Abstract
Lung cancer causes more deaths than any other cancer. Sphingolipids encompass metabolically interconnected species whose balance has pivotal effects on proliferation, migration, and apoptosis. In this study, we paralleled quantification of sphingolipid species with quantitative (q)PCR analyses of metabolic enzymes in order to identify dysregulated routes of sphingolipid metabolism in different subtypes of lung cancers. Lung samples were submitted to histopathological reexamination in order to confirm cancer type/subtype, which included adenocarcinoma histological subtypes and squamous cell and neuroendocrine carcinomas. Compared with benign lesions and tumor-free parenchyma, all cancers featured decreased sphingosine-1-phosphate and SMs. qPCR analyses evidenced differential mechanisms leading to these alterations between cancer types, with neuroendocrine carcinomas upregulating SGPL1, but CERT1 being downregulated in adenocarcinomas and squamous cell carcinomas. 2-Hydroxyhexosylceramides (2-hydroxyHexCers) were specifically increased in adenocarcinomas. While UDP-glycosyltransferase 8 (UGT8) transcript levels were increased in all cancer subtypes, fatty acid 2-hydroxylase (FA2H) levels were higher in adenocarcinomas than in squamous and neuroendocrine carcinomas. As a whole, we report differing mechanisms through which all forms of lung cancer achieve low SM and lysosphingolipids. Our results also demonstrate that FA2H upregulation is required for the accumulation of 2-hydroxyHexCers in lung cancers featuring high levels of UGT8.
Keywords: 2-hydroxyhexosylceramide, sphingolipid, squamous, neuroendocrine, cancer, fatty acids, gene expression, mass spectrometry, fatty acid 2-hydroxylase, uridine 5′-diphosphate-glycosyltransferase 8
Lung cancer is the leading cause of cancer-related deaths worldwide, with an estimated 1.8 million deaths per year (1). The overall prognosis of lung cancer patients is poor, with an 18% survival rate after 5 years (1). Given the clinical advantage of cancer type-specific therapies, it is required to enhance our understanding of differential mechanisms underlying lung cancers.
Sphingolipid alterations are increasingly associated with oncogenesis (2). Sphingolipids encompass a wide array of bioactive molecules centrally involved in the determination of cell fate (3). The balance between the various members of this family has a decisive impact on proliferation (4), migration (5), cell death (6), and angiogenesis (7). Several sphingolipid metabolites/enzymes, including ceramides (8), glycosphingolipids (9), SMs (10), and sphingosine kinases (11), are dysregulated in malignant neoplastic lesions. The study of isolated actors has provided useful yet limited cues on the involvement of sphingolipids in oncogenesis. In fact, these bioactive lipids are part of a complex and dynamic metabolic pathway containing enzymatically interconnected metabolites with pleiotropic and sometimes opposite effects on cell biology (12). Single target/single analyte-based approaches have led to a significant, yet partial, success for this class of agents in clinical trials (13–15). Considering the dynamic metabolic links between these bioactive lipids, a more global pathway analysis should enhance our understanding of their involvement in different types of cancer and potentially lead to the identification of new targets, or combinations of targets, to interfere with lesion-specific mechanisms of oncogenesis.
Histological types of lung cancer are associated with specific biological processes (16), which could be specifically targeted if better understood. In the last decade, the development of several targeted lung cancer therapies based on histological characteristics increased progression-free survival (17), but the need for identifying alterations with therapeutic potential for specific histological types of lung cancers remains. In this study, we compared sphingolipid species and the transcript levels of selected sphingolipid-metabolizing enzymes across adenocarcinoma and squamous and neuroendocrine lung cancers classified using the latest recommendations of the World Health Organization (18). We found SMs, sphingosine, sphinganine, and sphingosine-1-phosphate (S1P) to be decreased in all of the investigated types of lung cancer, while hexosylceramides (HexCers) and 2-hydroxyhexosylceramides (2-hydroxyHexCers) were specifically increased in adenocarcinomas.
MATERIALS AND METHODS
Subjects
The patients included in this study were diagnosed with low stage lung cancers and underwent surgical resection between the years 2000 and 2016 at the Institut Universitaire de Cardiologie et de Pneumologie de Québec (IUCPQ). The patients signed a consent form allowing the utilization of their tissues for research purposes. Samples were collected and stored using methods that minimize degradation (19). Samples were stored at the IUCPQ site of the Respiratory Health Network Tissue Bank of the Fonds de recherche du Québec-Santé (www.tissuebank.ca). Availability of both tumor and nontumor tissue as well as access to medical history were required for inclusion. Exclusion criteria included previous use of chemotherapeutics that might alter sphingolipid levels (12) and sphingolipid modifiers or sphingolipid-modifying genetic disease. Using this procedure, we obtained samples from 97 exsmoker patients (Table 1) whose histological types were reclassified by a thoracic pathologist (P.J.) according to the 2015 World Health Organization guidelines (18) (Table 1). Adenocarcinomas were classified as high-grade (micropapillary and solid predominance) or low-grade (lepidic, acinar, or papillary predominance). Nineteen never-smoker adenocarcinoma patients were also included. In addition, 22 exsmoker and 14 never-smoker patients operated for benign lung diseases were analyzed. Chronic obstructive pulmonary disease (COPD) GOLD stages were determined according to the 2017 recommendations (20). The study was approved by the IUCPQ Research Ethics Committee (Project 20852) and abided by the Declaration of Helsinki principles.
TABLE 1.
Characteristics of patients
| Exsmokers | Never-Smokers | ||||||
| Benign (n = 22) | AD high (n = 25) | AD low (n = 25) | Squamous (n = 25) | Neuro (n = 22) | Benign (n = 14) | AD (n = 19) | |
| Age, years | 59.2 (43–78) | 64 (51–80) | 68.8 (50–80)a | 66.1 (50–78)a | 55.2 (28–75) | 58.3 (36–78) | 64.6 (26–83) |
| Sex, male/female | 11/11 | 11/14 | 11/14 | 11/14 | 9/13 | 4/10 | 5/14 |
| Packs per year | 31.8 | 37.2a | 32.7 | 42.43a | 23.3 | 0 | 0 |
| Predominance (adenocarcinoma) | |||||||
| Lepidic | — | 0 | 5 | — | — | — | 2 |
| Acinar | — | 0 | 17 | — | — | — | 7 |
| Papillary | — | 0 | 4 | — | — | — | 1 |
| Micropapillary | — | 6 | 0 | — | — | — | 5 |
| Solid | — | 18 | 0 | — | — | — | 3 |
| T Stage | |||||||
| T1 | — | 6 (24%)b | 12 (48%) | 3 (12%)b | 12 (60%) | — | 4 (22%) |
| T2 | — | 16 (64%) | 12 (48%) | 19 (76%) | 8 (40%) | — | 12 (67%) |
| T3 | — | 2 (8%) | 1 (4%) | 3 (12%) | 0 (0%) | — | 0 (0%) |
| T4 | — | 1 (4%) | 0 (0%) | 0 (0%) | 0 (0%) | — | 2 (11%) |
| Stage | |||||||
| I | — | 13 (57%) | 21 (84%) | 11 (48%) | 17 (94%) | — | 9 (60%) |
| II | — | 6 (26%) | 3 (12%) | 8 (35%) | 1 (6%) | — | 3 (20%) |
| III | — | 4 (17%) | 1 (4%) | 4 (17%) | 0 (0%) | — | 3 (20%) |
| V | — | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Lymph node | |||||||
| Positive | — | 7 (28%) | 2 (8%) | 9 (36%) | 3 (15%) | — | 5 (28%) |
| Negative | — | 18 (72%) | 23 (92%) | 16 (64%) | 17 (85%) | — | 13 (72%) |
| FEV1, percent predicted | 94 (45–131) | 89.4 (54–140) | 92.1 (47–130) | 80.5 (43–130) | 97.5 (72–129) | 100.9 (75–139) | 101.4 (61–137) |
| DLCO, percent | 91 (41–125)a | 91.3 (53–153)a | 89.2 (34–135)a | 91.9 (42–138)a | 110.7 (75–144) | 103.7 (60–146) | 102.3 (71–139) |
| With COPD, percent | 8 (36.3) | 17 (68) | 15 (60) | 12 (48) | 7 (32) | 1 (7) | 4 (21) |
| GOLD stage (1/2/3/4) | 6/1/1/0 | 14/3/0/0 | 7/7/1/0 | 5/6/1/0 | 6/1/0/0 | 0/1/0/0 | 2/2/0/0 |
| Died | 4 (16%) | 9 (36%) | 5 (20%) | 12 (48%) | 4 (16%) | 0 (0%) | 7 (28%) |
AD, adenocarcinoma; Neuro, neuroendocrine; DLCO, diffusing capacity of the lung for carbon monoxide.
