Abstract
Previous data have suggested that the large nerve terminals present in the synaptosomal fraction from squid optic lobe are capable of protein synthesis (Crispino et al., 1993a,b). We have further examined this issue by comparing the translation products of synaptosomal and microsomal polysomes. Both preparations programmed an active process of translation, which was completely abolished by their previous treatment with EDTA. After immunoabsorption of the newly synthesized neurofilament (NF) proteins, the labeling ratio of the 60 and 70 kDa NF proteins was found to differ, in agreement with comparable differences obtained with intact synaptosomes. These observations indicate that the set of mRNAs translated by synaptosomes differs from that translated by nerve cell bodies. Hence, because NF proteins are neuron-specific, they support the view that the active synaptosomal polysomes are mostly localized in the large nerve terminals that represent the most abundant neuronal component of the fraction. This hypothesis was confirmed (1) by electron spectroscopic data demonstrating the presence of ribosomes and polysomes within the large nerve endings of the synaptosomal fraction, as well as in the carrot-like nerve endings of the retinal photoreceptors that constitute the only large terminals in the optic lobe, and (2) by light and high resolution autoradiography of synaptosomal samples incubated with [3H]leucine, showing that most labeled proteins are associated with the large nerve endings. This response was abolished by cycloheximide. Taken together, the data provide the first unequivocal demonstration that presynaptic nerve terminals are capable of protein synthesis.
Keywords: nerve terminals, synaptosomes, protein synthesis, polysomes, neurofilament proteins, squid
The existence of a local system of protein synthesis in the axon compartment has long been proposed and investigated (for reviews, see Koenig, 1984; Giuditta et al., 1990) but is not widely accepted. Thus, the opinion that axons and nerve terminals are entirely reliant on proteins transported from the perikaryon is still the prevailing view. Nonetheless, the local synthesis of axonal proteins has been reported in several animal species, notably in the squid giant axon (Giuditta et al., 1977, 1980,1986, 1991; Perrone Capano et al., 1987; Kaplan et al., 1992; Gioio et al., 1994; Chun et al., 1995, 1996, 1997) and the goldfish Mauthner axon (Koenig, 1979, 1991; Koenig and Martin, 1996). More controversial data are available with regard to the issue of protein synthesis in nerve terminals. This topic was actively investigated in the 60s and early 70s (Austin and Morgan, 1967; Gambetti et al., 1972; Gilbert, 1972; Ramirez et al., 1972), but lost momentum in later years as a result of growing concern over the purity of the synaptosomal fractions (Rao and Steward, 1991).
A few considerations will help in presenting the main experimental questions. The protein synthetic activity of the synaptosomal fraction is generally determined by the incorporation of a radiolabeled amino acid into protein. Under these conditions, extramitochondrial protein synthesis may only be attributed to sheared cellular fragments still enclosing polysomes and the entire complement of soluble factors required for protein synthesis. These fragments may include presynaptic terminals, postsynaptic elements, and glial processes sealed up by membrane fusion soon after shearing. Discrimination between these possible cellular sources has been attempted often but has not yet yielded a comprehensive picture. Notably, the occurrence of active polysomes in dendritic regions is now well accepted, whereas their presence in nerve terminals is questioned (Steward, 1983; Steward and Banker, 1992).
Several reasons suggest, however, that the issue of presynaptic protein synthesis merits additional investigation. First, the theoretical difficulties and experimental data suggesting that axonal proteins may not be imported exclusively from the perikaryon (Nixon, 1980; Alvarez, 1992) apply even more cogently to the proteins of the presynaptic terminals that are farthest from the nerve cell body and are frequently undergoing long-term plastic changes requiring modifications of their protein complement (Crispino et al., 1993a). Second, the demonstration of local protein synthesis in squid and goldfish axons (see above) makes the hypothesis of the existence of a similar synthetic system in nerve terminals more plausible. Last, studies of the synaptosomal fraction from squid optic lobes support the view that proteins may be synthesized in the large presynaptic endings present in that fraction (Crispino et al., 1993a,b; 1994).
On the basis of these considerations, we have extended our investigations of the optic lobe synaptosomal fraction, and now report that active polysomes are indeed present in the large presynaptic terminals of that fraction. In addition, these terminals have been found to derive from the carrot-shaped endings of the retinal photoreceptor neurons.
MATERIALS AND METHODS
Animals and tissue fractionation. Adult squid (Loligo pealii) were provided by the Marine Biological Laboratory, Woods Hole, MA, and kept in large tanks with running seawater. They were used within a day or two of their capture. Optic lobes were dissected from decapitated heads, washed in ice-cold filtered seawater, and processed as described previously (Crispino et al., 1993a). Briefly, a 10% tissue homogenate in 0.7 msucrose, 20 mm Tris-Cl, pH 7.3, was cleared of nuclei and gross particles by centrifugation in rotor JA-20 of a model J2–21M Beckman centrifuge (3000 rpm, 11 min, 4°C), and the supernatant fraction was centrifuged again in the same rotor at higher speed (12,000 rpm, 30 min, 4°C). The latter step yielded a sedimented pellet (mitochondrial fraction), a floating particulate layer (synaptosomal fraction), and an opaque supernatant (microsomal fraction) that contained free and membrane-bound polysomes and the cytosol. The synaptosomal fraction was collected by decantation, washed several times with homogenizing medium, gently resuspended in the same medium, and stored in ice.
