuhmann and Adamska, 2012; Nishimura et al., 2016).
According to sequence-based predictions and previous experimental evidence, eight of the Deg proteases of Arabidopsis may be localized in mitochondria: DEG3, DEG4, DEG6, DEG7, DEG10, DEG11, DEG12, and DEG14 (Schuhmann and Adamska, 2012; Basak et al., 2014; Tanz et al., 2014). For DEG3, DEG4, and DEG12, evidence of gene expression has been obtained only by microarray analyses, while DEG6 transcripts were hardly ever detected even in RNA-seq experiments [eFP Browser (Winter et al., 2007), data not shown]. A previous phylogenetic study of Deg proteases in photosynthetic eukaryotes revealed a group of eight Deg proteases present in all analyzed organisms, suggesting the existence of a core set of Deg proteases essential for protein quality control (Schuhmann et al., 2012). Among these eight Deg proteases, DEG10 is the only representative with predicted mitochondrial localization, suggesting an important function of DEG10 in the mitochondria of plants and algae.
For Arabidopsis DEG10 (At5g36950), dual targeting to mitochondria and plastids has been proposed on the basis of transient overexpression of a DEG10:GFP fusion protein (Tanz et al., 2014). However, based on the synthesis of results from 20 sequence-based prediction algorithms, the ARAMEMNON 8.1 database calculates a 3-fold higher score for localization in mitochondria than in plastids (Schwacke et al., 2003; Supplementary Fig. S1W). Of 11 prediction programs that consider both organelles, one predicts nuclear localization, seven predict exclusive mitochondrial localization, two predict equal probability for import into mitochondria or plastids, and only one favors plastid localization. Transcripts encoding DEG10 and a second mitochondrial Deg protease in Arabidopsis, DEG14, were more abundant after heat stress (Sinvany-Villalobo et al., 2004; Larkindale and Vierling, 2008; Basak et al., 2014). While overexpression of DEG14 suggested a function in thermotolerance, the native physiological function of DEG10 had not been analyzed previously (Larkindale and Vierling, 2008; Basak et al., 2014). In general, not much is known about the physiological functions of mitochondrial Deg proteases in plants.
In this study, we settled the ambiguity of the localization and showed by green fluorescent protein (GFP) expression analysis and cell fractionation the exclusive localization of DEG10 in mitochondria. Promoter:GUS fusion analysis revealed predominant expression of DEG10 in trichomes and, to a lesser extent, in aboveground and root vascular tissue. Loss of DEG10 impaired root development, particularly at elevated temperatures, and resulted in changes in the abundance of electron transport chain complex subunits and mitochondrial stomatin-like proteins. Field trials showed reduced seed production of deg10 mutants, suggesting a contribution of DEG10 to plant evolutionary fitness.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh. ecotype Columbia (Col-0), hereafter referred to as wild type (WT), and mutant lines carrying T-DNA insertions in DEG10 (see Supplementary Table S1 at JXB online) and catr3-1 (SALK_036082) were obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003) and from the GABI-KAT project (Kleinboelting et al., 2012). Heterozygous loss-of-function deg10-1, deg10-2, and catr3-1 plants were backcrossed with WT plants for three consecutive generations. Homozygous deg10-1, deg10-2, and catr3-1 mutant lines were selected by PCR (Supplementary Fig. S5) and the positions of the T-DNA insertions were confirmed by sequencing. For developmental analyses, WT plants and backcrossed deg10-1, deg10-2, and catr3-1 mutants were raised for two generations side by side in a glasshouse.
For the generation of plants expressing a DEG10:GFP:6x-His fusion protein under the control of the CaMV 35S promoter, the full-length DEG10 open reading frame was amplified (primers 38 and 39; Supplementary Table S2) from cDNA clone RAFL09-10-L10 (R21313; Seki et al., 2002; Sakurai et al., 2005), inserted into pENTR/D-TOPO (Life Technologies), and transferred into the vector pEG103 (Earley et al., 2006). For the generation of the GFP-GUS double reporter line (PrDEG10:GFP:GUS), the 854 bp intergenic region between CATR3 and DEG10 was amplified (primers 40 and 41) and inserted via pDONR221 (Life Technologies) into the plant transformation vector pHGWFS7 (Karimi et al., 2002). For a second reporter construct (PrDEG10:GUS) expressing β-glucuronidase (GUS) with an N-terminal fusion of the first three amino acids of DEG10, primers 42 and 43 were used and the amplicon was inserted into the XbaI site of pCB308 (Xiang et al., 1999). All final constructs were verified by sequencing. Agrobacterium tumefaciens strains LBA4404 and GV3101 were used to transform WT plants by floral dip (Clough and Bent, 1998). Transformants were selected on half-strength Murashige and Skoog (MS, Duchefa) agar plates supplemented with 20 µg ml−1 hygromycin B, 150 µg ml−1 cefotaxime, 150 µg ml−1 ticarcillin, and 2% sucrose, or by spraying soil-grown seedlings with 50 µg ml−1 BASTA (Bayer) solution.
Soil-grown plants were kept in a glasshouse at a photon flux density of approximately 150 µmol m−2 s−1 under short-day conditions (9 h light at 22 °C; 15 h dark at 20 °C) unless stated otherwise. For proteomic analysis of roots, 3-week-old seedlings were transferred to an aerated hydroponic culture system with constant renewal (1 l day–1) of the nutrient solution [1 mM Ca(NO3)2, 0.5 mM MgSO4, 0.5 mM K2HPO4, 0.1 mM KCl, 20 μM Fe(III)-EDDHA, 10 μM H3BO3, 0.1 μM MnSO4, 0.2 μM Na2MoO4, 0.5 μM NiSO4, 0.1 μM CuSO4, 0.1 μM ZnSO4, 1 mM MES pH 5.8; adapted from (Küpper et al., 2007)]. After 3 additional weeks, the nutrient solution was heated to 30 °C for 2 days before the roots were harvested, rinsed briefly, and snap-frozen in liquid nitrogen.
For growth in axenic culture, seeds were surface sterilized by incubation in 70% ethanol for 5 min, followed by 1.25% sodium hypochlorite and 0.01% (v/v) Triton X-100 for 20 min under agitation, and finally washed twice with deionized water. To synchronize germination, all seeds were incubated in 0.1% agarose for 3 days at 4 °C. Seedlings were grown on agar plates (half-strength MS salts, pH 5.8, 1% agar, with or without 2% sucrose). Short-day conditions were 9 h light with a photon flux density of 90 µmol m−2 s−1 followed by 15 h dark; long-day conditions were 16 h light at a photon flux density of 180–200 µmol m−2 s−1 followed by 8 h dark. Growth temperatures varied between experiments as detailed in the Results. Liquid cultures were grown in gently agitated flasks containing half-strength MS salts and 2% sucrose at 22 °C at a continuous photon flux density of 100 µmol m−2 s−1.
Isolation and fractionation of organelles
Mitochondria were purified from 10–60 g of Arabidopsis seedlings grown in axenic liquid cultures. The procedure, consisting of differential centrifugation and a continuous Percoll density gradient, was adapted from Eubel et al. (2007) and Millar et al. (2007). Intact chloroplasts were isolated from 3-week-old WT plants grown on soil and kept in darkness on the day of preparation. The procedure, consisting of a two-step Percoll density protocol, was adapted from Lohscheider et al. (2015).
Protein overexpression and antibody production
Primer pairs 35 and 37 or 36 and 37 (see Supplementary Table S3) were used to amplify truncated versions of the DEG10 cDNA; products were inserted into the pET151/D-TOPO vector (Life Technologies). Recombinant DEG10 fusion proteins with the N-terminal 51 or 92 amino acids replaced by a 6xHis-tag were expressed in Escherichia coli BL21 (DE3) Star and purified under native or denaturing conditions by metal affinity chromatography followed by size exclusion chromatography (1 ml HisTrap FF and Superdex 200 10/300 GL columns, GE Healthcare) using an FPLC system (Äkta Purifier, GE Healthcare). A polyclonal rabbit antiserum was raised at the Animal Research Facility of the University of Konstanz. The sensitivity of the antiserum was determined with known amounts of 6xHis:DEG10ΔN51 in E. coli lysates.
Protein detection
Proteins were purified by precipitation in 80% (v/v) acetone at –20 °C, solubilized in protein extraction buffer [104 mM Tris–HCl pH 6.8, 3.3% SDS, 1.67× protease inhibitor (complete EDTA free, Roche), 83.3 mM DTT, 83.3 mM EDTA] and the concentration was determined with a kit (RC/DC assay, BioRad). Proteins were separated by SDS-PAGE according to Laemmli (1970) on 10–15% polyacrylamide gels, and stained with Coomassie brilliant blue or blotted semi-dry onto PVDF membranes (0.2 µm pore size, Roche). The membranes were blocked for 1 h at room temperature with blocking solution [1× Roti Block (Roth) in PBS-T (137 mM NaCl, 2.7 mM KCl, 10.2 mM Na2HPO4, 2 mM NaH2PO4, 0.1% (v/v) Tween-20, pH 7.4)] and incubated in antibody solution (see Supplementary Table S3) for 1 h at room temperature or overnight at 4 °C. Proteins were detected with an enhanced chemiluminescence assay (ECL, GE Healthcare Life Sciences).
Proteome preparation and mass spectrometry data acquisition
Deep-frozen roots were ground with a mortar and pestle, transferred into 2 ml reaction cups, and thawed in protein extraction buffer [6 M guanidine-HCl, 5 mM EDTA, 100 mM HEPES pH 7.5, 1× Complete protease inhibitor (Roche)]. Protein concentration was determined with the Pierce BCA assay (Thermo Fisher) and adjusted to 1.5 µg/µl. A 200 µl volume of each sample was reduced with 12.5 mM DTT for 30 min at 55 °C and alkylated with 37.5 mM iodoacetic acid for 40 min at 22 °C. Excess iodoacetic acid was quenched with 25 mM DTT for 20 min at 37 °C before purification by chloroform/methanol precipitation (Wessel and Flügge, 1984). Protein pellets were dissolved in 100 mM HEPES pH 7.5, proteomics-grade trypsin (Thermo) at a ratio of 1:125 protease/protein (w/w) and incubated at 37 °C for 16 h. Tryptic peptides were desalted using C18 reverse-phase cartridges (Sep-Pak, Waters) before analysis in duplicate injections using an Impact II QTOF (Bruker) nano-LC-MS/MS system as described by Soares et al. (2019).
