Abstract
Specialized associations between interacting species fundamentally determine the diversity and distribution of both partners. How the specialization of guilds of organisms varies along environmental gradients underpins popular theories of biogeography and macroecology, whereas the degree of specialization of a species is typically considered fixed. However, the extent to which environmental context impacts specialization dynamics is seldom examined empirically. In this study, we examine how specialization within a bipartite network consisting of three co-occurring plant species and their foliar fungal endophyte symbionts changes along a 1000-meter elevation gradient where host species were held constant. The gradient, along the slope of Mauna Loa shield volcano, represents almost the entire elevational range of two of the three plants. Network and plant specialization values displayed a parabolic relationship with elevation, and were highest at middle elevations, whereas bipartite associations were least specific at low and high elevations. Shannon’s diversity of fungal endophytes correlated negatively with specificity, and was highest at the ends of the transects. Although plant host was a strong determinant of fungal community composition within sites, fungal species turnover was high among sites. There was no evidence of spatial or elevational patterning in fungal community compositon. Our work demonstrates that specificity can be a plastic trait, which is influenced by the environment and centrality of the host within its natural range.
Subject terms: Microbial ecology, Microbial ecology
Introduction
Plant–microbe symbioses vary in the degree to which one or more partners “specializes” on the other. Some hosts only associate with specific symbiont species, such as mycoheterotrophic plant species from the subfamily Monotropoideae, where each associates with a different, but specific, ectomycorrhizal fungal partner [1]. Other hosts are specific at higher taxonomic ranks such as the legume–rhizobia symbiosis [2] or species of Pinaceae that are the only hosts of the ectomycorrhizal fungal genus Rhizopogon [3]. Other plant hosts and fungal symbionts form more general associations, such as most ectomycorrhizal fungi, which form symbioses with many woody host plants [4] and arbuscular mycorrhizal fungi, where ~300 species associate with ~70% of land plant families [5, 6]. Foliar pathogens, as a group, lie somewhere in between, as many species encompass the ability to infect a broad diversity of hosts in some circumstances, although the probability of two different hosts both being susceptible to the same pathogen diminishes as a function of host phylogenetic distance as seen in tropical systems [7].
In nature these species interactions act on a continuum ranging from complete generalization to full specialization [8]. Placement along this continuum presents a series of tradeoffs. For example, in the case of mutualisms, if symbionts are optimally suited for each other, co-located, and abundant, specialization can confer a fitness benefit, since energy is not wasted maintaining associations with inferior partners. Interestingly, microbial systems appear to be dominated by specialist taxa, with strong positive correlations observed between habitat specificity and abundance [9] and where dominance of specialist taxa has been linked to their preference for specific abiotic factors such as rainfall [10]. However, for microbial symbionts, if one symbiotic partner is sparse or absent, specialization comes at a cost, since it might limit the distribution or fitness of the other partner.
Network specialization is a measure of specialization between multiple hosts and microbial symbionts, and can be quantified by enumerating the frequency of the interaction between two species relative to other community members. In this way, network metrics can be used to characterize the interaction dynamics of multiple co-occurring species at once, accounting for the fact that some species are likely specialized while others are more generalist. The degree to which network specialization varies across systems and environments has long been considered an important component of latitudinal diversity clines [11, 12]. In fact, analyses of interactions across broad geographic scales provide some support for this idea [13, 14]. Broad-scale measures of species interactions, however, are minimally informative for interactions happening at the local scale, and it is unclear how environmental gradients impact specificity within a given species or community [15]. This gap is likely attributable to the fact that species composition typically covaries as a function of environment, such that specialization as a function of the environment cannot be disentangled from specialization as a function of species identity.
Elevation gradients are an ideal system in which to examine how specialization between plant hosts and microbial symbionts covaries with changes in environmental conditions, as the same host species can be found along the entirety of a gradient along with coinciding large variations in abiotic conditions [16]. In this study, we took advantage of a steep elevation gradient on the Island of Hawaiʻi to examine how foliar fungal endophyte host specialization is influenced by environmental conditions. Specifically, we examined foliar fungal endophyte hosts along Mauna Loa, a shield volcano on Hawaiʻi Island. The volcano forms a steep environmental gradient as it rises from sea level to 4200 meters above sea level (masl) over ~20 km. Additionally, the young geological age and isolation of the Hawaiian archipelago supports a relatively species poor (ca. 1100 spp.) but primarily endemic flora, some of which encompass wide geographical niches and elevational distributions [17]. For example, Metrosideros polymorpha, an ecologically important and endemic tree species, can be found near sea level up to 2500 m along the eastern slope of Mauna Loa [18]. In addition to M. polymorpha, there are several other woody species that co-occur along the gradient, enabling us to examine wide environmental variance while keeping host identity constant.
To examine host specialization dynamics we focus on the symbiosis between plants and their foliar fungal endophytes. This relationship, defined here as all fungi living within leaf tissue but not causing any outward signs of disease [19], are effectively invisible but represent one of the most ubiquitous symbioses in nature. Researchers have yet to find a plant lineage lacking these cryptic microbial associates. Endophytes can play important roles in plant biochemistry (reviewed in ref. [20]), water conductance [21], heat and drought tolerance [22], and disease resistance [23]. Despite a presumed horizontal transmittance [24] foliar fungal endophytes demonstrate varied degrees of specialization among plant hosts. In one study spanning North America [25] fungal community dissimilarity correlated with phylogenetic dissimilarity of hosts, and in another, plant species in lowland tropical forests in New Guinea [26] showed that the composition of fungal communities within the same host species were more similar to each other than they were to other fungal communities of a different host species. Within controlled experiments, foliar fungal endophyte composition can vary among host plants at the population level [27, 28]. However, the extent to which the environment affects the strength of specialization in this host–microbe symbiosis is largely unknown.
We combined the unique characteristics of the Hawaiian flora with the steep elevation gradient of Mauna Loa to examine how changes in environmental conditions can affect host specialization, local richness, and community composition. Because environmental conditions along Mauna Loa become more stressful with increasing elevation (e.g. decreased precipitation, increased solar radiation, and greater diurnal temperature differences), we expected to observe a decrease in fungal species richness with increasing elevation, as fewer species would be able to tolerate and persist under conditions at higher elevations. Based on previous studies examining host-associated microbes, we hypothesized that elevational gradients would structure foliar fungal endophyte community composition, such that communities located at more similar elevations will be more similar in composition. Furthermore, we predicted that host identity would play an important role within and among sites in determining fungal community composition. Finally, we predicted that network and host specificity would decrease as a linear function of elevation, analogous to observations of mutualistic interaction network specialization patterns along latitudinal gradients [29].
