Abstract
Aims
Outcomes of colorectal cancer (CRC) treatment and survival have steadily improved during the past decades, accompanied by an increased risk of developing second primary tumours and metastatic tumours at unusual sites. Metastatic CRC can show mucosal colonisation, thereby mimicking a second primary tumour. This potential confusion could lead to incorrect diagnosis and consequently inadequate treatment of the patient. The aim of this study was to differentiate between metastatic CRC and a second primary (gallbladder cancer, GBC) using a combination of standard histopathology and molecular techniques.
Methods and results
Ten consecutive patients with both CRC and GBC were identified in our region using the Dutch National Pathology Archive (PALGA). Two patients served as negative controls. Histology of GBC was reviewed by nine pathologists. A combination of immunohistochemistry, microsatellite analysis, genomewide DNA copy number analysis and targeted somatic mutation analysis was used to aid in differential diagnosis. In two patients, CRC and GBC were clonally related, as confirmed by somatic mutation analysis. For one case, this was confirmed by genomewide DNA copy number analysis. However, in both cases, pathologists initially considered the GBC as a second primary tumour.
Conclusions
Metastatic CRC displaying mucosal colonisation is often misinterpreted as a second primary tumour. A combination of traditional histopathology and molecular techniques improves this interpretation, and lowers the risk of inadequate treatment.
Keywords: clonality, colorectal cancer, gallbladder cancer, metastasis, next‐generation sequencing
Introduction
During the past several decades outcomes of cancer treatment and surveillance have steadily improved, in particular for colorectal cancer (CRC), due to earlier diagnosis, more accurate staging techniques and better treatment options.1, 2 Death rates of CRC in the United States decreased by half in 2015 (14 per 100 000) compared to 1975 (28 per 100 000).3 One of the concerns of (long‐term) cancer survivorship is the increased risk of developing a new primary tumour, but also metastases in more uncommon sites. Approximately 25% of CRC patients present with metastatic disease at presentation (stage IV), and an additional 18% of patients will develop metastases metachronously after treatment of the primary CRC.4 Metastatic CRC (mCRC) mainly develops in liver and lung,5 but rare metastatic sites include, for example, the small bowel in 0.7%6 of cases and the gallbladder in 0.8% of cases.6 Especially in these rare locations, it is important to distinguish metastatic disease from new primaries, as this is essential for therapeutic planning and determining prognosis. The priority for a primary tumour is early diagnosis followed by therapy, mainly surgery with curative intent, while a metastatic tumour requires a different approach.
Traditionally, primary tumours are histologically discriminated from metastatic tumours based on the presence of dysplasia. However, distinguishing primary and metastatic tumour can occasionally be difficult. Studies by Shepherd et al.7 and Estrella et al.8 demonstrated that metastatic gastrointestinal tumours can show significant mucosal colonisation, which cannot be distinguished from precursor lesions, suggesting that the presence of dysplasia does not automatically imply a primary tumour. Other studies have shown this phenomenon in the urinary bladder, lung and liver.9, 10, 11
Various additional techniques can help to establish the origin of a tumour and distinguish primary from metastatic cancer. Immunohistochemistry (IHC) is often inconclusive,12 but in the era of next‐generation molecular pathology novel techniques such as somatic mutation analysis and DNA copy number analysis have been proven to be valuable tools for clonality analysis in solid tumours.13
We hypothesised that metastatic spread from CRC might be unnoticed due to mucosal colonisation, leading to the incorrect diagnosis of a second primary. We used a combination of histological and molecular techniques to determine the extent of the problem and determine diagnostic strategies, using gallbladder cancer (GBC) as a model.
Patients and Methods
Below is an overall description; additional details on experimental procedures and (statistical) analyses are provided as online Data S1.
Patient Selection
Patients were identified using a search question (LZV‐1152) in the PALGA‐database (the nationwide network and registry of histo‐ and cytopathology in the Netherlands).14 We combined patient data from the Radboud University Medical Center (UMC) (Nijmegen), Rijnstate (Arnhem) and Jeroen Bosch (’s‐Hertogenbosch) hospitals to select patients with both GBC and CRC. In the period between 1989 and 2015 we identified 10 consecutive patients with both tumours, two of which served as negative controls for the current study: the first patient developed a primary GBC 5 years before the CRC and the second patient presented with a neuroendocrine carcinoma of the colon with a simultaneous gallbladder adenocarcinoma.
Information on gender, age at time of diagnosis and location of the CRC and GBC were collected retrospectively via review of the pathology report and medical history. This study was approved by the Radboud UMC medical ethics committee (2014/306).
Histological Examination
Tumour blocks were selected based on the pathology reports and review of histology. One haematoxylin‐and eosin (H&E)‐stained slide of the gallbladder of each patient was analysed by nine independent pathologists: six non‐academic pathologists of the Jeroen Bosch Hospital and three academic pathologists of the Radboud UMC. Histological typing was performed using the World Health Organisation (WHO) histological classification of tumours of the gallbladder and extrahepatic bile ducts.15 T‐classification for CRC and GBC was performed using the American Joint Committee on Cancer (AJCC) tumour–node–metastasis (TNM) classification system (7th edn).16 Furthermore, slides were assessed on the presence of dysplasia and pathologists were asked if the gallbladder tumour should be considered a primary or metastatic tumour based on histology. Presence or absence of dysplasia and primary/metastasis were indicated when more than half of the pathologists agreed.