Mass spectrometry data analysis
Peptides were identified, quantified, and matched to corresponding proteins using the Max-Quant software package, version 1.6.0.16 (Tyanova et al., 2016a), with generic settings for Bruker Q-TOF instruments and the Arabidopsis Uniprot proteome database (release 2017-07-09). Trypsin was set as digestion protease allowing for up to three missed cleavages. N-terminal protein acetylation and methionine oxidation were considered as variable modifications, and carbamidomethylation of cysteine as a fixed modification. Peptide-spectrum match and protein false discovery rates were set to 0.01. For label-free protein quantification (LFQ), the ‘match between runs’ algorithm was enabled with standard settings. The Perseus software package, version 1.6.1.1 (Tyanova et al., 2016b), was used for statistical evaluation. Due to inefficient digestion, one replicate sample of the WT Col-0 correlated poorly with the other three and was excluded from further analysis. Proteins identified as contaminants, reverse hits, or ‘only identified by site’ were excluded, as were proteins not quantified in at least three biological replicates of one genotype. Missing values were imputed using standard settings before determination of proteins with significantly altered abundance by ANOVA of log2-transformed LFQ intensities with a permutation-based false discovery rate of 0.05. Tukey’s honestly significant difference post-hoc test was used to determine significant pairwise differences among the four genotypes. Fold differences were calculated from the log2-transformed LFQ intensities.
Quantitative reverse transcription PCR
Relative transcript levels were determined by quantitative reverse transcription PCR (qRT–PCR) as described previously (Mishra et al., 2019), using the primers listed in Supplementary Table S2. RNA was extracted from the seedlings that were used to determine the impact of DEG10 and temperature on root growth. Cq values were obtained by using CFX Maestro software (BioRad) and relative transcript levels are reported as 2(−ΔΔCq ±SD). ΔΔCq values were analyzed with linear models (R base function lm) including genotype, temperature, and their interaction as fixed effects. P-values for the model effects were obtained using the Anova function in the car package (Fox and Weisberg, 2011).
Confocal microscopy
Protoplasts were isolated from leaves of DEG10:GFP-expressing or WT plants by overnight incubation in protoplast medium (0.45 M sorbitol, half-strength MS salt mixture) supplemented with 10 mg ml−1 cellulase and 2.5 mg ml−1 macerozyme. Mitochondria were stained by incubating protoplasts with 0.33 µM MitoTracker Orange (Life Technologies) for 20–30 min in the dark before embedding in protoplast medium supplemented with 0.1% agarose. Photographs were taken with a Zeiss LSM 510 Meta confocal microscope with a ×40 oil immersion lens. GFP, MitoTracker, and chlorophyll fluorescence signals were sequentially captured with the Meta detector (GFP: 497–561 nm, MitoTracker: 572–615 nm, chlorophyll: 657–690 nm). Fluorescence was excited with an argon ion laser at 488 nm and a He-Ne laser at 543 nm. For spectral deconvolution, a Zeiss LSM 880 with a ×63 water immersion lens was used. The samples were excited at 488 nm and images with 20 spectral channels (9 nm bandwidth) were recorded between 490 and 669 nm. A chlorophyll spectrum was obtained from WT protoplasts and a GFP spectrum from root protoplasts of a DEG10:GFP-expressing plant. Linear unmixing with background subtraction was performed with ZEN software (Zeiss). Channel overlay, false coloring, adjustment to identical gain, offset, and contrast settings were performed in ImageJ and Adobe Photoshop.
TEM analysis
For TEM analysis, 5-day-old seedlings of two parental lines per genotype were dissected into cotyledon, hypocotyl, and root, fixed in 2% glutaraldehyde [(EM Grade, Agar Scientific), 0.1 M Na-cacodylate pH 7.0, 1 mM CaCl2, 1% sucrose] and incubated at 4 °C for 2 h. The material was rinsed several times in fixation solution without glutaraldehyde and postfixed with 1% osmium tetroxide in 0.1 M Na-cacodylate pH 7.0 at 4 °C for 2 h. After dehydration in a graded series of ethanol (15%, 30%, and 50%), the material was incubated for several hours at 4 °C in Spurr’s epoxy resin (Spurr, 1969) at increasing concentrations, ranging from 10% to 100%, before the resin was polymerized at 65 °C for 3 days. Ultra-thin sections (80–100 nm) were made with an ultramicrotome (Ultracut R, Leica), stained with 2% uranyl acetate and 0.4% lead citrate according to standard methods, and analyzed using a 912 Omega transmission electron microscope (Zeiss). Brightness and contrast were adjusted with AxioVision 4.8 software (Zeiss).
Histochemical GUS assay
For the histochemical GUS assay, leaves were incised at the tip and siliques were carefully pricked using a needle to facilitate vacuum infiltration with staining solution [1 mM X-Gluc (Apollo Scientific Ltd) from a stock in dimethylformamide, 100 mM Na-phosphate buffer pH 7.2, 10 mM EDTA, 0.2% Triton-X-100]. Samples were incubated for 20 h at 37 °C. The reaction was stopped by replacing the staining solution with 80% ethanol followed by incubation for 30 to 40 min at 60 °C until all endogenous pigments were completely eluted. Photographs were taken with a stereomicroscope (Stemi 2000-C, Zeiss) with an external light source (KL 1500 LCD, Zeiss).
Analysis of development and seed production
For the analysis of seedling development, surface-sterilized seeds were placed individually on agar plates, sealed with Parafilm, and incubated vertically in light thermostats (Ernst Schütt Laborgerätebau, Göttingen) under long-day conditions at a photon flux density of 180–200 µmol m−2 s−1 (Osram Powerstar HQI-E/P 400 W/D light bulbs). Plate positions were rotated every other day. The seedlings were grown with three different temperature regimes: 21 °C/17 °C light/dark (Fig. 6, Supplementary Fig. S6), 22 °C/20 °C light/dark (Supplementary Fig. S7), and 30 °C/25 °C light/dark (Fig. 6, Supplementary Figs S6 and S7). Four parameters of seedling development were scored visually: germination, seedling establishment (a seedling was considered to have established when it had formed a radicle and had two fully unfolded cotyledons with an angle between the cotyledon blades ≥180°), number of true leaves present at day 10, and the proportion of plants that survived and continuously developed until day 18. As a fifth growth parameter, the length of the primary root was measured 13 or 18 days after plating using ImageJ (version 1.46f).
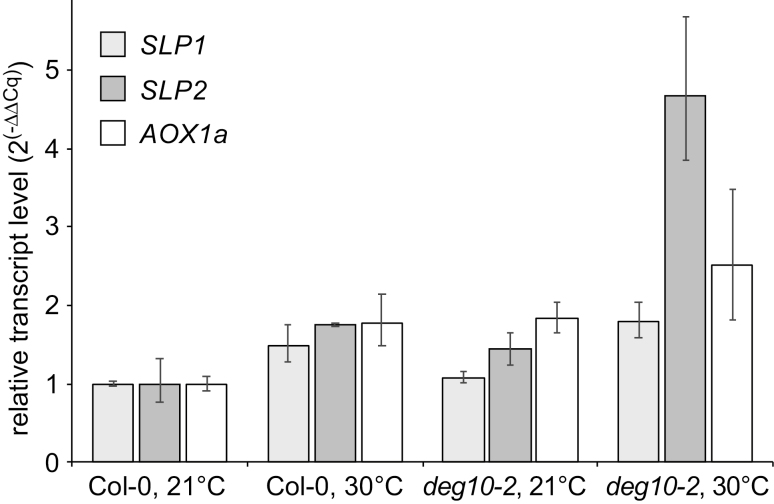
Fig. 6.
Relative transcript levels of mitochondrial proteins with altered expression in deg10 mutants. RNA was isolated from 13-day-old seedlings grown for 3 days at ambient temperature (21 °C day/17 °C night) followed by 10 days’ incubation at ambient temperature or elevated temperature (30 °C day/25 °C night) on medium with 2% sucrose (see Supplementary Fig. S6). Transcript levels were normalized to TUBULINα1 and UBIQUITIN5 and to transcript levels in Col-0 plants grown at 21 °C. Columns represent 2(−ΔΔCq± SD). Statistical analysis of ΔΔCq values confirmed a significant effect of the deg10-2 mutation on SLP2 transcript levels, which was confirmed in an independent experiment and was similar in the comparison of deg10-1 with catr3-1 mutants (Supplementary Fig. S8 and Table S4).
For reproduction studies in the glasshouse, plants were cultivated in soil (Einheitserde, type P, Gebr. Patzer, Sinntal-Altengronau, Germany). Seedlings were transplanted to individual pots 10 days after sowing. From the beginning of the reproductive phase, plants were cultivated under long-day conditions. During the entire growth period, plants of the different lines were placed alternatingly in common trays to guarantee similar growth conditions. When plants reached the senescence phase, seeds in 10 siliques harvested from the main shoot were counted per plant.
To analyze development and fertility under semi-natural field conditions, WT and mutant lines were cultivated as described in Frenkel et al. (2008) and Wagner et al. (2011) in a garden lot at the Wallenberg laboratories in Umeå, Sweden, during the summer period (June to September) of the years 2011, 2012, and 2014. Vegetative growth was assessed by measuring the rosette diameter after 3, 4, and 5 weeks of growth. When plants stopped producing new flowers, seed production was analyzed by counting the seeds, whenever possible, in five siliques per plant and by counting all siliques per plant. Weather data were obtained from the Department of Applied Physics and Electronics, Umeå University, Sweden (http://www.tfe.umu.se), and were collected at a station 650 m from the growth site.
