Abstract
Background
Aeromonas are ubiquitous bacteria causing many clinical conditions including acute diarrhea. Diarrheagenic Aeromonas harbors aerolysin gene secreting virulent enterotoxin, aerolysin.
Objectives
To develop a molecular and immunological based method for detection of Aeromonas.
Methods
Diarrheal Aeromonas strains were identified from stool samples using culture, enterotoxicity testing using mice model. During immune magnetic polymerase chain reaction IM-PCR protocol, aerolysin specific antibodies were bound with immuno magnetic binding. Sensitivity and specificity tests for IM-PCR were conducted.
Results
There was high detection of Aeromonas using IM-PCR (12.4 %) technique when compared to low isolation with culture (5.1%). Our study confirmed that some strains of enterotoxic Aeromonas strains were uncultivable. Enterotoxicity tests on culture isolates revealed many strains were negative. IM-PCR detected high, (62/500) rate of identification of Aeromonas with aerolysin toxin gene. Aeromonas species identified after IM-PCR were A. hydrophila (40.3% ), A. veronii (17.7 %), A. caviae (14.5 %), A. trota (11.2 %), A. jandei (9.6 %) and A. schuberti (6.4%). All A. trota strains were undetected by cultivation.
Conclusion
High sensitivity and specificity of IM-PCR are due to preparation of aerolysin antibodies and immuno magnetic binding, prior to PCR. Since diseases due to Aeromonas are increasingly reported, IM-PCR is recommended for detection from clinical specimens.
Keywords: Aeromonas, IM-PCR, acute diarrhea, aerolysin, enterotoxicity
Introduction
The members of the genus Aeromonas are Gram negative bacilli similar to Vibrionaceae1,2. Importance of aeromonad diarrhea has been established by many microbiologists3–6. In developing nations, numerous food and water related epidemics due to Aeromonas were reported7–9. Even in developed countries variety of infections due to Aeromonas have been noted10–13. Aeromonas is also known to cause other infections ranging from meningitis, pneumonia, wound associated sepsis to ocular disease14–16. It was noticed that aeromonads can cause diseases in healthy individuals and very severe infections in immune compromised patients17,18. Aeromonad diarrhea in humans is caused by six species of Aeromonas consisting of A. hydrophila, A. veronii (biotype Sobria and Veronii), A. caviae, A. trota, A. jandei and A. schuberti19. Many virulent determinants of this bacterium such as pili adhesins, enterotoxin (aerolysin), cytolytic toxin, hemoytic toxin, lipases, and proteases have been studied20,21. Various bacteriological data revealed that aerolysin toxin gene is similar to cholera toxin (CT) gene and Aeromonas strains containing aerolysin toxin gene are highly associated with diarrhea22. This article brings out our study based on immuno magnetic polymerase chain reaction (IM-PCR) technique to detect pathogenic Aeromonas strains harboring aerolysin gene from diarrheal stool samples. Current diagnostic methods in diarrheal Aeromonas in laboratories and research organizations include microscopy, staining, culture using selective media, biotyping and serotyping. These routinely used procedures are time consuming, inaccurate and insensitive in detection. Current proposed method is novel and unique because it uses both immunological based binding and molecular based PCR set up for the diagnosis of diarrheal Aeromonas.
Materials and methods
Cultivation of Aeromonas using conventional culture techniques
A total of 500 diarrheal stool specimens were analyzed over a period of twelve months in our Diarrheal Active Surveillance Unit (DASU), Jimma University, Ethiopia. Isolation of Aeromonas was performed using alkaline peptone water and ampicillin sheep blood agar (ASBA) for 24 hours at 37°C23.
Enterotoxic potential of isolated Aeromonas strains
Aeromonas cultures grown in peptone water were centrifuged at 6000 x g for 10 minutes to obtain cell free supernatant. Aerolysin present in the supernatant was furtherconcentrated and purified by sterile filtration (0.22 µm filters, Millipore, Billerica, USA). 50 µL doses of purified aerolysin toxin were injected through sub-plantar route into paw of Swiss male mice (7 week old with weight 30 – 35 g). Severity of inflammatory responses and edema were recorded starting from 1 hour up to 96 hours24.
