Abstract
The LIM domain is a zinc-binding amino acid motif that characterizes various proteins which function in protein–protein interactions and transcriptional regulation. Expression patterns of several LIM protein genes are compatible with roles in vertebrate CNS development, but little is known about the expression, regulation, or function of LIM proteins in the mature CNS. Lmo1, Lmo2,and Lmo3 are LIM-only genes originally identified as putative oncogenes that have been implicated in the control of cell differentiation and are active during CNS development. Using in situ hybridization for mRNA and immunohistochemical detection of reporter protein expression in transgenic mice, we found thatLmo1, Lmo2, and Lmo3 show individually unique but partially overlapping patterns of expression in several regions of the adult mouse forebrain, including hippocampus, caudate putamen, medial habenula, thalamus, amygdala, olfactory bulb, hypothalamus, and cerebral cortex. In the hippocampal formation,Lmo1, Lmo2, and Lmo3 show different combinatorial patterns of expression levels in CA pyramidal and dentate granule neurons, and Lmo1 is present in topographically restricted subpopulations of astrocytes. Kainic acid-induced limbic seizures differentially regulated Lmo1, Lmo2, andLmo3 mRNA levels in hippocampal pyramidal and granule neurons, such that Lmo1 mRNA increased, whereasLmo2 and Lmo3 mRNAs decreased significantly, with maximal changes at 6 hr after seizure onset and return to baseline by 24 hr. These findings show that Lmo1, Lmo2, and Lmo3 continue to be expressed in the adult mammalian CNS in a cell type-specific manner, are differentially regulated by neuronal activity, and may thus be involved in cell phenotype-specific regulatory functions.
Keywords: LIM-only proteins, transcriptional regulation, limbic seizures, gene expression, cell phenotype, hippocampus, brain
LIM proteins are a diverse group of transcription factors, proto-oncogene products, cytoskeletal proteins, and constituents of adhesion plaques that contain one or more cysteine-rich amino acid sequence motifs known as LIM domains (Way and Chalfie, 1988;Freyd et al., 1990; Karlsson et al., 1990; Sanchez-Garcia and Rabbitts, 1994). More than 40 different eukaryote LIM proteins have been identified in various tissues across a wide range of species, and the LIM domain sequence is highly conserved among these (Sanchez-Garcia and Rabbitts, 1994; Dawid et al., 1995). The LIM domain binds zinc ions and shows structural similarities to DNA-binding zinc finger proteins (Michelsen et al., 1993; Archer et al., 1994); however, most evidence favors a role for the LIM domain in protein–protein interactions (Rabbitts and Boehm, 1990; Schmeichel and Beckerle, 1994).
Several classes of LIM proteins have been identified in which one or more LIM motifs are present either alone (LIM-only proteins) or in association with recognized protein regions that may confer specific functions, such as DNA-binding homeodomain regions, cytoskeletal proteins, protein kinase domains, and regions that bind other proteins (Freyd et al., 1990; Crawford et al., 1994; Sanchez-Garcia and Rabbitts, 1994; Dawid et al., 1995; Taira et al., 1995; Weiskirchen et al., 1995). The functions of LIM-only proteins are less well understood. Homology between LIM domains suggests that they may be involved in LIM–LIM interactions (Rabbitts and Boehm, 1990) through which they may indirectly affect transcription (Sanchez-Garcia et al., 1993), as suggested by their putative roles in the regulation of cell differentiation or tumorigenesis (Boehm et al., 1991a; Warren et al., 1994).
Lmo1, Lmo2, and Lmo3 comprise a family of genes encoding LIM-only proteins (Foroni et al., 1992; Sanchez-Garcia and Rabbitts, 1994). Lmo1 and Lmo2 (originallyrbtn1, -2 or tgt1, -2) were identified as highly expressed transcription units near chromosomal translocation breakpoints associated with T-cell tumors (Boehm et al., 1988; McGuire et al., 1989; Boehm et al., 1991a). The LMO1 and LMO2 proteins have 50% amino acid identity, and LMO3 was identified subsequently on the basis of sequence homology (Foroni et al., 1992). Lmo1, Lmo2, and Lmo3 are all highly expressed in the developing mouse CNS (Boehm et al., 1991b; Foroni et al., 1992).Lmo1 has an unusual structure, with two promoter regions designated 1 and 1a (Boehm et al., 1990, 1991b). Transgenic mice expressing the bacterial lacZ gene fromLmo1-promoter 1 show β-galactosidase (β-gal) activity in cells in various regions of the developing CNS (Greenberg et al., 1990).
Considerable evidence now indicates that a number of LIM protein genes, including LIM-only genes, are active during development of the vertebrate CNS (Foroni et al., 1992; Tsuchida et al., 1994; Lumsden, 1995; Shawlot and Behringer, 1995). Less is known about the extent of expression, regulation, or function of LIM proteins in the mature vertebrate CNS. In this study we have compared the regional distribution and cellular patterns of Lmo1, Lmo2, andLmo3 expression in the hippocampus and other forebrain regions of adult mice by in situ hybridization using radiolabeled antisense oligonucleotide probes. To investigate whether expression of these LIM protein genes might be regulated by changes in neuronal activity, we examined the effects of kainic acid (KA)-induced limbic seizures (Ben-Ari, 1985) on relative levels of their mRNAs in the hippocampus, a site of considerable plasticity of gene expression in response to seizures in adult rodents (Bendotti et al., 1994; Morgan et al., 1987; Gall and Isackson, 1989). In addition, the hippocampal distribution of Lmo1 mRNA was compared with that of staining for β-gal in adult mice expressing the bacterial lacZ gene from the Lmo1-promoter 1 (Greenberg et al., 1990).
MATERIALS AND METHODS
Animals. Wild-type BKTO mice weighing 25–30 gm (Bantin-Kingman, Freemont, CA) were used for evaluation by in situ hybridization with and without KA-induced limbic seizures. The generation of transgenic mice expressing the bacteriallacZ gene from the Lmo1-promoter 1 has been described (Greenberg et al., 1990); these mice were used for histochemical evaluation with and without KA-induced limbic seizures. Animals were housed in small groups, maintained in a constant 12 hr light/dark cycle with controlled temperature and humidity, and allowed free access to food and water. All experiments were conducted in accordance with the United Kingdom Animal Scientific Procedures Act, 1986.
Drug administration. Animals were placed in the experimentation room 24 hr before drug injection to allow them to acclimatize. KA (30 mg/kg in 0.9% NaCl) (Sigma, St. Louis, MO) or vehicle (0.9% NaCl) were administered intraperitoneally. Animals were kept under observation for 1 hr and then returned to the home cage. Wild-type animals analyzed by in situ hybridization were killed by decapitation at set time intervals. The brains were rapidly frozen in n-pentane at −30°C and stored at −70°C before sectioning. Transgenic animals analyzed by histochemistry were killed by terminal barbiturate anesthesia followed by transcardiac perfusion with fixative, as described below.
In situ hybridization. Sections (10 μm, three sections/slide) were cut at −20°C using a cryostat (Bright Instruments) and thaw-mounted onto sterile poly-l-lysine-coated slides. Sections were air-dried and fixed using 4% paraformaldehyde in PBS for 5 min, rinsed twice in 1× PBS for 1 min, dehydrated in 70% ethanol for 5 min, and then stored in 95% ethanol at 4°C until use. Slides containing control and experimental sections were hybridized overnight in hybridization buffer, and [35S]dATP-labeled probes were diluted to a concentration of 3000 cpm/ml hybridization buffer. Sections were covered with parafilm and hybridized at 42°C in a humid atmosphere. Excess unbound probe was removed using the following stringency washes: 1× standard saline citrate buffer (SSC) at room temperature for 30 min and then 1× SSC at 55°C for 30 min. Slides were finally rinsed in 1× SSC, dehydrated in ethanol, and air-dried. Sections from control and experimental animals were apposed to the same sheet of Biomax MS film for a period of time before the film was developed under standard conditions. Nonspecific binding was determined using 100-fold excess of unlabeled probe. The length of time that sections were apposed to film was dependent on the mRNA under study, but once determined this time was kept constant for that particular probe. Care was taken to standardize procedures for control and experimental section at all stages. A selection of slides were coated with emulsion (LM1; Amersham, Arlington Heights, IL), and after 6 weeks in a light-proof box were developed, stained, and counterstained with hematoxylin and eosin for microscopic examination.
