Abstract
In the last decade, extensive application of hydraulic fracturing technologies to unconventional low-permeability hydrocarbon-rich formations has significantly increased natural-gas production in the United States and abroad. The injection of surface-sourced fluids to generate fractures in the deep subsurface introduces microbial cells and substrates to low-permeability rock. A subset of injected organic additives has been investigated for their ability to support biological growth in shale microbial community members; however, to date, little is known on how complex xenobiotic organic compounds undergo biotransformations in this deep rock ecosystem. Here, high-resolution chemical, metagenomic, and proteomic analyses reveal that widely-used surfactants are degraded by the shale-associated taxa Halanaerobium, both in situ and under laboratory conditions. These halotolerant bacteria exhibit surfactant substrate specificities, preferring polymeric propoxylated glycols (PPGs) and longer alkyl polyethoxylates (AEOs) over polyethylene glycols (PEGs) and shorter AEOs. Enzymatic transformation occurs through repeated terminal-end polyglycol chain shortening during co-metabolic growth through the methylglyoxal bypass. This work provides the first evidence that shale microorganisms can transform xenobiotic surfactants in fracture fluid formulations, potentially affecting the efficiency of hydrocarbon recovery, and demonstrating an important association between injected substrates and microbial growth in an engineered subsurface ecosystem.
Subject terms: Environmental chemistry, Microbial ecology
Introduction
The integration of horizontal drilling and hydraulic fracturing (HF) technologies for the recovery of hydrocarbons from unconventional low permability (“tight”) formations has increased the production of natural-gas in the United States and abroad [1]. In 2017, the United States became a net exporter of natural-gas for the first time since 1957, producing on average more than 70 billion cubic feet per day [2]. During HF, fluids are injected at high pressures to induce new and reopen existing fractures in extremely low-permeability (nano-Darcy conductivities) rock formations, increasing production of petroleum resources [1, 3]. Injected fluids are comprised primarily of water, proppant (e.g. fine sand), and organic chemical additives, with formulations optimized by industry to increase natural-gas yields, while concurrently protecting well infrastructure [3]. Once the petroleum well has been completed and natural-gas production commences, injected fluids continue to mix with highly saline formation pore waters, generating produced fluid brines that return to the surface over the course of several months to years [3–8].
The interconnected fractures generated by HF create a new microbial ecosystem through an infusion of surface-derived water, nitrogen, and carbon sources to the shale [4, 7, 9, 10]. Microbial community dynamics in samples from HF natural-gas wells have been described for several different U.S. black shale formations [9]. Within months, the microbial communities converge, and are characterized by low species-level diversity and predominance of halotolerant, fermentative Bacteria and Archaea [4, 9–18]. This microbial community shift occurs as HF-injected carbon and electron acceptors are depleted, and salinity becomes enriched from formational brine and dissolution of solid phase salts [4–8, 11, 12, 15, 17, 19–21]. Recent studies have begun to describe the unique metabolisms and adaptations enabling microbial persistence in this engineered subsurface [11, 12, 18, 22–24], with some influenced by the lability of chemical additives used by the petroleum industry to protect the petroleum well and enhance its production [11, 18, 22, 23, 25].
Organic chemical components of HF fluids play an important, yet under-characterized role in sustaining microbial communities in shales. For example, urea is a naturally-derived additive that reduces friction in fracturing fluids [26], and may be consumed as a carbon and nitrogen source through conversion to CO2 and NH3 by taxa present in the first few months in HF shale-gas wells (e.g. Arcobacter, Marinobacter) [18]. Additionally, the vitamin choline (added as choline chloride) is the most frequently disclosed clay stabilizer used by the industry, and may fuel a community metabolism culminating in biogenic production of osmoprotectants and ultimately methanogenesis [11, 25]. Furthermore, the gelling agent guar gum, a polysaccharide derived from plants, is readily degraded by Halanaerobium, which couples guar gum fermentation to sulfide production months to years after HF occurs [23]. The above referenced studies focus on the biotransformation of labile or naturally-derived compounds, underscoring the need to better understand metabolisms influencing xenobiotic compound degradation during and after HF operations [25].
Xenobiotic polyglycols are common and essential components of HF fluids in natural-gas wells (up to 49.7%) as crosslinkers, scale inhibitors, solvents, and surfactants (Supplementary Table 1) [25, 27]. Specifically, alkyl polyethoxylates (AEOs) containing a branched or linear alkyl hydrocarbon connected to repeating ethoxylate chains have been identified in produced fluid samples from petroleum wells that underwent HF [28–30]. In one study examining fluids produced from the Niobrara Formation, AEO relative concentrations decreased as the petroleum well matured, but the mechanism(s) responsible for AEO changes were not thoroughly explored [28]. One possible explanation for surfactant losses in shale wells stimulated by HF is biological attenuation. Anaerobic degradation of glycols is accomplished via stepwise cleavage of ethoxylate units catalyzed by the propanediol dehydratase gene cluster, generating alcohols, carboxylic acids, and ketone metabolites [31–33]. This biotransformation pathway has not been investigated in situ during HF operations, but was described in groundwater microcosms simulating spills of fracturing and produced fluids, highlighting environmental impacts that might arise from an accidental release of these constituents [33, 34]. Moreover, this surfactant degradation mechanism may have a negative effect on hydrocarbon recovery due to unintended changes in produced fluid chemistry, as the transformation pathway can (1) reduce concentrations of injected chemicals resulting in a loss of chemical efficacy, and (2) produce corrosive organic acids that damage well infrastructure [35]. Biological degradation of surfactants may help explain the aforementioned surfactant trends; nonetheless, the environmental and industrial consequences of this pathway are significant.
To determine whether xenobiotic polymers are attenuated by microorganisms in HF systems, we analyzed changes in unsubstituted and alkylated polyglycols in fluid samples collected up to 204 days after production began in a Utica-Point Pleasant Formation natural-gas well in Ohio, U.S.A. Temporal changes in surfactant chemistry were associated with putative biotransformation genes from produced fluid metagenomes. To confirm the capacity of relevant taxa to enzymatically transform these xenobiotic compounds, we applied genome sequencing, proteomics, and metabolite analysis in laboratory batch experiments, uncovering a previously uncharacterized, co-metabolic pathway for surfactant chain shortening by the halotolerant bacterial strain Halanaerobium congolense WG10. Our results show that key microbial strains are capable of transforming xenobiotic organic additives in natural-gas wells stimulated by HF.
Materials and methods
Produced water sampling and pre-processing
Fluid samples were recovered from a natural-gas well drilled in the Utica-Point Pleasant Formation (2.6 km depth) in eastern Ohio between July 2014 and February 2015 as reported previously [12]. Due to a ~3 month “shut-in”, the first fluid sampling occurred 86 days after HF. Samples were collected from the gas-water separator in 1 L sterile Nalgene HDPE bottles (Thermo-Fisher Scientific, Waltham, MA) with no headspace. The flow rate in the separator ranged from 190,000 to 380,000 L day−1, resulting in a maximum 8 h residence time before any sampling event [12]. Biomass from produced fluid samples (300–1000 mL) was concentrated onto 0.22 µm PES filters (Nalgene, Thermo-Fisher Scientific, Waltham, MA) within 24 h of sampling and stored at −80 °C until further use [12]. Filtrate was collected and preserved for ion (4 °C, with or without nitric acid), dissolved carbon analysis (4 °C, HCl), and surfactants (−20 °C) as described below and in the Supplementary Methods.
