Abstract
Microbial community data are commonly subjected to computational tools such as correlation networks, null models, and dynamic models, with the goal of identifying the ecological processes structuring microbial communities. A major assumption of these methods is that the signs and magnitudes of species interactions and vital rates can be reliably parsed from observational data on species’ (relative) abundances. However, we contend that this assumption is violated when sample units contain any underlying spatial structure. Here, we show how three phenomena—Simpson’s paradox, context-dependence, and nonlinear averaging—can lead to erroneous conclusions about population parameters and species interactions when samples contain heterogeneous mixtures of populations or communities. At the root of this issue is the fundamental mismatch between the spatial scales of species interactions (micrometers) and those of typical microbial community samples (millimeters to centimetres). These issues can be overcome by measuring and accounting for spatial heterogeneity at very small scales, which will lead to more reliable inference of the ecological mechanisms structuring natural microbial communities.
Subject terms: Microbial ecology, Microbial ecology, Community ecology
Common “pattern-to-process” inferential methods yield erroneous results
Advances in sequencing technology offer microbiologists unprecedented access to the composition and dynamics of microbial communities [1]. Marker gene and metagenomic surveys regularly chronicle hundreds to thousands of taxa, many previously unknown, all seemingly co-occurring within their respective habitats. In possession of these large observational datasets, we have adapted theory and methods developed from plant and animal ecology to investigate how species interactions—such as competition, predation, and facilitation—structure microbial communities [2, 3].
Without experimental systems in which competition (or any other interaction) may be directly manipulated and detected, researchers often employ randomization-based null models, correlation networks, and population dynamic models to identify and quantify putative interspecific interactions from observational sequence data [4–7]. Here, negative covariation between the (relative) abundances of taxa are assumed to result from negative interspecific interactions such as competition, though practitioners generally agree that precisely parsing ecological mechanisms from observational data remains unreliable. In fact, the utility of these methods for reliably isolating and quantifying signals of competition from alternative community assembly processes such as habitat filtering and trophic interactions has been disputed in the community ecology literature for decades [8].
Recently, a number of studies have challenged null model and correlation-based methods to recapitulate known interactions in well-studied marine intertidal habitats [9–11]. In all cases, these tests revealed troubling inaccuracies and discrepancies among the various methods, calling into question their ability to reliably identify true ecological interactions. For microbial communities, the only successful validations of these methods have occurred in simple, well-mixed liquid cultures [7] or in situations where direct, inter-kingdom chemical inhibition occurs [12]. Taken in concert, these studies highlight potential pitfalls in our ability to correctly identify species interactions when communities are sampled over underlying spatial heterogeneity. Most natural microbial communities are spatially structured and exhibit marked heterogeneity at multiple spatial scales. We argue that failure to account for this underlying spatial heterogeneity in environmental samples can undermine our conclusions about the ecological processes structuring microbial assemblages [13].
Causes and consequences of heterogeneity in microbial samples
Typical sample volumes used for environmental marker gene and metagenomics studies are rarely smaller than 0.1 ml, but can be as large as 100 l of seawater and 100 g of soil in low-DNA habitats. Unless these samples come from a perfectly mixed, completely homogeneous medium, they will contain at least some amount of spatial structure. For example, a typical 0.25 g sample of soil containing particles 1 mm in diameter (i.e., a very coarse sand) will inevitably contain hundreds to thousands of discrete granules on which microbial communities can assemble. These discrete habitats can represent a heterogeneous array of environments or resources, each selecting for their own unique local microbial communities [13]. However, even a physicochemically homogeneous collection of particles can contain a mosaic of distinct microbial communities owing to the effects of limited or asymmetric dispersal, priority effects, and successional turnover.
Fine-scale heterogeneity in microbial communities appears to be a general property of environmental samples, having been repeatedly documented in aquatic, soil, fecal, leaf surface, and wastewater habitats [14–19]. Owing to this, marker gene samples commonly represent sums or averages of sequence reads made over underlying environmental heterogeneity, leaving us with a bulk inventory of operational taxonomic units (OTUs) and their (often relative) abundances without information on spatial context. Because microbial interactions such as resource competition, phage predation, DNA transfer, and syntrophy are hypothesized to take place at spatial scales much smaller than that of the typical bulk sample [20], it can be argued that many marker gene samples actually measure the metacommunity—a collection of semi-autonomous communities linked through dispersal [21]. In the following sections, we illustrate how collecting samples at the metacommunity scale can introduce errors into computational estimates of interspecific interactions by virtue of three phenomena: Simpson’s paradox, context-dependence, and nonlinear averaging. Note that although we present total abundance data throughout our scenarios, these phenomena also apply to compositional (i.e., relative abundance) data, which are more commonly collected in environmental marker gene surveys.
