Abstract
Regulator of G-protein signaling 4 (RGS4) is a potent modulator of G-protein-coupled receptor signal transduction that is expressed throughout the pain matrix. Here, we use genetic mouse models to demonstrate a role of RGS4 in the maintenance of chronic pain states in male and female mice. Using paradigms of peripheral inflammation and nerve injury, we show that the prevention of RGS4 action leads to recovery from mechanical and cold allodynia and increases the motivation for wheel running. Similarly, RGS4KO eliminates the duration of nocifensive behavior in the second phase of the formalin assay. Using the Complete Freud's Adjuvant (CFA) model of hindpaw inflammation we also demonstrate that downregulation of RGS4 in the adult ventral posterolateral thalamic nuclei promotes recovery from mechanical and cold allodynia. RNA sequencing analysis of thalamus (THL) from RGS4WT and RGS4KO mice points to many signal transduction modulators and transcription factors that are uniquely regulated in CFA-treated RGS4WT cohorts. Ingenuity pathway analysis suggests that several components of glutamatergic signaling are differentially affected by CFA treatment between RGS4WT and RGS4KO groups. Notably, Western blot analysis shows increased expression of metabotropic glutamate receptor 2 in THL synaptosomes of RGS4KO mice at time points at which they recover from mechanical allodynia. Overall, our study provides information on a novel intracellular pathway that contributes to the maintenance of chronic pain states and points to RGS4 as a potential therapeutic target.
SIGNIFICANCE STATEMENT There is an imminent need for safe and efficient chronic pain medications. Regulator of G-protein signaling 4 (RGS4) is a multifunctional signal transduction protein, widely expressed in the pain matrix. Here, we demonstrate that RGS4 plays a prominent role in the maintenance of chronic pain symptoms in male and female mice. Using genetically modified mice, we show a dynamic role of RGS4 in recovery from symptoms of sensory hypersensitivity deriving from hindpaw inflammation or hindlimb nerve injury. We also demonstrate an important role of RGS4 actions in gene expression patterns induced by chronic pain states in the mouse thalamus. Our findings provide novel insight into mechanisms associated with the maintenance of chronic pain states and demonstrate that interventions in RGS4 activity promote recovery from sensory hypersensitivity symptoms.
Keywords: genetic mouse models, inflammatory pain, neuropathic pain, RGS, RNA sequencing, viral gene transfer
Introduction
Chronic pain promotes robust neurochemical and cellular adaptations in peripheral and central circuits (Ossipov et al., 2010; Kuner and Flor, 2016). Prolonged inflammation or nerve injury leads to central sensitization, characterized by increased neuronal excitability at spinal and supraspinal sites (Latremoliere and Woolf, 2009; Ji et al., 2018) and the expression of a wide range of sensory and affective symptoms (Turner et al., 2004; Hummel et al., 2008; Mathey et al., 2015). A better understanding of the synaptic maladaptations that contribute to pain chronicity may provide novel avenues for the treatment of chronic pain conditions (Reichling and Levine, 2009; Megat et al., 2019).
Regulator of G-protein signaling 4 (RGS4) is a potent G-protein-coupled receptor (GPCR) regulator (Gold et al., 1997; Bastin et al., 2012; Gerber et al., 2016) expressed in brain circuits mediating nociception, mood, and motivation. Aberrant RGS4 function has been linked to severe human disorders including schizophrenia, Parkinson's disease, and addiction (Ding et al., 2006; Terzi et al., 2009; Lerner and Kreitzer, 2012). This 28 kDa protein modulates responses to chronic stress and the efficacy of monoamine-targeting drugs, such as psychostimulants and antidepressants (Gold et al., 2002; Schwendt and McGinty, 2007; Stratinaki et al., 2013). Furthermore, several studies have demonstrated a role of RGS4 in functional responses to opioid receptor agonists (Georgoussi et al., 2006; Ajit et al., 2007; Han et al., 2010). RGS4 is expressed in many structures involved in the transmission and maintenance of chronic pain, including the dorsal horn of the spinal cord, the periaqueductal gray (PAG), the thalamus, and the basal ganglia (Ni et al., 1999; Gold et al., 2003; Terzi et al., 2009; Taccola et al., 2016). Based on the distribution pattern of RGS4 and its potent modulatory role in signal transduction events triggered by Gα and Gβγ proteins, we expect that changes in RGS4 activity may affect functional responses of several GPCRs and ion channels throughout the pain matrix. Understanding the functional significance of RGS4 in the development and maintenance of chronic pain states may contribute essential knowledge to the cellular determinants of sensory hypersensitivity and pain chronicity. Recent reports propose that intrathecal application of RGS4 inhibitors in rodents alleviates mechanical allodynia and pain-like behaviors (Bosier et al., 2015; Yoon et al., 2015; Taccola et al., 2016). Here, we apply constitutive and conditional knock-out models to understand the role of RGS4 in the induction, intensity, and maintenance of chronic pain symptoms.
Our data reveal a prominent role of RGS4 in the maintenance of mechanical and cold allodynia. The role of RGS4 in the maintenance of central sensitization is further supported by formalin assay studies, showing that knockout of RGS4 attenuates nocifensive behaviors in the second phase of the formalin test. The ventral posterolateral thalamus (VPL-THL) receives spinothalamic input, which plays a prominent role in central sensitization and relays nociceptive information to other brain regions in response to peripheral inflammation or injury (Craig and Burton, 1981; Cliffer et al., 1991; Ossipov et al., 2000). Using a mouse line carrying a floxed RGS4 gene (RGS4fl\fl) and adeno-associated virus (AAV) vectors expressing Cre recombinase, we downregulated RGS4 expression in the VPL-THL and demonstrated that this intervention promotes recovery from mechanical allodynia in a model of peripheral inflammation. RNA-sequencing (Seq) analysis reveals that the prevention of RGS4 action affects a number of receptors and intracellular pathways in the THL, with major impact on glutamatergic plasticity and on the activity of G-protein-regulated networks. Furthermore, recovery from chronic pain states in RGS4 knock-out (RGS4KO) groups is associated with increased expression of metabotropic glutamate receptor 2 (mGluR2) in THL synaptosomes. In summary, our study provides insight into the intracellular mechanisms contributing to pain chronification and points to promising avenues for novel pain treatments.
Materials and Methods
Subjects.
For all behavioral experiments, we used 2- to 3-month-old male and female mice from the RGS4 knock-out or RGS4fl/fl lines (Han et al., 2010; Stratinaki et al., 2013). For quantitative PCR (qPCR) experiments, we used male C57BL/6 mice (The Jackson Laboratory). Mice were group housed (four to five per cage) on a 12 h light/dark cycle, and provided with food and water ad libitum. Animal handling was in accordance with the animal care and use committee guidelines of the Icahn School of Medicine at Mount Sinai.
Stereotaxic surgery and viral localization.
Stereotaxic coordinates for bilateral viral vector injections into the VPL-THL were as follows: anteroposterior (AP), −1.70 mm; mediolateral (ML), ±2.20 mm; and dorsoventral (DV) −3.50 mm, at an angle of 10° from the midline. Coordinates for the MD-THL were as follows: AP −1.58 mm; ML, ±0.44 mm; and DV, −3.20 mm. Coordinates for the nucleus accumbens (NAc) were as follows: AP, 1.60 mm; ML, ±1.50 mm; DV, −4.40 mm; at an angle of 10° from the midline. AAV-Cre and AAV-GFP vectors (serotype 2) were purchased from the Vector Core Facility at the University North Carolina (Chapel Hill, NC; Gaspari et al., 2018). Viral expression was evaluated at the end of each experiment using GFP fluorescence or Western blot analysis for RGS4.
Spared nerve injury operation.
Spared nerve injury (SNI) was performed in the left sciatic nerve, as previously described (Shields et al., 2003; Mitsi et al., 2015; Descalzi et al., 2017). Briefly, using a stereomicroscope, a skin and muscle incision of the left hindlimb at mid-thigh level was performed to reach the sciatic nerve. The common peroneal and the sural nerves were carefully ligated with 6.0 silk suture (Johnson & Johnson International), transected and 1–2 mm sections of these nerves were removed, while the tibial nerve was left intact. Skin and muscle were then closed with 4.0 silk suture (Johnson & Johnson International). In sham-operated mice, the same procedure was followed, but the nerves were left untouched.
Complete Freud's Adjuvant treatment.
Left hindpaw inflammation was induced by intraplantar injection of 25 μl of Complete Freud's Adjuvant (CFA; Sigma-Aldrich), diluted 1:1 in saline as described previously (Minett et al., 2012; Gaspari et al., 2018).
Paclitaxel treatment.
Paclitaxel (Tocris Bioscience) was dissolved in a 50% solution of absolute ethanol and 50% Cremophor EL to a concentration of 10 mg/ml and stored at 4°C, and then diluted to the necessary concentration with saline just before administration (Masocha, 2014; Toma et al., 2017). Control animals received vehicle, prepared the same way as the drug solution. Paclitaxel or vehicle was administered intraperitoneally at a volume of 100 μl four times, every other day.
Von Frey test for mechanical allodynia.
For the assessment of mechanical allodynia, we used Von Frey filaments (Mitsi et al., 2015; Descalzi et al., 2017; Tsantoulas et al., 2018) with ascending forces expressed in grams (Stoetling). Each filament was applied five times in a row against the ipsilateral hindpaw. Hindpaw withdrawal or licking was marked as a positive allodynia response. A positive response in three of five repetitive stimuli was defined as the allodynia threshold.
Hargreaves test for thermal hyperalgesia.
Mice were placed in Plexiglas boxes on top of a glass surface (IITC Life Science), and the latency to withdraw the injured (CFA-injected) hindpaw was measured after a high-intensity heat beam (30%) was applied to the mid-plantar area (IITC Life Science). Two measurements were obtained with a 10 min interval, and the average was defined as the thermal nociceptive threshold. A cutoff time of 20 s was used to avoid potential tissue damage (Patwardhan et al., 2009; Gaillard et al., 2014; Terzi et al., 2014; Gaspari et al., 2018).
Cold plate test.
Nocifensive behavior was assessed at 0°C using a cold plate (IITC-Life Science). Mice were habituated in the room for 1 h, and then placed on the cold surface, and behavioral responses were recorded using a video camera (VIXIAHF R600, Canon). Two blinded experimenters monitored latency to paw flinching (Roeckel et al., 2017).
Formalin test.
The left hindpaw was injected with 25 μl of 4% formaldehyde (Sigma-Aldrich), as previously described (Papachatzaki et al., 2011; Gaillard et al., 2014; Rodriguez et al., 2017). Mice were immediately placed into Plexiglas boxes and nocifensive behavior was recorded for 1 h, using a video camera (VIXIAHF R600, Canon). Two blinded investigators monitored licking and flinching behaviors over the 1 h period.
Wheel running.
We used a wireless running wheel activity monitoring system (Low-Profile Wireless Running Wheel for Mouse, Med Associates). Mice were habituated for 2 d in their home cage with the running wheel apparatus (Cobos et al., 2012). On testing days, each mouse was monitored for 1 h. Mice that ran <100 cycles per hour were excluded from the study. Activity was calculated as a percentage of naive RGS4 wild-type (RGS4WT) baseline activity.
Western blotting and subcellular fractionation.
