Abstract
Drosophila melanogaster males perform a series of courtship behaviors that, when successful, result in copulation with a female. For over a century, mutations in the yellow gene, named for its effects on pigmentation, have been known to reduce male mating success. Prior work has suggested that yellow influences mating behavior through effects on wing extension, song, and/or courtship vigor. Here, we rule out these explanations, as well as effects on the nervous system more generally, and find instead that the effects of yellow on male mating success are mediated by its effects on pigmentation of male-specific leg structures called sex combs. Loss of yellow expression in these modified bristles reduces their melanization, which changes their structure and causes difficulty grasping females prior to copulation. These data illustrate why the mechanical properties of anatomy, not just neural circuitry, must be considered to fully understand the development and evolution of behavior.
Research organism: D. melanogaster
eLife digest
More than 100 years ago, Nobel-prize winning geneticist Thomas Hunt Morgan and his colleagues discovered that some fruit flies inherited genetic mutations that caused their body color to change. The yellow flies had a mutation in one specific gene and these mutants did not only look different from normal flies, they behaved differently too. Specifically, yellow males were far less successful at mating than normal males, demonstrating for the first time that some behaviors had a genetic basis.
Since then it has remained a mystery how the genetic mutations that cause yellow coloration in fruit flies lead to unsuccessful mating attempts. Geneticists have long suggested that mutations in insect pigment genes cause changes in the fly’s brain because these pigments are made from dopamine, a chemical messenger that acts in the brain. They proposed that yellow flies must have altered levels of dopamine in their brains which was causing them to fail at mating.
To solve this mystery, Massey et al. used a series of genetic experiments and high speed-videos to assess how mutations in male yellow fruit flies affected their mating behavior. The experiments showed that yellow fruit flies mated poorly not because of changes in their brain but because of changes in specialized structures on their legs called sex combs. The yellow males lack melanin pigments in their sex combs, which changes their structure. As a result, the yellow males would court female flies but were then unable to grab and mount them. This explains why yellow flies often fail to mate and why fruit flies have sex combs in the first place.
The study reveals the importance of scientists considering that genes that affect behavior may do so by changing anatomy rather than by altering the brain. The results also may benefit those working to control insect pests. For example, they could help insect pest managers to develop strategies that prevent reproduction in other insects that spread disease or destroy crops.
Introduction
“The form of any behavior depends to a degree on the form of the morphology performing it.” – West-Eberhard (2003)
Over 100 years ago in Thomas Hunt Morgan’s fly room, Alfred Sturtevant described what is often regarded as the first example of a single gene mutation affecting behavior (Sturtevant, 1915; reviewed in Drapeau et al., 2003; Cobb, 2007; Greenspan, 2008): he noted that yellow mutant males, named for their loss of black pigment that gives their body a more yellow appearance (Figure 1A), mated successfully with wild-type females much less often than wild-type males. In 1956, in what is often considered the first ethological study (reviewed in Cobb, 2007; Greenspan, 2008), Margaret Bastock compared courtship of yellow mutant and wild-type males and concluded that despite all courtship actions being present, loss of yellow function likely reduces courtship vigor or drive, leading to copulation inhibition (Bastock, 1956). Despite more recent data consistent with this hypothesis (Drapeau et al., 2003), the precise mechanism by which the yellow gene affects male mating success in D. melanogaster has remained a mystery. Consequently, Bastock’s statement about yellow from her 1956 paper is equally true today: “It seemed worthwhile therefore to examine more closely one example of a gene mutation affecting behavior and to ask two questions, (1) how does it bring about its effect? [and], (2) what part might it play in evolution?”
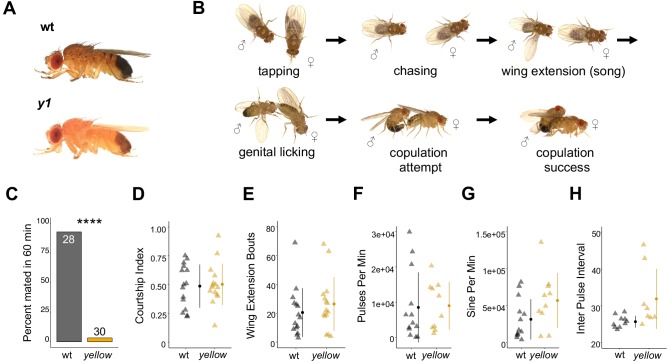
Figure 1. The Drosophila melanogaster yellow gene is required for male mating success.
(A) Photographs comparing wild-type and yellow (y1) body pigmentation [Reprinted from Atlas of Drosophila Morphology, 1 st Edition, Sylwester Chyb and Nicolas Gompel, Body Markers, pp.173, 175, Copyright (2013), with permission from Elsevier] This panel is not covered under the CC-BY 4.0 licence. (B) Snapshots from videos illustrating D. melanogaster courtship behaviors. (C) y1 males (yellow) showed significantly lower mating success levels compared to wild-type males (black) in non-competitive, one-hour trials. Sample sizes are shown at the top of each barplot. (D–H) y1 males showed similar levels of courtship activity and song compared to wild-type males. (D) Courtship index: the proportion of time a male engages in courtship activity divided by the total observation period. (E) Wing extension bouts: the number of unilateral wing extensions during the observation period. (F) Pulses per minute. (G) Sine per minute. (H) Inter pulse interval. (D–H) Show individual points that represent single fly replicates. Circles represent means and lines SD. Significance was measured using Fisher’s exact test in (C) and Welch’s Two Sample t-test in (D–H). Comparisons that were statistically (p<0.05) are indicated (****p<0.0001).
The D. melanogaster yellow gene encodes a protein hypothesized to act either structurally (Geyer et al., 1986) or enzymatically (Wittkopp et al., 2002) in the synthesis of dopamine melanin, and a Yellow homolog has been shown to bind dopamine and other biogenic amines in the sand fly Lutzomyia longipalpis (Xu et al., 2011). The interaction between Yellow and dopamine might explain the protein’s effects on male mating success because dopamine acts as a modulator of male courtship drive in D. melanogaster (Zhang et al., 2016). These effects of dopamine are mediated by neurons expressing the gene fruitless (fru) (Zhang et al., 2016), which is a master regulator of sexually dimorphic behavior in D. melanogaster that can affect every component of courtship and copulation (reviewed in Villella and Hall, 2008). fru has also been shown to regulate expression of yellow in the central nervous system (CNS) of male D. melanogaster larvae (Drapeau et al., 2003). These observations suggest that the pleiotropic effects of yellow on male mating success might result from effects of yellow in the adult CNS, particularly in fru-expressing neurons. Consistent with this hypothesis, functional links between the pigment synthesis pathway and behavior mediated by the nervous system have previously been reported for other pigmentation genes (Hotta and Benzer, 1969; Heisenberg, 1971; Borycz et al., 2002; Richardt et al., 2002; True et al., 2005; Suh and Jackson, 2007).
Results and discussion
Fruitless-expressing cells do not mediate the effect of yellow on male mating success
D. melanogaster males perform multiple behaviors, including tapping, chasing, singing, and genital licking, before attempting to copulate with females by curling their abdomen and grasping the female (Figure 1B, Video 1). In one-hour trials, we found that virgin males homozygous for a null allele of the yellow gene (y1) successfully mated with wild-type virgin females only 3% of the time, whereas wild-type males mated with wild-type virgin females 93% of the time (Fisher’s exact test, p=6×10−13; Figure 1C). Videos of mating trials (e.g., Videos 1 and 2) indicated that the difference in mating success between wild-type and yellow males did not come from differences in the amount of time spent courting (courtship index, t-test, p=0.81; Figure 1D) or the number of wing extensions during the trial period (t-test, p=0.37; Figure 1E). Courtship song analysis also indicated similar amounts of pulse (t-test, p=0.90; Figure 1F), sine song (t-test, p=0.07; Figure 1G), and interpulse interval (t-test, p=0.07; Figure 1H). Watching the courtship videos showed that copulation initiation was most strikingly different between the two genotypes, with copulation initiation reduced in yellow males compared to wild-type (compare Videos 3 and 4).
Video 1. Wild-type courtship and copulation.
Video 2. y1 courtship with wild-type female.
Video 3. Wild-type copulation.
Video 4. Copulation attempts between y1 male and wild-type female after 3 hr of courtship.
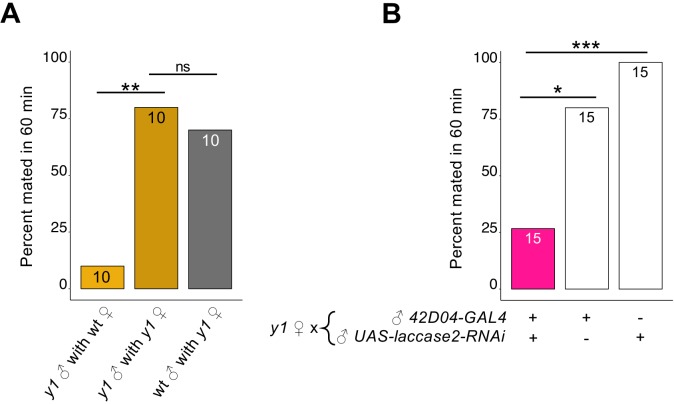
To determine whether yellow activity in fru-expressing cells was responsible for this difference in mating success, we used the UAS-GAL4 system (Brand and Perrimon, 1993) to drive expression of yellow-RNAi (Dietzl et al., 2007) with fruGAL4 (Stockinger et al., 2005), knocking down native yellow expression in these cells. We also used fruGAL4 to drive yellow expression in y1 mutants. In both cases, when the experimental genotype was compared to the (1) GAL4 only and (2) UAS only control genotypes using a Fisher’s exact test (FET) with p-values adjusted (Bonferroni) for the (n = 2) control comparisons, we found no significant effect on male mating success (Figure 2A, p=1 for both tests; Figure 2B, p=0.07 and 0.2), suggesting that expression of yellow in fru-expressing cells is neither necessary nor sufficient for yellow’s effect on male mating success.
