Abstract
Intratumoral hypoxia causes the formation of dysfunctional blood vessels, which contribute to tumor metastasis and reduce the efficacy of therapeutic treatments. Blood vessels are embedded in the tumor stroma of which cancer-associated fibroblasts (CAFs) constitute a prominent cellular component. We found that hypoxic human mammary CAFs promoted angiogenesis in CAF-endothelial cell co-cultures in vitro. Mass spectrometry-based proteomic analysis of the CAF secretome unraveled that hypoxic CAFs contributed to blood vessel abnormalities by altering their secretion of various pro- and anti-angiogenic factors. Hypoxia induced pronounced remodeling of the CAF proteome, including proteins that have not been previously related to this process. Among those, the uncharacterized protein NCBP2-AS2 that we renamed HIAR (hypoxia-induced angiogenesis regulator) was the protein most increased in abundance in hypoxic CAFs. Silencing of HIAR abrogated the pro-angiogenic and pro-migratory function of hypoxic CAFs by decreasing secretion of the pro-angiogenic factor VEGFA and consequently reducing VEGF/VEGFR downstream signaling in the endothelial cells. Our study has identified a regulator of angiogenesis and provides a map of hypoxia-induced molecular alterations in mammary CAFs.
Introduction
Intratumoral hypoxia favors tumor aggressiveness and is associated with risk of metastasis, limited response to therapies and poor clinical outcome. The tumor vasculature plays key roles in these processes. The uncontrolled growth of the cancer cells results in the rapid consumption of oxygen released by the adjacent vasculature, leading to the formation of intratumoral hypoxic regions. To counteract the lack of oxygen, hypoxic cancer and stromal cells secrete factors that stimulate the recruitment of blood vessels, a process that is called angiogenesis (1). However, rather than promoting the formation of a functional vasculature, excessive amounts of those factors result in the recruitment of blood vessels that are leaky and nonfunctional. This phenomenon in turn leads to the expansion of hypoxic areas and the formation of a positive feedback loop between hypoxia and non-functional blood vessels that is detrimental and challenging to disrupt (2–4). So far, most of the effort has been devoted to targeting the vascular endothelial growth factor (VEGF) pathway (5), which is a potent inducer of blood vessel growth and is transcriptionally regulated by the hypoxia inducible factor 1α (HIF1α) (6). However, VEGF controls also the growth of blood vessels in normal tissues (7); therefore therapies that block VEGF signaling result in unwanted adverse effects (8). The identification of mechanisms that regulate blood vessel recruitment in hypoxic tumors may provide useful hints to develop strategies to block this feedback loop without interfering with the normal vasculature. Tumor blood vessels are typically embedded within the stroma, where cancer-associated fibroblasts (CAFs) are an abundant cell population (9). CAFs can originate from activation of resident normal fibroblasts within the tumor stroma and secrete factors that alter the composition and physical properties of the extracellular matrix (ECM) and signal to adjacent cells (10–12). CAFs play crucial roles in the pathogenesis of cancer, including the recruitment of blood vessels within the tumor (13, 14). How CAFs respond to oxygen deprivation is largely unknown and rather controversial. Intratumoral hypoxia induces CAF activation and HIF1α-activated fibroblasts co-transplanted in an MDA-MB-231 breast cancer xenograft are pro-tumorigenic and pro-metastatic (15). Conversely, tumorigenesis in a PyMT breast cancer model is accelerated by fibroblast-specific depletion of Hif1α before tumor onset, but not by depletion of prolyl-hydroxylase 2 (Phd2), which stabilizes HIF1α (16, 17). Furthermore, decreasing PHD2 levels in CAFs reduces their activated phenotype and their pro-metastatic activity in a 4T1 breast cancer orthotopic model (18). A better understanding of the molecular mechanisms underpinning the response of CAFs to hypoxia could bring useful insights on the role of these cells in hypoxic tumors.
Hypoxia-induced signaling has been mostly studied at the genome-wide gene expression level, and there is little information on how it affects the proteome of the cells. Modern mass spectrometry (MS) technology and robust platforms for data analysis allow in-depth and highly accurate quantitative analysis of cellular and sub-cellular proteomes (19–21), including those shaped by hypoxia (22, 23). Proteome-wide analysis of breast cancer cell-derived conditioned media (CM) has shown that high levels of lysyl oxidase (LOX) are induced by hypoxia and associated with bone metastasis, leading to the discovery of a role for LOX in bone metastasis formation (23).
Here we have used three dimensional (3D) co-cultures of human primary endothelial cells with mammary CAFs and established that the secretome of hypoxic CAFs promotes endothelial cell sprouting, which is one of the initial steps of angiogenesis (24). We have identified proteomic alterations induced by hypoxia in CAFs using high resolution MS and stable isotope labelling with amino acids in cell culture (SILAC) (25). We found hundreds of regulated molecules that may support hypoxic CAFs to drive angiogenesis, and discovered a regulatory mechanism of VEGF signaling through which hypoxic but not normoxic CAFs control endothelial sprouting.
Results
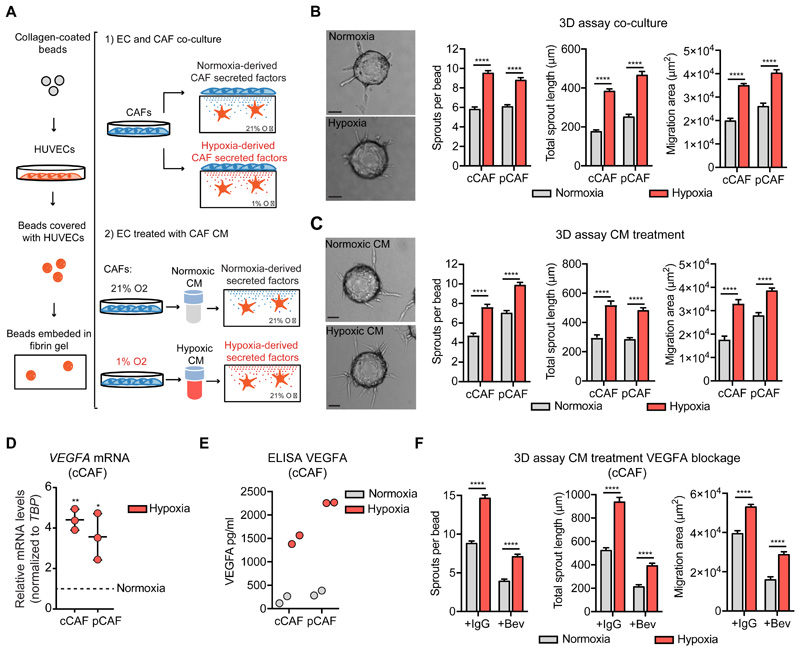
The secretome of hypoxic CAFs promotes endothelial sprouting
To determine how the hypoxic environment influences the ability of CAFs to regulate angiogenesis, we used an established three dimensional (3D) co-culture model in which collagen IV-coated beads are covered with endothelial cells and embedded into fibrin gel. CAFs are then plated on top of the solidified gel and secrete factors that influence the sprouting and invasion of the endothelial cells though the 3D matrix (fig. 1A) (26). We used human umbilical vein endothelial cells (referred to as ECs) because they are a well-characterized model to study endothelial functions and angiogenesis (27, 28). For fibroblasts, we used two human mammary CAF cell lines: cancer cell-induced CAFs (cCAFs) that we and others have previously characterized for their pro-angiogenic function in vitro and in vivo (13, 29) and patient-derived CAFs (pCAFs), which were isolated from a triple-negative invasive ductal carcinoma. The co-culture was exposed to either normoxia or hypoxia for 72h. Endothelial sprouting was enhanced by hypoxia in both co-cultures compared with their normoxic counterpart, as measured by the total number and length of EC sprouts and their migration area surrounding the bead (fig. 1B). To determine whether the factors secreted by hypoxic CAFs were the major contributor of this phenotype, normoxic ECs embedded in fibrin gel were treated with conditioned medium (CM) derived from cCAFs or pCAFs cultured under hypoxia or normoxia for 72h (fig. 1A). CAFs readily responded to hypoxia, because HIF1α was stabilized and accumulated into the nucleus after 4h of oxygen deprivation (fig. S1A,B). Moreover, after 72h of hypoxia, CAFs were still proliferative and not apoptotic, as measured by EdU incorporation (fig. S1C) and TUNEL assay (fig. S1D). Using this set-up, where there are no CAFs in the 3D culture and ECs are stimulated by CAF-derived CM, we observed a significant increase in sprouting when ECs were treated with the hypoxic CM (fig. 1C). Hence, hypoxic CAFs secrete factors that promote endothelial sprouting.
Figure 1. The secretome of hypoxic CAFs induces endothelial sprouting angiogenesis.
(A) Scheme of the 3D fibrin-based sprouting assay for ECs co-cultured with CAFs or alone and treated with CAF-derived CM. (B) Representative images and quantification of the sprouting of ECs co-cultured with CAFs under normoxia and hypoxia for 72h. Scale bar = 50 μm. N = beads assessed in 3 biological replicates; cCAF normoxia n = 120, cCAF hypoxia n = 135, pCAF normoxia n = 128, pCAF hypoxia n = 134. (C) Representative image and quantification of the sprouting of ECs treated with CM derived from CAFs cultured under normoxia or hypoxia for 72 h. Scale bar = 50 μm. N = beads assessed in 3 biological replicates; cCAFnormoxia n = 201, cCAF hypoxia n = 195, pCAF normoxia n = 112, pCAF hypoxia n = 135. (D) RT-qPCR analysis showing VEGFA transcription in cCAFs and pCAFs under hypoxic condition (normalized to the VEGFA levels in normoxia condition. VEGFA normoxia = 1 = dashed line). VEGFA mRNA levels were normalized to TBP. N = 3 biological replicates, one-sample t-test. (E) ELISA showing VEGFA levels in the CM from cCAFs and pCAFs. N = 2 biological replicates. (F) Quantification of the sprouting of ECs treated with the CM from normoxic and hypoxic CAFs in the presence or 1 μg/ml of bevacizumab or IgGK (control). N = beads assessed in 3 biological replicates; CM normoxia + IgGK n = 178, CM hypoxia + IgGK n = 161, CM normoxia + bevacizumab n = 176, CM hypoxia + bevacizumab n = 179. T-test. * p < 0.05, ** p < 0.01**** p < 0.0001.
Hypoxia increases the expression of the potent pro-angiogenic factor VEGFA, by activating the HIF pathway. As expected, hypoxia enhanced the mRNA levels and secretion of VEGFA in both cCAFs and pCAFs (fig 1D,E). To assess whether the pro-angiogenic effect of the CM from hypoxic CAFs was due to increased VEGFA levels, we blocked VEGFA/VEGFR signaling with the anti-VEGFA antibody bevacizumab. The addition of bevacizumab to the CM from hypoxic and normoxic CAFs reduced the sprouting of ECs by 60%, indicating that VEGF is a major contributor of the pro-angiogenic function of CAFs under either condition (fig. 1F). This finding was confirmed by the reduced sprouting observed upon HIF1α silencing in hypoxic CAFs (fig. S1E). Factors other than VEGF may also contribute to the pro-angiogenic effect of the hypoxic CM, because even in the presence of bevacizumab, the CM from hypoxic CAFs enhanced endothelial sprouting (fig. 1F).