Statistical analysis of growth and reproduction
In the developmental analysis, only the fraction of surviving plants was considered (for sample size per trait, see Supplementary Table S4). To test whether genotypes differed in their responses to high temperature, and whether the genotypes’ responses were different in the presence or absence of sucrose, we used linear mixed models as implemented in the lmer function in the lme4 package (Bates et al., 2014) of R software version 3.1.0 (R Core Team, 2013). The fixed part of the model included temperature, medium, genotype, and all possible interactions. The random part of the model included plate and thermostat. Tukey’s honestly significant difference post-hoc test was used to test for differences between each of the genotype–treatment combinations through the glht function in the multcomp package of the R software (Hothorn et al., 2008). For the analysis of reproduction, we used linear mixed models, using the lme function in the nlme package (Pinheiro et al., 2015), in which the fixed part of the model was genotype and the random part was plant individuals. Where appropriate, data were transformed to improve residual structure.
Results
DEG10 is exclusively found in mitochondria
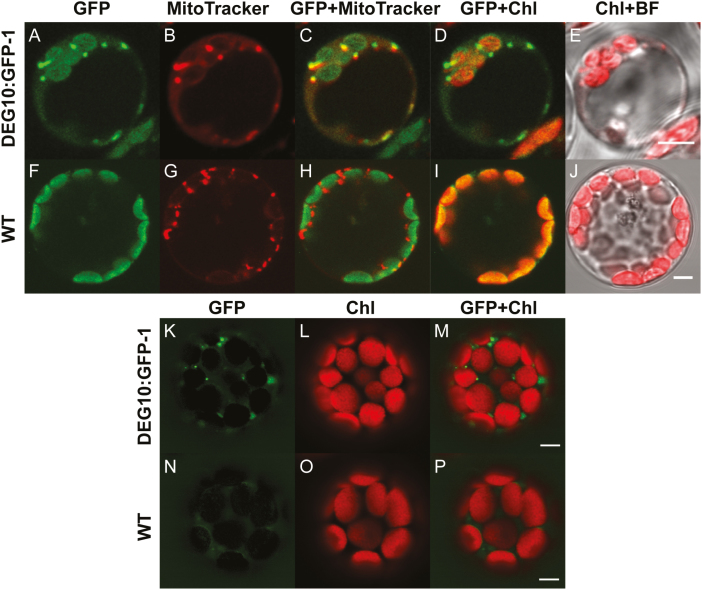
Sequence-based algorithms predominantly predict mitochondrial localization of DEG10 (Schwacke et al., 2003), whereas overexpression of DEG10:GFP in Arabidopsis cell cultures suggested dual targeting to mitochondria and chloroplasts (Tanz et al., 2014). To resolve this ambiguity, we generated transgenic plants stably expressing a DEG10:GFP fusion protein and analyzed three independent lines by confocal laser scanning microscopy. The brightest fluorescence signals in the GFP channel in DEG10:GFP-expressing cells formed small dots that co-localized with the fluorescence signals of MitoTracker, a cell-permeant mitochondria-selective dye (Fig. 1A–J, Supplementary Fig. S1). Additionally, fainter signals in the GFP channel matched chlorophyll autofluorescence of chloroplasts. We used linear unmixing of spectral images covering the range between 490 nm and 669 nm to demonstrate that the signal obtained from chloroplasts was entirely derived from chlorophyll and that GFP fluorescence was exclusively detected in mitochondria (Fig. 1K–P, Supplementary Fig. S1).
Fig. 1.
Mitochondrial localization of DEG10:GFP. Confocal fluorescence images of protoplasts isolated from DEG10:GFP-expressing (A–E, K–M) or wild-type (WT) (F–J, N–P) Arabidopsis plants. (A–J) protoplasts stained with MitoTracker Orange: (A, F) green fluorescence (497–561 nm), (B, G) MitoTracker Orange (572–615 nm, depicted in red); (C, H) merge of GFP and MitoTracker images; (D, I) merge of GFP and chlorophyll (Chl) autofluorescence (657–690 nm, depicted in red); (E, J) merge of Chl autofluorescence and a bright-field (BF) image. (K–P) Images obtained by spectral deconvolution using chlorophyll and GFP reference spectra: (K, N) GFP-specific fluorescence; (L, O) chlorophyll-specific fluorescence; (M, P) overlay of GFP and chlorophyll fluorescence. The images are false-colored; both MitoTracker and Chl signals are depicted in red for better visualization of co-localization in the merged images. Scale bars=5 µm.
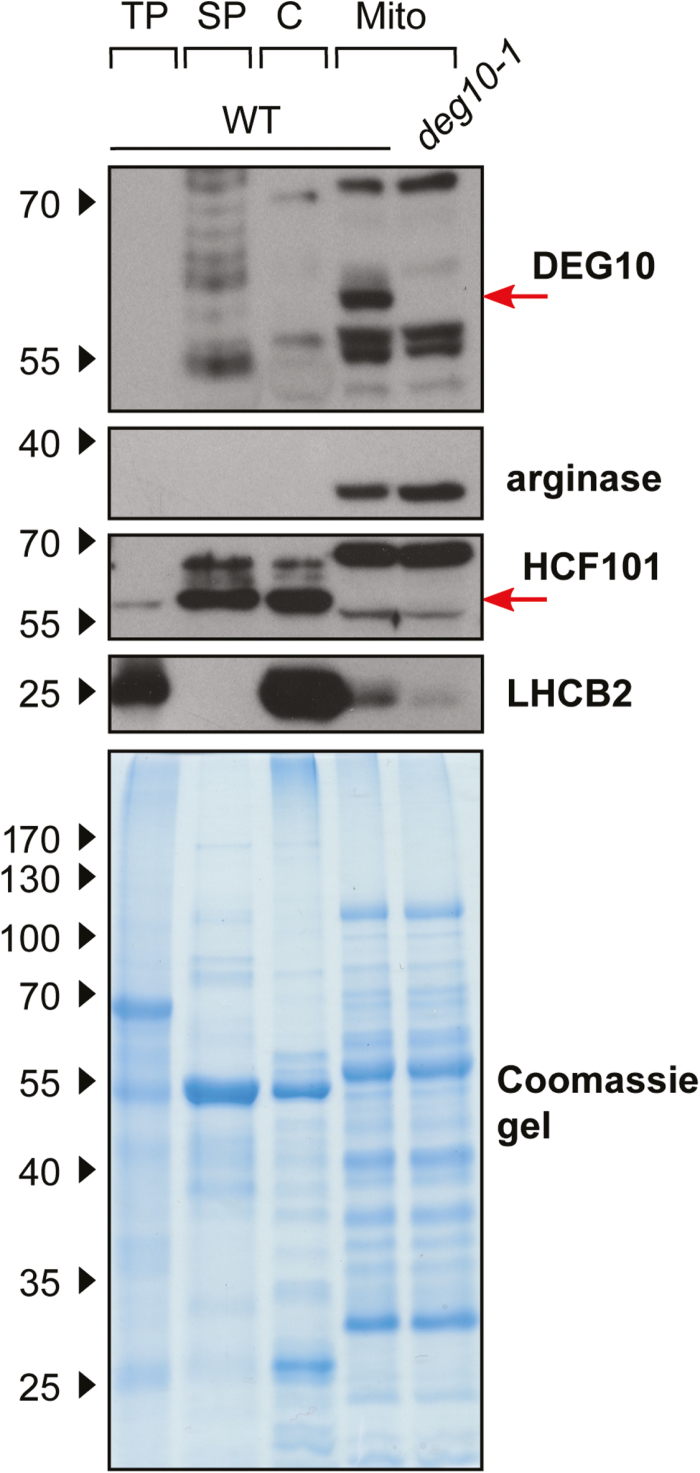
To analyze the localization of DEG10 in WT plants, we raised a polyclonal antiserum against a denatured, recombinant form of DEG10 in rabbits. The antiserum allowed sensitive immunoblot detection of recombinant DEG10 (≤0.2 ng) but cross-reacted with several other Arabidopsis proteins, particularly in soluble protein extracts (Fig. 2, Supplementary Fig. S2). After cell fractionation and immunodetection, comparison of mitochondrial protein extracts from WT plants and from the T-DNA insertion line deg10-1 (see below) allowed the unambiguous identification of a signal at ~60 kDa as originating from DEG10, which is in good agreement with the theoretical molecular mass of 62 kDa for mature DEG10 without the predicted transit peptide. DEG10 was below the detection limit in total protein extracts and was exclusively found in highly purified mitochondrial fractions, whereas no protein with the expected molecular mass was detected in preparations of intact chloroplasts (Fig. 2). The identity and purity of the isolated organelles were confirmed using arginase, HCF101 (high chlorophyll fluorescence protein 101), and LHCB2 (light-harvesting complex protein b2) as markers for mitochondria, chloroplast stroma, and thylakoid membranes, respectively. The distribution of HCF101, LHCB2, and arginase indicated that traces of thylakoid membranes were present in the mitochondrial preparation, while soluble proteins from chloroplasts and mitochondria were strictly separated. HCF101, but neither arginase nor DEG10, was detected in the soluble protein fraction, indicating that a substantial fraction of the chloroplasts was ruptured during organelle preparation, whereas the majority of the mitochondria stayed intact.
Fig. 2.
Analysis of the localization of DEG10 by cell fractionation. Arabidopsis wild-type (WT) plants or deg10-1 seedlings were separated into total protein (TP), soluble protein (SP), chloroplasts (C), and mitochondria (Mito). The following proteins were detected by immunoblotting: DEG10, arginase (mitochondrial, 38 kDa), HCF101 (high chlorophyll fluorescence protein 101, stroma, 57 kDa), LHCB2 (light-harvesting complex protein b2, thylakoids, 25 kDa). 8 µg protein per lane was separated on 10% polyacrylamide gels. A Coomassie-stained gel is shown as a loading control. The positions of molecular weight markers are indicated on the left.
