Abstract
In recent years, research in the field of Microbial Ecology has revealed the tremendous diversity and complexity of microbial communities across different ecosystems. Microbes play a major role in ecosystem functioning and contribute to the health and fitness of higher organisms. Scientists are now facing many technological and methodological challenges in analyzing these complex natural microbial communities. The advances in analytical and omics techniques have shown that microbial communities are largely shaped by chemical interaction networks mediated by specialized (water-soluble and volatile) metabolites. However, studies concerning microbial chemical interactions need to consider biotic and abiotic factors on multidimensional levels, which require the development of new tools and approaches mimicking natural microbial habitats. In this review, we describe environmental factors affecting the production and transport of specialized metabolites. We evaluate their ecological functions and discuss approaches to address future challenges in microbial chemical ecology (MCE). We aim to emphasize that future developments in the field of MCE will need to include holistic studies involving organisms at all levels and to consider mechanisms underlying the interactions between viruses, micro-, and macro-organisms in their natural environments.
Subject terms: Metabolomics, Microbial ecology
Background
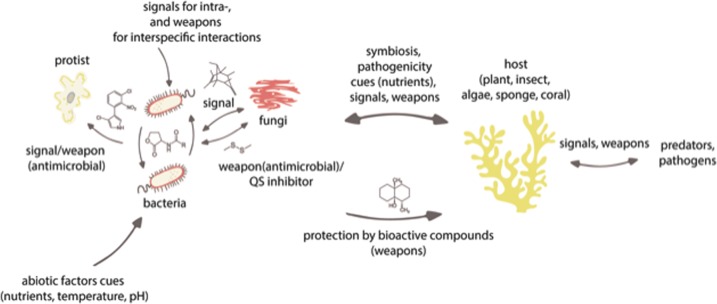
Chemical ecology first appeared as a keystone discipline in the early 1950s, advancing our understanding of insect communication and plant chemical defenses [1]. However, chemical communication is not restricted to plant–insect and plant–plant interactions. In fact, chemically mediated relationships are now being recognized as common in the microbial world across terrestrial and aquatic ecosystems (Fig. 1). Bonnie Bassler is one of the pioneers of microbial chemical communication being amongst the first to discover bacterial intra-specific quorum sensing via autoinducing chemical compounds. This mechanism is now proving to play a fundamental role in both intraspecific and interspecific interactions [2, 3]. Prof. Bassler coined the term “microbial language” and it was her initial work and the numerous follow-up studies that brought chemical communication between microbes into the spotlight. Researchers in the field of microbial ecology are recognizing the important roles that chemical communication and interactions play across all ecosystems (reviewed in ref. [4]). In fact, the oldest form of communication is probably the chemical communication between microorganisms and only later evolved in plants, insects, and other higher organisms [5]. Thus, by deciphering the chemical language, we will be able to better understand how species interact in their ecosystems. However, understanding the theoretical foundations of chemical language (its origin and diversity) is challenging and has been rarely studied.
Fig. 1.
Patterns of microbial communication across terrestrial and aquatic ecosystems. Cues—provide unintentional information; signals—provide intentional information and chemical weapons/antimicrobial—are produced targeted
Until now, the topic of microbial chemical ecology (MCE) has been largely neglected by microbiologists. The reason stems from methodological constraints concerning the analysis of microbiological communities under natural conditions. Furthermore, most of the research for natural products is focused on chemical and biochemical approaches and drug discovery with a less of an emphasis on ecological aspects. The traditional separation of disciplines limits our understanding and ultimately hinders scientific advances. Recent developments in genome sequencing and chemico-analytical tools enabling us to uncover the chemical communication networks of the microbial world, as well as cross-disciplinary collaborations between research fields will make MCE a central field within microbial ecology.
In order to raise awareness of the importance of MCE in the field of microbial ecology, we hosted a roundtable session entitled “Microbial chemical ecology: intra- and interspecies communication” during the ISME17 meeting in Leipzig, Germany (August 2018). The discussion raised several crucial points that will be addressed in this paper. We will also address recent breakthrough discoveries, methodological challenges, and future perspectives in this rapidly evolving field.
Microbial chemical diversity
Microorganisms produce a wide array of secondary metabolites with a variety of physico-chemical and biological properties. In recent years, these molecules have been increasingly referred to as specialized metabolites (SMs) in order to emphasize their important role in microbial ecology (Table 1 keyword definition) [6]. Most microorganisms produce both, volatile and water-soluble (nonvolatile) compounds (reviewed in ref. [7]). However, so far, most studies have focused on either volatile or soluble compounds and have ignored the fact that these compounds are usually produced simultaneously, sometimes by enzymes encoded in the same biosynthetic gene cluster [8]. In addition, the same molecule can be functional in both, gas and liquid phases. For example, naphthalene acts as an attractant for Pseudomonas putida bacteria in liquid media, while air-born naphthalene acts as a repellent for the same strain [9].
Table 1.