Materials and methods
Sites/fieldwork
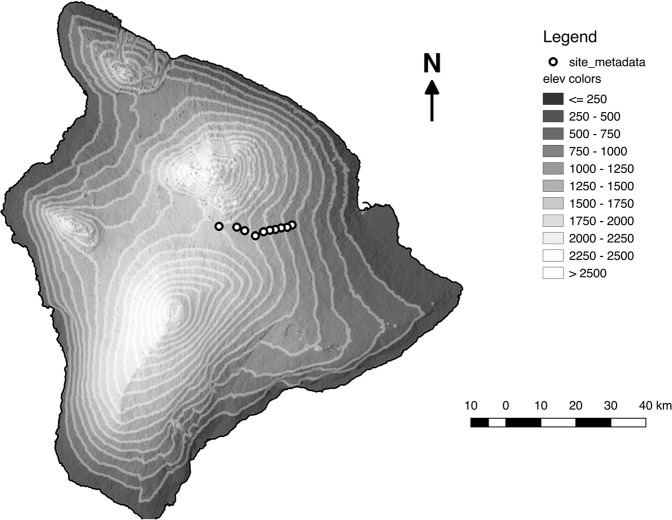
Sampling was conducted along the eastern slope of Mauna Loa on Hawaiʻi Island (19.4721°N, 155.5922°W; Fig. 1). Along this steep slope the volcano encompasses an environmental gradient over which temperature, rainfall, and solar irradiance vary rapidly (Fig. S1). To examine the influence of the abiotic environment on foliar endophyte host specialization we sampled three native Hawaiian plants that co-occur along the gradient. Hosts were sampled from 1100 to 2000 masl at 100 m intervals and included: Leptecophylla tameiameiae (pūkiawe) an indigenous species found throughout the Pacific, M. polymorpha (ʻōhiʻa) an endemic Hawaiian species common throughout the Hawaiian archipelago, and Vaccinium reticulatum (ʻōhelo) also a Hawaiian endemic. For both L. tameiameiae and V. reticulatum, our sampling range encompassed nearly the entire extent of their elevation distribution along this portion of Mauna Loa. M. polymorpha is the dominant tree species along the entire gradient, and its range extends both above and below the limits of sampling conducted for this study. Within each of the 10 plots sampled, we collected leaf samples from four different individuals of each host species (120 sampled individuals, 40 per host species total).
Fig. 1.
Sampling locations on the Island of Hawaiʻi. Circle points indicate sampling locations, shades of gray indicate elevation in meters above sea level (masl). Starting at 1100 masl, sampling locations were established in 100 m increments spanning ~20 km to our highest elevation site at 2000 masl. At each site, we collected leaf samples from four individuals for each of our target hosts (L. tameiameiae, M. polymorpha, and V. reticulatum)
Sampling was based on approximate leaf mass. We haphazardly collected five apparently healthy leaves from M. polymorpha and V. reticulatum individuals, and 40 leaves from L. tameiameiae individuals (as its leaves are much smaller). Leaf samples were placed in Ziploc bags and stored on ice in the field, before being placed in −20 °C storage in the lab. At each elevation, we collected a voucher specimen from each host species which were deposited in the Joseph F. Rock Herbarium at the University of Hawaiʻi at Mānoa.
Molecular analysis
Surface sterilization
For each replicate we collected 40 leaf-disks per individual by punching leaves using a surface sterilized standard paper single hole punch (~0.5 cm diameter). Because L. tameiameiae leaves are roughly 0.7 cm long, entire leaves of this species were processed. Leaf disks and L. tameiameiae leaves were placed inside loose-leaf tea bags, and surface-sterilized by submerging in 1% NaClO for 2 min, 70% EtOH for 2 min, and finally two rinses in sterile water for 2 min each. Rinse water was retained for use as a negative control in subsequent analyses.
DNA extraction
For DNA extraction, 10 surface-sterilized leaf-disks per sample were haphazardly chosen from tea bags and placed in MP Biomedical Lysing Matrix A tubes (MP Biomedical, Santa Ana, CA, USA) containing Solution PD1, Solution PD2, Phenolic Separation Solution, and RNase A Solution from the MoBio PowerPlant Pro DNA Isolation kit (MO Bio, Carlsbad, CA, USA). Leaf disks were homogenized using a Mini-Beadbeater 24 (BioSpecs Inc. OK, USA) at 3000 oscillations per minute for 2 min. Genomic DNA was then isolated using the MoBio PowerPlant Pro DNA Isolation kit protocol following the manufacturer’s instructions.
Amplification and Illumina Library Prep
We amplified the ITS1 region of the ribosomal RNA gene (rDNA) using the fungal-specific primers ITS1f and ITS2, along with Illumina adaptors and Golay barcodes following the protocols of Smith and Peay [30], but with the addition of a second 12 bp nucleotide index on the ITS1f primer construct to enable a dual indexed approach. Reaction conditions were as follows: initial denaturation at 95 °C for 180 s followed by 35 cycles of 95 °C for 20 s, 53 °C for 15 s, and 72 °C for 20 s, with a final elongation at 72 °C for 60 s. PCRs were carried out in 25 μL PCR reactions containing 1X KAPA3G Plant PCR kit master mix (KAPA Biosystems, Wilmington, MA, USA), 1.5 mg/mL BSA, 2 mM MgCl2, 0.3 μM forward primer, 0.3 μM reverse primer, 0.2 μL KAPA3G enzyme, and 9 μL DNA template. PCR amplification was verified via gel electrophoresis. PCR products were purified and normalized using Just-a-PlateTM 96 PCR Purification and Normalization Kit (Charm Biotech, San Diego, CA, USA) and concentrated using a streptavidin magnetic bead solution. The resulting library was sequenced by GENEWIZ (GENEWIZ, South Plainfield, NJ, USA) using the 2 x 300 paired-end sequencing chemistry on an Illumina MiSeq sequencing platform (Illumina Inc., San Diego, CA, USA).
Bioinformatics
Bioinformatic analyses were conducted using Quantitative Insights Into Microbial Ecology (QIIME v1.0) [31] and Mothur v1.39 [32]. Sequencing produced 23,531,047 sequences. After removal of sequence with an average Quality score < 25 or a sequence length < 75, 18,370,578 were retained. Because reverse reads contained a high proportion of low-quality reads, only forward reads were utilized for subsequent processing and analyses.