Significantly different from neuroendocrine patients (P < 0.05).
Significantly different from adenocarcinoma low and neuroendocrine patients (P < 0.05).
Lung lipid quantification
Lipid extraction from tissue samples was performed as described (21). Briefly, 15–20 mg of frozen lung was homogenized in 600 μl isopropanol-water-butylated hydroxytoluene (BHT) (75:25:0.01 v/v/w) using glass tissue grinders. For each sample, 25 μl of the homogenate was stored for DNA quantification and 900 μl of ethyl acetate was added to the remainder of the sample for a final concentration of ethyl acetate-isopropanol-water-BHT (60:30:10:0.01 v/v/v/w). A cocktail of internal standards dissolved in methanol was added to each sample at the start of the extraction process. Internal standards included 17:1 sphingosine (Cayman Chemical, Ann Arbor, MI), 17:1 S1P, 18:1/17:0 ceramide, 18:1/12:0 glucosylceramide, 18:1/12:0 lactosylceramide (LacCer), and 18:1/12:0 SM (Avanti Polar Lipids, Alabaster, AL). BHT, HPLC-grade isopropanol, ethyl acetate, and methanol were from Thermo Fisher (Waltham, MA). Lipids were resolved on a 3 × 150 mm XDB-C8 column (Agilent, Santa Clara, CA) and detected using a TSQ Altis triple quadrupole mass spectrometer (Thermo Fisher) as described previously (22). Precursor ions were the precise [M + H] species for each lipid. The m/z 184.1 product ion was used for SM species; the m/z 264.3 product ion was used for all other sphingolipids containing a sphingosine backbone; and the m/z 284.3 product ion was used for sphinganine.
DNA was extracted using Extracta DNA Prep (QuantaBio, Beverly, MA) and quantified using EvaGreen DNA binding dye (Biotium, Fremont, CA) against a standard curve of salmon sperm DNA (Thermo Fisher). Four samples were excluded from analysis due to DNA levels below the limit of detection.
Real-time PCR
RNA was extracted from 10–15 mg of frozen lung samples with an RNeasy extraction kit (Qiagen, Hilden, Germany), and cDNA was prepared with an iScript kit (Bio-Rad, Hercules, CA) according to manufacturers’ instructions. Predesigned quantitative (q)PCR primers (IDT, Coralville, IA) (Table 2) were validated for linearity using SsoAdvanced (Bio-Rad) master mix in a CFX 384 thermocycler (Bio-Rad) and normalized to the geometric mean of three reference genes validated for stability across tissue types.
TABLE 2.
qPCR primer sequences
| Gene | Forward | Reverse |
| B4GALT6 | ACA TTA TAC CTC GAG CTT GTA CC | CTT CTC CCT CTC TTC GTC CT |
| CERT | CCT GAG CAC GAA GAT ACC AAA | TGA AGA TGA AAC AGA GTA TGG CT |
| FA2H | GTA CTC GAT GAG GCT CCA GA | GCA ACG TCC GAC TCT TCA |
| GNB2L1 | TGG TCT TCA GCT TGC AGT TAG | GCA AAT ACA CTG TCC AGG ATG |
| HPRT1 | GTA TTC ATT ATA GTC AAG GGC ATA TCC | AGA TGG TCA AGG TCG CAA |
| SGMS1 | GTA CAG ATA GTC CCC ACA CAT | GCA TTT CAA CTG TTC TCC GAA G |
| SGMS2 | GTG TGC TAC AAG AAT GCA GAT G | CTT CCT CTT CAG CGG TCA C |
| SGPL1 | ATA TGA GTT TGT CTT CCA GCC A | CTT GGT CTT GTT CAA CTT GTC TTG |
| SMPD1 | GAG AGA GAT GAG GCG GAG A | CTG GCT CTA TGA AGC GAT GG |
| SMPD3 | GGT CCT GAG GTG TGC TTC | TCT TTG CCA GCC GCT AC |
| SPHK1 | TTC ACG CTG ATG CTC ACT G | CCG TTC ACC ACC TCG TG |
| SPHK2 | CCT TCA ACC TCA TCC AGA CAG | CCC GTT CAG CAC CTC AT |
| RPLP0 | TGT CTG CTC CCA CAA TGA AAC | TCG TCT TTA AAC CCT GCG TG |
| UGCG | GCA TTG CAA CTT GAG TGG AC | GAT GTG TTG GAT CAA GCA GGA |
| UGT8 | AAG ACA CCA AGA CAA AGC CA | GAA TTC CCA AGA CCC ACT CTG |
GNB2L1, G protein subunit beta 2; HPRT1, hypoxanthine phosphoribosyltransferase 1; RPLP0, ribosomal protein lateral stalk subunit P0; UGCG, UDP-glucose ceramide glucosyltransferase.
Statistical analyses
Continuous and nominal variables are expressed using mean ± SEM and percentages, respectively. Categorical variables expressed as a percentage were analyzed using chi-square or Fisher’s exact test. Continuous variables were analyzed using one-way ANOVA with heterogeneous variances, as the model could not be reduced to a one-way analysis with the same variance among groups. The Satterthwaite’s degree of freedom statement was added for unequal variance structures. For variables for which normality assumption was not fulfilled, analyses were performed using log-transformed data. The normality assumption was verified with the Shapiro-Wilk tests after a Cholesky factorization on residuals from the statistical model. All analyses were conducted using the statistical package SAS, version 9.4 (SAS Institute Inc., Cary, NC.) and R [R Core Team (2016), Foundation for Statistical Computing, Vienna, Austria].
