Abstract
Cardiovascular diseases are the number one cause of death worldwide and greatly impact quality of life and medical costs. Enormous effort has been made in research to obtain new tools for efficient and quick diagnosis and predicting the prognosis of these diseases. Discoveries of epigenetic mechanisms have related several pathologies, including cardiovascular diseases, to epigenetic dysregulation. This has implications on disease progression and is the basis for new preventive strategies. Advances in methodology and big data analysis have identified novel mechanisms and targets involved in numerous diseases, allowing more individualized epigenetic maps for personalized diagnosis and treatment. This paves the way for what is called pharmacoepigenetics, which predicts the drug response and develops a tailored therapy based on differences in the epigenetic basis of each patient. Similarly, epigenetic biomarkers have emerged as a promising instrument for the consistent diagnosis and prognosis of cardiovascular diseases. Their good accessibility and feasible methods of detection make them suitable for use in clinical practice. However, multicenter studies with a large sample population are required to determine with certainty which epigenetic biomarkers are reliable for clinical routine. Therefore, this review focuses on current discoveries regarding epigenetic biomarkers and its controversy aiming to improve the diagnosis, prognosis, and therapy in cardiovascular patients.
Keywords: epigenetics, biomarker, microRNA, cardiovascular diseases, myocardial infarction, heart failure, atherosclerosis, hypertension
Introduction
Cardiovascular diseases (CVDs) are one of the leading causes of mortality in developed countries. Cardiovascular diseases refer to disorders affecting the structures or function of the heart and blood vessels, including hypertension, atherosclerosis, myocardial infarction (MI), ischemia/reperfusion injury, stroke, and heart failure (HF), among others (Wang et al., 2016a; Thomas et al., 2018). Mechanisms underlying the complex pathophysiology that leads to CVDs are of great interest but still far from clear. Progress in the field of epigenetics have opened a new world for the comprehension and management of human diseases, including the prevalence of CVDs, based on the role of genetics and its environmental interaction in pathological conditions (Jaenisch and Bird, 2003). Significant evidence suggests that the environment and lifestyle can define epigenetic patterns throughout life. These epigenetic patterns are a cellular memory of further environmental exposure. Epigenetic modifications are reversible, different among cell types, and can potentially lead to disease susceptibility by producing long-term changes in gene transcription (Fraga et al., 2005; Beekman et al., 2010).
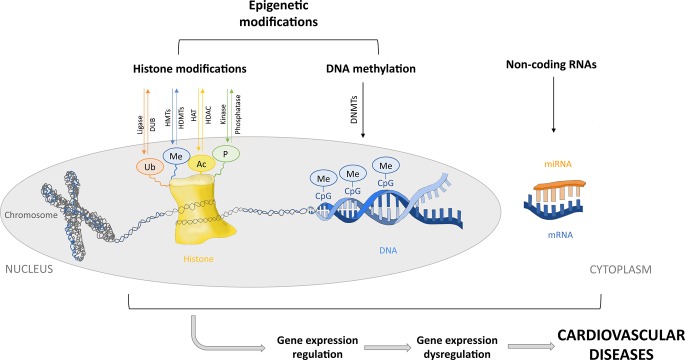
Epigenetic modifications include DNA methylation and posttranslational modifications of histone tails. However, in this review, posttranscriptional regulation of gene expression by noncoding RNAs (ncRNAs) is also considered a part of the epigenetic machinery. MicroRNAs (miRNAs) are small ncRNAs that contribute to regulation of the expression of different epigenetic regulators such as DNA methyltransferases (DNMTs) and histone deacetylases (HDACs), among others. Similarly, DNA methylation and histone modifications can regulate the expression of some miRNAs, forming a feedback loop. Thus, miRNAs and epigenetic regulators cooperate to modulate the expression of mutual targets. Therefore, although miRNAs are not strictly considered epigenetic factors, they contribute to the modulation of gene expression through epigenetics. Disruption of this complex regulation may participate in the development of different diseases (Iorio et al., 2010; Hoareau-Aveilla and Meggetto, 2017; Moutinho and Esteller, 2017; Wang et al., 2017a) ( Figure 1 ). DNA and histone proteins comprise the chromatin, which can be remodeled into a tightly condensed state (heterochromatin) or an open conformation (euchromatin) that would allow access to transcription factors or DNA binding proteins, allowing the regulation of gene expression (Kouzarides, 2007). Thus, epigenetics involves changes in gene expression due to chromatin adjustments that change the accessibility of DNA without changing its sequence, leading to silencing or downregulation/upregulation of gene expression (Baccarelli et al., 2010). Chromatin modifications, such as DNA methylation, consist of the transfer of a methyl group to carbon 5 of the cytosine residues [5-methylcytosine (5mC)] in CpG dinucleotides sites. CpG dinucleotides are localized throughout the genome but are more abundant in certain regions, such as gene promoters, forming so-called CpG islands. CpG methylation causes transcriptional repression by directly blocking transcription factor access to the DNA or indirectly via chromatin-modifying proteins (methyl-binding proteins) that recognize the methylated regions and recruit corepressors. DNA methyltransferases catalyze DNA methylation by recognizing and maintaining hypermethylated DNA during replication (DNMT1) or by de novo methylation (DNMT3a and DNMT3b). Moreover, gene bodies of actively transcribed genes normally show slightly higher DNA methylation levels as compared to gene bodies of nontranscribed genes. In contrast, hypomethylation is usually found in enhancer regions and promoters (Costantino et al., 2018). Posttranslational modification of histone tails is another epigenetic modification that regulates gene expression by chromatin remodeling. Histone acetylation, deacetylation, methylation, phosphorylation and ubiquitination change DNA accessibility, regulating gene transcription. The acetylation of histone tails is regulated by histone acetyltransferases (HATs) and HDACs. Histone acetyltransferase enzymes acetylate the lysine residues of the histones, whereas HDACs deacetylate them, promoting gene activation or silencing, respectively. Histone methylation is regulated by histone methyltransferases (HMTs) and histone demethylases (HDMT). Methylation occurs at the lysine or arginine residues and can activate or repress gene transcription depending on the degree of methylation and which residue is methylated (Li et al., 2017c; Sabia et al., 2017). The serine, threonine, and tyrosine residues of histone tails can also be phosphorylated and dephosphorylated by protein kinases and phosphatases, respectively. Histone tail phosphorylation modulates chromatin structure, taking part in transcription, DNA repair, and chromatin compaction in cell division and apoptosis (Rossetto et al., 2012). Lastly, histone tail ubiquitination is sequentially catalyzed by ligases enzymes, which attach ubiquitin to lysine residues. Ubiquitination and deubiquitination are involved in the activation of transcription and are usually associated with histone methylation. Their effect on repressing or activating transcription generally depends on what histone is modified (Cao and Yan, 2012). Finally, miRNAs regulate gene expression via degradation of the transcript or repression of translation when binding to the 3′-untranslated region of the target mRNA. Thus, miRNA represses mRNA translation without changing the DNA sequence of the gene. MicroRNA binding to mRNA is imperfect, so each miRNA has multiple targets. This allows the regulation of a great part of the human genome (Bartel, 2009). The miRNAs are 19-25 nucleotides in length, encoded in the genome and transcribed into primary miRNA (pri-miRNA). Pri-miRNAs derive into miRNAs precursors (pre-miRNA) by the nuclear RNase III called Dorsha and are transferred to the cytoplasm and processed by the endonuclease Dicer to generate a double-stranded miRNA duplex. This product is incorporated into an RNA-induced silencer complex (RISC)–loading complex. Then, one strand is removed from the complex, and the other strand forms a mature RISC, serving as a template for target mRNAs (Sato et al., 2011; Nishiguchi et al., 2015).
Figure 1.
Epigenetic regulatory mechanisms. Posttranslational modifications of histone tails by acetylation, deacetylation, ubiquitination, methylation, and phosphorylation. DNA methylation by DNA methyltransferases (DNMTs). Posttranscriptional regulation of gene expression by microRNAs. Epigenetic modifications involve silencing or downregulation/upregulation of gene expression. Dysregulation of the epigenetic machinery could lead to gene expression dysregulation and cardiovascular diseases. Ubiquitin (Ub), methionine (Me), acetyl group (Ac), phosphate (P), deubiquitinating enzyme (DUB), histone methyltransferase (HMTs), histone demethylase (HDMTs), histone acetyltransferase (HAT), histone deacetylase (HDAC), a cytosine followed by a guanine (CpG), microRNAs (miRNAs), and messenger RNA (mRNA).
Due to this important function in gene regulation, epigenetic modifications and miRNA may play a crucial role in the development of pathological conditions, including CVDs. Understanding the epigenetic machinery underlying cardiac disorders and how these epigenetic mechanisms can be introduced into diagnostics (i.e., biomarkers) and therapies is fundamental to improving the quality of life of patients. In medicine, a biomarker is defined as a measurable characteristic that indicates a particular physiological or pathological state or a response to a therapeutic treatment (Strimbu and Tavel, 2010). Ideally, biomarkers should have easy accessibility, predictable detection, and reliability (Sun et al., 2017). It is mandatory to present a specific measurable change that clearly associates with a diagnosis or a predictable outcome. Thus, biomarkers provide information to physicians when evaluating the probability of developing a disease, making a diagnosis, evaluating the severity of a disease and its progression; during therapeutic decision making; or when monitoring a patient’s response and may result in significant cost reduction (Baccarelli et al., 2010). Their classification can be based on their application (predisposition, diagnosis, monitoring, safety, prognostic, or predictive biomarkers). Predisposition biomarkers determine how likely it is for a patient to develop a certain disease and are usually utilized when there is a personal or family history that indicates a disease risk, and the results can help guide medical care. Diagnostic biomarkers are used to detect or confirm the existence of a health disorder and may assist its early detection. Monitoring biomarkers evaluate the status of a disease or determine exposure to an environmental agent or medical product. Safety biomarkers indicate the probability, presence, or extent of toxicity of a certain medical product or environmental agent. Prognostic biomarkers indicate how a disease may progress in patients who already have the particular disease. These biomarkers do not predict the treatment response but can be useful when selecting patients for treatment. Predictive biomarkers identify patients who are most likely to have a favorable or unfavorable response to a specific treatment. Thus, they can predict treatment success or undesired side effects in a particular patient. A particular disease can have different biological mechanisms in different patients. Predictive biomarkers can be associated with the specific mechanism of a health disorder. This facilitates a targeted therapy, which uses drugs specific for a particular biological mechanism associated with a disease, increasing its effectiveness (FDA-NIH Biomarker Working Group, 2016). Specifically, epigenetic biomarkers belonging to most of these classifications are discussed in this review, with a focus on CVDs. Among the epigenetic biomarkers, miRNAs are the most attractive, as they can be detected in small sample volumes, are stable, and can be obtained from plasma, serum, saliva, and urine. Interestingly, they are highly conserved, and this allows a reliable comparison between patients and animal models of disease (Matsumoto et al., 2013). Therefore, although all epigenetic mechanisms are being intensively investigated, miRNAs are evaluated the most for their use as predictive biomarkers. This review presents an overview of current research on epigenetic biomarkers in CVDs and how this knowledge can benefit the diagnosis, prognosis, and therapy for cardiovascular patients.
Epigenetic Biomarkers in CVDs
Over the last few years, numerous studies have linked cardiovascular risk factors to epigenetic modifications in human patients. Modification of the epigenetic environment alters cardiovascular homeostasis and impacts cardiovascular disorders. The function of epigenetic mechanisms in the regulation of gene expression is well known, although the role of epigenetic marks in CVDs is not clearly understood. Thus, the exploration of epigenetic biomarkers may lead to a deep comprehension of the molecular mechanisms and pathways associated with CVDs. In this section, we focus on major CVDs, such as hypertension, atherosclerosis, MI, and HF, and the epigenetic biomarkers associated with them.
Hypertension
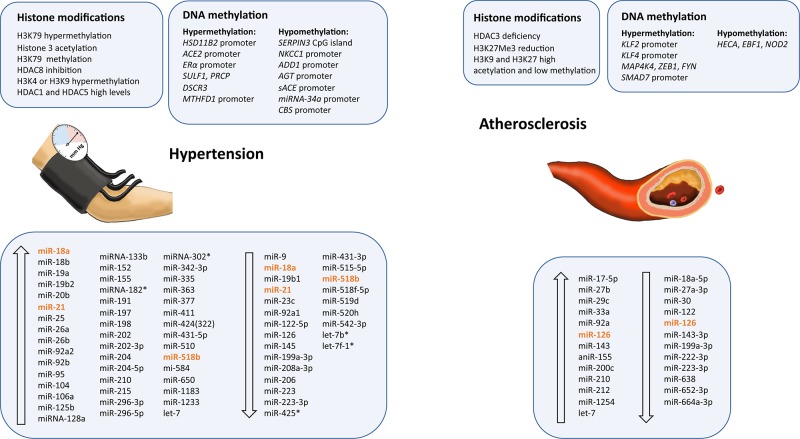
Arterial hypertension is a multifactorial disease with several mechanisms and metabolic systems involved in its pathogenesis. Genetic factors and environmental background may lead to alterations in multiple pathways that can eventually trigger development of the disease (Franceschini and Le, 2014). Intrauterine alterations, such as malnutrition, starvation, obesity, alcohol, drugs, nicotine, or environmental toxins, are some of the environmental factors directly related to hypertension development in the progeny (Bogdarina et al., 2007; Nuyt and Alexander, 2009). In addition, individuals who have aerobic training present with lower blood pressure than nontrained individuals (Fagard, 2006). This has an important impact on CVD risk factor control and is a nonpharmacological way to treat patients. There are also epigenetic factors that can influence the appearance of hypertension in adults, such as hypermethylation of genes, including superoxide dismutase-2 (SOD2) or Granulysin, or increased levels of histone acetylation at the promoter of the endothelial oxide synthetase gene (eNOS) (Wang et al., 2018b). Environmental factors are important to determining an individual’s predisposition to developing major cardiovascular risk factors by means of epigenetic modifications, and identification of the epigenetic mechanisms that participate in hypertension development may help generate new treatments. This is of great interest because hypertension is a key risk factor for CVDs, including MI, HF, stroke, and end-stage renal disease ( Table 1 and Figure 2 ).
Table 1.
Epigenetic biomarkers in hypertension.
| Epigenetic modification | Biomarker | Regulation in hypertension | Sample source | Study type | References |
|---|---|---|---|---|---|
| DNA methylation | HSD11B2 promoter | Highly methylated | Rat’s urine and tissues and human cell lines | Experimental: in vitro and rat model | (Alikhani-Koopaei et al., 2004) |
| SERPIN3 CpG island | Hypomethylation | Placental tissue | Clinical | (Chelbi et al., 2007) | |
| HSD11B2 promoter | Highly methylated | Blood and urine | Clinical | (Friso et al., 2008) | |
| 5mC | Lower levels | Blood | Clinical | (Smolarek et al., 2010) | |
| NKCC1 promoter | Hypomethylation | Aorta, heart and kidney | Experimental: spontaneously hypertensive rodent model | (Lee et al., 2010; Cho et al., 2011) | |
| sACE promoter | Hypermethylation | Blood | Clinical and experimental: in vitro | (Rivière et al., 2011) | |
| ERα promoter | Methylation | Uterine arteries | Clinical | (Dasgupta et al., 2012) | |
| SULF1, PRCP | SULF1: hypermethylation; PRCP: hypomethylation | Blood | Clinical | (Wang et al., 2013b) | |
| ADD1 promoter | Hypomethylation | Plasma | Clinical | (Zhang et al., 2013a) | |
| 5mC, 5hmC | Higher levels | Tissue | Experimental: Dahl salt-sensitive rats | (Liu et al., 2014) | |
| AGT promoter | Demethylation | H295R cells and visceral adipose tissue | Experimental: in vitro and rat model | (Wang et al., 2014a) | |
| DSCR3 | Hypermethylation | Maternal blood and placental tissue | Clinical | (Kim et al., 2015) | |
| miRNA-34a gene promoter | Hypomethylation | Placental tissue | Clinical | (Rezaei et al., 2018) | |
| ACE2 promoter | Hypermethylation | Plasma | Clinical | (Fan et al., 2017) | |
| CBS promoter | Hypermethylation | Maternal blood and placental tissue | Clinical | (Kim et al., 2015) | |
| MTHFD1 promoter | Hypermethylation | Plasma | Clinical | (Xu et al., 2019) | |
| Histone modifications | H3K79 | Hypermethylation | NA | Clinical | (Rodriguez-Iturbe, 2006; Duarte et al., 2012) |
| Histone 3 | Acetylation | Germ cells | Review | (Irmak and Sizlan, 2006) | |
| H3K79 | DNA methylation | Bibliography | Review | (Zhang et al., 2009) | |
| HDAC8 | Inhibition | mDCT cells and tissues | Experimental: rat models of salt-sensitive hypertension | (Mu et al., 2011) | |
| H3K4 or H3K9 | Hypermethylation | Tissue, plasma, and urine | Experimental: LSD1 knockout mice with a high-salt diet | (Pojoga et al., 2011) | |
| HDAC1, HDAC5 | High levels | Lung tissue and adventitial fibroblasts | Clinical and experimental: in vitro and hypoxic rat | (Zhao et al., 2012) | |
| miRNA | miR-18a, miR-210, miR-152, miR-363, miR-377, miR-411, miR-518b, miR-542-3p | miR-18a, miR-363, miR-377, miR-411, miR-542-3p: underexpression; miR-210, miR-152, miR-518b: overexpression | Placental tissue | Clinical | (Zhu et al., 2009) |
| 22 miRNAs | 15 upregulated and 7 downregulated | Serum | Clinical | (Yang et al., 2011) | |
| let-7b, miR-302*, miR-104, miR-128a, miR-182*, miR-133b | Overexpression | Placental tissue | Clinical | (Noack et al., 2011) | |
| miR-92b, miR-197, miR-342-3p, miR-296-5p, miR-26b, miR-25, miR-296-3p, miR-26a, miR-198, miR-202, miR-191, miR-95, miR-204, miR-21, miR-223 | miR-92b, miR-197, miR-342-3p, miR-296-5p, miR-26b, miR-25, miR-296-3p, miR-26a, miR-198, miR-202, miR-191, miR-95, miR-204: overexpression; miR-21, miR-223: underexpression | Placental tissue | Clinical | (Choi et al., 2013) | |
| miR-9, miR-126 | Lower levels | Peripheral blood mononuclear cells | Clinical | (Kontaraki et al., 2014) | |
| miR1233 | Higher levels | Serum | Clinical | (Ura et al., 2014) | |
| miR-18a, miR-19b1, miR-92a1, miR-210 | miR-210: upregulation; miR-18a, miR-19b1, and miR-92a1: downregulation | Plasma and placental tissue | Clinical | (Xu et al., 2014) | |
| miR-505 | Upregulation | Plasma | Clinical | (Yang et al., 2014) | |
| miR-106a, miR-18b, miR-20b, miR-19b-2, miR-92a-2, miR-363 | Dysregulation | Placental tissue | Clinical | (Zhang et al., 2015a) | |
| miR-515-5p, miR-518b, miR-518f-5p, miR-519d, miR-520h | Downregulation | Placental tissue | Clinical | (Hromadnikova et al., 2015) | |
| miR-335, miR-584 | Upregulation | Placental tissue and HTR8/Svneo cells | Clinical and experimental: in vitro | (Jiang F. et al., 2015) | |
| miR-125b | Overexpression | Plasma and placental tissue | Clinical | (Yang et al., 2016b) | |
| miR-215, miR-155, miR-650, miR-210, miR-21, miR-18a, miR-19b1 | MiR-215, miR-155, miR-650, miR-210, miR-21: upregulation; miR-18a, miR-19b1: downregulation | Plasma | Clinical | (Jairajpuri et al., 2017) | |
| miR-204-5p | Higher levels | Serum | Clinical | (Mei et al., 2017) | |
| let-7b*, let-7f-1*, miR-1183, miR-23c, miR-425* | miR-1183: upregulation; let-7b*, miR-23c, miR-425*, let-7f-1*: downregulation | Plasma and placental tissue | Clinical | (Gunel et al., 2017) | |
| miR-145 | Downregulation | Placental tissue | Clinical | (Han et al., 2017) | |
| miR-202-3p | Upregulation | Placental tissue | Clinical | (Singh et al., 2017) | |
| let-7 | Higher | Plasma | Clinical | (Huang et al., 2017b) | |
| miRNA | Dysregulation | Bibliography: Maternal serum and placental tissue | Bibliography review | (Laganà et al., 2018) | |
| miR-19a | Upregulation | Plasma and lung tissue | Clinical | (Chen and Li, 2017) | |
| miR-21 | Upregulation | Peripheral blood mononuclear cells | Clinical | (Parthenakis et al., 2017) | |
| miR-21 | Upregulation | Bibliography review | Bibliography review | (Sekar et al., 2017) | |
| miR-510 | Upregulation | Serum | Clinical | (Krishnan et al., 2017) | |
| miR-206 | Lower levels | Serum | Clinical | (Jin et al., 2017) | |
| miR-424(322) | Upregulation | Plasma | Clinical | (Baptista et al., 2018) | |
| miR-199a-3p, miR-208a-3p, miR-122-5p, miR-223-3p | Downregulation | Serum | Clinical | (Zhang et al., 2018c) | |
| miR-431-5p | Upregulation | Tissue | Experimental: mice made hypertensive and in vitro | (Huo et al., 2019) | |
| miR-143, NR_034083, NR_104181, | miR-143: upregulation; NR_034083: downregulation and NR_104181 and | Peripheral blood leucocytes | Clinical | (Chen et al., 2018b) |
NA, not available.
Figure 2.
Epigenetic modifications and microRNAs biomarkers dysregulated in atherosclerosis and hypertension. Ascending arrows indicate higher levels or upregulation, and descending arrows denote lower levels or downregulation, both compared to control conditions. Those miRNAs presenting opposite results are shown in orange.
Essential hypertension is a multifactorial disease with no identifiable cause that is affected by environmental and epigenetic factors. Environmental stressors cause acetylation of histone 3 in the neurons of the area postrema, leading to an increase in pressure that results in hypertension (Irmak and Sizlan, 2006). Low activity of the 11 beta-hydroxysteroid dehydrogenase 2 (HSD11B2) induces hypertension. In a study performed in patients with essential hypertension or glucocorticoid-induced hypertension, the HSD11B2 promoter was highly methylated. These changes may reflect a global status, with methylation of gene promoter being a potentially useful molecular biomarker to characterize hypertensive patients (Alikhani-Koopaei et al., 2004; Friso et al., 2008). Moreover, a polymorphism in the disruptor of telomeric silencing-1 gene (DOT1L), which encodes a methyltransferase that enhances methylation of histone 3 (H3K79) in the renal epithelial sodium channel gene (ENaC) promoter, is associated with blood pressure regulation (Duarte et al., 2012). It has also been reported that a DOT1A and ALL1 (fused gene from chromosome 9 [Af9]) interaction is associated with H3K79 hypermethylation of the ENaC promoter, suppressing its transcriptional activity. This interaction is disrupted by aldosterone and causes hypomethylation of H3K79 at specific regions, disinhibiting the ENaC promoter and leading to hypertension. Thus, the Dot1a-Af9 pathway may also be involved in the control of genes implicated in hypertension (Zhang et al., 2009). Hypomethylation of the α-adducin gene (ADD1) promoter has been found to be connected to the risk of essential hypertension. However, differences between females and males have been found (Zhang et al., 2013a). Moreover, histone 3 (H3K4 or H3K9) demethylation is induced by lysine-specific demethylase-1 (LSD1), which modifies gene transcription. Hypermethylation of histone 3 has been associated with hypertension, increased vascular contraction, and decreased relaxation via the nitric oxide-cGMP (NO-cGMP) pathway in heterozygous LSD1 knockout mice fed a high-salt diet (Pojoga et al., 2011). Histone deacetylation is also important in the development of pulmonary arterial hypertension. HDAC1 and HDAC5 protein levels have been demonstrated to be elevated in the lungs of patients and hypoxic rats. Inhibition of these proteins by valproic acid and suberoylanilide hydroxamic acid diminished the development of hypoxia-induced pulmonary hypertension in rats. Thus, HDAC1 and HDAC5 levels could be useful predictive biomarkers for the treatment of pulmonary hypertension in patients (Zhao et al., 2012).
In a study evaluating alterations in the global DNA methylation status of patients with essential hypertension, the level of the epigenetic marker 5mC was lower in hypertensive patients than in healthy people (Smolarek et al., 2010). In an in vivo model of hypertension using Dahl salt-sensitive rats, the levels of 5mC and 5-hydroxymethylcytosine (5hmC) were evaluated in the outer renal medulla. In response to salt administration, the 5mC levels were significantly higher for genes with low transcription and 5hmC levels higher in genes with higher expression. This study revealed important features of 5mC and 5hmC for understanding the role of epigenetic modifications in the regulation of hypertension (Liu et al., 2014).