Northern blot analysis. Wild-type animals for Northern analysis were killed by decapitation, and RNA was extracted from mouse brain areas pooled from five animals as described (Sambrooke et al., 1989). Brains were homogenized (Polytron homogenizer) in guanidium thiocyanate buffer, and the total RNA was pelleted by centrifugation on a cesium chloride cushion at 32,000 rpm for 24 hr at 20°C (Beckman L8–70M ultracentrifuge with SW41Ti swinging-bucket rotor). After resuspension in Tris-EDTA buffer containing 0.1% SDS, the RNA was reprecipitated in 100% ethanol and sodium acetate at 4°C overnight and collected by centrifugation at 12,000 × g at 4°C for 10 min. The RNA was diluted to 20 μg/4.5 μl in diethyl pyrocarbonate-treated water, and then 20 μg of RNA was loaded per lane. This was separated using a denaturing 1% agarose/formaldehyde gel (Sambrooke et al., 1989) in 1× 3-[N-morpholino]propanesulfonic acid buffer along with standard RNA markers. Ethidium bromide was added to the gel to visualize the RNA after electrophoresis. The samples were electrophoresed at 60 V for 2.5 hr (Bio-Rad submarine gel electrophoresis tank; Bio-Rad, Hercules, CA), and the resulting gel was photographed under ultraviolet light against a fluorescent ruler (Polaroid Land camera and Polaroid type 57 film). The separated RNA was transferred overnight from the gel onto a nitrocellulose membrane by capillary elution, using 10× SSC buffer. The membrane was baked under vacuum for 2 hr at 80°C to fix the RNA to the membrane. After membranes were prehybridized in hybridization buffer overnight (Hybaid mini-hybridization oven), [32P]-labeled oligonucleotide probe was added at a concentration of 3 × 106 cpm/ml hybridization buffer and hybridized at 42°C overnight. Membranes were washed in 1× standard sodium chloride EDTA (SSPE)/0.1% SDS for 30 min at room temperature, followed by 1× SSPE/0.1% SDS for 30 min at 55°C. After they were wrapped in clingfilm, membranes were exposed to x-ray film (Kodak X-OMAT AR) at −70°C overnight. The size of the transcripts was calculated by reference to standard RNA markers.
Oligonucleotide probes. Oligonucleotide probes were made on an Applied Biosystems (Foster City, CA) synthesizer. After deprotection at 55°C for 16 hr, the probes were dried in a Speed Vac centrifuge, resuspended, purified, and dissolved in sterile water to give a stock solution of 20 ng/ml. They were labeled at the 3′ end using terminal deoxynucleotidyl transferase (New England Nuclear) and [35S]dATP (1000 Ci/mmol) or [32P]dATP (300Ci/mmol). The radiolabeled probe was purified using Sephadex columns (Biospin6), and dithiothreitol was added to a final concentration of 50 mm for [35S]dATP-labeled probes. Sequences of the antisense oligonucleotide probes used in this study were selected from nonoverlapping regions of the Lmo1, Lmo2, andLmo3 genes (Boehm et al., 1991a; Foroni et al., 1992). The following antisense oligonucleotide sequences (kindly provided by Dr. T. H. Rabbitts, Cambridge) were used: Lmo1, 5′-GCA GAC GGA CAG ATG GAC CTG GAG GCC AGA TGG TGG GCG TTA CTG-3′, complimentary to nucleotides 586–631; Lmo2, 5′-GAC TCT GGG CTA GAT GAT CCC ATT GAT CTT GGT CCA CTC GTA GAT-3′, complimentary to nucleotides 810–855; and Lmo3, 5′-ATG TAG TGC TTT GCA TTG TTG GGG AGA CGC TGC TGC CCC CTC ACC-3′, complimentary to nucleotides 705–750.
Analysis of autoradiographical data. Analysis of in situ hybridization autoradiograms was carried out using an MCID image analyzer (model M4) to measure relative optical density (ROD) values. Measurements were taken within the linear range of optical density levels, and in situ hybridization results are expressed in arbitrary units (ROD values ×100). Measurements were taken from at least five animals with three to six sections measured per animal, and the measurements from each animal were processed independently on the MCID. The final data were statistically analyzed using the nonparametric Kruskal–Wallis test for multiple measures, followed by Dunn’s post test for post hoc pair-wise comparisons (GraphPad Software). p < 0.05 was taken as the minimum level of statistical significance.
Histochemical analysis. Perfusion-fixed transgenic brains were allowed to sink in 25% buffered sucrose, and frozen sections were prepared at 40 μm. For routine immunohistochemical detection of β-gal, the fixative used was 4% paraformaldehyde. Free-floating tissue sections were incubated in a polyclonal rabbit antiserum against β-gal (Cappell, West Chester, PA) followed by the avidin–biotin–peroxidase method (Vectastain ABC kit; Vector Laboratories, Burlingame, CA). Immunoreactivity was visualized using diaminobenzidine (Sigma) as the chromagen. For double-labeling immunohistochemistry of β-gal (brown) and glial fibrillary acidic protein (GFAP) (blue), β-gal was stained first as just described, using diaminobenzidine as the brown chromogen. Sections were then washed briefly and stained using the same protocol with a primary rabbit antiserum against GFAP (Dako, Carpinteria, CA), and Vector-SG (Vector) as a blue chromogen. Timm staining of zinc-containing fibers was conducted as described (Geneser et al., 1993). For double Timm/immunohistochemical staining, the fixative used was that for Timm staining, which gave somewhat poorer but identifiable immunohistochemical staining for β-gal. After fixation, the sections were immunohistochemically stained first, followed by the Timm stain procedure. All stained sections were mounted on gelatin-subbed slides, air-dried, cleared in xylene, and coverslipped in DePe-X.
RESULTS
Oligonucleotide probe specificity
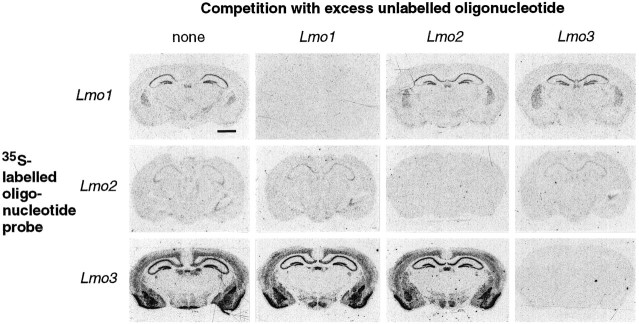
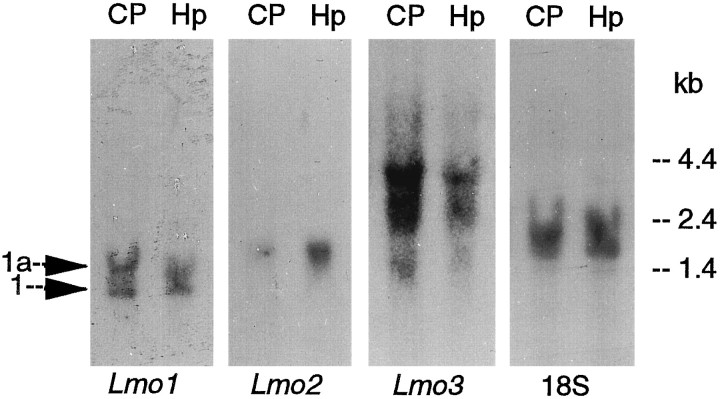
Lmo1, Lmo2, and Lmo3 share considerable sequence homology (Foroni et al., 1992), and the specificity of the in situ hybridization reaction obtained with the antisense oligonucleotide probes used in this study was tested and verified in several ways. The hybridization patterns obtained with each probe in the mouse forebrain were unique and included a number of nonoverlapping areas (Figs. 1, 2; Table1). In addition, the hybridization signal obtained with each radiolabeled probe was eliminated by competition with excess amounts of its corresponding unlabeled antisense oligonucleotide, whereas competition with excess amounts of the other two unlabeled oligonucleotides did not diminish the hybridization signal (Fig. 1). Northern blot analysis further showed that each of the probes bound to transcripts of different sizes, in agreement with previous studies that used probes derived directly from cDNA (Fig. 3).
Fig. 1.
Specificity of the oligonucleotide probes forLmo1, Lmo2, and Lmo3 was confirmed byin situ hybridization competition studies. Sections through the forebrain of mice injected with KA (6 hr survival) were processed for in situ hybridization such that each of the three 35S-labeled sequences was incubated either alone or in competition with 100-fold excess of each of the three unlabeled oligonucleotide sequences. Autoradiographs of the hybridization signals demonstrated that specific hybridization signal was prevented only by coincubation of labeled probes with unlabeled oligonucleotides of the same sequence. Scale bar, 1500 μm.
Fig. 2.