RESULTS
Patient characteristics
Baseline characteristics of patients (Table 1) were not significantly different between cancer types, with the exception of lower pack-year (23.3 ± 15.7, P = 0.001) and younger age of patients with neuroendocrine carcinomas, as well as a higher diffusing capacity of the lung for carbon monoxide (DLCO) in this group (P = 0.007). As expected (23), high-grade adenocarcinomas and squamous cell carcinoma groups featured the highest shift toward high pathological T stages and overall clinical stage. Neuroendocrine lesions featured mostly T1 typical (G1) lesions (24). The never-smoker adenocarcinoma group featured all adenocarcinoma types with a dominance of the acinar and micropapillary types. As expected, few never-smoker patients presented with COPD, while the frequency of COPD ranged from 32% to 68% between the exsmoker cancer subgroups and mainly featured GOLD 1 and 2 patients. Thus, our inclusion criteria allowed us to select subgroups of patients whose clinical features were aligned with previous studies (23–25).
Common and differential sphingolipid alterations between lung cancers
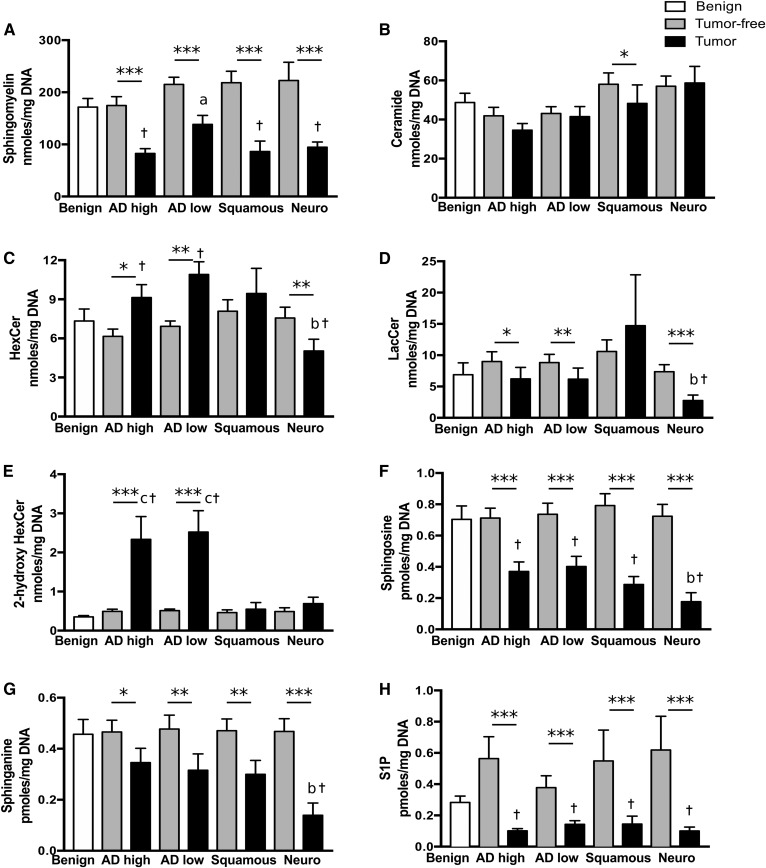
Given the direct relationship between DNA (as a surrogate indicator of cell number) and phospholipid content in lung tissue (26), quantified lipids were expressed relative to the DNA content in order to enable comparisons between samples with differing cellularity (supplemental Fig. S1). Total levels of each lipid class were compared between groups (Fig. 1), as there were strong correlations between chain lengths within each lipid class (R of 0.5–0.9; representative example in supplemental Fig. S2) with the exception of HexCer that did not correlate with 2-hydroxyHexCer (average R was 3.9 times higher within classes than between classes, data not shown). In agreement with documented sphingolipid levels in lung tissue (27), we found that the most prevalent species was SM (Fig. 1A, range 83–222 nmol·mg−1 DNA). These were followed by ceramides (Fig. 1B, 35–58 nmol·mg−1 DNA), glycosylceramides (Fig. 1 C–E, 0.4–11 nmol·mg−1 DNA), sphingosine/sphinganine and S1P (0.1–0.8 pmol·mg−1 DNA, Fig. 1 F–H).
Fig. 1.
Sphingolipids are modulated in different types of lung cancer. Tissue lipids were extracted and sphingolipids were quantified by LC-MS/MS in lung parenchyma of exsmoker patients with benign lung diseases; in tumor and tumor-free lung tissues from patients with high-grade adenocarcinomas (AD high), low-grade adenocarcinomas (AD low), and squamous and neuroendocrine (Neuro) lung carcinomas. The sums of metabolites per lipid class per milligram of DNA are shown for SMs (A), ceramides (B), HexCers (C), LacCers (D); 2-hydroxyHexCers (E), sphingosine (F), sphinganine (G), and S1P (H). Bars represent the average ± SEM, n = 22–25. *P < 0.05, **P < 0.01, ***P < 0.001 tumor compared with tumor-free tissue. aP < 0.05 versus AD high and squamous, bP < 0.05 versus AD high, low, and squamous, cP < 0.05 versus squamous and neuro, †P < 0.05 versus benign.
In our whole population, neither age, sex, COPD status, functional lung measures, tumor size, nor survival correlated with the total level of each sphingolipid class. We found no significant modulation of sphingolipid classes between benign lung disease samples and nontumor parenchyma from cancer patients (Fig. 1 A–H). The amount of ceramides did not differ between tumor and tumor-free tissues except for squamous cell carcinoma, where ceramides were decreased (17%) relative to their nontumor tissue counterpart. In contrast, tumor tissue contained lower levels of S1P (62–83% decrease), sphingosine (45–76% decrease), sphinganine (26–70% decrease), and SMs (35–60% decrease), when compared with tumor-free counterparts, regardless of the cancer type.
Sphingolipid modulations that discriminated cancer types were also noted. Relative to tumor-free tissue, 2-hydroxyHexCers were increased 7.8-fold in adenocarcinoma tumors (P < 0.0001, Fig. 1E), and this was observed in both low- and high-grade adenocarcinomas with 72% and 76% of patients having more 2-hydroxyHexCers in their tumor than in nontumor tissue (supplemental Fig. S3). This dysregulation was not observed in any of the other cancer types (Fig. 1E, supplemental Fig. S3). Neuroendocrine lesions showed the lowest levels of HexCers, LacCers, sphingosine, and sphinganine (34, 70, 76, and 62% decreases, respectively; Fig. 1C, D, F, G) with LacCer levels significantly different from the other cancer types. Thus, sphingolipid levels are modified in tumors when compared with nontumor tissue; and specific sphingolipidomic alterations were detected in adenocarcinomas and squamous cell and neuroendocrine lung cancers.
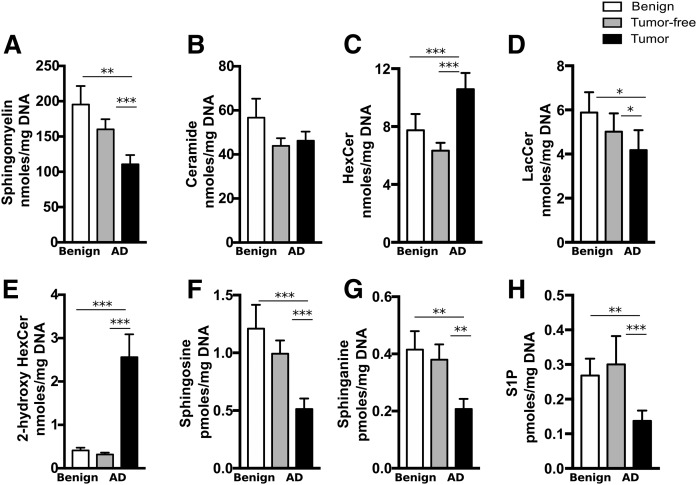
The effect of cigarette smoke exposure
Up to 25% of adenocarcinomas occur in patients who have never smoked, and lung cancers in this population have distinct clinical and biological features (28) that might translate into different sphingolipidomic alterations. First, our results suggest that smoking did not have a long-lasting impact on lipid levels in lung tissue, as exsmoker and never-smoker benign patients had similar levels of all lipid classes (Figs. 1, 2). The lipid alterations detected in tumor tissues of exsmoker adenocarcinoma patients were also observed in tumor tissues from adenocarcinoma patients who had never smoked, including the increase in 2-hydroxyHexCer (6.1-fold, P < 0.0001, Fig. 2E). Thus, smoking status did not have an impact at the sphingolipid level in adenocarcinoma patients.