Rivière et al. (2011) analyzed the regulation of somatic angiotensin-converting enzyme gene (sACE) expression by promoter methylation. sACE regulates blood pressure by catalyzing the conversion of angiotensin I into angiotensin II, a potent vasopressor. Hypermethylation of sACE promoter in cultures of human endothelial cells and rats was associated with transcriptional repression, suggesting an epigenetic mechanism in hypertension regulation (Rivière et al., 2011). More recently, Fan et al. (2017) demonstrated opposite results in patients with essential hypertension. The authors indicated that hypermethylation of the ACE2 promoter may increase essential hypertension risk, with variabilities in CpG islands methylation in males and females (Fan et al., 2017).
Moreover, a genome-wide methylation study on essential hypertension revealed that changes in the DNA methylation of leukocytes are involved in the pathogenesis of hypertension. They found increased methylation in the gene encoding sulfatase 1 (SULF1), which is involved in apoptosis, and decreased methylation in the gene encoding prolylcarboxypeptidase (PRCP), a regulator of angiotensin II and III cleavage (Wang et al., 2013b). Another genome-wide study of blood pressure characteristics found new genetic variants that influence blood pressure and are strongly associated with local CpG island methylation. This study demonstrated the role of DNA methylation in the regulation of blood pressure (Kato et al., 2015).
The pathogenesis of hypertension is affected by alterations in ion flux mechanisms. Hypomethylation of the Na/K/2Cl cotransporter 1 gene (NKCC1) promoter results in overexpression in a rodent model with spontaneous hypertension (Lee et al., 2010). DNA methyltransferase activity maintained hypomethylation in the NKCC1 promoter, playing an important role in NKCC1 upregulation during the course of the disease. This encourages evaluation of the NKCC1 methylation status in hypertensive patients (Cho et al., 2011). Furthermore, WNK4 is a serine-threonine kinase that negatively regulates the Na(+)-Cl(−)-cotransporter (NCC) and ENaC. This would affect the distal nephron, increasing the reabsorption of sodium. Stimulation of β(2)-adrenergic receptor (β(2)AR) in salt intake conditions would reduce WNK4 transcription, resulting in inhibition of HDAC8 activity and increased histone acetylation. In the rat models of salt-sensitive hypertension, salt diet repressed renal WNK4 expression, activating the NCC and inducing salt-dependent hypertension. Thus, WNK4 transcription is epigenetically modulated in the course of salt-sensitive hypertension, with the β(2)AR-WNK4 pathway as a potential therapeutic target for this disease (Mu et al., 2011).
Goyal et al. (2010) demonstrated that a low protein diet in pregnant mice leads to alterations in DNA methylation, miRNA, and gene expression in the brain renin–angiotensin system, a key regulator of hypertension in adults (Goyal et al., 2010). Along the same lines, in a study carried out in vitro and in a rat model, DNA demethylation of the angiotensinogen gene (AGT) promoter activated its expression. AGT is an important substrate of the renin–angiotensin–aldosterone system and an important target in hypertension research. Elevated concentrations of circulating aldosterone and high consumption of salt stimulate the AGT gene expression in adipose-induced hypertension (Wang et al., 2014a). In addition, cystathionine β-synthase (CBS), an important enzyme in the metabolism of plasma homocysteine, is associated with hypertension and stroke. Hypermethylation of the CBS promoter has been demonstrated to increase the risk of both diseases, especially in male patients (Wang et al., 2019a). Similarly, hypermethylation of the methylenetetrahydrofolate dehydrogenase 1 gene (MTHFD1) promoter, which is also associated with homocysteine metabolism, was observed in hypertensive patients, and proposed as a potential diagnostic biomarker in patients with essential hypertension (Xu et al., 2019).
In addition to the previous classic epigenetic modifications, miRNAs often regulate hypertension and are attractive biomarkers for the disease. The miR-9 and miR-126 expression levels are significantly lower in hypertensive patients than healthy individuals and are related to hypertension prognosis and organ damage. Thus, miR-9 and miR-126 may be possible biomarkers in essential hypertension (Kontaraki et al., 2014). Moreover, ncRNAs, such as miR-143, miR-145, and NR_104181, are significantly higher in essential hypertensive patients than controls, whereas NR_027032 and NR_034083 are significantly reduced. After evaluating cardiovascular risk factors, they concluded that lower expression levels of NR_034083 and higher expression levels of NR_104181 and miR-143 were risk factors for essential hypertension (Chen et al., 2018b). Another study evaluated the correlation between miRNA let-7 expression and subclinical atherosclerosis in untreated patients with newly diagnosed essential hypertension and found increased levels in hypertensive patients, suggesting that plasma let-7 could be an indicator for monitoring end-organ damage and a biomarker for atherosclerosis in these patients (Huang et al., 2017b). Similarly, upregulation of miR-505, miR-19a, miR-21, miR-510, or miR-424(322) in blood from hypertensive patients suggests a possible use for miR-510 as a diagnostic biomarker and therapeutic target (Yang et al., 2014; Chen and Li, 2017; Krishnan et al., 2017; Parthenakis et al., 2017; Sekar et al., 2017; Baptista et al., 2018). Lower levels of the combination of miR-199a-3p, miR-208a-3p, miR-122-5p, and miR-223-3p have also been shown to be suitable for diagnosis of hypertension (Zhang et al., 2018c). Decreased miR-206 levels might also be especially useful in the detection of pulmonary hypertension in patients with left heart disease (Jin et al., 2017). Furthermore, a study in hypertensive mice produced by infusion of angiotensin II concluded that miR-431-5p knockdown delays the increase in blood pressure induced by angiotensin II and reduces vascular injury. This demonstrates its potential as a target for the treatment of hypertension and vascular injury (Huo et al., 2019).
Preeclampsia is an important pregnancy-induced syndrome characterized by hypertension and proteinuria. Chronic hypoxia is a common pregnancy stress that increases the risk of preeclampsia and is associated with changes in methylation of the estrogen receptor α gene (ERα) promoter. ERα is involved in adjustments to the uterine blood flow, and promoter methylation results in gene repression in uterine arteries, increasing blood pressure (Dasgupta et al., 2012). Preeclampsia also modifies the expression profile of several serine protease inhibitors (SERPINs) in the placenta. Specifically, SERPIN3 CpG islands have a significantly low level of methylation in preeclampsia, providing a new potential marker for early diagnosis (Chelbi et al., 2007). Another study demonstrated a positive association between placenta global DNA methylation and hypertension in preeclampsia (Kulkarni et al., 2011). Next-generation sequencing technology and microarray assay analyses of the miRNA expression pattern in preeclamptic placentas versus healthy placentas have revealed that miRNAs expression is dysregulated in preeclampsia (Zhu et al., 2009; Noack et al., 2011; Yang et al., 2011; Choi et al., 2013; Xu et al., 2014; Hromadnikova et al., 2015; Zhang et al., 2015a; Gunel et al., 2017; Han et al., 2017). These results were in agreement with those found in the miRNA database from cell and tissue analyses. Thus, circulating miRNAs in the serum of pregnant women could be used as biomarkers for the diagnosis and prognosis of preeclampsia. To further demonstrate that miRNAs could be good predictors of preeclampsia, as well as its severity, circulating miRNA signatures were evaluated in women divided into groups based on preeclampsia severity. MiR-21, miR-29a, miR-125b, miR-155, miR-202-3p, miR-204-5p, miR-210, miR-215, miR-335, miR-518b, miR-584, miR-650, and miR-1233 were upregulated, whereas miR-15b, miR-18a, miR-19b1, and miR-144 were downregulated in women with severe preeclampsia compared to mild preeclampsia (Ura et al., 2014; Jiang et al., 2015; Yang et al., 2016b; Jairajpuri et al., 2017; Mei et al., 2017; Singh et al., 2017). In addition, a recent data recompilation supported a direct association between high or low expression of miRNAs in pregnancy serum and placenta in preeclamptic pregnancies (Laganà et al., 2018). Interestingly, an association has also been demonstrated between hypomethylation of the miR-34a promoter and preeclampsia severity (Rezaei et al., 2018). Another study analyzed the concentrations of Down syndrome critical region 3 (DSCR3), Ras association domain family 1 isoform A (RASSF1A), and sex-determining region Y (SRY) cell-free fetal DNA in maternal plasma from preeclamptic pregnancies and found that all of the markers significantly correlated with gestational age. The authors demonstrated that DSCR3 is a novel epigenetic biomarker and an alternative to RASSF1A for the prediction of early-onset preeclampsia (Kim et al., 2015). However, no association was found between the methylation status of the cortisol-controlling gene (HSD11B2), tumor suppressor gene (RUNX3), or long interspersed nucleotide element-1 gene (LINE-1) and hypertensive disorders of pregnancy when placental DNA methylation was analyzed (Majchrzak-Celińska et al., 2017).
Atherosclerosis
Atherosclerosis is a chronic inflammatory disease characterized by the accumulation of cholesterol in the walls of large- and medium-sized arteries, the accumulation of extracellular matrix and lipids, and smooth muscle cell proliferation. This process leads to the infiltration of immune cells (mostly macrophages) and endothelial dysfunction, forming a plaque, and eventually developing into acute cardiovascular events, such as MI, peripheral vascular disease, aneurysms, and stroke (Wissler, 1991). Proatherogenic stimuli, such as low-density lipoprotein (LDL) cholesterol and oxidized LDL, have been suggested to stimulate a long-term epigenetic reprogramming of innate immune system cells. This induces a constant activation, even after the removal of atherosclerotic stimuli (Bekkering et al., 2016). Emerging evidence supports epigenetic modifications being involved in the initiation and progression of atherosclerosis, playing an important role in plaque development and vulnerability, and highlighting the importance of epigenetic biomarkers as predictors of CVDs ( Table 2 and Figure 2 ) (Xu et al., 2018).
Table 2.
Epigenetic biomarkers in atherosclerosis.
| Epigenetic modification | Biomarker | Regulation in atherosclerosis | Sample source | Study type | References |
|---|---|---|---|---|---|
| DNA methylation | KLF2 promoter | Methylation | HUVEC cells | Experimental: in vitro | (Kumar et al., 2013) |
| KLF4 promoter | Methylation | HAEC cells | Experimental: in vitro | (Jiang et al., 2014) | |
| HECA, EBF1, NOD2, MAP4K4, ZEB1, FYN | HECA, EBF1, NOD2: Hypomethylated; MAP4K4, ZEB1, FYN: Hypermethylated | Human aortic intima and HEK293 cells | Clinical and experimental: in vitro | (Yamada et al., 2014) | |
| TIMP1, ABCA1, ACAT1 promoters | Altered methylation status | Peripheral blood | Clinical | (Ma et al., 2016) | |
| SMAD7 promoter | Hypermethylation | Peripheral blood and atherosclerotic plaques | Clinical | (Wei et al., 2018) | |
| 5mC, 5-hmC | Higher levels | Peripheral blood | Clinical | (Jiang et al., 2019) | |
| Histone modifications | HDAC3 | Deficiency | Aorta and HUVEC cells | Experimental: apoE−/− mice and in vitro | (Zampetaki et al., 2010) |
| H3K27Me3 | Reduction in H3K27Me3 modification | Perirenal aortic tissue patches | Clinical | (Wierda et al., 2015) | |
| H3K9, H3K27 | Higher histone acetylation and lower histone methylation | Carotid tissue | Clinical | (Greißel et al., 2016) | |
| miRNA | miR-130a, miR-27b, miR-210 | Higher levels | Serum and intima tissue | Clinical | (Li et al., 2011) |
| miR-17-5p | Higher levels | Plasma | Clinical | (Chen et al., 2015a) | |
| miR-143-3p, miR-222-3p | Lower levels | Microparticles | Clinical | (de Gonzalo-Calvo et al., 2016) | |
| miR-30 | Lower levels | Plasma | Clinical | (Huang et al., 2016b) | |
| miR-92a | Higher levels | Plasma | Clinical | (Huang et al., 2017a) | |
| miR-18a-5p, miR-27a-3p, miR-199a-3p, miR-223-3p, miR-652-3p | Lower levels | Plasma | Clinical | (Vegter et al., 2017) | |
| miR-33a | Higher levels | Plasma | Clinical | (Kim et al., 2017) | |
| miR-126 | Lower levels | Plasma | Experimental: apoE−/− mice | (Hao and Fan, 2017) | |
| miR-212 | Overexpression | Serum | Clinical | (Jeong et al., 2017) | |
| miRNA let-7 | Higher levels | Plasma | Clinical | (Huang et al., 2017b) | |
| miR-1254 | Higher levels | Plasma | Clinical | (de Gonzalo-Calvo et al., 2018) | |
| miR-200c | Overexpression | Carotid plaques and plasma | Clinical | (Magenta et al., 2018) | |
| miR-29c | Higher levels | Plasma | Clinical | (Huang et al., 2018) | |
| miR-221, miR-222 | Lower expression levels | Serum | Clinical | (Yilmaz et al., 2018) | |
| miR-638 | Lower levels | Serum | Clinical | (Luque et al., 2018) | |
| miR-122 | Higher levels | Serum | Clinical | (Wang and Yu, 2018) | |
| miR-221, miR-222 | Higher levels in tissue samples and lower levels in whole blood | Coronary artery atherosclerotic plaques, and internal mammary arteries and whole blood | Clinical | (Bildirici et al., 2018) | |
| miR-664a-3p | Downregulation | Serum | Clinical | (Li et al., 2018b) | |
| miR-155 | Higher levels | Serum | Clinical | (Qiu and Ma, 2018) | |
| miR-19A, miR-19B, miR-126, miR-155 | Differential levels | GEO dataset | High throughput | (Mao et al., 2018) | |
| miR-126, miR-143 | Higher levels | Plasma | Clinical | (Gao et al., 2019) |
Regarding histone modifications, HDAC3 is reported to have a protective effect in apolipoprotein E deficient (apoE−/−) mice. HDAC3 maintains the endothelial integrity, and its deficiency results in atherosclerosis (Zampetaki et al., 2010). Similarly, increased histone acetylation has been proposed to play some role in the progression of atherogenesis by modulating the expressions of proatherogenic genes (Choi et al., 2005). Histone deacetylases are upregulated in aortic smooth muscle cells when they were stimulated with mitogens. In contrast, inhibition of HDACs reduces aortic smooth muscle cell proliferation by changing cell cycle genes expression. This suggests a protective effect against atherosclerosis (Findeisen et al., 2011). Investigations of the association between changes in lysine 27 trimethylation of histone 3 (H3K27Me3), and atherosclerotic plaque development revealed a reduction in global levels of H3K27Me3 modification in vessels with advanced atherosclerotic plaques. This does not correlate with a reduction in the corresponding HMT, enhancer of zeste homolog 2 (EZH2). There was a relationship between the repression of H3K27Me3 mark in the vessels with advanced atherosclerotic plaques and the dynamic differentiation and proliferation of smooth muscle cells associated with atherosclerotic disease (Wierda et al., 2015). Histone acetylation, methylation, and the expression of their corresponding transferases in the atherosclerotic plaques of patients with carotid artery stenosis have been analyzed. Greißel et al. (2016) analyzed the expression of HATs GCN5L, P300, MYST1, and MYST2 and HMTs MLL2/4, SET7/9, hSET1A, SUV39H1, SUV39H2, ESET/SETDB1, EHMT1, EZH2, and G9a and described an enhancement in histone acetylation on H3K9 and H3K27 in the smooth muscle cells from severe atherosclerotic lesions that correlated with plaque severity. In addition, H3K9 and H3K27 methylation were significantly lower in atherosclerotic plaques and significantly associated with disease severity (Greißel et al., 2016).
DNA methylation is also involved in atherosclerosis. To identify CpG methylation profiles in the progression of atherosclerosis in the human aorta, Valencia-Morales et al. (2015) performed DNA methylation microarray analyses. They detected a correlation between histological pathology and the differential methylation of numerous autosomal genes in vascular tissue, providing potential biomarkers of damage severity and treatment targets (Valencia-Morales et al., 2015). Genes such as Drosophila headcase (HECA), early B-cell factor 1 (EBF1), and nucleotide-binding oligomerization domain containing 2 (NOD2) are significantly hypomethylated, whereas mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4), zinc finger E-box binding homeobox 1 (ZEB1), and proto-oncogene tyrosine-protein kinase (FYN) are hypermethylated in atheromatous plaque lesions compared to the plaque-free intima (Yamada et al., 2014). Another study described differentially methylated regions in genes associated with atherosclerosis in swine aorta endothelial cells (Jiang et al., 2015). Low-density lipoprotein cholesterol risk factor upregulates DNMT1, which methylates and represses the Krüppel-like factor 2 gene (KLF2) promoter. KLF2 is a transcription factor essential for endothelium homeostasis, and its repression results in endothelial dysfunction (Kumar et al., 2013). Similarly, DNMT3a upregulation in human aortic endothelial cells exposed to disturbed flow induces the methylation and repression of the Krüppel-like factor 4 gene (KLF4) promoter, increasing regional atherosusceptibility (Jiang et al., 2014). In an attempt to determine biomarkers of atherosclerosis in the primary stages, the DNA methylation status was determined in a selection of gene promoters associated with the disease. They analyzed the promoter methylation of ATP binding cassette subfamily A member 1 (ABCA1), TIMP metallopeptidase inhibitor 1 (TIMP1), and acetyl-CoA acetyltransferase 1 (ACAT1) and observed significant alterations in the peripheral blood of atherosclerosis patients (Ma et al., 2016). A recent study found that SMAD7 expression is decreased and its promoter highly methylated in atherosclerotic plaques compared to normal artery walls. There was also increased DNA methylation of the SMAD7 promoter in the peripheral blood of atherosclerosis patients. Thus, the SMAD7 promoter is hypermethylated in atherosclerosis patients and their atherosclerotic plaques, with a positive association with homocysteine levels (Wei et al., 2018). Moreover, increased 5mC and 5-hmC levels, which indicate DNA methylation and hydroxymethylation, respectively, have been demonstrated in peripheral blood mononuclear cells from elderly patients with coronary heart disease. These results positively correlate with the severity of coronary atherosclerosis (Jiang et al., 2019).
MicroRNAs have also been identified as attractive epigenetic biomarkers for atherosclerosis. Li et al. (2011) examined miRNA levels in serum samples and the intima of atherosclerosis obliterans patients and compared them to controls. They observed increased levels of miR-27b, miR-130a, and miR-210 in serum and sclerotic tissue from patients, proposing these miRNAs as epigenetic biomarkers for early stages of the disease (Li et al., 2011). Later, a study with a reduced number of patients suggested that elevated levels of circulating miR-17-5p may be a useful biomarker in the diagnosis of coronary atherosclerosis (Chen et al., 2015a).
Microparticles secreted by human coronary artery smooth muscle cells are a different source of cardiovascular biomarkers. These extracellular vesicles can contain miRNAs, such as miR-21-5p, miR-143-3p, miR-145-5p, miR-221-3p, and miR-222-3p. Lower levels of miR-143-3p and miR-222-3p have been found in microparticles derived from atherosclerotic plaque areas compared to nonatherosclerotic areas (de Gonzalo-Calvo et al., 2016).
Huang et al. (2016b) evaluated the expression of miR-30 in patients with essential hypertension compared to control individuals. They observed a reduction in miR-30 levels in the hypertensive patients and in the increased carotid intima-media thickness group. Thus, the authors suggested that circulating miR-30 may be a useful noninvasive atherosclerosis biomarker for patients with essential hypertension (Huang et al., 2016b). Later, the authors also identified higher levels of miR-92a as a possible biomarker of atherosclerosis in the same type of patients (Huang et al., 2017a).With the aim of investigating correlations between circulating miRNAs specific for HF and atherosclerosis in HF patients, Vegter et al. (2017) assessed miRNAs levels and related them to biomarkers associated with atherosclerotic disease and rehospitalizations of cardiovascular patients. They demonstrated a consistent trend between a high number of atherosclerosis manifestations and lower levels of miR-18a-5p, miR-27a-3p, miR-199a-3p, miR-223-3p, and miR-652-3p. Thus, lower levels of circulating miRNAs in HF patients with atherosclerotic disease and an elevated probability of cardiovascular-related rehospitalization were described (Vegter et al., 2017). High levels of miR-33a have also been demonstrated to be a potential cause of cholesterol accumulation and to exacerbate vessel walls inflammation in atherosclerotic disease. Thus, plasma miR-33a has been proposed as a suitable biomarker in atherosclerosis (Kim et al., 2017).
In an attempt to identify more atherosclerosis biomarkers, Hao and Fan (2017) performed microarray analysis using the plasma from apoE−/− mice and discovered that a reduction in miR-126 levels is a good indicator of atherosclerotic disease. They also determined that miR-126 is involved in the mitogen-associated protein kinase (MAPK) signaling pathway, reducing cytokine release and progressing atherosclerotic pathogenesis (Hao and Fan, 2017). In contrast, Gao et al. (2019) determined that higher expression levels of miR-126 and miR-143 correlate with the presence and severity of cerebral atherosclerosis (Gao et al., 2019). In another study, the authors evaluated the synergy of circulating miRNAs with cardiovascular risk factors to estimate the presence of atherosclerosis in ischemic stroke patients. They identified miR-212 as a novel marker that enhances the estimation of atherosclerosis presence in combination with hemoglobin A1c, high-density lipoprotein cholesterol, and lipoprotein(a) (Jeong et al., 2017). Another candidate biomarker for atherosclerosis is miR-200c. The authors analyzed plaque instability in the carotid arteries of patients undergoing carotid endarterectomy by examining the expression of miR-200c. Higher expression of miR-200c positively correlated with instability biomarkers, such as monocyte chemoattractant protein-1, cyclooxygenase-2, interleukin 6 (IL-6), metalloproteinases, and miR-33a/b, and negatively correlated with stability biomarkers, such as ZEB1, endothelial nitric oxide synthase, forkhead boxO1, and Sirtuin1. Thus, miR-200c could be a biomarker of atherosclerotic plaque progression and clinically useful for identifying patients at high embolic risk (Magenta et al., 2018). Along the same lines, lower serum levels of miR-638 may be a suitable biomarker of plaque vulnerability and ischemic stroke in individuals with high cardiovascular risk (Luque et al., 2018). With the intention to explore the role of miRNAs associated with carotid atherosclerosis, Mao et al. (2018) analyzed the genes differentially expressed between primary and advanced atherosclerotic plaques using two public datasets from the Gene Expression Omnibus (GEO) databases. The authors found a total of 23 miRNAs and focused on miR-19A, miR-19B, miR-126, and miR-155, which may be considered biomarkers of carotid atherosclerosis (Mao et al., 2018). In addition, Li et al. (2018b) identified downregulation of specific circulating miR-664a-3p as a biomarker of atherosclerosis in patients with obstructive sleep apnea and enlarged maximum carotid intima-media thickness (Li et al., 2018b).
Circulating miR-221 and miR-222 could also be suitable biomarkers for the diagnosis of atherosclerosis, as lower levels of these miRNAs correlate with the disease (Bildirici et al., 2018; Yilmaz et al., 2018). However, higher levels have been found in samples from coronary atherosclerotic plaques and internal mammary arteries (Bildirici et al., 2018). On the other hand, higher circulating levels of miR-29c, miR-122, and miR-155 in coronary atherosclerosis patients might allow noninvasive detection of the disease and its severity (Huang et al., 2018; Qiu and Ma, 2018; Wang and Yu, 2018). In another interesting study that assessed whether atherosclerosis of different arterial territories, not including the coronary artery, is associated with specific circulating miRNAs, the investigators were able to identify specific miRNA profiles for each territory with atherosclerotic disease. These findings may provide a pathophysiological understanding and be useful for selecting potential biomarkers for clinical practice (Pereira-da-Silva et al., 2018).