We also used the recombinant DEG10 for biochemical analyses. The best yield of soluble recombinant DEG10 was obtained when the N-terminus was truncated by 92 amino acids (DEG10ΔN92, Supplementary Fig. S2A). Analysis by size exclusion chromatography indicated that the majority of the protein formed an oligomeric complex of ~400 kDa as estimated by comparison to globular marker proteins (Supplementary Fig. S3). We tested DEG10ΔN92 for proteolytic activity against β-casein, a typical model substrate for Deg/HtrA proteases (Sun et al., 2007; Ströher and Dietz, 2008; Sun et al., 2012), and a commercial resorufin-labeled casein substrate under a variety of buffer conditions (pH 5.5, 7.0, and 8.5), temperatures (5 °C, 10 °C, 30 °C, and 50 °C) and incubation times (0, 1, 2, 17 h), but did not detect unambiguous proteolytic activity compared with blanks or incubations with an inactive variant of DEG10 in which Ser265 of the catalytic triad was replaced by Ala (data not shown).
DEG10 is expressed predominantly in trichomes but also in the vascular tissue, and is induced by temperature stress
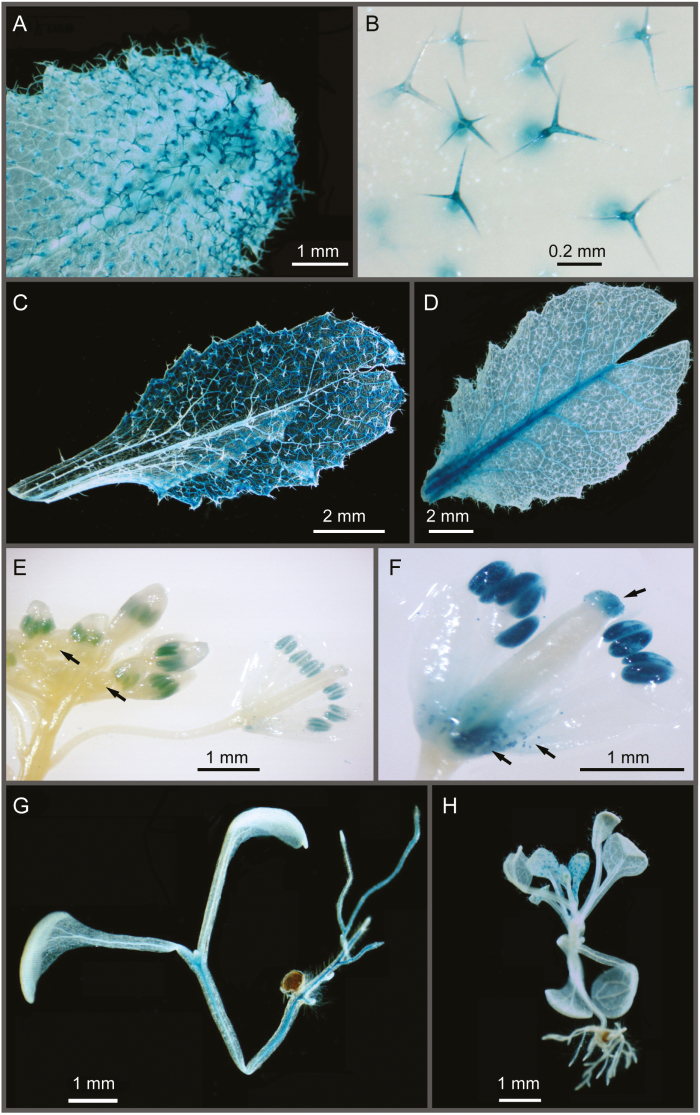
We analyzed the activity of GUS under the control of the DEG10 promoter to determine the organ and tissue specificity of DEG10 expression. At 6 weeks of age, approximately 20% of the 159 and 49 primary transformants carrying a PrDEG10:GUS or a PrDEG10:GFP:GUS construct, respectively, showed GUS activity in rosette leaves, independent of the construct used. GUS activity was observed in the trichomes of 67% (PrDEG10:GFP:GUS) and 83% (PrDEG10:GUS) of the GUS-positive plants (Fig. 3A, B, Supplementary Fig. 4B). With a lower frequency, GUS activity was detected in the entire leaf blade and in the vascular tissue (Fig. 3C, D, Supplementary Fig. 4B). Staining of inflorescences revealed GUS expression in ~12% of the plants with GUS activity in the leaves. Only in plants carrying the PrDEG10:GUS construct was a consistent pattern observed, with GUS activity in the male germline and in mature pollen as well as in stigmata (Fig. 3E, F). Otherwise, the tissue specificity and the intensity distributions of GUS activity were very similar for both reporter constructs.
Fig. 3.
Organ and tissue specificity of DEG10 promoter activity in Arabidopsis. Histochemical GUS staining was performed on T1 (A–F) and T2 (G, H) transgenic plants carrying either a PrDEG10:GFP:GUS (B, D, G, H) or a PrDEG10:GUS (A, C, E, F) expression cassette. (A–D) Different patterns of GUS expression observed in leaves of 6-week-old plants cultivated on soil in a glasshouse. (E, F) GUS expression in reproductive organs of approximately 3-month-old plants, showing GUS activity in pollen and at the stigma. Arrows in E indicate very young buds, in which no GUS activity was detected. Arrows in F indicate staining in the stigma and in pollen grains shattered from the anthers and caught in the base of the flower. (G, H) Staining of 18-day-old, whole T2 seedlings. Images were taken with a stereomicroscope.
Furthermore, we analyzed 18-day-old T2 seedlings from eight independent GUS-positive T1 plants carrying the PrDEG10:GFP:GUS construct. Neither staining pattern nor intensity were affected by the developmental stage (data not shown). In more than half of the lines, almost all seedlings showed GUS staining in trichomes of rosette leaves and occasionally in the vascular tissue of cotyledons, rosette leaves, and hypocotyls (Fig. 3G, H). In some lines, we additionally observed GUS expression in the vascular cylinder in roots, whereas in root tips expression was virtually absent (Fig. 3G, Supplementary Fig. S4A).
Expression of DEG14, another mitochondrial Deg protease, was reported to be induced selectively by heat stress (Basak et al., 2014). We analyzed the influence of elevated temperature (30 °C) on transcript levels of DEG10 and on GUS activity in plants carrying the PrDEG10:GFP:GUS construct (Supplementary Fig. S4C–E). Both the transcript level of DEG10 in entire seedlings and the frequency of GUS activity detection in roots were slightly increased at elevated temperature, indicating that DEG10 may contribute to acclimation.
DEG10 but not CATR3 is needed for optimal root growth, especially at elevated temperature
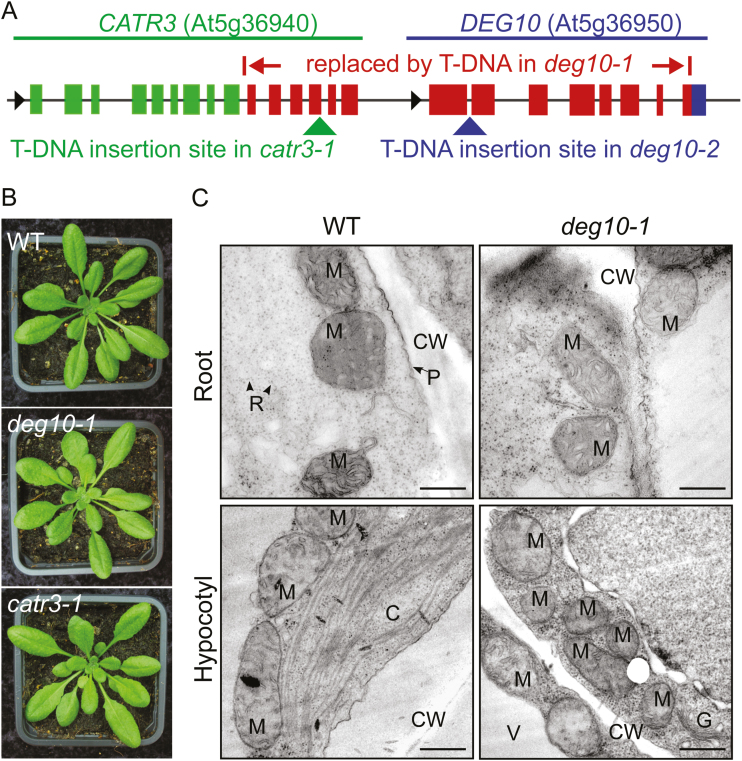
In order to analyze the physiological function of the DEG10 protease, we characterized deg10 T-DNA insertion lines. We screened 12 candidate lines of the SALK (Alonso et al., 2003), SAIL (Sessions et al., 2002), and GABI-KAT (Kleinboelting et al., 2012) collections (see Supplementary Table S1). Among these lines, native DEG10 transcripts were undetectable by RT–PCR in SALK_135850, SALK_127867, and GABI_088E04 (Supplementary Fig. S5). Analysis of flanking sequences showed that the two SALK lines carry the same T-DNA insertion replacing 5395 bp between intron 9 of the Cationic Amino Acid Transporter 3 gene (CATR3; At5g36940, previously referred to as CAT3, although this is commonly used for Catalase 3) and exon 8 of DEG10 (Fig. 4A). Since we did not find any phenotypic changes that result from a deletion of CATR3 (see below), we named this line deg10-1. Sequences corresponding to the 3′-end of CATR3 and the 5′-part of DEG10 could not be amplified by PCR from homozygous deg10-1 mutants (Supplementary Fig. S5A), excluding insertion of the deleted fragment elsewhere in the genome. RT–PCR analysis confirmed that deg10-1 is a deg10/catr3 double mutant (Supplementary Fig. S5B). In the line GABI_088E04 (deg10-2), the T-DNA junction towards the 3′-end was confirmed in the first intron, while we were unable to identify the opposite end of the T-DNA. The 3′-end of CATR3 and the 5′-end of DEG10 were unaffected in homozygous deg10-2 mutants, and no changes in CATR3 transcript levels were observed (Supplementary Fig. S5A, B). DEG10 protein was not detected in mitochondria of deg10-1 or deg10-2 mutants (Fig. 2A, Supplementary Fig. S2C). To attribute phenotypic alterations of the deg10-1 mutant specifically to the loss of DEG10 expression, we isolated a catr3 knockout line, SALK_036082, renamed catr3-1, as a control. In the catr3-1 line, the T-DNA is inserted in exon 13 (3467 bp downstream of the start codon) of the CATR3 gene (Fig. 4A, Supplementary Fig. S5C). Expression of DEG10 was slightly reduced in homozygous catr3-1 mutants in some experiments, while no native CATR3 transcripts were detected downstream of the T-DNA insertion site (Supplementary Fig. S4C and S5B, Table S4). Hence, any phenotypic or metabolic alterations found in deg10-1 and deg10-2 but not in catr3-1 are due to the loss of DEG10 expression.