Keyword definitions
| Keyword | Definition |
|---|---|
| Infochemicals |
Chemical compounds released by microbes, animals and plants into their environment and used as signals. The term “infochemical” generally indicates low-weight SMs. However, macromolecules, such as DNA, can also serve as an information carrier in a form of mobile genetic elements (plasmids, transposons, and bacteriophages) via horizontal gene transfer. These elements, especially plasmids, carrying genes for antibiotic resistance, virulence, or nitrogen fixation contribute to microbial community fitness and interaction with a host. |
| Microbial chemical interaction | Process in which a chemical signal (“infochemical”) from one organism has an effect on the counterpart behavior and physiology. The interaction can occur directly cell-to-cell, or signals can be spread on short and long distances. The signal may or may not activate the “feedback” signal production in the counterpart. |
| Microbial chemical communication | An active exchange of (targeted) chemical signals, where signals of one organism activate response in the counterpart. |
| Secondary (specialized) metabolites | Historical name for metabolites produced by microorganisms mostly in the stationary phase of growth in laboratory cultivations and considered to be nonessential for survival (in contrast to primary metabolites). However, the term “secondary” does neither reflect the real function nor the timing of production of several of these metabolites in nature. For this reason the term “specialized” is increasingly used in connection with metabolites functioning as signals in microbial interactions. |
| Volatile organic compound | Small molecular weight compounds with low boiling points and a high vapor pressure. |
| Quorum sensing | Mechanism how microorganisms sense community and coordinate its behavior by production of chemical compounds (autoinducers, peptides, and microbial hormones). |
| Hormesis | A process in a cell or organism that exhibits biphasic dose response to an environmental compound—low dose has stimulating or beneficial effect and a high dose inhibitory or toxic effect. |
Water-soluble compounds from terrestrial and aquatic microorganisms are increasingly gaining attention as compared to volatiles, mostly due to relatively simple extraction and detection methodology, and due to the fact that many of the soluble compounds have potent bioactive properties. Soluble compounds serve as antimicrobial weapons in antagonistic interactions, as well as signaling compounds within the same or between different species of free-living or host-associated microbial communities. In contrast to soluble compounds, volatile organic compounds can diffuse easily through air- and gas-filled pores and play an important role in long-distance interactions between microorganisms [10]. Recently, Schulz-Bohm et al. [11] have shown that volatile compounds can diffuse within 20 min over distance of >12 cm, which is a veritable distance for most soil microorganisms. Despite their mostly hydrophobic nature, volatiles are widely produced in both terrestrial and aquatic environments by marine plankton, algae, animals, and marine bacteria [12–15].
Interestingly, although the ability of microorganisms to produce structurally diverse volatile compounds has been known for decades [16], their antimicrobial activities have only recently attracted attention making them potential candidates for future drug development (reviewed in ref. [17]). In addition, volatiles can have synergistic effects with soluble antimicrobials. For example, hydrophilic antibiotics such as vancomycin and β-lactams that have marginal inhibitory effects on Gram-negative bacteria, exhibit enhanced antibacterial activity when the exposed strains are pre-treated with the volatile phenylpropanoid eugenol [18]. Due to their lipophilic nature, volatiles may interfere with membrane structures causing depolarization of the cell membrane thus, leading to a higher sensitivity toward the more polar antibiotics.
The microbial dialog may also involve small inorganic molecules such as HCN, ammonia, others. For example, stimulation of NO production in Streptomyces by fungal bacteriostatic compound followed by NO-mediated transcriptional activation of fungistatic heronapyrrole biosynthesis [19]. Another study reported that nitrite produced in nitrogen oxide cycle functioned as an intercellular communication molecule in Streptomyces coelicolor [20].
Factors affecting the production of SMs
The production of both, soluble and volatile SMs, is influenced by various environmental biotic and abiotic factors. Playing with abiotic factors such as nutrients, light, temperature, pH, moisture, salinity, and others, one can trigger the expression of genes leading to the production of diverse and novel SMs in terrestrial and marine microorganisms. There are several examples revealing chemical diversity of single isolate by applying different cultivation parameters using so-called OSMAC (one strain-many compounds) approach [21, 22]. Molecular mechanisms of SM regulation by nutrients are best-described for major nutrient sources, such as carbon, nitrogen, phosphate, and a few selected micronutrients, such as the trace metals like iron, copper, and zinc (reviewed in ref. [23, 24]). However, these molecular mechanisms have been mostly studied in isolated microorganisms cultivated as pure cultures and little is known about how nutrients and other abiotic factors influence SM production in microbial communities under natural conditions. As an example, a higher proportion of bioactive actinomycetes strains were repeatedly reported in alkaline soils [25, 26]. However, in a later study, actinomycetes isolated from the acidic soil samples produced a higher number of low-molecular-weight compounds as compared to alkaline sites. This result indicates that acidic soils may be a reservoir for novel actinobacterial strains [26]. Yet, so far, little is known about the selective pressure pH plays on SM evolution.
Interspecific interactions and competitor sensing are considered the main biotic factors affecting the production of SMs [7]. The nonantibiotic producing soil bacteria can be triggered to produce broad-spectrum antibiotics when confronted with unrelated bacterial species. For example, when Pseudomonas fluorescens Pf0–1 is confronted with taxonomically different bacterial species, it can produce broad-spectrum antimicrobial compounds with activity against a range of plant pathogenic fungi, making fungi the victim of this particular bacterial–bacterial interaction [27].