Myocardial Infarction
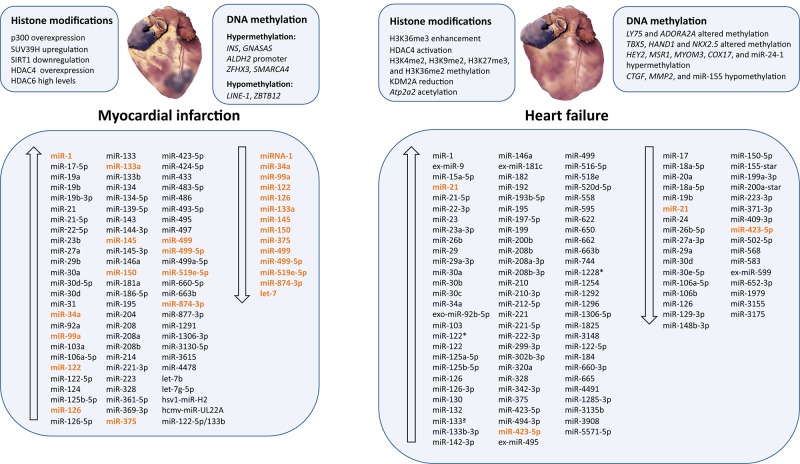
Acute MI (AMI) is a threatening disease worldwide. Early and accurate differential diagnosis is critical for immediate medical intervention and improved prognosis (Reed et al., 2017). In particular, it is important to notice that patients with ST-segment elevation MI (STEMI) have different requirements than patients with non-STEMI (NSTEMI). For the first group, reperfusion therapy should be administered quickly to reduce infarct size and mortality (Authors/Task Force members et al., 2014). However, in NSTEMI patients, revascularization strategies are recommended based on individual clinical characteristics (Reed et al., 2017). Therefore, biomarkers with the capacity to diagnose and personalize a therapeutic schedule in AMI would be of great interest. Currently, the favored diagnostic biomarkers of AMI are cardiac troponin I (cTnI) and T (cTnT), both of which are released from necrotic cardiomyocytes within 2 to 4 h post-MI (Babuin and Jaffe, 2005), with maximum levels at 24 to 48 h and lasting for more than 1 week (Jaffe et al., 2006). For this reason, small repeat infarctions after the main infarction are difficult to detect. Thus, it is fundamental to identify biomarkers for very early diagnosis of STEMI and for monitoring the entire pathological process of AMI ( Table 3 and Figure 3 ).
Table 3.
Epigenetic biomarkers in myocardial infarction.
| Epigenetic modification | Biomarker | Regulation in myocardial infarction | Sample source | Study type | References |
|---|---|---|---|---|---|
| DNA methylation | INS, GNASAS | Hypermethylation | Leukocytes | Clinical | (Talens et al., 2012) |
| LINE-1, ZBTB12 | Hypomethylation | White blood cells | Clinical | (Guarrera et al., 2015) | |
| ALDH2 promoter | Hypermethylation | Experimental: rat model of MI | Experimental: rat model of MI | (Wang et al., 2015) | |
| ZFHX3, SMARCA4 | Methylation | Whole blood | Clinical | (Nakatochi et al., 2017) | |
| Histone modifications | p300 | Overexpression | Myocardium | Experimental: mouse model of MI in HATmut p300-Tg mice | (Miyamoto et al., 2006) |
| SUV39H, SIRT1 | SUV39H upregulation and SIRT1 downregulation | H9C2 cells primary rat neonatal ventricular myocytes | Experimental: mouse model of MI in SUV39H−/− mice | (Yang et al., 2017a) | |
| HDAC4 | Overexpression | Myocardium | Experimental: mouse model of MI in MHC-HDAC4-Tg mice | (Zhang et al., 2018b) | |
| HDAC6 | Higher levels | Myocardium | Experimental: rat model of MI | (Nagata et al., 2019) | |
| miRNA | miR-1 | Higher levels | Plasma | Clinical | (Ai et al., 2010) |
| miR-31, miR-126, miR-214, miR-499-5p | miR-31, miR-214: upregulation; miR-126, miR-499-5p: downregulation | Myocardium | Experimental: rat model of MI | (Shi et al., 2010) | |
| miR-499 | Higher levels | Tissues and plasma | Clinical | (Adachi et al., 2010) | |
| miR-1, miR-133a, miR-133b, miR-499-5p, miR-122, miR-375 | miR-1, miR-133a, miR-133b, miR-499-5p: upregulation; miR-122, miR-375: downregulation | Plasma | Clinical and experimental: mouse model of MI | (D’Alessandra et al., 2010) | |
| miR-1, miR-126 | miR-1: increased; miR-126: decreased | Plasma | Clinical | (Long et al., 2012a) | |
| miR-133a | Higher levels | Plasma | Clinical | (Eitel et al., 2012) | |
| miR-30a, miR-195, let-7b | miR-30a, miR-195: increased; let-7: decreased | Plasma | Clinical | (Long et al., 2012b) | |
| miR-499-5p | Higher levels | Plasma | Clinical | (Olivieri et al., 2013) | |
| miR-1, miR-133a, miR-208b, miR-499 | Higher levels | Plasma | Clinical | (Li et al., 2013) | |
| miR-150 | Downregulation | plasma | Clinical | (Devaux et al., 2013) | |
| miR-133a | Higher levels | Plasma | Clinical | (Wang et al., 2013a) | |
| miR-21-5p, miR-361-5p, miR-519e-5p | miR-21-5p, miR-361-5p: increased; miR-519e-5p: reduced | Plasma | Clinical | (Wang et al., 2014b) | |
| miR-208a, miR-499 | Higher levels in serum; miR-499: lower levels in scar, miR-208a: unchanged in scar | Serum and heart tissues | Experimental: mouse model of MI | (Xiao et al., 2014) | |
| miR-208b, miR-34a | Higher levels | Plasma | Clinical | (Lv et al., 2014) | |
| miR-328, miR-134 | Higher levels | Plasma | Clinical | (He et al., 2014) | |
| miR-133, miR-1291, miR-663b | Higher levels | Plasma | Clinical | (Peng et al., 2014) | |
| miR-497 | Upregulation | Plasma | Clinical | (Li et al., 2014b) | |
| miR-1 | Higher levels | Plasma | Clinical | (Li et al., 2014a) | |
| miR-19a | Higher levels | Plasma | Clinical | (Zhong et al., 2014) | |
| miR-486-3p, miR-150-3p, miR-126-3p, miR-26a-5p, and miR-191-5p | miR-486-3p, miR-150-3p: upregulation; miR-126-3p, miR-26a-5p, miR-191-5p: downregulation | Serum | Clinical | (Hsu et al., 2014) | |
| miR-145 | Higher levels | Serum | Clinical | (Dong et al., 2015) | |
| hsa-miR-493-5p, hsa-miR-369-3p, hsa-miR-495, hsa-miR-3615, hsa-miR-433, hsa-miR-877-3p, hsa-miR-1306-3p, hsv1-miR-H2, hsa-miR-3130-5p, hcmv-miR-UL22A | hsa-miR-493-5p, hsa-miR-369-3p, hsa-miR-495, hsa-miR-3615, hsa-miR-433: upregulation, hsa-miR-877-3p, hsa-miR-1306-3p, hsv1-miR-H2, hsa-miR-3130-5p, hcmv-miR-UL22A: downregulation | Plasma | Clinical | (Liang et al., 2015) | |
| miR-499 | Higher levels | Plasma | Clinical | (Zhang et al., 2015b) | |
| miR-486, miR-150 | Higher levels | Plasma | Clinical | (Zhang et al., 2015c) | |
| miR-499 | Higher levels | Plasma | Clinical | (Chen et al., 2015b) | |
| miR-146a, miR-21 | Higher levels | Plasma | Clinical | (Liu et al., 2015a) | |
| miR-1, miR-208, miR-499 | Higher levels | Plasma | Clinical | (Liu et al., 2015b) | |
| miR-208a | Higher levels | Plasma | Clinical | (Białek et al., 2015) | |
| miR-208 | Overexpression | Plasma | Clinical | (Han et al., 2015) | |
| miR-122-5p | Higher levels | Plasma | Clinical | (Yao et al., 2015) | |
| miR-21 | Higher levels | Plasma | Clinical | (Zhang et al., 2016) | |
| miR-99a | Downregulation | Plasma | Clinical | (Yang et al., 2016a) | |
| miR-19b-3p, miR-134-5p and miR-186-5p | Higher levels | Plasma | Clinical | (Wang et al., 2016b) | |
| miR-106a-5p, miR-424-5p, let-7g-5p, miR-144-3p, miR-660-5p | Higher levels | Blood | Clinical | (Bye et al., 2016) | |
| miR-19b-3p, miR-134-5p and miR-186-5p | Overexpression | Plasma | Clinical | (Wang et al., 2016b) | |
| miR-125b-5p, miR-30d-5p | Overexpression | Plasma | Clinical | (Jia et al., 2016) | |
| miR-423-5p, miR-30d | Overexpression | Plasma | Clinical | (Eryılmaz et al., 2016) | |
| miR-221-3p | Overexpression | Plasma | Clinical | (Coskunpinar et al., 2016) | |
| miR-208a | Overexpression in myocardium and high levels in serum | Myocardium and serum | Experimental: rat model of MI | (Feng et al., 2016) | |
| miR-133b, miR-22-5p | Upregulation | Serum/plasma | Clinical | (Maciejak et al., 2016) | |
| miR-103a | Higher levels in plasma | Plasma and peripheral blood mononuclear cells | Clinical and experimental: in vitro | (Huang et al., 2016a) | |
| miR-122-5p/133b | High ratio | Serum | Clinical | (Cortez-Dias et al., 2016) | |
| miR499a-5p | Higher levels | Plasma | Clinical | (O’Sullivan et al., 2016) | |
| miR-181a | Higher levels | Plasma | Clinical | (Zhu et al., 2016) | |
| miR-145 | Decreased | Plasma | Clinical | (Zhang et al., 2017b) | |
| miR-133a | Higher levels | Plasma | Clinical | (Yuan et al., 2016) | |
| miR-208b | Higher levels | Plasma | Clinical | (Liu et al., 2017) | |
| miR-1, miR-92a, miR-99a, miR-143, miR-223 | miR-143: increased; miR-1, miR-92a, miR-99a, miR-223: decreased | Monocytes | Clinical | (Parahuleva et al., 2017) | |
| miR-92a | Higher levels | Plasma | Clinical | (Zhang et al., 2017c) | |
| miR-208b | Overexpression | Plasma | Clinical | (Zhang and Xie, 2017) | |
| miR-124 | Higher levels | Peripheral blood | Clinical | (Guo et al., 2017) | |
| miR-1, miR-21, miR-29b and miR-92a | miR-1, miR-21, miR-29b: increased | Plasma | Clinical | (Grabmaier et al., 2017) | |
| miR-874-3p | Downregulation | Plasma | Clinical | (Yan et al., 2017) | |
| pmiR-126 | Lower levels | Platelet | Clinical | (Li et al., 2017b) | |
| miR-133a | Lower levels | Serum/Plasma | Clinical | (Zhu et al., 2018) | |
| miR-21 | Upregulation | Serum | Clinical | (Wang et al., 2017b) | |
| miR-4478 | Higher levels | Serum | Clinical | (Gholikhani-Darbroud et al., 2017) | |
| miR-23b | Higher levels | Plasma | Clinical | (Zhang et al., 2018a) | |
| MiR-27a, miR-31, miR-1291, miR-139-5p, miR-204, miR-375 | Higher levels | GEO database | High throughput | (Wu et al., 2018a) | |
| miR-1, miR-133a, miR-34a | Lower levels | Myocardium | Experimental: mouse model of MI | (Qipshidze Kelm et al., 2018) | |
| miR-19b, miR-223, miR-483-5p | Higher levels | Plasma | Clinical | (Li et al., 2019) | |
| miR-17-5p, miR-126-5p, miR-145-3p | Higher levels | Plasma | Clinical | (Xue et al., 2019) | |
| miR-150 | Lower levels | Serum | Clinical | (Lin et al., 2019) | |
| miR-208b, miR-499 | Higher levels | Plasma | Clinical | (Devaux et al., 2012) |
Figure 3.
Epigenetic modifications and microRNAs biomarkers dysregulated in myocardial infarction and heart failure. Ascending arrows indicate higher levels or upregulation, and descending arrows denote lower levels or downregulation, both compared to control conditions. Those miRNAs presenting opposite results are shown in orange.
Regarding methylation as an indicator of MI, Talens et al. (2012) investigated the association between MI and DNA methylation at six loci described to be sensitive to prenatal nutrition. As a result, the researchers demonstrated that the risk of MI in women is associated with DNA hypermethylation at INS and GNASAS-specific loci (Talens et al., 2012). Moreover, microarray analyses investigating whole-genome DNA methylation using cases from the EPICOR study and EPIC-NL cohort (Fiorito et al., 2014) identified a hypomethylated region in the zinc finger and BTB domain-containing protein 12 (ZBTB12) and LINE-1, concluding that it is possible to detect specific methylation profiles in white blood cells a few years before MI occurs. This provides a promising early biomarker of MI (Guarrera et al., 2015). Another example is the hypermethylation of the aldehyde dehydrogenase 2 gene (ALDH2) promoter, which is associated with myocardial injury after MI in rats. The hypermethylation downregulates ALDH2, inhibiting its cardioprotective role (Wang et al., 2015). Rask-Andersen et al. (2016) performed an epigenome-wide association study to identify disease-specific alterations in DNA methylation. The authors observed differential DNA methylation at 211 CpG sites in individuals with MI, and some of these sites represented genes related to cardiac function, CVD, cardiogenesis, and recovery after ischemic injury. Their results highlight genes that might be important in the pathogenesis of MI or in recovery (Rask-Andersen et al., 2016). Along the same lines, a genome-wide DNA methylation and gene ontology analysis of white blood cells from a population-based study identified four differentially methylated sites in individuals who had a previous MI. Interestingly, they found a correlation between differences in DNA methylation in blood cells and the levels of growth differentiation factor 15 (GDF-15), which was overexpressed in the myocardium of MI patients (Ek et al., 2016). Later, a genome-wide DNA methylation study of whole blood samples from MI patients and controls identified two methylated regions in zinc finger homeobox 3 (ZFHX3) and SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 (SMARCA4) that were independently related to MI (Nakatochi et al., 2017).
Histone modifications are also involved in the pathological process of MI. To investigate the role of the HAT p300 in adverse left ventricular (LV) remodeling, Miyamoto et al. (2006) generated transgenic mice overexpressing wild-type p300 or its mutant in the heart. They subjected these mice to surgical MI and demonstrated that cardiac overexpression of p300 stimulated adverse LV remodeling. They concluded that the HAT activity of p300 is fundamental for the pathological course of MI (Miyamoto et al., 2006). Moreover, the class III deacetylase sirtuin 1 (SIRT1) is well known to confer a cardioprotective effect and is downregulated after cardiac injury. To understand the underlying mechanism, primary rat neonatal ventricular myocytes were exposed to ischemic or oxidative stress, leading to upregulation of the histone H3K9 methyltransferase SUV39H and downregulation of SIRT1. In addition, inhibition of SUV39H activity by chaetocin in wild-type mice and SUV39H-knockout mice protected against induced MI. SUV39H and heterochromatin protein 1 gamma cooperate to methylate the SIRT1 promoter and repress its transcription. Thus, the authors described a role for SUV39H linking SIRT1 repression to MI (Yang et al., 2017a). To examine the role of HDAC4 in the modulation of cardiac function after an MI, Zhang et al. (2018b) generated a myocyte-specific activated HDAC4-transgenic mouse. They found that HDAC4 overexpression increases myocardial fibrosis and hypertrophy, leading to cardiac dysfunction. Furthermore, the overexpression of activated HDAC4 aggravated cardiac dysfunction and increased adverse remodeling and apoptosis in the infarcted myocardium. Thus, HDAC4 is an indicator of heart injury (Zhang et al., 2018b). More recently, the role of HDAC6 in the development of HF following MI was investigated using a rat model. The authors found that the deacetylase activity of HDAC6 is increased after MI (Nagata et al., 2019).
Abundant research has focused on miRNAs as novel biomarkers for MI. MiR-1 levels have been analyzed in plasma from patients with AMI and found to be significantly elevated, but decreased to normal levels with medication (Ai et al., 2010; Long et al., 2012a). MiR-1, miR-126, and cTnI expression levels exhibited a similar tendency. Thus, circulating miR-1 and miR-126 may be useful indicators of AMI (Long et al., 2012a). However, when miR-1 was compared to cTnT, the authors found that cTnT was more specific and sensitive than miR-1 (Li et al., 2014a). Experiments performed in a rat model of MI revealed dysregulation of several miRNAs in the myocardium. Specifically, miR-31, miR-208, and miR-214 were upregulated, and miR-126 and miR-499-5p were downregulated in infarcted rats compared to sham-operated animals (Ji et al., 2009; Shi et al., 2010). MiR-499 has been widely analyzed as a possible biomarker of MI. MiR-499 has been reported to be produced almost exclusively in the heart and plasma and is significantly increased in individuals with AMI (Adachi et al., 2010; Devaux et al., 2012). MiR-499 positively correlates with serum creatine kinase-MB (CK-MB) and cTnI increasing their diagnostic accuracy (Chen et al., 2015b; Zhang et al., 2015b). Thus, miR-499 might be a suitable biomarker for MI and a predictor of myocardial ischemia risk (Adachi et al., 2010; Chen et al., 2015b; Zhang et al., 2015b). These results were confirmed in the mouse model of MI, with elevated serum miR-208a levels. However, the expression of miR-499 was significantly reduced in the MI region, whereas miR-208a remained unchanged in the same area. One explanation is that the damaged heart might release miR-499 into the circulation (Xiao et al., 2014). Other authors observed a high correlation between circulating miRNA-208a in STEMI patients and the levels of cTnI and CK-MB mass liberated from the infarcted zone (Białek et al., 2015). Thus, cardiac miR-208 and miR-499 seemed to be better biomarkers for predicting AMI than miR-1 (Liu et al., 2015b; Liu et al., 2018a). Another study analyzed the expression of miR-208a in the myocardium and serum of infarcted rats compared to control groups, as well as the expression of cAMP-PKA to evaluate the effect of this signaling pathway in the primary stages of MI; they found increased expression of miR-208a and cAMP-PKA. Moreover, the transfection of human myocardial cells with the miR-208a analog significantly increased the amount of cAMP-PKA protein. Thus, higher expression of miR-208a in the infarcted myocardium and serum may play a role in MI by affecting the cAMP-PKA signaling pathway (Feng et al., 2016).
D’Alessandra et al. (2010) investigated plasma levels of miRNAs in acute STEMI patients and infarcted mice and found higher levels of miR-1, miR-133a, miR-133b, and miR-499-5p compared to controls, whereas miR-122 and miR-375 levels were lower only in STEMI patients. Peak miR-1, miR-133a, and miR-133b expression correlated with cTnI levels in time, whereas the time course of miR-499-5p was slower (D’Alessandra et al., 2010). This was later confirmed in an exhaustive meta-analysis of relevant publications (Cheng et al., 2014). Similarly, geriatric patients with acute NSTEMI had greater miR-499-5p levels, exhibiting greater precision in diagnosis than cTnT in patients with mild ST elevation (Olivieri et al., 2013). On the other hand, increased levels of miR-1, miR-133a, miR-208b, and miR-499 in patients with AMI have been demonstrated to not be superior to cTnT (Li et al., 2013). The use of miR-133a as a biomarker in reperfused STEMI has been evaluated and compared to cardiovascular magnetic resonance imaging; high levels of miR-133a correlated with an increased infarct scar size, worse myocardial recovery, and prominent reperfusion injury. Nevertheless, miR-133a did not add further predictive information to cardiovascular magnetic resonance and conventional markers used in clinical practice in high-risk STEMI patients (Eitel et al., 2012). Moreover, the circulating levels of miR-133a were significantly enhanced in AMI patients compared to coronary heart disease and myocardial ischemia patients, presenting a similar trend as plasma cTnI concentration. Remarkably, we found a positive correlation between circulating miR-133a levels and the severity of coronary artery stenosis. Thus, circulating miR-133a may be a suitable tool for AMI diagnosis and predicting the presence and severity of coronary damage in coronary heart disease patients (Wang et al., 2013a). These results were later confirmed (Yuan et al., 2016; Zhu et al., 2018). Nevertheless, in another study analyzing miR-133a and miR-423-5p and their relationship with cardiac biomarkers, such as B-type natriuretic peptide (BNP), C-reactive protein, and cTnI in MI patients, an increase in circulating levels of both miRNAs was observed, but these changes were not associated with LV remodeling or BNP. The authors claimed that miR-133a and miR-423-5p are not useful biomarkers of LV remodeling after MI (Bauters et al., 2013). Another controversial pair of biomarkers is miR-423-5p and miR-30d, which were found to be higher in STEMI patients without a significant correlation with cTnI (Eryılmaz et al., 2016). In addition, the analysis of circulating miR-124a and miR-133 in STEMI and cardiogenic shock patients revealed a significant upregulation of both molecules. A negative correlation was found between miR-133 and MMP-9 levels, and a relationship between miR-124 and soluble ST2 levels, a marker associated with cardiac damage. Surprisingly, this study did not connect any of the miRNAs to the extent of the injury, disease progression, or the prognosis of patient outcomes. In this case, miRNAs would not bring any benefit compared to current markers (Goldbergova et al., 2018). Moreover, elevated circulating miR-1254 was described as predicting adverse LV remodeling in STEMI patients when compared to magnetic resonance imaging. However, the diagnosis and prognosis values of miR-1254 require further research (de Gonzalo-Calvo et al., 2018). Other investigations have described miR-150-3p and miR-486-3p as being upregulated, whereas miR-26a-5p, miR-126-3p, and miR-191-5p were significantly downregulated in STEMI patients (Hsu et al., 2014). In the same manner, circulating miR-19b-3p, miR-134-5p, and miR-186-5p have been reported to be significantly elevated in the initial stages of AMI. The expression of miR-19b-3p and miR-134-5p in the plasma reached a maximum earlier than miR-186-5p. However, all three positively correlated with cTnI and achieved peak expression before cTnI, which was 8 h after admission. Interestingly, the expression of these circulating miRNAs was not altered by heparin and medications for AMI, and the combination of all three miRNAs increased their diagnostic efficacy (Wang et al., 2016b). Moreover, a higher miR-122-5p/133b ratio was found in serum from STEMI patients (Cortez-Dias et al., 2016). The NSTEMI patients presented higher serum levels of miR-4478, soluble leptin receptor, cTnI, CKMB, urea, creatinine, glucose, cholesterol, TG, and ALP but lower levels of ALT compared to healthy individuals (Gholikhani-Darbroud et al., 2017). Moreover, there was an increase in miR-143 expression in monocytes from STEMI patients, whereas miR-1, miR-92a, miR-99a, and miR-223 expression was significantly reduced. Also, monocytic expression of miR-143 positively correlated with high-sensitivity C-reactive protein (hs-CRP), but not cTnT. These findings demonstrated that circulating monocytes could also be suitable biomarkers (Parahuleva et al., 2017).
Interestingly, cell-specific miRNA patterns are able to distinguish STEMI and NSTEMI patients. A correlation was found between miRNA 30d-5p and plasma, platelets, and leukocytes in patients with STEMI and NSTEMI. Furthermore, miR-221-3p and miR-483-5p were associated with plasma and platelets, but only in NSTEMI patients (Ward et al., 2013).
High levels of plasma miR-134 and miR-328 are described as being possible AMI biomarkers, as they correlate with a superior risk of developing HF and mortality. However, the miRNA levels were not superior to high-sensitivity cTnT (hs-cTnT) concentrations (He et al., 2014). In addition, elevated levels of miR-19a, miR-22-5p, miR-27a, miR-30a, miR-30a-5p, miR-30d-5p, miR-31, miR-34a, miR-122-5p, miR-125b-5p, miR-133, miR-133b, miR-139-5p, miR-150, miR-181a, miR-195, miR-204, miR-208, miR-208b, miR-221-3p, miR-375, miR-486, miR-497, miR-499a-5p, miR-663b, miR-1291, and let-7b can be potential biomarkers for AMI, increased risk of mortality, or HF (Devaux et al., 2012; Long et al., 2012b; Devaux et al., 2013; Li et al., 2014b; Lv et al., 2014; Peng et al., 2014; Zhong et al., 2014; Han et al., 2015; Yao et al., 2015; Zhang et al., 2015c; Coskunpinar et al., 2016; Jia et al., 2016; Maciejak et al., 2016; O’Sullivan et al., 2016; Zhu et al., 2016; Liu et al., 2017; Zhang and Xie, 2017; Alavi-Moghaddam et al., 2018; Maciejak et al., 2018; Wu et al., 2018a; Wang et al., 2019b). Other potential biomarkers for AMI are downregulated in patients’ plasma, such as miR-99a, miR-122-5p, and miR-874-3p (Yang et al., 2016a; Yan et al., 2017; Wang et al., 2019b). Interestingly, high levels of the combination of miR-21-5p, miR-361-5p, and miR-519e-5p or the reduction of miR-519e-5p correlates with cTnI concentrations, significantly increasing the diagnostic accuracy in AMI patients (Wang et al., 2014b;Liu et al., 2015a). Similarly, miR-21 and miR-124 have similar diagnostic ability compared to CK, CK-MB, and cTnI (Zhang et al., 2016; Guo et al., 2017).