Fig. 2.
Never-smoker patients with benign lung disease or adenocarcinoma display lipid alterations similar to exsmoker patients. Lipids were extracted and sphingolipids were quantified by LC-MS/MS in tumor and tumor-free lung tissues from never-smoker patients with benign lung disease as well as patients with adenocarcinoma. The sums of metabolites per lipid class per milligram of DNA are shown for SMs (A), ceramides (B), HexCers (C), LacCers (D), 2-hydroxyHexCers (E), sphingosine (F), sphinganine (G), and S1P (H). Bars represent the average ± SEM, n = 14–19. *P < 0.05, **P < 0.01, ***P < 0.001.
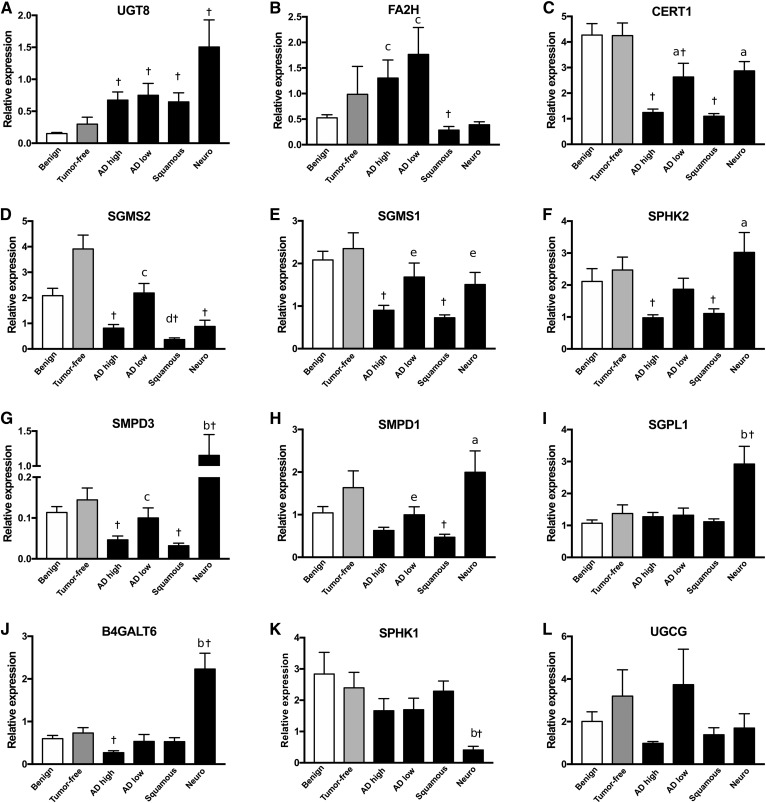
Transcript level alterations of sphingolipid-metabolizing enzymes
Sphingolipids belong to a family of lipids interconverted by specific enzymes (12). We quantified mRNA of sphingolipid-metabolizing enzymes and transport protein genes related to the observed cancer-associated lipid modulations (Fig. 3). Relative levels of transcripts for the studied genes were not significantly different between benign tissue and nontumor tissue from adenocarcinoma patients. UDP-glycosyltransferase 8 (UGT8), which catalyzes HexCer synthesis from ceramide, was the only gene significantly increased in all cancer types (4.3- to 10-fold increases, Fig. 3A). Fatty acid 2-hydroxylase (FA2H) was significantly higher in both types of adenocarcinomas compared with the other cancer types (Fig. 3B). Compared with benign lesions and nontumor tissues, the majority of the investigated genes featured decreased transcript levels, including ceramide transporter 1 (CERT1), which was profoundly decreased in squamous cell carcinoma and high-grade adenocarcinomas (Fig. 3C). We also observed genes with decreased expression specifically in the most severe cancer types, i.e., squamous carcinomas and high-grade adenocarcinomas, including SM synthases (SGMS2, SGMS1), sphingosine kinase 2 (SPHK2), and SM phosphodiesterase 3 (SMPD3) (Fig. 3D–G). Neuroendocrine carcinomas featured the most distinct dysregulations, with unique increases in SMPD3, S1P lyase 1 (SGPL1), and β-1,4-galactosyltransferase 6 (B4GALT6), and a decrease in SPHK1 (Fig. 3I–K). Therefore, although there appears to be consistency between transcript levels and the general decrease of sphingolipids in the several cancer types, distinct patterns of sphingolipid-metabolizing gene transcript levels were observed between cancer types.
Fig. 3.
Sphingolipid-associated gene expression differs between tumor-free tissues and histological cancer types. mRNA was extracted from lung samples from patients with benign lung diseases, from tumor-free tissues of adenocarcinoma patients, and from the tumors from patients with high-grade adenocarcinomas (AD high), low-grade adenocarcinomas (AD low), and squamous and neuroendocrine (Neuro) lung carcinomas. Shown is average expression relative to the geometric mean of three reference genes ± SEM for UGT8 (A), FA2H (B), CERT1 (C), SGMS2 (D), SGMS1 (E), SPHK2 (F), SMPD3 (G), SMPD1 (H), SGPL1 (I), B4GALT6 (J), SPHK1 (K), and UDP-glucose ceramide glucosyltransferase (UGCG) (L). aP < 0.05 versus AD high and squamous, bP < 0.05 all other cancer types, cP < 0.05 versus squamous and Neuro, dP < 0.05 versus AD high, eP < 0.05 versus squamous; †P < 0.05 versus benign; n = 14–22.
