Abstract
We analyzed the effects of two growth factors that regulate oligodendrocyte progenitor (O-2A) development on the expression of glutamate receptor (GluR) subunits in cortical O-2A cells. In the absence of growth factors, GluR1 was the AMPA subunit mRNA expressed at the lowest relative level. Basic fibroblast growth factor (bFGF) caused an increase in GluR1 and GluR3 steady-state mRNA levels. Platelet-derived growth factor (PDGF) did not modify the mRNA levels for any of the AMPA subunits but selectively potentiated the effects of bFGF on GluR1 mRNA (4.5-fold increase). The kainate-preferring subunits GluR7, KA1, and KA2 mRNAs were increased by bFGF, but these effects were not modified by cotreatment with PDGF. Nuclear run-on assays demonstrated that PDGF+bFGF selectively increased the rate of GluR1 gene transcription (2.5-fold over control). Western blot analysis showed that GluR1 protein levels were increased selectively (sixfold over control) by PDGF+bFGF. Functional expression was assessed by rapid application of AMPA to cultured cells. AMPA receptor current densities (pA/pF) were increased nearly fivefold in cells treated with PDGF+bFGF, as compared with untreated cells. Further, AMPA receptor channels in cells treated with PDGF+bFGF were more sensitive to voltage-dependent block by intracellular polyamines, as expected from the robust and selective enhancement of GluR1 expression. Our combined molecular and electrophysiological findings indicate that AMPA receptor function can be regulated by growth factor-induced changes in the rate of gene transcription.
Keywords: platelet-derived growth factor, basic fibroblast growth factor, oligodendrocytes, gene transcription, glutamate-gated channels, rectification
Glutamate receptors (GluRs) are widely expressed in both neurons and glia (Mayer and Westbrook, 1987; Gallo and Russell, 1995; Steinhauser and Gallo, 1996). GluRs exist as a mosaic of hetero-oligomeric and possibly homo-oligomeric glutamate-activated channels with different molecular and functional properties that depend on subunit composition (Nakanishi, 1992; Seeburg, 1993; Hollmann and Heinemann, 1994; McBain and Mayer, 1994). Three separate families of non-NMDA GluR genes encode the AMPA-preferring subunits GluR1–4 (or GluRA–D) and the kainate-preferring subunits GluR5–7 and KA1–2 (Nakanishi, 1992; Seeburg, 1993; Hollmann and Heinemann, 1994). mRNA transcripts for all of these subunits are expressed in a developmentally regulated pattern in the rat embryo (Monyer et al., 1991; Pellegrini-Giampietro et al., 1991; Bahn et al., 1994), and functional glutamate-activated channels are known to exist in immature brain cells (Wyllie et al., 1991; Konig et al., 1992; Walton et al., 1993; Patneau et al., 1994) (for review, see Gallo et al., 1995).
The cellular factors that determine and regulate the expression of GluR genes are of great interest because of the broad effects that these receptors have in synaptic transmission and in cellular development in the brain as well as in neurodegenerative conditions. In particular, non-NMDA GluRs have been shown to regulate cell proliferation and differentiation of neural progenitor cells (LoTurco et al., 1995; Gallo et al., 1996), but the growth factors that in turn regulate GluR expression primarily are unknown. This is attributable not only to the heterogeneity of the cell types expressing GluRs in the brain but also to the complexity of the network of growth factors that regulate different stages of neural development (Jacobson, 1991).
The levels of RNA transcripts for the AMPA-preferring subunits GluR1–4 increase after birth (Monyer et al., 1991; Pellegrini-Giampietro et al., 1991; Durand and Zukin, 1993), and basic fibroblast growth factor (bFGF) has been implicated in the regulation of the AMPA receptor subunits GluR1 and GluR4 in developing neurons and glia, respectively (Cheng et al., 1995; Gallo et al., 1994). Recent molecular and electrophysiological studies have demonstrated that, in both neurons and glia, a correlation exists between steady-state RNA levels for distinct GluR subunits and functional properties of glutamate-activated channels within the same cell (Lambolez et al., 1992; Geiger et al., 1995). These findings suggest that regulation and plasticity of GluRs may occur, at least in part, via modifications of the RNA levels for individual subunits. These are determined, ultimately, not only by the rate of gene transcription but also by RNA stability.
In the present study, we have analyzed the effects of platelet-derived growth factor (PDGF) and bFGF on non-NMDA receptor subunit gene expression in oligodendrocyte progenitor (O-2A) cells. We have addressed directly the question of whether modifications in transcriptional rate of GluR genes can cause changes in glutamate-activated channel activity. O-2A cells coexpress functional AMPA- and kainate-preferring receptors (Patneau et al., 1994; Puchalski et al., 1994), and their proliferation and differentiation are regulated by PDGF and bFGF (Raff et al., 1988; Besnard et al., 1989;Bogler et al., 1990; McKinnon et al., 1990; Gard and Pfeiffer, 1993;Gallo and Armstrong, 1995). Here we report that these two factors act in concert to upregulate expression of the AMPA receptor subunit GluR1 selectively via an increase in gene transcription. This molecular event results in upregulation of functional AMPA receptors in the O-2A cell plasma membrane. Our analysis not only identifies two cellular factors that regulate GluR gene expression during glial development but also establishes a direct link between transcriptional regulation of a GluR gene and functional plasticity of the corresponding membrane channels.
MATERIALS AND METHODS
Cell cultures. Purified cortical O-2A progenitor cultures were prepared by modifications of previously described methods (Armstrong et al., 1990a; McKinnon et al., 1990). Briefly, E20–P1 Sprague Dawley rats were decapitated, and cortices were removed, mechanically dissociated, suspended in DMEM (Life Technologies, Gaithersburg, MD) containing 10% fetal bovine serum (FBS; HyClone, Logan, UT), and plated in plastic T75 flasks. After 12 d in culture, O-2A progenitor cells growing on top of a confluent monolayer of astrocytes were detached by overnight shaking (McCarthy and de Vellis, 1980). Contaminating microglial cells were eliminated further by plating this fraction on plastic culture dishes for 1 hr. The O-2A progenitor cells, which do not attach well to plastic, were collected by gently washing the dishes, were replated (18–25 × 103 cells/cm2) onto poly-d-ornithine-coated plates (0.1 mg/ml), and were cultured in DME-N1 biotin-containing medium with 0.5% FBS (HyClone). After 2 hr, either PDGF (human AB, heterodimer form; 10 ng/ml) or bFGF (human; 10 ng/ml), or PDGF+bFGF (both from Upstate Biotechnology, Lake Placid, NY) or 30% B104-conditioned medium (B104-CM) was added to the culture medium. O-2A cells were cultured for 1–3 d and treated every 24 hr with PDGF and/or bFGF or B104-CM.
CG-4 cells were cultured as previously described by Louis et al. (1992a) and Patneau et al. (1994). Briefly, CG-4 progenitor cells were grown on poly-d-ornithine-coated dishes in DME-N1 biotin-containing medium supplemented with 30% B104-CM. B104-CM was prepared as described by Louis et al. (1992a). CG-4 cells were passaged (1:3) every 3–4 d.
Cortical type-1 astrocyte cultures were prepared as previously described (McCarthy and de Vellis, 1980). Briefly, tissue was dissociated with trypsin in the presence of DNase, and cells were cultured in DMEM containing 10% FBS (Life Technologies), 2 mm glutamine, and 2.5 μg/ml amphotericin B (Fungizone). Cells were plated in T75 flasks and maintained at 37°C in 5% CO2. The culture medium was changed every 3 d. Then cultures were shaken overnight to eliminate the majority of oligodendrocyte lineage cells and microglia. Cells were replated in T75 flasks and harvested for RNA preparations after 10 d in culture.
Cultures enriched in different cell types were characterized immunocytochemically by using cell specific markers (see below). Both in CG-4 and cortical cultures enriched in O-2A progenitors, 90–95% of the cells were LB1+/nestin+ (Gallo and Armstrong, 1995). Cell cultures used for immunostaining were grown on glass coverslips precoated with poly-d-ornithine.
Immunofluorescence. Indirect immunofluorescence experiments were performed as previously described (Gallo and Armstrong, 1995). For staining with the monoclonal antibody LB1 (anti-GD3 ganglioside; Levi et al., 1986; Curtis et al., 1988), live cells were incubated for 30 min with the primary antibody diluted (1:1) in DMEM, followed by fluorescein-conjugated goat anti-mouse IgG (Cappel-Organon Teknika, Durham, NC) for 20 min. After extensive washing in DMEM, cells were fixed in 4% paraformaldehyde and 0.2% glutaraldehyde, pH 7.4, in PBS for 15 min and mounted in Vectashield (Vector Laboratories, Burlingame, CA). For staining with the anti-GluR1 antibody, cells were fixed in 4% paraformaldehyde and 0.2% glutaraldehyde, pH 7.4, in PBS for 15 min, permeabilized in 0.25% Triton X-100 for 5 min, and incubated with anti-GluR1 antibodies for 1 hr (2 μg/ml; gift of Dr. Robert Wenthold, National Institutes of Health, Bethesda, MD). After incubation for 45 min with biotinylated goat anti-rabbit antibodies (1:200; Vector), cells were washed extensively and incubated with fluorescein-avidin (1:150; Vector) for 20 min and mounted in Vectashield.
All immunofluorescence experiments were analyzed with 25, 40, or 100× objectives on a Zeiss Photoscope III. In all experiments, controls for antibody specificity were performed by omitting the primary antibodies in the immunostaining protocols. The immunofluorescence pictures presented are representative of at least three experiments.
RNA isolation and Northern blot analysis. Total RNA was isolated from cortical O-2A progenitors and type-1 astrocytes by a single-step procedure (RNAzolB; Tel-Test, Friendswood, TX) (Chomczynski and Sacchi, 1987). Total RNA was isolated from CG-4 cells with the guanidine thiocyanate procedure (Chirgwin et al., 1979). To prepare RNA samples from purified cortical O-2A progenitor cells, we pooled four sets of cultures for each experiment. All of the treatments analyzed (N1, PDGF, bFGF, PDGF+bFGF, B104-CM) were represented in each set of cell cultures. The purified RNA samples were used to run nine parallel Northern blots, each containing RNAs for the five treatments tested. For AMPA receptor subunit RNA analysis, a total of three independent experiments was performed (see Fig. 2). All RNA samples (15 μg/lane) were resolved by electrophoresis through a 1.5% agarose-formaldehyde denaturing gel, electrotransferred onto Nytran (Schleicher & Schuell, Keene, NH) membranes, cross-linked to the membranes by UV irradiation (Stratalinker, Stratagene, La Jolla, CA), and hybridized with cDNA probes. The following cDNA internal fragments were used for random priming with [α-32P]dCTP: KpnI (GluR1),XbaI–XhoI (GluR2, 3, and 5), PstI (GluR4), EcoRI (GluR6), EcoRI–ApaI (GluR7), ClaI (KA1), XbaI–SalI (KA2), and pT7 18S ribosomal RNA template linearized with HindIII (Ambion, Austin, TX). Each probe was labeled at ∼108 cpm/μg DNA. For analysis of cellular RNA, blots were hybridized in 50% formamide at 42°C, washed at high stringency with 0.1× SSC at 60°C, and exposed to film for 3–4 d. GluR and 18S RNA transcripts were quantified with a PhosphorImager system (Molecular Dynamics, Sunnyvale, CA). At the stringency level used, with totally homologous RNA probes, there was no cross-hybridization with ribosomal RNA.