Western blotting and subcellular fractionation were performed as previously described (Charlton et al., 2008; Gaspari et al., 2018). Two 12 gauge thalamus punches were used per sample for all assays. For subcellular fractionation, samples were homogenized using a pestle in 100 μl of hypotonic cell lysis buffer (HCLB) plus protease and proteasome inhibitors (Sigma-Aldrich), incubated on ice for 45 min, and centrifuged at 371 × g at 4°C for 10 min. Supernatants were then collected as the crude cytoplasmic fractions. To obtain the synaptosomal fractions, the crude cytoplasmic fractions were centrifuged twice to remove the remaining nuclei (371 × g at 4°C for 10 min). The supernatants were collected and centrifuged at 9361 × g at 4°C for 10 min. Supernatants were collected as the cytoplasmic fractions, and the synaptosomal pellets were resuspended in 20 μl of HCLB. The following antibodies were used: rabbit anti-β-actin (catalog #4970, Cell Signaling Technology); rabbit anti-Gβ5 (from W. Simmonds, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD); rabbit anti-RGS4 (catalog #ABT17, Millipore); and mouse anti-metabotropic glutamate receptor 2 (catalog #ab15672, Abcam). Gβ5 or β-actin antibodies were used as loading controls. Data are calculated as the optical density (OD) ratio of sample/loading control and are expressed as the percentages of the control group. Bands were quantified using ImageJ software (Mitsi et al., 2015).
RNA sequencing and qPCR.
Four biological replicates per group were used for the RNA-Seq studies. Bilateral thalamus punches from two male mice were pooled per biological replicate (four 12 gauge punches per sample). Total RNA was isolated with TRIzol, and the integrity was confirmed with an Agilent 2100 Bioanalyzer (Mitsi et al., 2015; Descalzi et al., 2017). mRNA-Seq libraries were prepared using a TruSeq RNA sample preparation kit version 2 (Illumina). Sequencing was performed using the Illumina HiSeq 2000 apparatus. qPCR was performed using SYBR green and analyzed using the ΔΔCT method and GAPDH as a normalization control (Gaspari et al., 2018). Information on primers used for biological validation of gene expression and in RGS4 regulation studies is provided in Table 5.
Table 5.
Primer pairs used for real-time-PCR experiments
| Gene/primer | Forward 5′ → 3′ | Reverse 5′ → 3′ |
|---|---|---|
| Rgs4 | GTTTTGATGAAGCCCAGAAGAAG | CAGAGCATAGAGTCTTGGCA |
| Gapdh | GGTCGGTGTGAACGGATTTGG | TGATGTTAGTGGGGTCTCGC |
| Gng11 | AAGTTGCAGAGACAACAGGTATC | CACTTTGGCTTCATTTGGGGG |
| Glp1r | GCCAGTAGTGTGCTACAAGG | GACTCTTCACACTCCGACAGG |
| Ppp3r2 | GTCCTGGATAAGGATGGCGA | AGTCTTCTTGACCGTGTTCTACAAA |
| Atf4 | CTTGATGTCCCCCTTCGACC | GGAAAAGGCATCCTCCTTGC |
Bioinformatics analysis.
Read alignment, read counting, and differential analysis were performed using HISAT2 (Kim et al., 2015a), HTSSeq (Anders et al., 2015), and the DESeq2 R package, respectively (Love et al., 2014). Differential analysis aimed to dissect the CFA versus naive treatment effect in WT and KO was performed using a 2 × 2 factorial design with the following linear model: log(exp) ∼ Genotype + Treatment + Genotype:Treatment. The differential lists were defined by a p value cutoff of <0.05 and log2 (fold change) of <−0.25 or >0.25. Inclusion of the interaction term “Genotype:Treatment” to the model facilitated in studying the KO-specific treatment effect. Significantly contraregulated genes were defined as the genes exhibiting opposite directionality and the interaction term p value <0.05. Gene Ontology analysis was performed using DAVID 6.8. Only terms with p value <0.05 were reported. Venn diagrams and heat maps were generated using BioVenn-web application (http://www.biovenn.nl/) and Morpheus (https://software.broadinstitute.org/morpheus) software respectively. Pathway analysis was conducted using ingenuity pathway analysis (IPA). For pathway analysis, a cutoff p value of <0.003 was applied to the output pathways (Love et al., 2014; Descalzi et al., 2017; Gaspari et al., 2018).
Statistical analysis.
Data were analyzed using GraphPad Prism 8 software. For the experiments monitoring behavioral responses over time, we used two-way repeated-measures ANOVAs followed by Bonferroni's post hoc tests. For two-factor designs, we used two-way ANOVA followed by Bonferroni's post hoc test. For data containing a single independent variable, we used unpaired two-tailed t tests. Error bars depict ±SEM. F and t values for each dataset are provided in the figure legends.
Results
RGS4KO mice show accelerated recovery from mechanical and cold allodynia in models of peripheral inflammation
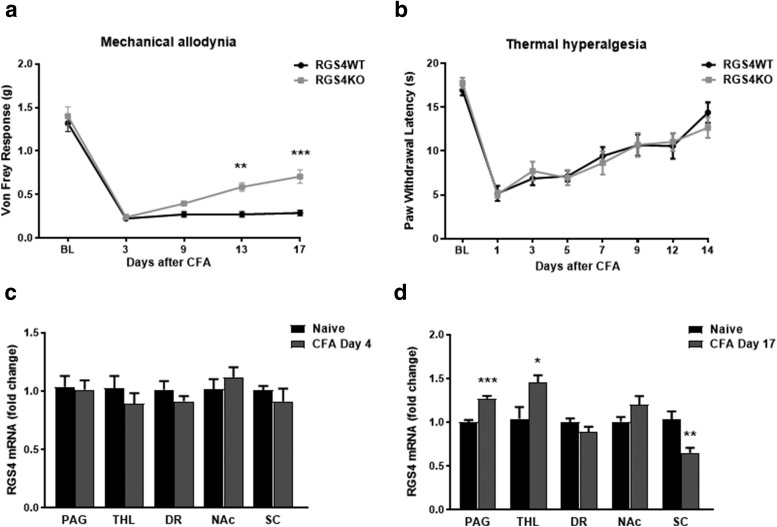
We applied the CFA model to gain insight on the role of RGS4 in hypersensitivity responses associated with prolonged hindpaw inflammation. We monitored mechanical allodynia and thermal hyperalgesia at several time points after CFA treatment. Male RGS4KO mice start recovering from mechanical allodynia at 9 d post-CFA injection, and they show a significant antiallodynic response on day 13, whereas the RGS4WT mice maintain the same allodynia levels throughout the monitoring period (Fig. 1a). Notably, there were no significant differences in thermal hypersensitivity in male mice between genotypes in the Hargreaves assay (Fig. 1b). To determine whether the expression of RGS4 is regulated by peripheral inflammation, we used qPCR and monitored RGS4 mRNA levels in several regions along the pain matrix at early and later time points after CFA treatment. PAG, THL, dorsal raphe (DR), NAc, and ipsilateral dorsal spinal cord (SC) from C57BL/6 mice were collected 4 or 17 d after CFA injection to the plantar surface of the hindpaw. Although there is no change in RGS4 expression at 4 d after CFA treatment (Fig. 1c), at 17 d there is a significant upregulation of RGS4 transcript in the PAG and in the THL. At this time point, RGS4 mRNA levels are decreased in the ipsilateral dorsal horn (Fig. 1d). These data further support the hypothesis that while there is no physiological role of RGS4 in the expression and intensity of sensory hypersensitivity, RGS4 activity at later time points promotes the maintenance of chronic pain states. There is no effect of RGS4 on hindpaw edema size between genotypes (DV: RGS4WT = 3.56 ± 0.03 mm; RGS4KO = 3.55 ± 0.06 mm; ML: RGS4WT = 4.57 ± 0.02 mm, RGS4KO = 4.42 ± 0.04 mm).
Figure 1.
RGS4KO mice rapidly recover from mechanical allodynia in a model of long-term peripheral inflammation. a, Male RGS4WT and RGS4KO mice show mechanical allodynia in the Von Frey assay in response to CFA injection to the left hindpaw, but RGS4KO mice recover by day 13 (two-way ANOVA followed by Bonferroni post hoc tests: F(1,28) = 16.39, **p = 0.001, ***p < 0.001; n = 15). b, RGS4KO mice show identical responses to their RGS4WT controls in the Hargreaves assay of thermal hyperalgesia (two-way ANOVA: F(1,20) = 0.0009, n = 11/group). c, qPCR analysis reveals that RGS4 mRNA expression in the PAG, THL, DR, NAc, and ipsilateral SC does not change at early time points (4 d) after the induction of peripheral inflammation (CFA injection to the left hindpaw). Unpaired t test: PAG, t(20) = 0.1893; THL, t(9) = 0.966; DR, t(10) = 1.153; NAc, t(10) = 0.8572; SC ipsilateral, t(18) = 0.8891; n = 5–11/group; for all comparisons, p > 0.05). d, At later time points (17 d post-CFA), there is an increase in Rgs4 transcript in the PAG and in the THL (unpaired t test: PAG: t(11) = 6.629, ***p < 0.001, n = 6–7/group; THL: t(10) = 2.64, *p = 0.02, n = 6/group; DR: t(11) = 1.521, n = 6–7/group, NAc: t(13) = 1.778, n = 7–8/group; SC: t(24) = 3.604 **p = 0.001, n = 13/group).
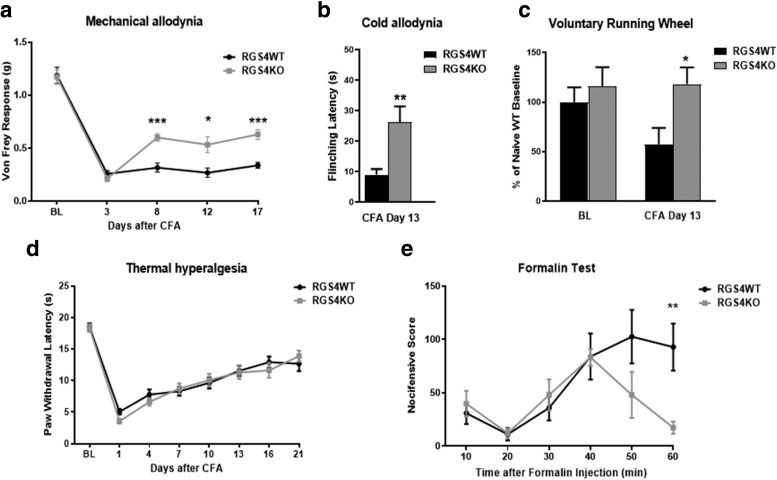
We followed up with studies in female mice to determine whether RGS4 has a sex-specific role in pain modulation. As shown in Figure 2 RGS4 plays a similar modulatory role in the maintenance of chronic pain symptoms in female mice. Female RGS4KO mice show attenuated allodynia in the Von Frey assay 8 d after CFA treatment, whereas the RGS4WT group maintained the same Von Frey thresholds throughout the monitoring period (Fig. 2a). Notably, 9 d after the induction of peripheral inflammation, female RGS4KO mice also show attenuated responses in the 0°C cold plate assay (Fig. 2b). Chronic pain states have been shown to reduce voluntary wheel running in rodents (Cobos et al., 2012), reflecting reduced motivational states. We applied the running wheel paradigm in female mice to determine the impact of RGS4 on running activity 2 weeks after the induction of peripheral inflammation. Under pain-free states (baseline), both RGS4WT and RGS4KO mice show similar running wheel activity. However, at 13 d post-CFA, RGS4KO mice exhibit higher activity compared with RGS4WT controls, further supporting the role of RGS4 in the maintenance of chronic pain states (Fig. 2c). Notably, knockout of RGS4 does not affect Hargreaves responses in female mice (Fig. 2d). The formalin assay for subchronic inflammatory pain was also used to assess the impact of RGS4 in nocifensive behaviors. The prevention of RGS4 action does not affect licking and flinching during the first phase of the formalin test (first 10 min), reflecting acute nociception. However, while the number of licking/flinching occurrences was not different between genotypes throughout the first 30 min of the second phase, RGS4KO mice showed minimal nocifensive behavior during the last 20 min of the second phase (Fig. 2e). This phenotype supports a role for RGS4 in the maintenance of central sensitization.