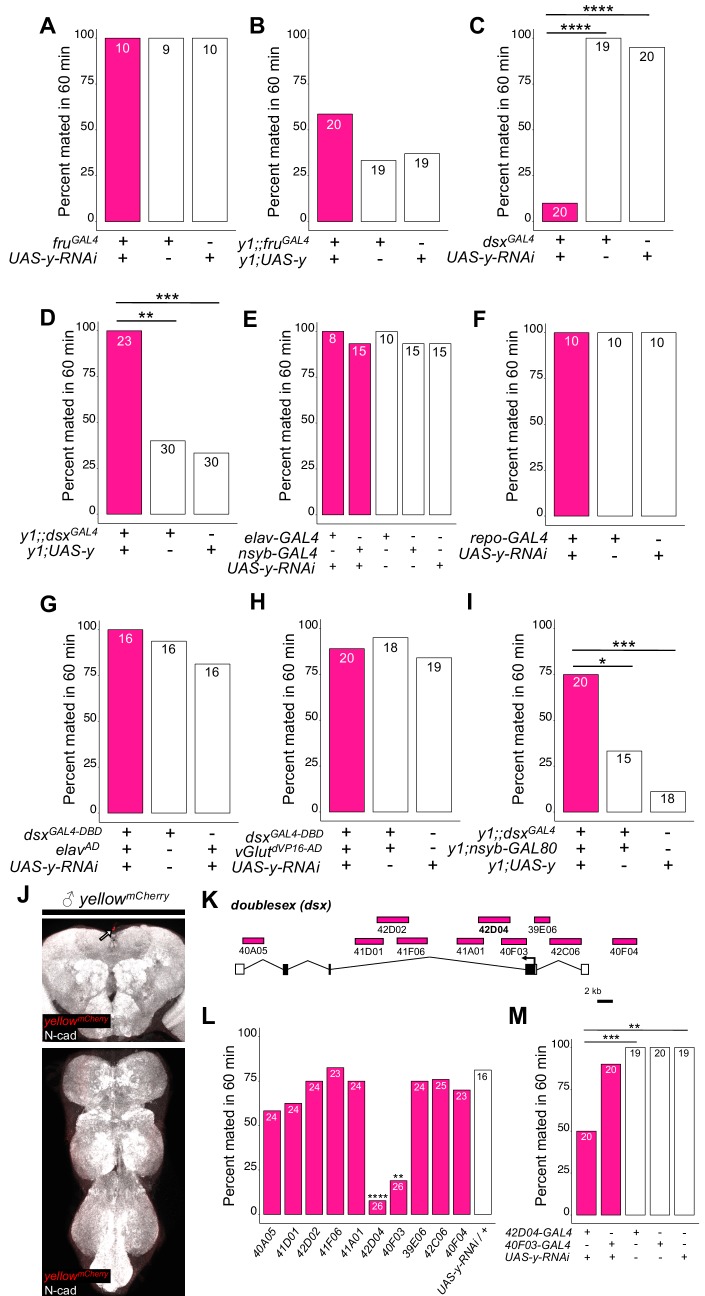
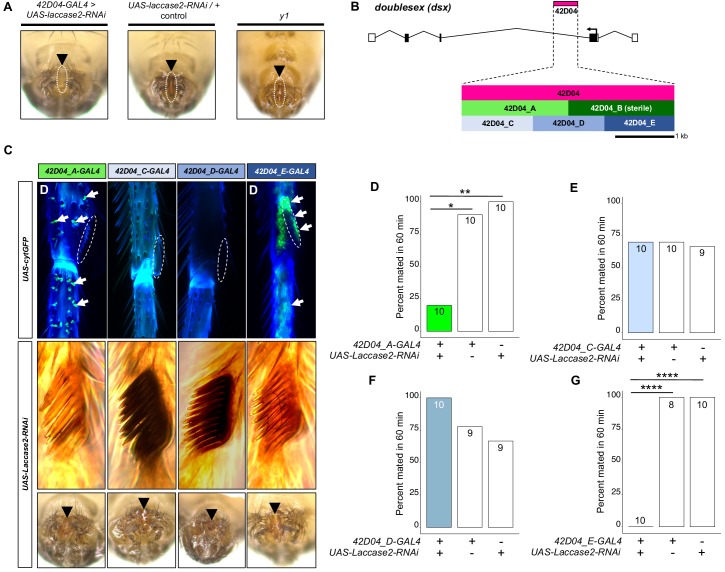
Figure 2. yellow expression in non-neuronal doublesex-expressing cells, but not fruitless-expressing cells, is necessary and sufficient for male mating success.
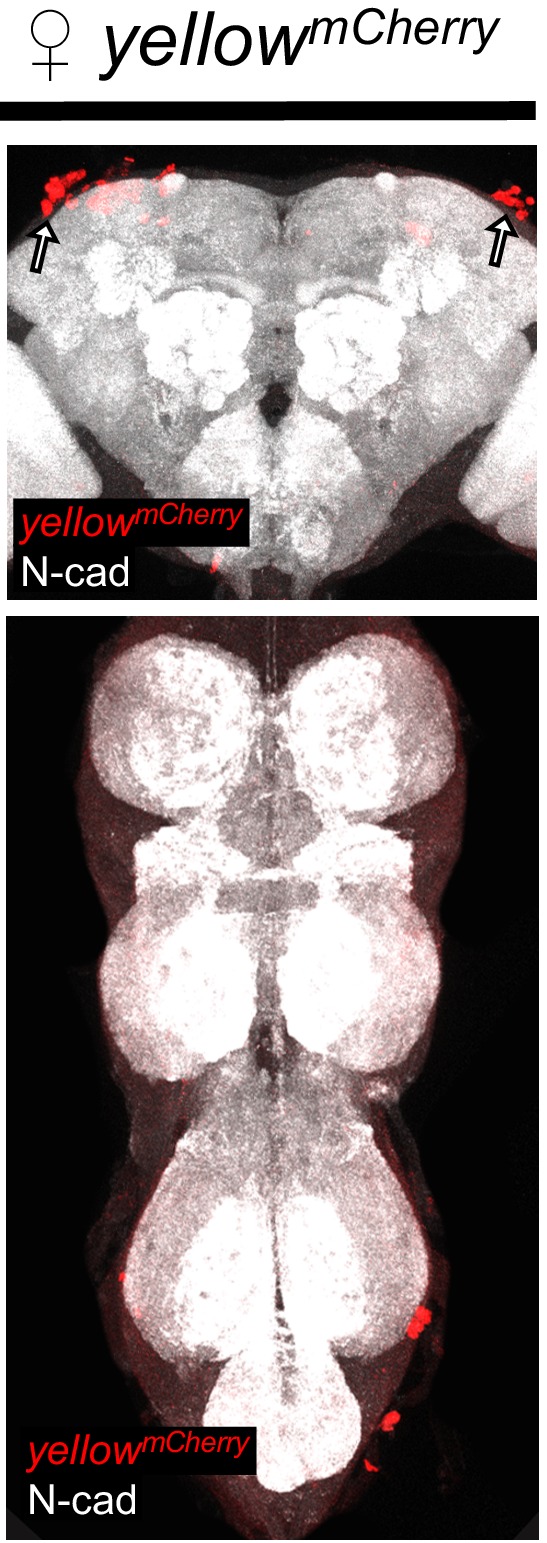
(A,B) Neither expressing yellow-RNAi nor yellow-cDNA in fru-expressing cells using fruGAL4 (Stockinger et al., 2005) affected male copulation. (C) Expressing yellow-RNAi in dsx-expressing cells using dsxGAL4 (Robinett et al., 2010) significantly inhibited male mating success. (D) Expressing yellow in dsx-expressing cells using dsxGAL4 in a y1 mutant background was sufficient to restore male mating success. (E,F) Expressing yellow-RNAi using pan-neuronal (elav-GAL4 and nsyb-GAL4) and pan-glia (repo-GAL4) drivers did not affect male mating success. (G) Restricting yellow-RNAi expression to dsx-expressing neurons using the split-GAL4 technique, combining dsxGAL4-DBD (Pavlou et al., 2016) with elavVP16-AD (Luan et al., 2006), did not affect male mating success. (H) Restricting yellow-RNAi expression to dsx-expressing glutamatergic neurons using the split-GAL4 technique, combining dsxGAL4-DBD (Pavlou et al., 2016) with vGlutdVP16-AD (Gao et al., 2008) did not affect male mating success. (I) Expressing yellow in dsx-expressing cells restricted outside the CNS using dsxGAL4 and nsyb-GAL80 (courtesy of Julie Simpson) in a y1 mutant background significantly increased male mating success. (J) Brain and ventral nerve cord of adult male ymCherry flies stained with anti-N-Cadherin (N-cad) antibody labeling neuropil (white) and anti-DsRed antibody labeling Yellow::mCherry (red). We observed sparse, inconsistent signal outside the CNS at the top of the brain in males (white arrow), but we were unable to confirm a previous report that ymCherry is expressed in the adult brain (Hinaux et al., 2018). (K) Diagram of the male exon structure of the dsx locus highlighting 10 genomic fragments between 1.7 and 4 kb used to clone Janelia enhancer trap GAL4 drivers (Pfeiffer et al., 2008). Black boxes indicate coding exons. White boxes indicate 5’ and 3’ UTRs, and the arrow in exon two denotes the transcription start site. (L) Expressing yellow-RNAi using each Janelia dsx-GAL4 driver identified 42D04-GAL4 and 40 F03-GAL4 as affecting male mating success when compared with the yellow-RNAi control. (M) A replicate experiment comparing 42D04-GAL4 and 40F03-GAL4 effects on male mating success with both GAL4 and UAS parental controls confirmed the significant effect of 42D04-GAL4 but not 40F03-GAL4. We attribute differences in the 40F03-GAL4 effect between (L) and (M) to between experiment variability in the levels of male mating success; each common genotype tested in (L), for example, mated at higher levels in (M), but 42D04-GAL4 consistently showed a significant effect relative to controls. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
Figure 2—figure supplement 1. yellow expression in dsx-expressing cells is necessary and sufficient for male mating success.
Figure 2—figure supplement 2. The mating regulatory sequence (MRS) from Drapeau et al. (2006) does not affect male mating success.
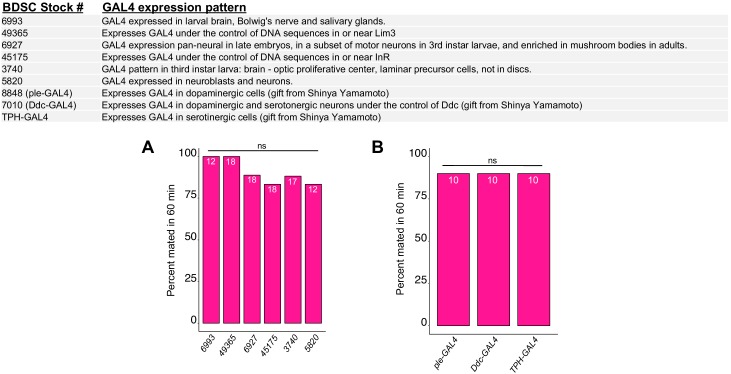
Figure 2—figure supplement 3. Expressing yellow-RNAi in subsets of CNS tissue does not affect male mating success.