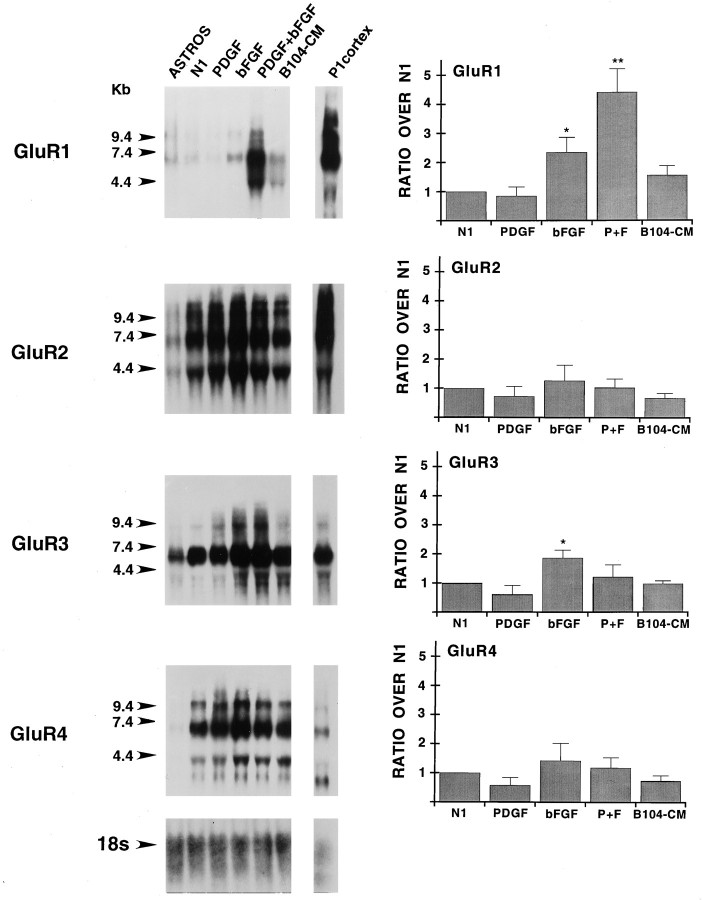
Fig. 2.
Expression and regulation by growth factors of AMPA receptor subunit mRNAs in purified cortical O-2A progenitor cells. Northern blot analysis was performed by using total RNA;lanes show, from left toright: Type-1 astrocyte RNA (ASTROS; 15 μg/lane) isolated from one set of cultures; RNA (15 μg/lane) pooled from four separate sets of O-2A cultures, with each set of cultures tested with all of the conditions analyzed; cerebral cortex RNA (P1 cortex; 15 μg/lane) isolated from postnatal day 1 rat cerebral cortex. O-2A cells and type-1 astrocytes were cultured for 3 and 10 d, respectively. Conditions for O-2A cultures included the following: N1, N1 medium + 0.5% FBS;PDGF, same medium supplemented with 10 ng/ml of PDGF;bFGF, same medium supplemented with 10 ng/ml of bFGF;PDGF+bFGF, same medium supplemented with 10 ng/ml of PDGF and bFGF each. Numbers on the leftrepresent molecular size in kilobases, as derived from an RNA standard. Blots were rehybridized with a cDNA probe for the 18S ribosomal RNA (rRNA) to control for uniform RNA loading; the bottom panel shows the result obtained with rehybridization of the GluR1 blot. All blots were exposed to film for 4 d to maximize signal detection. Histograms in the right panels were derived from Phosphor-Imager (Molecular Dynamics) analysis of the corresponding blots. All data are presented as ratios over untreated cells (N1) and represent averages (±SEM) of three to five independent experiments. *p < 0.05; **p < 0.005, compared with N1(Student’s t test). For GluR1, PDGF+bFGF was also significantly different from bFGF (p < 0.05).
Nuclear run-on assays. This was based on a procedure that has been described previously in detail (Murphy and Carter, 1990). Nuclei from 2 × 107 cells pooled from four different sets of cultures were used in each group. All of the treatments analyzed (N1, 3 hr with PDGF+bFGF and 20 hr with PDGF+bFGF) were represented in each cell culture. For the in vitrotranscription reaction, 160 μl of nuclei were thawed on ice, and [32P]UTP was incorporated into nascent RNA chains by the addition of 30 μl of 10× transcription buffer (1.5 mNaCl; 25 mm MgCl2; 50 mm Mg acetate; 10 mm DTT; 1.25 mm EDTA, pH 8.0; 5 mm each of ATP, CTP, and GTP; 20 mm creatine phosphate; 30 U/ml creatine phosphokinase; and 6 mg/ml heparin), RNase inhibitor to 500 U/ml, 330 μCi [32P]UTP, and DEPC water to a final volume of 300 μl. After termination of the transcription reaction and subsequent extraction and precipitation, the pellet was resuspended in 1.5 ml of Northern hybridization buffer (see above). Equal counts of run-off transcripts, determined by measuring incorporated [32P]UTP on DE81 filters (Sambrook et al., 1989), were hybridized to 3 μg of cloned cDNAs immobilized on nylon membranes and applied with a slot blot apparatus. The membranes were incubated at 65°C for 60 hr and then washed with 1× SSC/0.1%SDS twice at room temperature for 10 min, twice at 65°C for 10 min, incubated with 10 μg/ml RNase A in 2× SSPE for 1 hr, and then washed twice with 1× SSC/1% SDS at 65°C for 15 min. Autoradiography was performed for 3 d.
Ribonuclease protection assays. The GluR1 template used in RNA probe generation was constructed by PCR amplification of bases 2541–2741 (Hollmann et al., 1989), inserted between theEcoRI and the BamHI sites of pSP72 (Promega, Madison, WI), and sequenced to check for mutations. After linearization with BglII, 264 nucleotide (NTD) antisense run-off transcripts were produced by in vitro transcription with SP6 RNA polymerase. The GluR2 template was generated by allowing theEcoRI-digested cDNA to self-ligate. Linearization withEcoRI and transcription with T7 RNA polymerase produced a 296 NTD probe corresponding to bases 2924 and 3190 (Boulter et al., 1990). The 18S ribosomal RNA template was purchased from Ambion. Antisense 100 NTD 18S probes were generated by HindIII linearization and transcription with T7 RNA polymerase. The sizes of the protected fragments for GluR1, GluR2, and 18S were 200, 265, and 80 bp, respectively. Cells (2 × 106) were rinsed twice with ice-cold PBS before extraction of total RNA by using RNazol B (Tel-Test). Ribonuclease protection assays were performed according to the protocol provided with the RPAII kit (Ambion). Briefly, RNA probes were labeled to the following specific activities (cpm/μg): GluR1, 7 × 108; GluR2, 7 × 108; and 18S, 3 × 106. Gel-purified riboprobes were hybridized with 20 μg of total RNA in 20 μl of hybridization buffer at 45°C overnight. The hybridization mixtures were treated with a 1:100 dilution of A/T1 mixture (final concentrations, 2.5 U/ml RNase A; 100 U/ml RNase T1) at 37°C for 30 min, and the resulting samples were precipitated and resolved on a 6% denaturing polyacrylamide gel. The dried gel was exposed to film for 7 d for GluR1 and GluR2 and 5 hr for 18S products. Autoradiographs were scanned, and GluR1, GluR2, and 18S RNA levels were determined by densitometry (Microtek ScanWizard Plug-In, Redondo Beach, CA). Data for GluR1 and GluR2 RNAs were normalized with 18S RNA levels, as determined within the same samples.
Reverse transcriptase-PCR. For each Reverse Transcriptase (RT) reaction, 0.5–2.0 μg of total RNA, isolated as outlined above, was combined with 100 pmol of random hexamers and denatured at 80°C for 10 min. Then the reactions were assembled by adding 1× PCR buffer (1 mm MgCl2, 333 mm each of dNTPs, 10 mm DTT, and 100 mg/ml BSA), containing 1 unit/ml RNasin (Promega) and 200 units of pSuperscript II RT (Life Technologies) in a volume of 30 μl. Reactions were incubated at 25°C for 10 min and then at 42°C for 50 min. After heat inactivation, 30 units of RNase H (Life Technologies) were added, and incubation was continued at 37°C for 20 min.
For PCR reactions, 10 μl of each RT reaction was added to a tube containing 1× PCR buffer, 50 pmol of upstream primer (5′-TGGTGGTTCTTCACCCTGATCAT-3′, bases 1854 to 1877 of the GluR1 cDNA when the adenosine of the initiator codon is set to +1), and 50 pmol of downstream primer (5′-TATGGCTTCATTGATGGATTGC-3′, bases 1394 to 1373), yielding a 709 bp product (Jensen and Chiu, 1993), 200 mmeach of dNTPs, and 2.5 units of AmpliTaq Polymerase (Perkin-Elmer, Foster City, CA) in a volume of 100 μl. The PCR cycling profile was as follows: 95°C for 5 min and then 30 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min with a 5 sec time extension added per cycle and a final extension at 72° for 7 min. Products were separated on 1.5% agarose gels and visualized by ethidium bromide staining.
Western blot analysis. After treatment with growth factors, 2 × 106 cells were washed twice and harvested in cold PBS. The cells were resuspended and lysed in 300 μl of sample buffer (62.5 mm Tris, pH 6.8, 1% SDS, 10% glycerol, 10 μg/ml leupeptin, 1 μg/ml aprotinin, and 1 mm AEBSF). The lysate was centrifuged at 5000 × g for 15 min, and the supernatant was collected. An aliquot was taken for protein determination with the Bio-Rad DC protein assay kit (Hercules, CA). Forty micrograms of the cell extracts were resolved on a 9% mini SDS polyacrylamide gel and transferred to Immobilon P membranes (Millipore, Bedford, MA). Blots were blocked with 5% nonfat dry milk in TBST (10 mm Tris, pH 7.4, 150 mm NaCl, and 0.05% Tween 20) for 1 hr at room temperature and then incubated at room temperature for 1 hr in TBST and 5% nonfat dry milk containing GluR antibodies: GluR1 (1 μg/ml; R. Wenthold), GluR2/3 (0.1 μg/ml; R. Wenthold), and GluR4 (1 μg/ml; Upstate Biotechnology). The anti-GluR2/3 antibody also detected the GluR4 splice isoform GluR4c (Gallo et al., 1992;Puchalski et al., 1994). Protein bands were detected by the Amersham ECL kit (Arlington Heights, IL) with horseradish peroxidase-conjugated secondary antibodies. Relative intensities of the protein bands were quantified by scanning densitometry (Microtek ScanWizard Plug-In).
Electrophysiological analysis of AMPA receptor expression.Whole-cell recordings from cortical progenitor cells were obtained 2–3 d after plating in medium with 0.5% serum or the same medium containing PDGF+bFGF as described above. Phase-contrast video photomicrographs were obtained from individual cells before recording to establish a record of their morphology as multipolar or bipolar cells, which were generated after growth in low (0.5%) serum medium or medium with PDGF+bFGF, respectively. Thin-walled borosilicate glass microelectrodes (TW150F-6, World Precision Instruments, Sarasota, FL) had resistances of 3–5 MΩ when filled with intracellular recording solution (in mm): 85 CsMeSO3, 50 CsCl, 10 CsF, 10 HEPES, 5 EGTA, 1 MgCl2, and 0.5 CaCl2, pH 7.2, osmolarity 310 mOsm. The intracellular solution also included 30 μm spermine to compensate for loss of cytoplasmic polyamines as a result of dialysis during whole-cell recording (Bowie and Mayer, 1995; Donevan and Rogawski, 1995; Kamboj et al., 1995; Koh et al., 1995). The extracellular solution contained (in mm): 160 NaCl, 2.5 KCl, 10 HEPES, 2 CaCl2, 1 MgCl2, 10 glucose, 0.01 mg/ml phenol red, pH to 7.3 with NaOH, osmolarity 325 mOsm, and 400 nm tetrodotoxin. Cells were voltage-clamped at −60 mV with an Axopatch-1C amplifier (Axon Instruments, Foster City, CA). The series resistance was <10 MΩ and compensated by at least 80%; cell capacitance was read from the setting on the capacitance neutralization potentiometer of the amplifier. The time constant of the capacitive current for bipolar cells in PDGF+bFGF-treated cultures (82 ± 9.9 μsec;n = 9) was much briefer than that of multipolar cells in untreated cultures (449 ± 53 μsec; n = 8), but in both cell populations it was reasonably well fit by a single exponential function; as a result there was good agreement between values for the series resistance and cell capacitance calculated from the amplitude and time constant of the capacitive current and the settings of the series resistance and capacitance neutralization potentiometers of the amplifier (Marty and Neher, 1995).