Bacterial growth experiments
Surfactant biodegradation experiments were performed using H. congolense WG10, a strain isolated from this Utica-Point Pleasant natural-gas well 140 days after HF as previously described [12]. H. congolense WG10 was grown in SWDM medium (Supplementary Methods) containing 10 mM D-Glucose and amended with a commercial surfactant additive “Revert Flow”, which contains C6-C12 alcohol ethoxylates, isopropyl alcohol, orange terpenes, and a polyethylene glycol–polypropylene glycol (PEG–PPG) co-block polymer (Supplementary Table 2). Revert Flow was added to culture samples at concentrations of ~150 mg/L total organic carbon (TOC), with AEOs and PEG-PPGs each between 15–45% of total Revert Flow mass according to the manufacturer (Supplementary Table 2). Biotic samples (containing Revert Flow and 10 mM D-glucose) and glucose controls (10 mM D-glucose) were cultured in triplicate using a 10% inoculum. Killed controls, conducted in duplicate, were initially grown to mid-log phase with 10 mM D-glucose, heat killed using an autoclave, then transferred to fresh medium (10%). Abiotic controls (in duplicate) contained a 10% transfer of sterile SWDM. All transfers were performed under anoxic conditions (80% N2, 20% CO2) and were incubated at 37 °C in the dark. Growth was monitored using optical density readings (OD, 610 nm) on a Hach DR900 Colorimeter (Loveland, Colorado). Samples were collected aseptically immediately after inoculation/transfer (t0), at mid-log phase (tm) when OD reached ~0.3 (25.5 h for glucose controls, 43 h for surfactant amended cultures), stationary phase (ts) (51 h for both treatments), and death phase (td) (66 h for both treatments). Dissolved constituents were filtered (0.22 μM PES, EMD Millipore, Burlington, MA) for GC and LC analyses (Supplementary Methods, Supplemental Table 3), while unfiltered samples were used for ATP analysis (Supplementary Methods).
Shotgun proteomic analysis
Biomass for shotgun proteomic analysis was collected from biological triplicates at mid-log phase and pelleted by centrifugation at 10,000×g for 10 min at 4 °C. Pellets were immediately flash frozen and stored at −80 °C until shipment to Pacific Northwest National Laboratory for analysis. Protein extraction, digestion into peptides, and analysis on a 2D ACQUITY ultra high-performance liquid chromatography (UPLC) M-class system (Waters, Milford, MA) was performed as previously described [12, 36]. Measured peptides were searched against predicted peptides derived from the H. congolense WG10 genome. Resulting peptide identifications were filtered via MS-GF+ using a Q value of ≤0.01 [37] and a reverse decoy database search using the reported filters generated an FDR of 0.9%. For comparative analyses of replicates, protein spectral counts were normalized by the normalized spectral abundance frequency (NSAF) method [38] and Z-score values were calculated from the mean protein abundance across all conditions as previously reported [12]. Values between glucose controls and surfactant grown cells were considered significantly different if the difference in the protein z-score was ≥1.65 (90% confidence).
Polyglycol analysis
AEOs (C4, nC6, C6, C8) and polyethylene glycols (PEGs) from field samples were analyzed using a high-performance liquid chromatograph coupled to a high-resolution mass spectrometer (LTQ-Orbitrap Velos with positive polarity electrospray ionization (ESI)) (Thermo-Fisher Scientific, Waltham, MA) at Duke University. Chromatographic separation of branched and linear AEOs (e.g. nC6 vs. C6) was achieved (Supplementary Fig. 1). Sample preparation, instrument methodology, and identification methods have been previously reported [39]. Limits of detection and quantification were determined for each measured species (Supplementary File 1).
C8 AEOs and PPGs in laboratory samples were analyzed using UPLC with quadrupole time-of-flight mass spectrometry (qTOF-MS) at the Colorado State University Central Instrument Facility (Supplementary Fig. 2) [33]. The separation and identification methods have been previously reported [40] with the following exceptions: C8 alkyl polyethoxylates (C8 AEOn) and polypropylene glycols (PPGn) were manually quantified on the most abundant adduct [M+NH4]+, except for PPGs with propoxymers n = 4–8, which were quantified on the more abundant adduct [M+Na]+ (Supplementary Table 4). A calibration curve (R2 = 0.9879) was constructed using a C10EO8 standard (Sigma Aldrich) for quality assurance and to establish a linear range for similar species for accurate semi-quantitation based on relative abundances (see Supplementary Table 4, Supplementary Table 5). For both field and culture sample analysis, external calibrants for individual species or mixtures with known species concentrations were unavailable, therefore a semi-quantitative approach was employed which normalized all AEO, PEG, and PPG species to the starting concentrations of each individual polyglycol, as has been reported previously (Supplementary Table 6) [28, 33, 34, 40, 41].
Metagenomic sequencing and analysis
DNA was extracted from produced fluid filters and sequenced as previously reported [12]. Metagenomes in IMG/M were assembled by the Joint Genome Institute using MEGAHIT v. 1.0.3 and annotated using the IMG Annotation Pipeline v.4.10.0 [42]. Gene inquiry for isolate genomes and metagenomes were performed using IMG/M from JGI [43]. Genes in H. congolense WG10 were identified using annotated EC numbers and by comparison to characterized genes in Acetobacterium woodii (Supplementary Methods). Genes in assembled metagenomes were identified through comparison to H. congolense WG10 using BLASTp (within IMG/M) with quality cutoffs (≥90% identity and bitscore ≥200) (Supplementary Files 2, 3) [43, 44].
Data accession
Metagenomic sequence data from this study is available under NCBI BioProject ID PRJNA433267 and the Joint Genome Institute (JGI) Integrated Microbial Genomes and Microbiomes (IMG/M) database. Genomic sequence data is available under the JGI-IMG/M database under project number Ga0073285. Proteomic data is available in Supplementary information (Supplementary File 4).