Figure 2.
A role of RGS4 in symptoms of peripheral inflammation in female mice. a, Female RGS4KO mice show attenuated mechanical allodynia by day 8 after CFA injection to the ipsilateral hindpaw (two-way ANOVA followed by Bonferroni post hoc tests: F(1,27) = 14.08, *p = 0.03, ***p < 0.001, n = 14–15/group). b, Female RGS4KO mice show attenuated responses to cold (0°C) at 9 d after CFA treatment compared with the RGS4WT group (unpaired t test: t(27) = 3.219, **p = 0.003, n = 14–15/group). c, Prevention of RGS4 action improves voluntary wheel running. Female RGS4KO mice show higher running wheel activity compared with RGS4WT controls at 13 d after left hindpaw CFA injection, suggesting recovery from chronic pain states. Baseline responses are indistinguishable between phenotypes. Bars represent the percentage of naive baseline responses (two-way ANOVA followed by Bonferroni post-tests: F(1,26) = 5.106, *p = 0.04, n = 7–8/group). d, In contrast, both RGS4WT and RGS4KO female mice show identical responses in the Hargreaves assay for thermal hyperalgesia (two-way ANOVA: F(1,27) = 0.2289, n = 14–15/group). e, Female RGS4WT and RGS4KO mice show similar licking/flinching levels during the first 40 min of the formalin test, but after this time point RGS4KO mice show recovery from nocifensive states (two-way ANOVA followed by Bonferroni post hoc tests: F(1,14) = 3.256, **p = 0.008, n = 7–9/group).
A role of RGS4 in the maintenance of mechanical allodynia in models of nerve injury
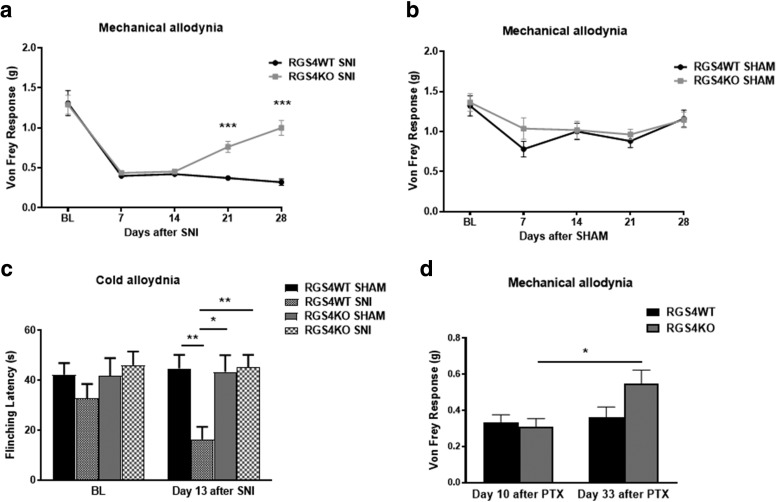
We next investigated whether RGS4 is required to promote and maintain mechanical hypersensitivity in response to peripheral nerve injury. We applied the SNI model of neuropathic pain (Shields et al., 2003) to determine the consequences of RGS4 gene inactivation in mechanical and cold allodynia developed under long-term nerve injury. SNI leads to mechanical allodynia, which is observed for several months after nerve injury (Descalzi et al., 2017). As shown in Figure 3a, female RGS4KO mice show levels of mechanical allodynia in the Von Frey assay similar to those of their RGS4WT controls throughout the first 2 weeks of testing; however, at 3 weeks after SNI, RGS4KO mice show a significant increase in Von Frey thresholds compared with their RGS4WT controls. Sham-operated female mice show similar Von Frey responses (Fig. 3b). Recovery from mechanical allodynia is also observed in male groups of mice [Von Frey force (g) 3.5 weeks post-SNI: RGS4WT = 0.35 ± 0.07; RGS4KO = 0.9 ± 0.05; ****p = 0.0001). At 2 weeks post-SNI, female RGS4KO mice also show recovery from cold hypersensitivity, as assessed in the cold plate assay (Fig. 3c). We observed a similar phenotype when we used the paclitaxel model of chemotherapy-induced peripheral nerve injury (Masocha, 2014; Toma et al., 2017). This model of chemotherapy-induced peripheral nerve injury is known to promote long-term mechanical hypersensitivity. Female RGS4WT and RGS4KO mice show indistinguishable responses in the Von Frey assay 10 d after the onset of paclitaxel treatment, but RGS4KO mice show higher Von Frey thresholds at 33 d (Fig. 3d).
Figure 3.
A role of RGS4 in the maintenance of neuropathic states. a, Using the SNI model of neuropathic pain, we show that female RGS4KO mice recover from mechanical allodynia at 21 d after nerve injury, while their RGS4WT controls show maximal allodynia in the Von Frey test throughout the 1 month monitoring period (two-way ANOVA followed by Bonferroni post-tests: F(1,18) = 17.28, ***p < 0.001, n = 9–11/group). b, Mechanical thresholds are indistinguishable in sham-treated (control) female groups, which do not develop mechanical allodynia (two-way ANOVA: F(1,19) = 0.6013, n = 10–11/group). c, SNI-treated female RGS4KO mice show attenuated responses to cold when tested at 13 d post SNI (two-way ANOVA followed by Bonferroni post hoc tests: F(3,35) = 5.817; RGS4WT SNI vs sham, **p = 0.005; RGS4KO sham vs RGS4WT SNI, *p = 0.01; RGS4KO SNI vs RGS4WT SNI, **p = 0.003; n = 9–11 female mice/group). d, Paclitaxel-treated female RGS4WT and RGS4KO mice show identical responses in the Von Frey assay at 10 d after the onset of paclitaxel (PTX) injections, but RGS4KO mice show reduced allodynia on day 33 (two-way ANOVA followed by Bonferroni post hoc tests: F(1,13) = 2.225 for RGS4KO PTX day 10 vs day 33; *p = 0.019; n = 7–8/group).
RGS4 actions in the VPL-THL contribute to the maintenance of mechanical allodynia
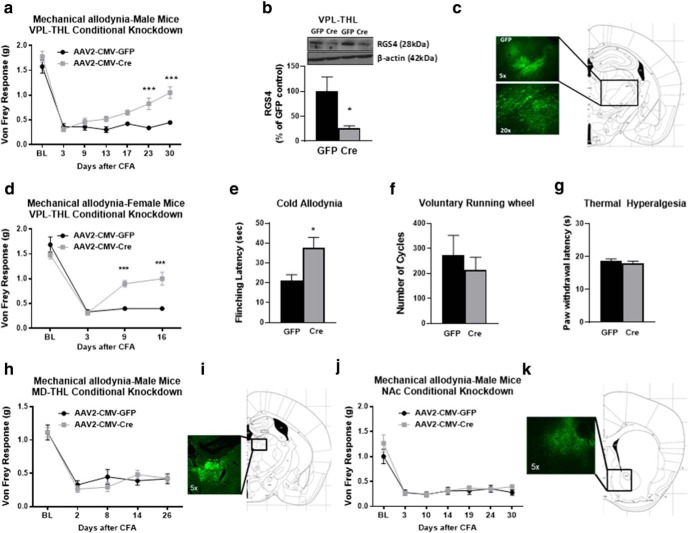
Based on findings from our qPCR analysis demonstrating the upregulation of RGS4 expression in the THL in response to long-term hindpaw inflammation, we hypothesized that RGS4 actions in the VPL-THL play a critical role in the maintenance of central sensitization. We used a conditional gene knock-down (KD) approach to assess the consequences of RGS4 downregulation in the VPL-THL in sensory hypersensitivity deriving from left hindpaw inflammation. KD of RGS4 was achieved by stereotaxic infection of the VPL-THL of RGS4fl\fl mice with AAV vectors (AAV2) expressing Cre recombinase (AAV-Cre; Stratinaki et al., 2013). As shown in Figure 4a, and, consistent with our findings from experiments using constitutive RGS4KO mice, male VPL-THL-RGS4KD mice recover from mechanical allodynia by 10 d after CFA treatment. These findings point to a key role of RGS4 actions in VPL-THL in pain chronicity. For the validation of RGS4 knockdown, GFP-positive THL tissue was collected using a fluorescent microscope and was used for Western blot analysis. Figure 4b demonstrates that there is a 75 ± 12% reduction in RGS4 protein expression in the THL of AAV-Cre- versus AAV-GFP-infected groups. Representative GFP immunofluorescence images (Fig. 4c) show the distribution of the AAV-Cre vector in the VPL-THL. This phenotype was also observed in female cohorts of VPL-THL-RGS4KD mice (Fig. 4d). The downregulation of RGS4 in the VPL-THL also leads to recovery from cold hypersensitivity (Fig. 4e), while it does not affect running wheel activity (recorded on day 13 after CFA; Fig. 4f) and Hargreaves responses (recorded on day 17 after CFA; Fig. 4g).
Figure 4.
RGS4 knockdown in VPL-THL neurons induces earlier recovery from mechanical allodynia. a, Conditional RGS4 downregulation in neurons of the VPL-THL of male RGS4fl/fl mice by stereotaxic infection of AAV2-CMV-CRE vectors led to recovery from mechanical allodynia in the Von Frey assay (two-way ANOVA followed by Bonferroni post hoc tests: F(1,14) = 19.32, ***p < 0.001; n = 8/group). b, Western blot analysis verifying RGS4 downregulation in the VPL-THL of mice infected with AAV2-CMV-Cre-EGFP compared with AAV2-CMV-EGFP vectors (t test unpaired two-tailed: t(6) = 2.528, *p = 0.04; n = 4/group). c, Representative image showing GFP fluorescence 5 weeks after AAV2-CMV-Cre-EGFP infection of the VPL-THL. d, A similar phenotype was observed in female RGS4fl/fl mice injected with AAV2-CMV-CRE vectors in the VPL-THL. e, Downregulation of RGS4 in the female VPL-THL leads to recovery from mechanical allodynia in the Von Frey assay (two-way ANOVA followed by Bonferroni post hoc tests: F(1,13) = 12.33; ***p < 0.001; n = 7–8/group) and alleviates cold hypersensitivity (unpaired two-tailed t test: t(13) = 2.552; *p = 0.0241, n = 7–8/group). f, g, Notably, the downregulation of RGS4 in the VPL-THL does not affect wheel-running activity (unpaired two-tailed t test: t(10) = 0.6497; n = 6/group; f) and Hargreaves responses (unpaired two-tailed t test: t(13) = 0.7550; n = 7–8/group; g). h, Downregulation of RGS4 in the MD-THL of male mice does not promote recovery from mechanical allodynia in the CFA model (two-way ANOVA: F(1,12) = 0.1690; n = 7/group). i, Representative image showing GFP fluorescence in the MD-THL of mice infected with AAV2-CMV-EGFP. j, Downregulation of RGS4 in the NAc of male mice does not affect the trajectory of mechanical allodynia in the CFA model (two-way ANOVA: F(1,13) = 0.8012; n = 6–9/group). k, Representative image showing GFP fluorescence in the NAc of mice infected with AAV2-CMV-EGFP.
We also investigated whether downregulation of RGS4 in the medial dorsal THL (MD-THL) is sufficient to promote recovery from symptoms of sensory hypersensitivity. The MD-THL is involved in the processing of cognitive/emotional information in the modulation of pain-related aversion as well as in the modulation of nociception, but it projects to different areas than the VPL-THL (Price and Slotnick, 1983; Wang et al., 2004; Parnaudeau et al., 2018; Meda et al., 2019). In contrast to our observations on the VPL-THL knockdown groups, downregulation of RGS4 in the MD-THL of male mice did not affect Von Frey responses in the CFA model of peripheral inflammation (Fig. 4h). RGS4 is also expressed in the NAc, where it has been shown to modulate responses to psychoactive drugs (Han et al., 2010; Stratinaki et al., 2013). The NAc is implicated in the processing of sensory and affective pain symptoms (Baliki and Apkarian, 2015; Mitsi and Zachariou, 2016). The downregulation of RGS4 in the NAc of male mice did not affect mechanical allodynia in the CFA model of peripheral inflammation (Fig. 4j), highlighting the regional specificity of RGS4 actions.