Solutions were applied via a stepper motor-based fast perfusion system, as described previously (Vyklicky et al., 1990). Agonist-evoked currents were evoked by using 500 μm (R,S) AMPA to selectively activate AMPA versus kainate-preferring glutamate receptors. To allow an accurate estimation of the maximum amplitude of AMPA receptor responses, we preincubated cells for at least 30 sec with the AMPA receptor-selective allosteric modulator cyclothiazide (100 μm) before application of agonist; this reliably blocked desensitization. Cyclothiazide (20 mm) was dissolved in dimethylsulfoxide (DMSO) before dilution with extracellular solution; an equivalent final concentration of DMSO (0.5%) was added to the remaining extracellular solution. Responses were filtered at 2 kHz (8-pole Bessel filter), digitized as required, and stored on a MacIntosh IIfx computer with an ITC-16 interface (Instrutech) under control of the data acquisition and analysis program Synapse (Synergistic Research, Silver Spring, MD). To examine the rectification properties of AMPA receptor responses, we ramped the membrane potential from −105 to +100 mV at 0.41 V/sec during steady-state responses to application of AMPA and cyclothiazide. The resulting current–voltage (I–V) relationships were leak-subtracted and fit with a ninth-order polynomial to estimate the reversal potential (Vrev); then the chord conductance was calculated from the equation G =I/(V−Vrev). Data points surrounding the reversal potential were masked during subsequent fits to a modified Woodhull equation:
where
in which GR is the component of the response to AMPA-resistant to voltage-dependent block by polyamines;GV the voltage-dependent, polyamine-sensitive component of the AMPA response; [S], the internal spermine concentration (assumed to be 30 μm);KD(0 mV), the dissociation constant for binding of spermine at 0 mV membrane potential; z, the valence of spermine; and δ, the fraction of the membrane electric field at the polyamine binding site, with F, R, andT having their standard values (Woodhull, 1973). Data are presented as mean ± SEM.
RESULTS
Proliferation and morphology of O-2A cells cultured with PDGF and bFGF
The strongly mitogenic effects of PDGF and bFGF on O-2A progenitor cells are well documented (Raff et al., 1988; Bogler et al., 1990; McKinnon et al., 1990; Gard and Pfeiffer, 1993; Gallo and Armstrong, 1995; Gallo et al., 1996). We have demonstrated previously that both growth factors significantly stimulated O-2A cell proliferation (three- to fourfold increase after 24 hr treatment), as measured by [3H]thymidine or bromodeoxyuridine incorporation (Gallo and Armstrong, 1995; Gallo et al., 1996). Additionally, continuous treatment with PDGF and bFGF for 3 d produced distinct effects on O-2A cell morphology, as revealed after staining with the monoclonal antibody LB1 (Fig. 1). Cells cultured in the absence of growth factors (N1; untreated cells) had round cell bodies and a complex stellate morphology with several processes that covered a large surface area (Fig. 1). O-2A cells cultured in PDGF displayed a typical bipolar morphology with long processes, whereas bFGF-treated cells were multipolar and had radially oriented short processes (Fig. 1). O-2A progenitors cultured with both PDGF and bFGF displayed a wider range of morphologies (Fig. 1). Clusters of cells with a more multipolar morphology also were found in the cultures (see also Fig. 8). Finally, O-2A cells cultured with conditioned medium from the neuroblastoma cell line B104 (B104-CM) displayed a proliferation index and morphology similar to cells cultured in PDGF+bFGF (data not shown).
Fig. 1.
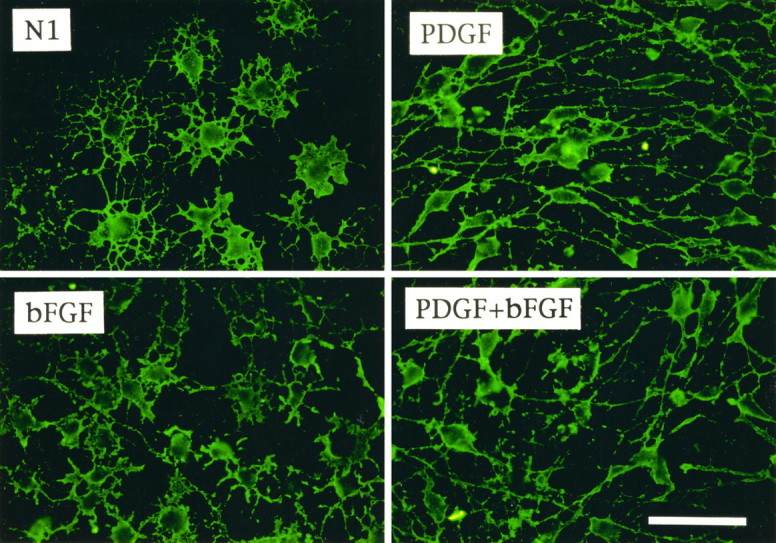
O-2A progenitor cells express a different phenotype when cultured in the presence or absence of PDGF and/or bFGF. Immunofluorescence of rat cortical O-2A cells cultured in N1 medium + 0.5% FBS without growth factors (N1) or with PDGF (10 ng/ml), bFGF (10 ng/ml), or PDGF+bFGF (10 ng/ml each) for 3 d. Cells were stained with the monoclonal antibody LB1 and a fluorescein-conjugated anti-mouse secondary antibody. Representative fields are shown for each culture condition. All O-2A progenitors were LB1+, but their morphology differed according to culture condition. Cells cultured without mitogens were larger and multipolar; PDGF, smaller and bipolar; bFGF, smaller and multipolar with short processes; PDGF+bFGF, mostly bipolar (but also other morphologies, see Fig. 7). Scale bar, 100 μm.
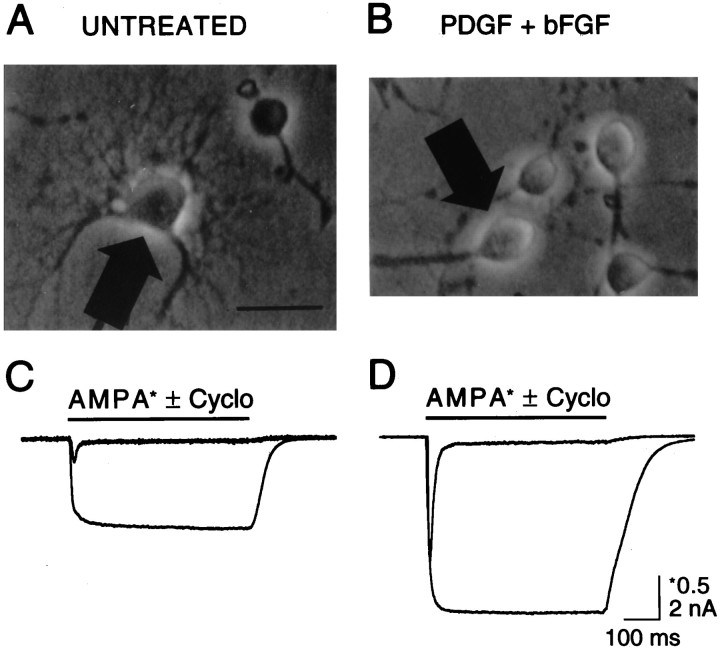
Fig. 8.
Whole-cell currents mediated by AMPA receptors are enhanced after treatment of O-2A cells with PDGF+bFGF.A, Untreated cells grown in low serum medium for 2–3 d displayed multipolar morphology with highly branched processes.B, Cells in parallel cultures supplemented with PDGF+bFGF (10 ng/ml each) maintained a bipolar morphology. C, D, Application of 500 μm AMPA by fast perfusion to the cells marked by arrows produced rapidly desensitizing responses in both untreated (C) and in PDGF+bFGF-treated cultures (D); the capacitance of the cells was 56 pF (C) and 9 pF (D). In the presence of 100 μm cyclothiazide, which blocks AMPA receptor desensitization, both the peak and equilibrium responses increased in amplitude; the vertical scale bar for control responses to AMPA (*0.5 nA) is four times that for responses in the presence of cyclothiazide (2 nA).
PDGF and bFGF in combination selectively increase GluR1 RNA levels
We have demonstrated previously that primary cortical O-2A progenitor cells cultured with B104-CM express both AMPA- and kainate-preferring subunit RNA transcripts (Patneau et al., 1994). Figure 2 shows that the mitogenic factors present in B104-CM did not modify AMPA receptor subunit expression significantly in O-2A cells, as measured by Northern blot analysis of total RNA isolated from cells cultured in B104-CM for 3 d versus cells cultured without any growth factor (N1 lanes). PDGF also did not modify the RNA levels of any AMPA receptor subunit, whereas bFGF caused a significant increase of GluR1 and GluR3 RNAs (Fig. 2).
Treatment of O-2A cells with PDGF+bFGF for 3 d resulted in a selective, 4.5-fold increase of GluR1 RNAs (Fig. 2). The synergistic effect of the two growth factors seemed to be unrelated to the proliferative effects of the growth factors, because treatment of the cells with B104-CM, which was shown previously to stimulate O-2A cell proliferation similarly to PDGF+bFGF (Gallo and Armstrong, 1995), did not modify GluR1 expression significantly (Fig. 2).
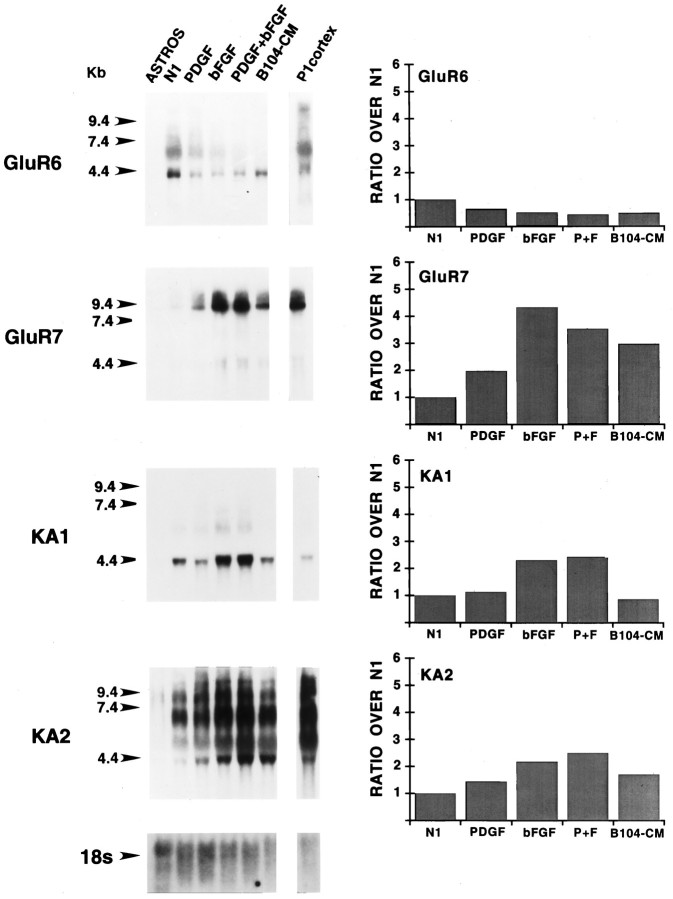
Among the kainate-preferring subunits, GluR7, KA1, and KA2 RNAs were upregulated by bFGF, whereas PDGF only increased GluR7 RNAs and did not modify the effects of bFGF on kainate-preferring receptor RNAs (Fig.3). GluR5 RNAs were not detected in O-2A progenitor cells (Patneau et al., 1994) and were not induced by PDGF and/or bFGF (data not shown).
Fig. 3.
