Abstract
Plant genomes sustain various forms of DNA damage that stall replication forks. Translesion synthesis (TLS) is one of the pathways to overcome stalled replication in which specific polymerases (TLS polymerase) perform bypass synthesis across DNA damage. This article gives a brief overview of plant TLS polymerases. In Arabidopsis, DNA polymerase (Pol) ζ, η, κ, θ, and λ and Reversionless1 (Rev1) are shown to be involved in the TLS. For example, AtPolη bypasses ultraviolet (UV)-induced cyclobutane pyrimidine dimers in vitro. Disruption of AtPolζ or AtPolη increases root stem cell death after UV irradiation. These results suggest that AtPolζ and ATPolη bypass UV-induced damage, prevent replication arrest, and allow damaged cells to survive and grow. In general, TLS polymerases have low fidelity and often induce mutations. Accordingly, disruption of AtPolζ or AtRev1 reduces somatic mutation frequency, whereas disruption of AtPolη elevates it, suggesting that plants have both mutagenic and less mutagenic TLS activities. The stalled replication fork can be resolved by a strand switch pathway involving a DNA helicase Rad5. Disruption of both AtPolζ and AtRAD5a shows synergistic or additive effects in the sensitivity to DNA-damaging agents. Moreover, AtPolζ or AtRev1 disruption elevates homologous recombination frequencies in somatic tissues. These results suggest that the Rad5-dependent pathway and TLS are parallel. Plants grown in the presence of heat shock protein 90 (HSP90) inhibitor showed lower mutation frequencies, suggesting that HSP90 regulates mutagenic TLS in plants. Hypersensitivities of TLS-deficient plants to γ-ray and/or crosslink damage suggest that plant TLS polymerases have multiple roles, as reported in other organisms.
Keywords: translesion synthesis, UV, mutation, DNA damage, genome stability
Introduction
Accurate replication of genomic DNA is vital for maintaining genome integrity. However, genomic DNA sustains various forms of damage caused by internal and external agents. Ultraviolet (UV) light is a major cause of DNA damage for land plants. It induces the formation of covalent bonds between the two adjacent pyrimidines. The two major products of UV damage, cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidone photoproducts [(6-4)PPs], are quickly repaired by the action of CPD and 6-4 photolyases in plant cells (Britt, 1999; Li et al., 2010; Hitomi et al., 2012). In addition, nucleotide excision repair (NER) plays an important role in removing UV damage (Kimura et al., 2004; Kunz et al., 2005; Canturk et al., 2016). Nevertheless, the remaining damage is toxic for cells because it distorts the template structure and prevents replication. This stalled replication creates a fragile single-strand region that easily leads to double-strand breaks (DSBs), so organisms have multiple pathways to solve the stalled replication fork. Translesion synthesis (TLS) is one such pathway in which specific polymerases (TLS polymerase) are recruited to the replication machinery and perform the bypass synthesis across the DNA damage (Vaisman and Woodgate, 2017). Figure 1A illustrates the concept of TLS activity. When encountering DNA damage, the replicative polymerase (replicase) stalls because of distorted helix geometry. TLS polymerase, carrying a flexible active site, replaces the replicase and inserts one or more nucleotide(s) opposite the damage. Because of the relaxed constraints of these active sites, the TLS polymerase has a low fidelity and often incorporates one or more incorrect nucleotide(s) that can be removed by the exonuclease activity of replicases or the mismatch repair mechanism. However, unremoved errors result in base substitutions, frameshifts, or other types of mutation. This mutagenic nature of TLS has been linked to the senescence, carcinogenesis, and evolution of organisms.
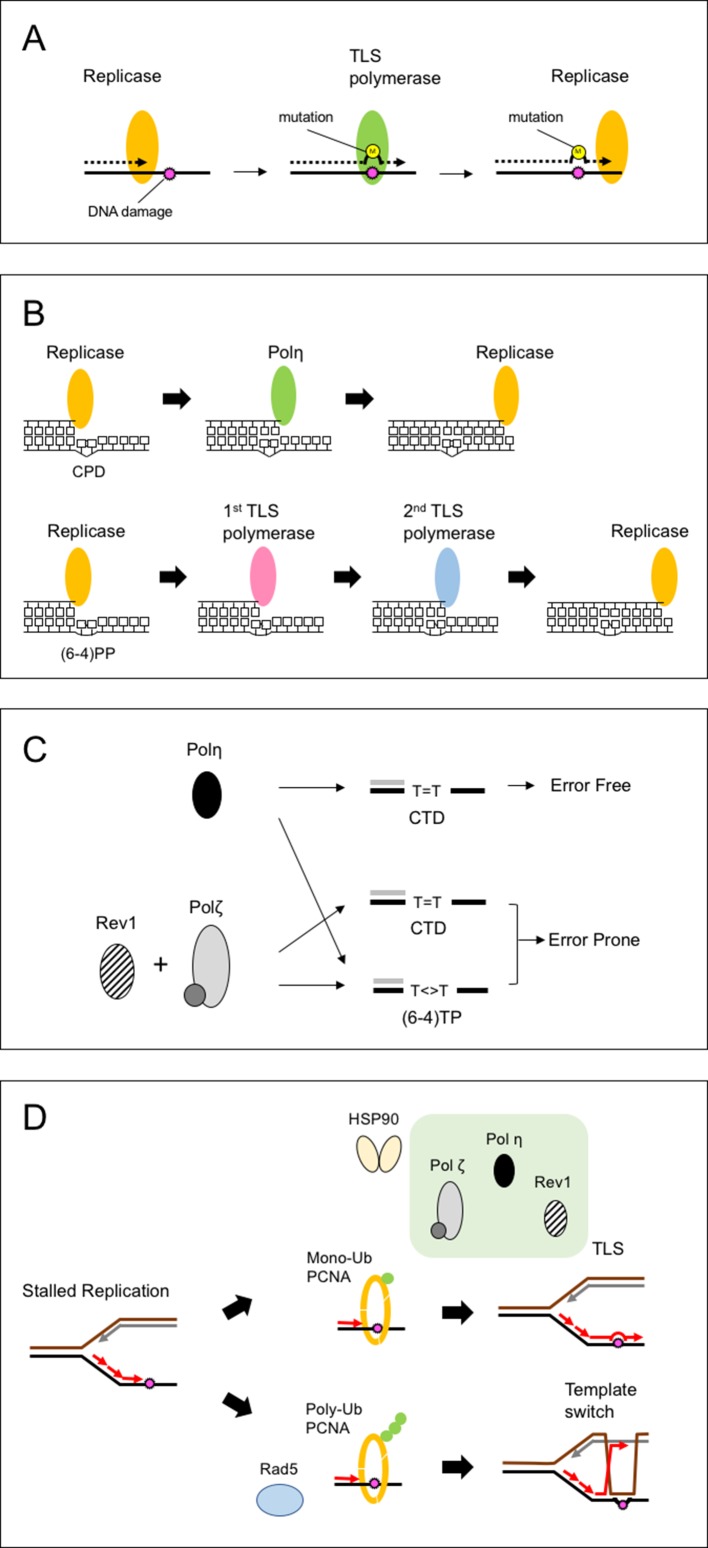
Figure 1.
