Abstract
The proteolipid protein gene products DM-20 and PLP are adhesive intrinsic membrane proteins that make up ≥50% of the protein in myelin and serve to stabilize compact myelin sheaths at the extracellular surfaces of apposed membrane lamellae. To identify which domains of DM-20 and PLP are positioned topologically in the extracellular space to participate in adhesion, we engineered N-glycosylation consensus sites into the hydrophilic segments and determined the extent of glycosylation. In addition, we assessed the presence of two translocation stop-transfer signals and, finally, mapped the extracellular and cytoplasmic dispositions of four antibody epitopes. We find that the topologies of DM-20 and PLP are identical, with both proteins possessing four transmembrane domains and N and C termini exposed to the cytoplasm. Consistent with this notion, DM-20 and PLP contain within their N- and C-terminal halves independent stop-transfer signals for insertion into the bilayer of the rough endoplasmic reticulum during de novo synthesis. Surprisingly, the conformation (as opposed to topology) of DM-20 and PLP may differ, which has been inferred from the divergent effects that many missense mutations have on the intracellular trafficking of these two isoforms. The 35 amino acid cytoplasmic peptide in PLP, which distinguishes this protein from DM-20, imparts a sensitivity to mutations in extracellular domains. This peptide may normally function during myelinogenesis to detect conformational changes originating across the bilayer from extracellular PLP interactions in trans and trigger intracellular events such as membrane compaction in the cytoplasmic compartment.
Keywords: protein topology, protein conformation, protein misfolding, proteolipid protein, PLP, DM-20, 1A9 monoclonal antibody, O10 monoclonal antibody, myelin, missense mutations, transfection, central nervous system, evolution, transmembrane signaling
In the CNS of tetrapods, compact myelin is maintained as a multilamellar sheath by the combined action of two sets of highly abundant membrane proteins, the myelin basic proteins and the proteolipid proteins (DM-20/PLP). The myelin basic proteins are extrinsic membrane proteins that function to bring into close apposition the cytoplasmic aspects of the myelin membrane bilayer (Omlin et al., 1982; Roach et al., 1985). On the other hand, DM-20 and its more abundant insertion isoform PLP are polytopic integral membrane proteins that act to stabilize appositions of the extracellular surfaces of the myelin membrane (Duncan, 1990; Boison and Stoffel, 1994; Boison et al., 1995) (Dr. K.-A. Nave, personal communication); DM-20 and PLP arise by alternative splicing of a single genomic transcript and differ by a hydrophilic peptide segment 35 amino acids in length, the presence of which generates the PLP product (Macklin et al., 1987; Nave et al., 1987; Simons et al., 1987). Indirect evidence suggests that DM-20 and PLP form homo- and heteropolymers in vitro (Sinoway et al., 1994; Jung et al., 1995; Gow and Lazzarini, 1996), but precisely how these molecules may interact and how they may influence the conformation of one another is at present unknown.
Subsequent to the elucidation of the primary structure of PLP, several topologies for this protein have been proposed based on mathematical modeling, chemical labeling, limited protease digestion, and immunolabeling. These data have yielded contradictory models (for review, see Popot et al., 1991). However, recent data provide evidence favoring an N-terminus-in/C-terminus-in model (N-in/C-in) of PLP with four intervening transmembrane segments. This topology has been deduced by mapping disulfide-bonded cysteine residues, thioester-linked fatty acids, and chemically glycosylated lysine residues and by immunocytochemical studies (Shaw et al., 1989; Weimbs and Stoffel, 1992, 1994; Greer et al., 1996). In other studies, Kitagawa et al. (1993), Yan et al. (1993), and Yoshida and Colman (1996) have identified a family of closely related brain proteolipids, the primary structure of each containing within it functional N-glycosylation sequences, at least one of which is located in a hydrophilic segment that is presumably exposed on the extracellular surface.
In this study, we have examined PLP and DM-20 topologies by using specific antibodies and by engineering N-glycosylation sites into hydrophilic segments in each of these molecules and determining the extent of glycosylation. We find that the transmembrane topologies of DM-20 and PLP are identical, with both proteins possessing four transmembrane domains and both N and C termini located within the cytoplasmic compartment. However, the precise conformations of DM-20 and PLP may differ, because they each respond differently to many identical missense mutations in their primary sequences. We also tested for the presence of translocation signals that enable insertion of these proteins into rough endoplasmic reticulum (RER) membranes. We find that DM-20 and PLP contain at least two such signals that are active during de novo synthesis. Finally, we find that the IgM monoclonal antibodies 1A9 and 010, widely used as markers for the oligodendrocyte lineage, bind to an extracellular hydrophilic segment common to both proteolipid proteins.
MATERIALS AND METHODS
cDNA constructs. A 1.4 kb EcoRI fragment made up of human PLP cDNA (Puckett et al., 1987) was used as the starting point for all of the constructs. The DM-20 cDNAs were constructed by replacing the 330 bp BglII/NcoI fragment from the PLP cDNA with a 225 bpBglII/NcoI fragment from murine DM-20 cDNA (Timsit et al., 1992b). Neither DM-20 nor PLP is a glycoprotein; glycosylation sites were introduced into the cDNAs encoding these proteins by PCR-based site-directed mutagenesis as described previously (Gow et al., 1994b). A single codon was inserted into cDNAs to generate each of the consensus N-glycosylation recognition sequences (Table1). Thereafter, the cDNAs were subcloned into pSP64 polyA (Promega, Madison, WI) for subsequent use in in vitrotranscription/translation reactions (see below).
Table 1.
The positions and flanking amino acids in the primary sequences of DM-20 and PLP where N-glycosylation consensus sites were engineered for in vitro translation experiments (see Fig.2)
| Designation for N-glycosylation consensus site (see Fig.1) | The number of the amino acid preceding the N-glycosylation consensus sequence1-a | Amino acids surrounding the N-glycosylation consensus sequence1-b | |
|---|---|---|---|
| PLP | DM-20 | ||
| α | 46 | 46 | …iNety… |
| β | 91 | 91 | …yNktt… |
| γ | 221 | 186 | …snlTl… |
| δ | 272 | 237 | …rNgtk… |
The residue numbers are for the mature DM-20/PLP proteins.
Single letter codes are used to specify amino acids. Small capital letters indicate the wild-type amino acid sequence, and capital letters show the amino acids inserted by site-directed mutagenesis to generate N-glycosylation sequences conforming to the NxT/S consensus.