Expression and regulation by growth factors of kainate receptor subunit RNAs in purified cortical O-2A progenitor cells. Northern blot analysis was performed by using total RNA;lanes show, from left toright: Type-1 astrocyte RNA (ASTROS; 15 μg/lane) isolated from one set of cultures; RNA (15 μg/lane) pooled from four separate sets of O-2A cultures, each set tested with all of the treatments analyzed; cerebral cortex RNA (P1 cortex; 15 μg/lane) isolated from postnatal day 1 rat cerebral cortex. O-2A cells and type-1 astrocytes were cultured for 3 and 10 d, respectively. N1, N1 medium + 0.5% FBS;PDGF, same medium supplemented with 10 ng/ml of PDGF;bFGF, same medium supplemented with 10 ng/ml of bFGF;PDGF+bFGF, same medium supplemented with 10 ng/ml of PDGF and bFGF each. Numbers on the leftrepresent molecular size in kilobases, as derived from an RNA standard. Blots were rehybridized with a cDNA probe for 18S rRNA to control for uniform RNA loading; the bottom panel shows the result obtained with rehybridization of the KA1 blot. All blots were exposed to film for 4 d to maximize signal detection. No expression of GluR5 was detected by Northern blot in cultured glia under any culture condition, even after longer exposure times (see also Patneau et al., 1994). Histograms in the right panels were derived for PhosphorImager analysis of the corresponding blots. The experiments was repeated by using an independent set of cultures with similar results. All data are presented as ratios over untreated cells (N1).
AMPA receptor subunits were also detectable in purified cortical type-1 astrocyte cultures, with GluR3 being expressed at the highest relative levels (Fig. 2). Kainate-preferring subunit transcripts were not detectable in type-1 astrocytes by Northern blot analysis (Fig. 3).
PDGF and bFGF increase the rate of GluR1 transcription within 20 hr
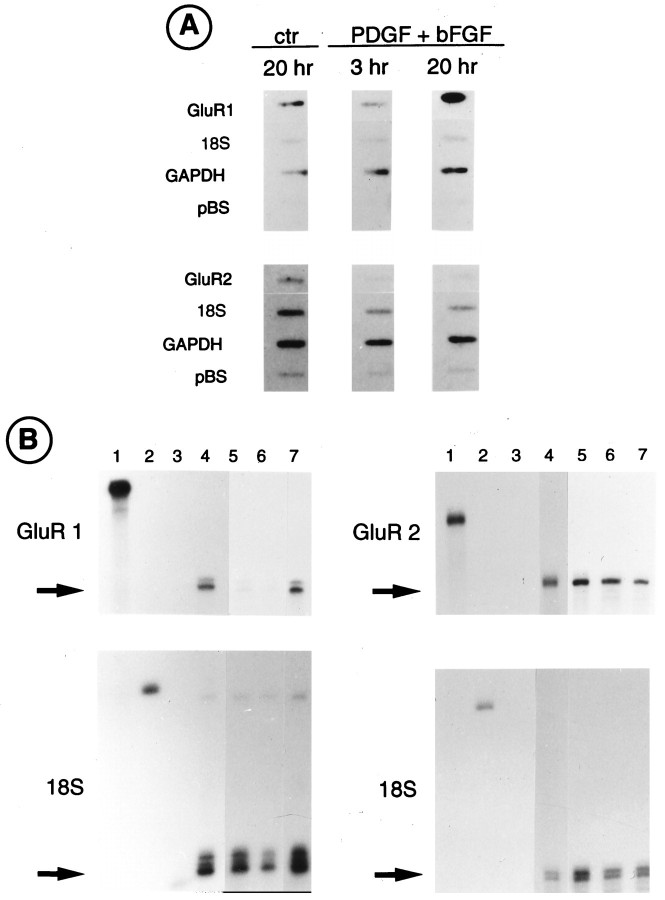
The increase in GluR1 mRNA expression induced by treatment of O-2A cells with PDGF+bFGF could be attributable to an increase in the stability of the GluR1 message, an increase in the transcriptional activity of the GluR1 subunit gene, or a combination of both. To distinguish between these mechanisms, we performed nuclear run-on assays to measure levels of newly synthesized GluR transcripts after treatment with the two growth factors. Nuclei were obtained from O-2A cells grown in the absence of growth factors (untreated cells) or in the presence of PDGF+bFGF for 3 and 20 hr (Fig.4A). Data for GluR1 and GluR2 transcripts were normalized with respect to 18S ribosomal transcript levels. Treatment with the growth factors for 3 hr caused a modest decrease in GluR1 transcription, but after 20 hr PDGF+bFGF caused a 2.5-fold increase in GluR1 transcription rate (Fig.4A). GluR2 transcription rate was not increased by treatment with the two growth factors at both time points (Fig.4A).
Fig. 4.
PDGF and bFGF increase the rate of GluR1 gene transcription in O-2A cells. A, Nuclear run-on transcription assays. Nuclei isolated from six separate cultures were pooled. O-2A cells were cultured either in N1+0.5% FBS (ctr) for 20 hr or in the same medium supplemented with PDGF and bFGF (10 ng/ml each) for 3 and 20 hr. Labeled RNAs were hybridized to the GluR1, GluR2, 18S ribosomal (18S), and the glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) cDNAs, the latter as an internal control for metabolic integrity of nuclear preparations. pBluescript vector DNA (pBS) was used to determine background hybridization. The hybridization signals were quantified by PhosphorImager analysis and normalized with respect to the 18S RNA. GluR1 probe showed a 2.5-fold increase in the hybridization signal after 20 hr of treatment with PDGF+bFGF. No increase was observed in GluR2 or in 18S signals. B, Ribonuclease protection assay. Twenty micrograms of total RNA isolated from O-2A progenitor cells were hybridized with the indicated RNA probes (GluR1, GluR2, and 18S).Left panels, Lane 1, Full-length GluR1 RNA probe;lane 2, full-length 18S ribosomal RNA probe; lane 3, tRNA negative control; lane 4, cerebellar RNA (positive control); lane 5, O-2A cells cultured in N1 + 0.5% FBS; lane 6, O-2A cells treated with PDGF+bFGF for 3 hr; lane 7, O-2A cells treated with PDGF+bFGF for 24 hr. Right panels, Lane 1, Full-length GluR2 RNA probe;lanes 2, 3, as above; lane 4, rat cerebellar RNA (10 μg); lanes 5–7, as above.Arrows indicate protected RNA fragments.
A time course to study the effects of PDGF+bFGF on GluR1 and GluR2 mRNA accumulation within the first 24 hr of treatment was, therefore, performed and analyzed by RNase protection assays. Data for GluR1 and GluR2 RNAs were normalized with respect to 18S RNA levels, as determined within the same samples (Fig. 4B). No changes in GluR1 or GluR2 steady-state RNA levels were detected after 3 hr of treatment with PDGF+bFGF (Fig. 4B, lanes 5, 6). The two growth factors stimulated a threefold increase in GluR1 RNA expression after 24 hr, although no change in GluR2 could be detected (Fig. 4B, lane 7). The close agreement between the transcription assay and the RNase protection data in both the magnitude and temporal pattern of stimulation indicates that elevated GluR1 RNA levels, at least in the first 20 hr of growth factor treatment, are primarily the result of increased transcriptional activity. A further increase in GluR1 RNA levels (fivefold over control) was found after 3 d of treatment (data not shown). The subunit-specific effect of PDGF+bFGF on GluR1 gene expression in both nuclear run-on and RNase protection assays is consistent with the Northern blot data (Fig. 2).
PDGF and bFGF do not stimulate GluR1 expression in the oligodendrocyte cell line CG-4
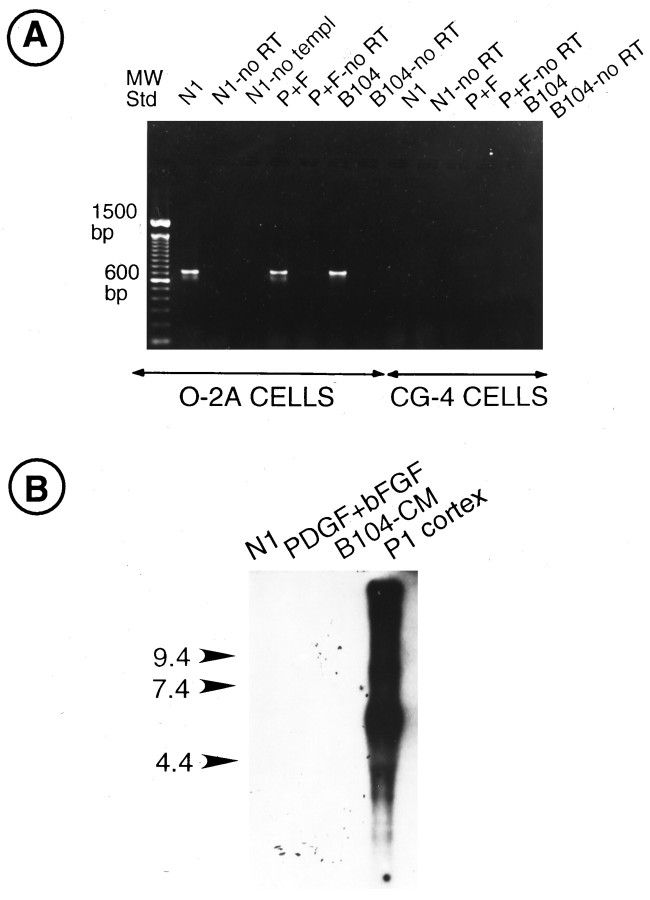
We have demonstrated previously by Northern blot analysis that the oligodendrocyte cell line CG-4 expresses all AMPA receptor subunit mRNAs, except for GluR1 (Patneau et al., 1994). Because CG-4 cells are sensitive to the mitogenic action of PDGF and bFGF (Louis et al., 1992b), we analyzed whether the two growth factors also would regulate GluR1 expression in this oligodendrocyte cell line. Figure5A shows that GluR1 transcripts could not be detected by RT-PCR in CG-4 cells maintained under the culture conditions previously used (Patneau et al., 1994), i.e., in B104-CM. Furthermore, treatment of CG-4 cells with PDGF+bFGF did not induce detectable GluR1 gene expression, as demonstrated by RT-PCR (Fig.5A) and Northern blot analysis (Fig. 5B).
Fig. 5.
GluR1 mRNAs are neither expressed nor regulated by growth factors in CG-4 cells. A, RT-PCR analysis of GluR1 transcripts in O-2A and CG-4 cells cultured in N1 + 0.5% FBS (N1), in PDGF+bFGF (P+F; 10 ng/ml each), and in B104-conditioned medium (B104). Negative controls included no RT reaction to exclude genomic DNA contamination (no RT) and no template in the RT reaction to exclude reagent contamination (no templ). The expected RT products (700 bp) were obtained from O-2A cells cultured inN1, PDGF+bFGF (P+F), or B104, but not from CG-4 cells. B, Northern blot analysis of total RNA isolated from CG-4 cells cultured in N1 + 0.5% FBS (N1), in PDGF+bFGF (P+F; 10 ng/ml each), and in B104-conditioned medium (B104). The blot was hybridized with a GluR1 KpnI cDNA probe. P1 cortex, Positive control. Treatment with PDGF+bFGF, which strongly upregulates GluR1 RNA expression in O-2A cells, did not stimulate GluR1 expression in CG-4 cells. This blot was overexposed (2 weeks) to detect low levels of GluR1 transcripts.
PDGF and bFGF increase the levels of GluR1 protein
We analyzed whether the increase in GluR1 gene transcription rate triggered by PDGF and bFGF resulted in a parallel increase in GluR1 protein. Western blot analysis and immunocytochemical staining were used to determine whether AMPA receptor protein levels were modified by the growth factors after 3 d in culture. Consistent with the RNA analysis, basal levels of GluR1 protein in untreated cells were relatively lower than GluR2, 3, 4c, and GluR4 (Fig. 6A). Figure 6, A andB, shows that GluR1 protein levels were increased moderately by PDGF (1.2 ± 0.04-fold increase over untreated cells; average ± SEM; n = 4) and by bFGF (2.1 ± 0.3-fold increase; n = 6). The combined treatment of PDGF+bFGF caused a synergistic 6.0 ± 1.4-fold increase in total GluR1 protein over untreated cells (Fig. 6A,B;n = 6), but GluR1 protein levels in O-2A cells cultured in B104-CM were similar to those of untreated cells (data not shown). As predicted from the RNA analysis, GluR1 protein could not be detected in CG-4 cells (Fig. 6A).