Schematic of translesion synthesis (TLS). (A) Concept of TLS. When encountering DNA damage, the replicase stalls before the damage. TLS polymerase replaces the replicase and inserts one or more nucleotides opposite the damage. Because of the low fidelity, TLS polymerase incorporates one or more incorrect nucleotides, resulting in base substitutions, frameshifts, or other types of mutation. (B) Proposed model for the bypass of two major forms of ultraviolet (UV) damage. The model was proposed from the biochemical activities of TLS polymerases. The cyclobutane pyrimidine dimer (CPD) is efficiently bypassed by Polη (upper). However, no polymerase can complete the bypass of (6-4) photoproducts [(6-4)PP] by itself. Thus, (6-4)PPs may be bypassed by two polymerases, incorporating nucleotides one after the other (lower). (C) A model for UV-induced mutagenesis at the TT site in plants. The cyclobutane TT dimer (CTD) is efficiently bypassed by Polη in an error-free manner; any misincorporation is removed by replicases. In contrast, Polζ and Rev1 are involved in the error-prone bypass for both CTD and (6-4) TT photoproducts [(6-4)TP]. Polη cannot complete the bypass of (6-4)TP, so error-prone bypass is achieved by Polζ. (D) A model for damage tolerance mechanism in plants. The stalled replication fork is processed by either of two pathways: mutagenic synthesis by specific TLS polymerases or accurate synthesis using an intact template (template switch). The stalled replication fork signals the modification of PCNA. When PCNA is monoubiquitinated, the TLS polymerases interact with the Ub-PCNA and are recruited to the replication fork. The stalled replication fork also signals the transfer of Polη and Rev1. The 90-kDa heat shock protein (HSP90) promotes TLS activity through interaction with TLS polymerases. When TLS is deficient or reduced by depletion of HSP90, Rad5-dependent polyubiquitination of PCNA leads to a template switch, which causes genome instability.
It is more than a decade since the first report of TLS in plants. The accumulation of reports from multiple groups has clarified the roles and importance of TLS not only in UV resistance but also in the maintenance of genome stability in plants. This mini-review aims to summarize 1) TLS activity in plants in comparison with that in other organisms, 2) the contribution of TLS activity to plant responses to DNA-damaging stresses, and 3) possible other functions of TLS polymerases, which may unveil novel damage-resistant mechanisms in plants.
DNA Polymerase Family Members in Plants
DNA polymerases are classified into seven families based on their amino acid sequence similarity (Ishino and Ishino, 2014). Eukaryotes have Family A, B, X, and Y polymerases, whereas Family C polymerases are only seen in bacteria and Family D and E polymerases only in archaea. Arabidopsis has at least 11 polymerases classified into five families based on comparisons with human and yeast homologs ( Table 1 ). The representative member of Family A polymerases is Escherichia coli polymerase I, which was the first DNA polymerase to be identified (Kornberg et al., 1956). Eukaryotic members of this group are polymerase γ (Polγ) and DNA polymerase θ (Polθ). Arabidopsis also has homologs of two prokaryotic-type DNA polymerases, PolI-like A and B (Parent et al., 2011), as well as AtPolθ, which was originally isolated as the causative gene of the short-root mutant tebichi (Inagaki et al., 2006). Family B polymerases include E. coli Pol II and eukaryotic polymerases α (Polα), δ (Polδ), and ε (Polε), which are involved in the replication of nuclear DNA. Polα, δ, and ε are conserved in Arabidopsis (Ronceret et al., 2005; Shultz et al., 2007; Liu et al., 2010; Iglesias et al., 2015; Pedroza-Garcia et al., 2016). This family includes DNA polymerase ζ, the first identified TLS polymerase that is also conserved in Arabidopsis (Sakamoto et al., 2003). Family X is only conserved in eukaryotes: its representative polymerase is Polymerase β, which is involved in base excision repair. Humans have four members in Family X (Polβ, Polλ, Polμ, and terminal deoxytransferase), whereas plants only have Polλ, which is phylogenetically distant from the Polλ of other organisms (Filée et al., 2002; Pavlov et al., 2006). Family Y carries the largest number of TLS polymerases, including E. coli Pol IV and V; eukaryotic Polη, Polκ, Polι; and Rev1 (Ohmori et al., 2001). Rev1 was originally isolated as a responsible gene for yeast reversionless1 mutant, which carries a deoxycytidyl transferase activity (Nelson et al., 1996). Homologs of Polη, Polκ, and Rev1 are found in Arabidopsis (Takahashi et al., 2007).
Table 1.
DNA polymerases in Arabidopsisa,b.
| Family | Category | Subunit |
A. thaliana Gene ID |
Reference | Function |
|---|---|---|---|---|---|
| A | DNA polymerase IA DNA polymerase IB |
POLIA POLIB |
At3g20540 At1g50840 |
Parent et al., 2011 | Replication of organellar DNA, TLS Replication of organellar DNA, TLS |
| DNA polymerase θ | POLQ | At4g32700 | Inagaki et al., 2006 | Repair of crosslink damage DSB repair TLS |
|
| B | DNA polymerase α | POLA1 POLA2 POLA3 POLA4 |
At5g67100 At1g67630 At1g67320 At5g41880 |
Shultz et al., 2007; Liu et al., 2010 | Replication |
| DNA polymerase δ | POLD1 POLD2 POLD3 POLD4 |
At5g63960 At2g42120 At1g78650 At1g09815 |
Shultz et al., 2007; Iglesias et al., 2015 | Replication | |
| DNA polymerase ε | POLE1 POLE2 POLE3 POLE4 |
At1g08260 At2g27120 At5g22110 At1g07980 At5g43250 At2g27470 |
Ronceret et al., 2005; Pedroza-Garcia et al., 2016 | Replication | |
| DNA polymerase ζ | REV3 REV7 |
At1g67500 At1g16590 | Sakamoto et al., 2003; Takahashi et al., 2005 | TLS, Repair of crosslink damage DSB repair |
|
| X | DNA polymerase λ | POLL | At1g10520 | Uchiyama et al., 2004 | Repair synthesis TLS |
| Y | DNA polymerase η | POLH | At5g44740 | Santiago et al., 2006 | TLS, Repair of crosslink damage |
| DNA polymerase κ | POLK | At1g49980 | García-Ortiz et al., 2004 | TLS | |
| Rev1 | REV1 | At5g44750 | Takahashi et al., 2005 | TLS, Repair of crosslink damage |
aHomologs for DNA polymerase σ are not listed here because opinions are divided whether Polσ has a DNA polymerase activity or not. bOrganellar DNA primases are not listed here.
Most recently, it has been shown that some members of the Archea-Eucaryotic Primase superfamily, such as human PrimPol, perform bypass synthesis across DNA damage (Iyer et al., 2005; Bianchi et al., 2013; Guilliam et al., 2015). Arabidopsis has a herpes-pox type primase (Iyer et al., 2005), although its function has not yet been investigated.
Isolation of Translesion Synthesis Polymerases Based on Ultraviolet Resistance
A UVB-sensitive mutant rev3 was isolated in Arabidopsis by screening ion-beam mutagenized seedlings under non-photoreactivating conditions (Sakamoto et al., 2003). The responsible gene, AtREV3, encodes a homolog of the catalytic subunit of DNA polymerase ζ (Polζ). DNA replication in the rev3 root meristem was reduced after UVB irradiation (Sakamoto et al., 2003). AtREV7 and AtREV1 encode a regulatory subunit of Polζ and a Family Y polymerase, respectively (Takahashi et al., 2005). The rev7 and rev1 plants showed reduced growth compared with wild-type plants under chronic UVB irradiation (Takahashi et al., 2005). AtPOLH encodes a homolog of DNA polymerase η (Polη) that complements the yeast rad30 mutant (Santiago et al., 2006). Disruption of Polζ and Polη had an additive effect on Arabidopsis root growth after UVB treatment (Anderson et al., 2008). Moreover, cell death was induced at root stem cells, and the number of mitotic cells was reduced severely in the UV-irradiated Polζ- and Polη-deficient plants (Curtis and Hays, 2007). This series of studies showed that these polymerases are important in plant UV resistance. The polymerases allow DNA replication to continue, saving the stem cell from cell death and maintaining growth in the presence of harmful UV irradiation.