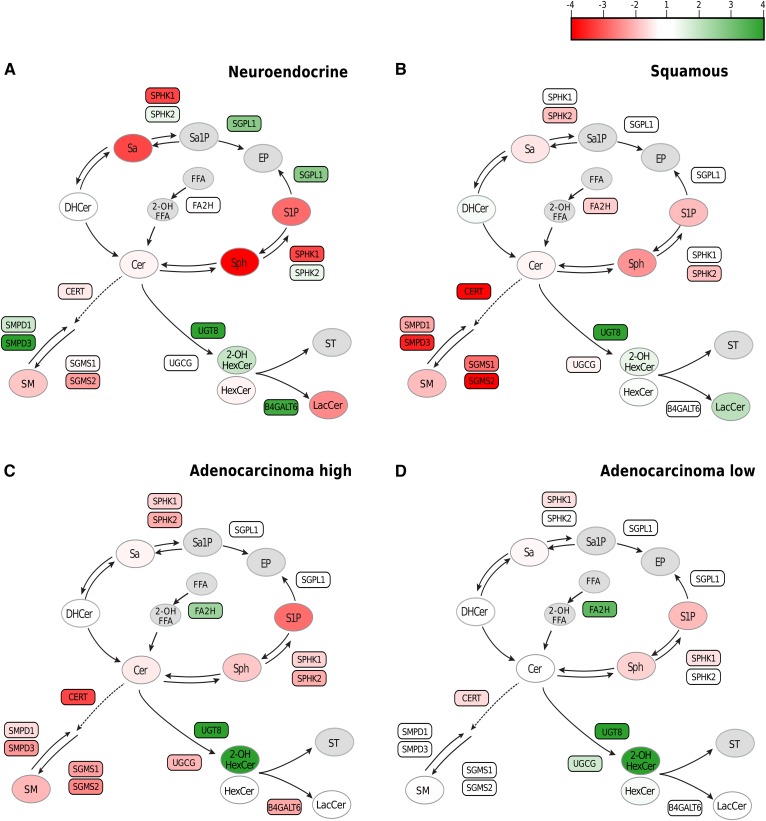
Combining lipidomic and transcriptomic data identifies cancer-specific sphingometabolic dysregulated nodes
Cancer to benign tissue ratios were used to study the interplay between sphingometabolic genes and the metabolites (Fig. 4). Neuroendocrine carcinomas featured the most important decreases in sphinganine, sphingosine, and S1P, all of which could be related by a large increase in SGPL1, the enzyme responsible for irreversible degradation of sphingolipids (Fig. 4A). All cancer types were characterized by a decrease in SMs, which could be explained by a decrease in mRNA level for CERT1 (Fig. 4), necessary to the transport of ceramide to the Golgi for SM production (29), and a decrease in SGMSs, especially in squamous and high-grade adenocarcinomas (Fig. 4B, C). Neuroendocrine carcinomas featured decreased SMs and significantly increased mRNA expression of the SMPD3 enzyme that catabolizes SMs (10-fold increase vs. benign, Fig. 3G) that could participate to reduce SM levels. Lastly, adenocarcinomas were the only cancer types that overexpressed FA2H (Fig. 4C, D), the enzyme that catalyzes hydroxylation of FFAs (30), which are precursors of 2-hydroxyHexCer.
Fig. 4.
Dysregulated metabolite levels are supported by altered gene expression in tumor tissue. The ratio of tumor tissue over tissues from patients with benign diseases was computed for group averages of lipid classes (ovals) and for mRNA levels (rectangles) for neuroendocrine carcinomas (A), squamous cell carcinomas (B), high-grade adenocarcinomas (C), and low-grade adenocarcinomas (D). The data ranges from −4- to +4-fold changes. Red indicates a decrease of the ratio; white indicates a ratio of 1; and green indicates an increased ratio. Dashed line indicates transport of ceramide. Absence of the data is indicated by the gray color (n = 14–25). Cer, ceramide; DHCer, dihydroceramide; Sa, sphinganine; Sa1P, sphinganine-1-phosphate; Sph, sphingosine; ST, sulfatide.
FA2H determines 2-hydroxyHexCer levels in UGT8-expressing lung cancers
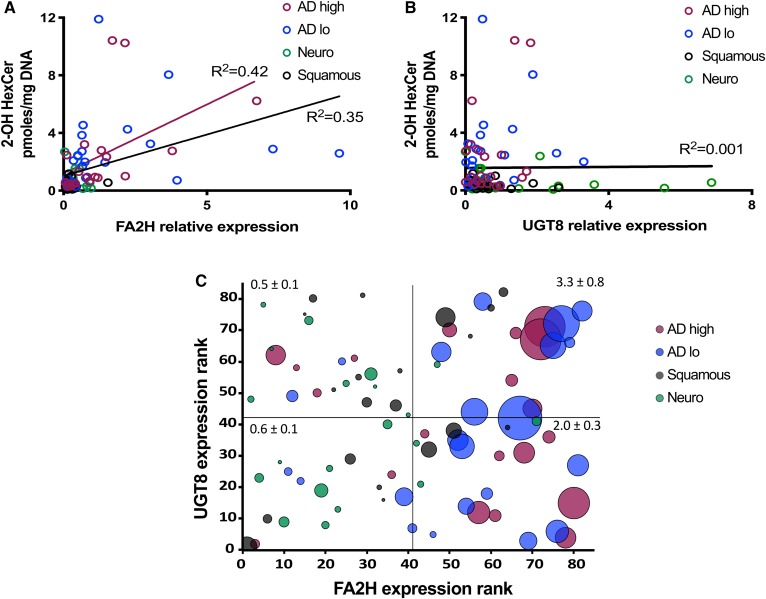
In this study, we found weak to null correlation between 2-hydroxyHexCer and the mRNA levels of UGT8 across all cancer types or for adenocarcinomas specifically (Fig. 5B), despite high levels of 2-hydroxyHexCer in the latter. Given that FA2H and UGT8 cooperate in order to produce 2-hydroxyHexCer, we also studied the relationship between FA2H and 2-hydroxyHexCer levels (Fig. 5A). Multiple regression analyses reveal that the variance of 2-hydroxyHexCer is explained at 53% by the combination of cancer types and the transcript levels of FA2H and UGT8. When addressing specifically how FA2H impacted on 2-hydroxyHexCer levels in cancer subtypes, we determined that FA2H alone had a minimal impact in neuroendocrine (R2 0.02) and squamous cell carcinoma (R2 0.016). However, the FA2H transcript level accounted by itself for 43% and 18% of 2-hydroxyHexCer variance, respectively, in high-grade and low-grade adenocarcinomas. Accordingly, 2-hydroxyHexCers were on average 4.8-fold higher in adenocarcinomas with FA2H transcript levels above the median (Fig. 5C), indicating that FA2H determines the accumulation of 2-hydroxyHexCers in UGT8-expressing cancer tissues independently of UGT8 expression levels. Conversely, there was no relationship between FA2H and 2-hydroxyHexCer levels in neuroendocrine carcinomas, which might relate to their very low overall level of sphingolipids and, more specifically, HexCer (Fig. 1).
Fig. 5.
High FA2H expression is associated with increased 2-hydroxyHexCers in lung cancer. Linear regression of the level of 2-hydroxyHexCer (2-OH HexCer) versus FA2H (A) and UGT8 (B) mRNA relative expression in adenocarcinoma (AD) high (red), AD low (blue), squamous (black), or neuroendocrine (Neuro) (green) lung cancer tissues. C: mRNA expression rank for FA2H and UGT8 among lung cancer samples are plotted on the x and y axes. Circle diameter indicates the 2-OH HexCer levels. Average 2-OH HexCer ± SEM for each quadrant is indicated. Black lines are the regression curves for all samples; red line is for AD high only (n = 20–22).
DISCUSSION
Sphingolipids are produced in a biosynthetic pathway of interconverted species known to regulate numerous processes that are critical for oncogenesis (2). Patients with lung cancer have a low life expectancy and a better understanding of cancer-associated changes could lead to treatment or diagnostic solutions (1). In the current study, we uncovered alterations of metabolites that are common to several types of lung cancers, including a strong decrease in SMs, S1P, sphingosine, and sphinganine. These findings could have important clinical implications because low CERT1 and SM levels were shown to contribute to the progression of breast cancer (31). In addition, we unraveled the specific increase of HexCer and 2-hydroxyHexCer in lung adenocarcinomas. This metabolic dysregulation is of interest given that, in breast cancer, increased UGT8 and galactosylceramides are associated with tumor aggressiveness and lung metastasis (32) and low galactosylceramide levels in UGT8-silenced breast cancer cells lead to decreased proliferation and metastasis potential and increased apoptosis (33).