Microbial communication by autoinducers and autoregulatory factors/microbial hormones was initially considered to be an intra-specific microbial communication mechanism, which influenced a range of physiological responses to microbial density environmental changes, such as antibiotic and toxin production, biofilm formation, etc. (reviewed in ref. [28, 29]). However, it has been demonstrated that inter-specific communication between closely related and distant microbial species using species-specific signaling molecule is possible under laboratory conditions [30] (reviewed in ref. [31]). Thus, such interspecies signaling may also take place in nature.
Another interesting example of interspecific interaction is cell-to-cell contact between mycolic acid-containing actinomycete and other nonmycolic actinomycete species in a combined culture. This direct interaction induces SM production in nonmycolic actinomycete by an unknown mechanism [32]. However, it has been found that mycolic acid-containing bacteria need to be alive since dead cells do not induce compound production in combined culture [33]. In addition to the ecological aspect, the understanding of factors affecting SM production has also an applicational impact. The compounds acting as signal molecules can be used as elicitors of silent natural product biosynthetic gene clusters that might have potential applications as drugs [34]. Similarly, co-cultivation with other microorganisms and modification of abiotic cultivation factors is an important tool for natural product discovery (reviewed in ref. [35]).
Transport in the natural environment
To elicit an effect, chemicals need to physically reach their potential recipients, i.e., need to become accessible and available at sufficient concentrations [36]. Hence, transport and accessibility of chemical signals is an important and often overlooked factor in chemical ecology. Following the definitions used in the risk assessment of environmental chemicals [37], the term bioavailability refers to the degree of interaction of chemicals with living organisms and includes two major exposure scenarios. First, if a chemical gets transformed by the recipient, the bioavailability is a dynamic feature and bioavailable (steady-state) concentrations are determined by the rate of mass transfer of a compound to the recipient and the recipient’s intrinsic catabolic activity to degrade the compound [38]. Second, if chemicals act by nonconsumptive processes, their equilibrium concentration at the recipient will be effect determining. In either of the scenarios, the transport of the chemical from the source to the recipient is driven by its molecular reactivity and physical–chemical properties as well the prevailing environmental conditions. Hence, the bioavailability of any chemical should be perceived as a habitat-specific rather than solely a compound property. For chemical communication to develop, microbes should be within communication distances. For instance in soil, typical inter-cell distances of 10–20 µm [39], and cell-to-cell communication distances of soluble chemicals of up to 78 µm have been described [40]. The soil structure and its complex pore space are another driver of cell-to-cell communication and microbial functioning. The diffusion rate of volatile compounds throughout the porous network of the soil is influenced by the physical properties of the soil, including shape and size of soil aggregates as well as chemical parameters, such as soil moisture, pH, and temperature. Arrangement, size, and composition of particles influence the retention capacity of water and nutrients [41] and provide pathways for the exchange of cells and vapor- or water-bound communication signals. Compound molecules are typically transported by diffusion, advection, or by biological transport vectors. While volatile chemicals have been considered as the “lingua franca” [42] for long distance signaling through the air-phase, diffusive transport of water-born chemicals is often restricted to short distances, as molecular diffusion coefficients generally are 103–104 lower in water than in air. Moreover, transport of nonvolatile water-soluble chemicals requires continuous liquid phases and thus, may be restricted by air-filled pores. However, a study by Barto et al. [43] has shown that information-carrying chemicals may be transmitted at long distances by mycorrhizal networks acting as below ground information networks between plants. Efficient resource translocation at velocities up to 600 µm min−1 in their mycelia enables fungi to grow even in air-filled, heterogeneous habitats. Thereby, mycelia also enable bacterial activity by cm-range metabolite, nutrient, and water transfer to bacteria in the hyphosphere as was shown by a combination of stable isotope probing and chemical microscopy [44]. Via their hyphal transport (“hyphal pipelines”) [45], they may also transport hydrophobic chemicals to distant bacteria up to 100-fold better than diffusion would do.
Another option for the exchange of information carriers and microbial chemical interaction is the transport of microorganisms themselves. Microorganisms may contain information carriers such as plasmids, prophages, or endobacteria [46], and interact with neighboring recipients as agents of horizontal gene transfer (HGT) or by the exchange of smaller signals. As for chemicals microbial dispersal may take place via (i) advective or quasi-diffusive transport in air or water, (ii) intrinsic random or targeted cellular motility, or (iii) by deposition to abiotic or biotic transport vectors such as colloidal particles or the micro or macro fauna. For instance, research on bacterial fungal interactions has highlighted the role of hyphae and the mycosphere as a hotspot of microbial transport and activity [47]. Hyphae enable the directed and random transport of less immobilized bacteria in heterogeneous (soil) habitats. Hyphae also serve as scaffolds for bacterial transport [48], as well as presumed habitat for preferential HGT [49–51].
