Abstract
Although the involvement of pathological tau in neurodegenerative dementias is indisputable, its physiological roles have remained elusive in part because its abrogation has been reported without overt phenotypes in mice and Drosophila. This was addressed using the recently described Drosophila tauKO and Mi{MIC} mutants and focused on molecular and behavioral analyses. Initially, we show that Drosophila tau (dTau) loss precipitates dynamic cytoskeletal changes in the adult Drosophila CNS and translation upregulation. Significantly, we demonstrate for the first time distinct roles for dTau in adult mushroom body (MB)-dependent neuroplasticity as its downregulation within α′β′neurons impairs habituation. In accord with its negative regulation of translation, dTau loss specifically enhances protein synthesis-dependent long-term memory (PSD-LTM), but not anesthesia-resistant memory. In contrast, elevation of the protein in the MBs yielded premature habituation and depressed PSD-LTM. Therefore, tau loss in Drosophila dynamically alters brain cytoskeletal dynamics and profoundly affects neuronal proteostasis and plasticity.
SIGNIFICANCE STATEMENT We demonstrate that despite modest sequence divergence, the Drosophila tau (dTau) is a true vertebrate tau ortholog as it interacts with the neuronal microtubule and actin cytoskeleton. Novel physiological roles for dTau in regulation of translation, long-term memory, and footshock habituation are also revealed. These emerging insights on tau physiological functions are invaluable for understanding the molecular pathways and processes perturbed in tauopathies.
Keywords: Drosophila, habituation, memory, proteome, tau
Introduction
Tau is a microtubule-associated protein (MAP), a neuronal protein enriched in axons. Through its interaction with tubulin, tau is involved in the regulation of neuronal polarity, axon outgrowth, and axonal transport mediated by kinesin and dynein motor proteins. Apart from its axonal function, several studies indicate alternative cellular functions of tau proteins at the synapse and in the nucleus, as well as interactions with the plasma membrane and the actin cytoskeleton (Sotiropoulos et al., 2017).
Under pathological conditions, tau undergoes post-translational modifications that trigger its pathogenicity (Wang and Mandelkow, 2016). The prevalence of tauopathies and the current lack of prevention or treatment mandate elucidation of the physiological functions of tau, which are requisite to understanding the molecular etiology of its pathogenicity. Transgenic expression of human tau isoforms in Drosophila has contributed to identification of novel tau phosphorylation sites (Nishimura et al., 2004; Papanikolopoulou and Skoulakis, 2011, 2015) and molecular pathways contributing to neuronal dysfunction and toxicity (Shulman and Feany, 2003; Chatterjee et al., 2009). However, the physiological function of the endogenous Drosophila protein has not been fully elucidated.
Drosophila possesses a single tau gene encoding multiple transcripts and potential protein isoforms ostensibly via alternative splicing (http://flybase.org/reports/FBgn0266579). It contains the characteristic conserved tubulin binding repeats with 46% identity and 66% similarity to the corresponding human tau sequences (Heidary and Fortini, 2001). Despite the similarity, the presence of unique and divergent sequences outside the repeats, and the presence of an apparent fifth repeat (Gistelinck et al., 2012) have led to questions as to whether the fly protein functions as a physiological tau ortholog (Chen et al., 2007).
Drosophila tau (dTau) is expressed in the developing and adult CNS, prominently in photoreceptors (Heidary and Fortini, 2001), cell bodies, and neuropils of the visual system and the central brain (Bolkan and Kretzschmar, 2014). Functional regulation by phosphorylation seems conserved in flies, since dTau possesses multiple SKXGS motifs and has been shown to be phosphorylated (Doerflinger et al., 2003; Burnouf et al., 2016). Examination of its physiological functions was attempted with the generation of a knock-out (tauKO) mutant lacking exons 2–6 including the tubulin-binding repeats (Burnouf et al., 2016). However, obvious phenotypes were not reported for these mutants, and apparently this was not a consequence of the upregulation of other microtubule-associated proteins, as in mice (Harada et al., 1994), furthering the notion that dTau may not be an ortholog of the vertebrate protein. This prompted us to determine whether dTau loss affects the neuronal cytoskeleton, as would be expected if functionally conserved with its vertebrate homolog.
Reduced tau characterizes humans with variants of frontotemporal lobar degeneration with granulin (GRN) mutations (Zhukareva et al., 2001, 2003; Mackenzie et al., 2009; Papegaey et al., 2016). In addition, a deletion encompassing the tau gene is linked to mental retardation, although it also removes adjacent genes (Koolen et al., 2006; Shaw-Smith et al., 2006), making assignment of the pathological phenotype to tau loss difficult. Therefore, we also investigated whether dTau loss also precipitates phenotypes in learning and memory as upon human tau expression in the fly CNS (Papanikolopoulou et al., 2010; Papanikolopoulou and Skoulakis, 2015; Sealey et al., 2017).
Materials and Methods
Drosophila culture and strains.
Flies were cultured on standard wheat-flour-sugar food supplemented with soy flour and CaCl2, at 25°C in 50–70% relative humidity in a 12 h light/dark cycle. tauKO mutant flies (Burnouf et al., 2016) were a gift from Dr. L. Partridge (Max Planck Institute for Biology of Aging, Cologne, Germany). The mutant was backcrossed into the resident Cantonised-w1118control isogenic background for six generations. The transposon insertion mutant Mi{MIC} tau[MI03440] was obtained from the Bloomington Stock Center (catalog #BL37602), and Mi{PT-GFSTF.0}tau[MI03440-GFSTF.0] (catalog #BL60199) was a gift from Dr. Carla Sofia Lopes, Universidade do Porto (Porto, Portugal). The Gal4 driver lines used in this work were as follows: elav[C155]-Gal4 (Robinow and White, 1988), LeoMB-Gal4 (Messaritou et al., 2009), dnc-Gal4 (BL48571; Aso et al., 2009), and MB247-Gal4 and c739-Gal4 (Zars et al., 2000). The Gal4 α′/β′ c305a driver was a gift of Dr. S. Waddell (University of Oxford, Oxford, UK). The c739-Gal4;TubGal80ts line was from Dr. G. Roman (University of Mississippi, Oxford, MS). The TubGal80ts transgene (McGuire et al., 2004) was introduced into all other Gal4 strains through standard genetic crosses.
To generate UAS-Flag-dtau a NotI/XbaI fragment containing the RA dtau cDNA (Mershin et al., 2004) was subcloned into pUAST-FLAG vector (Kosmidis et al., 2010). The dtau RNA interference (RNAi) target region was selected to be a 632 bp BglII-BamHI fragment from the entire dtau cDNA to target all tau splice forms. UAS-dTauRNAi was designed as a genomic-cDNA hybrid consisting of the BglII-BamHI fragment cloned into pUAST vector in forward and reverse orientations. Germline transformants were obtained in the Canton S-w1118 genetic background using standard methods. A second dtau RNAi line was obtained from the Bloomington Stock Center (catalog #BDSC-40875).
Proteomic analysis.
Three to four biological and two technical replicas from each genotype (w1118 vs backcrossed tauKO and Elav-Gal4;TubGal80ts >+ vs Elav-Gal4; TubGal80ts >dtauRNAi induced for 3 d at 30°C) were used for this experiment. Briefly, 10 fly brains per genotype were dissected in PBS and, after removal of the optic lobes, were lysed by boiling for 3 min in 50 μl of a solution containing 4% SDS, 100 mm fresh DTT, and 10 mm Tris, pH 7.6. The lysates were processed according to the filter-aided sample preparation (FASP) protocol using spin filter devices with 10 kDa cutoff (catalog # VN01H02, Sartorius; Wis̀niewski et al., 2009). Proteins were subsequently subjected to alkylation and trypsin digestion (1 μg trypsin/ LysC mix mass spectrometry grade; Promega). Peptide products were analyzed by nano-LC-MS/MS (liquid chromatography with tandem mass spectrometry) using a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to a nano-LC high-performance liquid chromatography (RSLCnano, Thermo Fisher Scientific) as described in the study by Terzenidou et al. (2017).
The raw files were analyzed using MaxQuant 1.5.3.30 (Tyanova et al., 2016a) against the complete Uniprot proteome of Drosophila melanogaster (Downloaded 1 April 2016/42,456 entries) and a common contaminants database by the Andromeda search engine. Protein abundance was calculated on the basis of the normalized spectral protein intensity as label-free quantitation (LFQ intensity). The statistical analysis was performed with Perseus (version 1.5.3.2) using a two-sample t test with a false discovery rate (FDR) value of 0.05 (Tyanova et al., 2016b).
Western blot analyses.
Single female fly heads at 1–3 d posteclosion were homogenized in 1× Laemmli buffer (50 mm Tris, pH 6.8, 100 mm DTT, 5% 2-mercaptoethanol, 2% SDS, 10% glycerol, and 0,01% bromophenol blue). Proteins were transferred to PVDF membrane and probed with the following monoclonal antibodies: N2 7A1 Armadillo [Developmental Studies Hybridoma Bank (DSHB)] at 1:200, E7 β-tubulin 97EF (DSHB) at 1:250, ADL84.12 Lamin Dm0 (DSHB) at 1:100, acetyl-α-tubulin (Cell Signaling Technology) at 1:2000, FLAGM2 (Sigma-Aldrich) at 1:1000, and 8C3 syntaxin (DSHB) at 1:3000. Anti-αTub84B+D guinea pig polyclonal antibody (1:300) was a gift from Stefan Baumgartner (Lund University, Sweden) (Fahmy et al., 2014), rabbit polyclonal anti-βTub56D (1:1000) was from Dr. Detlev Buttgereit (Philipps Universität Marburg, Germany) (Buttgereit et al., 1991) and rabbit polyclonal anti-dTau was from Dr. Nick Lowe (Cambridge University, UK) (1:2000). Appropriate HRP-conjugated secondary antibodies were applied at 1:5000. Proteins were visualized with chemiluminescence (ECL Plus, GE Healthcare), and signals were quantified by densitometry with the Image Lab 5.2 program (Bio-Rad). Results were plotted as mean ± SEM values from two or three independent experiments. The data were analyzed by standard parametric statistics (t tests) as indicated in the text.
Confocal microscopy.
The protein-trap Drosophila strain Mi{PT-GFSTF.0}tau[MI03440-GFSTF.0] was used to examine the expression pattern of dTau in the adult brain. Flies were dissected in cold PBS, fixed in 4% paraformaldehyde for 15 min, and imaged by laser-scanning confocal microscopy (TCS SP8, Leica). F-actin levels were determined in adult fly brains from three independent experiments as described previously (Fulga et al., 2007; Kotoula et al., 2017).
F-actin and microtubule precipitation assay.
Total F-actin has been isolated as in (Fulga et al., 2007). Briefly four brains from each genotype (1- to 3-d-old flies) were dissected in cold PBS and transferred in 25 μl of homogenization buffer (100 mm Na2HPO4–NaH2PO4 at pH 7.2, 2 mm ATP, 2 mm MgCl2), supplemented with phosphatase (Sigma-Aldrich) and protease (Thermo Fisher Scientific) inhibitor cocktails. After homogenization, Invitrogen biotinylated phalloidin (Thermo Fisher Scientific) was added to a final concentration of 0.15 units per brain followed by precipitation with streptavidin-coupled Invitrogen Dynabeads (Thermo Fisher Scientific). The precipitated material and supernatant were probed for dTau (1:2000) and actin (1:1000; Sigma-Aldrich). To control for nonspecific binding, the same protocol was followed without the addition of biotinylated phalloidin to the lysate. Microtubule-binding experiments were based on established methods (Feuillette et al., 2010; Gorsky et al., 2017). Briefly, Taxol-stabilized microtubules have been isolated from head extracts by ultracentrifugation at 100,000 × g for 1 h. Soluble and insoluble fractions were probed with E7 and acetyl-α-tubulin.
Puromycin assay.
The protocol was adapted from the studies by Belozerov et al. (2014) and Deliu et al. (2017). Briefly, flies were starved for 4 h and transferred onto 5% sucrose-1% low-melting agarose supplemented with 600 μm puromycin (Santa Cruz Biotechnology) for 16 h. Six female fly heads were homogenized in 20 μl of buffer containing 20 mm Tris-HCl pH 8.0, 137 mm NaCl, 1 mm EDTA, 25% glycerol, 1% NP-40, and a protease inhibitor cocktail (Thermo Fisher Scientific). Samples were run on SDS 10% PAGE gels, proteins were transferred to PVDF membrane and probed sequentially with anti-puromycin (Millipore) at 1:2000 and anti-syntaxin (8C3, DSHB) at 1:3000. Anti-mouse HRP-conjugated secondary antibody was applied at 1:5000. Proteins were visualized with chemiluminescence (ECL Plus, GE Healthcare), and signals corresponding to the molecular weight region 30–125 kDa were quantified by densitometry with Image Lab 5.2 (Bio-Rad). Results were plotted as mean ± SEM values from two independent experiments. The data were analyzed by standard parametric statistics (t tests), as indicated in the text.
Behavioral analyses.
All experiments were performed in mixed sex populations. Animals bearing TubG80ts were raised at 18°C until adulthood, and transgenes were induced maximally by placing 2- to 5-d-old flies at 30°C for 72 h. Flies were kept at the training temperature (25°C) for 30 min before the behavioral assays. Other flies or mutants were raised at 25°C throughout development, adulthood, and behavioral testing. Olfactory memory in the negatively reinforced paradigm coupling aversive odors as conditioned stimuli (CS) with the electric shock unconditioned stimulus (US; Tully and Quinn, 1985) was performed essentially as described previously (Pavlopoulos et al., 2008). The aversive odors used were benzaldehyde and 3-octanol, diluted in isopropylmyristate (Fluka). Electric foot shock avoidance and habituation to electric shock experiments were performed under the conditions described in the study by Acevedo et al. (2007). Each genotype was tested in yoked experiments whereby estimation of the avoidance was immediately followed by habituation training and testing of the same genotype. Hence, all data consist of yoked pairs per genotype. The avoidance fraction was calculated by dividing the number of naive flies preferring shock by the total number of flies. After exposure of the flies to several 1.2 s electric shocks at 45 V, the habituation fraction was calculated by dividing the number of flies preferring shock by the total number of flies. Finally, the habituation index was calculated by subtracting the avoidance fraction from the habituation fraction and multiplying by 100. Cycloheximide treatment has been performed as in the study by Plaçais et al. (2017). Briefly, flies were treated with 35 mm CXM (cycloheximide) in 5% sucrose for 16 h before training at 30°C. After training and until memory test, flies were kept on regular food.