Hypoxia remodels the proteome and secretome of CAFs to promote angiogenesis
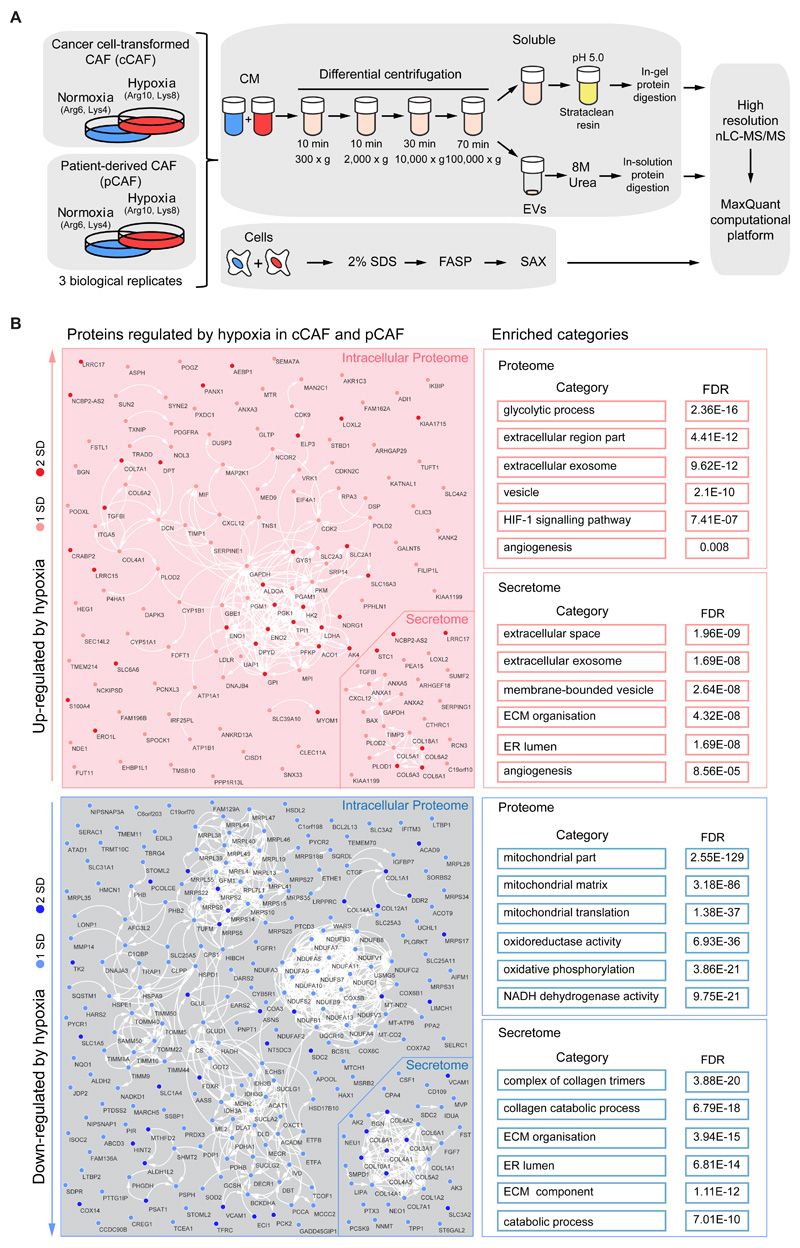
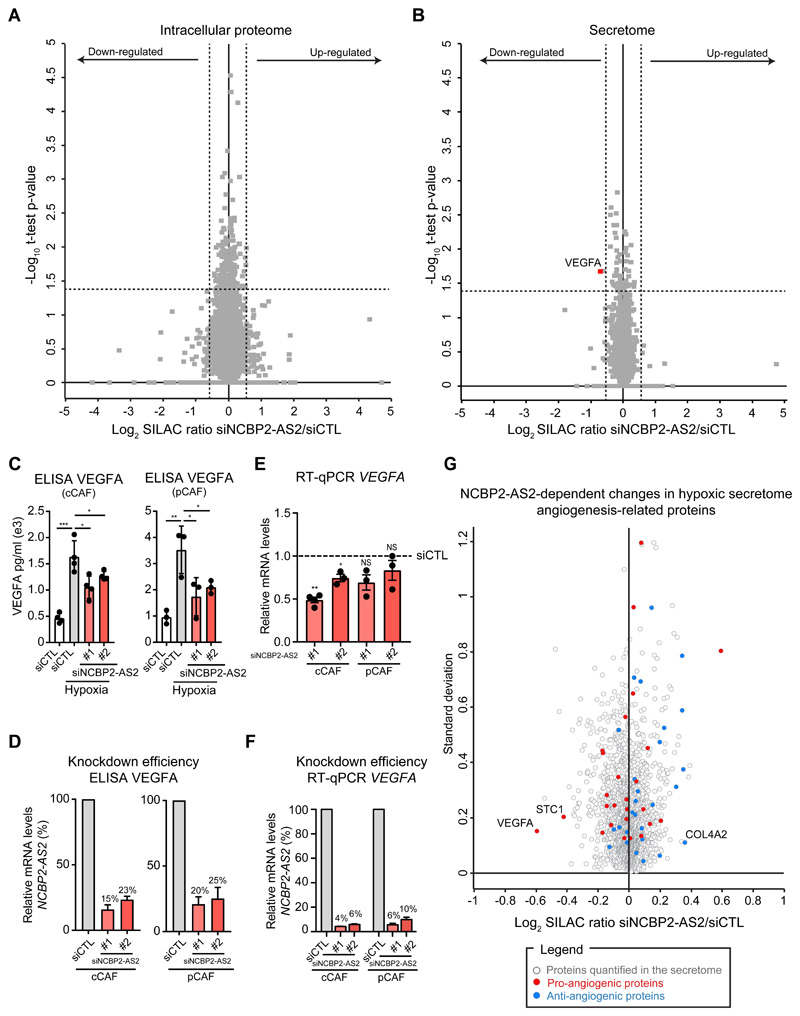
To determine the molecular events underpinning the pro-angiogenic function of hypoxic CAFs we used MS-based proteomics to compare the proteome of cCAFs and pCAFs cultured under normoxia or hypoxia for 72h, using SILAC for accurate protein quantification. We additionally analyzed the secretome of these cells to map the composition of the pro-angiogenic secretome of hypoxic CAFs. We measured the total cell proteome by fractionating with strong anion exchange chromatography tryptic peptides obtained from SDS-extracted proteins (30). We separated the CM into two fractions using multistep centrifugation: a fraction containing soluble factors (soluble) and a fraction containing extracellular vesicles (EVs). All samples were measured on an Orbitrap mass spectrometer followed by analysis with the MaxQuant computational platform (31) (fig. 2A). Overall 3,513 proteins were robustly quantified in the cell proteome (in at least two out of three biological replicates in both cCAFs and pCAFs), 681 proteins in the soluble secretome and 165 proteins in the EV fraction (Data File S1). Because we found that replicates strongly correlated with each other (fig. S1F), for each protein we averaged the hypoxia/normoxia SILAC ratio between replicates; proteins were defined as hypoxia-regulated based on the standard deviation of their ratio from the mean SILAC ratio calculated for the entire dataset. We classified regulated proteins as strongly regulated when the hypoxia/normoxia ratio was greater than two standard deviations (SD) from the mean, or as modestly regulated when the hypoxia/normoxia ratio was between one and two SDs. The latter group contained well-established HIF1α-target genes, such as CXCL12, lysyl oxidase-like 2 (LOXL2), procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 (PLOD2) and prolyl 4-hydroxylase subunit alpha-1 (P4HA1), among others (32–34) (Data File S1,S2), supporting the choice of this threshold to identify hypoxia-controlled proteins in our study. We required proteins to be regulated in both cCAFs and pCAFs. With these criteria, we identified 124 upregulated and 221 downregulated proteins by hypoxia in the intracellular proteome, and 27 upregulated and 34 downregulated proteins in the secretomes (fig. S2A). The physical and functional interactions between these proteins and the enriched functional categories showed that a vast proportion mirrored the metabolic reprogramming required for cellular adaptation to reduced oxygen availability (fig. 2B). Hypoxia increased the levels of proteins involved in glycolysis, while decreased levels of those involved in oxidative phosphorylation. Additionally, this analysis revealed a significant regulation of extracellular proteins, such as those located in the ECM or in exosomes and extracellular vesicles (fig. 2B), thus underlining the impact that hypoxia exerts on the ability of CAFs to alter the surrounding environment. In addition, we pinpointed the angiogenesis-regulating factors that were secreted by hypoxic CAFs. These included staniocalcin 1 (STC1), LOXL2 (32, 35), CXCL12 (14), annexin 1 and 2 (36, 37) and the endotrophin fragment of collagen 6-alpha (38), all of which were upregulated by hypoxia (fig. S2A,B). We confirmed VEGFA, LOXL2 and STC1 upregulation by RT-qPCR analysis (fig. S2C). Conversely, collagen fragments with anti-angiogenic properties, such as vastatin (COL8A1) (39), arresten (COL4A2) (40) and canstatin (COL4A1) (41), were down-regulated (fig. S2A,B).
Figure 2. Hypoxia-induced remodeling of the proteome and secretome of mammary CAFs.
(A) SILAC-based workflow used to characterize the total proteome and secretome of hypoxic cCAFs and pCAFs. (B) Physical and functional relationship among the hypoxia-regulated proteins in the intracellular proteome and secretome, as determined by STRING analysis and visualized with Cytoscape. GO categories and KEGG pathways enriched in the subset of proteins up-regulated and down-regulated by hypoxia are indicated on the right. FASP: Filter-aided sample preparation; SAX: strong anion exchange. Results from n=3 biological replicates for both cCAF and pCAF.
NCBP2-AS2 is a hypoxia-induced factor in CAFs
Next, we exploited our data to identify mechanisms that underpin hypoxic CAF functions. We compared our hypoxia-induced proteomic signature with known HIF1α-responsive genes and publicly available hypoxia-related gene expression signatures (fig. S3A and Data File S2). More than 75% of our signature (300 proteins) was not identified as hypoxia-regulated at the gene expression level by any of the analyzed datasets. This result suggests that the extent of gene expression regulation was below the statistical threshold, or that these proteins are regulated by hypoxia either post-transcriptionally or specifically in CAFs. Supporting the hypothesis of post-transcriptional regulation, hypoxia down-regulated pentraxin-related protein 3 (PTX3) and carboxypeptidase A4 (CPA4) at the protein but not at the gene expression level (fig. S3B,C). Moreover, we identified 134 mitochondrial proteins with decreased levels in hypoxic CAFs (fig. S3A and Data File S2), indicating that hypoxia may also induce extensive mitochondrial damage in CAFs (22, 42).
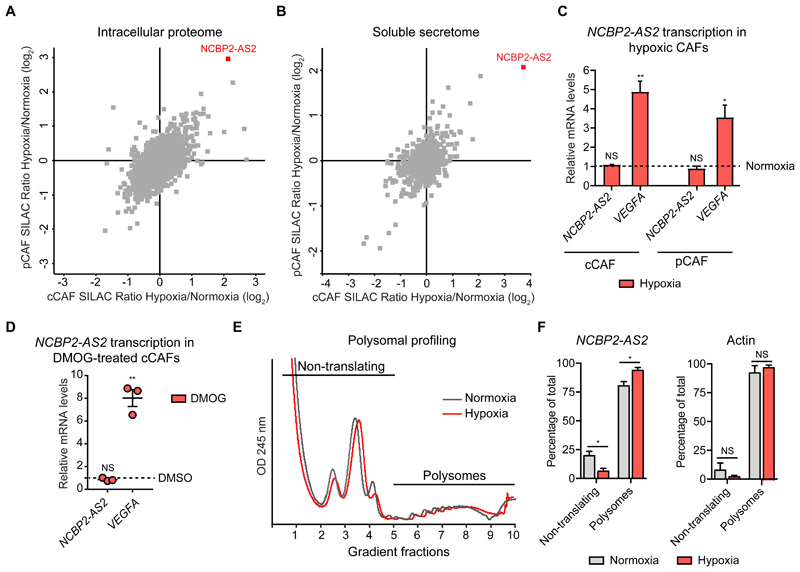
Within our signature, there were proteins concordantly regulated in both the proteome and secretome: COL1A1, COL6A2, COL14A1, LRRC17, NCBP2-AS2, SDC2 and VCAM1 (fig. S3A and Data File S2). These proteins could be important because they may have a strong impact not only on CAF functions, but also as paracrine factors on other cell types; moreover they could potentially be used as biomarkers for hypoxia. Of these, NCBP2-AS2 was strongly up-regulated by hypoxia in the proteome and soluble secretome of cCAFs and pCAFs (fig. 3A,B, Data File S2). Fewer peptides of NCBP2-AS2 were identified in the secretome than in the proteome (Data File S2), suggesting that NCBP2-AS2 is predominantly intracellular but also released as soluble factor. Increased levels of NCBP2-AS2 in total CAF lysate were confirmed using a targeted MS-based approach (fig. S4A-C). Conversely, we did not detect a correspondent increase in NCBP2-AS2 at the mRNA levels in hypoxic CAFs (fig. 3C, and fig S5A,B) or upon treatment with the HIF1α-stabilizing prolyl hydroxylase domain-containing protein inhibitor dimethyloxaloylglycine (DMOG) (fig. 3D). Conversely, NCBP2-AS2 translation increased with hypoxia. Ribosomal profiling analysis showed that normoxic and hypoxic CAFs had similar levels of total protein translation (fig. 3E). Furthermore, the amount of NCBP2-AS2 mRNA associated with polysomes, which are the actively translating subset of ribosomes, was significantly higher in hypoxic than in normoxic CAFs (Fig. 3F). We confirmed an increase in NCBP2-AS2 translation upon stimulation with hypoxia also using a pulsed-SILAC approach coupled to targeted MS-proteomics (fig. S6A). Thus, our findings indicate that hypoxia induces NCBP2-AS2 levels mostly post-transcriptionally, by enhancing its translation.