Adenocarcinomas, whether low-grade or high-grade, from exsmokers or nonsmokers, featured a striking increase in HexCer levels, and especially 2-hydroxyHexCer, which was not observed in the other investigated cancer types. The FA2H enzyme hydroxylates the FFAs required for the generation of 2-hydroxyceramides, the preferred substrate for UGT8 to generate 2-hydroxygalactosylceramides (34). Because only adenocarcinoma tumors exhibited both high FA2H and UGT8, this suggests that combined FA2H and UGT8 expression could drive increased 2-hydroxyHexCer levels in adenocarcinomas. Our results strongly argue that a combination of FA2H and UGT8 expression drives the synthesis of 2-hydroxyHexCer, the exact mechanism accounting for the accumulation of these metabolites specifically in adenocarcinomas, which should be confirmed by ulterior investigations of sulfatides and gangliosides, which were also linked with cancer metastasis (35, 36) and multi-drug resistance (37). Aligning with the theory that complex metabolic alterations, rather than single molecule levels, should be taken into account for delineating mechanisms of disease, low FA2H was associated with tumor growth and patient survival in gastric tumors (38), but UGT8 is not overexpressed in gastric tumors in two publicly available gene expression datasets (39, 40), suggesting that the modulation of FA2H in gastric tumor is part of a different cancer-associated metabolic change. Our results thus suggest that, in addition to UGT8, FA2H could be a target to counteract 2-hydroxyHexCer accumulation and the associated oncogenic processes.
Our observations that total SMs are decreased in lung cancer are consistent with a recent study where phospholipid profiling was performed on combined non-small-cell lung cancer types (27). SMs and their metabolizing enzymes are involved in apoptosis (41), and the mechanism of action of a chemotherapeutic agent currently in clinical trials (42), 2-hydroxyoleic acid (Minerval), involves an increase of SM levels, which in turn triggers cell cycle arrest and apoptosis (43). Correcting the low sphingolipid levels in lung cancers could thus be a potentially successful therapeutic approach. For that matter, our results suggest that the type of cancer could dictate the choice of sphingolipid modifier to be used. For instance, the decreased SM accumulation appears to relate to a combined defect in ceramide transfer and SM synthesis in the high-grade adenocarcinomas, which could mitigate the effect of SGMS-promoting drugs, compared with low-grade adenocarcinomas where CERT1 is less downregulated, and thus, SM precursors are available at the Golgi apparatus. Altogether, our results support a critical role for CERT1 in the dysregulation of SM levels in squamous cell carcinoma and high-grade adenocarcinomas.
Neuroendocrine carcinomas featured the most distinctive profile of mRNA alterations, which might relate to the ontogeny of this type of tumors (24). Cells from grade 1 typical carcinoid, which are the majority of our cohort, consist generally of neuron-like small cells with low proliferation rates and secretory abilities (24), and this could have an impact on sphingolipid and enzyme levels. Neuroendocrine carcinomas presented the greatest decreases of HexCer, sphinganine, sphingosine, and LacCer levels, when compared with the other lung cancer histological types. In fact, the global shift toward glycosylceramides appears to be of less importance in this type of cancer when considering the overall decrease of sphingolipid levels. Indeed, SGPL1, the enzyme responsible for irreversible degradation of sphingolipids, was significantly more expressed in neuroendocrine tumors. Our observation of low LacCer levels in this cancer type, despite increased B4GALT6 mRNA, is also consistent with the theory that this cancer type features a generalized reduction of sphingolipids and, thus, of LacCer precursors.
Adenocarcinomas often share histopathological features, but S1P levels were shown to be decreased in pancreas (44), unchanged in colon (45), and increased in breast (46) tumors, when compared with nontumor tissues. For many types of studies, it appears practical to normalize lipids according to tissue weight (44–46). However, we determined that in lung tissue, the amount of DNA per milligram of tissue is consistently increased in tumors by approximately 45% (supplemental Fig. S1A), stressing the importance of using a surrogate for cell numbers to resolve the sphingolipid alterations on a per-cell basis (supplemental Fig. S1B vs. Fig. 1H). Similar to our study, Marien et al. (27) observed a profound decrease in SM in tumor nests by 2D-imaging, a method independent of mass or DNA, supporting that modulations in homogenized tissues normalized by DNA can adequately represent tumor-associated change in situ. In addition to the methods of normalization, comorbidities could mitigate the interpretations of lung tissue sample analyses. As such, COPD was shown to increase sphingolipid levels in the sputum (47). Yet, COPD status did not have an impact on sphingolipid levels in our lung tissues, which might either relate to the average low GOLD stages of the patients included in the current study or to the fact that sputum (47) and parenchyma are inherently different in their composition.
Although SPHK1 levels were previously shown to be increased in adenocarcinoma and squamous lung cancers (48), we did not observe significant modulations at the mRNA level in our qPCR results. The possibility exists that SPHK1 expression is influenced by patient characteristics. Indeed, the Song et al. (48) study concluded that severe non-small-cell lung cancer cases had higher SPHK1 levels, but the patients in their cohort had lung cancers of higher severity (higher T, N, and clinical stages). Of note, S1P levels were often associated with SPHK1 expression in different tissues (49, 50) and our finding of no SPHK1 overexpression in tissues featuring low levels of S1P is consistent with this contention.
CONCLUSIONS
We conclude that lung cancers feature common and distinct sphingolipid dysregulations. While single chained sphingolipids and SMs are generally decreased, distinct patterns could be observed for HexCers. Although we observed a generalized upregulation of UGT8 in all cancers, only adenocarcinomas featured high levels of 2-hydroxyHexCer, which is explained by preferential overexpression of FA2H in this cancer type. Neuroendocrine carcinoma featured the most profound decrease in several sphingolipids, including S1P, which was associated with the strongest upregulation of SGPL1 compared with all the other cancer types. As a whole, our study identifies distinct regulatory nodes associated with cancer-specific sphingolipid alterations.
Supplementary Material
Acknowledgments
The authors thank Serge Simard for the statistical analyses, Christine Racine and the team at the IUCPQ site of the Tissue Bank of the Quebec Respiratory Health Network (www.tissuebank.ca), and Anick Langlois for technical help.