Long-term peripheral inflammation promotes distinct gene expression patterns in the thalamus of RGS4WT and RGS4KO mice
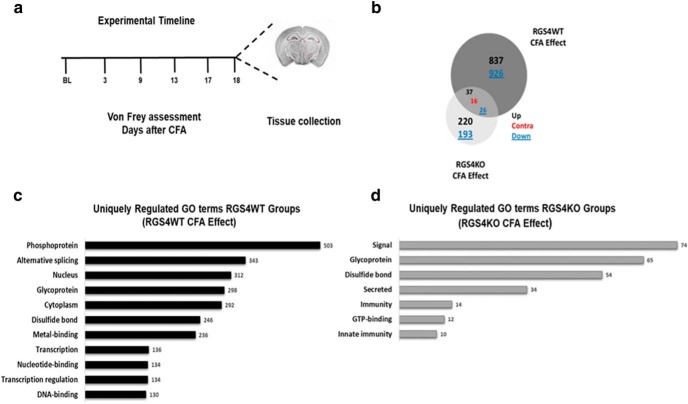
Based on the observation that RGS4KO is sufficient to promote recovery from chronic pain states, we hypothesized that RGS4 affects the expression of many genes that contribute to central sensitization and pain chronicity. To gain insight into the genes and pathways affected by RGS4 activity in the THL under chronic pain states, we applied RNA-Seq analysis and investigated the impact of peripheral inflammation and/or RGS4 knockout in gene expression. We used adult male cohorts of mice, and we selected a time point of 18 d post-CFA treatment, at which RGS4KO mice show recovery from mechanical allodynia while RGS4WT controls still display maximal allodynia, and we collected thalamic tissue from CFA and naive groups of RGS4WT and RGS4KO mice (Fig. 5a). Dissections were performed 24 h after the last Von Frey assessment. In particular, 1763 genes were differentially expressed only in the RGS4WT group, while 413 genes were uniquely regulated in the RGS4KO group. As shown in the Venn diagram in Figure 5b, 837 genes were upregulated and 926 genes were downregulated in the RGS4WT group, while only 220 genes were upregulated and 193 downregulated in the RGS4KO group. Gene ontology analysis indicated distinct functional roles for genes regulated by CFA treatment under each genotype. The list of differentially expressed genes between CFA and naive in the RGS4WT group were enriched in: phosphoproteins, transcription factors, genes expressed in the nucleus, and DNA-binding molecules, reflecting high levels of transcriptional activity (Fig. 5c). Notably, at this time point, these GO term enrichments were not observed in the RGS4KO group (Fig. 5d). The GO categorization suggests a role of RGS4 in the CFA-induced transcriptional profile. On the other hand, genes associated with signal transduction, glycoproteins, and molecules forming disulfide bonds were differentially expressed in both genotypes (Fig. 5c,d). Our findings suggest that the maintenance of chronic pain states requires a number of gene expression adaptations in the THL, and several of these events are prevented by knockout of the RGS4 gene.
Figure 5.
THL-specific RNA-Seq profiling of RGS4KO in groups of naive and CFA-treated mice. a, Experimental time line showing that THL tissue from male groups of mice was collected 18 d after the induction of peripheral inflammation (a time point that RGS4KO mice show recovery from mechanical allodynia). b–d, Venn diagram depicting the number of genes affected in each group and their direction of regulation suggest that long-term peripheral inflammation promotes distinct gene expression adaptations between genotypes (b) and associated gene ontology (GO) categories (c, d). Bar graphs show GO of genes uniquely regulated by pain (CFA) in the THL of RGS4WT (black color) and RGS4KO (gray color) mice. Only GO terms with p < 0.05 are depicted.
Recovery from mechanical allodynia in RGS4KO mice is associated with unique gene expression changes
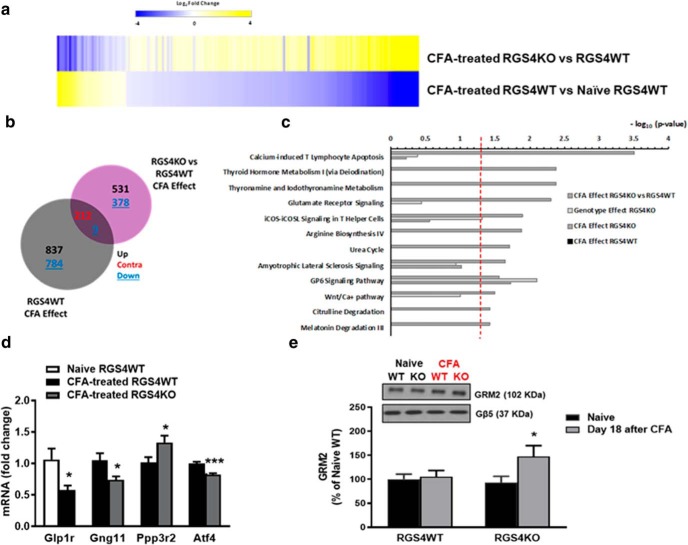
A heat map analysis comparing CFA-treated RGS4WT versus naive RGS4WT and CFA-treated RGS4KO versus RGS4WT reveals that a large number of genes are contraregulated between RGS4WT and RGS4KO groups under states of inflammatory pain (Fig. 6a) and in particular 212 genes are contraregulated between genotypes (Fig. 6b). To further validate the contraregulation gene pattern, we applied a multifactor differential analysis. This analysis permits the extraction of genes that are significantly contraregulated. A specific set of 62 genes are upregulated by pain in the RGS4WT group, whereas these genes are downregulated in the RGS4KO group (Table 1). Similarly, a set of 27 genes that are downregulated by peripheral inflammation in the THL of RGS4WT mice are upregulated in the THL of RGS4KO groups (Table 2). We applied IPA to gain insight into the signaling pathways affected by long-term peripheral inflammation in each genotype. Pathways differentially modulated in RGS4KO versus RGS4WT at 18 d post-CFA treatment include glutamate receptor signaling, calcium signaling, and metabolic pathways (p < 0.003; Fig. 6c). We followed up with qPCR validation of the RNA-Seq findings. THL tissue from naive and 18 d CFA groups of male RGS4WT and RGS4KO mice was collected from separate cohorts from those used in the RNA-Seq analysis. Figure 6d shows qPCR validation of RNA-Seq findings for Gng11 (G-protein subunit gamma 11), Ppp3r2 (protein phosphatase 3 regulatory subunit B, beta), Atf4 (activating transcription factor 4), and Glp1r (glucagon-like peptide-1 receptor). Previous studies have shown that the upregulation of mGluR2 is correlated with attenuated sensory hypersensitivity (Chiechio et al., 2010; Notartomaso et al., 2017). Based on our RNA-Seq findings, we hypothesized that increased expression of mGluR2s in the THL of RGS4KO mice contributes to synaptic remodeling necessary for recovery from sensory hypersensitivity. Using cellular fractionation and Western blot analysis from THL tissue of naive and 18 d CFA RGS4WT and RGS4KO mice, we show that the expression of mGluR2 is increased in the THL synaptosomal fraction of the RGS4KO group (Fig. 6e). Interestingly, cytoplasmic mGluR2 levels were not different between CFA-treated RGS4WT and RGS4KO mice (% OD of naive RGS4WT control: naive RGS4WT = 100 ± 9.5; naive RGS4KO = 107 ± 11.3; CFA RGS4WT = 138.8 ± 18; CFA RGS4KO = 138.2 ± 15). These findings further support our hypothesis that the prevention of RGS4 action permits rearrangement of mGluR2 expression that is necessary for synaptic remodeling and recovery from sensory hypersensitivity.
Figure 6.
RGS4 in the THL modulates gene expression patterns of peripheral inflammation. a, b, Heat map analysis (a) and Venn diagrams (b) showing the effect of peripheral inflammation in RGS4WT and RGS4KO THL (cutoff p value <0.05; cutoff log2fold <−0.25 and >0.25). The overall patterns of gene expression regulation were distinct between RGS4KO and RGS4WT groups. Interestingly, the commonly regulated genes are contraregulated by peripheral inflammation between genotypes (212 contraregulated genes). c, IPA of the set of genes affected by CFA in the RGS4KO group compared with RGS4WT predicts pathways that are selectively modulated upon RGS4 gene ablation in the RGS4KO group (cutoff: p value <0.003). d, qPCR validation of a subset of genes in a separate cohort of male animals (naive RGS4WT and CFA-treated RGS4WT: Glp1r (unpaired t test: t(11) = 2.735, *p = 0.02); CFA-treated RGS4WT and CFA-treated RGS4KO (unpaired t test: Gng11 t(24) = 2.486, *p = 0.02; Ppp3r2 t(10) = 2.364, *p = 0.04; Atf4 t(9) = 5.236, ***p < 0.001; n = 5–13/group. e, Western blot analysis in THL synaptosomes from naive and CFA groups of RGS4WT and RGS4KO mice reveals an upregulation of mGluR2 at 18 d after the induction of peripheral inflammation in the RGS4KO group. Two-way ANOVA followed by Bonferroni post hoc tests: F(1,59) = 3.505, *p = 0.04, n = 15–16/group.
Table 1.