Simpson’s paradox
Simpson’s paradox refers to the reversal or negation of a statistical association between two variables, X and Y, when conditioned on a third variable, Z [22–25]. In ecology, this Z variable might include information on spatial variation among local patches, which, if accounted for, changes the direction of a trend at larger spatial scales [26]. Computational approaches to inferring microbial interactions can be sensitive to the effects of Simpson’s paradox. For instance, the inferred signs of interspecific correlation coefficients might change when comparing analytic results obtained from bulk community samples with results that have statistically accounted for underlying variation in microhabitats or resource availability within bulk samples.
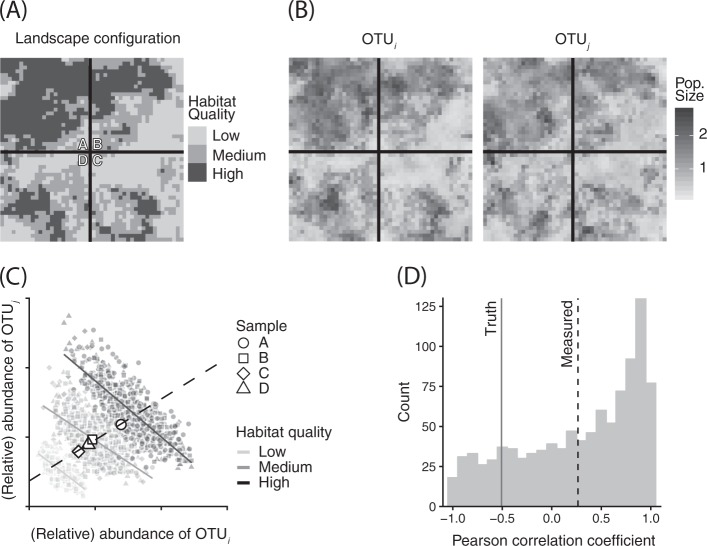
To illustrate this point, consider a hypothetical study that uses data obtained from bulk soil samples to infer the sign of interspecific interaction between two microbial OTUs. If the true nature of this interaction is competitive, then our results are anticipated to reveal a negative correlation between the abundances of the OTUs. To add some realism to this scenario, let us assume that each of our samples represent collections of discrete microhabitats on which our focal taxa grow (Fig. S1). Finally, we might also make the realistic assumption that both OTUs respond similarly to these discrete microhabitats such that sub-optimal habitats support fewer individuals of both species. If we create bulk soil samples by subsampling a particulate, heterogeneous soil habitat containing populations of our two competitors (Fig. 1a), we find that these samples’ OTU abundances reflect both local habitat composition, as well as negative interspecific interactions (Fig. 1b). As a result, our interspecific correlation estimates from bulk samples predict positive correlations between our two OTUs, whereas their true, competitive interactions result in negative correlations when the local environment is accounted for (Fig. 1c). Furthermore, by repeating this experiment many times using the same parameters but different habitat configurations (Fig. S2), each time re-assembling our bulk samples, we encounter an overwhelming majority of cases where the inferred sign of interaction between our two OTUs (positive) is the opposite of its true sign (negative), leading us to erroneously conclude that these species are not strong competitors when, in truth, they are (Fig. 1d; details in supplemental information).
Fig. 1.
Example of how Simpson’s paradox can influence the identification of interspecific interactions. a We randomly generated microbial landscapes with underlying spatial heterogeneity in habitat quality and then simulated competition between two OTUs using a spatially explicit gLV model with local dispersal. b After 60 generations, the populations had reached global equilibria. c From these gridded populations, we can see that the interspecific Pearson’s correlation coefficients, when controlled for habitat type, show the true negative correlations. However, correlations measured using samples taken from these landscapes (here, the solid lines bisecting panel a), result in erroneous estimates of interspecific correlations. d To assess the generality of this finding, we repeated this simulation exercise 1000 times across a variety of different habitat configurations and found that, on average, the sample-scale interspecific correlation coefficients (dashed line) do not have the same sign as the true, local-scale coefficients (solid line). Methods and code are provided in the supplementary information
Crucially, detection methods relying on false-discovery rate correction will fail to identify this error as such, as statistical significance is decoupled from the effects of Simpson’s Paradox [27]. Because of these issues, we contend that unless the assumption of homogeneity within and among microbial community samples is justified, interspecific interaction coefficients derived from correlation or model-based approaches should be interpreted with extreme caution, and should always include a statement concerning the spatial context of the sample including potential sources of underlying spatial heterogeneity.