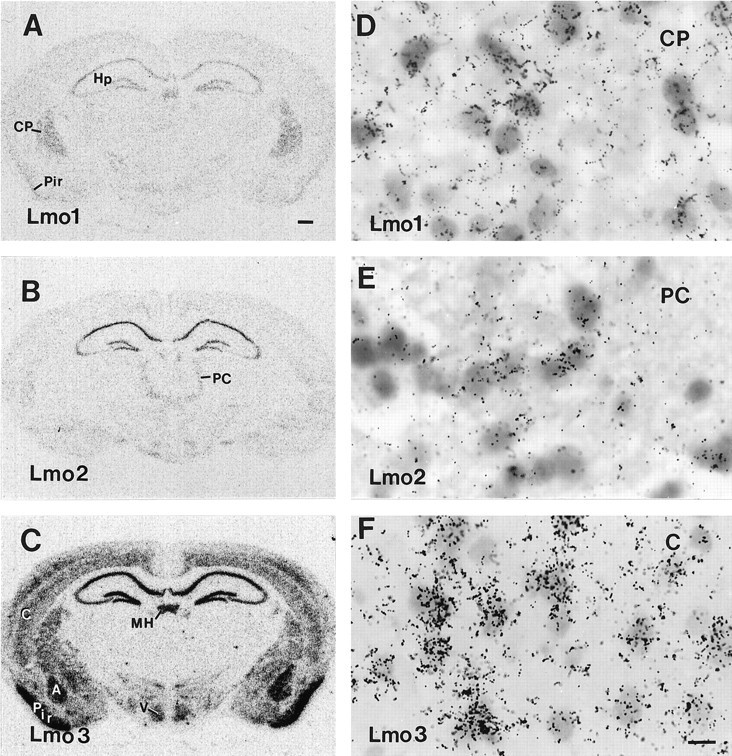
Photomicrographs showing the distribution ofLmo1 (A, D), Lmo2(B, E), and Lmo3 (C, F) mRNAs in the forebrain of untreated mice as detected by in situ hybridization with three35S-labeled oligonucleotide probes and exposure of tissue sections to radiographic film (A–C) or dipping in photographic emulsion (D–F).A–C show the different but partially overlapping patterns of labeling obtained for Lmo1,Lmo2, and Lmo3 in prominent forebrain structures. D–F show that labeling clusters over cells in the caudate putamen (D), paracentral nucleus of the thalamus (E), and layer V of parietal neocortex (F). Scale bar (shown inF): A–C, 400 μm;D–F, 12 μm. A, Basolateral nucleus of the amygdala; C, cerebral neocortex; CP, caudate putamen; Hp, hippocampal formation;MH, medial habenula; PC, paracentral nucleus of the thalamus; Pir, piriform cortex;V, ventromedial nucleus of the hypothalamus.
Table 1.
Qualitative estimates of the relative distributions ofLmo1, Lmo2, and Lmo3 mRNA in various prominent forebrain regions of untreated adult mice as determined by in situ hybridization
| Forebrain region | Lmo1 | Lmo2 | Lmo3 |
|---|---|---|---|
| Hippocampal formation | |||
| CA1, pyramidal neurons | +++ | ++++ | ++++ |
| CA2, pyramidal neurons | ++++ | ++++ | ++++ |
| CA3, pyramidal neurons | ++ | ++ | ++ |
| Dentate gyrus, granule neurons | ++++ | +++ | +++ |
| Subiculum, pyramidal neurons | +++ | ++++ | ++++ |
| Interneurons | + | + | |
| Caudate putamen | ++++ | ++++ | |
| Thalamus | |||
| Intralaminar nucleus | ++ | ||
| Parafasicular nucleus | ++ | ||
| Reticular nucleus | ++ | ||
| Lateral geniculate nucleus | ++ | ||
| Medial habenula | +++ | + | ++++ |
| Septum | +++ | +++ | |
| Hypothalamus | + | + | +++ |
| Amygdaloid complex | ++++ | ||
| Olfactory bulb | + | + | |
| Piriform cortex | + | + | ++++ |
| Cerebral neocortex | +++ |
+ to ++++, Qualitative estimates of increasing levels of hybridization signal for each individual probe relative to itself. Note that comparisons of levels are made across anatomical regions for each individual probe and that no comparison of the expression levels is inferred among the three different probes in any of the anatomical regions.
Fig. 3.
Northern blot analysis ofLmo1, Lmo2, and Lmo3 mRNAs in forebrain tissue of untreated mice. RNA was extracted from the caudate putamen (CP) or hippocampus (Hp) and hybridized with each of the three 32P-labeled oligonucleotide sequences. Autoradiographs of the hybridization signals showed that the Lmo1 oligonucleotide probe bound to two species of RNA (1.4 and 1.2 kb) derived from promoters1a and 1 (Boehm et al., 1990, 1991b), theLmo2 probe to a single species of RNA (1.7 kb), and theLmo3 probe to three RNA species (4.3, 2.7, and 1.5 kb) in agreement with previous studies using cDNA probes (Foroni et al., 1992). Note that striatal tissue shows approximately equal regulation from promoters 1 and 1a ofLmo1, whereas promoter 1 predominates somewhat in hippocampal tissue. Rehybridization with an 18S rRNA probe indicated approximately equal lane loading.
Lmo1, Lmo2, and Lmo3 mRNA in mouse forebrain
Lmo1, Lmo2, and Lmo3 showed individually unique but partially overlapping patterns of expression in many regions of the adult mouse forebrain. Prominent sites of expression included various portions of the hippocampal formation, caudate putamen, medial habenula, thalamus, amygdala, olfactory bulb, hypothalamus, and cerebral cortex (Figs. 1, 2). Table 1 shows qualitative estimates of the relative intensity of the labeling obtained with individual Lmo mRNA probes in various forebrain regions, prepared by comparison of five untreated mice hybridized with all three probes in an alternate series of sections. The comparison is only between the expression levels of each individual gene relative to itself in different anatomical regions. It is not possible using in situ hybridization to compare the expression levels of different genes with one another, and no attempt has been made to do so. Thus, for example, the level of Lmo1expression is compared across various anatomical regions, but the expression levels of Lmo1, Lmo2, and Lmo3 within an anatomical region are not compared with one another.
The forebrain distribution patterns varied for the threeLmo genes such that Lmo3 showed the most widespread distribution, followed by Lmo1 and thenLmo2 (Figs. 1, 2; Table 1). The caudate putamen showed high relative levels of expression for both Lmo1 andLmo3, whereas Lmo2 was essentially undetectable, as determined by both in situ hybridization and Northern blot analysis (Figs. 1, 2, 3). Interestingly, Northern blot analysis showed approximately equal activities from Lmo1 promoters 1 and 1a in the caudate putamen (Fig. 3). In the amygdaloid complex,Lmo3 showed prominent expression in several nuclei, particularly the basolateral amygdala, whereas Lmo1 showed little and Lmo2 showed no expression (Figs. 1, 2; Table 1). In the thalamus, Lmo2 expression was prominent in the parafascicular nucleus, paracentral and other intralaminar nuclei;Lmo1 was prominent in the reticular and Lmo3 in the lateral geniculate nuclei (Figs. 1, 2; Table 1). In most of these areas, the size of the labeled cells in emulsion-dipped sections suggested that they were neuronal (Fig.2D–F). Nevertheless, some small cells that may have been glial cells were also labeled. For this report, the expression patterns of Lmo1, Lmo2, and Lmo3 in the hippocampal formation and their changes after limbic seizures are described in greater detail.
Lmo1, Lmo2, and Lmo3 expression in hippocampus
All three Lmo genes showed prominent expression in different portions of the hippocampal formation. Hippocampal principal cells, i.e., pyramidal neurons of CA1, CA2, CA3 and subiculum, and granule neurons of the dentate gyrus, showed substantial but different combinatorial patterns of Lmo1, Lmo2, andLmo3 mRNA expression by in situ hybridization (Figs. 2, 4; Table 1). Hippocampal expression was confirmed for all three Lmo genes by Northern blot analysis (Fig. 3). In situ hybridization showed high relative levels of expression of Lmo1 in pyramidal neurons of CA2 and in dentate granule neurons, and moderate levels in pyramidal neurons of CA1, CA3, and subiculum (Figs. 2, 4; Table 1). Northern blot analysis of hippocampal tissue showed evidence of activities from bothLmo1 promoters 1 and 1a, with a slight predominance of activity from promoter 1 (Fig. 3). In situ hybridization showed about equal relative levels of expression of Lmo2 in pyramidal neurons of CA1, CA2, and subiculum, with somewhat less expression in dentate granule neurons and CA3 pyramidal neurons (Figs.2, 4; Table 1). In situ hybridization showed high relative levels of expression of Lmo3 in pyramidal cells in CA1, CA2, and the subiculum, as well as by dentate granule neurons, with somewhat less expression in CA3 pyramidal neurons (Figs. 2, 4; Table 1). Microscopic examination of sections dipped in photographic emulsion confirmed at the cellular level the observations made on autoradiographic film, and showed that within a particular subgroup of principal neurons (i.e., CA1, CA2, etc.) the individual expression levels of Lmo1, Lmo2, or Lmo3 were broadly similar across most neurons, so that differences between subgroups reflected essentially the whole group (Fig.4D–F,H). Examination of emulsion-dipped sections also showed that Lmo1 and Lmo3, but notLmo2, were expressed by presumptive interneurons in various hippocampal regions (Fig. 4G).
