Abstract
During splicing of pre-mRNA, 5′ and 3′ splice sites are brought within proximity by interactions between the pre-mRNA bound U1 and U2 snRNPs, followed by recruitment of the tri-snRNP for assembly of the mature spliceosome. Previously, we identified an interaction between the U2 snRNP-specific protein SF3A1 and the stem–loop 4 (SL4) of the U1 snRNA that occurs during the early steps of spliceosome assembly. Although harboring many annotated domains, SF3A1 lacks a canonical RNA binding domain. To identify the U1-SL4 binding region in SF3A1, we expressed amino- and carboxy-terminal deletion constructs using a HeLa cell-based cell free expression system. UV-crosslinking of the truncated proteins with 32P-U1-SL4 RNA identified the carboxy-terminal ubiquitin-like (UBL) domain of SF3A1 as the RNA binding region. Characterization of the interaction between SF3A1-UBL and U1-SL4 by electrophoretic mobility shift assay and surface plasmon resonance determined high binding affinity (KD = ∼97 nM), and revealed the double-stranded G-C rich stem of U1-SL4 as an important feature for binding to the UBL domain. Further, mutations of two conserved tyrosine residues, Y772 and Y773, were found to cause a two- and fivefold decrease in the binding affinity for U1-SL4, respectively. Finally, we found that SF3A1-UBL can specifically pull down the U1 snRNP from HeLa nuclear extract, demonstrating its capacity to bind U1-SL4 in the context of the intact snRNP. Thus, the data show that the UBL domain of SF3A1 can function as an RNA binding domain and that mutations in this region may interfere with U1-SL4 binding.
Keywords: SF3A1, U1 snRNA, ubiquitin-like domain, pre-mRNA splicing, stem–loop 4
INTRODUCTION
Removal of introns and ligation of exons in pre-mRNAs is catalyzed by the spliceosome, a dynamic complex comprising five small nuclear ribonucleoproteins (snRNPs) (U1, U2, U4, U5, and U6) and many auxiliary proteins (Will and Lührmann 2011). Spliceosome assembly occurs de novo onto each intron and proceeds through a series of intermediate complexes. Stable binding of the U1 snRNP to the 5′ splice site and of the U2 snRNP to the branch point sequence forms the prespliceosomal A complex that interacts with the preformed U4/U6.U5 tri-snRNP to generate the spliceosomal pre-B complex. Subsequent to this, extensive structural and conformational remodeling leads to formation of at least six distinct complexes that are referred to as B, Bact, B*, C, C*, and P (Boesler et al. 2016; Galej et al. 2016; Bertram et al. 2017; Fica et al. 2017; Plaschka et al. 2017; Haselbach et al. 2018; Zhan et al. 2018; Zhang et al. 2018). The catalytic steps of splicing occur during the B* → C and C* → P transitions. Cryo-EM analyses have revealed in great detail the compositional and structural changes that occur in spliceosomal complexes after binding of the tri-snRNP and during the transitions that accompany the catalytic steps. However, interactions that occur during the very early steps of spliceosome assembly, prior to binding of the tri-snRNP, remain to be elucidated.
Spliceosome assembly begins with binding of the U1 snRNP to the 5′ splice site, splicing factor 1 (SF1) to the branch-point, and the U2 auxiliary factor 65 (U2AF65 or U2AF2) and U2AF35 (U2AF1) to the polypyrimidine tract and the 3′ splice site, respectively (Will and Lührmann 2011). The U2 snRNP has been reported to associate with this early (E) complex via interactions between SF3B1 and U2AF65 (Gozani et al. 1998; Das et al. 2000). Stable binding of the U2 snRNP to the branchpoint forms the prespliceosomal A complex. Cryo-EM analysis of the yeast prespliceosome has provided some insight into the U1–U2 interface and identified two regions of contact between components of pre-mRNA bound U1 and U2 snRNPs (Plaschka et al. 2018). The first interface forms after a stable interaction occurs between yeast U1 specific protein Prp39 and the core U2 protein U2A′ (Lea1 in yeast). The second interface involves interactions of yeast U1 snRNA stem–loop 3 (SL3) with the SF3B complex protein SF3B3 (SF3B130; Rse1 in yeast) and with the SF3A complex protein SF3A3 (Prp9 in yeast). However, the human U1 and U2 snRNPs differ significantly from those of yeast (Pomeranz Krummel et al. 2009; Li et al. 2017). The human U1 (164 nt) and U2 (188 nt) snRNAs are considerably shorter than their yeast orthologs, which are 568 and 1175 nt long, respectively. The human U1 snRNP consists of three particle specific proteins, U1-70k, U1C, and U1A. On the other hand, the yeast U1 contains seven additional particle specific proteins, namely, Prp39, Prp40 (human Prp40 or FBP11), Prp42, Nam8 (human TIA-1), Snu56, LUC7 (human LUC7L), and Snu71 (human RBM25). The human and yeast U2 snRNPs have similar numbers of particle specific proteins, 7 and 6, respectively. However, the primary structures of some of these proteins are significantly different. For example, the SF3A1 protein that is relevant to this study is 793 amino acids (aa) in humans, and its yeast counterpart, Prp21, is only 280 aa (Lin and Xu 2012). Thus, it is likely that the U1–U2 interface contacts during the early steps of human spliceosome assembly differ from those observed in yeast.
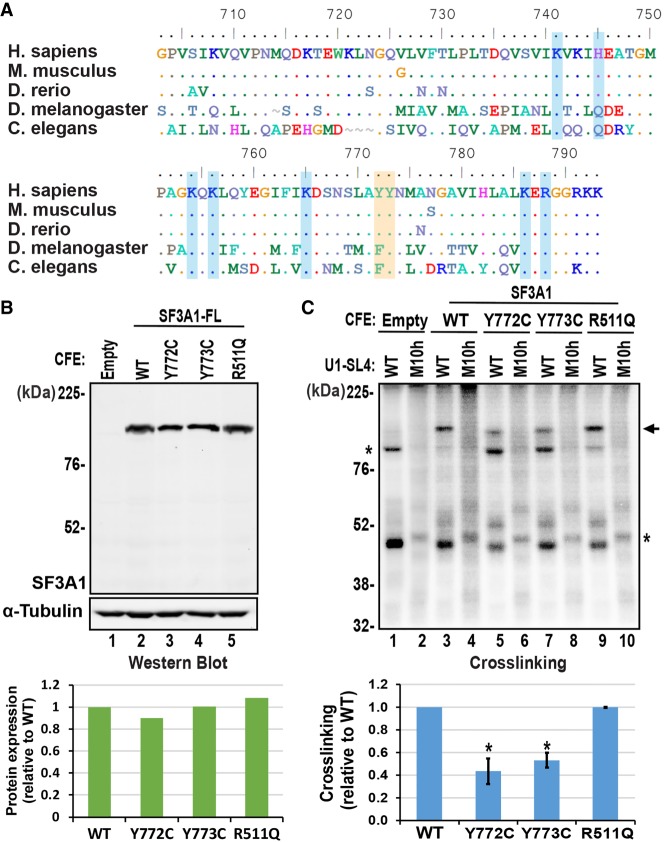
Previously, we reported that during spliceosome assembly, the SF3A complex engages in direct contact with the U1 snRNP via interactions between the SF3A1 protein and stem–loop 4 of the U1 snRNA (U1-SL4). This interaction occurs between pre-mRNA bound U1 and U2 snRNPs and was found to be critical for the formation of the prespliceosomal A complex (Sharma et al. 2014). The SF3A1 protein (120 kDa) interacts with SF3A2 (66 kDa) and SF3A3 (60 kDa) to create the SF3A complex, which is necessary for formation of the mature 17S U2 snRNP and for pre-mRNA splicing in vitro and in vivo (Brosi et al. 1993; Tanackovic and Krämer 2004; Crisci et al. 2015). Notably, SF3A1 lacks a conventional RNA-binding domain. At its amino terminus, SF3A1 contains two suppressor-of-white-apricot (SURP) domains and a short segment of charged residues (Fig. 1A). SF3A1 interacts with SF3A3 through a region harboring the SURP2 domain (aa 145–243) and with SF3A2 via a 26 residue region (aa 269–295) (Huang et al. 2011). The carboxy-terminal region of SF3A1 harbors a nuclear localization signal (NLS) and a ubiquitin-like (UBL) domain. The amino- and carboxy-terminal domains are separated by a long central region that contains two proline-rich segments. Integrity of all these regions is important for normal physiology as several point mutations that occur across the entire length of SF3A1 are known to be associated with hematological diseases including myelodysplastic syndromes (MDS), chronic myelomonocytic leukemia (CMML), and acute myeloid leukemia (AML) (Yoshida et al. 2011; Papaemmanuil et al. 2016).
