Abstract
Background:
The fetal fraction of cell-free DNA (cfDNA) in maternal plasma is decreased in obese women. The underlying mechanism is not well understood. The amount of cfDNA released from the placenta has not been directly examined in maternal obesity.
Objective:
We sought to quantify release of cfDNA from the placenta and fetal membranes in maternal diet-induced obesity using explant cultures in an established mouse model.
Study Design:
C57BL6/J females were fed either 60% high-fat diet or 10% fat-matched control diet for 14 weeks prepregnancy and throughout gestation. Placentas and fetal membranes were collected on e18 and randomly allocated to time 0-, 1-, or 6-hour culture times. The CfDNA was isolated from culture media, quantified, and normalized to tissue weight.
Results:
Placentas from obese dams released significantly less cfDNA compared to those of lean dams at time 0 (45.8 ± 4.3 ng/mg vs 65.6 ± 7.9 ng/mg, P = .02). Absolute cfDNA levels increased with longer placental culture, with no significant differences between obese and lean dams at 1 and 6 hours. Membranes released significantly less cfDNA than did placentas at every time point.
Conclusions:
Maternal obesity is associated with decreased release of cfDNA from the placenta compared to lean controls immediately after tissue harvest. This may provide an alternative explanation for the lower fetal fraction of cfDNA noted in maternal obesity.
Keywords: cell-free fetal DNA, maternal obesity, fetal fraction, placenta, mouse
Introduction
Cell-free DNA (cfDNA) is released into maternal plasma by cells undergoing apoptosis.1 In pregnancy, the placenta is the primary source of cell-free “fetal” DNA (cffDNA) in maternal plasma, releasing DNA via apoptosis of villous trophoblasts.2 The application of cffDNA for aneuploidy screening has become well integrated into prenatal care in the first and second trimesters. Recent investigations have also examined the association of cffDNA with adverse pregnancy outcomes at later gestational ages, including intrauterine growth retardation,3,4 severe preeclampsia,5 and preterm labor.6 In addition, the results of a small human study suggest that increased methylation ratios in total cfDNA at term may be associated with spontaneous labor.7
These clinical applications rely on the assessment of the fetal fraction of cfDNA, defined as the amount of cffDNA in a sample divided by the total cfDNA (maternal plus fetal cfDNA). Multiple studies have found that maternal obesity is associated with a decreased fetal fraction of cfDNA,8–13 but the underlying mechanism is poorly understood. It has been hypothesized that this may be due to a larger volume of maternal adipose tissue that undergoes apoptosis, thereby adding to the pool of maternal cfDNA in circulation and diluting the fetal fraction.14,15 This hypothesis relies on the assumption that lean and obese pregnant women release the same amount of cfDNA from the placenta, which has not been evaluated. We sought to test an alternative hypothesis that placental release of cfDNA is diminished in the setting of maternal obesity using placental explant cultures in our established mouse model of maternal diet-induced obesity.16
Fetal membranes are biologically active and respond to changes in the intrauterine environment.17 Studies in human and animal models have noted increased senescence and apoptosis of fetal membranes at term17–19 and in the setting of preterm premature rupture of membranes.17,20,21 However, the effect of maternal obesity on membrane cfDNA release is poorly understood, and little is known about the relative contribution of the fetal membranes to cffDNA. We thus also sought to investigate the effect of maternal obesity on cfDNA released by fetal membranes and to compare the relative contribution of membranes versus placenta to the total amount of cfDNA released by fetal tissues at term.
Materials and Methods
Animal Model
The Tufts Medical Center Institutional Animal Care and Use Committee approved this protocol (B2016-69); all institutional guidelines for animal care and use were followed. From 4 weeks of age, C57BL/6J females (Jackson Laboratories, Bar Harbor, Maine) were fed ad libitum either a lard-based, high-fat diet (HFD) containing 60% calories from fat (n = 10; Research Diets, D12492, New Brunswick, New Jersey) or a matched control diet (CD) containing 10% calories from fat (n = 10; Research Diets, D12450J) for 14 weeks prebreeding to ensure maternal diet-induced obesity and throughout the pregnancy. The diets were matched for fiber, protein, sucrose, and micronutrient content (Supplemental Table 1). Previous studies have validated the use of these diets in BL/6 dams to establish maternal diet-induced obesity.16,22–24 C57BL/6J breeder males were fed the CD. Dams were weighed weekly until breeding. Maternal obesity was defined as at least 30% increase in weight compared to age-matched controls.16,25 Dams were paired with breeder males and examined daily for vaginal plugs. Males were removed when vaginal plugs were noted and pregnant dams were singly housed. Dams were weighed during pregnancy on day P0 (day of mating), P10, and P15.