Figure 2—figure supplement 4. ymCherry expression in adult female central nervous system.
Doublesex-expressing cells require yellow for normal male mating success
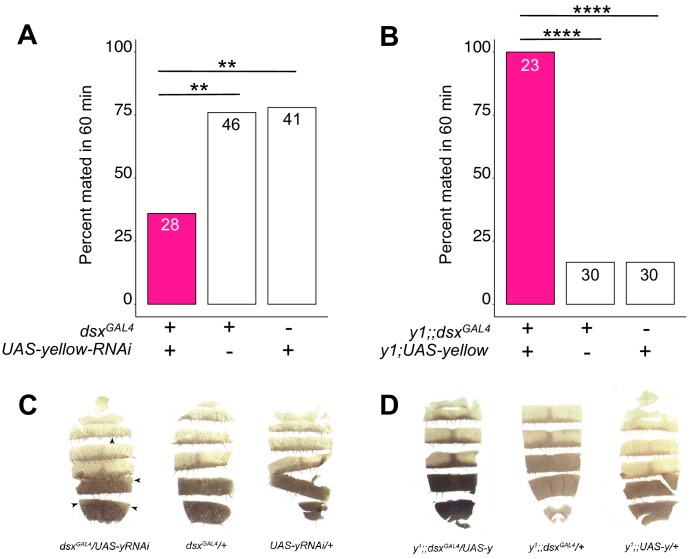
To continue searching for cells responsible for yellow’s effects on mating, we examined a 209 bp sequence 5’ of the yellow gene called the ‘mating-success regulatory sequence’ (MRS) in a prior study that reported it was required for male mating success (Drapeau et al., 2006). We hypothesized that the MRS might contain an enhancer driving yellow expression and found that ChIP-seq data indicate the Doublesex (Dsx) transcription factor binds to this region in vivo (Clough et al., 2014). Like fru, dsx expression is required to specify sex-specific behaviors in D. melanogaster (Rideout et al., 2010; Robinett et al., 2010; reviewed in Villella and Hall, 2008; Yamamoto and Koganezawa, 2013), suggesting that yellow expression regulated by Dsx through the MRS enhancer might be responsible for its effects on male mating behavior. We found that reducing yellow expression in dsx-expressing cells with either of two different dsxGAL4 drivers (Robinett et al., 2010; Rideout et al., 2010) strongly reduced male mating success (Figure 2C, FET, p=7×10−9 and 1 × 10−7; Figure 2—figure supplement 1A, FET, p=0.002 and 0.002), whereas restoring yellow activity in cells expressing dsxGAL4 in y1 mutants significantly increased male mating success compared with y1 controls (Figure 2D, FET, p=0.001 and 0.0004; Figure 2—figure supplement 1B, FET, p=5×10−10 and 5 × 10−10). Video recordings of male flies with reduced yellow expression in dsx-expressing cells showed the same mating defect observed in y1 mutants: males seem to perform all courtship actions normally, but repeatedly failed to copulate (Video 5). We therefore conclude that yellow expression is required in dsx-expressing cells for normal male mating behavior.
Video 5. Copulation attempts between male expressing yellow-RNAi in dsxGAL4-expressing cells and wild-type female.
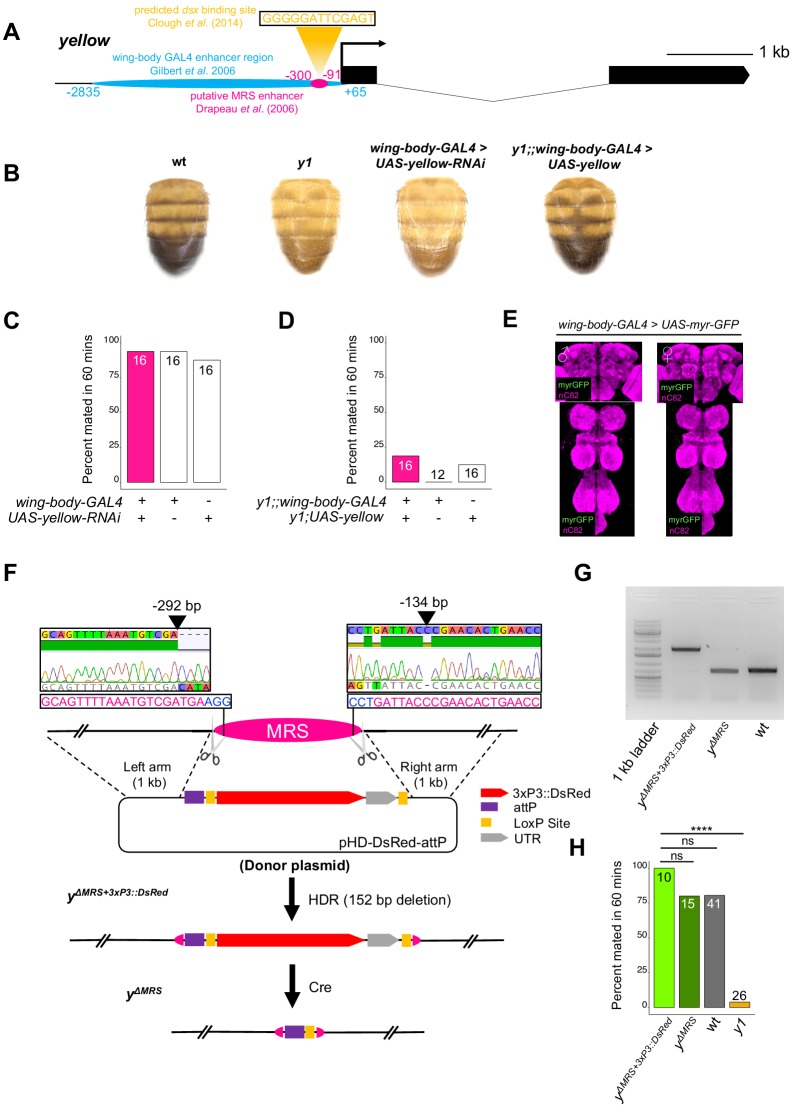
To determine whether the MRS sequence might be the enhancer mediating yellow expression in dsx-expressing cells that affects male mating success, we manipulated yellow expression with GAL4 driven by a 2.7 kb DNA region located 5’ of yellow that includes the wing, body, and putative MRS enhancers (Gilbert et al., 2006, Figure 2—figure supplement 2A). Altering yellow expression with this GAL4 driver modified pigmentation as expected but did not affect male mating success (Figure 2—figure supplement 2B–D), possibly because this GAL4 line did not show any detectable expression in the adult CNS (Figure 2—figure supplement 2E). To test more directly whether the MRS was necessary for male mating success, we deleted 152 bp of the 209 bp MRS sequence using CRISPR/Cas9 gene editing (Bassett et al., 2013) (Figure 2—figure supplement 2F,G). We found that this deletion had no significant effect on male mating success (Figure 2—figure supplement 2H, FET, p=0.99 compared to wild-type (CS)), contradicting the previous deletion mapping data (Drapeau et al., 2006). We conclude therefore that effects of yellow expression in dsx-expressing cells on mating behavior are likely mediated by other cis-regulatory sequences associated with the yellow gene.
dsx-expressing cells outside the CNS require yellow for normal male mating success
Although dsx is expressed broadly throughout the fly (Robinett et al., 2010; Rideout et al., 2010), we hypothesized that its expression in the nervous system would be responsible for yellow’s effects on mating because yellow has been reported to be expressed in the adult brain (Hinaux et al., 2018) and behavioral effects of other pigmentation genes are mediated by neurons (Hotta and Benzer, 1969; Heisenberg, 1971; Borycz et al., 2002; True et al., 2005). However, we found that suppressing yellow expression in the larval CNS, dopaminergic neurons, or serotonergic neurons (Figure 2—figure supplement 3, FET, P values ranging from 0.45 to 1), or in all neurons (Figure 2E, FET, p=1 in all cases) or all glia (Figure 2F, FET, p=1), had no significant effect on male mating success. Specifically reducing yellow expression in either all dsx-expressing neurons (Figure 2G, FET, p=1 and 0.45) or all dsx-expressing glutamatergic neurons that are required for genital coupling (Pavlou et al., 2016) (Figure 2H, FET, p=1 and 0.68) also had no significant effect on male mating success. In addition, when we examined yellow expression in adult brains, we were only able to observe non-specific signal at the anterior of the adult brain in females (Figure 2J, Figure 2—figure supplement 4). Given this lack of evidence that yellow is required in neuronal cells for normal male mating behavior, we limited dsxGAL4 activation of yellow expression in y1 mutants to non-neuronal cells and found that these flies exhibited an increase in male mating success compared with y1 mutant males (Figure 2I, FET, p=0.04 and 0.0002), showing that yellow expression in non-neuronal dsx-expressing cells is required for normal male mating behavior.
To identify which non-neuronal dsx-expressing cells require yellow expression for normal male mating success, we screened ten dsx-enhancer GAL4 lines that each contains a different ~ 3 kb region of dsx noncoding sequence (Figure 2K; Pfeiffer et al., 2008). Two of these lines, 42D04-GAL4 and 40F03-GAL4, significantly decreased male mating success when driving yellow-RNAi (Figure 2L, FET, p=0.001 and 2 × 10−5). These two GAL4 drivers contain overlapping sequences from intron 2 of dsx (Figure 2K), suggesting that their similar effects result from reduction of yellow expression in the same cells. Line 42D04-GAL4 had stronger effects than 40 F03-GAL4 (Figure 2M, FET, p=0.0009 for both controls for 42D04-GAL4 versus p=0.97 for both controls for 40 F03-GAL4), so we performed all further analyses with 42D04-GAL4. Males with yellow reduced by 42D04-GAL4 performed courtship behavior in a pattern similar to y1 mutant males: males performed all precopulatory courtship behaviors normally, but repeatedly failed to copulate, even after hours of attempts (Video 6). These data indicate that some or all cells in which 42D04-GAL4 drives expression require yellow expression for normal male mating behavior.
Video 6. Copulation attempts between male expressing yellow-RNAi in 42D04-GAL4-expressing cells and wild-type female.