Context dependence
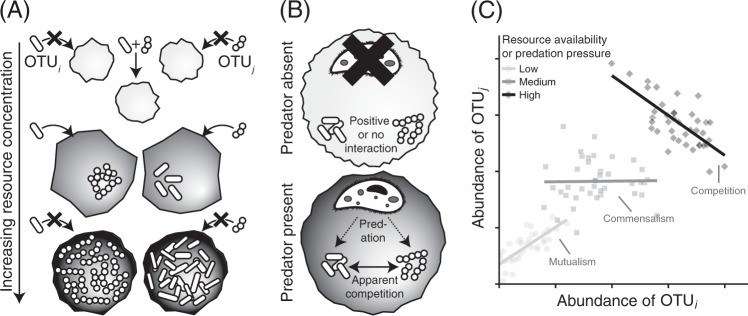
A common assumption of computational models for identifying species interactions is that the sign and strength of interactions are immutable across time and space, yet this simplifying assumption is widely acknowledged not to hold. Often invoked for statistical convenience, this assumption reduces the sample sizes required for estimating correlation coefficients or population parameters, and permits the use of graph theoretic descriptors of network structure (connectance, nestedness, etc.). However, numerous laboratory experiments have documented context-dependent interactions arising from variation in population densities, community composition, or environmental context, such that interactions measured at one place and time cannot reliably be extrapolated across habitats [28–31]. For instance, a recent study documented predictable shifts in the sign of species interactions with changing resource concentrations in experimental yeast communities as cross-feeding gave way to competition [32] (Fig. 2a). The presence of predators can also mediate the sign of interspecific interactions through a variety of mechanisms [33] (e.g., Fig. 2b). Likewise, a meta-analysis of hundreds of experiments uncovered a strong effect of spatial heterogeneity on context-dependent species interactions [34]. Consequently, it is not unreasonable to expect the signs of microbial interactions to change across gradients of resource density, predation pressure, or other indicators of habitat quality (Fig. 2c). While temporal correlation network approaches might be used to circumvent the static interactions assumption at larger spatial scales or in well-mixed samples, they cannot account for variable interactions arising from underlying, unmeasured spatial heterogeneity within individual samples.
Fig. 2.
Examples of context-dependent species interactions. a Resource availability can modulate the sign of interspecific interactions. For instance, local resource limitation can weaken the strength of competition when (i) it selects for cross-feeding or another mutualistic, resource-concentrating behavior, or (ii) when it limits the strength of interspecific negative density dependence. b Likewise, in situations where a shared predator is present, species that do not compete for shared resources can experience apparent competition by supplementing the predator densities. The dashed lines denote trophic interactions, solid line denotes competitive interaction. c These context-dependent interactions can lead to highly variable estimates of the signs of OTU interactions, depending on the spatial distribution of resources or predators within the sample
From a theoretical perspective, context-dependence is hypothesized to be a critical factor for maintaining diversity in spatially structured communities. For instance, the abilities of two competing microbial strains to coexist will be enhanced if the negative impacts of competition experienced by each strain are stronger in more favorable habitat patches [35]. Given that microbial species richness appears to peak in particulate, heterogeneous habitats (soil, sediments) [1], context-dependent interactions within these habitats may be quite common and important in promoting high levels of diversity. Currently, the extent of context-dependent interactions in spatially structured microbial communities remains largely unknown. We note, though, that correlation network approaches have been successfully used to identify context-dependent interactions robust to experimental ground-truthing [12, 36]. However, until the prevalence and magnitude of context-dependent microbial interactions are better understood, we encourage researchers to exercise discretion when making general statements concerning any local estimates of interspecific interactions, ideally contextualizing results to the specific environment and scale at which measurements were taken.