Forward sequences were clustered into operational taxonomic units (OTUs) using a chain-clustering protocol. We chose this method as it tends to be more accurate compared with using a single OTU clustering algorithm [33]. Sequences were first grouped into de novo OTUs using a sequence similarity of ≥ 96% using USEARCH [34]. Chimera checking referenced the UNITE v7 database [35]. A second round of de novo clustering was then performed using UCLUST [34], again using ≥ 96% sequence similarity. Representative sequences from OTUs were then assigned taxonomy using the BLAST v2.6 algorithm [36] and the UNITE v7 database within QIIME. Query sequences aligning to <85% of the length of a fungal database sequence were removed from the OTU table. No OTUs were detected in PCR-negative controls after OTU clustering. We then exported our OTU table to R (v3.3.2 [37]) for further processing.
To reduce the likelihood of mis-attribution due to tag switching during the sequencing run, we removed OTUs from samples in which their abundance was < 0.1% of the maximum number of reads found in another sample. We then filtered all OTUs with < 10 reads in the dataset, as well as OTUs not identified to the kingdom of Fungi. This resulted in 2058 OTUs. We then down-sampled our OTU table to 2300 reads per sample to account for uneven sequencing depth across the dataset. While there is no consensus on how best to account for differences in sequencing depth across samples, given a mean per sample richness of fewer than 60 OTUs in our study, we believe that down-sampling to achieve equal sequencing coverage minimally impacts statistical sensitivity.
Statistical analyses
Effect of elevation on local fungal richness and Shannon diversity (alpha-diversity)
We examined local alpha-diversity of foliar fungal endophytes using both observed species richness and Shannon’s diversity index as our metrics. Both diversity metrics were calculated within individual plants. Species richness was calculated using the specnumber function and Shannon diversity was calculated using index = “shannon” in the diversity function, both of which are available in the vegan package [38]. Changes in alpha-diversity with increasing elevation were examined along our gradient using Pearson’s moment correlation with the cor.test function in base R [38].
Effects of elevation and host identity on fungal community composition (beta-diversity)
To examine differences in community composition among samples we first calculated Bray–Curtis [39] dissimilarity among samples using log-transformed OTU abundances. To determine the changes in fungal community composition along our gradient, we calculated Mantel correlations [40] between elevational and community compositional dissimilarities using the mantel function with 10,000 permutation in the vegan package [38]. To further quantify changes in beta-diversity along our gradient, we determined the relative contribution of species nestedness vs. species turnover separately for all three hosts [41]. Changes in beta-diversity were partitioned into the nestedness and turnover components using the beta.multi.abund function in the betapart package [42].
To determine the extent to which host identity predicts community composition within each site along the gradient and to determine how the interaction between host and elevation affect community composition, we performed a permutational multivariate analysis of variance (PERMANOVA) [43] using the adonis function in the Vegan package [38].
Host specialization and selectivity
To determine how host specialization of foliar fungal endophytes changes as a function of elevation, we implemented a network approach. We assembled bipartite networks by aggregating fungal OTUs by host species within each elevation. We calculated the H2′, a measure of specialization [8] generalized across the entire network, for each elevation. The index describes the extent to which observed interactions deviate from those that would be expected given the species and sample abundance distributions of species [8]. H2′ values range from 0 (no network specialization) to 1 (perfect network specialization) based on potential associations given OTU abundance totals. H2′ was calculated using the H2fun function in the bipartite package [44]. Next, we examined plant host specialization for foliar fungal endophytes for our three target hosts separately. Host specialization was determined using the d′ specialization index using the dfun function [8]. The d′ index calculates how strongly a species deviates from a random sampling of interacting partners available, and like H2′ it ranges from 0 (no specialization) to 1 (perfect specialist) [8]. Both indices consider interaction frequency (abundance) and are standardized to account for heterogeneity in the interaction strength and taxon richness. To examine the relationship between elevation and H2′ and d′, we modeled both indices with increasing elevation using quadratic polynomial regression. For each polynomial regression, model assumptions were verified using the gvlma function in the gvlma package [45].
We used a null model approach to test whether observed networks exhibited non-random patterns. To generate null networks, we randomized observed networks 10,000 times using the swap.web algorithm and the nullmodel function in the bipartite package and compared empirical values to the randomized distribution using a one-sample t-test.
Data and code availability
All amplicon sequencing data generated in this study can be accessed upon publication on National Center for Biotechnology Information’s Sequence Read Archive under Foliar Fungal Endophytes along Mauna Loa Elevation Gradient—BioProject (PRJNA474551). All metadata, OTU tables, and code for data analysis are available at figshare [46].
Results
Effect of elevation on local fungal richness and Shannon diversity (alpha-diversity)
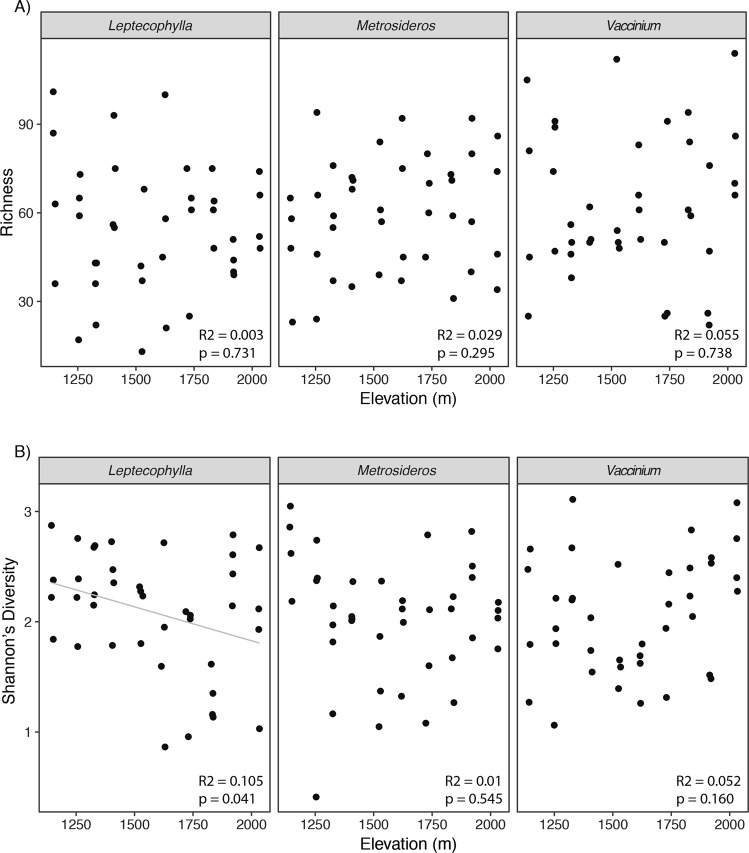
With the exception of endophytic communities associated with L. tameiameiae, which showed a negative correlation between diversity and elevation explaining 11% of the variation (Fig. 2, r = −0.325, p = 0.041), we observed no significant relationship between fungal species richness or diversity and elevation among sampled hosts (Fig. 2).