At embryonic day 18 (e18.0, reflective of term gestation in humans), embryos and placentas were rapidly dissected from the uterine horns using sterile surgical technique and immediately washed in ice-cold sterile Dulbecco phosphate-buffered saline (DPBS-1×; Gibco, Grand Island, Nebraska). The membranes were carefully separated from the placenta, with care taken to minimize tissue trauma. Tissues were rinsed in ice-cold DPBS, and placentas and membranes were individually placed into sterile tissue culture media (45% Dulbecco modified Eagle medium [DMEM]/45% Ham F12/10% fetal bovine serum [FBS] + Pen/Strep) for transfer to the explant cultures. Fetal sex genotyping was performed on tail snip DNA using real-time polymerase chain reaction (PCR) with specific probes for the Sry gene (Transnetyx, Cordova, Tennessee).
Placental Explant Cultures
In order to avoid the contribution of maternal cfDNA inherent in studies of maternal plasma and evaluate only placental (fetal) cfDNA release, placental explant cultures were performed using an established and validated method,26 as depicted in Supplemental Figure 1. Briefly, under sterile conditions, each placenta was quartered with a scalpel into 4 equal pieces, and all 4 pieces were placed into a single culture well with 1 mL of room temperature tissue culture media (45% DMEM/45% Ham’s F12/10% FBS + Pen/Strep). Placentas were randomly allocated to time 0 (immediately after tissue retrieval, approximately 10-15 minutes elapsed between euthanasia of the mouse and plating the explant cultures), 1-hour, or 6-hour culture times and incubated at 37°C, 5% CO2, and 95% room air (ambient O2 levels of 21%). Although normoxic conditions for placental physiology are described to be 8%,27 prior placental explant studies have demonstrated that cfDNA release does not differ between 8% and 21% O2.26,28 One to 3 placentas were used from each litter for each time point, depending on the litter size.
Membrane Explant Cultures
Evaluation of membrane cfDNA release was performed in a similar fashion as the placental explant cultures as described earlier, with the exception that the membranes were not quartered, due to their small size. All membranes from each individual gestation were placed into a well containing 500 µL of media and incubated in the conditions described earlier. Fetal membranes were assigned to the same time point as their corresponding placentas, with 1 to 3 fetal membranes utilized per litter for each time point, depending on the litter size.
Cell-Free DNA Extraction
At the designated time point, the culture media were collected and centrifuged at 14 600 rpm for 20 minutes to remove cellular debris. The CfDNA was subsequently extracted from 300 µL aliquots of supernatant using the Roche High Pure PCR Template Preparation Kit (Roche, Indianapolis, Indiana) to a final volume of 50 µL. Concentration of cfDNA was determined with NanoDrop spectrophotometry (Thermo Scientific, Waltham, Massachusetts) and the Qubit dsDNA HS Assay Kit (Thermo Scientific). Levels of CfDNA were normalized to placental tissue weight by calculating the total nanogram amount of cfDNA in the extracted volume (based on the measured cfDNA concentration in ng/µL) and then dividing the nanogram amount by the placental tissue wet weight in milligrams. Quantification assays were performed in duplicate for each sample.
Statistics
Differences between cfDNA released by placentas and membranes of lean and obese dams were evaluated with Student t test (normally distributed data) or Mann-Whitney U testing (nonnormally distributed data). Two-way analysis of variance (ANOVA; sex × maternal obesity status) was performed to examine interactions between maternal obesity, fetal sex, and cfDNA release. Data are reported as mean ± standard error of the mean unless otherwise noted. All statistical analyses were performed using GraphPad Prism 7 (GraphPad Software, San Diego, California).
Results
Dams on the 60% HFD gained significantly more weight and ended the prebreeding feeding period significantly heavier than dams on the 10% fat CD (26.06 ± 0.22 g vs 19.31 ± 0.34 g, P < .0001). The HFD dams were significantly heavier than CD dams at the time of e18 placental retrievals (46.12 ± 1.62 g vs 36.08 ± 0.79 g, P < .0001).
In total, 60 placentas and membranes from obese dams (10 litters) and 51 placentas/membranes from lean dams (10 litters) were retrieved. Litter size did not differ significantly between maternal obese and lean groups. One to three placentas/membranes per litter were allocated to each time point. Table 1 describes the characteristics of the study population and tissue samples at each time point.
Table 1.