Nonlinear averaging
The previous two sections concerned issues that arise when quantifying local microbial interactions from heterogeneous samples. However, we also face difficulties when using microbial community data collected at very small scales to quantify the aggregate behavior of aggregate microbial communities. Imagine that we are now able to obtain measurements of microbial populations at the scale of the individual microhabitat patches. Importantly, these data are collected at the spatial scale over which intraspecific interactions play out, which, in a heterogeneous sample experiencing dispersal among particles, is at the scale of individual microhabitat patches or particles. Called the characteristic scale, it is the scale which maximizes the ratio of deterministic signal to the influences of stochasticity and spatial heterogeneity [37], making it the optimal scale for measuring and characterizing the effects of deterministic species interactions.
Let us now envision a scenario where we wish to quantify whether a microbial OTU’s competitive ability is a function of its local soil habitat. Since accurately estimating the strength of competition in our samples is of paramount importance, suppose we have conducted our sequencing surveys at appropriately small characteristic scales and have generated time series data from this assortment of individual particles. We then fit a population dynamic model to these data in order to estimate our OTU’s growth rate and competitive interactions among different soil types, adequately replicated within each type. The generalized Lotka-Volterra (gLV) population dynamic model is increasingly being utilized for this purpose [38]. Fitting such a differential equation model requires estimating parameters describing a focal species’ growth rates and interspecific interactions. The gLV model commonly takes the form
| 1 |
where Ni is the abundance of OTU i, μi is its maximum per capita growth rate, and αij is a parameter describing the proportional change in its growth rate with conspecific or heterospecific densities. Values of αij greater than zero imply that OTU j has a positive effect on OTU i, which might stem from interactions such as syntrophy, whereas values less than zero can signify interactions such as competition or chemical inhibition.
For illustrative purposes, let us simplify our problem of estimating competition among soil types by assuming that only our single focal OTU occupies our habitats, and so is only capable of experiencing intraspecific competition. This permits us to simplify our model to the case where (i = j), and define αij = −μiKi−1, where Ki represents the local carrying capacity of our OTU i. This results in the familiar logistic population growth model describing decelerating microbial population growth with increasing population density. Expanding this model across a spatially structured array of n individual particles, we obtain the equation
| 2 |
where Nx are the local sub-populations of our focal OTU on habitat particle x.
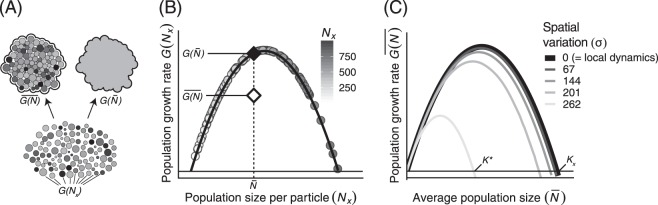
With a collection of population dynamic equations describing our individual particles, we can now aggregate these local dynamics to obtain general growth parameters to compare growth across soil types. This scaling-up process requires a spatial averaging of local population dynamics (Fig. 3a). Crucially, because the average of a nonlinear function is not equal to the function of its averaged covariates (i.e., when f″(N) ≠ 0), to scale up microbial population dynamics—which are almost unanimously nonlinear—by averaging across spatially variable local populations will result in biases proportional to the spatial population variation and model’s nonlinearity. This principle, called Jensen’s inequality, has important consequences for our abilities to accurately estimate scaled-up model parameters and make predictions from any gLV model when the local populations contain spatial heterogeneity, as is often the case.
Fig. 3.