Fig. 2.
Top three panels (a) display fungal richness associated with our three target hosts plotted against elevation. Fungal richness was not affected by elevation. Bottom three panels (b) display fungal community Shannon’s diversity associated with our three target hosts plotted against elevation. Fungal Shannon’s diversity for both M. polymorpha and V. reticulatum were not significantly influenced by elevation. Shannon’s diversity of fungal communities associated with L. tameiameiae declined significantly with increasing elevation. Solid gray line represents linear regression
Effects of elevation and host identity on fungal community composition (beta-diversity)
Overall, elevational dissimilarity was a poor predictor of endophyte compositional similarity (Fig. S2; Mantel test for correlation between two distance matrices; r = 0.023; p = 0.119). Neither M. polymorpha (Fig. S2; r = 0.099; p = 0.054), nor V. reticulatum (Fig. S2; r = 0.089; p = 0.056) associated communities demonstrated a significant relationship between elevational dissimilarity and foliar endophytic fungal community composition. Communities associating with L. tameiameiae were significantly structured by elevation, although this relationship only explained 1% of the variation (Fig. S2; r = 0.105; p = 0.018).
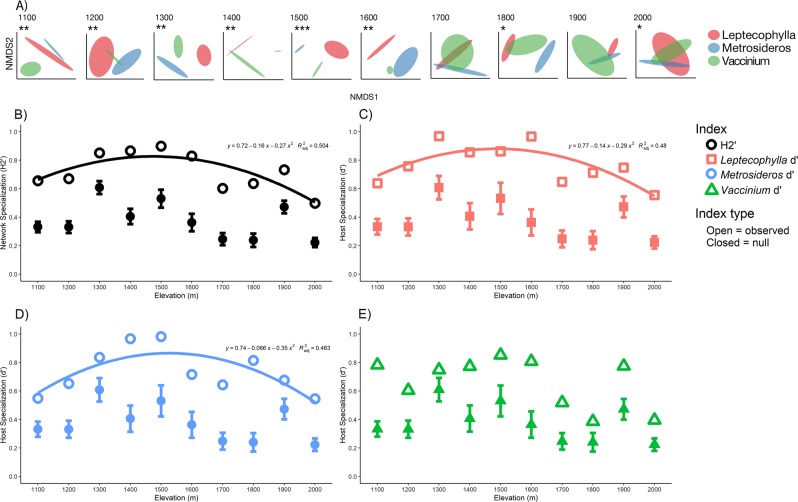
In contrast, host identity strongly determined foliar fungal endophytic community composition at all elevations sampled, with the exception of 1700 and 2000 masl (Fig. 3a, Table 1). Host identity explained the greatest amount of variance at mid-elevations (1300–1600 m), and then decreased at lower and higher elevations (Table 1). When all hosts along the elevation were considered, host, elevation, and their interaction were all significant predictors of fungal community composition. Their interaction explained the greatest amount of variation (Table S1; Fig. S3; ADONIS: R2 = 0.176, p = 0.001) followed by host (Table S1; Fig. S3; ADONIS: R2 = 0.101, p < 0.001) then elevation (Table S1; Fig. S3; ADONIS: R2 = 0.094, p < 0.001). When we examined the relative contribution of species turnover vs. nestedness in determining changes in community composition, we observed that changes along our gradient were driven primarily by species turnover for all hosts (Table 2).
Fig. 3.
Top panel (a) displays NMDS plots for each elevation site and illustrates how endophyte communities are structured by host identity. Shaded ellipses represent standard error of the mean (95%) and are colored by host (red = L. tameiameiae, blue = M. polymorpha, green = V. reticulatum). Asterisks above NMDS plots show significance values of PERMANOVA tests, where *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001). Lower four panels (b–e) display observed; network specialization (H2′; b, open black circles), and host specialization (d′) for L. tameiameiae (c, open red squares), M. polymorpha (d, open blue circles), and V. reticulatum (e, open green triangles), respectively, plotted against elevation. For all network indices, significant trends (at p < 0.05) with elevation are illustrated with a polynomial regression curve fitted to the graph (polynomial order = 2), along with associated quadratic equation and adjusted-R2 value. Observed network indices were compared to random expectation by assembling null networks. Null means for each network index at each site are denoted by closed shapes, along with standard deviations around the null mean (error bars around closed shapes). Observed H2′ and d′ for all three species were significantly more specialized than expected by chance at all sites (see Tables S2 and S3 for t-test results)
Table 1.
