Abstract
Many cellular and physiological processes are coordinated by regulatory networks that produce a remarkable complexity of transcript isoforms. In the mammalian nervous system, alternative pre-mRNA splicing generates functionally distinct isoforms that play key roles in normal physiology, supporting development, plasticity, complex behaviors, and cognition. Neuronal splicing programs controlled by RNA-binding proteins, are influenced by chromatin modifications and can exhibit neuronal subtype specificity. As highlighted in recent publications, aberrant alternative splicing is a major contributor to disease phenotypes. Therefore, understanding the underlying mechanisms of alternative splicing regulation and identifying functional splicing isoforms with critical phenotypic roles are expected to provide a comprehensive resource for therapeutic development, as illuminated by recent successful interventions of spinal muscular atrophy. Here, we discuss the latest progress in the study of the emerging complexity of alternative splicing mechanisms in neurons, and how these findings inform new therapies to correct and control splicing defects.
Keywords: alternative splicing, CACNA1B, spinal muscular atrophy (SMA), autism spectrum disorder (ASD), axonogenesis, chromatin
Introduction
Alternative pre-mRNA splicing expands the mammalian transcriptome in the nervous system, paving the way for neuronal differentiation and plasticity. The transcriptome and proteome are customized by alternative splicing based on neuronal subtype, and developmental and physiological states. Recent publications show that disruption of alternatively spliced isoform levels, rather than disruption of gene expression levels, is a major source of pathological effects in psychiatric and neurological diseases (Gandal et al., 2018); splicing-correcting therapies are being successfully applied to alleviate disease symptoms in both animal models and in humans (Hua et al., 2011; Finkel et al., 2016; Sinha et al., 2018).
Alternative splicing dictates exon composition of mRNAs that will be translated into the final protein. This mechanism generates numerous exon combinations and, consequently, functionally different isoforms from a single multiexon gene. The best-studied mechanisms for determining alternative exon usage involve recognition of cis-regulatory elements on pre-mRNAs by RNA-binding proteins (for reviews, see Fu and Ares, 2014 and Vuong et al., 2016). RNA-binding proteins act as trans-acting splicing factors that inhibit or promote exon usage during alternative splicing. But epigenetic factors also act as molecular regulators of splicing (for review, see Luco et al., 2011) (Fig. 1). The RNA polymerase II (Pol II) can interact with DNA-binding proteins and histone post-translational modifications, altering transcription elongation rate or recruiting RNA splicing factors, to promote or inhibit alternative exon usage (Luco et al., 2010; Shukla et al., 2011; Ding et al., 2017; Maslon et al., 2019). The fundamentally different nature of these mechanisms calls for different design principles for therapeutics intended to alter splicing.
Figure 1.

Key mechanisms of alternative pre-mRNA splicing. A, Histone modifications and DNA-binding proteins associated with alternative exons (light blue rectangles) can interact with the RNA Pol II and regulate exon usage by recruiting RNA-binding proteins that recognize pre-mRNA-binding motifs or by slowing down RNA Pol II elongation rate and favoring the inclusion of alternative exons. B, RNA-binding proteins regulate alternative splicing by binding pre-mRNA and driving inclusion or exclusion of alternative exons (light blue rectangles). Gray rectangles represent constitutive exons.
The advances in transcriptome and epigenome profiling technologies, together with sophisticated bioinformatic analyses, have vastly improved the detection and quantification of mRNA isoforms in the nervous system. Assigning functional roles for these splicing isoforms is more challenging, but isoforms of some proteins, including REST, PSD-95, and NRXN1, have been thoroughly characterized, and the progress on this front is accelerating.
Rest encodes the RE1-silencing transcription factor, a master repressor of neuronal-specific genes during cell differentiation. During development, the activation of neuronal genes depends on the release of REST binding from neuronal gene loci (Ballas et al., 2005). This can be achieved by REST post-translational degradation, transcriptional repression, or inactivation through alternative splicing (Ballas et al., 2005; Westbrook et al., 2008; Raj et al., 2011; Nakano et al., 2012). The RNA-binding protein SRRM4 promotes Rest exon 4 inclusion, triggering a frameshift in Rest mRNA that truncates REST, impairing its gene-silencing function and thus derepressing many neuronal genes (Raj et al., 2011; Nakano et al., 2012). Splicing-inactivated REST in the mechanosensory hair cells is required for normal hearing in humans and mice. Mutations causing either loss of function of SRRM4 in mice or inhibition of the frameshifting splicing event in humans are associated with dominantly inherited deafness (Nakano et al., 2012, 2018).
Alternative splicing of Psd-95 exemplifies the power of splicing control in shaping neuronal differentiation. PSD-95 is an excitatory postsynaptic protein (Cho et al., 1992); and although it is widely considered to be a neural-specific protein, its mRNA is readily detectable outside the nervous system (Zheng, 2016). The underlying mechanism for neural-specific expression of PSD-95 is attributed to different isoforms expressed in neurons versus non-neuronal cells. In neurons, Psd-95 exon 18 is specifically included to enable PSD-95 translation; whereas in non-neuronal cells, exon 18 is excluded, leading to a frameshift and nonsense-mediated mRNA decay without productive translation (Zheng et al., 2012). During development, the splicing switch occurs before synaptogenesis and is regulated by polypyrimidine tract binding proteins 1 and 2 (PTBP1 and PTBP2), RBM10, and Puf60 (Zheng et al., 2012, 2013). Therefore, neuronal-specific alternative splicing of Psd-95 enforces neuronal-restricted expression of PSD-95 and ensures the timing of synapse formation.