A role of RGS4 in CFA-induced gene upregulation
| Gene | Description | RGS4WT day 18 post-CFA | RGS4KO day 18 post-CFA | Interaction padj |
|---|---|---|---|---|
| Ghsr | Growth hormone secretagogue receptor | 0.96 | −0.12 | 2.63E-08 |
| Cbln1 | Cerebellin 1 precursor protein | 0.58 | −0.34 | 5.10E-07 |
| Hcn1 | Hyperpolarization-activated, cyclic nucleotide-gated K + 1 | 0.47 | −0.02 | 2.11E-05 |
| Prss12 | Protease, serine 12 neurotrypsin (motopsin) | 0.55 | −0.13 | 2.86E-05 |
| Pcdh9 | Protocadherin 9 | 0.34 | −0.10 | 0.0001 |
| Gm13339 | Predicted gene 13339 | 0.17 | −0.24 | 0.0003 |
| Ndst4 | N-Deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 | 0.39 | −0.11 | 0.0009 |
| Slit3 | Slit homolog 3 | 0.41 | −0.07 | 0.0010 |
| Nefl | Neurofilament, light polypeptide | 0.40 | −0.09 | 0.0012 |
| Caln1 | Calneuron 1 | 0.33 | −0.03 | 0.0045 |
| Sspo | SCO-spondin | 1.52 | −0.90 | 0.0045 |
| Cela1 | Chymotrypsin-like elastase family, member 1 | 0.91 | −0.66 | 0.0045 |
| Lrrc38 | Leucine-rich repeat containing 38 | 0.88 | −0.18 | 0.0049 |
| Esrrb | Estrogen-related receptor, β | 1.07 | −0.32 | 0.0053 |
| Igf1 | Insulin-like growth factor 1 | 0.53 | −0.19 | 0.0053 |
| Gm11413 | Predicted gene 11413 | 1.00 | −0.41 | 0.0053 |
| Nr4a2 | Nuclear receptor subfamily 4, group A, member 2 | 0.56 | −0.49 | 0.0053 |
| Gm10222 | Predicted gene 10222 | 0.07 | −0.25 | 0.0055 |
| Sim1 | Single-minded homolog 1 | 0.84 | −0.13 | 0.0062 |
| Krt222 | Keratin 222 | 0.21 | −0.10 | 0.0064 |
| Anxa4 | Annexin A4 | 0.86 | −0.24 | 0.0067 |
| Ntng1 | Netrin G1 | 0.21 | −0.07 | 0.0067 |
| Tfrc | Transferrin receptor | 0.07 | −0.24 | 0.0069 |
| Ugcg | UDP-glucose ceramide glucosyltransferase | 0.05 | −0.12 | 0.0070 |
| Mfap3l | Microfibrillar-associated protein 3-like | 0.25 | −0.14 | 0.0104 |
| Postn | Periostin, osteoblast-specific factor | 2.20 | −0.16 | 0.0111 |
| Gm13340 | Predicted gene 13340 | 0.13 | −0.11 | 0.0123 |
| Calcb | Calcitonin-related polypeptide, β | 2.29 | −0.01 | 0.0131 |
| Scrg1 | Scrapie responsive gene 1 | 0.42 | −0.09 | 0.0131 |
| 4632404H12Rik | RIKEN cDNA 4632404H12 gene | 0.30 | −0.12 | 0.0131 |
| Pax6 | Paired box 6 | 0.82 | −0.44 | 0.0131 |
| Cpox | Coproporphyrinogen oxidase | 0.16 | −0.25 | 0.0131 |
| Slc35a5 | Solute carrier family 35, member A5 | 0.10 | −0.19 | 0.0131 |
| Gm11408 | Predicted gene 11408 | 0.07 | −0.27 | 0.0131 |
| Cartpt | CART prepropeptide | 3.06 | −0.10 | 0.0134 |
| Paqr8 | Progestin and adipoQ receptor family member VIII | 0.36 | −0.10 | 0.0171 |
| Gfod1 | Glucose-fructose oxidoreductase domain containing 1 | 0.40 | −0.06 | 0.0171 |
| Cep97 | Centrosomal protein 97 | 0.11 | −0.37 | 0.0171 |
| Dner | Delta/notch-like EGF repeat containing | 0.14 | −0.05 | 0.0182 |
| Kcng4 | Potassium voltage-gated channel, subfamily G, member 4 | 1.09 | −0.17 | 0.0183 |
| Adamtsl5 | ADAMTS-like 5 | 0.67 | −0.20 | 0.0197 |
| Pappa | Pregnancy-associated plasma protein A | 0.55 | −0.06 | 0.0204 |
| Abhd10 | Abhydrolase domain containing 10 | 0.11 | −0.22 | 0.0225 |
| Ppm1e | Protein phosphatase 1E (PP2C domain containing) | 0.28 | −0.03 | 0.0244 |
| Mbnl2 | Muscleblind-like splicing factor 2 | 0.16 | −0.04 | 0.0266 |
| Kcnn1 | Potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 | 0.26 | −0.13 | 0.0266 |
| Mbnl1 | Muscleblind-like splicing factor 1 | 0.10 | −0.07 | 0.0266 |
| Nell1 | NEL-like 1 | 0.13 | −0.09 | 0.0267 |
| Spp1 | Secreted phosphoprotein 1 | 1.41 | −0.26 | 0.0292 |
| Ube2g1 | Ubiquitin-conjugating enzyme E2G 1 | 0.12 | −0.14 | 0.0301 |
| Chodl | Chondrolectin | 1.45 | −0.23 | 0.0305 |
| 1700020I14Rik | RIKEN cDNA 1700020I14 gene | 0.32 | −0.06 | 0.0306 |
| Heatr5b | HEAT repeat containing 5B | 0.17 | −0.01 | 0.0308 |
| Ppargc1a | Peroxisome proliferative activated receptor, gamma, coactivator 1 α | 0.29 | −0.12 | 0.0308 |
| Fblim1 | Filamin binding LIM protein 1 | 0.83 | −0.34 | 0.0321 |
| Grin3b | Glutamate receptor, ionotropic, NMDA3B | 0.90 | −0.19 | 0.0336 |
| Nrip3 | Nuclear receptor-interacting protein 3 | 0.24 | −0.06 | 0.0404 |
| 9330132A10Rik | RIKEN cDNA 9330132A10 gene | 0.22 | −0.07 | 0.0404 |
| Spp2 | Secreted phosphoprotein 2 | 1.71 | −0.70 | 0.0404 |
| Vdr | Vitamin D (1,25-dihydroxyvitamin D3) receptor | 2.43 | −0.12 | 0.0479 |
| Eif5a2 | Eukaryotic translation initiation factor 5A2 | 0.23 | −0.08 | 0.0490 |
| Inpp5j | Inositol polyphosphate 5-phosphatase J | 0.39 | −0.07 | 0.0496 |
Multifactor analysis reveals that a set of 62 genes are upregulated by CFA treatment in the RGS4WT group, whereas these genes are downregulated in the RGS4KO group. Table 1 contains genes with interaction-adjusted p < 0.05.
Table 2.
A role of RGS4 in CFA-induced gene downregulation
| Gene | Description | RGS4WT day 18 post-CFA | RGS4KO day 18 post-CFA | Interaction padj |
|---|---|---|---|---|
| Nup210 | Nucleoporin 210 | −0.297 | 0.046 | 0.0112 |
| Rps6ka2 | Ribosomal protein S6 kinase, polypeptide 2 | −0.212 | 0.060 | 0.0053 |
| Ngfr | Nerve growth factor receptor (TNF receptor superfamily, member 16) | −0.776 | 0.305 | 0.0017 |
| Pde2a | Phosphodiesterase 2A, cGMP-stimulated | −0.429 | 0.234 | 0.0003 |
| Muc6 | Mucin 6, gastric | −1.319 | 0.132 | 0.0479 |
| Rab36 | RAB36, member RAS oncogene family | −0.239 | 0.040 | 0.0404 |
| Wdfy4 | WD repeat and FYVE domain containing 4 | −0.525 | 0.113 | 0.0411 |
| Hdac6 | Histone deacetylase 6 | −0.178 | 0.072 | 0.0104 |
| Cplx2 | Complexin 2 | −0.213 | 0.108 | 0.0045 |
| Megf6 | Multiple EGF-like-domains 6 | −0.449 | 0.250 | 0.0055 |
| Cml5 | NA | −0.961 | 0.459 | 0.0190 |
| Id4 | Inhibitor of DNA binding 4 | −0.351 | 0.164 | 0.0171 |
| Pygl | Liver glycogen phosphorylase | −0.338 | 0.145 | 0.0371 |
| Stk30 | NA | −0.255 | 0.128 | 0.0329 |
| Cryba2 | Crystallin, β A2 | −1.248 | 1.126 | 0.0053 |
| Scn7a | Sodium channel, voltage-gated, type VII, α | −0.279 | 0.173 | 0.0267 |
| Gpc4 | Glypican 4 | −0.425 | 0.330 | 0.0175 |
| Tpra1 | Transmembrane protein, adipocyte-associated 1 | −0.130 | 0.148 | 0.0066 |
| Esr2 | Estrogen receptor 2 (β) | −0.681 | 0.675 | 0.0171 |
| Rgag1 | Retrotransposon gag domain containing 1 | −0.333 | 0.315 | 0.0306 |
| Stk32b | Serine/threonine kinase 32B | −0.320 | 0.303 | 0.0308 |
| Gpr15 | G-protein-coupled receptor 15 | −1.075 | 1.402 | 0.0183 |
| Cdh23 | Cadherin 23 (otocadherin) | −0.411 | 0.710 | 0.0046 |
| Sfi1 | Sfi1 homolog, spindle assembly associated (yeast) | −0.097 | 0.163 | 0.0092 |
| Crhr2 | Corticotropin-releasing hormone receptor 2 | −0.717 | 1.204 | 0.0171 |
| Adhfe1 | Alcohol dehydrogenase, iron containing, 1 | −0.061 | 0.259 | 0.0485 |
| Hjurp | Holliday junction recognition protein | −0.028 | 0.870 | 5.87E-19 |
Multifactor analysis reveals that a set of 27 genes are downregulated by CFA treatment in the THL of RGS4WT mice while they are upregulated in the THL of RGS4KO groups. Table 2 contains genes with interaction adjusted p < 0.05. NA, Not available.
Discussion
Our study reveals an essential role of RGS4 in the maintenance of chronic pain states. Using murine models of peripheral inflammation and nerve injury, we show that knockout of the RGS4 gene leads to recovery from tactile and cold hypersensitivity and improves voluntary wheel running. RGS4 is widely expressed in pain-modulating circuits, but the downregulation of RGS4 in the VPL-THL of adult mice is sufficient to promote recovery from mechanical and cold hypersensitivity.
A role of RGS4 in the maintenance of sensory hypersensitivity
Given that GPCRs play a major role in pain sensitivity and pain chronicity (Stone and Molliver, 2009; Veldhuis et al., 2015), we expect that potent modulators of GPCR signaling and desensitization such as RGS proteins affect the development of pathological pain symptoms and the maintenance of chronic pain states. We selected RGS4 for study as this protein is expressed at various levels of the pain matrix, and recent evidence has highlighted its modulatory role in the function of opioid and monoamine GPCRs (Gold et al., 2003; Georgoussi et al., 2006; Han et al., 2010; Stratinaki et al., 2013; Yoon et al., 2015). Using genetic tools that we have available in our laboratory, we assessed the consequences of RGS4 gene inactivation in hypersensitivity behaviors associated with long-term peripheral inflammation or nerve injury. These tools permit the investigation of the functional significance of RGS4 in sensory hypersensitivity at early and later time points after the induction of pain-like states. Interestingly, RGS4 gene knockout has no effect on Von Frey or cold plate responses during the first week of peripheral inflammation or during the first 3 weeks of nerve injury. These findings highlight the role of RGS4 pathways in the maintenance, but not in the induction or the intensity, of mechanical/cold allodynia. Studies using the formalin assay also support a role for RGS4 in the maintenance of sensory hypersensitivity.
While we expect that RGS4 controls central sensitization at several pain-modulating circuits, our findings demonstrate a critical role of RGS4 in the VPL-THL in pain chronicity. The VPL is a component of the spinothalamic and trigeminothalamic tracts with major impact on central sensitization. Notably, the downregulation of RGS4 in the MD-THL did not have any influence on the trajectory of mechanical allodynia associated with prolonged hindpaw inflammation, highlighting the specificity of RGS4 actions within THL nuclei. The NAc has also been shown to play a prominent role in the processing of sensory and affective pain symptoms and in the modulation of the antiallodynic efficacy of antidepressants (Baliki et al., 2010; Mitsi et al., 2015; Mitsi and Zachariou, 2016); however, the downregulation of RGS4 in this brain region was not sufficient to prevent the maintenance of mechanical allodynia. Future studies will further investigate the impact of chronic pain states on RGS4 expression in other brain regions mediating nociceptive processing, including the medial prefrontal cortex, the anterior cingulate, somatosensory cortices, and the locus ceruleus. To this extent, we will assess the circuit-specific actions of RGS4 in sensory, cognitive, and affective symptoms of chronic pain. We will also cross the RGS4fl\fl mice with lines expressing Cre recombinase under the control of specific promoters to understand the function of RGS4 in specific cell types and circuits.
RGS4 knockout affects the expression of several GPCRs involved in central sensitization, including noradrenergic, metabotropic glutamate, and GABA receptors (Ding et al., 2006; Fowler et al., 2007; McGinty et al., 2008; Shen et al., 2015). Future work will apply pharmacology and electrophysiology approaches to identify receptors that are regulated by RGS4 under chronic pain states.