FIGURE 1.
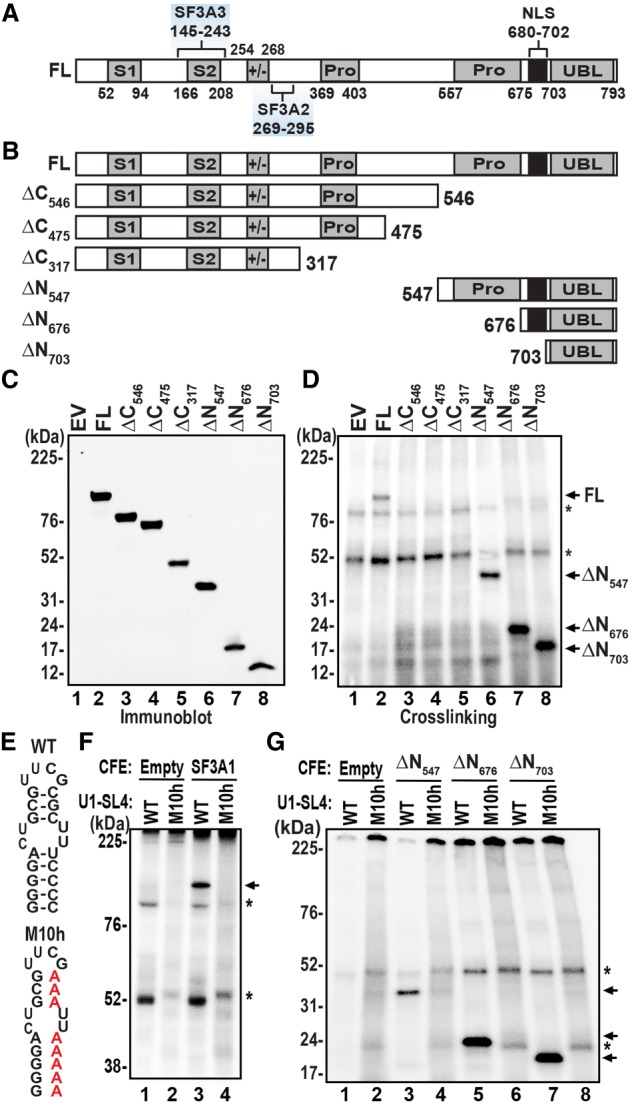
The carboxy-terminal UBL domain of SF3A1 binds to U1-SL4 RNA. (A) Schematic diagram of human SF3A1 protein depicting the known domain organization and the regions that interact with SF3A2 and SF3A3. (B) Schematic of all amino- and carboxy-terminal deletion constructs. (C) Western blot of 6×His-tagged protein expressed in cell-free expression (CFE) reactions containing the control empty vector (EV), or expression vectors with full-length (FL) protein, carboxy-terminal truncations (ΔC546, ΔC475, ΔC317), and amino-terminal truncations (ΔN547, ΔN676, ΔN703). Proteins were detected using anti-6×His primary antibody. (D) 32P-U1-SL4 was added to CFE extracts containing SF3A1 proteins, UV-crosslinked, and then separated on SDS-PAGE gels and visualized by phosphor imaging. (E) Schematic of wild-type (WT) and mutant (M10h) U1-SL4 RNAs. (F) Crosslinking of 32P-U1-SL4-WT and -M10h RNAs in CFE extracts expressing EV or FL SF3A1. (G) Crosslinking of WT or M10h 32P-U1-SL4 in CFE extracts expressing EV or ΔN SF3A1 constructs. Arrows indicate specific crosslinked products while asterisks indicate nonspecific products that were observed in all reactions including the EV control.
In this study, we report that the UBL domain of SF3A1 is a noncanonical RNA-binding domain. Applying a combination of techniques including UV-crosslinking, electrophoretic mobility shift assays (EMSAs), and surface plasmon resonance (SPR), we demonstrate that the SF3A1-UBL domain binds U1-SL4 with high affinity (KD = ∼97 nM). Investigations of the impact of an MDS-linked residue Y772, and an adjacent highly conserved residue Y773, revealed that Y772C and Y773C mutations decrease the affinity of SF3A1-UBL for U1-SL4 and reduced its ability to interact with the U1 snRNP. Since Prp21, the yeast ortholog of SF3A1, lacks the carboxy-terminal region that harbors the UBL domain (Lin and Xu 2012), we propose that the U1-SL4/SF3A1 contact may be unique to the U1–U2 interface during the early steps of human spliceosome assembly.
RESULTS
SF3A1 interacts with U1-SL4 through the carboxy-terminal ubiquitin-like domain
Human SF3A1 is a 793 aa protein that lacks a canonical RNA binding domain (Fig. 1A). To identify the U1-SL4 binding domain in SF3A1, we created carboxy-terminally 6×His-tagged constructs for expression of full-length (FL) SF3A1, and a series of amino-terminal (ΔN) and carboxy-terminal (ΔC) deletions (Fig. 1B). These constructs were expressed by the HeLa cell lysate based in vitro cell-free expression (CFE) system (Thermo Fisher Scientific) that uses coupled transcription-translation reactions. Western analysis with an anti-6×His antibody demonstrated efficient expression of all FL, ΔN, and ΔC SF3A1 proteins (Fig. 1C).
To identify the U1-SL4 interacting domain in SF3A1, binding reactions were assembled by adding uniformly 32P-labeled U1-SL4 RNA to the expressed proteins under splicing conditions. The reactions were UV-crosslinked and separated by SDS-PAGE. This analysis showed that U1-SL4 crosslinked to FL SF3A1, but not to carboxy-terminal truncations, ΔC546, ΔC475, or ΔC317 (Fig. 1D, compare lane 2 with lanes 3–5). All amino-terminal truncations, however, retained the capacity to crosslink to U1-SL4, including ΔN703 that only contains the UBL domain (lanes 6–8). There was some nonspecific crosslinking in all reactions, which was also observed in the EV control (lane 1). However, specific binding of SF3A1-FL and ΔN proteins to 32P-U1-SL4 was clearly identifiable by the appearance of appropriate size bands. To check the specificity of binding, we used the U1-SL4/M10h mutant that was previously found to reduce U1 snRNP splicing activity (Fig. 1E). Crosslinking analysis with FL protein demonstrated binding of SF3A1 to wild-type U1-SL4, but not to M10h (Fig. 1F, compare lanes 3 and 4). Similarly, all amino-terminal truncations ΔN547, ΔN676, and ΔN703 crosslinked to wild-type U1-SL4 (Fig. 1G lanes 3,5,7), but not to the mutant M10h (lanes 4,6,8). Thus, the crosslinking analysis demonstrated that the U1-SL4 interacting domain resides in the carboxy-terminal region of SF3A1, from residues 703–793 that harbors the UBL domain.