Figure 3. Hypoxic CAFs increase NCBP2-AS2 protein levels through enhanced translation.
(A, B) Scatter plot of the average SILAC ratio, hypoxia/normoxia, measured by MS in the pCAF and cCAF proteome (A) and secretome (B) highlighting the induction of NCBP2-AS2 by hypoxia in both cell types. Results from n=3 biological replicates. (C) RT-qPCR showing mRNA levels of NCBP2-AS2 and VEGFA normalized to TBP in CAFs upon exposure to hypoxia for 72 h (normalized to the levels in CAFs in normoxia. Normoxia = 1 = dashed line). VEGFA was used as a positive control. N = 3 biological replicates. One sample t-test. (D) RT-qPCR showing the mRNA levels of NCBP2-AS2 and VEGFA upon treatment with 1 mM DMOG for 24 h (normalized to the levels in DMSO control-treated cells. DMSO = 1 = dashed line). The mRNA levels were normalized to TBP. VEGFA was used as a positive control. N = 3 biological replicates. One-sample t-test. (E) Polysomal profiling of cCAFs exposed to hypoxia and normoxia for 24 h. Representative of 3 biological replicates. (F) RT-qPCR analysis for NCBP2-AS2 and actin mRNAs, which was used as control, on the non-translating and polysomal ribosomal fractions. N = 3 biological replicates. T-test. * p < 0.05, ** p < 0.01, NS: not significant.
Hypoxic CAFs promote angiogenesis through NCBP2-AS2
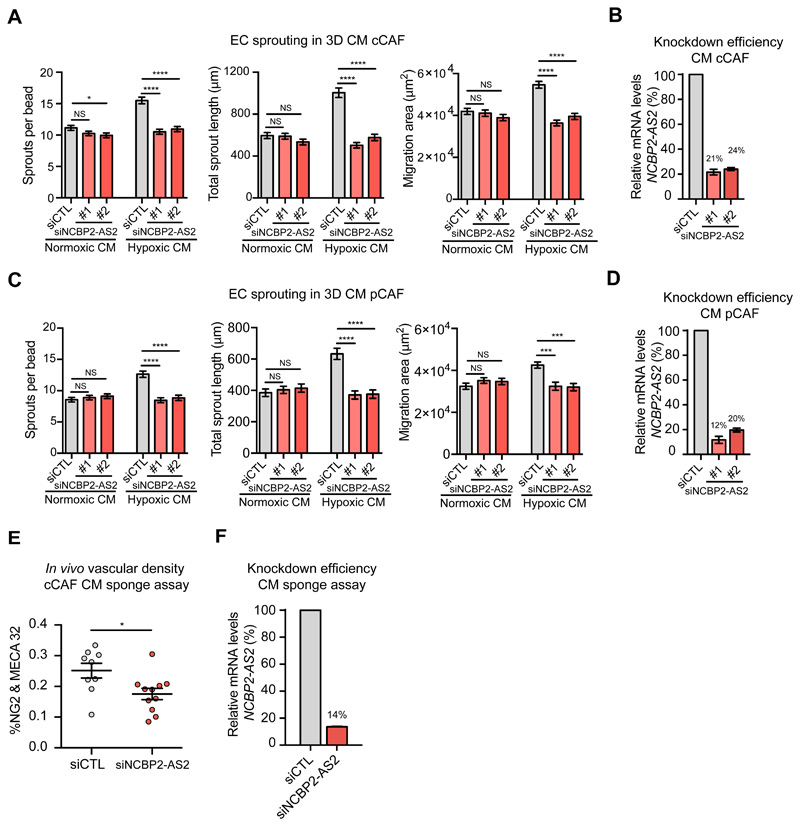
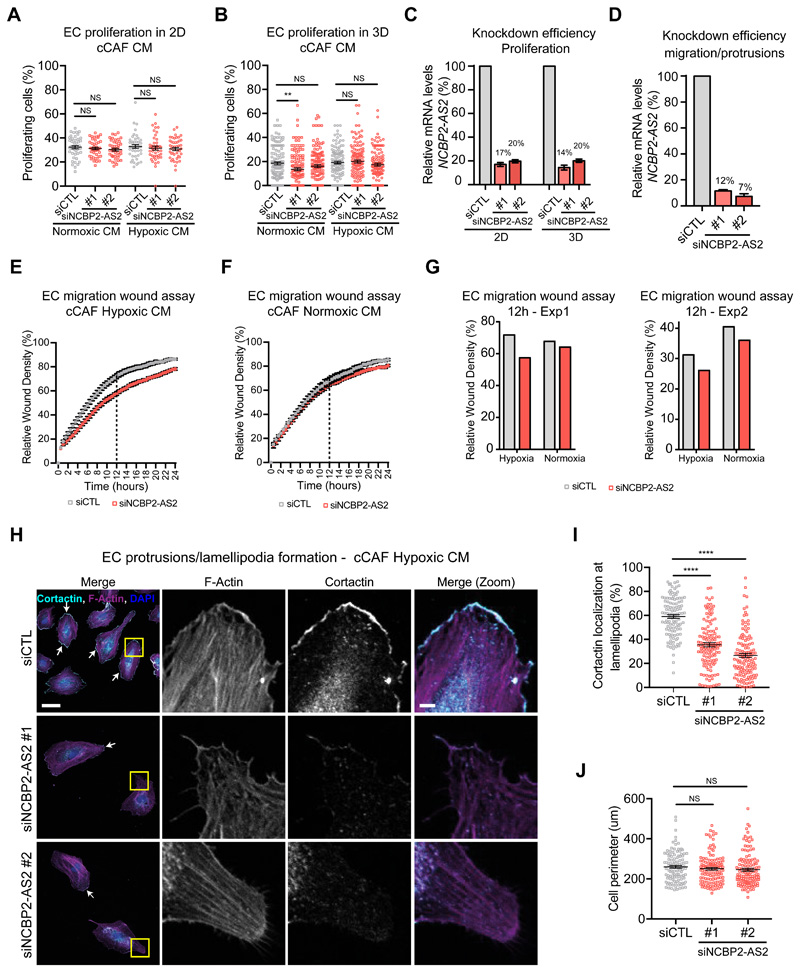
To determine whether NCBP2-AS2 contributed to the pro-angiogenic function of hypoxic CAFs, we silenced NCBP2-AS2 in CAFs and assessed the pro-angiogenic function of their CM. ECs treated with the CM from hypoxic CAFs with NCBP2-AS2 silencing sprouted significantly less than those treated with the CM from control CAFs. Conversely, the CM from normoxic CAFs control or those with NCBP2-AS2 silencing promoted endothelial sprouting to a similar extent (Fig. 4A-D and fig. S6B). Of note, the sprouting induced by normoxic and hypoxic CAFs with NCBP2-AS2 silencing was comparable. Since we did not detect a significant increase in sprouting when the recombinant purified GST-tagged NCBP2-AS2 was added to the medium of ECs cultured in 3D fibrin gel (fig. S7A), NCBP2-AS2 is likely to control endothelial sprouting by regulating intracellular CAF functions rather than as a paracrine factor. Next, we confirmed that NCBP2-AS2 supports the pro-angiogenic function of hypoxic CAFs also in vivo in a subcutaneous sponge assay typically used to investigate angiogenesis in vivo (43). In this assay, a sponge is subcutaneously implanted to initiate an inflammatory reaction that induces endothelial cell infiltration. This leads to the formation of a functional capillary network, whose extension is determined by the factors or conditioned medium contained in the sponge. The vessel density in sponges that were embedded with the CM from CAFs with NCBP2-AS2 silencing was significantly lower than in those containing the CM from control CAFs (fig. 4 E,F).
Figure 4. The CM from hypoxic CAFs silenced for NCBP2-AS2 show a decreased pro-angiogenic activity in vitro and in vivo.
(A, C) Sprouting quantification of ECs treated with CM from hypoxic and normoxic cCAFs (A) or pCAFs (C) transfected with siCTL or siNCBP2-AS2 (2 different siRNAs). N = beads assessed in 3 biological replicates; cCAF Hypoxic CM: siCTL n = 129, siNCBP2-AS2 #1 n = 124 and siNCBP2-AS2 #2 n = 116. Normoxic CM: siCTL n = 121, siNCBP2-AS2 #1 n = 131 and siNCBP2-AS2 #2 n = 122. pCAF Hypoxic CM: siCTL n= 119, siNCBP2-AS2 #1 = 107 and siNCBP2-AS2 #2 = 97. Normoxic CM: siCTL n= 111 beads, siNCBP2-AS2 #1 = 127 and siNCBP2-AS2 #2 = 104. (B, D) RT-qPCR showing the remaining mRNA levels of NCBP2-AS2 normalized to TBP in (B) cCAFs and (D) pCAFs used to produce CM. (E) Vascular density quantification, as measured by Meca32+ and Ng2+ staining of the blood vessels (y axis), of subcutaneous sponges which were embedded with CM derived from hypoxic CAFs transfected with siCTL or siNCBP2-AS2. N = ½ sponges derived from 6 mice per group. T-test. (F) RT-qPCR showing the remaining mRNA levels of NCBP2-AS2 normalized to TBP in CAFs producing the CM used in the sponge assay in (E). * p < 0.05, *** p < 0.001, **** p < 0.0001, NS: not significant.
Because sprouting angiogenesis is regulated by a combination of EC proliferation and migration, we investigated how NCBP2-AS2 silencing affected these functions. When treated with the CM from CAFs with or without NCBP2-AS2 silencing, EC proliferation in 3D and 2D culture was comparable, as measured by EdU incorporation (fig 5A-C, fig. S7B,C). Next, we established that the CM from hypoxic CAFs with NCBP2-AS2 silencing had reduced pro-migratory properties compared with the CM from hypoxic CAFs control. Wound closure in an endothelial monolayer was slower in ECs treated with CM from NCBP2-AS2-silenced hypoxic CAFs. Conversely, silencing of NCBP2-AS2 in normoxic CAFs had minor impact on the pro-migratory function of their secretome (fig. 5D-G). To confirm the role of NCBP2-AS2 in cell migration, we evaluated the ability of ECs to form lamellipodia, because pro-angiogenic factors such as VEGF induce the formation of cell protrusions (44). To monitor lamellipodia, we performed immunostaining for cortactin, which is a cortical actin remodeling protein and major mediator of lamellipodia formation (45). The presence or absence of NCBP2-AS2 in hypoxic CAFs did not affect the total perimeter of ECs, but when treated with the CM from hypoxic CAFs with NCBP2-AS2 silencing the ECs had significantly fewer cortactin-enriched protrusions (fig. 5H-J). These data show that NCBP2-AS2 in CAFs is required to support the pro-angiogenic activity of their secretome.
Figure 5. ECs treated with the CM from hypoxic CAFs silenced for NCBP2-AS2 have a decreased migratory capacity.