a Illustration of the concept of scaling-up local microbial community dynamics to quantify the behavior of an aggregate sample. Degree of shading denotes an OTU’s population sizes across a heterogeneous collection of particles governed by the shared, nonlinear dynamics, G(Nx), from Eq. 2. Note the conceptual differences between aggregating these data by averaging over the local nonlinear dynamics, , and by fitting our small-scale dynamical model to the average population density, . b The differences in these aggregation procedures result in differing estimates for scaled-up population dynamics. The black curve shows the logistic governing dynamics, G(Nx), of populations on individual particles (shaded circles). Note the difference in growth rates between the correctly spatially averaged growth function (white diamond) and growth function fit to the spatial average population density (black diamond). c Increasing the spatial variation of local populations results in vastly different spatially averaged population dynamics. Here again, the black line denotes the local dynamics, G(Nx), which equals the spatially averaged dynamics when there is no variation among subpopulations. For this concave-down function, increasing the spatial variation causes the scaled-up carrying capacity, K*, to be smaller than the local carrying capacity, Kx
The consequences of this spatial averaging process are illustrated in Fig. 3b. For notational simplicity, we replace the growth function in Eq. 2, μNx(1−Nx/K), with G(Nx). The spatially averaged dynamical equation that we wish to obtain is . Calculating our population dynamic model using the spatial averages of the populations we have measured, , overestimates the correctly scaled-up population growth function, . In Fig. 3c, we generated four collections of particles in which spatially explicit populations have been randomly drawn from lognormal distributions having equal means but different variances (σ2). We then used these simulated data to fit four spatially averaged population growth functions, . Our results demonstrate how increasing the spatial variation among local populations has the effect of changing our scaled-up estimates of carrying capacity. The challenge for microbiologists is to correctly estimate using their measured population densities, Nx. Fortunately, if we have already collected these values, and if they can be reasonably fit to a population dynamic model, we can use the tools of scale transition theory [39, 40] to correctly obtain scaled-up population parameters. We introduce these methods in the following section and provide a more thorough overview in the supplemental information.
Recommendations moving forward
Despite the various ways in which spatial heterogeneity can subvert our interpretation or complicate our assessment of microbial community interactions and dynamics, we are optimistic that these issues can be surmounted with prudent data collection, analysis, and interpretation. The lurking effects of habitat heterogeneity are most effectively mitigated by quantifying microbial populations or communities at the spatial scales over which cell-cell interactions occur, which is on the scale of micrometers to millimeters. Micron-scale sampling has successfully been accomplished using individual grains of sand [14], aquatic organic particles [41], and sludge granules [42, 43]— all of which encountered marked heterogeneity among particles. Sampling at this scale is facilitated by technologies such as fluorescence-activated cell sorting and laser-assisted microdissection, which offer opportunities to precisely and efficiently capture individual microscopic particles for sequencing, as well as fluorescence in situ hybridization microscopy to determine the spatial associates of individual populations. A noteworthy example of such an approach is the in situ microscopic validation of a network-predicted mutualistic interaction [44]. However, as we have seen, even measurements made at the appropriate characteristic scales can be challenging to generalize due to the issues of nonlinear averaging and context-dependence.
The restrictive assumptions of most correlation network and null models hinder our reliable assessment of microbial interactions in all but the most homogeneous samples. However, the influence of Simpson’s paradox and context-dependence may be surmounted by measuring and statistically accounting for the confounding effects of environmental and/or community variation among samples. Though methods to control for environmental factors using partial correlations or joint species distribution models are available [4, 45, 46], unless environmental characterization is made at the appropriate scales, they cannot account for the spatial variation contained within a single heterogeneous sample. These methods have also delivered underwhelming results when challenged to predict an empirically measured interaction web, after accounting for the confounding effects of environmental variation [10]. Though it will be challenging to collect environmental data at such fine spatial grains, such data could be used to test the alternative hypotheses of habitat filtering, competition, and dispersal limitation—all of which can feasibly manifest as identical metacommunity patterns in the presence of spatial variation.
While creative new statistical and mechanistic modeling approaches for identifying nonlinear and context-dependent species interactions are becoming available [46–50], we suggest these methods be ground-truthed with more complex and realistic data than are currently in use. For example, rather than using time series simulated from equilibrial, non-spatial Lotka-Volterra equations to benchmark a new method, a more powerful validation routine could use data simulated from spatially explicit agent-based models, which can test methods’ robustness to spatial heterogeneity, scale-dependence, and demographic stochasticity. We also encourage the inclusion of dynamic parameters in generalized Lotka-Volterra models. While it is challenging to estimate these parameters from observational data, experiments consistently show that microbial growth rate, carrying capacity, and interaction parameters are functions of their underlying environments. A benefit of including environmentally dependent growth parameters in gLV models is that these models can then be used to quantify the effects of various coexistence-promoting mechanisms [51]. Context-dependent parameters also allow us to investigate the effects of environmental change on microbial populations and communities.
The increasing use of gLV models in microbial ecology also prompts us to account for the effects of nonlinear spatial averaging on scaled up population dynamics (see section Nonlinear averaging). Chesson’s scale transition theory [34, 35] provides a mathematical framework for tackling the issues of spatial heterogeneity and nonlinearities in gLV models. We introduce the scale transition using two simple models, but refer interested readers to the original papers [39, 40] and the supplementary information section for general scale transition approaches. Continuing from section Nonlinear averaging, we can calculate the scaled-up population dynamics, , by accounting for the nonlinearity in G(Nx) using its second derivative, G″(Nx), as well as the spatial variation in Nx, measured by the spatial variance, Var(N). The full, spatially averaged population model can be approximated as
| 3 |
Where g(N) = μ[1−N/K] and . This approximation is exact when the growth function is quadratic (as is the case for logistic growth).
