Abstract
Antibiotic resistance is a rapidly growing health care problem globally and causes many illnesses and deaths. Bacteria can acquire antibiotic resistance genes (ARGs) by horizontal transfer mediated by mobile genetic elements, where the role of phages in their dissemination in natural environments has not yet been clearly resolved. From metagenomic studies, we showed that the mean proportion of predicted ARGs found in prophages (0–0.0028%) was lower than those present in the free viruses (0.001–0.1%). Beta-lactamase, from viruses in the swine gut, represented 0.10 % of the predicted genes. Overall, in the environment, the ARG distribution associated with viruses was strongly linked to human activity, and the low dN/dS ratio observed advocated for a negative selection of the ARGs harbored by the viruses. Our network approach showed that viruses were linked to putative pathogens (Enterobacterales and vibrionaceae) and were considered key vehicles in ARG transfer, similar to plasmids. Therefore, these ARGs could then be disseminated at larger temporal and spatial scales than those included in the bacterial genomes, allowing for time-delayed genetic exchanges.
Subject terms: Metagenomics, Antimicrobials
Introduction
Globally, antibiotic resistance is a rapidly growing health care problem. The World Health Organization estimated that, in 2010, foodborne illnesses affected 600 million people and caused 420,000 deaths globally [1]. Some bacteria display intrinsic resistance [2]. In others, resistance is acquired by mutations in different chromosomal loci or by the horizontal acquisition of antibiotic resistance genes (ARGs), which is mediated by mobile genetic elements (MGEs). The majority of MGEs, such as plasmids, genomic islands, transposons, and integrative conjugative elements, are transferred through cell-cell contact by a conjugation mechanism [3]. Other mechanisms do not require cell contact between microorganisms, but the persistence of the DNA in the environment is then critical. Thus, DNA transformation is unlikely a reason for the ARG transfer, and the most suitable vehicle for the transfer between noncontiguous cells could be phages or, more generally, all vehicles protecting the nucleic acids as gene transfer agents or vesicles [4]. Viruses are the most abundant biological entities on earth, with an estimated abundance ranging from 109 to 1010 per liter of seawater (e.g., [5]) and from 108 to 109 per gram of human feces [6]. In addition, some studies show that in certain environments, the transduction frequencies are several orders of magnitude greater than what was previously thought [7, 8]. Phages may, therefore, act as vectors for genetic exchange via generalized or specialized transduction. In the first mechanism, some host DNA is erroneously packaged in the capsid, whereas in the second phenomenon, the DNA prophage is excised with a small part of the host chromosome. An important characteristic of transduction is that gene transfer does not require that the donor and recipient bacteria be present in the same biome at the same time. In addition, phages can survive in the environment for long periods of time, allowing for a time-delayed transfer of genetic information [9].
The acquisition of antimicrobial resistance by transduction has already been demonstrated in clinically relevant bacterial species. For example, prophages of Staphylococcus aureus are believed to be responsible for the spread of some antibiotic resistance genes [10]. Of the 243 coliphages, 24.7% are able to transduce one or more antibiotic genes, encoding for ampicillin, tetracycline, kanamycin and chloramphenicol, to the laboratory strain Escherichia coli ATCC 13706 [11]. The transfer of the ampicillin resistance gene between E. coli cells is done at a surprisingly high frequency (ranging between 10−4 and 10−3) [7]. Finally, phage DNA may constitute 20% of the bacterial genomes, and some cryptic forms help bacteria (E. coli) to resist sublethal concentrations of antibiotics or, more generally, to resist various stresses [12]. The importance of generalized or specialized transductions are, therefore, rather well described for foodborne bacteria, mainly among the Gammaproteobacteria. In this regard, the role of phages in the dissemination of antibiotic resistance genes among bacterial hosts in natural environments has not yet been clearly resolved, since the results seem conflicting. Surprisingly, the relative abundance of ARGs in the phage DNA fraction (0.26%) was higher than in the bacterial DNA fraction (0.18%) [13]. However, by qPCR, higher copy numbers of ARGs were detected in the bacterial DNA fraction than in the phage fraction [14]. Another example of the intense debate within the scientific community about this topic is the new analysis [15] of the metagenomics results obtained by Modi et al. [16]. In the original paper, the authors shed light on the fact that antibiotic treatment leads to the enrichment of phage-encoded genes that confer resistance to the antibiotics. However, the ARG detection from the reads is challenging, and false results can be obtained by using too relaxed or explanatory thresholds for the sequence analysis. A more stringent analysis of the proteins in contigs does not allow for the detection of ARGs among dominant viruses. Finally, if the viromes built by Modi and collaborators were not contaminated by cellular components, any ARG enrichment can be evidenced, since the percentage of ARGs was correlated with the gene content found in bacteria, suggesting, rather, a generalized transduction. The metagenomics approach represents, therefore, the standard method for studying the gene contents of viruses that cannot be isolated without their host. However, ARG detection is very sensitive to the following: (i) the thresholds used to the similarity search among the public databases; (ii) the kind of data (short reads vs contigs), and (iii) the reference databases [17].
To the best of our knowledge, ARG detection in the viruses from various environments have mainly been realized from short reads [13, 18, 19], and their importance has to be confirmed by a more robust approach, namely, assembling and protein affiliation with stringent thresholds against a curated database, as recommended [15, 17]. In this paper, we (i) analyzed the virome data generated by high-throughput sequencing and (ii) compared the role of the viruses to plasmids as ARG vehicles in the biomes by a network approach. This work allows therefore to decipher the role of viruses in the dissemination of the ARGs in environments compared to plasmids and could contribute to limit the spread of such resistances in the future.