Ubiquitin is a 76 aa protein consisting of two α-helices and five β-sheets that fold into a ββαββαβ topology (Vijay-kumar et al. 1987). A previous analysis showed that the carboxyl terminus of SF3A1 (aa 714–790) shares ∼29.6% identity and ∼54.9% similarity with ubiquitin (Krämer et al. 1995). However, the region of SF3A1 that folds into a UBL motif is 90 aa, encompassing residues 703–793 (Fig. 2A). A comparison of the structures and sequences of ubiquitin and SF3A1-UBL revealed that the first β strand in SF3A1-UBL is located further upstream starting at aa P704. In SF3A1-UBL, the linker between the β1 and β2 strands is longer (15 aa) than the 4 aa linker in ubiquitin. Additionally, at its carboxy-terminal end, SF3A1-UBL contains three positively charged residues (RKK) downstream from an RGG sequence, which is present in ubiquitin (Fig. 2A), and also conserved in SF3A1-UBL domains from other species (see Fig. 5A).
FIGURE 2.
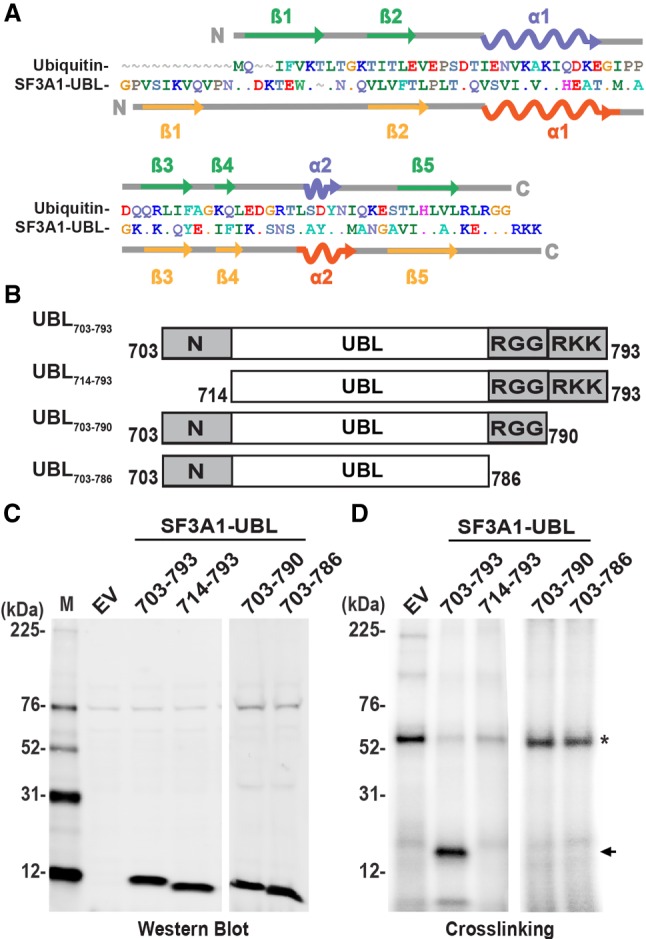
Full-length SF3A1-UBL domain is required for RNA binding. (A) Alignment of the SF3A1 UBL domain (aa 703–793) to ubiquitin. Graphical representations of secondary structure in relation to primary structure were created based on PDB entries for ubiquitin (PDB ID: 1UBQ) and SF3A1-UBL (PDB ID: 1ZKH). In the alignment, a dot indicates the presence of an identical residue to the reference sequence used in the alignment (ubiquitin), and a tilde indicates a gap. (B) Schematic of all amino- and carboxy-terminal deletion constructs made to study the SF3A1-UBL domain. (C) Western blot of 6×His-tagged proteins in CFE reactions containing the EV control, or expression vectors for FL (UBL703–793), amino-terminal truncated (UBL714–793), and carboxy-terminal truncated (UBL703–790 and UBL703–786) SF3A1-UBL. The expressed UBL domain proteins were detected with anti-6×His primary antibody. (D) UV-crosslinking of 32P-U1-SL4 in CFE reactions expressing EV or UBL proteins. Arrows indicate specific crosslinked products, and asterisks indicate nonspecific products that were also observed in reactions containing the EV control.
FIGURE 5.
Mutation of conserved tyrosine residues affects U1-SL4 binding by SF3A1. (A) Alignment of SF3A1-UBL domains from human, mouse, zebrafish, flies, and worms. In the aligned sequences, a dot indicates the presence of an identical residue and a tilde indicates a gap. (B) Western analysis using anti-SF3A1 primary antibody demonstrating expression of SF3A1-WT and point-mutants Y772C, Y773C, and R511Q in CFE reactions. The expression of mutant proteins relative to WT, normalized to α-Tubulin, is graphed below the western blot. (C) Crosslinking of WT and mutant SF3A1 proteins to 32P-U1-SL4-WT and -M10h RNAs. Crosslinking efficiency, relative to WT SF3A1, is represented in the graph below (* = P < 0.05, Student's t-test). For gels, arrows indicate specific crosslinked products and asterisks indicate nonspecific products that were seen in reactions containing the EV control.
To determine if these unique terminal features of the SF3A1-UBL domain are required for RNA binding, we created three additional constructs that involved deleting the first β strand in UBL714–793, the RKK motif in UBL703–790, and the RGGRKK sequence in UBL703–786 (Fig. 2B). Expression of 6×His-tagged UBL domain deletion constructs in CFE reactions was confirmed by immunoblotting (Fig. 2C). UV-crosslinking analysis demonstrated that the UBL703–793 construct bound 32P-U1-SL4 RNA, whereas UBL714–793, UBL703–790, and UBL703–786 did not (Fig. 2D). Thus, these results confirmed that the unique amino- and carboxy-terminal features of the SF3A1-UBL domain are required for RNA binding.