Characteristics of Study Population and Tissue Samples.
| Time 0 | Time 1 hour | Time 6 hour | ||||
|---|---|---|---|---|---|---|
| Obese | Lean | Obese | Lean | Obese | Lean | |
| Number of placentas | 24 | 20 | 24 | 20 | 12 | 11 |
| Placental weight,a mg, mean ± SEM | 74.16 ± 3.01 | 77.45 ± 3.12 | 69.49 ± 3.38 | 74.33 ± 2.44 | 71.60 ± 6.60 | 81.82 ± 5.03 |
| Number of litters | 10 | 10 | 10 | 10 | 8 | 9 |
| Fetal sexb | ||||||
| Male | 12 | 11 | 9 | 7 | 7 | 6 |
| Female | 9 | 6 | 12 | 10 | 4 | 2 |
Abbreviation: SEM, standard error of the mean.
a P = .03 for placental weight of obese versus lean placentas overall.
b Fetal sex was not determined for the initial 3 litters, so sex is unknown for 1 to 3 placentas per group.
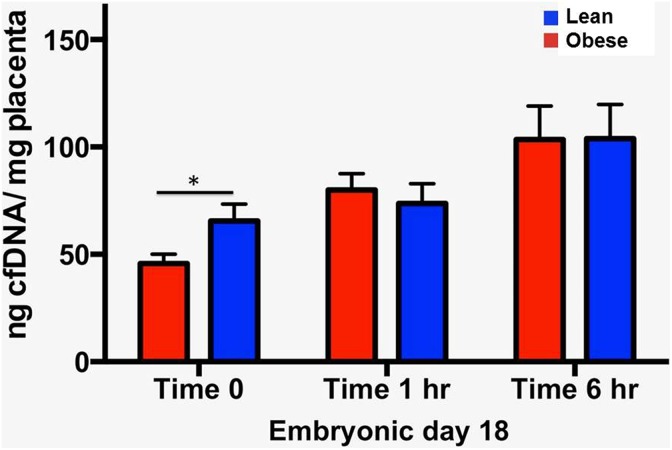
At time 0, placentas from obese dams released significantly less cfDNA compared to those of lean dams (45.77 ± 4.27 ng/mg vs 65.56 ± 7.87 ng/mg placenta, P = .02; Figure 1). The CfDNA levels in both obese and lean groups increased with longer placental culture, with no significant differences between obese and lean groups at 1 and 6 hours (Figure 1; time 1 hour: 80.06 ± 7.60 ng/mg vs 73.70 ± 9.22 ng/mg, P = .63; time 6 hour: 103.50 ± 15.54 ng/mg vs 103.90 ± 15.91 ng/mg, P = .98). Placentas from obese dams weighed significantly less than those of lean dams (71.80 mg vs 78.80 mg, P = .03). Given that tissue weight was accounted for by normalizing cfDNA concentration to placental weight, the weight difference was not the etiology for the diminished release of cfDNA from obese placentas.
Figure 1.
Placental cell-free DNA release in obese versus lean dams by time in culture. Placentas from obese dams release significantly less cell-free DNA at time 0 compared to those from lean dams. While cell-free DNA release rises in both groups as time in culture increases, there are no significant differences between obese and lean placentas at 1 hour and 6 hours. Lean: Control dams. Obese: Maternal obesity. *P < .05.
Similar to the placentas, cfDNA release by the fetal membrane explants from the obese dams was lower than that from the lean dams at time 0, although this finding did not achieve statistical significance (mean cfDNA release by fetal membranes 2.12 ± 0.58 ng/mg in obese vs 4.56 ± 1.61 ng/mg in lean, P = .12). Within both obese and lean study groups, membranes released significantly less cfDNA than placentas at every time point (Table 2).
Table 2.
Placental and Membrane Cell-Free DNA Release in Obese Versus Lean Dams by Time in Culture.
| Time Point | Amount of cfDNA Released, ng/mga, mean ± SEM | |||||
|---|---|---|---|---|---|---|
| Lean | Obese | |||||
| Placenta | Membrane | P Value | Placenta | Membrane | P Value | |
| 0 | 65.56 ± 7.87 | 4.56 ± 1.61 | <.0001 | 45.77 ± 4.27 | 2.12 ± 0.58 | <.0001 |
| 1 hour | 73.70 ± 9.22 | 15.50 ± 3.87 | <.0001 | 80.06 ± 7.60 | 16.70 ± 3.11 | <.0001 |
| 6 hours | 103.90 ± 15.91 | 43.90 ± 10.10 | .005 | 103.50 ± 15.54 | 44.70 ± 7.19 | .002 |
Abbreviations: cfDNA, cell-free DNA; SEM, standard error of the mean.
aAll cell-free DNA values normalized to tissue wet weight. P values reflect differences in cfDNA release between placenta and membranes within obese and lean study groups.