Fig. 4.
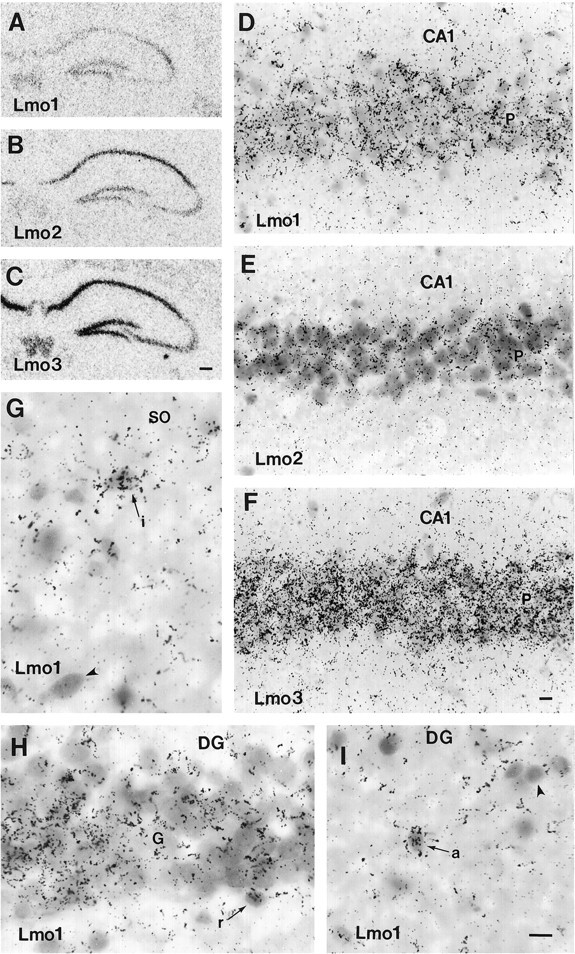
Photomicrographs of the distribution ofLmo1 (A, D, G–I),Lmo2 (B, E), andLmo3 (C, F) mRNAs in the hippocampus of untreated mice as detected by in situhybridization with three 35S-labeled oligonucleotide probes and exposure of tissue sections to radiographic film (A–C) or by dipping in photographic emulsion (D–I). D–F, Note the heavy and even labeling of pyramidal neurons with all three probes in the CA1 region of the hippocampus. G, Lmo1-labeled (arrow) as well as Lmo1-unlabeled (arrowhead) presumptive interneurons (i) are present in the stratum oriens (SO). H, Lmo1-labeled granule cells (G) in the dentate gyrus (DG). Arrow indicates aLmo1-labeled, small, potential radial astrocyte (r) at the base of the granule cell layer. I, Arrow indicates a Lmo1-labeled small cell in the inner molecular layer of the dentate gyrus (DG) likely to be an astrocyte (a), compared with a small, unlabeled cell (arrowhead). Scale bars: A–C, 200 μm; D–F, 15 μm; G–I, 12 μm.
The distribution of Lmo1 mRNA was also compared with the distribution of immunoreactive β-gal in six untreated adult transgenic mice expressing lacZ fromLmo1-promoter 1. In these transgenic mice, most pyramidal neurons in the subiculum and CA1 and nearly all pyramidal neurons in CA2 were β-gal-positive (Fig. 5A,D,E). These findings correlated well with observations of Lmo1mRNA as detected by in situ hybridization (Figs. 2, 4). In contrast, no pyramidal neurons in CA3 and no granule neurons in the dentate gyrus were β-gal-positive in transgenic mice (Fig.5A,E,F), suggesting that Lmo1 promoter 1 was not active in these neurons and that the moderate expression levels of Lmo1 in CA3 pyramidal neurons and the more robust expression observed in dentate granule neurons by in situhybridization were attributable to activity of promoter 1a. This possibility is further strengthened by observations from Northern blot analysis indicating that although activity from Lmo1promoter 1 predominates somewhat, promoter 1a is also active in the adult hippocampus (Fig. 3). In the transgenic mice, scattered interneurons were also β-gal-positive throughout the hippocampus (Fig. 5A,D,E). In addition, numerous cells that were stained positively for β-gal had the morphological appearance (Kosaka and Hama, 1986) of astrocytes in the molecular layer and hilus of the dentate gyrus or of radial astrocytes in the subgranular zone (Fig.5A,F,I–L). β-Gal-stained presumptive astrocytes had a tufted appearance, with a small central cell body and many fine branches (Fig. 5I). These cells, as well as radial glia, stained positively for both GFAP and β-gal in double-labeled sections (Fig. 5J–L), confirming their identity as astrocytes. Some small β-gal-positive cells, which may have been progenitor cells that are known to exist in this region, were observed in the dentate subgranular zone (Suhonen et al., 1996); further analysis will be required to establish the identity of all β-gal-positive cell types. All cell types identified in the transgenic animals could be identified in emulsion-dipped sections processed for in situ hybridization detection ofLmo1, including pyramidal neurons in subiculum, CA1, and CA2, as well as presumptive interneurons and small potential glia cells (Fig. 4D,G–I), strongly supporting the likelihood that β-gal expression in these transgenic mice reflects sites of endogenous expression of Lmo1 from promoter 1. In addition, previous analysis has shown that progeny of five different transgenic founder mice generated with this fusion gene construct all showed similar developmental patterns of β-gal staining, further indicating that the transgene expression patterns observed are not artifact (Greenberg et al., 1990). In comparing the pattern of labeling obtained by in situ hybridization with that of reporter gene expression in these transgenic mice, it should be noted that the immunohistochemical reaction product for β-gal was often deposited not only in the cell body but also throughout its processes, particularly in astrocytes. Thus, when viewed at low power as in Figure5A,F, the degree of reporter protein expression visualized considerably overestimates the number of expressing cells.
Fig. 5.
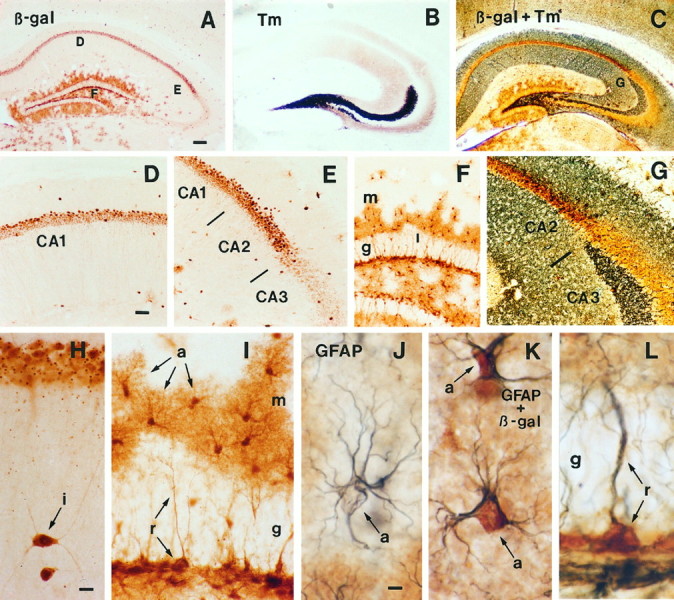
Immunohistochemical staining for β-galactosidase (β-gal) in transgenic mice expressing lacZ from the Lmo1-promoter 1.A, Survey of hippocampus stained for β-gal.B, Survey of hippocampus stained by the Timm method to show zinc-containing fibers, particularly in mossy fibers projecting from the dentate gyrus to CA3. C, Survey of hippocampus stained by both the Timm method and immunohistochemistry for β-gal illustrating the sharp border between Timm-stained fibers in CA3 and β-gal-positive neurons in CA2. D, E, Details ofA showing β-gal-positive pyramidal cells in CA1 and CA2 but not CA3, as well as scattered interneurons. F,Detail of A showing numerous β-gal-positive astrocytes in the dentate gyrus molecular layer (m) and hilus, and the absence of staining in granule layer (g) neurons. G, Detail ofC showing that β-gal-positive pyramidal neurons are present in CA2 but not in CA3 as delineated by the CA2–CA3 border demarcated by Timm-stained mossy fibers in CA3. H,Detail showing two β-gal-positive interneurons (i) in the stratum radiatum below CA1 pyramidal neurons. I,Detail of F showing the morphology of β-gal-positive astrocytes (a) in the molecular layer of the dentate gyrus. Note that in addition to the darkly stained cell bodies, the finely branched processes of the astrocytes are also stained and radiate out from the cell bodies to cover a large area and give the cells a tufted appearance (a). Not all astrocytes in the molecular layer are β-gal positive. Note also the β-gal-positive radial astrocytes (r) whose processes span the granule layer (g), and that the granule neurons are not stained. J, Detail showing an astrocyte (a) immunohistochemically stained only for GFAP using a blue chromogen. Note that blue staining for GFAP is present in the astrocyte processes and outlines but does not fill the cell body. This cell is located in a portion of the molecular layer similar to that shown in I, in between astrocytes stained positively for both β-gal and GFAP, as in K. K, Two astrocytes (a) located in the molecular layer that are immunohistochemically double-stained for both GFAP (blue) and β-gal (brown). Note that thebrown-stained cell bodies (β-gal) and dark blue-stained processes (GFAP) clearly belong to the same cells and compare in appearance with the single-stained astrocytes in I and J. L,Radial astrocyte (r) located in the granule layer that is immunohistochemically double-stained for both GFAP (blue) and β-gal (brown). Note that thebrown-stained cell body (β-gal) and dark blue-stained process (GFAP) clearly belong to the same cell, which compares in appearance with the single-stained radial astrocytes in I. Scale bars: A–C, 180 μm;D–G, 70 μm; H, I, 10 μm;J–L, 5 μm.