Learning experiments described in Figure 4 were performed as in the study by Cervantes-Sandoval et al. (2016). Briefly, training for 3 min learning experiments consisted of 1 single session of 12 US/CS pairings of 90 V electric shocks (US) with one odor (CS+) over 1 min, followed after a 30 s purge with air, by the presentation of the second odor (CS−) without shocks for 1 min. Training for reversal learning consisted of a standard conditioning protocol (CS+ = OCT, CS− = BNZ; reciprocal, CS+ = BNZ, CS− = OCT) followed by 1 min of air and then the reverse odor–shock contingency (CS+ = BNZ, CS− = OCT; reciprocal, CS+ = OCT, CS− = BNZ). Testing was performed immediately after reversal training.
Figure 4.
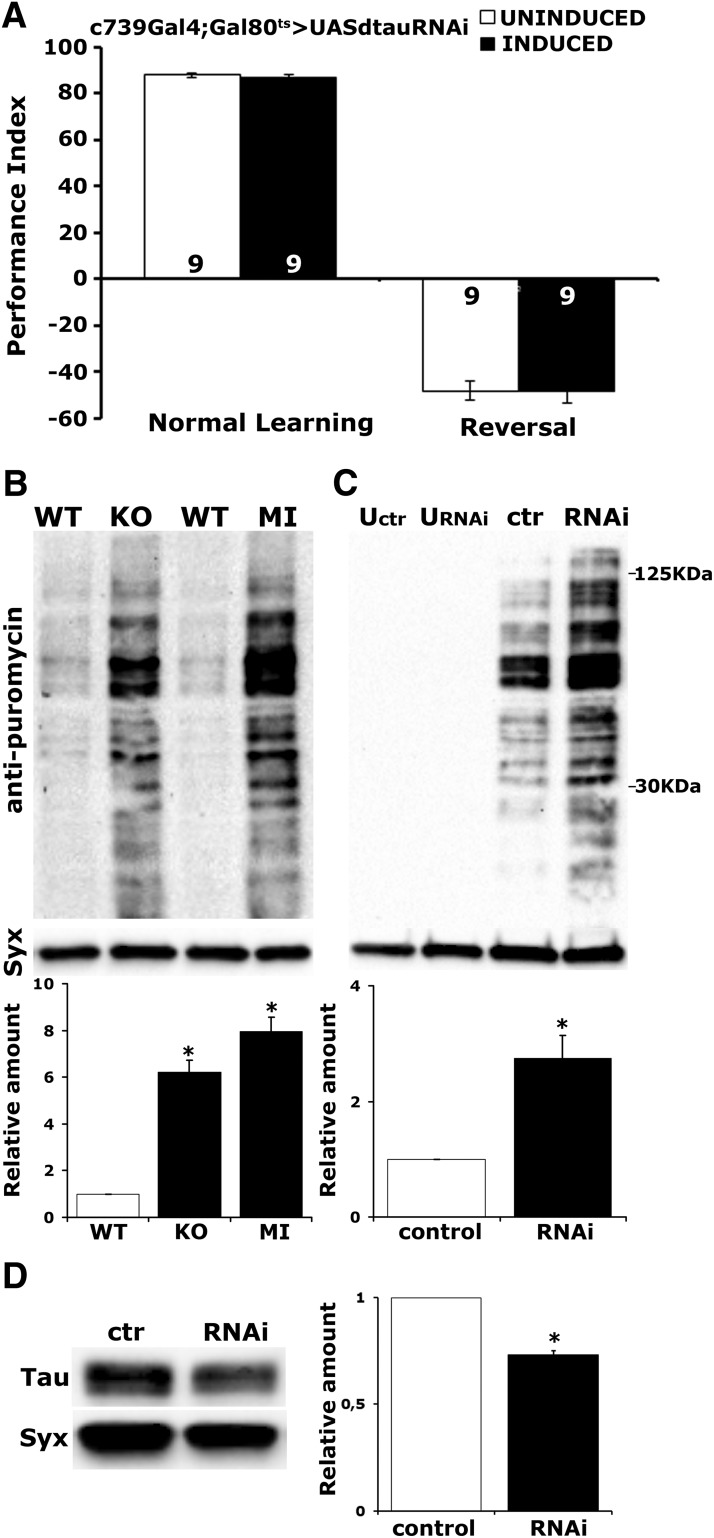
dTau is not required for forgetting of olfactory memories, and its abrogation increases protein synthesis levels. A, Flies expressing dTauRNAi within αβ MB neurons under c739-Gal4;TubGal80ts performed at levels similar to the control group when they were trained with a reversed contingency. Both groups expressed considerable memory to the more recent learning event. No significant difference in 3 min memory was observed between the experimental and control groups when using the typical learning protocol. Animals were raised at 18°C and shifted to 30°C for 3 d, while uninduced animals were kept at 18°C for these 3 d and were used as controls. The number of experimental replicates (n) is indicated within the bars. B, To measure protein synthesis levels flies were treated with 600 μm puromycin for 16 h. Representative blots of head lysates from WT, tauKO, and tauMI flies probed with either anti-puromycin or anti-syntaxin (Syx). For quantifications, levels of the signal corresponding to molecular weight region 30–125 Da in the mutants were normalized using the Syx loading control and are shown as a ratio of their mean ± SEM values relative to their respective level in WT flies, which is arbitrarily set to 1. Stars indicate significant differences (p < 0.0001) from control (open bars) for tauKO and tauMI. C, Representative Western blot of head lysates from flies expressing UAS-dtauRNAi using Elav-Gal4;TubG80ts and probed with anti-puromycin antibody. Animals were raised at 18°C and shifted to 30°C for 3 d, while uninduced animals (U) were kept at 18°C for these 3 d. The genotype of control animals was Elav-Gal4;TubG80ts/+. For the quantification, levels of the signal corresponding to molecular weight region 30–125 kDa were normalized using the Syx loading control and are shown as a ratio of their mean ± SEM values relative to their respective level in control flies, which is arbitrarily set to 1. The star indicates significant differences (p = 0.0044) from control (open bar), indicative of increased protein synthesis upon dTau loss. D, Representative Western blot of head lysates from flies expressing UAS-dtauRNAi with Elav-Gal4;TubG80ts using an anti-dTau antibody. The genotype of control animals was Elav-Gal4;TubG80ts/+ shifted to 30°C for 3 d. Compared with its levels in control animals, dTau was significantly reduced. For the quantification, tau levels were normalized using the Syx loading control and are shown as a ratio of their mean ± SEM values relative to respective levels in control flies, which was set to 1. The star indicates significant differences from the control indicative of reduced dTau levels (*p < 0.0001).
Experimental design and statistical analysis.
For all experiments, controls and experimental genotypes were tested in the same session in balanced design. The order of training and testing these genotypes was randomized. We required an experimental result to be significantly different from both genetic controls. Data are shown as the mean ± SEM. Data were analyzed parametrically with the JMP 7.1 statistical software package (SAS Institute) as described previously (Gouzi et al., 2018). Following initial ANOVA, planned multiple comparisons were performed, using a p value = 0.05. The level of significance was adjusted for the experimentwise error rate. Detailed results of all ANOVA and planned comparisons using the Least Squares Means (LSM) approach are reported in the text.
Results
dTau loss alters microtubule polymerization and stability
Because the Drosophila genome contains only one homolog of the tau/MAP2/MAP4 family (Heidary and Fortini, 2001), and given the involvement of tau in multiple vital functions (Sotiropoulos et al., 2017), it was highly surprising that null mutants of the protein were viable in Drosophila (Burnouf et al., 2016). Potential explanations for this could be that, unlike its apparent vertebrate homologs, dTau is not involved in essential cytoskeletal functions, or that, like in mice, its activities are compensated at least in part by other MAPs (Harada et al., 1994; Bettencourt da Cruz et al., 2005; Barlan et al., 2013). If dTau acted like its vertebrate homolog, then its absence should affect the cytoskeleton and/or the levels of other microtubule-associated proteins, perhaps in compensation for its absence.
To address this hypothesis, we adopted a comparative proteomic approach using LC-MS/MS and label-free quantitation in protein extracts from the brains of null tauKO (Fig. 1A; Burnouf et al., 2016) versus control (WT) flies (Table 1). As expected, dTau was present in the CNS of WT flies and absent in the mutants. In agreement with a previous report (Burnouf et al., 2016), dTau loss did not affect the levels of Futsch and Ensconsin, the known fly homologs of MAP1 and MAP7, respectively, or those of Map205 (Table 1). Since Drosophila does not contain a MAP2 homolog, these are the MAPs that could presumably compensate dTau loss and account for the viability of the mutants as suggested for tau loss in mice (Harada et al., 1994; Bettencourt da Cruz et al., 2005; Barlan et al., 2013). Furthermore, although the centrosomal MAP60 was elevated, previous studies suggested that it cannot functionally replace dTau (Bolkan and Kretzschmar, 2014). The atypical MAP Jupiter (Karpova et al., 2006) was found significantly reduced in tauKO animals. In addition, the spectraplakin Short Stop (Shot), a large actin-microtubule linker molecule, which could in principle functionally overlap tau for microtubule stabilization (Alves-Silva et al., 2012), remained unaltered upon dTau deletion (Table 1). Only the highly divergent Mapmodulin (Goldstein and Gunawardena, 2000) was highly upregulated in the mutant. However, Mapmodulin is leucine rich, unlike the proline rich-dTau, and this, along with its divergent sequence, strongly suggests that it is rather unlikely to functionally compensate for loss of the latter. Hence, there is no obvious upregulation of one of the major Drosophila MAPs under chronic dTau loss (Table 1), similar to that reported to account for the viability and lack of gross mutant phenotypes in tauKO mice (Harada et al., 1994). Therefore, functional compensation of tau loss by MAP upregulation may characterize vertebrates, but divergent molecular mechanisms appear able to overcome the deficit in Drosophila.
Figure 1.
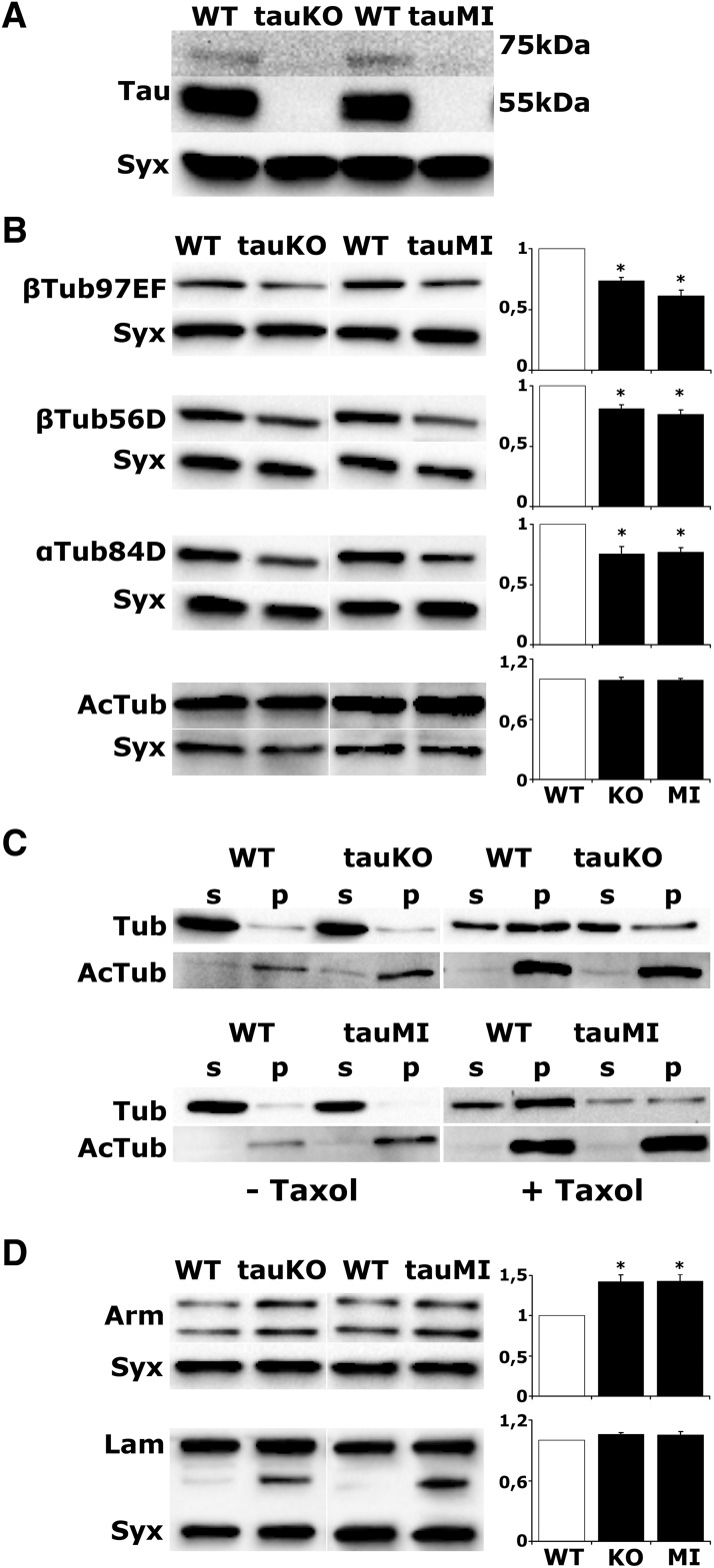
dTau loss precipitates changes in microtubule cytoskeleton. A, Western blot analysis of head lysates from WT, tauKO, and tauMI flies probed with anti-dTau. B, D, Representative blots of head lysates probed with the indicated antibodies. For quantifications, levels of the indicated protein in the mutants were normalized using the syntaxin (Syx) loading control and are shown as a ratio of their mean ± SEM values relative to their respective level in WT flies, which is arbitrarily set to 1. Stars indicate significant differences (p < 0.05) from control (open bars) for tauKO and tauMI. C, Endogenous microtubules were purified from fly head lysates in the absence or presence of Taxol. p, Pellet fraction; s, supernatant fraction. Fractions were analyzed by Western blotting using antibodies against total tubulin (Tub) and acetylated tubulin (AcTub).
Table 1.