SF3A1-UBL domain binds G-C rich stem–loop RNA
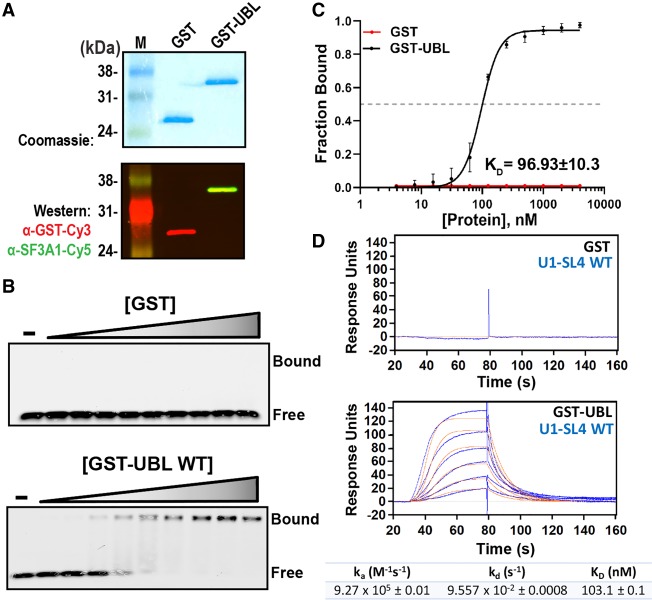
To characterize the affinity and specificity of RNA binding by the SF3A1-UBL domain, we created a fusion construct of GST and the UBL domain (GST-UBL). GST and GST-UBL proteins were expressed in Escherichia coli, purified using glutathione agarose beads, and confirmed by Coomassie blue staining, and western blotting using antibodies to both GST and SF3A1 (Fig. 3A). To determine the dissociation constant (KD) for the interaction between SF3A1-UBL and U1-SL4, we applied two independent quantitative methods: EMSA and SPR. For EMSAs, binding reactions consisting of varying concentrations of GST or GST-UBL protein and Cy5-labeled U1-SL4 RNA were prepared and then separated on native gels as described previously (Fig. 3B; Pagano et al. 2011). Dose-response curves created from the fraction of RNA–protein complexes formed indicated dose-dependent assembly of GST-UBL/U1-SL4 complexes with a KD = 96.93 ± 10 nM, whereas GST alone did not exhibit any RNA binding (Fig. 3B,C).
FIGURE 3.
GST-UBL fusion protein binds to free U1-SL4. (A) Expression and purification of GST alone and GST-UBL fusion proteins were confirmed by Coomassie blue staining and western blotting using anti-GST and anti-SF3A1 antibodies. (B) EMSAs for binding of GST and GST-UBL proteins to 5′-Cy5-labeled U1-SL4 RNA. The concentrations of GST and GST-UBL were 0, 0.02, 0.03, 0.06, 0.13, 0.25, 0.5, 1, 2, and 4 µM. (C) Dose-response curves generated by plotting the fraction of Cy5-U1-SL4 bound versus GST or GST-UBL protein concentration. The KD value was determined from triplicate experiments. (D) SPR analysis for binding of GST and GST-UBL to immobilized 5′-biotinylated U1-SL4. Blue lines in the sensorgrams represent the raw curve generated by association and dissociation of GST injected at 1 µM and GST-UBL injected at 0.02, 0.03, 0.06, 0.13, 0.25, and 0.5 nM. Orange traces represent the global fit to the raw data by the Langmuir binding model assuming a 1:1 stoichiometry. Binding kinetics and dissociation constant values are shown below the sensorgram.
For SPR experiments, biotinylated U1-SL4 RNA was immobilized on Neutravidin coated biosensors. Sensorgrams recorded during association and dissociation phases demonstrated a concentration-dependent response upon injection of GST-UBL and lack of a response with GST (Fig. 3D). Binding parameters were calculated by nonlinear regression analysis of the data assuming a 1:1 stoichiometry by the Langmuir binding model. Kinetics of the SF3A1-UBL/U1-SL4 interaction are characterized by an association rate of 9.27 ± 0.01 × 105 M−1 sec−1 and a dissociation rate of 9.56 ± 0.0008 × 10−2 sec−1, resulting in a KD = 103.1 ± 0.1 nM. The agreement of the KD values obtained by EMSA and SPR indicates that immobilization of the U1-SL4 RNA on the biosensor does not perturb the binding reaction and that dissociation constants for the UBL–SL4 interaction can be reliably determined by either technique.
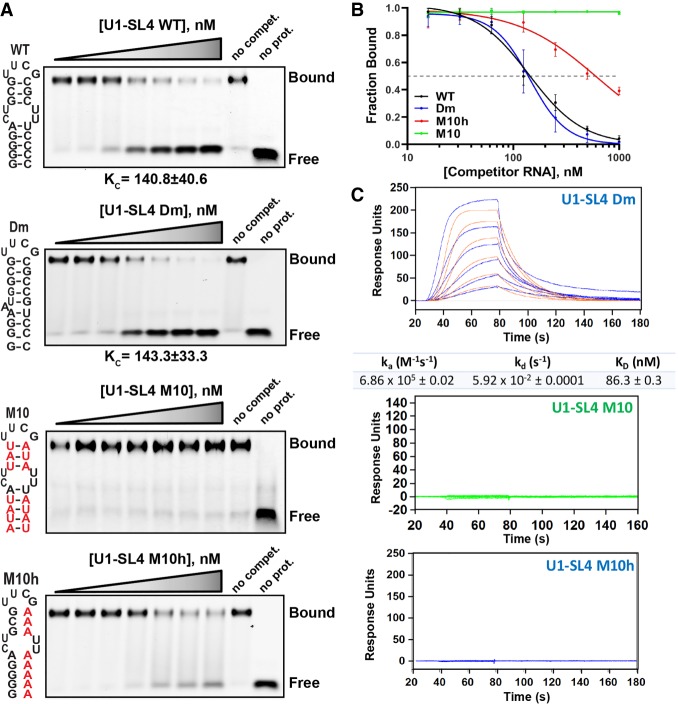
To determine the specificity of the UBL–SL4 interaction, we performed competitive EMSAs and SPR. To reactions containing preformed GST-UBL/Cy5-U1-SL4 complexes, increasing concentrations of competitor RNAs were added. The RNA–protein complexes were separated, quantified, and the dissociation constants for competitor RNAs (KC) were determined from plots of fraction of Cy5-U1-SL4 RNA bound versus competitor RNA concentration (Fig. 4A,B; Ryder et al. 2008). As expected, U1-SL4/WT strongly competed out Cy5-U1-SL4 with a KC = 140.8 ± 40.6 nM. Like human SL4, the Drosophila melanogaster U1-SL4 RNA (U1-SL4/Dm) forms a G-C rich stem ending in a tetra-loop. We previously demonstrated that U1-SL4/Dm can be functionally substituted for human U1-SL4 in U1 complementation assays and that it binds SF3A1 (Sharma et al. 2014). U1-SL4/Dm was able to compete out GST-UBL/U1-SL4 complexes as efficiently as WT human U1-SL4 with a similar KC = 143.3 ± 33.3 nM and also exhibited the capacity to bind the SF3A1-UBL protein by SPR with an association rate of 6.86 ± 0.02 × 105 M−1 sec−1 and a dissociation rate of 5.92 ± 0.0001 × 10−2 sec−1, resulting in a KD = 86.3 ± 0.3 nM (Fig. 4B,C). On the other hand, minimal to no competition was observed when using two mutant RNAs. In the U1-SL4/M10 mutant, all G-C base pairs were replaced with A-U to retain the secondary structure of U1-SL4 while lowering the G-C content, and in the U1-SL4/M10h mutant, base-pairing was disrupted to obtain a single-stranded RNA (ssRNA) by changing guanine or cytosine nucleotides of one of the strands to adenines. The M10 RNA did not exhibit any UBL-binding by competitive EMSAs or SPR (Fig. 4B,C). While the M10h mutant did not crosslink to SF3A1 (Fig. 1F,G) or demonstrate binding to GST-UBL by SPR (Fig. 4C), it showed some capacity to compete out GST-UBL/SL4 bound complexes by EMSA but the binding did not reach saturation (Fig. 4A). This nonspecific binding might be because of a short stem–loop structure that the M10h mutant may form as predicted by the mfold algorithm (Supplemental Fig. S1; Zuker 2003). Taken together, EMSA and SPR analyses demonstrate that the UBL domain of SF3A1 binds to a G-C base-paired stem–loop RNA, but not to an A-U rich stem–loop or to ssRNA.