Fig. 4.
Characterization of Arabidopsis T-DNA insertion mutants. (A) Schematic illustration of the genomic loci of DEG10 and CATR3 with the T-DNA insertion sites of the mutants deg10-1, deg10-2, and catr3-1. Exons are represented as boxes, and introns, untranslated regions, and intergenic regions as black lines. Black arrows indicate the direction of transcription. In the deg10-1 mutant, the T-DNA replaces 5395 bp from intron 9 of CATR3 to a part of exon 8 of DEG10 (indicated by red boxes for exons). While a larger part of the CATR3 gene (green boxes) is intact in deg10-1 mutants, only a fragment of exon 8 of DEG10 is retained (blue box). In the deg10-2 mutant, the T-DNA is inserted in intron 1 of Deg10 (blue arrow), while the catr3-1 mutant carries a T-DNA within exon 13 of CATR3 (green arrow). (B) Photographs of representative 6-week-old wild-type (WT), deg10-1, and catr3-1 plants grown under short-day conditions in a glasshouse. (C) TEM images of mitochondria in root and hypocotyl tissue of 5-day-old WT and deg10-1 seedlings. C, chloroplast; CW, cell wall; G, Golgi apparatus; M, mitochondrion; P, plasma membrane; R, ribosome; V, vacuole. Scale bars=500 nm.
Phenotypic analysis of WT plants and deg10-1 and catr3-1 mutants showed no overt differences in vegetative growth under common glasshouse conditions (Fig. 4B). In addition, the mitochondrial ultrastructure was not altered in deg10-1 (Fig. 4C), indicating that neither DEG10 nor CATR3 plays an indispensable role for growth or mitochondrial organization.
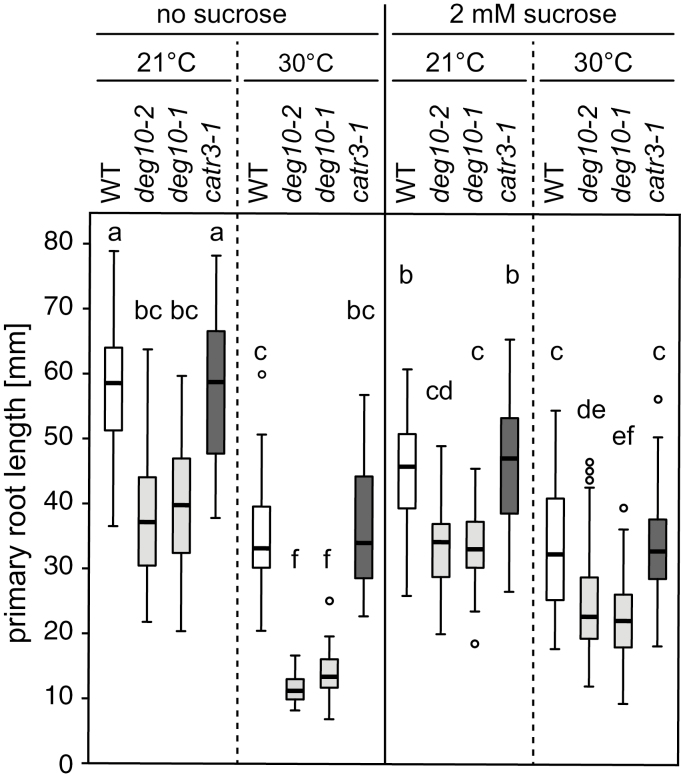
Publicly accessible microarray data indicated that DEG10 transcripts accumulated most strongly in imbibed seeds (eFP Browser; Winter et al., 2007). We therefore monitored seeds of WT plants and deg10-1 and catr3-1 mutants in axenic culture to test whether loss of DEG10 affects germination and early seedling development. Since Deg proteases are often associated with responses to protein-folding stress, the seedlings were grown under four different conditions: absence or presence of 2% sucrose (to promote mitochondrial respiration) and ambient temperature (22 °C/20 °C or 21 °C/17 °C light/dark) or elevated temperature (30 °C/25 °C light/dark). Germination and seedling establishment did not differ significantly between the analyzed genotypes, while cultivation at elevated temperature or in the presence of sucrose generally promoted seedling establishment slightly, by up to 10% (data not shown). Elongation of the primary root was not different between WT plants and catr3-1 mutants, but was reduced in deg10 mutants under all conditions tested (Fig. 5, Supplementary Figs S6 and S7, Table S5). The addition of sucrose to the medium reduced the root growth of WT plants and catr3-1 mutants at ambient temperature, while it had no significant effect on root growth of deg10 mutants. Elevated temperature reduced the root growth in all genotypes, and this effect was most pronounced in deg10 mutants in the absence of sucrose. At 30 °C day temperature, the root growth of WT plants and catr3-1 mutants was not affected by the addition of sucrose, but under the same regime of heat stress the root growth of deg10 mutants was slightly enhanced by sucrose. In a previous experiment with freshly harvested seeds and slightly different temperature regimes and plate geometries, elevated temperature stimulated root elongation; this effect was much stronger in WT plants and catr3-1 mutants, and depended on sucrose in deg10-1 mutants (Supplementary Fig. S7, Table S5). In contrast, leaf number at day 10 and continuous development until day 18 were not affected by the loss of DEG10, as no significant differences in these parameters between WT seedlings and either the deg10-1 or catr3-1 mutants were observed. For each line, leaf number at day 10 was higher at the elevated temperature, and this effect was stronger when sucrose was added to the medium. In the absence of sucrose, ~15% of the seedlings grown at room temperature were arrested in development during the first week in all experiments, and at elevated temperature 30–50% of the seedlings stopped growing. With sucrose, nearly all plants developed continuously, regardless of temperature (Supplementary Fig. S7, Table S5).
Fig. 5.
Effect of temperature and sucrose on primary root growth of Arabidopsis wild-type (WT), deg10-2, deg10-1, and catr3-1 seedlings. Per genotype and condition, 54 seeds (from three different parental plants) were plated on medium with or without 2% sucrose. All plates were incubated at ambient temperature (21 °C day/17 °C night) for 3 days to allow germination and seedling establishment. Thereafter, the plates for temperature stress were incubated at elevated temperature (30 °C day/25 °C night), and after 13 days the length of the primary root was measured. Between 27 and 54 plants per genotype and condition showed continuous growth during this period; all others were excluded from the analysis. Boxes show the interquartile range with medians, whiskers extend to the most extreme data point within 1.5 times the interquartile range, and open circles indicate data points outside this range. Boxes that are not marked by a common letter have significantly different means (P<0.05 in all-pairwise comparisons of log-transformed values with Tukey’s honestly significant difference post-hoc test). Data are from one out of three experiments with similar results. See Supplementary Table S5 for the analysis of interactions between genotype and treatment.
DEG10 deficiency affects mitochondrial proteostasis at elevated temperatures
To identify proteins affected by DEG10 deficiency that may be linked to the reduced root growth at elevated temperature, we compared the root proteomes of WT, deg10-1, deg10-2, and catr3-1. Plants were hydroponically grown and exposed to an elevated temperature of 30 °C for 2 days before extraction of total proteins from roots. For each genotype, four biological replicate samples consisting of pooled roots from two or three plants were analyzed by mass spectrometry. Overall, this label-free shotgun proteome analysis identified 2125 proteins that were quantified in at least three biological replicates of one line (Supplementary Table S6). Among these, only 16 proteins exhibited significantly altered abundance in the comparison between all genotypes. Ten of these proteins, all nuclear encoded but with predicted mitochondrial location, showed significantly and consistently altered protein abundance in deg10-2 compared with Col-0, and in deg10-1 compared with catr3-1 (because deg10-1 is actually a deg10/catr3 double mutant, as described above; Table 1, Supplementary Table S7). In both DEG10-deficient mutants, the mitochondrial stomatin-like scaffold proteins SLP1 and SLP2 and a mitochondrial Hsp23.5 heat shock protein were more abundant than in the corresponding control line, while the Complex III subunit 4-2, four mitochondrial ATP synthase subunits, and the two subunits of the mitochondrial processing peptidase were moderately lower in abundance (Table 1).
Table 1.
Proteins with significantly altered abundance in deg10-2 compared with Col-0 wild-type (WT) and deg10-1 compared with catr3-1 roots stressed for 3 days at 30 °C
| UniProt protein IDs | Protein names | Gene locus | log2 (deg10-1/ catr3-1)b | log2 (deg10-2/ Col-0)b | Peptides | Unique peptides | Sequence coverage (%) |
|---|---|---|---|---|---|---|---|
| Q9LVW0 | SLP2 | At5g54100 | 1.58 | 1.48 | 6 | 3 | 21.9 |
| Q9FGM9 | HSP23.5 | At5g51440 | 0.95 | 1.08 | 4 | 3 | 27.6 |
| Q93VP9 | SLP1 | At4g27585 | 0.63 | 0.69 | 10 | 7 | 32.4 |
| Q9ZU25 | Probable MPP subunit alpha-1 | At1g51980 | –0.30 | –0.24 | 26 | 18 | 78.1 |
| Q42290 | Probable MPP subunit beta | At3g02090 | –0.33 | –0.24 | 19 | 19 | 49 |
| P83484 a; P83483 a; Q9C5A9 a | ATP synthase subunit beta-1,-2,-3 | At5g08670; At5g08680; At5g08690 | –0.41 | –0.36 | 27 | 26 | 67.3 |
| Q96250 | ATP synthase subunit gamma | At2g33040 | –0.42 | –0.45 | 15 | 15 | 57.8 |
| P92549 | ATP synthase subunit alpha | AtMg01190 | –0.55 | –0.48 | 27 | 6 | 59.6 |
| Q96251 | ATP synthase subunit O | At5g13450 | –0.28 | –0.48 | 11 | 11 | 52.1 |
| Q9FKS5 | Complex III subunit 4-2 | At5g40810 | –0.49 | –0.55 | 7 | 3 | 43.5 |
a These three highly homologous proteins could not be distinguished based on the identified peptides.
b Significance was determined by ANOVA with correction for multiple testing by Tukey’s honestly significant difference post-hoc test on data from three (WT) or four (all mutants) biological replicates per genotype.