Results
ARGs predicted in free viruses and prophages
The predicted ARGs in the virus genomes (free and prophages) represented 0.02% of the total predicted genes (Fig. 1). Surprisingly, the mean proportions of the predicted ARGs found in prophages (0–0.0028%) were lower than those present in the free viruses (0.001–0.1%) (P < 0.001, Chi-squared test). The prophages were certainly under-sampled compared to the free viruses. The genomes of the viruses from the swine guts integrated the most ARGs, with a value of 0.10%. These genes were also well represented in the viruses inhabiting oceans, freshwater ecosystems and human guts.
Fig. 1.

Antibiotic Resistance Genes predicted in the viromes (i.e., viruses) and microbiomes (i.e., prophages) expressed in percentage of the genes predicted
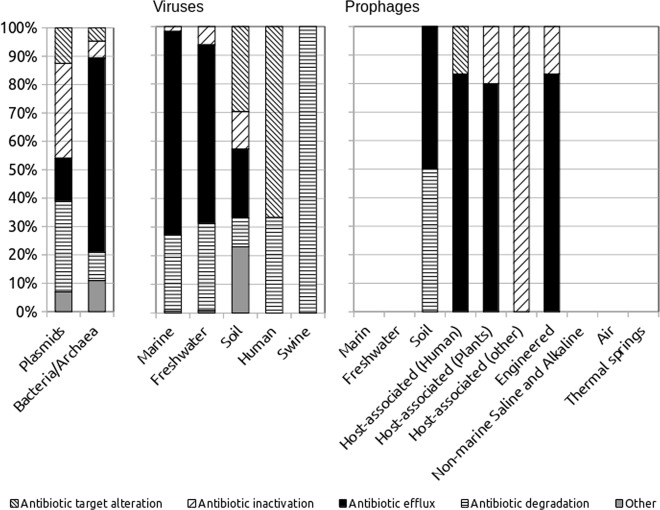
The resistance mechanisms differed greatly between the vehicles analyzed, including bacteria, archaea, plasmids and viruses (Fig. 2). The greatest richnesses in the ARGs were detected in the chromosomes, plasmids and soil viruses. Antibiotic efflux seemed, therefore, the main mechanism in the viruses, with the exception of the gut microbiota, where only antibiotic degradation and target alterations were found. The swine gut consisted only of genes coding for Beta-lactamase.
Fig. 2.
Mechanisms of the resistance to the antibiotics detected in the environments for the viruses (free and prophages) and the prophages
Interactions between microorganisms and viruses inferred from networks
To decipher the putative gene transfer between the vehicles, two networks were built. The first, a bipartite network (Supplementary Information Fig. S1a), allowed us to discriminate the main associations within and between the genomics units (GUs) defined as bacteria, archaea, plasmids and viruses in the various environments. These GUs were linked by protein clusters of ARGs named Homologous protein Clusters (HpCs). An HpC, including at least 2 ARGs could be then linked to another GU or within the GU by an edge. There were few edges (i.e., HpC) between archaea and bacteria unlike with the marine viruses, showing that both domains did not share many ARGs, whereas viruses shared numerous ARGs with bacteria. The second network (Fig. 3) was built from the first but with various distances (from proteins, genes and phylogenies in a same HpC) allowing us (i) to identify the vehicles linking the GUs at a finer level and (ii) to select the best vehicle involved in the interaction (i.e., ARG transfer). The best vehicles were defined as those with the lowest evolutionary distance. The most interesting results were the associations between the GUs, including viruses, plasmids and bacteria/archaea. Indeed, a protein cluster within a GU of viruses no make sense here, since (i) viruses from different species can share a same ARGs, because they infected a same host or (ii) an edge between viruses can also correspond to a cluster consisting of closed viruses.
Fig. 3.
a Molecular network built with the best vehicles of the ARGs inferred from the gene phylogenies (patristic distances). b Main network topological indices computed from the network
The first network was built with 15937 HpCs, with a strong identity between each other (Supplementary Information Fig. S1b), since the median value was 99% and more specifically 99.3% by taking account only viruses and prophages. A total of 403 of these HpCs allowed us to define an edge between at least 2 GUs and at the most 7 GUs. The bacteria (no archaea) were involved in all the edges defined. From these 403 edges, 210 involved plasmids and almost the same number (205) of viruses and prophages. The most important viruses in this network were those sampled from the oceans and freshwater ecosystems with 160 and 29 edges with bacteria, respectively.
From the HpC described, patristic distances (branch lengths) from the phylogenies were computed and a new network was built allowing to visualize the interactions between bacteria and putative vehicles consisting in 40 plasmids and 180 viruses and prophages (Fig. 3a). Finally, this network recruited significantly more viruses than plasmids (P < 0.001, Chi-squared test). As expected plasmids appeared as central in this network but also viruses. Both of the indices, betweenness and closeness, that can measure this centrality, were used to determine the keystone nodes and, therefore, the main vehicles involved in the ARG flux (Fig. 3). A high closeness meant that the node was near all other nodes and had a central position in the network, and a high betweenness allowed us to detect the nodes that acted as bridges between the nodes or modules. Overall, the viruses detected in the viromes, more numerous in this network, had the highest closeness and betweenness values among the mobile genetic elements (Fig. 3b, Supplementary Information Fig S2). Nevertheless, the statistical tests (Table 1) show that among the vehicles these indices were rather similar with slightly differences. For example, the betweenness computed from plasmids were significantly different from marine viruses but not significant with freshwater viruses.
Table 1.