Damage Bypass Activities of Translesion Synthesis Polymerases
TLS activity has been investigated in vitro using purified or recombinant polymerases and synthetic damage-inducing templates, such as cyclobutane TT dimer (CTD) and (6-4)TT photoproducts [(6-4)TP]. These analyses revealed that the bypass efficiency is dependent on both the type of damage and the polymerases involved ( Figure 1B ). For example, yeast and human Polη bypasses CTD efficiently (Johnson et al., 1999; Masutani et al., 1999), but Polη only inefficiently bypasses (6-4)TP (Johnson et al., 2001). In humans, DNA polymerase ι (Polι) inserts a nucleotide opposite 3′-T in the (6-4)TP (Vaisman et al., 2003). The 3′-end is thought to be elongated by the second polymerase (Polζ, Polκ, or Polθ), which has 3′-end elongation activity (Prakash et al., 2005, Seki and Wood, 2008). The subsequent in vivo analyses suggest that the UV damage at CC or CT sequence are also bypassed by a similar one- or two-step mechanism. Thus, TLS involves the multiple switching of polymerases at the replication site ( Figure 1B ; Prakash and Prakash, 2002; Bebenek and Kunkel, 2004).
The bypass activity of AtPolη for the major UV damage was examined by two groups who showed that AtPolη bypasses the CTD in vitro (Anderson et al., 2008; Hoffman et al., 2008). The activity of AtPolη is comparable to that of human Polη when examined at optimum salt concentration and temperature, and HsPolη, ScPolη, and AtPolη do not bypass (6-4)TP (Hoffman et al., 2008). Research has also been done on other types of DNA damage: AtPolκ inserted an A/C opposite 8-oxoG, a common form of oxidative damage induced by reactive oxygen species (García-Ortiz et al., 2007). Deletion of the C-terminal domain elevates the processivity and fidelity of AtPolκ, suggesting that the C-terminal domain regulates the activities of this polymerase through interactions with other proteins (García-Ortiz et al., 2004; García-Ortiz et al., 2007). DNA polymerase λ bypassed 8-oxoG in both error-free (dC insertion) and error-prone (dA insertion) manners (Amoroso et al., 2011). AtRev1 inserted a C opposite an apurin/apyrimidine (AP) site (Takahashi et al., 2007), which is formed by spontaneous depurination or occurs as an intermediate in the base excision repair process (Boiteux and Guillet, 2004). AtPolIA and AtPolIB have also been shown to bypass the AP site in vitro (Baruch-Torres and Brieba, 2017).
Detection of Mutations Induced by Translesion Synthesis
Mutations induced by TLS have been investigated in in vivo assay systems (Lawrence and Christensen, 1978; Lawrence and Christensen, 1979; Roche et al., 1994; Harfe and Jinks-Robertson, 2000; Yu et al., 2001; Bresson and Fuchs, 2002; Kozmin et al., 2003; Gibbs et al., 2005; Szüts et al., 2008; Yoon et al., 2009; Yoon et al., 2010). In yeast, the deletion of Polζ or Rev1 reduces the UV-induced mutation frequency (Lawrence and Christensen, 1978, Lawrence and Christensen, 1979), whereas the deletion of Polη increases the frequency (Yu et al., 2001; Kozmin et al., 2003). These observations are not consistent with the in vitro characteristics of Polζ and Polη because Polη is less accurate than Polζ when replicating undamaged DNA (McCulloch et al., 2007; Zhong et al., 2006). Comprehensive analysis of in vitro and in vivo data suggested that Polη bypasses CTD with some errors, which are removed by the exonuclease activity of other polymerase(s) (McCulloch and Kunkel, 2008). Prakash et al. (2005) suggest that yeast Polη bypasses CPD at CC or CT sequence in an error-free manner. However, Pol η seems to induce C to T transition by inserting dA opposite deaminated C or mC in CPD (Ikehata and Ono, 2011). Yeast and mammalian Polη bypass (6-4)TP in an error-prone manner (Bresson and Fuchs, 2002; Yoon et al., 2010). It is suggested that Polζ contributes to the mutagenic bypass of (6-4)PP by extending the mismatched primer end caused by the action of Polη or other polymerases (Prakash et al., 2005; Hirota et al., 2010). Thus, the mutation frequency depends on the polymerases available, damage type, sequence context, and the assay system, and so on.
In plants, the reversion frequencies in Arabidopsis plants were measured using β-glucuronidase (GUS)-based markers (Kovalchuk et al., 2000; Nakagawa et al., 2011a; Nakagawa et al., 2011b).
The markers carry a G-T mutation, which corresponds to the 3’-T of TT sequence, a possible target of UV dimer. A misincorporation of dC opposite 3’-T leads to detect a reversion (a T to G transversion). When irradiated with UVB, the Polζ- and Rev1-deficient plants made fewer reversions in somatic cells compared with wild-type plants. By contrast, the Polη-deficient plant showed higher reversion frequencies than wild-type plants, which were reduced in Polζ and Polη double-deficient plants. From these results, the authors proposed a model in which Arabidopsis has two TLS pathways for responding to UV damage: a more mutagenic pathway involving Polζ and Rev1 and a less mutagenic pathway involving Polη (Nakagawa et al., 2011a). Polη bypasses CTD in an error-free manner ( Figure 1C ). Polζ and Rev1 bypass both CTD and (6-4)TP in an error-prone manner. The Polη inserts a nucleotide opposite (6-4)TP, which is extended by Polζ and causes the mutation. Since the bypass activity across (6-4)TP is low anyway, the minor dC insertions would be detected in this assay system. However, other explanations are possible, for example, when UV induces a double-strand break near the TT sequence, which is wrongly repaired and causes a mutation. Also, further analysis by employing a C-containing marker is necessary to profile UV-induced mutations in plants.
Regulation of Translesion Synthesis
Maintenance of the replication fork is crucial because stalled replication forks easily lead to strand breaks. It has been suggested that a stalled replication fork signals the modification of proliferating cell nuclear antigen (PCNA), which triggers the switching of replicase to TLS polymerase (Stelter and Ulrich, 2003; Kanao and Masutani, 2017). That is, when the PCNA is monoubiquitinated, the replicase detaches from PCNA and TLS polymerases are recruited to the replication site to perform the bypass of damaged DNA, whereas polyubiquitinated PCNA leads to the strand switch pathway. The mammalian Polη, Polκ, and Rev1 have been shown to interact with monoubiquitinated PCNA through the UBZ or UBM motif located in the C-terminal (Bienko et al., 2005; Wood et al., 2007). Moreover, Rev1 has also been shown to interact with other TLS polymerases (Guo et al., 2003) and is suggested to function as a bridge through which the best polymerase for TLS is selected (Boehm et al., 2016).