Ecological function of microbial natural products
The chemical diversity of microbial natural products is so immense, yet most of them still remain unknown. Widespread soil bacteria like Streptomyces or myxobacteria might encode >30 biosynthetic gene clusters for the production of several structurally different polyketides, peptides, or terpenes in a single strain (not counting SM derivatives derived from the same biosynthetic gene cluster) [52]. While the number of putative natural product families correlates with the number of biosynthetic gene clusters that can easily be predicted from the bacterial genome sequence, in most cases, only a small fraction of these natural products have been identified. Even for the natural products that have been well-known for decades, we often know more about their potential use (as anti-infectives or other drugs) than about their original ecological function. Several clinically used antibiotics of microbial origin have been shown to act as signaling molecules at sub-inhibitory concentrations [53, 54]. Assuming that the true target is addressed clinically (and not an off-target effect), these examples show that the metabolite concentration matters. The phenomena of low-dose stimulation/signaling and high-dose toxicity by the same molecule is called hormesis and is very common for microbial natural products [55]. In contrast to the much higher concentrations that are often used in the clinical situations, these low concentrations might be more relevant in nature. For example, in terrestrial ecosystems, microbial biomass can be triggered by trace concentrations of low-molecular weight compounds, so-called “trigger solutions” [56].
Bacteria always live in a complex environment surrounded by several other organisms, including other bacteria, fungi, protozoa, as well as complex multicellular organisms such as insects, mammals, and plants. Assuming that many of the required organismic interactions are being mediated by natural products, we can expect toxic or beneficial compounds, signals or metallophores, along with compounds enabling ultraviolet-protection, swarming motility or sporulation [7]. If we look into bacterial quorum sensing enabling the communication within but also among microbial species [57], it is obvious that we have identified only a small fraction of the natural communication systems in some model systems that often have not been analyzed with respect to other microbes present in these environments [58]. Moreover, we need more information concerning the regulatory mechanisms and triggers (signals/elicitors) that are required for the production of natural products. Transcription factors (often encoded in the respective biosynthetic gene clusters) that mediate the activation or repression of biosynthetic gene clusters often require specific ligands, which might be difficult to identify due to their low abundance. With respect to other regulatory elements as regulatory sRNAs, riboswitches or DNA-binding proteins that interfere with transcription, we have hardly started to identify them.
New tools to address methodological challenges in MCE
Understanding the natural metabolites that mediate interactions between organisms is key to deciphering chemical communication and interactions. Unfortunately, the detection and identification of the compounds that mediate these interactions still remains challenging. The two principal methods in metabolomics used to detect and structurally elucidate metabolites are mass spectrometry (MS) and nuclear magnetic resonance (NMR). However, NMR is difficult to use in an ecological context and one must distinguish between the analysis of ecologically relevant mixtures (done by MS) and NMR used for pure compounds, but being the ultimate proof for compound structure. The emerging MS imaging (MSI) provides new opportunities to study environmentally relevant metabolites in their spatial and temporal context [59]. This approach helps to overcome limitations in traditional MS-based metabolomics techniques that require extraction and ample amounts of sample preparation. MSI techniques are excellent tools for monitoring metabolic processes and for studying chemical communication in an ecological context. For example, MALDI-IMS analysis of S. coelicolor staged with other actinomycetes revealed the production of many interaction-specific metabolites that were not produced in monoculture [60].
The biggest methodological challenge in MCE is to mimic natural environmental conditions in the laboratory. Recent approaches in creating optically transparent microcosms for long-term observations of cell–cell interactions [61] or mesocosms to test the SM effect on the microbial community [62] have opened up new opportunities for carrying out microbial interaction studies.
Artificial microcosm systems (“designer” ecosystems) bring the advantage of studying microbial interactions on a molecular level while creating controlled environments that mimic environmental conditions [63]. As such, the 3D printing of soil structures or microfluidic techniques prove to be promising approaches to studying microbial chemical interactions [64]. Borer et al. developed glass-etched pore networks based on soil-aggregate cross sections that are used to study microbial interactions in response to O and C gradients [65]. The “lab-on-a-chip” technology is another promising platform to study microbial chemical interactions due to its compatibility with flow cytometry and MS tools [66]. A range of model microbiome systems have been developed that have the capability of mimicking the complexity of natural environments while testing hypotheses with statistical power in a controlled setting [67].
Future trends and perspectives
Great progress has been made in understanding unidirectional chemical responses without considering the dialogs and bidirectional interaction between organisms. Current studies are often focused on SM(s) produced by a single organism and the responses of a perceiving organism. However, chemical communications taking place in nature are complex and may play a role in almost every possible interaction between the member of the community. Most microorganisms produce a multitude of metabolites into their environment but probably only a few of these have a true communicative function. Nevertheless, substances emitted for noncommunicative purposes can provide multiple starting points for the evolution of chemical communication.
Several compounds, such as terpenoids, sulfur compounds, indole, others are commonly produced by different microorganisms and even plants and insects. Analyses of such chemical compounds in a phylogenetic context could be very helpful for understanding the evolution of chemical communication. In addition, important factor to improve our understanding of the evolution of chemical communication is the expansion of our current knowledge of receptors and olfactory systems that are responsible for signal perception.