(A) Proliferation of ECs cultured in 2D and treated with the CM from normoxic and hypoxic CAFs transfected with siCTL or siNCBP2-AS2. N = 45 fields assessed per condition from 3 biological replicates. (B) Proliferation of ECs cultured in 3D fibrin gel and treated with the CM from normoxic and hypoxic CAFs transfected with siCTL or siNCBP2-AS2. N = beads assessed from 3 biological replicates. Normoxic CM: siCTL n = 124, siNCBP2-AS2 #1 n = 132 and siNCBP2-AS2 #2 n = 136. Hypoxic CM: siCTL n = 127, siNCBP2-AS2 #1 n = 120 and siNCBP2-AS2 #2 n = 134. (C) RT-qPCR showing remaining mRNA levels of NCBP2-AS2 normalized to TBP. (D) RT-qPCR showing remaining mRNA levels of NCBP2-AS2 normalized to TBP. (E,F) Scratch wound migration assay using ECs treated with the CM from hypoxic (E) or normoxic (F) cCAFs transfected with siCTL or siNCBP2-AS2. N = 2 biological replicates. (G) Relative wound density measured at 12h in the two independent replicate experiments (dashed line in E and F). (H) Representative images of lamellipodia formation in ECs treated with the CM from cCAFs silenced or not for NCBP2-AS2. (I) Quantification of cortactin at the cell membrane. (J) Quantification of the total cell perimeter. N= cells measured in 3 biological replicates. siCTL n = 111, siNCBP2-AS2 #1 n = 126 and siNCBP2-AS2 #2 n = 125. Scale bar = 10μm, scale bar in zoomed inset = 5μm. P-value based on Kruskal-Wallis test corrected for multiple testing using Dunn’s test. T-test was used for the other panels. ** p < 0.01, **** p < 0.0001, NS: not significant.
NCBP2-AS2 regulates VEGFA expression
To understand how NCBP2-AS2 controls the pro-sprouting function of the CAF secretome, we used SILAC MS proteomics to identify molecular alterations induced upon silencing of NCBP2-AS2 in hypoxic CAFs. The knockdown of NCBP2-AS2 did not significantly alter the intracellular proteome of the CAFs (fig. 6A and Data File S3). Conversely, the levels of VEGFA in the secretome of CAFs silenced for NCBP2-AS2 were down-regulated (fig. 6B and Data File S4), as also confirmed by ELISA (fig 6C, D). RT-qPCR analysis showed that VEGFA downregulation occurred also at transcriptional level (fig. 6E, F). VEGF levels in CAFs were not regulated in an autocrine manner by secreted NCBP2-AS2, because the amount of VEGFA in the secretome of hypoxic CAFs silenced for NCBP2-AS2 could not be rescued upon treatment with conditioned media from hypoxic control CAFs (fig S7D-F). Next, we investigated how other hypoxia-regulated pro- and anti-angiogenic factors were affected by NCBP2-AS2 silencing in CAFs. The levels of the pro-angiogenic protein STC1 decreased upon NCBP2-AS2 silencing, although at a lower extent compared to VEGFA, whereas the anti-angiogenic factor COL4A2 was modestly increased upon NCBP2-AS2 silencing (fig 6G). Thus STC1 and COL4A2 may also contribute to the decreased pro-angiogenic function of hypoxic CAFs. Together, these results show that VEGFA could account for most of the reduced pro-angiogenic effect of the secretome of CAFs with NCBP2-AS2 silencing.
Figure 6. NCBP2-AS2 regulates the levels of CAF-derived VEGFA.
(A,B) Volcano plot showing the SILAC ratio of the proteins quantified in the total proteome (A) and secretome (B) of hypoxic cCAFs transfected with siCTL and siNCBP2-AS2. The dashed lines show the regulation cut-off of at least 1-fold change and p-value < 0.05. Results from 3 biological replicates. (C) ELISA of VEGFA in the CM from hypoxic CAFs silenced or not for NCBP2-AS2 and normoxic CAFs siCTL. One-tailed t-test. N = biological replicates. (D) RT-qPCR showing remaining mRNA levels of NCBP2-AS2 normalized to TBP in cells used in (C). (E) RT-qPCR for VEGFA in hypoxic cCAFs and pCAFs transfected with siCTL or siNCBP2-AS2. The mRNA levels were normalized to TBP. One-sample t-test. N = biological replicates. (F) RT-qPCR for NCBP2-AS2 showing the silencing efficiency in cells used in (E). NCBP2-AS2 mRNA levels were normalized to TBP. N = biological replicates. (G) Influence of NCBP2-AS2 silencing on the angiogenic secretome of hypoxic CAFs. * p < 0.05, ** p < 0.01, *** p < 0.001, **** p<0.0001.
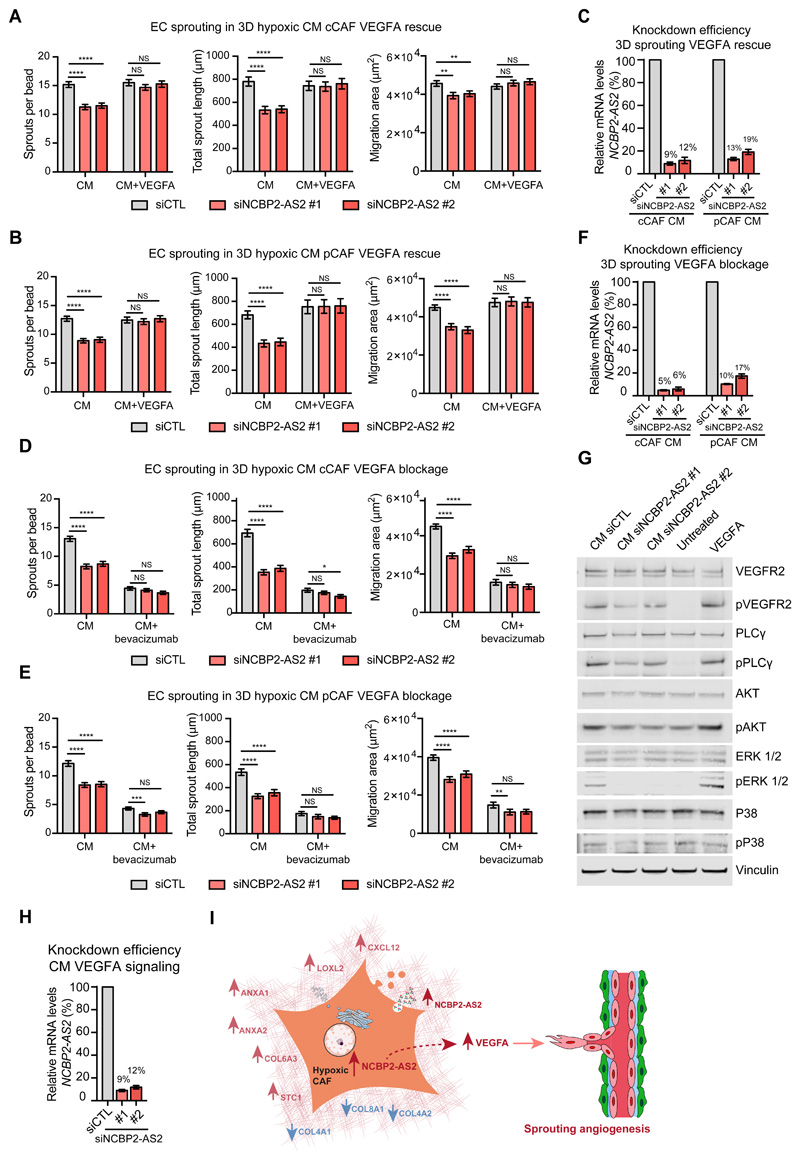
We reasoned that reduced amounts of VEGFA in the CM from NCBP2-AS2-silenced hypoxic CAFs caused the reduction in sprouting observed in ECs treated with this CM. Defective sprouting in ECs was rescued by supplementing the CM from hypoxic NCBP2-AS2-silenced CAFs with VEGFA (fig 7A-C), thus confirming our hypothesis. Likewise, the addition of bevacizumab in the CM from hypoxic CAFs with NCBP2-AS2 silencing did not have an additive inhibitory effect on endothelial sprouting (fig. 7D-F). Moreover, ECs treated with hypoxic CAF-derived CM had activated VEGFR2 and canonical downstream signaling, as shown by phosphorylation of the regulatory site of PLCγ (Tyr783), AKT (Ser473) and ERK1/2 (Thr202/Tyr204). Conversely, the extent of VEGFR2 activation and the downstream signaling were reduced in ECs treated with CM from hypoxic CAFs silenced for NCBP2-AS2 (Fig. 7G,H and Fig. S8). Thus, we show that NCBP2-AS2 controls the pro-angiogenic function of CAF secretome by regulating VEGFA levels and that this, in turn, controls VEGF-dependent sprouting in ECs (Fig 7I).
Figure 7. NCBP2-AS2 regulates EC sprouting through VEGFA signaling.
(A) Sprouting quantification of ECs treated with the CM from hypoxic cCAFs transfected with siCTL or siNCBP2-AS2 (two different siRNAs) with or without VEGFA. N = beads assessed from 3 biological replicates. CM without VEGFA: siCTL n = 149, siNCBP2-AS2 #1 n = 128 and siNCBP2-AS2 #2 n = 110. CM with VEGFA: siCTL n=132, siNCBP2-AS2 #1 n = 130 and siNCBP2-AS2 #2 n = 126. (B) Sprouting quantification of ECs treated with the CM from hypoxic pCAFs transfected with siCTL or siNCBP2-AS2 (two different siRNAs) with or without VEGFA. N = beads assessed from 3 biological replicates. CM without VEGFA: siCTL n = 162 beads, siNCBP2-AS2 #1 n = 132 and siNCBP2-AS2 #2 n = 124. CM with VEGFA: siCTL n=127, siNCBP2-AS2 #1 n = 133 and siNCBP2-AS2 #2 n = 136. (C) RT-qPCR showing remaining mRNA levels of NCBP2-AS2 normalized to TBP. (D) Sprouting quantification of ECs treated with the CM from hypoxic cCAFs transfected with siCTL or siNCBP2-AS2 (two different siRNAs) in the presence of bevacizumab or IgGK (control). N = beads assessed from 3 biological replicates. CM with IgG: siCTL n = 135, siNCBP2-AS2 #1 n = 129 and siNCBP2-AS2 #2 n = 107. CM with bevacizumab: siCTL n = 127, siNCBP2-AS2 #1 n = 147 and siNCBP2-AS2 #2 n = 122. (E) Sprouting quantification of ECs treated with the CM from hypoxic pCAFs transfected with siCTL or siNCBP2-AS2 (two different siRNAs) in the presence of bevacizumab or IgGK (control). N = beads assessed from 3 biological replicates. CM with IgG: siCTL n = 121, siNCBP2-AS2 #1 n = 108 and siNCBP2-AS2 #2 n = 107. CM with bevacizumab: siCTL n = 126, siNCBP2-AS2 #1 n = 130 and siNCBP2-AS2 #2 n = 110. (F) RT-qPCR showing the remaining mRNA levels of NCBP2-AS2 normalized to TBP. (G) Representative western blot for proteins downstream of VEGFR signaling in ECs treated with the CM from cCAFs silenced or not for NCBP2-AS2. N = 3 biological replicates. (H) RT-qPCR showing remaining mRNA levels of NCBP2-AS2 normalized to TBP. (I) Working model. T-test. * p < 0.05, ** p < 0.01, **** p < 0.0001, NS: not significant.