Various metrics inferred from the second network built (Fig. 3), with distances computed from the phylogenies (patristic distances) for each HpC
| Bacteria | Plasmids | Freshwater viruses | Marine viruses | Prophages | Pa | |
|---|---|---|---|---|---|---|
| Betweenness | 0.040 | 0.487bc | 0.342ab | 0.286a | 0.725c | < 0.001 |
| Closeness | 0.508 | 0.721bde | 0.687abc | 0.641ad | 0.843ce | < 0.001 |
| Neighbors | 1.716 | 4.875a | 4.450ab | 4.221a | 3.141a | < 0.001 |
Because the low abundances in the second network, prophages were grouped together and swine viruses were not take account in this statistical test
aANOVA one way (ddl: 4, 748)
Tukey test: values with the same subscripts (a, b c, d, e) did not differ significantly (P > 0.05)
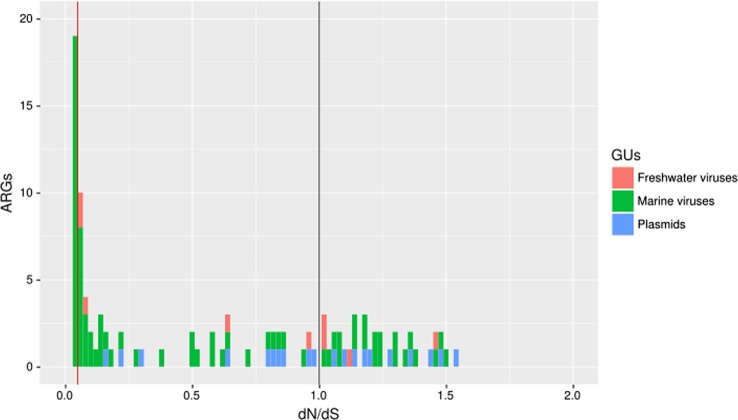
From the ARGs linking the viruses or plasmids to bacteria, the dN/dS ratios were computed for estimating the ratio between the nonsynonymous and synonymous substitutions by using the bacteria in each cluster (HpC) as a gene reference (Fig. 4). The median values for the plasmids, freshwater and marine viruses were 0.99, 0.03, and 0.05, respectively. Few prophages were found in this second network, but interestingly the prophages that originated from the human gut and human engineering had a low dN/dS ratio (0.02).
Fig. 4.
Selective pressure acting on the ARGs included in the second network (Fig. 3) evaluated by the dN/dS ratio
Geographical distribution of the ARGs
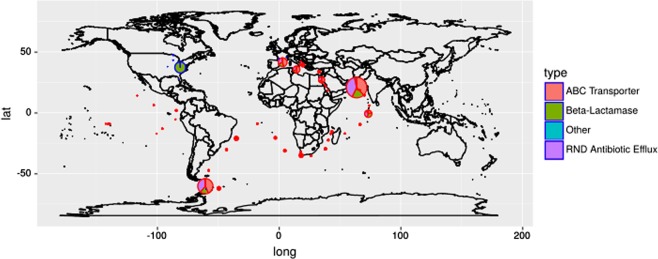
The results show therefore the importance of the aquatic viruses in the ARG dissemination. These ecosystems can considered by their watershed as integrating all the human activities and ultimately, it reflects the pollution. Thus, the study of the spatial distribution of ARGs in this part was focused to aquatic environments. In Fig. 5, the main ARG categories are displayed in the map, and the pie size is proportional to the quantity of ARGs (i.e., bases mapped against genes) among the ecosystem considered (ocean or lakes). In the few lakes studied, the ARGs, mostly represented by Beta-lactamase, were mainly found in the eastern part of the USA. From the TARA-Ocean experiments [20], “ABC Transporter”, “Gene Modulating Resistance” and “RND Antibiotic Efflux” were significantly different (P < 0.05) between biomes. Thus, a more precise geographical distribution was generated, and the ARGs were less numerous in the open ocean than along the coasts. More precisely, the ARGs in viruses were the most abundant in the close seas (Mediterranean and Red Seas) and the Indian coast. Surprisingly, an ARG spot was also detected in the southern ocean close to the Cape Horn passage and far away from dense populations.
Fig. 5.
Relative importance of the ARGs and the main resistance mechanisms in the aquatic ecosystems in the earth. The pie size is proportional to the reads mapped on the viral contigs for each environment: oceans and lakes
Taxonomies of the viruses involved in the ARG flux
After sampling down the sequences (i.e. 1000 contigs) among the various GUs defined (total viruses or prophages inhabited various environments) for avoiding sampling bias, the proteomic trees generated by VipTree evidenced that the taxonomy of GUs were significantly different (ANOSIM, P = 0.001). The prophages, whatever the environments, were also significantly different from the free viruses (ANOSIM, P = 0.001).
More precisely, among the viruses involved in the ARG fluxes, some remained mainly unclassified because no landmark viruses or cellular genes were found in the contig (Fig. 6). This category remained rather weak (15%), and two categories dominated the viral community, including caudovirales and Leptospira phages. Surprisingly, the taxonomy of the ocean viruses (Supplementary Information Fig. S3) was quite similar with the freshwater one. This result sheds light on the fact that the close viruses harbored ARGs in their genomes in both ecosystems or that this taxonomy reflected the paucity of the virus databases. However, the results obtained from the proteomic trees showed no significant differences (ANOSIM, P = 0.23) between the viral communities (Supplementary Information Fig. S4) harboring ARGs in their genomes. Likely, the first hypothesis should be retained.
Fig. 6.