Results and discussion
Surfactants attenuate in situ during the months following hydraulic fracturing
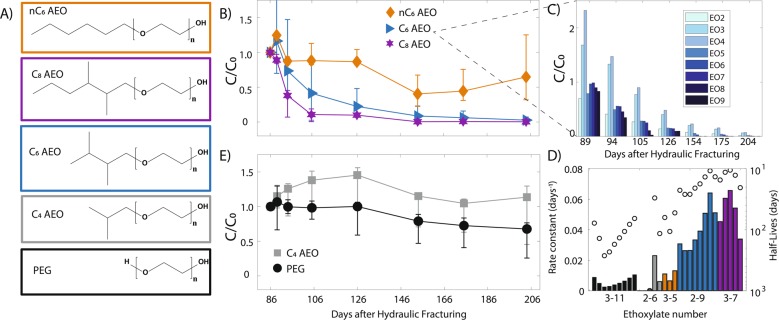
Five different polyglycols were detected in produced fluids from this Utica-Point Pleasant natural-gas well, with relative abundances varying through time based on surfactant class and chain length (Fig. 1a). Branched C6 AEOs, containing two to nine ethoxylate units (EO2 to EO9) decreased by 97% on average between the first day of flowback (day 86) and later production on day 204 (Fig. 1b). During earlier flowback (days 86–92 after HF), branched C6 AEOs with short ethoxylate chain lengths (EO3, EO4) doubled in abundance concurrent with ~50% loss for longer chains by day 94 (EO5-EO9) (Fig. 1c). This trend is consistent with enzymatic chain shortening of linear AEOs under anaerobic conditions [31, 32]. Pseudo-first order rate constants based on temporal changes in surfactant relative abundances were used to estimate half-lives for individual ethoxylate lengths and ranged from 10 and 26 days for branched C6 AEOs (Fig. 1d). In contrast to the trends described for branched C6 AEOs, linear structures with the same alkyl length (nC6 AEO) decreased only 35% between days 86 and 204, resulting in half-lives on the order of 52–106 days (Fig. 1b, d). AEOs with shorter alkyl chain lengths (C4 AEOs) were removed more slowly (Fig. 1e) resulting in significantly longer half-lives than AEOs with longer alkyl chain lengths (115 to 990 days) (Fig. 1d). On the other hand, C8 AEOs decreased the quickest, with 99.7% reduction by day 204, and exhibited the shortest half-life of all detected AEO species (10–20 days) (Fig. 1b, d). Here, branched C6 and C8 AEOs attenuated at a considerably faster rate than linear C6 or C4 AEO structures, indicating a clear preference for the longer, branched alkyl chains over shorter or linear structures in this system. To further support this point, we also tracked PEGs through time, which contain an ethoxylate backbone without an alkyl chain. PEGs decreased only 32% by day 204, with estimated half-lives between 65 and 266 days (Fig. 1d, e). Similarly, monomeric ethylene glycol only decreased 17% between days 86 and 204, while monomeric propylene glycol decreased 55% during this time (Supplementary Fig. 3). Earlier studies have revealed similar temporal declines in specific AEOs compared to PEGs, yet little attention has been given to examining potential causes other than sorption or mixing effects [28].
Fig. 1.
a Structures of polyglycol compounds observed in this Utica-Point Pleasant natural-gas well. b Produced fluid temporal change in total AEO species for linear nC6, branched C6, and C8 AEOs where C0 is the sum of all ethoxylates of a particular alkyl species at Day 86. Error bars correspond to the standard deviation of the average change experienced by the sum of all ethoxylates for a given alkyl chain. c Branched C6 AEO temporal trends relative to Day 86, with each bar representing a different C6 ethoxylate. d First-order rate constants (bars, left y-axis) and half-lives (circles, right y-axis) for PEGs, C4, nC6, C6, and C8 AEOs. Colors correspond to legend given in b, e. e Produced fluid temporal change in total AEO for C4 AEOs and PEGs where C0 is the sum of all ethoxylates of a particular alkyl species at Day 86. Error is the same as described in b
Geochemical mixing model discerns physical from biochemical surfactant trends
To elucidate whether observed field trends in AEOs and PEGs were a result of geochemical mixing, we applied an end-member mixing model to inorganic elements and ions measured in the input and produced fluid samples using input fluid and day 204 as end-members. Mg2+, Ca2+, Sr2+, and Br− were strongly related to Cl- in formation waters (Spearman’s > 0.94), suggesting a conservative mixing model was appropriate to discern physical (i.e. dilution) effects in this system (Supplementary Table 7) [20]. Ten days after flowback began (day 96), Li+/Cl− molar ratios approached a plateau near 2 × 10−3 (Supplementary Fig. 4), indicating fluid mixing effects within the formation had diminished. In addition, pH and conductivity measurements were consistent for the first 12 days after flowback began (Supplementary Table 7), suggesting solubility variations from redox changes were minimal during this period of time. Throughout the temporal flowback series, Cl− and Sr2+ increased by 1.5 and 2.8 fold, respectively (Supplementary Fig. 4). By comparison, the abundance of branched C6 and C8 AEOs decreased monotonically through time by 2 orders of magnitude, while PEGs, C4 and nC6 AEOs remained steady or increased during the flowback period (Fig. 1). If trends in surfactant abundance were due solely to dilution, we would expect to see the same magnitude of change in both inorganic ions and organic constituents in the absence of appreciable pH/redox driven changes in anticipated adsorption, which was not the case. These combined trends indicate dilution and/or mixing could not completely explain surfactant temporal trends, and that another attenuation mechanism occurred.
We considered whether decreases in branched C6 and C8 AEOs could be due to sorption effects from interactions with shale [28], but ruled this out as the primary loss mechanism based on two possible explanations. Firstly, as observed here, AEOs with shorter ethoxylate chains increased in abundance before decreasing (e.g. branched C6 EO3, EO4 between day 86 and 89, Fig. 1c), while other chain lengths for the same alkyl group monotonically decreased (e.g. branched C6 EO2, and EO5-EO9). A system dominated by sorption should show similar trends for ethoxylate units of the same alkyl structure (e.g. decreases, not increases). Secondly, if we consider partitioning of surfactants from an aqueous phase (fracture fluid) into an organic phase (e.g. kerogen within source rock), we would expect the more hydrophobic surfactant (higher alkyl chain lengths, linear structures) to have a greater attraction for other organic phases than the less hydrophobic surfactant (lower alkyl chain lengths, branched structures). Sorption may explain the rapid losses in C8 AEOs relative to C4 AEOs or PEGs; however, the opposite trend is observed in the linear versus branched isomers of the C6 AEOs. Specifically, the slightly less hydrophobic branched C6 AEOs decreased to a greater extent than the more hydrophobic linear nC6 AEOs at all observed time points (Fig. 1b, Supplementary File 1). It is therefore unlikely the surfactant trends described here are dictated solely by differences in hydrophobicity or sorption to an organic phase. Polymers such as guar gum are known to form filter cakes in situ, which may be one possible explanation for the observed decrease in polyglycol concentrations; however, polyglycols are used to prevent and disrupt filter cake formation during drilling [45], rendering filter cake formation an unlikely mechanism explaining these trends. Sorption to cell biomass is also unlikely to dictate surfactant trends as viable cells rarely exceed 106 cells mL−1 in produced fluid [11]. Altogether these data suggest a third mechanism, such as biotransformation, may govern surfactant fate in this system.
Metagenomic identification of surfactant degrading genes
Enzymatic chain shortening of AEOs, PEGs, and ethylene glycol can occur under anaerobic conditions through the propanediol dehydratase gene family (pduCDE). The biochemical reaction generates acetaldehyde, which can be dismutated to ethanol and acetate by an aldehyde dehydrogenase (pduP) [31, 46] (Fig. 2a). Although there is currently no characterized enzyme for the biotransformation of PPGs under anaerobic conditions, propylene glycol, the monomer of PPGs, can be degraded by a diol dehydratase under both aerobic and anaerobic conditions, generating propionaldehyde, n-propanol, propionate, and acetone as products [33, 47–49]. We therefore mined metagenomes from Utica-Point Pleasant produced fluid samples for genes encoding enzymes known to degrade polyglycols to determine whether the aforementioned microbial biotransformation pathway existed in this system.