Alternative splicing of the Nrxn1 gene generates thousands of neurexin isoforms (Südhof, 2017). Neurexins are presynaptic cell-adhesion molecules essential for synapse formation and synaptic transmission. Learning-induced changes in Nrxn1's exon usage determine binding affinity of neurexins to postsynaptic partners (e.g., Neuroligin 1), impacting synaptic strength (Aoto et al., 2013; Traunmüller et al., 2016; Ding et al., 2017; Dai et al., 2019). A well-studied splicing event is the inclusion of the Nrxn1 alternative exon 22. In hippocampal neurons, neuronal activation drives the inclusion of Nrxn1 exon 22; the resulting neurexin isoform is essential for memory consolidation (Ding et al., 2017). In hippocampus, a specific form of histone 3 modification (trimethylation at lysine 9; H3K9me3) cotranscriptionally regulates exon 22 inclusion by slowing the elongation rate of RNA Pol II (Ding et al., 2017); whereas in cerebellum, the RNA-binding protein SAM68 guides the activity-dependent exclusion of exon 22 after Nrxn1 transcription (Iijima et al., 2011). These observations suggest that alternative splicing of Nrxn1 is temporally and spatially controlled by different mechanisms in the brain to establish precise synaptic connections.
The aforementioned examples illustrate how neuronal-specific and dynamically regulated splicing events underlie neuronal differentiation and plasticity. Despite the recent progress in the characterization of individual alternative exons, our understanding of the functional roles for the vast majority of spliced isoforms that have been detected in the last years by transcriptome profiling studies, remains limited. A major future goal is to characterize all splicing events with important phenotypic roles, and especially those associated with pathologies. In this review, we summarize recently identified neuronal-specific splicing programs, focusing on individual splicing events and their implications in neurophysiology. We also discuss how disruption of these programs contributes to disease pathology and how understanding these mechanisms can facilitate the development of therapeutic tools to correct mis-splicing events. This review presents recent advances on these fronts as presented at the Mini-Symposium Novel Mechanisms of Neuronal Alternative Splicing and Strategies to Correct Aberrant-Splicing during the Neuroscience 49th Annual Meeting.
Alternative splicing of Cacna1b pre-mRNA modifies CaV2.2 channel properties across different neurons
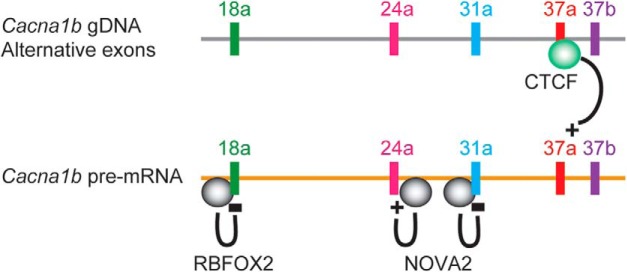
Transcriptome profiling across neuronal populations reveals substantial differences in the pattern of alternative splicing of ion channels. The vast majority of large multiexon ion channel genes contain alternatively spliced exons that are expressed in neuronal subtype-specific patterns and that modify protein function (for review, see Lipscombe et al., 2013; Allen et al., 2017; Szczot et al., 2017; Thalhammer et al., 2017; Regan et al., 2018; Bunda et al., 2019). A well-studied example of this is Cacna1b, which encodes the functional core of CaV2.2 voltage-gated calcium channels, and controls presynaptic calcium entry that triggers exocytosis. The Lipscombe laboratory has shown that exon usage during Cacna1b pre-mRNA splicing varies across different neurons to generate distinct patterns of CaV2.2 isoforms with unique properties and functions (Bell et al., 2004; Lipscombe and Andrade, 2015; Allen et al., 2017). Subsequent research in the laboratory has focused on identifying the mechanisms that regulate alternative splicing of Cacna1b. For example, the neuronal splicing-factor families RBFOX and NOVA bind Cacna1b pre-mRNA to regulate the usage of alternative exons 18a, 24a, and 31a (Allen et al., 2010, 2017) (Fig. 2). In superior cervical ganglia, RBFOX2 binding upstream of Cacna1b exon 18a suppresses its inclusion early in development, but in adults this suppression is reduced and exon 18a is included (Allen et al., 2017). As a result, CaV2.2 channel trafficking to plasma membrane is promoted in adult neurons because of sequences encoded by exon 18a.
Figure 2.
Different mechanisms of Cacna1b alternative splicing. The DNA-binding protein CTCF, when bound to genomic DNA locus 37a, promotes exon 37a inclusion during alternative splicing. The RNA-binding proteins RBFOX2 and NOVA2, when bound to Cacna1b pre-mRNA, inhibit inclusion of exons 18a and 31a, respectively, but NOVA2 promotes inclusion of exon 24a.
The best studied example of cell-specific alternative splicing of Cacna1b involves a pair of mutually exclusive exons (37a and 37b) that encode a 32 amino acid segment in the proximal region of the CaV2.2 C-terminus. CaV2.2 channels are located at nociceptor terminals in the dorsal horn of the spinal cord, and this is the site of action of drugs and neurotransmitters that activate G-protein-coupled receptors to downregulate nociception. The neuronal-specific inclusion of exon 37a replaces the more common exon 37b in a subset of nociceptors, resulting in more efficient CaV2.2 channel membrane trafficking and enhanced G-protein inhibition of CaV2.2 by μ-opioid receptors (Bell et al., 2004; Castiglioni et al., 2006; Raingo et al., 2007; Macabuag and Dolphin, 2015). This splicing event enhances intrathecal morphine analgesia in nonpathological pain conditions, but it is disrupted following peripheral nerve injury that results in neuropathic pain (Altier et al., 2007; Andrade et al., 2010; Jiang et al., 2013).
The Lipscombe laboratory has identified the factors that promote inclusion of exon 37a during Cacna1b splicing. Unexpectedly, Cacna1b exon 37a inclusion is controlled by the DNA-binding protein CTCF: CTCF binding to exon 37a promotes its inclusion in nociceptors. CTCF is a ubiquitously expressed chromatin regulator that has been shown to influence alternative exon usage by pausing RNA Pol II (Shukla et al., 2011). In a subset of nociceptors, epigenetic modulation of CpG methylation in locus 37a controls CTCF binding and thus controls Cacna1b exon 37a usage. Peripheral nerve injury alters the epigenetic marks in Cacna1b exon 37a, reducing CTCF binding and limiting exon 37a usage, thus contributing to the reduced morphine analgesia in neuropathic pain. This study underscores the importance of identifying the factors that control cell-specific alternative pre-mRNA splicing that impact drug action in disease states, and could inform the development of therapies to increase drug efficiency.