Immunohistochemistry (IHC)
IHC was performed on 4 µm formalin‐fixed paraffin‐embedded (FFPE) tissue sections of CRC and GBC with standard chromogenic horseradish peroxidase‐diaminobenzidine (HRP‐DAB) detection method (Table S1). CK7/CK20/CDX2/SATB2 staining was applied and immunoexpression examined as negative, weak (weak intensity/limited cells) and strong (strong intensity/extensive positive cells). MMR protein staining was scored as described previously.17 Two independent observers (N.K., I.N.) evaluated all sections. Inconsistencies in scoring were re‐evaluated to obtain agreement.
Molecular Analyses
For microsatellite, mutation and copy number analysis, DNA was isolated from FFPE‐derived CRC and GBC and matched normal colon tissue, as described previously.18 Microsatellite analysis was performed with a pentaplex polymerase chain reaction (PCR) for five quasimonomorphic mononucleotide markers.
For mutation analysis, the single‐molecule Molecular Inversion Probe (smMIP) target enrichment technology18 was used. Two custom gene panels (Table S2) were designed using the MIPgen pipeline.19 SmMIP library preparations were performed manually, in essence as described previously.18 BCL to FASTQ conversion and de‐multiplexing of barcoded reads was performed at the Radboud UMC Genomics Technology Center. Mapping of reads to the reference genome and consensus read building was performed with SeqNext software from JSI medical systems (version 4.3.0, build 505; JSI Medical Systems, Ettenheim, Germany). Specific settings and variant filtering strategy are provided in Figure S1.
Shallow whole genome sequencing for copy number analysis was performed at The Tumour Genome Analysis Core, Department of Pathology, Amsterdam UMC, Vrije Universiteit Amsterdam, Cancer Center Amsterdam, as described previously.20 In short, approximately 10 million SR50 sequence reads were generated on the Illumina HiSeq 2500 sequencing platform. Sequence reads were aligned to the reference genome (GRCh37/hg19) with BWA (version 0.5.9) and downstream analyses were performed with R‐packages QDNAseq20 and DNAcopy.21
Statistical Analyses
Clonality analysis was based on somatic mutations and DNA copy number aberrations. For somatic mutations, a statistical test strategy developed by Ostrovnaya et al. was used.22 DNA copy number profiles were statistically compared using Pearson's correlation and the log‐likelihood ratio measure, also developed by Ostrovnaya et al.23
Results
Patient and Tumour Characteristics
Patient and tumour characteristics and results of IHC/MSI analysis are shown in Table 1. In total, eight cases with both a GBC and CRC and two controls were selected. Control 1 developed a GBC before developing a CRC and control 2 had a neuroendocrine CRC together with a gallbladder adenocarcinoma. In five patients the diagnosis of CRC and GBC was synchronous. Three patients had a history of CRC and developed GBC in an average time of 4 years. All CRCs were classified as adenocarcinoma not otherwise specified, except for control 2 (neuroendocrine carcinoma). Dysplasia in the gallbladder was observed in both negative controls and in six of eight cases (Figure 1A), and correlated with the suggested diagnosis of primary GBC based on histological evaluation (Figure 1B). In two cases (2 and 7), pathologists suspected mCRC to the gallbladder (Figure 1B), and no dysplasia was observed.
Table 1.
Patient and tumour characteristics
| Patient | Gender | Tumour site | Age at diagnosis | Stage | Immunohistochemistry results | MSI analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CK7 | CK20 | CDX2 | SATB2 | MLH1 | MSH2 | MSH6 | PMS2 | ||||||
| Case 1 | Male | Gallbladder | 72 | NA | +/− | + | + | + | + | + | + | + | NA |
| Colon (descending) | 66 | T4N1 | +/− | + | + | + | + | + | + | + | NA | ||
| Case 2 | Female | Gallbladder | 51 | T3N0 | + | − | − | − | +/− | + | + | +/− | MSS |
| Rectum | 51 | T3N1 | − | +/− | + | + | + | + | + | + | MSS | ||
| Case 3 | Male | Gallbladder | 64 | T3N1 | − | + | + | +/− | + | + | + | + | NA |
| Colon (sigmoid) | 64 | T4N1 | − | + | + | + | + | + | + | + | NA | ||
| Case 4 | Male | Gallbladder | 69 | T3N0 | − | +/− | + | − | + | + | + | + | NA |
| Colon (sigmoid) | 74 | T2N0 | − | + | + | + | + | + | + | + | NA | ||
| Case 5 | Female | Gallbladder | 69 | T2Nx | +/− | − | + | − | + | + | + | + | MSS |
| Colon (sigmoid) | 69 | T3N1 | +/− | +/− | + | + | NA | NA | NA | NA | MSS | ||
| Case 6 | Female | Gallbladder | 80 | T3Nx | + | + | − | − | + | + | + | + | NA |
| Rectum | 78 | T3N1 | − | + | + | + | + | + | + | + | NA | ||
| Case 7* | Male | Gallbladder | 57 | NA | + | − | − | − | +/− | + | + | − | MSS |
| Colon (caecum) | 53 | T3N1 | − | +/− | + | + | − | + | + | − | MSI | ||
| Colon (hepatic flexure) | 53 | T3N0 | − | − | + | + | − | + | + | − | MSI | ||
| Case 8 | Male | Gallbladder | 67 | T3Nx | +/− | +/− | +/− | − | + | + | + | + | NA |
| Colon (ascending) | 67 | T3N0 | +/− | +/− | +/− | − | + | + | + | + | NA | ||
| Control 1 † | Male | Gallbladder | 50 | T2Nx | + | + | + | − | + | + | + | + | MSS |
| Colon (ascending) | 55 | NA | + | − | − | NA | NA | NA | NA | NA | MSS | ||
| Control 2 ‡ | Male | Gallbladder | 64 | T1Nx | + | − | +/− | NA | + | + | + | + | NA |
| Rectum | 64 | T3N0 | − | − | − | − | + | + | + | + | NA | ||
NA, Not assessable/not assesed; MSS, Microsatellite stability; MSI, Microsatellite instability.
