Abstract
Multiple signaling pathways are thought to control the selective expression of genes over the course of neuronal differentiation. One approach to elucidating these pathways is to identify specificcis-acting elements that serve as the final targets for these signaling pathways in neural-specific genes. We now identify a novel repressive element from the growth-associated protein 43 (GAP-43) gene that can contribute to neuron-specific gene expression by inhibiting transcription in a wide range of non-neuronal cell types. This repressive element is located downstream of the GAP-43 TATA box and is highly position-dependent. When transferred to viral promoters this element preferentially inhibits transcription in non-neuronal cells. Electrophoretic mobility shift assays show that the repressive element comprises at least two protein recognition sites. One of these is a novel sequence motif that we designate the SNOG element, because it occurs downstream of the TATA boxes of the synaptosomal-associated protein of 25 kDa and neuronal nitric oxide synthase genes, as well as the GAP-43 gene. The GAP-43 repressive element is distinct in sequence and position dependence from the repressor element 1/neuron-restrictive silencer element previously described in other neural genes and therefore is a likely target for a distinct set of signaling pathways involved in the control of neuronal differentiation.
Keywords: GAP-43, gene, transcription, neuron, repressive element, SNOG element, TATA box
The differentiation of neurons and other distinct cell types during embryonic development requires the selective activation or repression of many different sets of genes, and a large number of extracellular and intracellular signaling events are thought to control this process. The transcriptional regulation of individual genes is ultimately mediated by short DNA sequences that serve as the binding sites for transcription factors that can either stimulate or repress the activity of the general transcriptional machinery (Mitchell and Tjian, 1989), and the expression pattern of an individual gene can be determined by the integration of the effects of a very large number of these cis-acting elements (Yuh and Davidson, 1996). The identification of individual cis-acting elements involved in neuron-specific gene expression therefore provides a powerful approach to identifying signaling pathways that govern the development of the nervous system.
The gene for the axonal growth cone protein growth-associated protein 43 (GAP-43) is one of a cohort of genes activated in neurons during periods of axon outgrowth (Skene, 1989) and this gene is also expressed in glial cells under some circumstances (Deloulme et al., 1993;Plantinga et al., 1993). GAP-43 is expressed in the great majority of differentiating neurons (Jacobson et al., 1986), suggesting that the regulation of this gene is tied to important differentiation signals common to many neurons.
Transcriptional regulation of the GAP-43 gene involves multiple regulatory elements. Sequences within the first 1000 bases 5′ to the protein coding region can confer a strong preference for neural expression on a reporter gene, both in cell culture systems (Nedivi et al., 1992; Starr et al., 1994) and in transgenic zebra fish (Reinhard et al., 1994). This region contains two potential promoters, a TATA-less promoter proximal to the protein coding region (Ortoft et al., 1993; Eggen et al., 1994; Starr et al., 1994) and a more distal promoter that contains CCAAT and TATA box consensus sequences (Nedivi et al., 1992; Ortoft et al., 1993; Eggen et al., 1994). Although the roles of these putative promoters are not fully resolved, it is clear that regulatory elements within each region can influence transcription of the gene. Results from stable lines of transgenic mice are consistent with the ability of the 5′ flanking sequences to confer preferential expression in the nervous system and have also been taken to suggest that additional sequences within the first intron further refine this neural specificity by suppressing expression in non-neural tissues (Vanselow et al., 1994).
Taken together, these studies indicate that the GAP-43 gene is likely to be a target for multiple signaling pathways involved in the differentiation of the nervous system. One approach to dissecting these signaling pathways is to isolate individual regulatory elements and then to define the response properties of these elements. One small region of the GAP-43 gene that likely contains important neuron-specific regulatory elements is a 386 base pair (bp) DNA sequence that includes the distal promoter. In mammalian cell culture systems, this promoter drives expression of a reporter gene much more strongly in neurons than in other cell types (Nedivi et al., 1992). Moreover, the rat and human promoters have long sequences in the TATA box region with 100% sequence identity (Ortoft et al., 1993; Groen et al., 1995), and the rat 386 bp promoter is preferentially expressed in the developing nervous system of transgenic zebra fish, indicating a high degree of functional phylogenetic conservation (Reinhard et al., 1994). These findings indicate that this DNA sequence contains at least one target site for a signaling pathway(s) critical for development of the vertebrate nervous system.
Here we show that the 386 bp GAP-43 promoter contains a tissue-specific repressive element that inhibits transcription in a wide range of non-neuronal cell types. We also present evidence suggesting that a related element may be present in at least two other neuron-specific genes.
MATERIALS AND METHODS
DNA constructs. The pGL2-Basic plasmid (Promega, Madison, WI), which contains the luciferase reporter gene downstream of a polylinker region, was modified by inserting two simian virus 40 polyadenylation signals upstream of the polylinker region (in addition to the single polyadenylation sequence already present) to reduce cryptic promoter activity driven by vector sequences. The simian virus 40 polyadenylation signal sequences were copied by PCR from the pSVoATCAT vector (Gorman et al., 1982) and cloned into the pGL2-Basic vector using the KpnI and XhoI restriction sites. The resulting plasmid is referred to as pGL3A-Basic. The GAP-43 promoter constructs (Nedivi et al., 1992) were cloned into theHindIII site of the pGL3A-Basic plasmid. The 386 bp GAP-43 promoter comprises sequences from 358 to 742 bp upstream of the ATG start codon of the protein coding region. Modified versions of the 386 bp promoter were produced using a PCR technique in which convenient restriction sites were added to the 5′ ends of the PCR primers (Scharf et al., 1986). Mutations were introduced by including mismatches in PCR primers, and the mutations are reported in the figures or figure legends. The cloning strategy for mutations downstream of the GAP-43 TATA box took advantage of a naturally occurring XbaI site in this region. Modified viral promoters were produced by a similar PCR strategy.
The wild-type adenovirus E1b promoter was made by extending overlapping synthetic oligonucleotides with the Klenow fragment of DNA polymerase I and includes bases −49 to +10 of the sequence published by Wu et al. (1987). The E1b promoter was cloned into the pGL3A-Basic vector using its BglII and HindIII sites. The herpes simplex virus thymidine kinase (TK) promoter was obtained as anEcoRI–PstI restriction fragment (Balkan et al., 1992), and synthetic linkers were added to each end to clone the promoter into the BglII and HindIII sites of pGL3A-Basic. The thymidine kinase promoter is composed of bases −110 to +19 of the sequence published by McKnight and Kingsbury (1982). Because there is a naturally occurring MluI site downstream of the TATA box of the TK promoter, the GAP-43 sequences added to this promoter were synthesized as MluI–HindIII fragments and cloned directly into pGL3A-TK. All constructs made by PCR or direct synthesis of small fragments were confirmed by DNA sequencing.
Cell cultures and transfections. Primary cultures of dissociated rat cerebral cortex from embryonic day 18 were produced as described previously (Nedivi et al., 1992) and cultured in DMEM with 10% supplemented calf serum (Life Technologies, Gaithersburg, MD). Three days after plating, the cortical cultures were treated with 5 μm arabinose C for 24 hr to kill the majority of proliferating cells. HTC hepatoma cells (Thompson et al., 1966) were cultured in DMEM with 10% donor calf serum. RAT2 cells (Topp, 1981), B1.1 cells (the B1.1 cell line was generated by immortalizing primary mouse Schwann cells from neonatal sciatic nerve with a transforming retrovirus; B. Yankner and W. D. Matthew, unpublished observations), and C6 cells (Benda et al., 1968) were cultured in DMEM with 10% fetal bovine serum. PC12 cells (Greene and Tischler, 1976) were cultured in 5% horse serum and 10% fetal bovine serum. CAD Cells (Qi et al., 1997) were cultured in DMEM/F12 medium (BioWhittaker, Walkersville, MD) with 10% fetal bovine serum. The cortical cultures were plated at a density of 2 times 106 cells per 35 mm dish. Cell lines were plated at densities that resulted in ∼1–2 times 106 cells per 35 mm dish at the time of harvest. All cultures were maintained in a humidified 37°C incubator at 5% CO2, except for PC12 cells, which were maintained at 10% CO2. 1× penicillin–streptomycin–fungisome (Life Technologies) was included in all culture media, except during liposome-mediated transfection.
Cortical cultures were transiently transfected 5 d after plating. All cultures were transfected using 0.5 ml of serum-free DMEM containing 18 μg of lipofectin reagent (Life Technologies), 6 μg of plasmid with the promoter–luciferase construct, and 3 μg of an internal control, the Rous sarcoma virus promoter-chloramphenicol acetyltransferase plasmid (pRSVcat) (Gorman et al., 1982). Plasmid DNA and lipofectin reagent were diluted separately into equal volumes of serum free medium and then mixed briefly. The plasmid DNA–lipofectin mixture was incubated at room temperature for a minimum of 40 min before adding to cultures, because shorter incubation times will give much lower and more variable transfection efficiencies, which, along with the lack of an internal control plasmid for monitoring transfection efficiency, likely explains the high degree of experimental variability and the overestimation of the average activity of the 386 bp GAP-43 promoter, relative to the 1 kilobase (kb) GAP-43 promoter, in previous work (Nedivi et al., 1992). Cultures were rinsed once with serum-free medium before adding the plasmid DNA–lipofectin solution. After 5 hr at 37°C, transfection was stopped by changing to medium with serum. All cultures were harvested 2 d after transfection with 250 μl of Promega reporter lysis buffer (catalog #E397A)/35 mm dish.