A human PLP cDNA, previously engineered to create a BstEII site beginning at amino acid G146, was linearized withBstEII, filled in with Klenow (NEB, Beverly, MA), and ligated, thereby bringing into frame a translation-termination signal eight codons downstream. Thus, the amino acid sequence after G146 is HVTPTRG, and the resulting truncated protein contains amino acids 1–147 of PLP. A human cDNA, modified to encode the C-terminal 177 amino acids of PLP, was generated by inserting a polycloning site into an existing BstXI site across the translation start signal for the protein. The first restriction site in the polylinker isXbaI. The resulting cDNA was restricted with XbaI and BglII to remove the 5′ 300 bp of coding sequence, then filled in with Klenow. Ligation of the DNA preserved the reading frame with the amino acid sequence at the N terminus becoming MGSRIFGD. Thus, the resulting truncated protein comprises amino acids 99–276 of PLP. The synthesis of all other mutant cDNAs used is described elsewhere (Gow et al., 1994b; Gow and Lazzarini, 1996).
In vitro transcription and translation reactions. Plasmids encoding DM-20s and PLPs, which had been engineered to contain N-glycosylation consensus sequences within the open reading frame, were linearized with HindIII or SalI (NEB) and transcribed in vitro using the Promega Riboprobe kit following the protocols supplied by the manufacturers. SP6 RNA polymerase was used in all reactions to generate sense-strand mRNAs, which were uncapped at the 5′ ends but polyadenylated at the 3′ ends because of the presence of a poly(A+) stretch in the polylinker immediately upstream of the HindIII site. Plasmids linearized with SalI did not give rise to polyadenylated mRNAs, but these performed equally well to poly(A+)-tailed transcripts in subsequent reactions.
The mRNAs were purified free from plasmid DNA and translatedin vitro using Promega reticulocyte lysates in the presence or absence of dog pancreas microsomes to effect carbohydrate addition to any N-glycosylation sites in the nascent polypeptides, which were cotranslationally translocated. Protein molecules synthesized in these reactions were radiolabeled with [35S]methionine (NEN), which was included in accordance with the manufacturers recommendations. The cotranslational addition of carbohydrates to polypeptide chains in the presence of microsomes was verified through enzymatic cleavage. After in vitro translation, the reactions were divided into two and incubated in an equal volume of buffer containing 100 mm Tris, pH 8.6, 50 mmEDTA, 1% Triton X-100, and 1% 2-mercaptoethanol and PMSF for 1 hr at 37°C (Johnson et al., 1989) in the presence or absence of 0.2 units of endoglycosidase F (Boehringer Mannheim, Indianapolis, IN).
SDS-polyacrylamide gels. The discontinuous gel system devised by Laemmli (Laemmli, 1970) was used for electrophoresis of proteins on 12% polyacrylamide gels using a BioRad miniprotean electrophoresis unit (Richmond, CA). The in vitrotranslation reaction products were mixed with equal volumes of a 2× stock of sample buffer containing the standard concentrations of Tris salts but with 5% SDS 0.5% 2-mercaptoethanol added fresh. The samples were heated at 50°C for 10 min, then loaded onto gels alongside125I-labeled 14.4–220 kDa Mrmarkers (NEN). Gels were infiltrated with Amplify (Amersham, Arlington Heights, IL), vacuum-dried onto Whattman 3MM paper (Fisher, Agawam, MA), and exposed to x-ray film.
Immunofluorescence staining. COS-7 cells were cultured and transfected using CaPO4–DNA precipitates as described previously (Gow et al., 1994a). Twenty-four hours after commencing the transfections, culture dishes of cells were prepared for immunofluorescence staining. Cells were washed twice for 1 min in HEPES-containing modified Eagle’s medium (MEM, Life Technologies, Gaithersburg, MD), warmed to 37°C, and then fixed for 30 min in the same medium containing 2% paraformaldehyde. For live cell staining, dishes were gradually cooled to room temperature, then to 4°C over 10 min. The dishes were then placed in a water-ice slurry for 30 min and the cells incubated in 100 μl of culture medium containing 25 mm HEPES buffer, pH 7.2, and antibody (the mouse monoclonal IgM antibody 1A9 was diluted 200-fold; the hybridoma supernate for the mouse monoclonal IgM antibody O10, a kind gift from M. Schachner, ETH Zurich, was undiluted; and the rabbit polyclonal 81-11 antiserum, a kind gift from J. Benjamins, Wayne State University, was diluted 100-fold). The cells were washed thrice for 1 min each in ice-cold MEM, then fixed in the same buffer containing 2% paraformaldehyde (Sigma, St. Louis, MO). The fixative was initially at 4°C but warmed to room temperature during the fixation. Thereafter, the cells were immunostained as described previously (Gow et al., 1994b) and viewed using a Leica TCS 4D laser scanning microscope. Confocal images of transfected cells were obtained as a series of optical sections covering a distance of 2–5 μm on the z-axis, and these sections were superimposed to generate extended-focus images. Additional primary antibodies used for immunocytochemistry were rat anti-DM-20/PLP (AB3) (Yamamura et al., 1991); rabbit anti-PLP (PLP130) (Sinoway et al., 1994); mouse anti-vimentin monoclonal IgM (Sigma). Goat anti-IgM μ-chain-specific antibodies were obtained from Jackson ImmunoResearch Labs (West Grove, PA), and all other immunochemical reagents were obtained through Amersham or Molecular Probes (Eugene, OR). The confocal images presented below are representative examples of at least 80% of the transfectants in our experiments.