Taxonomies of the vehicles of the ARGs, bacteria (a) and viruses (b), present in the second network displayed in Fig. 3
In the network, these viruses interacted with bacteria that were represented mostly by Gammaproteobacteria and Alphaproteobacteria (Fig. 6). Enterobacterales and vibrionaceae, within which human pathogens are found, accounted for 6% and 0.7% of the bacterial community, respectively.
Discussion
Viruses are known as the most abundant and diverse biological entities on earth, and their main role in ecosystems was identified in the first time as the microbial population regulation. The innovation in microbial studies though “omic” approaches allows us to now decipher intriguing virus-host interactions in the environment, such as the auxiliary metabolic genes or HGT [21]. However, the study of viruses is technical challenged, the abundance may be overestimated in the environment [22] and the gene content may be biased by cellular contaminants [23]. In addition, the gene annotation, and more particularly the ARG annotation, is sensitive to the database and bioinformatic procedures used [17]. Finally, this reanalysis of the Modi’s study [16] allowed to define the major pitfalls in ARG identification [15]. The bioinformatic pipeline used in this study (briefly gene annotation from contigs with RESFAM) represents certainly the most accurate procedure right now. However, similar to microbial annotation, the ARG activity cannot be deduced, undoubtedly, since changes of a few amino acids in a gene can alter its substrate preference or binding site. However, there was a strong identity between the microbial and virus proteins within an HpC, and the low dN/dS ratio, found in this study advocate for a negative selection (i.e. amino acid sequences not modified) of the ARGs detected in the virus genomes and likely a preservation of the function.
Our study allowed us to confirm part of the results found by Lekunberri et al. [19] from metagenomic reads analysis. Most of the viromes harbored ARGs, and the pig sewage showed the highest relative abundance dominated by a resistance mechanism, namely Beta-lactamase. However, our work sheds light on the fact that a lower relative abundance of the ARGs was estimated compared to the maximum at ~0.45% in the rare previous studies [18, 19]. For the reasons given above (i.e., bioinformatic procedures), our estimation is certainly more accurate. Intriguingly, the ARGs were dominant within the pig sewage but not in the human feces, while the antibiotic pressure was also strong. As underlined by Colombo et al. [24], ARGs may, therefore, be mobilized even in the absence of antibiotic treatment in some environments. To support this hypothesis, [25] also found a high proportion of ARGs in a pristine pond of the Mauritanian Sahara. In addition, the microbial genes mobilized in the genome could be the result of past transfer events rather than a picture of the current microbial diversity. For example, ARGs were evidenced in viromes from fossilized fecal material from the 14th century [26]. However, our study on the geographical distribution of the ARGs in the aquatic ecosystem, which are considered through the watershed as a summarize of the human activities, showed that, globally, the hot spot of the ARGs in the viromes corresponded to the most anthropized systems and/or a closed sea (Mediterranean sea). In contrast, viruses from open oceans included few ARGs in their genomes, with the exception of one sample close to Cap Horn. Overall, in the environment, the ARG distribution associated with the viruses seemed to be strongly linked to human activity.
Another intriguing result was the lower proportion of ARGs in the prophages compared to the free viruses, since from the reference genomes [27] concluded that the ARGs were 10-fold less abundant in phages than in prophages. These statistics were determined from the RefSeq database [28], which is known to be enriched in isolates from agriculture and medicine fields. On the other hand, (i) our results were partly biased by the sampling effort, since the proteins predicted from the viruses were therefore approximately four times more abundant than those analyzed from prophages (Supplementary Information Table S1), and (ii) the cellular contaminants might still be present in the viromes despite the precautions taken to exclude such contigs. This last aspect should be minimal since we reanalyzed entirely or checked the data already appraised when the contigs were available. In addition, the viromes originated from the various sources, minimizing the methodological bias, and this conclusion was true for each ecosystem, with the exception of the soil. Since the specialized transduction is associated with a lysogenic cycle (prophage), we then considered, in a first approximation, that this mechanism was a minority compared to the general transduction. This mechanism is indeed evidenced, for example, in freshwater ecosystems [29]. Enault et al. [15] hypothesized that the main factor explaining ARG increases is that the antibiotic-treatment-inducing prophages, with some subset, performed a generalized transduction. Nevertheless, generalized transducing particles completely lack DNA originating from the viral vector, containing instead only bacterial sequences. With the exception of the unclassified viruses, the virus contig harbored viral genes, and we excluded general transduction as the main mechanism for transferring the ARGs. Finally, the free-reference approach (i.e., VipTree) highlighted a significant difference between the viruses from viromes and prophages. Thus, the few studies on the gene contents from prophages from various environments may be the best explanation for understanding the low proportion of ARGs in their genomes.