Polysome isolation and cell-free translation. The synaptosomal and microsomal fractions were homogenized in polysomal buffer (0.32 m sucrose, 50 mm Tris-Cl, pH 7.4, 0.1 m KCl, 10 mm MgCl2, 10 mm dithiothreitol, 25 μg/ml tRNA, and 10 U/ml RNasin), brought to 0.5% (v/v) with respect to both Triton X-100 and sodium deoxycholate, and clarified by centrifugation at 13,000 ×g for 10 min. Polysomes were subsequently collected by sedimentation through 2 m sucrose in a SW50.1 rotor (40,000 rpm, 4 hr, 2°C) (Perrone Capano et al., 1986; Giuditta et al., 1991). The purified polysomal pellets were resuspended directly in the translation medium (25 μl) containing 10 μl rabbit reticulocyte lysate (NEN, Boston, MA), 5.5 μl translation cocktail, 96 mm potassium acetate, 1.4 mm magnesium acetate, and 10–25 μCi [35S]methionine (1175 Ci/mmol; NEN), and were routinely incubated at 37°C for 60 min. A sample not containing polysomes was treated similarly and served as a measure of the endogenous activity of the lysate (blank values). To determine protein radioactivity, 1–5 μl aliquots were taken at different times of incubation (up to 1 hr), and acid-precipitable, alkali-resistant material was collected onto GF/C filter disks (Giuditta et al., 1991).
Immunoabsorption analyses. Suitable aliquots of the translation products were treated with 0.5 ml lysis buffer (10 mm Tris-Cl, pH 7.4, 30 mm NaCl, 1 mm EDTA, 0.25% NP-40) and immunoabsorbed on protein A-Sepharose CL-4B (Pharmacia Biotech, Uppsala, Sweden) using rabbit antisera prepared against squid neurofilament (NF) proteins (a generous gift from Dr. H. Gainer, National Institute of Neurological Disorders and Stroke) or nonimmune sera (Szaro et al., 1991; Way et al., 1992). After incubation, the antigen-antibody complex was washed in Ripa buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1% NP-40, 0.5% DOC, 0.1% SDS), and the immunoabsorbed proteins were separated by SDS-PAGE and visualized by fluorography (Harlow and Lane, 1988; Sambrook et al., 1989).
Autoradiography. Two 4 ml aliquots of the synaptosomal fraction were incubated at 18–20°C for 1 hr in a medium containing 0.84 m sucrose, 20 mm NaCl, 10 mmKCl, 10 mm Tris-Cl, pH 7.4, and 10 μCi/ml [3H]leucine (121 Ci/mmol; ICN Pharmaceuticals, Costa Mesa, CA). Protein concentration was kept at 55 μg/ml, within the linear range of the incorporation reaction (Crispino et al., 1993a). One of the two reaction mixtures contained 100 μg/ml cycloheximide. At the end of the incubation period, after the addition of cold synaptosomes (675 μg protein), synaptosomes were collected again by centrifugation (Crispino et al., 1993a). The addition of unlabeled synaptosomes allowed the recovery of a sizable synaptosomal layer that was gently washed with cold medium until excess [3H]leucine was fully removed. Washed synaptosomes were then fixed in 4% paraformaldehyde, 460 mm NaCl, 10 mm KCl, and 20 mm Tris-Cl, pH 7.4, and stored in the same medium. This procedure prevents the retention of free tracers (Bergeron and Droz, 1968), preserves the integrity of synaptosomes, and increases the distance between radiolabeled synaptosomes, which permits the unambiguous identification of the radioactive source in high resolution autoradiograms.
The fixed synaptosomes were treated with 1% osmium tetroxide for 2 hr, dehydrated in acetone, and embedded in Epon. The autoradiograms were produced as described previously (Alvarez and Benech, 1983). Briefly, for light autoradiography, sections were placed on glass slides, sensitized with Ilford K5 emulsion, and developed with D19 after suitable exposure times. Silver grains were counted in five fields of 2240 μm2 with the aid of an ocular reticle. Because grains were present in clusters or appeared randomly dispersed throughout the pellet (see Results), the number of grains was counted separately in the two populations. Background level was determined in regions located just outside the sections and subtracted from grain counts. To estimate the relative contribution of the underlying structures to the overall synaptosomal mass, the diameter and number of the clusters were also determined.
The regions of the synaptosomal fraction exhibiting the most intense autoradiographic response under the light microscope were chosen for high resolution autoradiography. Thin sections were placed on Parlodion-coated slides, stained with lead citrate, coated with carbon, sensitized with Ilford L4 emulsion, exposed at 4°C, and developed with Phenidon. Parlodion films were floated, sections were recovered on grids, and the film was thinned with amyl acetate.
Electron spectroscopic imaging (ESI). Synaptosomes were fixed in filtered seawater (0.2 μm cellulose nitrate filters) containing 2% glutaraldehyde, adjusted to pH 7.4 with NaOH. An optic lobe was fixed and stored in 100 mm sodium cacodylate-HCl buffer, pH 7.4, 348 mm sucrose, 169 mm NaCl, 5% glutaraldehyde diluted 1:1 with filtered seawater. Pieces of the optic lobe were dissected and cut into thin slices.
For conventional electron microscopy, fixed samples were rinsed in artificial seawater (ASW) and osmicated for 1 hr in 2% OsO4 dissolved in half-strength ASW. ESI specimens were not osmicated. Both types of specimens were dehydrated in a graded series of ethanol concentrations, observing long dehydration times and using absolutely dry ethanol in the last three steps. After further infiltration with propylene oxide and propylene oxide/epon mixtures, tissue samples were embedded in Epon. Favorable areas were selected in semithin sections in the light microscope and trimmed for ultramicrotomy.
For ESI, extremely thin sections (10–20 nm) of the unosmicated specimens were collected on 700-mesh thin bar grids. The thickness of the sections was determined by electron energy loss spectroscopy (Door et al., 1991). The energy-filtering transmission electron microscope was model CEM 902 of Carl Zeiss. Phosphorus distribution images were taken on Kodak SO 163 electron image film at ΔE = 150 ± 10 eV, the phosphorus edge being at ΔE = 132 eV. Control images recorded at ΔE = 110 ± 10 eV contained much weaker signals.