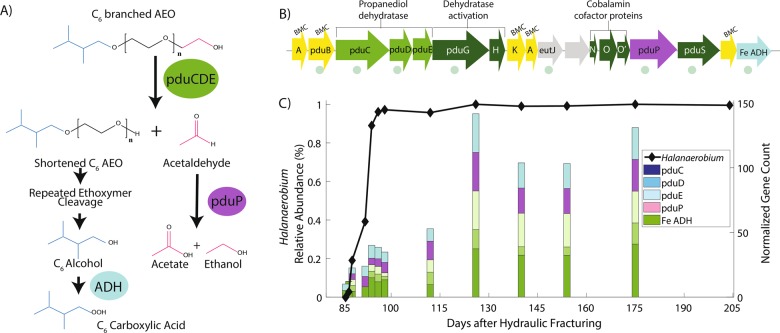
Fig. 2.
a Scheme for enzymatic biotransformation of a branched C6 alcohol ethoxylate based on Huber et al. [31] and Trifunović et al. [46] . b H. congolense WG10 genomic contig containing pdu and related genes with homologous genes to Acetobacterium woodii denoted by green circles (bitscore > 200, identity > 35%) c Relative abundance of Halanaerobium through time and the normalized gene copies ((gene count/assembled metagenome size) × 108) present in produced fluid metagenomes using blastp (>90% identity and bit-score > 250) compared to the genes present in the genome of H. congolense WG10
We detected the surfactant biotransformation gene, pduC, in produced fluid metagenomes throughout the lifetime of the Utica-Point Pleasant natural-gas well (Supplementary File 2) [44]. Observed pduC genes were highly similar (top hit, bit score > 100, identity > 90%) to dominant Halanaerobium, Thermoanaerobacter, and Clostridiales (Supplementary File 2), previously identified as key microbial community members in this natural-gas well [12]. Despite the association of pduC to these three different bacterial genera, 86.5% of all pduC genes mapped with high homology (bit-score > 70, identity % > 70) to Halanaerobium, and all pduC genes detected after day 112 were matched to Halanaerobium (Supplementary File 2). The halotolerant taxon Halanaerobium, which can grow at NaCl concentrations greater than 25%, but grows optimally between 10 and 15% NaCl [50], commonly dominates the microbial community in later produced fluids from natural-gas wells stimulated by HF [4, 11, 12, 14, 23, 24]. As such, Halanaerobium is an important model organism for understanding biogeochemical changes in the highly saline, HF stimulated shale ecosystem. Since Halanaerobium was persistent in this natural-gas well and its reconstructed genome contained putative surfactant degrading genes, we focused our search to genes within metagenomic data having high homology to this genus.
We compared metagenomic data to a genome sequenced from a Halanaerobium strain isolated from the Utica-Point Pleasant natural-gas well on day 140 [12]. The isolate, H. congolense WG10, contained several important surfactant degrading genes with homology (bit score > 200, identity > 30%) to a known glycol-degrading bacterium, Acetobacterium woodii (Fig. 2b) (Supplementary File 3) [46]. The normalized metagenomic gene counts for the propanediol dehydratase gene family (pduCDE), aldehyde dehydrogenase (pduP), and iron-containing alcohol dehydrogenase (Fe-ADH) paralleled the increasing abundance of Halanaerobium as the natural-gas well matured, except on day 204 where gene absence may be related to viral predation of Halanaerobium (Fig. 2c) [11, 51]. The decreasing abundance of branched C6 and C8 AEOs in this natural-gas well corresponds with higher pduCDE gene abundances (Fig. 1b, Fig. 2c), suggesting Halanaerobium may be primarily responsible for surfactant chain shortening in this system. In addition to detecting the putative enzymes in the most dominant bacterial taxa, we also identified four possible metabolites that could result from polyglycol chain shortening in produced fluid using LC-MS and the MetFish approach [52], including aldehydes, carboxylic acids, alcohols, and a ketone (Supplementary Fig. 3). Although these metabolites could result from the transformation of other natural and additive compounds, given the portion of fracture fluid chemical mass accounted for by polyglycols in this shale-gas well (~0.007%, Supplementary Table 8) their presence further supports the possibility that this biotransformation pathway is occurring in situ. Metabolite data generated by LC-MS analysis was further supported by NMR analysis of produced fluid samples (Supplementary Fig. 3).
Surfactant degradation observed in isolate cultures
Given the likely presence of a surfactant degrading metabolism in Halanaerobium, we tested the ability of the isolate H. congolense WG10 to metabolize a commercially available surfactant mixture (Revert Flow, Supplementary Table 2) containing PPGs and C6-C12 AEOs by tracking changes in surfactant chemistry, protein abundances, and metabolite production during batch growth over 60 h (Supplementary Fig. 5). As this strain did not grow on the commercially available surfactant mixture (Revert Flow), PEGs, ethylene glycol, or propylene glycol as a sole carbon source, surfactant treatments were amended with D-glucose and compared with a glucose control (Supplementary Fig. 7).
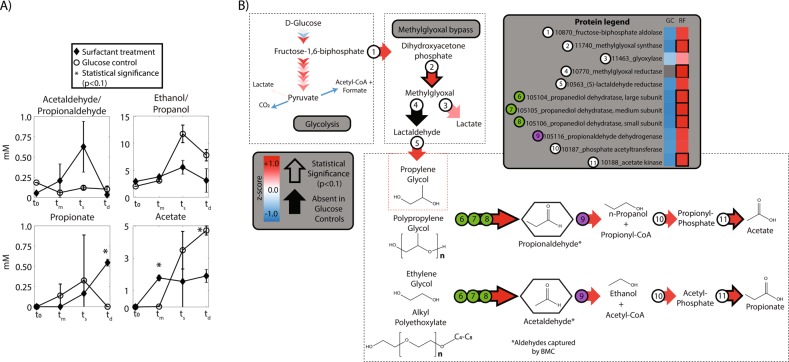
Total PPGs decreased in surfactant treatments by 19% with no PPG losses observed in killed or abiotic controls (Fig. 3a). PPGs containing higher propoxylate (PO) chain lengths (PO6 through PO10) displayed more significant decreases in relative concentration (26–47%) as compared with shorter propoxylate lengths (PO4 and PO5) (8 and 12%, respectively), a trend which may be attributed to the shortening of longer PPG chains (Fig. 3b). Metabolite data showed significantly higher propionate concentrations in surfactant treatments (0.54 mM) compared to glucose controls during the death phase (Fig. 4a), supporting the enzymatic production of carboxylic acids from PPG chain shortening. To our knowledge, H. congolense WG10 represents the first report of a microbial isolate transforming PPG under anaerobic conditions.
Fig. 3.
