Abstract
We experienced two cases of hepatitis C virus (HCV) eradication failure in patients with a history of non-responsiveness to previous treatments with direct-acting antiviral agents (DAAs) who were subsequently treated with the combination of glecaprevir and pibrentasvir (GLE/PIB). Direct sequencing at commencement of GLE/PIB therapy showed non-structural protein (NS) 5A-P32 deletion in the first patient and NS5A-R30E/Q54H/A92K in the second patient (both genotype 1b). The common point was that L31/Y93 was double wild-type, and the IL28B polymorphism was non-TT type. Even when L31/Y93 is double wild-type, other NS5A mutations may affect the DAA re-treatment outcome. We analyzed the transition of amino acid mutations at NS5A by ultra-deep sequencing.
Keywords: direct-acting antivirals, glecaprevir, hepatitis C virus, pibrentasvir, resistance-associated variants, ultra-deep sequencing
Introduction
The treatment outcome of chronic hepatitis C has changed radically following the development of direct-acting antiviral agents (DAAs). Glecaprevir (GLE), a non-structural protein (NS) 3/4 protease inhibitor, and pibrentasvir (PIB), a NS5A inhibitor, are the latest approved hepatitis C virus (HCV) inhibitors in Japan. Both drugs have potent in vivo antiviral activities against genotypes 1 to 6 with little or no loss of potency against common single resistance-associated amino acid substitutions (1, 2). In reality, however, the overall sustained virological response rate at 12 weeks after the end of treatment (SVR12) for these two inhibitors is not 100%. In this regard, the evolution of NS5A resistance-associated variants (RAVs) and treatment efficacy in patients who are treated with GLE/PIB after more than one failure of treatment with other DAAs is unknown at present.
We experienced two cases of failure of combination therapy of GLE/PIB among 105 cases with a history of DAA failures at our hospital. In both cases, the HCV genotype was 1b. We conducted a comprehensive analysis of host and viral factors in these two failed cases. In particular, we analyzed the amino acid substitutions in the NS5A regions by direct sequencing and ultra-deep sequencing at both the commencement of each DAA treatment and re-elevation of the viral load.
Assessment of amino acid substitutions
The RAVs were viruses with amino acid substitutions of NS5A-L31 M/V, NS5A-P32 deletion, and NS5A-Y93H, as detected by direct sequencing at the commencement of each DAA treatment and re-elevation of viral loads. Direct sequencing was performed by the dye terminator method using the dideoxynucleotide termination sequencing FS Ready Reaction kit (Life Technologies, Carlsbad, USA). Using the HCV-J (accession no. D90208) as a reference (3), we determined the sequence of 1-140 amino acids in the NS5A protein of HCV 1b. In particular, RAVs in the NS5A region were evaluated for amino acid substitutions of L28, R30, L31, P32, Q54, A92, and Y93 (4-6).
An ultra-deep sequencing analysis was performed as described previously in detail (7). Sequencing was performed using the MiSeqⓇ sequencing platform (Illumina, San Diego, USA) using 150-bp paired-end reads, according to the instructions provided by the manufacturer. Libraries were prepared using NexteraⓇ XT DNA, according to the Library Preparation Guide (Illumina). Based on the results of a control experiment using plasmid encoding the HCV NS5A sequences, amino acid mutations were defined as amino acid substitutions detected at frequencies exceeding 0.1% of the total coverage, in order to exclude potential putative errors caused by the ultra-deep sequencing method.
Case Reports
Case 1
The first patient was a 57-year-old woman with HCV-related compensated liver cirrhosis (genotype 1b, IL28B polymorphism TG). She underwent treatment with interferon (IFN) plus ribavirin (RBV) in 2003, pegylated interferon (PEG IFN) plus RBV in 2011, and her first DAA treatment with daclatasvir (DCV) (60 mg/day) and asunaprevir (ASV) (200 mg/day) for 24 weeks in November 2014 at the original medical facility. However, all treatments were considered failures based on non-responsiveness.