RGS4 in acute pain and actions of analgesic drugs
RGS4 activity does not affect acute pain responses. Knockout of RGS4 did not affect nociceptive behaviors during the first phase of the formalin test, whereas the Von Frey and Hargreaves latencies of naive RGS4KO mice were indistinguishable from those of RGS4WT controls. While the blockade of RGS4 action promotes recovery from chronic pain symptoms, RGS4 acts as a positive modulator of many analgesic drugs: in models of acute pain, RGS4KO mice show lower responses to the opioids fentanyl and methadone, while they respond to morphine in a similar manner to that of RGS4WT controls (Han et al., 2010). In studies evaluating the impact of RGS4 in the antiallodynic effects of the tricyclic antidepressant desipramine, the ablation of the RGS4 gene reduced the antiallodynic efficacy of the drug within the first 3 weeks after SNI induction (when no genotype differences are observed in Von Frey assay; Stratinaki et al., 2013). These findings highlight the complex role of RGS4 and downstream pathways in physiological modulation of long-term pain states and in the efficacy of pain-alleviating drugs.
Regulation of chronic pain by RGS proteins
While numerous studies demonstrate the implication of RGS proteins in physiological processes and pathological conditions (Zachariou et al., 2003; Ding et al., 2006; Levitt et al., 2006; Terzi et al., 2014; Gerber et al., 2016), there is limited information on the role of RGS proteins in pain transmission, perception, and modulation. Earlier studies in our group revealed that the prevention of RGS9–2 action delayed the development of maximal mechanical allodynia the first week after the induction of nerve injury (Terzi et al., 2014). However, after this time point, RGS9–2 did not have any influence on sensory hypersensitivity. Another component of RGS9–2 and RGS7 protein complexes, the adaptor molecule R7BP, has been reported to modulate sensitivity to acute thermal stimuli (Zhou et al., 2012). Previous studies have reported that intrathecal application of RGS4 antagonists in rats attenuated the development of mechanical allodynia (Bosier et al., 2015; Yoon et al., 2015; Taccola et al., 2016). These studies support a role of RGS4 in nociceptive processing in the dorsal root ganglia and spinal cord. The presence of RGS4 in both spinal and supraspinal sites further highlights the importance of this molecule in the modulation of chronic pain symptoms and suggests that the inhibition of RGS4 may provide a powerful therapeutic tool. Notably, our data reveal that long-term peripheral inflammation downregulates RGS4 in the lumbar spinal cord. Future studies will delineate the functional role of RGS4 in the dorsal spinal cord and dorsal root ganglia. As many of the brain regions expressing RGS4 also control mood and motivation, future work will determine the role of RGS4 in anxiodepressive symptoms and motivation-related behaviors.
The role of RGS4 in signal transduction and gene expression under chronic pain states
We demonstrate that long-term pain states promote the expression of RGS4, and that this effect contributes to the maintenance of long-term pain symptoms. By controlling the availability of Gα and Gβγ subunits to effector molecules, RGS4 may affect the function of several phosphoproteins, ion channels, and transcription factors mediating sensory hypersensitivity.
We used next-generation sequencing to gain insight into genes and pathways affected by long-term peripheral inflammation and RGS4 knockout in the mouse THL. These experiments highlighted genes and intracellular pathways implicated in long-term pain states, and supported our hypothesis that RGS4 activity promotes synaptic events that contribute to sensory hypersensitivity. It is well established that chronic pain states promote the function of NMDA, AMPA, and mGluRs in various regions of the pain matrix (Miyata et al., 2003; Qiu et al., 2014). Our RNA-Seq analysis demonstrates that genes related to glutamatergic activity and signal transduction, including ionotropic glutamate receptors GRIK1, GRIK2, GRIN2D, and GRIA1; metabotropic glutamate receptors GRM2, GRM4, and GRM8; and homer scaffolding protein 1 (homer1) are differentially regulated between RGS4WT and RGS4KO groups. Many of these genes are downregulated in the absence of RGS4, indicating altered glutamatergic signaling in the thalamus of RGS4KO groups at time points that correlate with recovery from sensory hypersensitivity symptoms (Tables 3). Recovery from chronic pain states has been associated with increased expression of the Gαi/o-coupled mGluR2s (Chiechio et al., 2010). Indeed, our Western blot analysis demonstrates that recovery from mechanical allodynia in RGS4KO groups is associated with elevated mGluR2 levels in THL synaptosomes.
Table 3.
IPA predicts a prominent effect of RGS4 in the expression of genes involved in glutamatergic signaling
| Gene | Description | Naive RGS4KO vs RGS4WT | RGS4WT CFA-treated vs naive | RGS4KO CFA-treated vs naive | CFA-treated RGS4KO vs RGS4WT |
|---|---|---|---|---|---|
| Calm3 | Calmodulin 1 | 0.491 | |||
| Camk4 | Calcium/calmodulin-dependent protein kinase IV | −0.679 | −0.458 | ||
| Gng2 | G-Protein subunit gamma 2 | −0.378 | |||
| Gria2 | Glutamate ionotropic receptor AMPA type subunit 2 | −0.456 | −0.359 | ||
| Gria3 | Glutamate ionotropic receptor AMPA type subunit 3 | −0.310 | −0.257 | ||
| Grik1 | Glutamate ionotropic receptor kainate type subunit 1 | −0.336 | −0.366 | ||
| Grik2 | Glutamate ionotropic receptor kainate type subunit 2 | −0.414 | |||
| Grik5 | Glutamate ionotropic receptor kainate type subunit 5 | 0.641 | |||
| Grin1 | Glutamate ionotropic receptor NMDA type subunit 1 | 0.571 | |||
| Grin2d | Glutamate ionotropic receptor NMDA type subunit 2D | 0.578 | |||
| Grm2 | Glutamate metabotropic receptor 2 | 0.877 | |||
| Grm4 | Glutamate metabotropic receptor 4 | 0.832 | |||
| Grm8 | Glutamate metabotropic receptor 8 | −0.373 | −0.413 | ||
| Homer1 | Homer scaffolding protein 1 | −0.258 | |||
| Pick1 | Protein interacting with PRKCA 1 | 0.429 | |||
| Gng11 | G-Protein subunit gamma 11 | −0.258 | |||
| Gria1 | Glutamate ionotropic receptor AMPA type subunit 1 | 0.286 | |||
| Slc17a8 | Solute carrier family 17 member 8 | 0.894 | |||
| Slc1a6 | Solute carrier family 1 member 6 | 0.333 |
Several ionotropic subunits and metabotropic receptors are downregulated in the RGS4KO group. Only genes with p values <0.05 are shown.
In addition to the critical role of long-term potentiation (LTP), alterations in synaptic long-term depression (LTD) also contribute to the establishment of chronic pain states (Ikeda et al., 2006; Schwartz et al., 2014; Pradier et al., 2018). In particular, LTD in the GABAergic spinal cord interneurons drives LTP in projection neurons, resulting in mechanical pain hypersensitivity (Kim et al., 2015b). Also, electrophysiology data note the role of LTD in the anterior cingulate cortex in sensory hypersensitivity (Toyoda et al., 2005, 2009). Interestingly, our IPA suggests the modulation of synaptic LTD components, G-protein-coupled receptor, Gαi, and inflammation-related pathways such as STAT3 and nuclear factor (NF)-κB in CFA groups of RGS4WT cohorts, which show maximal allodynia (Table 3, 4). Human studies also document that the loss of GABAergic inhibition in thalamocortical circuits affects pain perception (Henderson et al., 2013).
Table 4.
Top differentially regulated molecular pathways by peripheral inflammation in RGS4WT
| Canonical pathway | CFA effect RGS4KO vs RGS4WT | Genotype effect RGS4KO | CFA effect RGS4KO | CFA effect RGS4WT |
|---|---|---|---|---|
| STAT3 pathway | ND | 1.9 | 0.3 | 3.2 |
| Thyroid cancer signaling | 0.6 | 2.3 | ND | 3.1 |
| Synaptic long-term depression | 1.2 | 0.7 | 0.5 | 3.1 |
| Hepatic fibrosis/hepatic stellate cell activation | 0.8 | 1.3 | 2.1 | 2.9 |
| VEGF family ligand-receptor interactions | 0.3 | 0.7 | 0.4 | 2.9 |
| Gap junction signaling | 0.3 | 1.7 | ND | 2.7 |
| Agranulocyte adhesion and diapedesis | ND | ND | 0.3 | 2.7 |
| NF-κB signaling | ND | 1.6 | ND | 2.6 |
| Antiproliferative role of somatostatin receptor 2 | 0.4 | 1.0 | ND | 2.5 |
| HIF1α signaling | ND | 1.7 | ND | 2.5 |
| IL-6 signaling | ND | 1.7 | ND | 2.4 |
| Axonal guidance signaling | 1.9 | 6.6 | ND | 2.4 |
| Relaxin signaling | 0.3 | 0.6 | ND | 2.4 |
| PAK signaling | ND | 3.3 | ND | 2.4 |
| Role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis | 1.1 | 1.5 | ND | 2.3 |
| Cellular effects of sildenafil (Viagra) | 1.0 | 0.6 | ND | 2.3 |
| Cardiac hypertrophy signaling | 0.3 | 3.2 | ND | 2.3 |
| UVC-induced MAPK signaling | ND | 1.8 | 0.3 | 2.3 |
| Endothelin-1 signaling | ND | 0.8 | 1.1 | 2.3 |
| BMP signaling pathway | 0.2 | 2.2 | 0.4 | 2.2 |
| Gβγ signaling | 0.8 | 1.0 | ND | 2.2 |
| IL-8 signaling | ND | 2.7 | ND | 2.1 |
| Gαi signaling | 0.5 | 0.8 | 0.5 | 2.1 |
| G-protein-coupled receptor signaling | 1.4 | 0.4 | ND | 2.1 |
IPA points to several signaling pathways affected by CFA in RGS4WT but not in RGS4KO. Pathways are ranked -log10 p value for the CFA-treated RGS4WT group in descending order. ND, Pathways that remained unchanged; HIF1α, hypoxia-inducible factor-1α. UVC-induced MAPK signaling: UVC stands for UV (ultraviolet) radiation in a specific spectrum C (200–280 nm).
Our bioinformatics analysis suggests that inflammatory pain states in RGS4WT affect the expression transcription factors, such as CREB1 (log2fold change = −0.26, p = 0.02), Jun (log2fold change = 0.39, p = 0.04), and Sirt1 (log2fold change = −0.37, p = 0.01) have been shown by various studies to promote transcriptional events that facilitate chronic pain states (Doya et al., 2005; Descalzi et al., 2012; Zhou et al., 2012). Several ion channels are contraregulated by CFA treatment in RGS4WT versus RGS4KO, including HCN1 (Resta et al., 2018) the potassium voltage-gated Kcng4, and calcium-gated Kcnn1 channels (Table 1). Furthermore, our analysis reveals that peripheral inflammation affects the expression of a number of nucleotide binding proteins in the RGS4WT cohort, including Eph receptors, inositol-trisphosphate 3-kinase A, and guanylate cyclase isoforms, which are well documented contributors of central sensitization (Duan et al., 2012; Yu et al., 2012; Lee et al., 2015). Components of the nitric oxide/cGMP pathway, which is a key mediator of central sensitization (Lee et al., 2018) are also identified in the list of genes contraregulated between genotypes at 18 d after CFA treatment (Table 2).
Conclusions
In summary, our findings reveal a prominent role of RGS4 in the maintenance of long-term pain states. Using genetically modified mice, we show that while the onset of pathological pain symptoms is not affected by RGS4 activity, the maintenance of mechanical and cold allodynia is dynamically modulated by RGS4 actions. Our findings support the notion that distinct intracellular mechanisms promote sensory hypersensitivity at early versus later time points after nerve injury or peripheral inflammation, and point to RGS4 as a critical mediator of pain chronicity. Our findings from RNA sequencing analysis suggest that the inhibition of RGS4 activity restores gene expression maladaptations associated with central sensitization and point to novel intracellular pathways that can be targeted for the management of chronic pain.