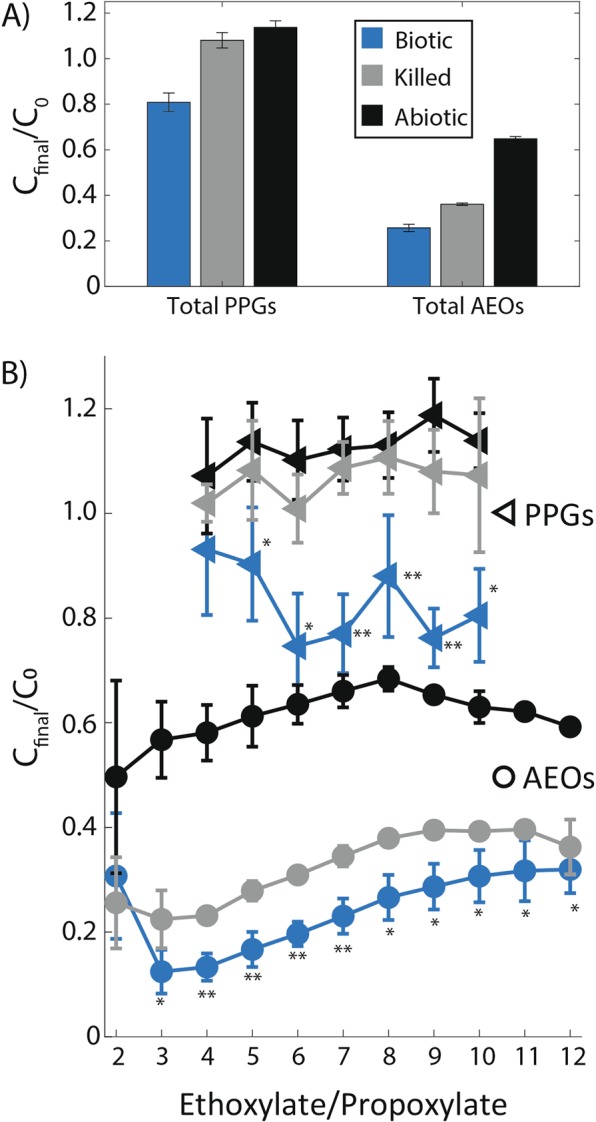
Relative change in initial and final concentrations of polypropylene glycols (PPGs) and C8 alkyl polyethoxylates (AEOs) in surfactant amended cell cultures between inoculation (t0) and stationary phase (ts, 51 h). a The sum of all PPG and C8 AEO polyglycol species. b Relative change in each individual ethoxylate/propoxylate. Error bars correspond to standard deviations between triplicate measurements, and statistical significance for biotic cells (p < 0.1) is denoted by (*) compared to abiotic controls only and (**) compared to both killed and abiotic controls
Fig. 4.
a Relevant metabolite concentrations for surfactant grown cultures and glucose controls at t0, tm (mid-log), ts (stationary phase), and td (death phase), and statistical significance (p < 0.1) is denoted by (*) next to the measurement. b Proposed surfactant metabolic reconstruction of Halanaerobium congolense WG10 cultured with surfactants compared to glucose controls. Arrow color represents average z-score in proteins from surfactant cultures. Each arrow denotes one biochemical step, whereas each number corresponds to individual proteins. Statistical significance (p < 0.1) for differences in protein relative abundance in surfactant grown cultures are denoted by a black outline around the corresponding arrow. Blacked out arrows indicate the protein was solely found in surfactant grown cultures and not in glucose controls. The protein legend includes the gene number, name, and z-score differences between glucose controls and surfactant grown cultures
Biotransformation of AEOs also occurred during growth of H. congolense WG10 on surfactants. Total AEOs decreased in relative concentration by 74%, with losses in several ethoxylate chain lengths (EO4 through EO7) significantly lower than killed and abiotic controls (Fig. 3a, b). Acetaldehyde/propionaldehyde, the initial product(s) of AEO/PPG chain shortening, had a higher concentration in the surfactant treatment (>0.6 mM) at stationary phase compared to glucose controls, which never surpassed 0.2 mM at any time point (Fig. 4a). Acetate concentrations were significantly higher during mid-log phase in surfactant treatments; however, acetate continued to increase in glucose controls during the death phase whereas concentrations plateaued in surfactant treatments (Fig. 4a). No significant differences in concentration of ethanol/propanol were detected between the two treatments (Fig. 4a). It is important to note that we measured considerable losses in the relative concentration of AEOs in abiotic and killed controls (35 and 64%, respectively, Fig. 3a), which we attributed to interaction with growth medium reagents (abiotic) and/or sorption to dead biomass (killed) estimated at 109 cells/mL. It is also possible high salt concentrations caused matrix suppression during analysis of AEO species using mass spectrometry [30]; however, medium conditions were held constant, therefore these effects are similar across treatments.
Using shotgun proteomics, we sought to determine which biochemical pathway was responsible for surfactant transformations in H. congolense WG10. Protein relative abundance for each of the three genes in the propanediol dehydratase gene cluster pduCDE (Gene 6–8 in Fig. 4b/Ga0072835_105104-105106) was significantly higher in surfactant grown cultures as compared with glucose controls (Fig. 4b, Supplementary Fig. 6). The activity of pduCDE generates one mole of an aldehyde per ethoxylate (AEO) or propoxylate (PPG) cleaved. As aldehydes are toxic to bacterial cells, they are immediately captured by a bacterial microcompartment (BMC) before conversion to propionyl/acetyl CoA using an aldehyde dehydrogenase (pduP, Gene 9/105116) [47]. Here, we saw a significant increase in the relative abundance of proteins associated with BMCs (105102, 105103, 105110) as well as overall increases in pduP for surfactant grown cells (Gene 9/105116) (Supplementary Fig. 6). In addition, proteins responsible for converting propionyl/acetyl-phosphate to their corresponding carboxylic acids (propionate/acetate) were identified at significantly higher relative abundances in surfactant grown cultures compared to glucose controls (Gene 11/10188) (Fig. 4b, Supplementary Fig. 6). Furthermore, at least eleven proteins associated with Vitamin B12 synthesis, a key cofactor during pduCDE activity, were observed at significantly higher levels for cells grown on surfactants (Supplementary Fig. 6) [47].
In addition to increased abundance of proteins associated with AEO and PPG chain shortening, we also observed clear differences in methylglyoxal synthase (Gene 2/11740), methylglyoxal reductase (Gene 4/10770), and (S)-lactaldehyde reductase (Gene 5/10563) protein abundance for surfactant grown cultures (Fig. 4b, Supplementary Fig. 6). These three enzymes convert dihydroxyacetone phosphate derived from glucose to methylglyoxal, next reducing it to lactaldehyde, and finally producing propylene glycol. Propylene glycol is then taken into the propanediol dehydratase gene cluster (pduCDE) for conversion to carboxylic acids and alcohols as described previously. When grown in the presence of glycol-containing compounds, we infer that H. congolense WG10 utilizes the methylglyoxal bypass during glycolysis, thereby “turning up” the pathway for propanediol dehydratase. Consequently, surfactant grown cells likely shuttle more glucose through the methylglyoxal bypass relative to glucose controls. This would culminate in decreased glycolytic activity and increased production of aldehydes (acetaldehyde/propionaldehyde) and carboxylic acids (acetate/propionate) from the generation of propylene glycol and from shortening of AEO and PPG chains (Fig. 4a).