Distribution of fetal sex in the obese and lean groups at the 3 time points is described in Table 1. Two-way ANOVA demonstrated no significant effect of fetal sex on placental cfDNA release and no significant sex–diet interaction. The CfDNA release by fetal sex and diet group is depicted in Supplemental Figure 2.
Comment
In our established mouse model of maternal diet-induced obesity,16 we demonstrated that placentas from obese dams released significantly less cfDNA compared to placentas from lean dams at time 0, the time point most likely to reflect in vivo physiology of placental cfDNA release just prior to parturition. Later time points (time 1 hour and 6 hours) may be more reflective of cellular changes/apoptosis of fetal tissues in vitro. Placentas from obese dams were smaller than those of lean dams, but even after accounting for this difference by normalizing cfDNA release to placental weight, the quantity of cfDNA released was still significantly decreased in the setting of maternal obesity. The amount of cfDNA released by the placenta was not influenced by fetal sex in our study. These data suggest a refined hypothesis for the lower fetal fraction of cfDNA observed in maternal obesity: There is an intrinsic difference in placental cfDNA release in the setting of maternal obesity compared to normal maternal weight. The CfDNA release by fetal membranes trended in the same direction as the placental explants (reduced in the setting of maternal obesity at time 0), albeit not statistically significant. Fetal membranes released significantly less cfDNA compared to placentas in both lean and obese study groups at every time point. The placenta, not the fetal membranes, was the primary source of cffDNA released in this model system.
After Lo and colleagues described the presence of cffDNA in human maternal plasma,1 multiple studies have since described the presence of cffDNA in maternal plasma in various animal models.29–32 Studies by Tjoa and colleagues determined that the “fetal” source of cfDNA in maternal circulation is the placenta, with increased cfDNA released by villous trophoblasts in the setting of apoptosis and oxidative stress.2 However, studies of cffDNA in maternal plasma are challenging due to the relatively small contribution from fetal compared to maternal cfDNA in maternal circulation (fetal DNA comprises only 3%-20% of cfDNA in maternal circulation, depending on gestational age and study).15,33–36 Phillippe and Adeli have demonstrated that murine placental explant cultures provide a valid and useful model for fetal/placental cfDNA release into maternal circulation.26 They demonstrated the feasibility of isolating and quantifying placental cfDNA in culture media at multiple time points from time 0 through 21 hours and that these explant cultures at time 0 were an appropriate model for in vivo physiology and could serve as a proxy for cffDNA in maternal plasma.26 They also demonstrated that apoptosis was the likely mechanism releasing cfDNA from the placentas into media, with increasing biomarkers of apoptosis seen with increasing time in culture.26 Thus, in our experimental protocol, time 0 best represents the in vivo physiology of placental tissue release of cfDNA into maternal circulation, while the 1- and 6-hour time points may be more reflective of apoptosis of the placenta in vitro.
To date, no studies have examined the release of cffDNA in an animal model of maternal diet-induced obesity. In addition, given that the initial focus on cffDNA in maternal plasma was for prenatal screening,1,37,38 the data on cffDNA in the setting of maternal obesity have been primarily derived from studies evaluating first- and early second-trimester samples. We are not aware of any study that has examined cffDNA in maternal circulation of obese versus lean women at term. In contrast, a few studies have examined total cfDNA in maternal circulation of pregnant women at term, and these have been limited by their inability to distinguish fetal from maternal cfDNA or to account for the effect of maternal body mass index (BMI) on cfDNA in maternal circulation.7,14,39 Evaluation of cffDNA in maternal plasma is difficult due to the low fetal fraction of cfDNA in maternal plasma. Given that all the cfDNA collected in the explant cultures comes directly from fetal tissues, this model has the advantage of definitively answering the question about quantity of fetal cfDNA release, without needing to simultaneously consider the contribution of maternal cfDNA. Thus, the data in the current study fill a knowledge gap regarding the effect of maternal obesity on fetal (placental) DNA release at term. The decreased cfDNA released by placentas from obese dams in our model suggests that the placental release of cfDNA is itself modified by maternal obesity.
Our model system also allows for examining the amount of cfDNA released by fetal membranes. We are not aware of any study that has examined cfDNA release by membranes, nor one that has compared the quantity of cfDNA released by the fetal membranes versus the placenta. Our results demonstrate that while the fetal membranes do release increasing amounts of cfDNA over time in culture, the amount of cfDNA released by the placenta is significantly greater at every time point. Therefore, the majority of cffDNA in maternal circulation likely originates from the placenta, not fetal membranes.