Genomewide interrogation of microexon regulators reveals genetic links to autism spectrum disorder (ASD)
ASD is a neurodevelopmental and neuropsychiatric disorder characterized by impaired social interactions and repetitive patterns of behavior that affects >1% of the population. Technological advances in genomics have revealed that ASD is not a single disease but rather a heterogeneous spectrum of etiologically distinct conditions, which result in common behavioral manifestations. Despite the profound genetic heterogeneity of ASD, this disorder is characterized by the disruption of convergent molecular and cellular mechanisms, such as synaptic function, activity-dependent transcription, and localized translation (de la Torre-Ubieta et al., 2016; Quesnel-Vallières et al., 2019). A major advancement in our understanding of the underlying causes of ASD has emerged through transcriptome profiling studies, which revealed that alternative pre-mRNA splicing is often mis-regulated in ASD patients (Irimia et al., 2014; Parikshak et al., 2016; Gandal et al., 2018; Quesnel-Vallières et al., 2019).
Alternative microexons (3–27 nucleotides) represent the most highly conserved class of splicing events and tend to preserve the open reading frame, often overlapping domains that mediate protein–protein interactions (Black, 1995; Irimia et al., 2014; Li et al., 2015; Ustianenko et al., 2017). The Blencowe laboratory recently discovered that an extensive network of neuronal microexons is frequently disrupted in the brains of ASD patients (Irimia et al., 2014). Approximately 120 microexons, enriched in synaptic genes, display reduced inclusion in the brains of >30% of analyzed individuals with idiopathic autism. Notably, the expression level of neuronal-specific splicing factor SRRM4, which is critical for the splicing of most neuronal microexons, is reduced in the brains of ASD subjects that display attenuated microexon inclusion (Irimia et al., 2014). SRRM4 is a vertebrate-conserved RNA-binding protein critical for nervous system development (Calarco et al., 2009; Raj et al., 2011; Quesnel-Vallières et al., 2015). Importantly, mice haploinsufficient for Srrm4 recapitulate reduced microexon inclusion and display multiple ASD-like features, including altered social behavior, increased sensitivity to environmental stimuli, and altered synaptic transmission, strongly suggesting that microexon mis-regulation is causally linked to autism (Quesnel-Vallières et al., 2015, 2016).
Despite the importance of microexons in nervous system development and disorders, the molecular mechanisms that regulate microexon usage during splicing are not well understood. This was addressed by developing a CRISPR-based genomewide screening strategy that identified >200 microexon regulators, including chromatin, protein turnover, and RNA processing factors enriched for genetic links to autism (Gonatopoulos-Pournatzis et al., 2018). These results revealed that SRRM4 regulates neuronal microexon usage by forming interactions with the SR-related proteins SRSF11 and RNPS1, as well as an intronic enhancer element, and that these interactions are required for early splicing complex assembly. Furthermore, critical chromatin regulators, including EP300 and CREBBP, impact microexon splicing by controlling SRRM4 expression. Interestingly, mutations and/or alterations in the expression of SRSF11, RNPS1, EP300, and CREBP are linked to neurodevelopmental disorders (Nguyen et al., 2013; Korzus, 2017; Lim et al., 2017; Yuen et al., 2017), providing additional genetic evidence that disruption of microexon splicing underlies nervous system development disorders.
A major challenge that lies ahead is to determine the functions of individual microexons, especially in relation to autistic phenotypes, and to explore their mechanisms of action. Recent studies have revealed that individual microexons can impact animal behavior (Rusconi et al., 2017; Parras et al., 2018). The Cytoplasmic Polyadenylation Element Binding (CPEB) proteins are RNA-binding proteins that promote cytoplasmic mRNA polyadenylation regulating several biological processes, including learning and memory (Ivshina et al., 2014; Si and Kandel, 2016). Interestingly, inclusion of a neuron-specific microexon in CPEB4 controls the deadenylation and translation of ASD-risk genes, and imbalance in the expression of the Cpeb4 microexon causes ASD-like phenotypes in mice (Parras et al., 2018). In addition, a microexon in the Lysine Demethylase 1A (KDM1A) controls activity-dependent transcriptional programs and impacts emotional behavior and seizure susceptibility (Rusconi et al., 2015, 2016). Collectively, these studies provide evidence that individual microexons underlie distinct phenotypic outcomes and that disruption of the SRRM4-dependent microexon network contributes to ASD-related phenotypes. The next challenge is to develop high-throughput screening strategies that systematically identify alternative exons, including neuronal microexons, with critical roles in development and whose mis-regulation contributes to disease and disorders. Identification of such splicing events is expected to facilitate the development of therapeutic approaches to revert specific disease phenotypes.
Chromatin-mediated alternative splicing is linked to reward pathophysiology
Chromatin state regulates alternative pre-mRNA splicing, underlying remodeling events by which mature neurons could mount a stable response to environmental stimuli (Ding et al., 2017) (for review, see Luco et al., 2011). The Heller laboratory posits that, in postmitotic neurons, alternative exon usage maintained by stably associated histone modifications underlies chronic disease states, such as addiction. This hypothesis is based on two findings: (1) cocaine exposure changes isoform expression to a far greater extent than it affects total gene expression levels (Feng et al., 2014); and (2) alternative splicing is functionally coupled to specific histone modifications (Auboeuf et al., 2002; Luco et al., 2010; Kim et al., 2011), which are also altered by cocaine exposure (Kumar et al., 2005; Feng et al., 2014). While many neurological diseases, including frontotemporal dementia, Rett syndrome, bipolar disorder, ASD, and schizophrenia, have been linked to large disruptions in isoform expression levels (Licatalosi and Darnell, 2006; Gandal et al., 2018), this mechanism is understudied in the context of drug abuse and addiction. Therefore, the Heller laboratory is currently investigating the hypothesis that cocaine alters alternative splicing in brain reward regions by remodeling chromatin states. To examine this, they analyze the genomewide association between a suite of seven histone modifications and alternative exon usage in nucleus accumbens, a structure central to reward seeking, from mice treated with cocaine or saline. Using novel computational approaches, classical statistical methods combined with machine learning, they found that two histone modifications (trimethylation at lysine 36 of histone 3, [H3K36me3], and methylation of lysine 4 of histone 3 [H3K4me1]) have the strongest association with alternative splicing (Hu et al., 2017).