A similar, albeit more complicated scale transition can be calculated for a multispecies gLV model (Eq. 1) [34]. This model is commonly used to identify interspecific interactions, denoted by the αij parameters. By defining and g(Wi) = μi+Wi, the scaled up version of Eq. 1 can be written as a function of mean field terms, a nonlinearity term, and spatial variances and covariances:
| 4 |
where . Once again, we see that the spatially averaged population dynamics are not simply a function of average populations across space. However, the only extra information needed to calculate the scale transition are the spatial variances and covariances of the populations, which we can approximate by measuring local population densities across a sufficient number of particles within a sample. Thus, the calculation of scale transition terms is straightforward once they are defined for a particular dynamic model.
Given the potential for biases and errors stemming from the joint effects of underlying spatiotemporal heterogeneity and other methodological choices (e.g., relative abundance transformations, normalization techniques) [52], it may seem like the inference of species interactions from observational microbial data represents an underdetermination problem. That is, there may be multiple, or even infinite potential mechanisms capable of generating an observed community pattern. However, this problem, like many in ecology and evolution, can more precisely be described as an example of contrast failure [53]. Instead of a solution-free, underdetermined system, we instead have one where our failure to parse competing hypotheses is a transient consequence of data insufficiency. Access to better, more contrastive data, derived either experimentally or observationally at the appropriate spatiotemporal scales, will refine our ability to discriminate among alternative hypotheses. To clarify, we do not advocate for the abandonment of ‘pattern-to-process’ approaches for parsing microbial communities, which have already proven useful for identifying the ‘keystone taxa’ associated with alternative community configurations and ecosystem functioning [12, 36]. On the contrary, we are optimistic about continued methodological development in this area—particularly with regard to the spatial scale of sample characterization. In the meantime, we implore researchers to consider and confront the lurking effects of spatial structure on their inferred microbial interaction networks and growth parameters. At minimum, this could simply comprise a comment on the spatiotemporal scale over which the results are anticipated to hold and a description of the spatial structure contained within a sample unit.
Supplementary information
Acknowledgements
The authors thank members of the Jones Lab, J. Prosser, and three anonymous referees for helpful discussion and feedback. Financial support was provided by the US National Science Foundation (DEB-1442246).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41396-019-0463-3) contains supplementary material, which is available to authorized users.
References
- 1.Thompson LR, Sanders JG, McDonald D, Amir A, Ladau J, Locey KJ, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551:457–63. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prosser JI, Bohannan BJM, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, et al. The role of ecological theory in microbial ecology. Nat Rev Microbiol. 2007;5:384–92. doi: 10.1038/nrmicro1643. [DOI] [PubMed] [Google Scholar]
- 3.Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77:342–56. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Micro. 2012;10:538–50. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 5.Bálint M, Bahram M, Eren AM, Faust K, Fuhrman JA, Lindahl B, et al. Millions of reads, thousands of taxa: microbial community structure and associations analyzed via marker genes. FEMS Microbiol Rev. 2016;40:686–700. doi: 10.1093/femsre/fuw017. [DOI] [PubMed] [Google Scholar]
- 6.Layeghifard M, Hwang DM, Guttman DS. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 2017;25:217–28. doi: 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Angulo MT, Friedman J, Waldor MK, Weiss ST, Liu Y-Y. Mapping the ecological networks of microbial communities. Nat Commun. 2017;8:2042. doi: 10.1038/s41467-017-02090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roughgarden J. Competition and theory in community ecology. Am Natural. 1983;122:583–601. doi: 10.1086/284160. [DOI] [Google Scholar]