Chemical interaction processes are not restricted to prokaryotes and eukaryotes only. Recent studies revealed that viruses (phages) use phage-produced communication peptide or host-produced quorum sensing autoinducer to control phage lysis-lysogeny decisions [68, 69]. To counteract, bacteria developed a natural product-based defense mechanism against phage infections [70]. However, the role of viruses in microbial chemical communication has been rarely tackled and so far, largely unexplored. Thus, future directions of MCE will ideally involve studies on all organismal levels, and consider mechanisms underlying the communication including viruses, micro- and macro-organisms in their natural environment.
Another important direction of MCE is to study how climate change (e.g., low/high temperatures and drought/flooding) will affect SM production and their function in the changing natural environment. A final, yet important question is “How to promote chemical studies in the course of microbial ecological work and vice versa?” Traditionally, microbiology and microbial ecology have been separated from the field of chemical ecology, with the latter focusing mainly on above-ground communication. However, since recent advance have shown the importance of chemical interactions in the microbial world as part of a bigger communication network with their host, we argue for a merge of disciplines and integrate functional, evolutionary, physiological and ontogenetic levels [71, 72].
Understanding the various chemical interactions between microbes and their plant host will have important implications for agriculture to counteract drought and increased pathogen pressure. One promising solution stems from microbial engineering of the holobiont—the inseparable unit of the host and its microbiome [73]. Moreover, volatiles can play important roles in suppressing pathogens in disease suppressive soils [74, 75]. Thus, future studies could usefully address the underlying mechanisms of microbial communication and pathogen control via volatiles in the plant holobiont, which will help linking genes to enzymes and metabolites and set the basis for microbial engineering strategies.
Finally, advances in MCE will help to uncover mechanisms driving human–microbiome interactions that influence our health. Till today, only a small fraction of chemistry carried out in this microbial habitat has been characterized [76, 77]. A critical step in understanding human gut microbial interactions is linking metabolites with specific microbial genes and enzymes. Artificial systems that mimic gut conditions, such as the “Robogut” [78] or microfluidic devices such as the HuMiX (human–microbial crosstalk) [79] system combined with metabolomics and transcriptomics approaches will be essential tools to close the knowledge gap and to develop strategies for improved health and treatment of infectious diseases.
Whether in human or any other environment, deep understanding of the complex microbially mediated chemical interactions remains a large and intricate puzzle that will require efficient collaborative effort.
Acknowledgements
The authors would like to thank all participants of the roundtable session “Microbial chemical ecology: intraspecies and interspecies communication” at ISME 17 for the fruitful discussion. We acknowledge funding from the Education, Culture, Sports, Science, and Technology Ministry of Japan special project costs Four-Dimensional Kuroshio Marine Science (4D-KMS) Project and Japan Society for the Promotion of Science (JSPS KAKENHI Grant Number 16K18678) to DU. HBB acknowledges support from the State of Hesse for the LOEWE TBG research center. LYW acknowledges support from the Collaborative Research Centre AquaDiva (CRC 1076 AquaDiva) at the Friedrich Schiller University Jena and the Helmholtz Centre for Environmental Research—UFZ, funded by the Deutsche Forschungsgemeinschaft (DFG). PG acknowledges the Netherlands Organization for Scientific Research (NWO), VIDI personal grant (864.11.015). This is publication 6750 of the NIOO-KNAW.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ruth Schmidt, Dana Ulanova
References
- 1.Hartmann T. The lost origin of chemical ecology in the late 19th century. Proc Natl Acad Sci USA. 2008;105:4541–6. doi: 10.1073/pnas.0709231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–7. doi: 10.1016/S1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 3.Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–5. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson S, Williams P. Quorum sensing and social networking in the microbial world. J R Soc Interface. 2009;6:959–78. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mithofer A, Boland W. Do you speak chemistry? Small chemical compounds represent the evolutionary oldest form of communication between organisms. EMBO Rep. 2016;17:626–9. doi: 10.15252/embr.201642301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies J. Specialized microbial metabolites: functions and origins. J Antibiot. 2013;66:361–4. doi: 10.1038/ja.2013.61. [DOI] [PubMed] [Google Scholar]