To confirm that the differences in the staining observed between pyramidal neurons in different CA regions genuinely reflect anatomically recognized borders, we combined immunohistochemical analysis of β-gal with Timm staining. Timm stain identifies projections of the mossy fibers that pass from the dentate granule cells to CA3 cells and provides an accurate and well characterized means of identifying the CA2–CA3 border (Haug et al., 1971; Amaral and Dent, 1981; Swanson et al., 1987). Combined detection of β-gal and Timm in neighboring (Fig. 5A,B) and the same sections (Fig.5C) clearly demonstrated that Lmo1 expression from promoter 1 observes the CA2–CA3 border and is high in CA2 and absent from CA3 pyramidal neurons.
Regulation of Lmo1, Lmo2, and Lmo3 mRNA by seizure activity
To investigate whether hippocampal Lmo1, Lmo2, and Lmo3 expression levels might be regulated by changes in neuronal activity, we examined the effects of limbic seizure activity induced by intraperitoneal injections of KA. KA injections were dosed so that they resulted in behavioral activity such as freezing and rearing (Lothman and Collins, 1981; Ben-Ari, 1985) but did not cause generalized seizures and did not lead to hippocampal neuronal degeneration visible in tissue sections stained for cresyl violet after 7 d survival times. These KA-induced limbic seizures caused differential and statistically significant changes, as compared with vehicle-injected control animals, in the relative levels of Lmo1, Lmo2, and Lmo3 mRNA in hippocampal pyramidal and granule neurons as determined by densitometric analysis of sections processed by in situ hybridization (Figs. 6,7). Lmo1 mRNA increased and Lmo2and Lmo3 mRNAs decreased in both of these cell types in various hippocampal regions over a time course of 3–24 hr (Figs. 6,7). Few statistically significant changes were seen at 1 hr, and all changes were maximal at 6 hr after onset of seizure activity and had returned approximately to baseline by 24 hr. The changes can be summarized as follows. Lmo1 showed a very pronounced increase in expression in CA1 and particularly in CA2 pyramidal neurons, with a substantial increase in dentate granule neurons and little change in CA3 (Figs. 6A, 7A).Lmo2 showed a pronounced drop in expression to very low relative levels in CA1–CA3 pyramidal neurons and to essentially undetectable levels in dentate granule neurons (Figs.6B, 7B). Lmo3 showed a substantial decrease in expression but was still easily detectable in CA1, CA2, and CA3 pyramidal neurons and dentate gyrus granule neurons (Figs. 6C, 7C). Microscopic examination of sections dipped in photographic emulsion confirmed observations made on autoradiographic film and showed that within a particular subgroup of principal neurons (i.e., CA1, CA2, etc.), the individual expression levels of Lmo1, Lmo2, or Lmo3 were similar across most neurons, and that changes induced by limbic seizure activity in different subgroups reflected essentially the whole group. No changes in relative levels of Lmo1, Lmo2, and Lmo3 mRNA were observed qualitatively or quantitatively in the habenula, striatum, or cerebral cortex over this same time period (Figs. 6, 7). Cortical measurements were taken from the entire cortex overlying the hippocampus and may not have detected changes in particular layers or subregions. Although limbic seizure activity modulated expression levels of all three of these genes, seizure activity did not alter the basic pattern of cells expressing these genes; in particular, seizure activity did not induce Lmo gene expression in novel groups of CNS cells not already observed in untreated mice. In addition, four transgenic mice expressing lacZ from Lmo1promoter 1 were examined at 12 hr (to allow time for translation of protein) after KA-induced limbic seizures. The pattern of positive cell types in these animals was identical to that seen in untreated transgenic mice, and no novel sites of β-gal staining were observed.
Fig. 6.
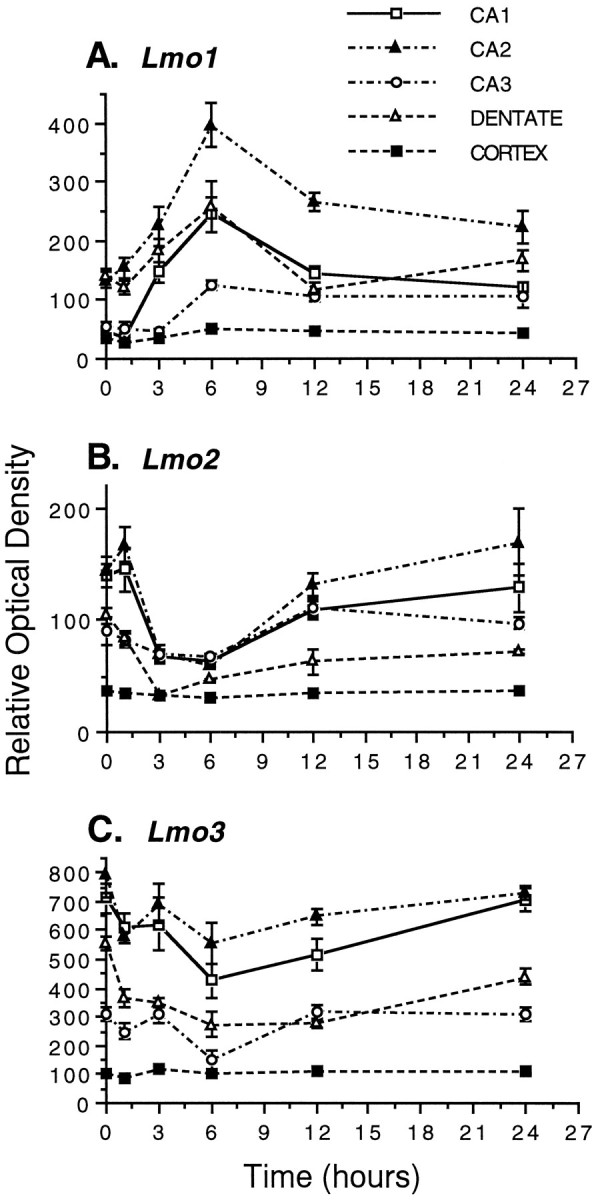
Effects of KA-induced limbic seizures on the relative mRNA levels of Lmo1 (A),Lmo2 (B), and Lmo3(C) in hippocampal subdivisions CA1, CA2, CA3, and dentate gyrus, and cerebral cortex. Mice were administered 30 mg/kg KA or saline vehicle intraperitoneally, and the levels of mRNA for the three Lmo genes were examined at 1, 3, 6, 12, and 24 hr after injection by in situ hybridization. Quantitation was performed on autoradiographs, representative examples of which are shown in Figure 7. The graphs show the ROD measurements taken from five animals with three to six measurements per animal and are expressed as the mean ± SEM. No statistically significant changes were observed at any time point for any probe in the cerebral cortex. In contrast, statistically significant changes of varying magnitude were observed for all three probes in all regions of the hippocampus, and these changes were maximal at 6 hr after onset of seizure activity.