Sex combs require yellow expression for normal male mating success
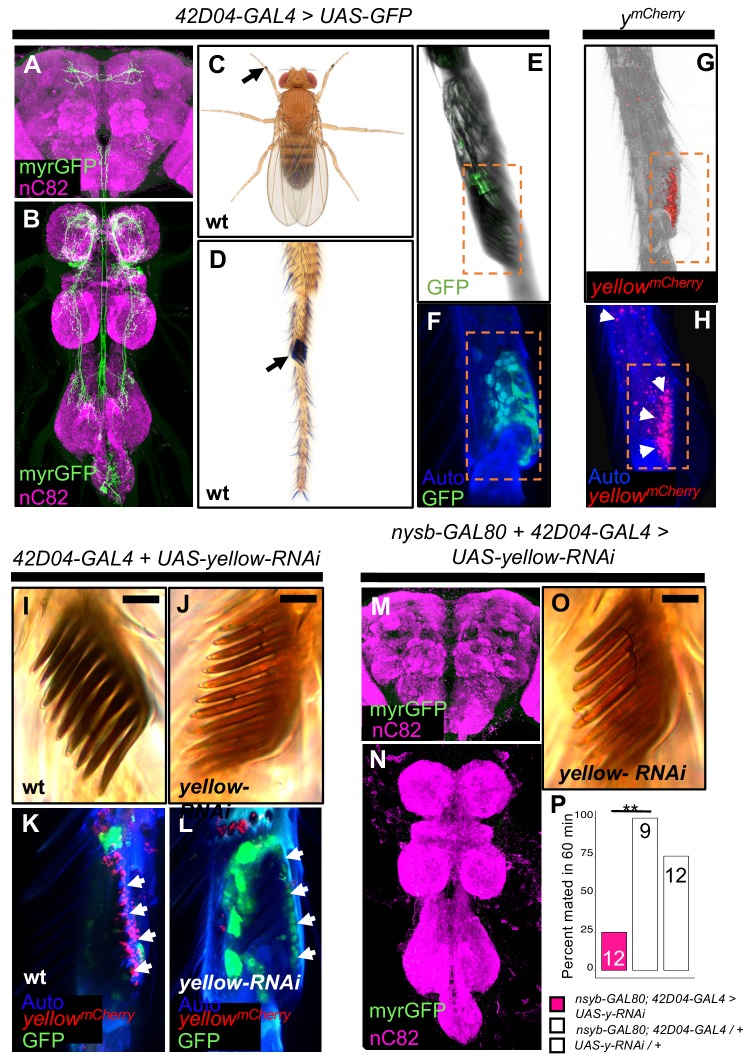
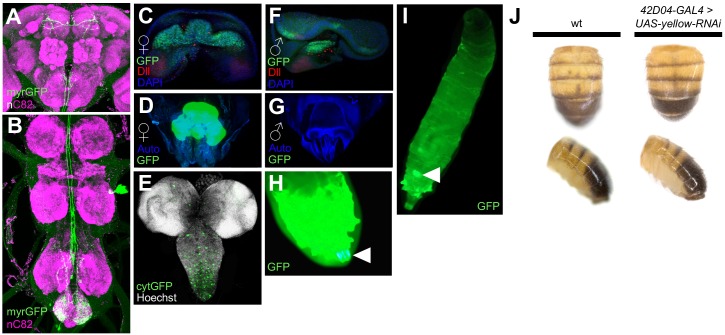
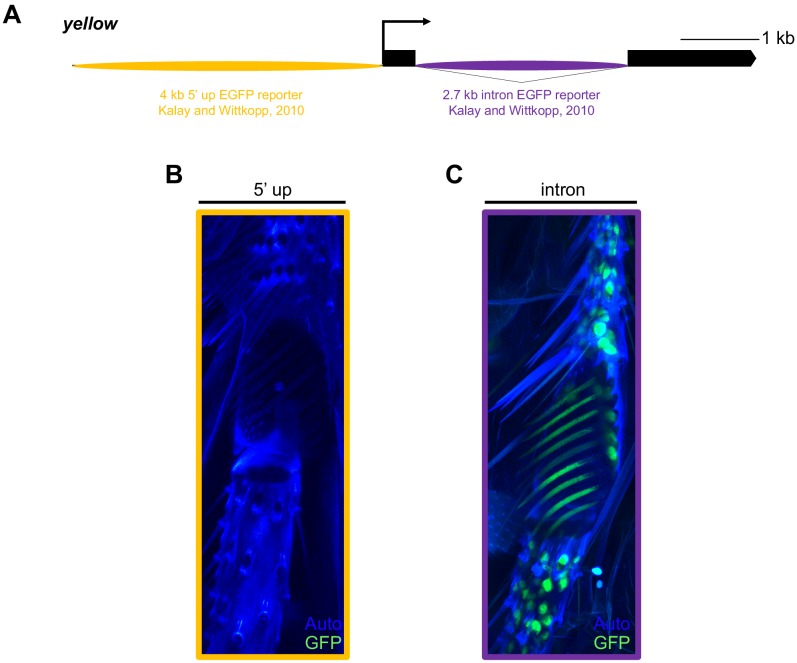
42D04-GAL4 drives expression in a sexually dimorphic pattern in multiple neurons of the adult male (Figure 3A,B) and female CNS (Figure 3—figure supplement 1A,B), consistent with previously described dsxGAL4 expression in the posterior cluster, the abdominal cluster, and, in males, in the prothoracic TN1 neurons (Robinett et al., 2010). 42D04-GAL4 also drives expression in male and female larval CNS and genital discs, with expression in the genital tissues persisting into the adult stage only in females (Figure 3—figure supplement 1C–G). Finally, we observed 42D04-GAL4 expression at the base of the sex combs (also observed by Robinett et al., 2010 and Rice et al., 2019), which are modified bristles used during mating (Cook, 1975; Ng and Kopp, 2008; Hurtado-Gonzales et al., 2015) that are present only on the first tarsal segment of adult male forelegs (Figure 3C–F). Yellow protein is expressed in sex combs (Hinaux et al., 2018, Figure 3G,H), where it is presumably required for synthesis of black dopamine melanin in the sex comb ‘teeth’. This expression of yellow in sex comb cells is driven by enhancer sequences in the yellow intron (Figure 3—figure supplement 2), potentially explaining why manipulating yellow expression using GAL4 driven by sequences 5’ of the yellow gene failed to affect mating (Figure 2—figure supplement 2A–D). Driving expression of yellow-RNAi with 42D04-GAL4 eliminated expression of an mCherry tagged version of the native Yellow protein in sex combs and strongly reduced black melanin in the sex combs (Figure 3I–L) but not the abdomen (Figure 3—figure supplement 1J).
Figure 3. yellow expression in non-neuronal 42D04-GAL4 expressing cells is necessary for sex comb melanization and male mating success.
(A,B) Brain and ventral nerve cord of adult male fly stained with anti-GFP (green) antibody for myrGFP expressed using 42D04-GAL4 and counterstained with anti-nC82 (magenta) for neuropil. (C) Wild-type (wt) D. melanogaster adult male fly highlighting the location of sex combs (Nicolas Gompel). (D) Close up of a wild-type (wt) sex comb on the first tarsal segment (ts1) of the front leg (courtesy of Nicolas Gompel). (E) Bright field illumination of a male front leg expressing cytGFP (green) in sex-comb cells using 42D04-GAL4. (F) Confocal image of the sex comb cells expressing cytGFP (green) with 42D04-GAL4 and leg cuticle autofluorescence (blue). (G) Confocal image of a ymCherry male leg highlighting native ymCherry sex comb expression (red). (H) Zoomed in confocal image shown in (G) with leg cuticle autofluorescence (blue) and native ymCherry sex comb expression (red). (I) Wild-type (wt) sex comb. (J) Loss of black melanin in sex combs in males expressing yellow-RNAi using 42D04-GAL4. (K) Co-localization of ymCherry (red) at the base of the sex comb cells expressing cytGFP (green) with 42D04-GAL4. (L) Loss of ymCherry (red) at the base of the sex comb cells expressing cytGFP (green) and yellow-RNAi using 42D04-GAL4. (M,N) Brain and ventral nerve cord of adult male expressing nsyb-GAL80 to block GAL4 activity in the CNS, stained with anti-GFP (green) antibody for myrGFP expressed using 42D04-GAL4, and counterstained with anti-nC82 (magenta) for neuropil. (O) Loss of black melanin in sex combs in nsyb-GAL80 males expressing yellow-RNAi using 42D04-GAL4. (P) Expressing yellow-RNAi using 42D04-GAL4 in males expressing nsyb-GAL80 significantly inhibited male mating success. Scale bars in (I), (J), and (O) measure 12.5 μm. Sample sizes are shown at the top of each barplot. Significance was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (**p<0.01).
Figure 3—figure supplement 1. Expression pattern of 42D04-GAL4.
Figure 3—figure supplement 2. yellow EGFP reporters localize yellow sex comb expression to the intronic bristle enhancer.
To test the impact of yellow expression in sex combs on male mating behavior, we used 42D04-GAL4 to drive yellow-RNAi, but inhibited the function of 42D04-GAL4 in the CNS with nysb-GAL80 (courtesy of Julie Simpson). These flies showed no GAL4 activity in the CNS (Figure 3M,N), but lost black melanin in the sex combs (Figure 3O) and had reduced male mating success (Figure 3P, FET, p=0.002 and 0.08). High-speed videos (1000 frames per second) revealed that yellow mutant (y1) males fail repeatedly to grasp the female abdomen with their sex combs when attempting to mount and copulate (Video 7), whereas wild-type males more readily grasp the female with their melanized sex combs and initiate copulation efficiently (Video 8). These observations suggest that yellow expression in sex combs affects their melanization, which in turn affects their function.
Video 7. High-speed (1000 fps) video capture of copulation attempts between y1 male and wild-type female.
Video 8. High-speed (1000 fps) video capture of wild-type copulation.