Variation in foliar fungal endophyte community composition (beta (β) diversity) among hosts within each elevation sampled, as determined by permutational multivariate analysis of variance using distance matrices (PERMANOVA)
| Elevation (masl) | DF | SS | MS | PsudoF/F | R 2 | p-value | |
|---|---|---|---|---|---|---|---|
| 1100 | |||||||
| Host | 2 | 1.155 | 0.578 | 1.849 | 0.291 | 0.006** | |
| Residuals | 9 | 2.813 | 0.313 | 0.709 | |||
| 1200 | |||||||
| Host | 2 | 1.119 | 0.375 | 1.493 | 0.249 | 0.033* | |
| Residuals | 9 | 3.372 | 0.751 | ||||
| 1300 | |||||||
| Host | 2 | 1.518 | 0.759 | 2.085 | 0.317 | 0.002** | |
| Residuals | 9 | 3.277 | 0.364 | 0.683 | |||
| 1400 | |||||||
| Host | 2 | 1.858 | 0.929 | 3.642 | 0.477 | 0.001*** | |
| Residuals | 9 | 2.040 | 0.255 | 0.523 | |||
| 1500 | |||||||
| Host | 2 | 2.136 | 1.068 | 3.751 | 0.455 | 9.999E−05*** | |
| Residuals | 9 | 2.563 | 0.285 | 0.545 | |||
| 1600 | |||||||
| Host | 2 | 1.779 | 0.890 | 3.084 | 0.407 | 0.002** | |
| Residuals | 9 | 2.595 | 0.288 | 0.593 | |||
| 1700 | |||||||
| Host | 2 | 0.710 | 0.355 | 0.850 | 0.159 | 0.623 | |
| Residuals | 9 | 3.756 | 0.417 | 0.841 | |||
| 1800 | |||||||
| Host | 2 | 1.408 | 0.704 | 2.283 | 0.337 | 0.013* | |
| Residuals | 9 | 2.776 | 0.308 | 0.663 | |||
| 1900 | |||||||
| Host | 2 | 1.230 | 0.615 | 1.658 | 0.269 | 0.019* | |
| Residuals | 9 | 3.339 | 0.371 | 0.731 | |||
| 2000 | |||||||
| Host | 2 | 0.734 | 0.367 | 0.949 | 0.174 | 0.520 | |
| Residuals | 9 | 3.480 | 0.387 | 0.826 | |||
Values shown include degrees of freedom (DF), sum of squares (SS), and mean squares (MS). Asterisks associated with p-values indicate significance values where: *≤ 0.05, **≤ 0.01, ***≤ 0.001
Table 2.
Partitioned abundance-based beta (β) diversity metrics for foliar fungal endophyte community composition within each host examined along the gradient
| Measure of compositional dissimlarity (beta diversity) | Host | ||
|---|---|---|---|
| L. taeiameiae | M. polymorpha | V. reticulatum | |
| Turnover (βBC-bal) | 0.975 | 0.960 | 0.965 |
| Nestedness (βBC-gra) | 0.000 | 0.000 | 0.000 |
| Overall (βBC) | 0.975 | 0.960 | 0.965 |
βBC-bal indicates changes in community composition caused by balanced variation in abundance, whereby the individuals of some species in one site are substituted by the same number of individuals of different species on another site (i.e. species turnover) [41]. βBC-gra indicates the changes in community composition caused by abundance gradients, whereby some individuals are lost from one site to the other (i.e. community nestedness) [41]. βBC indicates changes in community composition as determined by Bray–Curtis dissimilarity [39]
Host specialization and selectivity
Network specialization displayed a unimodal pattern along the elevation gradient (Fig. 3b, Table S2), where highest network specialization values were observed at mid-elevations, and then decreased at higher and lower elevations. Network specialization peaked at 0.9 at 1500 masl and then was lowest at 0.5 at 2000 masl (Fig. 3b, Table S2). Network specialization could be well fit to a polynomial regression equation (Fig. 3b; polynomial order = 2; R2adj = 0.504; F-statistic = 5.572; df = 2,7; p = 0.036), indicating that elevation is a strong predictor of network specialization between plants and foliar fungal endophytes.
Like network specialization, host specialization on foliar fungal endophytes for all three hosts displayed a unimodal pattern with elevation, where specialization for all three species peaked between 1400 and 1500 masl and decreased at lower and higher elevations (Table S3). The lowest host specialization for all three-host species was observed at the highest elevation sampled (Table S3). Both L. tameiameiae (Fig. 3c; red curve; Polynomial order = 2; R2adj = 0.480; F-statistic = 5.157; df = 2,7; p = 0.040) and M. polymorpha (Fig. 3d; blue curve; Polynomial order = 2; R2adj = 0.462; F-statistic = 4.877; df = 2,7; p = 0.047) specialization fit a polynomial regression equation. In contrast, V. reticulatum was poorly fit to a polynomial regression equation (Fig. 3e; Polynomial order = 2; R2adj = 0.174; F-statistic = 0.1732; df = 2,7; p = 0.21), indicating that elevation strongly influenced L. tameiameiae and M. polymorpha specialization on fungal foliar endophytes, but not V. reticulatum. Null model analysis revealed that within all sites along our gradient observed network specialization (Fig. 3b, Table S2), as well as observed specialization for all hosts (Fig. 3c–e, Table S3) were significantly higher than expected by chance (p < 0.001).
Discussion
Host specificity is typically considered a static trait, although environmental impacts on biotic interaction patterns and strength are seldom evaluated directly. Here, we provide evidence for elevational clines in specialization. We observed that both total network specialization (H2′), as well as individual host specializations (d′) peaked at mid-elevations and then decreased towards the ends of the elevation gradient we examined. This pattern ran contrary to our prediction that specificity would decrease linearly as a function of elevation, which is analogous to patterns found for other species interactions along latitudinal gradients [29]. Although future empirical work will be needed to examine the mechanisms of host specificity along elevational gradients, we consider two plausible and non-exclusive mechanisms that might explain the pattern observed.
First, we hypothesize that fungal endophyte specialization is influenced by host species density. Host density-dependent disease dynamics predict a higher occurrence of host-specific disease when hosts are more abundant, and specificity of disease decreases as host species become more scarce [12, 47]. We presume that factors influencing the association with woody plants among horizontally transferred foliar fungal endophytes are similar to those of fungal pathogens: mainly involving unrestricted dispersal of gametes and sexual reproduction prior to infecting a new host [48]. These factors are highly likely to be affected by host density. In our transect host species proportional occurrences are lower at the edges of their ranges, compare to mid-elevations (Fig. S4). Many fungi exhibit a tradeoff between dispersal and competitive ability [49]. Assuming that specialized interactions confer a competitive advantage, the higher host specialization found at mid-elevations might be due to decreased reliance on dispersal ability exhibited by more generalist fungi at low and high elevations.
Additionally, fungal endophyte communities may be less specialized at lower and higher elevations because these represent the limits of the host species range. Populations at the margins of their species’ distributions tend to be at lower abundances than populations in the middle of their species range distributions [50, 51]. As such, marginal populations of some plant species also tend to have elevated physiological stress and lower genetic diversity [52] compared to populations near the middle of their range distribution, which in turn leads to lower population growth rates and higher mortality [53]. Because our gradient constituted almost the entirety of the elevational range of two of the host species examined (and the majority of the third), we suspect that sampled plants experience increased stress at either end of the elevation gradient, which concurrently could be influencing endophyte specialization. This perception is supported by our data, as we did not observe specialization for V. reticulatum, a species which has been shown to be tolerant to abiotic stress, especially at higher elevations in Hawaii [54, 55]. Although physiological mechanisms governing host control of microbial symbionts, particularly leaf symbionts, is poorly understood, plant innate immunological responses are metabolically expensive and require the (re)allocation of resources from other plant functions, such as growth, reproduction, and stress responses [56]. Thus, hosts would likely have a better ability to allocate resources to select for and against symbionts when they are central in their ranges, whereas plants located at the margins will lack resources to be selective.