Arabidopsis has two copies of PCNA, but only AtPCNA2 complements the yeast pol30 mutant (Anderson et al., 2008). The AtPolη has a UBM motif and two PIP repeats but does not have a UBZ motif conserved in animal and yeast Polηs. The mutant AtPolη disrupted in PIP1, PIP2, or UBM still interacts with Arabidopsis PCNA2 but does not fully complement yeast rad30 cells (Anderson et al., 2008). Both Arabidopsis PCNAs interact with ubiquitin in N. benthamiana cells and are ubiquitinated in vitro (Strzalka et al., 2013). The AtREV1 interacts with PCNA2, AtPolη, and AtREV7, a regulatory subunit of AtPolζ in yeast (Sakamoto et al., 2018). The processivity of rice Polλ is stimulated in the presence of PCNA (Uchiyama et al., 2004). Moreover, when Arabidopsis Polλ bypasses 8-oxoG, the ratio of error-free (dC insertion) to error-prone (dA insertion) bypass changed depending on its interaction with PCNA2 (Amoroso et al., 2011). These results suggest that the modification of PCNA leads to the switching from replicase to the appropriate TLS polymerase in plants.
It has been suggested that stalled replication in plants is also resolved by a Rad5-dependent strand switch pathway (Wang et al., 2011). The rev3 and rad5a mutations caused synergistic or additive effects on root growth in plants exposed to UV, MMS, or crosslink agents compared with plants containing each single mutation (Wang et al., 2011). The rad5a plant failed to induce homologous recombination events after bleomycin treatment (Chen et al., 2008). By contrast, rev3 and rev1 plants induced significantly more recombination events after UV irradiation (Sakamoto et al., 2018). If AtRAD5a and AtREV3 work via two alternative pathways, the elevation of recombination activities in rev3 and rev1 plant could be due to the activation of a RAD5-dependent pathway ( Figure 1D ).
Translesion Synthesis and Heat Shock Protein 90
The 90-kDa heat shock protein (HSP90) is an evolutionarily conserved molecular chaperone that stabilizes and activates various proteins involved in homeostasis, transcriptional regulation, chromatin remodeling, and DNA repair (Pennisi et al., 2015). The Arabidopsis genome has four copies of cytosolic HSP90 and three copies of organellar HSP90 (Krishna and Gloor, 2001). Queitsch et al. (2012) reported that the application of geldanamycin, a specific inhibitor of HSP90, to Arabidopsis plants elevated homologous recombination (HR) frequencies, suggesting that the HSP90s are involved in genome maintenance in plants.
Human HSP90 interacts with HsPolη and HsRev1 and regulates the TLS activities (Sekimoto et al., 2010; Pozo et al., 2011). The frequency of UV-induced supF mutation in hPolη-proficient cells is elevated by applying 17-AAG, an HSP90 inhibitor, due to the inhibition of error-free bypass of UV damage (Sekimoto et al., 2010). Conversely, in HsPolη-deficient cells, 17-AAG treatment reduces mutation due to the inhibition of the REV1-dependent error-prone bypass (Pozo et al., 2011). In contrast with the results in mammals, treatment with geldanamycin reduces mutation frequencies in wild-type plants, which are AtPolη-proficient (Sakamoto et al., 2018). This suggests that HSP90 mainly regulates the error-prone TLS pathway, involving AtRev1, in Arabidopsis.
Translesion Synthesis and Cell Cycle Checkpoint
In Arabidopsis, UVB or gamma irradiation induces programmed cell death of stem and progenitor (StPr) cells in the root meristem that depends on ataxia-telangiectasia mutated (ATM), ataxia-telangiectasia and Rad3-related (ATR), and SUPPRESSOR OF GAMMA RESPONSE1 (Curtis and Hays, 2011; Furukawa et al., 2010). Curtis and Hays (2011) investigated the time course of cell death in UV-irradiated Polζ-deficient (rev3) and Polη-deficient (polh) roots as well as the roots of damage checkpoint kinase atm and atr mutants. They found that the cells in polh plants started dying at around 16 h after UV treatment, but the cells in rev3 plant started to die at around 20 h. The time courses of cell death in atr and rev3 atr plants were similar to that in rev3 plants, whereas the UV dose-dependency plots of atr, rev3 atr, and rev3 fitted similar slopes. Thus, they hypothesized that there are two types of TLS in Arabidopsis StPr cells: rapid TLS involving Polη and slow TLS involving Polζ. No Polη or a failure of rapid TLS results in the accumulation of single-stranded DNA, which activates a damage checkpoint and Polζ bypasses the damage (slow TLS). If both Polη and Polζ are absent, or if Polη and ATR are absent, then the stalled replication fork collapses to produce DSB, and the ATM activates DSB repair pathways. A similar epistatic relationship between ATR and Polζ was observed in yeast; the Polζ-dependent mutation requires the yeast ATR homolog Mec1 (Pagès et al., 2009). Therefore, some, but not all, of the TLS activities appear to be controlled by checkpoint activation in both plants and microorganisms.
REV7, the regulatory subunit of Polζ, contains a HORMA (Hop1, Rev7, and MAD2) domain (Aravind and Koonin, 1998). Based on its homology to MAD2, the key component of the mitotic-spindle-assembly checkpoint, and Hop1, a meiotic-synaptonemal complex component, it has been speculated that REV7 acts as an adaptor for DNA repairs and the spindle assembly checkpoint (Aravind and Koonin, 1998). Human REV7 makes a homodimer, and REV7-MAD2 a heterodimer, in vitro (Murakumo et al., 2000). In the absence of REV7, human cells arrest in the G2/M-phase and display increased monoastral and abnormal spindles with misaligned chromosomes (Bhat et al., 2015). Crystal structure and NMR analyses showed that two copies of REV7 bind to the canonical REV7-binding motifs (RBMs) of REV3 (Rizzo et al., 2018). In plants, Arabidopsis REV7 makes a homodimer in both the nucleus and the cytosol (Sakamoto et al., 2018). However, there is only one repeat of RBM in AtREV3 sequences, which is similar to yeast REV3 (Tomida et al., 2015). Therefore, the conformation of active Polζ in plants could be different from that in mammals.
Translesion Synthesis Dna Polymerase in the Repair of Double-Strand Breaks or Crosslink Damage
Substantial evidence points to the involvement of TLS polymerases in the DSB repair pathway. For example, chicken REV3(−/−) cells and Arabidopsis rev3 plants are sensitive to ionizing radiation (Sakamoto et al., 2003; Sonoda et al., 2003). Arabidopsis Polλ-disruption plants are hypersensitive to ionizing radiation and bleomycin (Furukawa et al., 2016). In yeast, Polζ and Rev1 are associated with the homing endonuclease (HO)-induced DSB end (Hirano and Sugimoto, 2006). Moreover, ScREV3 is responsible for mutations near the HO-induced cleavage site (Holbeck and Strathern, 1997; Rattray et al., 2002). These results show that some TLS polymerases, at least, have a role in DSB-repair processes in both animals and plants. Recently, DNA polymerase θ was shown to be involved in the alternative end-joining (Alt-EJ) pathway in animals (Chan et al., 2010; Wood and Doublié, 2016; Schimmel et al., 2017) and in moss (Mara et al., 2019). Polθ-deficient Arabidopsis cannot integrate T-DNA, suggesting that the Polθ stabilizes two minimally paired 3’ overhanging DNA ends during the T-DNA integration process (van Kregten et al., 2016). It is possible that TLS polymerases work in DSB repair pathway in some context.