Sex comb melanization is required for efficient grasping, mounting and copulation
To test whether sex comb melanization (as opposed to some other unknown effect of losing yellow expression in sex combs) is critical for male sexual behavior, we suppressed expression of Laccase2 (Arakane et al., 2005; Riedel et al., 2011) in sex combs using 42D04-GAL4 and Laccase2-RNAi (Dietzl et al., 2007). Laccase2 is required to oxidize dopamine into dopamine quinones and thus acts upstream of Yellow in the melanin synthesis pathway (Figure 4A; Riedel et al., 2011). Males with Laccase2 suppressed in sex combs lacked both black and brown dopamine melanin, making these sex combs appear translucent (Figure 4B). These males displayed strongly reduced mating success compared with wild-type males (Figure 4C, FET, p=1×10−7 and 8 × 10−6) and behavioral defects similar to those observed for y1 mutants (Videos 9 and 10), including inefficient grasping of the female for mounting and copulation. We noticed, however, that flies with Laccase2-RNAi driven by 42D04-GAL4 also showed a loss of melanin in the aedeagus (Figure 4—figure supplement 1A), which is the main part of the male genitalia used for copulation, despite no visible expression of 42D04-GAL4 in the adult male genitalia (Figure 3—figure supplement 1G) nor changes in aedeagus pigmentation in y1 mutants (Figure 4—figure supplement 1A). We therefore used subsets of the 42D04 enhancer (Figure 4—figure supplement 1B) to drive expression of Laccase2-RNAi, separating the effects of expression in the sex combs from expression in the genitalia (Figure 4—figure supplement 1C). Male mating success was reduced when Laccase2 suppression reduced melanization in the sex combs, but not the genitalia (Figure 4—figure supplement 1D–G).
Figure 4. Sex comb melanization is specifically required for male mating success.
(A) Simplified version of the insect melanin synthesis pathway. (B) Light microscopy images of sex combs from wild-type (wt), y1, and 42D04-GAL4; UAS-Laccase2-RNAi males. Expressing Laccase2-RNAi in sex combs completely blocked melanin synthesis. (C) Expressing Laccase2-RNAi using 42D04-GAL4 in males significantly inhibited male mating success. (D) Scanning Electron Microscopy (SEM) of sex combs from wild-type (wt), y1, and Laccase2-RNAi males (expressed using 42D04-GAL4). Compared to wild-type, sex comb teeth in y1 mutants appeared thinner and smoother, whereas Laccase2-RNAi sex comb teeth appeared even smoother than y1 mutants, and one comb tooth had a visible crack in the cuticle (white rectangle, enlarged on the right). Scale bars in (B) measure 12.5 μm. Sample sizes are shown at the top of each barplot. Significance in was measured using Fisher’s exact tests with Bonferroni corrections for multiple comparisons. Comparisons that were statistically (p<0.05) are indicated (****p<0.0001).
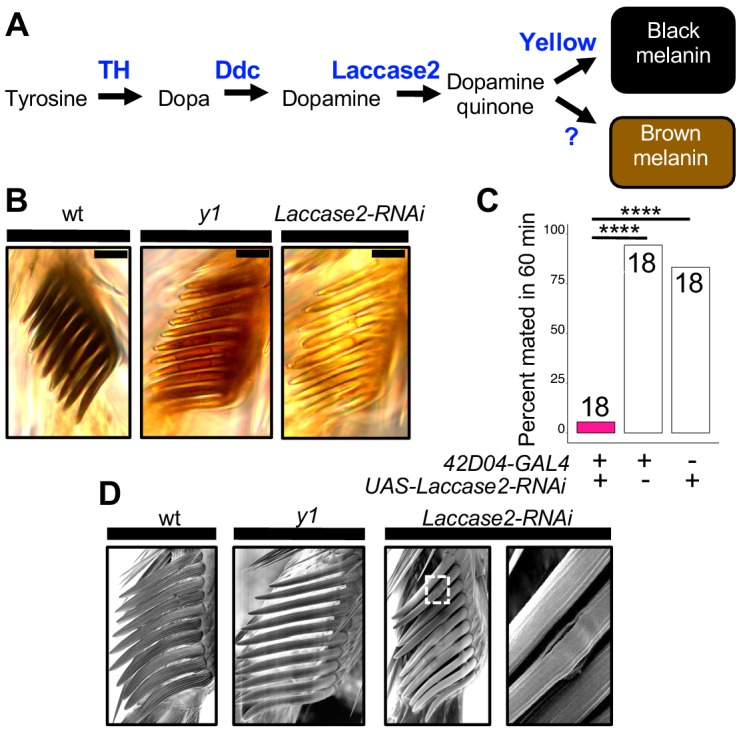
Figure 4—figure supplement 1. Genetic dissection of the 42D04-GAL4 enhancer confirms the specific role of sex comb melanization, and not the aedeagus, in male mating success.
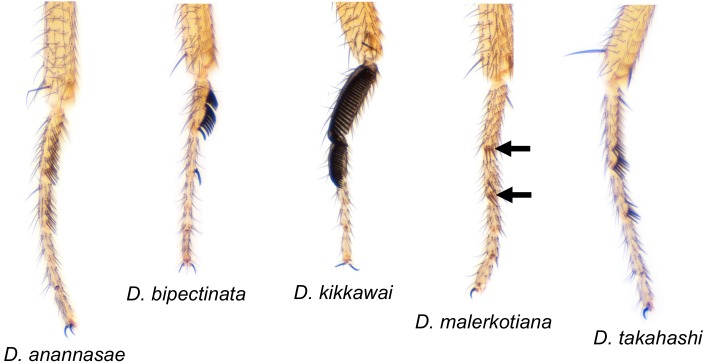
Figure 4—figure supplement 2. Drosophila species with varying sex comb morphology used for high-speed video assays.
Figure 4—figure supplement 3. Sex comb melanization is required for male mating success with y1 females.
Video 9. Copulation attempts between male expressing Laccase2-RNAi in 42D04-GAL4-expressing cells and wild-type female.
Video 10. High-speed (1000 fps) video capture of copulation attempts between male expressing Laccase2-RNAi in 42D04-GAL4-expressing cells and wild-type female.
How can sex comb melanization affect sex comb function? In insects, melanization impacts not only the color of the adult cuticle but also its mechanical stiffness (Xu et al., 1997; Kerwin et al., 1999; Vincent and Wegst, 2004; Andersen, 2005; Arakane et al., 2005; Suderman et al., 2006; Riedel et al., 2011; Noh et al., 2016). For example, expressing Laccase2-RNAi in D. melanogaster wings softens the cuticle to such a degree that the wings collapse (Riedel et al., 2011). Butterflies lacking dopamine melanin due to loss of yellow or another gene required for melanin synthesis, Dopa decarboxylase, also show changes in the fine structure of their wing scales (Matsuoka and Monteiro, 2018). Consistent with these observations, we observed structural changes in D. melanogaster sex comb teeth lacking yellow or Laccase2 expression using scanning electron microscopy (SEM), with a crack appearing in one of the Laccase2-RNAi comb teeth (Figure 4D). We thus conclude that these structural changes in sex combs are responsible for inhibiting the yellow mutant male’s ability to grasp a female for mounting and copulation (Video 10). In 1976, Wilson et al. (1976) speculated about this very hypothesis based on their own observations of behavior in yellow mutant males.
Data from other Drosophila species are also consistent with this structural hypothesis. Specifically, yellow mutants in D. subobscura, D. pseudoobscura, and D. gaucha, all of which have sex combs, show reduced male mating success with wild-type females (Rendel, 1944; Tan, 1946; Frias and Lamborot, 1970; Pruzan-Hotchkiss et al., 1992) whereas yellow mutants in Drosophila willistoni, a species that lacks sex combs (Kopp, 2011; Atallah et al., 2014), do not (da Silva et al., 2005). Sex comb morphology is highly diverse among species that have sex combs (Kopp, 2011), but these structures generally seem to be melanized (Figure 4—figure supplement 2; Tanaka et al., 2009) and used to grasp females (Videos 11–15). (Our high-speed video recordings of mating in D. anannasae, D. bipectinata, D. kikkawai, D. malerkotiana, and D. takahashi show that differences in sex comb morphology (Figure 4—figure supplement 2) correspond with differences in how (where on the female and with which part of the male leg) the male grasps the female prior to copulation (Videos 11–15).
Video 11. Drosophila anannasae wild-type copulation.
Video 12. Drosophila bipectinata wild-type copulation.
Video 13. Drosophila kikkawai wild-type copulation.
Video 14. Drosophila malerkotiana wild-type copulation.
Video 15. Drosophila takahashi wild-type copulation.
It remains unclear how D. willistoni males (and males of other species without sex combs) are able to efficiently grasp females prior to copulation (Video 16). However, differences in females might be part of the answer, as D. melanogaster y1 mutant males are able to mate with y1 mutant females at rates similar to wild-type males (Bastock, 1956; Dow, 1976; Heisler, 1984; Liu et al., 2019; Figure 4—figure supplement 3A, FET, p=1). That said, removing all melanin from D. melanogaster sex combs by knocking down Laccase-2 reduced mating efficiency with y1 females (Figure 4—figure supplement 3B, FET, p=0.02 and 0.0001), suggesting that the brown melanin remaining in y1 sex-combs (Figure 4B) played a role in the mating success of y1 males with y1 females.
Video 16. Drosophila willistoni wild-type copulation.
Conclusion
Taken together, our data show that melanization of a secondary sexual structure affects mating in D. melanogaster. Specifically, we find that the reduced mating success of D. melanogaster yellow mutant males, which was perceived as a behavioral defect for decades, is caused by changes in the morphology of the structures used during mating. Other recent studies have also shown the importance of morphological structures for stickleback schooling (Greenwood et al., 2015), water strider walking (Santos et al., 2017), and cricket singing (Pascoal et al., 2014) behaviors. These observations all underscore that behavior cannot be understood by studying the nervous system alone; anatomy and behavior function and evolve as an interconnected system.
Materials and methods
We have included a Key Resources Table as Supplementary file 4.