The decreased placental weights observed in the obese dams is consistent with other studies demonstrating decreased embryo weights16,40 and smaller placentas41 in obese dams using this 60% HFD model of diet-induced obesity. In addition, metabolic studies of C57BL6/J dams in this model have demonstrated hyperlipidemia and glucose intolerance.16,25,42,43 Given that vasculopathy and morbid obesity can result in placental insufficiency and fetal growth restriction in human pregnancies,44,45 and that long-standing exposure to HFD was similarly noted to result in both smaller placentas and smaller fetuses41 in a mouse model, there is both precedent and biologic plausibility for our observation of decreased placental weights in our obese dams.
Given that obesity is often considered a chronic state of inflammation46 and that increased inflammation leads to increased cell turnover, our observation of reduced cfDNA release in obese placentas seems counterintuitive. However, a small human study demonstrated decreased placental apoptosis and reduced cell turnover in the setting of maternal obesity.47 In addition, analysis of amniotic fluid samples demonstrates gene expression patterns consistent with reduced apoptosis in multiple fetal organ systems in the setting of maternal obesity.48 Therefore, a small number of prior studies demonstrate biologic plausibility for our finding that the obese placenta releases less cfDNA.
Our findings may have implications for prenatal aneuploidy screening using cffDNA. The accuracy of noninvasive prenatal screening in assessing the risk of aneuploidy relies on a sufficient fetal fraction,33,49 and studies in patients with an elevated maternal BMI have demonstrated increased rates of “no calls” for noninvasive prenatal testing50,51 and a diminished fetal fraction.8–13 Our results suggest that the diminished fetal fraction in the setting of maternal obesity may not be attributable solely to increased maternal adipose tissue apoptosis but also to decreased release of cfDNA by the placenta itself. This nuanced understanding of the low fetal fraction seen in the setting of maternal obesity may spur modifications in the analysis of cfDNA testing to account for these differences and to improve interpretation of results in obese patients. In addition, this novel observation may aid in a better understanding of pregnancy outcomes at term. Further studies are needed to evaluate the effect of cffDNA release at term.
Limitations of our study include the use of in vitro culture techniques to gain insight into cffDNA in maternal serum in vivo. The strength of using placental explant cultures in this setting is that we were able to ensure quantification of fetal/placental, rather than maternal cfDNA, which other studies examining the effects of maternal obesity on cffDNA have been unable to do. We did not attempt to directly measure cffDNA in maternal plasma in our model. While the ability to extrapolate cfDNA released by placenta in culture to cffDNA in maternal circulation is limited, experiments quantifying cffDNA in maternal plasma in a mouse model may be confounded by the multiple gestations with varying number of pups that necessarily exist in murine pregnancy. In human pregnancies, studies have demonstrated an apparent homeostatic pool of cffDNA in maternal circulation which is slightly increased but not proportional to the number of gestations.52 Moreover, studies have shown that dizygotic twins often contribute different amounts of cffDNA into the maternal circulation.53,54 In addition, our study is also limited in its power to detect sex differences in placental and membrane release of cfDNA in the setting of obesity, if such differences exist. Sex-specific placental adaptations have been noted in humans and in mice in the setting of maternal medical comorbidities, including obesity.55–57 We assessed relatively small numbers of male and female placentas/membranes, and it was not possible to achieve equal number of each sex at a given time point, as the fetal sex was not known at the time the tissue was allocated to culture wells. More males and females would be needed in each group at each time point to draw definitive conclusions about whether there is an impact of fetal sex on placental cfDNA release in maternal obesity. Finally, while it is plausible that the mechanism underlying our observation is reduced placental apoptosis in the setting of maternal obesity, markers of placental apoptosis were not specifically quantified in these placentas of obese dams. The release of cfDNA in placental explant cultures has been demonstrated to correlate with placental apoptosis in normal-weight dams, however.24
In conclusion, our mouse model demonstrates that maternal diet-induced obesity is associated with diminished release of cfDNA by the placenta. These findings offer an alternative hypothesis for the decreased fetal fraction of cfDNA seen in obese pregnancies.
Supplemental Material
ReproSci_supplemental_figure_1 for The Effect of Maternal Obesity on Placental Cell-Free DNA Release in a Mouse Model by Mohak Mhatre, Sharareh Adeli, Errol Norwitz, Sabrina Craigo, Mark Phillippe and Andrea Edlow in Reproductive Sciences
Supplemental Material
ReproSci_supplemental_figure_2 for The Effect of Maternal Obesity on Placental Cell-Free DNA Release in a Mouse Model by Mohak Mhatre, Sharareh Adeli, Errol Norwitz, Sabrina Craigo, Mark Phillippe and Andrea Edlow in Reproductive Sciences
Supplemental Material
Supplemental_Material for The Effect of Maternal Obesity on Placental Cell-Free DNA Release in a Mouse Model by Mohak Mhatre, Sharareh Adeli, Errol Norwitz, Sabrina Craigo, Mark Phillippe and Andrea Edlow in Reproductive Sciences
Footnotes
Authors’ Note: Study was conducted at Tufts Medical Center, Massachusetts General Hospital.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the 2K12HD000849 (Edlow); March of Dimes (#4-FY17, Edlow), Natalie V. Zucker Research Grant (Mhatre).