- 9.Sander EL, Wootton JT, Allesina S. Ecological network inference from long-term presence-absence data. Sci Rep. 2017;7:7154. doi: 10.1038/s41598-017-07009-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barner AK, Coblentz KE, Hacker SD, Menge BA. Fundamental contradictions among observational and experimental estimates of non-trophic species interactions. Ecology. 2018;99:557–66. doi: 10.1002/ecy.2133. [DOI] [PubMed] [Google Scholar]
- 11.Freilich MA, Wieters E, Broitman BR, Marquet PA, Navarrete SA. Species co-occurrence networks: Can they reveal trophic and non-trophic interactions in ecological communities? Ecology. 2018;99:690–9. doi: 10.1002/ecy.2142. [DOI] [PubMed] [Google Scholar]
- 12.Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, et al. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell. 2018;175:973–83.e14. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry D, Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5:219. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Probandt D, Eickhorst T, Ellrott A, Amann R, Knittel K. Microbial life on a sand grain: from bulk sediment to single grains. ISME J. 2018;12:623–33. doi: 10.1038/ismej.2017.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–5. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 16.Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microbiol. 2001;67:4215–24. doi: 10.1128/AEM.67.9.4215-4224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swidsinski A, Loening–Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008;135:568–79.e2. doi: 10.1053/j.gastro.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 18.Remus-Emsermann MNP, Tecon R, Kowalchuk GA, Leveau JHJ. Variation in local carrying capacity and the individual fate of bacterial colonizers in the phyllosphere. ISME J. 2012;6:756–65. doi: 10.1038/ismej.2011.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Gil G, Holliger C. Aerobic granules: microbial landscape and architecture, stages, and practical implications. Appl Environ Microbiol. 2014;80:3433–41. doi: 10.1128/AEM.00250-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordero OX, Datta MS. Microbial interactions and community assembly at microscales. Curr Opin Microbiol. 2016;31:227–34. doi: 10.1016/j.mib.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leibold MA, Holyoak M, Mouquet N, Amarasekare P, Chase JM, Hoopes MF, et al. The metacommunity concept: a framework for multi-scale community ecology. Ecol Lett. 2004;7:601–13. doi: 10.1111/j.1461-0248.2004.00608.x. [DOI] [Google Scholar]
- 22.Thorndike EL. On the fallacy of imputing the correlations found for groups to the individuals or smaller groups composing them. Am J Psychol. 1939;52:122–4. doi: 10.2307/1416673. [DOI] [Google Scholar]
- 23.Simpson EH. The Interpretation of interaction in contingency tables. J R Stat Soc Ser B. 1951;13:238–41. [Google Scholar]
- 24.Blyth CR. On Simpson’s paradox and the sure-thing principle. J Am Stat Associat. 1972;67:364–6. doi: 10.1080/01621459.1972.10482387. [DOI] [Google Scholar]
- 25.Appleton DR, French JM, Vanderpump MPJ. Ignoring a covariate: an example of Simpson’s paradox. Am Stat. 1996;50:340–1. [Google Scholar]
- 26.Scheiner SM, Cox SB, Willig MR, Mittelbach GG, Osenberg CW, Kaspari M. Species richness, species–area curves and Simpson’s paradox. Evol Ecol Res. 2000;2:791–802. [Google Scholar]
- 27.Heydtmann M. The nature of truth: Simpson’s paradox and the limits of statistical data. QJM. 2002;95:247–9. doi: 10.1093/qjmed/95.4.247. [DOI] [PubMed] [Google Scholar]
- 28.de Muinck EJ, Stenseth NC, Sachse D, Roost J, vander, Rønningen KS, Rudi K, et al. Context-dependent competition in a model gut bacterial community. PLoS ONE. 2013;8:e67210. doi: 10.1371/journal.pone.0067210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu A, Archer AM, Biggs MB, Papin JA. Growth-altering microbial interactions are responsive to chemical context. PLoS ONE. 2017;12:e0164919. doi: 10.1371/journal.pone.0164919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tecon R, Ebrahimi A, Kleyer H, Levi SE, Or D. Cell-to-cell bacterial interactions promoted by drier conditions on soil surfaces. PNAS. 2018;115:9791–6. doi: 10.1073/pnas.1808274115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gould AL, Zhang V, Lamberti L, Jones EW, Obadia B, Gavryushkin A, et al. High-dimensional microbiome interactions shape host fitness. Proc Nat Acad Sci USA. 2018;115 E11951–E11960. [DOI] [PMC free article] [PubMed]