Fig. 6.
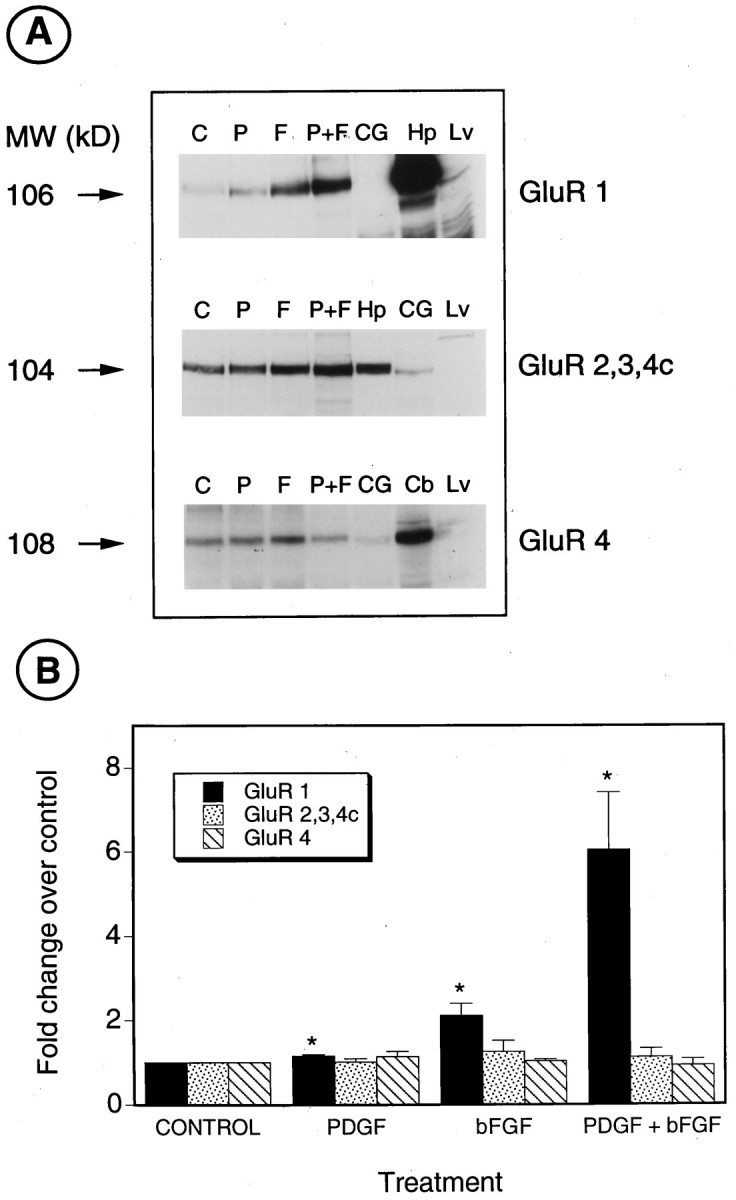
PDGF and bFGF increase the levels of GluR1 protein in O-2A progenitor cells. A, Western blot analysis of total proteins isolated from O-2A cells cultured for 3 d in N1 medium + 0.5% FBS (C), 10 ng/ml PDGF (P), 10 ng/ml bFGF (F), and PDGF+bFGF (P+F; 10 ng/ml each). CG, CG-4 cells cultured in B104-conditioned medium; Hp, hippocampus; Lv, liver; Cb, cerebellum. Antibodies used were anti-GluR1, anti-GluR2, 3, 4c, and anti-GluR4 in the top, middle, and bottom panels, respectively. The small increase in GluR2, 3, 4c observed in cells treated with bFGF and PDGF+bFGF (middle panel) was not reproducible (3 experiments; seeB). Numbers on the leftindicate molecular sizes in kilodaltons, as derived from protein standards. B, Relative levels of the AMPA receptor subunits were determined by densitometric analysis of Western blots. Values are expressed as ratios over untreated cells (N1 + 0.5% FBS) and are mean ± SEM of three separate experiments, except for GluR1, in which n = 6 for control, bFGF, and PDGF+bFGF, and n = 4 for PDGF. *p < 0.05, as compared with control (Student’st test).
A similar analysis with anti-GluR2, 3, 4c, or anti-GluR4 antibodies demonstrated that neither PDGF nor bFGF significantly modified the levels of these AMPA receptor proteins (Fig. 6A,B). The relative levels of GluR2, 3, 4c, and 4 proteins were lower in CG-4 than in primary O-2A cells (Fig. 6A), in agreement with previous findings at the RNA level (Patneau et al., 1994).
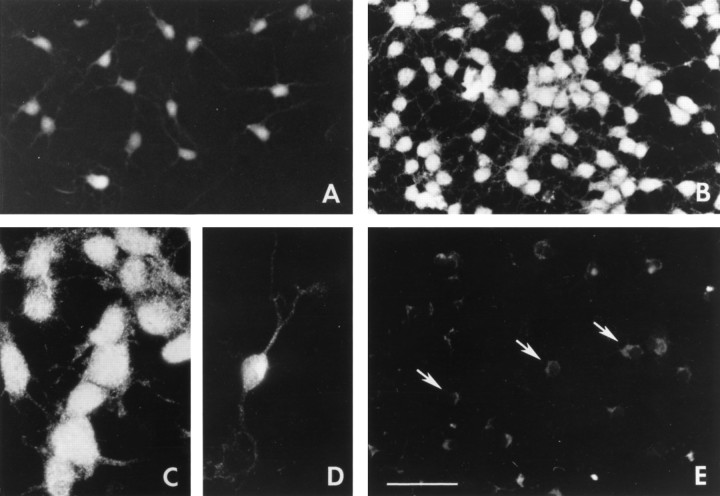
Oligodendrocyte lineage cells maintained in culture for 3 d were stained specifically with anti-GluR1 antibodies (Fig.7). Cells cultured in the absence of growth factors displayed low levels of anti-GluR1 staining (Fig. 7A), which significantly increased in cells treated with PDGF+bFGF for 3 d (Fig. 7B). Interestingly, the increase in GluR1 immunoreactivity was not confined to a subpopulation of cells but was found in virtually all cells (see representative field inB), independent of their morphology (Fig. 7C,D). GluR1 staining was found not only in the cell bodies but also in the processes of all cells (Fig. 7B–D).
Fig. 7.
GluR1 protein is expressed in all O-2A progenitor cells and is upregulated uniformly after treatment with PDGF+bFGF. Immunofluorescence staining of cortical O-2A cells with anti-GluR1 antibodies in cells cultured for 3 d. A, Untreated cells (N1 + 0.5% FBS). B–D, PDGF+bFGF (10 ng/ml each). E, PDGF+bFGF, negative control; cells stained after omitting the anti-GluR1 antibody. Arrows indicate background cytoplasmic fluorescence. PDGF+bFGF-treated cells were strongly stained independently of their morphology, and GluR1 staining was found both on the cell bodies and on the processes (C, D). Scale bar: A, B, E, 200 μm; C, D, 100 μm.
Taken together, our biochemical and immunocytochemical analyses unequivocally demonstrate that changes in AMPA receptor subunit RNA expression, in particular GluR1, result in parallel changes in AMPA protein levels.
PDGF and bFGF increase the density of functional AMPA receptors
Next we sought to determine whether treatment with PDGF+bFGF also resulted in increased functional expression of AMPA-preferring receptors. Using the whole-cell recording configuration, with agonist applied by fast perfusion, we compared the magnitude of AMPA-evoked currents in cells grown in low serum medium (untreated cells) with those of cells grown in the same medium containing PDGF+bFGF. Whole-cell recordings were obtained from cells selected on the basis of morphology (see Fig. 8A). The majority of untreated cells was multipolar, whereas PDGF+bFGF-treated cells were predominantly bipolar (see Figs. 1, 7). All of the cells chosen for electrophysiological recordings were either multipolar (untreated; Fig.8A) or bipolar (PDGF+bFGF-treated; Fig.8B). Figure 8, C and D, compares the responses of these two cell types to application of 500 μm AMPA in the absence and presence of 100 μm cyclothiazide, used to block AMPA receptor desensitization selectively. As previously reported (Patneau et al., 1994), application of AMPA to multipolar cells produced a rapidly desensitizing response well fit by a single exponential of the time constant, 9.31 ± 0.56 msec (n = 13). AMPA responses of bipolar cells from cultures grown in the presence of PDGF+bFGF desensitized with a similar time constant, 8.82 ± 0.34 msec (n = 18). The rapidity of AMPA receptor desensitization and the difficulty of ensuring rapid solution exchange over the surface of multipolar cells would be expected to contribute to an underestimate of the total number of functional AMPA receptors. Consistent with this, whole-cell responses to AMPA were greatly enhanced after preincubation with 100 μm cyclothiazide, and, at steady-state, AMPA currents in untreated multipolar cells were increased >50-fold in the presence of cyclothiazide. However, even after block of desensitization, AMPA responses were larger in cells treated with PDGF+bFGF (5.76 ± 0.57 nA; n = 39), as compared with untreated cells grown in low serum medium (4.94 ± 0.59 nA, n = 31). Because these culture conditions generate cells with strikingly different morphology and size and AMPA receptor currents varied 5- to 10-fold in amplitude between cells grown in both low serum media as well as in PDGF+bFGF, amplitude measurements alone are unable to quantify accurately the possible changes in AMPA receptor density that result from treatment with PDGF+bFGF.
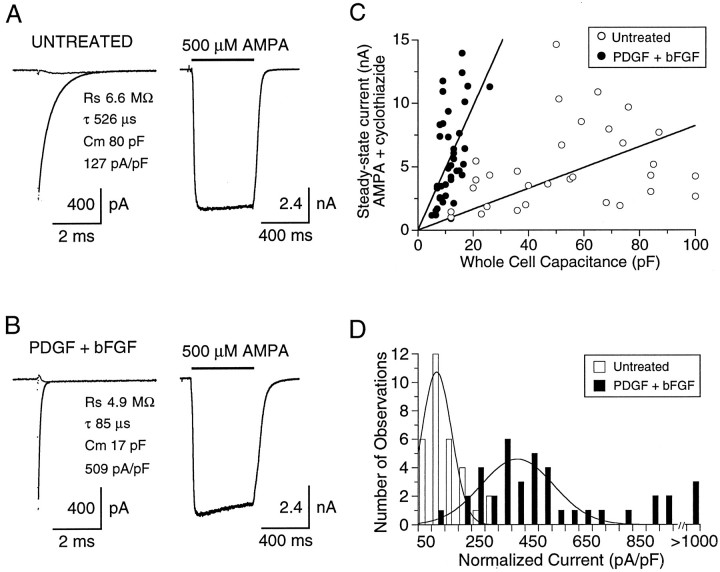
To estimate the effects of growth factors on receptor density more effectively, AMPA receptor responses recorded in the presence of cyclothiazide were normalized relative to whole-cell capacitance measurements (Fig. 9). As would be expected from morphological analysis, the multipolar cells in untreated cultures had a significantly larger capacitance than bipolar growth factor-treated cells (53.1 ± 4.7 pF vs 11.7 ± 0.7 pF; p < 0.001, two-tailed t test), although smaller multipolar cells purposely were selected for recording to minimize this difference (in addition, larger differentiated cells did not adhere well during rapid perfusion and expressed currents too big to adequately voltage clamp). Although in both culture conditions AMPA receptor current amplitude and cell capacitance varied among cells, the slope of the relationship between cell capacitance and AMPA-evoked current amplitude (Fig.9A) was much steeper for cells grown with PDGF+bFGF (488 pA/pF) than for untreated cells (82 pA/pF). Analysis of the range of steady-state current densities, derived by normalization to whole-cell capacitance (Fig. 9B), revealed two dissimilar distributions with a mean current density approximately fivefold higher in cells treated with growth factors (496 ± 47 pA/pF; n = 39), as compared with untreated cells (106 ± 12 pA/pF;n = 31; p < 0.001, one-tailedt test).