In an attempt to predict HF and cardiovascular death after AMI, circulating miR-145, the N-terminal fragment of the precursor BNP, myocardial-band CK, and cTnI concentrations were analyzed for short- and long-term clinical outcomes. As a result, the authors concluded that miR-145 was a significant independent predictor of cardiac events, predicting long-term outcomes after AMI (Dong et al., 2015). Later, another group found that miR-145 levels were significantly lower in AMI patients and correlate with increased serum BNP and cTnT and decreased LV ejection fraction (Zhang et al., 2017b).
An miRNA array revealed differences in the miRNA expression patterns in patients with different phases of HF after MI. Specifically, human miR-369-3p, miR-433, miR-493-5p, miR-495, and miR-3615 were overexpressed, whereas miR-877-3p, miR-1306-3p, hsv1-miR-H2, miR-3130-5p, and hcmv-miR-UL22A were underexpressed in these patients. Thus, these circulating miRNAs are novel candidates as biomarkers of MI and HF (Liang et al., 2015).
An important aspect of circulating miRNAs as biomarkers is their temporal release, source, and transportation. Using the ischemia–reperfusion injury model, Deddens et al. (2016) showed that the ischemic myocardium releases extracellular vesicles. They also demonstrated that these extracellular vesicles transported specific miRNAs from the heart and muscle and were quickly detected in plasma. Interestingly, these vesicles had a high miRNAs content and rapid detection compared to traditional injury markers. This makes them a promising tool for the early detection of MI (Deddens et al., 2016). Along the same lines, microparticles and the expression levels of miR-92a were investigated in AMI and stable coronary artery disease patients and compared to cTnI. The number of microparticles and expression levels of miR-92a were higher in AMI patients than in the stable coronary artery disease patients and control groups, with a positive correlation between the levels of microparticles and cTnI. Thus, microparticles containing miR-92a may be suitable for MI diagnosis and possibly regulate dysfunctional endothelial tissue in AMI patients (Zhang et al., 2017c). However, according to Grabmaier et al. (2017), miR-92a seems to not be a good biomarker of adverse ventricular remodeling in post-AMI patients. The authors evaluated circulating miR-1, miR-21, miR-29b, and miR-92a from the SITAGRAMI trial population and found that miR-1, miR-21, and miR-29b expression was higher in AMI patients. The levels of miR-1 and miR-29b in plasma post-AMI correlated with variations in infarct volume, and the levels of miR-29b and changes in LV ejection fraction over time were also associated (Grabmaier et al., 2017).
Investigation of the expression of miR-103a in AMI patients with and without high blood pressure and the effect on endothelial cell function revealed increased levels of miR-103a in all patients but no changes in peripheral blood mononuclear cells. Moreover, miR-103a suppressed the expression of Piezo1 protein, which diminished the capacity to produce capillary tubes and the viability of human umbilical vein endothelial cells (HUVECs). Thus, miR-103a may take part in the development of high blood pressure and the initiation of AMI via regulation of Piezo1 expression (Huang et al., 2016a).
In a study based on samples from the HUNT study biobank, Bye et al. (2016) analyzed the utility of circulating miRNAs to predict future fatal AMI in healthy participants. MiR-424-5p and miR-26a-5p were associated exclusively with risk in men and women, respectively, suggesting a gender-specific association. They discovered that the best model for predicting future AMI consisted of miR-106a-5p, miR-424-5p, let-7g-5p, miR-144-3p, and miR-660-5p, and these miRNAs were proposed as a panel to enhance the prediction of AMI risk in healthy individuals (Bye et al., 2016).
Platelet activation is critical for AMI pathogenesis, but the role of platelet miRNAs (pmiRNAs) as biomarkers in AMI and their correlation with indices of platelet activity are unclear. Assessment of pmiR-126 expression in STEMI patients revealed reduced levels and a correlation with plasma cTnI. However, pmiR-126 expression did not correlate well with platelet activity indices, and its potential diagnostic utility is limited (Li et al., 2017b).
MiR-1, miR-133a, and miR-34a induce adverse structural remodeling to impair cardiac contractile function. Increased levels of all three miRNAs have been shown in the hearts of old MI mice compared to young MI mice, and the miR-1 increase was more prolonged and corresponded to LV wall thinning. This suggests that significantly increased levels of miR-1 in the aged post-MI heart could be a biomarker for high-risk prediction (Qipshidze Kelm et al., 2018). In addition, miRNA-21 has been reported to be overexpressed in the serum of ancient patients with AMI and to positively correlate with serum levels of CK-MB and cTnI. In vitro experiments with human cardiomyocytes transfected with the miR-21 mimic short hairpin RNA have shown that, following tumor necrosis factor α (TNF-α) induction, apoptosis rates are downregulated. The upregulation of miR-21 expression in the serum of elderly patients with AMI inhibited apoptosis induced by TNF-α in human cardiomyocytes via activation of the JNK/p38/caspase-3 signaling pathway (Wang et al., 2017b). Along the same lines, cardiomyocyte apoptosis and hypoxic reduction of cell growth can be promoted by miR-23b overexpression, suggesting that it could be a potential biomarker for STEMI (Zhang et al., 2018a).
A recent study explored the diagnostic use of circulating miRNAs in patients with acute chest pain in the emergency department. They found that higher circulating miR-19b, miR-223, and miR-483-5p levels may be clinically useful for AMI diagnosis in early phases (Li et al., 2019). Similarly, circulating miR-17-5p, miR-126-5p, and miR-145-3p levels are elevated in plasma from AMI patients. Combining these three miRNAs achieves a more precise AMI diagnosis (Xue et al., 2019). Interestingly, next-generation miRNA sequencing from whole blood samples has been useful for identifying new biomarkers of MI (Kanuri et al., 2018).
Heart Failure
Heart failure is a chronic and progressive condition that hampers the ability of the heart to pump enough blood to the body and fulfill its needs. Heart failure is caused by multiple disorders, such as hypertension, cardiomyopathy, MI, arrhythmias, or valvular diseases, among others (Khatibzadeh et al., 2013). Numerous scientific reports connect HF and epigenetic modifications ( Table 4 and Figure 3 ). High-density epigenome-wide mapping of DNA methylation in the myocardium and blood from dilated cardiomyopathy patients and healthy individuals has been analyzed. This technology has been used to find regions of epigenetic susceptibility and new biomarkers related to HF and heart dysfunction; they recognized different patterns of epigenetic methylation that were preserved through tissues—the CpGs regions identified as novel biomarkers of HF (Meder et al., 2017; Rau and Vondriska, 2017). Differentially methylated DNA regions were also identified in blood leukocytes from HF patients (Li et al., 2017a). Dilated cardiomyopathy is an important cause of HF. Genome-wide cardiac DNA methylation in idiopathic dilated cardiomyopathy patients revealed abnormal DNA methylation, which was related to important variations in the expression of lymphocyte antigen 75 (LY75) and adenosine receptor A2A (ADORA2A) mRNA (Haas et al., 2013). Similarly, genome-wide maps of DNA methylation and enrichment of histone 3 lysine-36 trimethylation (H3K36me3) in pathological and healthy hearts were analyzed. Differences in DNA methylation were found in promoter CpG islands, genes, intragenic CpG islands, and H3K36me3-rich regions of the genome. The promoters of upregulated genes had altered DNA methylation, but not the promoters of downregulated genes. In particular, an abundance of DUX4 transcripts was associated with differences in DNA methylation and H3K36me3 enrichment. Although further studies need to be carried out, there is evidence that the expression of genes critical for the development of cardiomyopathies may be controlled by the epigenome (Movassagh et al., 2011). Moreover, in patients with dilated cardiomyopathy, there is an altered methylation pattern in the regulatory regions of cardiac development genes, such as T-box protein 5 (TBX5), heart and neural crest derivatives expressed 1 (HAND1), and NK2 homeobox 5 (NKX2.5) (Jo et al., 2016). Koczor et al. (2013) also studied the differential methylation patterns in patients with dilated cardiomyopathy, which is characterized by congestive HF. Computational analysis detected few differentially methylated gene promoters (AURKB, BTNL9, CLDN5, and TK1). This study provides relevant information on DNA methylation and altered expression in dilated cardiomyopathy that would help in treatment (Koczor et al., 2013).
Table 4.
Epigenetic biomarkers in heart failure.
| Epigenetic modification | Biomarker | Regulation in heart failure | Sample source | Study type | References |
|---|---|---|---|---|---|
| DNA methylation | LY75 and ADORA2A | Aberrant DNA methylation | Left ventricle myocardium and zebrafish | Clinical and experimental: zebrafish | (Haas et al., 2013) |
| TBX5, HAND1, and NKX2.5 | Altered DNA methylation | Myocardium | Clinical | (Jo et al., 2016) | |
| HEY2, MSR1, MYOM3, COX17, miR-24-1, CTGF, MMP2, miR-155 | HEY2, MSR1, MYOM3, COX17, and miR-24-1: hypermethylation; CTGF, MMP2, and miR-155: hypomethylation | Myocardium | Clinical | (Glezeva et al., 2019) | |
| Histone modifications | H3K36me3 | H3K36me3 enhancement | Myocardium | Clinical | (Movassagh et al., 2011) |
| HDAC4 | HDAC4 activation | Myocardium | Clinical and experimental: mouse model of pressure overload | (Hohl et al., 2013) | |
| H3K4me2, H3K9me2, H3K27me3, H3K36me2, KDM2A | H3K4me2, H3K9me2, H3K27me3, and H3K36me2 methylation and KDM2A reduction | Myocardium | Experimental: mouse model of pressure overload | (Angrisano et al., 2014) | |
| Atp2a2 | Atp2a2 acetylation | Ventricular myocytes and myocardium | Clinical and experimental: mouse model of pressure overload in MHC-SIRT1−/− Tg mice and swine model of MI | (Gorski et al., 2019) | |
| miRNA | miR423-5p | Higher levels | Plasma | Clinical | (Tijsen et al., 2010) |
| miR-192 | Upregulation | Serum | Clinical | (Matsumoto et al., 2013) | |
| miR-122*, miR-200b, miR-520d-5p, miR-622, miR-1228*, miR-558 | miR-122*, miR-200b, miR-520d-5p, miR-622, miR-1228*: upregulation; miR-558: downregulation | Whole peripheral blood | Clinical | (Vogel et al., 2013) | |
| miR-103, miR-142-3p, miR-30b, miR-342-3p | Differentially expressed | Plasma | Clinical | (Ellis et al., 2013) | |
| miR-210, miR-30a | Upregulation | Serum | Clinical | (Zhao et al., 2013) | |
| miR-210 | Higher levels | Plasma, mononuclear cells, and skeletal muscles | Clinical and experimental: Dahl salt-sensitive rats | (Endo et al., 2013) | |
| miR-1 | Higher levels | Plasma | Clinical | (Zhang et al., 2013b) | |
| miR-423-5p | Positive transcoronary gradients | Transcoronary gradients | Clinical | (Goldraich et al., 2014) | |
| miR-423-5p | Lower levels | Plasma | Clinical | (Seronde et al., 2015) | |
| MiR-30c, miR-146a, miR-221, miR-328, miR-375 | Downregulation | Serum | Clinical | (Watson et al., 2015) | |
| miR-21, miR-650, miR-744, miR-516-5p, miR-1292, miR-182, miR-1228, miR-595, miR-663b, miR-1296, miR-1825, miR-299-3p, miR-662 miR-122, miR-3148, miR-518e, miR-129-3p, miR-3155, miR-3175, miR-583, miR-568, miR-30d, miR-200a-star, miR-1979, miR-371-3p, miR-155-star, miR-502-5p | miR-21, miR-650, miR-744, miR-516-5p, miR-1292, miR-182, miR-1228, miR-595, miR-663b, miR-1296, miR-1825, miR-299-3p, miR-662 miR-122, miR-3148, miR-518e: increased; miR-129-3p, miR-3155, miR-3175, miR-583, miR-568, miR-30d, miR-200a-star, miR-1979, miR-371-3p, miR-155-star, miR-502-5p: decreased | Serum | Clinical | (Cakmak et al., 2015) | |
| miR-1233, miR-671-5p, miR-183-3p, miR-190a, miR-193b-3p, miR-193b-5p, miR-211-5p, miR-494 | miR-1233, miR-671-5p: Upregulation; miR-183-3p, miR-190a, miR-193b-3p, miR-193b-5p, miR-211-5p, miR-494: downregulation | Whole blood and plasma | Clinical | (Wong et al., 2015) | |
| miR-1, miR-21 | miR-1: downregulation; miR-21: upregulation | Serum | Clinical | (Sygitowicz et al., 2015) | |
| miR-126 | Downregulation | Serum | Clinical | (Wei et al., 2015) | |
| miR-1, miR-21, miR-23, miR-29, miR-130, miR-195, miR-199 | Upregulation | Myocardial biopsy | Clinical | (Lai et al., 2015) | |
| miR-106a-5p, miR-223-3p, miR-652-3p, miR-199a-3p, miR-18a-5p | Downregulation | Plasma | Clinical | (Vegter et al., 2016) | |
| miR-148b-3p, miR-409-3p | Downregulation | Serum and left atrial tissue | Clinical | (Chen et al., 2016) | |
| miR-122-5p, miR-184 | Upregulation | H9C2 cells and blood and myocardium | Experimental: in vitro and rat model of post-MI HF | (Liu et al., 2016) | |
| miR-660-3p, miR-665, miR-1285-3p, miR-4491 | Upregulation | Plasma and heart | Clinical | (Li et al., 2016) | |
| miR-18a-5p, miR-26b-5p, miR-27a-3p, miR-30e-5p, miR-106a-5p, miR-199a-3p, miR-652-3p | Lower levels | Plasma | Clinical | (Ovchinnikova et al., 2016) | |
| miR-19b | Lower levels | Serum and myocardial | Clinical and experimental: in vitro | (Beaumont et al., 2017) | |
| miR-30d | Lower levels | Serum | Clinical | (Xiao et al., 2017) | |
| miR-195-3p | Higher levels | Plasma | Clinical | (He et al., 2017) | |
| miR-22-3p | Higher levels | Blood | Clinical | (van Boven et al., 2017) | |
| miR-150-5p | Downregulation | Blood | Clinical | (Scrutinio et al., 2017) | |
| miR-133b-3p, miR-208b-3p, miR-125a-5p, miR-125b-5p, miR-126-3p, miR-21-5p, miR-210-3p, miR-29a-3p, miR-320a, miR-494-3p | Upregulation | Blood | Experimental: sheep model of HF | (Wong et al., 2017) | |
| miR-146a | Upregulation | Exosomal and total plasma | Clinical and experimental: in vitro | (Beg et al., 2017) | |
| miR-9, miR-495, miR-599, miR-181c | ex-miR-9, ex-miR-181c, ex-miR-495: increased; ex-miR-599: decreased | Exosomal and total plasma | Experimental: dogs with myxomatous mitral valve disease, mitral valve prolapse | (Yang et al., 2017b) | |
| miR-21-5p, miR-23a-3p, miR-222-3p | Higher levels | Plasma | Clinical and experimental: rat model of post-MI HF | (Dubois-Deruy et al., 2017) | |
| miRNA-21 | Higher levels | Serum | Clinical | (Zhang et al., 2017a) | |
| miR-132 | Higher levels | Plasma | Clinical | (Masson et al., 2018) | |
| miR-1254, miR-1306-5p | Higher levels | Blood | Clinical | (Bayés-Genis et al., 2018) | |
| miR-423, miR-34a, miR-21-3p, miR-30a | miR-21-3p, miR-30a: Positive transcoronary gradient in non-ischemic HF; miR-423, miR-34a: Negative transcoronary gradient in ischemic HF | Transcoronary gradients | Clinical | (De Rosa et al., 2018) | |
| miR-3135b, miR-3908, miR-5571-5p | Upregulation | Plasma | Clinical | (Chen et al., 2018a) | |
| miR-302b-3p | Higher levels | Plasma | Clinical | (Li et al., 2018a) | |
| exo-miR-92b-5p | increased | Serum | Clinical | (Wu et al., 2018c) | |
| miR-26b, miR-208b, miR-499 | Higher levels | Peripheral blood mononuclear cells | Clinical | (Marketou et al., 2018) | |
| miR-423-5p, miR-221-5p, miR-212-5p, miR-193b-5p, miR-15a-5p, miR-208a-3p | Upregulation | Plasma, mouse myocardium and NRVMs cells | Clinical and experimental: in vitro, and murine model of hypertrophy and HF | (Shah et al., 2018b) | |
| miR-192 | Upregulation | Serum | Clinical | (Klenke et al., 2018) | |
| miR-34a, miR-208b, miR-126, miR-24, miR-29a | miR-34a, miR-208b, miR-126: upregulation; miR-24, miR-29a: downregulation | Serum | Clinical | (Lakhani et al., 2018) | |
| miR-17, miR-20a, miR-106b | Lower levels | Plasma | Clinical | (Shah et al., 2018a) | |
| miR-197-5p | Upregulation | Plasma | Clinical | (Liu et al., 2018b) | |
| miR-133a, miR-221 | Higher levels | Plasma | Clinical | (Guo et al., 2018) | |
| exo-miR-92b-5p | Higher levels | Serum | Clinical | (Wu et al., 2018b; Wu et al., 2018c) |
Furthermore, epigenetic modifications have been proposed to play an important role in HF progression in the murine model of pressure overload. The researchers observed a reduction in sarcoplasmic reticulum Ca2+ATPase (Atp2a2) levels and a significant induction of β-myosin-heavy chain (Myh7) mRNA levels. They also detected H3K4me2, H3K9me2, H3K27me3, and H3K36me2 and a reduction in the lysine-specific demethylase KDM2A after 8 weeks of transverse aortic constriction (Angrisano et al., 2014). Atp2a2 is a determinant of cardiac function, and its reduced activity is a clear feature of HF. Gorski et al. (2019) investigated the role of lysine acetylation in Atp2a2 function in HF patients and found that acetylation at lysine 492 is regulated by SIRT1 and HAT p300 and significantly reduced the gene activity (Gorski et al., 2019). All of this knowledge would be fundamental to identifying potential biomarkers and new epigenetic drugs in HF therapy. Interestingly, an association has been reported between epigenetic remodeling in the atrial natriuretic peptide (ANP) and BNP promoters and reactivation of the fetal gene program in HF. Their reported upregulation in HF patients did not respond to an increase in histone acetylation but HDAC4, which is exported from the nucleus. In contrast, demethylation of H3K9 and dissociation of heterochromatin protein 1 from gene promoters were regulated by HDAC4. Thus, HDAC4 is fundamental to histone methylation in HF caused by increased cardiac load and a potential target for treatment (Hohl et al., 2013). More recently, Glezeva et al. (2019) performed targeted DNA methylation sequencing to detect DNA methylation alterations in coding and ncRNA in cardiac interventricular septal tissue from HF patients. They found hypermethylation in HEY2, MSR1, MYOM3, COX17, and miR-24-1 and hypomethylation in CTGF, MMP2, and miR-155. Therefore, they defended a unique cohort of loci useful as diagnostic and therapeutic targets in HF (Glezeva et al., 2019).
More than 10 years ago, few reports suggested that specific miRNAs are differentially regulated in the failing heart (Divakaran and Mann, 2008; Small and Olson, 2011). Since then, an extensive evidence base has been published in the literature regarding the use of miRNAs as possible biomarkers for HF diagnosis and prognosis. In evaluating whether miRNAs can differentiate clinical HF from healthy individuals and from non-HF dyspnea, miRNA arrays have revealed miR423-5p enrichment in the blood of HF patients (Tijsen et al., 2010). However, criticisms have been raised in this study regarding age differences between groups, reduced sample size, and statistics (Kumarswamy et al., 2010). Moreover, patients with HF of different etiologies presented with different expression levels of circulating miRNAs. Ischemic HF patients were found to have a positive transcoronary gradient for miR-423-5p, miR-423, and miR-34a, but the nonischemic HF group was positive only for miR-21-3p and miR-30a. The transcoronary concentration gradient suggests that the failing heart may selectively release the miRNAs into the coronary circulation. These miRNAs could be useful for discriminating different etiologies of HF (Goldraich et al., 2014; De Rosa et al., 2018).
Circulating miRNAs have been screened in an attempt to identify any that could be used for the prognosis of ischemic HF in post-AMI patients. Knowing that p53 has been involved in HF development in mice (Sano et al., 2007), the authors took great interest in p53-responsive miRNAs. The serum levels of miR-34a, miR-192, and miR-194 were significantly and coordinately upregulated in AMI patients with ischemic HF progression, and all three were p53-responsive. Interestingly, these miRNAs were contained in extracellular vesicles, suggesting that they are circulating regulators of HF. Furthermore, there was a significant correlation between the LV end-diastolic dimension 1 year after AMI and the miR-194 and miR-34a expression levels. Thus, although further investigations are needed, these results suggest the usefulness of miR-34a, miR-192, and miR-194 in predicting the risk of HF progression after AMI (Evans and Mann, 2013; Matsumoto et al., 2013; Klenke et al., 2018).
Vogel et al. (2013) assessed the genome-wide miRNA expression profiles in HF patients with reduced ejection fraction (HFrEF). They demonstrated that dysregulated levels of miRNAs, such as miR-122*, miR-200b, miR-520d-5p, miR-622, miR-1228* (upregulated), or miR-558 (downregulated) significantly correlate with disease severity, as indicated by LV ejection fraction (Vogel et al., 2013). Moreover, Ellis et al. (2013) tried to find differences between HF patients and non–HF-related breathlessness, and between HFrEF and HF with preserved ejection fraction (HFpEF); although they found a differential expression of miR-103, miR-142-3p, miR-30b, and miR-342-3p in HF and breathless patients, individually, classical biomarkers such as NT-proBNP and hs-cTnT exhibited greater sensitivity and specificity. However, the combination of miRNAs with NT-proBNP significantly improved prediction performance (Ellis et al., 2013). Similarly, elevated plasma levels of miR-210 were reported in congestive HF patients, although no significant correlation was observed with BNP. However, patients with an improved BNP profile presented with low plasma miR-210 levels. MiR-210 might reflect a mismatch between heart contraction and oxygen demand in the peripheral tissues (Endo et al., 2013). Interestingly, miR-210 and miR-30a expression is upregulated in HF patients, with a tendency toward fetal values (Zhao et al., 2013). Moreover, changes in myocardial miRNA in patients with stable and end-stage HF partially resemble the fetal myocardium. Target mRNA levels negatively correlate with changes in highly expressed miRNAs in HF and fetal hearts. The circulation is dominated by miRNAs, fragments of tRNAs, and small cytoplasmic RNAs. Heart- and muscle-specific circulating miRNAs (myomirs) are also increased in advanced HF, correlating with cTnI levels. These findings support miRNA-based therapies and the use of circulating miRNAs as biomarkers for heart injury (Akat et al., 2014). Cardiac fibroblast–derived miRNAs, such as miR-660-3p, miR-665, miR-1285-3p, and miR-4491, have also been found to be significantly upregulated in heart and plasma during HF, discriminating patients from controls (Li et al., 2016). However, miRNAs in the pericardial fluid are not related to cardiovascular pathologies or clinically assessed stages of HF. MicroRNAs may be paracrine signaling factors that intervene in cardiac cells crosstalk (Kuosmanen et al., 2015).
In another study performed in patients with chronic congestive HF, microarray profiling demonstrated increased expression of miR-21, miR-122, miR-182, miR-299-3p, miR-516-5p, miR-518e, miR-595, miR-650, miR-662, miR-663b, miR-744, miR-1228, miR-1292, miR-1296, miR-1825, and miR-3148 and decreased expression of miR-30d, miR-129-3p, and miR-502-5p, miR-155-star miR-200a-star, miR-371-3p, miR-583, miR-568, miR-1979, miR-3155, and miR-3175. Among these miRNAs, miR-182 seemed to have a better prognostic value than hs-CRP (Cakmak et al., 2015). Furthermore, miR-30c, miR-146a, miR-221, miR-328, and miR-375 had different expression levels in HFrEF and HFpEF. The combination of two or more miRNAs with BNP could significantly improve the discrimination of these pathological conditions compared to BNP alone (Watson et al., 2015). Additional miRNAs have been identified as promising biomarkers to discriminate HF from healthy individuals and to differentiate HFrEF from HFpEF: miR-125a-5p, miR-183-3p, miR-190a, miR-193b-3p, miR-193b-5p, miR-211-5p, miR-494, miR-545-5p, miR-550a-5p, miR-638, miR-671-5p, miR-1233, miR-3135b, miR-3908, and miR-5571-5p. The use of a combination of miRNAs and NT-proBNP increases its discernment capacity (Schulte et al., 2015; Wong et al., 2015; Chen et al., 2018a). Similarly, increased levels of miR-133a and miR-221 can be used as suitable HF diagnostic biomarkers in elderly HF patients, and the combination of NT-proBNP and miR-133a can improve the diagnostic accuracy (Guo et al., 2018). Serum levels of miR-1, miR-21, and miR-208a have also been analyzed in symptomatic HF patients. Expression of miR-1 is reduced in symptomatic HF patients, with decreasing levels correlating with increasing severity. In contrast, miR-21 has been shown to be overexpressed with no relation to HF severity. No circulating miR-208a has been observed in symptomatic HF patients. A negative correlation between miR-1 expression and NT-proBNP has been reported in HF patients, whereas miR-21 and galectin-3 have been positively correlated. Therefore, dysregulated levels of miR-1 and miR-21 may be fundamental for HF progression (Sygitowicz et al., 2015). An inverse correlation between miR-1 levels and ejection fraction has also been reported. Thus, elevated levels of miR-1 may inhibit cardiac function and be a predictor of the onset of HF secondary to AMI (Zhang et al., 2013b).