Footnotes
Abbreviations:
- B4GALT6
- β-1,4-galactosyltransferase 6
- BHT
- butylated hydroxytoluene
- CERT1
- ceramide transporter 1
- COPD
- chronic obstructive pulmonary disease
- FA2H
- fatty acid 2-hydroxylase
- HexCer
- hexosylceramide
- 2-hydroxyHexCer
- 2-hydroxyhexosylceramide
- IUCPQ
- Institut Universitaire de Cardiologie et de Pneumologie de Québec
- LacCer
- lactosylceramide
- SGMS
- SM synthase
- SGPL1
- sphingosine-1-phosphate lyase 1
- SMPD
- SM phosphodiesterase
- S1P
- sphingosine-1-phosphate
- SPHK
- sphingosine kinase
- UGCG
- UDP-glucose ceramide glucosyltransferase
- UGT8
- UDP-glycosyltransferase 8
This study was funded using peer-reviewed grants from the Cancer Research Society (Grant 22339) (70%) and from the Faculty of Medicine of Université Laval using funds from a donation by Merck Sharpe and Dohme (30%). The latter company did not have any involvement in any step of this project. D.M. is a Fonds de Recherche du Québec J2 scholarship awardee and a member of the Quebec Respiratory Health Network. Y.B. holds a Canada Research Chair in Genomics of Heart and Lung Diseases. T.A.C. is supported by a National Health and Medical Research Council/Australian Research Council Dementia Research Fellowship (1110400). A.S.D. is supported by National Health and Medical Research Council Project Grant 1100626.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Torre L. A., Siegel R. L., and Jemal A.. 2016. Lung cancer statistics. In Lung Cancer and Personalized Medicine: Current Knowledge and Therapies. A. Ahmad and S. Gadgeel, editors. Springer International Publishing, Cham, Switzerland. 1–19. [Google Scholar]
- 2.Don A. S., Lim X. Y., and Couttas T. A.. 2014. Re-configuration of sphingolipid metabolism by oncogenic transformation. Biomolecules. 4: 315–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goetzl E. J., and An S.. 1998. Diversity of cellular receptors and functions for the lysophospholipid growth factors lysophosphatidic acid and sphingosine 1-phosphate. FASEB J. 12: 1589–1598. [PubMed] [Google Scholar]
- 4.Zhang H., Desai N. N., Olivera A., Seki T., Brooker G., and Spiegel S.. 1991. Sphingosine-1-phosphate, a novel lipid, involved in cellular proliferation. J. Cell Biol. 114: 155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadahira Y., Ruan F., Hakomori S., and Igarashi Y.. 1992. Sphingosine 1-phosphate, a specific endogenous signaling molecule controlling cell motility and tumor cell invasiveness. Proc. Natl. Acad. Sci. USA. 89: 9686–9690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuvillier O., Pirianov G., Kleuser B., Vanek P. G., Coso O. A., Gutkind S., and Spiegel S.. 1996. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature. 381: 800–803. [DOI] [PubMed] [Google Scholar]
- 7.Kimura T., Watanabe T., Sato K., Kon J., Tomura H., Tamama K., Kuwabara A., Kanda T., Kobayashi I., Ohta H., et al. 2000. Sphingosine 1-phosphate stimulates proliferation and migration of human endothelial cells possibly through the lipid receptors, Edg-1 and Edg-3. Biochem. J. 348: 71–76. [PMC free article] [PubMed] [Google Scholar]
- 8.Karahatay S., Thomas K., Koybasi S., Senkal C. E., ElOjeimy S., Liu X., Bielawski J., Day T. A., Gillespie M. B., Sinha D., et al. 2007. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C18-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 256: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoda Y., Gasa S., Makita A., Fujioka Y., Kikuchi Y., and Hashimoto M.. 1979. Glycolipids in human lung carcinoma of histologically different types. J. Natl. Cancer Inst. 63: 1153–1160. [PubMed] [Google Scholar]
- 10.Merchant T. E., Meneses P., Gierke L. W., Den Otter W., and Glonek T.. 1991. 31P magnetic resonance phospholipid profiles of neoplastic human breast tissues. Br. J. Cancer. 63: 693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruckhäberle E., Rody A., Engels K., Gaetje R., von Minckwitz G., Schiffmann S., Grösch S., Geisslinger G., Holtrich U., Karn T., et al. 2008. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res. Treat. 112: 41–52. [DOI] [PubMed] [Google Scholar]
- 12.Truman J-P., García-Barros M., Obeid L. M., and Hannun Y. A.. 2014. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochim. Biophys. Acta. 1841: 1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adan-Gokbulut A., Kartal-Yandim M., Iskender G., and Baran Y.. 2013. Novel agents targeting bioactive sphingolipids for the treatment of cancer. Curr. Med. Chem. 20: 108–122. [PubMed] [Google Scholar]
- 14.Dickson M. A., Carvajal R. D., Merrill A. H., Gonen M., Cane L. M., and Schwartz G. K.. 2011. A phase I clinical trial of safingol in combination with cisplatin in advanced solid tumors. Clin. Cancer Res. 17: 2484–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pal S. K., Drabkin H. A., Reeves J. A., Hainsworth J. D., Hazel S. E., Paggiarino D. A., Wojcjak J., Woodnutt G., and Bhatt R. S.. 2017. A phase 2 study of the sphingosine-1-phosphate antibody sonepcizumab in patients with metastatic renal cell carcinoma. Cancer. 123: 576–582. [DOI] [PubMed] [Google Scholar]
- 16.Li L., Wei Y., To C., Zhu C. Q., Tong J., Pham N. A., Taylor P., Ignatchenko V., Ignatchenko A., Zhang W., et al. 2014. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat. Commun. 5: 5469. [DOI] [PubMed] [Google Scholar]
- 17.Chan B. A., and Hughes B. G. M.. 2015. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl. Lung Cancer Res. 4: 36–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Travis W. D., Brambilla E., Nicholson A. G., Yatabe Y., Austin J. H., Beasley M. B., Chirieac L. R., Dacic S., Duhig E., Flieder D. B., et al. 2015. The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J. Thorac. Oncol. 10: 1243–1260. [DOI] [PubMed] [Google Scholar]
- 19.Bossé Y., Postma D. S., Sin D. D., Lamontagne M., Couture C., Gaudreault N., Joubert P., Wong V., Elliott M., van den Berge M., et al. 2012. Molecular signature of smoking in human lung tissues. Cancer Res. 72: 3753–3763. [DOI] [PubMed] [Google Scholar]
- 20.Vogelmeier C. F., Criner G. J., Martinez F. J., Anzueto A., Barnes P. J., Bourbeau J., Celli B. R., Chen R., Decramer M., Fabbri L. M., et al. 2017. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am. J. Respir. Crit. Care Med. 195: 557–582. [DOI] [PubMed] [Google Scholar]
- 21.Hejazi L., Wong J. W., Cheng D., Proschogo N., Ebrahimi D., Garner B., and Don A. S.. 2011. Mass and relative elution time profiling: two-dimensional analysis of sphingolipids in Alzheimer’s disease brains. Biochem. J. 438: 165–175. [DOI] [PubMed] [Google Scholar]
- 22.Wong J. W., Abuhusain H. J., McDonald K. L., and Don A. S.. 2012. MMSAT: automated quantification of metabolites in selected reaction monitoring experiments. Anal. Chem. 84: 470–474. [DOI] [PubMed] [Google Scholar]
- 23.Yoshizawa A., Motoi N., Riely G. J., Sima C. S., Gerald W. L., Kris M. G., Park B. J., Rusch V. W., and Travis W. D.. 2011. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod. Pathol. 24: 653–664. [DOI] [PubMed] [Google Scholar]
- 24.Fisseler-Eckhoff A., and Demes M.. 2012. Neuroendocrine tumors of the lung. Cancers (Basel). 4: 777–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ou S. H. I., Zell J. A., Ziogas A., and Anton-Culver H.. 2007. Prognostic factors for survival of stage I nonsmall cell lung cancer patients. Cancer. 110: 1532–1541. [DOI] [PubMed] [Google Scholar]