RESULTS
In vitro translation of microsomal and synaptosomal polysomes
The synaptosomal fraction from squid optic lobe catalyzes an active process of translation inhibited by cycloheximide, the products of which differ substantially from those of nerve perikarya and glial cells (Crispino et al., 1993a,b). These results indicate that the protein synthetic activity of the synaptosomal fraction cannot be attributed to the latter cells.
The nerve perikarya and glial cells used in the above experiments, however, were not derived from the optic lobe. Hence, the possibility that cells of the optic lobe might synthesize a set of proteins similar to that of the synaptosomal fraction was not excluded. This possibility, although unlikely, was examined directly by purifying polysomes from optic lobe microsomes (a subcellular fraction derived from nerve and glial cell bodies) and comparing their translation products with those of purified synaptosomal polysomes.
As shown in Figure 1, polysomes isolated from the synaptosomal fraction of the optic lobe programmed a massive increase (50-fold) in methionine incorporation over the endogenous activity of the lysate. At 37°C, [35S]methionine incorporation into protein was linear for ∼20 min. Similar kinetics were observed with purified microsomal polysomes. In three independent experiments, polysomes isolated from the synaptosomal fraction yielded 3.8 ± 1.2% (SD) of the activity obtained from total microsomal polysomes. This should be considered a minimal figure because the percentage recovery of synaptosomes is not known, and polysomes released from damaged synaptosomes are likely to be recovered with the microsomal fraction.
Fig. 1.

Kinetics of the translation reaction programmed by synaptosomal and microsomal polysomes from the squid optic lobe. Blank values represent the endogenous activity of the reticulocyte lysate. The different rates displayed by synaptosomal and microsomal polysomes merely reflect the different amounts of polysomes added to each reaction mixture.
To establish that the cell-free translation system was programmed by bona fide polysomes, the putative synaptosomal polysomes were translated in a ribosome-free lysate in which the endogenous ribosomes were removed by previous centrifugation (150,000 × g, 3 hr, 4°C). As shown in Table 1, the ribosome-free lysate retained a significant amount of its polysome translation activity (60–70%) but did not support the translation of exogenous mRNA. As shown in Figure 2, translation of synaptosomal polysomes in the clarified lysate markedly increased (25-fold) methionine incorporation into protein over the endogenous lysate activity (blank values).
Table 1.
Translation activity of the complete and clarified reticulocyte lysatea
| Sample controlb | Cpm/reaction × 104 | % | |
|---|---|---|---|
| Complete | Clarified | ||
| OL polysomes | 285.3 ± 12.08 | 180.3 ± 3.12 | 63.2 |
| BMV mRNA | 120.8 ± 5.54 | 1.8 ± 0.18 | 1.5 |
| Luciferase mRNA | 45.3 ± 3.82 | 0.1 ± 0.07 | 0.3 |
| OL mRNA | 17.5 ± 1.26 | 0.1 ± 0.05 | 0.7 |
aPolysomes isolated from squid optic lobe (OL; 0.1 A260 U) and mRNA standards (0.25 μg each) were translated in standard reactions containing either complete or clarified lysate as detailed in Materials and Methods. All reactions contained 10 μCi[35S]methionine and were incubated for 60 min at 37°C. Values are the mean (±SD) of triplicate determinations. BMV, Brome mosaic virus.
Clarified/complete × 100.
Fig. 2.

Translation activity of synaptosomal polysomes prepared in the presence of either Mg2+ (10 mm) or EDTA (20 mm). Values represent the mean (±SEM) of triplicate determinations of each reaction. *p < 0.001; NS, nonsignificant (two-tailed t test).
In a second control experiment, polysomes from the synaptosomal fraction were isolated in buffers in which Mg2+ was replaced with EDTA (20 mm), an agent known to disrupt polysome structure. Because EDTA-treated polysomes will not sediment through 2.0 m sucrose, samples prepared under these conditions should yield little or no translation activity. As shown in Figure 2, exposure of synaptosomal polysomes to EDTA completely eliminated the translational activity of the samples in the clarified lysate. Identical results were obtained with microsomal polysomes (data not shown).
NF proteins synthesized by synaptosomal polysomes
The fluorograph in Figure 3indicates that on gross inspection the electrophoretic patterns of the translation products of synaptosomal and microsomal polysomes are similar. In contrast, the patterns of the NF proteins isolated by immunoabsorption from the two translation mixtures display differences comparable to those observed previously (Crispino et al., 1993b). In fact, the 60 and 70 kDa NF proteins translated by synaptosomal polysomes show a similar degree of labeling, whereas in microsomal polysomes the 70 kDa form is much less labeled. Densitometric scanning of the fluorograph shown in Figure 3 indicated that the ratio of labeling between the 70 and 60 kDa NF protein was 4.75 times higher in the synaptosomal translation products than in the microsomal translation products. In two additional experiments, the same ratio was found to be 1.6 and two times higher in the synaptosomal translation product.
Fig. 3.

Electrophoretic patterns of the translation products of synaptosomal and microsomal polysomes from the squid optic lobe. Lanes 1 and 4, synaptosomal and microsomal translation products, respectively; lanes 2and 5, after immunoabsorption with a rabbit anti-squid NF antiserum; lanes 3 and 6, after immunoabsorption with a nonimmune antiserum.
The above data indicate that synaptosomal polysomes translate a partially different set of mRNAs than microsomal polysomes. Hence, the synaptosomal protein synthetic activity may not derive entirely from contaminating microsomal polysomes. Rather, it seems to reflect the existence of an intrinsic eukaryotic system of protein synthesis, likely to be localized in the large presynaptic endings that abound in the synaptosomal fraction (Crispino et al., 1993a). To further support the latter hypothesis, additional analyses were aimed at investigating synaptosomal protein synthesis using autoradiographic methods and assessing the localization of synaptosomal polysomes using the ESI procedure (see Materials and Methods).