Luciferase and CAT assays. Luciferase assays were conducted in a Turner luminometer (Promega) using either the Promega luciferase assay system (catalog #E1500) and 20 μl of cell lysate or 50 μl of cell lysate, 180 μl of assay buffer (Brasier and Fortin, 1995), and injection of 100 μl of 2 mm luciferin. CAT assays were done using tritiated acetate (Nordeen et al., 1987). Promoter activity for each construct was determined using duplicate plates and a minimum of three independent experiments. The promoterless luciferase construct pGL3A-Basic was included in every experiment so that the minor luciferase signal driven by vector sequences could be subtracted from activity driven by promoter constructs. CAT enzyme activity from the pRSVcat vector was used to monitor transfection efficiency for each cell culture dish, and luciferase activity was normalized to the CAT activity. To compare promoter activities between cell types, luciferase expression for each promoter construct was normalized to a modified version of the adenovirus E1b promoter that has a deletion of a single G residue upstream of the TATA box (GGGGCGGGGC to GGGGCGGGC). This control promoter has similar activity in neurons and hepatoma cells (signal-to-noise ratios of 8:1 and 9:1, respectively), but is approximately eightfold less active than the wild-type E1b promoter. Unless otherwise indicated, activities of experimental promoter constructs are expressed as a percent of the luciferase activity obtained with this control promoter.
Western blot. Cell lysates were harvested using 0.25% Triton X-100, 1 mm EDTA, and 10 mm Tris-Cl, pH 7.9. Lysates from nontransfected cell cultures were harvested at the same time as the cell cultures for transfection assays. For each cell lysate, 40 μg of total protein was run on a 12% acrylamide-SDS gel (Laemmli, 1970) and then electrotransferred to an Immobilon-N membrane (Millipore, Bedford, MA) in a semidry apparatus using 20% methanol, 150 mm glycine, and 20 mm Tris-Cl, pH 9.0. GAP-43 was detected using monoclonal antibody 9-1E12 as described previously (Schreyer and Skene, 1991).
Nuclear extracts and electrophoretic mobility shift assays.Nuclear extracts from cell cultures were prepared using a modified version of the method of Dignam and coworkers (Abmayr and Workman, 1993) with 300 mm NaCl and a protease inhibitor mixture (catalog #1697498; Boehringer Mannheim, Indianapolis, IN) included in the nuclear extraction buffer. Arabinose C-treated cortical neuronal cultures were harvested 7 d after plating (same as for cultures used in transfections). The nuclear extracts were dialyzed against 20 mm HEPES, pH 7.9, 100 mm KCl, 0.1 mm EDTA, 0.01 mm ZnCl2, 20% glycerol, and 0.5 mm dithiolthreitol. Total nuclear protein for all extracts was quantitated according to the Bradford method (Smith, 1987).
Electrophoretic mobility shift assays were conducted using 5% polyacrylamide (29:1 ratio of acrylamide to bisacrylamide) gels in low ionic strength buffer (Chodosh, 1988). Oligonucleotide probes were synthesized with an Applied Biosystems (Foster City, CA) 391 DNA synthesizer and purified with Applied Biosystems oligonucleotide purification cartridges. Complementary strands were annealed and then purified on nondenaturing 15% polyacrylamide gels. T4 polynucleotide kinase was used to end label probes with 32-phosphate. Binding reactions were conducted for 40 min on ice using 50,000 cpm of radiolabeled probe (∼0.5 ng of DNA), 7 μg of nuclear extract, 2 μg of poly(dI-dC), 15 mm HEPES, pH 7.9, 60 mmKCl, 1 mm MgCl2, 12% glycerol, and 1 mm dithiolthreitol in a reaction volume of 50 μl. Samples were loaded directly onto gels and run at 35 mA for ∼1.5 hr in a 4°C room. The gels were dried and then exposed to film overnight at −80°C with an intensifying screen.
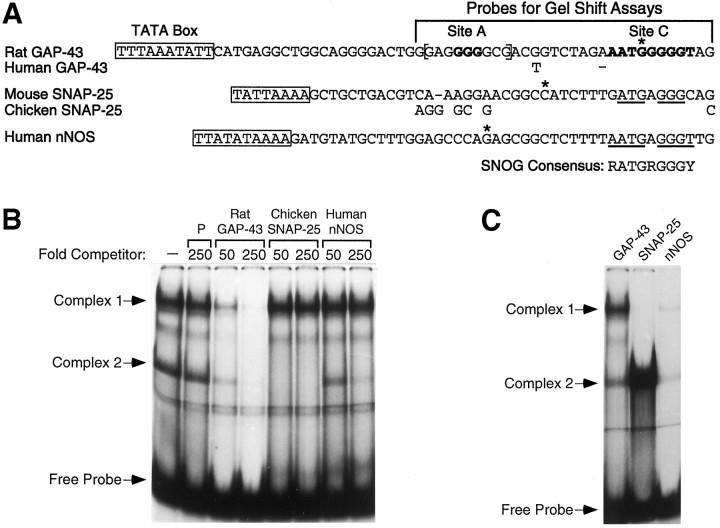
DNA database searches. The GenBank DNA database was searched using the program Stringsearch (University of Wisconsin Genetics Computer Group) for vertebrate entries annotated as having known or putative TATA boxes (TATA_signal). The list produced by the Stringsearch program (2040 sequences) was then searched using the program Findpatterns for any occurrence of the SNOG element consensus sequence (G/A)ATG(G/A)GGG(C/T) located with 60 bp of a (T/A)(T/A)(T/A)(T/A)(T/A)(T/A) sequence. The GenBank annotations of each find were then checked to determine which finds involved proposed TATA boxes. The genes identified by this search were rat GAP-43 (Nedivi et al., 1992), mouse major histocompatibility complex (MHC) Q10-K gene for class I antigen (Watts et al., 1989), mouse MHC Q2-k gene for class I antigen (Watts et al., 1989), mouse MHC D2d gene for class I antigen (Hedley et al., 1989), rat MHC RT1.A gene for class I antigen (Lambracht and Wonigeit, 1995), mouse proteasome Lmp-7 (Zanelli et al., 1993), and chicken myelomonocytic growth factor (Sterneck et al., 1992). The synaptosomal-associated protein of 25 kDa (SNAP-25) and neuronal nitric oxide synthase (nNOS) sequences given in Figure 8 did not show up in this search, because their TATA boxes were not listed in the annotations for their GenBank entries.
Fig. 8.
Phylogenetic conservation of the GAP-43 repressive element and potentially related repressive elements in other neuronal genes. A, Sequences downstream of the TATA boxes for GAP-43, SNAP-25, and neuronal NOS are shown. Human GAP-43 sequences are shown only where different from the rat sequence, and chicken SNAP-25 sequences are shown only where different from the mouse sequence. Adash indicates a gap used for alignment. The bases that were mutated in previous figures to define Site A andSite C of the GAP-43 repressive element are inbold face type (the wild-type sequence is shown), and a sequence similar to the NGFI-A/EGR consensus sequence is inbrackets. SNAP-25 and nNOS sequences that are identical to sequences in Site C are underlined, and a consensus sequence for a potential regulatory element in all three promoter is listed as the SNOG consensus.Asterisks indicate transcription start sites. The mouse SNAP-25 promoter has another major transcription start site located further downstream of the sequence shown (Ryabinin et al., 1995), and there is evidence of multiple GAP-43 transcription start sites (see Fig. 2). B, SNAP-25 and nNOS SNOG element probes were tested as competitors for Complex 2 in an electrophoretic mobility shift assay that used hepatoma nuclear extracts and a radiolabeled GAP-43 repressive element probe (Complex 2 was the site C-specific band in Fig. 6).P, Polylinker DNA. C, GAP-43, SNAP-25, and nNOS probes were radiolabeled and compared for direct binding to the protein(s) in Complex 2. The nNOS band of the same mobility as the GAP-43 complex 1 is discussed in Results.
