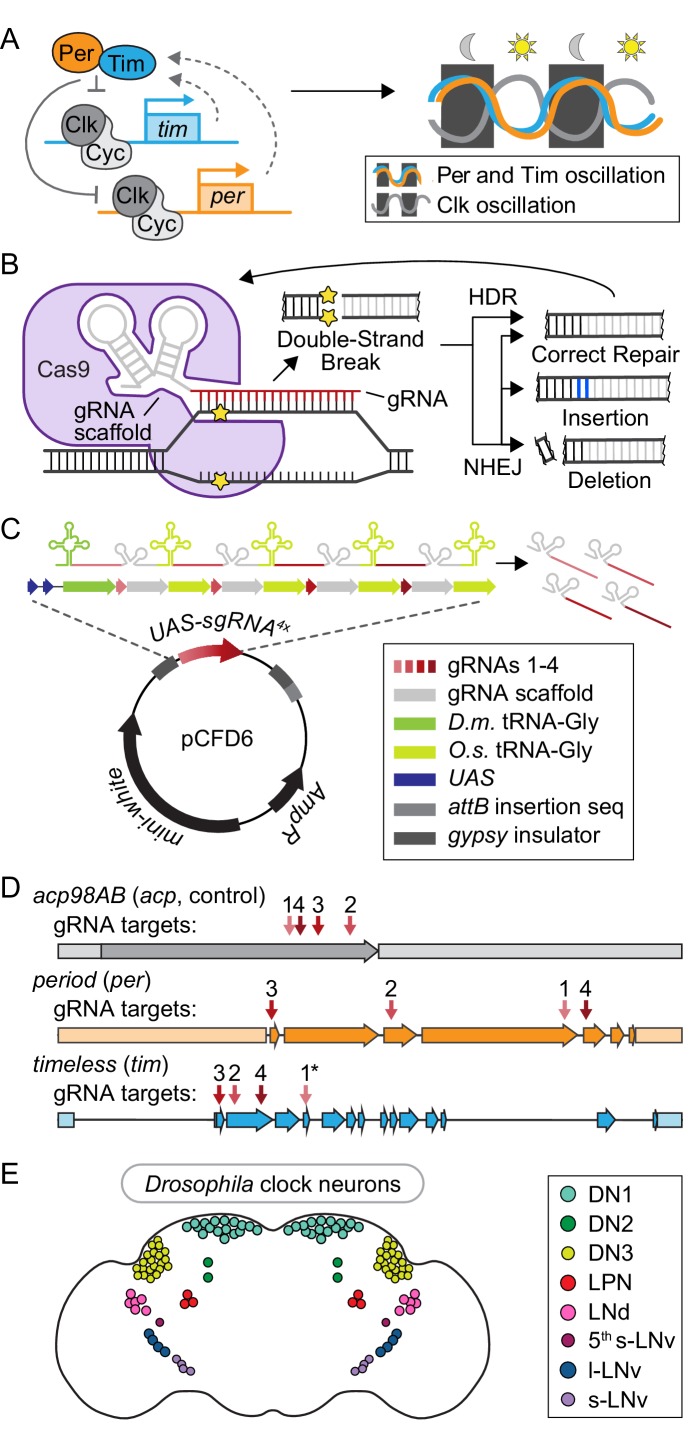
Figure 1. Toolbox for cell-specific, CRISPR-mediated disruption of core circadian regulators.
(A) Schematic of the transcriptional/translational negative feedback loop that drives rhythmic expression and activity of the four core circadian regulators: Period (Per), Timeless (Tim), Clock (Clk), and Cycle (Cyc). (B) Diagram of CRISPR-Cas9 mediated DNA damage and repair pathways. (C) Diagram of plasmid (pCFD6, adapted from Port and Bullock, 2016) used to generate UAS-sgRNA4x transgenic flies. D.m. = Drosophila melanogaster. O.s. = Oryza sativa, Asian rice. (D) Diagram showing sgRNA target sites for acp98AB (acp, gray), period (per, orange), and timeless (tim, blue), numbered in order of 5’−3’ position in the respective UAS-sgRNA4x construct. Arrows = exons; shaded rectangles = promoters and UTRs. *tim sgRNA one has a single base pair deletion in the Cas9-binding scaffold region (see Materials and methods). (E) Diagram of ~150 clock neurons organized into the following anatomical and functional clusters in the Drosophila brain: dorsal neurons (DN1, DN2, DN3), lateral posterior neurons (LPN), dorsal lateral neurons (LNd), and small and large ventral lateral neurons (s-LNv, 5th s-LNv, l-LNv).