The higher abundance of mitochondrial proteins may be caused directly by the absence of DEG10 activity in mitochondria, but also by altered gene expression. Therefore, we quantified the transcript levels of SLP1, SLP2, and Alternative Oxidase 1a (AOX1a) as marker gene for mitochondrial retrograde signaling in seedlings cultivated at either 21 °C or 30 °C daytime temperature (Fig. 6; Supplementary Fig. S8, Table S4). Transcript levels of all three genes were significantly increased at elevated temperature, indicating that the applied stress induced mitochondrial retrograde signaling. In agreement with the strongest accumulation of SLP2 in the mitochondria of deg10 mutants, SLP2 transcript levels were consistently higher in deg10 mutants compared with the corresponding control line, especially at elevated temperature. In addition, AOX1a transcripts were more abundant in deg10 mutants compared with the corresponding controls, but this effect was not consistently observed in all replicate experiments. Transcript levels of SLP1 were not affected in deg10 mutants. These findings indicate that DEG10 activity counteracts temperature stress in mitochondria and/or dampens retrograde signaling. Taken together, both the proteome and the transcript data indicate a small but significant contribution of DEG10 to mitochondrial proteostasis under temperature stress conditions, in agreement with the absence of obvious mutant phenotypes in deg10 mutants under laboratory conditions.
Decreased fecundity of deg10-1 mutants under natural conditions
Exposure to multiple simultaneous stresses, as frequently encountered in nature, places greater demands on plant protein quality control mechanisms. Therefore, we hypothesized that the role of DEG10 might become more apparent under natural conditions, and performed field trials in Umeå, Sweden, during the summers of 2011, 2012, and 2014. The deg10-2 mutant line had not been isolated at that time and was therefore not included in these field trials.
Initial growth and final rosette size were lower for all genotypes in 2012 compared with 2011, presumably due to lower temperatures and less precipitation (Supplementary Fig. S9, Table S8; weather data: http://www.tfe.umu.se). Extremely challenging environmental conditions in 2014 resulted in very poor growth, low survival rates, low seed yield, and a high variability among WT plants and insertion mutants alike. In 2011 and 2012, after 3 weeks in the field, the average rosette diameter was 13% and 25% bigger, respectively, for the deg10-1 mutants compared with WT plants; this difference was significant in 2012. After the fourth and fifth weeks of growth, rosettes of WT plants and deg10-1 mutants were no longer different in size. No further obvious phenotypic differences between the vegetative rosettes of WT plants and deg10-1 mutants were observed in the field.
However, in all 3 years the deg10-1 plants produced 3–20% less seeds per silique compared with WT plants (Table 2). This decreased seed production was statistically significant in 2011 and 2012. In 2014, the fertility of all genotypes was reduced by approximately 64%, with fewer siliques per plant and fewer seeds per silique compared with previous years. In all three years, the total number of siliques per plant was not significantly different between WT plants and deg10-1 mutants (Table 3), indicating that there was no compensation for the reduced number of seeds per silique. Consequently, the deg10-1 mutants produced fewer seeds per plant under natural conditions in the field. In addition, under controlled conditions in the glasshouse in Konstanz, the deg10-1 mutants showed in two of three passes a tendency to produce fewer seeds per silique (Supplementary Table S9), although the differences were not statistically significant (P≥0.05). In the field trial in 2014 and in several glasshouse experiments, we never observed consistent differences in growth or fertility between WT plants and catr3-1 mutants. Therefore, we conclude that the impaired fertility of the deg10-1 mutants can be attributed to the loss of DEG10 expression.
Table 2.
Seed production of Arabidopsis WT plants, deg10-1 and catr3-1 mutants under field conditions
| Year | Genotype | Seeds per silique (mean ±SE)a | t-valueb | P-valueb | N siliques (Nplants)c |
|---|---|---|---|---|---|
| 2011 | WT | 54.5±1.1 | 150 (30) | ||
| deg10-1 | 43.5±2.0* | –4.79 | 0.000 | 150 (30) | |
| 2012 | WT | 55.2±0.8 | 140 (29) | ||
| deg10-1 | 52.8±0.9* | –2.03 | 0.047 | 150 (30) | |
| 2014 d | WT | 18.8±1.8 | 143 (31) | ||
| deg10-1 | 18.3±1.3 | –0.33 | 0.739 | 184 (39) | |
| catr3-1 | 20.4±1.5 | 0.93 | 0.355 | 130 (29) |
a Based on plant mean.
b For comparison to WT reference (model intercept).
c Number (N) of analyzed siliques and plants.
d Statistical analysis performed on square-root-transformed data.
* Significantly lower values for deg10-1 mutants compared with WT plants.
Table 3.
Analysis of the number of siliques per plant of Arabidopsis wild-type (WT) plants and deg10-1 and catr3-1 mutants under field conditions
| Year | Genotype | Siliques per plant (mean ±SE) | t-valuea | P-valuea | Number of analyzed plants |
|---|---|---|---|---|---|
| 2011 b | WT | 324.9±23.8 | 30 | ||
| deg10-1 | 309.7±24.1 | –0.24 | 0.815 | 30 | |
| 2012 b | WT | 318.1±26.6 | 29 | ||
| deg10-1 | 332.7±32.2 | 0.31 | 0.756 | 30 | |
| 2014 b | WT | 141.2±20.9 | 41 | ||
| deg10-1 | 179.4±27.7 | 0.90 | 0.371 | 48 | |
| catr3-1 | 132.6±30.6 | –0.93 | 0.355 | 38 |
a For comparison to WT reference (model intercept).
b Statistical analysis performed on log-transformed data.
Discussion
Proteases of the Deg/HtrA family contribute to the eukaryotic protein quality control system in several cellular compartments, particularly those of endosymbiotic origin. The data obtained in this study reveal that DEG10 in Arabidopsis is exclusively localized in mitochondria, contributes to mitochondrial proteostasis at elevated temperature, and is important for root growth and reproduction.
Plant Deg proteases are predominantly localized in mitochondria or chloroplasts, with some ambiguous predictions and individual reports of dual localization (Schuhmann and Adamska, 2012; Tanz et al., 2014). A DEG10:GFP fusion protein overexpressed in cell cultures was detected in mitochondria as well as in plastids (Tanz et al., 2014). In contrast, we consistently found DEG10 exclusively in the mitochondria using two independent experimental approaches, stable expression of a DEG10:GFP fusion protein and cell fractionation in combination with immunoblotting (Figs 1 and 2; Supplementary Fig. S1). In our experiments, the green fluorescence in chloroplasts of both DEG10:GFP-expressing and WT protoplasts resulted exclusively from endogenous pigments, presumably chlorophyll. In immunoblot analyses, we detected DEG10 in isolated mitochondria but not in chloroplasts. We further determined the tissue specificity of DEG10 expression using the GUS reporter system and found a high level of expression of DEG10 in trichomes (Fig. 3). This is in agreement with microarray data, which showed 2-fold higher levels of DEG10 transcript in trichomes compared with the leaf blade, while in gl3-sst/nok-1 double mutant trichomes the transcript levels were 10-fold higher than in the WT leaf blade [eFP Browser; Marks et al., 2009). In contrast, our detection of DEG10 promoter activity exclusively in the central cylinder of mature roots (Supplementary Fig. S4) is in conflict with microarray data, which showed the highest DEG10 transcript levels in the root tip (Birnbaum et al., 2003). This might be due either to missing promoter elements in our GUS expression construct or to post-transcriptional regulation of DEG10 expression in roots. In summary, DEG10 is exclusively localized in mitochondria, with strongest expression in trichomes, pollen, and the vascular tissue.
The main aim of this study was to characterize the physiological functions of DEG10 through analysis of loss-of-function deg10 mutants. However, the expression of a putative endoplasmic reticulum-localized Cationic Amino acid Transporter 3 (CATR3) (Yang et al., 2014) is also compromised in the deg10-1 mutant (Fig. 4, Supplementary Fig. S5). We therefore isolated a catr3-1 single mutant and included this in our experiments as a control. None of the phenotypic alterations of deg10-1 mutants were detected in catr3-1 mutants, while our key findings were confirmed with a second, independent DEG10-deficient mutant line, deg10-2, which was identified at a later stage in the study. Therefore, we conclude that the reduced root growth and lower seed production of deg10 mutants described in this paper are due to loss of DEG10 and were not influenced by the presence or absence of CATR3.