FIGURE 4.
SF3A1-UBL binds to G-C rich stem–loop RNA. (A) All competitive EMSA reactions contained 500 nM GST-UBL and 10 nM WT Cy5-U1-SL4. Final concentrations of competitor RNAs for WT U1-SL4, Drosophila melanogaster (Dm), and two mutant RNAs, M10 and M10h, were 0.02, 0.03, 0.06, 0.13, 0.25, 0.5, and 1 µM. The dissociation constants of competitor RNAs (KC) that competed out Cy5-U1-SL4 are provided (n = 3). (B) Dose-response curves plotted from the fraction of Cy5-U1-SL4 bound versus the concentration of competitor RNA are shown. (C) Sensorgrams from SPR experiments for binding of GST-UBL to Dm, M10, and M10h RNAs. GST-UBL concentrations were 0.02, 0.03, 0.06, 0.13, 0.25, and 0.5 µM. Orange traces represent the global fit to the raw data by the Langmuir binding model, assuming a 1:1 stoichiometry. Binding kinetics and dissociation constant values are summarized below sensorgrams where binding was detected.
Tyrosines 772 and 773 are important for U1-SL4 binding by SF3A1-UBL
The UBL domain of SF3A1 shares a ∼98% and ∼93% sequence identity with the mouse and the zebrafish domains, respectively, and <50% with flies and worms (Supplemental Table S1). Multiple alignment of these sequences identified several highly conserved residues (Fig. 5A). Notable among these are tyrosine residues 772 (Y772) and 773 (Y773). Y772 has been reported to be mutated to a cysteine in individuals with MDS and CMML (Yoshida et al. 2011). It is conserved between humans, mice, and fish, and has a conservative substitution to phenylalanine in flies and worms, while being replaced by an aspartate in ubiquitin (Figs. 2A, 5A). Y773, on the other hand, is conserved from humans to worms and is even present in ubiquitin. To examine if these residues are involved in U1-SL4 binding, we created SF3A1 plasmid constructs carrying the Y772C and Y773C mutations and expressed them by the HeLa CFE system. We made an additional mutant, SF3A1-R511Q, which has been reported to be associated with AML (Papaemmanuil et al. 2016). Western analysis confirmed the expression of WT and mutant FL SF3A1 proteins in the CFE reactions (Fig. 5B lanes 2–5). UV-crosslinking of binding reactions containing the SF3A1 proteins and 32P-U1-SL4 revealed that the Y772C and Y773C mutations reduced the binding capacity of SF3A1 by approximately twofold in comparison to SF3A1-WT, whereas the R511Q mutation did not have any effect (Fig. 5C).
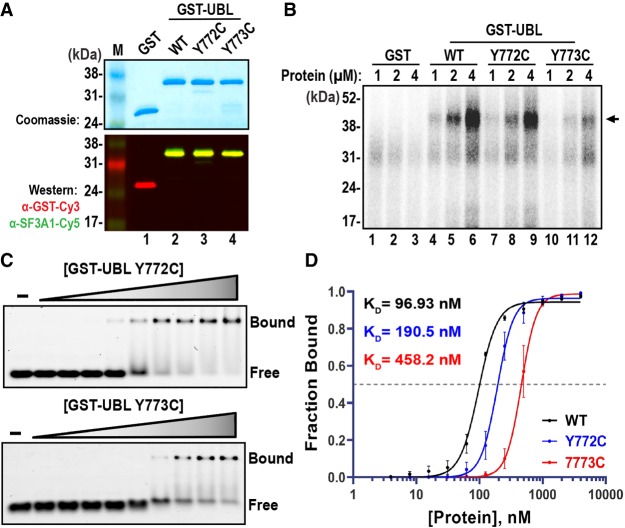
To further characterize the impact of the Y772C and Y773C mutations on U1-SL4 binding, we introduced these mutations into the GST-UBL fusion construct. Wild-type and mutant GST-UBL proteins were purified using glutathione agarose beads and their purity was confirmed by Coomassie blue staining and western blotting (Fig. 6A). Initial analysis by UV-crosslinking indicated that the Y772C mutation caused a moderate reduction in U1-SL4 binding, whereas the Y773C mutation led to a drastic reduction in crosslinking efficiency (Fig. 6B, compare lanes 7–9 and 10–12 to 4–6). Binding analysis by EMSAs supported these initial observations; GST-UBL proteins carrying either mutation formed complexes with Cy5-U1-SL4, however, not as efficiently as the WT protein (Fig. 6C). The dose-response curves exhibited a clear rightward shift and quantification revealed approximately twofold (KD = 190.5 ± 35.1) and approximately fivefold (KD = 458.2 ± 101.4) increase in KD values for Y772C and Y773C mutations, respectively (Fig. 6D). Thus, Y772C and Y773C mutations cause a decrease in binding affinity. These results indicate that Y772 and Y773 may have a role in U1-SL4 binding by the SF3A1-UBL domain.
FIGURE 6.
Y772C and Y773C mutations reduce the UBL domain binding affinity for U1-SL4. (A) Expression and purification of WT and Y772C/Y773C GST-UBL variants as assessed by Coomassie blue staining and western analysis. (B) Crosslinking of 32P-U1-SL4 RNA to WT and mutant GST-UBL proteins. (C) EMSA analysis for binding of GST-UBL carrying Y772C and Y773C mutants to Cy5-U1-SL4. The final concentrations for mutant GST-UBL proteins were 0, 0.02, 0.03, 0.06, 0.13, 0.25, 0.5, 1, 2, and 4 µM. (D) Dose-response curves were created by plotting the fraction of bound Cy5-U1-SL4 versus GST-UBL/WT, Y772C, and Y773C protein concentrations, and KD values were determined from triplicate experiments.
GST-UBL protein selectively interacts with the U1 snRNP in HeLa nuclear extract
To confirm that the UBL domain of SF3A1 can interact with U1-SL4 in the context of an intact U1 snRNP, we performed GST affinity pull-down assays. First, HeLa cell nuclear extracts were pre-cleared of GST-binding proteins, as previously described (Masuda et al. 2005). Then, GST and GST-UBL/WT proteins were added in the absence or presence of U1-SL4/WT or U1-SL4/M10 competitor RNAs, followed by pull-down using glutathione agarose beads. Spliceosomal components associated with bound complexes were analyzed by northern and western blotting (Fig. 7A,B). Northern analysis confirmed the enrichment of the U1 snRNA, but not the U2, U4, U5, or U6 snRNAs, in GST-UBL/WT complexes (Fig. 7A, lane 2). Preincubation with the U1-SL4/WT RNA competed out the U1 snRNA from the GST-UBL/WT complexes (Fig. 7A, compare lanes 2 and 4), whereas preincubation with the U1-SL4/M10 RNA did not have any impact (Fig. 7A, lane 6). Western analysis confirmed the presence of U1 snRNP-specific proteins U1C and U1-70k in the pull-down complexes and the absence of U2 specific protein SF3A3 (Fig. 7B, lane 2). The U1 proteins were similarly competed out by preincubation with the U1-SL4/WT, but not the U1-SL4/M10 RNA (Fig. 7B lanes 4 and 6). GST alone did not pull down any snRNP-specific RNAs or proteins under any of these conditions (Supplemental Fig. S2A,B).
FIGURE 7.