RESULTS
Two hydrophilic segments common to DM-20 and PLP are exposed on the luminal surface of rough microsomes during de novo protein synthesis
We engineered N-glycosylation sites in the first, second, and third internal hydrophilic loops and adjacent to the C terminus in both DM-20 and PLP by insertion into the primary structure of a single amino acid (see Table 1 and Fig. 1). We then used these mutated cDNAs containing the consensus N-glycosylation sites to programin vitro transcription/translation systems in the presence or absence of dog pancreas microsome membranes, which are competent to perform N-glycosylation. Glycosylation sites α and γ in thea–b and c–d loops were efficiently glycosylated, whereas sites β and δ in theb–c loop and at the C terminus were not glycosylated in either protein. Glycosylation was demonstrated by a retardation in electrophoretic mobility equivalent to ∼3 kDa, consistent with the N-glycosylation of a single site in each molecule (Fig. 2a). In some in vitro translations in the presence of membranes, the glycosylation reaction, although clearly apparent, was not complete; in these reaction mixtures, nonglycosylated product could be detected in the same reaction mixture (Fig.2a). When the glycosylated products were treated with endoglycosidase F, the labeled DM-20 or PLP reverted to the same electrophoretic mobility as the nonglycosylated product (Fig.2b). These data are consistent with the prediction that the two hydrophilic a–b andc–d loops (see Fig. 1) are translocated during synthesis on RER membranes into the lumen where N-glycosylation can occur. On transport and assembly into the oligodendrocyte plasma membrane, therefore, these segments in both DM-20 and PLP are exposed on the topologically equivalent extracellular membrane surface. In contrast, the b–c loop, which contains the PLP-specific segment, is likely to be cytoplasmically disposed in both molecules, because the engineered N-glycosylation site in this loop was incapable of becoming glycosylated. In fact, immunolabeling studies (see below) confirm this prediction.
Fig. 1.
Hydrophobicity plot calculated for PLP.a, The GES algorithm (Engelman et al., 1986) with a window size of 17 amino acids identifies four potential transmembrane domains (labeled a–d). Although amino acids 116–150 of PLP are absent in DM-20 (bidirectional arrow), the shape of the hydrophobicity plots for the two proteins is almost identical. In the current study, four consensus N-glycosylation sites (see Table 1) were inserted into cDNAs encoding PLP and DM-20 at the positions denoted α, β, γ, and δ. The positions of two peptides used to raise the antibodies, PLP130 (Sinoway et al., 1994) and AB3 (Yamamura et al., 1991), are indicated. b, Bidirectional arrowsindicate regions of full-length PLP, which is encoded by the truncated PLP cDNAs used for transfection in Figure 3.
Fig. 2.
SDS-PAGE of 35S-labeled DM-20 and PLP. The proteins were synthesized by in vitro translation of mRNAs containing engineered N-glycosylation consensus sites (see Fig.1a) in reticulocyte lysates in the presence of dog pancreas microsomes. a, Decreased mobilities of DM-20 and PLP in some lanes indicate that these proteins have been co- or post-translationally modified. b, Digestion of the modified proteins with endoglycosidase F indicates that the mobility shifts are attributable to N-glycosylation.
There are at least two signals for membrane insertion in PLP
The foregoing glycosylation experiments tested for exposure of each hydrophilic loop on the luminal or extracellular membrane surface. It was concluded from these experiments that the hydrophilica–b and c–d loop domains (see Fig. 1) are both predicted to be at the extracellular apposition of the myelin membrane, i.e., in the wild-type DM-20/PLP molecules; these hydrophilic loops should be exposed in the lumen of the RER during biosynthesis on membrane-bound ribosomes. Taken together with the fact that the b–c loop in PLP, which has a net charge of +6 and, therefore, is almost certainly cytoplasmically disposed (and also cannot be glycosylated), we may conclude that either hydrophobic segments c or d, or both, are membrane-embedded. This necessitates that the signal sequences for insertion into membranes are present in either segments c ord, or both, and, therefore, it was worth testing whether such insertion signals are located in these segments.
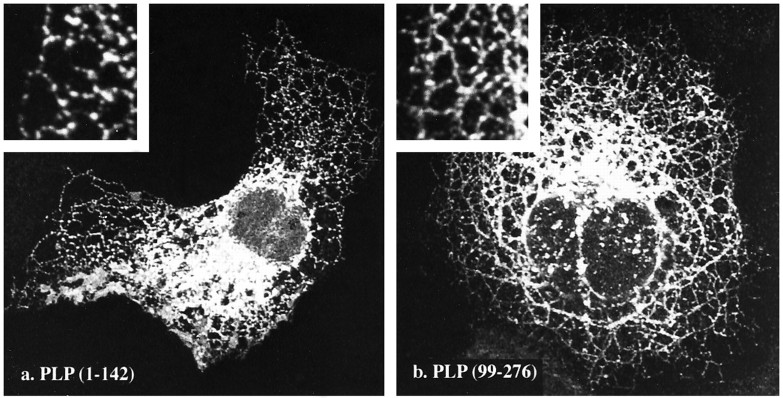
Accordingly, we engineered two truncated PLP cDNAs, PLP 1–147 and PLP 99–276, which encode the first two or second two hydrophobic segments, respectively, and use the natural methionine for translation initiation. These cDNAs were then transfected individually into cells and the intracellular distributions of the truncated molecules mapped using appropriate antibodies. In both cases, a reticular pattern of labeling (Fig. 3), highly concentrated in the perinuclear region but extending out into the cell periphery, was observed. Curiously, the reticular pattern strongly resembles that reported previously for some cells expressing wild-type DM-20 and PLP and is remarkable in that the intracellular membranes in which the truncated PLPs are found are not vesiculated (A. Gow, unpublished data) (Timsit et al., 1992b) as is normally the case for several intracellular organelles after formaldehyde fixation (Sabatini et al., 1963). This suggests that when present at high levels, these very hydrophobic segments may confer a certain rigidity to the membranes of the RER, Golgi, and perhaps lysosomal membranes. Furthermore, the absence of the truncated forms in the plasma membrane and their retention in intracellular compartments are consistent with previous observations that alterations in the PLP primary sequence causes retention in these compartments, most notably the RER (Gow et al., 1994b; Gow and Lazzarini, 1996; Tosic et al., 1996).
Fig. 3.
Subcellular distributions of two truncated PLPs (see Fig. 1b) in transfected COS-7 cells. Immunofluorescence staining, using an antibody raised against amino acids 107–130 of PLP (PLP130) (Sinoway et al., 1994), reveals the truncated PLPs that are present in the endoplasmic reticulum.a, The N-terminal two potential transmembrane domains of PLP (amino acids 1–147). b, The C-terminal two potential transmembrane domains of PLP (amino acids 99–276). Theinsets show, at higher power, reticular staining that is consistent with the localization of PLP in the endoplasmic reticulum.