Polθ is the best understood polymerase involved in the repair of interstrand crosslink (ICL) damage (Harris et al., 1999; Shima et al., 2003; Beagan et al., 2017). Other TLS polymerases have also been suggested to work in the process of ICL damage repair. For example, REV3(−/−) cells or organisms are sensitive to ICL-inducing treatments in mammals, chickens, yeast, and plants (Grossmann et al., 2000; Sakamoto et al., 2003; Nojima et al., 2005; Takahashi et al., 2005; Sarkar et al., 2006; Sharma et al., 2012). Disruption of Rev1 and Polη makes the cell or organism hypersensitive to ICL treatment (Takahashi et al., 2005; Sharma et al., 2012), and hPolη can bypass the ICL adduct in vitro (Vaisman et al., 2000). It has been suggested that ICL damage is processed by the Fanconi anemia complementation group A (FANCA)-dependent pathway, which includes nucleolytic incision, TLS, and HR (Kim and D’Andrea, 2012). Several TLS polymerases have been shown to bypass ICL damage if the DNA around the ICL is appropriately trimmed (Ho et al., 2011; Roy et al., 2016). These data suggest that TLS activities are important in overcoming ICL damage. In conclusion, TLS polymerases have multiple roles, which are critical for the genome stability of animals, plants, and microorganisms.
Conclusion and Future Perspectives
The deletion of TLS polymerase often causes lethal or severe phenotypes in animals (Esposito et al., 2000; O-Wang et al., 2002; Wittschieben et al., 2006; Dumstorf et al., 2006; Stallons and McGregor, 2010). By contrast, almost all TLS polymerase activities can be disrupted in plants without severe reduction of fertility. Therefore, the plant system is ideal for analyzing the function, regulation, and interaction of TLS polymerases. Information on the structure and catalytic fidelity of TLS polymerases can assist us to build a novel genome-editing enzyme with elaborate specificities. Plant cells can provide a good platform for developing these upcoming technologies.
Author Contributions
ANS wrote all the parts of this mini-review.
Funding
This work was partially supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant number 24241028, 25440147, and 17K00561 to AS.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author thanks Shinya Takahashi for his critical reading of the manuscript. She also thanks Ann Seward and Sarah Williams, ELS, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
References
- Amoroso A., Concia L., Maggio C., Raynaud C., Bergounioux C., Crespan E., et al. (2011). Oxidative DNA damage bypass in Arabidopsis thaliana requires DNA polymerase λ and proliferating cell nuclear antigen 2. Plant Cell. 23, 806–822. 10.1105/tpc.110.081455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson H. J., Vonarx E. J., Pastushok L., Nakagawa M., Katafuchi A., Gruz P., et al. (2008). Arabidopsis thaliana Y-family DNA polymerase eta catalyses translesion synthesis and interacts functionally with PCNA2. Plant J. 55, 895–908. 10.1111/j.1365-313X.2008.03562.x [DOI] [PubMed] [Google Scholar]
- Aravind L., Koonin E. V. (1998). The HORMA domain: a common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 23, 284–286. 10.1016/S0968-0004(98)01257-2 [DOI] [PubMed] [Google Scholar]
- Baruch-Torres N., Brieba L. G. (2017). Plant organellar DNA polymerases are replicative and translesion DNA synthesis polymerases. Nucleic Acids Res. 45, 10751–10763. 10.1093/nar/gkx744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagan K., Armstrong R. L., Witsell A., Roy U., Renedo N., Baker A. E., et al. (2017). Drosophila DNA polymerase theta utilizes both helicase-like and polymerase domains during microhomology-mediated end joining and interstrand crosslink repair. PLoS Genet. 13, e1006813. 10.1371/journal.pgen.1006813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebenek K., Kunkel T. A. (2004). Function of DNA polymerases. Adv. Protein Chem. 69, 137–165. 10.1016/S0065-3233(04)69005-X [DOI] [PubMed] [Google Scholar]
- Bhat A., Wu Z., Maher V. M., McCormick J. J., Xiao W. (2015). Rev7/Mad2B plays a critical role in the assembly of a functional mitotic spindle. Cell Cycle 14, 3929–3938. 10.1080/15384101.2015.1120922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi J., Rudd S. G., Jozwiakowski S. K., Bailey L. J., Soura V., Taylor E., et al. (2013). PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell. 52, 566–573. 10.1016/j.molcel.2013.10.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., et al. (2005). Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310, 1821–1824. 10.1126/science.1120615 [DOI] [PubMed] [Google Scholar]
- Boehm E. M., Spies M., Washington M. T. (2016). PCNA tool belts and polymerase bridges form during translesion synthesis. Nucleic Acids Res. 44, 8250–8260. 10.1093/nar/gkw563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiteux S., Guillet M. (2004). Abasic sites in DNA: repair and biological consequences in Saccharomyces cerevisiae. DNA Repair 3, 1–12. 10.1016/j.dnarep.2003.10.002 [DOI] [PubMed] [Google Scholar]
- Bresson A., Fuchs R. P. (2002). Lesion bypass in yeast cells: Pol Z participates in a multi-DNA polymerase process. EMBO J. 21, 3881–3887. 10.1093/emboj/cdf363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt A. B. (1999). Molecular genetics of DNA repair in higher plants. Trends Plant. Sci. 4, 20–25. 10.1016/S1360-1385(98)01355-7 [DOI] [PubMed] [Google Scholar]
- Canturk F., Karaman M., Selby C. P., Kemp M. G., Kulaksiz-Erkmen G., Hu J., et al. (2016). Nucleotide excision repair by dual incisions in plants. Proc. Natl. Acad. Sci. U.S.A. 113, 4706–4710. 10.1073/pnas.1604097113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. H., Yu A. M., McVey M. (2010). Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 6, e1001005. 10.1371/journal.pgen.1001005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. P., Mannuss A., Orel N., Heitzeberg F., Puchta H. (2008). A homolog of ScRAD5 is involved in DNA repair and homologous recombination in Arabidopsis. Plant Physiol. 146, 1786–1796. 10.1104/pp.108.116806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. J., Hays J. B. (2007). Tolerance of dividing cells to replication stress in UVB-irradiated Arabidopsis roots: requirements for DNA translesion polymerases eta and zeta. DNA Repair 6, 1341–1358. 10.1016/j.dnarep.2007.03.004 [DOI] [PubMed] [Google Scholar]
- Curtis M. J., Hays J. B. (2011). Cooperative responses of DNA-damage-activated protein kinases ATR and ATM and DNA translesion polymerases to replication-blocking DNA damage in a stem-cell niche. DNA Repair 10, 1272–1281. 10.1016/j.dnarep.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Dumstorf C. A., Clark A. B., Lin Q., Kissling G. E., Yuan T., Kucherlapati R., et al. (2006). Participation of mouse DNA polymerase iota in strand-biased mutagenic bypass of UV photoproducts and suppression of skin cancer. Proc. Natl. Acad. Sci. U.S.A. 103, 18083–18088. 10.1073/pnas.0605247103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Godindagger I., Klein U., Yaspo M. L., Cumano A., Rajewsky K. (2000). Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Curr. Biol. 10, 1221–1224. 10.1016/S0960-9822(00)00726-0 [DOI] [PubMed] [Google Scholar]
- Filée J., Forterre P., Sen-Lin T., Laurent J. (2002). Evolution of DNA polymerase families: evidences for multiple gene exchange between cellular and viral proteins. J. Mol. Evol. 54, 763–773. 10.1007/s00239-001-0078-x [DOI] [PubMed] [Google Scholar]