Immunization and preparation of aerolysin specific antibodies
Rabbits were immunized with aerolysin recombinant protein (aerA, MyBioSource, Canada) in divided, increasing doses over a period of 1, 2, 3 and 6 months.
Immuno magnetic binding of aerolysin immunoglobulins 50 µL of aerolysin specific immunoglobulins were allowed to bind with anti-immunoglobulin coated on magnetic chromium oxide for 30 minutes at 25°C in a shaker.
Immune magnetic binding of aerolysin secreting Aeromonas in stool
Stool specimens were reacted with magnetic particles containing aerolysin specific immunoglobulins at 25°C for 1 hour. Magnetic particles were centrifuged at 1000 g for 5 minutes. Clear supernatant was subjected to PCR.
Molecular detection using Immuno magnetic PCR technique (IM-PCR)
Using AeroFr -CCAAGGGGTCTGTGGCGACA-(forward primer) and AeroRv-TTTCACCGGTAACAGGATTG-(reverse primer) (Toyoba, Japan) a 209 bp part of aerolysin gene was amplified using thermal cycler (Perkin Elmer, US). Denatured DNA sample was obtained by heating the supernatant for 15 minutes and treating it with ice. Reaction mixture contained 2.0 µl of 2 mM dATP, dGTP, and dCTP, 15 nM of denatured DNA and 2.0 µl of 10 X amplification buffer. PCR thermal conditions were set as 32 cycles at 93° C for 1 minute, 54°C for 1minute and 74°C for 10 minutes.
Specificity and sensitivity of Immuno magnetic PCR (IM-PCR)
An overall number of 150 stool specimens from diarrheal cases were included for assessment of the specificity and sensitivity of Immuno magnetic PCR. Specimens from various sources were included. Statistical analysis were conducted using IBM — SPSS version 21.0.
Results
Comparison of Aeromonas isolates with other established pathogens using conventional culture techniques
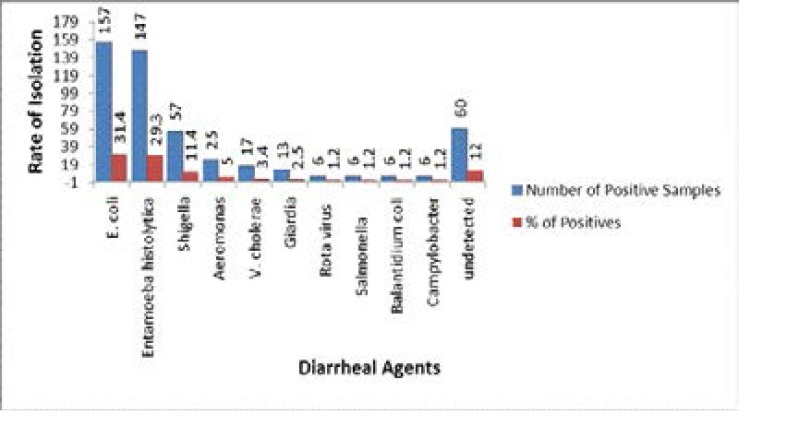
Our results showed that the isolation rate of Aeromonas using culture was low when compared with established pathogens. (Figure 1). A. trota was not isolated by culture method. (Table 1).
Figure 1.
Isolation rates of different diarrheal agents using conventional methods (n=500).
Table 1.
Identification of Aeromonas species using culture and immune magnetic polymerase chain reaction (IM-PCR) and comparison with the enterotoxicity of isolates
|
Aeromonas species identified |
Rate of isolation by culture, % |
Enterotoxicity test of Aeromonas isolates after culture |
Rate of detection after IM-PCRa, % |
Number of strains showing enterotoxicity after IM-PCRa |
| A. hydrophila | 12, (38.7) | 10/12 | 25, (40.3) | 25/25 |
| A. caviae | 8, (25.8) | 7/8 | 9, (14.5) | 9/9 |
| A. veronii | 5, (16.1) | 5/5 | 11, (17.7) | 11/11 |
| A. schuberti | 3, (9.7) | 2/3 | 4, (6.4) | 4/4 |
| A. Jandei | 3, (9.7) | 1/3 | 6, (9.6) | 6/6 |
| A. trota | NDb | NDb | 7, (11.2) | 7/7 |
Key to abbreviations:
a: Immune magnetic polymerase chain reaction.
b: Not detected.