MiR-126 has also been studied in atrial fibrillation and/or HF patients, with downregulated expression in patients and positive correlation with LV ejection fraction but a negative association with the cardiothoracic ratio and NT-proBNP. Thus, the reduction in miR-126 expression is a potential indicator of severity in atrial fibrillation and HF (Wei et al., 2015). A significant negative correlation has also been found between several miRNAs and classical clinical biomarkers indicative of a worse clinical outcome in HF patients. MiR-16-5p has been correlated to CRP, miR-106a-5p to creatinine, miR-223-3p to growth differentiation factor 15, miR-652-3p to soluble ST-2, miR-199a-3p to procalcitonin and galectin-3, and miR-18a-5p to procalcitonin (Vegter et al., 2016). Furthermore, an analysis of myocyte and fibroblast-related miRNAs and mRNAs in myocardium samples from HF patients and control individuals revealed that miR-1, miR-21, miR-23, miR-29, miR-130, miR-195, and miR-199 are significantly upregulated in HF patients, whereas miR-30, miR-133, miR-208, and miR-320 do not significantly change. Related mRNAs, such as caspase 3, collagenase I, collagenase III, and transforming growth factor (TGF), are also upregulated in HF patients. MicroRNAs involved in apoptosis, hypertrophy, and fibrosis are upregulated in the myocardium of HF patients and may be suitable biomarkers in the early stages of chronic HF and future therapeutic targets (Lai et al., 2015).
Evaluation of miR-148b-3p and miR-409-3p in mitral regurgitation patients, asymptomatic mitral regurgitation patients, and controls revealed that circulating and tissue miR-148b-3p and circulating miR-409-3p are significantly downregulated in mitral regurgitation patients with HF, and miR-148b-3p is significantly downregulated only in the mitral regurgitation patients without HF. Notably, the mRNAs of target genes of both miRNAs have been shown to be upregulated in HF patients with mitral regurgitation. Thus, circulating miR-148b-3p may be used as a biomarker of HF and miR-409-3p for incident HF in mitral regurgitation patients (Chen et al., 2016).
Specific overexpression of miR-221 in the hearts of transgenic mice has been shown to induce cardiac dysfunction and HF by impairing autophagy. In addition, in vitro miR-221 upregulation inhibits autophagic vesicle formation. Thus, autophagy balance and cardiac remodeling are regulated by miR-221 levels through modulation of the p27/CDK2/mTOR axis, and miR-221 might be a therapeutic target in HF (Su et al., 2015). Furthermore, high-throughput sequencing has been used to determine the differential miRNA pattern in a rat model of post-MI HF. Upregulation of miR-122-5p and miR-184 was found in HF rats, describing a proapoptotic role of both miRNAs (Liu et al., 2016). In another study using the same model, the authors identified a significant increase in miR-21-5p, miR-23a-3p, and miR-222-3p and their target SOD2 in the plasma and myocardium of HF rats. They showed a direct interaction between miR-222-3p and SOD2. An inhibition or increase in SOD2 expression was found when human cardiomyocytes were transfected with miR-222-3p mimic or inhibitor, respectively (Dubois-Deruy et al., 2017).
Myocardial fibrosis–related miRNAs, such as miR-19b, are reduced in the myocardium and serum of HF patients with aortic stenosis. Inhibition of miR-19b in cultured human fibroblasts increases the expression of connective tissue growth factor protein and the enzyme lysyl oxidase (LOX). This could lead to excessive collagen fibril cross-linking and a subsequent increase in LV stiffness in aortic stenosis patients, particularly those with HF. Thus, miR-19b could be a biomarker of alterations in the myocardial collagen network (Beaumont et al., 2017).
Numerous studies have been performed to find miRNAs with a predictive value in HF patients. Increased levels of miR-1, miR-21, miR-21-5p, miR-22-3p, miR-29a-3p, miR30d, miR-125a-5p, miR-125b-5p, miR-126-3p, miR-133b-3p, miR-195-3p, miR-197-5P, miR-208b-3p, miR-210-3p, miR-302b-3p, miR-320a, and miR-494-3p (Zhang et al., 2013b; He et al., 2017; van Boven et al., 2017; Wong et al., 2017; Xiao et al., 2017; Zhang et al., 2017a; Li et al., 2018a; Liu et al., 2018b;) or decreased levels of miR-17, miR-18a-5p, miR-20a, miR-150, miR-26b-5p, miR-27a-3p, miR-30e-5p, miR-106a-5p, miR-106b, miR-150-5p, miR-199a-3p, miR-423-5p, and miR-652-3p (Seronde et al., 2015; Ovchinnikova et al., 2016; Scrutinio et al., 2017; Shah et al., 2018a; Lin et al., 2019) have been described as potential biomarkers in HF patients. These discoveries may serve to develop miRNA-based therapies and to identify new pharmacological targets.
Beg et al. (2017) measured exosomal and total plasma miRNAs separately in HF patients to distinguish between the transfer of biological materials for signaling alteration in distant organs (exosomal) and the level of tissue damage (plasma). They found that the circulating exosomal miR-146a/miR-16 ratio was higher in HF patients, with miR-146a induced in response to inflammation. These results suggest circulating exosomal miR-146a as a biomarker of HF (Beg et al., 2017). Moreover, elevation of exosomal miRNA exo-miR-92b-5p has been suggested as a potential biomarker for the diagnosis of HF (Wu et al., 2018b; Wu et al., 2018c). In a preclinical study in dogs with myxomatous mitral valve disease, dysregulation of exosomal miR-9, miR-495, and miR-599 was observed as the dogs aged. In addition, levels of miR-9, miR-599, miR-181c, and miR-495 changed in myxomatous mitral valve disease. Thus, the exosomal miRNA expression level appears to be more specific to disease states than total plasma miRNA (Yang et al., 2017b). Furthermore, the downregulation of miR-425 and miR-744 in the plasma exosomes has been shown to induce cardiac fibrosis by suppressing TGFβ1 expression (Wang et al., 2018a).
Circulating miR-132 levels increased in chronic HF with disease severity, and lower levels improve risk prediction for HF readmission beyond traditional risk factors, but not for mortality. MiR-132 may be useful for finding strategies that would reduce rehospitalization in HF patients (Masson et al., 2018; Panico and Condorelli, 2018). Moreover, in an exhaustive analysis of two independent cohorts using a strict quality evaluation for miRNA testing, an association was found between high levels of miR-1254 and miR-1306-5p and mortality and HF hospitalization in HF patients. However, these two circulating miRNAs were not shown to improve standard predictors of prognostication, such as age, sex, hemoglobin, renal function, and NT-proBNP (Bayés-Genis et al., 2018).
MiR-26b, miR-208b, and miR-499 expression levels have been assessed in peripheral blood mononuclear cells from hypertensive HFpEF patients to evaluate their association with their exercise capacity. All three miRNAs were expressed at higher levels in the patients group, but miR-208b showed the strongest correlations with cardiopulmonary exercise test parameters, including oxygen uptake, exercise duration, and the minute ventilation–carbon dioxide production relationship (Marketou et al., 2018). In a study performed in patients and a mice model of hypertrophy and HF, miRNAs dysregulation was shown to occur during HF development in animals, with downregulation of target genes. These miRNAs were associated with adverse LV remodeling in humans, suggesting coordinated regulation of miRNA-mRNA. They also revealed target clusters of genes, such as autophagy, metabolism, and inflammation, implicated in HF mechanisms, (Shah et al., 2018b).
With the intention to establish a biomarkers panel useful for early detection of HF resulting from MI, Lakhani et al. (2018) found significant upregulation of miR-34a, miR-208b, miR-126, TGFβ-1, TNF-α, IL-6, and MMP-9 and reduced miR-24 and miR-29a levels. A positive association between IL-10 and ejection fraction in MI patients also suggested an important role of IL-10 in predicting HF (Lakhani et al., 2018).
Systems biology analyses of LV remodeling after MI allow molecular comprehensions; for example, miRNA modulation may be used as a marker of HF evolution. Two systems biology strategies were used to define an miRNA mark of LV remodeling in MI. They integrated either multiomics data (proteins and ncRNAs) produced from post-MI plasma or proteomic data generated from a rat model of MI. As a result, several miRNAs were associated with LV remodeling: miR-21-5p, miR-23a-3p, miR-222-3p, miR-17-5p, miR-21-5p, miR-26b-5p, miR-222-3p, miR-335-5p, and miR-375. These outcomes support the use of integrative systems biology analyses for the definition of miRNA marks of HF evolution (Charrier et al., 2019).
Limitations and Perspectives of the Epigenetic Biomarkers
Limitations of the current field include the lack of large multicenter studies to provide convincing evidence for clinical applicability. Rather than a single ncRNA, it is likely that there will be patterns of different ncRNAs and other biomarkers (e.g., protein-based) that, together with machine-learning algorithms, will provide more sensitive and specific diagnostic and prognostic approaches to CVDs. Also, several technical challenges must be overcome before CE-marked ncRNA biomarkers will enter the clinical realm. DNA methylation and histone modifications are epigenetic mechanisms that have been reported to be sources of potential biomarkers useful in clinical practice. However, each CVD is regulated by multiple epigenetic pathways, and different CVDs are regulated by the same epigenetic mechanism, most of which are still under study. For example, hypermethylation of H3K79 (Rodriguez-Iturbe, 2006; Duarte et al., 2012) and ACE2 promoter (Fan et al., 2017) in hypertensive patients has been described. Moreover, H3K4 and H3K9 were also hypermethylated in both mouse models of hypertension (Pojoga et al., 2011) and HF (Angrisano et al., 2014). This makes it difficult to select and implement a set of biomarkers for a particular CVD. Another potential problem is the quality of the samples, especially those obtained from collections in the pathology department. These samples are usually preserved in formaldehyde and paraffin, which highly degrades DNA. The stability, size, and integrity of a sample depend on the duration of fixation and storage (Kristensen et al., 2009). Thus, assessment of the quality of DNA is fundamental. However, the DNA methylation analysis can be performed successfully using polymerase chain reaction (PCR) methods with small amplicons in old samples (Tournier et al., 2012; Wong et al., 2014). In other cases, it is important to carefully adjust the protocol. It is also important to consider that frozen and paraffin-preserved samples may have different results, and they should not be compared without appropriate correction (García-Giménez et al., 2017).
Among the epigenetic biomarkers, miRNAs are the most promising, and numerous studies have been carried out in the last few years. The relatively easy detection and accessibility to samples in fluids, such as blood, urine, or saliva, make them very attractive. However, a few issues should be solved before their implementation in the clinical practice. The main problem is that miRNAs usually target multiple mRNAs from different genes, and one gene can be targeted by several miRNAs. This complex network should be deeply investigated before determining the use of a specific miRNA as a biomarker for the diagnosis or treatment of a particular disease (Akhtar et al., 2016). Regarding sample preparation, it is highly recommended to use plasma instead of whole blood, because if it is hemolyzed, the circulating miRNA content can be altered. Increasing the centrifugation time is also important in order to reduce platelet contamination (de Gonzalo-Calvo et al., 2017; García-Giménez et al., 2017).
Recently, great advances have been made to implement the new technology in the detection of new epigenetic biomarkers. However, a few concerns should be alleviated before their clinical implementation. Studies with big cohorts in different independent laboratories, using the same experimental design, sample preparation, methodology, and disease specifications, are necessary. Small patient cohorts should be considered as pilot studies before the validation of results in bigger sample analysis. The method of detection should be standardized for clinical application, and the clinical trials have to be randomized and prospective. It is also important to compare the new biomarkers with the classical biomarkers in order to validate them and determine their usefulness. The sensitivity and specificity for a certain disease also have to be determined for each biomarker (Engelhardt, 2012; García-Giménez et al., 2017). Regarding the method of DNA methylation detection, the luminometric methylation assay and the methylation analysis of CpG islands in repeatable elements (LINE-1) are widely used. Although there is a certain correlation with the measurements obtained with both methods, the comparison is not recommended, since a consistent bias between the results has been described (Knothe et al., 2016). Interestingly, a large multicenter study comparing DNA methylation assays compatible with routine clinical use has been performed. According to the authors, good agreement was observed between DNA methylation assays, which can be implemented in large-scale validation studies, development of new biomarkers, and clinical diagnostics (BLUEPRINT Consortium, 2016). The most used system to detect miRNAs is quantitative PCR, being the normalization protocol critical. Most laboratories use housekeeping genes or miRNAs as normalizers, changing their expression levels within serums. Another approach employs identical volumes of serum for all samples, generating different amounts of total RNA (Chen et al., 2008; Wang et al., 2009; Rockenbach et al., 2012). Both approaches include spike-in normalization, which consists of adding RNA of known sequence and quantity to calibrate measurements. However, spike-in normalization does not consider internal variation in circulating miRNA between different individuals. Thus, a combination of both methods should always be performed to guarantee results reliability (van Empel et al., 2012). Polymerase chain reaction technology has to be performed with rigorous controls to avoid artifacts in the amplification step. To overcome this problem, digital PCR based on the amplification of one single molecule per reaction constitutes a valuable option (Hindson et al., 2013). Another attractive alternative for accurate measuring RNAs is the direct nucleic acid sequencing, although it is still expensive when considering large screening analysis (Kozomara and Griffiths-Jones, 2011). Finally, it is also important to understand the processes controlling miRNAs release and stability. The correlation between circulating and tissue miRNAs is not clear, and several studies indicate that miRNA levels in blood are not a reflection of changes in the tissue of origin. The reason is that miRNAs can also be produced by immune cells (Zheng et al., 2018).
Concluding Remarks
Over the past few years, a great amount of research has focused on epigenetics and its dynamic cross-talk with genetics. Unveiling a personalized epigenetic pattern can provide a large amount of information on epigenetic machinery that could be employed to tailor diagnosis and therapeutic strategies in CVDs. Recent advances in technology and data analysis have made it possible to create detailed epigenetic maps, which may represent a new tool in the clinical practice to discern cardiovascular risk beyond traditional risk determinants. Epigenetic information can also help in predicting individual drug responses. Importantly, epigenetic biomarkers are gaining ground in the scientific community as tools for the diagnosis and prognosis of CVDs. However, discrepancies in specific diagnostic biomarkers make replication of the current results in independent laboratories, with multiple research centers and a big sample size, mandatory. All of this will lead to a standardized clinical application in the near future.
Author Contributions
CS-B and AB-G conceived the idea and wrote the manuscript with support from CG-M. CG-M performed the drawings and structure of the figures. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
This work was supported by grants from the Spanish Ministry of Economy and Competitiveness-MINECO (SAF2017-84324-C2-1-R), the Instituto de Salud Carlos III (PIC18/0014, PI18/00256), the Red de Terapia Celular–TerCel (RD16/0011/0006) and the CIBER Cardiovascular (CB16/11/00403) projects, as part of the Plan Nacional de I+D+I, and it was co-funded by ISCIII-Sudirección General de Evaluación y el Fondo Europeo de Desarrollo Regional (FEDER). This work was also funded by the Fundació La MARATÓ de TV3 (201516-10, 201502-20), the Generalitat de Catalunya (SGR2017 00483, SLT002/16/00234), the CERCA Programme/Generalitat de Catalunya, and “la Caixa” Banking Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AMI, acute myocardial infarction; ApoE, apolipoprotein E; BNP B-type natriuretic peptide; CK, creatine kinase; cTnI, cardiac troponin I; cTnT, cardiac troponin T; DOT1L, disruptor of telomeric silencing-1; ENaC, epithelial sodium channel; EZH2, enhancer of zeste homolog 2; GEO, Gene Expression Omnibus; HDAC, histone deacetylase; HF, heart failure; HFrEF, heart failure with reduced ejection fraction; HFpEF, heart failure with preserved ejection fraction; hs-cTnT, high-sensitivity cardiac troponin T; hs-CRP, high-sensitivity C-reactive protein; lncRNAs, long noncoding RNAs; LV, left ventricular; MI, myocardial infarction; miRNAs, microRNAs; ncRNAs, noncoding RNAs; NSTEMI, non- ST-segment elevation myocardial infarction; STEMI, ST-segment elevation myocardial infarction; pmiRNAs, platelet miRNAs; piRNAs, p-element-induced wimpy testis (PIWI)-interacting RNAs; tRNA, transfer RNA; ZEB1, zinc finger E-box binding homeobox 1.
Acknowledgments
We apologize to all authors whose work could not be mentioned because of space limitations or inadvertent omissions. We are greatly grateful to Sonia V Forcales for her comments and discussion on epigenetic regulation.
References
- Adachi T., Nakanishi M., Otsuka Y., Nishimura K., Hirokawa G., Goto Y., et al. (2010). Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin. Chem. 56, 1183–1185. 10.1373/clinchem.2010.144121 [DOI] [PubMed] [Google Scholar]
- Ai J., Zhang R., Li Y., Pu J., Lu Y., Jiao J., et al. (2010). Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem. Biophys. Res. Commun. 391, 73–77. 10.1016/j.bbrc.2009.11.005 [DOI] [PubMed] [Google Scholar]
- Akat K. M., Moore-McGriff D., Morozov P., Brown M., Gogakos T., Correa Da Rosa J., et al. (2014). Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc. Natl. Acad. Sci. U. S. A 111, 11151–11156. 10.1073/pnas.1401724111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M. M., Micolucci L., Islam M. S., Olivieri F., Procopio A. D. (2016). Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 44, 24–44. 10.1093/nar/gkv1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alavi-Moghaddam M., Chehrazi M., Alipoor S. D., Mohammadi M., Baratloo A., Mahjoub M. P., et al. (2018). A preliminary study of microRNA-208b after acute myocardial infarction: impact on 6-month survival. Dis. Markers 2018, 2410451–7. 10.1155/2018/2410451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alikhani-Koopaei R., Fouladkou F., Frey F. J., Frey B. M. (2004). Epigenetic regulation of 11 beta-hydroxysteroid dehydrogenase type 2 expression. J. Clin. Investig. 114, 1146–1157. 10.1172/JCI21647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angrisano T., Schiattarella G. G., Keller S., Pironti G., Florio E., Magliulo F., et al. (2014). Epigenetic switch at atp2a2 and myh7 gene promoters in pressure overload-induced heart failure. PLoS One 9, e106024. 10.1371/journal.pone.0106024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Authors/Task Force members. Windecker S., Kolh P., Alfonso F., Collet J.-P., Cremer J., et al. (2014). 2014 ESC/EACTS guidelines on myocardial revascularization: the task force on myocardial revascularization of the European Society of Cardiology (ESC) and the European Association for Cardio-Thoracic Surgery (EACTS) developed with the special contribution of the European Association of Percutaneous Cardiovascular Interventions (EAPCI). Eur. Heart J. 35, 2541–2619. 10.1093/eurheartj/ehu278 [DOI] [PubMed] [Google Scholar]
- Babuin L., Jaffe A. S. (2005). Troponin: the biomarker of choice for the detection of cardiac injury. CMAJ 173, 1191–1202. 10.1503/cmaj/051291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A., Rienstra M., Benjamin E. J. (2010). Cardiovascular epigenetics: basic concepts and results from animal and human studies. Circ. Cardiovasc. Genet. 3, 567–573. 10.1161/CIRCGENETICS.110.958744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista R., Marques C., Catarino S., Enguita F. J., Costa M. C., Matafome P., et al. (2018). MicroRNA-424(322) as a new marker of disease progression in pulmonary arterial hypertension and its role in right ventricular hypertrophy by targeting SMURF1. Cardiovasc. Res. 114, 53–64. 10.1093/cvr/cvx187 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauters C., Kumarswamy R., Holzmann A., Bretthauer J., Anker S. D., Pinet F., et al. (2013). Circulating miR-133a and miR-423-5p fail as biomarkers for left ventricular remodeling after myocardial infarction. Int. J. Cardiol. 168, 1837–1840. 10.1016/j.ijcard.2012.12.074 [DOI] [PubMed] [Google Scholar]
- Bayés-Genis A., Lanfear D. E., de Ronde M. W. J., Lupón J., Leenders J. J., Liu Z., et al. (2018). Prognostic value of circulating microRNAs on heart failure–related morbidity and mortality in two large diverse cohorts of general heart failure patients. Eur. J. Heart Fail. 20, 67–75. 10.1002/ejhf.984 [DOI] [PubMed] [Google Scholar]
- Beaumont J., López B., Ravassa S., Hermida N., José G. S., Gallego I., et al. (2017). MicroRNA-19b is a potential biomarker of increased myocardial collagen cross-linking in patients with aortic stenosis and heart failure. Sci. Rep. 7, 40696. 10.1038/srep40696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman M., Nederstigt C., Suchiman H. E. D., Kremer D., van der Breggen R., Lakenberg N., et al. (2010). Genome-wide association study (GWAS)–identified disease risk alleles do not compromise human longevity. Proc. Natl. Acad. Sci. U. S. A 107, 18046–18049. 10.1073/pnas.1003540107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg F., Wang R., Saeed Z., Devaraj S., Masoor K., Nakshatri H. (2017). Inflammation-associated microRNA changes in circulating exosomes of heart failure patients. BMC Res. Notes 10, 751. 10.1186/s13104-017-3090-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekkering S., van den Munckhof I., Nielen T., Lamfers E., Dinarello C., Rutten J., et al. (2016). Innate immune cell activation and epigenetic remodeling in symptomatic and asymptomatic atherosclerosis in humans in vivo . Atherosclerosis 254, 228–236. 10.1016/j.atherosclerosis.2016.10.019 [DOI] [PubMed] [Google Scholar]
- Białek S., Górko D., Zajkowska A., Kołtowski Ł., Grabowski M., Stachurska A., et al. (2015). Release kinetics of circulating miRNA-208a in the early phase of myocardial infarction. Kardiologia Polska 73, 613–619. 10.5603/KP.a2015.0067 [DOI] [PubMed] [Google Scholar]