Footnotes
This study was supported by National Institute of Neurological Disorders and Stroke Grants NS-086444 and NS-093537 (to V.Z.); and National Institute on Drug Abuse Grants PPG-POIDAO8227 (to V.Z.) and T32-5T32-DA007135-34 (to K.D.P.). C.P. is a recipient of the Irene and Eric Simon Brain Research Foundation Summer Fellowship (2016).
The authors declare no competing financial interests.
References
- Ajit SK, Ramineni S, Edris W, Hunt RA, Hum WT, Hepler JR, Young KH (2007) RGSZ1 interacts with protein kinase C interacting protein PKCI-1 and modulates mu opioid receptor signaling. Cell Signal 19:723–730. 10.1016/j.cellsig.2006.09.008 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics 32:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliki MN, Apkarian AV (2015) Nociception, pain, negative moods, and behavior selection. Neuron 87:479–491. 10.1016/j.neuron.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliki MN, Geha PY, Fields HL, Apkarian AV (2010) Predicting value of pain and analgesia: nucleus accumbens response to noxious stimuli changes in the presence of chronic pain. Neuron 66:149–160. 10.1016/j.neuron.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin G, Singh K, Dissanayake K, Mighiu AS, Nurmohamed A, Heximer SP (2012) Amino-terminal cysteine residues differentially influence RGS4 protein plasma membrane targeting, intracellular trafficking, and function. J Biol Chem 287:28966–28974. 10.1074/jbc.M112.345629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosier B, Doyen PJ, Brolet A, Muccioli GG, Ahmed E, Desmet N, Hermans E, Deumens R (2015) Inhibition of the regulator of G protein signalling RGS4 in the spinal cord decreases neuropathic hyperalgesia and restores cannabinoid CB1receptor signalling. Br J Pharmacol 172:5333–5346. 10.1111/bph.13324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton JJ, Allen PB, Psifogeorgou K, Chakravarty S, Gomes I, Neve RL, Devi LA, Greengard P, Nestler EJ, Zachariou V (2008) Multiple actions of spinophilin regulate mu opioid receptor function. Neuron 58:238–247. 10.1016/j.neuron.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiechio S, Copani A, Zammataro M, Battaglia G, Gereau RW 4th, Nicoletti F (2010) Transcriptional regulation of type-2 metabotropic glutamate receptors: an epigenetic path to novel treatments for chronic pain. Trends Pharmacol Sci 31:153–160. 10.1016/j.tips.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Cliffer KD, Burstein R, Giesler GJ Jr (1991) Distributions of spinothalamic, spinohypothalamic, and spinotelencephalic fibers revealed by anterograde transport of PHA-L in rats. J Neurosci 11:852–868. 10.1523/JNEUROSCI.11-03-00852.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos EJ, Ghasemlou N, Araldi D, Segal D, Duong K, Woolf CJ (2012) Inflammation-induced decrease in voluntary wheel running in mice: a nonreflexive test for evaluating inflammatory pain and analgesia. Pain 153:876–884. 10.1016/j.pain.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AD Jr, Burton H (1981) Spinal and medullary lamina I projection to nucleus submedius in medial thalamus: a possible pain center. J Neurophysiol 45:443–466. 10.1152/jn.1981.45.3.443 [DOI] [PubMed] [Google Scholar]
- Descalzi G, Fukushima H, Suzuki A, Kida S, Zhuo M (2012) Genetic enhancement of neuropathic and inflammatory pain by forebrain upregulation of CREB-mediated transcription. Mol Pain 8:90. 10.1186/1744-8069-8-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descalzi G, Mitsi V, Purushothaman I, Gaspari S, Avrampou K, Loh YH, Shen L, Zachariou V (2017) Neuropathic pain promotes adaptive changes in gene expression in brain networks involved in stress and depression. Sci Signal 10:eaaj1549. 10.1126/scisignal.aaj1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Guzman JN, Tkatch T, Chen S, Goldberg JA, Ebert PJ, Levitt P, Wilson CJ, Hamm HE, Surmeier DJ (2006) RGS4-dependent attenuation of M4autoreceptor function in striatal cholinergic interneurons following dopamine depletion. Nat Neurosci 9:832–842. 10.1038/nn1700 [DOI] [PubMed] [Google Scholar]
- Doya H, Ohtori S, Fujitani M, Saito T, Hata K, Ino H, Takahashi K, Moriya H, Yamashita T (2005) C-jun N-terminal kinase activation in dorsal root ganglion contributes to pain hypersensitivity. Biochem Biophys Res Commun 335:132–138. 10.1016/j.bbrc.2005.07.055 [DOI] [PubMed] [Google Scholar]
- Duan B, Liu DS, Huang Y, Zeng WZ, Wang X, Yu H, Zhu MX, Chen ZY, Xu TL (2012) PI3-kinase/Akt pathway-regulated membrane insertion of acid-sensing ion channel 1a underlies BDNF-induced pain hypersensitivity. J Neurosci 32:6351–6363. 10.1523/JNEUROSCI.4479-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CE, Aryal P, Suen KF, Slesinger PA (2007) Evidence for association of GABAB receptors with Kir3 channels and regulators of G protein signalling (RGS4) proteins. J Physiol 580:51–65. 10.1113/jphysiol.2006.123216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard S, Lo Re L, Mantilleri A, Hepp R, Urien L, Malapert P, Alonso S, Deage M, Kambrun C, Landry M, Low SA, Alloui A, Lambolez B, Scherrer G, Le Feuvre Y, Bourinet E, Moqrich A (2014) GINIP, a Gαi-interacting protein, functions as a key modulator of peripheral GABABreceptor-mediated analgesia. Neuron 84:123–136. 10.1016/j.neuron.2014.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspari S, Purushothaman I, Cogliani V, Sakloth F, Neve RL, Howland D, Ring RH, Ross EM, Shen L, Zachariou V (2018) Suppression of RGSz1 function optimizes the actions of opioid analgesics by mechanisms that involve the Wnt/β-catenin pathway. Proc Natl Acad Sci U S A 115:E2085–E2094. 10.1073/pnas.1707887115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgoussi Z, Leontiadis L, Mazarakou G, Merkouris M, Hyde K, Hamm H (2006) Selective interactions between G protein subunits and RGS4 with the C-terminal domains of the mu- and delta-opioid receptors regulate opioid receptor signaling. Cell Signal 18:771–782. 10.1016/j.cellsig.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Gerber KJ, Squires KE, Hepler JR (2016) Roles for regulator of G protein signaling proteins in synaptic signaling and plasticity. Mol Pharmacol 89:273–286. 10.1124/mol.115.102210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Ni YG, Dohlman HG, Nestler EJ (1997) Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci 17:8024–8037. 10.1523/JNEUROSCI.17-20-08024.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold SJ, Heifets BD, Pudiak CM, Potts BW, Nestler EJ (2002) Regulation of regulators of G protein signaling mRNA expression in rat brain by acute and chronic electroconvulsive seizures. J Neurochem 82:828–838. 10.1046/j.1471-4159.2002.01002.x [DOI] [PubMed] [Google Scholar]
- Gold SJ, Han MH, Herman AE, Ni YG, Pudiak CM, Aghajanian GK, Liu RJ, Potts BW, Mumby SM, Nestler EJ (2003) Regulation of RGS proteins by chronic morphine in rat locus coeruleus. Eur J Neurosci 17:971–980. 10.1046/j.1460-9568.2003.02529.x [DOI] [PubMed] [Google Scholar]
- Han MH, Renthal W, Ring RH, Rahman Z, Psifogeorgou K, Howland D, Birnbaum S, Young K, Neve R, Nestler EJ, Zachariou V (2010) Brain region specific actions of regulator of G protein signaling 4 oppose morphine reward and dependence but promote analgesia. Biol Psychiatry 67:761–769. 10.1016/j.biopsych.2009.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LA, Peck CC, Petersen ET, Rae CD, Youssef AM, Reeves JM, Wilcox SL, Akhter R, Murray GM, Gustin SM (2013) Chronic pain: lost inhibition? J Neurosci 33:7574–7582. 10.1523/JNEUROSCI.0174-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Lu P, Cummons TA, Whiteside GT (2008) The persistence of a long-term negative affective state following the induction of either acute or chronic pain. Pain 140:436–445. 10.1016/j.pain.2008.09.020 [DOI] [PubMed] [Google Scholar]
- Ikeda H, Stark J, Fischer H, Wagner M, Drdla R, Jäger T, Sandkühler J (2006) Synaptic amplifier of inflammatory pain in the spinal dorsal horn. Science 312:1659–1662. 10.1126/science.1127233 [DOI] [PubMed] [Google Scholar]
- Ji RR, Nackley A, Huh Y, Terrando N, Maixner W (2018) Neuroinflammation and central sensitization in chronic and widespread pain. Anesthesiology 129:343–366. 10.1097/ALN.0000000000002130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015a) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Tillu DV, Quinn TL, Mejia GL, Shy A, Asiedu MN, Murad E, Schumann AP, Totsch SK, Sorge RE, Mantyh PW, Dussor G, Price TJ (2015b) Spinal dopaminergic projections control the transition to pathological pain plasticity via a D1/D5-mediated mechanism. J Neurosci 35:6307–6317. 10.1523/JNEUROSCI.3481-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuner R, Flor H (2016) Structural plasticity and reorganisation in chronic pain. Nat Rev Neurosci 18:20–30. 10.1038/nrn.2016.162 [DOI] [PubMed] [Google Scholar]
- Latremoliere A, Woolf CJ (2009) Central sensitization: a generator of pain hypersensitivity by central neural plasticity. J Pain 10:895–926. 10.1016/j.jpain.2009.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Xu Z, Ashpole NM, Hudmon A, Kulkarni PM, Thakur GA, Lai YY, Hohmann AG (2015) Small molecule inhibitors of PSD95-nNOS protein-protein interactions as novel analgesics. Neuropharmacology 97:464–475. 10.1016/j.neuropharm.2015.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Li LL, Chawla A, Hudmon A, Lai YY, Courtney MJ, Hohmann AG (2018) Disruption of nNOS–NOS1AP protein–protein interactions suppresses neuropathic pain in mice. Pain 159:849–863. 10.1097/j.pain.0000000000001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Kreitzer AC (2012) RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron 73:347–359. 10.1016/j.neuron.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt P, Ebert P, Mirnics K, Nimgaonkar VL, Lewis DA (2006) Making the case for a candidate vulnerability gene in schizophrenia: convergent evidence for regulator of G-protein signaling 4 (RGS4). Biol Psychiatry 60:534–537. 10.1016/j.biopsych.2006.04.028 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masocha W. (2014) Paclitaxel-induced hyposensitivity to nociceptive chemical stimulation in mice can be prevented by treatment with minocycline. Sci Rep 4:6719. 10.1038/srep06719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathey EK, Park SB, Hughes RA, Pollard JD, Armati PJ, Barnett MH, Taylor BV, Dyck PJ, Kiernan MC, Lin CS (2015) Chronic inflammatory demyelinating polyradiculoneuropathy: from pathology to phenotype. J Neurol Neurosurg Psychiatry 86:973–985. 10.1136/jnnp-2014-309697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty JF, Shi XD, Schwendt M, Saylor A, Toda S (2008) Regulation of psychostimulant-induced signaling and gene expression in the striatum. J Neurochem 10:1440–1449. 10.1111/j.1471-4159.2008.05240.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda KS, Patel T, Braz JM, Malik R, Turner ML, Seifikar H, Basbaum AI, Sohal VS (2019) Microcircuit mechanisms through which mediodorsal thalamic input to anterior cingulate cortex exacerbates pain-related aversion article microcircuit mechanisms through which mediodorsal thalamic input to anterior cingulate cortex exacerbates pain-related. Neuron 102:944–959.e3. 