Autoradiographic localization of newly synthesized synaptosomal proteins
After incubation of the synaptosomal fraction with [3H]leucine, the resulting pellet (see Materials and Methods) examined by transmission EM revealed the presence of a large majority of round profiles ranging in size from <0.1 μm to 2–3 μm diameter (see Fig. 5A). As a rough approximation, the size of the profiles was inversely proportional to the frequency of their occurrence. Small profiles were generally empty, medium profiles contained a variable amount of synaptic vesicles, and large profiles were usually filled with a sizable amount of synaptic vesicles and sometimes damaged mitochondria. Some of the large profiles contained dense core vesicles (not shown). Profiles containing synaptic vesicles were identified as nerve terminals. Interspersed between profiles, clusters of vesicles and additional material could be observed occasionally, indicating damage to some nerve terminals.
Fig. 5.
A, Electron micrograph of the synaptosomal fraction after its incubation with [3H]leucine (see Materials and Methods). Several presynaptic bodies of different size and degree of preservation are identified by their content of synaptic vesicles and/or mitochondria. Partially empty profiles inside presynaptic bodies are remnants of swollen mitochondria, identified by their remaining cristae (synaptosome on the bottom left). A postsynaptic spine attached to a presynaptic body is shown (arrow). Partially empty profiles outside presynaptic terminals are likely to derive from damaged presynaptic bodies (medium-size profiles) or postsynaptic spines (small-size profiles). B–D, High-resolution autoradiograms of the synaptosomal pellet presented inA. Compact clusters of silver grains overlay large presynaptic terminals. Labeling of smaller terminals is shown clearly in B. A high content of synaptic vesicles and a cortical localization of silver grains is present in the large terminal shown inD. Magnification, 16,000×. Scale bar, 1 μm.
Light autoradiograms of the synaptosomal fraction incubated with [3H]leucine exhibited a dual response, i.e., it showed the presence of numerous clusters of silver grains with an average diameter of 4.3 μm and single randomly dispersed silver grains. Clustered and single grains were distributed throughout the pellet (Fig. 4A). Cycloheximide completely suppressed both responses: the grain density over the pellet was barely above background and the clusters of silver grains were conspicuously absent (Fig. 4B). Grain counts indicated that almost two thirds of the protein radioactivity was associated with the clusters (64.5 ± 3.0% as compared with 35.6 ± 3.0% for the dispersed grains; average values with SEM). Because the clusters were found to overlay an area corresponding to 2.9% of the total area, the labeling density of the underlying structures was calculated to be more than 60-fold greater than that of the structures responsible for the disperse labeling (93.4 ± 8.0 as compared with 1.4 ± 0.2 grains/100 μm2).
Fig. 4.

A, Autoradiogram of a synaptosomal fraction after incubation with [3H]leucine. Silver grains have both a clustered and a dispersed distribution. The background of the photographic emulsion is shown in the bottom left corner for comparison. B, Cycloheximide abolishes the response, because clusters and dispersed silver grains are virtually absent. The background is shown in the bottom right corner. In both A and B the underfocused synaptosomes appear as ghosts. Magnification, 600×. Scale bar, 20 μm.
Under the electron microscope, clusters of silver grains were frequent, and they consistently overlaid nerve endings (Fig.5B–D). Sometimes the clusters could be arrayed as a crown of grains sitting on the periphery of the synaptosome (Fig. 5D). Grains were never found to be associated with intraterminal mitochondria. Single dispersed grains were also present, but they did not consistently associate with a particular profile class, although the underlying structure was often a small nerve terminal. A sizable number of presynaptic endings were devoid of silver grains. Most of these structures corresponded to the unlabeled synaptosomes added as carrier to the labeled synaptosomal fraction after the pulse (see Materials and Methods). Taken together, these data indicate that most protein radioactivity was associated with nerve terminals, i.e., almost two thirds with the large nerve terminals, a smaller fraction with terminals of smaller size, and an even smaller amount with material not enclosed in nerve terminals, but presumably derived from damaged presynaptic bodies.
Identification of intraterminal polysomes by ESI analyses
The localization of polysomes and ribosomes in the synaptosomal fraction was examined using the method of ESI. This technique allows the selective visualization of a cluster of as few as 50 phosphorus atoms on the basis of the degree of element-specific energy loss suffered by inelastically scattered electrons (Ottensmeyer, 1986; Wang et al., 1992). Because ribosomes contain several thousand phosphate groups in a limited volume, they may be identified as bright signals of 25 nm diameter on a dark background (Korn et al., 1983). These signals may be distinguished from those emitted by phosphoproteins on the basis of their spectral properties (Martin et al., 1993). Using the ESI method, polysomes have been detected in postsynaptic and distal regions of the squid giant axon (Martin et al., 1989; Giuditta et al., 1991) and more recently in the cortical regions of the goldfish Mauthner axon (Koenig and Martin, 1996).
As shown in Figure6B,C, ESI analyses of the squid synaptosomal fraction indicated that phosphorus signals corresponding in size and spectroscopic properties to those emitted by bona fide polysomes and ribosomes (Fig.7A) were enclosed within the large presynaptic terminals identified by their content of synaptic vesicles and mitochondria. Similar signals were sometimes observed outside the nerve endings, presumably associated with damaged pre- or postsynaptic elements or microsomes.
Fig. 6.

Transmission (A) and electron energy loss spectroscopic micrographs (B, C, ΔE = 150 ± 10 eV) of the synaptosomal fraction. A large nerve terminal with many mitochondria and synaptic vesicles is shown inA. The arrows point to dense spots that may be interpreted as ribosomes. In B andC, many single and clustered ribosome-like signals occur within large nerve terminals. The small arrows inB point to profiles of synaptic vesicles. Magnification, 40,000×. Scale bar, 0.5 μm.
Fig. 7.