Enterotoxicity test using paw of mice
Enterotoxicity tests using paw of mice on Aeromonas isolates exhibited that not all strains were enterotoxigenic. Positive results of out of the total aeromonad culture isolates were A. hydrophila (10/12), A. caviae (7/8), A. veronii (5/5), A. schubertii (2/3) and A. jandaei (1/3) (Table 1).
Immune magnetic binding of aerolysin secreting Aeromonas in stool
Immune diffusion method showed specific antigen-antibody reaction suggesting antibodies produced in rabbits was highly specific. This recommended the application of aerolysin specific antibodies in immune magnetic binding. Molecular detection of all aerolysin genes containing aeromonads in stool were detected using IM-PCR technique.
Identification of pathogenic Aeromonas by IM-PCR
The total number of Aeromonas strains identified using this technique was 62/500 (12.4%) (Figure 2). In addition, species of Aeromonas detected after IM-PCR were, A. hydrophila (40.3%), A. veronii (17.7%), A. caviae (14.5%), A. trota (11.2%), A. jandei (9.6%) and A. schuberti (6.4%) (Table 1).
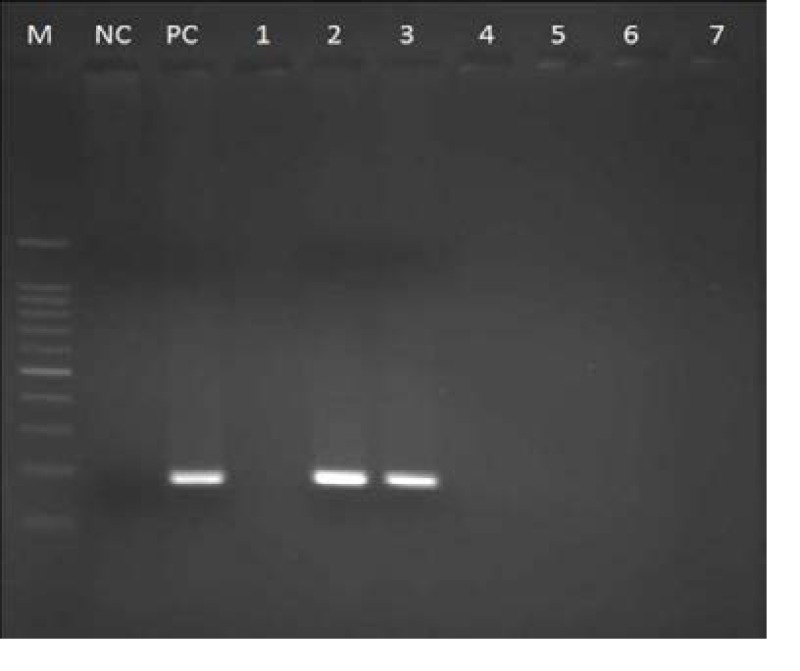
Figure 2.
Agarose gel electrophoresis (2% agarose) after IM -PCR. M- Molecular marker (100 bp DNA ladder), NC- Negative control, PC- Positive control, Lane 1-. negative sample, Lane 2 & 3 - AEROMONAS isolates with aerolysin gene amplicons. Lane 4 to 7 — negative samples.
Specificity and sensitivity of IM-PCR
Specificity and sensitivity assessments revealed that all the 40 stool samples from established Aeromonas diarrhea were confirmed as positives by IM-PCR technique. Surprisingly we observed that 30 cholera samples subjected for Aeromonas testing by culture revealed negative whereas 1/30 of these V. cholerae containing sample shown positive for IM-PCR. Interestingly IM-PCR technique detected 2 out of 15 as positives for Aeromonas possessing aerolysin toxin gene from these environmental isolates (Table 2).
Table 2.