Fig. 9.
AMPA receptor density is higher in cells treated with PDGF+bFGF. Capacitive currents evoked by 10 mV hyperpolarizing voltage steps (left) and inward currents evoked by 500 μm AMPA (right) recorded from cells in untreated (A) and PDGF+bFGF-treated (B) cultures. Capacitive currents are shown before and after compensation; the line drawn through the uncompensated response is a single exponential fit of indicated time constant (τ); the series resistance (Rs) was calculated from the amplitude of the capacitive current extrapolated to the start of the voltage jump; the cell capacitance (Cm) was calculated from the time constant of the capacitive current divided byRs. After the capacitive current was compensated, the calculated values for Rsand Cm (A, 6.6 MΩ, 80 pF;B, 4.9 MΩ, 17 pF) were in excellent agreement with the settings of the series resistance and capacitance neutralization controls of the amplifier (A, 6.5 MΩ, 76 pF;B, 5.5 MΩ, 15 pF). Note that the AMPA receptor current density in the PDGF+bFGF-treated cell (509 pA/pF) is four times larger than that in the untreated cell (127 pA/pF). C, The amplitude of steady-state responses to AMPA recorded in the presence of cyclothiazide is plotted versus whole-cell capacitance. The slope of linear regressions forced through the origin is steeper for cells treated with growth factors (488 pA/pF, r = 0.54) than for untreated cells (82 pA/pF, r = 0.30).D, Histograms of the distribution of whole-cell AMPA receptor current densities (pA/pF) fit with Gaussian functions. Mean current densities were 106 ± 12 pA/pF in untreated cells (n = 31) versus 496 ± 46 pA/pF in cells treated with growth factors (n = 39).
Effects of growth factors on AMPA receptor subunit composition
To determine further whether the enhanced functional expression of AMPA receptors in PDGF+bFGF-treated progenitors is selective for GluR1, as indicated by our molecular analysis at the RNA and protein levels, we examined the current–voltage relationship of AMPA receptor responses in PDGF+bFGF-treated and untreated cells. We reasoned that a selective enhancement of the expression of GluR1 should either generate homomeric AMPA receptors assembled from GluR1 or dilute the relative contribution of GluR2 subunits to heteromeric AMPA receptors generated by coassembly with GluR1 and perhaps other subunits. In either case enhanced sensitivity to voltage-dependent channel block by cytoplasmic polyamines would be expected (Bowie and Mayer, 1995; Donevan and Rogawski, 1995; Kamboj et al., 1995; Koh et al., 1995). Current–voltage (I–V) relationships were generated from voltage ramps from −100 to +100 mV during the steady-state component of the response to combined application of AMPA and cyclothiazide. To prevent loss of cytoplasmic polyamines during whole-cell recording, we included 30 μm spermine in the intracellular solution. I–V relationships were recorded from 23 untreated and 23 growth factor-treated cells. Examples are shown in Figure 10A and illustrate pronounced rectification for PDGF+bFGF-treated, but not untreated, cells. An index of rectification (IR), calculated from the ratio of slope conductances at +30/−60 mV, averaged 0.77 ± 0.06 and 0.32 ± 0.04 for untreated and growth factor-treated cells, respectively (p < 0.0001, one-tailedt test; n = 23 cells per group).
Fig. 10.
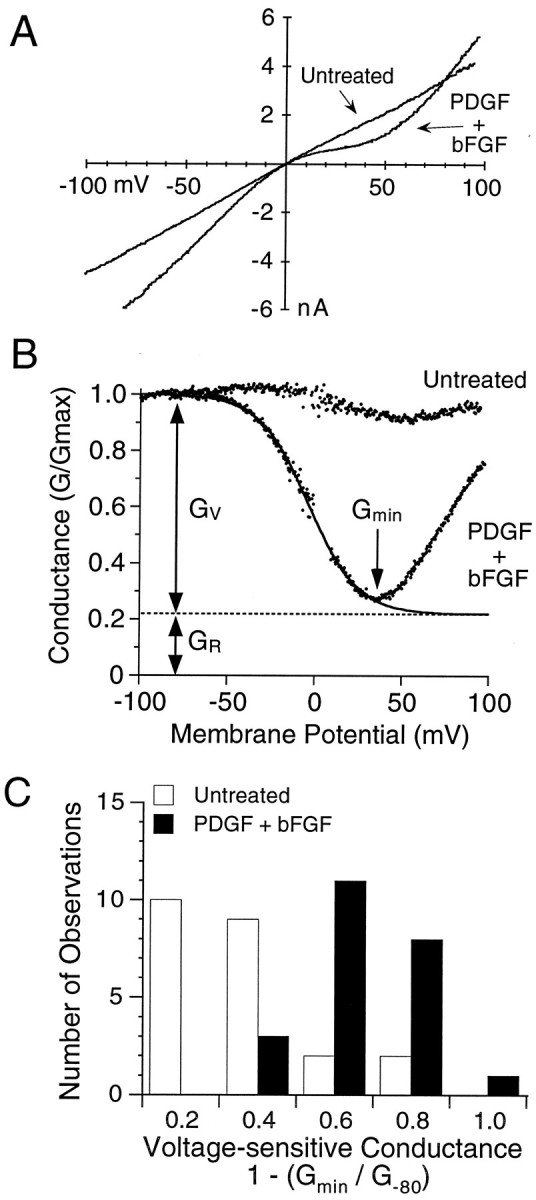
Increased sensitivity to voltage-dependent block by cytoplasmic polyamines in O-2A cells treated with PDGF+bFGF.A, Current–voltage plots of AMPA responses from representative cells in untreated and growth factor-treated cultures showing the enhanced inward rectification between 0 and +50 mV produced by treatment with PDGF+bFGF. B, Conductance–voltage plots from the same pair of cells showing pronounced voltage-dependent block by intracellular polyamines in the cell treated with PDGF+bFGF. The response from the treated cell was well fit by Equation 1 (see Materials and Methods) with KD(0 mV) = 23.4 μm and zδ = 1.79;Gv and Gr, the voltage-dependent and voltage-resistant components of the response to AMPA, were 78 and 22% of total, respectively. C, Histogram of the distribution of the relative amplitude of the voltage-sensitive component of responses to AMPA expressed as 1 − (Gmin/G−80 mV); note that cells treated with PDGF+bFGF showed responses with much greater voltage sensitivity than untreated cells.
To analyze more precisely the effect of growth factor treatment on the expression of voltage-dependent AMPA receptor responses, we determined reversal potentials, and we transformed the I–V data to conductance–voltage plots (Fig. 10B). After being normalized for whole-cell capacitance, conductance values at −80 mV (G−80) were 5.7 ± 0.77 nS/pF (n = 23) versus 1.8 ± 0.45 nS/pF (n = 23) for PDGF+bFGF-treated versus untreated cells, respectively (p < 0.001, one-tailedt test). The proportion of the AMPA-activated conductance-sensitive to voltage-dependent block by polyamines was estimated initially from the relationship 1 − (Gmin/G−80), in whichGmin is the minimum conductance recorded over the membrane potential range +10 to +90 mV (Fig.10B). In PDGF+bFGF-treated cells (n = 23) this ratio averaged 0.56 ± 0.04, whereas in untreated cells the ratio was 0.27 ± 0.05 (n = 23). Analysis of the distribution ofGmin/G−80 values revealed two discrete populations (Fig. 10C), with PDGF+bFGF-treated cells expressing strongly voltage-dependent responses to AMPA more frequently than untreated cells.
By fitting equation 1 (see Materials and Methods) to conductance–voltage plots obtained from cells with well defined voltage-dependent responses to AMPA, we were able to estimateKD(0 mV) and zδ values for block by spermine in both untreated (n = 9) and PDGF+bFGF-treated cells (n = 12). In the 9 of 23 untreated cells selected for this analysis, GV, the voltage-sensitive component of the response to AMPA, was 44 ± 7.8% of GTotal, withKD(0 mV) 48.8 ± 3.7 μm andzδ 1.7 ± 0.16; the remaining 14 of 23 cells showed too little voltage-dependent block for accurate analysis. In the cells treated with PDGF+bFGF and selected for analysis,GV was 78 ± 3.0% ofGTotal, with KD(0 mV) 49.3 ± 4.4 μm and zδ 1.4 ± 0.11; conductance–voltage plots from 11 of 23 growth factor-treated cells were rejected for this analysis, because the large amplitude of AMPA receptor currents saturated the amplifier at −100 mV, preventing accurate analysis of GTotal.
DISCUSSION
The functional implications of GluR diversity in the brain are beginning to be elucidated, but the molecular mechanisms that control GluR expression are not yet understood. The major finding of the present study is that changes in the transcriptional rate of GluR1 triggered by the growth factors PDGF and bFGF in oligodendrocyte progenitor cells are associated with a large increase in agonist-generated responses at AMPA receptors. Furthermore, the formation of new receptor channels functionally detected after growth factor treatment indicated a change in subunit composition from pre-existing receptors.
Previous studies with recombinant GluRs have analyzed whether the functional properties of native glutamate-gated channels are correlated with expression of RNAs for different subunits (Lambolez et al., 1992;Ruano et al., 1995). Geiger et al. (1995) used electrophysiological analysis and single-cell RT-PCR to demonstrate that differential expression of GluR subunit genes accounted for the gating properties and the Ca2+-permeability of GluRs in nine distinct brain cell types. Our analysis in O-2A cells (1) establishes a direct correlation between transcriptional rate of a GluR gene and activity of functional cell membrane channels, (2) reveals a pathway of regulation of GluRs in a well identified population of neural cells, and (3) demonstrates that the genes encoding these receptors are potential targets of growth factors that have widespread distribution in the developing brain.
PDGF and bFGF exert their biological effects on a variety of cell types in the mammalian CNS (for review, see Ross et al., 1986; Deuel et al., 1987; Raff, 1989; Pfeiffer et al., 1993; Temple and Qian, 1995). The effects of the two growth factors on O-2A cell development include stimulation of proliferation and migration and timing of differentiation (Raff et al., 1988; Besnard et al., 1989; Armstrong et al., 1990b; McKinnon et al., 1990; Gard and Pfeiffer, 1993). bFGF upregulates PDGF α-receptor expression in O-2A cells, and the combination of the two growth factors prevents O-2A differentiation (Bogler et al., 1990; McKinnon et al., 1990; Gallo and Armstrong, 1995;Gallo et al., 1996). Our data now indicate that PDGF and bFGF cooperate to upregulate GluR1 gene expression in O-2A cells. The effects of the growth factors on GluR1 gene expression seemed to be specific, because treatment of O-2A cells with B104-CM, shown previously to stimulate cell proliferation and to prevent differentiation to the same extent as treatment with PDGF+bFGF (Gallo and Armstrong, 1995; Gallo et al., 1996), did not affect GluR1 gene expression. The lack of effect of B104-CM on GluR1 RNA levels also indicated that upregulation of GluR1 by PDGF+bFGF was not simply a consequence of increased cell division.
An important aspect of our study is that different molecular approaches consistently demonstrated that PDGF+bFGF triggered GluR1 transcription, with parallel accumulation of GluR1 RNA transcripts within 24 hr. Three days of treatment with PDGF+bFGF produced an even larger increase in GluR1 RNA accumulation, in agreement with a sixfold increase in the levels of GluR1 protein (Fig. 6) and an overall fivefold increase in the density of functional AMPA receptor channels (Figs. 8, 9, 10). At present, additional post-translational effects of PDGF and bFGF cannot be excluded. However, our results indicate that the increase in AMPA receptor density triggered by the growth factors is primarily attributable to changes in GluR1 transcription rate.