Although there are likely multiple mechanisms that reinforce endophyte specialization, numerous studies have indicated some degree of host control. Because endophytic fungi are contained within host leaves, they are in intimate contact with their hosts and receive continual chemical feedback. For example, endophytic fungi grow faster on media containing host leaf extract than non-host leaf extract [24, 57]. Our observance of hosts being a strong determinant of endophyte community across our gradient, combined with previous lab and greenhouse studies, suggest that hosts provide unique biotic environments that select for specific fungal partners, and that host selection remains a strong determinant of community composition regardless of local abiotic conditions.
In this study, we observed that host species significantly influenced fungal endophyte community composition both within and among elevations, contributing to the growing body of literature showing that endophytic communities are strongly selected for by host species [25, 26, 58, 59]. That host species is a stronger determinant of endophyte community composition than elevation was a surprising result, particularly when coupled with high species turnover across sites. That is to say, whereas host species was the primary determinant of fungal community composition within elevations, hosts associate with a largely unique set of fungal endophytes at each elevation that are dissimilar from both communities associated with co-occurring plant species and from conspecific plants at other elevations.
In contrast, multiple studies show that local environmental conditions influence foliar fungal endophyte communities. For example, endophyte communities associated with two different grass species were structured by rainfall [60]. Similarly, Zimmerman and Vitousek [61] observed that foliar endophyte communities within M. polymorpha were structured by elevation and aspect (but not soil age) along a steep volcanic gradient on the Island of Hawaiʻi. Despite starkly different values of solar radiation, temperature, precipitation, humidity, and cloud cover at the ends of our gradients, we did not find evidence that endophyte communities were structured significantly by local abiotic conditions. One reason for this discrepancy may be geographic scale. Even though our study was conducted along a portion of the same gradient as Zimmerman and Vitousek, and with one of the same host species (M. polymorpha), our data did not replicate their strong elevation partitioning. However, in their study Zimmerman and Vitousek observed that endophyte communities sampled from similar elevations and aspect (1100 and 1800 masl) were less differentiated compared to samples collected at higher and lower elevations along the transect. The niche breadth of M. polymorpha is remarkably wide, and our sampling, limited by the elevational distributions of the other two plant species, did not capture the most divergent communities. Similarly, the large effect sizes noted by Giauque and Hawkes [60] considered hosts sampled across 400 km, nearly an order of magnitude greater than the distance sampled in this study. This increased scale may lead to greater effects as well an increased likelihood that both host genetic variation and/or fungal dispersal limitation, in addition to environmental variance, might also contribute to their patterns noted. In fact, in a larger archipelago-wide survey of endophytes associated with >100 host plant species and spanning 2300 m in elevation, we found that numerous environmental factors, principally elevation and plot level evapotranspiration, correlated with endophyte community composition [62].
Alpha diversity was similarly unaffected by elevation. For larger organisms, such as plants and animals, species richness tends to decrease with increasing elevation. However, correlations between elevation and diversity are less consistent among microbial communities. In this study, we observed no relationship between elevation and fungal endophyte species richness or diversity, congruent with other studies examining soil fungi [63], soil bacteria [64], or leaf bacterial communities [65], but in contrast to others [66, 67]. Collectively these results indicate that mechanisms structuring ‘micro’ organismal groups are not uniform, resulting in different diversity patterns along elevation gradients.
Foliar fungal endophytes form intimate associations with their plant hosts. While previous research has shown that both environment and host identity strongly influence endophyte community composition, we observed that the local abiotic environment has little influence on fungal richness or community composition compared with host species. This highlights the need for research spanning a diverse array of ecosystem types to generalize our knowledge of the factors structuring these ubiquitous plant-associated microbes. We observed that host and network specialization, as well as community dissimilarity peak at mid-elevations indicating that the strength of biotic interactions between plant and symbiont vary as a function of elevation. Collectively, our results show a strong interaction between host and endophyte, and that the environment influences fungal communities, indirectly, by modulating host specialization.
Supplementary information
Acknowledgements
The authors wish to thank Erin Datlof at UH Hilo and Tomoko Sakishima at UNLV for field assistance, Nhu Nguyen for bioinformatics assistance, and members of the Amend and Hynson labs for critical review of earlier versions of the manuscript. We appreciate Dr. Heather Sahli for contributing data on plant cover. The authors also wish to thank the National Science Foundation for their generous support as NSF grant #1255972 to ASA and NSF grant #1329626 to GMC.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0470-4) contains supplementary material, which is available to authorized users.
References
- 1.Hynson NA, Bruns TD. Fungal hosts for mycoheterotrophic plants: a nonexclusive, but highly selective club. New Phytol. 2010;185:598–601. doi: 10.1111/j.1469-8137.2009.03152.x. [DOI] [PubMed] [Google Scholar]
- 2.Bohlool BB, Schmidt EL. Lectins: a possible basis for specificity in the Rhizobium—legume root nodule symbiosis. Science. 1974;185:269–71. doi: 10.1126/science.185.4147.269. [DOI] [PubMed] [Google Scholar]
- 3.Grubisha LC, Trappe JM, Molina R, Spatafora JW. Biology of the ectomycorrhizal genus Rhizopogon. VI. Re-examination of infrageneric relationships inferred from phylogenetic analyses of ITS sequences. Mycologia. 2002;94:607–19. doi: 10.1080/15572536.2003.11833189. [DOI] [PubMed] [Google Scholar]
- 4.Bruns TD, Bidartondo MI, Taylor DL. Host specificity in ectomycorrhizal communities: what do the exceptions tell us? Integr Comp Biol. 2002;42:352–9. doi: 10.1093/icb/42.2.352. [DOI] [PubMed] [Google Scholar]
- 5.Klironomos JN. Host-specificity and functional diversity among arbuscular mycorrhizal fungi. Micro Biosyst New Front. 2000;1:845–51. [Google Scholar]
- 6.Smith SE, Read DJ. Mycorrhizal symbiosis. 3rd ed. London, UK: Academic Press; 2008.