Correlation of culture method and immune magnetic polymerase chain reaction (IM-PCR) in the identification of enterotoxic Aeromonas
| Source of sample causative agent |
Number of samples analyzed |
Number of positive samples for Aeromonas by culture |
Number of positive Enterotoxicity samples for paw Aeromonas by IM-PCRa |
test using of mice |
| Aeromonas | 40 | 40 | 40 | 40 |
| E. coli | 15 | -veb | -ve | -c |
| Plesiomonas | 10 | -ve | -ve | - |
| Healthy volunteers | 40 | -ve | -ve | - |
| Vibrio cholerae | 30 | -ve | 1/30 | - |
| Environmental Aeromonas isolates |
15 | 15 | 2/15 | -ve |
Key to abbreviations:
a: Immune magnetic polymerase chain reaction.
b: Negative.
c: Samples not tested for enterotoxicity test using paw of mice.
Discussion
Developing countries employ culture based identification strategy which is time consuming and results may delay the initiation of appropriate treatment25. There were some molecular based detection systems are developed for detection of aeromonad infections but unfortunately these protocols were found to be unaffordable to all diagnostic laboratories26,27. To overcome these disadvantages, our team has developed a novel and rapid PCR based method for detecting diarrhea causing Aeromonas directly from stool samples. Our IM-PCR based detection method is one of the first of its kind to detect Aeromonas directly from stool by coupling antigen — antibody based followed by PCR protocol.
Routine culture and identification for established pathogens along with Aeromonas resulted in high percentage of isolation rates for E. coli (31.4%), Entamoeba histolytica (29.3%) and Shigella (11.4%). It was observed that the isolation rate for Aeromonas was only 5.1%. In addition we have found that the undetected pathogens were also high with 12% (Table 1). Reasons for low isolation rate of Aeromonas and inability to detect pathogens may be attributed to possible presence of antibiotics in the samples, ampicillin sensitivity of some Aeromonas strains, significantly low number of pathogens in the samples or otherstressful factors that may influence the retardation of growth of pathogens in culture media. These issues during conventional culture methods were already faced by many microbiologists23,28,29.
PCR based IM-PCR method detected significantly high, (12.4%) identification of Aeromonas from stool specimens. Culture media identification of Aeromonas species resulted in the detection of five species whereas IM-PCR technique identified six species with, A. trota (Table 1). The failure of growth of A. trota on selective media culture may be due to ampicillin sensitivity of this species.
Enterotoxicity test using paw of mice revealed that, 2 strains of A. hydrophila, 1 from A. caviae, 1 strain from A. schubertii and 2 strains of A. jandei were non-enterotoxic. IM-PCR method detected significantly high, 62/500 (12.4%) species of Aeromonas with aerolysin gene (Table 1).
IM-PCR method expressed sensitivity of 100% and specificity of 99%. IM-PCR detected one V. cholerae as positive and may be due to genetic similarity between cholera toxin (CT) gene and aerolysin gene22. Moreover two environmental samples (fresh water sources) have also become positives for IM-PCR and this suggests that environment is the main reservoir and source for pathogenic Aeromonas30–33.
When compared to culture methods, IM-PCR method showed high accuracy, efficacy, quality, ease in application and rapidity. Increased sensitivity and specificity of IM-PCR method may be due to its novel procedures like, preparation of aerolysin specific antibodies and Immuno magnetic binding of aerolysin immunoglobulins, prior to actual PCR protocol.
Due to vast number of aeromonad cases are increasingly reported in humans, it is suggested that IM-PCR may be useful in the diagnosis.
Acknowledgments
This work was supported by USAID to Jimma University, Ethiopia. (Grant No. External-USAID-ET-2396725281-2016-18).
Conflict of interest statement
We declare that we have no conflict of interest.