NCBP2-AS2 is expressed in human tumors
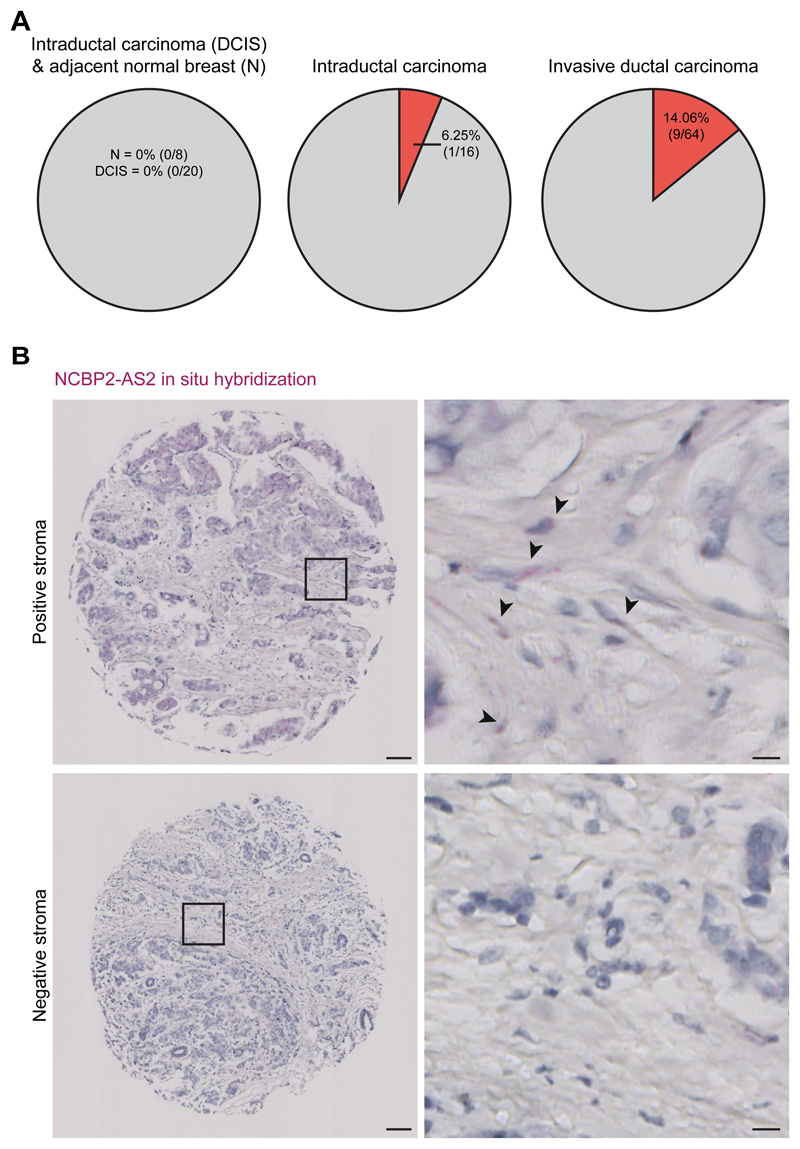
Finally, we determined NCBP2-AS2 expression in publicly available TCGA datasets of breast cancer patient samples. While this analysis showed that NCBP2-AS2 is expressed in tumors, giving that NCBP2-AS2 regulation by hypoxia occurs predominantly at the post-transcriptional level, we could not find a positive correlation between NCBP2-AS2 gene expression levels and hypoxia (fig. S9). We then looked at NCBP2-AS2 expression in tissue microarray (TMA). Because antibodies that recognize NCBP2-AS2 by immunohistochemistry staining are not available, we assessed its expression by in situ hybridization. In a breast cancer TMA containing 108 patient samples, we observed NCBP2-AS2 positive staining in the stroma of 10% of the tumors. In particular, ductal carcinomas positively stained for NCBP2-AS2 whereas no staining was detected in the stroma of lobular carcinomas. We found that 3% of intraductal carcinoma and 14% of invasive ductal carcinoma positively stain for NCBP2-AS2 in the stroma (fig. 8A,B). These findings indicate that the frequency of NCBP2-AS2 expression in the stroma increases with the progression of the disease, suggesting a possible role in advanced disease.
Figure 8. NCBP2-AS2 is expressed in the stroma of breast tumors.
(A) Pie chart showing for each type of breast cancer contained in a breast cancer TMA of 100 patient samples, how many showed a positive staining (as assessed by in situ hybridization with RNAscope) for NCBP2-AS2 in the tumor stroma. (B) Representative images of breast cancer tissues with positive or negative staining (as assessed by in situ hybridization with RNAscope) for NCBP2-AS2 in the tumor stroma. Scale bar = 100 μm for the panels on the left and 10 μm for the panels on the right.
Discussion
The ability of hypoxia to create an aberrant and dysfunctional tumor vasculature is underpinned by secreted pro-angiogenic factors that are downstream of HIF1α signaling, of which VEGF plays a key role. CAFs are highly abundant in invasive carcinomas where they create an aberrant microenvironment permissive for blood vessel growth. Notably, hypoxic and pro-angiogenic gene expression signatures found in the stroma of breast cancer patients correlate with poor clinical outcome (46), highlighting the importance of better understanding the role of hypoxia in the tumor stroma. Here we showed that hypoxia exacerbated the pro-angiogenic functions of CAFs, and MS-proteomic analyses pinpointed potential molecular mechanisms responsible for this function. VEGF is the major, but not the only, contributor of the enhanced pro-angiogenic activity of hypoxic CAFs, and we unveiled a mechanism through which NCBP2-AS2 controls VEGFA expression in hypoxic CAFs in vitro.
The CAF secretome is unique for its complexity and ability to remodel the tumor microenvironment including recruiting blood vessels. The cCAF model used in this study has been previously exploited by us and others to determine that TGFβ and CXCL12 dependent pathways support the pro-angiogenic activity of these cells (13) and to uncover a pro-angiogenic function for the CAF-secreted protein chloride intracellular channel 3 (CLIC3) (29). Here we deployed cCAFs and patient-derived CAFs to draw an intracellular and extracellular molecular portrait of hypoxic mammary CAFs. We did not find significant changes in CAF activation marker αSMA (ACTA2 in Data File S1), indicating that in our cell models and experimental conditions hypoxia does not inactivate CAFs, as previously shown in head and neck and vulvar CAFs (18). Conversely, reduced oxygen availability remodels the CAF secretome. Hypoxic CAFs secrete increased amounts of pro-angiogenic factors and decreased amounts of anti-angiogenic factors, conferring enhanced pro-angiogenic properties to their CM. We asked whether our datasets could be exploited to identify mechanisms that promote the pro-angiogenic effect of hypoxic CAFs. These could be potential targets to block the formation of a dysfunctional vasculature in hypoxic cancers. Proteins up-regulated by hypoxia and which were found also secreted by CAFs may be useful not only as targets but also as potential biomarkers in body fluids. We discovered the antisense gene protein NCBP2-AS2 (transcribed from the antisense DNA strand of the gene NCBP2-AS2) as a hypoxia-responsive molecule. Hypoxia controls protein levels and regulates both transcription and translation mechanisms (47, 48). Notably, the latter promotes a swifter response to environmental stimuli. NCBP2-AS2 translation was enhanced by hypoxia, thus indicating that it may play key roles to counteract hypoxic stress in the cells, for example, boosting VEGFA signaling for the recruitment of blood vessels. We showed that targeting NCBP2-AS2 in hypoxic CAFs reduced VEGFA expression and secretion to the levels of VEGFA expressed in normoxic CAFs, but that this manipulation did not affect the pro-angiogenic activity of normoxic CAFs in our in vitro setup. These results make it tempting to speculate that NCBP2-AS2 could offer a therapeutic opportunity to normalize the dysfunctional vasculature typical of hypoxic malignant cancers without altering physiological angiogenesis. Indeed, silencing NCBP2-AS2 in hypoxic CAFs significantly reduced the pro-angiogenic activity of their CM in vivo. In situ hybridization for NCBP2-AS2 in breast cancer patient samples showed that it is expressed in the tumor stroma. This prompts investigation into the functional consequences of modulating the levels of stromal NCBP2-AS2 in cancer pathology in vivo.
How NCBP2-AS2 controls VEGFA levels remains to be elucidated. STC1 may play a role because it was also found down-regulated in the CAF secretome in the absence of NCBP2-AS2. In gastric cancer, STC1 promotes angiogenesis in vitro and in vivo by increasing VEGF expression through the activation of protein kinase C βII (PKCβII) and ERK1/2 pathways (49). Additional players are suggested by unbiased MS-based interactome analyses in HeLa cells (50) which identified the GTPase KRAS, the Ras-related protein Rab-5C (RAB5C), the nuclear cap-binding protein subunit 2 (NCBP2), the serine/threonine-protein phosphatase 2A regulatory subunit B” subunit gamma (PPP2R3C), the kinesin-like protein KIF19 (KIF19), the RNA-binding protein 8A (RBM8A) and the survival motor neuron protein (SMN1) as potential binding partners of NCBP2-AS2. Of note, SMN1 and VEGF are upregulated in ischemic rat hippocampus (51) and VEGF expression is up-regulated by wild type KRAS in human colon carcinoma cells (52) and by mutant KRAS in human colon carcinoma (53), pancreatic epithelial (54) and NIH3T3 (55) cells.
In conclusion, here we provide new insight into how mammary CAFs adapt to and behave in a low oxygen environment and provide a resource to discover molecular mechanisms underpinning CAF functions. Furthermore, we unraveled a mechanism through which hypoxic CAFs control the expression of the most potent pro-angiogenic factor, VEGF, through NCBP2-AS2, which we propose to rename to a more functional name, HIAR (hypoxia-induced angiogenesis regulator).
Materials and Methods
Cell culture
Human umbilical vein endothelial cells (HUVECs) were isolated in house from umbilical cords and cultured in EGM-2 medium (Lonza) on plastic dishes coated in 1% gelatin. Patient-derived mammary cancer-associated fibroblasts (pCAFs) were isolated in house from a grade 3 triple negative breast cancer patient sample obtained through NHS Greater Glasgow and Clyde Biorepository. All participants gave specific consent to use their tissue samples for research. The pCAFs were cultured on plastic dishes coated in 35 μg/ml collagen I (Gibco). The cancer cell-derived human mammary CAFs (cCAFs) cell line was kindly provided by Professor Akira Orimo (Paterson Institute, Manchester). The pCAFs, cCAFs and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS, Life technologies), 1% glutamine (Life technologies) and 1% penicilin/streptomycin (Life technologies). For quantitative proteomics experiments, pCAFs and cCAFs were cultured in SILAC DMEM (Life Technologies) supplemented with 2% FBS, 8% 10 kDa dialysed FBS (PAA), 1% glutamine and 1% penicillin/streptomycin. SILAC DMEM used for the ‘medium’ labelled cells contained 84 mg/l 13C6 L-arginine and 175 mg/l D4 L-lysine, whereas the medium for the ‘heavy’ labelled cells contained 84 mg/l 13C615N4 L-arginine and 175 mg/l 13C615N2 L-lysine (Cambridge Isotope Laboratories). All the cell lines described above were kept in standard tissue culture incubators at 37 °C, 5% CO2 in 21% O2 (normoxic condition) or in a hypoxic chamber at 37°C, 5% CO2 in 1% O2 (hypoxic condition). The hypoxic chamber utilized was a Whitley H30/H35 Hypoxystation (Don Whitley Scientific). All cell lines were routinely tested for mycoplasma.
pCAFs isolation
Tumour tissue was minced and transferred into a 10 cm culture dish. 10 ml of RPMI supplemented with 10% FCS, 1% fungizone (Life Technologies), 1% penicillin/streptomycin (Invitrogen) and activated collagenase 1A (1 mg/ml, Sigma) were added and incubated overnight at 37 °C. The digested tissue was then filtered through a cell strainer and centrifuged. The CAF-containing cell pellet was washed in PBS, resuspended in DMEM, 10% FBS, 1% P/S, 1% fungizone, and plated on collagen coated (35 μg/ml, rat tail collagen 1, Gibco) culture dish. Patient samples were obtained through NHS Greater Glasgow and Clyde Biorepository (project #305). All participants gave specific consent to use their tissue samples for research.
pCAF immortalization
The pCAFs were immortalized using a human telomerase reverse transcriptase (hTERT)-expressing plasmid (pIRES2-hygro), kindly provided by Dr. Fernando Calvo (ICR, London). Lentivirus-containing the hTERT plasmid were generated in HEK293T cells. Two rounds of viral transduction were performed on consecutive days. Cells were selected using 10 μg/ml hygromycin (Sigma).