- Furukawa T., Curtis M. J., Tominey C. M., Duong Y. H., Wilcox B. W., Aggoune D., et al. (2010). A shared DNA-damage-response pathway for induction of stem-cell death by UVB and by gamma irradiation. DNA Repair (Amst.) 9, 940–948. 10.1016/j.dnarep.2010.06.006 [DOI] [PubMed] [Google Scholar]
- Furukawa T., Angelis K. J., Britt A. B. (2016). Arabidopsis DNA polymerase lambda mutant is mildly sensitive to DNA double strand breaks but defective in integration of a transgene. Front. Plant Sci. 6, 357. 10.3389/fpls.2015.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ortiz M. V., Ariza R. R., Hoffman P. D., Hays J. B., Roldán-Arjona T. (2004). Arabidopsis thaliana AtPOLK encodes a DinB-like DNA polymerase that extends mispaired primer termini and is highly expressed in a variety of tissues. Plant J. 39, 84–97. 10.1111/j.1365-313X.2004.02112.x [DOI] [PubMed] [Google Scholar]
- García-Ortiz M. V., Roldán-Arjona T., Ariza R. R. (2007). The noncatalytic C-terminus of AtPOLK Y-family DNA polymerase affects synthesis fidelity, mismatch extension and translesion replication. FEBS J. 274, 3340–3350. 10.1111/j.1742-4658.2007.05868.x [DOI] [PubMed] [Google Scholar]
- Gibbs P. E. M., McDonald J., Woodgate R., Lawrence C. W. (2005). The Relative Roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 Protein and Pol32 in the Bypass and Mutation Induction of an Abasic Site, T-T (6-4) Photoadduct and T-T cis-syn Cyclobutane Dimer. Genetics 169, 575–582. 10.1534/genetics.104.034611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann K. F., Ward A. M., Moses R. E. (2000). Saccharomyces cerevisiae lacking Snm1, Rev3 or Rad51 have a normal S-phase but arrest permanently in G2 after cisplatin treatment. Mutat. Res. 461, 1–13. 10.1016/S0921-8777(00)00035-5 [DOI] [PubMed] [Google Scholar]
- Guilliam T. A., Keen B. A., Brissett N. C., Doherty A. J. (2015). Primase-polymerases are a functionally diverse superfamily of replication and repair enzymes. Nucleic Acids Res. 43, 6651–6664. 10.1093/nar/gkv625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C., Fischhaber P. L., Luk-Paszyc M. J., Masuda Y., Zhou J., Kamiya K., et al. (2003). Mouse rev1 protein interacts with multiple DNA polymerases involved in translesion DNA synthesis. EMBO J. 22, 6621–6630. 10.1093/emboj/cdg626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B. D., Jinks-Robertson S. (2000). DNA polymerase zeta introduces multiple mutations when bypassing spontaneous DNA damage in Saccharomyces cerevisiae. Mol. Cell. 6, 1491–1499. 10.1016/S1097-2765(00)00145-3 [DOI] [PubMed] [Google Scholar]
- Harris P. V., Mazina O. M., Leonhardt E. A., Case R. B., Boyd J. B., Burtis K. C. (1999). Molecular cloning of Drosophila mus308, a gene involved in DNA cross-link repair with homology to prokaryotic DNA polymerase I genes. Mol. Cell. Biol. 16, 5764–5771. 10.1128/MCB.16.10.5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y., Sugimoto K. (2006). ATR homolog Mec1 controls association of DNA polymerase zeta-Rev1 complex with regions near a double-strand break. Curr. Biol. 16, 586–590. 10.1016/j.cub.2006.01.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Sonoda E., Kawamoto T., Motegi A., Masutani C., Hanaoka F., et al. (2010). Simultaneous disruption of two DNA polymerases, Polη and Polζ, in avian DT40 cells unmasks the role of Polη in cellular response to various DNA lesions. PLoS Genet. 6, e1001151. 10.1371/journal.pgen.1001151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi K., Arvai A. S., Yamamoto J., Hitomi C., Teranishi M., Hirouchi T., et al. (2012). Eukaryotic class II cyclobutane pyrimidine dimer photolyase structure reveals basis for improved ultraviolet tolerance in plants. J. Biol. Chem. 287, 12060–12069. 10.1074/jbc.M111.244020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. V., Guainazzi A., Derkunt S. B., Enoiu M., Schärer O. D. (2011). Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic Acids Res. 39, 7455–7464. 10.1093/nar/gkr448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman P. D., Curtis M. J., Iwai S., Hays J. B. (2008). Biochemical evolution of DNA polymerase eta: properties of plant, human, and yeast proteins. Biochem. 47, 4583–4596. 10.1021/bi701781p [DOI] [PubMed] [Google Scholar]
- Holbeck S. L., Strathern J. N. (1997). A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147, 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias F. M., Bruera N. A., Dergan-Dylon S., Marino-Buslje C., Lorenzi H., Mateos J. L., et al. (2015). The Arabidopsis DNA polymerase δ has a role in the deposition of transcriptionally active epigenetic marks, development and flowering. PLoS Genet. 11, e1004975. 10.1371/journal.pgen.1004975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehata H., Ono T. (2011). The mechanisms of UV mutagenesis. J. Radiat. Res. 52, 115–125. 10.1269/jrr.10175 [DOI] [PubMed] [Google Scholar]
- Inagaki S., Suzuki T., Ohto M. A., Urawa H., Horiuchi T., Nakamura K., et al. (2006). Arabidopsis TEBICHI, with helicase and DNA polymerase domains, is required for regulated cell division and differentiation in meristems. Plant Cell. 18, 879–892. 10.1105/tpc.105.036798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino S., Ishino Y. (2014). DNA polymerases as useful reagents for biotechnology - the history of developmental research in the field. Front. Microbiol. 5, 465. 10.3389/fmicb.2014.00465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L. M., Koonin E. V., Leipe D. D., Aravind L. (2005). Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33, 3875–3896. 10.1093/nar/gki702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Haracska L., Prakash S., Prakash L. (2001). Role of DNA polymerase eta in the bypass of a (6-4) TT photoproduct. Mol. Cell. Biol. 21, 3558–3563. 10.1128/MCB.21.10.3558-3563.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Prakash S., Prakash L. (1999). Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science 283, 1001–1004. 10.1126/science.283.5404.1001 [DOI] [PubMed] [Google Scholar]
- Kanao R., Masutani C. (2017). Regulation of DNA damage tolerance in mammalian cells by post-translational modifications of PCNA. Mutat. Res. 803–805, 82–88. 10.1016/j.mrfmmm.2017.06.004 [DOI] [PubMed] [Google Scholar]
- Kim H., D’Andrea D. (2012). Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 26, 1393–1408. 10.1101/gad.195248.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Tahira Y., Ishibashi T., Mori Y., Mori T., Hashimoto J., et al. (2004). DNA repair in higher plants; photoreactivation is the major DNA repair pathway in non-proliferating cells while excision repair (nucleotide excision repair and base excision repair) is active in proliferating cells. Nucleic Acids Res. 32, 2760–2767. 10.1093/nar/gkh591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A., Lehman I. R., Bessman M. J., Simms E. S. (1956). Enzymic synthesis of deoxyribonucleic acid. Biochim. Biophysl. Acta 21, 197–198. 10.1016/0006-3002(56)90127-5 [DOI] [PubMed] [Google Scholar]