Supplemental Material: Supplemental material is available for this article online.
References
- 1. Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. [DOI] [PubMed] [Google Scholar]
- 2. Tjoa ML, Cindrova-Davies T, Spasic-Boskovic O, Bianchi DW, Burton GJ. Trophoblastic oxidative stress and the release of cell-free feto-placental DNA. Am J Pathol. 2006;169(2):400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alberry MS, Maddocks DG, Hadi MA, et al. Quantification of cell free fetal DNA in maternal plasma in normal pregnancies and in pregnancies with placental dysfunction. Am J Obstet Gynecol. 2009;200(1):98.e91–e96. [DOI] [PubMed] [Google Scholar]
- 4. Al Nakib M, Desbriere R, Bonello N, et al. Total and fetal cell-free DNA analysis in maternal blood as markers of placental insufficiency in intrauterine growth restriction. Fetal Diagn Ther. 2009;26(1):24–28. [DOI] [PubMed] [Google Scholar]
- 5. Sifakis S, Koukou Z, Spandidos DA. Cell-free fetal DNA and pregnancy-related complications (review). Mol Med Rep. 2015;11(4):2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dugoff L, Barberio A, Whittaker PG, Schwartz N, Sehdev H, Bastek JA. Cell-free DNA fetal fraction and preterm birth. Am J Obstet Gynecol. 2016;215(2):231 e231–237. [DOI] [PubMed] [Google Scholar]
- 7. Herrera CA, Stoerker J, Carlquist J, et al. Cell-free DNA, inflammation, and the initiation of spontaneous term labor. Am J Obstet Gynecol. 2017;217(5):583.e581–e588. [DOI] [PubMed] [Google Scholar]
- 8. Scott FP, Menezes M, Palma-Dias R, et al. Factors affecting cell-free DNA fetal fraction and the consequences for test accuracy. J Matern Fetal Neonatal Med. 2018;31(14):1865–1872. [DOI] [PubMed] [Google Scholar]
- 9. Poon LC, Musci T, Song K, Syngelaki A, Nicolaides KH. Maternal plasma cell-free fetal and maternal DNA at 11-13 weeks’ gestation: relation to fetal and maternal characteristics and pregnancy outcomes. Fetal Diagn Ther. 2013;33(4):215–223. [DOI] [PubMed] [Google Scholar]
- 10. Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13(11):913–920. [DOI] [PubMed] [Google Scholar]
- 11. Ashoor G, Syngelaki A, Poon LC, Rezende JC, Nicolaides KH. Fetal fraction in maternal plasma cell-free DNA at 11-13 weeks’ gestation: relation to maternal and fetal characteristics. Ultrasound Obstet Gynecol. 2013;41(1):26–32. [DOI] [PubMed] [Google Scholar]
- 12. Zhou Y, Zhu Z, Gao Y, et al. Effects of maternal and fetal characteristics on cell-free fetal DNA fraction in maternal plasma. Reprod Sci. 2015;22(11):1429–1435. [DOI] [PubMed] [Google Scholar]
- 13. Wataganara T, Peter I, Messerlian GM, Borgatta L, Bianchi DW. Inverse correlation between maternal weight and second trimester circulating cell-free fetal DNA levels. Obstet Gynecol. 2004;104(3):545–550. [DOI] [PubMed] [Google Scholar]
- 14. Vora NL, Johnson KL, Basu S, Catalano PM, Hauguel-De Mouzon S, Bianchi DW. A multifactorial relationship exists between total circulating cell-free DNA levels and maternal BMI. Prenat Diagn. 2012;32(9):912–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canick JA, Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat Diagn. 2013;33(7):667–674. [DOI] [PubMed] [Google Scholar]
- 16. Edlow AG, Guedj F, Pennings JL, Sverdlov D, Neri C, Bianchi DW. Males are from Mars, and females are from Venus: sex-specific fetal brain gene expression signatures in a mouse model of maternal diet-induced obesity. Am J Obstet Gynecol. 2016;214(5):623 e621–623 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menon R. Human fetal membranes at term: dead tissue or signalers of parturition?. Placenta. 2016;44:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumagai K, Otsuki Y, Ito Y, Shibata MA, Abe H, Ueki M. Apoptosis in the normal human amnion at term, independent of Bcl-2 regulation and onset of labour. Mol Hum Reprod. 2001;7(7):681–689. [DOI] [PubMed] [Google Scholar]