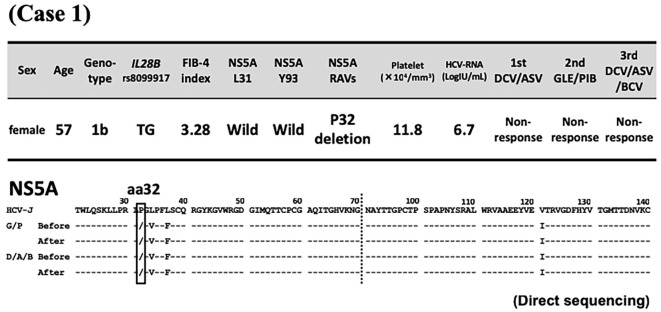
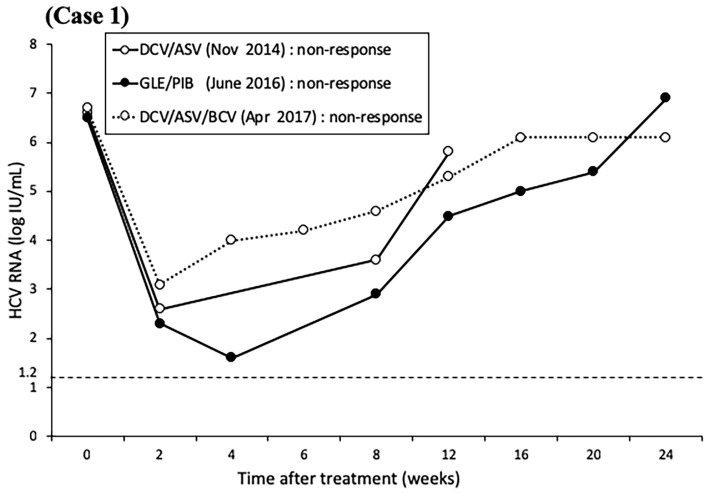
At the commencement of GLE/PIB therapy at our hospital, liver function tests showed compensated cirrhosis (Fibrosis-4 index 3.28), and both L31/Y93 of NS5A were the double wild-type. Direct sequencing showed deletion of P32 (Fig. 1). The combination treatment of 300 mg/day GLE and 120 mg/day PIB was started in April 2016 and continued for 12 weeks as a clinical trial. A close clinical follow-up showed non-responsiveness to the treatment. In April 2017, the patient was treated with the triple regimen of 60 mg/day DCV, 400 mg/day ASV, and 150 mg/day beclabuvir (BCV) for 12 weeks. However, as with the previous regimen, the treatment was considered a failure (Fig. 2). Currently, the patient is being treated with 3 million units of IFN-α (NAMALWA) 3 times a week (since October 2017). In the 2.5 years since the commencement of GLE/PIB, ultra-deep sequencing has shown P32 deletion alone (≥99%), lasting L31 wild dominance, and maintenance of Y93 wild alone in NS5A (Table).
Figure 1.
Case 1. Patient background characteristics at the commencement of glecaprevir/pibrentasvir and sequences of amino acids 21-140 in the non-structural protein 5A region at the commencement of direct-acting antiviral agent treatment and re-elevation of viral loads. Daclatasvir/asunaprevir treatment was provided at another medical facility, and unfortunately, no samples were available from before or after the analysis. Dashes indicate amino acids identical to the sequence of HCV-J (accession no. D90208) (3). Substituted amino acids are indicated by standard single-letter codes. aa: amino acids, DCV/ASV: daclatasvir/asunaprevir, D/A/B, DCV/ASV/BCV: daclatasvir/asunaprevir/beclabuvir, FIB-4: fibrosis-4, G/P, GLE/PIB: glecaprevir/pibrentasvir, NS: non-structural protein, RAVs: resistance-associated variants
Figure 2.
The levels of hepatitis C virus RNA over time in Case 1 treated with direct-acting antiviral agents. The patient was treated for 24 weeks with daclatasvir/asunaprevir, for 12 weeks with glecaprevir/pibrentasvir, and for 12 weeks with daclatasvir/asunaprevir/beclabuvir. Non-responsiveness was the outcome of all treatments.
Table.