Case 7 developed two colorectal cancers (CRCs) and one gallbladder cancer (GBC).
Control 1 developed a GBC first.
The CRC of control 2 was a neuro‐endocrine tumour.
Figure 1.
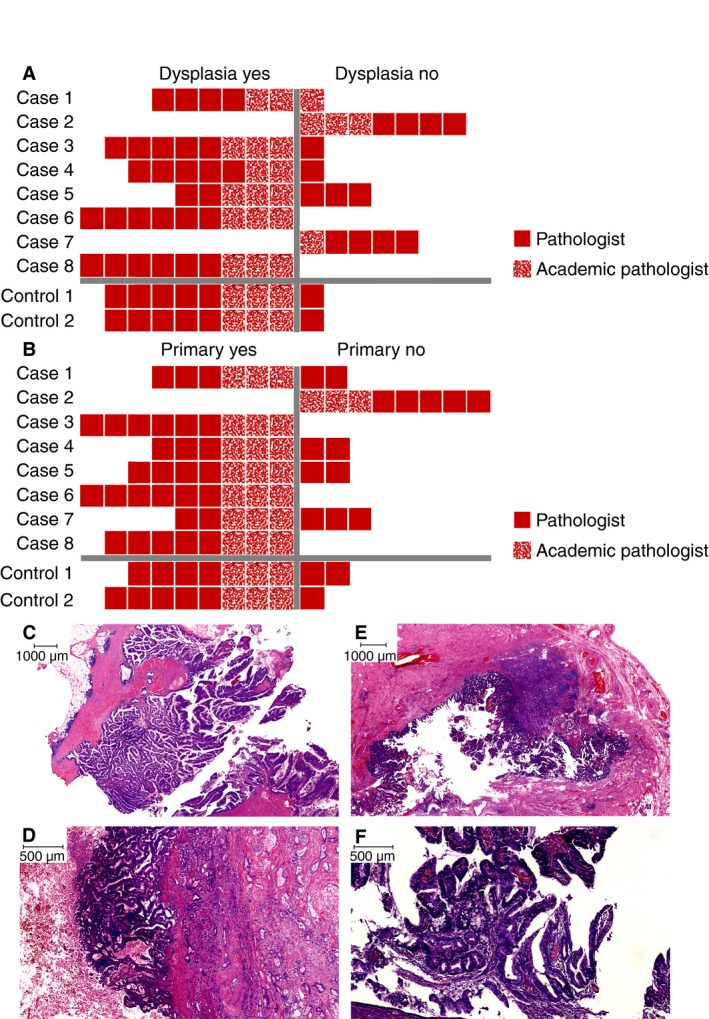
Review results of histopathology of gallbladder cancers (GBCs) of eight cases and two controls (not all features were examined by each pathologist). A, Presence of dysplasia; B, primary GBC or metastatic CRC; C–E, dysplasia in the gallbladder; Country, case 1; D, case 3; E, control 1; F, intestinal metaplasia in the gallbladder (control 1).
Examples of gallbladder dysplasia are shown in Figure 1C (case 1), 1D (case 3) and 1E (control 1). Figure 1F shows intestinal metaplasia of the gallbladder (control 1).
Immunohistochemical Results and MSI Status
Three of the cases showed a concordant CK7/CK20/CDX2/SATB2 profile in both CRC and GBC (Table 1), whereas the other five cases and both controls showed a discordant pattern (Table 1 and Figure 2). In one patient (case 7), both CRCs showed microsatellite instability (MSI), with loss of MLH1 and PMS2 expression. The GBC also showed loss of PMS2 expression, but MSI could not be demonstrated by pentaplex PCR. Other cases were microsatellite stable (MSS).
Figure 2.
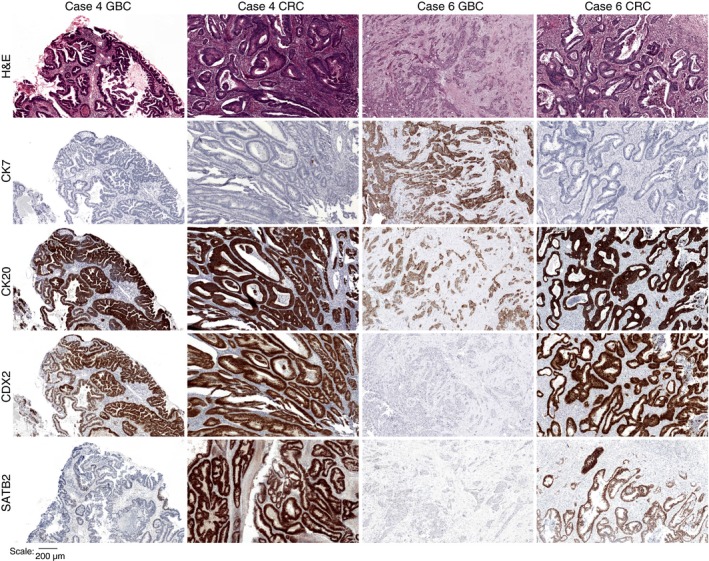
Immunohistochemical staining for cases 4 and 6. Case 4 showed discordance for SATB2 between gallbladder cancer (GBC) and colorectal cancer (CRC), whereas case 6 showed discordance for CK7, CDX2 and SATB2 between GBC and CRC. H&E, haematoxylin and eosin; CK7, cytokeratin 7; CK20, cytokeratin 20; CDX2, caudal type homeobox 2; SATB, homeobox 2.