RESULTS
Identification of a tissue-specific repressive element downstream of the GAP-43 TATA box
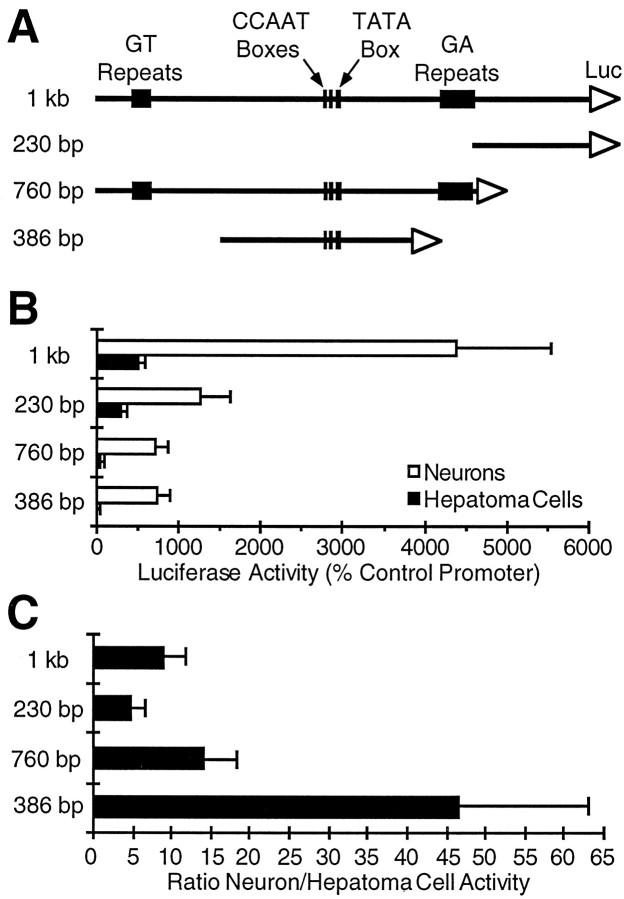
As an initial screen for sequences involved in neuron-specific gene expression, we have assayed the abilities of various GAP-43 promoter fragments to direct expression of a reporter gene in cell cultures highly enriched for neurons from embryonic rat cerebral cortex, compared with a hepatoma cell line. Consistent with previous results (Nedivi et al., 1992), we found that the first 1 kb of DNA 5′ to the coding region of the GAP-43 gene drove reporter gene expression several times more strongly in neuronal cultures than in non-neuronal cells (Fig. 1). We then tested two subdivisions of this 1 kb sequence, a 230 bp fragment containing regulatory sequences and a putative promoter proximal to the GAP-43 coding region (Eggen et al., 1994) and a more distal 760 bp fragment, which contains the promoter with the TATA box (Nedivi et al., 1992;Eggen et al., 1994). Each fragment had only a fraction (<30%) of the activity of the full 1 kb region, suggesting that the transcriptional activity of the 1 kb sequence depends on a synergistic interaction between elements in the two subdivisions. The highly reproducible reduction in activity after subdividing the 1 kb region contrasts with previous results (Nedivi et al., 1992) in which the average activity of the more distal promoter region was comparable to the 1 kb sequence, but with a high degree of variability. The difference appears to reflect improvements in the establishment of the neuronal cultures and in the transfection procedures (see Materials and Methods).
Fig. 1.
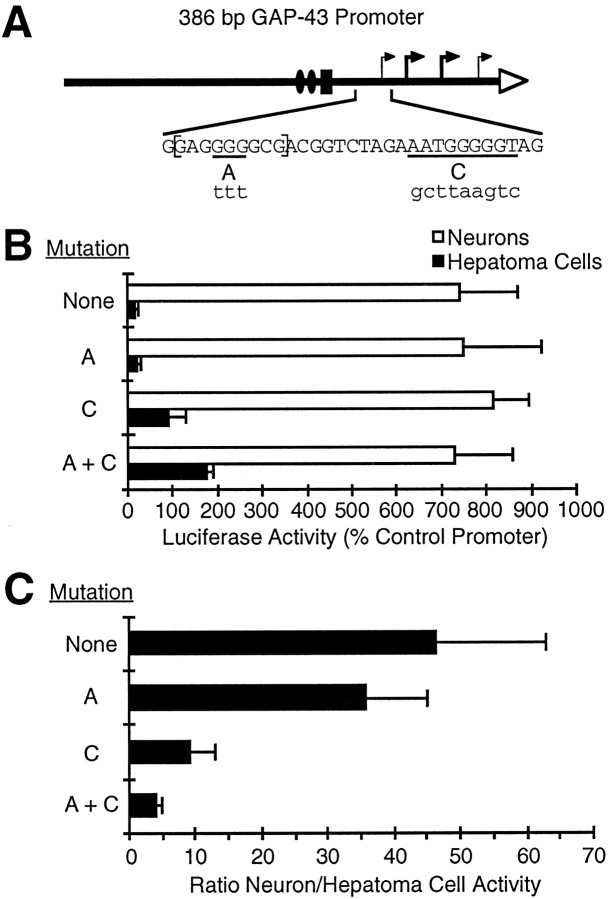
A 386 bp promoter is the most neuron-specific subdivision of the 5′ flanking region of the GAP-43 gene. The rat GAP-43 promoter constructs shown schematically in A were tested for the ability to drive the expression of a luciferase reporter gene (Luc) in primary neuronal cultures or hepatoma cells. Luciferase activity in each cell type is reported inB, and the ratio of activity in neurons to activity in hepatoma cells is reported in C. Promoter activity in each cell type is normalized to the activity of a modified adenovirus promoter that has similar activity in neurons and hepatoma cells (see Materials and Methods). SEs of the mean are based on at least three experiments. If normalized to the activity of an RSV promoter instead, the 1 kb promoter would have an activity of 13.1% RSV in neurons and 2.5% RSV in hepatoma cells. The neuronal cultures are dissociated cells from rat embryonic cerebral cortex treated with an antimitotic agent to kill the majority of non-neuronal cells. The 1 kb promoter construct shown in A comprises the first 1000 bases 5′ to the ATG start codon of the protein coding region of the GAP-43 gene. Consensus CCAAT and TATA sequences are labeled, as are long GT and GA repeating sequences.
Both subdivisions of the 1 kb region directed expression preferentially in neurons compared with non-neuronal cells, suggesting that regulatory elements within each of these subdivisions are used during the course of neuronal differentiation. In our cell culture systems, the highest degree of neuron-specific expression was displayed by a 386 bp fragment containing the distal GAP-43 promoter (Fig. 1). This promoter was expressed more than 40 times more strongly in neurons than in hepatoma cells, making this small region an attractive place to search for regulatory elements involved in neuron-specific gene expression.
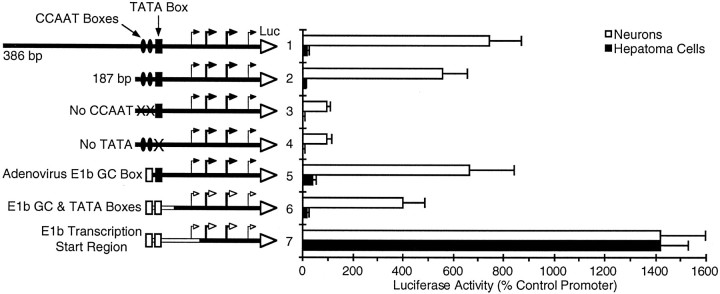
To locate neuron-specific regulatory elements within the 386 bp promoter region, we examined the effects of systematic deletions and substitutions on the activity of this promoter construct in neurons and hepatoma cells (Fig. 2). The promoter contained in the 386 bp sequence includes consensus CCAAT and TATA sequences. Deleting 200 bp upstream of the CCAAT boxes resulted in a small decrease in activity in neurons but did not reduce the ratio of activity in neurons to activity in hepatoma cells (Fig. 2, compare constructs 1, 2). Mutations that disrupted either the CCAAT boxes or the TATA box (Fig. 2, constructs 3, 4, respectively) resulted in an even more dramatic loss of activity in neurons, indicating that the CCAAT and TATA sequences are important for transcriptional activity.
Fig. 2.
A 38 bp sequence located downstream of the GAP-43 TATA box can determine tissue-specific expression. The promoter constructs shown schematically were tested for their ability to drive the expression of a luciferase reporter gene (Luc) in neurons or hepatoma cells (same as Fig. 1). In construct3 both CCAAT consensus sequences have been mutated to C ACCT, and in construct 4 the TATA box sequence has been changed from TTTAAATATT to TT GCAA GCTT. Sequences borrowed from the adenovirus E1b promoter (constructs 5–7) are shown as open boxes or lines. The transcription start sites (bent arrows) for the rat 386 bp GAP-43 promoter are based on RNase protection analysis of human transcripts (Ortoft et al., 1993), because the rat and human promoters are highly homologous in this region. Heavier arrowsdesignate the more dominant transcription start sites. For the hybrid promoters with the adenovirus TATA box (constructs 6, 7), the open arrows are intended only as reference points, because we have not determined where transcription actually starts. The wild-type adenovirus E1b promoter normally initiates transcription from a single site 23 bp downstream of its TATA box (Wu et al., 1987).
To determine whether the CCAAT or TATA sequences might play a critical role in governing the tissue specificity of this promoter, we replaced these elements with equivalent sequences from a promoter that is expressed in a wide range of cell types, the adenovirus E1b promoter (Wu et al., 1987). Replacing the GAP-43 CCAAT boxes with an adenovirus GC box resulted in a small change in neuronal specificity by increasing expression in hepatoma cells (Fig. 2, construct 5). Subsequent replacement of the GAP-43 TATA box and several surrounding bases with an adenovirus TATA box resulted in no further change in tissue specificity, although there was a slight decrease in activity in both cell types (Fig. 2, construct 6).
In contrast, replacing an additional 38 bp downstream of the TATA box resulted in a strong promoter that expressed equally well in both neurons and non-neuronal cells (Fig. 2, construct 7). Although replacement of this small region caused activity in neurons to increase 3.5-fold, the activity in hepatoma cells increased 75-fold (Fig. 2, compare constructs 6, 7). These dramatic changes in activity, which result in a promoter with no tissue specificity, suggest that a region located downstream of the TATA box plays a key role in determining the neuron-specific activity of the 386 bp promoter by preferentially repressing transcription in non-neuronal cells.