Fly stocks and maintenance
The following lines were used for this work: y1 [which was backcrossed into a wild-type (Canton-S) line for six generations before starting our experiments; the y1 allele contains an A to C transversion in the ATG initiation and is considered a null allele (Geyer et al., 1990)]; Canton-S as wild-type (courtesy of Scott Pletcher); UAS-yellow-RNAi obtained from the Vienna Drosophila Resource Centre (VDRC) (Dietzl et al., 2007, KK106068); y1;UAS-y (BDSC 3043); elav-GAL4 (BDSC 49226); nsyb-GAL4 (BDSC 39171); repo-GAL4 (BDSC 7415); dsxGAL4 (Robinett et al., 2010) (courtesy of Bruce Baker); dsxGAL4 (Rideout et al., 2010) (courtesy of Stephen Goodwin); fruGAL4 (Stockinger et al., 2005) (courtesy of Barry Dickson); the following Janelia enhancer trap GAL4 lines (Pfeiffer et al., 2008): 40A05-GAL4 (BDSC 48138), 41D01-GAL4 (BDSC 50123), 42D02-GAL4 (BDSC 41250), 41 F06-GAL4 (BDSC 47584), 41A01-GAL4 (BDSC 39425), 42D04-GAL4 (BDSC 47588), 40 F03-GAL4 (BDSC 47355), 39E06-GAL4 (BDSC 50051), 42 C06-GAL4 (BDSC 50150), 40 F04-GAL4 (BDSC 50094); ymCherry (courtesy of Nicolas Gompel); nsyb-GAL80 (courtesy of Julie Simpson); UAS-Laccase2-RNAi obtained from the VDRC (Dietzl et al., 2007, KK101687); dsxGAL4-DBD (Pavlou et al., 2016) (courtesy of Stephen Goodwin); vGlutdVP16-AD (Gao et al., 2008) (courtesy of Stephen Goodwin); BDSC 6993; BDSC 49365; BDSC 6927; BDSC 45175; BDSC 3740; BDSC 5820; BDSC 8848; BDSC 7010; TPH-GAL4 (courtesy of Shinya Yamamoto); wing-body-GAL4 (BDSC 44373); D. melanogaster yellow 5’ up EGFP reporter (Kalay and Wittkopp, 2010) (courtesy of Gizem Kalay); D. melanogaster yellow intron EGFP reporter (Kalay and Wittkopp, 2010) (courtesy of Gizem Kalay); vasa-Cas9 (BDSC 51324); UAS-cytGFP (courtesy of Janelia Fly Core); pJFRC12-10XUAS-IVS-myr::GFP (courtesy of Janelia Fly Core). All flies were grown at 23°C with a 12 hr light-dark cycle with lights on at 8AM and off at 8PM on standard corn-meal fly medium.
Behavior
Mating assays
Virgin males and females were separated upon eclosion and aged for 3–8 d before each experiment. Experiments were carried out at 23°C on a 12 hr light dark cycle with lights on at 8 AM and off at 8 PM on standard corn-meal fly medium. Males were isolated in glass vials, and females were group housed in standard plastic fly vials at densities of 20–30 flies. All mating assays were performed at 23°C between 8-11AM or 6-9PM. For each assay replicate, a single virgin male and female fly were gently aspirated into a 35 mm diameter Petri dish (Genesee Scientific, catalog #32–103) placed on top of a 17 inch LED light pad (HUION L4S) and immediately monitored for 60 min for courtship and copulation activity. All genotypes tested initiated courtship (including tapping, chasing, wing extension, genital licking, and attempted copulation) towards the female. Any genotype that copulated within the 60 min window was scored as a successful mating. Except for the experiment described in Figure 4—figure supplement 3 in which y1 females were used, all females in mating assays were wild-type (Canton-S). The percent mated in 60 min values shown in figures were calculated as the number of replicates that mated divided by the total number of replicates and multiplied by 100.
Courtship analysis
For courtship analysis, 60 min videos were recorded using Canon VIXIA HF R500 camcorders mounted to Manfrotto (MKCOMPACTACN-BK) aluminum tripods. To calculate courtship indices in Figure 1 between wild-type and y1 males, the amount of time males spent engaged in courtship: tapping, chasing, wing extension, genital licking, or attempted copulation was quantified for the first 10 min of the assay and divided by the total 10 min period. We chose to quantify courtship activity within the first 10 min of the assay, because wild-type (Canton-S) males will often begin copulating after this window, while y1 males will continue to court throughout the entire 60 min period. Wing extension bouts were quantified by noting every unilateral wing extension bout for each genotype within the first 10 min of the assay.
Song analysis
Courtship song was recorded as described previously (Arthur et al., 2013). All genotypes were recorded simultaneously. Song data were segmented (Arthur et al., 2013) and analyzed (http://www.github.com/dstern/BatchSongAnalysis) without human intervention. Values for pulse per minute, sine per minute, and interpulse interval were then extracted from the software.
High-speed video capture
For high-speed video capture of attempted mounting and copulation events, virgin males and females were isolated upon eclosion and aged for 4–7 d before each assay. Using a Fascam Photron SA4 (courtesy of Gwyneth Card) mounted with a 105 mm AF Micro Nikkor Nikon lens (courtesy of Gwyneth Card), we recorded individual pairs of males and females that were gently aspirated into a single well of a 96 well cell culture plate (Corning 05-539-200) partially filled with 2% agarose and covered with a glass coverslip. We recorded mounting and copulation attempts at 1000 frames per second (fps) and played back at 30 fps. Most wild-type males attempted mounting 3–5 times before copulating, whereas y1, yellow-RNAi, and Laccasse2-RNAi males repeatedly attempted mounting without engaging in copulation, mirroring the videos we captured on the Canon VIXIA HF R500 at 30 fps.
Imaging sex combs and genitalia
Sex comb images highlighting different melanization states (Figure 3I,J,O; Figure 4B) were taken using a Zeiss Axio Cam ERc 5 s mounted on a Zeiss Axio Observer A1 Inverted Microscope. Front legs were cut and placed sex comb side down on a microscope slide (Fisher brand 12-550-123) and imaged through a 40x objective. Images were processed using AxioVision LE software. Abdomens and genitalia images highlighting different melanization states of the aedeagus and female genital bristles were captured using a Canon EOS Rebel T6 camera mounted with a Canon MP-E 65 mm macro lens. Genitalia images were processed in Adobe Photoshop (version 19.1.5) (Adobe Systems Inc, San Jose, CA).
Focus Ion Beam Scanning Electron Microscope (FIB-SEM) images (Figure 4D) were taken by placing individual, dissected legs on carbon tape adhered to a scanning electron microscope pin stud mount with sex combs facing up. The samples were then coated with a 20 nm Au layer using a Gatan 682 Precision Etching and Coating System, and imaged by scanning electron microscopy in a Zeiss Sigma system. The samples were imaged using a 3-nA electron beam with 1.5 kV landing energy at 2.5MHz.
Immunohistochemistry and confocal imaging
Central Nervous System
Dissections, immunohistochemistry, and imaging of fly central nervous systems were done as previously described (Aso et al., 2014). In brief, brains and VNCs were dissected in Schneider’s insect medium and fixed in 2% paraformaldehyde (diluted in the same medium) at room temperature for 55 min. Tissues were washed in PBT (0.5% Triton X-100 in phosphate buffered saline) and blocked using 5% normal goat serum before incubation with antibodies. Tissues expressing GFP were stained with rabbit anti-GFP (ThermoFisher Scientific A-11122, 1:1000) and mouse anti-BRP hybridoma supernatant (nc82, Developmental Studies Hybridoma Bank, Univ. Iowa, 1:30), followed by Alexa Fluor 488-conjugated goat anti-rabbit and Alexa Fluor 568-conjugated goat anti-mouse antibodies (ThermoFisher Scientific A-11034 and A-11031), respectively. Tissues expressing mCherry-tagged Yellow protein (ymCherry) were stained with rabbit anti-dsRed (Clontech 632496, 1:1000) and rat anti-DN-Cadherin (DN-Ex #8, Developmental Studies Hybridoma Bank, Univ. Iowa, 1:100) as neuropil marker, followed by Cy3-conjugated goat anti-rabbit and Cy5-conjugated goat anti-rat antibodies (Jackson ImmunoResearch 111-165-144 and 112-175-167), respectively. After staining and post-fixation in 4% paraformaldehyde, tissues were mounted on poly-L-lysine-coated cover slips, cleared, and embedded in DPX as described. Image z-stacks were collected at 1 μm intervals using an LSM710 confocal microscope (Zeiss, Germany) fitted with a Plan-Apochromat 20x/0.8 M27 objective. Images were processed in Fiji (http://fiji.sc/) and Adobe Photoshop (version 19.1.5) (Adobe Systems Inc, San Jose, CA).
Sex combs and genitalia
Adult flies were 2–7 d old and pupae were 96 hr old after pupal formation (APF) for the EGFP reporter experiment summarized in Figure 3—figure supplement 2. Flies were anesthetized on ice, submerged in 70% ethanol, rinsed twice in phosphate buffered saline with 0.1% Triton X-100 (PBS-T), and fixed in 2% formaldehyde in PBS-T. Forelegs and genitalia/abdomen tips were removed with fine scissors and mounted in Tris-buffered (pH 8.0) 80% glycerol. Serial optical sections were obtained at 1.5 µm or 0.5 µm intervals on a Zeiss 880 confocal microscope with a LD-LCI 25x/0.8 NA objective (genitalia) or a Plan-Apochromat 40x/1.3 NA objective (appendages/tarsal sex combs). The native fluorescence of GFP, mCherry and autofluorescence of cuticle were imaged using 488, 594 and 633 lasers, respectively. Images were processed in Fiji (http://fiji.sc/), Icy (http://icy.bioimageanalysis.org/) and Adobe Photoshop (version 19.1.5) (Adobe Systems Inc, San Jose, CA).
Generation of the mating regulatory sequence (MRS) deletion line
Using the 209 bp region mapped in Drapeau et al. (2006) between −300 and −91 bp upstream of yellow’s transcription start site, we designed two single guide RNA (gRNA) target sites at −291 bp and −140 bp that maximized the MRS deletion region, given constraints of identifying NGG PAM sites required for CRISPR/Cas9 gene editing (Figure 2—figure supplement 2). We in-vitro transcribed these gRNAs using a MEGAscript T7 Transcription Kit (Invitrogen) following the PCR-based protocol from Bassett et al. (2013). Two 1 kb homology arms were PCR amplified from the yellow locus immediately upstream and downstream of the gRNA target sites using forward and reverse primers with NcoI and BglII tails, respectively, for the Left Arm (5’-TTACCATGGGGGATCAAGTTGAACCAC-3’, 5’-GGAGATCTGGCCTTCATCGACATTTA-3’) and the forward and reverse primers with Bsu36I and MluI tails, respectively, for the Right Arm (5’-TACATCCCTAAGGCCTGATTACCCGAACACT-3’, 5’-TATACGCGTTGCCATGCTATTGGCTTC-3’) and cloned into pHD-DsRed-attp (Gratz et al., 2014; Addgene Plasmid # 51019) in two steps, digesting first with NcoI and BglII (Left Arm) to transform the Left Arm and second with Bsu36I and MluI (Right Arm) to transform the Right Arm, flanking the 3xP3::DsRed, attP, and LoxP sites. Homology arms were ligated into pHD-DsRed-attp using T4 DNA Ligase (ThermoFisher Scientific), and products were transformed into One Shot TOP10 (Invitrogen) DH5 alpha competent cells. Purified donor plasmid was then co-injected at 500 ng/uL with the two gRNAs at 100 ng/uL total concentration into a vasa-Cas9 (BDSC 51324) line. Flies were then screened for DsRed expression in the eyes, and Sanger sequenced verified for a 3xP3::DsRed replacement of the MRS region (Figure 2—figure supplement 2). We confirmed that we deleted 152 bp of the 209 bp region based on Sanger sequencing the CRISPR/Cas9 cut sites (Figure 2—figure supplement 2). Next, we crossed yΔMRS+3xP3::DsRed with a Cre-expressing fly line (courtesy of Bing Ye, University of Michigan) to excise 3xP3::DsRed and screened for flies that lost DsRed expression in the eyes. Finally, we PCR-gel verified that DsRed was indeed removed in creation of the yΔMRS line using the forward and reverse primers, respectively (5’-CAGTCGCCGATAAAGATGAACACTG-3’, 5’-CAAGGTGATCAGGGTCACAAGGATC-3’) (Figure 2—figure supplement 2).