DNA COPY NUMBER PROFILES
All DNA copy number profiles were first inspected visually, expected and observed noise were calculated, and gender was checked. All profiles were of high quality, as shown by the small difference between the observed and the expected ratio. All the genders, as observed in the data, matched the annotation in the patient records.
The detection of numerical chromosomal copy number aberrations was limited for a few of the samples. The small number of aberrations detected decreased their interpretation, such that for case 1 both CRC and GBC and case 7 no clonality score could be made based on CNA profiling (Figure 5, Figure S2).
Figure 5.

Summary of findings. Green fill, suspected metastasis/clonally related (concordance); red fill, suspected primary (discordance); grey fill, not assessable. [Colour figure can be viewed at http://wileyonlinelibrary.com]
Profiles were compared with Pearson's correlation and subsequent hierarchical clustering of the correlation matrix. This showed that the CRC and GBC of patient 3 were highly similar, suggestive of clonality (Figure 3). Furthermore, we observed clustering of the biopsy and resection of the CRC of case 2 as a validation which serves as an attractive internal control in this analysis. Clonal relatedness was also evaluated with a log‐likelihood ratio measure, confirming clonality in case 3 [log2(LR2) = 36.27; P < 0.0001].
Figure 3.

Correlation matrix of genome‐wide copy number data, hierarchically clustered. Colorectal cancer (CRC) and gallbladder cancer (GBC) of case 3 cluster together, as well as the biopsy and resection specimen of CRC of case 2. Case 7: CRC‐1, caecum; CRC‐2, hepatic flexure.
Mutation Profiles
DNA quality was insufficient for somatic mutation and clonality analysis in three patients (cases 2 and 5 and control 1). In two cases (1 and 3), all mutations were shared between the CRC and GBC (Figure 4) and tumours were clonally related (P < 0.0001). In the other cases (4, 6, 7 and 8), and in control 2, tumours did not share identical mutations and were not clonally related.
Figure 4.
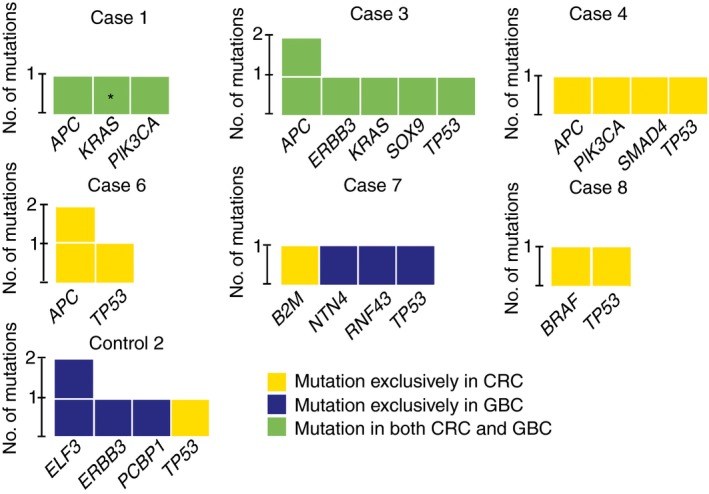
Summary of mutation and clonality analysis. For cases 2 and 5 and control 1, mutation analysis failed/was not performed because of insufficient DNA quantity and quality. In cases 1 and 3, identical mutations were present both in colorectal cancer (CRC) and gallbladder cancer (GBC) (green fill) and tumours were clonally related P < 0.0001. In the other cases and in control 2, mutations were exclusively present in either CRC (yellow fill) or GBC (blue fill) and tumours were unrelated. *Confirmed retrospectively in CRC, with coverage lower than threshold of >5 mut. reads and 5% mut. reads. As this variant was also identified in a diagnostic setting using an Ion Torrent platform with 40% mut. allele, it was included in clonality analysis. [Colour figure can be viewed at http://wileyonlinelibrary.com]
An overview of all findings is shown in Figure 5. In cases 1 and 3, clonal relatedness of both tumours was supported by a combination of concordant IHC, DNA copy number and DNA mutation profiles. In contrast, based on histology, they were regarded as independent primary tumours. Although a concordant IHC profile was observed in case 8, tumours were presumed unrelated, indicated by histological and clonality analyses.
Discussion and Conclusion
In a consecutive series, we describe two patients with metastatic spread of CRC to the gallbladder. Based on histopathological similarity to primary GBC, these cases would have gone unnoticed and patients would not have received the optimal treatment.