- 7.Gilbert GS, Webb CO. Phylogenetic signal in plant pathogen-host range. Proc Natl Acad Sci USA. 2007;104:4979–83. doi: 10.1073/pnas.0607968104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blüthgen N, Menzel F, Blüthgen N. Measuring specialization in species interaction networks. BMC Ecol. 2006;6:9. doi: 10.1186/1472-6785-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariadassou M, Pichon S, Ebert D. Microbial ecosystems are dominated by specialist taxa. Ecol Lett. 2015;18:974–82. doi: 10.1111/ele.12478. [DOI] [PubMed] [Google Scholar]
- 10.Waring B, Hawkes CV. Ecological mechanisms underlying soil bacterial responses to rainfall along a steep natural precipitation gradient. FEMS Microbiol Ecol. 2018;94:1–10. [DOI] [PubMed]
- 11.Connell J. On the role of natural enemies in preventing competitive exclusion in some marine animals and in rain forest trees. Dyn Numbers Popul Proc Adv Study Inst Dyn Numbers Popul Oosterbeck. 1971;298–312.
- 12.Janzen DH. Herbivores and the number of tree species in tropical forests. Am Nat. 1970;104:501–28. doi: 10.1086/282687. [DOI] [Google Scholar]
- 13.Forister ML, Novotny V, Panorska AK, Baje L, Basset Y, Butterill PT, et al. The global distribution of diet breadth in insect herbivores. Proc Natl Acad Sci USA. 2015;112:442–7. doi: 10.1073/pnas.1423042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schemske DW, Mittelbach GG, Cornell HV, Sobel JM, Roy K. Is there a latitudinal gradient in the importance of biotic interactions? Annu Rev Ecol Evol Syst. 2009;40:245–69. doi: 10.1146/annurev.ecolsys.39.110707.173430. [DOI] [Google Scholar]
- 15.Poisot T, Stouffer DB, Gravel D. Beyond species: why ecological interaction networks vary through space and time. Oikos. 2015;124:243–51. doi: 10.1111/oik.01719. [DOI] [Google Scholar]
- 16.Körner C. The use of ‘altitude’ in ecological research. Trends Ecol Evol. 2007;22:569–74. doi: 10.1016/j.tree.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Wagner WL. Manual of the flowering plants of Hawaiʻi, Rev. ed. Honolulu, HI: University of Hawaiʻi Press: Bishop Museum Press; 1999.
- 18.Vitousek PM, Matson PA, Turner DR. Elevational and age gradients in Hawaiian montane rainforest: foliar and soil nutrients. Oecologia. 1988;77:565–70. doi: 10.1007/BF00377275. [DOI] [PubMed] [Google Scholar]
- 19.Stone JK, Bacon CW, White Jr JF. Endophytism defined. In: Bacon CW, White Jr JF, editors. An overview of endophytic microbes. New York, NY: Marcel Dekker, Inc.; 2000. p. 3–29.
- 20.Arnold AE. Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev. 2007;21:51–66. doi: 10.1016/j.fbr.2007.05.003. [DOI] [Google Scholar]
- 21.Arnold AE, Engelbrecht BMJ. Fungal endophytes nearly double minimum leaf conductance in seedlings of a neotropical tree species. J Trop Ecol. 2007;23:369–72. doi: 10.1017/S0266467407004038. [DOI] [Google Scholar]
- 22.Kannadan S, Rudgers JA. Endophyte symbiosis benefits a rare grass under low water availability. Funct Ecol. 2008;22:706–13. doi: 10.1111/j.1365-2435.2008.01395.x. [DOI] [Google Scholar]
- 23.Zahn G, Amend AS. Foliar microbiome transplants confer disease resistance in a critically-endangered plant. PeerJ. 2017;5:e4020. doi: 10.7717/peerj.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnold AE, Herre EA. Canopy cover and leaf age affect colonization by tropical fungal endophytes: ecological pattern and process in Theobroma cacao (Malvaceae) Mycologia. 2003;95:388–98. doi: 10.1080/15572536.2004.11833083. [DOI] [PubMed] [Google Scholar]
- 25.U’Ren JM, Lutzoni F, Miadlikowska J, Laetsch AD, Arnold AE. Host and geographic structure of endophytic and endolichenic fungi at a continental scale. Am J Bot. 2012;99:898–914. doi: 10.3732/ajb.1100459. [DOI] [PubMed] [Google Scholar]
- 26.Vincent JB, Weiblen GD, May G. Host associations and beta diversity of fungal endophyte communities in New Guinea rainforest trees. Mol Ecol. 2016;25:825–41. doi: 10.1111/mec.13510. [DOI] [PubMed] [Google Scholar]
- 27.Bálint M, Tiffin P, Hallström B, O’Hara RB, Olson MS, Fankhauser JD, et al. Host genotype shapes the foliar fungal microbiome of balsam poplar (Populus balsamifera) PLoS One. 2013;8:e53987. doi: 10.1371/journal.pone.0053987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saunders M, Kohn LM. Evidence for alteration of fungal endophyte community assembly by host defense compounds. New Phytol. 2009;182:229–38. doi: 10.1111/j.1469-8137.2008.02746.x. [DOI] [PubMed] [Google Scholar]
- 29.Schleuning M, Fründ J, Klein A-M, Abrahamczyk S, Alarcón R, Albrecht M, et al. Specialization of mutualistic interaction networks decreases toward tropical latitudes. Curr Biol. 2012;22:1925–31. doi: 10.1016/j.cub.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 30.Smith DP, Peay KG. Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS One. 2014;9:e90234. doi: 10.1371/journal.pone.0090234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen NH, Smith D, Peay K, Kennedy P. Parsing ecological signal from noise in next generation amplicon sequencing. New Phytol. 2015;205:1389–93. doi: 10.1111/nph.12923. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Team RC. R language definition. Vienna Austria R Found Stat Comput 2000.
- 38.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. vegan: community ecology package; 2017.