Results of Ultra-deep Sequencing Showing the Evolution of NS5A Resistance-associated Variants over Time.
| Case | point | aa28 | aa30 | aa31 | aa32 | aa54 | aa92 | aa93 |
|---|---|---|---|---|---|---|---|---|
| 1 | Before GLE/PIB | L(99.8%) · M(0.2%) | - | - | deletion (99.8%) | - | - | - |
| After GLE/PIB | - | - | L(99.6%) · V(0.2%) | deletion (99.6%) | - | - | - | |
| Before DCV/ASV/BCV | - | - | L(99.7%) · V(0.2%) | deletion (99.6%) | Q(99.3%) · L(0.4%) | - | - | |
| After DCV/ASV/BCV | L(98.9%) · M(1.0%) | - | L(95.7%) · V(4.2%) | deletion (99.7%) | - | - | - | |
| Before IFN† | L(98.9%) · M(1.0%) | - | L(95.7%) · V(4.2%) | deletion (99.7%) | - | - | - | |
| During IFN | L(99.4%) · M(0.5%) | - | L(91.1%) · V(8.8%) | deletion (99.5%) | - | - | - | |
| 2 | Before DCV/ASV | L(49.9%) · M(49.8%) | Q(99.9%) | - | - | Q(66%) · H(33.8%) | T(99.7%) | - |
| After DCV/ASV | T(98.6%) · M(1.4%) | Q(99.8%) · R(0.2%) | - | - | H(99.8%) | K(99.9%) | - | |
| Before LDV/SOF | T(28.7%) · M(71.2%) | E(49.9%) · Q(46.6%) · R(2.1%) · L(1.2%) | - | - | H(98.3%) · Y(1.7%) | K(40.3%) · E(35%) · T(21.7%) · N(2.3%) · Q(0.3%) | - | |
| After LDV/SOF | M(99.7%) | E(99.4%) · G(0.3%) | - | - | H(99.8%) | K(99.6%) · A(0.1%) | - | |
| Before GLE/PIB | M(99.8%) | E(98.8%) · D(0.8%) · G(0.1%) | L(99%) · C(0.8%) | - | H(99.9%) | K(70.4%) · T(29.5%) | - | |
| After GLE/PIB | M(99.8%) | E(99.4%) · G(0.3%) | - | - | H(99.7%) | K(99.6%) · A(0.2%) · R(0.2%) | - |
Substituted amino acids are shown by standard single-letter codes, and frequencies relative to the total coverage by ultra-deep sequencing are also presented.
Dashes indicate amino acids of L28, R30, L31, P32, Q54, P58, A92, and Y93.
†The result before IFN was similar to that after DCV/ASV/BCV.
aa: amino acid, ASV: asunaprevir, BCV: beclabuvir, DCV: daclatasvir, GLE: glecaprevir, IFN: interferon, LDV: ledipasvir, NS: non-structural protein, PIB: pibrentasvir, SOF: sofosbuvir
Case 2
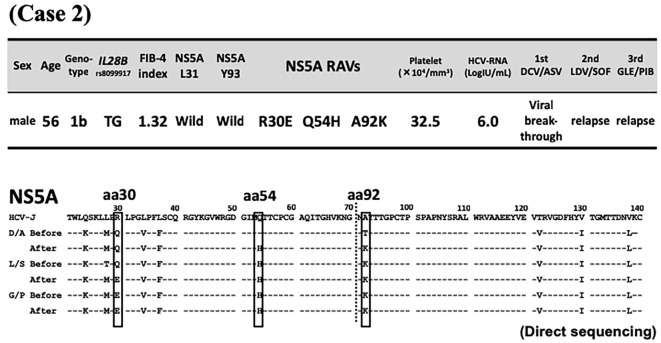
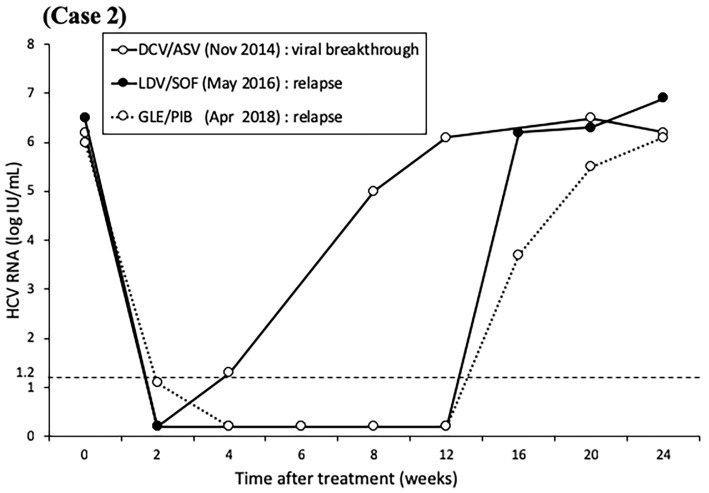
The second patient was a 56-year-old man with chronic hepatitis C (genotype 1b, IL28B polymorphism TG). In November 2014, he received dual DAA therapy of 60 mg/day DCV and 120 mg/day ASV, but treatment was discontinued after 11 weeks due to viral breakthrough. Subsequently, he was treated in May 2016 with 90 mg/day ledipasvir (LDV) combined with 400 mg/day sofosbuvir (SOF) for 12 weeks but later showed relapse. Just before the commencement of GLE/PIB, liver function tests indicated chronic hepatitis (Fibrosis-4 index 1.32). Both L31/Y93 were double wild-type, and RAVs of R30E/Q54H/A92K were detected in NS5A by direct sequencing (Fig. 3). In April 2018, treatment with the dual regimen of 300 mg/day GLE and 120 mg/day PIB was started and continued for 12 weeks, but relapse eventually developed (Fig. 4). The RAVs shifted from R30Q to E (≥99%) and from Q54 wild to H (≥99%) during the course of treatment, Furthermore, A92K increased at the time of each treatment relapse but decreased with time, as determined by ultra-deep sequencing (Table).