The PLP-specific peptide is exposed on the cytoplasmic side of the membrane bilayer
To probe directly the membrane orientation of the PLP-specific peptide segment, which, as we demonstrated above, cannot be glycosylated, PLP expressors were fixed and treated with an antibody raised against this region of the rat PLP sequence (Sinoway et al., 1994) encompassing amino acids 107–130 that form part of the PLP-specific segment. Nonpermeabilized cells do not react with the peptide antiserum (absence of red fluorescence in Fig.4a); in contrast, when cells are permeabilized, PLP immunofluorescence is clearly observed (red fluorescence in Fig. 4b). Clearly, the PLP-specific peptide, and therefore the b–c loop of this protein, is located cytoplasmically where it is in a favorable position to participate in the formation of the major dense line, perhaps by contributing positive charges to the membrane interface (Yoshida and Colman, 1996). As a control for the fixation and permeabilization procedures, an IgM monoclonal vimentin antibody was used on either live or fixed PLP expressors in conjunction with the AB3 antibody. As expected, vimentin antibody treatment of live cells revealed no labeling (Fig. 4c); these same cells when fixed, permeabilized, and treated with AB3 revealed surface and intracellular pools of PLP (green). In contrast, fixed and permeabilized cells labeled with AB3 (green) and vimentin (red) revealed intracellular but nonoverlapping distributions for both proteins (Fig. 4d). The asterisks in Figure 4d denote COS-7 cell processes in which vimentin intermediate filaments are clearly visible extending away from the perinuclear region toward the tips of these processes.
Fig. 4.

The epitopes in PLP for the PLP130 polyclonal antibody and the AB3 monoclonal antibody (see Fig. 1a) are cytoplasmic. In each panel, the antibody used for live staining of transfected cells is shown in brackets. The antibody used after fixation/permeabilization is underlined. Ina and b, neither of the antibodies incubated with the live cells binds to their epitopes, whereas both of these antibodies bind their epitopes after fixation/permeabilization. In c and d, the vimentin antibody is used as a negative control to indicate that the plasmalemma of the transfected cells was not compromised during live cell staining.
Antibodies against common sites in DM-20 and PLP yield identical staining patterns in transfected cells
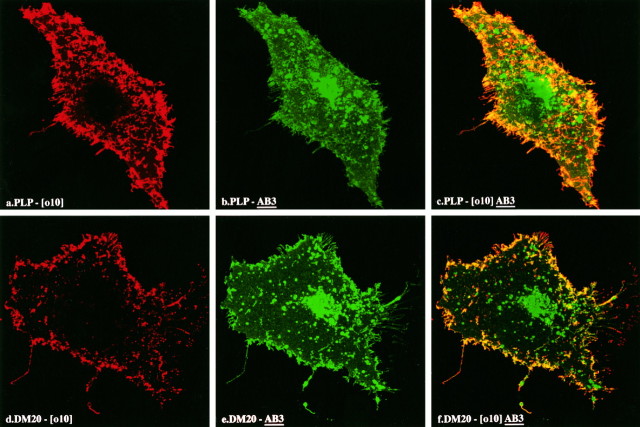
In the course of screening COS-7 cells transfected with cDNAs encoding either DM-20 or PLP, we used three monoclonal antibodies [1A9 (Gard and Dutton, 1987), O10 (Schachner, 1982), and AB3 (Yamamura et al., 1991)] that have been shown to strongly label oligodendrocytes, which naturally express DM-20 and PLP simultaneously. When the two proteins were expressed individually in transfectants, the following observations were made. The 1A9 antibody (red) labels the surface of either live (Fig. 5a,d) or paraformaldehyde-fixed (data not shown) DM-20 and PLP expressors. On the other hand, the AB3 antibody (green), which is directed against the C-terminal sequence common to both DM-20 and PLP, does not label live cells expressing DM-20 or PLP (absence of green fluorescence in Fig.4b) but heavily labels surface and internal pools (asterisk in Fig. 5b) of both proteins in fixed and permeabilized expressors (see Figs. 4a,5b,e). In these studies, the 010 antibody yields immunofluorescence staining that is identical to that obtained with the 1A9 reagent (Fig. 6).
Fig. 5.
Double-immunofluorescence staining shows that the epitope for the 1A9 monoclonal antibody is extracellular. Transfected COS-7 cells are expressing PLP (a–c) or DM-20 (d–f). Live cell staining with 1A9 (red) is shown in a andd, whereas AB3 staining (green) after fixation/permeabilization is shown in b ande. Overlays of the 1A9 with AB3 staining are shown inc and f. The arrow ina indicates a microvillus on the surface of the cell, which is brightly stained for PLP.
Fig. 6.
Double-immunofluorescence staining shows that the epitope for the O10 monoclonal antibody is extracellular. Transfected COS-7 cells are expressing PLP (a–c) or DM-20 (d–f). Live cell staining with O10 (red) is shown in a andd, whereas AB3 staining (green) after fixation/permeabilization is shown in b ande. Overlays of the O10 with AB3 staining are shown inc and f.
Missense mutations have disparate effects on the trafficking of DM-20 and PLP
The biochemical and immunolabeling studies described above, as well as evidence from other sources (Kitagawa et al., 1993; Yan et al., 1993; Yoshida and Colman, 1996), argue strongly for identical topologies with respect to the phospholipid bilayer for DM-20 and PLP. In this light, it is of particular interest that certain missense mutations exact differential effects on the intracellular trafficking of these isoforms in oligodendrocytes and transfected fibroblasts (Gow and Lazzarini, 1996). An example of this is shown in Figure7, in which the missense mutation L223I was expressed in transfected cells and labeled live (Fig. 7a,c) with the polyclonal antibody 81-11, which recognizes amino acids 209–215 in PLP. The cells were then fixed/permeabilized and labeled with the AB3 monoclonal antibody (Fig. 7b,d). As shown previously, a wild-type pattern of immunofluorescence staining was observed for the mutant DM-20 (compare Figs. 7b and5b) but not with the cognate mutant PLP (Fig.7d). Interestingly, the disparate effects of the mutation at leucine 223 on the trafficking of DM-20/PLP are dependent on the identity of the substituted amino acid, because the expression of cDNAs encoding the L223P mutation results in the retention of both isoforms in the RER (data not shown). Perhaps the capacity of a particular mutation to disrupt the trafficking of both DM-20 and PLP depends on the relatedness of the side chains between the wild-type and substituted amino acid. In this regard, L223I is a conservative substitution, whereas L223P is a nonconservative change. However, not all nonconservative substitutions behave the same. In this regard, D202H is a nonconservative mutation also in the c–d loop domain that nevertheless does not impede DM-20 trafficking (see Table2) while arresting the trafficking of PLP (Gow et al., 1994b). Remarkably, therefore, although DM-20 and PLP have a common transmembrane topology in terms of the disposition of sequential hydrophobic and hydrophilic segments, more subtle conformational differences between these proteins almost certainly exist.