Differentially regulated cytoskeletal proteins upon dTau deletion
| Gene | Identifier | Log 2 fold change | p Value |
|---|---|---|---|
| tau | FBgn0266579 | −7.66922013 | 0* |
| futsch | FBgn0259108 | −0.098929167 | 0.23816 |
| Map205 | FBgn0002645 | 0.172052622 | 0.163753 |
| Map60 | FBgn0010342 | 0.447441737 | 0.012972* |
| Jupiter | FBgn0051363 | −0.281016827 | 0.013477* |
| ens | FBgn0264693 | −0.176790555 | 0.411188 |
| Mapmodulin | FBgn0034282 | 0.385273457 | 0.00035* |
| βTub56D | FBgn0003887 | −0.467973868 | 0.000265* |
| βTub97EF | FBgn0003890 | −0,399647633 | 0.004722* |
| αTub84D | FBgn0003885 | −0.209974686 | 0.01011* |
| HDAC6 | FBgn0026428 | −0.393520594 | 0.014513* |
| Dhc64C | FBgn0261797 | 0.173733711 | 0.00421* |
| Dlic | FBgn0030276 | 0.268746932 | 0.001894* |
| BicD | FBgn0000183 | 0.389154911 | 0.013152* |
| Klc | FBgn0010235 | 0.258972645 | 0.008748* |
| Khc | FBgn0001308 | 0.284088135 | 0.000915* |
| Klp10A | FBgn0030268 | 0.30295078 | 0.000371* |
| Mtor | FBgn0013756 | 0.234465 | 0.000624* |
| CLIP-190 | FBgn0020503 | 0.197493394 | 0.001698* |
| shot | FBgn0013733 | 0.026000439 | 0.729461 |
| Actin-5C | FBgn0000042 | 0.101068815 | 0.097274 |
| Arm | FBgn0000117 | 0.287528 | 0.005765* |
| α-Cat | FBgn0010215 | 0.194383144 | 0.008364* |
| sqh | FBgn0003514 | 0.302096923 | 0.011639* |
| sn | FBgn0003447 | 0.478138049 | 0.000003* |
| cpb | FBgn0011570 | 0.188986699 | 0.013467* |
| tsr | FBgn0011726 | 0.160349607 | 0.000611* |
| cpa | FBgn0034577 | 0.211062511 | 0.00578* |
| zip | FBgn0265434 | 0.229537 | 0.018528* |
| vib | FBgn0267975 | −0.180737 | 0.015157* |
| flr | FBgn0260049 | 0.207514842 | 0.000992* |
| Lam | FBgn0002525 | 0.170005878 | 0.016684* |
Selected proteins, p values, and average log2 fold differences from three biological and two technical replicas have been calculated as described in Materials and Methods. The log2 fold change becomes positive when mutant > control and negative when control > mutant. The t test was performed with a permutation-based FDR (0.05) calculation, and the p value determines the statistical significance (*p < 0.05). Proteins are functionally grouped into Drosophila MAP family proteins, tubulin and microtubule-associated proteins, actin, and actin associated proteins.
However, dTau depletion resulted in significant reductions in both α- and β-tubulin and, interestingly, elevation in the major microtubule-associated motor proteins (Table 1). We aimed to validate independently the reduction of tubulins in tauKO and a second Mi{MIC} transposon insertion mutant (tauMI) by Western blots. Both mutants are null as they lack the 50 and 75 kDa dTau isoforms (Fig. 1A), and both harbored reduced levels of both tubulin (Tub) isoforms in head lysates (Fig. 1B; for tauKO and tauMI, respectively, βTub97EF p = 0.004, p = 0.009, n = 4, βTub56D p < 0.0001, n = 6 and n = 8, αTub84D p = 0.0003, n = 5 and n = 7), in agreement with the results in Table 1. Given the lack of obvious phenotypes despite the significant reduction, we wondered whether tubulin attenuation affected its partitioning between monomer and polymer pools.
Neuronal-specific dTau reduction has been reported to affect microtubule morphology and density, resulting in fewer, but larger axonal microtubules (Bolkan and Kretzschmar, 2014). A hallmark of long-lived, stably polymerized microtubules is tubulin acetylation, which is negatively regulated by the tubulin-histone deacetylase HDAC6 (Hubbert et al., 2002). Importantly, HDAC6 levels were reduced in tauKO flies (Table 1), suggesting that tubulin acetylation could be elevated in the mutants. As total tubulin acetylation did not differ among genotypes (Fig. 1B; for tauKO and tauMI, respectively, AcTub p = 0.678, p = 0.875, n = 4), it raised the possibility that it is the nonacetylated tubulin, which is lower in the mutants and is reflected in the reduced α- and β-tubulin isoforms (Fig. 1B).
To validate this notion, we extracted intact microtubules from the same number of Drosophila heads in the presence of the microtubule-stabilizing paclitaxel (Taxol; Feuillette et al., 2010). Microtubules were then sedimented by ultracentrifugation, and the pellet and supernatant fractions were probed for total and acetylated tubulin. In the absence of the microtubule-stabilizing Taxol, polymerized acetylated tubulin appeared more abundant in the mutants. However, the addition of Taxol revealed a significant increase in pelleted tubulin in control, but not the mutant lysates, while the fraction of acetylated tubulin remained equivalent (Fig. 1C). Together, the data indicate that whereas total polymerized tubulin is reduced in the mutants, its acetylation appears unaltered. Because acetylation enhances flexibility and confers resilience against mechanical stresses (Portran et al., 2017), the data suggest that the microtubule lattice in dTau mutants is likely less rigid. Collectively, the results underscore the essential role of dTau in microtubule cytoskeleton dynamics, supporting its functional role as a true ortholog of its vertebrate counterpart.
Loss of dTau destabilizes F-actin
Although significant changes in the ubiquitous actin 5C levels were not detected, dTau loss resulted in elevation of both α-catenin and β-catenin (Armadillo-Arm) and a number of other major actin-binding proteins, as well as the nucleoskeletal protein Lamin (Table 1). These results were selectively verified for Arm and Lamin due to reagent availability (Fig. 1D; for tauKO and tauMI, respectively: Arm, p = 0.007, p = 0.001, n = 5; Lam, p = 0.231, p = 0.817, n = 4). Notably, although the full-length 75 kDa Lamin was present in equal quantities in both mutants and controls, a 45–50 kDa band, representing a cleavage product (Martin and Baehrecke, 2004), was detected only in the tauKO and tauMI flies, which is suggestive of excess Lamin degradation. In accord with these results, transgenic tau elevation by the expression of human tau in the fly CNS promotes F-actin stabilization leading to disruption of the Lamin nucleoskeleton and reduction of Lamin levels in flies (Fulga et al., 2007; Frost et al., 2016).
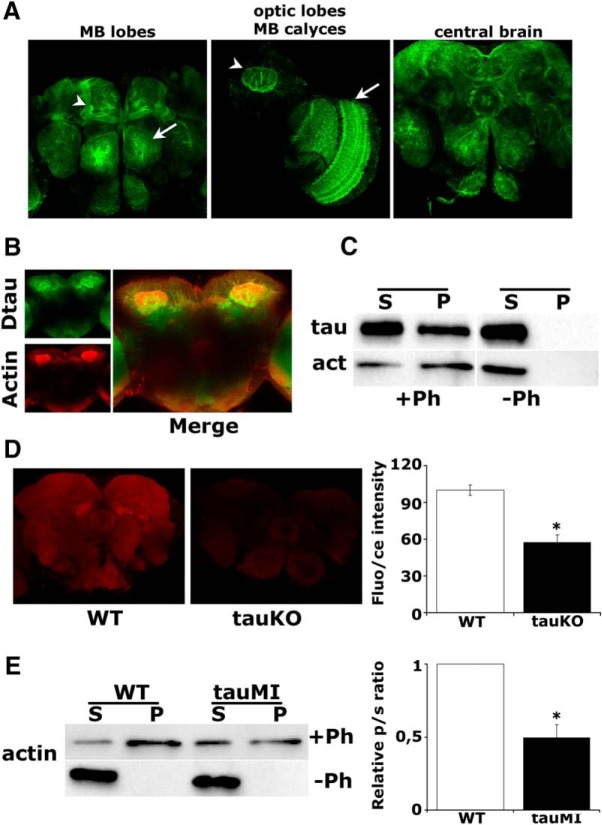
Given the broad upregulation of actin-binding proteins (Table 1) and its interaction with tau (Henríquez et al., 1995; Kempf et al., 1996), we investigated whether the actin cytoskeleton is altered upon dTau loss. We capitalized on a protein-trap fly strain expressing a GFP::dTau fusion protein, which recapitulated the distribution of dTau in the visual system and central brain, including the mushroom bodies (MBs; Fig. 2A). The MBs are bilateral clusters of neurons in the dorsal posterior cortex of the brain, which is essential for olfactory learning and memory in Drosophila (Davis, 2005). Notably, the GFP-marked dTau colocalized with rhodamine-phalloidin within these neurons (Fig. 2B). In accord, total F-actin isolated from brain lysates with biotinylated phalloidin coprecipitated a significant amount of dTau (Fig. 2C). Because dTau was absent from the pellet if phalloidin was omitted, this validates the specificity of dTau interaction with polymerized filamentous actin.
Figure 2.
dTau loss affects the actin cytoskeleton. A, Expression pattern of a GFP::dTau protein-trap in the adult brain at the level of the MB lobes (arrowhead) and antennal lobe (arrow; left), MB calyx (arrowhead) and optic lobe (arrow), (middle), and the central brain (right). B, Prominent colocalization of rhodamine-phalloidin-stained F-actin with the GFP::dTau fusion protein in adult MBs. C, Coprecipitation of phalloidin-bound F-actin and dTau from WT fly brains. D, Confocal images in the central fly brain following rhodamine-phalloidin staining of whole-mount brains from WT and tauKO flies (arrow, ellipsoid body). The mean relative fluorescence intensities ± SEM are shown as a percentage of control. E, Phalloidin-bound F-actin was isolated from fresh brain extracts of WT and tauMI mutants, and its levels were assessed by probing for actin. The ratio of precipitated actin in the pellet (p) to the actin in the supernatant (s) was used for quantification and was significantly different in the mutant, as indicated by the star.
Furthermore, immunofluorescence microscopy of tauKO mutant brains revealed a substantial decrease in total F-actin levels (Fig. 2D; p < 0.0001, n = 10). This was independently verified in tauMI flies, where pelleted, phalloidin-bound F-actin levels were also significantly reduced (p = 0.01, n = 3), while total actin was unaltered (Fig. 2E, Table 1). Collectively therefore, dTau promotes actin polymerization and cytoskeletal dynamics in vivo. It follows that the upregulation of actin-binding proteins upon dTau loss (Table 1) is likely a reflection of homeostatic compensatory responses consequent to decreased F-actin levels (Fig. 2D,E).
dTau is a negative regulator of translation and protein synthesis-dependent long-term memory
Because cytoskeletal proteins are essential for multiple neuronal properties, including plasticity and metabolism (Matamoros and Baas, 2016), in conjunction with its presence within the MBs, neurons essential for learning and memory in the fly, prompted us to investigate whether dTau loss affects behavioral plasticity.
To examine whether dTau is involved in associative learning and memory, we used the olfactory classical conditioning paradigm and examined controls and mutants immediately after training to assess 3 min memory/learning and 24 h later to probe consolidated memories (Fig. 3A,B). Two forms of consolidated memories can be assayed in Drosophila, the protein synthesis-dependent long-term memory (PSD-LTM), induced after multiple rounds of spaced training and the protein synthesis-independent anesthesia-resistant memory (ARM), elicited after repeated massed training cycles (Tully et al., 1994). Whereas 3 min memory was not affected (tauKO, ANOVA: F(2,29) = 0.1721, p = 0.8428; tauMI, ANOVA: F(2,31) = 0.7133, p = 0.4984), both mutants surprisingly presented enhanced LTM (tauKO, ANOVA: F(2,31) = 11.2031, p = 0.0002; subsequent LSM: p = 7.4 × 10−5 vs WT; tauM, ANOVA: F(2,31) = 23.8424, p < 0.0001; subsequent LSM: p = 9.6 × 10−7 vs WT). In contrast, ARM remained at control levels (tauKO, ANOVA: F(2,29) = 1.9692, p = 0.1591; tauMI, ANOVA: F(2,31) = 1.7708, p = 0.1881).
Figure 3.
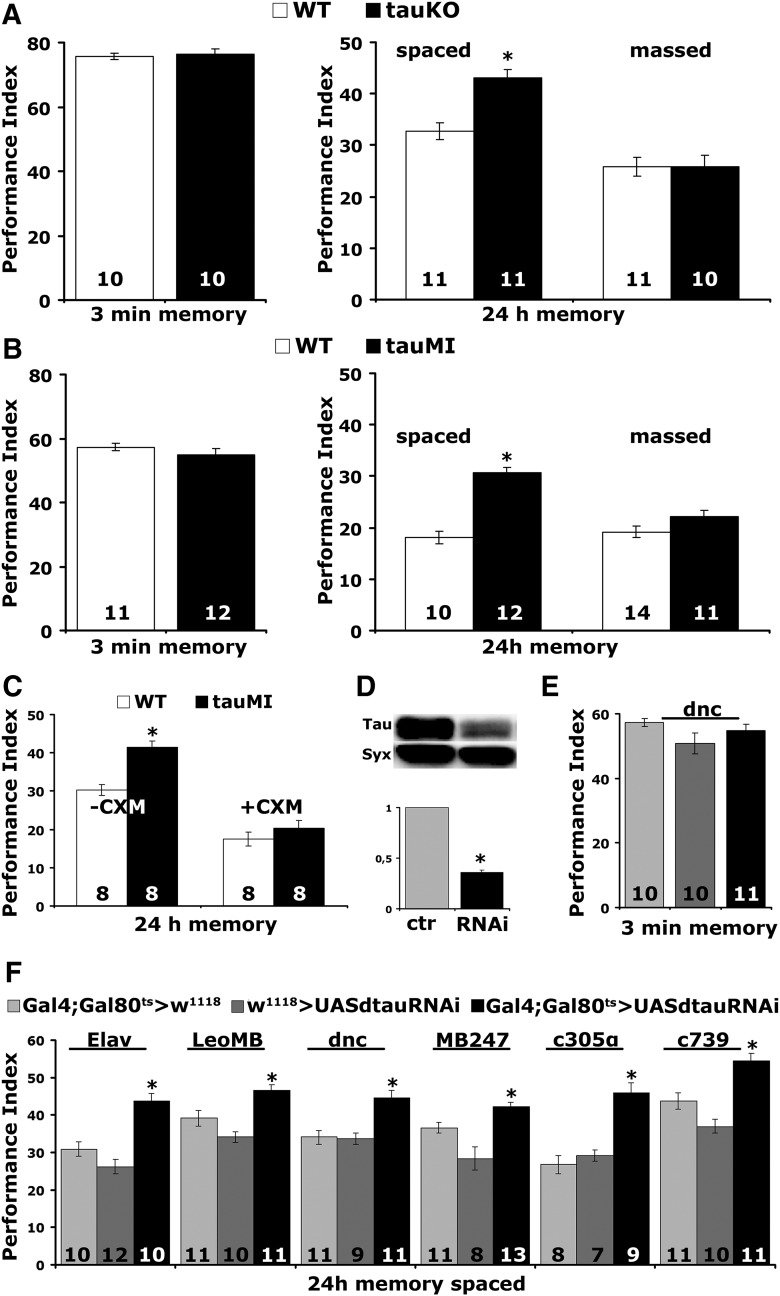
dTau abrogation affects long term memory. A, B, tauKO and tauMI mutants present enhanced LTM (*p < 0.0001), whereas 3 min memory and ARM are not affected (p > 0.05). The number of experimental replicates (n) is indicated within the bars. C, CXM administration eliminated the enhanced LTM of tauMI flies (*p < 0.0001). D, Representative Western blot of head lysates from flies expressing UAS-dtauRNAi with Elav-Gal4 using an anti-dTau antibody. The genotype of control animals was Elav-Gal4/+. Compared with its levels in control animals, dTau was significantly reduced. For the quantification, tau levels were normalized using the syntaxin (Syx) loading control and shown as a ratio of their mean ± SEM values relative to its respective levels in control flies, which was set to 1. The star indicates significant differences from the control indicative of reduced dTau levels (*p < 0.0001). E, Three minute memory is not affected after the downregulation of dTau in the adult MBs using dnc compared with driver and transgene heterozygotes. F, Enhanced LTM performance upon abrogation of dTau during adulthood using Elav (*p < 0.0001), LeoMB (*p = 0.0027), MB247 (*p = 0.0199), dnc (*p = 0.0001), c305α (*p = 0.00001), and c739 (*p = 0.0005).