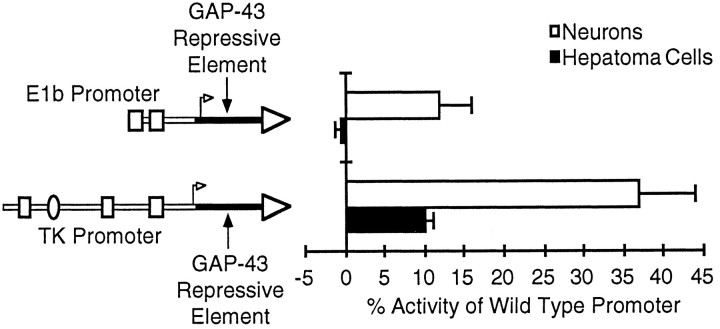
To test whether this region is sufficient to confer tissue-specific expression, we inserted the 38 bp GAP-43 sequence downstream of the TATA boxes of two viral promoters that express well in both neurons and hepatoma cells (Fig. 3). In both cases, the GAP-43 sequence was able to confer a substantial degree of neuronal specificity by preferentially repressing activity in hepatoma cells. For the adenovirus E1b promoter, activity in neurons was reduced eightfold compared with a wild-type E1b promoter, but activity in hepatoma cells was blocked entirely. For the herpes simplex virus thymidine kinase promoter, activity in neurons was reduced 2.8-fold, but activity in hepatoma cells was reduced by >10-fold. It is unclear whether the reduction in activity in neurons means that the repressive element has some effect in neurons, or whether it is an artifact resulting from disturbing the initiator region of the viral promoters. Nevertheless, for both viral promoters the GAP-43 sequence caused a greater reduction in activity in hepatoma cells, resulting in promoters that expressed more strongly in neurons. These findings demonstrate that a 38 bp sequence downstream of the GAP-43 TATA box contains a repressive element, which can preferentially inhibit transcription in non-neuronal cells.
Fig. 3.
The GAP-43 repressive element can confer a preference for expression in neurons on heterologous promoters. The GAP-43 repressive element (solid black line) was inserted downstream of the TATA boxes of two viral promoters, the adenovirus E1b promoter and the herpes simplex virus TK promoter. These hybrid promoters were tested for activity in primary neuronal cultures and hepatoma cells (same as Fig. 1), and in each case promoter activity is reported as a percent of the activity of the wild-type viral promoter with its own transcription initiation region. Replacement of the transcription start region of either viral promoter with the GAP-43 repressive element results in a substantial loss of promoter activity in neurons (discussed in Results) but has an even greater repressive effect in the non-neuronal cells, thereby resulting in a hybrid promoter with a preference for expression in neurons. The wild-type E1b promoter has average signal-to-noise ratios of 61:1 and 65:1 in neurons and hepatoma cells, respectively, and the TK promoter has average signal-to-noise ratios of 58:1 and 100:1 in neurons and hepatoma cells, respectively. The TATA box and upstream GC box of the E1b promoter are shown as open rectangles. The TATA box and two upstream GC boxes of the TK promoter are shown as open rectangles, and its single CCAAT box is shown as an open oval. The wild-type E1b and TK promoters initiate transcription 23 and 20 bp downstream of their TATA boxes, respectively (McKnight and Kingsbury, 1982; Wu et al., 1987). The transcription start site (open arrow) for each hybrid promoter is intended only as a reference point, because we have not determined where transcription actually starts.
Identification of sequence mutations that disrupt the GAP-43 repressive element
The GAP-43 repressive element contains the sequence GAGGGGGCG, which resembles the consensus binding site for the NGFI-A/EGR family of transcription factors (Madden and Rauscher, 1993; Swirnoff and Milbrandt, 1995). At least one member of this family can function as a repressive factor (Werner et al., 1994). We therefore tested whether a mutation that disrupts this sequence could eliminate the ability of the GAP-43 repressive element to preferentially inhibit transcription in non-neuronal cells. Disruption of this sequence (Fig.4A, site A) partially, but not entirely, eliminated the effects of the repressive element on the thymidine kinase promoter (Fig. 4B,C). This partial effect raised the possibility that the repressive element might contain a second binding site, which contributes to preferential expression in neurons. We therefore tested the site A mutation in combination with a series of mutations spanning the 38 bp region. Mutation of site A and a nonadjacent site (Fig. 4, site C) fully eliminated the effects of the GAP-43 repressive element on the thymidine kinase promoter, whereas mutation of either site B or site D in combination with site A had no greater effect on tissue specificity than mutation of site A alone. These results suggest that the repressive element is less than 30 bp in size and may contain binding sites for more than one negative-acting factor.
Fig. 4.
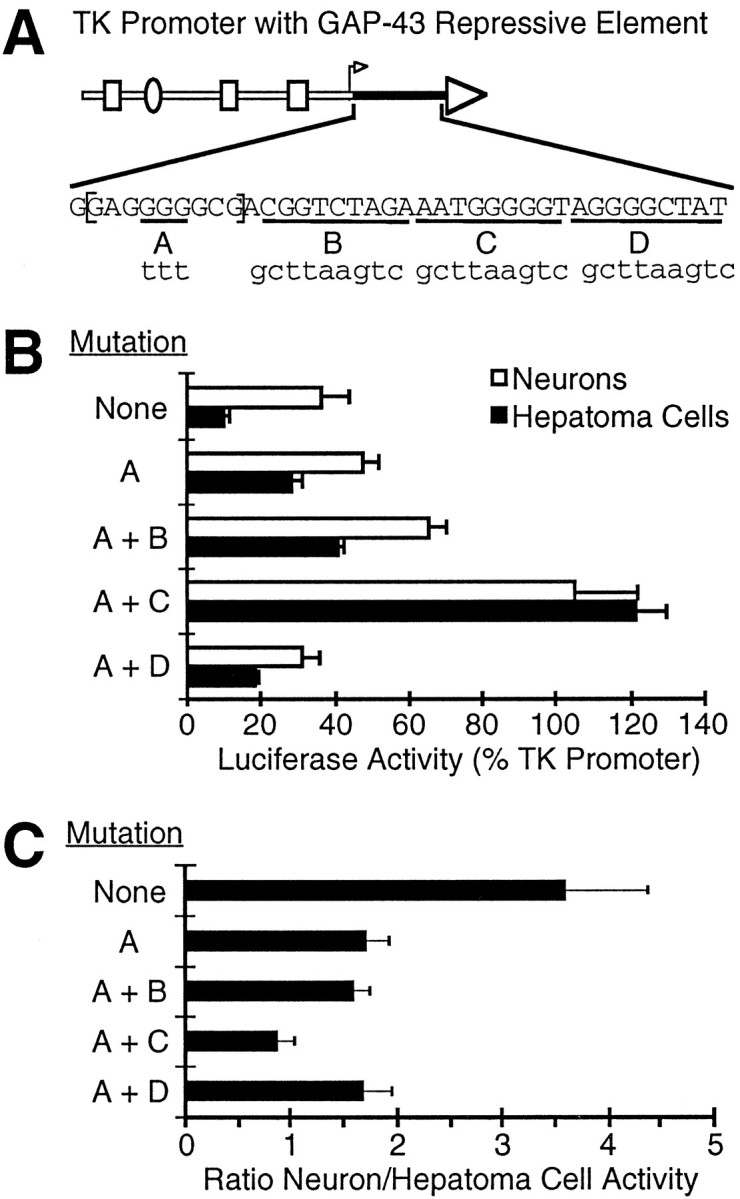
Identification of mutations that disrupt the negative effects of the GAP-43 repressive element. A, The GAP-43 repressive element is shown downstream of the TK promoter. Sequences that were mutated to produce new promoter constructs areunderlined (sites A-D), and the mutant sequences are given in lower-case letters.Brackets enclose a 9 bp sequence with similarity to the NGFI-A/EGR consensus sequence. B, TK promoters with the repressive element and the indicated mutations were tested for activity in neurons and hepatoma cells (same as Fig. 1), and activity in each cell type is reported a percent of the activity of the entirely wild-type TK promoter. Mutation C was not tested on its own in this context. C, The ratio of activity in neurons to activity in hepatoma cells is reported for each promoter construct.
We also mutated sites A and C in the context of the original 386 bp GAP-43 promoter (Fig. 5A), and the greatest effect on tissue specificity was achieved by mutating sites A and C in combination (Fig. 5B). Surprisingly, mutation of site A alone had no significant effect on tissue specificity in the context of the 386 bp promoter. However, mutation of site C alone resulted in a fivefold change in tissue specificity, and mutation of the combination of sites A and C resulted in a 10-fold change in tissue specificity, from a wild-type promoter that expresses over 40 times more strongly in neurons to a mutated promoter that expresses only four times more strongly in neurons. These results demonstrate that the repressive element can account for the majority, although not all, of the tissue specificity of the 386 bp promoter.
Fig. 5.
The repressive element can account for the majority of the tissue specificity of the 386 bp GAP-43 promoter.A, Mutations in sites A andC, which were defined in Figure 4, were introduced into the 386 bp GAP-43 promoter. B, The GAP-43 promoters with the indicated mutations were tested for activity in neurons and hepatoma cells (same as Fig. 1). C, The ratio of activity in neurons to activity in hepatoma cells is reported for each promoter construct.