Fig. 7.
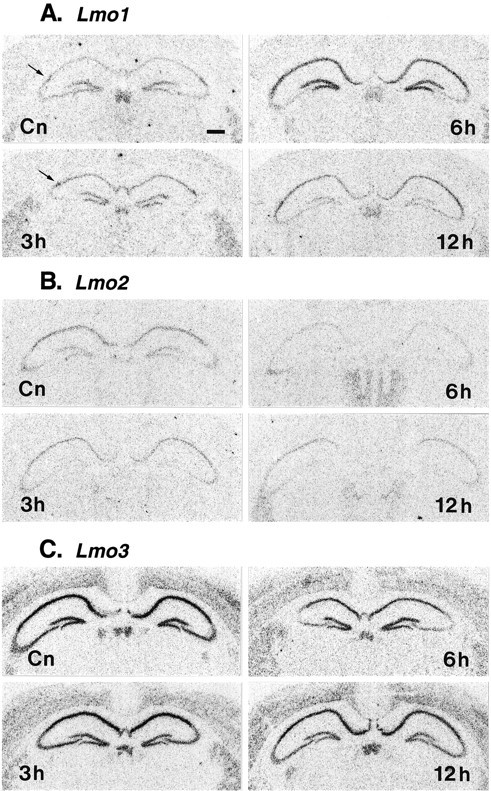
Effects of KA-induced limbic seizures on the relative mRNA levels of Lmo1 (A),Lmo2 (B), and Lmo3(C) in hippocampus as described in Figure 6. Photomicrographs show representative autoradiographs taken from vehicle-injected (Cn) and KA-injected mice at 3, 6, and 12 hr. Photomicrographs have been printed to achieve similar levels of background to allow comparison of the changes in relative hybridization levels in hippocampal principal neurons in CA1, CA2 (arrow), CA3, and dentate gyrus.
DISCUSSION
Lmo1, Lmo2, and Lmo3 are a family of LIM-only genes originally identified as putative oncogenes (Boehm et al., 1991a) that are active during mammalian CNS development (Greenberg et al., 1990; Boehm et al., 1991b; Foroni et al., 1992). In this study we have shown that Lmo1, Lmo2, and Lmo3 continue to be expressed in a number of forebrain regions in the adult mouse, particularly in the hippocampal formation where they show different combinatorial patterns of expression in pyramidal neurons of CA1, CA2, CA3, and dentate granule neurons, as well as in restricted subpopulations of astrocytes. We also found that expression levels of these LIM-only genes in hippocampal principal neurons were differentially regulated in response to limbic seizure activity. Together these observations suggest that Lmo1, Lmo2, andLmo3 may be involved in cell phenotype-specific regulatory functions in the adult CNS.
LIM proteins and the adult CNS
LIM proteins represent a broad family of molecules whose functions in the CNS are not well understood. With their potential for protein–protein interactions and associations with various different functional domains such as DNA binding regions, cytoskeletal regions, or kinase domains, LIM proteins are candidates to be important regulatory molecules. A number of LIM proteins, including LIM-only proteins, seem to be involved in the control of cell differentiation in neural as well as non-neural tissues and in vertebrates and invertebrates (Way and Chalfie, 1988; Freyd et al., 1990;Sanchez-Garcia and Rabbitts, 1994; Warren et al., 1994). Such observations are consistent with findings that various LIM protein genes are expressed during, and may be involved in controlling, particular aspects of CNS development. In a well documented example, the combinatorial expression patterns of a family of LIM homeobox genes have been shown to define distinct subclasses of motor neurons in the spinal cord, and it has been suggested that these expression patterns play a prominent role in determining the topographic organization of motor projections in the developing vertebrate CNS (Tsuchida et al., 1994; Lumsden, 1995).
Less is known about LIM proteins in the adult CNS. The LIM-homeodomain protein ISL-1 is present in various regions of the adult rat brain (Thor et al., 1991), related Isl genes have been identified in mature fish brain (Gong et al., 1995), andKiz-1, which encodes a LIM-kinase protein, has been identified in mature mouse brain (Bernard et al., 1994). Factors affecting the regulation of these genes have not been reported, and the functions of these or other LIM proteins in the mature CNS are not known.
In this study we found prominent expression of the LIM-only genesLmo1, Lmo2, and Lmo3 in numerous regions of the adult mouse forebrain, including the hippocampal formation, caudate putamen, medial habenula, thalamus, amygdala, olfactory bulb, hypothalamus, and cerebral cortex. The results were confirmed with various specificity tests and were consistent across the three different types of techniques used: in situ hybridization, Northern blot analysis, and evaluation of transgenic mice. In most areas Lmo gene expression seemed to be neuronal. In the transgenic mice, some areas contained Lmo1-β-gal-positive cells that had the morphological appearance of astrocytes and were double-labeled with GFAP.
The role of LIM-only proteins in the adult CNS is not yet clear. LIM-only proteins have been proposed to function by protein–protein interactions, possibly also with other LIM proteins (Rabbitts and Boehm, 1990; Sanchez-Garcia et al., 1993; Sanchez-Garcia and Rabbitts, 1994). If this proves to be the case, then the widespread expression of LIM-only genes in the adult CNS suggests that other LIM protein genes may be active in the adult CNS as well. Additional studies will be required to determine this. Previous studies have shown thatLmo1, Lmo2, and Lmo3 are expressed in specific and restricted populations of cells in the developing mouse CNS, in the hindbrain, and in cerebellum and various forebrain regions, including olfactory bulb, caudate putamen, hippocampus, thalamus, hypothalamus, and neocortex (Greenberg et al., 1990; Boehm et al., 1991b; Foroni et al., 1992), in many of the same areas in which expression was demonstrated in adult mice in the present study. Although detailed comparisons have not yet been made, preliminary observations suggest that Lmo1, Lmo2, and Lmo3 are expressed in the same cell types in some brain regions, such as the hippocampus, from early development through to and persisting in the adult. Thus, these LIM-only genes may be involved not only in the development but also in the subsequent maintenance in the adult of specific cell phenotypes in the CNS. The combinatorial pattern of LIM-only gene expression in hippocampal neurons favors this possibility.
Lmo1, Lmo2, and Lmo3 and the phenotype of adult hippocampal cells
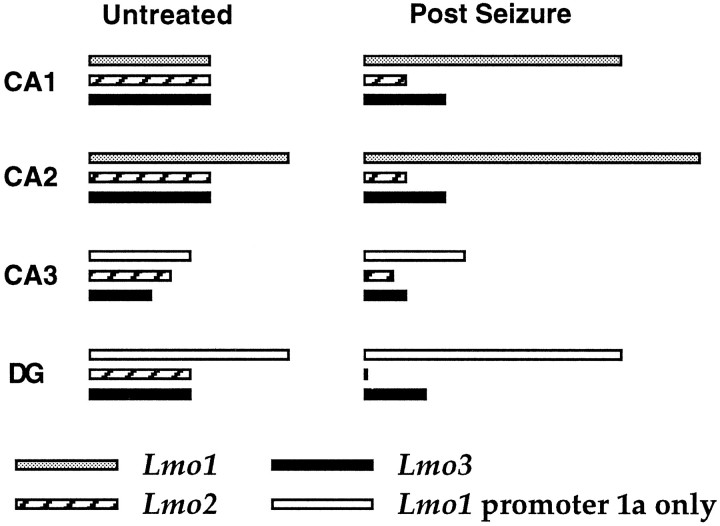
The function of the hippocampal formation depends on a precisely defined topographic arrangement of groups of neurons that exhibit well documented differences not only in cellular structure but also in afferent and efferent projections, intrinsic chemistry (e.g., of transmitters, growth factors, and receptors, etc.), and electrophysiological characteristics (Swanson et al., 1987; Amaral and Witter, 1989; Gall et al., 1991). In this study we found that the different combinatorial patterns of relative expression levels ofLmo1, Lmo2, and Lmo3 defined the well recognized subgroups of hippocampal principal neurons (Fig. 8). Pyramidal neurons of CA2 showed prominent expression of all three LIM-only genes, whereas CA1 pyramidal neurons showed somewhat lower relative expression levels. Pyramidal neurons of CA3 showed considerably lower relative expression levels of Lmo2, Lmo3, and Lmo1 from promoter 1a and no expression ofLmo1 from promoter 1. Granule neurons of the dentate gyrus show prominent expression of Lmo3 and Lmo1 from promoter 1a, but little expression of Lmo2 and no expression of Lmo1 from promoter 1 (Fig. 8). Importantly, major anatomical and functional boundaries that exist between well recognized groups of hippocampal pyramidal neurons, such as that between CA2 and CA3, were also precisely, and in some cases dramatically, defined by differences in relative expression levels of LIM-only genes. In addition, expression of Lmo1 from promoter 1 defined several topographically restricted subpopulations of astrocytes in the dentate gyrus. These findings suggest that LIM-only genes may be involved in defining subgroups of hippocampal cells. Interestingly, most of theLmo-expressing groups of principal neurons in the hippocampus did not seem to be identified by the expression of a singleLmo gene, but by different combinations of relative levels of Lmo gene expression. These findings are consistent with suggestions that combinatorial genetic events underlie the diversity of cell phenotype in the CNS (He and Rosenfeld, 1991; Struhl, 1991), and with reports that combinatorial expression patterns of a family of LIM homeobox genes distinguishes subclasses of developing motor neurons (Tsuchida et al., 1994).