To determine whether the enhanced 24 h performance is indeed PSD-LTM, we took advantage of its requirement for de novo protein synthesis (Tully et al., 1994) and fed control and mutant flies with the protein synthesis inhibitor CXM. CXM-fed mutants did not present elevated 24 h spaced training-induced memory (ANOVA: F(3,31) = 35.9177, p < 0.0001; subsequent LSM: p = 0.2981, tauMI + CXM vs WT + CXM), but their performance was reduced equally with the expected (Krashes et al., 2009) PSD-LTM reduction of drug-fed controls (Fig. 3C). Therefore, the enhanced memory of the mutants (ANOVA: F(3,31) = 35.9177, p < 0.0001; subsequent LSM: p = 1.6 × 10−4, tauMI-CXM vs WT-CXM) is the PSD-LTM form of consolidated memory.
To confirm these surprising results independently and to determine whether they are a consequence of altered development upon dTau loss, we used RNAi and the TARGET system (McGuire et al., 2004). Adult-specific pan-neuronal RNAi-mediated abrogation of dTau in the CNS reduced its level by ∼65% (Fig. 3D; p < 0.0001, n = 4). As shown in Figure 3E, this did not alter learning, but resulted in strong PSD-LTM enhancement (Fig. 3F; Elav, ANOVA: F(2,31) = 23.2665, p < 0.0001; subsequent LSM: p = 6.4 × 10−5 and p = 2.5 × 10−7 vs controls, respectively). Hence, the elevated memory is not developmental in origin, but reflects an acute requirement for dTau-engaging processes to limit negatively reinforced olfactory PSD-LTM.
Given their essential role for LTM (Davis, 2005), we limited dTau abrogation to adult MBs (Fig. 3F). The LeoMB and dnc-Gal4 are pan-mushroom body drivers (Aso et al., 2009; Messaritou et al., 2009), MB247-Gal4 drives expression mainly in α/β and γ neurons, whereas c305α and c739 are restricted to α′/β′ and α/β, respectively (Aso et al., 2009). To verify that pan-MB dTau attenuation under dnc-Gal4 did not result in enhanced learning, we limited the number of odor/shock pairings from 6 to 3, conditions conducive to revealing such properties (Pavlopoulos et al., 2008; Gouzi et al., 2011). However, enhanced learning was not detectable even under such limited training (Fig. 3E; ANOVA: F(2,30) = 1.9220, p = 0.1651), but, in contrast, PSD-LTM was significantly enhanced (LeoMB, ANOVA: F(2,31) = 14.6273, p < 0.0001; subsequent LSM: p = 0.0027 and p = 9.5 × 10−6 vs controls, respectively; dnc, ANOVA: F(2,30) = 12.8673, p < 0.0001; subsequent LSM: p = 0.0001 and p = 0.0002 vs controls respectively; MB247, ANOVA: F(2,31) = 14.8900, p < 0.0001; subsequent LSM: p = 0.0199 and p = 7.2 × 10−6 vs controls, respectively) under these drivers (Fig. 3F). These results were confirmed with an independent RNAi-mediating transgene (dTau levels for control set to 1, pan-neuronally expressed dtauRNAi40875 = 0.674 ± 0.0497, p < 0.0001, n = 6), which also yielded elevated PSD-LTM (dnc >+ = 23.32 ± 1.51; dtauRNAi40875 >+ = 27.72 ± 1.10; dnc >dtauRNAi40875 = 32.36 ± 1.26; n = 9; ANOVA: F(2,23) = 12.4614, p = 0.0003; subsequent LSM: p = 6.2 × 10−5 and p = 0.0224 vs controls respectively) under dnc-Gal4;TubG80ts.
Significantly, the attenuation of dTau within α′/β′ and α/β MB neurons (Fig. 3F, c305α and c739, respectively), yielded strong PSD-LTM memory improvement (c305α, ANOVA: F(2,23) = 18.8052, p < 0.0001; subsequent LSM: p = 0.00001 and p = 0.00011 vs controls, respectively; c739, ANOVA: F(2,31) = 20.0470, p < 0.0001; subsequent LSM: p = 0.0005 and p = 8 × 10−7 vs controls, respectively). Both subtypes of neurons are known to be essential for 24 h memory with apparently distinct roles in olfactory memory processing (Isabel et al., 2004; Yu et al., 2006; Krashes et al., 2007; Gouzi et al., 2018). Output from the α′/β′ neurons is required for olfactory memory acquisition and stabilization (Krashes et al., 2007), whereas neurotransmission from the α/β neurons is required for its retrieval (Dubnau et al., 2001; McGuire et al., 2001; Pascual and Préat, 2001; Akalal et al., 2006). Collectively, the data strongly indicate that dTau acts as a negative regulator of PSD-LTM within MB neurons in accord with cumulative evidence on the role of these neurons in the process (McGuire et al., 2001; Krashes et al., 2007).
The CXM treatment experiment in Figure 3C suggested that elevated memory upon dTau loss could be due to improved consolidation, whereas the equivalent performance of control and dTauRNAi flies immediately after training with a limited number of US/CS pairings (Fig. 3E) indicated that the increased memory observed at later time points was not due to improved acquisition. However, the enhanced memory observed upon the attenuation of dTau could also arise from decreased forgetting (Cervantes-Sandoval et al., 2016). To test whether forgetting is defective upon dTau attenuation, we performed reversal learning in which we trained flies to associate an aversive odor to footschock and 1 min later to the opposite contingency. Control flies typically avoid the odor most recently associated with shock, whereas flies with decreased forgetting keep the memory of the initial contingency (Cervantes-Sandoval et al., 2016). As shown in Figure 4A, dTau attenuation within αβ MB neurons yielded equal learning as the in-genotype controls both in the typical learning paradigm (learning, ANOVA: F(1,17) = 0.7532, p = 0.3983) and upon reverse training (reversal, ANOVA: F(1,17) = 0.0067, p = 0.9356). The reversal Performance Indexes are negative because the performance was scored as if the initial contingency was the correct choice. This would be expected to be positive if dTau loss impaired forgetting, in essence eliminating the effect of the second contingency in favor of the initial odor shock pairing. Therefore, the increased memory upon dTau loss is unlikely to be due to impaired forgetting.
The proteomic results suggested an elevation of proteins essential for translation in the mutant (Table 2), and this included a number of proteins involved in memory formation or recall (Table 3), in accord with the PSD-LTM dependence on translation. To independently confirm that protein synthesis is in fact elevated upon dTau loss, we used a functional assay, that of puromycin incorporation. Puromycin acts as an aminoacyl-tRNA analog becoming incorporated into nascent peptides causing termination (Nathans, 1964), but also labeling newly synthesized proteins, whose levels are readily measured with an anti-puromycin antibody.
Table 2.
Upregulation of proteins that function in translation upon dTau loss
| Gene | Identifier | Log 2 fold change | p Value |
|---|---|---|---|
| RpL3 | FBgn0020910 | 0.29769754 | 0.015227* |
| RpL4 | FBgn0003279 | 0.28274552 | 0.017456* |
| RpL10 | FBgn0024733 | 0.38811628 | 0.019366* |
| RpL10Ab | FBgn0036213 | 0.41334375 | 0.010486* |
| RpL11 | FBgn0013325 | 0.47628427 | 0.008492* |
| RpL12 | FBgn0034968 | 0.22205798 | 0.008651* |
| RpL13 | FBgn0011272 | 0.42983913 | 0.005514* |
| RpL18A | FBgn0010409 | 0.46355637 | 0.000564* |
| RpL23 | FBgn0010078 | 0.44537226 | 0.022022* |
| RpL26 | FBgn0036825 | 0.33991647 | 0.020167* |
| RpL30 | FBgn0086710 | 0.33723116 | 0.011247* |
| RpLP2 | FBgn0003274 | 0.38862832 | 0.005669* |
| RpS2 | FBgn0004867 | 0.32947826 | 0.000604* |
| RpS3 | FBgn0002622 | 0.33969498 | 0.002684* |
| RpS3A | FBgn0017545 | 0.29286544 | 0.014919* |
| RpS4 | FBgn0011284 | 0.44215218 | 0.000334* |
| RpS6 | FBgn0261592 | 0.35850573 | 0.013199* |
| RpS7 | FBgn0039757 | 0.34377821 | 0.008244* |
| RpS8 | FBgn0039713 | 0.28432178 | 0.007677* |
| RpS10b | FBgn0261593 | 0.30869754 | 0.020561* |
| RpS11 | FBgn0033699 | 0.47895050 | 0.001293* |
| RpS12 | FBgn0260441 | 0.45860616 | 0.005995* |
| RpS13 | FBgn0010265 | 0.35336463 | 0.012484* |
| RpS14b | FBgn0004404 | 0.37592173 | 0.000738* |
| RpS15Aa | FBgn0010198 | 0.3678902 | 0.00019* |
| RpS16 | FBgn0034743 | 0.36565614 | 0.00736* |
| RpS17 | FBgn0005533 | 0.34326967 | 0.011799* |
| RpS18 | FBgn0010411 | 0.34453487 | 0.003018* |
| RpS23 | FBgn0033912 | 0.30110852 | 0.005235* |
| RpS27 | FBgn0039300 | 0.3223815 | 0.010737* |
| Rbp2 | FBgn0262734 | 0.26778308 | 0.002853* |
| sta | FBgn0003517 | 0.32786949 | 0.005807* |
| Ef1beta | FBgn0028737 | 0.23767066 | 0.015910* |
| Elf | FBgn0020443 | 0.31957237 | 0.002937* |
| eEF1delta | FBgn0032198 | 0.20101547 | 0.007045* |
| eIF2gamma | FBgn0263740 | 0.33932988 | 0.009390* |
| eIF-3p66 | FBgn0040227 | 0.36954904 | 0.005558* |
| eIF3-S5–1 | FBgn0037270 | 0.43084621 | 0.005214* |
| eIF3-S8 | FBgn0034258 | 0.38344447 | 0.000346* |
| eIF3-S9 | FBgn0034237 | 0.25910266 | 0.005049* |
| eIF3-S10 | FBgn0037249 | 0.37967841 | 0.001322* |
| eIF4G | FBgn0023213 | 0.31082757 | 0.011195* |
| eRF1 | FBgn0036974 | 0.34124954 | 0.002162* |
| bel | FBgn0263231 | 0.37415822 | 0.000987* |
| bol | FBgn0011206 | 0.36492339 | 0.000232* |
| glo | FBgn0259139 | 0.31756496 | 0.002210* |
| Trip1 | FBgn0015834 | 0.41510550 | 0.000583* |
| Tango7 | FBgn0033902 | 0.37924329 | 0.003916* |
| AGO1 | FBgn0262739 | 0.20476087 | 0.011386* |
| U2af50 | FBgn0005411 | 0.25440500 | 0.017200* |
| Dp1 | FBgn0027835 | 0.27406081 | 0.009374* |
| Fmr1 | FBgn0028734 | 0.32938814 | 0.000069* |
| Hrb27C | FBgn0004838 | 0.29338336 | 0.000018* |
| Hrb98DE | FBgn0001215 | 0.18872627 | 0.002404* |
| kra | FBgn0250753 | 0.36629526 | 0.005815* |
| Not1 | FBgn0085436 | 0.27462665 | 0.002692* |
| Not3 | FBgn0033029 | 0.33431784 | 0.011705* |
| pAbp | FBgn0265297 | 0.32483967 | 0.000818* |
| tyf | FBgn0026083 | 1.42353360 | 0.000399* |
Average log2 fold differences and p values for the indicated proteins calculated from three biological and two technical replicas. As the log2 fold changes denote, all listed proteins were upregulated in the tauKO mutant. The t tests were performed with a permutation-based FDR (0.05) calculation, and the p value determines the statistical significance of the difference (*significant p value <0.05).
Table 3.
Differentially regulated proteins upon dTau deletion that affect memory formation
| Gene | Identifier | Log2 fold change | p Value | References |
|---|---|---|---|---|
| A2bp1 | FBgn0052062 | 0.242381 | 0.01749* | 1 |
| arm | FBgn0000117 | 0.287528 | 0.00576* | 2 |
| cer | FBgn0034443 | 0.352768 | 0.0016* | 3 |
| emb | FBgn0020497 | 0.187307 | 0.005865* | 4 |
| Fmr1 | FBgn0028734 | 0.329388 | 0.00007* | 5 |
| Hop | FBgn0024352 | 0.358213 | 0.0002* | 6 |
| lig | FBgn0020279 | 0.360493 | 0.0066* | 4 |
| Pdk | FBgn0017558 | 0.392657 | 0000001* | 4 |
| Pkc53E | FBgn0003091 | 0.207134 | 0.0022* | 7 |
| CG4612 | FBgn0035016 | 0.332883 | 0.00988* | 8 |
| Ugt | FBgn0014075 | 0.248749 | 0.0097* | 4 |
| Ugt35b | FBgn0026314 | 4.02456 | 0* | 4 |
Selected proteins, p values, and average log2 fold differencess (four biological and three technical replicas) have been calculated as described in Materials and Methods. As the log2 fold change denotes all proteins are upregulated in the tauKO mutant. The t test was performed with a permutation-based FDR (0.05) calculation, and the p value determines the statistical significance (*p value <0.05). References: 1, Guven-Ozkan et al. (2016); 2, Tan et al. (2013); 3, Comas et al. (2004); 4, Walkinshaw et al. (2015); 5, Kanellopoulos et al. (2012); 6, Copf et al. (2011); 7, Colomb and Brembs (2016); and 8, Khan et al. (2015). The two proteins highlighted in gray are family members of a protein identified in 4.