- Bildirici A. E., Arslan S., Özbilüm Şahin N., Berkan Ö., Beton O., Yilmaz M. B. (2018). MicroRNA-221/222 expression in atherosclerotic coronary artery plaque versus internal mammarian artery and in peripheral blood samples. Biomarkers 23, 670–675. 10.1080/1354750X.2018.1474260 [DOI] [PubMed] [Google Scholar]
- BLUEPRINT Consortium (2016). Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat. Biotechnol. 34, 726–737. 10.1038/nbt.3605 [DOI] [PubMed] [Google Scholar]
- Bogdarina I., Welham S., King P. J., Burns S. P., Clark A. J. L. (2007). Epigenetic modification of the renin–angiotensin system in the fetal programming of hypertension. Circ. Res. 100, 520–526. 10.1161/01.RES.0000258855.60637.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bye A., Røsjø H., Nauman J., Silva G. J. J., Follestad T., Omland T., et al. (2016). Circulating microRNAs predict future fatal myocardial infarction in healthy individuals—the HUNT study. J. Mol. Cell Cardiol. 97, 162–168. 10.1016/j.yjmcc.2016.05.009 [DOI] [PubMed] [Google Scholar]
- Cakmak H. A., Coskunpinar E., Ikitimur B., Barman H. A., Karadag B., Tiryakioglu N. O., et al. (2015). The prognostic value of circulating microRNAs in heart failure: preliminary results from a genome-wide expression study. J. Cardiovasc. Med. (Hagerstown) 16, 431–437. 10.2459/JCM.0000000000000233 [DOI] [PubMed] [Google Scholar]
- Cao J., Yan Q. (2012). Histone ubiquitination and deubiquitination in transcription, DNA damage response, and cancer. Front. Oncol. 2, 26. 10.3389/fonc.2012.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier H., Cuvelliez M., Dubois-Deruy E., Mulder P., Richard V., Bauters C., et al. (2019). Integrative system biology analyses identify seven microRNAs to predict heart failure. Noncoding RNA 5, E22–E30. 10.3390/ncrna5010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi S. T., Mondon F., Jammes H., Buffat C., Mignot T.-M., Tost J., et al. (2007). Expressional and epigenetic alterations of placental serine protease inhibitors: SERPINA3 is a potential marker of preeclampsia. Hypertension 49, 76–83. 10.1161/01.HYP.0000250831.52876.cb [DOI] [PubMed] [Google Scholar]
- Chen F., Yang J., Li Y., Wang H. (2018. a). Circulating microRNAs as novel biomarkers for heart failure. Hellenic J. Cardiol. 59, 209–214. 10.1016/j.hjc.2017.10.002 [DOI] [PubMed] [Google Scholar]
- Chen J., Xu L., Hu Q., Yang S., Zhang B., Jiang H. (2015. a). MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int. J. Cardiol. 197, 123–124. 10.1016/j.ijcard.2015.06.037 [DOI] [PubMed] [Google Scholar]
- Chen M.-C., Chang T.-H., Chang J.-P., Huang H.-D., Ho W.-C., Lin Y.-S., et al. (2016). Circulating miR-148b-3p and miR-409-3p as biomarkers for heart failure in patients with mitral regurgitation. Int. J. Cardiol. 222, 148–154. 10.1016/j.ijcard.2016.07.179 [DOI] [PubMed] [Google Scholar]
- Chen S., Chen R., Zhang T., Lin S., Chen Z., Zhao B., et al. (2018. b). Relationship of cardiovascular disease risk factors and noncoding RNAs with hypertension: a case-control study. BMC Cardiovasc. Disord. 18, 58. 10.1186/s12872-018-0795-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Li S. (2017). Circulating microRNA as a novel biomarker for pulmonary arterial hypertension due to congenital heart disease. Pediatr. Cardiol. 38, 86–94. 10.1007/s00246-016-1487-3 [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., et al. (2008). Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- Chen X., Zhang L., Su T., Li H., Huang Q., Wu D., et al. (2015. b). Kinetics of plasma microRNA-499 expression in acute myocardial infarction. J. Thorac. Dis. 7, 890–896. 10.3978/j.issn.2072-1439.2014.11.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., Wang Q., You W., Chen M., Xia J. (2014). MiRNAs as biomarkers of myocardial infarction: a meta-analysis. PLoS ONE 9, e88566. 10.1371/journal.pone.0088566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.-M., Lee H.-A., Kim H. Y., Han H. S., Kim I. K. (2011). Expression of Na+-K+ -2Cl- cotransporter 1 is epigenetically regulated during postnatal development of hypertension. Am. J. Hypertens. 24, 1286–1293. 10.1038/ajh.2011.136 [DOI] [PubMed] [Google Scholar]
- Choi J.-H., Nam K.-H., Kim J., Baek M. W., Park J.-E., Park H.-Y., et al. (2005). Trichostatin A exacerbates atherosclerosis in low density lipoprotein receptor-deficient mice. Arteriosclerosis, Thrombosis, and Vascular Biol. 25, 2404–2409. 10.1161/01.ATV.0000184758.07257.88 [DOI] [PubMed] [Google Scholar]
- Choi S. Y., Yun J., Lee O. J., Han H. S., Yeo M. K., Lee M. A., et al. (2013). MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta 34, 799–804. 10.1016/j.placenta.2013.06.006 [DOI] [PubMed] [Google Scholar]
- Cortez-Dias N., Costa M. C., Carrilho-Ferreira P., Silva D., Jorge C., Calisto C., et al. (2016). Circulating miR-122-5p/miR-133b ratio is a specific early prognostic biomarker in acute myocardial infarction. Circ. J. 80, 2183–2191. 10.1253/circj.CJ-16-0568 [DOI] [PubMed] [Google Scholar]
- Coskunpinar E., Cakmak H. A., Kalkan A. K., Tiryakioglu N. O., Erturk M., Ongen Z. (2016). Circulating miR-221-3p as a novel marker for early prediction of acute myocardial infarction. Gene 591, 90–96. 10.1016/j.gene.2016.06.059 [DOI] [PubMed] [Google Scholar]
- Costantino S., Libby P., Kishore R., Tardif J.-C., El-Osta A., Paneni F. (2018). Epigenetics and precision medicine in cardiovascular patients: from basic concepts to the clinical arena. Eur. Heart J. 39, 4150–4158. 10.1093/eurheartj/ehx568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessandra Y., Devanna P., Limana F., Straino S., Di Carlo A., Brambilla P. G., et al. (2010). Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 31, 2765–2773. 10.1093/eurheartj/ehq167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta C., Chen M., Zhang H., Yang S., Zhang L. (2012). Chronic hypoxia during gestation causes epigenetic repression of the estrogen receptor-α gene in ovine uterine arteries via heightened promoter methylation. Hypertension 60, 697–704. 10.1161/HYPERTENSIONAHA.112.198242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gonzalo-Calvo D., Cediel G., Bär C., Núñez J., Revuelta-Lopez E., Gavara J., et al. (2018). Circulating miR-1254 predicts ventricular remodeling in patients with ST-segment-elevation myocardial infarction: a cardiovascular magnetic resonance study. Sci. Rep. 8, 15115. 10.1038/s41598-018-33491-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gonzalo-Calvo D., Cenarro A., Civeira F., Llorente-Cortés V. (2016). microRNA expression profile in human coronary smooth muscle cell–derived microparticles is a source of biomarkers. Clin. Investig. Arterioscler. 28, 167–177. 10.1016/j.arteri.2016.05.005 [DOI] [PubMed] [Google Scholar]
- de Gonzalo-Calvo D., Iglesias-Gutiérrez E., Llorente-Cortés V. (2017). Biomarcadores epigenéticos y enfermedad cardiovascular: los microARN circulantes. Rev. Española Cardiol. 70, 763–769. 10.1016/j.recesp.2017.02.027 [DOI] [PubMed] [Google Scholar]
- De Rosa S., Eposito F., Carella C., Strangio A., Ammirati G., Sabatino J., et al. (2018). Transcoronary concentration gradients of circulating microRNAs in heart failure. Eur. J. Heart Fail. 20, 1000–1010. 10.1002/ejhf.1119 [DOI] [PubMed] [Google Scholar]
- Deddens J. C., Vrijsen K. R., Colijn J. M., Oerlemans M. I., Metz C. H. G., van der Vlist E. J., et al. (2016). Circulating extracellular vesicles contain miRNAs and are released as early biomarkers for cardiac injury. J. Cardiovasc Transl. Res. 9, 291–301. 10.1007/s12265-016-9705-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaux Y., Vausort M., Goretti E., Nazarov P. V., Azuaje F., Gilson G., et al. (2012). Use of circulating microRNAs to diagnose acute myocardial infarction. Clin. Chem. 58, 559–567. 10.1373/clinchem.2011.173823 [DOI] [PubMed] [Google Scholar]
- Devaux Y., Vausort M., McCann G. P., Zangrando J., Kelly D., Razvi N., et al. (2013). MicroRNA-150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ. Cardiovasc. Genet. 6, 290–298. 10.1161/CIRCGENETICS.113.000077 [DOI] [PubMed] [Google Scholar]
- Divakaran V., Mann D. L. (2008). The emerging role of microRNAs in cardiac remodeling and heart failure. Circ. Res. 103, 1072–1083. 10.1161/CIRCRESAHA.108.183087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y.-M., Liu X.-X., Wei G.-Q., Da Y.-N., Cha L., Ma C.-S. (2015). Prediction of long-term outcome after acute myocardial infarction using circulating miR-145. Scand. J. Clin. Lab. Invest. 75, 85–91. 10.3109/00365513.2014.981855 [DOI] [PubMed] [Google Scholar]
- Duarte J. D., Zineh I., Burkley B., Gong Y., Langaee T. Y., Turner S. T., et al. (2012). Effects of genetic variation in H3K79 methylation regulatory genes on clinical blood pressure and blood pressure response to hydrochlorothiazide. J. Transl. Med. 10, 56. 10.1186/1479-5876-10-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois-Deruy E., Cuvelliez M., Fiedler J., Charrier H., Mulder P., Hebbar E., et al. (2017). MicroRNAs regulating superoxide dismutase 2 are new circulating biomarkers of heart failure. Sci. Rep. 7, 14747. 10.1038/s41598-017-15011-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitel I., Adams V., Dieterich P., Fuernau G., de Waha S., Desch S., et al. (2012). Relation of circulating MicroRNA-133a concentrations with myocardial damage and clinical prognosis in ST-elevation myocardial infarction. Am. Heart J. 164, 706–714. 10.1016/j.ahj.2012.08.004 [DOI] [PubMed] [Google Scholar]
- Ek W. E., Hedman Å. K., Enroth S., Morris A. P., Lindgren C. M., Mahajan A., et al. (2016). Genome-wide DNA methylation study identifies genes associated with the cardiovascular biomarker GDF-15. Hum. Mol. Genet. 25, 817–827. 10.1093/hmg/ddv511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis K. L., Cameron V. A., Troughton R. W., Frampton C. M., Ellmers L. J., Richards A. M. (2013). Circulating microRNAs as candidate markers to distinguish heart failure in breathless patients. Eur. J. Heart Fail. 15, 1138–1147. 10.1093/eurjhf/hft078 [DOI] [PubMed] [Google Scholar]
- Endo K., Naito Y., Ji X., Nakanishi M., Noguchi T., Goto Y., et al. (2013). MicroRNA 210 as a biomarker for congestive heart failure. Biol. Pharm. Bull. 36, 48–54. 10.1248/bpb.b12-00578 [DOI] [PubMed] [Google Scholar]
- Engelhardt S. (2012). Small RNA biomarkers come of age. J. Am. Coll. Cardiol. 60, 300–303. 10.1016/j.jacc.2012.04.018 [DOI] [PubMed] [Google Scholar]
- Eryılmaz U., Akgullu C., Beser N., Yıldız Ö., Kurt Ömürlü İ., Bozdogan B. (2016). Circulating microRNAs in patients with ST-elevation myocardial infarction. Anatol. J Cardiol 16, 392–396. 10.5152/AnatolJCardiol.2015.6603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S., Mann D. L. (2013). Circulating p53-responsive microRNAs as predictive biomarkers in heart failure after acute myocardial infarction: the long and arduous road from scientific discovery to clinical utility. Circ. Res. 113, 242–244. 10.1161/CIRCRESAHA.113.301951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard R. H. (2006). Exercise is good for your blood pressure: effects of endurance training and resistance training. Clin. Exp. Pharmacol. Physiol. 33, 853–856. 10.1111/j.1440-1681.2006.04453.x [DOI] [PubMed] [Google Scholar]
- Fan R., Mao S.-Q., Gu T.-L., Zhong F.-D., Gong M.-L., Hao L.-M., et al. (2017). Preliminary analysis of the association between methylation of the ACE2 promoter and essential hypertension. Mol. Med. Rep. 15, 3905–3911. 10.3892/mmr.2017.6460 [DOI] [PubMed] [Google Scholar]
- FDA-NIH Biomarker Working Group (2016). BEST (Biomarkers, EndpointS, and other Tools) Resource. [PubMed]
- Feng G., Yan Z., Li C., Hou Y. (2016). microRNA-208a in an early stage myocardial infarction rat model and the effect on cAMP-PKA signaling pathway. Mol. Med. Rep. 14, 1631–1635. 10.3892/mmr.2016.5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen H. M., Gizard F., Zhao Y., Qing H., Heywood E. B., Jones K. L., et al. (2011). Epigenetic regulation of vascular smooth muscle cell proliferation and neointima formation by histone deacetylase inhibition. Arteriosclerosis, Thrombosis, and Vascular Biol. 31, 851–860. 10.1161/ATVBAHA.110.221952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorito G., Guarrera S., Valle C., Ricceri F., Russo A., Grioni S., et al. (2014). B-vitamins intake, DNA-methylation of one carbon metabolism and homocysteine pathway genes and myocardial infarction risk: the EPICOR study. Nutr. Metab. Cardiovasc. Dis. 24, 483–488. 10.1016/j.numecd.2013.10.026 [DOI] [PubMed] [Google Scholar]
- Fraga M. F., Ballestar E., Paz M. F., Ropero S., Setien F., Ballestar M. L., et al. (2005). Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl. Acad. Sci. U. S. A 102, 10604–10609. 10.1073/pnas.0500398102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini N., Le T. H. (2014). Genetics of hypertension: discoveries from the bench to human populations. Am. J. Physiol. Renal Physiol. 306, F1–F11. 10.1152/ajprenal.00334.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S., Pizzolo F., Choi S.-W., Guarini P., Castagna A., Ravagnani V., et al. (2008). Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis 199, 323–327. 10.1016/j.atherosclerosis.2007.11.029 [DOI] [PubMed] [Google Scholar]
- Gao J., Yang S., Wang K., Zhong Q., Ma A., Pan X. (2019). Plasma miR-126 and miR-143 as potential novel biomarkers for cerebral atherosclerosis. J. Stroke Cerebrovasc. Dis. 28, 38–43. 10.1016/j.jstrokecerebrovasdis.2018.09.008 [DOI] [PubMed] [Google Scholar]
- García-Giménez J. L., Mena-Mollá S., Beltrán-García J., Sanchis-Gomar F. (2017). Challenges in the analysis of epigenetic biomarkers in clinical samples. Clin. Chem. Lab. Med. 55, 1474–1477. 10.1515/cclm-2016-1162 [DOI] [PubMed] [Google Scholar]
- Gholikhani-Darbroud R., Khaki-Khatibi F., Mansouri F., Hajahmadipoorrafsanjani M., Ghojazadeh M. (2017). Decreased circulatory microRNA-4478 as a specific biomarker for diagnosing nonn–ST-segment elevation myocardial infarction (NSTEMI) and its association with soluble leptin receptor. Bratisl. Lek. Listy 118, 684–690. 10.4149/BLL_2017_129 [DOI] [PubMed] [Google Scholar]
- Glezeva N., Moran B., Collier P., Moravec C. S., Phelan D., Donnellan E., et al. (2019). Targeted DNA methylation profiling of human cardiac tissue reveals novel epigenetic traits and gene deregulation across different heart failure patient subtypes. Circ. Heart Fail. 12, e005765. 10.1161/CIRCHEARTFAILURE.118.005765 [DOI] [PubMed] [Google Scholar]
- Goldbergova M. P., Lipkova J., Fedorko J., Sevcikova J., Parenica J., Spinar J., et al. (2018). MicroRNAs in pathophysiology of acute myocardial infarction and cardiogenic shock. Bratisl. Lek. Listy 119, 341–347. 10.4149/BLL_2018_064 [DOI] [PubMed] [Google Scholar]
- Goldraich L. A., Martinelli N. C., Matte U., Cohen C., Andrades M., Pimentel M., et al. (2014). Transcoronary gradient of plasma microRNA 423-5p in heart failure: evidence of altered myocardial expression. Biomarkers 19, 135–141. 10.3109/1354750X.2013.870605 [DOI] [PubMed] [Google Scholar]
- Gorski P. A., Jang S. P., Jeong D., Lee A., Lee P., Oh J. G., et al. (2019). Role of SIRT1 in modulating acetylation of the sarco-endoplasmic reticulum Ca2+-ATPase in heart failure. Circ. Res. 124, e63–e80. 10.1161/CIRCRESAHA.118.313865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal R., Goyal D., Leitzke A., Gheorghe C. P., Longo L. D. (2010). Brain renin–angiotensin system: fetal epigenetic programming by maternal protein restriction during pregnancy. Reprod. Sci. 17, 227–238. 10.1177/1933719109351935 [DOI] [PubMed] [Google Scholar]
- Grabmaier U., Clauss S., Gross L., Klier I., Franz W. M., Steinbeck G., et al. (2017). Diagnostic and prognostic value of miR-1 and miR-29b on adverse ventricular remodeling after acute myocardial infarction—the SITAGRAMI-miR analysis. Int. J. Cardiol. 244, 30–36. 10.1016/j.ijcard.2017.06.054 [DOI] [PubMed] [Google Scholar]
- Greißel A., Culmes M., Burgkart R., Zimmermann A., Eckstein H.-H., Zernecke A., et al. (2016). Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc. Pathol. 25, 79–86. 10.1016/j.carpath.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Guarrera S., Fiorito G., Onland-Moret N. C., Russo A., Agnoli C., Allione A., et al. (2015). Gene-specific DNA methylation profiles and LINE-1 hypomethylation are associated with myocardial infarction risk. Clin. Epigenet. 7, 133. 10.1186/s13148-015-0164-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunel T., Hosseini M. K., Gumusoglu E., Kisakesen H. I., Benian A., Aydinli K. (2017). Expression profiling of maternal plasma and placenta microRNAs in preeclamptic pregnancies by microarray technology. Placenta 52, 77–85. 10.1016/j.placenta.2017.02.019 [DOI] [PubMed] [Google Scholar]
- Guo M., Luo J., Zhao J., Shang D., Lv Q., Zang P. (2018). Combined use of circulating miR-133a and NT-proBNP improves heart failure diagnostic accuracy in elderly patients. Med. Sci. Monit. 24, 8840–8848. 10.12659/MSM.911632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M.-L., Guo L.-L., Weng Y.-Q. (2017). Implication of peripheral blood miRNA-124 in predicting acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 21, 1054–1059. [PubMed] [Google Scholar]
- Haas J., Frese K. S., Park Y. J., Keller A., Vogel B., Lindroth A. M., et al. (2013). Alterations in cardiac DNA methylation in human dilated cardiomyopathy. EMBO Mol. Med. 5, 413–429. 10.1002/emmm.201201553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Zhao Y., Luo Q. Q., Liu X. X., Lu S. S., Zou L. (2017). The significance of miR-145 in the prediction of preeclampsia. Bratisl. Lek. Listy 118, 523–528. 10.4149/BLL_2017_101 [DOI] [PubMed] [Google Scholar]
- Han Z., Zhang L., Yuan L., Liu X., Chen X., Ye X., et al. (2015). Change of plasma microRNA-208 level in acute myocardial infarction patients and its clinical significance. Ann. Transl. Med. 3, 307. 10.3978/j.issn.2305-5839.2015.10.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X.-Z., Fan H.-M. (2017). Identification of miRNAs as atherosclerosis biomarkers and functional role of miR-126 in atherosclerosis progression through MAPK signalling pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 2725–2733. [PubMed] [Google Scholar]
- He F., Lv P., Zhao X., Wang X., Ma X., Meng W., et al. (2014). Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol. Cell Biochem 394, 137–144. 10.1007/s11010-014-2089-0 [DOI] [PubMed] [Google Scholar]
- He X., Ji J., Wang T., Wang M.-B., Chen X.-L. (2017). Upregulation of circulating miR-195-3p in heart failure. Cardiology 138, 107–114. 10.1159/000476029 [DOI] [PubMed] [Google Scholar]
- Hindson C. M., Chevillet J. R., Briggs H. A., Gallichotte E. N., Ruf I. K., Hindson B. J., et al. (2013). Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 10, 1003–1005. 10.1038/nmeth.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau-Aveilla C., Meggetto F. (2017). Crosstalk between microRNA and DNA methylation offers potential biomarkers and targeted therapies in ALK-positive lymphomas. Cancers (Basel) 9, E100–E114. 10.3390/cancers9080100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M., Wagner M., Reil J.-C., Müller S.-A., Tauchnitz M., Zimmer A. M., et al. (2013). HDAC4 controls histone methylation in response to elevated cardiac load. J. Clin. Invest. 123, 1359–1370. 10.1172/JCI61084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromadnikova I., Kotlabova K., Ondrackova M., Pirkova P., Kestlerova A., Novotna V., et al. (2015). Expression profile of C19MC microRNAs in placental tissue in pregnancy-related complications. DNA Cell Biol. 34, 437–457. 10.1089/dna.2014.2687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A., Chen S.-J., Chang Y.-S., Chen H.-C., Chu P.-H. (2014). Systemic approach to identify serum microRNAs as potential biomarkers for acute myocardial infarction. Biomed. Res. Int. 2014, 418628–418613. 10.1155/2014/418628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Li L., Chen X., Zhang H., Shi Z. (2016. a). MiR-103a targeting Piezo1 is involved in acute myocardial infarction through regulating endothelium function. Cardiol. J. 23, 556–562. 10.5603/CJ.a2016.0056 [DOI] [PubMed] [Google Scholar]
- Huang Y., Chen J., Zhou Y., Yu X., Huang C., Li J., et al. (2016. b). Circulating miR-30 is related to carotid artery atherosclerosis. Clin. Exp. Hypertens. 38, 489–494. 10.3109/10641963.2016.1163370 [DOI] [PubMed] [Google Scholar]
- Huang Y., Tang S., Ji-yan C., Huang C., Li J., Cai A.-P., et al. (2017. a). Circulating miR-92a expression level in patients with essential hypertension: a potential marker of atherosclerosis. J. Hum. Hypertens. 31, 200–205. 10.1038/jhh.2016.66 [DOI] [PubMed] [Google Scholar]
- Huang Y.-Q., Huang C., Chen J.-Y., Li J., Feng Y.-Q. (2017. b). Plasma expression level of miRNA let-7 is positively correlated with carotid intima-media thickness in patients with essential hypertension. J. Hum. Hypertens. 31, 843–847. 10.1038/jhh.2017.52 [DOI] [PubMed] [Google Scholar]
- Huang Y.-Q., Li J., Huang C., Feng Y.-Q. (2018). Plasma microRNA-29c levels are associated with carotid intima-media thickness and is a potential biomarker for the early detection of atherosclerosis. Cell. Physiol. Biochem. 50, 452–459. 10.1159/000494158 [DOI] [PubMed] [Google Scholar]
- Huo K.-G., Richer C., Berillo O., Mahjoub N., Fraulob-Aquino J. C., Barhoumi T., et al. (2019). miR-431-5p knockdown protects against angiotensin II–induced hypertension and vascular injury. Hypertension 73, 1007–1017. 10.1161/HYPERTENSIONAHA.119.12619 [DOI] [PubMed] [Google Scholar]
- Iorio M. V., Piovan C., Croce C. M. (2010). Interplay between microRNAs and the epigenetic machinery: an intricate network. Biochim. Biophys. Acta 1799, 694–701. 10.1016/j.bbagrm.2010.05.005 [DOI] [PubMed] [Google Scholar]
- Irmak M. K., Sizlan A. (2006). Essential hypertension seems to result from melatonin-induced epigenetic modifications in area postrema. Med. Hypotheses 66, 1000–1007. 10.1016/j.mehy.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Bird A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 Suppl, 245–254. 10.1038/ng1089 [DOI] [PubMed]
- Jaffe A. S., Babuin L., Apple F. S. (2006). Biomarkers in acute cardiac disease: the present and the future. J. Am. Coll. Cardiol. 48, 1–11. 10.1016/j.jacc.2006.02.056 [DOI] [PubMed] [Google Scholar]
- Jairajpuri D. S., Malalla Z. H., Mahmood N., Almawi W. Y. (2017). Circulating microRNA expression as predictor of preeclampsia and its severity. Gene 627, 543–548. 10.1016/j.gene.2017.07.010 [DOI] [PubMed] [Google Scholar]
- Jeong H. S., Kim J.-Y., Lee S. H., Hwang J., Shin J. W., Song K. S., et al. (2017). Synergy of circulating miR-212 with markers for cardiovascular risks to enhance estimation of atherosclerosis presence. PLoS One 12, e0177809. 10.1371/journal.pone.0177809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Takahashi R., Hiura Y., Hirokawa G., Fukushima Y., Iwai N. (2009). Plasma miR-208 as a biomarker of myocardial injury. Clin. Chem. 55, 1944–1949. 10.1373/clinchem.2009.125310 [DOI] [PubMed] [Google Scholar]
- Jia K., Shi P., Han X., Chen T., Tang H., Wang J. (2016). Diagnostic value of miR-30d-5p and miR-125b-5p in acute myocardial infarction. Mol. Med. Rep. 14, 184–194. 10.3892/mmr.2016.5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Sun M., You L., Lu K., Gao L., Hu C., et al. (2019). DNA methylation and hydroxymethylation are associated with the degree of coronary atherosclerosis in elderly patients with coronary heart disease. Life Sci. 224, 241–248. 10.1016/j.lfs.2019.03.021 [DOI] [PubMed] [Google Scholar]
- Jiang F., Li J., Wu G., Miao Z., Lu L., Ren G., et al. (2015). Upregulation of microRNA-335 and microRNA-584 contributes to the pathogenesis of severe preeclampsia through downregulation of endothelial nitric oxide synthase. Mol. Med. Rep. 12, 5383–5390. 10.3892/mmr.2015.4018 [DOI] [PubMed] [Google Scholar]