A second study tested the hypothesis that specific histone modifications could drive development by regulating expression of alternative spliced isoforms during differentiation (Hu et al., 2017). To address this idea, they undertook a comprehensive genomewide analysis that related histone modifications to alternative exon usage during mammalian tissue development. They found that H3K36me3 and H3K4me1 play a dominant role in exon exclusion across all the tissues and developmental time points examined. Specifically, H3K36me3 positioning in alternative exons is the strongest predictor for exon exclusion during alternative splicing in all tissues.
Given the evidence that H3K36me3 positioning strongly predicts alternative exon usage across various tissues and development, Xu and Heller (2018) next analyzed the role of H3K36me3 in influencing alternative splicing in adult mouse nucleus accumbens. Analysis was performed on ChIP/RNA-Seq datasets from nucleus accumbens following either cocaine (or saline) self-administration or overexpression of SET2, the histone methyltransferase that catalyzes H3K36me3. The authors found an overlapping set of alternative exons that are differentially expressed, are enriched for H3K36me3, and contain the binding motif of the Serine and Arginine Rich Splicing Factor 11 (SRSF11). Unexpectedly, Srsf11 mRNA is itself differentially spliced and enriched in H3K36me3 in the corresponding alternative exon in cocaine- or SET2-treated tissue relative to controls. Then, they used a targeted epigenetic editing tool to distinguish the direct causal relevance of the H3K36me3 at the alternative exon from pleiotropic, genomewide effects. In this approach, a histone methyltransferase catalytic domain linked to an engineered sequence-specific DNA-binding protein enables methylation of histones located only in the vicinity of the target alternative exon. Interestingly, they found that H3K36me3 enrichment targeted to the Srsf11 alternative exon promotes exon inclusion during alternative splicing but has no effect on overall Srsf11 expression.
Together, the combination of machine learning and epigenetic editing tools suggests that H3K36me3 enrichment influences alternative exon usage. The Heller laboratory is currently working to determine the relevance of SRSF11 isoform expression to cocaine-mediated behavior and the precise mechanism by which this histone modification contributes to alternative splicing, either via direct recruitment of RNA-binding proteins or regulation of the kinetics of RNA Pol II.
PTBP2 orchestrates alternative splicing programming required for axon formation
Neurons become polarized in phases: neurites form, then begin to grow before acquiring axonal characteristics (Arimura and Kaibuchi, 2007). Despite significant progress in understanding the cell biology of neuronal polarization, many basic questions remain. For example, although many genes that control axonogenesis, the first step in neuronal polarization, have been identified (Liu and Szaro, 2011; Zollinger et al., 2015; Rao et al., 2017; Tilot et al., 2018), how they are coordinated is unclear. In a quest to identify genetic determinants of cortical neuronal polarization, the Zheng laboratory generated the first transcriptome datasets of cortical neurons before and after axon specification (Zhang et al., 2019). Multipronged analyses revealed a surprisingly strong association between neural-specific splicing events and axonogenesis. The expression levels of previously identified axonogenesis-controlling genes were similar before and after axon extension, but many showed changes in isoform expression. This finding suggests that neuronal polarization is executed in part by regulating polarity genes through alternative splicing mechanisms. Importantly, the authors found that the alternative exons that are expressed during early axonogenesis are neural-specific, suggesting that they may be part of regulatory mechanisms that distinguish neuronal polarization from planar cell polarity.
The Zheng laboratory found that the RNA-binding protein PTBP2 governs axonogenesis-associated alternative splicing (Zhang et al., 2019). Specifically, Ptbp2 depletion results in shorter axons of cortical neurons both in vitro and in vivo. Interestingly, some Ptbp2 null neurons can produce surplus axons marked by axonal markers Tau1 and AnkG. Therefore, axonal growth is impeded, but axon specification appears to be enhanced in the absence of PTBP2. The unusual phenotype caused by Ptpb2 knockout likely reflects that the initial period of neurite growth builds the structural prerequisites for axon formation. If the alternative splicing program changes too soon, as happens when Ptb2 is knocked out, those prerequisites are not met, so the axon cannot be specified. The Zheng laboratory data suggest that the hierarchy of cellular events leading to axon formation is coordinated by PTBP2 regulation of alternative exon usage (Zhang et al., 2019).
Since the polarity genes alter their isoform expression during early axon formation, it is tempting to speculate that alternative splicing gives rise to functionally different isoforms involved in axon formation. Future studies are directed toward identifying the critical PTBP2-regulated events that underlie axonogenesis. Considering that structural and functional impairment of axons is a common manifestation of neurodegenerative, neuroinflammatory, and neurodevelopmental disorders (Medana and Esiri, 2003), this research provides a unique perspective to better understand the genetic, molecular, and cellular control of disease processes.
Novel insights from splicing regulation of the spinal muscular atrophy (SMA) genes
SMA is a devastating autosomal-recessive disorder associated with infant mortality, with an incidence of ∼1 in 10,000 live births. SMA is caused by low expression levels of a ubiquitous protein, survival motor neuron (SMN) protein, which plays an essential role in transcription, splicing, translation, RNA trafficking, and cell signaling (Singh et al., 2017b). Homologous deletion or disruption of SMN1, the main gene that encodes SMN protein, accounts for >90% of SMA cases (Lefebvre et al., 1995; Wirth, 2000).
Humans have another gene, SMN2, which is an almost identical copy of SMN1. Due to the predominant skipping of exon 7 during splicing, SMN2 produces low levels of SMN (for review, see Khoo and Krainer, 2009; Singh and Singh, 2018). Importantly, promoting inclusion of this exon has been exploited for rescuing SMA phenotypes (Hua et al., 2008, 2010, 2011, 2015; Passini et al., 2011; Rigo et al., 2012; Sahashi et al., 2013). Nusinersen (Spinraza), the first drug approved by the FDA for treatment of SMA, is an antisense oligonucleotide that promotes SMN2 exon 7 inclusion (Singh et al., 2017a). Nusinersen blocks the Intronic Splicing Silencer N1 (ISS-N1) located downstream of SMN2 exon 7 (Singh et al., 2006; Singh and Singh, 2018). Of note, ISS-N1 has been shown to be a highly inhibitory sequence for SMN2 exon 7 usage (Singh et al., 2006). In addition to several antisense oligonucleotides that have been successfully used to restore SMN protein, small molecules, and engineered small nuclear ribonucleoproteins have shown promise for correcting SMN2 exon 7 splicing (Singh et al., 2017a). Recent work by the Singh laboratory has demonstrated the utility of small-nuclear-ribonucleoprotein based approaches in correcting splicing that cannot be corrected by antisense oligonucleotides and small compounds (Singh and Singh, 2019).