3D fibrin gel-based sprouting assay
To assess the sprouting ability of ECs in 3D we have used a previously described protocol (26) with minor modifications (29). Around 3.3x105 ECs embedded in the fibrin gel were either treated with CAF-derived conditioned medium (CM, prepared from CAFs cultured in 2D on plastic dish) or co-cultured with 2x104 CAFs in a 24-well plate. For the experiments where the CAF-derived CM was used, 900 μl of freshly collected CM derived from 8x104 CAFs cultured in EGM-2 for 72h under normoxia or hypoxia were admixed with 100 μl of fresh EGM-2. The CM was replaced with fresh CM every 24h. For experiments using VEGFA neutralizing antibody, 1 μg/ml of bevacizumab (Roche) or IgGK (Sigma) was added in the CM. For the rescue experiments, 50 ng/ml of VEGFA (Sigma) was added. For the 3D experiments, the images used for quantification were acquired using the Axiovert 25 microscope (Zeiss) with a Retiga EXi Fast 1394 camera (Imaging) two days after the beads were embedded in the fibrin gel. Three technical replicates were performed for each biological replicate. A total of 15 images were acquired and quantified for each biological replicate. In total n=3 biological replicates (n=45 images) were performed for each experiment.
EdU proliferation assay
5-ethynyl-2'-deoxyuridine (EdU, Life technologies) was added to the culture medium to a final concentration of 10 μM and cells were allowed to incorporate for 6 h, when cultured in 3D, and 1.5 h, when cultured in 2D. Following EdU incorporation, cells were fixed with 4% PFA for 15 min, washed twice with 3% BSA and permeabilized with 0.5% Triton-X for 20 min. EdU positive cells were fluorescently labelled using a click reaction following the manufactures’ instructions (Life technologies), whilst all cells were stained with DAPI. After the staining, images were acquired in the Nikon A1R confocal microscope for the 3D assay and in the Zeiss 710 confocal for 2D cultures. The percentage of cells that entered the S phase of the cell cycle was determined by dividing the number of EdU positive cells by the total number of cells positive for DAPI.
TUNEL assay
Click-iT TUNEL Alexa Fluor 488 Imaging Assay (Life technologies) was used to monitor DNA degradation in apoptotic cells. In a 24-well plate, 8x104 cells were seeded on a 13 mm coverslip. CAFs were exposed to hypoxia and fixed with 4% PFA for 15 min. Cells were permeabilized with 0.25% Triton X-100 in PBS for 20 min and the TUNEL assay was performed according to manufacturer’s instructions.
Secretome production and analysis
For the cCAF- and pCAF-derived secretome, 6x106 ‘medium’ and ‘heavy’ SILAC labelled cells were seeded in 15 cm dishes. The following day, cells were washed six times with pre-warmed PBS to remove FBS residuals, serum-free SILAC medium was added and cells were allowed to secrete for 72 h in either normoxic or hypoxic conditions. Subsequently, the same volume of conditioned media from 3 dishes from each condition was mixed and subjected to sequential centrifugations at 4°C: 300 x g for 10 min, 2,000 x g for 10 min and 10,000 x g for 30 min. An ultracentrifugation step of 100,000 x g for 70 min was carried out to separate the soluble secretome from the extracellular vesicle (EV) fraction. The pellet containing the EVs was lysed with 8 M Urea buffer and in-solution digested with trypsin (Promega). The soluble fraction was acidified using 10% trifluoroacetic acid (TFA) to pH 5.0 and proteins enriched using Strataclean-resin (Agilent Technologies) as previously described (29). The eluted proteins from the resin were loaded in a 4-12% gradient NuPAGE Novex Bis-Tris gel, stained with instant blue and in-gel digested (56) with trypsin (Promega). For the secretome of hypoxic cCAFs silenced for NCBP2-AS2, ‘medium’ and ‘heavy’ SILAC labelled cells were transfected with siRNA CTL and siRNA NCBP2-AS2. 2x106 cells from each condition were seeded in a 10 cm dish and the CM prepared as described above. In this experiment, all secreted proteins and EVs were enriched with Strataclean-resin and in-gel digested with trypsin.
Total proteome sample preparation
For the analysis of the total proteome, total lysates from the same cells used for the isolation of the secretome were collected using 2% SDS, 100 mM TrisHCl pH 7.4 lysis buffer. Proteins were digested into peptides using filter-aided sample preparation using filter with 30 KDa cutoff (FASP,(57)). For the proteomes of cCAFs and pCAFs exposed to hypoxia, 50 μg of FASP-derived peptides were fractionated using strong anion exchange chromatography (SAX,(30)). For the proteome of hypoxic cCAFs silenced for NCBP2-AS2, 50 μg of FASP-derived peptides were fractionated using high-pH reverse phase StageTip fractionation as previously described (58), with minor modifications.
nLC-MS/MS analysis
After trypsin digestion, peptides were desalted using StageTip (59). Peptides were resuspended in 1% TFA, 0.2% acetic acid or formic acid buffer and injected on an EASY-nLC or 1200 system coupled online to an LTQ-Orbitrap Pro Elite or Q-Exactive HF, respectively, via nanoelectrospray ion source (Thermo Fisher scientific). Peptides were separated on a 20 cm fused silica emitter (New Objective) packed in house with reverse phase Reprosil Pur Basic 1.9 μm (Dr. Maisch GmbH). For peptide elution, buffer A (0.1% formic acid) and Buffer B (80% ACN, 0.1% formic acid) were used. In the Orbitrap Elite, peptides were eluted with a flow of 200 nl/min from 5% to 30% of buffer B, in a 90 min linear gradient (for in-gel digested peptides) and 190 min gradient (for SAX fractions and extracellular vesicles). In the Q-Exactive HF, peptides were eluted with a flow of 300 nl/min from 5% to 30% of buffer B in a 60 min linear gradient. In the Orbitrap Elite, the mass range acquired for the full MS scan was 300-1,650 m/z with a resolution of 120,000 at 400 Th. The top ten most intense ions in the full MS were isolated for higher-energy collision dissociation fragmentation with a target of 40,000 ions at a resolution of 15,000 at 400 Th). For the Q-Exactive HF, the mass range acquired for the full MS scan was 375-1,500 m/z with a resolution of 60,000 at 250 Th. The top ten most intense ions in the full MS were isolated for fragmentation with a target of 50,000 ions at a resolution of 15,000 at 250 Th. MS data were acquired using the XCalibur software (Thermo Fisher Scientific).
Parallel Reaction Monitoring and data analysis
The mass spectrometer was operated in positive ion mode and used in parallel reaction monitoring (PRM). A data dependent experiment with inclusion list was set-up together with a full scan. The full scan was performed at a target value of 1E6 ions with resolution of 60,000 at m/z 200 over mass range of 375-1600 m/z followed by 6 PRM scans. The inclusion mass list included light or medium and heavy SILAC labelled NCBP2-AS2 peptides 5-18, 70-77 and 82-92. Targeted peptides were constantly fragmented in the HCD cell (Q-Exactive HF) or linear trap (Orbitrap Elite) and acquired into the Orbitrap analyzer or ion trap, respectively, at a resolution of 15,000 at 200 m/z (Q-Exactive HF). Quadrupole Isolation width was set to 1.2 m/z (Q-Exactive HF) or 1 m/z (Elite) and normalized collision energy of 28 (Q-Exactive HF) or 35 (Orbitrap Elite) were used to fragment a maximum target of 1E5 ions (or injection time of 25 ms for the Q-Exactive HF) or 7.5E3 ions in the Elite.
The PRM data traces of NCBP2-AS2 tryptic peptides were used for MS/MS based quantification. Raw data were imported into Skyline (version 4.1.0.18169), and y and b product ions chromatograms were extracted, summed and integrated. Only fragment ion peaks with the same retention time were used for quantification. In the Transition settings menu, the “MS/MS filtering tool” was applied to all replicates using “Targeted” acquisition method. The product mass analyzer was set to “Orbitrap” and resolution to 15,000 (Q-Exactive HF) or to “QIT” with tolerance of 0.5 m/z (Elite). All matching scans were included in data extraction. Ion series extracted were compared to the 5 most intense product ions of a NCBP2-AS2 SILAC medium and heavy peptides library built with previously acquired data. For the siRNA, .raw files were analyzed with MaxQuant; protein quantification was according to the MaxQuant label-free quantification (LFQ) algorithm.
MS data analysis
The MS .raw files were processed with the MaxQuant software (31) version 1.5.0.36 and searched with the Andromeda search engine (60) with the following settings: minimal peptide length 7 amino acids, multiplicity 2, where the light labels were Arg6 and Lys4 and the heavy labels were Arg10 and Lys8; variable modifications Acetyl (Protein N-term) and Oxidation (M); fixed modification Carbamidomethyl (C); specificity for trypsin cleavage was required and maximum 2 missed cleavages were allowed. For the search of the parent mass and fragment ions, an initial mass deviation of 4.5 ppm and 20 ppm, respectively, was required. The re-quantify parameter was enabled and match between runs was used between similar fractions. For identification, the false discovery rates (FDRs) at the protein and peptide level were set to 1%. For accurate protein quantification, only unique peptides with at least 2 ratio counts were used. Ratio count of 1 was used for the total proteome of CAFs silenced for NCBP2-AS2, to allow the quantification of NCBP2-AS2. The MaxQuant output table ‘ProteinGroups.txt’ was uploaded in Perseus (61) (version 1.5.2.11) for downstream analysis. The data were filtered to remove potential contaminants, reverse peptides which match a decoy database, and proteins only identified by site, which corresponds to proteins identify through peptides which were identified only in their modified version. To assure an unambiguous identification, only proteins identified with at least one unique peptide were considered. The normalized SILAC Ratio was used for the analysis. Ratios from the ‘Reverse’ experiment were inverted. Then, ratios and intensities were logarithmized, log2 and log10, respectively. When the distribution of the SILAC ratios was not centered on the zero, data were normalized by subtracting the median of the all SILAC ratios calculated for that experiment. Annotation from Gene Ontology Cellular Compartment (GOCC), Gene Ontology Biological Processes (GOBP), Gene Ontology Molecular Function (GOMF), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Protein families (Pfam) databases were added. For the STRING analysis, the list of the regulated proteins was uploaded in STRING version 10.0 (62) and the analysis was conducted using the default settings. The output files were visualized with Cystoscape (Version 3.4.0) (63).
Knockdown with siRNA
cCAFs and pCAFs 90% confluent were harvested and 2x106 cells were used in each transfection accordingly to the manufacturer's protocol using the program T-20 and the Amaxa kit R (Lonza). Cells were transfected with 1-3 nM of the non-targeting siRNA as a control (D-001810-10-05, GE Healthcare Dharmacon) or with siRNAs targeting NCBP2-AS2 (NCBP2-AS2 #1: CTGGTATAGCAGGACAGTTAA; NCBP2-AS2 #2: CAGGCAAGATGTGTTAAGAAT) (Qiagen). Cells were used for experiments 24h after silencing.
Scratch wound migration assay
6x104 HUVECs were seeded in each well of an image lock 96-well plate (Essen Bioscience). The following day, wounds were made using the wound maker (Essen Bioscience) and the CM derived from CAFs silenced or not for NCBP2-AS2 was added. The images were acquired every 30 min for 24 h using Incucyte Zoom (Essen Bioscience). Relative wound density over the time was measured using the Incucyte Zoom software (Essen Bioscience).
Protrusions and lamellipodia formation assay
1x104 HUVECs were seeded on a 13 mm coverslip coated with gelatin. The following day, cells were starved for 4 h in EBM2. Then, cells were treated for 1 h with CM derived from hypoxic CAFs siCTL, siNCBP2-AS2 #1 or siNCBP2-AS2 #2. Cells were fixed in 4% PFA for 10 min, permeabilised with 0.2% Triton X-100 in PBS for 5 min, blocked in 5% BSA for 5 min and incubated with anti-Cortactin antibody for 1 h (1:200, Millipore 4F11). For F-actin staining we used Alexa Fluor 594 Phalloidin (1:200, Molecular Probes, A12381). For nuclei staining DAPI was used (1:1,000 Sigma). Images were acquired on a Nikon A1R confocal microscope.