- Jiang Y.-Z., Jiménez J. M., Ou K., McCormick M. E., Zhang L.-D., Davies P. F. (2014). Hemodynamic disturbed flow induces differential DNA methylation of endothelial Kruppel-like factor 4 promoter in vitro and in vivo . Circ. Res. 115, 32–43. 10.1161/CIRCRESAHA.115.303883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y.-Z., Manduchi E., Stoeckert C. J., Davies P. F. (2015). Arterial endothelial methylome: differential DNA methylation in athero-susceptible disturbed flow regions in vivo . BMC Genomics 16, 506. 10.1186/s12864-015-1656-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Gu W., Lai Y., Zheng W., Zhou Q., Wu X. (2017). The circulating microRNA-206 level predicts the severity of pulmonary hypertension in patients with left heart diseases. Cell. Physiol. Biochem. 41, 2150–2160. 10.1159/000475569 [DOI] [PubMed] [Google Scholar]
- Jo B.-S., Koh I.-U., Bae J.-B., Yu H.-Y., Jeon E.-S., Lee H.-Y., et al. (2016). Methylome analysis reveals alterations in DNA methylation in the regulatory regions of left ventricle development genes in human dilated cardiomyopathy. Genomics 108, 84–92. 10.1016/j.ygeno.2016.07.001 [DOI] [PubMed] [Google Scholar]
- Kanuri S. H., Ipe J., Kassab K., Gao H., Liu Y., Skaar T. C., et al. (2018). Next generation MicroRNA sequencing to identify coronary artery disease patients at risk of recurrent myocardial infarction. Atherosclerosis 278, 232–239. 10.1016/j.atherosclerosis.2018.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Loh M., Takeuchi F., Verweij N., Wang X., Zhang W., et al. (2015). Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat. Genet. 47, 1282–1293. 10.1038/ng.3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatibzadeh S., Farzadfar F., Oliver J., Ezzati M., Moran A. (2013). Worldwide risk factors for heart failure: a systematic review and pooled analysis. Int. J. Cardiol. 168, 1186–1194. 10.1016/j.ijcard.2012.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J., Kim S. Y., Lim J. H., Kwak D. W., Park S. Y., Ryu H. M. (2015). Quantification and application of potential epigenetic markers in maternal plasma of pregnancies with hypertensive disorders. IJMS 16, 29875–29888. 10.3390/ijms161226201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Kim G. J., Umemura T., Lee S. G., Cho K. J. (2017). Aberrant expression of plasma microRNA-33a in an atherosclerosis-risk group. Mol. Biol. Rep. 44, 79–88. 10.1007/s11033-016-4082-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenke S., Eul S., Peters J., Neumann T., Adamzik M., Frey U. H. (2018). Circulating miR-192 is a prognostic marker in patients with ischemic cardiomyopathy. Future Cardiol. 14, 283–289. 10.2217/fca-2017-0108 [DOI] [PubMed] [Google Scholar]
- Knothe C., Shiratori H., Resch E., Ultsch A., Geisslinger G., Doehring A., et al. (2016). Disagreement between two common biomarkers of global DNA methylation. Clin. Epigenet. 8, 60–17. 10.1186/s13148-016-0227-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koczor C. A., Lee E. K., Torres R. A., Boyd A., Vega J. D., Uppal K., et al. (2013). Detection of differentially methylated gene promoters in failing and nonfailing human left ventricle myocardium using computation analysis. Physiol. Genomics 45, 597–605. 10.1152/physiolgenomics.00013.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontaraki J. E., Marketou M. E., Zacharis E. A., Parthenakis F. I., Vardas P. E. (2014). MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: potential markers of target-organ damage. J. Am. Soc. Hypertens. 8, 368–375. 10.1016/j.jash.2014.03.324 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128, 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2011). miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39, D152–D157. 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R., Mani P., Sivakumar P., Gopinath V., Sekar D. (2017). Expression and methylation of circulating microRNA-510 in essential hypertension. Hypertens. Res. 40, 361–363. 10.1038/hr.2016.147 [DOI] [PubMed] [Google Scholar]
- Kristensen L. S., Wojdacz T. K., Thestrup B. B., Wiuf C., Hager H., Hansen L. L. (2009). Quality assessment of DNA derived from up to 30 years old formalin fixed paraffin embedded (FFPE) tissue for PCR-based methylation analysis using SMART-MSP and MS-HRM. BMC Cancer 9, 453. 10.1186/1471-2407-9-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A., Chavan-Gautam P., Mehendale S., Yadav H., Joshi S. (2011). Global DNA methylation patterns in placenta and its association with maternal hypertension in pre-eclampsia. DNA Cell Biol. 30, 79–84. 10.1089/dna.2010.1084 [DOI] [PubMed] [Google Scholar]
- Kumar A., Kumar S., Vikram A., Hoffman T. A., Naqvi A., Lewarchik C. M., et al. (2013). Histone and DNA methylation–mediated epigenetic downregulation of endothelial Kruppel-like factor 2 by low-density lipoprotein cholesterol. Arteriosclerosis, Thrombosis, and Vascular Biol. 33, 1936–1942. 10.1161/ATVBAHA.113.301765 [DOI] [PubMed] [Google Scholar]
- Kumarswamy R., Anker S. D., Thum T. (2010). MicroRNAs as circulating biomarkers for heart failure: questions about MiR-423-5p. Circ. Res. 106, e8–author reply e9. 10.1161/CIRCRESAHA.110.220616 [DOI] [PubMed] [Google Scholar]
- Kuosmanen S. M., Hartikainen J., Hippeläinen M., Kokki H., Levonen A.-L., Tavi P. (2015). MicroRNA profiling of pericardial fluid samples from patients with heart failure. PLoS One 10, e0119646. 10.1371/journal.pone.0119646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganà A. S., Vitale S. G., Sapia F., Valenti G., Corrado F., Padula F., et al. (2018). miRNA expression for early diagnosis of preeclampsia onset: hope or hype? J. Matern. Fetal. Neonatal. Med. 31, 817–821. 10.1080/14767058.2017.1296426 [DOI] [PubMed] [Google Scholar]
- Lai K.-B., Sanderson J. E., Izzat M. B., Yu C.-M. (2015). Micro-RNA and mRNA myocardial tissue expression in biopsy specimen from patients with heart failure. Int. J. Cardiol. 199, 79–83. 10.1016/j.ijcard.2015.07.043 [DOI] [PubMed] [Google Scholar]
- Lakhani H. V., Khanal T., Gabi A., Yousef G., Alam M. B., Sharma D., et al. (2018). Developing a panel of biomarkers and miRNA in patients with myocardial infarction for early intervention strategies of heart failure in West Virginian population. PLoS One 13, e0205329. 10.1371/journal.pone.0205329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-A., Baek I., Seok Y. M., Yang E., Cho H.-M., Lee D.-Y., et al. (2010). Promoter hypomethylation upregulates Na+-K+-2Cl– cotransporter 1 in spontaneously hypertensive rats. Biochem. Biophys. Res. Commun. 396, 252–257. 10.1016/j.bbrc.2010.04.074 [DOI] [PubMed] [Google Scholar]
- Li B., Feng Z.-H., Sun H., Zhao Z.-H., Yang S.-B., Yang P. (2017. a). The blood genome-wide DNA methylation analysis reveals novel epigenetic changes in human heart failure. Eur. Rev. Med. Pharmacol. Sci. 21, 1828–1836. [PubMed] [Google Scholar]
- Li G., Song Y., Li Y.-D., Jie L.-J., Wu W.-Y., Li J.-Z., et al. (2018. a). Circulating miRNA-302 family members as potential biomarkers for the diagnosis of acute heart failure. Biomark. Med 12, 871–880. 10.2217/bmm-2018-0132 [DOI] [PubMed] [Google Scholar]
- Li H., Fan J., Yin Z., Wang F., Chen C., Wang D. W. (2016). Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget 7, 33–45. 10.18632/oncotarget.6631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Chen Z., Qin Y., Wei Y. (2018. b). MiR-664a-3p expression in patients with obstructive sleep apnea: a potential marker of atherosclerosis. Medicine (Baltimore) 97, e9813. 10.1097/MD.0000000000009813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li S., Wu M., Chi C., Hu D., Cui Y., et al. (2019). Early diagnostic value of circulating microRNAs in patients with suspected acute myocardial infarction. J. Cell Physiol. 234, 13649–13658. 10.1002/jcp.28045 [DOI] [PubMed] [Google Scholar]
- Li L.-M., Cai W.-B., Ye Q., Liu J.-M., Li X., Liao X.-X. (2014. a). Comparison of plasma microRNA-1 and cardiac troponin T in early diagnosis of patients with acute myocardial infarction. World. J. Emerg. Med. 5, 182–186. 10.5847/wjem.j.issn.1920-8642.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Guo L. Z., Kim M. H., Han J.-Y., Serebruany V. (2017. b). Platelet microRNA for predicting acute myocardial infarction. J. Thromb. Thrombolysis 44, 556–564. 10.1007/s11239-017-1537-6 [DOI] [PubMed] [Google Scholar]
- Li T., Cao H., Zhuang J., Wan J., Guan M., Yu B., et al. (2011). Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin. Chim. Acta 412, 66–70. 10.1016/j.cca.2010.09.029 [DOI] [PubMed] [Google Scholar]
- Li Y., Du W., Zhao R., Hu J., Li H., Han R., et al. (2017. c). New insights into epigenetic modifications in heart failure. Front. Biosci. (Landmark Ed) 22, 230–247. 10.2741/4483 [DOI] [PubMed] [Google Scholar]
- Li Y.-Q., Zhang M.-F., Wen H.-Y., Hu C.-L., Liu R., Wei H.-Y., et al. (2013). Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics (Sao Paulo) 68, 75–80. 10.6061/clinics/2013(01)OA12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Lu J., Luo Y., Li S., Chen M. (2014. b). High association between human circulating microRNA-497 and acute myocardial infarction. Sci. World J. 2014, 931845–931847. 10.1155/2014/931845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Bai S., Su L., Li C., Wu J., Xia Z., et al. (2015). A subset of circulating microRNAs is expressed differently in patients with myocardial infarction. Mol. Med. Rep. 12, 243–247. 10.3892/mmr.2015.3422 [DOI] [PubMed] [Google Scholar]
- Lin X., Zhang S., Huo Z. (2019). Serum circulating miR-150 is a predictor of post–acute myocardial infarction heart failure. Int. Heart J. 60, 280–286. 10.1536/ihj.18-306 [DOI] [PubMed] [Google Scholar]
- Liu G., Niu X., Meng X., Zhang Z. (2018. a). Sensitive miRNA markers for the detection and management of NSTEMI acute myocardial infarction patients. J. Thorac. Dis. 10, 3206–3215. 10.21037/jtd.2018.05.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Zheng J., Dong J., Bai R., Song D., Ma X., et al. (2018. b). Association of miR-197-5p, a circulating biomarker for heart failure, with myocardial fibrosis and adverse cardiovascular events among patients with stage C or D heart failure. Cardiology 141, 212–225. 10.1159/000493419 [DOI] [PubMed] [Google Scholar]
- Liu X., Dong Y., Chen S., Zhang G., Zhang M., Gong Y., et al. (2015. a). Circulating microRNA-146a and microRNA-21 predict left ventricular remodeling after ST-elevation myocardial infarction. Cardiology 132, 233–241. 10.1159/000437090 [DOI] [PubMed] [Google Scholar]
- Liu X., Fan Z., Zhao T., Cao W., Zhang L., Li H., et al. (2015. b). Plasma miR-1, miR-208, miR-499 as potential predictive biomarkers for acute myocardial infarction: an independent study of Han population. Exp. Gerontol. 72, 230–238. 10.1016/j.exger.2015.10.011 [DOI] [PubMed] [Google Scholar]
- Liu X., Meng H., Jiang C., Yang S., Cui F., Yang P. (2016). Differential microRNA Expression and regulation in the rat model of post-infarction heart failure. PLoS One 11, e0160920. 10.1371/journal.pone.0160920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yuan L., Chen F., Zhang L., Chen X., Yang C., et al. (2017). Circulating miR-208b: a potentially sensitive and reliable biomarker for the diagnosis and prognosis of acute myocardial infarction. Clin. Lab. 63, 101–109. 10.7754/Clin.Lab.2016.160632 [DOI] [PubMed] [Google Scholar]
- Liu Y., Liu P., Yang C., Cowley A. W., Liang M. (2014). Base-resolution maps of 5-methylcytosine and 5-hydroxymethylcytosine in Dahl S rats: effect of salt and genomic sequence. Hypertension 63, 827–838. 10.1161/HYPERTENSIONAHA.113.02637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G., Wang F., Duan Q., Chen F., Yang S., Gong W., et al. (2012. a). Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int. J. Biol. Sci. 8, 811–818. 10.7150/ijbs.4439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long G., Wang F., Duan Q., Yang S., Chen F., Gong W., et al. (2012. b). Circulating miR-30a, miR-195 and let-7b associated with acute myocardial infarction. PLoS One 7, e50926. 10.1371/journal.pone.0050926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque A., Farwati A., Krupinski J., Aran J. M. (2018). Association between low levels of serum miR-638 and atherosclerotic plaque vulnerability in patients with high-grade carotid stenosis. J. Neurosurg. 131, 1–8. 10.3171/2018.2.JNS171899 [DOI] [PubMed] [Google Scholar]
- Lv P., Zhou M., He J., Meng W., Ma X., Dong S., et al. (2014). Circulating miR-208b and miR-34a are associated with left ventricular remodeling after acute myocardial infarction. IJMS 15, 5774–5788. 10.3390/ijms15045774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S.-C., Zhang H.-P., Kong F.-Q., Zhang H., Yang C., He Y.-Y., et al. (2016). Integration of gene expression and DNA methylation profiles provides a molecular subtype for risk assessment in atherosclerosis. Mol. Med. Rep. 13, 4791–4799. 10.3892/mmr.2016.5120 [DOI] [PubMed] [Google Scholar]
- Maciejak A., Kiliszek M., Opolski G., Segiet A., Matlak K., Dobrzycki S., et al. (2016). miR-22-5p revealed as a potential biomarker involved in the acute phase of myocardial infarction via profiling of circulating microRNAs. Mol. Med. Rep. 14, 2867–2875. 10.3892/mmr.2016.5566 [DOI] [PubMed] [Google Scholar]
- Maciejak A., Kostarska-Srokosz E., Gierlak W., Dluzniewski M., Kuch M., Marchel M., et al. (2018). Circulating miR-30a-5p as a prognostic biomarker of left ventricular dysfunction after acute myocardial infarction. Sci. Rep. 8, 9883. 10.1038/s41598-018-28118-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magenta A., Sileno S., D’Agostino M., Persiani F., Beji S., Paolini A., et al. (2018). Atherosclerotic plaque instability in carotid arteries: miR-200c as a promising biomarker. Clin. Sci. 132, 2423–2436. 10.1042/CS20180684 [DOI] [PubMed] [Google Scholar]
- Majchrzak-Celińska A., Kosicka K., Paczkowska J., Główka F. K., Bręborowicz G. H., Krzyścin M., et al. (2017). HSD11B2, RUNX3, and LINE-1 methylation in placental DNA of hypertensive disorders of pregnancy patients. Reprod. Sci. 24, 1520–1531. 10.1177/1933719117692043 [DOI] [PubMed] [Google Scholar]
- Mao Z., Wu F., Shan Y. (2018). Identification of key genes and miRNAs associated with carotid atherosclerosis based on mRNA-seq data. Medicine (Baltimore) 97, e9832. 10.1097/MD.0000000000009832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marketou M. E., Kontaraki J. E., Maragkoudakis S., Patrianakos A., Konstantinou J., Nakou H., et al. (2018). MicroRNAs in peripheral mononuclear cells as potential biomarkers in hypertensive patients with heart failure with preserved ejection fraction. Am. J. Hypertens. 31, 651–657. 10.1093/ajh/hpy035 [DOI] [PubMed] [Google Scholar]
- Masson S., Batkai S., Beermann J., Bär C., Pfanne A., Thum S., et al. (2018). Circulating microRNA-132 levels improve risk prediction for heart failure hospitalization in patients with chronic heart failure. Eur. J. Heart Fail. 20, 78–85. 10.1002/ejhf.961 [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Sakata Y., Suna S., Nakatani D., Usami M., Hara M., et al. (2013). Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ. Res. 113, 322–326. 10.1161/CIRCRESAHA.113.301209 [DOI] [PubMed] [Google Scholar]
- Meder B., Haas J., Sedaghat-Hamedani F., Kayvanpour E., Frese K., Lai A., et al. (2017). Epigenome-wide association study identifies cardiac gene patterning and a novel class of biomarkers for heart failure. Circulation 136, 1528–1544. 10.1161/CIRCULATIONAHA.117.027355 [DOI] [PubMed] [Google Scholar]
- Mei Z., Huang B., Mo Y., Fan J. (2017). An exploratory study into the role of miR-204-5p in pregnancy-induced hypertension. Exp Ther Med 13, 1711–1718. 10.3892/etm.2017.4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Kawamura T., Morimoto T., Ono K., Wada H., Kawase Y., et al. (2006). Histone acetyltransferase activity of p300 is required for the promotion of left ventricular remodeling after myocardial infarction in adult mice in vivo . Circulation 113, 679–690. 10.1161/CIRCULATIONAHA.105.585182 [DOI] [PubMed] [Google Scholar]
- Moutinho C., Esteller M. (2017). MicroRNAs and epigenetics. Adv. Cancer Res. 135, 189–220. 10.1016/bs.acr.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Movassagh M., Choy M.-K., Knowles D. A., Cordeddu L., Haider S., Down T., et al. (2011). Distinct epigenomic features in end-stage failing human hearts. Circulation 124, 2411–2422. 10.1161/CIRCULATIONAHA.111.040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu S., Shimosawa T., Ogura S., Wang H., Uetake Y., Kawakami-Mori F., et al. (2011). Epigenetic modulation of the renal β-adrenergic-WNK4 pathway in salt-sensitive hypertension. Nat. Med. 17, 573–580. 10.1038/nm.2337 [DOI] [PubMed] [Google Scholar]
- Nagata S., Marunouchi T., Tanonaka K. (2019). Histone deacetylase inhibitor SAHA treatment prevents the development of heart failure after myocardial infarction via an induction of heat-shock proteins in rats. Biol. Pharm. Bull. 42, 453–461. 10.1248/bpb.b18-00785 [DOI] [PubMed] [Google Scholar]
- Nakatochi M., Ichihara S., Yamamoto K., Naruse K., Yokota S., Asano H., et al. (2017). Epigenome-wide association of myocardial infarction with DNA methylation sites at loci related to cardiovascular disease. Clin. Epigenet. 9, 54. 10.1186/s13148-017-0353-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi T., Imanishi T., Akasaka T. (2015). MicroRNAs and cardiovascular diseases. Biomed. Res. Int. 2015, 682857. 10.1155/2015/682857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack F., Ribbat-Idel J., Thorns C., Chiriac A., Axt-Fliedner R., Diedrich K., et al. (2011). miRNA expression profiling in formalin-fixed and paraffin-embedded placental tissue samples from pregnancies with severe preeclampsia. J. Perinat. Med. 39, 267–271. 10.1515/jpm.2011.012 [DOI] [PubMed] [Google Scholar]
- Nuyt A. M., Alexander B. T. (2009). Developmental programming and hypertension. Curr. Opin. Nephrol. Hypertens. 18, 144–152. 10.1097/MNH.0b013e328326092c [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan J. F., Neylon A., McGorrian C., Blake G. J. (2016). miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int. J. Cardiol. 224, 310–316. 10.1016/j.ijcard.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Olivieri F., Antonicelli R., Lorenzi M., D’Alessandra Y., Lazzarini R., Santini G., et al. (2013). Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int. J. Cardiol. 167, 531–536. 10.1016/j.ijcard.2012.01.075 [DOI] [PubMed] [Google Scholar]
- Ovchinnikova E. S., Schmitter D., Vegter E. L., ter J. M., Valente M. A. E., Liu L. C. Y., et al. (2016). Signature of circulating microRNAs in patients with acute heart failure. Eur. J. Heart Fail. 18, 414–423. 10.1002/ejhf.332 [DOI] [PubMed] [Google Scholar]
- Panico C., Condorelli G. (2018). microRNA-132: a new biomarker of heart failure at last? Eur. J. Heart Fail. 20, 86–88. 10.1002/ejhf.1044 [DOI] [PubMed] [Google Scholar]
- Parahuleva M. S., Euler G., Mardini A., Parviz B., Schieffer B., Schulz R., et al. (2017). Identification of microRNAs as potential cellular monocytic biomarkers in the early phase of myocardial infarction: a pilot study. Sci. Rep. 7, 15974. 10.1038/s41598-017-16263-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthenakis F., Marketou M., Kontaraki J., Patrianakos A., Nakou H., Touloupaki M., et al. (2017). Low levels of microRNA-21 are a marker of reduced arterial stiffness in well-controlled hypertension. J. Clin. Hypertens. (Greenwich) 19, 235–240. 10.1111/jch.12900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Chun-guang Q., Bei-fang L., Xue-zhi D., Zi-hao W., Yun-fu L., et al. (2014). Clinical impact of circulating miR-133, miR-1291 and miR-663b in plasma of patients with acute myocardial infarction. Diagn. Pathol. 9, 89. 10.1186/1746-1596-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-da-Silva T., Coutinho Cruz M., Carrusca C., Cruz Ferreira R., Napoleão P., Mota Carmo M. (2018). Circulating microRNA profiles in different arterial territories of stable atherosclerotic disease: a systematic review. Am J Cardiovasc Dis 8, 1–13. [PMC free article] [PubMed] [Google Scholar]
- Pojoga L. H., Williams J. S., Yao T. M., Kumar A., Raffetto J. D., Nascimento G. R. A., et al. (2011). Histone demethylase LSD1 deficiency during high-salt diet is associated with enhanced vascular contraction, altered NO-cGMP relaxation pathway, and hypertension. AJP: Heart Circ. Physiol. 301, H1862–H1871. 10.1152/ajpheart.00513.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qipshidze Kelm N., Piell K. M., Wang E., Cole M. P. (2018). MicroRNAs as predictive biomarkers for myocardial injury in aged mice following myocardial infarction. J. Cell Physiol. 233, 5214–5221. 10.1002/jcp.26283 [DOI] [PubMed] [Google Scholar]
- Qiu X.-K., Ma J. (2018). Alteration in microRNA-155 level correspond to severity of coronary heart disease. Scand. J. Clin. Lab. Invest. 78, 219–223. 10.1080/00365513.2018.1435904 [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M., Martinsson D., Ahsan M., Enroth S., Ek W. E., Gyllensten U., et al. (2016). Epigenome-wide association study reveals differential DNA methylation in individuals with a history of myocardial infarction. Hum. Mol. Genet. 25, 4739–4748. 10.1093/hmg/ddw302 [DOI] [PubMed] [Google Scholar]
- Rau C. D., Vondriska T. M. (2017). DNA methylation and human heart failure: mechanisms or prognostics. Circulation 136, 1545–1547. 10.1161/CIRCULATIONAHA.117.029840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed G. W., Rossi J. E., Cannon C. P. (2017). Acute myocardial infarction. Lancet 389, 197–210. 10.1016/S0140-6736(16)30677-8 [DOI] [PubMed] [Google Scholar]
- Rezaei M., Eskandari F., Mohammadpour-Gharehbagh A., Harati-Sadegh M., Teimoori B., Salimi S. (2018). Hypomethylation of the miRNA-34a gene promoter is associated with severe preeclampsia. Clin. Exp. Hypertens. 41, 1–5. 10.1080/10641963.2018.1451534 [DOI] [PubMed] [Google Scholar]
- Rivière G., Lienhard D., Andrieu T., Vieau D., Frey B. M., Frey F. J. (2011). Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics 6, 478–489. 10.4161/epi.6.4.14961 [DOI] [PubMed] [Google Scholar]
- Rockenbach G., De Melo Neto A. J., Barcellos N. T., Wolff F. H. (2012). Ethnic differences in viral dominance patterns in patients with hepatitis B virus and hepatitis C virus dual infection. Hepatology 55, 1640–author reply 1640. 10.1002/hep.24533 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Iturbe B. (2006). Arteriolar remodeling in essential hypertension: are connective tissue growth factor and transforming growth factor involved? Kidney Int. 69, 1104–1105. 10.1038/sj.ki.5000222 [DOI] [PubMed] [Google Scholar]
- Rossetto D., Avvakumov N., Côté J. (2012). Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 7, 1098–1108. 10.4161/epi.21975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabia C., Picascia A., Grimaldi V., Amarelli C., Maiello C., Napoli C. (2017). The epigenetic promise to improve prognosis of heart failure and heart transplantation. Transplant. Rev. (Orlando) 31, 249–256. 10.1016/j.trre.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Sano M., Minamino T., Toko H., Miyauchi H., Orimo M., Qin Y., et al. (2007). p53-Induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature 446, 444–448. 10.1038/nature05602 [DOI] [PubMed] [Google Scholar]
- Sato F., Tsuchiya S., Meltzer S. J., Shimizu K. (2011). MicroRNAs and epigenetics. FEBS J. 278, 1598–1609. 10.1111/j.1742-4658.2011.08089.x [DOI] [PubMed] [Google Scholar]