Generation of the 42D04-GAL4 enhancer sub-fragment pBPGUw lines
Enhancer sub-fragments (2 kb, 2 kb, 1.3 kb, 1.3 kb, and 1.3 kb for 42D04_A,B,C,D,E-GAL4, respectively) were synthesized as IDT gene blocks (sequences available in Supplementary file 1) based off of the 42D04 D. melanogaster dsx enhancer sequence (FBsf0000164494) (Figure 4—figure supplement 1). The gene blocks were designed with 5’ and 3’ Gibson tails to facilitate Gibson assembly (Gibson et al., 2009) into the GAL4 plasmid pBPGUw (Pfeiffer et al., 2008; Addgene Plasmid #17575) after digestion with FseI and AatII. Products were transformed into Mix and Go! DH5 alpha competent cells (Zymo). Clones were selected by ampicillin resistance on Amp-LB plates (60 mg/mL). Purified plasmids were injected at 500 ng/uL into the phiC31 integrase-expressing 86Fb landing site line BDSC 24749 (courtesy of Rainbow Transgenics) for phiC31 attP-attB integration and screened for using a mini-white marker.
Statistics
Supplementary file 2 is a Microsoft Excel file containing four worksheets with all of the data used for analysis. The worksheet labeled ‘Univar_Male_Mating_Success_Data’ contains a univariate description of each mating trial. The worksheet labeled ‘Summary of mating success data’ shows the number of successful and unsuccessful matings for each genotype tested (grouped by figure panel including the data) and was generated from the ‘Univar_Male_Mating_Success_Data’ worksheet using the Excel Pivot Table function. The worksheet labeled ‘Courtship_Data’ includes the data for courtship index and wing extensions shown in Figure 1D and E, respectively. The worksheet labeled ‘Song Data’ includes the measures of pulses per minute, sin per minute and interpulse interval (labeled ‘ModeEndToStartIPI’) exported from the software described in Arthur et al. (2013). R version 3.6.1 (2019-07-05) (R Development Core Team, 2013) was used for all statistical analyses using the code included in Source code 1. These analyses included t-tests comparing courtship index, number of wing extensions, pulses per minute, sine per minute, and interpulse interval that were run after exporting data in the Courtship Data and Song Data worksheets (separately) to tab delimited text files. Note that the default t-test parameters allowing for unequal variance between samples were used. Source code 1 also contains the R code for the Fisher’s Exact Tests, which were coded using data from the ‘Summary of mating success data’ worksheet. Supplementary file 3 contains a summary of all statistical tests. Whenever an experimental genotype was compared to two control genotypes, P-values were adjusted using a Bonferroni correction for N = 2 (see Supplementary file 3). We note that for N = 2, alternative adjustments available with the p.adjust function in R (‘holm’, ‘Hochberg’, ‘hommel’ and ‘fdr’) give the same adjusted P-value.
Acknowledgements
We thank members of the Wittkopp and Stern labs for helpful discussions. For fly strains, we thank Bruce Baker, Carmen Robinett, Stephen Goodwin, Barry Dickson, Scott Pletcher, Julie Simpson, Shinya Yamamoto, Bing Ye, Nicolas Gompel, Gizem Kalay, The Bloomington Drosophila Stock Center, The Vienna Drosophila RNAi Center, and the Janelia Fly Core. For fly injections, we thank Rainbow Transgenics Inc. For technical support with Scanning Electron Microscopy (SEM), we thank Harald Hess and Song Pang. For use of the Photron for high-speed video capture, we thank Gwyneth Card and W Ryan Williamson. CNS dissections, immunostaining, and imaging were performed by the Janelia Project Technical Resource team with special thanks to Gudrun Ihrke, Kari Close, and Christina Christoforou. We thank Nicolas Gompel, Abby Lamb, and Henry Ertl for comments on the manuscript. We also thank Shyama Nandakumar and Ajai Pulianmackal for help with dissections and confocal microscopy as well as Elena Kingston for capturing the Drosophila willistoni copulation video.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
David L Stern, Email: sternd@janelia.hhmi.org.
Patricia J Wittkopp, Email: wittkopp@umich.edu.
K VijayRaghavan, National Centre for Biological Sciences, Tata Institute of Fundamental Research, India.
K VijayRaghavan, National Centre for Biological Sciences, Tata Institute of Fundamental Research, India.
Funding Information
This paper was supported by the following grants:
National Institutes of Health T32GM007544 to Jonathan H Massey.
National Institutes of Health GM089736 to Patricia J Wittkopp.
National Institutes of Health 1R35GM118073 to Patricia J Wittkopp.
Additional information
Competing interests
Senior editor, eLife.
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Funding acquisition, Validation, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing.
Conceptualization, Data curation, Formal analysis, Validation, Investigation, Methodology, Writing—review and editing.
Resources, Data curation, Visualization, Methodology.
Supervision, Funding acquisition, Visualization, Writing—original draft, Project administration, Writing—review and editing.
Conceptualization, Supervision, Funding acquisition, Visualization, Writing—original draft, Project administration, Writing—review and editing.
Additional files
Data availability
All data generated or analyzed during this study are included in the manuscript and supporting files. Supplementary File 2 contains all source data and Supplementary File 3 contains R code for analyzing it.
References
- Andersen SO. Cuticular sclerotization and tanning. Comprehensive Molecular Insect Science. 2005:145–170. doi: 10.1016/b0-44-451924-6/00052-1. [DOI] [Google Scholar]
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. PNAS. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur BJ, Sunayama-Morita T, Coen P, Murthy M, Stern DL. Multi-channel acoustic recording and automated analysis of Drosophila courtship songs. BMC Biology. 2013;11:11. doi: 10.1186/1741-7007-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, Rubin GM. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah J, Vurens G, Mavong S, Mutti A, Hoang D, Kopp A. Sex-specific repression of dachshund is required for Drosophila sex comb development. Developmental Biology. 2014;386:440–447. doi: 10.1016/j.ydbio.2013.12.017. [DOI] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock M. A gene mutation which changes a behavior pattern. Evolution. 1956;10:421–439. doi: 10.1111/j.1558-5646.1956.tb02868.x. [DOI] [Google Scholar]
- Borycz J, Borycz JA, Loubani M, Meinertzhagen IA. tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. The Journal of Neuroscience. 2002;22:10549–10557. doi: 10.1523/JNEUROSCI.22-24-10549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Clough E, Jimenez E, Kim YA, Whitworth C, Neville MC, Hempel LU, Pavlou HJ, Chen ZX, Sturgill D, Dale RK, Smith HE, Przytycka TM, Goodwin SF, Van Doren M, Oliver B. Sex- and tissue-specific functions of Drosophila doublesex transcription factor target genes. Developmental Cell. 2014;31:761–773. doi: 10.1016/j.devcel.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb M. A gene mutation which changed animal behaviour: Margaret Bastock and the yellow fly. Animal Behaviour. 2007;74:163–169. doi: 10.1016/j.anbehav.2007.05.002. [DOI] [Google Scholar]
- Cook R. Courtship of Drosophila melanogaster: rejection without extrusion. Behaviour. 1975;52:155–171. doi: 10.1163/156853975X00010. [DOI] [Google Scholar]
- da Silva LB, Leite DF, Valente VL, Rohde C. Mating activity of yellow and sepia Drosophila willistoni mutants. Behavioural Processes. 2005;70:149–155. doi: 10.1016/j.beproc.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dow MA. The genetic basis of receptivity of yellow mutant Drosophila melanogaster females. Behavior Genetics. 1976;6:141–143. doi: 10.1007/BF01067144. [DOI] [PubMed] [Google Scholar]
- Drapeau MD, Radovic A, Wittkopp PJ, Long AD. A gene necessary for normal male courtship, yellow, acts downstream of fruitless in the Drosophila melanogaster larval brain. Journal of Neurobiology. 2003;55:53–72. doi: 10.1002/neu.10196. [DOI] [PubMed] [Google Scholar]
- Drapeau MD, Cyran SA, Viering MM, Geyer PK, Long AD. A cis-regulatory sequence within the yellow locus of Drosophila melanogaster required for normal male mating success. Genetics. 2006;172:1009–1030. doi: 10.1534/genetics.105.045666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias D, Lamborot M. Reproductive isolation between the yellow, white, and ‘‘wild’’ stocks of D. gaucha at two temperatures (in Spanish) Archivos De Biología Y Medicina Experimentales. 1970;7:67 [Google Scholar]
- Gao S, Takemura SY, Ting CY, Huang S, Lu Z, Luan H, Rister J, Thum AS, Yang M, Hong ST, Wang JW, Odenwald WF, White BH, Meinertzhagen IA, Lee CH. The neural substrate of spectral preference in Drosophila. Neuron. 2008;60:328–342. doi: 10.1016/j.neuron.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Spana C, Corces VG. On the molecular mechanism of gypsy-induced mutations at the yellow locus of Drosophila melanogaster. The EMBO Journal. 1986;5:2657–2662. doi: 10.1002/j.1460-2075.1986.tb04548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Green MM, Corces VG. Tissue-specific transcriptional enhancers may act in trans on the gene located in the homologous chromosome: the molecular basis of transvection in Drosophila. The EMBO Journal. 1990;9:2247–2256. doi: 10.1002/j.1460-2075.1990.tb07395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer PK, Corces VG. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes & Development. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gilbert MK, Tan YY, Hart CM. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics. 2006;173:1365–1375. doi: 10.1534/genetics.106.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O'Connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics. 2014;196:961–971. doi: 10.1534/genetics.113.160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan RJ. The origins of behavioral genetics. Current Biology. 2008;18:R192–R198. doi: 10.1016/j.cub.2008.01.015. [DOI] [PubMed] [Google Scholar]