A recent report from the Singh laboratory expands the diversity of alternative pre-mRNA splicing events of SMN genes. They found that SMN genes harbor a surprisingly high content, ∼39%, of the most common transposable elements in the human genome, the Alu elements (Ottesen et al., 2017). Multiple intronic repeats of inverted Alu elements in SMN genes likely influence the formation of secondary structures in their pre-mRNAs favoring the generation of circular RNAs during alternative splicing (Jeck et al., 2013; Liang and Wilusz, 2014; Wilusz, 2015; Zhang et al., 2016). The Singh laboratory confirmed the formation of a vast repertoire of circular RNAs generated by SMN genes during splicing in various human cell lines and tissues (Ottesen et al., 2019). Circular RNAs can affect various aspects of cellular metabolism, including sequestration of RNAs and proteins, and regulation of proteins synthesis and transcription (Barrett and Salzman, 2016). The findings of SMN circular RNAs expand the potential role of SMN genes in cells. Further studies will explore the function of these newly discovered SMN circular RNAs, whether they contribute to SMA pathology, and how they could be exploited for SMA therapeutic and diagnostic applications.
In conclusion, the discovery that alternative splicing is a major contributor to the core phenotypic properties in neurons has inspired investigation of splicing regulators and splicing mis-regulation in disease. Recent publications highlight the value of comprehensive analysis of alternative splicing in the context of neuronal development, plasticity, and pathology, including neuropathic pain, cognitive dysfunction, addiction, and genetic disorders. The emerging functional and regulatory complexity of alternative splicing generates new questions in the field: What are the main determinants that drive splicing factors to recognize RNA or DNA elements in some neurons, but not in others, even when those splicing factors are ubiquitously expressed? Is alternative exon usage influenced by one factor acting alone or in a coregulated fashion with other factors? What are the functional roles of individual splicing isoforms, and does their disruption underlie specific disease phenotypes? Is targeting these regulators a viable option to restore aberrant alternative splicing programs to baseline as a treatment for splicing-linked diseases? Identifying neuron-specific molecular drivers encompassing alternative pre-mRNA splicing and regulators offers new, attractive drug targets for cognitive disorders, addiction, pathological pain, and genetic disorders that are notoriously challenging pharmaceutical targets.
Footnotes
This work was supported by Warren Alpert Fellowship Award to E.J.L.S., Canadian Institutes of Health Research (CIHR) Postdoctoral Fellowship to T.G.-P., European Molecular Biology Organization Postdoctoral Fellowship to T.G.-P., Ontario Institute for Regenerative Medicine (OIRM) Postdoctoral Fellowship to T.G.-P., Charles E. Kaufman Foundation Young Investigator Award to E.A.H., Whitehall Foundation Grant to E.A.H., National Institutes of Health-National Institute on Drug Abuse Avenir Director's Pioneer Award to E.A.H., National Institutes of Health Grants NS055925 and R21NS101312 to Ravindra Singh (Diou Luo is a PhD student in Singh Laboratory), National Institutes of Health Grants R01MH116220 and R01NS104041 to S.Z., and SFARI Bridge to Independence Award to M.J.G. We thank Drs. Benjamin J. Blencowe, Diane Lipscombe, Eric W. Ottesen, Joonbae Seo, Natalia N. Singh, Ravindra N. Singh, and Kristin Webster for providing critical input on the review.
The authors declare no competing financial interests.
References
- Allen SE, Darnell RB, Lipscombe D (2010) The neuronal splicing factor nova controls alternative splicing in N-type and P-type CaV2 calcium channels. Channels 4:483–489. 10.4161/chan.4.6.12868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen SE, Toro CP, Andrade A, López-Soto EJ, Denome S, Lipscombe D (2017) Cell-specific RNA binding protein Rbfox2 regulates CaV2.2 mRNA exon composition and CaV2.2 current size. eNeuro 4:ENEURO.0332–16.2017. 10.1523/ENEURO.0332-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altier C, Dale CS, Kisilevsky AE, Chapman K, Castiglioni AJ, Matthews EA, Evans RM, Dickenson AH, Lipscombe D, Vergnolle N, Zamponi GW (2007) Differential role of N-type calcium channel splice isoforms in pain. J Neurosci 27:6363–6373. 10.1523/JNEUROSCI.0307-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade A, Denome S, Jiang YQ, Marangoudakis S, Lipscombe D (2010) Opioid inhibition of N-type Ca2+ channels and spinal analgesia couple to alternative splicing. Nat Neurosci 13:1249–1256. 10.1038/nn.2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J, Martinelli DC, Malenka RC, Tabuchi K, Südhof TC (2013) Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking. Cell 154:75–88. 10.1016/j.cell.2013.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura N, Kaibuchi K (2007) Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci 8:194–205. 10.1038/nrn2056 [DOI] [PubMed] [Google Scholar]
- Auboeuf D, Hönig A, Berget SM, O'Malley BW (2002) Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298:416–419. 10.1126/science.1073734 [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G (2005) REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121:645–657. 10.1016/j.cell.2005.03.013 [DOI] [PubMed] [Google Scholar]
- Barrett SP, Salzman J (2016) Circular RNAs: analysis, expression and potential functions. Development 143:1838–1847. 10.1242/dev.128074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TJ, Thaler C, Castiglioni AJ, Helton TD, Lipscombe D (2004) Cell-specific alternative splicing increases calcium channel current density in the pain pathway. Neuron 41:127–138. 10.1016/S0896-6273(03)00801-8 [DOI] [PubMed] [Google Scholar]
- Black DL. (1995) Finding splice sites within a wilderness of RNA. RNA 1:763–771. [PMC free article] [PubMed] [Google Scholar]