Metastases to the gallbladder are considered uncommon, and usually manifest as end‐stage of malignancy. Two large autopsy studies reported gallbladder involvement in 2.2–5.8% of all cancer patients,6, 24 but both studies are dated and it is unclear whether these metastases were part of widespread metastatic disease. One more recent imaging study, although not comparable in study size, confirmed that isolated metastasis to the gallbladder is rare.25 CRC metastasising to the gallbladder seems almost non‐existent, with only a few case reports in the English literature.26, 27, 28, 29 Metastases of other primary cancer types to the gallbladder, such as melanoma or renal cell carcinoma, are more frequently observed, presumably because of their distinct histological features.30, 31
Diagnostic confusion may arise because of mucosal colonisation by mCRC, i.e. growth along an intact basement membrane and colonisation of the existing epithelium, thereby mimicking a primary tumour. We also observed this phenomenon in case 3 (Figure 1D), where the GBC was initially considered a primary tumour. Mucosal colonisation has been described previously in lung,10 liver11 and urinary bladder,9 and seems more frequent in tumours originating from the GI tract.8 The mechanisms underlying mucosal colonisation are largely unknown. Perhaps it is the result of dynamic conversion of the epithelial–mesenchymal transition (EMT) required for migration and invasiveness of metastatic cells to mesenchymal–epithelial transition (MET) required for the cells to colonise their target site. Spatiotemporal regulation of EMT‐MET involves widespread reprogramming of gene expression by epigenetic and transcriptional regulators,32 and by microRNAs that regulate gene expression on post‐transcriptional level,33 both influenced by the tumour microenvironment.34, 35 Significant differential expression of microRNAs was observed in cancer cells cultured on matrigel basement membrane matrix, i.e. the in‐vitro substitute of a basal membrane, and on plastic.36
To differentiate mCRC from primary GBC, a panel of CK7, CK20, CDX2 and SATB2 IHC stains was used. CRC predominantly displays a CK7–/CK20+ immunophenotype (65.8% of cases),37 whereas GBC typically displays a CK7+/CK20– immunophenotype (47.6% of cases).38 CDX2 immunoreactivity has been observed in 72–100% of CRCs39, 40, 41 and in 36–45% of GBCs.42, 43 To our knowledge, there are no studies that specifically investigated the expression of SATB2 in primary GBC. In addition, gallbladders may display primary intestinal‐type adenocarcinoma and may also have intestinal metaplasia (Figure 1F), one of the precursor lesions of GBC, observed in 12–85% of cases,44, 45, 46, 47, 48, 49 which limits the usefulness of these IHC markers. In cases 1 and 3, concordant IHC profiles supported that tumours were clonally related.
Recent advances in molecular pathology are of great value in studying the molecular origin of a tumour. Microsatellite analysis, for example, was successfully applied in other studies50, 51 to determine the origin of synchronous and metachronous tumours from various organs. In this study, however, microsatellite analysis was less informative, as only one patient showed MSI in both CRCs. The GBC of the same case was MSS, suggesting that tumours were unrelated. With somatic mutation and clonality analysis, we concluded that the CRC and GBC of patients 1 and 3 were clonally related. Genes with shared variants included APC, ERBB3, KRAS, PIK3CA, SOX9 and TP53. Unfortunately, the amount and quality of the input DNA was limited. All DNA was derived from FFPE tissue, most was relatively old and some derived from biopsy material, significantly decreasing the quality and yield. DNA copy number analysis supported that both tumours of patient 3 were clonally related. For a few of the samples, low numbers of numerical chromosomal copy number aberrations were detected, limiting their interpretation. This could either be the result of lower tumour cell percentages than estimated or of intratumoural heterogeneity with subclonal copy number aberrations.
Discriminating primary from metastatic cancer is essential for adequate therapeutic planning and prognosis of the patient, as misdiagnosis will result in suboptimal treatment with detrimental consequences. Both timing and choice of chemotherapy treatment differ in CRC and GBC. Furthermore, evidence on the efficacy of adjuvant chemotherapy in GBC is limited.52 Chemotherapy is only indicated in unresectable and metastatic GBC, with gemcitabine and cisplatin (GemCis).53, 54 For unresectable mCRC, however, first‐line treatment involves a fluoropyrimidine (5‐fluorouracil or capecitabine), alone or in combination with a targeted therapy, in various combinations and schedules.55 A few small studies were performed in GBC using fluoropyrimidine‐based chemotherapeutics,56, 57 but schedules differed considerably from those used for CRC. In addition, GemCis treatment has not been widely investigated in (metastatic) CRC. Only a few studies have investigated the benefit of gemcitabine combined with 5‐fluorouracil as second‐ or third‐line therapy in a palliative setting with refractory CRC,58, 59 but not in combination with cisplatin. Proper distinction of mCRC and primary GBC is therefore essential to select appropriate treatment.
In conclusion, we described two patients with metastatic spread of CRC to the gallbladder in a consecutive series of eight patients and two controls with a CRC and a GBC. The majority of these GBCs were considered primary tumours based on the presence of dysplasia. As histological assessment alone is not sufficient for differential diagnosis, we recommend for all cases initial testing with immunohistochemistry, although this is of limited use when the stains are concordant. Molecular analyses are necessary in those cases to confirm clonal relations, which is imperative for an accurate diagnosis and optimal treatment.
Conflicts of interest
The authors declare no conflicts of interest.
Supporting information
Figure S1. Flowchart of variant filtering process. Grey boxes: exclusion steps.
Figure S2. Copy number profiles. CRC, colorectal cancer; GBC, gallbladder cancer. Case 7, CRC‐1 = caecum, CRC‐2 = hepatic flexure.
Table S1. Details of antibodies used for immunohistochemistry.
Table S2. Composition of the targeted Next‐Generation Sequencing panels. CDS, coding sequence.
Table S3. Variants identified with targeted Next‐Generation Sequencing.