The reduced fertility of deg10-1 mutants (Table 2) and the heat sensitivity of root elongation (Fig. 5, Supplementary Figs S6 and S7) in deg10-1 and deg10-2 seedlings correlated well with the observation of DEG10 expression in pollen and the vascular tissue of roots as important sink tissues. In contrast, mitochondrial morphology appeared normal in deg10-1 seedlings, and trichome appearance did not differ between WT and mutant leaves, indicating that DEG10 is not essential for mitochondria and trichome development. The developmental and fertility defects of the deg10-1 mutant were most pronounced under challenging environmental conditions, indicating that DEG10 contributes to the maintenance of mitochondrial functions under stress. At the proteome level, roots exposed to elevated temperatures showed a moderate but significant decrease in the abundance of several subunits of the mitochondrial electron transport chain complexes in both deg10-1 and deg10-2 mutants compared with catr3-1 and WT plants, respectively (Table 1). These included four subunits of the ATP synthase, as well as subunit 4-2 of complex III and the two subunits of the mitochondrial processing peptidase, which are integral components of the respiratory complex III in plants (Teixeira and Glaser, 2013). In contrast, a heat shock protein (HSP23.5) and the stomatin-like scaffold proteins SLP1 and SLP2 were more abundant in mitochondria of deg10 mutants. Both SLP1 and SLP2 are known to be regulated by mitochondrial retrograde signaling, with both up- and down-regulation reported depending on the agent used to induce mitochondrial stress (Willeke, 2011; Umbach et al., 2012; Ng et al., 2013). Alternative oxidases, the key markers for retrograde mitochondrial stress signaling, were not detected in our proteome analysis. We therefore used qRT–PCR analysis to assess the transcript levels of AOX1a, SLP1, and SLP2, and found higher expression in plants cultivated at 30 °C compared with 21 °C, indicating that retrograde signaling may have been induced by the temperature regime applied in our study (Fig. 6). In deg10 mutants, only the transcript level of SLP2 was reproducibly higher than in WT plants, and the difference was most pronounced at 30 °C, while no consistent DEG10-dependent differences were detected for AOX1a or SLP1 transcript levels. Interestingly, previous studies demonstrated that SLP1 and SLP2 affect the assembly of electron transport chain supercomplexes and interact with FtsH4 (Gehl et al., 2014; Opalinska et al., 2017). In addition, the human homologue HsSLP2 accumulated under conditions of mitochondrial stress and affected the stability of respiratory complexes I and IV (Da Cruz et al., 2008). Taken together, the DEG10-dependent transcript and proteome alterations indicate an altered abundance and/or composition of the mitochondrial electron transport chain complexes. This probably causes a mild impairment of the primary metabolism with associated altered retrograde signaling (da Cunha et al., 2015) in deg10 mutants, which in turn could explain the observed mild quantitative phenotypes. Overall, our data indicate a subtle influence of DEG10 on mitochondrial proteostasis, mitochondrial retrograde signaling, and the overall performance of mitochondria, resulting in mild developmental defects of deg10 mutants under stress conditions.
There may be several reasons for the impaired root growth of deg10 mutants. Either the transport to the root and utilization of photoassimilates may be compromised (Freixes et al., 2002; Durand et al., 2016; Kushwah and Laxmi, 2017), or the loss of DEG10 interferes with the regulation of root growth. Since exogenous sucrose supply rescued the elongation defect of deg10 mutants only partially, both scenarios may be true. The observation that the differences between WT plants and deg10 mutants became more pronounced after prolonged storage of the seeds may indicate that DEG10 contributes to the preservation of mitochondria in seeds or their reactivation during germination. While the requirement of mitochondrial energy production for root growth is obvious, the interplay of mitochondrial retrograde signaling with the regulation of root growth by polar auxin transport has only recently been revealed. In several studies, a reciprocal regulation of mitochondrial stress responses by auxin and an impact of mitochondrial stress on auxin signaling and root growth were observed (Ivanova et al., 2014; Kerchev et al., 2014; Zhang et al., 2014b; Van Aken et al., 2016; Wang and Auwerx, 2017).
In E. coli, it was shown that DegP generally removes or refolds damaged proteins, whereas DegS contributes to the generation of a stress-specific transcription factor (Clausen et al., 2011). Further studies are required to determine whether DEG10 mainly contributes to the maintenance of mitochondrial functions during stress, or whether it participates in the generation or transmission of specific stress signals from mitochondria. DEG14, another mitochondrial Deg protease of Arabidopsis, has also been implicated in stress tolerance, as its overexpression resulted in increased thermotolerance (Basak et al., 2014). It is conceivable that Deg proteases, as ATP-independent proteases, are more important under stress conditions, when ATP levels are low and partially unfolded or otherwise damaged proteins may accumulate. Further analyses will be required to determine whether DEG10 and DEG14 act synergistically or independently in mitochondria.
The essential function of mitochondria in fertility, in particular pollen fertility, has long been recognized, while the exact mechanisms by which mitochondrial defects cause cytoplasmic male sterility remain poorly understood (Horn et al., 2014). Two other proteases, the cysteine protease CEP1 in Arabidopsis and the aspartic protease AP65 in rice, have been reported to contribute to pollen development and male fertility, respectively (Huang et al., 2013; Zhang et al., 2014a). Our observations that DEG10 is essential for the maintenance of optimal fertility in variable and challenging environmental conditions are again consistent with a function of DEG10 in protein turnover to stabilize mitochondrial energy production during stress. So far, we have not been able to unambiguously detect proteolytic activity of heterologously expressed, N-terminally truncated DEG10 variants purified as soluble proteins. Recombinant DEG10ΔN92 exhibited a native molecular weight in agreement with the formation of a hexamer (Supplementary Fig. S3), an oligomerization state that is the inactive resting state of several Deg/HtrA proteases, including Arabidopsis DEG2 (Sun et al., 2012). At present, we do not know whether our variants missed essential parts, or whether the tested conditions or model substrates were unsuitable. Alternatively, DEG10 might primarily act as a chaperone for mitochondrial electron transport chain complexes, similar to the role of DEG1 in the assembly of photosystem II (Sun et al., 2010).
In summary, our findings suggest a function of DEG10 in mitochondrial electron transport chain proteostasis. DEG10 thereby contributes to optimal root development and maximal seed set under challenging environmental conditions, and thus to individual performance under variable environmental conditions and potentially the evolutionary fitness of plants. This may also explain why all plant genomes analyzed in detail contain a direct homolog of DEG10 (Schuhmann and Adamska, 2012).
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Mitochondrial localization of DEG10.
Fig. S2. Characterization of the polyclonal anti-DEG10 immune serum by immunoblot analyses.
Fig. S3. Apparent molecular mass of recombinant DEG10.
Fig. S4. Regulation of DEG10 promoter activity and localization in roots.
Fig. S5. Molecular characterization of the T-DNA insertion mutants deg10-1, deg10-2, and catr3-1.
Fig. S6. Effect of temperature and sucrose on root growth of Arabidopsis WT, deg10-2, deg10-1, and catr3-1 seedlings.
Fig. S7. Effect of temperature and sucrose on development of Arabidopsis WT, deg10-1, and catr3-1 seedlings.
Fig. S8. Transcript levels of SLP1, SLP2, and AOX1a in catr3-1 and deg10-1 mutants at different temperatures.
Fig. S9. Growth and development of WT plants and deg10-1 mutants under field conditions.
Table S1. Overview of T-DNA insertion lines analyzed for T-DNA insertions affecting DEG10 expression.
Table S2. Primers used for PCRs for the characterization of T-DNA insertion mutants and for qRT–PCR analysis of DEG10, SLP1, SLP2, and AOX1a transcript levels.
Table S3. Antibodies and conjugates used in this study.
Table S4. Statistical analyses of qRT–PCR data.
Table S5. Statistical analyses of seedling development.
Table S6. Proteins identified and quantified by comparative label-free proteome analysis of deg10-1, deg10-2, catr3-1, and Col-0 roots.
Table S7. Proteins with significant differences in abundance among deg10-1, deg10-2, catr3-1, and Col-0 WT roots.
Table S8. Analyses of rosette diameters of WT and deg10-1 plants grown in the field.
Table S9. Seed yield analyses of WT and deg10-1 plants grown in the glasshouse.
Author contributions
CVH, IA, CF, PFH, and DF planned and designed the research. CVH, BDJ, LSM, SN, GW, PFH, and DF performed the laboratory experiments. CF designed and performed the field experiments. CVH, BDJ, LSM, SN, GW, MS, CF, PFH, and DF analyzed the data. CVH wrote the initial draft of the paper, and PFH and DF wrote the final version. All authors contributed to the interpretation of the data, and edited and approved the final version of the manuscript.
Conflicts of interest
All authors declare that there are no conflicts of interest.
Data deposition
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [https://www.ebi.ac.uk/pride/archive/ (Vizcaíno et al., 2016)], with the dataset identifier PXD007204.
Acknowledgements
The authors thank Martin Stöckl and the Bioimaging Centre of the University of Konstanz for help with spectrally resolved confocal microscopy, Elzbieta Glaser and Beata Kmiec (Stockholm University, Sweden) for advice on promoter:GUS fusion constructs, Daniela Völker for assistance with recombinant protein assays, Eva Zeiser and the Animal Research Facility for the polyclonal anti-DEG10 immune serum, the team of the EM Center Konstanz and Carolina Rio Bartulos for help in preparation and interpretation of electron micrographs, the RIKEN Genomic Sciences Center for the RAFL cDNA clone, Patrik Storm (Umeå University, Sweden) for help with the field experiments, as well as our technicians and gardeners. This work was funded by the German Science Foundation (AD92/12-1; CRC969 Project C02), an EMBO short-term fellowship to CH, and a grant by the Swedish Energy Agency (2012-005889) and Umeå University to CF. PFH is in part supported by funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (starting grant 639905 ‘ProPlantStress’).