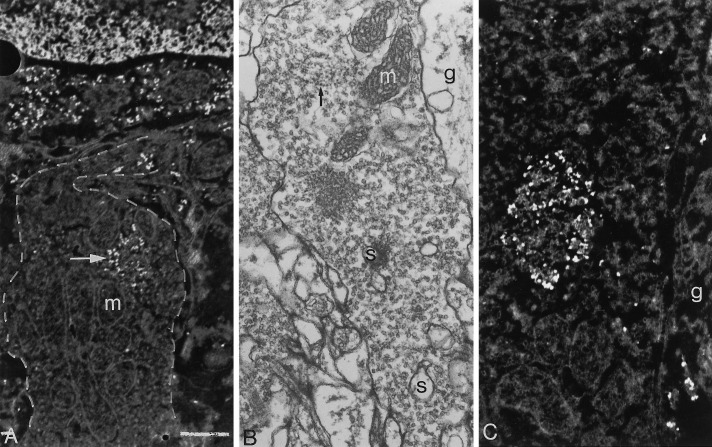
Transmission (B) and electron energy loss spectroscopic micrographs (A, C, ΔE = 150 ± 10 eV) of the cortical layer of the squid optic lobe (deep retina) showing carrot-shaped terminals of the optic nerve (bags). In A, several single and clustered ribosome-like signals (arrow) are seen in the bag outlined by adashed line. Magnification, 25,000×. Scale bar, 0.5 μm. The empty profiles within the bag are mitochondria (m). Identical ribosome-like signals are present in the rough endoplasmic reticulum of the nerve cell body located above the bag. A portion of the nucleus of the same neuron is shown at the top and is full of phosphorus signals derived from nucleosomal DNA. InC, a large aggregate of single and clustered ribosome-like signals is seen within a bag at a higher magnification (40,000×). By conventional EM (B), a carrot-shaped terminal of the optic nerve is characterized by the presence of several mitochondria, a host of synaptic vesicles, and small partially empty profiles representing the indentation of postsynaptic spines (s). Magnification, 25,000×. The arrow points to a long chain of dense spots resembling ribosomes. A glial process (g) is shown in B and C.
In contrast with the selective visualization of ribosomal signals with the ESI method, conventional micrographs of the synaptosomal fraction yielded little or no evidence of the presence of intraterminal ribosomes (Fig. 6A). This discrepancy may be attributed to the inadequacy of conventional EM methods for the identification of ribosomes or polysomes, notably if they occur in isolation or in low concentration, amid a host of additional subcellular components sharing a similar electron opacity.
The identification of ribosomes and polysomes in the large nerve endings of the synaptosomal fraction (Fig. 6) suggested the opportunity to perform similar ESI analyses of the optic lobe with the intent to identify the nerve terminals giving rise to the large endings of the synaptosomal fraction and to determine whether they contained ribosomes and polysomes. This search was greatly facilitated by knowledge that the outer plexiform layer of the optic lobe harbors unusually large, carrot-shaped terminals (bags) of the retinal photoreceptors (Cohen, 1973; Young, 1974). ESI analyses demonstrated that single and clustered ribosomal signals were indeed present in the bags, often in close association with intraterminal mitochondria (Fig. 7A,C). These signals were sometimes observed to form large aggregates (Fig.7C), reminiscent of the similar entities initially observed in the cortical layer of the squid giant axon (Martin et al., 1989; Giuditta et al., 1991) and studied in greater detail in the goldfish Mauthner axon (Koenig and Martin, 1996). As in the case of the synaptosomal fraction, conventional EM provided only ambiguous evidence of the presence of intraterminal ribosomes (Fig. 7B), presumably for the reason outlined above.
DISCUSSION
When the contribution of mitochondria is excluded by the addition of suitable inhibitors, the incorporation of a radiolabeled amino acid into proteins of the synaptosomal fraction (i.e., in the absence of exogenous soluble factors) may be attributed only to cell fragments enclosing eukaryotic polysomes and the requisite soluble factors. Conversely, polysomes located outside these enclosures are expected to remain inactive, because of the dilution of the native soluble factors during the homogenization and sedimentation steps. The complete inhibition of the protein synthetic reaction after hypo-osmotic treatment of the synaptosomal fraction and the insensitivity of the activity of the fraction to RNase are in agreement with this interpretation (Autilio et al., 1968; Morgan and Austin, 1968; Gambetti et al., 1972; Gilbert, 1972).
What has remained elusive, however, is the origin of the cellular fragments that synthesize protein. In principle, they could derive from pinched-off nerve terminals, dendrites, or glial processes, or from fragments of nerve or glial cell bodies, or even from non-neural cells (e.g., endothelial cells). Obviously, more than one type of fragment could contribute to the protein synthetic reaction.
Prompted by the increasing body of evidence indicating the presence of an extramitochondrial system of protein synthesis in several axonal types, notably in the Mauthner axon (Koenig, 1979, 1991; Koenig and Martin, 1996) and the squid giant axon (Giuditta et al., 1977, 1980,1986, 1991; Kaplan et al., 1992; Gioio et al., 1994; Chun et al., 1995,1996, 1997), we have approached the problem of local protein synthesis in presynaptic endings using the synaptosomal fraction of squid optic lobes. This model system offers several advantages in view of its preparation as a floating layer, its high rate of protein synthesis, and the predominance of axo-axonic-type synapses. As shown in previous publications (Crispino et al., 1993a,b), the protein synthetic activity of the squid synaptosomal fraction was completely inhibited by hypo-osmotic shock and strongly inhibited by cycloheximide but was insensitive to RNase. In addition, the pattern of newly synthesized synaptosomal proteins (notably NF proteins) showed substantial differences in comparison with model systems of nerve cell bodies (giant fiber lobe) and glial cells (stellate nerve).