The presence of the ARGs in the virus fraction was likely the result of a specialized transduction. These transduction events have been quantified in a few ecosystems. In the aquatic ecosystems, they vary from 0.3 × 10−3 transductants/plaque forming unit in freshwater ecosystems [29] to 5.33 × 10−9 in oceans [30]. These events are more frequent than expected when the methodology takes into account the noncultivable and cultivable bacteria [29]. Nevertheless, the presence of ARGs in metagenomes does not directly represent a risk for human health [31], and the gene transfer toward the pathogens is not straightforward to show. We, therefore, choose to compare these data with plasmids that are considered a reference vehicle for HGT and more particularly conferring some resistance/virulence factors to the bacteria [32]. This comparison was conducted mainly by combining both network approaches [33, 34]. Our network study showed that viruses are considered key vehicles in the ARG transfer similar to plasmids. Remarkably, this conclusion was drawn with the plasmids sequences that originated mainly from environments enriched in ARGs (i.e. medical and agricultural domains). In addition, they were linked to putative pathogens (Enterobacterales and vibrionaceae). From their study, Halary et al. [33] concluded that phages displayed lower betweenness centralities than plasmids and were on the periphery of the network, and thus, demonstrated that plasmids, not viruses, were key vectors of genetic exchange. However, the Halary’s study did not focus on the ARG transfer, and the network was built only with the DNA similarity between the vehicles (bacteria, plasmids and viromes). From a study based on a phylogenomic network between bacteria and phages, Popa et al. [35] revealed limited HGT events by transduction but highlighted transfer events of genes coding for a broad range of antibiotic resistance, demonstrating a putative role of phages in the spread of these resistances. Interestingly, this study was restricted to the reference genomes (bacteria and prophages) found in the RefSeq database, whereas our conclusions were drawn from a larger sampling of the viruses, not restricted to the prophages, and inhabited various environments. In addition, Popa et al. [35] showed that the barriers for gene transfer via transduction were primarily genetic, since the integration of the acquired DNA into the recipient genome was mediated by homologous recombination and, therefore, depended on the sequence similarity between the donor and recipient. In contrast, the ecological barriers played a minor role compared to the genetic recombination. However, a prophage, including a gene encoding for tetracycline resistance, was linked to a bacteria (Bacillus cereus) and an archaea (Methanobrevibacter smithii), shedding light on a transduction at the interdomain level [35]. Nevertheless, beyond the phylogenetic analysis, some experiments show that DNA exchange among bacteria via phage may occur in a more divergent range of bacteria than previously thought using cultural methods [29]. In addition, a significant proportion of the transferred genes (>20%) remain in viable recipient cells. Finally, these studies and the ours demonstrate that viruses are, therefore, a possible vehicle for ARGs at large temporal and spatial scales (i.e., biomes), and they transfer them between noncontiguous cells. These transfers could be directly involved in foodborn pathogens or indirectly because of the host specificity of the viruses. The transduction could be involved a first step, namely, specific bacteria in the environment (ocean, river or soil), and in a second step the ARG could be transferred by conjugation toward commensal bacteria and/or pathogens. In this model, the body waters, such as lakes or rivers, are considered hot-spots of the HGT [1].
The beneficial contribution of phage-mediated gene transfer to the host fitness has been documented in diverse environments as, for instance, the presence in the cyanophage genomes of the genes coding for components of the photosystems I and II (reviewed in [36]). There is now some evidence on the role of viruses in the dissemination of the antibiotic resistance by transduction, therefore requiring a selective pressure to maintain such genes in the phage (lytic or temperate) genomes. These genes have certainly the potential to be beneficial for the bacteria. Thus, the MazE/F toxin–antitoxin system encoded by prophages increases the persistence of Escherichia coli under antibiotic stress [37] or contributes significantly to the resistance to sublethal concentrations of some antibiotics [12].
Conclusion
This work contributes to deciphering the putative role of viruses as vehicles of the ARGs, whose dissemination represents a health care problem at the worldwide scale. These ARGs included in the viral genomes can be then disseminated at a larger temporal and spatial scales than those in bacterial genomes (included in the chromosomes or plasmids). This property can be correspond to the process of “gene externalization” predicted by Corel et al. [38]. This process is of sharing between chromosomes and extrachromosomal elements (plasmids, viruses). Understanding the prevalence, mechanisms and spread of such resistances are priorities from a heath perspective. However, in a first step, the ARG flux between bacteria, mainly the pathogens, and viruses must be quantified and the functionality of these ARGs assessed. If future studies confirmed the threat for the human health of such HGT in the environments, the elimination of viruses harboring ARGs will become a major challenge since they are known to persist more than bacteria after, for example, disinfection procedures in wastewaters from urban areas [39].
Methods
Data
The protein sequences of 32,188 bacteria and 535 archaea (without plasmids and phage protein sequences) and the plasmid protein sequences were downloaded from the NCBI RefSeq Protein Database (ftp://ftp.ncbi.nlm.nih.gov/refseq/release/bacteria/ and ftp://ftp.ncbi.nlm.nih.gov/refseq/release/archaea/, version 07/2017 - ftp://ftp.ncbi.nlm.nih.gov/refseq/release/plasmid/, version 03/2017).
For the viromes, the contigs (assembled data) or reads were downloaded from public databases. When only the reads were available, the assemblages were conducted with IDBA [40], with a k-mer size from 20 to 120 and a step of 20. The viromes corresponded to the sequencing of the viral DNA from various microbiotes and the data from prophages [41] (Supplementary Information Table S1). In this work, we called “virus”, the data from the assembly of the viromes (including of course the free state of the prophages) and prophages the public data contained based on the work by [41]. The protein sequences were predicted using the MetaGeneAnnotator tool [42]. The workflow for analyzing the contigs generated by the assembling is described thereafter, with the exception for the viromes from the freshwater ecosystems, where specific steps were processed to deal with multiple papers on this topic (Supplementary Information Fig. S5). The distribution of the contigs length therefore available for subsequent analysis are displayed in the supplementary information part (Supplementary Information Fig. S6).