In agreement with the proteomic results, protein synthesis levels were significantly elevated in both mutants relative to control (WT) flies (Fig. 4B; for tauKO and tauMI, respectively, p = 0.00004, p = 0.00002; n = 4). Moreover, qualitatively similar elevation of puromycin incorporation was obtained upon adult-specific pan-neuronal dTau abrogation (Fig. 4C; p = 0.0044, n = 4), suggesting rather acute effects on translation. It is also worth noting that protein synthesis increases in response to temperature elevation as expected [Fig. 4C, uninduced control (lane 1) vs control induced flies (lane 3)]. Given the dependence of PSD-LTM on translation, this protein synthesis upregulation could account, at least in part, for the enhanced memory in the mutants. The collective results in Tables 2 and 3, and Figure 4 support a role for dTau as an acute negative regulator of protein synthesis in the CNS.
Interestingly, acute abrogation of dTau yielded a memory enhancement similar to that in the null mutants. Therefore, we wondered whether the proteomic profiles would be similar or diverge, an indication of compensatory mechanisms dynamics in these situations of acute or chronic dTau attenuation. Therefore, proteomic profiling was performed after acute pan-neuronal dTau attenuation, which, as shown in Figure 4D, leads to 30% reduction of dTau expression levels (p < 0.0001, n = 6).
In accord with the results from chronic dTau reduction in the mutants, proteomic changes were also uncovered upon acute dTau attenuation. Although they represented the three main protein groups, cytoskeletal (Table 1), translation linked (Table 2), and neuronal function linked (Table 3), which were also altered in the mutants, few were in common (Tables 4, 5). For example, although tubulin levels and HDAC6 appeared unaltered upon acute attenuation, nevertheless proteins critical for the dynamics and function of the microtubule cytoskeleton were altered. These include the microtubule tip-localizing protein Eb1, which is critical for accelerating their dynamics (Li et al., 2012); the synaptic microtubule stabilization protein Ank2 (Pielage et al., 2008), which is downregulated upon acute dTau loss; and Arp2, an actin-related protein within the Arp2/3 complex, which is the basic actin nucleator in eukaryotes (Hudson and Cooley, 2002). In support of these results, Ank2 was recently suggested to interact with human tau expressed in Drosophila (Higham et al., 2019).
Table 4.
Differentially regulated proteins involved in cytoskeleton organization and translation upon acute pan-neuronal dTau downregulation
| Identifier | Log 2 fold change | p Value | |
|---|---|---|---|
| 14-3-3ζ | FBgn0004907 | −0.250421 | 0.023824* |
| Ank2 | FBgn0261788 | −0.16070 | 0.040059* |
| Eb1 | FBgn0027066 | −0.269836 | 0.038913* |
| dgt3 | FBgn0034569 | 1.62118 | 0.031084* |
| Klp35D | FBgn0267002 | −0.490059 | 0.016159* |
| Mtor | FBgn0013756 | −0.376771 | 0.040521* |
| mud | FBgn0002873 | 2.61944 | 0.010824* |
| Pka-C1 | FBgn0000273 | −0.229651 | 0.009899* |
| Pka-R1 | FBgn0259243 | −0.217834 | 0.006967* |
| Rab11 | FBgn0015790 | −0.172639 | 0.018478* |
| Rbp | FBgn0262483 | −0.173228 | 0.012758* |
| Strip | FBgn0035437 | −0.457799 | 0.016759* |
| Arp2 | FBgn0011742 | −0.410359 | 0.009826* |
| bt | FBgn0005666 | 0.361831 | 0.034574* |
| btsz | FBgn0266756 | −1.04969 | 0.049232* |
| didum | FBgn0261397 | 1.17976 | 0.001963* |
| mtm | FBgn0025742 | −1.24456 | 0.034627* |
| SelR | FBgn0267376 | −0.198595 | 0.039084* |
| vib | FBgn0267975 | 2.51317 | 0.001071* |
| WASp | FBgn0024273 | −0.415598 | 0.003013* |
| zip | FBgn0265434 | −0.16072 | 0.019419* |
| RpLP2 | FBgn0003274 | −0.22082 | 0.016296* |
| RpL9 | FBgn0015756 | 0.363787 | 0.014304* |
| Ef1β | FBgn0028737 | −0.18861 | 0.025385* |
| eIF-4B | FBgn0020660 | 1.22347 | 0.017551* |
| Elf | FBgn0020443 | 0.783829 | 0.029138* |
| EF2 | FBgn0000559 | −0.36835 | 0.001801* |
| eIF-2α | FBgn0261609 | −0.16968 | 0.027685* |
| LeuRs | FBgn0053123 | −1.95364 | 0.016296* |
| Fmr1 | FBgn0028734 | −0.8741 | 0.028043* |
| Dp1 | FBgn0027835 | −0.19861 | 0.03618* |
| U2af50 | FBgn0005411 | 0.37609 | 0.024622* |
| qkr54B | FBgn0022987 | 0.619797 | 0.049061* |
| nito | FBgn0027548 | −0.99659 | 0.045671* |
| La | FBgn0011638 | 0.505715 | 0.028674* |
| CG4612 | FBgn0035016 | −0.32493 | 0.011309* |
Selected proteins, p values, and average log2 fold differences from four biological and two technical replicas have been calculated as described in Materials and Methods. Control animals are Elav-Gal4;TubG80ts >+ vs Elav-Gal4;TubG80ts > dtauRNAi induced for 3 d at 30°C. The log2 fold change becomes positive when RNAi > control and negative when control > RNAi. The t test was performed with a permutation-based FDR (0.05) calculation, and the p value determines the statistical significance (*p < 0.05). In bold are proteins whose levels were also found to be changed in the mutant (Tables 1, 2).
Table 5.
Differentially regulated proteins upon acute dTau loss that affect memory formation
| Gene | Identifier | Log 2 fold change | p Value | References |
|---|---|---|---|---|
| Fmr1 | FBgn0028734 | −0.874096 | 0.028043* | 1 |
| lig | FBgn0020279 | −1.006 | 0.032011* | 2 |
| 14-3-3zeta | FBgn0004907 | −0.250421 | 0.023824* | 3 |
| Ank2 | FBgn0261788 | −0.1607 | 0.040059* | 4 |
| CG4612 | FBgn0035016 | −0.324925 | 0.011309* | 5 |
| SLC22A | FBgn0037140 | −0.850807 | 0.006731* | 6 |
| Fdh | FBgn0011768 | 0.29288 | 0.007593* | 7 |
| Pka-C1 | FBgn0000273 | −0.229651 | 0.009899* | 8 |
| Pka-R1 | FBgn0259243 | −0.217834 | 0.006967* | 9 |
| Sap47 | FBgn0013334 | −0.802852 | 0.003545* | 10 |
| Syn | FBgn0004575 | −0.150291 | 0.005463* | 11 |
Selected proteins, p value, and average log2 fold differences (four biological and three technical replicas) have been calculated as described in Materials and Methods. Control animals are Elav-Gal4;TubGal80ts >+ vs Elav-Gal4;TubGal80ts >dtauRNAi induced for 3 d at 30°C. The log2 fold change becomes positive when RNAi > control and negative when control > RNAi. The t test was performed with a permutation-based FDR (0.05) calculation, and the p value determines the statistical significance (*p < 0.05). In bold are proteins whose levels were also found to be changed in the mutant (Table 3). References: 1, Kanellopoulos et al. (2012); 2, Walkinshaw et al. (2015); 3, Philip et al. (2001); 4, Iqbal et al. (2013); 5, Khan et al. (2015); 6, Gai et al. (2016); 7, Hou et al. (2011); 8, Horiuchi et al. (2008); 9, Goodwin et al. (1997); 10, Saumweber et al. (2011); and 11, Godenschwege et al. (2004).
Similarly, few of the proteins involved in translation and memory formation such as Fmr1, lig, and CG4612 (Table 5), are shared between chronic and acute dTau attenuation. These similarities, but also the intriguing differences, suggest dynamic proteostatic adjustments of cytoskeletal and translation-linked proteins upon acute dTau loss, which evolve into steady-state long-term compensatory changes to support neuronal structure and function in the mutant. It is interesting that proteins such as 14-3-3ζ and the catalytic and regulatory subunits of protein kinase A, known to be involved in Drosophila learning and memory (Skoulakis and Grammenoudi, 2006), are significantly changed upon acute but not chronic dTau attenuation (Tables 3, 5; Walkinshaw et al., 2015). 14-3-3ζ has recently been reported to interact with human tau expressed pan-neuronally in Drosophila in support of this (Papanikolopoulou et al., 2018). This suggests that the molecular mechanisms underlying PSD-LTM enhancement upon acute and chronic dTau loss are also dynamically proteostatically adjusted, although the net effect may be similar. This is in accord with the notion that PSD-LTM formation or attenuation may result from the engagement of distinct, possibly parallel molecular pathways.
dTau is required for footshock habituation
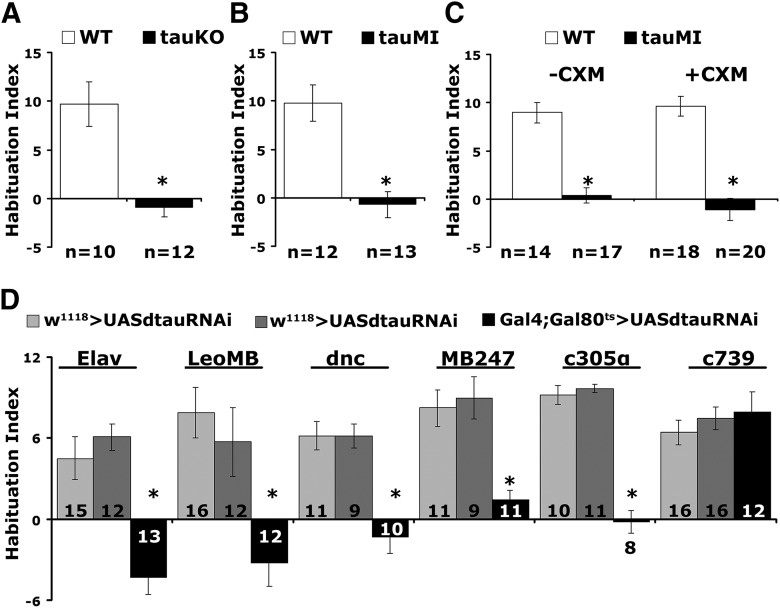
Apart from their established roles in olfactory learning and memory (Heisenberg, 2003), MBs are also involved in habituation to repeated footshocks (Acevedo et al., 2007). Habituation is a form of nonassociative plasticity manifested as a response attenuation to repetitive inconsequential stimuli. To investigate whether dTau is involved in mechanisms underlying habituation, both tauKO and tauMI mutants and their genetic background controls (w1118 and y1w1, respectively) were subjected to the established footshock habituation protocol (Acevedo et al., 2007) of repeated 45 V electric shocks (Fig. 5A,B).
Figure 5.
dTau abrogation impairs habituation. A, B, tauMI and tauKO mutants present strong habituation deficits (*p < 0.0001). The number of experimental replicates (n) is indicated below the graphs. C, CXM administration did not affect the defective habituation of tauMI flies. D, Deficient habituation upon dTau abrogation in adult animals using Elav (*p < 0.0001), LeoMB (*p = 0.0004), MB247 (*p = 0.0004), dnc (*p < 0.0001), and c305α (*p < 0.0001) drivers, but not under c739 (p = 0.3269). The number of experimental replicates (n) is indicated within the bars.
All genotypes avoided electric shock normally when naive (tauKO, ANOVA: F(2,34) = 5.5093, p = 0.0088; tauMI, ANOVA: F(2,40) = 1.8286, p < 0.1745), and the controls presented habituated responses after exposure to 15 such stimuli (Fig. 5A,B), as expected (Acevedo et al., 2007). In contrast, both dTau mutants failed to habituate to 15, 45 V shocks (tauKO, ANOVA: F(2,34) = 58.5474, p < 0.0001; subsequent LSM: p = 3 × 10−10 vs WT, tauMI, ANOVA: F(2,40) = 11.0184, p = 0.0002; subsequent LSM: p = 7.9 × 10−5 vs WT), indicating an inability to devalue inconsequential stimuli. The habituation deficit was not sensitive to CXM in the mutants (Fig. 5C; ANOVA: F(3,68) = 30.4059, p < 0.0001; subsequent LSM: p = 5 × 10−7, tauMI-CXM vs WT-CXM and ANOVA: F(3,68) = 30.4059, p < 0.0001; subsequent LSM: p = 8 × 10−11, tauMI + CXM vs WT + CXM), indicating that failure to habituate is not the result of elevated protein synthesis. Hence, dTau appears to physiologically engage neuronal mechanisms necessary to devalue the repeated footshock stimulation and to facilitate habituation.
To confirm that the habituation defect maps to the adult MBs, we used the same Gal4 drivers as described above to conditionally attenuate dTau via RNAi (Fig. 5D). Habituation deficits were uncovered upon pan-neuronal (Elav, ANOVA: F(2,39) = 18.8294, p < 0.0001; subsequent LSM: p = 1.3 × 10−5 and p = 2.2 × 10−6 vs controls, respectively) and adult MB limited attenuation (LeoMB, ANOVA: F(2,39) = 8.0835, p = 0.0012; subsequent LSM: p = 0.0004 and p = 0.0058 vs controls, respectively; dnc, ANOVA: F(2,29) = 16.7159, p < 0.0001; subsequent LSM: p = 2.2 × 10−5 and p = 4.3 × 10−5 vs controls respectively; MB247, ANOVA: F(2,30) = 11.5623, p = 0.0002; subsequent LSM: p = 0.0004 and p = 0.0002 vs controls, respectively). These results were recapitulated with an independent RNAi-mediating transgene driven pan-MB under dncGal4 (dnc-Gal4;TubG80ts >+ = 12.23 ± 1.34; dtauRNAi40875 >+ = 8.71 ± 1.88; dnc-Gal4;TubG80ts >dtauRNAi40875 = 1.99 ± 1.65; n = 14, ANOVA: F(2,42) = 9.8512, p = 0.0003; subsequent LSM: p = 0.0001 and p = 0.0056 vs controls, respectively).
Interestingly, dTau attenuation restricted to α′β′ neurons precipitated pronounced habituation defects (c305α, ANOVA: F(2,28) = 72.8606, p < 0.0001; subsequent LSM: p = 1.16 × 10−10 and p = 2.63 × 10−11 vs controls, respectively), in agreement with an accompanying report (Roussou et al., 2019). In contrast, habituation was normal if abrogation was limited to c739-Gal4-marked neurons (c739, ANOVA: F(2,43) = 0.5378, p = 0.5881). Collectively, the results indicate a distinct role for dTau specifically within the α′/β′ neurons in molecular mechanisms that facilitate footshock habituation, in addition to its role in limiting PSD-LTM within these and their α/β counterparts.
dTau elevation in adult MBs suppresses memory and results in premature habituation
Because LTM and footshock habituation appear sensitive to dTau levels within the MBs, we hypothesized that their elevation within these neurons may lead to the opposite phenotypes, akin to those observed upon overexpression of human tau (Sealey et al., 2017). In humans, duplication of the tau gene and, hence, presumably elevation of the protein levels, causes prominent neurofibrillary tangle pathology leading to early-onset dementia with an Alzheimer's disease (AD) clinical phenotype (Le Guennec et al., 2016).