- 19. Lei H, Furth EE, Kalluri R, et al. A program of cell death and extracellular matrix degradation is activated in the amnion before the onset of labor. J Clin Invest. 1996;98(9):1971–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Menon R, Boldogh I, Hawkins HK, et al. Histological evidence of oxidative stress and premature senescence in preterm premature rupture of the human fetal membranes recapitulated in vitro. Am J Pathol. 2014;184(6):1740–1751. [DOI] [PubMed] [Google Scholar]
- 21. Menon R. Oxidative stress damage as a detrimental factor in preterm birth pathology. Front Immunol. 2014;5:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kang SS, Kurti A, Fair DA, Fryer JD. Dietary intervention rescues maternal obesity induced behavior deficits and neuroinflammation in offspring. J Neuroinflammation. 2014;11:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gallou-Kabani C, Gabory A, Tost J, et al. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5(12):e14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krasnow SM, Nguyen ML, Marks DL. Increased maternal fat consumption during pregnancy alters body composition in neonatal mice. Am J Physiol Endocrinol Metab. 2011;301(6):E1243–E1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gallou-Kabani C, Vige A, Gross MS, et al. C57BL/6 J and A/J mice fed a high-fat diet delineate components of metabolic syndrome. Obesity (Silver Spring). 2007;15(8):1996–2005. [DOI] [PubMed] [Google Scholar]
- 26. Phillippe M, Adeli S. Cell-free DNA release by mouse placental explants. Plos One. 2017;12(6):e0178845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller RK, Genbacev O, Turner MA, Aplin JD, Caniggia I, Huppertz B. Human placental explants in culture: approaches and assessments. Placenta. 2005;26(6):439–448. [DOI] [PubMed] [Google Scholar]
- 28. Phillippe M. Cell-free fetal DNA—a trigger for parturition. N Engl J Med. 2014;370(26):2534–2536. [DOI] [PubMed] [Google Scholar]
- 29. de Leon PM, Campos VF, Dellagostin OA, Deschamps JC, Seixas FK, Collares T. Equine fetal sex determination using circulating cell-free fetal DNA (ccffDNA). Theriogenology. 2012;77(3):694–698. [DOI] [PubMed] [Google Scholar]
- 30. Kadivar A, Hassanpour H, Mirshokraei P, Azari M, Gholamhosseini K, Karami A. Detection and quantification of cell-free fetal DNA in ovine maternal plasma; use it to predict fetal sex. Theriogenology. 2013;79(6):995–1000. [DOI] [PubMed] [Google Scholar]
- 31. Khosrotehrani K, Wataganara T, Bianchi DW, Johnson KL. Fetal cell-free DNA circulates in the plasma of pregnant mice: relevance for animal models of fetomaternal trafficking. Hum Reprod. 2004;19(11):2460–2464. [DOI] [PubMed] [Google Scholar]
- 32. Wang G, Cui Q, Cheng K, Zhang X, Xing G, Wu S. Prediction of fetal sex by amplification of fetal DNA present in cow plasma. J Reprod Dev. 2010;56(6):639–642. [DOI] [PubMed] [Google Scholar]
- 33. Taglauer ES, Wilkins-Haug L, Bianchi DW. Review: cell-free fetal DNA in the maternal circulation as an indication of placental health and disease. Placenta. 2014;35(suppl):S64–S68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Edlow AG, Bianchi DW. Tracking fetal development through molecular analysis of maternal biofluids. Biochim Biophys Acta. 2012;1822(12):1970–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62(4):768–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105(42):16266–16271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Norton ME, Wapner RJ. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;373(26):2582. [DOI] [PubMed] [Google Scholar]
- 38. Bianchi DW, Platt LD, Goldberg JD, et al. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. [DOI] [PubMed] [Google Scholar]
- 39. Lapaire O, Volgmann T, Grill S, et al. Significant correlation between maternal body mass index at delivery and in the second trimester, and second trimester circulating total cell-free DNA levels. Reprod Sci. 2009;16(3):274–279. [DOI] [PubMed] [Google Scholar]