- Kovalchuk I., Kovalchuk O., Hohn B. (2000). Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J. 19, 4431–4438. 10.1093/emboj/19.17.4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmin S. G., Pavlov Y. I., Kunkel T. A., Sage E. (2003). Roles of Saccharomyces cerevisiae DNA polymerases Poleta and Polzeta in response to irradiation by simulated sunlight. Nucleic Acids Res. 31, 4541–4552. 10.1093/nar/gkg489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P., Gloor G. (2001). The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6, 238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A., Anderson H. J., Osmond M. J., Vonarx E. J. (2005). Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana. Environ. Mol. Mutagen. 45, 115–127. 10.1002/em.20094 [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. B. (1978). Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast I. rev1 mutant strains. L. Mol. Biol. 122, 1–21. 10.1016/0022-2836(78)90104-3 [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. B. (1979). Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast III. rev3 mutant strains. Genetics. 92, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Liu Z., Tan C., Guo X., Wang L., Sancar A., et al. (2010). Dynamics and mechanism of repair of ultraviolet-induced (6-4) photoproduct by photolyase. Nature 466, 887–890. 10.1038/nature09192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ren X., Yin H., Wang Y., Xia R., Wang Y., et al. (2010). Mutation in the catalytic subunit of DNA polymerase alpha influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J. 61, 36–45. 10.1111/j.1365-313X.2009.04026.x [DOI] [PubMed] [Google Scholar]
- Mara K., Charlot F., Guyon-Debast A., Schaefer D. G., Collonnier C., Grelon M., et al. (2019). POLQ plays a key role in the repair of CRISPR/Cas9-induced double-stranded breaks in the moss Physcomitrella patens . New Phytol. 222, 1380–1391. 10.1111/nph.15680 [DOI] [PubMed] [Google Scholar]
- Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., et al. (1999). The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399, 700–704. 10.1038/21447 [DOI] [PubMed] [Google Scholar]
- McCulloch S. D., Kunkel T. A. (2008). The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18, 148–161. 10.1038/cr.2008.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch S. D., Wood A., Garg P., Burgers P. M., Kunkel T. A. (2007). Effects of accessory proteins on the bypass of a cis-syn thyminethymine dimer by Saccharomyces cerevisiae DNA polymeraseη. Biochemistry 46, 8888–8896. 10.1021/bi700234t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakumo Y., Roth T., Ishii H., Rasio D., Numata S., Croce C. M., et al. (2000). A human REV7 homolog that interacts with the polymerase zeta catalytic subunit hREV3 and the spindle assembly checkpoint protein hMAD2. J Biol. Chem. 275, 4391–4397. 10.1074/jbc.275.6.4391 [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Takahashi S., Tanaka A., Narumi I., Sakamoto A. N. (2011. a). Role of AtPolζ, AtRev1, and AtPolη in UV light-induced mutagenesis in Arabidopsis. Plant Physiol. 155, 414–420. 10.1104/pp.110.166082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Takahashi S., Narumi I., Sakamoto A. N. (2011. b). Role of AtPolζ, AtRev1 and AtPolη in γ ray-induced mutagenesis. Plant Signal. Behav. 6, 728–731. 10.4161/psb.6.5.15124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J. R., Lawrence C. W., Hinkle D. C. (1996). Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382, 729–731. 10.1038/382729a0 [DOI] [PubMed] [Google Scholar]
- Nojima K., Hochegger H., Saberi A., Fukushima T., Kikuchi K., Yoshimura M., et al. (2005). Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 65, 11704–11711. 10.1158/0008-5472.CAN-05-1214 [DOI] [PubMed] [Google Scholar]
- Ohmori H., Friedberg E. C., Fuchs R. P., Goodman M. F., Hanaoka F., Hinkle D., et al. (2001). The Y-family of DNA polymerases. Mol. Cell. 8, 7–8. 10.1016/S1097-2765(01)00278-7 [DOI] [PubMed] [Google Scholar]
- O-Wang J., Kajiwara K., Kawamura K., Kimura M., Miyagishima H., Koseki H., et al. (2002). An essential role for REV3 in mammalian cell survival: absence of REV3 induces p53-independent embryonic death. Biochem. Biophys. Res. Commun. 293, 1132–1137. 10.1016/S0006-291X(02)00341-8 [DOI] [PubMed] [Google Scholar]
- Pagès V., Santa Maria S. R., Prakash L., Prakash S. (2009). Role of DNA damage-induced replication checkpoint in promoting lesion bypass by translesion synthesis in yeast. Genes Dev. 23, 1438–1449. 10.1101/gad.1793409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent J. S., Lepage E., Brisson N. (2011). Divergent roles for the two PolI-like organelle DNA polymerases of Arabidopsis. Plant Physiol. 156, 254–262. 10.1104/pp.111.173849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov Y. I., Shcherbakova P. V., Rogozin I. B. (2006). Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int. Rev. Cytol. 255, 41–132. 10.1016/S0074-7696(06)55002-8 [DOI] [PubMed] [Google Scholar]
- Pedroza-Garcia J. A., Domenichini S., Mazubert C., Bourge M., White C., Hudik E., et al. (2016). Role of the polymerase ϵ sub-unit DPB2 in DNA replication, cell cycle regulation and DNA damage response in Arabidopsis . Nucleic Acids Res. 44 (15), 7251–7266. 10.1093/nar/gkw449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi R., Ascenzi P., di Masi A. (2015). Hsp90: A New Player in DNA Repair? Biomolecules 5, 2589–2618. 10.3390/biom5042589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo F. M., Oda T., Sekimoto T., Murakumo Y., Masutani C., Hanaoka F., et al. (2011). Molecular chaperone Hsp90 regulates REV1-mediated mutagenesis. Mol. Cell. Biol. 31, 3396–3409. 10.1128/MCB.05117-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S., Prakash L. (2002). Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16, 1872–1883. 10.1101/gad.1009802 [DOI] [PubMed] [Google Scholar]
- Prakash S., Johnson R. E., Prakash L. (2005). Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74, 317–353. 10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- Queitsch C., Carlson K. D., Girirajan S. (2012). Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet. 8, e1003041. 10.1371/journal.pgen.1003041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A. J., Shafer B. K., McGill C. B., Strathern J. N. (2002). The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics 162, 1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo A. A., Vassel F. M., Chatterjee N., D’Souza S., Li Y., Hao B., et al. (2018). Rev7 dimerization is important for assembly and function of the Rev1/Polζ translesion synthesis complex. Proc. Natl. Acad. Sci. U.S.A. 115, E8191–E8200. 10.1073/pnas.1801149115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche H., Gietz R. D., Kunz B. A. (1994). Specificity of the yeast rev3 delta antimutator and REV3 dependency of the mutator resulting from a defect (rad1 delta) in nucleotide excision repair. Genetics 137, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronceret A., Guilleminot J., Lincker F., Gadea-Vacas J., Delorme V., Bechtold N., et al. (2005). Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 44 (2), 223–236. 10.1111/j.1365-313X.2005.02521.x [DOI] [PubMed] [Google Scholar]