These data supported the view that the active synaptosomal polysomes were not enclosed in contaminating fragments of nerve or glial cell bodies but might be localized in other subcellular structures present in the fraction, such as the large presynaptic endings. The proteins synthesized by the nerve and glial cells of the optic lobe (and hence by their fragments), however, might be more similar to those made by the synaptosomal fraction. If so, the synaptosomal translation activity might not be attributed conclusively to a discrete system of protein synthesis. Although unlikely, this possibility was tested directly by comparing the translation pattern of purified synaptosomal polysomes with that of microsomal polysomes, because microsomes are well known to derive from cell bodies.
The biochemical observations reported in this paper indicate that after immunoabsorption of the newly synthesized NF proteins, the labeling ratio of the 60 and 70 kDa NF proteins differs in the translation products of synaptosomal and microsomal polysomes, in agreement with the more conspicuous difference observed with intact synaptosomes (Crispino et al., 1993b). Because post-translational modifications are not supposed to occur in translation products of purified polysomes, the data indicate that the mRNAs translated by synaptosomal polysomes differ at least in part from those translated by microsomal polysomes. Hence, the translational activity of the synaptosomal fraction cannot be attributed to contamination by cell fragments. Rather, because squid NF proteins are neuron-specific (Szaro et al., 1991; Way et al., 1992), the data suggest that it is attributable to the major neuronal component of the synaptosomal fraction, i.e., the large nerve endings.
In view of these considerations, the gross similarity of the translation patterns of synaptosomal and microsomal polysomes (Fig. 3,lanes 1 and 4) may suggest that their complement of translated mRNAs differs only with regard to a few selected species, such as those coding for the NF proteins; however, two complementary mechanisms might blur additional differences. First, microsomal polysomes contaminating the synaptosomal fraction become active in the presence of exogenous soluble factors. Second, polysomes released from damaged synaptosomes would be recovered in the microsomal fraction. Neither mechanism would be active under conditions in which the translational activity of intact synaptosomes is determined by the incorporation of radiolabeled amino acids, without the supplement of exogenous soluble factors. This circumstance is likely to account for the much larger differences observed between the proteins synthesized by intact synaptosomes and model systems of nerve and glial cells (Crispino et al., 1993a).
The suggestion that the translational activity of the synaptosomal fraction is mostly attributable to the large nerve endings present in the fraction is strongly supported by the results of morphological analyses. ESI data demonstrate that ribosomes and polysomes are present in the large nerve terminals of the synaptosomal fraction. Their identification is based on the selective visualization of clusters of phosphorus atoms afforded by this technique (Korn et al., 1983;Ottensmeyer, 1986). The extremely high density of phosphorus atoms in ribosomes makes them essentially the only cytoplasmic components revealed by the ESI method. In fact, additional phosphorus compounds such as soluble metabolites and membrane phospholipids are mostly extracted out during the preparation of the sample. In this respect, it is relevant that the phosphorus signals detected inside the large presynaptic terminals of the synaptosomal fraction display the same size and spectroscopic properties of the phosphorus signals emitted by particles present in neuronal cytoplasm, which may safely be identified as ribosomes and polysomes (Fig. 7A).
The above observations were confirmed by the results of comparable ESI investigations of the outer plexiform layer of the squid optic lobe, which harbors the large carrot-like terminals of the retinal photoreceptors, the only terminals of such large size present in the optic lobe. As shown in Figure 7A,C, ribosomes and polysomes are present in these terminals examined in situ, because their phosphorus signals are indistinguishable from those emitted by similar particles present in the cytoplasm of a nearby neuron that are undoubtedly ribosomes and polysomes of the rough endoplasmic reticulum. The latter observation excludes the possibility that the ribosomes detected in the presynaptic bodies of the synaptosomal fraction may be attributed to an artifactual uptake of free or membrane-bound polysomes present in the homogenate. It is of more than passing interest that the large nerve terminals of the outer plexiform layer contain large aggregates of ribosomes and polysomes (Fig. 7C), because comparable aggregates are present in the cortical layer of the squid giant axon (Martin et al., 1989; Giuditta et al., 1991) and in the cortical layer of the goldfish Mauthner axon (Koenig and Martin, 1996).
Even more convincing data were obtained by autoradiographic analyses of the synaptosomal fraction after its incubation with [3H]leucine. This additional study demonstrated that approximately two thirds of the newly synthesized synaptosomal proteins are localized in the large nerve endings. This value is likely to be an underestimation, because part of the dispersed radioactivity of the pellet is associated with small nerve endings or with labeled protein set free from damaged presynaptic bodies. Because all autoradiographic signals were almost completely abolished by cycloheximide, they are to be attributed to the activity of eukaryotic polysomes.
Taken together, our previous observations (Crispino et al., 1993a,b) and the data presented in this paper provide strong biochemical and morphological evidence demonstrating that the large presynaptic terminals of the squid photoreceptor neurons contain polyribosomes that translate a particular set of mRNAs. To our knowledge, this is the first unequivocal demonstration that active polysomes are present in nerve endings. Whether this key feature extends to nerve endings of other animal species, notably of mammalian neurons, remains to be investigated. Additional relevant questions raised by these findings concern the role played by presynaptic polysomes in the maintenance and plastic modification of the nerve terminal, their cellular origin, and the neural and humoral mechanisms modulating their activity. Some of these questions may now be examined using the synaptosomal fraction of the squid optic lobe.
Footnotes
This work was supported by United States Public Health Service Grant NS30715, European Community Grant CI1*-CT93–0037; grants from Consiglio Nazionale delle Ricerche (PS Mezzogiorno), Ministero dell’Universita’ E della Ricerca Scientifica e Tecnologica, and the University of Naples; and Grant Fondo Nacional de Desarrollo Cientifico y Tecnologico 1950999 (Chile). We thank the staff of the Marine Biological Laboratory, Woods Hole, MA, for their kind and efficient cooperation during the completion of this work.