Taxonomic affiliation
The contigs were checked for removing the DNA from cellular origin [23]. The predicted proteins are aligned using the BLAST + tool [43] (e-value = 10−5) on a viral basis (database UniProtKB reduced to viral proteins) and on a “cellular” protein base (protein bases of bacteria, archaea and eukaryotes built from nonredundant UniProtKB: UNIREF100) [44]. Viral contigs were aligned using the BLASTn tool (e-value = 10−5) against the SILVA database [45], including the 16S/18S SSU rRNA and the 23S/28S rRNA. The presence of ribosomal RNA was confirmed if the length of the alignment was greater than 1200 bp or if the alignment was greater than 300 bp when the alignment was at one end of the contig. A contig was considered to be viral if the following criteria were met: (i) the absence of ribosomal RNA; (ii) not more than two proteins were affiliated with the “cellular organisms” base (protein databases of Bacteria, Archaea and eukaryotes), and (iii) the presence of viral proteins [23, 46]. If a contig fulfilled the first two conditions but had no alignment in the virus database, it was classified as an unclassified virus. The taxonomy of the viruses was deduced from an LCA (lowest common ancestor) analysis on, at most, the five best protein alignments of a contig on the viral protein base described above. A free-reference approach was used for assessing the distance between the contigs with VipTree [47]. This procedure was based on the normalized tBLASTx scores computed from the pairwise comparisons. A principal coordinate analysis (PcoA) and the statistical tests (ANalysis Of SIMilarity or ANOSIM) were computed from the distance matrix generated by VipTree with the vegan package [48] under the R environment [49].
Identification and quantification of the genes encoding antibiotic resistance
The predicted protein sequences were aligned using the HMMs (Hidden Markov Models) profiles based on the Resfams data [50] (version 1.2). The core Resfam consisted of 119 HMMs, whereas 47 additional HMMs profiles were collected from the Pfam databases [51] and TIGRFam [52] and corresponded to the full Resfams HMM Database. A protein sequence alignment against the Resfams HMM Database Core was performed using the HMMER tool set (version 3.1b2) [53], with the “-cut_ga” parameter, which defines the similarity threshold, to confirm the presence of antibiotic resistance in these sequences. By comparing our procedure applied to the bacterial genomes with the ARG predicted in the PATRIC database [54], we found the same proportions and concluded that the pipeline used was a reliable tool to predict ARGs (Supplementary Information Fig. S7). The Resfam database links the sequence to an identifier and a name corresponding to a family of antibiotics (e.g., AAC3) and its description, as well as an affiliation to an antibiotic resistance mechanism (e.g., Acetyltransferase).
For quantifying the genes encoding ARGs in marine and freshwater ecosystems, the reads were mapped against the ARG with bowtie2 [55]. The bases mapped against the ARGs were computed according to the procedure described by Sunagawa et al. [56]. The ARO features were compared between biomes such as defined by Longhurst [57] by using the package DESeq2 with R software [58].
Analysis of the ARG transfers by a network approach
Two networks were built from antibiotic resistance-related sequences, including a bipartite network to analyze the potential ARGs transferred between the viral entities and bacteria/archaea and a second network, derived from the first, representing the link between the best vehicles. The bipartite network describing the protein transfers between different “genomic units” or GUs is based on the Accessory Genome Constellation Network (AccNET) program [59]. The GUs corresponded to viruses and prophages in different ecosystems as well as plasmids, bacteria and archaea. These GUs were linked by protein clusters of ARGs named HpCs. An HpC was linked to at least two GUs when they had a protein sequence affiliated with a similar antibiotic resistance (Supplementary Information Fig. S8). Nevertheless, an HpC was also linked to a single GU when the protein sequence had no similarity with another GU. The construction of the bipartite network takes place in three stages as follows: (i) clustering proteins for defining the HpC; (ii) defining the distances between the proteins among each HpC, and (iii) calculation of the distances between the HpCs and the different GUs. The first step was realized with CD-HIT v4.8 [60] instead the kClust tool implemented in AccNET, which is an efficient program for grouping large protein or nucleotide sequence data according to a similarity threshold. The parameters used were a sequence identity of 90% and a coverage of at least 90% of the shortest sequence with respect to the representative sequence (-c 0.9 -n 5 -g 1 -aS 0.9). The second and third steps were described in the publication by Lanza et al. [59]. Briefly, these steps included the protdist program [61] for computing the protein distances within the various HpCs. The edge-weight was considered an attraction force between the nodes and thus was proportional to the inverse of the protein distances (the scripts used are available at the following address https://github.com/meb-team/AccNetPhylA).
The second network was focused on the putative vehicles of the ARGs by focusing on the clusters of the proteins (HpCs) linking the different GUs. First, the matrix of the distance between the proteins (protdist program) generated previously in step 2 was used for selecting the genes and computing both of the new distances. The gene distances were calculated by the dnadist program [61], and the patristic distances were from the phylogenies. More precisely, this last distance was computed from tree branch lengths describing the amount of genetic change represented by a tree. This tree was built with the maximum likelihood method using the PhyML tool [62] with the default settings and was rooted according to the midpoint rooting method (https://github.com/meb-team/HpC_to_vehicle). Finally, from an HpC including at least 2 GUs, only the best vehicles were selected on the basis of the minimal distance between the GUs (Supplementary Information Fig. S8). The network was then built with these vehicles, and the edges were equal to the inverse of the number of links. The pipelines used, the main command lines and the statistics can be found in the supplementary materials (Supplementary Information Fig. S8 and Table S2).