EC treatment for VEGF signaling assessment
2x106 HUVECs were seeded on a gelatin-coated 15 cm dish. The following day, cells were starved for 4 h in EBM2 and treated for 5 min with the CM derived from hypoxic CAFs siCTL, siNCBP2-AS2 #1 or siNCBP2-AS2 #2. As a negative control for VEGF signaling we used cells starved in EBM2; as positive control, following 4 h starvation, cells were treated for 5 min with 20 ng/ml of VEGFA (Sigma).
ELISA
VEGFA concentration in the conditioned medium was measured using Quantikine ELISA human VEGF Kit (DVE00, R&D systems) according to manufacturer instructions. Each sample was analyzed in three technical replicates.
Swap conditioned medium to assess VEGF levels
The cCAFs were transfected with NCBCP2-AS2 #1 or control siRNA as described above. The CM was derived from 8 x 104 transfected cells cultured for 72 hours under hypoxia. After centrifugation, the cleared CM was used to culture new NCBCP2-AS2 #1 siRNA transfected cCAFs. After 72 hours culture under hypoxia, VEGFA levels in the conditioned medium were measured by ELISA (see above). VEGFA secretion by the media conditioning cells was accounted for by incubating the CM in adjacent empty wells during the experiment and subtracted from the ELISA values to give the results shown.
Western blotting analysis
Total lysates were obtained using a lysis buffer containing 2% SDS, 100 mM TrisHCl pH 7.4, incubated at 95 °C for 5 min, sonicated using a metal tip (Soniprep 150, MSE) and centrifuged at 16,000 x g for 10 min. Protein concentration was determined using Optiblot Bradford reagent (Abcam). 20 μg of proteins were separated using 4-12% gradient NuPAGE Novex Bis-Tris gel (Life technologies). Protein transfer was performed on methanol-activated PVDF or Nitrocellulose membrane. Membrane was blocked overnight in 5% non-fat milk (Marvel) or in 3% BSA (Sigma) in TBST (tris-buffered saline polysorbate 20) at 4 °C. The following primary antibodies were used: HIF1α (1:500, 10006421, Cayman), Vinculin (1:2,000, V9131, Sigma), GAPDH (1:1,000, Santa Cruz, sc48167), total VEGFR2 (1:2,000, 55B11, CST), phospho VEGFR2 (Tyr 1175) (1:1,000, 2478S, CST), total PLC gamma (1:2,000, 2822S, CST), phospho PLC gamma (Tyr783) (1:1,000, 2821S, CST), total ERK 1/2 (p44/42 MAPK) (1:2,000, 9102, CST), phospho ERK 1/2 (p44/42 MAPK – Thr202/Tyr204) (1:1,000, 9106, CST), total p38 MAPK (1:2,000, 8690S, CST), phospho P38 MAPK (Thr180/Tyr182), (1:2,000, 9216S, CST), total AKT (1:1,000, D7D11, CST), phospho-Akt (Ser473) (1:1,000, 9271, CST). LI-COR secondary antibodies (1:10,000, LI-COR Biosciences) or HRP-conjugated (1:5,000, NEB) were incubated for 1h at RT. Western blot images were acquired using an Odyssey CLx Scanner (LI-COR Biosciences) or a myECL Imager (Thermo Scientific).
Reverse transcription polymerase chain reaction (RT-qPCR)
The primers for RT-qPCR used in this study were designed using Primer Blast (NCBI): NCBP2-AS2 (F: GCGCTGAGCGATTTAGAGAC, R: ACTGTAGAGCCCGGAAGGAA), VEGFA (F: GCTACTGCCATCCAATCGAG, R: CATCGCATCAGGGGCACAC), STC1: (F: TCTGGTGCTGGTGATCAGTG, R: CACTTCAGCTGAGTTTTGAGCC), CPA4 (F: ACCTGCAGGCCCTTTTAGACA, R: AGGAAAGTCTGCGGCAATGT), LOXL2 (F: CCCCCTGGAGACTACCTGTT, R: TCGCTGAAGGAACCACCTAT) and PTX3 (F: GCCGGCAGGTTGTGAAACAG, R: ACATCTGTGGCTTTGACCCAA). Tata-binding protein (TBP F: AGTGACCCAGCATCACTGTTT, R: TAAGGTGGCAGGCTGTTGTT) or 18S (F: AGGAATTGACGGAAGGGCA, R: GGACATCTAAGGGCATCACA) were used as housekeeping genes for normalization. For mRNA isolation, 5x105 cells were seeded in 6-well plate. After hypoxia (1% O2, 72 h) or DMOG (1 mM, 24h) treatment, mRNA was collected using RLT buffer (Qiagen). Total RNA extraction and DNase treatment were performed following the manufacturer’s instructions of the RNeasy kit (Qiagen). cDNA was synthesized using iScript kit (BioRad) using 500 ng to 1 μg of RNA. cDNA was diluted 1:10 and 2 μl were used in each RT-qPCR reaction (technical triplicates), with 10 μl of iTAQ Universal SYBR Green Supermix (BioRad) and 400 nM of primers forward and reverse diluted in 8 μl of water. Each plate contained the appropriate non-template controls. Runs were performed in a 7500 Fast Real Time PCR System (Applied Biosystems).
Polysomal RNA isolation
Cells exposed to normoxia or hypoxia for 24 h were treated with 100 μg/ml of cycloheximide for 3 min before harvesting. Cell pellets were lysed in lysis buffer (300 mM NaCl, 15 mM MgCl2 and 15 mM Tris/HCl pH 7.5, 1 mg/ml heparin sulfate, 100 μg/ml cycloheximide and 0.1% Triton X-100). Post-nuclear lysates were layered on 10 ml of 10%–50% (w/v) sucrose gradients of gradient buffer (lysis buffer without Triton X-100). Gradients were centrifuged at 256,824 x g for 2 h at 4°C in a SW40Ti rotor (Beckman Coulter) and separated through a live absorbance 254 nm UV spectrometer (Isco). Gradients were separated into 10 fractions, and RNA precipitated by addition of guanidine hydrochloride (2.9 M final concentration) and ethanol (50% v/v). Precipitated RNA was then ethanol precipitated for a second time prior to use. Data is represented as total RNA quantity in fractions from non-translating and polysomal sections of the gradients.
Sponge angiogenesis assay
All mouse procedures were in accordance with ethical approval from the Institutional Animal Care and Research Advisory Committee of the K.U. Leuven. We used the subcutaneous sponge assay described in details in the JoVe protocol (43), with minor modifications. A biopsy puncher was used to obtain 6 mm diameter sponges (Haromed, 1255889) for subcutaneous injection. Each sponge was embedded with 25 μl of conditioned medium produced for 48 h by hypoxic CAFs siCTL or siNCBP2-AS2. Four sponges per animal were subcutaneously transplanted and surgically dissected after 7 days. Each sponge was cut in two halves and embedded in OCT. Frozen sections of each sponge were stained for blood vessels. Meca32 (1:100, BD Biosciences, 550563) was used for the endothelial cells and Ng2 (1:100, Abcam, AB5320) for the pericytes. Whole sections were imaged as a tile scan and were analyzed by a macro used in ImageJ. The channels were split before the individual images were put together into individual mosaic images for each channel and subsequently merged. Regions of interest were included or excluded from the analysis generating a ‘zone’. Meca32 colocalization with Ng2 within the zone was measured.
RNA scope, IHC and TMA staining
In situ hybridization staining to detect mRNA expression in tissue sections was performed on tumors formalin-fixed for 24h at room temperature. The probe to detect the human NCBP2-AS2 was designed by Advanced Cell Diagnostics (Hs-NCBP2-AS2 gene 300031 ID: 152217). The staining of the paraffin-embedded sections was performed following manufacturer’s instructions (Advanced Cell Diagnostics). Breast cancer TMA was purchased and contained single core per case (BR2082b, Insight Biotechnology). 108 cases were analyzed, which include invasive ductal carcinoma, lobular carcinoma, intraductal carcinoma and normal tissues. Cores at the border of the slide were not included in the analysis because they were damaged. Cores were defined positive for NCBP2-AS2 staining if positive cells were detected in the stromal compartment. Slides and TMA were digitized and viewed using Slidepath Digital Image Hub (Leica Microsystems) and quantified using Halo software (ACD).
NCBP2-AS2 recombinant protein
Human NCBP2-AS2 was cloned in BamHI/EcoRI of pGEX-4T-1 vector. NCBP2-AS2 was expressed as GST fusion protein in BL21 Codon-plus (DE3). After overnight induction at 18°C cells were lysed and the protein was purified using a two-step chromatography procedure, including a GST affinity and size exclusion chromatography. Protein was aliquoted and stored in a buffer containing 150 mM NaCl, 20 mM Tris PH 7.5 and 2mM DTT.
Statistical analysis
GraphPad Prism version 7.0 was used for statistical analyses. For the quantification of the endothelial cell sprouting, first the distribution of the data was evaluated using the D’Agostino & Pearson normality test. For data with normal distribution, an unpaired two-tailed t-test with Welch’s correction was used. When the distribution was not normal, the Mann-Whitney t-test was used. For other experiments, the two-tailed unpaired t-test with Welch’s correction was used to compare samples. For cases in which samples were normalized to a control group, the normalized data were first log-transformed and then a one-sample t-test (against a comparison value of 0) was performed on the resulting log-transformed data. For the cortactin quantification, Kruskal-Wallis test with Dunn’s multiple comparison test was performed. P ≤ 0.05 was considered significant. Data are represented as mean ± standard error of the mean (SEM), unless stated otherwise. Data in the plot were obtained from three biological replicates, unless stated otherwise. For the MS proteomic data analysis the Perseus software was used: we required proteins to be quantified in at least 2 out of 3 biological replicates. Then, the SILAC ratios of these quantified proteins were averaged by the mean. For the cCAF and pCAF hypoxia-induced proteome and secretome, the threshold to define significantly up-regulated and down-regulated proteins was defined to be higher than one and two standard deviation from the mean of the SILAC ratios. This relaxed statistical threshold was chosen because known HIF1α responsive genes were found regulated within this threshold. For the NCBP2-AS2 knockdown experiment, one-sample t-test analysis was performed to ease visualization of the results with volcano plot.
Supplementary Material
Acknowledgements
We thank Colin Nixon, the BSU, the BAIR and the histology facilities at the CRUK Beatson Institute, Clare Orange for histopathology services, NHS Greater Glasgow and Clyde Biorepository for providing patient samples and the PRIDE team.
Funding
This work was funded by Cancer Research UK (F.G.K., S.A., L.J.N., S.L., D.M.B., E.K.M., CRUK Beatson Institute A17196, CRUK Glasgow Centre A18076 and Stand Up to Cancer campaign for Cancer Research UK A12935 for S.Z.; S.I. A19257; A.J. and L.M.M. A15673 for L.M.M., J.K. and O.J.S. A21139 and CRUK Science Committee Programme Award A24388 for O.J.S.). J.K. ERC Starting Grant ColonCan agreement 311301. M.M. and J.S. were funded by FWO (G0D1717N) and Stichting tegen Kanker.
Footnotes
Author contribution
F.G.K. and S.Z. conceptualization of the study. F.G.K., J.K., A.J. and S.Z. developed methodology. F.G.K., L.J.N., S.A., J.S., A.J. and J.K. performed investigation. S.I. and D.M.B. provided resources. F.G.K., J.K., A.J., E.K.M., and S.Z. performed data curation and formal analysis, F.G.K. and S.Z. wrote the original draft. F.G.K. and S.Z. visualized the data. S.I., O.J.S., L.M.M., M.M. and S.Z. supervised the work.
Competing interests
O.J.S. has received funding from Astra Zeneca, Novartis, Jansen and Cancer Research Technologies. The other authors declare that they have no competing interests.
Data and materials availability
The .raw MS files and search/identification files obtained with MaxQuant have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org/cgi/GetDataset) via the PRIDE partner repository (64) with the dataset identifier PXD006535 (hypoxia datasets) and PXD006539 (siNCBP2-AS2 datasets). All other data needed to evaluate the conclusions in the paper are presented in the paper or in the Supplementary Materials.