- Bunda A, LaCarubba B, Akiki M, Andrade A (2019) Tissue- and cell-specific expression of a splice variant in the II-III cytoplasmic loop of Cacna1b. FEBS Open Bio. Advance online publication. Retrieved Jul 17, 2019. doi: 10.1002/2211-5463.12701. 10.1002/2211-5463.12701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarco JA, Superina S, O'Hanlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, Zhen M, Ciruna B, Blencowe BJ (2009) Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell 138:898–910. 10.1016/j.cell.2009.06.012 [DOI] [PubMed] [Google Scholar]
- Castiglioni AJ, Raingo J, Lipscombe D (2006) Alternative splicing in the C-terminus of CaV2.2 controls expression and gating of N-type calcium channels. J Physiol 576:119–134. 10.1113/jphysiol.2006.115030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KO, Hunt CA, Kennedy MB (1992) The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9:929–942. 10.1016/0896-6273(92)90245-9 [DOI] [PubMed] [Google Scholar]
- Dai J, Aoto J, Südhof TC (2019) Alternative splicing of presynaptic neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron 102:993–1008.e5. 10.1016/j.neuron.2019.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH (2016) Advancing the understanding of autism disease mechanisms through genetics. Nat Med 22:345–361. 10.1038/nm.4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Liu S, Tian M, Zhang W, Zhu T, Li D, Wu J, Deng H, Jia Y, Xie W, Xie H, Guan JS (2017) Activity-induced histone modifications govern neurexin-1 mRNA splicing and memory preservation. Nat Neurosci 20:690–699. 10.1038/nn.4536 [DOI] [PubMed] [Google Scholar]
- Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C, Laitman B, Stockman V, LaPlant Q, Cahill ME, Nestler EJ, Shen L (2014) Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol 15:R65. 10.1186/gb-2014-15-4-r65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM (2016) Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet 388:3017–3026. 10.1016/S0140-6736(16)31408-8 [DOI] [PubMed] [Google Scholar]
- Fu XD, Ares M Jr (2014) Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 15:689–701. 10.1038/nrg3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal MJ, Zhang P, Hadjimichael E, Walker RL, Chen C, Liu S, Won H, van Bakel H, Varghese M, Wang Y, Shieh AW, Haney J, Parhami S, Belmont J, Kim M, Moran Losada P, Khan Z, Mleczko J, Xia Y, Dai R, et al. (2018) Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362:eaat8127. 10.1126/science.aat8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonatopoulos-Pournatzis T, Wu M, Braunschweig U, Roth J, Han H, Best AJ, Raj B, Aregger M, O'Hanlon D, Ellis JD, Calarco JA, Moffat J, Gingras AC, Blencowe BJ (2018) Genomewide CRISPR-Cas9 interrogation of splicing networks reveals a mechanism for recognition of autism-misregulated neuronal microexons. Mol Cell 72:510–524.e12. 10.1016/j.molcel.2018.10.008 [DOI] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR (2008) Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82:834–848. 10.1016/j.ajhg.2008.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR (2010) Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 24:1634–1644. 10.1101/gad.1941310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR (2011) Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature 478:123–126. 10.1038/nature10485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Liu YH, Sahashi K, Rigo F, Bennett CF, Krainer AR (2015) Motor neuron cell-nonautonomous rescue of spinal muscular atrophy phenotypes in mild and severe transgenic mouse models. Genes Dev 29:288–297. 10.1101/gad.256644.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Kim EJ, Feng J, Grant GR, Heller EA (2017) Histone posttranslational modifications predict specific alternative exon subtypes in mammalian brain. PLoS Comput Biol 13:e1005602. 10.1371/journal.pcbi.1005602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima T, Wu K, Witte H, Hanno-Iijima Y, Glatter T, Richard S, Scheiffele P (2011) SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell 147:1601–1614. 10.1016/j.cell.2011.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallières M, Tapial J, Raj B, O'Hanlon D, Barrios-Rodiles M, Sternberg MJ, Cordes SP, Roth FP, Wrana JL, Geschwind DH, Blencowe BJ (2014) A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159:1511–1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD (2014) Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol 30:393–415. 10.1146/annurev-cellbio-101011-155831 [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YQ, Andrade A, Lipscombe D (2013) Spinal morphine but not ziconotide or gabapentin analgesia is affected by alternative splicing of voltage-gated calcium channel CaV2.2 pre-mRNA. Mol Pain 9:67. 10.1186/1744-8069-9-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo B, Krainer AR (2009) Splicing therapeutics in SMN2 and APOB. Curr Opin Mol Ther 11:108–115. [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL (2011) Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A 108:13564–13569. 10.1073/pnas.1109475108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzus E. (2017) Rubinstein-Taybi syndrome and epigenetic alterations. Adv Exp Med Biol 978:39–62. 10.1007/978-3-319-53889-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ (2005) Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48:303–314. 10.1016/j.neuron.2005.09.023 [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165. 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28:2233–2247. 10.1101/gad.251926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Sanchez-Pulido L, Haerty W, Ponting CP (2015) RBFOX and PTBP1 proteins regulate the alternative splicing of micro-exons in human brain transcripts. Genome Res 25:1–13. 10.1101/gr.181990.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB (2006) Splicing regulation in neurologic disease. Neuron 52:93–101. 10.1016/j.neuron.2006.09.017 [DOI] [PubMed] [Google Scholar]