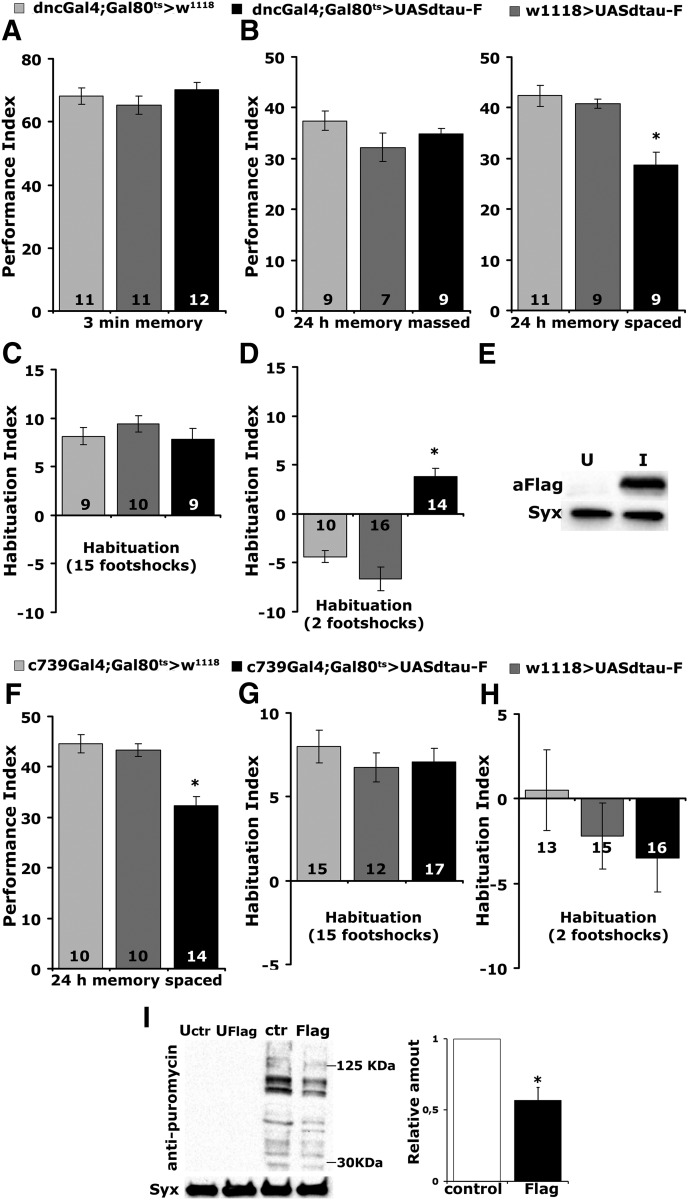
To elevate dTau, a UAS-Flag-dTau transgene was expressed specifically throughout the adult MBs under the pan-mushroom body driver dnc-Gal4;TubG80ts (Fig. 6E). The increase in dTau within the MBs did not affect learning (Fig. 6A; ANOVA: F(2,33) = 0.9706, p = 0.3901) or the protein synthesis-independent ARM (Fig. 6B; massed, ANOVA: F(2,23) = 1.9193, p = 0.1716), but PSD-LTM (Fig. 6B spaced) was deficient (ANOVA: F(2,28) = 13.7758, p < 0.0001; subsequent LSM: p = 4.1 × 10−5 and p = 0.0003 vs controls, respectively). In accord with the hypothesis that elevated translation is, at least in part, responsible for the increased PSD-LTM, adult-specific pan-neuronal accumulation of dTau resulted in an acute translation decrease (Fig. 6I; p = 0.0026, n = 4). Therefore, processes required for PSD-LTM are sensitive to dTau levels in a manner akin to that recently described for dAlk (Gouzi et al., 2018) and in accord with the interpretation that dTau participates in processes that limit LTM formation, storage, or recall.
Figure 6.
dTau elevation within the adult MBs leads to premature habituation and LTM deficits with a concomitant decrease in protein synthesis levels. A, A–D, Adult-specific expression of UAS-Flag dTau within the MBs under dnc-Gal4 results (A), normal 3 min memory (B), normal ARM after massed training but LTM deficits after spaced training (*p < 0.0001), normal habituation following 15-stimulus training (C), and premature habituation (*p < 0.0001) following 2-stimulus training (D). The number of experimental replicates (n) is indicated within the bars. E, Representative Western blot demonstrating accumulation of Flag-dTau in adult MBs using LeoMB-Gal4;TubG80ts of animals raised at 18°C and shifted to 30°C for 2 d (I), while uninduced animals (U) were kept at 18°C for these 2 d. dTau was revealed with an anti-Flag antibody. Syntaxin (Syx) was used as a loading control. F–H, Adult-specific expression of UAS-Flag dTau within αβ neurons under c739 yields significant LTM deficits (*p < 0.0001) compared with controls (F), and results in normal habituation following 15-stimuli (G) or 2-stimuli (H) training. The number of experimental replicates (n) is indicated within the bars. I, Representative Western blot of head lysates from flies expressing UAS-Flag-dTau using Elav-Gal4;TubG80ts and probed with anti-puromycin antibody. Animals were raised at 18°C and shifted to 30°C for 3 d, while uninduced animals (U) were kept at 18°C for these 3 d. The genotype of control animals was Elav-Gal4;Gal80ts/+. Flies were treated with 600 μm puromycin for 16 h. For the quantification, levels of the signal corresponding to molecular weight region 30–125 kDa were normalized using the Syx loading control and are shown as a ratio of their mean ± SEM values relative to their respective level in control flies, which is arbitrarily set to 1. The star indicates significant differences (p = 0.0026) from control (open bar), indicative of decreased protein synthesis upon dTau loss.
Interestingly, dTau elevation did not affect habituation (Fig. 6C) to 15 footshocks (ANOVA: F(2,27) = 0.7958, p = 0.4623), but onset of the habituated response was premature (Fig. 6D) as it occurred after only two stimuli (ANOVA: F(2,39) = 33.1018, p < 0.0001; subsequent LSM: p = 3.2 × 10−6 and p = 1.9 × 10−9 vs controls, respectively). Because silencing neurotransmission from α/β ΜΒ neurons results in premature habituation (Acevedo et al., 2007), we overexpressed dTau specifically in these neurons under c739-Gal4. Although this resulted in deficient (Fig. 6F) PSD-LTM (ANOVA: F(2,31) = 18.0720, p < 0.0001; subsequent LSM: p = 8.9 × 10−6 and p = 4 × 10−5 vs controls, respectively), it yielded normal habituation to 15 footshocks (ANOVA: F(2,43) = 0.5105, p = 0.6040) and did not result in premature habituation after two footshocks (ANOVA: F(2,43) = 0.9059, p = 0.4121; Fig. 6G,H). This suggests that its overaccumulation does not inhibit neurotransmission and that dTau is strongly implicated in a dosage-dependent manner in processes mediating footshock habituation within the MBs.
Discussion
Although our phenotypic search was not exhaustive, our results demonstrate, to our knowledge for the first time, robust mutant phenotypes upon dTau loss. In agreement with prior reports, we also find that both tauKO and tauMI mutants are viable and fertile (data not shown).
Proteostatic changes upon chronic and acute dTau loss
Significant changes in the adult CNS cytoskeletal proteome were uncovered by comparative proteomics and appear to underlie a global proteostatic adjustment to dTau abrogation. We have modeled two scenarios of dTau abrogation, chronic dTau loss as seen in the mutants, and a milder acute attenuation in the adult CNS. Although both situations elicited broad changes with certain proteins altered in common, they yielded differential proteomic signatures (Tables 1–5). Chronic changes appear to have resolved into a proteostatic steady state, presumably to minimize the effects of dTau loss. This is reflected, for example, by compensatory changes in HDAC levels, which stabilize the microtubule cytoskeleton, despite the ostensibly chronic reduction in tubulin (Fig. 1B, Table 1). On the other hand, acute dTau attenuation revealed the initial response of the CNS proteome to the insult, which included the downregulation of many tau interacting proteins (Table 4). We suggest that, with time, this acute proteostatic flux resolves to a steady state reflective of the level of dTau attenuation, and ongoing experiments are addressing this hypothesis.
Interestingly, the steady-state levels of all three tubulins were significantly reduced (Table 1), in accord with the notion that tau is essential for the maintenance of long labile domains of microtubules (Qiang et al., 2018) and may also be reflected in the acute downregulation of proteins such as labile end-organizing protein Eb1 (Li et al., 2012). dTau loss-dependent reduction of labile domain length is in effect reducing microtubule mass and hence the number of tubulins. The relative increase in acetylated tubulin (Fig. 1B,C) is consistent with the reduction in labile domains, which are expected to be underacetylated (Qiang et al., 2018). A potential consequence of the increased microtubule stability is the significant elevation in the subunits of both anterograde and retrograde moving motor proteins (Table 1).
Cytoskeletal homeostasis upon chronic dTau loss also benefits from the reduction in HDAC (Table 1), which may mediate the increase in acetylated tubulin and also account for the proposed reduction in the labile domains. Although how dTau loss affects HDAC levels is unclear at the moment, levels of HDAC are sensitive to tau dosage as HDAC6 is elevated in AD brains and tubulin acetylation is reduced in neurofibrillary tangle-bearing neurons (Hempen and Brion, 1996). Consistent with these observations, increased tubulin acetylation rescues human tau overexpression-induced defects in Drosophila (Xiong et al., 2013; Mao et al., 2017).
The functional role for dTau as a major regulator of cytoskeletal dynamics in vivo is also illustrated by the upregulation of the steady-state levels of actin binding proteins (Table 1), likely in response to the negative effects on actin polymerization upon dTau loss. Our data demonstrate that F-actin interacts directly with and is stabilized by dTau within the fly CNS (Fig. 2B–E). In congruence, perturbation of actin dynamics has also been reported upon pan-neuronal expression of human tau isoforms in Drosophila (Fulga et al., 2007) and may underlie some of the resultant neuropathologies. Therefore, our collective data strongly support the notion that dTau is a true MAP impacting the microtubule and actin cytoskeleton and, despite the sequence diversity, is an apparent ortholog of its vertebrate counterpart.
dTau translation regulation and neuroplasticity
Chronic, but surprisingly also acute, dTau abrogation resulted in the upregulation of translation-linked proteins (Tables 2, 4), strongly indicating that dTau is a negative regulator of translation (Fig. 4B). Congruently, vertebrate tau is a negative regulator of translation (Apicco et al., 2018) and was also recently reported to act as a negative regulator of ribosomal protein levels in mouse brains (Koren et al., 2019). Importantly, the fly brain comparative proteomics provide further validation of these results in a different system, as clearly dTau loss results in the broad elevation of proteins involved in regulation, initiation, and termination of translation, as well as most cellular ribosomal proteins (Tables 2, 4). In addition, tau is known to bind to ribosomes in the brain and to impair their function in reducing protein synthesis (Meier et al., 2016), an effect also observed in human tauopathy brains (Piao et al., 2002). Finally, this agrees with quantitative proteomics in a mouse model of tauopathy, which revealed a decrease in protein synthesis, specifically in neurons with high levels of pathological tau (Evans et al., 2019). This effect was recapitulated by acute dTau overexpression (Fig. 6I), demonstrating the sensitivity of translation to tau levels.
Interestingly, translational upregulation may be partially selective, because under chronic or acute dTau attenuation the levels of most MAPs, tubulins, HDACs, and other abundant proteins were not elevated, but rather reduced (Tables 1, 4). Consistent with this notion, the level of translational regulator dFmr1, whose loss results in LTM deficits (Kanellopoulos et al., 2012), is also elevated (Table 2), suggesting that dTau may be implicated in translational selectivity mechanisms. The elevation of proteins potentially involved in PSD-LTM in mutant brains (Tables 3, 5; Godenschwege et al., 2004; Hou et al., 2011; Iqbal et al., 2013) and probably within MB neurons, likely underlies, at least in part, the enhanced memory, in agreement with its CXM sensitivity (Fig. 3C). The enhanced 24 h memory is not ARM (Fig. 3A,B), or impaired forgetting (Fig. 4A). Furthermore, dTau is required within α/β and a′/β′neurons involved in recall and memory consolidation, respectively (Dubnau et al., 2001; McGuire et al., 2001; Pascual and Préat, 2001; Akalal et al., 2006; Krashes et al., 2007). Collectively then, the enhanced 24 h memory upon dTau abrogation is most likely due to enhanced consolidation of true PSD-LTM.
In contrast to PSD-LTM, the defective footshock habituation upon dTau loss is CXM insensitive, arguing that it is not consequent of excessive protein synthesis (Fig. 5). Is it possible that failure to devalue the electric footshock US results in better acquisition, resulting in better learning that eventually forms enhanced memory? This is unlikely because abrogation of dTau in the MBs did not result in better performance after limited training with three odor/shock pairings (Fig. 3E). Moreover, limiting dTau abrogation to the α′/β′ MB neurons resulted in enhanced PSD-LTM, as well as failure to devalue the shock stimulus, demonstrating that these effects are cell autonomous. Neurotransmission from these neurons is required for olfactory memory acquisition and stabilization (Krashes et al., 2007), but also to facilitate shock habituation (Roussou et al., 2019). Therefore, it is unlikely that dTau is implicated in a common mechanism affecting both processes, because the enhanced PSD-LTM mediated by these neurons (Fig. 3F) suggests increased neurotransmission, although failure to habituate its impairment.
A parsimonious explanation for this paradox would be that dTau functions in distinct mechanisms regulating conditional neurotransmitter traffic and release within these neurons, which is in line with the changes in actin binding proteins, and microtubule motor and associated proteins uncovered by the proteomics (Tables 1, 4). Changes upon dTau abrogation may selectively deregulate neurotransmitter levels and their regulated release, which are known to depend on presynaptic microtubule and cortical actin dynamics (Rust and Maritzen, 2015; Bodaleo and Gonzalez-Billault, 2016). This may selectively enhance neurotransmission upon associative (memory), but not upon nonassociative (habituation) stimulation. This is in line with the observation that dTau abrogation in αβ neurons, where neurotransmission is essential for LTM retrieval (Dubnau et al., 2001; McGuire et al., 2001; Pascual and Préat, 2001; Akalal et al., 2006), also enhances LTM (Fig. 3F). Interestingly, tau-null mice were also recently reported to perform better than WT littermates in a spatial navigation task (Ahmed et al., 2014) and showed enhanced exploration and recognition memory (Jara et al., 2018). Given these collective results, the effects of dTau loss on Drosophila PSD-LTM and the molecular targets offered by our comparative proteomic data, potential physiological roles for dTau in these processes are currently under investigation.