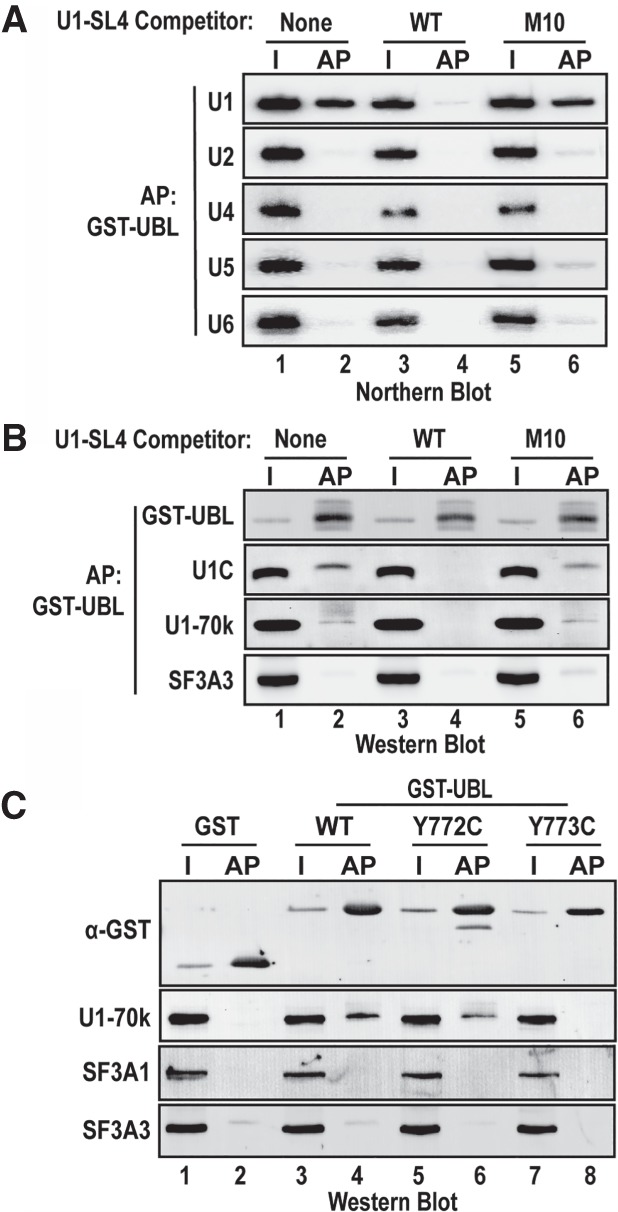
GST-UBL specifically binds to the U1 snRNP via U1-SL4. (A) Northern analysis for spliceosomal snRNAs in GST-UBL pull-down complexes. GST-UBL/WT protein was incubated in HeLa cell nuclear extracts in the absence or presence of U1-SL4/WT or M10 RNAs as competitors, followed by pull-down using glutathione agarose. (B) Western blot analysis of proteins in the pull-down complexes using antibodies to U1C, U1-70k, and SF3A3 proteins. (C) Western analysis of proteins present in GST-UBL/WT, Y772C, Y773 pull-down complexes. I and AP indicate input and affinity pull-down complexes, respectively.
To examine the impact of the tyrosine mutations, pull-down assays were performed with GST-UBL/Y772C and GST-UBL/Y773C fusion proteins. Western analysis confirmed the presence of the GST fusion proteins and revealed enrichment of the U1 snRNP-specific protein U1-70k in the pull-down complexes. In agreement with the mild impact of the Y772C mutation on binding affinity for U1-SL4, there was an approximately twofold reduction of U1-70k in the GST-UBL/Y772C complexes in comparison to the wild-type complexes. In the case of the more severe Y773C mutation, there was a drastic reduction of U1-70K in the pull-down complexes (Fig. 7C, compare lanes 6 and 8 to lane 4). U2 specific proteins SF3A1 and SF3A3 were not present in the complexes for wild-type or mutant GST-UBL proteins. Thus, these results demonstrate that the UBL domain of the SF3A1 protein is sufficient for binding to the U1 snRNA in the context of the U1 snRNP, and this interaction occurs through SL4. Results also indicate a more significant role for Y773 than Y772 in the interaction between SF3A1-UBL and the U1 snRNA.
Other human UBL domains with potential to bind nucleic acids
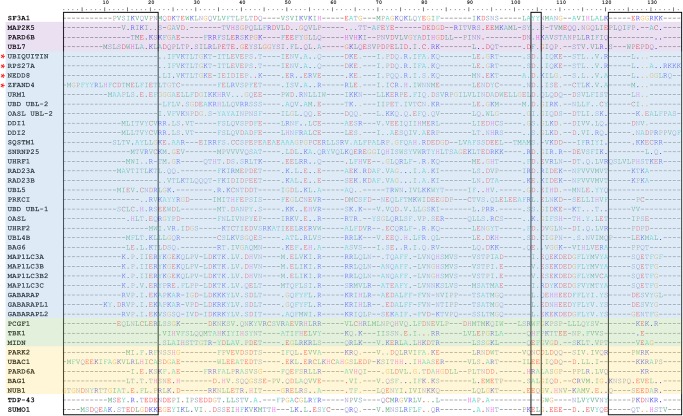
To identify other UBL domains with potential to bind RNA in humans, we searched databases of UBL domain-containing proteins for the salient features identified in SF3A1. A total of 955 proteins were scanned for the presence of a conserved tyrosine at positions analogous to 772 and/or 773, the carboxy-terminal RGG motif, or positively charged residues at the carboxyl terminus (see Fig. 2). This search identified 38 UBL domain-containing proteins that harbored one or more of these features. Alignment of the identified protein sequences is represented in Figure 8. Although not identified in our search, the UBL domains of transactivation response element (TAR) DNA-binding protein 43 (TDP-43), and the small ubiquitin-related modifier 1 (SUMO-1) are also included in the alignment as they have previously been demonstrated to bind nucleic acids (Smet-Nocca et al. 2011; Chang et al. 2012; Afroz et al. 2017).
FIGURE 8.
Alignment of human UBL domain sequences harboring features similar to SF3A1-UBL. Sequences were aligned by ClustalW, and key features that were used as criteria for performing the search for UBL domains are indicated by boxes. These features include the amino-terminal extension, conserved tyrosine residues at or near position 772 and/or 773 relative to SF3A1, and carboxy-terminal lysine and/or arginine residues. UBL domains with a tyrosine or a conservative phenylalanine substitution at position analogous to 772 are highlighted in purple. UBL domains with a tyrosine at position analogous to 773 are highlighted in blue, and domains with sequences containing a conservative phenylalanine substitution at this position are highlighted in green. Domains that lack a conserved tyrosine/phenylalanine or the RGG motif, but contain carboxy-terminal lysine and/or arginine residues, are highlighted in yellow. Sequences of UBL domains containing a conserved tyrosine and the RGG motif are indicated by an asterisk. In the aligned sequences, a dot indicates the presence of an identical residue to reference (SF3A1-UBL) and a dash indicates a gap. The UniProt accession numbers for all proteins included in the alignment are listed in Supplemental Table S2.