The order of magnitude change in tissue specificity caused by mutating sites A and C resulted entirely from an increase in activity in hepatoma cells, not from any loss of activity in neurons (Fig.5B). These results indicate that the mutations do not affect the ability of the promoter to initiate transcription in neurons, despite the fact that site C includes the most 5′ transcription start site of the promoter (Nedivi et al., 1992; Ortoft et al., 1993). These observations are consistent with evidence that there are multiple start sites downstream of the GAP-43 TATA box, and that the start site located within site C is a relatively minor transcription start site (Ortoft et al., 1993).
At least two different protein factors bind to the GAP-43 repressive element
The transfection experiments suggest that the repressive element may serve as a binding site for protein factors that preferentially inhibit transcription in non-neuronal cells. We therefore used electrophoretic mobility shift assays to search for factors that bind to the repressive element. Nuclear extracts from hepatoma cells contain at least two distinct binding activities, which specifically recognize the repressive element (Fig.6A, Complex 1 and Complex 2). Competition experiments showed that complex 2 is affected only by mutation of site C. Complex 1, on the other hand, is affected by mutations at sites A and C. An unlabeled repressive element probe with mutations in both sites failed entirely to compete for complex 1 binding, and mutation of either site alone caused an unlabeled probe to compete less effectively. A mutation located between sites A and C (Fig. 4A, siteB) appeared to have no effect on formation of complex 1 or 2 (data not shown). Either of the sequence-specific binding activities demonstrated in Figure 6 are good candidates for a negative-acting transcription factor.
Fig. 6.
Two different protein factors bind specific sequences in the GAP-43 repressive element. A, Nuclear extracts from hepatoma cells were tested in an electrophoretic mobility shift assay for the ability to bind a radiolabeled double-stranded DNA fragment containing the GAP-43 repressive element (the 31 bp sequence used is shown in Fig. 5A). The two most dominant sequence-specific bands are labeled as Complex 1 andComplex 2. The first lane has nuclear extract and labeled probe only, whereas the binding reactions for thenext eight lanes also included an unlabeled competitor probe added at 50- or 250-fold molar excess relative to the labeled probe. The wild-type repressive element (WT) probe competes very effectively against the labeled probe for bothComplex 1 and Complex 2, although probeP (polylinker DNA) fails altogether as a competitor. The competitors labeled as A, C, and A+Crefer to the repressive element probe with mutations in site A, site C, and both sites, respectively (same mutations as in Fig.5A). If a mutation disrupts protein binding, then the probe with that mutation competes less effectively than the WT probe.B, Nuclear extracts from neuronal cortical cultures were tested under conditions identical to those for the gel inA.
Although mutation of the repressive element in the context of the 386 bp GAP-43 promoter did not cause any increase in transcriptional activity in neurons, the putative negative-acting transcription factors are also present in nuclear extracts from neuronal cultures (Fig.6B). Complex 2 appears to be only slightly less abundant in neuronal nuclear extracts than in hepatoma extracts, whereas complex 1 is clearly much less abundant in the neuronal extracts. The relative amount of inhibitory factors to any positive-acting transcription factors might be critical for repressing promoter activity, or the putative negative-acting factors could be regulated by post-translational modifications or cofactors that differ between neurons and non-neuronal cells.
For both nuclear extracts, complex 2 is a single discrete band, but the heavy band that is labeled as complex 1 is surrounded by several minor bands that all appear to have the same binding specificity as complex 1 (although for the neuronal nuclear extracts these bands are quite faint and are not well resolved, even on much longer exposure of the autoradiographs). Multiple bands with the same or very similar DNA-binding specificity could reflect differences in post-translational modifications to a binding factor, the presence or absence of a cofactor, or perhaps binding by a family of related proteins. However, artifactual causes, such as protein damage, might also result in more than one band for the same binding activity.
The GAP-43 repressive element is effective in a wide range of non-neuronal cell types
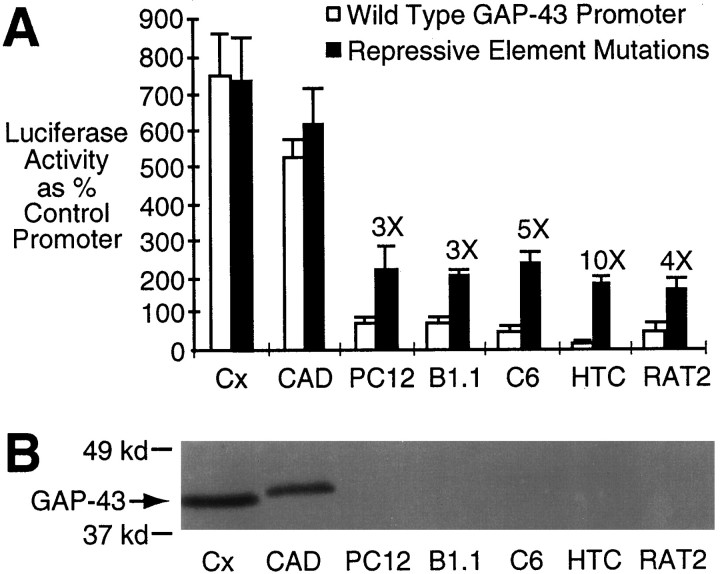
We have demonstrated that the repressive element inhibits the activity of the 386 bp GAP-43 promoter in hepatoma cells but has no effect in primary cultures of cortical neurons. To examine how this repressive element is used in a broader range of cell types, we compared the activity of the 386 bp GAP-43 promoter with mutations A and C to the wild-type promoter in several different cell cultures (Fig. 7A). The wild-type promoter drives expression most strongly in cortical neurons and in a neuronal cell line (CAD cells) (Qi et al., 1997), with a much weaker activity in five non-neuronal cell lines. Mutation of the repressive element in the context of the 386 bp promoter resulted in a 3- to 10-fold increase in the non-neuronal cell cultures but had no significant effect in the two neuronal cultures (primary cortical cultures and CAD cells).
Fig. 7.
The GAP-43 repressive element inhibits transcription in a wide range of non-neuronal cell types.A, Activity of the 386 bp GAP-43 promoter with repressive element mutations A and C (black columns) was compared with the wild-type promoter (white columns) in several different cell types (same methods as in Fig. 1). Significant changes in activity resulting from the repressive element mutations are reported as a fold increase (3×, 5×, 10×, 4×). The cell types are discussed in Results. Cx, Neurons from embryonic rat cerebral cortex; CAD, neuronal cell line;PC12, chromaffin cell derived line; B1.1, Schwannoma cells; C6, glioma cells; HTC, hepatoma cells; RAT2, fibroblast-like cell line.B, Western blot of cell lysates stained for GAP-43. Lysates were harvested from cells cultured under the same conditions as for the transfection assays, and equal amounts of total cellular protein were run in each lane. The slightly slower mobility for the CAD cell GAP-43 (compared with GAP-43 from cortical neuronal cultures) could be attributed to post-translational differences, because GAP-43 can be reversibly palmitoylated (Skene and Virag, 1989; Patterson and Skene, 1994) or phosphorylated (Spencer et al., 1992).
CAD cells constitute a CNS-derived cell line that displays a high level of spontaneous neurite outgrowth, and, like primary cultures from cerebral cortex, CAD cells express high levels of endogenous GAP-43 (Fig. 7B). The non-neuronal cells included three neural-derived lines: naive PC12 cells, which are derived from chromaffin cells of the adrenal medulla; B1.1 Schwannoma cells; and C6 glioma cells. Cells from these neural lineages have been shown to express endogenous GAP-43 under some circumstances (Deloulme et al., 1993; Plantinga et al., 1993, 1994; Costa et al., 1994; Moretto et al., 1995). However, under our culture conditions GAP-43 expression, if any, was below the level of detection for the chromaffin- and glial-derived lines. The hepatoma cell line and RAT2 fibroblast-like cell line do not express any endogenous GAP-43. In this survey, the repressive element was effective in all cell types except for the neurons, which were the only cells that expressed high levels of endogenous GAP-43.
The lack of activity by the wild-type 386 bp promoter in the chromaffin-like cell line (PC12 cells) is consistent with a published report that this promoter does not drive expression of a reporter gene in stably transfected PC12 cells, although sequences in the 230 bp proximal to the GAP-43 coding region could drive reporter expression (Starr et al., 1994). We have not tested our promoter constructs in NGF-treated PC12 cells, because Starr et al. (1994) have reported that NGF treatment did not increase activity from any of the GAP-43 promoter constructs they tested in stably transfected PC12 cells, and there is evidence that the NGF-mediated increase of GAP-43 mRNA in this cell line is mediated by post-transcriptional mechanisms (Perrone-Bizzozero et al., 1993; Starr et al., 1994).
The SNOG element: a potential tissue-specific repressive element in promoters for SNAP-25, neuronal NOS, and GAP-43
To assess the potential biological importance of the GAP-43 repressive element, we examined the phylogenetic conservation of this element and its occurrence in other genes. The repressive element defined by sites A and C is conserved between the rat and human GAP-43 genes. The sequences immediately downstream of the TATA box are shown in Figure 8A, and there is a perfect sequence identity between sites A and C. There are two nonconserved bases located between sites A and C, but the mutation series described in Figure 4 suggested that these intervening bases are not critical for the function of the repressive element. The perfect phylogenetic conservation of sites A and C suggests that the repressive element makes a critical contribution to the regulation of the endogenous GAP-43 gene.