Overexpression of human tau in the adult Drosophila CNS precipitates significant impairment in LTM, but not ARM (Sealey et al., 2017). This effect was recapitulated by adult-specific pan-neuronal or MB limited overexpression of dTau (Fig. 6). Again, relatively acute dTau elevation precipitated the complementary phenotype of premature habituation. Blocked neurotransmission from the MBs results in premature footshock habituation (Acevedo et al., 2007), which is similar to that observed upon dTau overexpression. Therefore, we hypothesize that excess dTau in the MBs might reduce neurotransmitter availability in accord with data from the larval neuromuscular junction (Chee et al., 2005).
In conclusion, it is apparent that dTau contributes in a dosage-dependent manner to a broad number of distinct processes involved in CNS function. Although it is a challenge of future work to understand how dTau can alter neuronal plasticity, the emerging insights into its physiological functions are expected to enhance our understanding of the molecular and cellular pathways perturbed in the various tauopathies.
Footnotes
This research was supported by a grant from the Stavros Niarchos Foundation to the Biomedical Sciences Research Center “Alexander Fleming,” as part of the initiative of the Foundation to support the Greek research center ecosystem. We thank L. Partridge, C.S. Lopes, and the Bloomington Stock Center for fly strains. We also thank S. Baumgartner, D. Buttgereit, N. Lowe, and DSHB for antibodies. Finally, we thank Babis Savakis for comments on the manuscript and helpful suggestions.
The authors declare no competing financial interests.
References
- Acevedo SF, Froudarakis EI, Kanellopoulos A, Skoulakis EM (2007) Protection from premature habituation requires functional mushroom bodies in Drosophila. Learn Mem 14:376–384. 10.1101/lm.566007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed T, Van der Jeugd A, Blum D, Galas MC, D'Hooge R, Buee L, Balschun D (2014) Cognition and hippocampal synaptic plasticity in mice with a homozygous tau deletion. Neurobiol Aging 35:2474–2478. 10.1016/j.neurobiolaging.2014.05.005 [DOI] [PubMed] [Google Scholar]
- Akalal DB, Wilson CF, Zong L, Tanaka NK, Ito K, Davis RL (2006) Roles for Drosophila mushroom body neurons in olfactory learning and memory. Learn Mem 13:659–668. 10.1101/lm.221206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves-Silva J, Sánchez-Soriano N, Beaven R, Klein M, Parkin J, Millard TH, Bellen HJ, Venken KJ, Ballestrem C, Kammerer RA, Prokop A (2012) Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins). J Neurosci 32:9143–9158. 10.1523/JNEUROSCI.0416-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apicco DJ, Ash PEA, Maziuk B, LeBlang C, Medalla M, Al Abdullatif A, Ferragud A, Botelho E, Ballance HI, Dhawan U, Boudeau S, Cruz AL, Kashy D, Wong A, Goldberg LR, Yazdani N, Zhang C, Ung CY, Tripodis Y, Kanaan NM, et al. (2018) Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat Neurosci 21:72–80. 10.1038/s41593-017-0022-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Grübel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H (2009) The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet 23:156–172. 10.1080/01677060802471718 [DOI] [PubMed] [Google Scholar]
- Barlan K, Lu W, Gelfand VI (2013) The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr Biol 23:317–322. 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belozerov VE, Ratkovic S, McNeill H, Hilliker AJ, McDermott JC (2014) In vivo interaction proteomics reveal a novel p38 mitogen-activated protein kinase/Rack1 pathway regulating proteostasis in Drosophila muscle. Mol Cell Biol 34:474–484. 10.1128/MCB.00824-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt da Cruz A, Schwärzel M, Schulze S, Niyyati M, Heisenberg M, Kretzschmar D (2005) Disruption of the MAP1B-related protein FUTSCH leads to changes in the neuronal cytoskeleton, axonal transport defects, and progressive neurodegeneration in Drosophila. Mol Biol Cell 16:2433–2442. 10.1091/mbc.e04-11-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodaleo FJ, Gonzalez-Billault C (2016) The presynaptic microtubule cytoskeleton in physiological and pathological conditions: lessons from Drosophila fragile X syndrome and hereditary spastic paraplegias. Front Mol Neurosci 9:60. 10.3389/fnmol.2016.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolkan BJ, Kretzschmar D (2014) Loss of tau results in defects in photoreceptor development and progressive neuronal degeneration in drosophila. Dev Neurobiol 74:1210–1225. 10.1002/dneu.22199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnouf S, Grönke S, Augustin H, Dols J, Gorsky MK, Werner J, Kerr F, Alic N, Martinez P, Partridge L (2016) Deletion of endogenous tau proteins is not detrimental in drosophila. Sci Rep 6:23102. 10.1038/srep23102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit D, Leiss D, Michiels F, Renkawitz-Pohl R (1991) During Drosophila embryogenesis the beta 1 tubulin gene is specifically expressed in the nervous system and the apodemes. Mech Dev 33:107–118. 10.1016/0925-4773(91)90077-J [DOI] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Chakraborty M, MacMullen C, Davis RL (2016) Scribble scaffolds a signalosome for active forgetting. Neuron 90:1230–1242. 10.1016/j.neuron.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Sang TK, Lawless GM, Jackson GR (2009) Dissociation of tau toxicity and phosphorylation: role of GSK-3beta, MARK and Cdk5 in a Drosophila model. Hum Mol Genet 18:164–177. 10.1093/hmg/ddn326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee FC, Mudher A, Cuttle MF, Newman TA, MacKay D, Lovestone S, Shepherd D (2005) Overexpression of tau results in defective synaptic transmission in Drosophila neuromuscular junctions. Neurobiol Dis 20:918–928. 10.1016/j.nbd.2005.05.029 [DOI] [PubMed] [Google Scholar]
- Chen X, Li Y, Huang J, Cao D, Yang G, Liu W, Lu H, Guo A (2007) Study of tauopathies by comparing Drosophila and human tau in drosophila. Cell Tissue Res 329:169–178. 10.1007/s00441-007-0401-y [DOI] [PubMed] [Google Scholar]
- Colomb J, Brembs B (2016) PKC in motorneurons underlies self-learning, a form of motor learning in Drosophila. Peer J 4:e1971. 10.7717/peerj.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comas D, Petit F, Preat T (2004) Drosophila long-term memory formation involves regulation of cathepsin activity. Nature 430:460–463. 10.1038/nature02726 [DOI] [PubMed] [Google Scholar]
- Copf T, Goguel V, Lampin-Saint-Amaux A, Scaplehorn N, Preat T (2011) Cytokine signaling through the JAK/STAT pathway is required for long-term memory in Drosophila. Proc Natl Acad Sci U S A 108:8059–8064. 10.1073/pnas.1012919108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL. (2005) Olfactory memory formation in drosophila: from molecular to systems neuroscience. Annu Rev Neurosci 28:275–302. 10.1146/annurev.neuro.28.061604.135651 [DOI] [PubMed] [Google Scholar]
- Deliu LP, Ghosh A, Grewal SS (2017) Investigation of protein synthesis in Drosophila larvae using puromycin labelling. Biol Open 6:1229–1234. 10.1242/bio.026294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger H, Benton R, Shulman JM, St Johnston D (2003) The role of PAR-1 in regulating the polarised microtubule cytoskeleton in the Drosophila follicular epithelium. Development 130:3965–3975. 10.1242/dev.00616 [DOI] [PubMed] [Google Scholar]
- Dubnau J, Grady L, Kitamoto T, Tully T (2001) Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature 411:476–480. 10.1038/35078077 [DOI] [PubMed] [Google Scholar]
- Evans HT, Benetatos J, van Roijen M, Bodea LG, Götz J (2019) Decreased synthesis of ribosomal proteins in tauopathy revealed by non-canonical amino acid labelling. EMBO J 38:e101174. 10.15252/embj.2018101174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmy K, Akber M, Cai X, Koul A, Hayder A, Baumgartner S (2014) αTubulin 67C and Ncd are essential for establishing a cortical microtubular network and formation of the Bicoid mRNA gradient in drosophila. PLoS One 9:e112053. 10.1371/journal.pone.0112053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillette S, Miguel L, Frébourg T, Campion D, Lecourtois M (2010) Drosophila models of human tauopathies indicate that tau protein toxicity in vivo is mediated by soluble cytosolic phosphorylated forms of the protein. J Neurochem 113:895–903. 10.1111/j.1471-4159.2010.06663.x [DOI] [PubMed] [Google Scholar]
- Frost B, Bardai FH, Feany MB (2016) Lamin dysfunction mediates neurodegeneration in tauopathies. Curr Biol 26:129–136. 10.1016/j.cub.2015.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Elson-Schwab I, Khurana V, Steinhilb ML, Spires TL, Hyman BT, Feany MB (2007) Abnormal bundling and accumulation of F-actin mediates tau-induced neuronal degeneration in vivo. Nat Cell Biol 9:139–148. 10.1038/ncb1528 [DOI] [PubMed] [Google Scholar]
- Gai Y, Liu Z, Cervantes-Sandoval I, Davis RL (2016) Drosophila SLC22A transporter is a memory suppressor gene that influences cholinergic neurotransmission to the mushroom bodies. Neuron 90:581–595. 10.1016/j.neuron.2016.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gistelinck M, Lambert JC, Callaerts P, Dermaut B, Dourlen P (2012) Drosophila models of tauopathies: what have we learned? Int J Alzheimers Dis 2012:970980. 10.1155/2012/970980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godenschwege TA, Reisch D, Diegelmann S, Eberle K, Funk N, Heisenberg M, Hoppe V, Hoppe J, Klagges BR, Martin JR, Nikitina EA, Putz G, Reifegerste R, Reisch N, Rister J, Schaupp M, Scholz H, Schwärzel M, Werner U, Zars TD, et al. (2004) Flies lacking all synapsins are unexpectedly healthy but are impaired in complex behaviour. Eur J Neurosci 20:611–622. 10.1111/j.1460-9568.2004.03527.x [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Gunawardena S (2000) Flying through the Drosophila cytoskeletal genome. J Cell Biol 150:F63–F68. 10.1083/jcb.150.2.F63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SF, Del Vecchio M, Velinzon K, Hogel C, Russell SR, Tully T, Kaiser K (1997) Defective learning in mutants of the Drosophila gene for a regulatory subunit of cAMP-dependent protein kinase. J Neurosci 17:8817–8827. 10.1523/JNEUROSCI.17-22-08817.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsky MK, Burnouf S, Sofola-Adesakin O, Dols J, Augustin H, Weigelt CM, Grönke S, Partridge L (2017) Pseudo-acetylation of multiple sites on human tau proteins alters tau phosphorylation and microtubule binding, and ameliorates amyloid beta toxicity. Sci Rep 7:9984. 10.1038/s41598-017-10225-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzi JY, Moressis A, Walker JA, Apostolopoulou AA, Palmer RH, Bernards A, Skoulakis EM (2011) The receptor tyrosine kinase Alk controls neurofibromin functions in Drosophila growth and learning. PLoS Genet 7:e1002281. 10.1371/journal.pgen.1002281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouzi JY, Bouraimi M, Roussou IG, Moressis A, Skoulakis EMC (2018) The Drosophila receptor tyrosine kinase alk constrains long-term memory formation. J Neurosci 38:7701–7712. 10.1523/JNEUROSCI.0784-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Busto GU, Schutte SS, Cervantes-Sandoval I, O'Dowd DK, Davis RL (2016) MiR-980 is a memory suppressor MicroRNA that regulates the autism-susceptibility gene A2bp1. Cell Rep 14:1698–1709. 10.1016/j.celrep.2016.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Oguchi K, Okabe S, Kuno J, Terada S, Ohshima T, Sato-Yoshitake R, Takei Y, Noda T, Hirokawa N (1994) Altered microtubule organization in small-calibre axons of mice lacking tau protein. Nature 369:488–491. 10.1038/369488a0 [DOI] [PubMed] [Google Scholar]
- Heidary G, Fortini ME (2001) Identification and characterization of the Drosophila tau homolog. Mech Dev 108:171–178. 10.1016/S0925-4773(01)00487-7 [DOI] [PubMed] [Google Scholar]
- Heisenberg M. (2003) Mushroom body memoir: from maps to models. Nat Rev Neurosci 4:266–275. 10.1038/nrn1074 [DOI] [PubMed] [Google Scholar]
- Hempen B, Brion JP (1996) Reduction of acetylated alpha-tubulin immunoreactivity in neurofibrillary tangle-bearing neurons in Alzheimer's disease. J Neuropathol Exp Neurol 55:964–972. 10.1097/00005072-199609000-00003 [DOI] [PubMed] [Google Scholar]
- Henríquez JP, Cross D, Vial C, Maccioni RB (1995) Subpopulations of tau interact with microtubules and actin filaments in various cell types. Cell Biochem Funct 13:239–250. 10.1002/cbf.290130404 [DOI] [PubMed] [Google Scholar]
- Higham JP, Malik BR, Buhl E, Dawson JM, Ogier AS, Lunnon K, Hodge JJL (2019) Alzheimer's disease associated genes ankyrin and tau cause shortened lifespan and memory loss in drosophila. Front Cell Neurosci 13:260. 10.3389/fncel.2019.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Yamazaki D, Naganos S, Aigaki T, Saitoe M (2008) Protein kinase A inhibits a consolidated form of memory in drosophila. Proc Natl Acad Sci U S A 105:20976–20981. 10.1073/pnas.0810119105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Q, Jiang H, Zhang X, Guo C, Huang B, Wang P, Wang T, Wu K, Li J, Gong Z, Du L, Liu Y, Liu L, Chen C (2011) Nitric oxide metabolism controlled by formaldehyde dehydrogenase (fdh, homolog of mammalian GSNOR) plays a crucial role in visual pattern memory in drosophila. Nitric Oxide 24:17–24. 10.1016/j.niox.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458. 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- Hudson AM, Cooley L (2002) A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J Cell Biol 156:677–687. 10.1083/jcb.200109065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z, Vandeweyer G, van der Voet M, Waryah AM, Zahoor MY, Besseling JA, Roca LT, Vulto-van Silfhout AT, Nijhof B, Kramer JM, Van der Aa N, Ansar M, Peeters H, Helsmoortel C, Gilissen C, Vissers LE, Veltman JA, de Brouwer AP, Frank Kooy R, Riazuddin S, et al. (2013) Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet 22:1960–1970. 10.1093/hmg/ddt043 [DOI] [PubMed] [Google Scholar]
- Isabel G, Pascual A, Preat T (2004) Exclusive consolidated memory phases in drosophila. Science 304:1024–1027. 10.1126/science.1094932 [DOI] [PubMed] [Google Scholar]