- Lim ET, Uddin M, De Rubeis S, Chan Y, Kamumbu AS, Zhang X, D'Gama AM, Kim SN, Hill RS, Goldberg AP, Poultney C, Minshew NJ, Kushima I, Aleksic B, Ozaki N, Parellada M, Arango C, Penzol MJ, Carracedo A, Kolevzon A, et al. (2017) Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat Neurosci 20:1217–1224. 10.1038/nn.4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Andrade A (2015) Calcium channel CaValpha(1) splice isoforms: tissue specificity and drug action. Curr Mol Pharmacol 8:22–31. 10.2174/1874467208666150507103215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscombe D, Andrade A, Allen SE (2013) Alternative splicing: functional diversity among voltage-gated calcium channels and behavioral consequences. Biochim Biophys Acta 1828:1522–1529. 10.1016/j.bbamem.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Szaro BG (2011) hnRNP K post-transcriptionally co-regulates multiple cytoskeletal genes needed for axonogenesis. Development 138:3079–3090. 10.1242/dev.066993 [DOI] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T (2010) Regulation of alternative splicing by histone modifications. Science 327:996–1000. 10.1126/science.1184208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T (2011) Epigenetics in alternative pre-mRNA splicing. Cell 144:16–26. 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macabuag N, Dolphin AC (2015) Alternative splicing in Ca(V)2.2 regulates neuronal trafficking via adaptor protein complex-1 adaptor protein motifs. J Neurosci 35:14636–14652. 10.1523/JNEUROSCI.3034-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslon MM, Braunschweig U, Aitken S, Mann AR, Kilanowski F, Hunter CJ, Blencowe BJ, Kornblihtt AR, Adams IR, Cáceres JF (2019) A slow transcription rate causes embryonic lethality and perturbs kinetic coupling of neuronal genes. EMBO J 38:e101244. 10.15252/embj.2018101244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medana IM, Esiri MM (2003) Axonal damage: a key predictor of outcome in human CNS diseases. Brain 126:515–530. 10.1093/brain/awg061 [DOI] [PubMed] [Google Scholar]
- Nakano Y, Jahan I, Bonde G, Sun X, Hildebrand MS, Engelhardt JF, Smith RJ, Cornell RA, Fritzsch B, Bánfi B (2012) A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx Waltzer mouse. PLoS Genet 8:e1002966. 10.1371/journal.pgen.1002966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Kelly MC, Rehman AU, Boger ET, Morell RJ, Kelley MW, Friedman TB, Bánfi B (2018) Defects in the alternative splicing-dependent regulation of REST cause deafness. Cell 174:536–548.e21. 10.1016/j.cell.2018.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LS, Kim HG, Rosenfeld JA, Shen Y, Gusella JF, Lacassie Y, Layman LC, Shaffer LG, Gécz J (2013) Contribution of copy number variants involving nonsense-mediated mRNA decay pathway genes to neuro-developmental disorders. Hum Mol Genet 22:1816–1825. 10.1093/hmg/ddt035 [DOI] [PubMed] [Google Scholar]
- Ottesen EW, Seo J, Singh NN, Singh RN (2017) A multilayered control of the human survival motor neuron gene expression by Alu elements. Front Microbiol 8:2252. 10.3389/fmicb.2017.02252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EW, Luo D, Seo J, Singh NN, Singh RN (2019) Human survival motor neuron genes generate a vast repertoire of circular RNAs. Nucleic Acids Res 47:2884–2905. 10.1093/nar/gkz034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta LT, Huang J, Lowe JK, Blencowe BJ, Horvath S, Geschwind DH (2016) Genomewide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540:423–427. 10.1038/nature20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parras A, Anta H, Santos-Galindo M, Swarup V, Elorza A, Nieto-Gonzalez JL, Picó S, Hernandez IH, Diaz-Hernandez JI, Belloc E, Rodolosse A, Parikshak NN, Peñagarikano O, Fernández-Chacón R, Irimia M, Navarro P, Geschwind DH, Méndez R, Lucas JJ (2018) Autism-like phenotype and risk gene mRNA deadenylation by CPEB4 mis-splicing. Nature 560:441–446. 10.1038/s41586-018-0423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, Hua Y, Rigo F, Matson J, Hung G, Kaye EM, Shihabuddin LS, Krainer AR, Bennett CF, Cheng SH (2011) Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med 3:72ra18. 10.1126/scitranslmed.3001777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnel-Vallières M, Irimia M, Cordes SP, Blencowe BJ (2015) Essential roles for the splicing regulator nSR100/SRRM4 during nervous system development. Genes Dev 29:746–759. 10.1101/gad.256115.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesnel-Vallières M, Dargaei Z, Irimia M, Gonatopoulos-Pournatzis T, Ip JY, Wu M, Sterne-Weiler T, Nakagawa S, Woodin MA, Blencowe BJ, Cordes SP (2016) Misregulation of an activity-dependent splicing network as a common mechanism underlying autism spectrum disorders. Mol Cell 64:1023–1034. 10.1016/j.molcel.2016.11.033 [DOI] [PubMed] [Google Scholar]
- Quesnel-Vallières M, Weatheritt RJ, Cordes SP, Blencowe BJ (2019) Autism spectrum disorder: insights into convergent mechanisms from transcriptomics. Nat Rev Genet 20:51–63. 10.1038/s41576-018-0066-2 [DOI] [PubMed] [Google Scholar]
- Raingo J, Castiglioni AJ, Lipscombe D (2007) Alternative splicing controls G protein-dependent inhibition of N-type calcium channels in nociceptors. Nat Neurosci 10:285–292. 10.1038/nn1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj B, O'Hanlon D, Vessey JP, Pan Q, Ray D, Buckley NJ, Miller FD, Blencowe BJ (2011) Cross-regulation between an alternative splicing activator and a transcription repressor controls neurogenesis. Mol Cell 43:843–850. 10.1016/j.molcel.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Rao AN, Patil A, Black MM, Craig EM, Myers KA, Yeung HT, Baas PW (2017) Cytoplasmic dynein transports axonal microtubules in a polarity-sorting manner. Cell Rep 19:2210–2219. 10.1016/j.celrep.2017.05.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MC, Grant T, McDaniel MJ, Karakas E, Zhang J, Traynelis SF, Grigorieff N, Furukawa H (2018) Structural mechanism of functional modulation by gene splicing in NMDA receptors. Neuron 98:521–529.e3. 10.1016/j.neuron.2018.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Hua Y, Chun SJ, Prakash TP, Krainer AR, Bennett CF (2012) Synthetic oligonucleotides recruit ILF2/3 to RNA transcripts to modulate splicing. Nat Chem Biol 8:555–561. 10.1038/nchembio.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi F, Paganini L, Braida D, Ponzoni L, Toffolo E, Maroli A, Landsberger N, Bedogni F, Turco E, Pattini L, Altruda F, De Biasi S, Sala M, Battaglioli E (2015) LSD1 neurospecific alternative splicing controls neuronal excitability in mouse models of epilepsy. Cereb Cortex 25:2729–2740. 10.1093/cercor/bhu070 [DOI] [PubMed] [Google Scholar]