We wanted to know whether other neuron-specific promoters might also use this repressive element. An observation that greatly reduced the difficulty of our search for potentially related elements is that the GAP-43 repressive element appears to be highly position-dependent. After eliminating the repressive element by mutating sites A and C in the context of the 386 bp GAP-43 promoter, we found that reinserting the repressive element in an upstream location did not produce any repression in hepatoma cells (data not shown). This lack of effect was true whether the repressive element was inserted as one or three copies in the forward or reverse orientation, and it did not matter whether the insertion site was 200 bp upstream of the CCAAT boxes or <20 bp upstream of the CCAAT boxes. Because the repressive element may need to be located in close proximity to a transcription start region, we searched for similar sequences located a short distance downstream of known or putative TATA boxes.
The sequences of 17 neuron-specific genes were retrieved from the GenBank database and searched for any similarities to the GAP-43 repressive element. These genes were selected based only on having a known or proposed TATA box and published evidence that the gene product, or in some cases the promoter region, has a preference for expression in neurons (list available on request). Although the combination of site A and C sequences together was not found downstream of a TATA box, the promoters for SNAP-25 and nNOS have a sequence very similar to site C located a short distance downstream of a TATA box (Fig. 8A). The transcription start region of the nNOS promoter (Hall et al., 1994) has a sequence with an 8 of 9 bp identity to site C, and the mouse (Ryabinin et al., 1995) and chicken (Bark, 1993) SNAP-25 promoters have a 6 of 7 bp identity to site C. A possible consensus sequence based on the promoters shown in Figure8A is (G/A)ATG(G/A)GGG(C/T). We have named this consensus sequence the SNOG element (SNAP-25, nNOS, and GAP-43).
We made a list of >2000 vertebrate GenBank entries containing a known or putative TATA box and then searched for the SNOG consensus sequence located within 60 bases downstream of the TATA box. We found only six additional promoters with this combination of sequences, and none included an NGFI-A/EGR-like sequence in close proximity to the SNOG consensus sequence. Four of the six were in genes coding for class I MHC antigen-presenting molecules, and one was in a gene for a proteasome subunit involved in antigen processing (see Materials and Methods for list of genes). The other find was in the gene for cMGF, a myeloid cell-specific growth factor. The results of this GenBank search raise the possibility that the SNOG sequence might also serve as a regulatory element in some non-neuronal promoters involved in immune responses.
The same protein factor binds the SNOG element in three neuronal promoters
The SNOG element alone can account for the binding of one of the two protein factors that recognize the GAP-43 repressive element (Fig.6, mutation of site C, which eliminates the SNOG consensus sequence, disrupted binding to Complex 2). We used electrophoretic mobility shift assays with a nuclear extract from hepatoma cells to test whether the proposed SNAP-25 or nNOS SNOG element could compete for the same binding factor as the GAP-43 SNOG element (Fig. 8B). When the rat GAP-43 repressive element was used as the radiolabeled probe, the chicken SNAP-25 sequence was an even better cold competitor than was the GAP-43 sequence, and the human nNOS sequence appeared to be a slightly less effective competitor than the GAP-43 sequence. Direct labeling of all three probes also indicated that the SNAP-25 SNOG element has a substantially higher affinity for complex 2 binding than does the GAP-43 SNOG element, and the nNOS SNOG element has the lowest affinity (Fig. 8C).
The SNAP-25 and nNOS unlabeled probes did not compete for binding to complex 1 (Fig. 8B). This result was expected, because complex 1 requires both sites A and C for strong binding, and the SNAP-25 and nNOS promoters do not have a sequence similar to the region that encompasses site A. An unexpected finding was that the radiolabeled nNOS probe produced a complex with virtually the same mobility as the GAP-43 complex 1 (Fig. 8C). Although this complex may be in some way related to the GAP-43 complex 1, the binding specificity of the nNOS complex is not identical to that of the GAP-43 complex 1. Under conditions in which cold nNOS probe entirely outcompetes the radiolabeled nNOS probe, a cold GAP-43 repressive element probe fails to compete (data not shown).
These electrophoretic shift assays demonstrate that the SNAP-25, nNOS, and GAP-43 proposed SNOG elements all compete for the same binding factor in hepatoma nuclear extracts, and the order of binding affinity appears to be SNAP-25 > GAP-43 > nNOS. These observations suggest that the SNAP-25 and nNOS promoters may contain a functional repressive element that is related to the GAP-43 repressive element.
DISCUSSION
Identification of a repressive element that inhibits transcription in non-neuronal cells
We have identified a novel repressive element that inhibits gene transcription in a wide range of non-neuronal cells. This ∼30 bp repressive element is sufficient to confer a preference for expression in neurons on two viral promoters, and it accounts for the majority of the neuronal specificity of a 386 bp GAP-43 promoter. In terms of both binding sequence and mechanism of transcriptional repression, the GAP-43 repressive element appears to be quite different from the only well characterized negative-acting element previously demonstrated to be involved in neural-specific gene regulation, the repressor element 1/neuron-restrictive silencer element (RE1/NRSE) that has been identified in several neural-specific genes (Kraner et al., 1992; Mori et al., 1992; Chong et al., 1995; Schoenherr and Anderson, 1995). Neither the NGFI-A/EGR-like site (site A) nor the novel SNOG element (site C) of the GAP-43 repressive element has any similarity to the RE1/NRSE consensus sequence. Furthermore, the GAP-43 repressive element is highly position-dependent. It is effective only if located downstream of a TATA box. In contrast, the RE1/NRSE, which is the binding site for a factor (RE1-silencing transcription factor/neuron-restrictive silencer factor, REST/NRSF) that represses transcription in non-neural cells, fits the formal definition of a silencer element in that it functions independently of orientation and distance from the promoter.
Because the proposed GAP-43 repressive element is located only a short distance downstream of a TATA box, it is possible that the structure of the repressive element sequence is inhibitory to the binding or activity of the factors involved in transcription initiation, but neurons have some specialized factors that override this problem. An alternative model is that the repressive element operates by binding negative-acting factors that repress transcription. The latter mechanism has clear precedent from work on other genes. Both the avian erythrocyte initiation binding receptor (Gomez-Cuadrado et al., 1992;Gomez-Cuadrado et al., 1995) and the cytomegalovirus IE2 protein (Macias et al., 1996) are negative-acting transcription factors that operate by binding downstream of TATA boxes. In principle, such proteins could operate by interfering with either the assembly of the general transcriptional machinery or the conversion of a transcription initiation complex into an elongation complex.
Electrophoretic mobility shift assays indicate that hepatoma cell nuclear extracts contain at least two distinct factors that bind to specific sequences in the GAP-43 repressive element. Either or both of these factors are good candidates for a transcriptional repressor. However, the actual relationship between these factors and the inhibitory activity of the repressive element remains to be established.
Neuronal nuclear extracts contain the same or very similar binding factors as hepatoma extracts, although one of these factors is present at much lower levels. There are several mechanisms by which proteins present in two cell types could inhibit transcription in only one of those cell types. The activity of transcription factors can be regulated by cofactors or post-translational modifications (Calkhoven and Ab, 1996). Protein phosphorylation, for example, is a common mechanism that allows transcription factor activity to respond directly to intracellular signaling pathways (Karin, 1994). The ability to repress transcription could also depend on a ratio of negative to positive factors. For example, the ability of the transcription factor Kruppel to block expression of the even-skipped gene inDrosophila embryos depends not on the simple presence or absence of Kruppel but rather on a delicate balance of limiting amounts of the negative-acting factor Kruppel and the positive-acting factor bicoid (Small et al., 1992).
One of the factors that binds to the GAP-43 repressive element also binds to similar sequences found downstream of the TATA boxes of promoters for two other neuronal genes, SNAP-25 and neuronal NOS. The consensus derived from all three promoters is (G/A)ATG(G/A)GGG(C/T), which we have named the SNOG element. The strong binding we detected with the SNAP-25 SNOG sequence is particularly intriguing, because this sequence is contained in a 241 bp mouse SNAP-25 promoter that Ryabinin and coworkers (1995) have previously shown is highly neuron-specific in transfections of cell cultures. The phylogenetic conservation of the mouse and chicken SNAP-25 SNOG sequence, along with the observation that a factor that binds the GAP-43 repressive element binds with an even higher affinity to the SNAP-25 SNOG element, suggests that this sequence will turn out to be an important regulatory element in the SNAP-25 gene. However, the SNOG element may not serve exactly the same role in regulation of the SNAP-25 and nNOS promoters as it does in the GAP-43 promoter, because the GAP-43 repressive element contains additional sequences not present in the SNAP-25 and nNOS promoters.
In addition to the putative SNOG factor, the GAP-43 repressive element also binds a second nuclear factor (complex 1), which recognizes sequences in both the SNOG element and a nearby NGFI-A/EGR-like sequence. The binding activity of this second factor is fairly well correlated with the inhibitory activity of the repressive element, because it appears to be much more elevated in hepatoma than neuronal nuclear extracts. Furthermore, the same combination of mutations that fully disrupt complex 1 binding also produced the greatest increase in GAP-43 promoter activity in hepatoma cells. At least one member of the NGFI-A/EGR family of transcription factors can function as a transcriptional inhibitor (Werner et al., 1994), and an interesting possibility is that one or more members of the NGFI-A/EGR family may form a complex with the factor that binds to the SNOG element. It remains to be determined, however, whether any of the binding activities we detected in the electrophoretic mobility shift assays actually mediate the functional effects observed for the GAP-43 repressive element.