Figure 3.
Case 2. Patient background characteristics at the commencement of glecaprevir/pibrentasvir and sequences of amino acids 21-140 in the non-structural protein 5A region at the commencement of direct-acting antiviral agent treatment and re-elevation of viral loads. Dashes indicate amino acids identical to the sequence of HCV-J (accession no. D90208) (3). Substituted amino acids are indicated by standard single-letter codes. aa: amino acids, DCV/ASV: daclatasvir/asunaprevir, D/A/B, DCV/ASV/BCV: daclatasvir/asunaprevir/beclabuvir, FIB-4: fibrosis-4, G/P, GLE/PIB: glecaprevir/pibrentasvir, NS: non-structural protein, RAVs: resistance-associated variants, L/S, LDV/SOF: ledipasvir/sofosbuvir
Figure 4.
The levels of hepatitis C virus RNA over time in Case 2 treated with direct-acting antiviral agents. The patient was treated for 11 weeks with daclatasvir/asunaprevir, for 12 weeks with ledipasvir/sofosbuvir, and for 12 weeks with glecaprevir/pibrentasvir. The result of the first treatment was viral breakthrough, while relapse followed the subsequent two treatments.
Discussion
Among DAA-treated GT1b-infected patients, baseline substitutions in NS5A, including those at L31 or Y93 in NS5A, are reported to have no impact on the SVR12 for GLE/PIB (8). The first of our two reported patients had compensated liver cirrhosis and a history of DAA failure, but L31/Y93 was double wild-type. P32 deletion alone or in combination with L31F in NS5A was reported to confer a high level of resistance to GLE/PIB (8). Therefore, P32 deletion in our patient was considered to have contributed to GLE/PIB failure. In another reported case, P32 deletion was recognized immediately after DCV/ASV treatment failure and persisted for at least two years, and it is still not clear how long this deletion persists after treatment failure (9). Indeed, we have confirmed the persistence of ≥99% P32 deletion in our case for 2.5 years by ultra-deep sequencing. Taken together, the findings from these two studies suggest that this resistance is long-term in nature.
Our second patient had chronic hepatitis, and L31/Y93 was double wild-type. P32 deletion was not detected in NS5A, but the patient showed failure of GLE/PIB therapy. Although we do not know the cause of failure at present, R30E/Q54H and A92K in NS5A showed different trends on ultra-deep sequencing during DAA treatment (Table), suggesting some associaation with treatment resistance. There are no reports of resistance in NS5A-R30E/Q54H/A92K, but the relationship with DAA resistance should be examined in the future. In contrast, the IL28B polymorphism non-TT type is reported to correlate with non-SVR12 in LDV/SOF treatment (10). Since the IL28B polymorphism was non-TT type in both of our cases, it may be a host resistance factor.