- Rusconi F, Grillo B, Ponzoni L, Bassani S, Toffolo E, Paganini L, Mallei A, Braida D, Passafaro M, Popoli M, Sala M, Battaglioli E (2016) LSD1 modulates stress-evoked transcription of immediate early genes and emotional behavior. Proc Natl Acad Sci U S A 113:3651–3656. 10.1073/pnas.1511974113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi F, Grillo B, Toffolo E, Mattevi A, Battaglioli E (2017) NeuroLSD1: splicing-generated epigenetic enhancer of neuroplasticity. Trends Neurosci 40:28–38. 10.1016/j.tins.2016.11.002 [DOI] [PubMed] [Google Scholar]
- Sahashi K, Ling KK, Hua Y, Wilkinson JE, Nomakuchi T, Rigo F, Hung G, Xu D, Jiang YP, Lin RZ, Ko CP, Bennett CF, Krainer AR (2013) Pathological impact of SMN2 mis-splicing in adult SMA mice. EMBO Mol Med 5:1586–1601. 10.1002/emmm.201302567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479:74–79. 10.1038/nature10442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Kandel ER (2016) The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb Perspect Biol 8:a021774. 10.1101/cshperspect.a021774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Singh NN, Androphy EJ, Singh RN (2006) Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol 26:1333–1346. 10.1128/MCB.26.4.1333-1346.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Howell MD, Androphy EJ, Singh RN (2017a) How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy. Gene Ther 24:520–526. 10.1038/gt.2017.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN, Howell MD, Ottesen EW, Singh NN (2017b) Diverse role of survival motor neuron protein. Biochim Biophys Acta Gene Regul Mech 1860:299–315. 10.1016/j.bbagrm.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN, Singh NN (2018) Mechanism of splicing regulation of spinal muscular atrophy genes. Adv Neurobiol 20:31–61. 10.1007/978-3-319-89689-2_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RN, Singh NN (2019) A novel role of U1 snRNP: splice site selection from a distance. Biochim Biophys Acta Gene Regul Mech 1862:634–642. 10.1016/j.bbagrm.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Kim YJ, Nomakuchi T, Sahashi K, Hua Y, Rigo F, Bennett CF, Krainer AR (2018) Antisense oligonucleotides correct the familial dysautonomia splicing defect in IKBKAP transgenic mice. Nucleic Acids Res 46:4833–4844. 10.1093/nar/gky249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. (2017) Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171:745–769. 10.1016/j.cell.2017.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczot M, Pogorzala LA, Solinski HJ, Young L, Yee P, Le Pichon CE, Chesler AT, Hoon MA (2017) Cell-type-specific splicing of Piezo2 regulates mechanotransduction. Cell Rep 21:2760–2771. 10.1016/j.celrep.2017.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalhammer A, Contestabile A, Ermolyuk YS, Ng T, Volynski KE, Soong TW, Goda Y, Cingolani LA (2017) Alternative splicing of P/Q-type Ca(2+) channels shapes presynaptic plasticity. Cell Rep 20:333–343. 10.1016/j.celrep.2017.06.055 [DOI] [PubMed] [Google Scholar]
- Tilot AK, Kucera KS, Vino A, Asher JE, Baron-Cohen S, Fisher SE (2018) Rare variants in axonogenesis genes connect three families with sound-color synesthesia. Proc Natl Acad Sci U S A 115:3168–3173. 10.1073/pnas.1715492115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traunmüller L, Gomez AM, Nguyen TM, Scheiffele P (2016) Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science 352:982–986. 10.1126/science.aaf2397 [DOI] [PubMed] [Google Scholar]
- Ustianenko D, Weyn-Vanhentenryck SM, Zhang C (2017) Microexons: discovery, regulation, and function. Wiley Interdiscip Rev RNA 8:4. 10.1002/wrna.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong CK, Black DL, Zheng S (2016) The neurogenetics of alternative splicing. Nat Rev Neurosci 17:265–281. 10.1038/nrn.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW, Elledge SJ (2008) SCFbeta-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452:370–374. 10.1038/nature06780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. (2015) Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements 5:1–7. 10.1080/2159256X.2015.1006109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth B. (2000) An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat 15:228–237. [DOI] [PubMed] [Google Scholar]
- Xu SJ, Heller EA (2018) Single sample sequencing (S3EQ) of epigenome and transcriptome in nucleus accumbens. J Neurosci Methods 308:62–73. 10.1016/j.jneumeth.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen R, Merico D, Bookman M, Howe JL, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N, Bingham J, Wang Z, Pellecchia G, Buchanan JA, Walker S, Marshall CR, Uddin M, Zarrei M, Deneault E, D'Abate L, Chan AJ, Koyanagi S, et al. (2017) Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci 20:602–611. 10.1038/nn.4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Ergin V, Lin L, Stork C, Chen L, Zheng S (2019) Axonogenesis is coordinated by neuron-specific alternative splicing programming and splicing regulator PTBP2. Neuron 101:690–706.e10. 10.1016/j.neuron.2019.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL (2016) The biogenesis of nascent circular RNAs. Cell Rep 15:611–624. 10.1016/j.celrep.2016.03.058 [DOI] [PubMed] [Google Scholar]
- Zheng S. (2016) Alternative splicing and nonsense-mediated mRNA decay enforce neural specific gene expression. Int J Dev Neurosci 55:102–108. 10.1016/j.ijdevneu.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O'Dell TJ, Black DL (2012) PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci 15:381–388, S1. 10.1038/nn.3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Damoiseaux R, Chen L, Black DL (2013) A broadly applicable high-throughput screening strategy identifies new regulators of Dlg4 (Psd-95) alternative splicing. Genome Res 23:998–1007. 10.1101/gr.147546.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollinger DR, Baalman KL, Rasband MN (2015) The ins and outs of polarized axonal domains. Annu Rev Cell Dev Biol 31:647–667. 10.1146/annurev-cellbio-100913-013107 [DOI] [PubMed] [Google Scholar]