- 7.Tyc O, Song C, Dickschat JS, Vos M, Garbeva P. The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends Microbiol. 2017;25:280–92. doi: 10.1016/j.tim.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Riclea R, Aigle B, Leblond P, Schoenian I, Spiteller D, Dickschat JS. Volatile lactones from streptomycetes arise via the antimycin biosynthetic pathway. Chembiochem. 2012;13:1635–44. doi: 10.1002/cbic.201200260. [DOI] [PubMed] [Google Scholar]
- 9.Hanzel J, Harms H, Wick LY. Bacterial chemotaxis along vapor-phase gradients of naphthalene. Environ Sci Technol. 2010;44:9304–10. doi: 10.1021/es100776h. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt R, Cordovez V, de Boer W, Raaijmakers J, Garbeva P. Volatile affairs in microbial interactions. ISME J. 2015;9:2329–35. doi: 10.1038/ismej.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulz-Bohm K, Gerards S, Hundscheid M, Melenhorst J, de Boer W, Garbeva P. Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 2018;12:1252–62. doi: 10.1038/s41396-017-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fink P. Ecological functions of volatile organic compounds in aquatic systems. Marine and Freshwater Behaviour and Physiology. 2007;40:155–68. doi: 10.1080/10236240701602218. [DOI] [Google Scholar]
- 13.Groenhagen U, Leandrini De Oliveira AL, Fielding E, Moore BS, Schulz S. Coupled Biosynthesis of Volatiles and Salinosporamide A in Salinispora tropica. Chembiochem. 2016;17:1978–85. doi: 10.1002/cbic.201600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harig T, Schlawis C, Ziesche L, Pohlner M, Engelen B, Schulz S. Nitrogen-containing volatiles from marine salinispora pacifica and roseobacter-group bacteria. J Nat Prod. 2017;80:3289–95. doi: 10.1021/acs.jnatprod.7b00789. [DOI] [PubMed] [Google Scholar]
- 15.Thiel V, Brinkhoff T, Dickschat JS, Wickel S, Grunenberg J, Wagner-Dobler I, et al. Identification and biosynthesis of tropone derivatives and sulfur volatiles produced by bacteria of the marine Roseobacter clade. Org Biomol Chem. 2010;8:234–46. doi: 10.1039/B909133E. [DOI] [PubMed] [Google Scholar]
- 16.Wheatley RE. The consequences of volatile organic compound mediated bacterial and fungal interactions. Antonie Van Leeuwenhoek. 2002;81:357–64. doi: 10.1023/A:1020592802234. [DOI] [PubMed] [Google Scholar]
- 17.Avalos M, van Wezel GP, Raaijmakers JM, Garbeva P. Healthy scents: microbial volatiles as new frontier in antibiotic research? Curr Opin Microbiol. 2018;45:84–91. doi: 10.1016/j.mib.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 18.Hemaiswarya S, Doble M. Synergistic interaction of eugenol with antibiotics against Gram negative bacteria. Phytomedicine. 2009;16:997–1005. doi: 10.1016/j.phymed.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Khalil ZG, Cruz-Morales P, Licona-Cassani C, Marcellin E, Capon RJ. Inter-kingdom beach warfare: microbial chemical communication activates natural chemical defences. ISME J. 2019;13:147–58. doi: 10.1038/s41396-018-0265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sasaki Y, Oguchi H, Kobayashi T, Kusama S, Sugiura R, Moriya K, et al. Nitrogen oxide cycle regulates nitric oxide levels and bacterial cell signaling. Sci Rep. 2016;6:22038. doi: 10.1038/srep22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bode HB, Bethe B, Hofs R, Zeeck A. Big effects from small changes: possible ways to explore nature’s chemical diversity. Chembiochem. 2002;3:619–27. doi: 10.1002/1439-7633(20020703)3:7<619::AID-CBIC619>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 22.Romano S, Jackson SA, Patry S, Dobson ADW. Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Mar Drugs. 2018;16:E244. doi: 10.3390/md16070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Locatelli FM, Goo KS, Ulanova D. Effects of trace metal ions on secondary metabolism and the morphological development of streptomycetes. Metallomics. 2016;8:469–80. doi: 10.1039/C5MT00324E. [DOI] [PubMed] [Google Scholar]
- 24.van der Heul HU, Bilyk BL, McDowall KJ, Seipke RF, van Wezel GP. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat Prod Rep. 2018;35:575–604. doi: 10.1039/C8NP00012C. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez I, Niebla A, Lemus M, Gonzalez L, Iznaga IO, Perez ME, et al. Ecological approach of macrolide-lincosamides-streptogramin producing actinomyces from Cuban soils. Lett Appl Microbiol. 1999;29:147–50. doi: 10.1046/j.1365-2672.1999.00580.x. [DOI] [PubMed] [Google Scholar]
- 26.Sagova-Mareckova M, Ulanova D, Sanderova P, Omelka M, Kamenik Z, Olsovska J, et al. Phylogenetic relatedness determined between antibiotic resistance and 16S rRNA genes in actinobacteria. BMC Microbiol. 2015;15:81. doi: 10.1186/s12866-015-0416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garbeva P, Tyc O, Remus-Emsermann MN, van der Wal A, Vos M, Silby M, et al. No apparent costs for facultative antibiotic production by the soil bacterium Pseudomonas fluorescens Pf0-1. PLoS One. 2011;6:e27266. doi: 10.1371/journal.pone.0027266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniel-Ivad M, Pimentel-Elardo S, Nodwell JR. Control of specialized metabolism by signaling and transcriptional regulation: opportunities for new platforms for drug discovery? Annu Rev Microbiol. 2018;72:25–48. doi: 10.1146/annurev-micro-022618-042458. [DOI] [PubMed] [Google Scholar]