Consistent with Western blot analysis, electrophysiological experiments demonstrated a fivefold increase in AMPA receptor current density after treatment with PDGF+bFGF. However, attributing this change entirely to an increase in receptor number is complicated by the effects of heteromerization of AMPA receptor subunits on single-channel conductance. For example, for heteromeric receptors assembled from GluR4 and GluR2, mean values for the weighted single-channel conductance can vary fivefold and are determined by subunit composition, RNA editing, and alternative splicing (Swanson et al., 1997). As a result, it is not possible to extrapolate from the effects of growth factors on whole-cell current density to changes in receptor density, except to note that the higher single-channel conductance of AMPA receptors lacking GluR2 could increase the amplitude of whole-cell responses independent of changes in channel density.
Electrophysiological data consistent with a selective increase in expression of GluR1, but not GluR2, are provided by the increase in rectification of AMPA receptor responses in cells treated with PDGF+bFGF. This almost certainly reflects a higher sensitivity to channel block by cytoplasmic polyamines (Bowie and Mayer, 1995; Donevan, and Rogawski, 1995; Kamboj et al., 1995; Koh et al., 1995). In the present experiments KD(0 mV) values for spermine block for both untreated and PDGF+bFGF-treated O-2A cells were ∼30 times higher than the value reported for homomeric GluR1 expressed in HEK293 cells (Bowie and Mayer, 1995). At present, there is no information available on how AMPA receptor affinity for spermine is regulated by heteromerization of GluR2 with other subunits, but given that edited glutamate receptor subunits are resistant to polyamine block, it is likely that the reduced sensitivity to spermine recorded in the present experiments reflects formation of GluR1/GluR2 heteromers with polyamine sensitivity lower than that for homomeric GluR1. The finding that KD(0 mV) values for spermine block obtained for untreated O-2A cells and PDGF+bFGF-treated cells were similar is possibly misleading, because 14 of 23 untreated cells were excluded because it was not possible to fit Boltzmann functions to conductance–voltage plots accurately for cells with weak inward rectification; thus, we cannot exclude the possibility that the affinity for spermine could be even lower for AMPA receptors in untreated O-2A cells with weakly voltage-sensitive AMPA receptor responses.
bFGF treatment of O-2A cells for 3 d increased the steady-state RNA levels of GluR1, GluR3, KA1, KA2, and GluR7 (see Figs. 2, 3). We also had reported previously a substantial induction of GluR4c RNA expression by bFGF after 5 d of treatment (Gallo et al., 1994); however, under our present treatment paradigm, corresponding changes in protein levels were not observed (see Fig. 6B). It is likely that very small changes in GluR4c after 3 d of treatment with the growth factor are undetectable by Western blotting. In our experiments on O-2A cells, bFGF affected both GluR1 RNA and protein levels, at variance with the results of Cheng et al. (1995), who reported an upregulation of GluR1 protein in cultured hippocampal neurons by bFGF in the absence of observable changes in RNA levels. This difference is likely to be attributable to the cell types analyzed, but the findings in O-2A cells and hippocampal neurons provide evidence that bFGF may regulate GluR expression in a variety of neural cell types via distinct mechanisms.
Basic fibroblast growth factor influences cortical neuroectoderm cell division and differentiation (Ghosh and Greenberg, 1995; Vicario-Abejon et al., 1995) and stimulates proliferation of committed progenitor cells derived from embryonic striatum (Vescovi et al., 1993) and olfactory epithelium (DeHamer et al., 1994). PDGF receptors are expressed in both neuronal and glial cells of virtually all regions of the brain (Smits et al., 1991; Yeh et al., 1993), and PDGF exerts neurotrophic effects on dopaminergic and GABAergic neurons (Nikkah et al., 1993; Smits et al., 1993). It is likely, therefore, that these growth factors or other factors that activate the same intracellular pathways may cooperate to regulate GluR expression also in neurons.
The intracellular molecular events that underlie the stimulation of GluR1 gene transcription by PDGF and bFGF are still unknown. Both PDGF and bFGF receptors are coupled to multiple signal transduction pathways that may lead to gene transcription (Hill and Treisman, 1995). One of these pathways involves Ras, activation and nuclear translocation of the MAP kinase to the cell nucleus with consequent phosphorylation of nuclear transcription factors (Hill and Treisman, 1995). We have found recently that PDGF and bFGF activate MAP kinase in O-2A cells (Pende and Gallo, unpublished data).
GluR1 transcripts are expressed abundantly in two white matter areas during development, the corpus callosum and the optic nerve (Jensen and Chiu, 1993; Matute and Miledi, 1993), and functional AMPA receptors presumably containing GluR1 have been characterized in oligodendrocyte lineage cells of these tissues (Barres et al., 1990; Wyllie et al., 1991; Fulton et al., 1992). Preliminary in situhybridization experiments from our laboratory indicate that the relative levels of GluR1 transcripts in cerebellar oligodendrocyte progenitor cells in vivo, identified as PDGF α-receptor-positive cells, are similar to those for GluR2 and GluR3 (W. P. Hayes, L.-J. Chew, V. Gallo, unpublished observations). These findings are strikingly different from our results in cultured O-2A cells, which express much lower levels of GluR1 RNAs relative to the other AMPA receptor subunits (see Fig. 2), and suggest that the GluR1 gene may be under the transcriptional control of factors (e.g., PDGF and bFGF) that are present in the intact tissue, but not in our basal culture medium. Thus, the low GluR1 content in primary O-2A progenitors offers a convenient model system in which potential factors that may act to maintain GluR1 levels in vivo can be identifiedin vitro.
In a previous study, we have demonstrated that GluR agonists regulate O-2A cell proliferation and lineage progression by activating AMPA receptors (Gallo et al., 1996). The experiments described in the present study indicate that the growth factors PDGF and bFGF can modulate O-2A responsiveness to glutamate by upregulating AMPA receptor density. It can be hypothesized that the proliferative potential of oligodendrocyte progenitors results from a complex relationship involving levels of extracellular mitogenic factors (e.g., PDGF and bFGF), levels of extracellular glutamate, and the density of cell surface GluRs.
Footnotes
We thank Peter Seeburg, Bernard Bettler, and Stephen Heinemann for providing cDNA clones; Robert Wenthold for the generous gift of the anti-GluR1 and anti-GluR2, -3, and -4c antibodies; Giulio Levi for the LB1 antibody; Jean Claude Louis for the CG-4 cell line; Stuart Cull-Candy for sharing results before publication; and Chris McBain and Mario Pende for discussion and for critically reading this manuscript.
Correspondence should be addressed to Dr. Vittorio Gallo, Laboratory of Cellular and Molecular Neurophysiology, National Institute of Child Health and Human Development, National Institutes of Health, Building 49, Room 5A78, Bethesda, MD 20892.
Dr. Wright’s present address: Biological Carcinogenesis and Development Program, National Cancer Institute, Frederick Cancer Research and Development Center, Frederick, MD 21702-1201.
REFERENCES
- 1.Armstrong RC, Friedrich VL, Holmes KV, Jr, Dubois-Dalcq M. In vitro analysis of the oligodendrocyte lineage in mice during demyelination and remyelination. J Cell Biol. 1990a;111:1183–1195. doi: 10.1083/jcb.111.3.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong RC, Harvath L, Dubois-Dalcq M. Type 1 astrocytes and oligodendrocyte-type 2 astrocyte glial progenitors migrate toward distinct molecules. J Neurosci Res. 1990b;27:400–407. doi: 10.1002/jnr.490270319. [DOI] [PubMed] [Google Scholar]
- 3.Bahn S, Volk B, Wisden W. Kainate receptor gene expression in the developing brain. J Neurosci. 1994;14:5525–5547. doi: 10.1523/JNEUROSCI.14-09-05525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barres BA, Koroshetz WJ, Swartz KJ, Chun LLY, Corey DP. Ion channel expression by white matter glia: the O-2A glial progenitor cell. Neuron. 1990;4:507–524. doi: 10.1016/0896-6273(90)90109-s. [DOI] [PubMed] [Google Scholar]
- 5.Besnard F, Perraud F, Sensenbrenner M, Labourdette G. Effects of acidic and basic fibroblast growth factors on proliferation and maturation of cultured rat oligodendrocytes. Int J Dev Neurosci. 1989;7:401–409. doi: 10.1016/0736-5748(89)90061-0. [DOI] [PubMed] [Google Scholar]
- 6.Bogler O, Wren D, Barnett SC, Land H, Noble M. Cooperation between two growth factors promotes extended self-renewal and inhibits differentiation of oligodendrocyte-type-2 astrocyte (O-2A) progenitor cells. Proc Natl Acad Sci USA. 1990;87:6368–6372. doi: 10.1073/pnas.87.16.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulter J, Hollmann M, O’Shea-Greenfield A, Hartley M, Deneris E, Maron C, Heinemann S. Molecular cloning and functional expression of glutamate receptor subunit genes. Science. 1990;249:1033–1037. doi: 10.1126/science.2168579. [DOI] [PubMed] [Google Scholar]
- 8.Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 9.Cheng B, Furukawa K, O’Keefe JA, Goodman Y, Kihiko M, Fabian T, Mattson MP. Basic fibroblast growth factor selectively increases AMPA-receptor subunit GluR1 protein level and differentially modulates Ca2+ responses to AMPA and NMDA in hippocampal neurons. J Neurochem. 1995;6:2525–2536. doi: 10.1046/j.1471-4159.1995.65062525.x. [DOI] [PubMed] [Google Scholar]
- 10.Chirgwin JM, Przybyla AE, Macdonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of mRNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Curtis R, Cohen J, Fok-Seang J, Hanley MR, Gregson NA, Reynolds R, Wilkin GP. Development of macroglial cells in rat cerebellum. I. Use of antibodies to follow early in vivo development and migration of oligodendrocytes. J Neurocytol. 1988;17:43–54. doi: 10.1007/BF01735376. [DOI] [PubMed] [Google Scholar]
- 13.DeHamer MK, Guevara JL, Hannon K, Olwin BB, Calof AL. Genesis of olfactory receptor neurons in vitro: regulation of progenitor cell divisions by fibroblast growth factors. Neuron. 1994;13:1083–1097. doi: 10.1016/0896-6273(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 14.Deuel TF. Polypeptide growth factors: roles in normal and abnormal cell growth. Annu Rev Cell Biol. 1987;3:443–492. doi: 10.1146/annurev.cb.03.110187.002303. [DOI] [PubMed] [Google Scholar]
- 15.Donevan SD, Rogawski MA. Intracellular polyamines mediate inward rectification of Ca2+-permeable α-amino-3-hydroxy-5-methyl-4-isoxazole-proprionic acid receptors. Proc Natl Acad Sci USA. 1995;92:9298–9302. doi: 10.1073/pnas.92.20.9298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durand GM, Zukin RS. Developmental regulation of mRNAs encoding rat brain kainate/AMPA receptors: a Northern analysis study. J Neurochem. 1993;61:2239–2246. doi: 10.1111/j.1471-4159.1993.tb07465.x. [DOI] [PubMed] [Google Scholar]