10.1016/j.neuron.2019.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megat S, Ray PR, Moy JK, Lou TF, Barragán-Iglesias P, Li Y, Pradhan G, Wanghzou A, Ahmad A, Burton MD, North RY, Dougherty PM, Khoutorsky A, Sonenberg N, Webster KR, Dussor G, Campbell ZT, Price TJ (2019) Nociceptor translational profiling reveals the Ragulator-Rag GTPase complex as a critical generator of neuropathic pain. J Neurosci 39:393–411. 10.1523/JNEUROSCI.2661-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minett MS, Nassar MA, Clark AK, Passmore G, Dickenson AH, Wang F, Malcangio M, Wood JN (2012) Distinct Nav1.7-dependent pain sensations require different sets of sensory and sympathetic neurons. Nat Commun 3:791. 10.1038/ncomms1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsi V, Zachariou V (2016) Modulation of pain, nociception, and analgesia by the brain reward center. Neuroscience 338:81–92. 10.1016/j.neuroscience.2016.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsi V, Terzi D, Purushothaman I, Manouras L, Gaspari S, Neve RL, Stratinaki M, Feng J, Shen L, Zachariou V (2015) RGS9–2-controlled adaptations in the striatum determine the onset of action and efficacy of antidepressants in neuropathic pain states. Proc Natl Acad Sci U S A 112:E5088–E5097. 10.1073/pnas.1504283112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Kashiwadani H, Fukaya M, Hayashi T, Wu D, Suzuki T, Watanabe M, Kawakami Y (2003) Role of thalamic phospholipase C[beta]4 mediated by metabotropic glutamate receptor type 1 in inflammatory pain. J Neurosci 23:8098–8108. 10.1523/JNEUROSCI.23-22-08098.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni YG, Gold SJ, Iredale PA, Terwilliger RZ, Duman RS, Nestler EJ (1999) Region-specific regulation of RGS4 in brain by stress and glucocorticoids: in vivo and in vitro studies. J Neurosci 19:3674–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notartomaso S, Mascio G, Bernabucci M, Zappulla C, Scarselli P, Cannella M, Imbriglio T, Gradini R, Battaglia G, Bruno V, Nicoletti F (2017) Analgesia induced by the epigenetic drug, L-acetylcarnitine, outlasts the end of treatment in mouse models of chronic inflammatory and neuropathic pain. Mol Pain 13:1744809176917009. 10.1177/1744806917697009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipov MH, Lai J, Malan TP Jr, Porreca F (2000) Spinal and supraspinal mechanisms of neuropathic pain. Ann N Y Acad Sci 909:12–24. 10.1111/j.1749-6632.2000.tb06673.x [DOI] [PubMed] [Google Scholar]
- Ossipov MH, Dussor GO, Porreca F (2010) Central modulation of pain. J Clin Invest 120:3779–3787. 10.1172/JCI43766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papachatzaki MM, Antal Z, Terzi D, Szücs P, Zachariou V, Antal M (2011) RGS9–2 modulates nociceptive behaviour and opioid-mediated synaptic transmission in the spinal dorsal horn. Neurosci Lett 501:31–34. 10.1016/j.neulet.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnaudeau S, Bolkan SS, Kellendonk C (2018) The mediodorsal thalamus: an essential partner of the prefrontal cortex for cognition. Biol Psychiatry 83:648–656. 10.1016/j.biopsych.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan AM, Scotland PE, Akopian AN, Hargreaves KM (2009) Activation of TRPV1 in the spinal cord by oxidized linoleic acid metabolites contributes to inflammatory hyperalgesia. Proc Natl Acad Sci U S A 106:18820–18824. 10.1073/pnas.0905415106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradier B, Shin HB, Kim DS, St Laurent R, Lipscombe D, Kauer JA (2018) Long-term depression induced by optogenetically driven nociceptive inputs to trigeminal nucleus caudalis or headache triggers. J Neurosci 38:7529–7540. 10.1523/JNEUROSCI.3032-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Slotnick BM (1983) Dual olfactory representation in the rat thalamus: an anatomical and electrophysiological study. J Comp Neurol 215:63–77. 10.1002/cne.902150106 [DOI] [PubMed] [Google Scholar]
- Qiu S, Zhang M, Liu Y, Guo Y, Zhao H, Song Q, Zhao M, Huganir RL, Luo J, Xu H, Zhuo M (2014) GluA1 phosphorylation contributes to postsynaptic amplification of neuropathic pain in the insular cortex. J Neurosci 34:13505–13515. 10.1523/JNEUROSCI.1431-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichling DB, Levine JD (2009) Critical role of nociceptor plasticity in chronic pain. Trends Neurosci 32:611–618. 10.1016/j.tins.2009.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resta F, Micheli L, Laurino A, Spinelli V, Mello T, Sartiani L, Di Cesare Mannelli L, Cerbai E, Ghelardini C, Romanelli MN, Mannaioni G, Masi A (2018) Selective HCN1 block as a strategy to control oxaliplatin-induced neuropathy. Neuropharmacology 131:403–413. 10.1016/j.neuropharm.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Rodriguez E, Sakurai K, Xu J, Chen Y, Toda K, Zhao S, Han BX, Ryu D, Yin H, Liedtke W, Wang F (2017) A craniofacial-specific monosynaptic circuit enables heightened affective pain. Nat Neurosci 20:1734–1743. 10.1038/s41593-017-0012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeckel LA, Utard V, Reiss D, Mouheiche J, Maurin H, Robé A, Audouard E, Wood JN, Goumon Y, Simonin F, Gaveriaux-Ruff C (2017) Morphine-induced hyperalgesia involves mu opioid receptors and the metabolite morphine-3-glucuronide. Sci Rep 7:10406. 10.1038/s41598-017-11120-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz N, Temkin P, Jurado S, Lim BK, Heifets BD, Polepalli JS, Malenka RC (2014) Decreased motivation during chronic pain requires long-term depression in the nucleus accumbens. Science 345:535–542. 10.1126/science.1253994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendt M, McGinty JF (2007) Regulator of G-protein signaling 4 interacts with metabotropic glutamate receptor subtype 5 in rat striatum: relevance to amphetamine behavioral sensitization. J Pharmacol Exp Ther 323:650–657. 10.1124/jpet.107.128561 [DOI] [PubMed] [Google Scholar]
- Shen W, Plotkin JL, Francardo V, Ko WK, Xie Z, Li Q, Fieblinger T, Wess J, Neubig RR, Lindsley CW, Conn PJ, Greengard P, Bezard E, Cenci MA, Surmeier DJ (2015) M4 muscarinic receptor signaling ameliorates striatal plasticity deficits in models of L-DOPA-induced dyskinesia. Neuron 88:762–773. 10.1016/j.neuron.2015.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields SD, Eckert WA 3rd, Basbaum AI (2003) Spared nerve injury model of neuropathic pain in the mouse: a behavioral and anatomic analysis. J Pain 4:465–470. 10.1067/S1526-5900(03)00781-8 [DOI] [PubMed] [Google Scholar]
- Stone LS, Molliver DC (2009) In search of analgesia: emerging poles of GPCRs in pain. Mol Interv 9:234–251. 10.1124/mi.9.5.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratinaki M, Varidaki A, Mitsi V, Ghose S, Magida J, Dias C, Russo SJ, Vialou V, Caldarone BJ, Tamminga CA, Nestler EJ, Zachariou V (2013) Regulator of G protein signaling 4 is a crucial modulator of antidepressant drug action in depression and neuropathic pain models. Proc Natl Acad Sci U S A 110:8254–8259. 10.1073/pnas.1214696110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taccola G, Doyen PJ, Damblon J, Dingu N, Ballarin B, Steyaert A, Rieux AD, Forget P, Hermans E, Bosier B, Deumens R (2016) A new model of nerve injury in the rat reveals a role of regulator of G protein signaling 4 in tactile hypersensitivity. Exp Neurol 286:1–11. 10.1016/j.expneurol.2016.09.008 [DOI] [PubMed] [Google Scholar]
- Terzi D, Stergiou E, King SL, Zachariou V (2009) Chapter 10 regulators of G protein signaling in neuropsychiatric disorders. Prog Mol Biol Transl Sci 86:299–333. 10.1016/S1877-1173(09)86010-9 [DOI] [PubMed] [Google Scholar]
- Terzi D, Gaspari S, Manouras L, Descalzi G, Mitsi V, Zachariou V (2014) RGS9–2 modulates sensory and mood related symptoms of neuropathic pain. Neurobiol Learn Mem 115:43–48. 10.1016/j.nlm.2014.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma W, Kyte SL, Bagdas D, Alkhlaif Y, Alsharari SD, Lichtman AH, Chen ZJ, Del Fabbro E, Bigbee JW, Gewirtz DA, Damaj MI (2017) Effects of paclitaxel on the development of neuropathy and affective behaviors in the mouse. Neuropharmacology 117:305–315. 10.1016/j.neuropharm.2017.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda H, Zhao MG, Zhuo M (2005) Roles of NMDA receptor NR2A and NR2B subtypes for long-term depression in the anterior cingulate cortex. Eur J Neurosci 22:485–494. 10.1111/j.1460-9568.2005.04236.x [DOI] [PubMed] [Google Scholar]
- Toyoda H, Zhao MG, Zhuo M (2009) Enhanced quantal release of excitatory transmitter in anterior cingulate cortex of adult mice with chronic pain. Mol Pain 5:4. 10.1186/1744-8069-5-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsantoulas C, Denk F, Signore M, Nassar MA, Futai K, McMahon SB (2018) Mice lacking Kcns1 in peripheral neurons show increased basal and neuropathic pain sensitivity. Pain 159:1641–1651. 10.1097/j.pain.0000000000001255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JA, Franklin G, Heagerty PJ, Wu R, Egan K, Fulton-Kehoe D, Gluck JV, Wickizer TM (2004) The association between pain and disability. Pain 112:307–314. 10.1016/j.pain.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Veldhuis NA, Poole DP, Grace M, McIntyre P, Bunnett NW (2015) The G protein-coupled receptor-transient receptor potential channel axis: molecular insights for targeting disorders of sensation and inflammation. Pharmacol Rev 67:36–73. 10.1124/pr.114.009555 [DOI] [PubMed] [Google Scholar]
- Wang JY, Zhang HT, Han JS, Chang JY, Woodward DJ, Luo F (2004) Differential modulation of nociceptive neural responses in medial and lateral pain pathways by peripheral electrical stimulation: a multichannel recording study. Brain Res 1014:197–208. 10.1016/j.brainres.2004.04.029 [DOI] [PubMed] [Google Scholar]
- Yoon SY, Woo J, Park JO, Choi EJ, Shin HS, Roh DH, Kim KS (2015) Intrathecal RGS4 inhibitor, CCG50014, reduces nociceptive responses and enhances opioid-mediated analgesic effects in the mouse formalin test. Anesth Analg 120:671–677. 10.1213/ANE.0000000000000607 [DOI] [PubMed] [Google Scholar]
- Yu LN, Zhou XL, Yu J, Huang H, Jiang LS, Zhang FJ, Cao JL, Yan M (2012) PI3K contributed to modulation of spinal nociceptive information related to ephrinBs/EPHBS. PLoS One 7:e40930. 10.1371/journal.pone.0040930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariou V, Georgescu D, Sanchez N, Rahman Z, DiLeone R, Berton O, Neve RL, Sim-Selley LJ, Selley DE, Gold SJ, Nestler EJ (2003) Essential role for RGS9 in opiate action. Proc Natl Acad Sci U S A 100:1365613661. 10.1073/pnas.2232594100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Chisari M, Raehal KM, Kaltenbronn KM, Bohn LM, Mennerick SJ, Blumer KJ (2012) GIRK channel modulation by assembly with allosterically regulated RGS proteins. Proc Natl Acad Sci U S A 109:19977–19982. 10.1073/pnas.1214337109 [DOI] [PMC free article] [PubMed] [Google Scholar]