Data S1. Materials and Methods.
Acknowledgements
The authors thank pathologists of the Jeroen Bosch Hospital (M. J. Koopmans, S. J. J. Mol, P. T. G. A. Nooijen, R. J. van Suylen and R. Rozendaal) and the Radboud University Medical Center (L. A. Brosens) for pathology review. The authors thank R. D. A. Weren and R. M. de Voer for designing the NGS panels. This study was funded by Foundation ADP. The views expressed in the submitted article are the authors’ own and not an official position of the institution or funder.
de Bitter T J J, van der Linden R L A, van Vliet S, Weren F, Sie D, Ylstra B, van der Linden H C, Knijn N, Ligtenberg M J L, van der Post R S, Simmer F & Nagtegaal I D (2019) Histopathology 75, 394–404. 10.1111/his.13892 Colorectal metastasis to the gallbladder mimicking a primary gallbladder malignancy: histopathological and molecular characteristics
References
- 1. Dutch Cancer Registry. Available at: http://www.cijfersoverkanker.nl (accessed 6 November 2018).
- 2. Toes‐Zoutendijk E, van Leerdam ME, Dekker E et al Real‐time monitoring of results during first year of Dutch colorectal cancer screening program and optimization by altering fecal immunochemical test cut‐off levels. Gastroenterology 2017; 152; 767–775 e762. [DOI] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J. Clin. 2018; 68; 7–30. [DOI] [PubMed] [Google Scholar]
- 4. van Gestel YR, de Hingh IH, van Herk‐Sukel MP et al Patterns of metachronous metastases after curative treatment of colorectal cancer. Cancer Epidemiol. 2014; 38; 448–454. [DOI] [PubMed] [Google Scholar]
- 5. Mariotto AB, Rowland JH, Ries LA, Scoppa S, Feuer EJ. Multiple cancer prevalence: a growing challenge in long‐term survivorship. Cancer Epidemiol. Biomark. Prev. 2007; 16; 566–571. [DOI] [PubMed] [Google Scholar]
- 6. Disibio G, French SW. Metastatic patterns of cancers: results from a large autopsy study. Arch. Pathol. Lab. Med. 2008; 132; 931–939. [DOI] [PubMed] [Google Scholar]
- 7. Shepherd NA, Hall PA. Epithelial–mesenchymal interactions can influence the phenotype of carcinoma metastases in the mucosa of the intestine. J. Pathol. 1990; 160; 103–109. [DOI] [PubMed] [Google Scholar]
- 8. Estrella JS, Wu TT, Rashid A, Abraham SC. Mucosal colonization by metastatic carcinoma in the gastrointestinal tract: a potential mimic of primary neoplasia. Am. J. Surg. Pathol. 2011; 35; 563–572. [DOI] [PubMed] [Google Scholar]
- 9. Silver SA, Epstein JI. Adenocarcinoma of the colon simulating primary urinary bladder neoplasia. A report of nine cases. Am. J. Surg. Pathol. 1993; 17; 171–178. [DOI] [PubMed] [Google Scholar]
- 10. Rosenblatt MB, Lisa JR, Collier F. Primary and metastatic bronciolo‐alveolar carcinoma. Dis. Chest 1967; 52; 147–152. [DOI] [PubMed] [Google Scholar]
- 11. Riopel MA, Klimstra DS, Godellas CV, Blumgart LH, Westra WH. Intrabiliary growth of metastatic colonic adenocarcinoma: a pattern of intrahepatic spread easily confused with primary neoplasia of the biliary tract. Am. J. Surg. Pathol. 1997; 21; 1030–1036. [DOI] [PubMed] [Google Scholar]
- 12. Anderson GG, Weiss LM. Determining tissue of origin for metastatic cancers: meta‐analysis and literature review of immunohistochemistry performance. Appl. Immunohistochem. Mol. Morphol. 2010; 18; 3–8. [DOI] [PubMed] [Google Scholar]
- 13. Begg CB, Ostrovnaya I, Geyer FC et al Contralateral breast cancers: independent cancers or metastases? Int. J. Cancer 2018; 142; 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casparie M, Tiebosch AT, Burger G et al Pathology databanking and biobanking in the Netherlands, a central role for PALGA, the nationwide histopathology and cytopathology data network and archive. Cell. Oncol. 2007; 29; 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Albores‐Saavedra J, Adsay NV, Crawford JM et al Tumours of the gallbladder and extrahepatic bile ducts In Bosman FT, Carneiro F, Hruban RH, Theise ND. eds. WHO classification of tumours of the digestive system. Lyon: IARC Press, 2010; 266–273. [Google Scholar]
- 16. Sobin LH, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. 7th ed. Chichester, UK/Hoboken, NJ: Wiley‐Blackwell, 2010; 309. [Google Scholar]
- 17. Koopman M, Kortman GA, Mekenkamp L et al Deficient mismatch repair system in patients with sporadic advanced colorectal cancer. Br. J. Cancer 2009; 100; 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eijkelenboom A, Kamping EJ, Kastner‐van Raaij AW et al Reliable next‐generation sequencing of formalin‐fixed, paraffin‐embedded tissue using single molecule tags. J. Mol. Diagn. 2016; 18; 851–863. [DOI] [PubMed] [Google Scholar]