Correspondence should be addressed to Professor Antonio Giuditta, Department of General and Environmental Physiology, University of Naples “Federico II,” Via Mezzocannone 8, 80134 Naples, Italy.
REFERENCES
- 1.Alvarez J. Maintenance of the axoplasm: can neurons accord with the accepted notions? Neurosci Lett. 1992;144:1–3. doi: 10.1016/0304-3940(92)90702-9. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez J, Benech CR. Axoplasmic incorporation of amino acids in a myelinated fiber exceeds that of its soma: a radioautographic study. Exp Neurol. 1983;79:283–286. doi: 10.1016/0014-4886(83)90240-6. [DOI] [PubMed] [Google Scholar]
- 3.Austin L, Morgan IG. Incorporation of 14-C-labelled leucine into synaptosomes from rat cerebral cortex “in vitro”. J Neurochem. 1967;14:377–387. doi: 10.1111/j.1471-4159.1967.tb09535.x. [DOI] [PubMed] [Google Scholar]
- 4.Autilio LA, Appel SH, Pettis P, Gambetti PG. Biochemical studies of synapses “in vitro”. I. Protein synthesis. Biochemistry. 1968;7:2615–2622. doi: 10.1021/bi00847a025. [DOI] [PubMed] [Google Scholar]
- 5.Bergeron M, Droz B. Analyse critique des conditions de fixation et de preparation des tissus pour la detection radioautographique des proteines neoformé en microscopie electronique. J Microsc (Paris) 1968;7:51–62. [Google Scholar]
- 6.Chun JT, Gioio AE, Crispino M, Giuditta A, Kaplan BB. Characterization of squid enolase mRNA: sequence analysis, tissue distribution, and axonal localization. Neurochem Res. 1995;20:923–930. doi: 10.1007/BF00970738. [DOI] [PubMed] [Google Scholar]
- 7.Chun JT, Gioio AE, Crispino M, Giuditta A, Kaplan BB. Differential compartmentation of mRNAs in squid giant axon. J Neurochem. 1996;67:1806–1812. doi: 10.1046/j.1471-4159.1996.67051806.x. [DOI] [PubMed] [Google Scholar]
- 8.Chun JT, Gioio AE, Crispino M, Eyman M, Giuditta A, Kaplan BB. Molecular cloning and characterization of a novel mRNA present in the squid giant axon. J Neurosci Res. 1997;49:144–153. [PubMed] [Google Scholar]
- 9.Cohen AI. An ultrastructural analysis of the photoreceptors of the squid and their synaptic connections. III. Photoreceptor terminations in the optic lobes. J Comp Neurol. 1973;147:399–426. doi: 10.1002/cne.901470306. [DOI] [PubMed] [Google Scholar]
- 10.Crispino M, Castigli E, Perrone Capano C, Martin R, Menichini E, Kaplan BB, Giuditta A. Protein synthesis in a synaptosomal fraction from squid brain. Mol Cell Neurosci. 1993a;4:366–374. doi: 10.1006/mcne.1993.1046. [DOI] [PubMed] [Google Scholar]
- 11.Crispino M, Perrone Capano C, Kaplan BB, Giuditta A. Neurofilament proteins are synthesized in nerve endings from squid brain. J Neurochem. 1993b;61:1144–1146. doi: 10.1111/j.1471-4159.1993.tb03632.x. [DOI] [PubMed] [Google Scholar]
- 12.Crispino M, Perrone Capano C, Kaplan BB, Giuditta A. Squid optic lobe synaptosomes: what can they tell us about presynaptic protein synthesis? J Neurochem. 1994;63:387–389. doi: 10.1046/j.1471-4159.1994.63010387.x. [DOI] [PubMed] [Google Scholar]
- 13.Door R, Frosch D, Martin R. Estimation of section thickness and quantification of iron standards with EELS. J Microsc. 1991;162:15–22. [Google Scholar]
- 14.Gambetti P, Autilio-Gambetti LA, Gonatas NK, Shafer B. Protein synthesis in synaptosomal fractions. J Cell Biol. 1972;52:526–535. doi: 10.1083/jcb.52.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert MJ. Evidence for protein synthesis in synaptosomal membranes. J Biol Chem. 1972;247:6541–6550. [PubMed] [Google Scholar]
- 16.Gioio AE, Chun JT, Crispino M, Perrone Capano C, Giuditta A, Kaplan BB. Kinesin mRNA is present in the squid giant axon. J Neurochem. 1994;63:13–18. doi: 10.1046/j.1471-4159.1994.63010013.x. [DOI] [PubMed] [Google Scholar]
- 17.Giuditta A, Metafora S, Felsani A, Del Rio A. Factors for protein synthesis in the axoplasm of the squid giant axons. J Neurochem. 1977;28:1393–1395. doi: 10.1111/j.1471-4159.1977.tb12339.x. [DOI] [PubMed] [Google Scholar]
- 18.Giuditta A, Cupello A, Lazzarini G. Ribosomal RNA in the axoplasm of the squid giant axon. J Neurochem. 1980;34:1757–1760. doi: 10.1111/j.1471-4159.1980.tb11271.x. [DOI] [PubMed] [Google Scholar]
- 19.Giuditta A, Hunt T, Santella L. Messenger RNA in squid axoplasm. Neurochem Intern. 1986;8:435–442. doi: 10.1016/0197-0186(86)90019-7. [DOI] [PubMed] [Google Scholar]
- 20.Giuditta A, Menichini E, Castigli E, Perrone Capano C. Protein synthesis in the axonal territory. In: Giuffrida Stella AM, de Vellis J, Perez Polo R, editors. Regulation of gene expression in the nervous system. Alan Liss; New York: 1990. pp. 205–218. [Google Scholar]
- 21.Giuditta A, Menichini E, Perrone Capano C, Langella M, Martin R, Castigli E, Kaplan BB. Active polysomes in the axoplasm of the squid giant axon. J Neurosci Res. 1991;28:18–28. doi: 10.1002/jnr.490280103. [DOI] [PubMed] [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. [Google Scholar]