Transcriptional regulation by a combinatorial code
The transcriptional regulation of the endogenous GAP-43 gene is likely to involve a large number of positive and negative regulatory elements scattered over a distance of several thousand bases. Transgenic zebra fish studies have been used to propose that somewhere in the 1–5 kb upstream of the GAP-43 coding region there is an enhancer recognized in ectodermal cells (Reinhard et al., 1994), and transgenic mouse studies have been used to propose that a region of the first intron suppresses expression in non-neural tissues (Vanselow et al., 1994). Even within the first 1 kb of the 5′ flanking region of the GAP-43 gene, there are important regulatory elements outside of the 386 bp region we have focused on. A 230 bp region located immediately adjacent to the protein coding region has been proposed as a TATA-less promoter which is active in neurons, glia, and chromaffin cells (Eggen et al., 1994; Plantinga et al., 1994; Starr et al., 1994), and an E-box sequence in this region can positively or negatively modulate promoter activity depending on what basic helix-loop-helix proteins bind this site (Chiaramello et al., 1996). Sequences upstream of the 230 bp region are responsive to the neurogenic transcription factor neuroD2, but it is not yet clear whether this activation is a direct effect (McCormick et al., 1996). Moreover, the 386 bp promoter must contain additional regulatory elements involved in tissue specificity, because after mutation of the repressive element this promoter is still expressed more strongly in neurons than in non-neuronal cell types. This high degree of regulatory complexity suggests that the GAP-43 gene may be targeted by a large number of signaling pathways.
Regulation by a combinatorial code involving multiple positive and negative elements may also be true of many other neuronal genes. For example, immediately upstream of the neuron-specific SNAP-25 promoter mentioned above there are sequences that confer responsiveness to the neuronal transcription factors Brn-3a, Brn-3b, and Brn-3c, but even further upstream is a negative element capable of countering the positive effects of Brn-3b (Morris et al., 1994). Also, some of the genes with RE1/NRSE elements have considerable differences in which populations of neurons they are expressed in, suggesting that additional regulatory elements must be involved in the control of their tissue-specific expression. In the case of the synapsin I gene, the RE1/NRSE element can only partially account for the neuron-specific activity of a 253 bp promoter, indicating that there must be at least one additional tissue-specific element in this region (Li et al., 1993).
One potential advantage of the use of a complex combinatorial code is that the regulation of individual genes can be fine-tuned by a large number of spatial and temporal cues in the environment of a cell. Another advantage is that a limited number of transcription factor families can be used to determine the expression patterns of a much larger number of target genes. With these advantages in mind, it is particularly interesting to note that RE1/NRSE elements, although originally identified in neural genes, have now been identified in some non-neural genes (Schoenherr et al., 1996). It is also unlikely that the SNOG element is used exclusively by neuronal genes. We found the SNOG consensus sequence downstream of the TATA boxes of a small number of genes involved in immune responses (see Results), suggesting that in the context of these genes the SNOG element may be used to modulate gene expression in non-neuronal cells under some conditions.
The repressive element in the GAP-43 gene comprises both the novel SNOG site and a second, neighboring site, which bears a similarity to the NGFI-A/EGR consensus sequence. Our present findings show that the repressive element can contribute to neuron-specific gene expression by inhibiting transcription in a wide range of non-neuronal cells. Deactivation of this repressive element, therefore, is likely to be one critical checkpoint in the selective activation of the GAP-43 gene and other SNOG-containing genes in the course of neuronal differentiation. Identification of this repressive element opens a new door to identification of signaling pathways involved in the control of neuronal differentiation.
Footnotes
This work was supported by National Institutes of Health Grant EY07397. The CAD cells were kindly provided by Dona Chikaraishi and Yanping Qi, and the B1.1 Schwannoma cells were kindly provided by Bill Matthew. We thank Eva Reinhard, Dona Chikaraishi, Yanping Qi, and Bill Matthew for critical reading of this manuscript.
Correspondence should be addressed to J. H. Pate Skene, Box 3209, Duke University Medical Center, Durham, NC 27710.
REFERENCES
- 1.Abmayr SM, Workman JL. Preparation of nuclear and cytoplasmic extracts from mammalian cells. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; 1993. pp. 12.1.1–12.1.3. [DOI] [PubMed] [Google Scholar]
- 2.Balkan W, Colbert M, Bock C, Linney E. Transgenic indicator mice for studying activated retinoic acid receptors during development. Proc Natl Acad Sci USA. 1992;89:3347–3351. doi: 10.1073/pnas.89.8.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bark IC. Structure of the chicken gene for SNAP-25 reveals duplicated exon encoding distinct isoforms of the protein. J Mol Biol. 1993;233:67–76. doi: 10.1006/jmbi.1993.1485. [DOI] [PubMed] [Google Scholar]
- 4.Benda P, Lightbody J, Sato G, Levine L, Sweet W. Differentiated rat glial cell strain in tissue culture. Science. 1968;161:370–371. doi: 10.1126/science.161.3839.370. [DOI] [PubMed] [Google Scholar]
- 5.Brasier AR, Fortin JJ. Nonisotopic assays for reporter gene activity. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; 1995. pp. 9.7.12–9.7.14. [DOI] [PubMed] [Google Scholar]
- 6.Calkhoven CF, Ab G. Multiple steps in the regulation of transcription-factor level and activity. Biochem J. 1996;317:329–342. doi: 10.1042/bj3170329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiaramello A, Neuman T, Peavy DR, Zuber MX. The Gap-43 gene is a direct downstream target of the basic helix-loop-helix transcription factors. J Biol Chem. 1996;271:22035–22043. doi: 10.1074/jbc.271.36.22035. [DOI] [PubMed] [Google Scholar]
- 8.Chodosh LA. Mobility shift DNA-binding assay using gel electrophoresis. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; 1988. pp. 12.2.1–12.2.10. [Google Scholar]
- 9.Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 10.Costa JJ, Averill S, Ching YP, Priestley JV. Immunocytochemical localization of a growth-associated protein (GAP-43) in rat adrenal gland. Cell Tissue Res. 1994;275:555–566. doi: 10.1007/BF00318824. [DOI] [PubMed] [Google Scholar]
- 11.Deloulme JC, Laeng P, Janet T, Sensenbrenner M, Baudier J. Expression of neuromodulin (GAP-43) and its regulation by basic fibroblast growth factor during the differentiation of O-2A progenitor cells. J Neurosci Res. 1993;36:147–162. doi: 10.1002/jnr.490360205. [DOI] [PubMed] [Google Scholar]
- 12.Eggen BJL, Nielander HB, Rensen-de Leeuw MG, Schotman P, Gispen WH, Schrama LH. Identification of two promoter regions in the rat B-50/GAP-43 gene. Mol Brain Res. 1994;23:221–234. doi: 10.1016/0169-328x(94)90229-1. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Cuadrado A, Rousseau S, Renaud J, Ruiz-Carrillo A. Repression of the H5 histone gene by a factor from erythrocytes that binds to the region of transcription initiation. EMBO J. 1992;11:1857–1866. doi: 10.1002/j.1460-2075.1992.tb05237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Cuadrado A, Martin M, Noel M, Ruiz-Carrillo A. Initiation binding repressor, a factor that binds to the transcription initiation site of the histone H5 gene, is a glycosylated member of a family of cell growth regulators. Mol Cell Biol. 1995;15:6670–6685. doi: 10.1128/mcb.15.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorman CM, Merlino GT, Willingham MC, Pastan I, Howard BH. The rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci USA. 1982;79:6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groen PC, Eggen BJL, Gispen WH, Schotman P, Schrama LH. Cloning and promoter analysis of the human B-50/GAP-43 gene. J Mol Neurosci. 1995;6:109–119. doi: 10.1007/BF02736770. [DOI] [PubMed] [Google Scholar]
- 18.Hall AV, Antoniou H, Wang Y, Cheung AH, Arbus AM, Olson SL, Lu WC, Kau CL, Marsden PA. Structural organization of the human neuronal nitric oxide synthase gene (NOS1). J Biol Chem. 1994;269:33082–33090. [PubMed] [Google Scholar]