- Greenwood AK, Ardekani R, McCann SR, Dubin ME, Sullivan A, Bensussen S, Tavaré S, Peichel CL. Genetic mapping of natural variation in schooling tendency in the threespine stickleback. G3: Genes, Genomes, Genetics. 2015;5:761–769. doi: 10.1534/g3.114.016519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg M. Separation of receptor and Lamina potentials in the electroretinogram of normal and mutant Drosophila. The Journal of Experimental Biology. 1971;55:85–100. doi: 10.1242/jeb.55.1.85. [DOI] [PubMed] [Google Scholar]
- Heisler IL. Inheritance of female mating propensities for yellow locus genotypes in Drosophila Melanogaster. Genetical Research. 1984;44:133–149. doi: 10.1017/S0016672300026343. [DOI] [Google Scholar]
- Hinaux H, Bachem K, Battistara M, Rossi M, Xin Y, Jaenichen R, Le Poul Y, Arnoult L, Kobler JM, Grunwald Kadow IC, Rodermund L, Prud'homme B, Gompel N. Revisiting the developmental and cellular role of the pigmentation gene yellow in Drosophila using a tagged allele. Developmental Biology. 2018;438:111–123. doi: 10.1016/j.ydbio.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Hotta Y, Benzer S. Abnormal electroretinograms in visual mutants of Drosophila. Nature. 1969;222:354–356. doi: 10.1038/222354a0. [DOI] [PubMed] [Google Scholar]
- Hurtado-Gonzales JL, Gallaher W, Warner A, Polak M. Microscale Laser Surgery Demonstrates the Grasping Function of the Male Sex Combs in Drosophila melanogaster and Drosophila bipectinata. Ethology. 2015;121:45–56. doi: 10.1111/eth.12316. [DOI] [Google Scholar]
- Kalay G, Lusk R, Dome M, Hens K, Deplancke B, Wittkopp PJ. Potential direct regulators of the Drosophila yellow gene identified by yeast one-hybrid and RNAi screens. G3: Genes|Genomes|Genetics. 2016;6:3419–3430. doi: 10.1534/g3.116.032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay G, Wittkopp PJ. Nomadic enhancers: tissue-specific cis-regulatory elements of yellow have divergent genomic positions among Drosophila species. PLOS Genetics. 2010;6:e1001222. doi: 10.1371/journal.pgen.1001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwin JL, Turecek F, Xu R, Kramer KJ, Hopkins TL, Gatlin CL, Yates JR. Mass spectrometric analysis of catechol-histidine adducts from insect cuticle. Analytical Biochemistry. 1999;268:229–237. doi: 10.1006/abio.1998.3069. [DOI] [PubMed] [Google Scholar]
- Kopp A. Drosophila sex combs as a model of evolutionary innovations. Evolution & Development. 2011;13:504–522. doi: 10.1111/j.1525-142X.2011.00507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Champer J, Langmüller AM, Liu C, Chung J, Reeves R, Luthra A, Lee YL, Vaughn AH, Clark AG, Messer PW. Maximum likelihood estimation of fitness components in experimental evolution. Genetics. 2019;211:1005–1017. doi: 10.1534/genetics.118.301893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M, Meng YB, Chia W. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. MGG Molecular & General Genetics. 1989;218:118–126. doi: 10.1007/BF00330574. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Monteiro A. Melanin pathway genes regulate color and morphology of butterfly wing scales. Cell Reports. 2018;24:56–65. doi: 10.1016/j.celrep.2018.05.092. [DOI] [PubMed] [Google Scholar]
- Ng CS, Kopp A. Sex combs are important for male mating success in Drosophila melanogaster. Behavior Genetics. 2008;38:195–201. doi: 10.1007/s10519-008-9190-7. [DOI] [PubMed] [Google Scholar]
- Noh MY, Koo B, Kramer KJ, Muthukrishnan S, Arakane Y. Arylalkylamine N-acetyltransferase 1 gene (TcAANAT1) is required for cuticle morphology and pigmentation of the adult red flour beetle, Tribolium castaneum. Insect Biochemistry and Molecular Biology. 2016;79:119–129. doi: 10.1016/j.ibmb.2016.10.013. [DOI] [PubMed] [Google Scholar]
- Pascoal S, Cezard T, Eik-Nes A, Gharbi K, Majewska J, Payne E, Ritchie MG, Zuk M, Bailey NW. Rapid convergent evolution in wild crickets. Current Biology. 2014;24:1369–1374. doi: 10.1016/j.cub.2014.04.053. [DOI] [PubMed] [Google Scholar]
- Pavlou HJ, Lin AC, Neville MC, Nojima T, Diao F, Chen BE, White BH, Goodwin SF. Neural circuitry coordinating male copulation. eLife. 2016;5:e20713. doi: 10.7554/eLife.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. PNAS. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzan-Hotchkiss A, Sato K, Thompson JF. Genetic and behavioral studies on yellow Drosophila. Behavior Genetics. 1992;22:747 [Google Scholar]
- R Development Core Team . Vienna, Austria: 2013. http://www.r-project.org/ [Google Scholar]
- Rendel JM. Genetics and cytology of Drosophila subobscura. II. Normal and selective matings in Drosophila subobscura. Journal of Genetics. 1944;46:287–302. doi: 10.1007/BF02989271. [DOI] [PubMed] [Google Scholar]
- Rice GR, Barmina O, Luecke D, Hu K, Arbeitman M, Kopp A. Modular tissue-specific regulation of doublesex underpins sexually dimorphic development in Drosophila. Development. 2019;146:dev178285. doi: 10.1242/dev.178285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardt A, Rybak J, Störtkuhl KF, Meinertzhagen IA, Hovemann BT. Ebony protein in the Drosophila nervous system: optic neuropile expression in glial cells. Journal of Comparative Neurology. 2002;452:93–102. doi: 10.1002/cne.10360. [DOI] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, Goodwin SF. Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nature Neuroscience. 2010;13:458–466. doi: 10.1038/nn.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel F, Vorkel D, Eaton S. Megalin-dependent yellow endocytosis restricts melanization in the Drosophila cuticle. Development. 2011;138:149–158. doi: 10.1242/dev.056309. [DOI] [PubMed] [Google Scholar]
- Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. there is a time and place for sex. PLOS Biology. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers WA, Grover S, Stringer SJ, Parks J, Rebeiz M, Williams TM. A survey of the trans-regulatory landscape for Drosophila melanogaster abdominal pigmentation. Developmental Biology. 2014;385:417–432. doi: 10.1016/j.ydbio.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Santos ME, Le Bouquin A, Crumière AJJ, Khila A. Taxon-restricted genes at the origin of a novel trait allowing access to a new environment. Science. 2017;358:386–390. doi: 10.1126/science.aan2748. [DOI] [PubMed] [Google Scholar]
- Siegal ML, Hartl DL. Transgene Coplacement and high efficiency site-specific recombination with the Cre/loxP system in Drosophila. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger P, Kvitsiani D, Rotkopf S, Tirián L, Dickson BJ. Neural circuitry that governs Drosophila male courtship behavior. Cell. 2005;121:795–807. doi: 10.1016/j.cell.2005.04.026. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. Experiments on sex recognition and the problem of sexual selection in Drosophila. Journal of Animal Behaviour. 1915;5:e366. doi: 10.1037/h0074109. [DOI] [Google Scholar]
- Suderman RJ, Dittmer NT, Kanost MR, Kramer KJ. Model reactions for insect cuticle sclerotization: cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochemistry and Molecular Biology. 2006;36:353–365. doi: 10.1016/j.ibmb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in Glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CC. Genetics of sexual isolation between DROSOPHILA PSEUDOOBSCURA and DROSOPHILA PERSIMILIS. Genetics. 1946;31:558–573. doi: 10.1093/genetics/31.6.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Barmina O, Kopp A. Distinct developmental mechanisms underlie the evolutionary diversification of Drosophila sex combs. PNAS. 2009;106:4764–4769. doi: 10.1073/pnas.0807875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Yeh SD, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou SR, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLOS Genetics. 2005;1:e63. doi: 10.1371/journal.pgen.0010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Hall JC. Neurogenetics of courtship and mating in Drosophila. Advances in Genetics. 2008;62:67–184. doi: 10.1016/S0065-2660(08)00603-2. [DOI] [PubMed] [Google Scholar]
- Vincent JF, Wegst UG. Design and mechanical properties of insect cuticle. Arthropod Structure & Development. 2004;33:187–199. doi: 10.1016/j.asd.2004.05.006. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. Oxford University Press; 2003. [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Burnet B, Eastwood L, Connolly K. Behavioural pleiotropy of the yellow gene in Drosophila melanogaster. Genetical Research. 1976;28:75–88. doi: 10.1017/s0016672300016748. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002;129:1849–1858. doi: 10.1242/dev.129.8.1849. [DOI] [PubMed] [Google Scholar]
- Xu R, Huang X, Hopkins TL, Kramer KJ. Catecholamine and histidyl protein cross-linked structures in sclerotized insect cuticle. Insect Biochemistry and Molecular Biology. 1997;27:101–108. doi: 10.1016/S0965-1748(96)00083-5. [DOI] [Google Scholar]
- Xu X, Oliveira F, Chang BW, Collin N, Gomes R, Teixeira C, Reynoso D, My Pham V, Elnaiem DE, Kamhawi S, Ribeiro JM, Valenzuela JG, Andersen JF. Structure and function of a "yellow" protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection. Journal of Biological Chemistry. 2011;286:32383–32393. doi: 10.1074/jbc.M111.268904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto D, Koganezawa M. Genes and circuits of courtship behaviour in Drosophila males. Nature Reviews Neuroscience. 2013;14:681–692. doi: 10.1038/nrn3567. [DOI] [PubMed] [Google Scholar]
- Zhang SX, Rogulja D, Crickmore MA. Dopaminergic Circuitry Underlying Mating Drive. Neuron. 2016;91:168–181. doi: 10.1016/j.neuron.2016.05.020. [DOI] [PubMed] [Google Scholar]