- 23.Kaplan BB, Gioio AE, Perrone Capano C, Crispino M, Giuditta A. β-actin and β-tubulin are components of a heterogeneous mRNA population present in the squid giant axon. Mol Cell Neurosci. 1992;3:133–144. doi: 10.1016/1044-7431(92)90017-v. [DOI] [PubMed] [Google Scholar]
- 24.Koenig E. Ribosomal RNA in Mauthner axon: implications for a protein synthesizing machinery in the myelinated axon. Brain Res. 1979;174:95–107. doi: 10.1016/0006-8993(79)90806-0. [DOI] [PubMed] [Google Scholar]
- 25.Koenig E. Local synthesis of axonal proteins. In: Lajtha A, editor. Handbook of neurochemistry, Vol 7, 2nd edition. Plenum; New York: 1984. pp. 315–340. [Google Scholar]
- 26.Koenig E. Evaluation of local synthesis of axonal proteins in the goldfish Mauthner cell axon and axons of dorsal and ventral roots of the rat in vitro. Mol Cell Neurosci. 1991;2:384–394. doi: 10.1016/1044-7431(91)90025-j. [DOI] [PubMed] [Google Scholar]
- 27.Koenig E, Martin R. Cortical plaque-like structures identify ribosome-containing domains in the Mauthner cell axon. J Neurosci. 1996;16:1400–1411. doi: 10.1523/JNEUROSCI.16-04-01400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korn AP, Spitnik-Elson P, Elson D, Ottensmeyer FP. Specific visualization of ribosomal RNA in the intact ribosome by electron spectroscopic imaging. Eur J Cell Biol. 1983;31:334–340. [PubMed] [Google Scholar]
- 29.Martin R, Fritz W, Giuditta A. Visualization of polyribosomes in the postsynaptic area of the squid giant synapse by electron spectroscopic imaging. J Neurocytol. 1989;418:11–18. doi: 10.1007/BF01188419. [DOI] [PubMed] [Google Scholar]
- 30.Martin R, Door R, Breitig D. High resolution imaging of protein phosphorylation in squid axons and synapse by electron energy loss spectroscopy. J Histochem Cytochem. 1993;41:1133–1134. doi: 10.1177/41.8.8331279. [DOI] [PubMed] [Google Scholar]
- 31.Morgan IG, Austin L. Synaptosomal protein synthesis in a cell-free system. J Neurochem. 1968;15:41–51. doi: 10.1111/j.1471-4159.1968.tb06172.x. [DOI] [PubMed] [Google Scholar]
- 32.Nixon RA. Protein degradation in the mouse visual system. I. Degradation of axonally transported and retinal proteins. Brain Res. 1980;200:69–83. doi: 10.1016/0006-8993(80)91095-1. [DOI] [PubMed] [Google Scholar]
- 33.Ottensmeyer FP. Elemental mapping by energy filtration: advantages, limitations and compromises. Ann NY Acad Sci. 1986;487:339–353. doi: 10.1111/j.1749-6632.1986.tb34541.x. [DOI] [PubMed] [Google Scholar]
- 34.Perrone Capano C, Gioio AE, Giuditta A, Kaplan BB. Complexity of nuclear and polysomal RNA from squid optic lobe and gill. J Neurochem. 1986;46:1517–1521. doi: 10.1111/j.1471-4159.1986.tb01770.x. [DOI] [PubMed] [Google Scholar]
- 35.Perrone Capano C, Giuditta A, Castigli E, Kaplan BB. Occurrence and sequence complexity of polyadenylated RNA in squid axoplasm. J Neurochem. 1987;49:698–704. doi: 10.1111/j.1471-4159.1987.tb00950.x. [DOI] [PubMed] [Google Scholar]
- 36.Ramirez G, Levitan IB, Mushynski E. Highly purified synaptosomal membranes from rat brain: incorporation of amino acids into membrane proteins in vitro. J Biol Chem. 1972;247:5382–5390. [PubMed] [Google Scholar]
- 37.Rao A, Steward O. Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized: analysis of proteins synthesized within synaptosomes. J Neurosci. 1991;11:2881–2895. doi: 10.1523/JNEUROSCI.11-09-02881.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 39.Steward O. Alterations in polyribosomes associated with dendritic spines during the reinnervation of the dentate gyrus of the adult rat. J Neurosci. 1983;3:177–188. doi: 10.1523/JNEUROSCI.03-01-00177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steward O, Banker GA. Getting the message from the gene to the synapse: sorting and intracellular transport of RNA in neurons. Trends Neurosci. 1992;15:180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- 41.Szaro BG, Pant HC, Way J, Battey J. Squid low molecular weight neurofilament proteins are a novel class of neurofilament protein. J Biol Chem. 1991;266:15035–15041. [PubMed] [Google Scholar]
- 42.Wang YY, Ho R, Shao Z, Somlyo AP. Optimization of quantitative electron energy loss spectroscopy in the low loss region: phosphorus L-edge. Ultramicroscopy. 1992;41:11–31. doi: 10.1016/0304-3991(92)90091-w. [DOI] [PubMed] [Google Scholar]
- 43.Way J, Hellmich MR, Jaffe H, Szaro B, Pant H, Gainer H : A high-molecular-weight squid neurofilament protein contains a lamin-like rod domain and a tail domain with lys-ser-pro repeats. Proc Natl Acad Sci USA. 1992;89:6963–6967. doi: 10.1073/pnas.89.15.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young JZ. The central nervous system of Loligo. I. The optic lobe. Philos Trans R Soc [Biol] 1974;267:263–302. doi: 10.1098/rstb.1974.0002. [DOI] [PubMed] [Google Scholar]