Research by others suggests the methylglyoxal bypass is important for bacteria grown under stress or nutrient limited conditions, including excess organic carbon [53–56]. Although carbon was not limited in our experiments, we observed significantly higher abundances of the universal stress protein (uspA, 10238) in surfactant amended cultures (Supplementary File 4). Moreover, in a recent study on a closely-related Halanaerobium strain isolated from the same natural-gas well (H. congolense WG8), the methylglyoxal bypass was initiated under growth at high pressures with glucose as the sole carbon source, possibly to dispose of excess reducing equivalents [48]. Here, we propose co-metabolism of glycols occurs either through cellular stress and/or high concentrations of glycols within the medium, activating both the methylglyoxal bypass and the propanediol dehydratase pathways.
Our combined field and laboratory data provide evidence for several important findings regarding bacteria living in highly saline anaerobic systems. Firstly, the halotolerant bacterial strain isolated from produced fluids obtained from this natural-gas well, H. congolense WG10, can co-metabolize two commonly used surfactant classes, namely, AEOs and PPGs, when grown on a labile substrate. Importantly, the pduCDE gene cluster in H. congolense WG10 acts on monomeric ethylene glycol, PPGs, and AEOs, but not PEGs (Supplementary Fig. 7), indicating a predilection for specific polymers. Our field data suggest that enzymatic preference extends to alkyl chain length (C8 and C6 over C4 AEOs) and structure (branching over linear AEOs, and AEOs over PEGs). This finding is surprising in light of previous work which described branched AEOs as significantly more recalcitrant to biodegradation than linear AEOs [57]. Here, the presence of glucose is key to surfactant biotransformation through presumed precursor activation of the propanediol dehydratase gene cluster after sending fructose-1,6-biphosphate through the methylglyoxal bypass. The propanediol dehydratase gene family (pduCDE) as well as the aldehyde and alcohol dehydrogenases (pduP, Fe-ADH) were detected in metagenomes assembled from fluids produced from the Utica-Point Pleasant natural-gas well (Fig. 2c). However, not all Halanaerobium strains contain these genes (Supplementary Table 9), suggesting that this pathway may be unique to certain strains occupying specific niches in the HF stimulated shale ecosystem.
During HF operations, both labile carbon and recalcitrant xenobiotic carbon compounds are injected into the subsurface along with microbial cells. Guar gum and other polysaccharides are frequently disclosed in fracture fluid formulations, and are readily metabolized by microorganisms in produced fluids [11, 23, 25]. In particular, Halanaerobium is capable of cleaving the mannose and galactose monomers from guar gum, yielding galactose which can be converted to glucose-1-phosphate for use in glycolysis [23]. Guar gum transformation may therefore initiate the methylglyoxal bypass in a similar way to glucose, triggering the propanediol dehydratase pathway and resulting in chain shortening of surfactants in produced fluids. The production of carboxylic acids and alcohols associated with this co-metabolic pathway in Halanaerobium may provide bottom-up support to other shale-associated microorganisms [22]. Moreover, the biological depletion of injected surfactants in parallel with the accumulation of metabolites may lead to sub-optimal hydrocarbon extraction, advanced petroleum well corrosion, and an overall lower rate of return for the petroleum industry [35].
Conclusion
Specific classes of AEOs were rapidly attenuated during the first few months after HF of a natural-gas well drilled in the Utica-Point Pleasant Formation. Although this trend has been observed in other petroleum wells stimulated by HF, the mechanism(s) behind surfactant losses were not well understood. In this Utica-Point Pleasant natural-gas well, Halanaerobium dominated the microbial community structure within months, despite the decreasing concentrations of labile carbon and increasing salinity. An isolate from this natural-gas well (H. congolense WG10) transformed AEOs and PPGs in the presence of glucose, utilizing the methylglyoxal bypass during glycolysis to initiate chain-shortening enzymes. The co-metabolic pathway described here may be initiated by other polymeric compounds that enter glycolysis (e.g. guar gum), suggesting this pathway may inadvertently occur until polymers are completely transformed. In contrast to the surfactant types attenuated in field and laboratory findings (AEOs, PPGs, monomer glycols), PEGs were not degraded in the Utica-Point Pleasant natural-gas well produced fluids nor by H. congolense WG10, suggesting that PEGs may be useful tracers of injected hydraulic fracturing fluid in this system. Our findings highlight the versatility of the halotolerant bacterial taxa, Halanaerobium, to metabolize xenobiotic organic additives injected during HF stimulation of natural-gas wells.
Supplementary information
Acknowledgements
This research was supported by funding from the National Science Foundation Dimensions of Biodiversity (award no. 1830742). We express thanks to Mikayla Borton, Kelly Wrighton, and Rebecca Daly for guidance on genomic analyses, and Desiree Plata and Nathaniel Warner for helpful conversations on chemical mixing analysis. We are especially grateful for cooperation from our industry partner for site access and sample support. Genomic and metagenomic sequencing for this research was performed by the Department of Energy’s Joint Genome Institute (JGI) via a large-scale sequencing award (no. 1931). NMR metabolite support was provided by the Environmental Molecular Sciences Laboratory (EMSL) via a JGI-EMSL Collaborative Science Initiative (award no. 48483). LC-MS metabolite analysis using MetFish was performed at the Pacific Northwest National Laboratory (PNNL) through support from the Department of Energy’s Genomic Science Program via the Metabolic and Spatial Interactions in Communities Scientific Focus Area. The JGI, EMSL, and PNNL are sponsored by the Office of Biological and Environmental Research and operated under contracts DE-AC02-05CH11231 (JGI) and DE-AC05-76L01830 (EMSL and PNNL).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0466-0) contains supplementary material, which is available to authorized users.
References
- 1.Arthur JD, Bohm B, Layne M. Hydraulic fracturing considerations for natural-gas wells of the Marcellus Shale. 2009. http://archives.datapages.com/data/gcags_pdf/2009/arthetal.htm.
- 2.U.S. Energy Information Administration (EIA). The United States exported more natural-gas than it imported in 2017—Today in Energy. https://www.eia.gov/todayinenergy/detail.php?id=35392.