References
- 1.Ruimy R, Breittmayer V, Elbaze P, Lafay B, Boussemart O, Gauthiery2 M, et al. Phylogenetic Analysis and Assessment of the Genera Vibrio, Photobacterium, Aeromonas, and Plesiomonas Deduced from Small-Subunit rRNA Sequences. Int J Syst Evol Microbiol. 1994;44(3):416–426. doi: 10.1099/00207713-44-3-416. [DOI] [PubMed] [Google Scholar]
- 2.Karolcek J, Urgeová E, Durisová A, Draskovicová M, Klokocníková L, Viteková K, et al. Ecology and importance of bacterial’ species of the family Vibrionaceae. J Hyg Epidemiol Microbiol Immunol [Internet] 1982;26(3):275–284. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7142689. [PubMed] [Google Scholar]
- 3.Vila J, Ruiz J, Gallardo F, Vargas M, Soler L, Figueras MJ, et al. Aeromonas spp. and traveler's diarrhea: Clinical features and antimicrobial resistance. Emerg Infect Dis. 2003;9(5):552–555. doi: 10.3201/eid0905.020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pablos M, Huys G, Cnockaert M, Rodríguez-Calleja JM, Otero A, Santos JA, et al. Identification and epidemiological relationships of Aeromonas isolates from patients with diarrhea, drinking water and foods. Int J Food Microbiol. 2011;147(3):203–210. doi: 10.1016/j.ijfoodmicro.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Von Graevenitz A. The role of Aeromonas in diarrhea: A review. Infection. 2007;35(2):59–64. doi: 10.1007/s15010-007-6243-4. [DOI] [PubMed] [Google Scholar]
- 6.Senderovich Y, Ken-Dror S, Vainblat I, Blau D, Izhaki I, Halpern M. A molecular study on the prevalence and virulence potential of Aeromonas spp. recovered from patients suffering from diarrhea in Israel. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0030070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isonhood JH, Drake M. Aeromonas species in foods. J Food Prot [Internet] 2002;65(3):575–582. doi: 10.4315/0362-028x-65.3.575. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11899061. [DOI] [PubMed] [Google Scholar]
- 8.Pablos M, Rodríguez-Calleja JM, Santos JA, Otero A, García-López ML. Occurrence of motile Aeromonas in municipal drinking water and distribution of genes encoding virulence factors. Int J Food Microbiol. 2009;135(2):158–164. doi: 10.1016/j.ijfoodmicro.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 9.Ghenghesh KS, Ahmed SF, El-Khalek RA, Al-Gendy A, Klena J. Aeromonas-associated infections in developing countries. Vol. 2. Journal of Infection in Developing Countries. 2008:81–98. [PubMed] [Google Scholar]
- 10.Ghenghesh KS, Rahouma A, Zorgani A, Tawil K, Al Tomi A, Franka E. Aeromonas in Arab countries: 1995–2014. Vol. 42. Comparative Immunology, Microbiology and Infectious Diseases. 2015:8–14. doi: 10.1016/j.cimid.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Egorov AI, Birkenhauer Best JM, Frebis CP, Karapondo MS. Occurrence of Aeromonas spp. in a random sample of drinking water distribution systems in the USA. J Water Health. 2011;9(4):785–798. doi: 10.2166/wh.2011.169. [DOI] [PubMed] [Google Scholar]
- 12.Janda JM, Abbott SL. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Vol. 23. Clinical Microbiology Reviews. 2010:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lamy B, Kodjo A, Laurent F. Prospective nationwide study of Aeromonas infections in France. J Clin Microbiol. 2009;47(4):1234–1237. doi: 10.1128/JCM.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay C, Chawla K, Sharma Y, Bairy I. Emerging extra-intestinal infections with Aeromonas hydrophila in coastal region of southern Karnataka. J Postgrad Med [Internet] 2008;54(3):199–202. doi: 10.4103/0022-3859.41801. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18626167. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee B, Madiyal M, Ramchandra L, Mukhopadhyay C, Garg R, Chawla K. Unusual severe extra-intestinal manifestations of a common enteric pathogen-aeromonas Spp. J Clin Diagnostic Res. 2017;11(5):DC01–DC03. doi: 10.7860/JCDR/2017/26600.9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Mukhopadhyay P, Chatterjee M, Bandyopadhyay MMK, Bandyopadhyay MMK, Ghosh T, et al. Necrotizing fasciitis caused by Aeromonas caviae. Avicenna J Med [Internet] 2012;2(4):94–96. doi: 10.4103/2231-0770.110740. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3696207&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yumoto T, Ichiba S, Umei N, Morisada S, Tsukahara K, Sato K, et al. Septic shock due to Aeromonas hydrophila bacteremia in a patient with alcoholic liver cirrhosis: A case report. J Med Case Rep. 2014;8(1) doi: 10.1186/1752-1947-8-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parker JL, Shaw JG. Aeromonas spp. clinical microbiology and disease. Journal of Infection. 2011;62:109–118. doi: 10.1016/j.jinf.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Chen PL, Tsai PJ, Chen CS, Lu YC, Chen HM, Lee NY, et al. Aeromonas stool isolates from individuals with or without diarrhea in southern Taiwan: Predominance of Aeromonas veronii. J Microbiol Immunol Infect. 2015;48(6):618–624. doi: 10.1016/j.jmii.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Chopra AK, Houston CW. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes and Infection. 1999;1:1129–1137. doi: 10.1016/s1286-4579(99)00202-6. [DOI] [PubMed] [Google Scholar]
- 21.Sen K, Rodgers M. Distribution of six virulence factors in Aeromonas species isolated from US drinking water utilities: a PCR identification. J Appl Microbiol [Internet] 2004;97(5):1077–1086. doi: 10.1111/j.1365-2672.2004.02398.x. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15479425. [DOI] [PubMed] [Google Scholar]
- 22.Rose JM, Houston CW, Coppenhaver DH, Dixon JD, Kurosky A. Purification and chemical characterization of a cholera toxin-cross-reactive cytolytic enterotoxin produced by a human isolate of Aeromonas hydrophila. Infect Immun. 1989;57(4):1165–1169. doi: 10.1128/iai.57.4.1165-1169.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perales I. Handbook of Culture Media Fof Food And Water Microbiology. 3rd Edition. 2012. Culture Media for the Isolation of Aeromonas spp. and Plesiomonas shigelloides; pp. 451–481. [Google Scholar]
- 24.Harne SD, Sharma VD, Rahman H. Paw oedema test for detection of Salmonella enterotoxin : modification and standardization. Indian J Exp Biol. 1990;28(12):1141–1144. [PubMed] [Google Scholar]
- 25.Archibald LK, Reller LB. Clinical microbiology in developing countries 14. Emerg Infect Dis. 2001;7(1080–6040):302–305. doi: 10.3201/eid0702.010232. (Print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cascón A, Anguita J, Hernanz C, Sánchez M, Fernández M, Naharro G. Identification of Aeromonas hydrophila hybridization group 1 by PCR assays. Appl Environ Microbiol. 1996;62(4):1167–1170. doi: 10.1128/aem.62.4.1167-1170.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tacão M, Alves A, Saavedra MJ, Correia A. BOX-PCR is an adequate tool for typing Aeromonas spp. Antonie van Leeuwenhoek. Int J Gen Mol Microbiol. 2005;88(2):173–179. doi: 10.1007/s10482-005-3450-9. [DOI] [PubMed] [Google Scholar]
- 28.Al-Awadhi H, Dashti N, Khanafer M, Al-Mailem D, Ali N, Radwan S. Bias problems in culture-independent analysis of environmental bacterial communities: A representative study on hydrocarbonoclastic bacteria. Springerplus. 2013;2(1):1–11. doi: 10.1186/2193-1801-2-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avarre J, Masseret E, Hartmann P, Mann W, Häfner N, Dürst M. PCR on slide : advantages in environmental and clinical microbiology. Formatex. 2010:1522–1532. [Google Scholar]
- 30.Rahim Z, Khan SI, Chopra AK. Biological characterization of Aeromonas spp. isolated from the environment. Epidemiol Infect. 2004;132(4):627–636. doi: 10.1017/s0950268804002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhowmik P, Bag PK, Hajra TK, De R, Sarkar P, Ramamurthy T. Pathogenic potential of Aeromonas hydrophila isolated from surface waters in Kolkata. India J Med Microbiol. 2009;58(12):1549–1558. doi: 10.1099/jmm.0.014316-0. [DOI] [PubMed] [Google Scholar]
- 32.Kannan S, Chattopadhyay UK, Pal D, Shimada T, Takeda Y, Bhattacharya SK, et al. Isolation and identification of Aeromonas from patients with acute diarrhoea in Kolkata, India. Indian J Med Microbiol. 2001;19(4):190–192. [PubMed] [Google Scholar]
- 33.Kannan S, Suresh Kanna P, Karkuzhali K, Chattopadhyay UK, Pal D. Direct detection of diarrheagenic Aeromonas from faeces by polymerase chain reaction (PCR) targeting aerolysin toxin gene. Eur Rev Med Pharmacol Sci. 2001;5(3):91–94. [PubMed] [Google Scholar]