- 17.Fulton BP, Burne JF, Raff MC. Visualization of O-2A progenitor cells in developing and adult rat optic nerve by quisqualate-stimulated cobalt uptake. J Neurosci. 1992;12:4816–4833. doi: 10.1523/JNEUROSCI.12-12-04816.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo V, Armstrong RC. Developmental and growth factor-induced regulation of nestin in oligodendrocyte lineage cells. J Neurosci. 1995;15:394–406. doi: 10.1523/JNEUROSCI.15-01-00394.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallo V, Russell JT. Excitatory amino acid receptors in glia: different subtypes for distinct functions? J Neurosci Res. 1995;42:1–8. doi: 10.1002/jnr.490420102. [DOI] [PubMed] [Google Scholar]
- 20.Gallo V, Upson JM, Hayes WP, Vyklicky LJ, Winters CA, Buonanno A. Molecular cloning and developmental analysis of a new glutamate subunit isoform in cerebellum. J Neurosci. 1992;12:1010–1023. doi: 10.1523/JNEUROSCI.12-03-01010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallo V, Wright PW, McKinnon RD. Expression and regulation of a glutamate receptor subunit by bFGF in oligodendrocyte progenitors. Glia. 1994;10:149–153. doi: 10.1002/glia.440100209. [DOI] [PubMed] [Google Scholar]
- 22.Gallo V, Pende M, Scherer S, Molne M, Wright P. Expression and regulation of kainate and AMPA receptors in uncommitted and committed neural progenitors. Neurochem Res. 1995;20:549–560. doi: 10.1007/BF01694536. [DOI] [PubMed] [Google Scholar]
- 23.Gallo V, Zhou JM, McBain CJ, Wright PW, Knutson PL, Armstrong RC. Oligodendrocyte progenitor cell proliferation and lineage progression are regulated by glutamate receptor-mediated K+ channel block. J Neurosci. 1996;16:2659–2670. doi: 10.1523/JNEUROSCI.16-08-02659.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gard AL, Pfeiffer SE. Glial cell mitogens bFGF and PDGF differentially regulate development of O4+GalC− oligodendrocyte progenitors. Dev Biol. 1993;159:618–630. doi: 10.1006/dbio.1993.1269. [DOI] [PubMed] [Google Scholar]
- 25.Geiger JRP, Meicher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh A, Greenberg M. Distinct roles for bFGF and NT-3 in the regulation of cortical neurogenesis. Neuron. 1995;15:89–103. doi: 10.1016/0896-6273(95)90067-5. [DOI] [PubMed] [Google Scholar]
- 27.Hill SH, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 28.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 29.Hollmann M, O’Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson M. Developmental neurobiology. Plenum; New York: 1991. [Google Scholar]
- 31.Jensen AM, Chiu SY. Expression of glutamate receptor genes in white matter: developing and adult optic nerve. J Neurosci. 1993;13:1664–1675. doi: 10.1523/JNEUROSCI.13-04-01664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamboj SK, Swanson GT, Cull-Candy SG. Intracellular spermine confers rectification on rat calcium-permeable AMPA and kainate receptors. J Physiol (Lond) 1995;486:297–303. doi: 10.1113/jphysiol.1995.sp020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh DS, Burnashev N, Jonas P. Block of native Ca2+-permeable AMPA receptors in rat brain by intracellular polyamines generates double rectification. J Physiol (Lond) 1995;486:305–312. doi: 10.1113/jphysiol.1995.sp020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konig N, Drian M-J, Pariat M, Manzoni O, Sladeczek F. Early functional glutamate receptors in acutely dissociated embryonic raphe cells. NeuroReport. 1992;3:738–740. doi: 10.1097/00001756-199209000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Lambolez B, Audinat E, Bochet P, Crepel F, Rossier J. AMPA receptor subunit expressed by single Purkinje cell. Neuron. 1992;9:247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 36.Levi G, Gallo V, Ciotti MT. Bipotential precursors of putative fibrous astrocytes and oligodendrocytes in rat cerebellar cultures express distinct surface features and “neuron-like” GABA transport. Proc Natl Acad Sci USA. 1986;83:1504–1508. doi: 10.1073/pnas.83.5.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LoTurco JJ, Owens DF, Heath MJS, Davis MBE, Kriegstein AR. GABA and glutamate depolarize cortical progenitor cells and inhibit DNA synthesis. Neuron. 1995;15:1287–1298. doi: 10.1016/0896-6273(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 38.Louis JC, Magal E, Muir D, Manthorpe M, Varon S. CG-4, a new bipotential cell line from rat brain, is capable of differentiating in vitro into either mature oligodendrocytes or type-2 astrocytes. J Neurosci Res. 1992a;31:193–204. doi: 10.1002/jnr.490310125. [DOI] [PubMed] [Google Scholar]
- 39.Louis JC, Muir D, Varon S. Autocrine inhibition of mitotic activity in cultured oligodendrocyte-type-2 astrocyte (O-2A) precursor cells. Glia. 1992b;6:30–38. doi: 10.1002/glia.440060105. [DOI] [PubMed] [Google Scholar]
- 40.Marty A, Neher E. Tight-seal whole-cell recording. In: Sakmann B, Neher E, editors. Single-channel recording. Plenum; New York: 1995. pp. 31–52. [Google Scholar]
- 41.Matute C, Miledi R. Neurotransmitter receptors and voltage-dependent Ca2+ channels encoded by mRNA from the adult corpus callosum. Proc Natl Acad Sci USA. 1993;90:3270–3274. doi: 10.1073/pnas.90.8.3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mayer ML, Westbrook G. The physiology of excitatory amino acids in the vertebrate central nervous system. Prog Neurobiol. 1987;28:197–276. doi: 10.1016/0301-0082(87)90011-6. [DOI] [PubMed] [Google Scholar]
- 43.McBain CJ, Mayer ML. N-Methyl-d-aspartic acid receptor structure and function. Physiol Rev. 1994;74:723–760. doi: 10.1152/physrev.1994.74.3.723. [DOI] [PubMed] [Google Scholar]
- 44.McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglia cell cultures from rat cerebral tissue. J Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKinnon RD, Matsui T, Dubois-Dalcq M, Aaronson SA. FGF modulates the PDGF-driven pathway of oligodendrocyte development. Neuron. 1990;5:603–614. doi: 10.1016/0896-6273(90)90215-2. [DOI] [PubMed] [Google Scholar]
- 46.Monyer H, Seeburg PH, Wisden W. Glutamate-operated channels, developmentally early and mature forms arise by alternative splicing. Neuron. 1991;6:799–810. doi: 10.1016/0896-6273(91)90176-z. [DOI] [PubMed] [Google Scholar]
- 47.Murphy D, Carter DA. Vasopressin gene expression in the rodent hypothalamus: transcriptional and post-transcriptional responses to physiological stimulation. Mol Endocrinol. 1990;4:1051–1059. doi: 10.1210/mend-4-7-1051. [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- 49.Nikkah G, Odin P, Smits A, Tingstrom A, Othberg A, Brundin P, Funa K, Lindvall O. Platelet-derived growth factor promotes survival of rat and human mesencephalic dopaminergic neurons in culture. Exp Brain Res. 1993;92:516–523. doi: 10.1007/BF00229041. [DOI] [PubMed] [Google Scholar]
- 50.Patneau DK, Wright PW, Winters C, Mayer ML, Gallo V. Glial cells of the oligodendrocyte lineage express both kainate- and AMPA-preferring subtypes of glutamate receptor. Neuron. 1994;12:357–371. doi: 10.1016/0896-6273(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 51.Pellegrini-Giampietro DE, Bennett MVL, Zukin RS. Differential expression of three glutamate receptor genes in developing brain: an in situ hybridization study. Proc Natl Acad Sci USA. 1991;88:4157–4161. doi: 10.1073/pnas.88.10.4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeiffer SE, Warrington AE, Bansal R. The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993;3:191–197. doi: 10.1016/0962-8924(93)90213-k. [DOI] [PubMed] [Google Scholar]
- 53.Puchalski RB, Louis JC, Brose N, Traynelis SF, Egebjerg J, Kukekov V, Wenthold RJ, Rogers SW, Lin F, Moran T, Morrison JH, Heinemann SF. Selective RNA editing and subunit assembly of native glutamate receptors. Neuron. 1994;13:131–146. doi: 10.1016/0896-6273(94)90464-2. [DOI] [PubMed] [Google Scholar]
- 54.Raff MC. Glial cell diversification in the rat optic nerve. Science. 1989;243:1509–1524. doi: 10.1126/science.2648568. [DOI] [PubMed] [Google Scholar]
- 55.Raff MC, Lillien LE, Richardson WD, Burne FJ, Noble MD. Platelet-derived growth factor from astrocytes drives the clock that times oligodendrocyte development in culture. Nature. 1988;333:562–565. doi: 10.1038/333562a0. [DOI] [PubMed] [Google Scholar]
- 56.Ross R, Raines EW, Bowen-Pope DF. The biology of platelet-derived growth factor. Cell. 1986;46:155–169. doi: 10.1016/0092-8674(86)90733-6. [DOI] [PubMed] [Google Scholar]
- 57.Ruano D, Lambolez B, Rossier J, Paternain AV, Lerma J. Kainate receptor subunits expressed in single cultured hippocampal neurons: molecular and functional variants by RNA editing. Neuron. 1995;14:1009–1017. doi: 10.1016/0896-6273(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd Ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 59.Seeburg PH. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993;9:359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- 60.Smits A, Kato M, Westermark B, Nister M, Heldin C-H, Funa K. Neurotrophic activity of platelet-derived growth factor (PDGF): rat neuronal cells possess functional PDGF β-type receptors and respond to PDGF. Proc Natl Acad Sci USA. 1991;88:8159–8163. doi: 10.1073/pnas.88.18.8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smits A, Ballagi AE, Funa K. PDGF-BB exerts trophic activity on cultured GABA interneurons from the newborn rat cerebellum. Eur J Neurosci. 1993;5:986–994. doi: 10.1111/j.1460-9568.1993.tb00950.x. [DOI] [PubMed] [Google Scholar]
- 62.Steinhauser C, Gallo V. News on glutamate receptors in glial cells. Trends Neurosci. 1996;19:339–345. doi: 10.1016/0166-2236(96)10043-6. [DOI] [PubMed] [Google Scholar]
- 63.Swanson GT, Kamboj SK, Cull-Candy SG. Single-channel properties of recombinant AMPA receptors depend on RNA editing, splice variation, and subunit composition. J Neurosci. 1997;17:58–69. doi: 10.1523/JNEUROSCI.17-01-00058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Temple S, Qian X. bFGF, neurotrophins, and the control of cortical neurogenesis. Neuron. 1995;15:249–252. doi: 10.1016/0896-6273(95)90030-6. [DOI] [PubMed] [Google Scholar]
- 65.Vescovi AL, Reynolds BA, Fraser DD, Weiss S. bFGF regulates the proliferative fate of unipotent (neuronal) and bipotent (neuronal/astroglial) EGF-generated CNS progenitor cells. Neuron. 1993;11:951–966. doi: 10.1016/0896-6273(93)90124-a. [DOI] [PubMed] [Google Scholar]
- 66.Vicario-Abejon C, Johe KK, Hazel TG, Collazo D, McKay RDG. Functions of basic fibroblast growth factor and neurotrophins in the differentiation of hippocampal neurons. Neuron. 1995;15:105–114. doi: 10.1016/0896-6273(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 67.Vyklicky L, Benveniste M, Mayer ML. Modulation of N-methyl-d-aspartic acid receptor desensitization by glycine in mouse cultured hippocampal neurones. J Physiol (Lond) 1990;428:313–331. doi: 10.1113/jphysiol.1990.sp018214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walton MK, Schaffner AE, Barker JL. Sodium channels, GABAA receptors, and glutamate receptors develop sequentially on embryonic rat spinal cord cells. J Neurosci. 1993;13:2068–2084. doi: 10.1523/JNEUROSCI.13-05-02068.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woodhull AM. Ionic blockage of sodium channels in nerve. J Gen Physiol. 1973;61:687–708. doi: 10.1085/jgp.61.6.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyllie DJA, Mathie A, Symonds CJ, Cull-Candy SG. Activation of glutamate receptors and glutamate uptake in identified macroglial cells in rat cerebellar cultures. J Physiol (Lond) 1991;432:235–258. doi: 10.1113/jphysiol.1991.sp018383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeh H-J, Silos-Santiago I, Wang Y-X, George RJ, Snider WD, Deuel TF. Developmental expression of the platelet-derived growth factor α-receptor gene in mammalian central nervous system. Proc Natl Acad Sci USA. 1993;90:1952–1956. doi: 10.1073/pnas.90.5.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]