- Schulte C., Westermann D., Blankenberg S., Zeller T. (2015). Diagnostic and prognostic value of circulating microRNAs in heart failure with preserved and reduced ejection fraction. WJC 7, 843–860. 10.4330/wjc.v7.i12.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrutinio D., Conserva F., Passantino A., Iacoviello M., Lagioia R., Gesualdo L. (2017). Circulating microRNA-150-5p as a novel biomarker for advanced heart failure: a genome-wide prospective study. J. Heart Lung Transplant. 36, 616–624. 10.1016/j.healun.2017.02.008 [DOI] [PubMed] [Google Scholar]
- Sekar D., Shilpa B. R., Das A. J. (2017). Relevance of microRNA 21 in different types of hypertension. Curr. Hypertens. Rep. 19, 57. 10.1007/s11906-017-0752-z [DOI] [PubMed] [Google Scholar]
- Seronde M.-F., Vausort M., Gayat E., Goretti E., Ng L. L., Squire I. B., et al. (2015). Circulating microRNAs and outcome in patients with acute heart failure. PLoS One 10, e0142237. 10.1371/journal.pone.0142237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R. V., Rong J., Larson M. G., Yeri A., Ziegler O., Tanriverdi K., et al. (2018. a). Associations of circulating extracellular RNAs with myocardial remodeling and heart failure. JAMA Cardiol. 3, 871–876. 10.1001/jamacardio.2018.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R., Ziegler O., Yeri A., Liu X., Murthy V., Rabideau D., et al. (2018. b). MicroRNAs associated with reverse left ventricular remodeling in humans identify pathways of heart failure progression. Circ. Heart Fail. 11, e004278. 10.1161/CIRCHEARTFAILURE.117.004278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi B., Guo Y., Wang J., Gao W. (2010). Altered expression of microRNAs in the myocardium of rats with acute myocardial infarction. BMC Cardiovasc. Disord. 10, 11. 10.1186/1471-2261-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Williams J., Brown J., Wang E. T., Lee B., Gonzalez T. L., et al. (2017). Up-regulation of microRNA-202-3p in first trimester placenta of pregnancies destined to develop severe preeclampsia, a pilot study. Pregnancy Hypertens. 10, 7–9. 10.1016/j.preghy.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E. M., Olson E. N. (2011). Pervasive roles of microRNAs in cardiovascular biology. Nature 469, 336–342. 10.1038/nature09783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolarek I., Wyszko E., Barciszewska A. M., Nowak S., Gawronska I., Jablecka A., et al. (2010). Global DNA methylation changes in blood of patients with essential hypertension. Med. Sci. Monit. 16, CR149–CR155. [PubMed] [Google Scholar]
- Strimbu K., Tavel J. A. (2010). What are biomarkers? Curr. Opin. HIV AIDS 5, 463–466. 10.1097/COH.0b013e32833ed177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M., Wang J., Wang C., Wang X., Dong W., Qiu W., et al. (2015). MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell Death Differ. 22, 986–999. 10.1038/cdd.2014.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Dong Y.-H., Du W., Shi C.-Y., Wang K., Tariq M.-A., et al. (2017). The role of MicroRNAs in myocardial infarction: from molecular mechanism to clinical application. IJMS 18, 745. 10.3390/ijms18040745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sygitowicz G., Tomaniak M., Błaszczyk O., Kołtowski Ł., Filipiak K. J., Sitkiewicz D. (2015). Circulating microribonucleic acids miR-1, miR-21 and miR-208a in patients with symptomatic heart failure: preliminary results. Arch. Cardiovasc. Dis. 108, 634–642. 10.1016/j.acvd.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Talens R. P., Jukema J. W., Trompet S., Kremer D., Westendorp R. G. J., Lumey L. H., et al. (2012). Hypermethylation at loci sensitive to the prenatal environment is associated with increased incidence of myocardial infarction. Int. J. Epidemiol. 41, 106–115. 10.1093/ije/dyr153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H., Diamond J., Vieco A., Chaudhuri S., Shinnar E., Cromer S., et al. (2018). Global atlas of cardiovascular disease 2000–2016: the path to prevention and control. Glob. Heart 13, 143–163. 10.1016/j.gheart.2018.09.511 [DOI] [PubMed] [Google Scholar]
- Tijsen A. J., Creemers E. E., Moerland P. D., de Windt L. J., van der Wal A. C., Kok W. E., et al. (2010). MiR423-5p as a circulating biomarker for heart failure. Circ. Res. 106, 1035–1039. 10.1161/CIRCRESAHA.110.218297 [DOI] [PubMed] [Google Scholar]
- Tournier B., Chapusot C., Courcet E., Martin L., Lepage C., Faivre J., et al. (2012). Why do results conflict regarding the prognostic value of the methylation status in colon cancers? The role of the preservation method. BMC Cancer 12, 12–12. 10.1186/1471-2407-12-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura B., Feriotto G., Monasta L., Bilel S., Zweyer M., Celeghini C. (2014). Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan J. Obstet. Gynecol. 53, 232–234. 10.1016/j.tjog.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Valencia-Morales M. D. P., Zaina S., Heyn H., Carmona F. J., Varol N., Sayols S., et al. (2015). The DNA methylation drift of the atherosclerotic aorta increases with lesion progression. BMC Med. Genomics 8, 7. 10.1186/s12920-015-0085-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boven N., Akkerhuis K. M., Anroedh S. S., Rizopoulos D., Pinto Y., Battes L. C., et al. (2017). Serially measured circulating miR-22-3p is a biomarker for adverse clinical outcome in patients with chronic heart failure: the Bio-SHiFT study. Int. J. Cardiol. 235, 124–132. 10.1016/j.ijcard.2017.02.078 [DOI] [PubMed] [Google Scholar]
- van Empel V. P. M., de Windt L. J., da Costa Martins P. A. (2012). Circulating miRNAs: reflecting or affecting cardiovascular disease? Curr. Hypertens. Rep. 14, 498–509. 10.1007/s11906-012-0310-7 [DOI] [PubMed] [Google Scholar]
- Vegter E. L., Ovchinnikova E. S., van Veldhuisen D. J., Jaarsma T., Berezikov E., van der Meer P., et al. (2017). Low circulating microRNA levels in heart failure patients are associated with atherosclerotic disease and cardiovascular-related rehospitalizations. Clin. Res. Cardiol. 106, 598–609. 10.1007/s00392-017-1096-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegter E. L., Schmitter D., Hagemeijer Y., Ovchinnikova E. S., van der Harst P., Teerlink J. R., et al. (2016). Use of biomarkers to establish potential role and function of circulating microRNAs in acute heart failure. Int. J. Cardiol. 224, 231–239. 10.1016/j.ijcard.2016.09.010 [DOI] [PubMed] [Google Scholar]
- Vogel B., Keller A., Frese K. S., Leidinger P., Sedaghat-Hamedani F., Kayvanpour E., et al. (2013). Multivariate miRNA signatures as biomarkers for non-ischaemic systolic heart failure. Eur. Heart J. 34, 2812–2822. 10.1093/eurheartj/eht256 [DOI] [PubMed] [Google Scholar]
- Wang C., Xu G., Wen Q., Peng X., Chen H., Zhang J., et al. (2019. a). CBS promoter hypermethylation increases the risk of hypertension and stroke. Clinics (Sao Paulo) 74, e630. 10.6061/clinics/2019/e630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Demura M., Cheng Y., Zhu A., Karashima S., Yoneda T., et al. (2014. a). Dynamic CCAAT/enhancer binding protein-associated changes of DNA methylation in the angiotensinogen gene. Hypertension 63, 281–288. 10.1161/HYPERTENSIONAHA.113.02303 [DOI] [PubMed] [Google Scholar]
- Wang F., Long G., Zhao C., Li H., Chaugai S., Wang Y., et al. (2013. a). Plasma microRNA-133a is a new marker for both acute myocardial infarction and underlying coronary artery stenosis. J. Transl. Med. 11, 222. 10.1186/1479-5876-11-222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Long G., Zhao C., Li H., Chaugai S., Wang Y., et al. (2014. b). Atherosclerosis-related circulating miRNAs as novel and sensitive predictors for acute myocardial infarction. PLoS One 9, e105734. 10.1371/journal.pone.0105734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Ma Y., Wang H., Qin H. (2017. a). Reciprocal regulation between microRNAs and epigenetic machinery in colorectal cancer. Oncol Lett. 13, 1048–1057. 10.3892/ol.2017.5593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Naghavi M., Allen C., Barber R. M., Bhutta Z. A., Carter A., et al. (2016. a). Articles Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544. 10.1016/S0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Zhang S., Marzolf B., Troisch P., Brightman A., Hu Z., et al. (2009). Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl. Acad. Sci. U. S. A 106, 4402–4407. 10.1073/pnas.0813371106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K.-J., Zhao X., Liu Y.-Z., Zeng Q.-T., Mao X.-B., Li S.-N., et al. (2016. b). Circulating MiR-19b-3p, MiR-134-5p and MiR-186-5p are promising novel biomarkers for early diagnosis of acute myocardial infarction. Cell. Physiol. Biochem. 38, 1015–1029. 10.1159/000443053 [DOI] [PubMed] [Google Scholar]
- Wang L., Liu J., Xu B., Liu Y.-L., Liu Z. (2018. a). Reduced exosome miR-425 and miR-744 in the plasma represents the progression of fibrosis and heart failure. Kaohsiung J. Med. Sci. 34, 626–633. 10.1016/j.kjms.2018.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Shen C., Diao L., Yang Z., Fan F., Wang C., et al. (2015). Aberrant hypermethylation of aldehyde dehydrogenase 2 promoter upstream sequence in rats with experimental myocardial infarction. Biomed. Res. Int. 2015, 503692–503613. 10.1155/2015/503692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Falkner B., Zhu H., Shi H., Su S., Xu X., et al. (2013. b). A genome-wide methylation study on essential hypertension in young African American males. PLoS One 8, e53938. 10.1371/journal.pone.0053938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chang W., Zhang Y., Zhang L., Ding H., Qi H., et al. (2019. b). Circulating miR-22-5p and miR-122-5p are promising novel biomarkers for diagnosis of acute myocardial infarction. J. Cell Physiol. 234, 4778–4786. 10.1002/jcp.27274 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yan L., Zhang Z., Prado E., Fu L., Xu X., et al. (2018. b). Epigenetic regulation and its therapeutic potential in pulmonary hypertension. Front. Pharmacol. 9, 241. 10.3389/fphar.2018.00241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-L., Yu W. (2018). Association of circulating microRNA-122 with presence and severity of atherosclerotic lesions. PeerJ 6, e5218. 10.7717/peerj.5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.-H., Sun X.-Y., Li C.-L., Sun Y.-M., Li J., Wang L.-F., et al. (2017. b). miRNA-21 expression in the serum of elderly patients with acute myocardial infarction. Med. Sci. Monit. 23, 5728–5734. 10.12659/MSM.904933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. A., Esa N., Pidikiti R., Freedman J. E., Keaney J. F., Tanriverdi K., et al. (2013). Circulating cell and plasma microRNA profiles differ between non–ST-segment and ST-segment-elevation myocardial infarction. Fam. Med. Med. Sci. Res. 2, 108. 10.4172/2327-4972.1000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C. J., Gupta S. K., O’Connell E., Thum S., Glezeva N., Fendrich J., et al. (2015). MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur. J. Heart Fail. 17, 405–415. 10.1002/ejhf.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Zhao S., Wang G., Zhang S., Luo W., Qin Z., et al. (2018). SMAD7 methylation as a novel marker in atherosclerosis. Biochem. Biophys. Res. Commun. 496, 700–705. 10.1016/j.bbrc.2018.01.121 [DOI] [PubMed] [Google Scholar]
- Wei X. J., Han M., Yang F. Y., Wei G. C., Liang Z. G., Yao H., et al. (2015). Biological significance of miR-126 expression in atrial fibrillation and heart failure. Braz. J. Med. Biol. Res. 48, 983–989. 10.1590/1414-431x20154590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierda R. J., Rietveld I. M., van Eggermond M. C. J. A., Belien J. A. M., van Zwet E. W., Lindeman J. H. N., et al. (2015). Global histone H3 lysine 27 triple methylation levels are reduced in vessels with advanced atherosclerotic plaques. Life Sci. 129, 3–9. 10.1016/j.lfs.2014.10.010 [DOI] [PubMed] [Google Scholar]
- Wissler R. W. (1991). Update on the pathogenesis of atherosclerosis. Am. J. Med. 91, 3S–9S. 10.1016/0002-9343(91)90050-8 [DOI] [PubMed] [Google Scholar]
- Wong L. L., Armugam A., Sepramaniam S., Karolina D. S., Lim K. Y., Lim J. Y., et al. (2015). Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur. J. Heart Fail. 17, 393–404. 10.1002/ejhf.223 [DOI] [PubMed] [Google Scholar]
- Wong L. L., Rademaker M. T., Saw E. L., Lew K. S., Ellmers L. J., Charles C. J., et al. (2017). Identification of novel microRNAs in the sheep heart and their regulation in heart failure. Sci. Rep. 7, 8250. 10.1038/s41598-017-08574-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. Q., Li J., Tan A. Y.-C., Vedururu R., Pang J.-M. B., Do H., et al. (2014). Sequence artefacts in a prospective series of formalin-fixed tumours tested for mutations in hotspot regions by massively parallel sequencing. BMC Med. Genomics 7, 23–10. 10.1186/1755-8794-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Zhao Q., Li Z., Li N., Xiao Q., Li X., et al. (2018. a). Bioinformatic screening for key miRNAs and genes associated with myocardial infarction. FEBS Open Bio. 8, 897–913. 10.1002/2211-5463.12423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Chen Y., Du Y., Tao J., Li W., Zhou Z., et al. (2018. b). Circulating exosomal miR-92b-5p is a promising diagnostic biomarker of heart failure with reduced ejection fraction patients hospitalized for acute heart failure. J. Thorac. Dis. 10, 6211–6220. 10.21037/jtd.2018.10.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Chen Y., Du Y., Tao J., Zhou Z., Yang Z. (2018. c). Serum exosomal MiR-92b-5p as a potential biomarker for acute heart failure caused by dilated cardiomyopathy. Cell. Physiol. Biochem. 46, 1939–1950. 10.1159/000489383 [DOI] [PubMed] [Google Scholar]
- Xiao J., Gao R., Bei Y., Zhou Q., Zhou Y., Zhang H., et al. (2017). Circulating miR-30d predicts survival in patients with acute heart failure. Cell. Physiol. Biochem. 41, 865–874. 10.1159/000459899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Shen B., Li J., Lv D., Zhao Y., Wang F., et al. (2014). Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int. J. Clin. Exp. Med. 7, 136–141. [PMC free article] [PubMed] [Google Scholar]
- Xu M., Li J., Chen X., Han L., Li L., Liu Y. (2019). MTHFD1 promoter hypermethylation increases the risk of hypertension. Clin. Exp. Hypertens. 41, 422–427. 10.1080/10641963.2018.1501057 [DOI] [PubMed] [Google Scholar]
- Xu P., Zhao Y., Liu M., Wang Y., Wang H., Li Y.-X., et al. (2014). Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 63, 1276–1284. 10.1161/HYPERTENSIONAHA.113.02647 [DOI] [PubMed] [Google Scholar]
- Xu S., Pelisek J., Jin Z. G. (2018). Atherosclerosis is an epigenetic disease. Trends Endocrinol. Metab. 29, 739–742. 10.1016/j.tem.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Liu D., Zhu W., Su Z., Zhang L., Zhou C., et al. (2019). Circulating MiR-17-5p, MiR-126-5p and MiR-145-3p are novel biomarkers for diagnosis of acute myocardial infarction. Front. Physiol. 10, 123. 10.3389/fphys.2019.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Nishida T., Horibe H., Oguri M., Kato K., Sawabe M. (2014). Identification of hypo- and hypermethylated genes related to atherosclerosis by a genome-wide analysis of DNA methylation. Int. J. Mol. Med. 33, 1355–1363. 10.3892/ijmm.2014.1692 [DOI] [PubMed] [Google Scholar]
- Yan Y., Song X., Li Z., Zhang J., Ren J., Wu J., et al. (2017). Elevated levels of granzyme B correlated with miR-874-3p downregulation in patients with acute myocardial infarction. Biomark. Med. 11, 761–767. 10.2217/bmm-2017-0144 [DOI] [PubMed] [Google Scholar]
- Yang G., Weng X., Zhao Y., Zhang X., Hu Y., Dai X., et al. (2017. a). The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction. Nat. Commun. 8, 14941. 10.1038/ncomms14941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Jia C., Wang P., Xiong M., Cui J., Li L., et al. (2014). MicroRNA-505 identified from patients with essential hypertension impairs endothelial cell migration and tube formation. Int. J. Cardiol. 177, 925–934. 10.1016/j.ijcard.2014.09.204 [DOI] [PubMed] [Google Scholar]
- Yang Q., Lu J., Wang S., Li H., Ge Q., Lu Z. (2011). Application of next-generation sequencing technology to profile the circulating microRNAs in the serum of preeclampsia versus normal pregnant women. Clin. Chim. Acta 412, 2167–2173. 10.1016/j.cca.2011.07.029 [DOI] [PubMed] [Google Scholar]
- Yang S.-Y., Wang Y.-Q., Gao H.-M., Wang B., He Q. (2016. a). The clinical value of circulating miR-99a in plasma of patients with acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 20, 5193–5197. [PubMed] [Google Scholar]
- Yang V. K., Loughran K. A., Meola D. M., Juhr C. M., Thane K. E., Davis A. M., et al. (2017. b). Circulating exosome microRNA associated with heart failure secondary to myxomatous mitral valve disease in a naturally occurring canine model. J. Extracell Vesicles 6, 1350088. 10.1080/20013078.2017.1350088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Wang A., Zhao C., Li Q., Pan Z., Han X., et al. (2016. b). miR-125b enhances IL-8 production in early-onset severe preeclampsia by targeting sphingosine-1-phosphate lyase 1. PLoS One 11, e0166940. 10.1371/journal.pone.0166940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.-L., Lu X.-L., Yan C.-Y., Wan Q.-L., Cheng G.-C., Li Y.-M. (2015). Circulating miR-122-5p as a potential novel biomarker for diagnosis of acute myocardial infarction. Int. J. Clin. Exp. Pathol. 8, 16014–16019. [PMC free article] [PubMed] [Google Scholar]
- Yilmaz S. G., Isbir S., Kunt A. T., Isbir T. (2018). Circulating microRNAs as novel biomarkers for atherosclerosis. In Vivo 32, 561–565. 10.21873/invivo.11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Liu X., Chen F., Zhang L., Chen X., Huang Q., et al. (2016). Diagnostic and prognostic value of circulating microRNA-133a in patients with acute myocardial infarction. Clin. Lab. 62, 1233–1241. 10.7754/Clin.Lab.2015.151023 [DOI] [PubMed] [Google Scholar]
- Zampetaki A., Zeng L., Margariti A., Xiao Q., Li H., Zhang Z., et al. (2010). Histone deacetylase 3 is critical in endothelial survival and atherosclerosis development in response to disturbed flow. Circulation 121, 132–142. 10.1161/CIRCULATIONAHA.109.890491 [DOI] [PubMed] [Google Scholar]
- Zhang C., Li Q., Ren N., Li C., Wang X., Xie M., et al. (2015. a). Placental miR-106a∼363 cluster is dysregulated in preeclamptic placenta. Placenta 36, 250–252. 10.1016/j.placenta.2014.11.020 [DOI] [PubMed] [Google Scholar]
- Zhang D., Yu Z.-Y., Cruz P., Kong Q., Li S., Kone B. C. (2009). Epigenetics and the control of epithelial sodium channel expression in collecting duct. Kidney Int. 75, 260–267. 10.1038/ki.2008.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Li Y., Zhao Q. (2018. a). Circulating miR-23b as a novel biomarker for early risk stratification after ST-elevation myocardial infarction. Med. Sci. Monit. 24, 1517–1523. 10.12659/MSM.908060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xing Q., Zhou X., Li J., Li Y., Zhang L., et al. (2017. a). Circulating miRNA-21 is a promising biomarker for heart failure. Mol. Med. Rep. 16, 7766–7774. 10.3892/mmr.2017.7575 [DOI] [PubMed] [Google Scholar]
- Zhang L. X., Du J., Zhao Y. T., Wang J., Zhang S., Dubielecka P. M., et al. (2018. b). Transgenic overexpression of active HDAC4 in the heart attenuates cardiac function and exacerbates remodeling in infarcted myocardium. J. Appl. Physiol. 125, 1968–1978. 10.1152/japplphysiol.00006.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen X., Su T., Li H., Huang Q., Wu D., et al. (2015. b). Circulating miR-499 are novel and sensitive biomarker of acute myocardial infarction. J. Thorac. Dis. 7, 303–308. 10.3978/j.issn.2072-1439.2015.02.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L.-N., Liu P.-P., Wang L., Yuan F., Xu L., Xin Y., et al. (2013. a). Lower ADD1 gene promoter DNA methylation increases the risk of essential hypertension. PLoS ONE 8, e63455. 10.1371/journal.pone.0063455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Cheng Y.-J., Sara J. D., Liu L.-J., Liu L.-P., Zhao X., et al. (2017. b). Circulating microRNA-145 is associated with acute myocardial infarction and heart failure. Chin. Med. J. 130, 51–56. 10.4103/0366-6999.196573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Lan C., Pei H., Duan G., Huang L., Li L. (2015. c). Expression of circulating miR-486 and miR-150 in patients with acute myocardial infarction. BMC Cardiovasc. Disord. 15, 51. 10.1186/s12872-015-0042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Niu H., Ban T., Xu L., Li Y., Wang N., et al. (2013. b). Elevated plasma microRNA-1 predicts heart failure after acute myocardial infarction. Int. J. Cardiol. 166, 259–260. 10.1016/j.ijcard.2012.09.108 [DOI] [PubMed] [Google Scholar]
- Zhang W.-Q., Xie B.-Q. (2017). A meta-analysis of the relations between blood microRNA-208b detection and acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 21, 848–854. [PubMed] [Google Scholar]
- Zhang X., Wang X., Wu J., Peng J., Deng X., Shen Y., et al. (2018. c). The diagnostic values of circulating miRNAs for hypertension and bioinformatics analysis. Biosci. Rep. 38, BSR20180525. 10.1042/BSR20180525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng J., Chen F., Wu C., Zhang J., Ren X., et al. (2017. c). Circulating endothelial microparticles and miR-92a in acute myocardial infarction. Biosci. Rep. 37, BSR20170047. 10.1042/BSR20170047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu Y.-J., Liu T., Zhang H., Yang S.-J. (2016). Plasma microRNA-21 is a potential diagnostic biomarker of acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 20, 323–329. [PubMed] [Google Scholar]
- Zhao D.-S., Chen Y., Jiang H., Lu J.-P., Zhang G., Geng J., et al. (2013). Serum miR-210 and miR-30a expressions tend to revert to fetal levels in Chinese adult patients with chronic heart failure. Cardiovasc. Pathol. 22, 444–450. 10.1016/j.carpath.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Zhao L., Chen C.-N., Hajji N., Oliver E., Cotroneo E., Wharton J., et al. (2012). Histone deacetylation inhibition in pulmonary hypertension: therapeutic potential of valproic acid and suberoylanilide hydroxamic acid. Circulation 126, 455–467. 10.1161/CIRCULATIONAHA.112.103176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B., Xi Z., Liu R., Yin W., Sui Z., Ren B., et al. (2018). The function of microRNAs in B-cell development, lymphoma, and their potential in clinical practice. Front. Immunol. 9, 936. 10.3389/fimmu.2018.00936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., He Y., Chen W., Shui X., Chen C., Lei W. (2014). Circulating microRNA-19a as a potential novel biomarker for diagnosis of acute myocardial infarction. IJMS 15, 20355–20364. 10.3390/ijms151120355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yao K., Wang Q., Guo J., Shi H., Ma L., et al. (2016). Circulating miR-181a as a potential novel biomarker for diagnosis of acute myocardial infarction. Cell. Physiol. Biochem. 40, 1591–1602. 10.1159/000453209 [DOI] [PubMed] [Google Scholar]
- Zhu L., Liu F., Xie H., Feng J. (2018). Diagnostic performance of microRNA-133a in acute myocardial infarction: a meta-analysis. Cardiol. J. 25, 260–267. 10.5603/CJ.a2017.0126 [DOI] [PubMed] [Google Scholar]
- Zhu X.-M., Han T., Sargent I. L., Yin G.-W., Yao Y.-Q. (2009). Differential expression profile of microRNAs in human placentas from preeclamptic pregnancies vs normal pregnancies. Am. J. Obstet. Gynecol. 200, 661.e1–661.e7. 10.1016/j.ajog.2008.12.045 [DOI] [PubMed] [Google Scholar]