In the newly identified sequences, a tyrosine at position analogous to 773 is the most conserved feature and is present in 29 UBL domains (highlighted blue) while three proteins (PCGF1, TBK1, and MIDN) have a conservative phenylalanine substitution at this position (highlighted green) (Fig. 8). A tyrosine or a conservative phenylalanine substitution at position analogous to 772 is present in three proteins: MAP2K5, PARD6B, and UBL7 (highlighted purple). Of the 29 domains harboring a conserved tyrosine, four (ubiquitin, RPS27A, NEDD8, and ZFAND4) also have the carboxy-terminal RGG motif (indicated by an asterisk). However, of these four, only RPS27A and NEDD8 contain positively charged residues downstream from RGG. Despite lacking the RGG motif, many UBL proteins have arginines and/or lysines at their carboxyl terminus. Five proteins (PARK2, UBAC1, PARD6A, BAG1, and NUB1) lack a conserved tyrosine(s), but are enriched in arginines and/or lysines at their carboxyl terminus (highlighted yellow). Although the residues involved in RNA-binding in the amino-terminal extension of SF3A1-UBL have yet to be determined, we observed that many of the UBL domains had similar amino-terminal extensions when compared with ubiquitin. Like SF3A1, the amino termini of most of these UBL domains contained at least one arginine or lysine residue. Notably, TDP43-UBL and SUMO-1 lack Y772 and Y773, but do have positively charged residues at their carboxyl and amino terminus, respectively.
A few proteins identified in our search are known to either bind RNA or be involved in an RNA processing step. These include seven members of the ATG8 family of UBL proteins (MAP1L3A, MAP1L3B, MAP1L3B2, MAP1L3C, GABARAP, GABARAPL1, and GABARAPL2), RPS27A, and SNRNP25. The ATG8 family proteins have a conserved tyrosine and an amino-terminal extension. Although lacking a positively charged carboxyl terminus, the amino-terminal half of these proteins are rich in arginine and lysine, and one of them, MAPILC3B, has been reported to bind RNA (Zhou et al. 1997; Ying et al. 2009). RPS27A is a ribosomal protein that is cleaved post-translationally to produce a free UBL domain and S27a, and may be involved in ribosome biogenesis (Finley et al. 1989; Kirschner and Stratakis 2000). SNRNP25 is of particular interest, as it is a unique component of the U11/U12 di-snRNP of the minor spliceosome. It is essential for cell viability and appears to be more associated with the U11 snRNP than the U12 snRNP (Will et al. 2004). Since the SF3A1-UBL directly binds the U1 snRNA via SL4, a similar function may be involved in the association of SNRNP25 with the U11 snRNA, which is structurally similar to U1 and folds into four distinct stem–loops. Thus, the sequence analysis indicates that many other UBL domains in humans have salient features that were found to be important for binding RNA by SF3A1-UBL, and the proteins harboring these domains may also have the capacity to bind RNA.
DISCUSSION
In this study, we identify a noncanonical RNA binding domain in the U2 snRNP-specific protein SF3A1. Results from experiments using UV-crosslinking, EMSA, and SPR demonstrate that the carboxy-terminal UBL domain of SF3A1 (aa 703–793) is necessary and sufficient for binding to the stem–loop 4 of the U1 snRNA (Figs. 1, 2). The UBL domain of SF3A1 specifically recognizes the double-stranded, G-C rich stem–loop features of this RNA substrate (Figs. 3, 4). Also, this domain specifically binds U1-SL4 in the context of the U1 snRNP in HeLa cell nuclear extracts (Fig. 7). Additionally, tyrosine residues Y772 and Y773 in the UBL domain likely play an important role in binding to U1-SL4 (Figs. 5–7). Finally, our search of human UBL domains for features that were found to be important for U1-SL4 binding by SF3A1-UBL identified 38 UBL domains that could potentially bind RNA (Fig. 8). The carboxy-terminal UBL domain of SF3A1 is unique to higher eukaryotes and is not present in the yeast ortholog, Prp21, which is only 280 aa. Prp21 is homologous to the amino-terminal region of SF3A1 that harbors the two SURP domains and the short charged region (Supplemental Fig. S3; Lin and Xu 2012). Thus, the absence of a UBL domain in Prp21 suggests that the potential role of this SF3A1 domain (and its capacity to bind U1-SL4) during the early steps of spliceosome assembly may be unique to higher eukaryotes.
The binding affinity of SF3A1-UBL for U1-SL4 is high (KD = ∼97 nM) in comparison to the canonical RNA recognition motifs (RRMs) of another U1-SL4 interacting protein, the polypyrimidine tract-binding protein P1 (PTBP1). PTBP1 contains four RRMs, and RRM1 and RRM2 bind U1-SL4 with dissociation constants of 850 and 390 nM, respectively, and to single-stranded CU-rich RNAs with KD values ≥ ∼1 µM (Auweter et al. 2007; Sharma et al. 2011). However, the affinity of the SF3A1-UBL/U1-SL4 interaction is significantly weaker than binding of the core U1 protein, U1A, to SL2 of the U1 snRNA. U1A very stably binds to SL2 via its amino-terminal RRM with a KD of ∼32 pM (Katsamba et al. 2001). Since the SF3A1–U1 snRNA interaction would need to be disrupted during the progression of spliceosome assembly (see below), the intermediate nM range affinity of the interaction reported here might be reflective of the transient nature of this prespliceosomal RNA–protein contact.
Mutation analysis of conserved tyrosines in SF3A1-UBL has revealed a critical role for Y773 in binding to the U1-SL4 RNA and the intact U1 snRNP. However, the MDS-associated mutation Y772C was found to have a weaker effect, and reduced U1-SL4 binding and U1 snRNP association by only approximately twofold. This small effect is similar to the MDS-mutation induced change in RNA-binding affinity observed for other splicing factors such as SRSF2 and U2AF1. SRSF2 mutations P95H/L/R were reported to cause ∼1.2- to ∼2.1-fold decrease in affinity for RNA sequences harboring the 5′-GGAG-3′ consensus (Kim et al. 2015; Zhang et al. 2015). Similarly, the S34F mutation was found to decrease the binding affinity of U2AF1 for consensus 3′ splice site sequences by ∼1.3- to approximately fourfold (Okeyo-Owuor et al. 2015; Fei et al. 2016). In both SRSF2 and U2AF1, the MDS-mutations also changed sequence specificity by causing an increase in the affinity for alternative sequences. However, any impact of the Y772C mutation on the binding specificity of SF3A1 remains to be determined. Overall, the analysis showed that the magnitude of decrease in binding affinity caused by the SF3A1-Y772C mutation is similar to that observed for other MDS-associated mutations and may be sufficient for induction of pathogenic molecular effects.
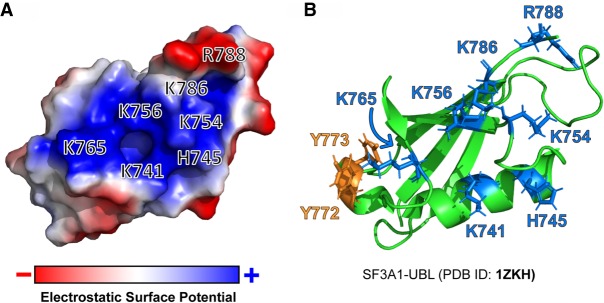
TDP-43-NTD and SUMO-1 have been shown to bind single-stranded (ss) and double-stranded (ds) DNA, respectively, but their capacity to bind RNA has not been reported (Smet-Nocca et al. 2011; Chang et al. 2012; Afroz et al. 2017). SUMO-1, but not TDP-43-NTD, appears to fold into the UBL ββαββαβ topology (Eilebrecht et al. 2010; Qin et al. 2014). Structural analysis of TDP-43-NTD in the presence of ssDNA and SUMO-1 in the presence of dsDNA have identified clusters of positively charged surface residues that facilitate DNA binding in both proteins. Electrostatic surface potential analysis of the available structure of the SF3A1-UBL domain (PDB ID:1ZKH) by PyMOL demonstrates the presence of a similarly highly positively charged surface that is formed by five lysine residues (K741, K754, K756, K765, and K786), histidine H745, and arginine R788 (Fig. 9A). These seven positively charged amino acids are conserved in SF3A1-UBL from humans to worms except for H745, which is replaced by a glutamine, in flies and worms. These residues are in the same plane as tyrosines 772 and 773 that were found to be important for binding to U1-SL4 (Figs. 5C, 9B). The carboxy-terminal RKK motif that was found to be critical for U1-SL4 binding could also contribute to the positively charged surface in SF3A1-UBL as it is immediately downstream from R788. Thus, this positive surface, in coordination with Y772 and Y773, likely confers the SF3A1-UBL with RNA-binding properties.