Fig. 7.
The L223I missense mutation, which is common to DM-20 and PLP, differentially affects the intracellular trafficking of these mutant proteins in transfected cells. Immunofluorescence staining of transfected COS-7 cells using the AB3 monoclonal antibody shows the wild-type distribution of DM-20L223I (b). Cell surface staining is clearly visible from 81-11 antibody staining (a), indicating that the mutation does not affect the intracellular trafficking of DM-20 through the secretory pathway. However, the identical mutation in PLP disrupts its trafficking; PLPL223I is not detectable on the cell surface (c) but rather accumulates in the RER of the cell (d).
Table 2.
Summary of immunofluorescence staining with the 1A9 and O10 monoclonal antibodies on transfected COS-7 cells expressing missense mutant DM-20s
| Missense mutation | Equivalent amino acid in DM-20 | Antibody used for live cell staining | Mutant DM-20 present on the cell surface2-e | |
|---|---|---|---|---|
| 1A9 | O10 | |||
| Wild type (human)2-a | ++ | ++ | ||
| H36Q (pt rabbit) | 36 | ++ | ++ | |
| T115K (PMD) | 115 | ++ | ++ | |
| T155I (PMD) | 120 | ++ | ++ | |
| I186T (rsh mouse) | 151 | + | + | |
| T198S (rabbit)2-b | 163 | ++ | ++ | |
| D202H (PMD) | 167 | − | − | yes |
| D202E (PMD)2-c | 167 | − | − | yes |
| P215S (PMD) | 180 | − | − | yes |
| V218F (PMD) | 183 | − | − | yes |
| L223I2-d | 188 | +/− | − | yes |
| S252P2-d | 217 | ++ | ++ | |
The amino acid sequence of human DM-20 is identical to the mouse protein.
Wild-type rabbit DM-20 differs from the human protein by the conservative substitution T198S, which would not be expected to affect protein function (Tosic et al., 1994) .
O. Boespflug-Tanguy, personal communication.
Denotes missense mutations that are not known to be naturally occurring.
Revealed by live cell 81-11 polyclonal antibody immunofluorescence staining (see Fig. 7a; data not shown).
Point mutations in the c–d loop of DM-20/PLP eradicate binding of the 1A9 and 010 reagents
Having established that both 1A9 and 010 IgM monoclonals react with epitopes in extracellularly disposed loops of DM-20 and PLP, we next attempted to localize the epitopes more precisely in these myelin proteolipids. To explore this, we took advantage of the fact that the binding of both of these monoclonal antibodies is sensitive to the conformation of their epitopes (Schachner, 1982; Gard and Dutton, 1987). Thus, we used a number of missense mutations and species-specific polymorphisms in DM-20 that, although changing the primary sequence, still allow the mutant DM-20s to reach the plasma membrane of host cells (see Fig. 7b). For these studies, 1A9 and 010 binding was detected by immunofluorescence staining and scored as either + or − in the summary in Table 2. Although the 1A9 and O10 immunoreactivities of the mutant proteins were unaffected by mutations located in the first two hydrophilic loops, labeling was abolished by mutations residing in the distal portion of thec–d loop (Fig. 8). Amino acid changes affecting the proximal region of the c–dloop (I186T and S198T) yielded protein products with, at most, slightly diminished reactivity to the 1A9 and 010 reagents when compared with controls expressing the wild-type proteins. These results suggest that the region of DM-20 (and by analogy, PLP as well) that is recognized by 1A9 and 010 lies between amino acids 200 and 223 of the PLP sequence (equivalent to amino acids 165–188 of the DM-20 sequence). It is of interest in this regard that the conformation of this segment of the c–d loop is almost certainly influenced by the presence of a disulfide linkage between amino acids 200 and 219, which may place this segment in an exposed position on the surface of the transfectants. In the myelin membrane, exposure of this segment at the extracellular interface may place it in a favorable position to interact with partner proteolipid molecules emanating from the opposite bilayer. In support of this notion, Stephens et al. (1996) have recently found that thec–d loop of PLP and, in particular, of amino acids 209–217, is a common B-cell determinant for many antimyelin and anti-PLP sera, which recognize the epitope on live and fixed mouse oligodendrocytes in culture and paraformaldehyde-fixed brain sections.
Fig. 8.
Topological representation of DM-20/PLP derived from the data in Figures 2, 3, 4. The biochemical and immunofluorescence staining analyses are consistent with a four-transmembrane, N-terminus-in/C-terminus-in topology for both DM-20 and PLP. The region of the protein in which missense mutations abrogate both 1A9 and O10 monoclonal antibody binding is indicated by the stippled stripe in the c–d loop domain.
DISCUSSION
DM-20 and PLP are members of an ancient family of proteolipid-type proteins that share common hydrophobicity profiles and contain certain identical elements in their primary sequences. In addition to their expression in white matter, these molecules are expressed in neurons, heart, kidney tubules, and choroid plexus (Baumrind et al., 1992;Campagnoni et al., 1992; Lagenaur et al., 1992), and all the gene products are predicted to be polytopic, containing noncleavable hydrophobic signals for insertion into the RER.
Although it is not known with any degree of precision how DM-20 and PLP function together in the establishment and maintenance of the myelin sheath, it is of interest that in all tetrapods studied to date (with the exception of Xenopus), the two proteins are found together in myelin and sometimes with P0 (see Yoshida and Colman, 1996). Their similar expression patterns across species, very similar biochemistries and hydrophobicities, apparent ability to interact with each other in vitro (Sinoway et al., 1994; Gow and Lazzarini, 1996), and possible evolutionary relationship with ligand-gated channels (Kitagawa et al., 1993) have lead to speculation that DM-20/PLP may form heteromeric complexes in myelin, perhaps with a defined stoichiometry. Furthermore, preliminary evidence indicates that when coexpressed on the surface of host cells, these two proteins induce rapid aggregation of expressing cells (M. P. Sinoway and D. R. Colman, unpublished data), providing evidence for the notion that the natural function of the DM-20/PLP complex in myelin is membrane adhesion (for review, see Braun, 1984; Duncan, 1990; Boison and Stoffel, 1994; Boison et al., 1995).