- 39.Bray JR, Curtis JT. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr. 1957;27:325–49. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 40.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209. [PubMed] [Google Scholar]
- 41.Baselga A. Partitioning abundance-based multiple-site dissimilarity into components: balanced variation in abundance and abundance gradients. Methods Ecol Evol. 2017;8:799–808. doi: 10.1111/2041-210X.12693. [DOI] [Google Scholar]
- 42.Baselga A, Orme CDL. betapart: an R package for the study of beta diversity: Betapart package. Methods Ecol Evol. 2012;3:808–12. doi: 10.1111/j.2041-210X.2012.00224.x. [DOI] [Google Scholar]
- 43.Anderson MJ. A new method for non-parametric multivariate analysis of variance: non-parametric manova for ecology. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 44.Dormann CF, Gruber B, Fründ J. Introducing the bipartite package: analysing ecological networks. Interaction. 2008;1:0–2413793. [Google Scholar]
- 45.Peña EA, Slate EH. Global validation of linear model assumptions. J Am Stat Assoc. 2006;101:341–54. doi: 10.1198/016214505000000637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cobian G, Amend A, Egan C. Plant–microbe specificity varies as a function of elevation. Figshare 2019. [DOI] [PMC free article] [PubMed]
- 47.Parker IM, Saunders M, Bontrager M, Weitz AP, Hendricks R, Magarey R, et al. Phylogenetic structure and host abundance drive disease pressure in communities. Nature. 2015;520:542–4. doi: 10.1038/nature14372. [DOI] [PubMed] [Google Scholar]
- 48.Schirrmann MK, Zoller S, Croll D, Stukenbrock EH, Leuchtmann A, Fior S. Genomewide signatures of selection in Epichloë reveal candidate genes for host specialization. Mol Ecol. 2018;27:3070–86. doi: 10.1111/mec.14585. [DOI] [PubMed] [Google Scholar]
- 49.Smith GR, Steidinger BS, Bruns TD, Peay KG. Competition–colonization tradeoffs structure fungal diversity. ISME J. 2018;12:1758–67. doi: 10.1038/s41396-018-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown JH. On the relationship between abundance and distribution of species. Am Nat. 1984;124:255–79. doi: 10.1086/284267. [DOI] [Google Scholar]
- 51.Caughley G, Grice D, Barker R, Brown B. The edge of the range. J Anim Ecol. 1988;57:771. doi: 10.2307/5092. [DOI] [Google Scholar]
- 52.Jump AS, Woodward FI, Burke T. Cirsium species show disparity in patterns of genetic variation at their range-edge, despite similar patterns of reproduction and isolation. New Phytol. 2003;160:359–70. doi: 10.1046/j.1469-8137.2003.00874.x. [DOI] [PubMed] [Google Scholar]
- 53.Sorte C, Hofmann G. Changes in latitudes, changes in aptitudes: Nucella canaliculata (Mollusca: Gastropoda) is more stressed at its range edge. Mar Ecol Prog Ser. 2004;274:263–8. doi: 10.3354/meps274263. [DOI] [Google Scholar]
- 54.Lipp CC, Goldstein G, Meinzer FC, Niemczura W. Freezing tolerance and avoidance in high-elevation Hawaiian plants. Plant Cell Environ. 1994;17:1035–44. doi: 10.1111/j.1365-3040.1994.tb02026.x. [DOI] [Google Scholar]
- 55.Vander Kloet SP, Avery TS. Vaccinium on the edge. Edinb J Bot. 2010;67:7–24. doi: 10.1017/S0960428609990199. [DOI] [Google Scholar]
- 56.Rauw WM. Immune response from a resource allocation perspective. Front Genet. 2012;3:1–14. [DOI] [PMC free article] [PubMed]
- 57.Lau MK, Arnold AE, Johnson NC. Factors influencing communities of foliar fungal endophytes in riparian woody plants. Fungal Ecol. 2013;6:365–78. doi: 10.1016/j.funeco.2013.06.003. [DOI] [Google Scholar]
- 58.David AS, Seabloom EW, May G. Plant host species and geographic distance affect the structure of aboveground fungal symbiont communities, and environmental filtering affects belowground communities in a coastal dune ecosystem. Microb Ecol. 2016;71:912–26. doi: 10.1007/s00248-015-0712-6. [DOI] [PubMed] [Google Scholar]
- 59.Lamit LJ, Lau MK, Sthultz CM, Wooley SC, Whitham TG, Gehring CA. Tree genotype and genetically based growth traits structure twig endophyte communities. Am J Bot. 2014;101:467–78. doi: 10.3732/ajb.1400034. [DOI] [PubMed] [Google Scholar]
- 60.Giauque H, Hawkes CV. Climate affects symbiotic fungal endophyte diversity and performance. Am J Bot. 2013;100:1435–44. doi: 10.3732/ajb.1200568. [DOI] [PubMed] [Google Scholar]
- 61.Zimmerman NB, Vitousek PM. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc Natl Acad Sci USA. 2012;109:13022–7. doi: 10.1073/pnas.1209872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darcy JL, Cobian GM, Swift SOI, Zahn GL, Perry BA, Amend AS. Fungal communities living within leaves of native Hawaiian dicots are structured by landscape-scale variables as well as by host plants. bioRxiv 2019;640029. [DOI] [PubMed]
- 63.Miyamoto Y, Nakano T, Hattori M, Nara K. The mid-domain effect in ectomycorrhizal fungi: range overlap along an elevation gradient on Mount Fuji, Japan. ISME J. 2014;8:1739–46. doi: 10.1038/ismej.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh D, Takahashi K, Kim M, Chun J, Adams JM. A hump-backed trend in bacterial diversity with elevation on Mount Fuji, Japan. Microb Ecol. 2012;63:429–37. doi: 10.1007/s00248-011-9900-1. [DOI] [PubMed] [Google Scholar]
- 65.Fierer N, McCain CM, Meir P, Zimmermann M, Rapp JM, Silman MR, et al. Microbes do not follow the elevational diversity patterns of plants and animals. Ecology. 2011;92:797–804. doi: 10.1890/10-1170.1. [DOI] [PubMed] [Google Scholar]
- 66.Bryant JA, Lamanna C, Morlon H, Kerkhoff AJ, Enquist BJ, Green JL. Microbes on mountainsides: contrasting elevational patterns of bacterial and plant diversity. Proc Natl Acad Sci USA. 2008;105:11505–11. doi: 10.1073/pnas.0801920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Egan CP, Callaway RM, Hart MM, Pither J, Klironomos J. Phylogenetic structure of arbuscular mycorrhizal fungal communities along an elevation gradient. Mycorrhiza. 2017;27:273–82. doi: 10.1007/s00572-016-0752-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All amplicon sequencing data generated in this study can be accessed upon publication on National Center for Biotechnology Information’s Sequence Read Archive under Foliar Fungal Endophytes along Mauna Loa Elevation Gradient—BioProject (PRJNA474551). All metadata, OTU tables, and code for data analysis are available at figshare [46].