It is widely known that mutations in NS5A, particularly Y93H, in patients with genotype 1b HCV infection are most frequently associated with treatment failure of NS5A inhibitors [e.g. DCV, LDV, ombitasvir (OBV)] (11). However, we herein showed the need to evaluate other NS5A mutations in DAA retreatment when L31/Y93 is double wild-type and the IL28B polymorphism is non-TT type, as in our two cases. Notably, triple therapy with SOF, velpatasvir (VEL), another NS5A inhibitor, plus RBV has been recently advocated for patients with NS5A-P32 deletion (considered to be DAAs tolerance) (12). Furthermore, a recent Japanese Phase III study reported an SVR12 rate for SOF/VEL + RBV treatment of 100% (2/2) at the end of 12-week treatment and 67% (2/3) at the end of 24-week treatment in patients infected with genotype 1 and NS5A-P32 deletion who had a history of DAA failure (13). New treatment regimens, including SOF/VEL + RBV, are expected to increase the rate of HCV eradication in GLE/PIB failure cases with high treatment resistance, as in our two cases.
Further research on the factors associated with GLE/PIB treatment failure may help identify factors that will aid in the selection of the most appropriate patients for such therapy and preventing its unnecessary use in others.
Author's disclosure of potential Conflicts of Interest (COI).
Norio Akuta: Honoraria, Bristol-Myers Squibb and AbbVie. Yoshiyuki Suzuki: Honoraria, Bristol-Myers Squibb and AbbVie. Hiromitsu Kumada: Honoraria, MSD, Bristol-Myers Squibb, Gilead Sciences, AbbVie and Dainippon Sumitomo Pharma.
Acknowledgement
The authors thank Ms. R. Mineta and Ms. Y. Suzuki for their assistance in the measurement of HCV resistant-associated variants.
References
- 1. Ng TI, Krishnan P, Pilot-Matias T, et al. . In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS5A inhibitor pibrentasvir. Antimicrob Agents Chemother 61: e02558, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ng T, Tripathi R, Dekyhyar T, et al. . In vitro antiviral activity and resistance profile of the next-generation hepatitis C virus NS3-4A protease inhibitor glecaprevir. Antimicrob Agents Chemother 62: e01620, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kato N, Hijikata M, Ootsuyama Y, et al. . Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA 87: 9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McPhee F, Hernandez D, Zhou N, et al. . Virological escape in HCV genotype-1-infected patients receiving daclatasvir plus ribavirin and peginterferon alfa-2a or alfa-2b. Antivir Ther 19: 479-490, 2014. [DOI] [PubMed] [Google Scholar]
- 5. Pawlotsky JM. Hepatitis C virus resistance to direct-acting antiviral drugs in interferon-free regimens. Gastroenterology 151: 70-86, 2016. [DOI] [PubMed] [Google Scholar]
- 6. Sarrazin C. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J Hepatol 64: 486-504, 2016. [DOI] [PubMed] [Google Scholar]
- 7. Köser CU, Fraser LJ, Ioannou A, et al. . Rapid single-colony whole-genome sequencing of bacterial pathogens. J Antimicrob Chemother 69: 1275-1281, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krishnan P, Schnell G, Tripathi R, et al. . Integrated resistance analysis of CERTAIN-1 and CERTAIN-2 studies in hepatitis C virus-infected patients receiving glecaprevir and pibrentasvir in Japan. Antimicrob Agents Chemother 62: e02217, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iio E, Ogawa S, Shimada N, et al. . Serial changes of NS5A P32 deletion mutant in HCV genotype 1b patients after daclatasvir/asunaprevir failure. Kanzo 59: 230-233, 2018(in Japanese, Abstract in English). [Google Scholar]
- 10. Akuta N, Sezaki H, Suzuki F, et al. . Ledipasvir plus sofosbuvir as salvage therapy for HCV genotype 1 failures to prior NS5A inhibitors regimens. J Med Virol 89: 1248-1254, 2017. [DOI] [PubMed] [Google Scholar]
- 11. Dietz J, Susser S, Vermehren J, European HCV Resistance Study Group, et al. . Patterns of resistance-associated substitutions in patients with chronic HCV infection following treatment with direct-acting antivirals. Gastroenterology 154: 976-988.e4, 2018. [DOI] [PubMed] [Google Scholar]
- 12. Gane EJ, Shiffman ML, Etzkorn K, et al. . Sofosbuvir-velpatasvir with ribavirin for 24 weeks in hepatitis C virus patients previously treated with a direct-acting antiviral regimen. Hepatology 66: 1083-1089, 2017. [DOI] [PubMed] [Google Scholar]
- 13. Izumi N, Takehara T, Chayama K, et al. . Sofosbuvir-velpatasvir plus ribavirin in Japanese patients with genotype 1 or 2 hepatitis C who failed direct-acting antivirals. Hepatol Int 12: 356-367, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]