- 3.Vidic RD, Brantley SL, Vandenbossche JM, Yoxtheimer D, Abad JD. Impact of shale-gas development on regional water quality. Science. 2013;340:1235009. doi: 10.1126/science.1235009. [DOI] [PubMed] [Google Scholar]
- 4.Cluff MA, Hartsock A, MacRae JD, Carter K, Mouser PJ. Temporal changes in microbial ecology and geochemistry in produced water from hydraulically fractured Marcellus shale-gas wells. Environ Sci Technol. 2014;48:6508–17. doi: 10.1021/es501173p. [DOI] [PubMed] [Google Scholar]
- 5.Barbot E, Vidic NS, Gregory KB, Vidic RD. Spatial and temporal correlation of water quality parameters of produced waters from Devonian-age shale following hydraulic fracturing. Environ Sci Technol. 2013;47:2562–9. doi: 10.1021/es304638h. [DOI] [PubMed] [Google Scholar]
- 6.Haluszczak LO, Rose AW, Kump LR. Geochemical evaluation of flowback brine from Marcellus gas wells in Pennsylvania, USA. Applied Geochemistry. 2013;28:55–61. doi: 10.1016/j.apgeochem.2012.10.002. [DOI] [Google Scholar]
- 7.Orem W, Tatu C, Varonka M, Lerch H, Bates A, Engle M, et al. Organic substances in produced and formation water from unconventional natural-gas extraction in coal and shale. Int J Coal Geol. 2014;126:20–31. doi: 10.1016/j.coal.2014.01.003. [DOI] [Google Scholar]
- 8.Engle MA, Reyes FR, Varonka MS, Orem WH, Ma L, Ianno AJ, et al. Geochemistry of formation waters from the Wolfcamp and “Cline” shales: Insights into brine origin, reservoir connectivity, and fluid flow in the Permian Basin, USA. Chem Geol. 2016;425:76–92. doi: 10.1016/j.chemgeo.2016.01.025. [DOI] [Google Scholar]
- 9.Mouser PJ, Borton M, Darrah TH, Hartsock A, Wrighton KC. Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol Ecol. 2016;92:fiw166. doi: 10.1093/femsec/fiw166. [DOI] [PubMed] [Google Scholar]
- 10.Hanson AJ, Lipp JS, Daly RA, Sheets JM, Welch SA, Sharma S et al. Microbial membrane lipids show de novo ecosystem transformation in hydraulically fractured Marcellus Shale. In preparation.
- 11.Daly RA, Borton MA, Wilkins MJ, Hoyt DW, Kountz DJ, Wolfe RA, et al. Microbial metabolisms in a 2.5-km-deep ecosystem created by hydraulic fracturing in shales. Nature Microbiology. 2016;1:16146. doi: 10.1038/nmicrobiol.2016.146. [DOI] [PubMed] [Google Scholar]
- 12.Booker AE, Borton MA, Daly RA, Welch SA, Nicora CD, Hoyt DW, et al. Sulfide generation by dominant Halanaerobium microorganisms in hydraulically fractured shales. mSphere. 2017;2:pii: e00257–17. doi: 10.1128/mSphereDirect.00257-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipus D, Roy D, Khan E, Ross D, Vikram A, Gulliver D, et al. Microbial communities in Bakken region produced water. FEMS Microbiol Lett 2018; 365:fny107. [DOI] [PubMed]
- 14.Mohan AM, Hartsock A, Bibby KJ, Hammack RW, Vidic RD, Gregory KB. Microbial community changes in hydraulic fracturing fluids and produced water from shale-gas extraction. Environ Sci Technol. 2013;47:13141–50. doi: 10.1021/es402928b. [DOI] [PubMed] [Google Scholar]
- 15.Struchtemeyer CG, Elshahed MS. Bacterial communities associated with hydraulic fracturing fluids in thermogenic natural-gas wells in North Central Texas, USA. FEMS Microbiol Ecol. 2012;81:13–25. doi: 10.1111/j.1574-6941.2011.01196.x. [DOI] [PubMed] [Google Scholar]
- 16.An BA, Shen Y, Voordouw G. Control of sulfide production in high salinity Bakken Shale oil reservoirs by halophilic bacteria reducing nitrate to nitrite. Front Microbiol. 2017;8:1164. doi: 10.3389/fmicb.2017.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akob DM, Cozzarelli IM, Dunlap DS, Rowan EL, Lorah MM. Organic and inorganic composition and microbiology of produced waters from Pennsylvania shale-gas wells. Appl Geochem. 2015;60:116–25. doi: 10.1016/j.apgeochem.2015.04.011. [DOI] [Google Scholar]
- 18.Evans MV, Panescu J, Hanson AJ, Welch SA, Sheets JM, Nastasi N, et al. Members of Marinobacter and Arcobacter influence system biogeochemistry during early production of hydraulically fractured natural-gas wells in the Appalachian Basin. Front Microbiol. 2018;9:2646. doi: 10.3389/fmicb.2018.02646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman EC, Capo RC, Stewart BW, Kirby CS, Hammack RW, Schroeder KT, et al. Geochemical and Strontium isotope characterization of produced waters from Marcellus Shale natural-gas extraction. Environ Sci Technol. 2012;46:3545–53. doi: 10.1021/es204005g. [DOI] [PubMed] [Google Scholar]
- 20.Warner NR, Christie CA, Jackson RB, Vengosh A. Impacts of shale-gas wastewater disposal on water quality in Western Pennsylvania. Environ Sci Technol. 2013;47:11849–57. doi: 10.1021/es402165b. [DOI] [PubMed] [Google Scholar]
- 21.Vengosh A, Kondash A, Harkness J, Lauer N, Warner N, Darrah TH. The geochemistry of hydraulic fracturing fluids. Procedia Earth and Planetary Science. 2017;17:21–24. doi: 10.1016/j.proeps.2016.12.011. [DOI] [Google Scholar]
- 22.Borton MA, Hoyt DW, Roux S, Daly RA, Welch SA, Nicora CD, et al. Coupled laboratory and field investigations resolve microbial interactions that underpin persistence in hydraulically fractured shales. PNAS. 2018;115:E6585. doi: 10.1073/pnas.1800155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang R, Davidova IA, Marks CR, Stamps BW, Harriman BH, Stevenson BS, et al. Metabolic capability of a predominant Halanaerobium sp. in hydraulically fractured gas wells and its implication in pipeline corrosion. Front Microbiol. 2016;7:988. doi: 10.3389/fmicb.2016.00988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipus D, Vikram A, Ross D, Bain D, Gulliver D, Hammack R, et al. Predominance and metabolic potential of Halanaerobium in produced water from hydraulically fractured Marcellus Shale wells. Appl Environ Microbiol. 2017;83:AEM.02659–16. doi: 10.1128/AEM.02659-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elsner M, Hoelzer K. Quantitative survey and structural classification of hydraulic fracturing chemicals reported in unconventional gas production. Environ Sci Technol. 2016;50:3290–314. doi: 10.1021/acs.est.5b02818. [DOI] [PubMed] [Google Scholar]
- 26.Ahrenst A, Lungwitz B, Fredd CN, Abad C, Gurmen N, Chen Y et al. Friction reduction fluids. 2015.