References
- 1.Folkman J. Role of angiogenesis in tumor growth and metastasis. Semin Oncol. 2002;29:15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- 2.Casazza A, Di Conza G, Wenes M, Finisguerra V, Deschoemaeker S, Mazzone M. Tumor stroma: a complexity dictated by the hypoxic tumor microenvironment. Oncogene. 2014;33:1743–1754. doi: 10.1038/onc.2013.121. [DOI] [PubMed] [Google Scholar]
- 3.Jain RK. Antiangiogenesis Strategies Revisited: From Starving Tumors to Alleviating Hypoxia. Cancer Cell. 2014;26:605–622. doi: 10.1016/j.ccell.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. Cancer-stromal cell interactions mediated by hypoxia-inducible factors promote angiogenesis, lymphangiogenesis, and metastasis. Oncogene. 2013;32:4057–4063. doi: 10.1038/onc.2012.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrara N. Role of vascular endothelial growth factor in physiologic and pathologic angiogenesis: therapeutic implications. Semin Oncol. 2002;29:10–14. doi: 10.1053/sonc.2002.37264. [DOI] [PubMed] [Google Scholar]
- 6.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nature Medicine. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 7.Carmeliet P, Ferreira V, Breier G, Pollefeyt S, Kieckens L, Gertsenstein M, Fahrig M, Vandenhoeck A, Harpal K, Eberhardt C, Declercq C, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 8.Kamba T, McDonald DM. Mechanisms of adverse effects of anti-VEGF therapy for cancer. Br J Cancer. 2007;96:1788–1795. doi: 10.1038/sj.bjc.6603813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sappino AP, Skalli O, Jackson B, Schurch W, Gabbiani G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int J Cancer. 1988;41:707–712. doi: 10.1002/ijc.2910410512. [DOI] [PubMed] [Google Scholar]
- 10.Desmouliere A, Guyot C, Gabbiani G. The stroma reaction myofibroblast: a key player in the control of tumor cell behavior. Int J Dev Biol. 2004;48:509–517. doi: 10.1387/ijdb.041802ad. [DOI] [PubMed] [Google Scholar]
- 11.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 12.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 13.Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg RA, Orimo A. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci U S A. 2010;107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 15.Chiavarina B, Whitaker-Menezes D, Migneco G, Martinez-Outschoorn UE, Pavlides S, Howell A, Tanowitz HB, Casimiro MC, Wang C, Pestell RG, Grieshaber P, et al. HIF1-alpha functions as a tumor promoter in cancer associated fibroblasts, and as a tumor suppressor in breast cancer cells: Autophagy drives compartment-specific oncogenesis. Cell Cycle. 2010;9:3534–3551. doi: 10.4161/cc.9.17.12908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JW, Evans C, Weidemann A, Takeda N, Lee YS, Stockmann C, Branco-Price C, Brandberg F, Leone G, Ostrowski MC, Johnson RS. Loss of fibroblast HIF-1alpha accelerates tumorigenesis. Cancer Res. 2012;72:3187–3195. doi: 10.1158/0008-5472.CAN-12-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuchnio A, Dewerchin M, Carmeliet P. The PHD2 oxygen sensor paves the way to metastasis. Oncotarget. 2015;6:35149–35150. doi: 10.18632/oncotarget.6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madsen CD, Pedersen JT, Venning FA, Singh LB, Moeendarbary E, Charras G, Cox TR, Sahai E, Erler JT. Hypoxia and loss of PHD2 inactivate stromal fibroblasts to decrease tumour stiffness and metastasis. EMBO Rep. 2015;16:1394–1408. doi: 10.15252/embr.201540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 20.Han X, Aslanian A, Yates JR., 3rd Mass spectrometry for proteomics. Curr Opin Chem Biol. 2008;12:483–490. doi: 10.1016/j.cbpa.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet. 2013;14:35–48. doi: 10.1038/nrg3356. [DOI] [PubMed] [Google Scholar]
- 22.Bousquet PA, Sandvik JA, Arntzen MO, Jeppesen Edin NF, Christoffersen S, Krengel U, Pettersen EO, Thiede B. Hypoxia Strongly Affects Mitochondrial Ribosomal Proteins and Translocases, as Shown by Quantitative Proteomics of HeLa Cells. Int J Proteomics. 2015 doi: 10.1155/2015/678527. 678527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cox TR, Rumney RM, Schoof EM, Perryman L, Hoye AM, Agrawal A, Bird D, Latif NA, Forrest H, Evans HR, Huggins ID, et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106–110. doi: 10.1038/nature14492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- 25.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 26.Nakatsu MN, Hughes CC. An optimized three-dimensional in vitro model for the analysis of angiogenesis. Methods Enzymol. 2008;443:65–82. doi: 10.1016/S0076-6879(08)02004-1. [DOI] [PubMed] [Google Scholar]
- 27.Goodwin AM. In vitro assays of angiogenesis for assessment of angiogenic and anti-angiogenic agents. Microvasc Res. 2007;74:172–183. doi: 10.1016/j.mvr.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kvietys PR, Granger DN. Endothelial cell monolayers as a tool for studying microvascular pathophysiology. Am J Physiol. 1997;273:G1189–1199. doi: 10.1152/ajpgi.1997.273.6.G1189. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez-Fernaud JR, Ruengeler E, Casazza A, Neilson LJ, Pulleine E, Santi A, Ismail S, Lilla S, Dhayade S, MacPherson IR, McNeish I, et al. Secreted CLIC3 drives cancer progression through its glutathione-dependent oxidoreductase activity. Nat Commun. 2017;8 doi: 10.1038/ncomms14206. 14206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wisniewski JR, Zougman A, Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 31.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 32.Bignon M, Pichol-Thievend C, Hardouin J, Malbouyres M, Brechot N, Nasciutti L, Barret A, Teillon J, Guillon E, Etienne E, Caron M, et al. Lysyl oxidase-like protein-2 regulates sprouting angiogenesis and type IV collagen assembly in the endothelial basement membrane. Blood. 2011;118:3979–3989. doi: 10.1182/blood-2010-10-313296. [DOI] [PubMed] [Google Scholar]
- 33.Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, Capla JM, Galiano RD, Levine JP, Gurtner GC. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- 34.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Zaffryar-Eilot S, Marshall D, Voloshin T, Bar-Zion A, Spangler R, Kessler O, Ghermazien H, Brekhman V, Suss-Toby E, Adam D, Shaked Y, et al. Lysyl oxidase-like-2 promotes tumour angiogenesis and is a potential therapeutic target in angiogenic tumours. Carcinogenesis. 2013;34:2370–2379. doi: 10.1093/carcin/bgt241. [DOI] [PubMed] [Google Scholar]
- 36.Maji S, Chaudhary P, Akopova I, Nguyen PM, Hare RJ, Gryczynski I, Vishwanatha JK. Exosomal Annexin II Promotes Angiogenesis and Breast Cancer Metastasis. Mol Cancer Res. 2017;15:93–105. doi: 10.1158/1541-7786.MCR-16-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yi M, Schnitzer JE. Impaired tumor growth, metastasis, angiogenesis and wound healing in annexin A1-null mice. Proc Natl Acad Sci U S A. 2009;106:17886–17891. doi: 10.1073/pnas.0901324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J, Scherer PE. Endotrophin - a novel factor linking obesity with aggressive tumor growth. Oncotarget. 2012;3:1487–1488. doi: 10.18632/oncotarget.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu R, Yao ZY, Xin L, Zhang Q, Li TP, Gan RB. NC1 domain of human type VIII collagen (alpha 1) inhibits bovine aortic endothelial cell proliferation and causes cell apoptosis. Biochem Biophys Res Commun. 2001;289:264–268. doi: 10.1006/bbrc.2001.5970. [DOI] [PubMed] [Google Scholar]
- 40.Nyberg P, Xie L, Sugimoto H, Colorado P, Sund M, Holthaus K, Sudhakar A, Salo T, Kalluri R. Characterization of the anti-angiogenic properties of arresten, an alpha1beta1 integrin-dependent collagen-derived tumor suppressor. Exp Cell Res. 2008;314:3292–3305. doi: 10.1016/j.yexcr.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamphaus GD, Colorado PC, Panka DJ, Hopfer H, Ramchandran R, Torre A, Maeshima Y, Mier JW, Sukhatme VP, Kalluri R. Canstatin, a novel matrix-derived inhibitor of angiogenesis and tumor growth. J Biol Chem. 2000;275:1209–1215. doi: 10.1074/jbc.275.2.1209. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Ren H, Xu C, Zhu C, Wu H, Liu D, Wang J, Liu L, Li W, Ma Q, Du L, et al. Hypoxic mitophagy regulates mitochondrial quality and platelet activation and determines severity of I/R heart injury. Elife. 2016;5 doi: 10.7554/eLife.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deskins DL, Ardestani S, Young PP. The polyvinyl alcohol sponge model implantation. J Vis Exp. 2012 doi: 10.3791/3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ren G, Crampton MS, Yap AS. Cortactin: Coordinating adhesion and the actin cytoskeleton at cellular protrusions. Cell Motil Cytoskeleton. 2009;66:865–873. doi: 10.1002/cm.20380. [DOI] [PubMed] [Google Scholar]
- 46.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 47.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, Cangiarella J, Arju R, Formenti SC, Schneider RJ. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 48.Liu L, Simon MC. Regulation of transcription and translation by hypoxia. Cancer Biol Ther. 2004;3:492–497. doi: 10.4161/cbt.3.6.1010. [DOI] [PubMed] [Google Scholar]
- 49.He LF, Wang TT, Gao QY, Zhao GF, Huang YH, Yu LK, Hou YY. Stanniocalcin-1 promotes tumor angiogenesis through up-regulation of VEGF in gastric cancer cells. J Biomed Sci. 2011;18:39. doi: 10.1186/1423-0127-18-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 51.Jin K, Mao XO, Eshoo MW, Nagayama T, Minami M, Simon RP, Greenberg DA. Microarray analysis of hippocampal gene expression in global cerebral ischemia. Ann Neurol. 2001;50:93–103. doi: 10.1002/ana.1073. [DOI] [PubMed] [Google Scholar]
- 52.Zeng M, Kikuchi H, Pino MS, Chung DC. Hypoxia activates the K-ras proto-oncogene to stimulate angiogenesis and inhibit apoptosis in colon cancer cells. PLoS One. 2010;5:e10966. doi: 10.1371/journal.pone.0010966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rak J, Mitsuhashi Y, Bayko L, Filmus J, Shirasawa S, Sasazuki T, Kerbel RS. Mutant ras oncogenes upregulate VEGF/VPF expression: implications for induction and inhibition of tumor angiogenesis. Cancer Res. 1995;55:4575–4580. [PubMed] [Google Scholar]
- 54.Matsuo Y, Campbell PM, Brekken RA, Sung B, Ouellette MM, Fleming JB, Aggarwal BB, Der CJ, Guha S. K-Ras promotes angiogenesis mediated by immortalized human pancreatic epithelial cells through mitogen-activated protein kinase signaling pathways. Mol Cancer Res. 2009;7:799–808. doi: 10.1158/1541-7786.MCR-08-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Figueras A, Arbos MA, Quiles MT, Vinals F, Germa JR, Capella G. The impact of KRAS mutations on VEGF-A production and tumour vascular network. BMC Cancer. 2013;13:125. doi: 10.1186/1471-2407-13-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 57.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 58.Dimayacyac-Esleta BR, Tsai CF, Kitata RB, Lin PY, Choong WK, Lin TD, Wang YT, Weng SH, Yang PC, Arco SD, Sung TY, et al. Rapid High-pH Reverse Phase StageTip for Sensitive Small-Scale Membrane Proteomic Profiling. Anal Chem. 2015;87:12016–12023. doi: 10.1021/acs.analchem.5b03639. [DOI] [PubMed] [Google Scholar]
- 59.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 60.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J Proteome Res. 2011 doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 61.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 62.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O'Kelly G, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.