- 19. Boyle EA, O'Roak BJ, Martin BK, Kumar A, Shendure J. Mipgen: optimized modeling and design of molecular inversion probes for targeted resequencing. Bioinformatics 2014; 30; 2670–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scheinin I, Sie D, Bengtsson H et al DNA copy number analysis of fresh and formalin‐fixed specimens by shallow whole‐genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014; 24; 2022–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array‐based DNA copy number data. Biostatistics 2004; 5; 557–572. [DOI] [PubMed] [Google Scholar]
- 22. Ostrovnaya I, Seshan VE, Begg CB. Using somatic mutation data to test tumors for clonal relatedness. Ann. Appl. Stat. 2015; 9; 1533–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ostrovnaya I, Olshen AB, Seshan VE, Orlow I, Albertson DG, Begg CB. A metastasis or a second independent cancer? Evaluating the clonal origin of tumors using array copy number data. Stat. Med. 2010; 29; 1608–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Abrams HL, Spiro R, Goldstein N. Metastases in carcinoma; analysis of 1000 autopsied cases. Cancer 1950; 3; 74–85. [DOI] [PubMed] [Google Scholar]
- 25. Barretta ML, Catalano O, Setola SV, Granata V, Marone U, D'Errico Gallipoli A. Gallbladder metastasis: spectrum of imaging findings. Abdom. Imaging 2011; 36; 729–734. [DOI] [PubMed] [Google Scholar]
- 26. Galvao FH, Pestana JO, Capelozzi VL. Fatal gemcitabine‐induced pulmonary toxicity in metastatic gallbladder adenocarcinoma. Cancer Chemother. Pharmacol. 2010; 65; 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen SC, Hsu CY, Wang SM, Tai TY. Adenocarcinoma of the transverse colon manifested as acute cholecystitis. Am. J. Emerg. Med. 1994; 12; 386. [DOI] [PubMed] [Google Scholar]
- 28. Abacherli C, Metzger J. Single metastasis in the gallbladder arising from adenocarcinoma of the rectum. South Med. J. 2008; 101; 1183–1184. [DOI] [PubMed] [Google Scholar]
- 29. Bukhari N, Abdulkader M. Metastatic cecal adenocarcinoma to the gallbladder presenting with acute cholecystitis. Case Rep. Oncol. Med. 2018; 2018; 5308585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Costa Neves M, Neofytou K, Giakoustidis A et al Two cases of gallbladder metastasis from renal cell carcinoma and review of literature. World J. Surg. Oncol. 2016; 14; 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giannini I, Cutrignelli DA, Resta L, Gentile A, Vincenti L. Metastatic melanoma of the gallbladder: report of two cases and a review of the literature. Clin. Exp. Med. 2016; 16; 295–300. [DOI] [PubMed] [Google Scholar]
- 32. Tam WL, Weinberg RA. The epigenetics of epithelial–mesenchymal plasticity in cancer. Nat. Med. 2013; 19; 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell 2009; 139; 871–890. [DOI] [PubMed] [Google Scholar]
- 34. Chen S, Chen X, Li W et al Conversion of epithelial‐to‐mesenchymal transition to mesenchymal‐to‐epithelial transition is mediated by oxygen concentration in pancreatic cancer cells. Oncol. Lett. 2018; 15; 7144–7152. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Rupaimoole R, Calin GA, Lopez‐Berestein G, Sood AK. MiRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016; 6; 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Price KJ, Tsykin A, Giles KM et al Matrigel basement membrane matrix influences expression of micrornas in cancer cell lines. Biochem. Biophys. Res. Commun. 2012; 427; 343–348. [DOI] [PubMed] [Google Scholar]
- 37. Bayrak R, Yenidunya S, Haltas H. Cytokeratin 7 and cytokeratin 20 expression in colorectal adenocarcinomas. Pathol. Res. Pract. 2011; 207; 156–160. [DOI] [PubMed] [Google Scholar]
- 38. Duval JV, Savas L, Banner BF. Expression of cytokeratins 7 and 20 in carcinomas of the extrahepatic biliary tract, pancreas, and gallbladder. Arch. Pathol. Lab. Med. 2000; 124; 1196–1200. [DOI] [PubMed] [Google Scholar]
- 39. Suh N, Yang XJ, Tretiakova MS, Humphrey PA, Wang HL. Value of CDX2, villin, and alpha‐methylacyl coenzyme a racemase immunostains in the distinction between primary adenocarcinoma of the bladder and secondary colorectal adenocarcinoma. Mod. Pathol. 2005; 18; 1217–1222. [DOI] [PubMed] [Google Scholar]
- 40. De Lott LB, Morrison C, Suster S, Cohn DE, Frankel WL. CDX2 is a useful marker of intestinal‐type differentiation: a tissue microarray‐based study of 629 tumors from various sites. Arch. Pathol. Lab. Med. 2005; 129; 1100–1105. [DOI] [PubMed] [Google Scholar]
- 41. Bayrak R, Haltas H, Yenidunya S. The value of CDX2 and cytokeratins 7 and 20 expression in differentiating colorectal adenocarcinomas from extraintestinal gastrointestinal adenocarcinomas: cytokeratin 7‐/20 + p henotype is more specific than CDX2 antibody. Diagn. Pathol. 2012; 7; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu XS, Akiyama Y, Igari T et al Expression of homeodomain protein CDX2 in gallbladder carcinomas. J. Cancer Res. Clin. Oncol. 2005; 131; 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li QL, Yang ZL, Liu JQ, Miao XY. Expression of CDX2 and hepatocyte antigen in benign and malignant lesions of gallbladder and its correlation with histopathologic type and clinical outcome. Pathol. Oncol. Res. 2011; 17; 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sakamoto H, Mutoh H, Ido K, Satoh K, Hayakawa H, Sugano K. A close relationship between intestinal metaplasia and cdx2 expression in human gallbladders with cholelithiasis. Hum. Pathol. 2007; 38; 66–71. [DOI] [PubMed] [Google Scholar]