- 29.Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–99. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen TB, Kitani S, Shimma S, Nihira T. Butenolides from Streptomyces albus J1074 act as external signals to stimulate avermectin production in Streptomyces avermitilis. Appl Environ Microbiol. 2018;84:e02791–17. doi: 10.1128/AEM.02791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiology. 2008;154:1845–58. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- 32.Onaka H, Mori Y, Igarashi Y, Furumai T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl Environ Microbiol. 2011;77:400–6. doi: 10.1128/AEM.01337-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asamizu S, Ozaki T, Teramoto K, Satoh K, Onaka H. Killing of mycolic acid-containing bacteria aborted induction of antibiotic production by streptomyces in combined-culture. PLoS One. 2015;10:e0142372. doi: 10.1371/journal.pone.0142372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdelmohsen UR, Grkovic T, Balasubramanian S, Kamel MS, Quinn RJ, Hentschel U. Elicitation of secondary metabolism in actinomycetes. Biotechnol Adv. 2015;33:798–811. doi: 10.1016/j.biotechadv.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhu H, Sandiford SK, van Wezel GP. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol. 2014;41:371–86. doi: 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- 36.Semple KT, Doick KJ, Wick LY, Harms H. Microbial interactions with organic contaminants in soil: Definitions, processes and measurement. Environmental Pollution. 2007;150:166–76. doi: 10.1016/j.envpol.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 37.Ortega-Calvo JJ, Harmsen J, Parsons JR, Semple KT, Aitken MD, Ajao C, et al. From bioavailability science to regulation of organic chemicals. Environ Sci Technol. 2015;49:10255–64. doi: 10.1021/acs.est.5b02412. [DOI] [PubMed] [Google Scholar]
- 38.Johnsen AR, Wick LY, Harms H. Principles of microbial PAH-degradation in soil. Environ Pollut. 2005;133:71–84. doi: 10.1016/j.envpol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 39.Raynaud X, Nunan N. Spatial ecology of bacteria at the microscale in soil. PLoS One. 2014;9:e87217. doi: 10.1371/journal.pone.0087217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gantner S, Schmid M, Durr C, Schuhegger R, Steidle A, Hutzler P, et al. In situ quantitation of the spatial scale of calling distances and population density-independent N-acylhomoserine lactone-mediated communication by rhizobacteria colonized on plant roots. FEMS Microbiol Ecol. 2006;56:188–94. doi: 10.1111/j.1574-6941.2005.00037.x. [DOI] [PubMed] [Google Scholar]
- 41.Tecon R, Or D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol Rev. 2017;41:599–623. doi: 10.1093/femsre/fux039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt R, Etalo DW, de Jager V, Gerards S, Zweers H, de Boer W, et al. Microbial small talk: volatiles in fungal–bacterial interactions. Front Microbiol. 2016;6:12. doi: 10.3389/fmicb.2015.01495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barto EK, Weidenhamer JD, Cipollini D, Rillig MC. Fungal superhighways: do common mycorrhizal networks enhance below ground communication? Trends Plant Sci. 2012;17:633–7. doi: 10.1016/j.tplants.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Worrich A, Stryhanyuk H, Musat N, Konig S, Banitz T, Centler F, et al. Mycelium-mediated transfer of water and nutrients stimulates bacterial activity in dry and oligotrophic environments. Nat Commun. 2017;8:15472. doi: 10.1038/ncomms15472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furuno S, Foss S, Wild E, Jones KC, Semple KT, Harms H, et al. Mycelia promote active transport and spatial dispersion of polycyclic aromatic hydrocarbons. Environ Sci Technol. 2012;46:5463–70. doi: 10.1021/es300810b. [DOI] [PubMed] [Google Scholar]
- 46.Torres-Cortes G, Ghignone S, Bonfante P, Schussler A. Mosaic genome of endobacteria in arbuscular mycorrhizal fungi: transkingdom gene transfer in an ancient mycoplasma-fungus association. Proc Natl Acad Sci USA. 2015;112:7785–90. doi: 10.1073/pnas.1501540112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deveau A, Bonito G, Uehling J, Paoletti M, Becker M, Bindschedler S, et al. Bacterial-fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol Rev. 2018;42:335–52. doi: 10.1093/femsre/fuy008. [DOI] [PubMed] [Google Scholar]
- 48.Kohlmeier S, Smits TH, Ford RM, Keel C, Harms H, Wick LY. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ Sci Technol. 2005;39:4640–6. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 49.Berthold T, Centler F, Hubschmann T, Remer R, Thullner M, Harms H, et al. Mycelia as a focal point for horizontal gene transfer among soil bacteria. Sci Rep. 2016;6:36390. doi: 10.1038/srep36390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pratama AA, van Elsas JD. A novel inducible prophage from the mycosphere inhabitant Paraburkholderia terrae BS437. Sci Rep. 2017;7:9156. doi: 10.1038/s41598-017-09317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang M, Visser S, Pereira e Silva MC, van Elsas JD. IncP-1 and PromA group plasmids are major providers of horizontal gene transfer capacities across bacteria in the mycosphere of different soil fungi. Microb Ecol. 2015;69:169–79. doi: 10.1007/s00248-014-0482-6. [DOI] [PubMed] [Google Scholar]