From an evolutionary standpoint (see Yoshida and Colman, 1996), the DM-20 protein seems to have been expressed in the first myelinated vertebrates, which presumably were shark-like organisms that, along with DM-20, expressed P0 as the primary compactor of myelin membranes. Significantly later in the course of evolution, coincident with the emergence of amphibia, the highly charged PLP-specific segment was inserted into the DM-20 gene to yield the current arrangement of exons and introns that constitutes the PLP gene. Our results demonstrate that incorporation of the charged segment into DM-20 to yield the new PLP molecule apparently did not alter the topology of these proteolipids but instead preserved what must have been a very ancient form of association of a polypeptide with a phospholipid bilayer. In particular, the absence of cleavable, N-terminal signal sequences in many polytopic proteins is consistent with the idea that polypeptides with permanent insertion signals arose in very ancient organisms, even before the development of the secretory apparatus (Sabatini et al., 1982). Several other myelin proteins with similar topologies have recently been described including PMP22 (Suter et al., 1992), MAL/MVP17 (Kim et al., 1995; Schaeren-Wiemers et al., 1995), OSP (J. M. Bronstein, unpublished data), and the connexins (Milks et al., 1988).
It is of great interest that although DM-20 and PLP share identical topologies, the presence of the PLP segment on the cytoplasmic aspect of the bilayer in some way renders PLP susceptible to conformational changes that prevent it from obtaining a stable conformation in the RER and reaching the cell surface (see Fig. 7). Many missense mutations, when introduced into the DM-20 sequence, allow this protein to reach the cell surface apparently unimpeded (see Table 2). On the other hand, these identical mutations, when introduced into PLP, cause a dramatic accumulation of the altered polypeptide in intracellular compartments from which it cannot exit. Presumably, these particular missense mutations change protein conformation sufficiently to yield misfolded PLP products that are recognized and retained in the RER by resident proteins (Gething and Sambrook, 1992; Hammond and Helenius, 1995), although other explanations cannot be presently discounted (see below). In any case, whatever the improvement or modification of function in the myelin membrane that is conferred by the PLP-specific peptide, which enabled the domination of plp gene products over P0 in the CNS of terrestrial vertebrates, this segment also introduced a sensitivity to even subtle changes in primary sequence in the protein that profoundly affects its intracellular trafficking afterde novo synthesis in several cell types.
Curiously, changes in protein conformation imparted by the PLP-specific peptide not only confer sensitivity to amino acid substitutions in or near cytoplasmic domains of PLP, such as that observed for theT155I mutation (Gow and Lazzarini, 1996), but also to mutations in extracellular domains (see Fig. 8). This implies that a transfer of information across the lipid bilayer occurs and suggests that our previous mutational analyses (Gow et al., 1994b; Gow and Lazzarini, 1996) may have revealed a property of the PLP-specific peptide that was selected during its evolution from the primitive DMα-like gene (Yoshida and Colman, 1996). Furthermore, the fact that the expression of the PLP-specific peptide is developmentally regulated by alternative splicing (Ikenaka et al., 1992; Timsit et al., 1992a) suggests an important role for this cytoplasmic domain in the normal function of DM-20/PLP. At present, we can only speculate about the nature of such a function and in this regard have drawn heavily on the results of others who have reported evidence of transbilayer signaling. For example, the deletion of cytoplasmic peptide segments from P0, E-cadherin, αIIbβ3integrin, or PECAM-1 dramatically influences the adhesion mediated by the extracellular domains of these proteins (Nagafuchi and Takeichi, 1988; Williams et al., 1994; Wong and Filbin, 1994; Yan et al., 1995).
A provocative feature arising from previous cotransfection experiments in fibroblasts (Sinoway et al., 1994; Gow and Lazzarini, 1996) is that DM-20 appears to facilitate the folding and trafficking of PLP through the secretory pathway to the cell surface, thereby suggesting that these proteins form heteropolymers. In this regard, we note that during the course of evolution, events that resulted in the appearance of the PLP-specific segment and subsequent expunction of P0 from CNS myelin have nevertheless preserved the splice site that enables the independent expression of DM-20 and that this splice site is present in all terrestrial vertebrates that have been studied except for frogs, in which, for whatever reason, this site did not survive the rigors of natural selection (Schliess and Stoffel, 1991). An important implication of this notion is revealed in the pathogenesis of Pelizaeus– Merzbacher disease in humans (Gow and Lazzarini, 1996); mutations within the coding region of plp, which arrest the intracellular trafficking of both DM-20 and PLP in the RER of oligodendrocytes, give rise to a disease phenotype that is more severe (connatal Pelizaeus–Merzbacher disease) than mutations that result either in null alleles at plp or arrest the trafficking of PLP but not of DM-20 (classical Pelizaeus– Merzbacher disease).
Footnotes
This work was supported by research grants awarded to R.A.L. from the National Multiple Sclerosis Society (RG2734-A-3) and National Institutes of Health (3P01NS33165-O1A1S1), and to D.R.C. from the National Multiple Sclerosis Society (RG2636-A-3) and the National Institute of Neurological and Communicative Disorders and Stroke (NS20147). This is manuscript number 225 from the Brookdale Center for Molecular Biology, Mount Sinai School of Medicine. We thank Christopher P. De Marco, Amanda Battisti, and Cherie Southwood for their excellent technical support and Charles ffrench-Constant for helpful discussions.
Correspondence should be addressed to Dr. Robert A. Lazzarini, Brookdale Center for Molecular Biology, P.O. Box 1126, One Gustave L. Levy Place, Mount Sinai School of Medicine, New York, NY 10029-6574.