- 32.Hoek TA, Axelrod K, Biancalani T, Yurtsev EA, Liu J, Gore J. Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 2016;14:e1002540. doi: 10.1371/journal.pbio.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chesson P, Kuang JJ. The interaction between predation and competition. Nature. 2008;456:235–8. doi: 10.1038/nature07248. [DOI] [PubMed] [Google Scholar]
- 34.Chamberlain SA, Bronstein JL, Rudgers JA. How context dependent are species interactions? Ecol Lett. 2014;17:881–90. doi: 10.1111/ele.12279. [DOI] [PubMed] [Google Scholar]
- 35.Chesson P. General theory of competitive coexistence in spatially-varying environments. Theor Population Biol. 2000;58:211–37. doi: 10.1006/tpbi.2000.1486. [DOI] [PubMed] [Google Scholar]
- 36.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim S-T, Weigel D, et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascual M, Levin SA. From individuals to population densities: searching for the intermediate scale of nontrivial determinism. Ecology. 1999;80:2225–36. doi: 10.1890/0012-9658(1999)080[2225:FITPDS]2.0.CO;2. [DOI] [Google Scholar]
- 38.Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, et al. Ecological modeling from time-series inference: Insight into dynamics and stability of intestinal microbiota. PLoS Comput Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chesson P, Donahue MJ, Melbourne BA, Sears ALW. Scale transition theory for understanding mechanisms in metacommunities. In: Holyoak M, Leibold MA, Holt RD, editors. Metacommunities: spatial dynamics and ecological communities. Chicago: University Of Chicago Press; 2005. pp. 279–306. [Google Scholar]
- 40.Chesson P. Scale transition theory: Its aims, motivations and predictions. Ecol Complex. 2012;10:52–68. doi: 10.1016/j.ecocom.2011.11.002. [DOI] [Google Scholar]
- 41.Bižić-Ionescu M, Ionescu D, Grossart H-P. Organic particles: heterogeneous hubs for microbial interactions in aquatic ecosystems. Front Microbiol. 2018;9:2569. doi: 10.3389/fmicb.2018.02569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuroda K, Nobu MK, Mei R, Narihiro T, Bocher BTW, Yamaguchi T, et al. A single-granule-level approach reveals ecological heterogeneity in an upflow anaerobic sludge blanket reactor. PLoS ONE. 2016;11:e0167788. doi: 10.1371/journal.pone.0167788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leventhal GE, Boix C, Kuechler U, Enke TN, Sliwerska E, Holliger C, et al. Strain-level diversity drives alternative community types in millimetre-scale granular biofilms. Nat Microbiol. 2018;3:1295. doi: 10.1038/s41564-018-0242-3. [DOI] [PubMed] [Google Scholar]
- 44.Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, et al. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 45.Ovaskainen O, Hottola J, Siitonen J. Modeling species co-occurrence by multivariate logistic regression generates new hypotheses on fungal interactions. Ecology. 2010;91:2514–21. doi: 10.1890/10-0173.1. [DOI] [PubMed] [Google Scholar]
- 46.Harris DJ. Inferring species interactions from co-occurrence data with Markov networks. Ecology. 2016;97:3308–14. doi: 10.1002/ecy.1605. [DOI] [PubMed] [Google Scholar]
- 47.Biswas S, McDonald M, Lundberg DS, Dangl JL, Jojic V. Learning microbial interaction networks from metagenomic count data. In: Przytycka TM editor. Research in computational molecular biology. Springer International Publishing; 2015. pp. 32–43. [DOI] [PubMed]
- 48.Momeni B, Xie L, Shou W. Lotka-Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. Elife. 2017;6:e25051. doi: 10.7554/eLife.25051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tackmann J, Rodrigues JFM, von Mering C. Rapid inference of direct interactions in large-scale ecological networks from heterogeneous microbial sequencing data. bioRxiv 2018;390195. 10.1101/390195. [DOI] [PubMed]
- 50.Ma B, Wang H, Dsouza M, Lou J, He Y, Dai Z, et al. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J. 2016;10:1891–901. doi: 10.1038/ismej.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chesson P. Mechanisms of maintenance of species diversity. Ann Rev Ecol Systemat. 2000;31:343–66. doi: 10.1146/annurev.ecolsys.31.1.343. [DOI] [Google Scholar]
- 52.Weiss S, Treuren WV, Lozupone C, Faust K, Friedman J, Deng Y, et al. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 2016;10:1669–81. doi: 10.1038/ismej.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Forber P. Spandrels and a pervasive problem of evidence. Biol Philos. 2008;24:247. doi: 10.1007/s10539-008-9144-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.