- 19.Hedley ML, Hunt SW, Brorson KA, Andris JS, Hood L, Forman J, Tucker PW. Analysis of D2d: a D-region class I gene. Immunogenetics. 1989;29:359–365. doi: 10.1007/BF00375863. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson RD, Virag I, Skene JHP. A protein associated with axon growth, GAP-43, is widely distributed and developmentally regulated in rat CNS. J Neurosci. 1986;6:1843–1855. doi: 10.1523/JNEUROSCI.06-06-01843.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karin M. Signal transduction from the cell surface to the nucleus through the phosphorylation of transcription factors. Curr Opin Cell Biol. 1994;6:415–424. doi: 10.1016/0955-0674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 22.Kraner SD, Chong JA, Tsay HJ, Mandel G. Silencing the type II sodium channel gene: a model for neural-specific gene regulation. Neuron. 1992;9:37–44. doi: 10.1016/0896-6273(92)90218-3. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lambracht D, Wonigeit K. Sequence analysis of the promoter regions of the classical class I gene RT1.A1 and two other class I genes of the rat MHC. Immunogenetics. 1995;41:375–379. doi: 10.1007/BF00163995. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Suzuki T, Mori N, Greengard P. Identification of a functional silencer element involved in neuron-specific expression of the synapsin I gene. Proc Natl Acad Sci USA. 1993;90:1460–1464. doi: 10.1073/pnas.90.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macias MP, Huang L, Lashmit PE, Stinski MF. Cellular or viral protein binding to a cytomegalovirus promoter transcription initiation site: effects on transcription. J Virol. 1996;70:3628–3635. doi: 10.1128/jvi.70.6.3628-3635.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madden SL, Rauscher FJ. Positive and negative regulation of transcription and cell growth mediated by the EGR family of zinc-finger gene products. Ann NY Acad Sci. 1993;684:75–84. doi: 10.1111/j.1749-6632.1993.tb32272.x. [DOI] [PubMed] [Google Scholar]
- 28.McCormick MB, Tamimi RM, Snider L, Asakura A, Bergstrom D, Tapscott SJ. NeuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol Cell Biol. 1996;16:5792–5800. doi: 10.1128/mcb.16.10.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKnight SL, Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982;217:316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 31.Moretto G, Xu RY, Monaco S, Rizzuto N, Kim SU. Expression and distribution of GAP-43 in human astrocytes in culture. Neuropathol Appl Neurobiol. 1995;21:362–367. doi: 10.1111/j.1365-2990.1995.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 32.Mori N, Schoenherr C, Vandenbergh DJ, Anderson DJ. A common silencer element in the SCG10 and type II Na+ channel genes binds a factor present in nonneuronal cells but not in neuronal cells. Neuron. 1992;9:45–54. doi: 10.1016/0896-6273(92)90219-4. [DOI] [PubMed] [Google Scholar]
- 33.Morris PJ, Theil T, Ring CJ, Lillycrop KA, Moroy T, Latchman DS. The opposite and antagonistic effects of the closely related POU family transcription factors Brn-3a and Brn-3b on the activity of a target promoter are dependent on differences in the POU domain. Mol Cell Biol. 1994;14:6907–6914. doi: 10.1128/mcb.14.10.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nedivi E, Basi GS, Akey IV, Skene JH. A neural-specific GAP-43 core promoter located between unusual DNA elements that interact to regulate its activity. J Neurosci. 1992;12:691–704. doi: 10.1523/JNEUROSCI.12-03-00691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordeen SK, Green PP, Fowlkes DM. A rapid, sensitive, and inexpensive assay for chloramphenicol acetyltransferase. DNA. 1987;6:173–178. doi: 10.1089/dna.1987.6.173. [DOI] [PubMed] [Google Scholar]
- 36.Ortoft E, Pahlman S, Andersson G, Parrow V, Betsholtz C, Hammerling U. Human GAP-43 gene expression: multiple start sites for initiation of transcription in differentiating human neuroblastoma cells. Mol Cell Neurosci. 1993;4:549–561. doi: 10.1006/mcne.1993.1068. [DOI] [PubMed] [Google Scholar]
- 37.Patterson SI, Skene JH. Novel inhibitory action of tunicamycin homologues suggests a role for dynamic protein fatty acylation in growth cone-mediated neurite extension. J Cell Biol. 1994;124:521–536. doi: 10.1083/jcb.124.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perrone-Bizzozero NI, Cansino VV, Kohn DT. Posttranscriptional regulation of GAP-43 gene expression in PC12 cells through protein kinase C-dependent stabilization of the mRNA. J Cell Biol. 1993;120:1263–1270. doi: 10.1083/jcb.120.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plantinga LC, Verhaagen J, Gispen WH. The expression of B-50/GAP-43 in Schwann cells. Ann NY Acad Sci. 1993;679:412–417. doi: 10.1111/j.1749-6632.1993.tb18331.x. [DOI] [PubMed] [Google Scholar]
- 40.Plantinga LC, Schrama LH, Eggen BJL, Gispen WH, Verhaagen J, Lemke G. B-50/GAP-43 mRNA expression in cultured primary Schwann cells is regulated by cyclic AMP. NeuroReport. 1994;5:2465–2468. doi: 10.1097/00001756-199412000-00015. [DOI] [PubMed] [Google Scholar]
- 41.Qi Y, Wang JKT, McMillian M, Chikaraishi DM. Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J Neurosci. 1997;17:1217–1225. doi: 10.1523/JNEUROSCI.17-04-01217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinhard E, Nedivi E, Wegner J, Skene JH, Westerfield M. Neural selective activation and temporal regulation of a mammalian GAP-43 promoter in zebrafish. Development. 1994;120:1767–1775. doi: 10.1242/dev.120.7.1767. [DOI] [PubMed] [Google Scholar]
- 43.Ryabinin AE, Sato TN, Morris PJ, Latchman DS, Wilson MC. Immediate upstream promoter regions required for neurospecific expression of SNAP-25. J Mol Neurosci. 1995;6:201–210. doi: 10.1007/BF02736765. [DOI] [PubMed] [Google Scholar]
- 44.Scharf SJ, Horn GT, Erlich HA. Direct cloning and sequence analysis of enzymatically amplified genomic sequences. Science. 1986;233:1076–1078. doi: 10.1126/science.3461561. [DOI] [PubMed] [Google Scholar]
- 45.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 46.Schoenherr CJ, Paquette AJ, Anderson DJ. Identification of potential target genes for the neuron-restrictive silencer factor. Proc Natl Acad Sci USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schreyer DJ, Skene JHP. Fate of GAP-43 in ascending spinal axons of DRG neurons after peripheral nerve injury: delayed accumulation and correlation with regenerative potential. J Neurosci. 1991;11:3738–3751. doi: 10.1523/JNEUROSCI.11-12-03738.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skene JHP. Axonal growth-associated proteins. Annu Rev Neurosci. 1989;12:127–156. doi: 10.1146/annurev.ne.12.030189.001015. [DOI] [PubMed] [Google Scholar]
- 49.Skene JH, Virag I. Posttranslational membrane attachment and dynamic fatty acylation of a neuronal growth cone protein, GAP-43. J Cell Biol. 1989;108:613–624. doi: 10.1083/jcb.108.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith JA. Quantitation of proteins. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. Wiley; 1987. p. 10.1.1. [Google Scholar]
- 52.Spencer SA, Schuh SM, Liu WS, Willard MB. GAP-43, a protein associated with axon growth, is phosphorylated at three sites in cultured neurons and rat brain. J Biol Chem. 1992;267:9059–9064. [PubMed] [Google Scholar]
- 53.Starr RG, Lu B, Federoff HJ. Functional characterization of the rat GAP-43 promoter. Brain Res. 1994;638:211–220. doi: 10.1016/0006-8993(94)90652-1. [DOI] [PubMed] [Google Scholar]
- 54.Sterneck E, Blattner C, Graf T, Leutz A. Structure of the chicken myelomonocytic growth factor gene and specific activation of its promoter in avian myelomonocytic cells by protein kinases. Mol Cell Biol. 1992;12:1728–1735. doi: 10.1128/mcb.12.4.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swirnoff AH, Milbrandt J. DNA-binding specificity of NGFI-A and related zinc finger transcription factors. Mol Cell Biol. 1995;15:2275–2287. doi: 10.1128/mcb.15.4.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson EB, Tomkins GM, Curran JF. Induction of tyrosine alpha-ketoglutarate transaminase by steroid hormones in a newly established tissue culture cell line. Proc Natl Acad Sci USA. 1966;56:296–303. doi: 10.1073/pnas.56.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Topp WC. Normal rat cell lines deficient in nuclear thymidine kinase. Virology. 1981;113:408–411. doi: 10.1016/0042-6822(81)90168-9. [DOI] [PubMed] [Google Scholar]
- 58.Vanselow J, Grabczyk E, Ping J, Baetscher M, Teng S, Fishman MC. GAP-43 transgenic mice: dispersed genomic sequences confer a GAP-43-like expression pattern during development and regeneration. J Neurosci. 1994;14:499–510. doi: 10.1523/JNEUROSCI.14-02-00499.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watts S, Davis AC, Gaut B, Wheeler C, Hill L, Goodenow RS. Organization and structure of the Qa genes of the major histocompatibility complex of the C3H mouse: implications for Qa function and class I evolution. EMBO J. 1989;8:1749–1759. doi: 10.1002/j.1460-2075.1989.tb03568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Werner H, Rauscher FJ, Sukhatme VP, Drummond IA, Roberts CT, Jr, LeRoith D. Transcriptional repression of the insulin-like growth factor I receptor (IGF-I-R) gene by the tumor suppressor WT1 involves binding to sequences both upstream and downstream of the IGF-I-R gene transcription start site. J Biol Chem. 1994;269:12577–12582. [PubMed] [Google Scholar]
- 61.Wu L, Rosser DS, Schmidt MC, Berk A. A TATA box implicated in E1A transcriptional activation of a simple adenovirus 2 promoter. Nature. 1987;326:512–515. doi: 10.1038/326512a0. [DOI] [PubMed] [Google Scholar]
- 62.Yuh CH, Davidson EH. Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development. 1996;122:1069–1082. doi: 10.1242/dev.122.4.1069. [DOI] [PubMed] [Google Scholar]
- 63.Zanelli E, Zhou P, Cao H, Smart MK, David CS. Genomic organization and tissue expression of the mouse proteasome gene Lmp-7. Immunogenetics. 1993;38:400–407. doi: 10.1007/BF00184520. [DOI] [PubMed] [Google Scholar]