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Basak I, Pal R, Patil KS, et al. 2014. Arabidopsis AtPARK13, which confers thermotolerance, targets misfolded proteins. Journal of Biological Chemistry 289, 14458–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1–7. 2014. https://cran.r-project.org/web/packages/lme4/index.html [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. 2003. A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. HTRA proteases: regulated proteolysis in protein quality control. Nature Reviews Molecular Cell Biology 12, 152–162. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Parone PA, Gonzalo P, Bienvenut WV, Tondera D, Jourdain A, Quadroni M, Martinou JC. 2008. SLP-2 interacts with prohibitins in the mitochondrial inner membrane and contributes to their stability. Biochimica et Biophysica Acta 1783, 904–911. [DOI] [PubMed] [Google Scholar]
- da Cunha FM, Torelli NQ, Kowaltowski AJ. 2015. Mitochondrial retrograde signaling: triggers, pathways, and outcomes. Oxidative Medicine and Cellular Longevity 2015, 482582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M, Porcheron B, Hennion N, Maurousset L, Lemoine R, Pourtau N. 2016. Water deficit enhances C export to the roots in Arabidopsis thaliana plants with contribution of sucrose transporters in both shoot and roots. Plant Physiology 170, 1460–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. The Plant Journal 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Eubel H, Heazlewood JL, Millar AH. 2007. Isolation and subfractionation of plant mitochondria for proteomic analysis. In: Thiellement H, Zivy M, Damerval C, Mechin V, eds. Plant protocols: methods and protocols. Methods in molecular biology Vol. 355 Totowa: Humana Press, 49–62. [DOI] [PubMed] [Google Scholar]
- Fox J, Weisberg S. 2011. An R companion to applied regression. Los Angeles: Sage. [Google Scholar]
- Freixes S, Thibaud M-C, Tardieu F, Muller B. 2002. Root elongation and branching is related to local hexose concentration in Arabidopsis thaliana seedlings. Plant, Cell & Environment 25, 1357–1366. [Google Scholar]
- Frenkel M, Jänkänpää HJ, Moen J, Jansson S. 2008. An illustrated gardener’s guide to transgenic Arabidopsis field experiments. New Phytologist 180, 545–555. [DOI] [PubMed] [Google Scholar]
- Gehl B, Lee CP, Bota P, Blatt MR, Sweetlove LJ. 2014. An Arabidopsis stomatin-like protein affects mitochondrial respiratory supercomplex organization. Plant Physiology 164, 1389–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibala M, Kicia M, Sakamoto W, Gola EM, Kubrakiewicz J, Smakowska E, Janska H. 2009. The lack of mitochondrial AtFtsH4 protease alters Arabidopsis leaf morphology at the late stage of rosette development under short-day photoperiod. The Plant Journal 59, 685–699. [DOI] [PubMed] [Google Scholar]
- Horn R, Gupta KJ, Colombo N. 2014. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 19(Pt B), 198–205. [DOI] [PubMed] [Google Scholar]
- Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biometrical Journal Biometrische Zeitschrift 50, 346–363. [DOI] [PubMed] [Google Scholar]
- Huang J, Zhao X, Cheng K, Jiang Y, Ouyang Y, Xu C, Li X, Xiao J, Zhang Q. 2013. OsAP65, a rice aspartic protease, is essential for male fertility and plays a role in pollen germination and pollen tube growth. Journal of Experimental Botany 64, 3351–3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, et al. 2014. A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates Alternative Oxidase1a expression in Arabidopsis. Plant Physiology 165, 1233–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kerchev PI, De Clercq I, Denecker J, Mühlenbock P, Kumpf R, Nguyen L, Audenaert D, Dejonghe W, Van Breusegem F. 2014. Mitochondrial perturbation negatively affects auxin signaling. Molecular Plant 7, 1138–1150. [DOI] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. 2012. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Research 40, D1211–D1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küpper H, Parameswaran A, Leitenmaier B, Trtílek M, Setlík I. 2007. Cadmium-induced inhibition of photosynthesis and long-term acclimation to cadmium stress in the hyperaccumulator Thlaspi caerulescens. New Phytologist 175, 655–674. [DOI] [PubMed] [Google Scholar]
- Kushwah S, Laxmi A. 2017. The interaction between glucose and cytokinin signaling in controlling Arabidopsis thaliana seedling root growth and development. Plant Signaling & Behavior 12, e1312241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Larkindale J, Vierling E. 2008. Core genome responses involved in acclimation to high temperature. Plant Physiology 146, 748–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohscheider JN, Rojas-Stütz MC, Rothbart M, Andersson U, Funck D, Mendgen K, Grimm B, Adamska I. 2015. Altered levels of LIL3 isoforms in Arabidopsis lead to disturbed pigment-protein assembly and chlorophyll synthesis, chlorotic phenotype and impaired photosynthetic performance. Plant, Cell & Environment 38, 2115–2127. [DOI] [PubMed] [Google Scholar]
- Marks MD, Wenger JP, Gilding E, Jilk R, Dixon RA. 2009. Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Molecular Plant 2, 803–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Liddell A, Leaver CJ. 2007. Isolation and subfractionation of mitochondria from plants. In: Pon LA, Schon EA, eds, Mitochondria, 2nd edn. Methods in cell biology, Vol. 80 San Diego, Elsevier, 65–90. [DOI] [PubMed] [Google Scholar]
- Mishra LS, Mielke K, Wagner R, Funk C. 2019. Reduced expression of the proteolytically inactive FtsH members has impacts on the Darwinian fitness of Arabidopsis thaliana. Journal of Experimental Botany 70, 2173–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, et al. 2013. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. The Plant Cell 25, 3450–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Kato Y, Sakamoto W. 2016. Chloroplast proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiology 171, 2280–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinska M, Parys K, Janska H. 2017. Identification of physiological substrates and binding partners of the plant mitochondrial protease FTSH4 by the trapping approach. International Journal of Molecular Sciences 18, 2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, Debroy S, Sarkar D, R Core Team 2015. Linear and nonlinear mixed effects models. https://cran.r-project.org/web/packages/nlme/index.html [Google Scholar]
- R Core Team 2013. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, Ito T, Konagaya A, Toyoda T, Shinozaki K. 2005. RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Research 33, D647–D650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmann H, Adamska I. 2012. Deg proteases and their role in protein quality control and processing in different subcellular compartments of the plant cell. Physiologia Plantarum 145, 224–234. [DOI] [PubMed] [Google Scholar]
- Schuhmann H, Huesgen PF, Adamska I. 2012. The family of Deg/HtrA proteases in plants. BMC Plant Biology 12, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R. 2003. ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiology 131, 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, et al. 2002. Functional annotation of a full-length Arabidopsis cDNA collection. Science 296, 141–145. [DOI] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, et al. 2002. A high-throughput Arabidopsis reverse genetics system. The Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvany-Villalobo G, Davydov O, Ben-Ari G, Zaltsman A, Raskind A, Adam Z. 2004. Expression in multigene families. Analysis of chloroplast and mitochondrial proteases. Plant Physiology 135, 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares A, Niedermaier S, Faro R, Loos A, Manadas B, Faro C, Huesgen PF, Cheung AY, Simoes I. 2019. An atypical aspartic protease modulates lateral root development in Arabidopsis thaliana. Journal of Experimental Botany 70, 2157–2171. [DOI] [PubMed] [Google Scholar]
- Spurr AR. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. Journal of Ultrastructure Research 26, 31–43. [DOI] [PubMed] [Google Scholar]
- Ströher E, Dietz KJ. 2008. The dynamic thiol-disulphide redox proteome of the Arabidopsis thaliana chloroplast as revealed by differential electrophoretic mobility. Physiologia Plantarum 133, 566–583. [DOI] [PubMed] [Google Scholar]
- Sun R, Fan H, Gao F, Lin Y, Zhang L, Gong W, Liu L. 2012. Crystal structure of Arabidopsis Deg2 protein reveals an internal PDZ ligand locking the hexameric resting state. Journal of Biological Chemistry 287, 37564–37569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Ouyang M, Guo J, Ma J, Lu C, Adam Z, Zhang L. 2010. The thylakoid protease Deg1 is involved in photosystem-II assembly in Arabidopsis thaliana. The Plant Journal 62, 240–249. [DOI] [PubMed] [Google Scholar]
- Sun X, Peng L, Guo J, Chi W, Ma J, Lu C, Zhang L. 2007. Formation of DEG5 and DEG8 complexes and their involvement in the degradation of photodamaged photosystem II reaction center D1 protein in Arabidopsis. The Plant Cell 19, 1347–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Small I, Millar AH. 2014. Using the SUBcellular database for Arabidopsis proteins to localize the Deg protease family. Frontiers in Plant Science 5, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira PF, Glaser E. 2013. Processing peptidases in mitochondria and chloroplasts. Biochimica et Biophysica Acta 1833, 360–370. [DOI] [PubMed] [Google Scholar]
- Todd CD, Gifford DJ. 2002. The role of the megagametophyte in maintaining loblolly pine (Pinus taeda L.) seedling arginase gene expression in vitro. Planta 215, 110–118. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols 11, 2301–2319. [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740. [DOI] [PubMed] [Google Scholar]
- Umbach AL, Zarkovic J, Yu J, et al. 2012. Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS One 7, e44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, De Clercq I, Ivanova A, Law SR, Van Breusegem F, Millar AH, Whelan J. 2016. Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiology 171, 2150–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, del-Toro N, et al. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Research 44, D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R, Aigner H, Pružinská A, Jänkänpää HJ, Jansson S, Funk C. 2011. Fitness analyses of Arabidopsis thaliana mutants depleted of FtsH metalloproteases and characterization of three FtsH6 deletion mutants exposed to high light stress, senescence and chilling. New Phytologist 191, 449–458. [DOI] [PubMed] [Google Scholar]
- Wang X, Auwerx J. 2017. Systems phytohormone responses to mitochondrial proteotoxic stress. Molecular Cell 68, 540–551.e5. [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Analytical Biochemistry 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Willeke C. 2011. Einfluss von Hypoxie auf das Transkriptom und das mitochondriale Proteom von Arabidopsis thaliana. PhD thesis. Christian-Albrechts-University Kiel, Kiel, 209. [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. 1999. A mini binary vector series for plant transformation. Plant Molecular Biology 40, 711–717. [DOI] [PubMed] [Google Scholar]
- Yang H, Krebs M, Stierhof YD, Ludewig U. 2014. Characterization of the putative amino acid transporter genes AtCAT2, 3 & 4: the tonoplast localized AtCAT2 regulates soluble leaf amino acids. Journal of Plant Physiology 171, 594–601. [DOI] [PubMed] [Google Scholar]
- Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H. 2014a. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. The Plant Cell 26, 2939–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wu J, Yuan D, Zhang D, Huang Z, Xiao L, Yang C. 2014b. Perturbation of auxin homeostasis caused by mitochondrial FtSH4 gene-mediated peroxidase accumulation regulates Arabidopsis architecture. Molecular Plant 7, 856–873. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.