- 26.Singer S. J., and Nicolson G. L.. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175: 720–731. [DOI] [PubMed] [Google Scholar]
- 27.Marien E., Meister M., Muley T., Fieuws S., Bordel S., Derua R., Spraggins J., Van de Plas R., Dehairs J., Wouters J., et al. 2015. Non-small cell lung cancer is characterized by dramatic changes in phospholipid profiles. Int. J. Cancer. 137: 1539–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thu K. L., Vucic E. A., Chari R., Zhang W., Lockwood W. W., English J. C., Fu R., Wang P., Feng Z., MacAulay C. E., et al. 2012. Lung adenocarcinoma of never smokers and smokers harbor differential regions of genetic alteration and exhibit different levels of genomic instability. PLoS One. 7: e33003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanada K., Kumagai K., Yasuda S., Miura Y., Kawano M., Fukasawa M., and Nishijima M.. 2003. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 426: 803–809. [DOI] [PubMed] [Google Scholar]
- 30.Alderson N. L., Rembiesa B. M., Walla M. D., Bielawska A., Bielawski J., and Hama H.. 2004. The human FA2H gene encodes a fatty acid 2-hydroxylase. J. Biol. Chem. 279: 48562–48568. [DOI] [PubMed] [Google Scholar]
- 31.Heering J., Weis N., Holeiter M., Neugart F., Staebler A., Fehm T. N., Bischoff A., Schiller J., Duss S., Schmid S., et al. 2012. Loss of the ceramide transfer protein augments EGF receptor signaling in breast cancer. Cancer Res. 72: 2855–2866. [DOI] [PubMed] [Google Scholar]
- 32.Dzięgiel P., Owczarek T., Plazuk E., Gomulkiewicz A., Majchrzak M., Podhorska-Okolów M., Driouch K., Lidereau R., and Ugorski M.. 2010. Ceramide galactosyltransferase (UGT8) is a molecular marker of breast cancer malignancy and lung metastases. Br. J. Cancer. 103: 524–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Owczarek T. B., Suchanski J., Pula B., Kmiecik A. M., Chadalski M., Jethon A., Dziegiel P., and Ugorski M.. 2013. Galactosylceramide affects tumorigenic and metastatic properties of breast cancer cells as an anti-apoptotic molecule. PLoS One. 8: e84191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaeren-Wiemers N., van der Bijl P., and Schwab M. E.. 1995. The UDP-galactose:ceramide galactosyltransferase: expression pattern in oligodendrocytes and Schwann cells during myelination and substrate preference for hydroxyceramide. J. Neurochem. 65: 2267–2278. [DOI] [PubMed] [Google Scholar]
- 35.Cao Q., Chen X., Wu X., Liao R., Huang P., Tan Y., Wang L., Ren G., Huang J., and Dong C.. 2018. Inhibition of UGT8 suppresses basal-like breast cancer progression by attenuating sulfatide-alphaVbeta5 axis. J. Exp. Med. 215: 1679–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia J., Callewaert N., and Borsig L.. 2007. P-selectin mediates metastatic progression through binding to sulfatides on tumor cells. Glycobiology. 17: 185–196. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y. Y., Gupta V., Patwardhan G. A., Bhinge K., Zhao Y., Bao J., Mehendale H., Cabot M. C., Li Y. T., and Jazwinski S. M.. 2010. Glucosylceramide synthase upregulates MDR1 expression in the regulation of cancer drug resistance through cSrc and beta-catenin signaling. Mol. Cancer. 9: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao Y., Yang X., Sun L., Sun S., Huang X., Zhou D., Li T., Zhang W., Abumrad N. A., Zhu X., et al. 2019. Fatty acid 2-hydroxylation inhibits tumor growth and increases sensitivity to cisplatin in gastric cancer. EBioMedicine. 41: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D’Errico M., de Rinaldis E., Blasi M. F., Viti V., Falchetti M., Calcagnile A., Sera F., Saieva C., Ottini L., Palli D., et al. 2009. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur. J. Cancer. 45: 461–469. [DOI] [PubMed] [Google Scholar]
- 40.Wang G., Hu N., Yang H. H., Wang L., Su H., Wang C., Clifford R., Dawsey E. M., Li . M., Ding T., et al. 2013. Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in china. PLoS One. 8: e63826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tepper A. D., Ruurs P., Wiedmer T., Sims P. J., Borst J., and van Blitterswijk W. J.. 2000. Sphingomyelin hydrolysis to ceramide during the execution phase of apoptosis results from phospholipid scrambling and alters cell-surface morphology. J. Cell Biol. 150: 155–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massalha W., Markovits M., Pichinuk E., Feinstein-Rotkopf Y., Tarshish M., Mishra K., Llado V., Weil M., Escriba P. V., and Kakhlon O.. 2019. Minerval (2-hydroxyoleic acid) causes cancer cell selective toxicity by uncoupling oxidative phosphorylation and compromising bioenergetic compensation capacity. Biosci. Rep. 39: doi:10.1042/BSR20181661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barceló-Coblijn G., Martin M. L., de Almeida R. F., Noguera-Salvà M. A., Marcilla-Etxenike A., Guardiola-Serrano F., Lüth A., Kleuser B., Halver J. E., and Escribá P. V.. 2011. Sphingomyelin and sphingomyelin synthase (SMS) in the malignant transformation of glioma cells and in 2-hydroxyoleic acid therapy. Proc. Natl. Acad. Sci. USA. 108: 19569–19574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Y., DiVittore N. A., Young M. M., Jia Z., Xie K., Ritty T. M., Kester M., and Fox T. E.. 2013. Altered sphingolipid metabolism in patients with metastatic pancreatic cancer. Biomolecules. 3: 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uranbileg B., Nishikawa T., Ikeda H., Kurano M., Sato M., Saigusa D., Aoki J., Watanabe T., and Yatomi Y.. 2018. Evidence suggests sphingosine 1-phosphate might be actively generated, degraded, and transported to extracellular spaces with increased S1P2 and S1P3 expression in colon cancer. Clin. Colorectal Cancer. 17: e171–e182. [DOI] [PubMed] [Google Scholar]
- 46.Nagahashi M., Tsuchida J., Moro K., Hasegawa M., Tatsuda K., Woelfel I. A., Takabe K., and Wakai T.. 2016. High levels of sphingolipids in human breast cancer. J. Surg. Res. 204: 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Telenga E. D., Hoffmann R. F., t’Kindt R., Hoonhorst S. J., Willemse B. W., van Oosterhout A. J., Heijink I. H., van den Berg M., Jorge L., Sandra P., et al. 2014. Untargeted lipidomic analysis in chronic obstructive pulmonary disease. Uncovering sphingolipids. Am. J. Respir. Crit. Care Med. 190: 155–164. [DOI] [PubMed] [Google Scholar]
- 48.Song L., Xiong H., Li J., Liao W., Wang L., Wu J., and Li M.. 2011. Sphingosine kinase-1 enhances resistance to apoptosis through activation of PI3K/Akt/NF-kappaB pathway in human non-small cell lung cancer. Clin. Cancer Res. 17: 1839–1849. [DOI] [PubMed] [Google Scholar]
- 49.Nagahashi M., Ramachandran S., Kim E. Y., Allegood J. C., Rashid O. M., Yamada A., Zhao R., Milstien S., Zhou H., Spiegel S., et al. 2012. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 72: 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Z., Ma J., Hu B., Zhang Y., and Wang S.. 2018. SPHK1 promotes metastasis of thyroid carcinoma through activation of the S1P/S1PR3/Notch signaling pathway. Exp. Ther. Med. 15: 5007–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.