- 40. Luzzo KM, Wang Q, Purcell SH, et al. High fat diet induced developmental defects in the mouse: oocyte meiotic aneuploidy and fetal growth retardation/brain defects. Plos One. 2012;7(11):e49217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kamimae-Lanning AN, Krasnow SM, Goloviznina NA, et al. Maternal high-fat diet and obesity compromise fetal hematopoiesis. Mol Metab. 2015;4(1):25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ferrari F, Facchinetti F, Ontiveros AE, et al. The effect of combined inositol supplementation on maternal metabolic profile in pregnancies complicated by metabolic syndrome and obesity. Am J Obstet Gynecol. 2016;215(4):503 e501–508. [DOI] [PubMed] [Google Scholar]
- 43. Murabayashi N, Sugiyama T, Zhang L, et al. Maternal high-fat diets cause insulin resistance through inflammatory changes in fetal adipose tissue. Eur J Obstet Gynecol Reprod Biol. 2013;169(1):39–44. [DOI] [PubMed] [Google Scholar]
- 44. Leguizamon G, Trigubo D, Pereira JI, Vera MF, Fernandez JA. Vascular complications in the diabetic pregnancy. Curr Diab Rep. 2015;15(4):22. [DOI] [PubMed] [Google Scholar]
- 45. Myatt L, Maloyan A. Obesity and placental function. Semin Reprod Med. 2016;34(1):42–49. [DOI] [PubMed] [Google Scholar]
- 46. Pendeloski KPT, Ono E, Torloni MR, Mattar R, Daher S. Maternal obesity and inflammatory mediators: a controversial association. Am J Reprod Immunol. 2017;77(5). [DOI] [PubMed] [Google Scholar]
- 47. Higgins L, Mills TA, Greenwood SL, Cowley EJ, Sibley CP, Jones RL. Maternal obesity and its effect on placental cell turnover. J Matern Fetal Neonatal Med. 2013;26(8):783–788. [DOI] [PubMed] [Google Scholar]
- 48. Edlow AG, Vora NL, Hui L, Wick HC, Cowan JM, Bianchi DW. Maternal obesity affects fetal neurodevelopmental and metabolic gene expression: a pilot study. Plos One. 2014;9(2):e88661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kang X, Xia J, Wang Y, et al. An advanced model to precisely estimate the cell-free fetal DNA concentration in maternal plasma. Plos One. 2016;11(9):e0161928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Livergood MC, LeChien KA, Trudell AS. Obesity and cell-free DNA “no calls”: is there an optimal gestational age at time of sampling? Am J Obstet Gynecol. 2017;216(4):413 e411–413 e419. [DOI] [PubMed] [Google Scholar]
- 51. Yared E, Dinsmoor MJ, Endres LK, et al. Obesity increases the risk of failure of noninvasive prenatal screening regardless of gestational age. Am J Obstet Gynecol. 2016;215(3):370 e371–376. [DOI] [PubMed] [Google Scholar]
- 52. Canick JA, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenat Diagn. 2012;32(8):730–734. [DOI] [PubMed] [Google Scholar]
- 53. Leung TY, Qu JZ, Liao GJ, et al. Noninvasive twin zygosity assessment and aneuploidy detection by maternal plasma DNA sequencing. Prenat Diagn. 2013;33(7):675–681. [DOI] [PubMed] [Google Scholar]
- 54. Qu JZ, Leung TY, Jiang P, et al. Noninvasive prenatal determination of twin zygosity by maternal plasma DNA analysis. Clin Chem. 2013;59(2):427–435. [DOI] [PubMed] [Google Scholar]
- 55. Anelli GM, Cardellicchio M, Novielli C, et al. Mitochondrial content and hepcidin are increased in obese pregnant mothers. J Matern Fetal Neonatal Med. 2018;31(18):2388–2395. [DOI] [PubMed] [Google Scholar]
- 56. Clifton VL. Review: sex and the human placenta: mediating differential strategies of fetal growth and survival. Placenta. 2010;31(suppl):S33–S39. [DOI] [PubMed] [Google Scholar]
- 57. Gabory A, Ferry L, Fajardy I, et al. Maternal diets trigger sex-specific divergent trajectories of gene expression and epigenetic systems in mouse placenta. Plos One. 2012;7(11):e47986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ReproSci_supplemental_figure_1 for The Effect of Maternal Obesity on Placental Cell-Free DNA Release in a Mouse Model by Mohak Mhatre, Sharareh Adeli, Errol Norwitz, Sabrina Craigo, Mark Phillippe and Andrea Edlow in Reproductive Sciences
ReproSci_supplemental_figure_2 for The Effect of Maternal Obesity on Placental Cell-Free DNA Release in a Mouse Model by Mohak Mhatre, Sharareh Adeli, Errol Norwitz, Sabrina Craigo, Mark Phillippe and Andrea Edlow in Reproductive Sciences
Supplemental_Material for The Effect of Maternal Obesity on Placental Cell-Free DNA Release in a Mouse Model by Mohak Mhatre, Sharareh Adeli, Errol Norwitz, Sabrina Craigo, Mark Phillippe and Andrea Edlow in Reproductive Sciences