The network was visualized by using Cytoscape software (version 3.2.1) [63]. The various parameters characterizing the networks were calculated using the Cytoscape Network Analyzer plugin [64]. This module allowed us to compute a set of topological parameters, such as the degree distribution, the betweenness and the closeness of the nodes. The betweenness centrality a node n is defined as follows: Σs ≠ n ≠ t (σst(n)/σst). In this formula, s and t are nodes in the network different from n, σst denotes the number of shortest paths from s to t, and σst (n) is the number of shortest paths from s to t. The betweenness value for each node n is normalized by dividing the number of node pairs excluding n: (N − 1)(N − 2)/2, where N is the total number of nodes in the connected component. The closeness centrality of a node n is the reciprocal of the average shortest path length. These values computed from each node are a number between 0 and 1 [64]. These both indices help to define the keystone nodes. The topological indices computed from the different vehicles were tested by rewiring the network with the igraph package [65]. Briefly, 10000 networks were determined and for each a F value was computed from an ANOVA test. This F distribution allowed to compare the F value obtained from the real network with the simulations.
dN/dS ratio
The selective pressure acting on the ARGs included in the second network was evaluated based the dN/dS ratio using the kaks calculator [66]. This ration corresponds to the rates of non-synonymous (Ka) to synonymous (Ks) substitution. For each HpC, the putative ARG within the viruses or plasmids was compared to each bacterial sequence considered as a reference in the cluster, and the median was then computed for each vehicle among an HpC.
Supplementary information
Author contributions
DD supervised research, conducted some bio-informatic analyses and prepared the paper with contributions from CS. CS performed bio-informatic analyses. Both authors read and approved the final paper.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0478-9) contains supplementary material, which is available to authorized users.
References
- 1.Colavecchio A, Cadieux B, Lo A, Goodridge LD. Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the enterobacteriaceae family? A Review. Front Microbiol. 2017;8. 10.3389/fmicb.2017.01108. [DOI] [PMC free article] [PubMed]
- 2.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown-Jaque M, Calero-Cáceres W, Muniesa M. Transfer of antibiotic-resistance genes via phage-related mobile elements. Plasmid. 2015;79:1–7. doi: 10.1016/j.plasmid.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Lossouarn J, Dupont S, Gorlas A, Mercier C, Bienvenu N, Marguet E, et al. An abyssal mobilome: viruses, plasmids and vesicles from deep-sea hydrothermal vents. Res Microbiol. 2015;166:742–52. doi: 10.1016/j.resmic.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–8. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 6.Kim M-S, Park E-J, Roh SW, Bae J-W. Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol. 2011;77:8062–70. doi: 10.1128/AEM.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenzaka T, Tani K, Sakotani A, Yamaguchi N, Nasu M. High-frequency phage-mediated gene transfer among escherichia coli cells, determined at the single-cell level. Appl Environ Microbiol. 2007;73:3291–9. doi: 10.1128/AEM.02890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muniesa M, Imamovic L, Jofre J. Bacteriophages and genetic mobilization in sewage and faecally polluted environments. Micro Biotechnol. 2011;4:725–34. doi: 10.1111/j.1751-7915.2011.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Touchon M, Moura de Sousa JA, Rocha EP. Embracing the enemy: the diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr Opin Microbiol. 2017;38:66–73. doi: 10.1016/j.mib.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, et al. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun. 2016;7:13333. doi: 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shousha A, Awaiwanont N, Sofka D, Smulders FJM, Paulsen P, Szostak MP, et al. Bacteriophages isolated from chicken meat and the horizontal transfer of antimicrobial resistance genes. Appl Environ Microbiol. 2015;81:4600–6. doi: 10.1128/AEM.00872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, et al. Cryptic prophages help bacteria cope with adverse environments. Nat Commun. 2010;1:1146. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subirats J, Sànchez-Melsió A, Borrego CM, Balcázar JL, Simonet P. Metagenomic analysis reveals that bacteriophages are reservoirs of antibiotic resistance genes. Int J Antimicrob Agents. 2016;48:163–7. doi: 10.1016/j.ijantimicag.2016.04.028. [DOI] [PubMed] [Google Scholar]
- 14.Marti E, Variatza E, Balcázar JL. Bacteriophages as a reservoir of extended-spectrum β -lactamase and fluoroquinolone resistance genes in the environment. Clin Microbiol Infect. 2014;20:O456–O459. doi: 10.1111/1469-0691.12446. [DOI] [PubMed] [Google Scholar]
- 15.Enault F, Briet A, Bouteille L, Roux S, Sullivan MB, Petit M-A. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2016;11:237. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–22. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengtsson-Palme J, Larsson DGJ, Kristiansson E. Using metagenomics to investigate human and environmental resistomes. J Antimicrob Chemother. 2017;72:2690–703. doi: 10.1093/jac/dkx199. [DOI] [PubMed] [Google Scholar]
- 18.Balcazar JL. Bacteriophages as vehicles for antibiotic resistance genes in the environment. PLoS Pathog. 2014;10:e1004219. doi: 10.1371/journal.ppat.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lekunberri I, Subirats J, Borrego CM, Balcázar JL. Exploring the contribution of bacteriophages to antibiotic resistance. Environ Pollut. 2017;220(Part B):981–4. doi: 10.1016/j.envpol.2016.11.059. [DOI] [PubMed] [Google Scholar]
- 20.Brum JR, Ignacio-Espinoza JC, Roux S, Doulcier G, Acinas SG, Alberti A, et al. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- 21.Parmar KM, Gaikwad SL, Dhakephalkar PK, Kothari R, Singh RP. Intriguing interaction of bacteriophage-host association: an understanding in the era of omics. Front Microbiol. 2017;8. 10.3389/fmicb.2017.00559. [DOI] [PMC free article] [PubMed]