Fig. 8.
Summary diagram of relative levels of expression of Lmo1, Lmo2, and Lmo3 in different groups of hippocampal principal neurons in untreated mice and in mice after limbic seizures. The approximate differences indicated are for the expression levels of each individual Lmo gene relative to itself in the two conditions and do not reflect differences in expression between the three different genes.
Regulation of gene expression in the adult hippocampus after limbic seizure activity
Changes in transcription may be fundamental to certain hippocampal functions, such as its role in the formation of long-term memory (Squire, 1992; Bourtchuladze et al., 1994; Huang et al., 1994;Stevens, 1994; Sossin, 1996). In agreement with this possibility, hippocampal neurons show considerable plasticity of gene expression in response to changes in neural activity (Gall et al., 1991; Berzaghi et al., 1993; Bendotti et al., 1994), and factors produced by hippocampal neurons affect the phenotype and function of afferent neurons (Sofroniew et al., 1990; Thoenen, 1995). Activity-dependent changes in hippocampal gene expression induced by limbic seizures have been well characterized. After seizure activity, expression levels of immediate early genes in hippocampal principal neurons increase rapidly, peak at 1–2 hr, and decline rapidly (Morgan et al., 1987; Gass et al., 1993). Over a more prolonged time course that varies from 3–6 hr to 1–3 d and in some cases longer, expression levels of numerous other genes change, including genes encoding neuropeptides, neurotrophins, and growth associated proteins (Gall, 1988; Gall and Isackson, 1989; Gall et al., 1991; Bendotti et al., 1994; Lauterborn et al., 1995). All of these genes are expressed in a cell phenotype-specific manner, show differential levels of expression in different subsets of hippocampal principal neurons, and show changes in levels of expression in some but not other neuronal subgroups after seizure activity. In some cases expression levels increase after seizures, whereas in others they decrease.
In this study we found that expression levels of the LIM-only genes Lmo1, Lmo2, and Lmo3 are regulated by neuronal activity in a cell-specific manner, such that relative levels of Lmo1 increased and relative levels of Lmo2 andLmo3 decreased in different subgroups of hippocampal principal neurons after limbic seizure activity (Fig. 8). It is important to note that although changes in neural activity modulated expression levels of these genes, neural activity did not alter the basic pattern of cell type expressing these genes, such that the changes seen after seizure activity were specific to neurons already showing Lmo gene expression. Thus, combinatorial patterns ofLmo gene expression not only defined specific subsets of hippocampal neurons in untreated animals, but seizure-related increases in neural activity differentially regulated the relative levels of expression of these LIM proteins in different subsets of hippocampal neurons.
The functional role of the activity-dependent changes in expression levels of LIM-only genes that we observed in hippocampal principal neurons is not certain. As described above, several lines of evidence suggest that LIM-only proteins may have intracellular regulatory functions, perhaps by interacting with other LIM proteins that have functional domains such as DNA binding regions, kinase regions, or cytoskeletal regions. Taken together these observations suggest that the combinatorial expression patterns of Lmo1, Lmo2, and Lmo3 may be involved in the specification of defining characteristics to hippocampal neurons of particular phenotypes in the adult, and may differentially influence transcription or other regulatory processes in response to changes in neuronal activity in a cell phenotype-specific manner.
Footnotes
This work was supported by Parke-Davis, The Wellcome Trust, and Medical Research Council. We thank Dr. T. H. Rabbitts for oligonucleotide sequences, J. Drew for technical assistance, and J. A. Bashford, I. Bolton, and A. P. Newman for photography.
Correspondence should be addressed to M. V. Sofroniew, Medical Research Council Cambridge Centre for Brain Repair, Forvie Site, Robinson Way, Cambridge CB2 2PY, UK.
REFERENCES
- 1.Amaral DG, Dent JA. Development of the mossy fibers of the dentate gyrus. I. A light and electron microscopic study of the mossy fibers and their expansions. J Comp Neurol. 1981;195:51–86. doi: 10.1002/cne.901950106. [DOI] [PubMed] [Google Scholar]
- 2.Amaral DG, Witter MP. The three dimensional organization of the hippocampal formation: a review of anatomical data. Neuroscience. 1989;31:571–591. doi: 10.1016/0306-4522(89)90424-7. [DOI] [PubMed] [Google Scholar]
- 3.Archer V, Breton J, Sanchez-Garcia I, Osada H, Forster A, Thomson AJ, Rabbitts TH. The cysteine-rich domains of LIM proteins RBTN and Isl-1 contain zinc but not iron. Proc Natl Acad Sci USA. 1994;91:316–320. doi: 10.1073/pnas.91.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Ari Y. Limbic seizure and brain damage produced by kainic acid: mechanisms and relevance to human temporal lobe epilepsy. Neuroscience. 1985;14:375–403. doi: 10.1016/0306-4522(85)90299-4. [DOI] [PubMed] [Google Scholar]
- 5.Bendotti C, Pende M, Samanin R. Expression of GAP-43 in the granule cells of rat hippocampus after seizure-induced sprouting of mossy fibres: in situ hybridization and immunocytochemical studies. Eur J Neurosci. 1994;6:509–515. doi: 10.1111/j.1460-9568.1994.tb00294.x. [DOI] [PubMed] [Google Scholar]
- 6.Bernard O, Ganiatsas S, Kannourakis G, Dringen R. Kiz-1, a protein with LIM zinc finger and kinase domains, is expressed mainly in neurons. Cell Growth Differ. 1994;5:1159–1171. [PubMed] [Google Scholar]
- 7.Berzaghi MP, Cooper JD, Castren E, Zafra F, Sofroniew MV, Thoenen H, Lindholm D. Cholinergic regulation of brain-derived neurotrophic factor (BDNF) and nerve growth factor (NGF) but not neurotrophin-3 (NT-3) mRNA levels in the developing rat hippocampus. J Neurosci. 1993;13:3818–3826. doi: 10.1523/JNEUROSCI.13-09-03818.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boehm T, Baer R, Lavenir I, Forster A, Waters JJ, Nacheva E, Rabbitts TH. The mechanism of chromosomal translocation t(11;14) involving the T-cell receptor C∂ locus on human chromosome 14q11 and a transcribed region of chromosome 11p15. EMBO J. 1988;7:385–394. doi: 10.1002/j.1460-2075.1988.tb02825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boehm T, Greenberg JM, Buluwela L, Lavenir I, Forster A, Rabbitts TH. An unusual structure of a putative T cell oncogene which allows production of similar proteins from distinct mRNAs. EMBO J. 1990;9:857–868. doi: 10.1002/j.1460-2075.1990.tb08183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boehm T, Foroni L, Kaneko Y, Perutz MF, Rabbitts TH. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc Natl Acad Sci USA. 1991a;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehm T, Spilantini MG, Sofroniew MV, Surani MA, Rabbitts TH. Developmentally regulated and tissue specific expression of mRNAs encoding the two alternative forms of the limb domain oncogene rhombotin: evidence for thymus expression. Oncogene. 1991b;6:695–703. [PubMed] [Google Scholar]
- 12.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 13.Crawford AW, Pino JD, Beckerle MC. Biochemical and molecular characterization of the chicken cysteine-rich protein, a developmentally regulated LIM-domain protein that is associated with the actin cytoskeleton. J Cell Biol. 1994;124:117–127. doi: 10.1083/jcb.124.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawid IB, Toyama R, Taira M. LIM domain proteins. C R Acad Sci Paris Life Sci. 1995;318:295–306. [PubMed] [Google Scholar]
- 15.Foroni L, Boehm T, White L, Forster A, Sherrington P, Liao XB, Brannan CI, Jenkins NA, Copeland NG, Rabbitts TH. The rhombotin gene family encode related LIM-domain proteins whose differin expression suggests multiple roles in mouse development. J Mol Biol. 1992;226:747–761. doi: 10.1016/0022-2836(92)90630-3. [DOI] [PubMed] [Google Scholar]
- 16.Freyd G, Kim SK, Horvitz HR. Novel cysteine-rich motif nd homeodomain in the product of the Caenorhabditis elegans cell lineage gene lin-II. Nature. 1990;344:876–882. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- 17.Gall C. Seizures induce dramatic and distinctly different changes in enkephalin, dynorphin, and CCK immunoreactivities in mouse hippocampal mossy fibers. J Neurosci. 1988;8:1852–1862. doi: 10.1523/JNEUROSCI.08-06-01852.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gall CM, Isackson PJ. Limbic seizures increase neuronal production of messenger RNA for nerve growth factor. Science. 1989;245:758–761. doi: 10.1126/science.2549634. [DOI] [PubMed] [Google Scholar]
- 19.Gall C, Lauterborn J, Bundman M, Murray K, Isackson P. Seizures and the regulation of neurotrophic factor and neuropeptide gene expression in brain. Epilepsy Res [Suppl] 1991;4:225–245. [PubMed] [Google Scholar]
- 20.Gass P, Herdegen T, Bravo R, Kiessling M. Spatiotemporal induction of immediate early genes in the rat brain after limbic seizures: effects of NMDA receptor antagonist MK-801. Eur J Neurosci. 1993;5:933–943. doi: 10.1111/j.1460-9568.1993.tb00944.x. [DOI] [PubMed] [Google Scholar]
- 21.Geneser FA, Holm IE, Slomianka L (1993) Application of the Timm and selenium methods to the central nervous system. Neurosci Protocols 93–050-15:1–14.