REFERENCES
- 1.Baumrind NL, Parkinson D, Wayne DB, Heuser JE, Pearlman AL. EMA: a developmentally regulated cell-surface glycoprotein of CNS neurons that is concentrated at the leading edge of growth cones. Dev Dyn. 1992;194:311–325. doi: 10.1002/aja.1001940407. [DOI] [PubMed] [Google Scholar]
- 2.Boison D, Stoffel W. Disruption of the compacted myelin sheath of axons of the central nervous system in proteolipid protein-deficient mice. Proc Natl Acad Sci USA. 1994;91:11709–11713. doi: 10.1073/pnas.91.24.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boison D, Bussow H, D’Urso D, Muller H-W, Stoffel W. Adhesive properties of proteolipid protein are responsible for the compaction of CNS myelin sheaths. J Neurosci. 1995;15:5502–5513. doi: 10.1523/JNEUROSCI.15-08-05502.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun PE. Molecular organization of myelin. In: Morrell P, editor. Myelin. Plenum; New York: 1984. pp. 97–116. [Google Scholar]
- 5.Campagnoni CW, Garbay B, Micevych P, Pribyl T, Kampf K, Handley VW, Campagnoni AT. DM20 mRNA splice product of the myelin proteolipid protein gene is expressed in the murine heart. J Neurosci Res. 1992;33:148–155. doi: 10.1002/jnr.490330119. [DOI] [PubMed] [Google Scholar]
- 6. Duncan ID. Dissection of the phenotype and genotype of the X-linked myelin mutants. Myelination and dysmyelination, Vol 605 Duncan I, Skoff R, Colman D. 1990. 110 121 New York Academy of Science; New York: s. [DOI] [PubMed] [Google Scholar]
- 7.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Chem. 1986;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 8.Gard AL, Dutton GR. Myelin-specific domain on the plasmalemma of oligodendroglia: differential expression in the rat and hypomyelinating mouse mutants jimpy and quaking. J Neurosci Res. 1987;17:329–343. doi: 10.1002/jnr.490170403. [DOI] [PubMed] [Google Scholar]
- 9.Gething M-J, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 10.Gow A, Lazzarini RA. A cellular mechanism governing the severity of Pelizaeus-Merzbacher disease. Nat Genet. 1996;13:422–428. doi: 10.1038/ng0896-422. [DOI] [PubMed] [Google Scholar]
- 11.Gow A, Friedrich VLJ, Lazzarini RA. Intracellular transport and sorting of the oligodendrocyte transmembrane proteolipid protein. J Neurosci Res. 1994a;37:563–573. doi: 10.1002/jnr.490370503. [DOI] [PubMed] [Google Scholar]
- 12.Gow A, Friedrich VLJ, Lazzarini RA. Many naturally occurring mutations of myelin proteolipid protein impair its intracellular transport. J Neurosci Res. 1994b;37:574–583. doi: 10.1002/jnr.490370504. [DOI] [PubMed] [Google Scholar]
- 13.Greer JM, Dyer CA, Pakaski M, Symonowicz C, Lees MB. Orientation of myelin proteolipid protein in the oligodendrocyte cell membrane. Neurochem Res. 1996;21:431–440. doi: 10.1007/BF02527707. [DOI] [PubMed] [Google Scholar]
- 14.Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 15.Ikenaka K, Kagawa T, Mikoshiba K. Selective expression of DM20, an alternatively spliced myelin proteolipid protein gene product in developing nervous system and nonglial cells. J Neurochem. 1992;58:2248–2253. doi: 10.1111/j.1471-4159.1992.tb10970.x. [DOI] [PubMed] [Google Scholar]
- 16.Johnson PW, Attia J, Richardson CD, Roder JC, Dunn RJ. Synthesis of soluble myelin-associated glycoproteins in insect and mammalian cells. Gene. 1989;77:287–296. doi: 10.1016/0378-1119(89)90076-0. [DOI] [PubMed] [Google Scholar]
- 17.Jung M, Schneider A, Nave K-A. Dominant negative action of mutations in the PLP/DM-20 gene and direct interaction of PLP polypeptides in vivo. J Neurochem. 1995;64:S101. [Google Scholar]
- 18.Kim T, Fiedler K, Madison DL, Krueger WH, Pfeiffer SE. Cloning and characterization of MVP17: a developmentally regulated myelin protein in oligodendrocytes. J Neurosci Res. 1995;42:413–422. doi: 10.1002/jnr.490420316. [DOI] [PubMed] [Google Scholar]
- 19.Kitagawa K, Sinoway MP, Yang C, Gould RM, Colman DR. A proteolipid protein gene family: expression in sharks and rays and possible evolution from an ancestral gene encoding a pore-forming polypeptide. Neuron. 1993;11:433–448. doi: 10.1016/0896-6273(93)90148-k. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lagenaur C, Kunemund V, Fischer G, Fushiki S, Schachner M. Monoclonal M6 antibody interferes with neurite extension of cultured neurons. J Neurobiol. 1992;23:71–88. doi: 10.1002/neu.480230108. [DOI] [PubMed] [Google Scholar]
- 22.Macklin WB, Campagnoni CW, Deininger PL, Gardinier MV. Structure and expression of the mouse myelin proteolipid protein gene. J Neurosci Res. 1987;18:383–394. doi: 10.1002/jnr.490180302. [DOI] [PubMed] [Google Scholar]
- 23.Milks LC, Kumar NM, Houghten R, Unwin N, Gilula NB. Topology of the 32 kDa liver gap junction protein determined by site-directed antibody localizations. EMBO J. 1988;7:2967–2976. doi: 10.1002/j.1460-2075.1988.tb03159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagafuchi A, Takeichi M. Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J. 1988;7:3679–3684. doi: 10.1002/j.1460-2075.1988.tb03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nave K-A, Lai C, Bloom FE, Milner RJ. Splice site selection in the proteolipid protein (PLP) gene transcript and primary structure of the DM-20 protein of central nervous system myelin. Proc Natl Acad Sci USA. 1987;84:5665–5669. doi: 10.1073/pnas.84.16.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Omlin F, Webster Hd F, Pulkovits CG, Cohen SR. Immunocytochemical localization of BP in major dense line regions of central and peripheral myelin. J Cell Biol. 1982;95:242–248. doi: 10.1083/jcb.95.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popot J-L, Pham-Dinh D, Dautigny A. Major myelin proteolipid: the 4-alpha-helix topology. J Membr Biol. 1991;120:233–246. doi: 10.1007/BF01868534. [DOI] [PubMed] [Google Scholar]