- 22.Forterre P, Soler N, Krupovic M, Marguet E, Ackermann H-W. Fake virus particles generated by fluorescence microscopy. Trends Microbiol. 2013;21:1–5. doi: 10.1016/j.tim.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Roux S, Krupovic M, Debroas D, Forterre P, Enault F. Assessment of viral community functional potential from viral metagenomes may be hampered by contamination with cellular sequences. Open Biol. 2013;3:130160. doi: 10.1098/rsob.130160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colombo S, Arioli S, Guglielmetti S, Lunelli F, Mora D. Virome-associated antibiotic-resistance genes in an experimental aquaculture facility. FEMS Microbiol Ecol. 2016;92:fiw003. doi: 10.1093/femsec/fiw003. [DOI] [PubMed] [Google Scholar]
- 25.Fancello L, Trape S, Robert C, Boyer M, Popgeorgiev N, Raoult D, et al. Viruses in the desert: a metagenomic survey of viral communities in four perennial ponds of the Mauritanian Sahara. ISME J. 2013;7:359–69. doi: 10.1038/ismej.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Appelt S, Fancello L, Bailly ML, Raoult D, Drancourt M, Desnues C. Viruses in a 14th-Century Coprolite. Appl Environ Microbiol. 2014;80:2648–55. doi: 10.1128/AEM.03242-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinheinz KA, Joensen KG, Larsen MV. Applying the ResFinder and VirulenceFinder web-services for easy identification of acquired antibiotic resistance and E. coli virulence genes in bacteriophage and prophage nucleotide sequences. Bacteriophage. 2014;4:e27943. doi: 10.4161/bact.27943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatusova T, Ciufo S, Fedorov B, O’Neill K, Tolstoy I. RefSeq microbial genomes database: new representation and annotation strategy. Nucleic Acids Res. 2015;43:3872. doi: 10.1093/nar/gkv278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenzaka T, Tani K, Nasu M. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J. 2010;4:648–59. doi: 10.1038/ismej.2009.145. [DOI] [PubMed] [Google Scholar]
- 30.Jiang SC, Paul JH. Gene transfer by transduction in the marine environment. Appl Environ Microbiol. 1998;64:2780–7. doi: 10.1128/aem.64.8.2780-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martínez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol. 2015;13:116–23. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 32.Popa O, Dagan T. Trends and barriers to lateral gene transfer in prokaryotes. Curr Opin Microbiol. 2011;14:615–23. doi: 10.1016/j.mib.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 33.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci. 2010;107:127–32. doi: 10.1073/pnas.0908978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corel E, Lopez P, Méheust R, Bapteste E. Network-thinking: graphs to analyze microbial complexity and evolution. Trends Microbiol. 2016;24:224–37. doi: 10.1016/j.tim.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popa O, Landan G, Dagan T. Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 2016 10.1038/ismej.2016.116. [DOI] [PMC free article] [PubMed]
- 36.Puxty RJ, Millard AD, Evans DJ, Scanlan DJ. Shedding new light on viral photosynthesis. Photosynth Res. 2015;126:71–97. doi: 10.1007/s11120-014-0057-x. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280:3143–50. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- 38.Corel E, Méheust R, Watson AK, McInerney JO, Lopez P, Bapteste E. Bipartite network analysis of gene sharings in the microbial world. Mol Biol Evol. 2018;35:899–913. doi: 10.1093/molbev/msy001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calero-Cáceres W, Muniesa M. Persistence of naturally occurring antibiotic resistance genes in the bacteria and bacteriophage fractions of wastewater. Water Res. 2016;95(Supplement C):11–18. doi: 10.1016/j.watres.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 41.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering Earth’s virome. Nature. 2016;536:425–30. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 42.Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res Int J Rapid Publ Rep Genes Genomes. 2008;15:387–96. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinforma. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. PeerJ. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishimura Y, Yoshida T, Kuronishi M, Uehara H, Ogata H, Goto S. ViPTree: the viral proteomic tree server. Bioinformatics. 2017;33:2379–80. doi: 10.1093/bioinformatics/btx157. [DOI] [PubMed] [Google Scholar]
- 48.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003;14:927–30. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 49.Team RC. R: a language and environment for statistical computing. 2018
- 50.Gibson MK, Forsberg KJ, Dantas G. Improved annotation of antibiotic resistance determinants reveals microbial resistomes cluster by ecology. ISME J. 2015;9:207–16. doi: 10.1038/ismej.2014.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res. 2003;31:371–3. doi: 10.1093/nar/gkg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eddy SR. Profile hidden Markov models. Bioinforma Oxf Engl. 1998;14:755–63. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 54.Antonopoulos DA, Assaf R, Aziz RK, Brettin T, Bun C, Conrad N, et al. PATRIC as a unique resource for studying antimicrobial resistance. Brief Bioinform. 2017. 10.1093/bib/bbx08. [DOI] [PMC free article] [PubMed]
- 55.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 57.Longhurst AR. Biomes: The Primary Partition. Ecological Geography of the Sea Elsevier: 2010. p. 89–99.
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lanza VF, Baquero F, de la Cruz F, Coque TM. AcCNET (Accessory Genome Constellation Network): comparative genomics software for accessory genome analysis using bipartite networks. Bioinformatics. 2017;33:283–5. doi: 10.1093/bioinformatics/btw601. [DOI] [PubMed] [Google Scholar]
- 60.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinforma Oxf Engl. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 61.Felsenstein J. PHYLIP—Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–6. [Google Scholar]
- 62.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 63.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinforma Oxf Engl. 2008;24:282–4. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 65.Csardi G, Nepusz T. The igraph software package for complex network research. 2006;9.
- 66.Zhang Z, Li J, Zhao X-Q, Wang J, Wong GK-S, Yu J. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom Proteom Bioinforma. 2006;4:259–63. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.