- 22.Gong Z, Hui C, Hew CL. Presence of isl-1-related LIM domain homeobox genes in teleost and their similar patterns of expression in brain and spinal cord. J Biol Chem. 1995;270:3335–3345. doi: 10.1074/jbc.270.7.3335. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg J, Boehm T, Sofroniew MV, Keynes RJ, Barton SC, Norris ML, Surani MA, Spilantini MG, Rabbitts TH. Segmental and developmental regulation of a presumptive T cell oncogene in the central nervous system. Nature. 1990;344:158–160. doi: 10.1038/344158a0. [DOI] [PubMed] [Google Scholar]
- 24.Haug FMS, Blackstad TW, Simonsen AH, Zimmer J. Timm’s sulfide silver reaction for zinc during experimental anterograde degeneration of hippocampal mossy fibers. J Comp Neurol. 1971;142:23–32. doi: 10.1002/cne.901420103. [DOI] [PubMed] [Google Scholar]
- 25.He X, Rosenfeld MG. Mechanisms of complex transcriptional regulation: implications for brain development. Neuron. 1991;7:183–196. doi: 10.1016/0896-6273(91)90257-z. [DOI] [PubMed] [Google Scholar]
- 26.Huang YY, Li XC, Kandel ER. cAMP contributes to mossy fiber LTP by initiating a covalently mediated early phase and macromolecular synthesis-dependent late phase. Cell. 1994;79:69–79. doi: 10.1016/0092-8674(94)90401-4. [DOI] [PubMed] [Google Scholar]
- 27.Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo- and a Cys-His domain. Nature. 1990;344:879–882. doi: 10.1038/344879a0. [DOI] [PubMed] [Google Scholar]
- 28.Kosaka T, Hama K. Three dimensional structure of astrocytes in the rat dentate gyrus. J Comp Neurol. 1986;249:242–260. doi: 10.1002/cne.902490209. [DOI] [PubMed] [Google Scholar]
- 29.Lauterborn J, Berschauer R, Gall C. Cell-specific modulation of basal and seizure-induced neurotrophin expression by adrenalectomy. Neuroscience. 1995;68:363–378. doi: 10.1016/0306-4522(95)00150-h. [DOI] [PubMed] [Google Scholar]
- 30.Lothman EW, Collins RC. Kainic acid induced limbic seizures: metabolic, behavioral, electroencephalographic and neuropathological correlates. Brain Res. 1981;218:299–318. doi: 10.1016/0006-8993(81)91308-1. [DOI] [PubMed] [Google Scholar]
- 31.Lumsden A. A “LIM code” for motor neurons? Curr Biol. 1995;5:491–495. doi: 10.1016/s0960-9822(95)00100-x. [DOI] [PubMed] [Google Scholar]
- 32.McGuire EA, Hockett RD, Pollock KM, Bartholdi MF, O’Brien SJ, Korsmeyer SJ. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts including Ttg-1, a gene encoding a potential zinc finger protein. Mol Cell Biol. 1989;9:2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michelsen JW, Schmeichel KL, Beckerle MC, Winge DR. The LIM motif defines a specific zinc-binding protein domain. Proc Natl Acad Sci USA. 1993;90:4404–4408. doi: 10.1073/pnas.90.10.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- 35.Rabbitts TH, Boehm T. LIM domains. Nature. 1990;346:418. doi: 10.1038/346418a0. [DOI] [PubMed] [Google Scholar]
- 36.Sambrooke J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 37.Sanchez-Garcia I, Rabbitts TH. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Garcia I, Osada H, Forster A, Rabbitts TH. The cysteine-rich LIM domains inhibit DNA binding by the associated homeodomain in Isl-1. EMBO J. 1993;12:4243–4250. doi: 10.1002/j.1460-2075.1993.tb06108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 40.Shawlot W, Behringer RR. Requirement for Lim1 in head-organizer function. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 41.Sofroniew MV, Galletly NP, Isacson O, Svendsen CN. Survival of adult basal forebrain cholinergic neurons after loss of target neurons. Science. 1990;247:338–342. doi: 10.1126/science.1688664. [DOI] [PubMed] [Google Scholar]
- 42.Sossin WS. Mechanism for the generation of synapse specificity in long-term memory: the implications of a requirement for transcription. Trends Neurosci. 1996;19:215–218. doi: 10.1016/0166-2236(96)20016-5. [DOI] [PubMed] [Google Scholar]
- 43.Squire LR. Memory and the hippocampus: a synthesis from findings with rat, monkeys, and humans. Psychol Rev. 1992;99:195–231. doi: 10.1037/0033-295x.99.2.195. [DOI] [PubMed] [Google Scholar]
- 44.Stevens CF. CREB and memory consolidation. Neuron. 1994;13:770–771. doi: 10.1016/0896-6273(94)90244-5. [DOI] [PubMed] [Google Scholar]
- 45.Struhl K. Mechanisms for diversity in gene expression patterns. Neuron. 1991;7:177–181. doi: 10.1016/0896-6273(91)90256-y. [DOI] [PubMed] [Google Scholar]
- 46.Suhonen JO, Peterson DA, Ray J, Gage FH. Differentiation of hippocampus-derived progenitors into olfactory neurons in vivo. Nature. 1996;383:624–627. doi: 10.1038/383624a0. [DOI] [PubMed] [Google Scholar]
- 47.Swanson LW, Kohler C, Bjorklund A. The limbic region. I: The septohippocampal system. In: Bjorklund A, Hokfelt T, Swanson LW, editors. Handbook of chemical neuroanatomy, Vol 5, Integrated systems of the CNS, Part I. Elsevier; Amsterdam: 1987. pp. 125–277. [Google Scholar]
- 48.Taira M, Evrard JL, Steinmetz A, Dawid IB. Classification of LIM proteins. Trends Genet. 1995;11:431–432. doi: 10.1016/s0168-9525(00)89139-8. [DOI] [PubMed] [Google Scholar]
- 49.Thoenen H. Neurotrophins and neuronal plasticity. Science. 1995;270:593–598. doi: 10.1126/science.270.5236.593. [DOI] [PubMed] [Google Scholar]
- 50.Thor S, Ericson J, Brannstrom T, Edlund T. The homeodomain LIM protein Isl-1 is expressed in subsets of neurons and endocrine cells in the adult rat. Neuron. 1991;7:881–889. doi: 10.1016/0896-6273(91)90334-v. [DOI] [PubMed] [Google Scholar]
- 51.Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 52.Warren AJ, Colledge WH, Carlton MBL, Evans MJ, Smith AJH, Rabbitts TH. The oncogenic cysteine-rich LIM domain protein Rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 53.Way JC, Chalfie M. mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 54.Weiskirchen R, Pino JD, Macalma T, Bister K, Beckerle MC. The cysteine-rich protein family of highly related LIM domain proteins. J Biol Chem. 1995;270:28946–28954. doi: 10.1074/jbc.270.48.28946. [DOI] [PubMed] [Google Scholar]