- Jara C, Aránguiz A, Cerpa W, Tapia-Rojas C, Quintanilla RA (2018) Genetic ablation of tau improves mitochondrial function and cognitive abilities in the hippocampus. Redox Biol 18:279–294. 10.1016/j.redox.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulos AK, Semelidou O, Kotini AG, Anezaki M, Skoulakis EM (2012) Learning and memory deficits consequent to reduction of the fragile X mental retardation protein result from metabotropic glutamate receptor-mediated inhibition of cAMP signaling in Drosophila. J Neurosci 32:13111–13124. 10.1523/JNEUROSCI.1347-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova N, Bobinnec Y, Fouix S, Huitorel P, Debec A (2006) Jupiter, a new Drosophila protein associated with microtubules. Cell Motil Cytoskeleton 63:301–312. 10.1002/cm.20124 [DOI] [PubMed] [Google Scholar]
- Kempf M, Clement A, Faissner A, Lee G, Brandt R (1996) Tau binds to the distal axon early in development of polarity in a microtubule- and microfilament-dependent manner. J Neurosci 16:5583–5592. 10.1523/JNEUROSCI.16-18-05583.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MR, Li L, Pérez-Sánchez C, Saraf A, Florens L, Slaughter BD, Unruh JR, Si K (2015) Amyloidogenic oligomerization transforms Drosophila Orb2 from a translation repressor to an activator. Cell 163:1468–1483. 10.1016/j.cell.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, Kooy RF, Reyniers E, Romano C, Fichera M, Schinzel A, Baumer A, Anderlid BM, Schoumans J, Knoers NV, van Kessel AG, Sistermans EA, Veltman JA, Brunner HG, de Vries BB (2006) A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet 38:999–1001. 10.1038/ng1853 [DOI] [PubMed] [Google Scholar]
- Koren SA, Hamm MJ, Meier SE, Weiss BE, Nation GK, Chishti EA, Arango JP, Chen J, Zhu H, Blalock EM, Abisambra JF (2019) Tau drives translational selectivity by interacting with ribosomal proteins. Acta Neuropathol 137:571–583. 10.1007/s00401-019-01970-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidis S, Grammenoudi S, Papanikolopoulou K, Skoulakis EM (2010) Differential effects of tau on the integrity and function of neurons essential for learning in Drosophila. J Neurosci 30:464–477. 10.1523/JNEUROSCI.1490-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoula V, Moressis A, Semelidou O, Skoulakis EMC (2017) Drk-mediated signaling to rho kinase is required for anesthesia-resistant memory in Drosophila. Proc Natl Acad Sci U S A 114:10984–10989. 10.1073/pnas.1704835114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S (2007) Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron 53:103–115. 10.1016/j.neuron.2006.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S (2009) A neural circuit mechanism integrating motivational state with memory expression in drosophila. Cell 139:416–427. 10.1016/j.cell.2009.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guennec K, Quenez O, Nicolas G, Wallon D, Rousseau S, Richard AC, Alexander J, Paschou P, Charbonnier C, Bellenguez C, Grenier-Boley B, Lechner D, Bihoreau MT, Olaso R, Boland A, Meyer V, Deleuze JF, Amouyel P, Munter HM, Bourque G, et al. (2016) 17q21.31 duplication causes prominent tau-related dementia with increased MAPT expression. Mol Psychiatry 22:1119–1125. 10.1038/mp.2016.226 [DOI] [PubMed] [Google Scholar]
- Li W, Moriwaki T, Tani T, Watanabe T, Kaibuchi K, Goshima G (2012) Reconstitution of dynamic microtubules with Drosophila XMAP215, EB1, and Sentin. J Cell Biol 199:849–862. 10.1083/jcb.201206101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, Ince PG, Kamphorst W, Revesz T, Rozemuller AJ, Kumar-Singh S, Akiyama H, Baborie A, Spina S, Dickson DW, Trojanowski JQ, et al. (2009) Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol 117:15–18. 10.1007/s00401-008-0460-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CX, Wen X, Jin S, Zhang YQ (2017) Increased acetylation of microtubules rescues human tau-induced microtubule defects and neuromuscular junction abnormalities in drosophila. Dis Model Mech 10:1245–1252. 10.1242/dmm.028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Baehrecke EH (2004) Caspases function in autophagic programmed cell death in Drosophila. Development 131:275–284. 10.1242/dev.00933 [DOI] [PubMed] [Google Scholar]
- Matamoros AJ, Baas PW (2016) Microtubules in health and degenerative disease of the nervous system. Brain Res Bull 126:217–225. 10.1016/j.brainresbull.2016.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Davis RL (2001) The role of Drosophila mushroom body signaling in olfactory memory. Science 293:1330–1333. 10.1126/science.1062622 [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL (2004) Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004:pl6. 10.1126/stke.2202004pl6 [DOI] [PubMed] [Google Scholar]
- Meier S, Bell M, Lyons DN, Rodriguez-Rivera J, Ingram A, Fontaine SN, Mechas E, Chen J, Wolozin B, LeVine H 3rd, Zhu H, Abisambra JF (2016) Pathological tau promotes neuronal damage by impairing ribosomal function and decreasing protein synthesis. J Neurosci 36:1001–1007. 10.1523/JNEUROSCI.3029-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mershin A, Pavlopoulos E, Fitch O, Braden BC, Nanopoulos DV, Skoulakis EM (2004) Learning and memory deficits upon TAU accumulation in Drosophila mushroom body neurons. Learn Mem 11:277–287. 10.1101/lm.70804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaritou G, Leptourgidou F, Franco M, Skoulakis EM (2009) A third functional isoform enriched in mushroom body neurons is encoded by the Drosophila 14-3-3zeta gene. FEBS Lett 583:2934–2938. 10.1016/j.febslet.2009.08.003 [DOI] [PubMed] [Google Scholar]
- Nathans D. (1964) Puromycin inhibition of protein synthesis: incorporation of puromycin into peptide chains. Proc Natl Acad Sci U S A 51:585–592. 10.1073/pnas.51.4.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura I, Yang Y, Lu B (2004) PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in drosophila. Cell 116:671–682. 10.1016/S0092-8674(04)00170-9 [DOI] [PubMed] [Google Scholar]
- Papanikolopoulou K, Skoulakis EM (2011) The power and richness of modelling tauopathies in drosophila. Mol Neurobiol 44:122–133. 10.1007/s12035-011-8193-1 [DOI] [PubMed] [Google Scholar]
- Papanikolopoulou K, Skoulakis EM (2015) Temporally distinct phosphorylations differentiate tau-dependent learning deficits and premature mortality in drosophila. Hum Mol Gen 24:2065–2077. 10.1093/hmg/ddu726 [DOI] [PubMed] [Google Scholar]
- Papanikolopoulou K, Kosmidis S, Grammenoudi S, Skoulakis EM (2010) Phosphorylation differentiates tau-dependent neuronal toxicity and dysfunction. Biochem Soc Trans 38:981–987. 10.1042/BST0380981 [DOI] [PubMed] [Google Scholar]
- Papanikolopoulou K, Grammenoudi S, Samiotaki M, Skoulakis EMC (2018) Differential effects of 14-3-3 dimers on tau phosphorylation, stability and toxicity in vivo. Hum Mol Gen 27:2244–2261. 10.1093/hmg/ddy129 [DOI] [PubMed] [Google Scholar]
- Papegaey A, Eddarkaoui S, Deramecourt V, Fernandez-Gomez FJ, Pantano P, Obriot H, Machala C, Anquetil V, Camuzat A, Brice A, Maurage CA, Le Ber I, Duyckaerts C, Buée L, Sergeant N, Buée-Scherrer V (2016) Reduced tau protein expression is associated with frontotemporal degeneration with progranulin mutation. Acta Neuropathol Commun 4:74. 10.1186/s40478-016-0345-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual A, Préat T (2001) Localization of long-term memory within the Drosophila mushroom body. Science 294:1115–1117. 10.1126/science.1064200 [DOI] [PubMed] [Google Scholar]
- Pavlopoulos E, Anezaki M, Skoulakis EM (2008) Neuralized is expressed in the alpha/beta lobes of adult Drosophila mushroom bodies and facilitates olfactory long-term memory formation. Proc Natl Acad Sci U S A 105:14674–14679. 10.1073/pnas.0801605105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip N, Acevedo SF, Skoulakis EM (2001) Conditional rescue of olfactory learning and memory defects in mutants of the 14-3-3z gene leonardo. J Neurosci 21:8417–8425. 10.1523/JNEUROSCI.21-21-08417.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao YS, Hayashi S, Wakabayashi K, Kakita A, Aida I, Yamada M, Takahashi H (2002) Cerebellar cortical tau pathology in progressive supranuclear palsy and corticobasal degeneration. Acta Neuropathol 103:469–474. 10.1007/s00401-001-0488-2 [DOI] [PubMed] [Google Scholar]
- Pielage J, Cheng L, Fetter RD, Carlton PM, Sedat JW, Davis GW (2008) A presynaptic giant ankyrin stabilizes the NMJ through regulation of presynaptic microtubules and transsynaptic cell adhesion. Neuron 58:195–209. 10.1016/j.neuron.2008.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaçais PY, de Tredern É, Scheunemann L, Trannoy S, Goguel V, Han KA, Isabel G, Preat T (2017) Upregulated energy metabolism in the Drosophila mushroom body is the trigger for long-term memory. Nat Commun 8:15510. 10.1038/ncomms15510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D, Schaedel L, Xu Z, Théry M, Nachury MV (2017) Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nat Cell Biol 19:391–398. 10.1038/ncb3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Sun X, Austin T, Muralidharan H, Jean DC, Liu M, Yu W, Baas PW (2018) Tau does not stabilize axonal microtubules but rather enables them to have long labile domains. Curr Biol 28:2181–2189.e4. 10.1016/j.cub.2018.05.045 [DOI] [PubMed] [Google Scholar]
- Robinow S, White K (1988) The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol 126:294–303. 10.1016/0012-1606(88)90139-X [DOI] [PubMed] [Google Scholar]
- Roussou IG, Papanikolopoulou K, Savakis C, Skoulakis E (2019) Drosophila Bruton's Tyrosine Kinase regulates habituation latency and facilitation in distinct mushroom body neurons. J Neurosci, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MB, Maritzen T (2015) Relevance of presynaptic actin dynamics for synapse function and mouse behavior. Exp Cell Res 335:165–171. 10.1016/j.yexcr.2014.12.020 [DOI] [PubMed] [Google Scholar]
- Saumweber T, Weyhersmüller A, Hallermann S, Diegelmann S, Michels B, Bucher D, Funk N, Reisch D, Krohne G, Wegener S, Buchner E, Gerber B (2011) Behavioral and synaptic plasticity are impaired upon lack of the synaptic protein SAP47. J Neurosci 31:3508–3518. 10.1523/JNEUROSCI.2646-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sealey MA, Vourkou E, Cowan CM, Bossing T, Quraishe S, Grammenoudi S, Skoulakis EMC, Mudher A (2017) Distinct phenotypes of three-repeat and four-repeat human tau in a transgenic model of tauopathy. Neurobiol Dis 105:74–83. 10.1016/j.nbd.2017.05.003 [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Pittman AM, Willatt L, Martin H, Rickman L, Gribble S, Curley R, Cumming S, Dunn C, Kalaitzopoulos D, Porter K, Prigmore E, Krepischi-Santos AC, Varela MC, Koiffmann CP, Lees AJ, Rosenberg C, Firth HV, de Silva R, Carter NP (2006) Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat Genet 38:1032–1037. 10.1038/ng1858 [DOI] [PubMed] [Google Scholar]
- Shulman JM, Feany MB (2003) Genetic modifiers of tauopathy in drosophila. Genetics 165:1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoulakis EM, Grammenoudi S (2006) Dunces and da vincis: the genetics of learning and memory in Drosophila. Cell Mol Life Sci 63:975–988. 10.1007/s00018-006-6023-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos I, Galas MC, Silva JM, Skoulakis E, Wegmann S, Maina MB, Blum D, Sayas CL, Mandelkow EM, Mandelkow E, Spillantini MG, Sousa N, Avila J, Medina M, Mudher A, Buee L (2017) Atypical, non-standard functions of the microtubule associated tau protein. Acta Neuropathol Commun 5:91. 10.1186/s40478-017-0489-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Yu D, Busto GU, Wilson C, Davis RL (2013) Wnt signaling is required for long-term memory formation. Cell Rep 4:1082–1089. 10.1016/j.celrep.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzenidou ME, Segklia A, Kano T, Papastefanaki F, Karakostas A, Charalambous M, Ioakeimidis F, Papadaki M, Kloukina I, Chrysanthou-Piterou M, Samiotaki M, Panayotou G, Matsas R, Douni E (2017) Novel insights into SLC25A46-related pathologies in a genetic mouse model. PLoS Genet 13:e1006656. 10.1371/journal.pgen.1006656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG (1985) Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 157:263–277. 10.1007/BF01350033 [DOI] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M (1994) Genetic dissection of consolidated memory in drosophila. Cell 79:35–47. 10.1016/0092-8674(94)90398-0 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Cox J (2016a) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319. 10.1038/nprot.2016.136 [DOI] [PubMed] [Google Scholar]
- Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J (2016b) The perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 13:731–740. 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, Davis RL (2015) Identification of genes that promote or inhibit olfactory memory formation in drosophila. Genetics 199:1173–1182. 10.1534/genetics.114.173575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Mandelkow E (2016) Tau in physiology and pathology. Nat Rev Neurosci 17:5–21. 10.1038/ncb3086 [DOI] [PubMed] [Google Scholar]
- Wis̀niewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Zhao K, Wu J, Xu Z, Jin S, Zhang YQ (2013) HDAC6 mutations rescue human tau-induced microtubule defects in Drosophila. Proc Natl Acad Sci U S A 110:4604–4609. 10.1073/pnas.1207586110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Akalal DB, Davis RL (2006) Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron 52:845–855. 10.1016/j.neuron.2006.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zars T, Wolf R, Davis R, Heisenberg M (2000) Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: in search of the engram. Learn Mem 7:18–31. 10.1101/lm.7.1.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukareva V, Vogelsberg-Ragaglia V, Van Deerlin VM, Bruce J, Shuck T, Grossman M, Clark CM, Arnold SE, Masliah E, Galasko D, Trojanowski JQ, Lee VM (2001) Loss of brain tau defines novel sporadic and familial tauopathies with frontotemporal dementia. Ann Neurol 49:165–175. [DOI] [PubMed] [Google Scholar]
- Zhukareva V, Sundarraj S, Mann D, Sjogren M, Blenow K, Clark CM, McKeel DW, Goate A, Lippa CF, Vonsattel JP, Growdon JH, Trojanowski JQ, Lee VM (2003) Selective reduction of soluble tau proteins in sporadic and familial frontotemporal dementias: an international follow-up study. Acta Neuropathol 105:469–476. 10.1007/s00401-002-0668-8 [DOI] [PubMed] [Google Scholar]