- 27.Rogers JD, Burke TL, Osborn SG, Ryan JN. A framework for identifying organic compounds of concern in hydraulic fracturing fluids based on their mobility and persistence in groundwater. Environ Sci Technol Lett. 2015;2:158–64. doi: 10.1021/acs.estlett.5b00090. [DOI] [Google Scholar]
- 28.Rosenblum J, Thurman EM, Ferrer I, Aiken G, Linden KG. Organic chemical characterization and mass balance of a hydraulically fractured well: from fracturing fluid to produced water over 405 days. Environ Sci Technol. 2017;51:14006–15. doi: 10.1021/acs.est.7b03362. [DOI] [PubMed] [Google Scholar]
- 29.Thurman EM, Ferrer I, Blotevogel J, Borch T. Analysis of hydraulic fracturing flowback and produced waters using accurate mass: Identification of ethoxylated surfactants. Anal Chem. 2014;86:9653–61. doi: 10.1021/ac502163k. [DOI] [PubMed] [Google Scholar]
- 30.Nell M, Helbling DE. Exploring matrix effects and quantifying organic additives in hydraulic fracturing associated fluids using liquid chromatography electrospray ionization mass spectrometry. Environ Sci: Processes Impacts. 2018;21:195–205. doi: 10.1039/c8em00135a. [DOI] [PubMed] [Google Scholar]
- 31.Huber M, Meyer U, Rys P. Biodegradation mechanisms of linear alcohol ethoxylates under anaerobic conditions. Environ Sci Technol. 2000;34:1737–41. doi: 10.1021/es9903680. [DOI] [Google Scholar]
- 32.Wagener S, Schink B. Fermentative degradation of nonionic surfactants and polyethylene glycol by enrichment cultures and by pure cultures of homoacetogenic and propionate-forming bacteria. Appl Environ Microbiol. 1988;54:561–5. doi: 10.1128/aem.54.2.561-565.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyob KM, Blotevogel J, Brooker M, Evans MV, Lenhart JJ, Wright J, et al. Natural attenuation of nonionic surfactants used in hydraulic fracturing fluids: degradation rates, pathways, and mechanisms. Environ Sci Technol. 2017;51:13985–94. doi: 10.1021/acs.est.7b01539. [DOI] [PubMed] [Google Scholar]
- 34.Rogers JD, Thurman EM, Ferrer I, Rosenblum JS, Evans MV, Mouser PJ, et al. Degradation of polyethylene glycols and polypropylene glycols in microcosms simulating a spill of produced water in shallow groundwater. Environ Sci: Processes Impacts. 2018;21:256–68. doi: 10.1039/c8em00291f. [DOI] [PubMed] [Google Scholar]
- 35.Fichter J, Wunch K, Moore R, Summer E, Braman S, Holmes P. How hot is too hot for bacteria? A technical study assessing bacterial establishment in downhole drilling, fracturing and stimulation operations. NACE International, Houston, TX. 2012;1310.
- 36.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6:359. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 37.Kim S, Pevzner PA. MS-GF makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, et al. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. PNAS. 2006;103:18928. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Getzinger GJ, O’Connor MP, Hoelzer K, Drollette BD, Karatum O, Deshusses MA, et al. Natural-gas residual fluids: Sources, endpoints, and organic chemical composition after centralized waste treatment in Pennsylvania. Environ Sci Technol. 2015;49:8347–55. doi: 10.1021/acs.est.5b00471. [DOI] [PubMed] [Google Scholar]
- 40.Hanson AJ, Luek JL, Tummings SS, McLaughlin MC, Blotevogel J, Mouser PJ. High total dissolved solids in shale-gas wastewater inhibit biodegradation of alkyl and nonylphenol ethoxylate surfactants. Sci Total Environ. 2019;668:1094–103. doi: 10.1016/j.scitotenv.2019.03.041. [DOI] [PubMed] [Google Scholar]
- 41.McLaughlin MC, Borch T, Blotevogel J. Spills of hydraulic fracturing chemicals on agricultural topsoil: biodegradation, sorption, and co-contaminant interactions. Environ Sci Technol. 2016;50:6071–8. doi: 10.1021/acs.est.6b00240. [DOI] [PubMed] [Google Scholar]
- 42.Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Tennessen K, et al. The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v.4) Standards in Genom Sci. 2016;11:17. doi: 10.1186/s40793-016-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen I-MA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–16. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 45.Bland RQ. Quality criteria in selecting glycols as alternatives to oil-based drilling fluid systems. SPE Health, Safety and Environment in Oil and Gas Exploration and Production Conference, 25-27 January, Jakarta, Indonesia. Society of Petroleum Engineers. 1994.
- 46.Trifunović D, Schuchmann K, Müller V. Ethylene glycol metabolism in the acetogen Acetobacterium woodii. J Bacteriol. 2016;198:1058–65. doi: 10.1128/JB.00942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol. 1999;181:5967–75. doi: 10.1128/jb.181.19.5967-5975.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Booker AE, Hoyt DW, Meulia T, Eder E, Nicora CD, Purvine SO, et al. Deep subsurface pressure stimulates metabolic plasticity in shale-colonizing Halanaerobium. Appl Environ Microbiol. 2019;85:AEM.00018–19. doi: 10.1128/AEM.00018-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuchmann K, Schmidt S, Lopez AM, Kaberline C, Kuhns M, Lorenzen W, et al. Nonacetogenic growth of the acetogen Acetobacterium woodii on 1,2-propanediol. J Bacteriol. 2015;197:382–91. doi: 10.1128/JB.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oren A. Halanaerobium. In: Whitman WB (ed). Bergey’s Manual of Systematics of Archaea and Bacteria. 2015. John Wiley & Sons, Inc. in association with Bergey's Manual Trust, Hoboken, NJ. p. 1–8.
- 51.Daly RA, Roux S, Borton MA, Morgan DM, Johnston MD, Booker AE, et al. Viruses control dominant bacteria colonizing the terrestrial deep biosphere after hydraulic fracturing. Nature Microbiology. 2018;4:352–61. doi: 10.1038/s41564-018-0312-6. [DOI] [PubMed] [Google Scholar]
- 52.Xu C, Couvillion SP, Sontag RL, Isern NG, Maezato Y, Lindemann SR, et al. MetFish: a metabolomics platform for studying microbial communities in chemically extreme environments. bioRxiv 2019; 518647. [DOI] [PMC free article] [PubMed]
- 53.Booth IR, Ferguson GP, Miller S, Li C, Gunasekera B, Kinghorn S. Bacterial production of methylglyoxal: a survival strategy or death by misadventure? Biochem SocTrans. 2003;31:1406–8. doi: 10.1042/bst0311406. [DOI] [PubMed] [Google Scholar]
- 54.Chandrangsu P, Dusi R, Hamilton CJ, Helmann JD. Methylglyoxal resistance in Bacillus subtilis: Contributions of bacillithiol-dependent and independent pathways. Mol Microbiol. 2014;91:706–15. doi: 10.1111/mmi.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tran-Din K, Gottschalk G. Formation of d(−)-1,2-propanediol and d(−)-lactate from glucose by Clostridium sphenoides under phosphate limitation. Arch Microbiol. 1985;142:87–92. doi: 10.1007/BF00409243. [DOI] [Google Scholar]
- 56.Cameron DC, Cooney CL. A novel fermentation: the production of R(–)–1,2–propanediol and acetol by Clostridium thermosaccharolyticum. Bio/Technology. 1986;4:651. [Google Scholar]
- 57.Mösche M. Anaerobic degradability of alcohol ethoxylates and related non-ionic surfactants. Biodegradation. 2004;15:327–36. doi: 10.1023/B:BIOD.0000042188.10331.61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.