- 28.Puckett C, Hudson L, Ono K, Friedrich V, Benecke J, Dubois-Dalcq M, Lazzarini RA. Myelin-specific proteolipid protein is expressed in myelinating Schwann cells but is not incorporated into myelin sheaths. J Neurosci Res. 1987;18:511–518. doi: 10.1002/jnr.490180402. [DOI] [PubMed] [Google Scholar]
- 29.Roach A, Takahashi N, Pravtcheva D, Ruddle F, Hood L. Chromosomal mapping of mouse myelin basic protein gene and structure and transcription of the partially deleted gene in shiverer mutant mice. Cell. 1985;42:149–155. doi: 10.1016/s0092-8674(85)80110-0. [DOI] [PubMed] [Google Scholar]
- 30.Sabatini DD, Bensch K, Barrnett RJ. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J Cell Biol. 1963;17:19–58. doi: 10.1083/jcb.17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabatini DD, Kreibich G, Morimoto T, Adesnik M. Mechanisms for the incorporation of proteins in membranes and organelles. J Cell Biol. 1982;92:1–22. doi: 10.1083/jcb.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schachner M. Cell type-specific surface antigens in the mammalian nervous system. J Neurochem. 1982;39:1–8. doi: 10.1111/j.1471-4159.1982.tb04694.x. [DOI] [PubMed] [Google Scholar]
- 33.Schaeren-Wiemers N, Valenzuela DM, Frank M, Schwab ME. Characterization of a rat gene, rMAL, encoding a protein with four hydrophobic domains in central and peripheral myelin. J Neurosci. 1995;15:5753–5764. doi: 10.1523/JNEUROSCI.15-08-05753.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schliess F, Stoffel W. Evolution of the myelin integral membrane proteins of the central nervous system. Biol Chem Hoppe Seyler. 1991;372:865–874. doi: 10.1515/bchm3.1991.372.2.865. [DOI] [PubMed] [Google Scholar]
- 35.Shaw SY, Laursen RA, Lees MB. Identification of thiol groups and a disulfide crosslink site in bovine myelin proteolipid protein. FEBS Lett. 1989;250:306–310. doi: 10.1016/0014-5793(89)80744-6. [DOI] [PubMed] [Google Scholar]
- 36.Simons R, Alon N, Riordan JR. Human myelin DM-20 proteolipid protein deletion defined by cDNA sequence. Biochem Biophys Res Commun. 1987;146:666–671. doi: 10.1016/0006-291x(87)90580-8. [DOI] [PubMed] [Google Scholar]
- 37.Sinoway MP, Kitagawa K, Timsit S, Hashim GA, Colman DR. Proteolipid protein interactions in transfectants: implications for myelin assembly. J Neurosci Res. 1994;37:551–562. doi: 10.1002/jnr.490370502. [DOI] [PubMed] [Google Scholar]
- 38.Stephens TS, Pakaski M, Lees MB, Potter NT. Identification and characterization of a B-cell determinant within the amphipathic domain (residues 178–238) of the myelin proteolipid protein. J Neurosci Res. 1996;43:545–553. doi: 10.1002/(SICI)1097-4547(19960301)43:5<545::AID-JNR4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.Suter U, Welcher AA, Ozcelik T, Snipes GJ, Kosaras B, Franke U, Billings-Gagliardi S, Sidman RL, Shooter EM. Trembler mouse carries a point mutation in a myelin gene. Nature. 1992;356:241–244. doi: 10.1038/356241a0. [DOI] [PubMed] [Google Scholar]
- 40.Timsit SG, Bally-Cuif L, Colman DR, Zalc B. DM20 mRNA is expressed during the embryonic development of the nervous system of the mouse. J Neurochem. 1992a;58:1172–1175. doi: 10.1111/j.1471-4159.1992.tb09378.x. [DOI] [PubMed] [Google Scholar]
- 41.Timsit S, Sinoway MP, Levy L, Allinquant B, Stempak J, Staugaitis SM, Colman DR. The DM-20 protein of myelin: intracellular and surface expression patterns in transfectants. J Neurochem. 1992b;58:1936–1942. doi: 10.1111/j.1471-4159.1992.tb10072.x. [DOI] [PubMed] [Google Scholar]
- 42.Tosic M, Dolivo M, Amiguet P, Domanska-Janik K, Matthieu J-M. Paralytic tremor (pt): a new allele of the proteolipid protein gene in rabbits. J Neurochem. 1994;63:2210–2216. doi: 10.1046/j.1471-4159.1994.63062210.x. [DOI] [PubMed] [Google Scholar]
- 43.Tosic M, Gow A, Dolivo M, Domanska-Janik K, Lazzarini RA, Matthieu J-M. Proteolipid/DM20 proteins bearing the paralytic tremor mutation in peripheral nerves and transfected Cos-7 cells. Neurochem Res. 1996;21:423–430. doi: 10.1007/BF02527706. [DOI] [PubMed] [Google Scholar]
- 44.Weimbs T, Stoffel W. Proteolipid protein (PLP) of CNS myelin: positions of free, disulfide-bonded, and fatty acid thioester-linked cysteine residues and implications for the membrane topology of PLP. Biochemistry. 1992;31:12289–12296. doi: 10.1021/bi00164a002. [DOI] [PubMed] [Google Scholar]
- 45.Weimbs T, Stoffel W. Topology of CNS myelin proteolipid protein: evidence for the nonenzymatic glycosylation of extracytoplasmic domains in normal and diabetic animals. Biochemistry. 1994;33:10408–10415. doi: 10.1021/bi00200a023. [DOI] [PubMed] [Google Scholar]
- 46.Williams MJ, Hughes PE, O’Toole TE, Ginsberg MH. The inner world of cell adhesion: integrin cytoplasmic domains. Trends Cell Biol. 1994;4:109–112. doi: 10.1016/0962-8924(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 47.Wong M-H, Filbin MT. The cytoplasmic domain of the myelin P0 protein influences the adhesive interactions of its extracellular domain. J Cell Biol. 1994;126:1089–1097. doi: 10.1083/jcb.126.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamamura T, Konola JT, Wekerle H, Lees MB. Monoclonal antibodies against myelin proteolipid protein: identification and characterization of two major determinants. J Neurochem. 1991;57:1671–1680. doi: 10.1111/j.1471-4159.1991.tb06367.x. [DOI] [PubMed] [Google Scholar]
- 49.Yan Y, Lagenaur C, Narayanan V. Molecular cloning of M6: identification of a PLP/DM20 gene family. Neuron. 1993;11:423–431. doi: 10.1016/0896-6273(93)90147-j. [DOI] [PubMed] [Google Scholar]
- 50.Yan H-C, Baldwin HS, Sun J, Buck CA, Albelda SM, DeLisser HM. Alternative splicing of a specific cytoplasmic exon alters the binding characteristics of murine platelet/endothelial cell adhesion molecule-1 (PECAM-1). J Biol Chem. 1995;270:23672–23680. doi: 10.1074/jbc.270.40.23672. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida M, Colman DR. Parallel evolution and coexpression of the proteolipid proteins and protein zero in vertebrate myelin. Neuron. 1996;16:1115–1126. doi: 10.1016/s0896-6273(00)80138-5. [DOI] [PubMed] [Google Scholar]