- 52.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–84. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011;111:5492–505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traxler MF, Kolter R. Natural products in soil microbe interactions and evolution. Nat Prod Rep. 2015;32:956–70. doi: 10.1039/C5NP00013K. [DOI] [PubMed] [Google Scholar]
- 55.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–53. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 56.De Nobili M, Contin M, Mondini C, Brookes PC. Soil microbial biomass is triggered into activity by trace amounts of substrate. Soil Biol Biochem. 2001;33:1163–70. doi: 10.1016/S0038-0717(01)00020-7. [DOI] [Google Scholar]
- 57.Abisado RG, Benomar S, Klaus JR, Dandekar AA, Chandler JR. Bacterial quorum sensing and microbial community interactions. MBio. 2018;8. 10.1128/mBio.02331-17. [DOI] [PMC free article] [PubMed]
- 58.Brameyer S, Bode HB, Heermann R. Languages and dialects: bacterial communication beyond homoserine lactones. Trends Microbiol. 2015;23:521–3. doi: 10.1016/j.tim.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Stasulli NM, Shank EA. Profiling the metabolic signals involved in chemical communication between microbes using imaging mass spectrometry. FEMS Microbiol Rev. 2016;40:807–13. doi: 10.1093/femsre/fuw032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. MBio. 2013;4:e00459–13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shank EA. Considering the lives of microbes in microbial communities. mSystems. 2018;3:e00155–17. doi: 10.1128/mSystems.00155-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patin NV, Schorn M, Aguinaldo K, Lincecum T, Moore BS, Jensen PR. Effects of actinomycete secondary metabolites on sediment microbial communities. Appl Environ Microbiol. 2017;83:e02331. [DOI] [PMC free article] [PubMed]
- 63.Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci USA. 2013;110:18380–5. doi: 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rangel DP, Superak C, Bielschowsky M, Farris K, Falconer RE, Baveye PC. Rapid prototyping and 3-D printing of experimental equipment in soil science research. Soil Sci Soc Am J. 2013;77:54–59. doi: 10.2136/sssaj2012.0196n. [DOI] [Google Scholar]
- 65.Borer BAM, Hatzimanikatis V, Or D. Integrating metabolic networks into an individual based model of bacterial life in soil (conference poster). ISME17. 2018.; pp 1.
- 66.Aleklett K, Kiers ET, Ohlsson P, Shimizu TS, Caldas VE, Hammer EC. Build your own soil: exploring microfluidics to create microbial habitat structures. ISME J. 2018;12:312–9. doi: 10.1038/ismej.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pessotti RC, Hansen BL, Traxler MF. In search of model ecological systems for understanding specialized metabolism. mSystems. 2018;3:e00175–17. doi: 10.1128/mSystems.00175-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Erez Z, Steinberger-Levy I, Shamir M, Doron S, Stokar-Avihail A, Peleg Y, et al. Communication between viruses guides lysis-lysogeny decisions. Nature. 2017;541:488–93. doi: 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Silpe JE, Bassler BL. A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell. 2019;176:268–80 e213. doi: 10.1016/j.cell.2018.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kronheim S, Daniel-Ivad M, Duan Z, Hwang S, Wong AI, Mantel I, et al. A chemical defence against phage infection. Nature. 2018;564:283–6. doi: 10.1038/s41586-018-0767-x. [DOI] [PubMed] [Google Scholar]
- 71.Traxler MF, Kolter R. A massively spectacular view of the chemical lives of microbes. Proc Natl Acad Sci USA. 2012;109:10128–9. doi: 10.1073/pnas.1207725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watrous J, Roach P, Alexandrov T, Heath BS, Yang JY, Kersten RD, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109:E1743–1752. doi: 10.1073/pnas.1203689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenberg E, Sharon G, Atad I, Zilber-Rosenberg I. The evolution of animals and plants via symbiosis with microorganisms. Environ Microbiol Rep. 2010;2:500–6. doi: 10.1111/j.1758-2229.2010.00177.x. [DOI] [PubMed] [Google Scholar]
- 74.Carrion VJ, Cordovez V, Tyc O, Etalo DW, de Bruijn I, de Jager VCL, et al. Involvement of Burkholderiaceae and sulfurous volatiles in disease-suppressive soils. ISME J. 2018;12:2307–21. doi: 10.1038/s41396-018-0186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cordovez V, Carrion VJ, Etalo DW, Mumm R, Zhu H, van Wezel GP, et al. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front Microbiol. 2015;6:1081. doi: 10.3389/fmicb.2015.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joice R, Yasuda K, Shafquat A, Morgan XC, Huttenhower C. Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab. 2014;20:731–41. doi: 10.1016/j.cmet.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.The Human Microbiome Project C. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McDonald JA, Fuentes S, Schroeter K, Heikamp-deJong I, Khursigara CM, de Vos WM, et al. Simulating distal gut mucosal and luminal communities using packed-column biofilm reactors and an in vitro chemostat model. J Microbiol Methods. 2015;108:36–44. doi: 10.1016/j.mimet.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 79.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]