- 45. Sakamoto H, Mutoh H, Ido K et al Intestinal metaplasia in gallbladder correlates with high amylase levels in bile in patients with a morphologically normal pancreaticobiliary duct. Hum. Pathol. 2009; 40; 1762–1767. [DOI] [PubMed] [Google Scholar]
- 46. Halder S, Kundu S, Chakraborty J, Chakrabarti S. Significance of HER2 and Ki‐67 in preneoplastic lesions and carcinoma of gallbladder. J. Gastrointest. Cancer 2018. 10.1007/s12029-018-0162-8. [DOI] [PubMed] [Google Scholar]
- 47. Khan MR, Raza SA, Ahmad Z et al Gallbladder intestinal metaplasia in Pakistani patients with gallstones. Int. J. Surg. 2011; 9; 482–485. [DOI] [PubMed] [Google Scholar]
- 48. Duarte I, Llanos O, Domke H, Harz C, Valdivieso V. Metaplasia and precursor lesions of gallbladder carcinoma. Frequency, distribution, and probability of detection in routine histologic samples. Cancer 1993; 72; 1878–1884. [DOI] [PubMed] [Google Scholar]
- 49. Fernandes JE, Franco MI, Suzuki RK, Tavares NM, Bromberg SH. Intestinal metaplasia in gallbladders: prevalence study. São Paulo Med. J. 2008; 126; 220–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaneki E, Oda Y, Ohishi Y et al Frequent microsatellite instability in synchronous ovarian and endometrial adenocarcinoma and its usefulness for differential diagnosis. Hum. Pathol. 2004; 35; 1484–1493. [DOI] [PubMed] [Google Scholar]
- 51. Tang M, Pires Y, Schultz M, Duarte I, Gallegos M, Wistuba II. Microsatellite analysis of synchronous and metachronous tumors: a tool for double primary tumor and metastasis assessment. Diagn. Mol. Pathol. 2003; 12; 151–159. [DOI] [PubMed] [Google Scholar]
- 52. Anderson C, Kim R. Adjuvant therapy for resected extrahepatic cholangiocarcinoma: a review of the literature and future directions. Cancer Treat. Rev. 2009; 35; 322–327. [DOI] [PubMed] [Google Scholar]
- 53. Valle J, Wasan H, Palmer DH et al Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010; 362; 1273–1281. [DOI] [PubMed] [Google Scholar]
- 54. Valle JW, Borbath I, Khan SA et al Biliary cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow‐up. Ann. Oncol. 2016; 27; v28–v37. [DOI] [PubMed] [Google Scholar]
- 55. Van Cutsem E, Cervantes A, Nordlinger B, Arnold D. Group EGW. Metastatic colorectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow‐up. Ann. Oncol. 2014; 25 (Suppl. 3); iii1–iii9. [DOI] [PubMed] [Google Scholar]
- 56. Ellis PA, Norman A, Hill A et al Epirubicin, cisplatin and infusional 5‐fluorouracil (5‐FU) (ECF) in hepatobiliary tumours. Eur. J. Cancer 1995; 31A; 1594–1598. [DOI] [PubMed] [Google Scholar]
- 57. Park SH, Park YH, Lee JN et al Phase II study of epirubicin, cisplatin, and capecitabine for advanced biliary tract adenocarcinoma. Cancer 2006; 106; 361–365. [DOI] [PubMed] [Google Scholar]
- 58. Merl M, Hoimes C, Pham T, Saif MW. Is there a palliative benefit of gemcitabine plus fluoropyrimidines in patients with refractory colorectal cancer? A review of the literature previously presented: Poster at the 2008 gastrointestinal cancer symposium (abstract no. 512). Expert Opin. Investig. Drugs 2009; 18; 1257–1264. [DOI] [PubMed] [Google Scholar]
- 59. Bitossi R, Sculli CM, Tampellini M et al Gemcitabine and protracted 5‐fluorouracil infusion as third‐line chemotherapy in refractory colorectal cancer patients. Anticancer Res. 2008; 28; 3055–3060. [PubMed] [Google Scholar]
- 60. Buhard O, Suraweera N, Lectard A, Duval A, Hamelin R. Quasimonomorphic mononucleotide repeats for high‐level microsatellite instability analysis. Dis. Markers 2004; 20; 251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lek M, Karczewski KJ, Minikel EV et al Analysis of protein‐coding genetic variation in 60,706 humans. Nature 2016; 536; 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ostrovnaya I, Seshan VE, Olshen AB, Begg CB. Clonality: an R package for testing clonal relatedness of two tumors from the same patient based on their genomic profiles. Bioinformatics 2011; 27; 1698–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flowchart of variant filtering process. Grey boxes: exclusion steps.
Figure S2. Copy number profiles. CRC, colorectal cancer; GBC, gallbladder cancer. Case 7, CRC‐1 = caecum, CRC‐2 = hepatic flexure.
Table S1. Details of antibodies used for immunohistochemistry.
Table S2. Composition of the targeted Next‐Generation Sequencing panels. CDS, coding sequence.
Table S3. Variants identified with targeted Next‐Generation Sequencing.
Data S1. Materials and Methods.