- Roy U., Mukherjee S., Sharma A., Frank E. G., Schärer O. D. (2016). The structure and duplex context of DNA interstrand crosslinks affects the activity of DNA polymerase η. Nucleic Acids Res. 44, 7281–7291. 10.1093/nar/gkw485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A., Lan V. T., Hase Y., Shikazono N., Matsunaga T., Tanaka A. (2003). Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and gamma-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell. 15, 2042–2057. 10.1105/tpc.012369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A. N., Kaya H., Endo M. (2018). Deletion of TLS polymerases promotes homologous recombination in Arabidopsis. Plant Signal. Behav. 13, e1483673. 10.1080/15592324.2018.1483673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago M. J., Alejandre-Durán E., Ruiz-Rubio M. (2006). Analysis of UV-induced mutation spectra in Escherichia coli by DNA polymerase eta from Arabidopsis thaliana . Mutat. Res. 601, 51–60. 10.1016/j.mrfmmm.2006.05.036 [DOI] [PubMed] [Google Scholar]
- Sarkar S., Davies A. A., Ulrich H. D., McHugh P. J. (2006). DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 25, 1285–1294. 10.1038/sj.emboj.7600993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel J., Kool H., van Schendel R., Tijsterman M. (2017). Mutational signatures of non-homologous and polymerase theta-mediated end-joining in embryonic stem cells. EMBO J. 6, 3634–3649. 10.15252/embj.201796948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Wood R. D. (2008). DNA polymerase θ (POLQ) can extend from mismatches and from bases opposite a (6–4) photoproduct. DNA Repair (Amst.) 7, 119–127. 10.1016/j.dnarep.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto T., Oda T., Pozo F. M., Murakumo Y., Masutani C., Hanaoka F., et al. (2010). The molecular chaperone Hsp90 regulates accumulation of DNA polymerase eta at replication stalling sites in UV-irradiated cells. Mol. Cell. 37, 79–89. 10.1016/j.molcel.2009.12.015 [DOI] [PubMed] [Google Scholar]
- Sharma S., Shah N. A., Joiner A. M., Roberts K. H., Canman C. E. (2012). DNA polymerase ζ is a major determinant of resistance to platinum-based chemotherapeutic agents. Mol. Pharmacol. 81, 778–787. 10.1124/mol.111.076828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima N., Hartford S. A., Duffy T., Wilson L. A., Schimenti K. J., Schimenti J. C. (2003). Phenotype-based identification of mouse chromosome instability mutants. Genetics 163, 1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz R. W., Tatineni V. M., Hanley-Bowdoin L., Thompson W. F. (2007). Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. Plant Physiol. 144, 1697–1714. 10.1104/pp.107.101105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Okada T., Zhao G. Y., Tateishi S., Araki K., Yamaizumi M., et al. (2003). Multiple roles of Rev3, the catalytic subunit of pol zeta in maintaining genome stability in vertebrates. EMBO J. 22, 3188–3197. 10.1093/emboj/cdg308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallons L. J., McGregor W. G. (2010). Translesion synthesis polymerases in the prevention and promotion of carcinogenesis. J. Nucleic Acids. 2010, 643857. 10.4061/2010/643857 [DOI] [PMC free article] [PubMed]
- Stelter P., Ulrich H. D. (2003). Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425 (6954), 188–191. 10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- Strzalka W., Bartnicki F., Pels K., Jakubowska A., Tsurimoto T., Tanaka K. (2013). RAD5a ubiquitin ligase is involved in ubiquitination of Arabidopsis thaliana proliferating cell nuclear antigen. J. Exp. Bot. 64, 859–869. 10.1093/jxb/ers368 [DOI] [PubMed] [Google Scholar]
- Szüts D., Marcus A. P., Himoto M., Iwai S., Sale J. E. (2008). REV1 restrains DNA polymerase ζ to ensure frame fidelity during translesion synthesis of UV photoproducts in vivo . Nucleic Acids Res. 36, 6767–6780. 10.1093/nar/gkn651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Sakamoto A., Sato S., Kato T., Tabata S., Tanaka A. (2005). Roles of Arabidopsis AtREV1 and AtREV7 in translesion synthesis. Plant Physiol. 138, 870–881. 10.1104/pp.105.060236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S., Sakamoto A. N., Tanaka A., Shimizu K. (2007). AtREV1, a Y-family DNA polymerase in Arabidopsis, has deoxynucleotidyl transferase activity in vitro . Plant Physiol. 145, 1052–1060. 10.1104/pp.107.101980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida J., Takata K., Lange S. S., Schibler A. C., Yousefzadeh M. J., Bhetawal S., et al. (2015). REV7 is essential for DNA damage tolerance via two REV3L binding sites in mammalian DNA polymerase ζ. Nucleic Acids Res. 43, 1000–1011. 10.1093/nar/gku1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y., Kimura S., Yamamoto T., Ishibashi T., Sakaguchi K. (2004). Plant DNA polymerase λ, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur. J. Biochem. 271, 2799–2807. 10.1111/j.1432-1033.2004.04214.x [DOI] [PubMed] [Google Scholar]
- Vaisman A., Masutani C., Hanaoka F., Chaney S. G. (2000). Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry 39, 4575–4580. 10.1021/bi000130k [DOI] [PubMed] [Google Scholar]
- Vaisman A., Frank E. G., Iwai S., Ohashi E., Ohmori H., Hanaoka F., et al. (2003). Sequence context-dependent replication of DNA templates containing UV-induced lesions by human DNA polymerase iota. DNA Repair (Amst.) 2, 991–1006. 10.1016/S1568-7864(03)00094-6 [DOI] [PubMed] [Google Scholar]
- Vaisman A., Woodgate R. (2017). Translesion DNA polymerases in eukaryotes: what makes them tick? Crit. Rev. Biochem. Mol. Biol. 52, 274–303. 10.1080/10409238.2017.1291576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kregten M., de Pater S., Romeijn R., van Schendel R., Hooykaas P. J., Tijsterman M., et al. (2016). T-DNA integration in plants results from polymerase-θ-mediated DNA repair. Nat. Plants 2, 16164. 10.1038/nplants.2016.164 [DOI] [PubMed] [Google Scholar]
- Wang S., Wen R., Shi X., Lambrecht A., Wang H., Xiao W. (2011). RAD5a and REV3 function in two alternative pathways of DNA-damage tolerance in Arabidopsis. DNA Repair 10, 620–628. 10.1016/j.dnarep.2011.04.009 [DOI] [PubMed] [Google Scholar]
- Wittschieben J. P., Reshmi S. C., Gollin S. M., Wood R. D. (2006). Loss of DNA polymerase ζ causes chromosomal instability in mammalian cells. Cancer Res. 66, 134–142. 10.1158/0008-5472.CAN-05-2982 [DOI] [PubMed] [Google Scholar]
- Wood A., Garg P., Burgers P. M. (2007). A ubiquitin-binding motif in the translesion DNA polymerase Rev1 mediates its essential functional interaction with ubiquitinated proliferating cell nuclear antigen in response to DNA damage. J. Biol. Chem. 282, 20256–20263. 10.1074/jbc.M702366200 [DOI] [PubMed] [Google Scholar]
- Wood R. D., Doublié S. (2016). DNA polymerase θ (POLQ), double-strand break repair, and cancer. DNA Repair 44, 22–32. 10.1016/j.dnarep.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Prakash L., Prakash S. (2009). Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U.S.A. 106, 18219–18224. 10.1073/pnas.0910121106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. H., Prakash L., Prakash S. (2010). Error-free replicative bypass of (6-4) photoproducts by DNA polymerase zeta in mouse and human cells. Genes Dev. 24, 123–128. 10.1101/gad.1872810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. L., Johnson R. E., Prakash S., Prakash L. (2001). Requirement of DNA polymerase eta for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21, 185–188. 10.1128/MCB.21.1.185-188.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Garg P., Stith C. M., Nick McElhinny S. A., Kissling G. E., Burgers P. M., et al. (2006). The fidelity of DNA synthesis by yeast DNA polymerase ζ alone and with accessory proteins. Nucleic Acids Res. 34, 4731–4742. 10.1093/nar/gkl465 [DOI] [PMC free article] [PubMed] [Google Scholar]