FIGURE 9.
The SF3A1-UBL domain has a positively charged surface that could potentially facilitate interactions with U1-SL4 RNA. (A) Electrostatic potential analysis of the SF3A1-UBL domain surface performed using the available NMR structure (SF3A1 aa 704–789; PDB ID: 1ZKH). General positions and identity of residues that contribute to the positively charged surface (blue) are highlighted. (B) Ribbon diagram of SF3A1-UBL represented in the same orientation as in A. All residues highlighted in the electrostatic surface potential analysis, and tyrosines Y772 and Y773, are shown.
Our biochemical analyses support the occurrence of SF3A1 contact with U1 snRNA during the transition of the E to A complex (Sharma et al. 2014). These observations are supported by other studies that have demonstrated an essential role for SF3A1 in early prespliceosome formation and the potential association of SF3A1 with prespliceosomal proteins such as SF1 (Nesic and Krämer 2002; Crisci et al. 2015). How long the SF3A1-UBL/SL4 interaction may persist after A complex formation is not clear. Structural and compositional analyses of higher order human pre-B, Bact, and B* complexes demonstrate the presence of SF3A1 in pre-B and Bact complexes, but not in B*, and that the SF3A complex is released through the action of the Prp2 helicase during the Bact → B* transition (Bessonov et al. 2010; Haselbach et al. 2018). Prior to the release of SF3A, the U1 snRNP is released by the Prp28 helicase during the conversion of the pre-B to Bact complex (Boesler et al. 2016). The cryo-EM structures of the human pre-B complex do not demonstrate any contacts between the U1–U2 snRNPs, indicating that the SF3A1/U1 snRNA contact may be disrupted during the A → pre-B complex transition (Zhan et al. 2018; Charenton et al. 2019). However, the carboxy-terminal half of SF3A1 is not visible in either of the pre-B complex structures, and therefore its orientation relative to U1-SL4 cannot be determined definitively. This difficulty in resolving the carboxy-terminal region of SF3A1 is probably due to the central, proline-rich region that separates the UBL domain from the amino terminus, and is likely unstructured (Fig. 1A). Thus, this study provides relevant functional insight into a critical prespliceosome interaction involving SF3A1 and U1 snRNA that has been challenging to evaluate by structural analyses, and may be uniquely occurring during human spliceosome assembly. In addition, it identifies UBL domains with RNA binding potential in several other proteins that are involved in RNA processing steps in essential cellular pathways.
MATERIALS AND METHODS
Cell-free protein expression and UV-crosslinking
Full-length and truncated SF3A1 proteins were expressed using the 1-Step Human Coupled In Vitro Translation Kit (Thermo Fisher Scientific). SF3A1 cDNA was cloned into NdeI/SalI restriction sites of pT7CFE-CHis expression vector and truncations were made by whole plasmid amplification via PCR. Constructs were added to CFE reactions and incubated for 6 h according to manufacturer's instructions. CFE reactions were then dialyzed against one liter of buffer DG (20 mM HEPES pH 7.9, 80 mM K-glutamate, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF, 20% glycerol) using Slide-A-Lyzer dialysis cassettes (Thermo Fisher Scientific) overnight at 4°C. Dialyzed reactions were aliquoted and stored at −20°C.
Radioactive 32P-UTP labeled wild-type and mutant U1-SL4 RNAs were in vitro transcribed by T7 RNA polymerase from annealed DNA templates, gel purified, and ethanol precipitated. Crosslinking reactions consisted of 100 nM 32P-U1-SL4, 2.2 mM MgCl2, and 60% dialyzed CFE reactions. After a 30-min incubation at room temperature (RT), reactions were UV-crosslinked with 1800 mJ total energy in a GS Gene Linker (Bio-Rad Laboratories). Reactions were separated on an SDS-PAGE gel that was dried and visualized by the Typhoon FLA 9500 Phosphor Imager.
Western and northern analysis
For protein analysis, all samples were boiled in 1× SDS-PAGE sample buffer, separated on a 10% or 12% SDS-PAGE gel and analyzed by Coomassie blue staining or western blotting using PVDF membranes. To analyze all SF3A1 truncations on single gels, precast 4%–20% gradient gels were used (Bio-Rad Laboratories). Polyclonal antibodies against SF3A1 have been described previously (Sharma et al. 2014). Anti-α-Tubulin mouse monoclonal antibody (Calbiochem; CP06-DM1A), anti-6×His rabbit monoclonal antibody (Thermo Fisher Scientific; MA1-21315), and anti-GST monoclonal antibody (Abcam; Ab92) were obtained commercially. Secondary anti-mouse and anti-rabbit antibodies conjugated to Cy3 and Cy5 fluorophores were purchased from GE Healthcare.
For northern blotting, RNA samples were separated on 10% Urea-PAGE gels and transferred onto Amersham Hybond Nylon Membrane (GE Healthcare) for 1 h at 15 V and 400 mA using the Trans-blot Turbo Semi-dry Transfer System (Bio-Rad). Transferred RNA was UV-crosslinked to nylon membranes for 10 min and prehybridized in 15 mL of ULTRAhyb Hybridization Buffer (Thermo Fisher Scientific). The membranes were probed with 32P-labeled oligo probes at 1.0 × 106 cpm/mL hybridization buffer overnight at 42°C. Sequences of oligonucleotides used to probe for target snRNAs are summarized in Supplemental Table S3. Membranes were washed at the temperature of hybridization once with 2× saline-sodium citrate buffer (SSC) containing 0.1% SDS for 10 min and twice with 2× SSC for 10 min. Northern blots were visualized using the Typhoon FLA 9500 Imager.
Glutathione agarose affinity chromatography and pull-down assay
GST-UBL fusion constructs were created by cloning the cDNA for the UBL domain of SF3A1 (aa 704–793) into the BamHI and XhoI restriction sites of plasmid pGEX-5x. GST and GST-UBL fusion proteins were expressed in E. coli (BL21-DE3) by induction with 0.5 mM IPTG for 4 h at 37°C. Induced proteins were isolated using glutathione agarose beads (Thermo Fisher Scientific) according to the manufacturer's protocol, separated by SDS-PAGE, and analyzed by Coomassie blue staining and western blotting. Purified proteins were dialyzed against two liters of buffer DG and stored at −80°C.
For GST pull-down assays, HeLa cell nuclear extracts were pre-cleared of GST-binding proteins, as described previously (Masuda et al. 2005). Purified GST and GST-UBL proteins were added to 100 µL of nuclear extract at a final concentration of 2–5 µM. After a 30 min incubation at 4°C, the reactions were added to a 30 µL packed volume of glutathione agarose beads and allowed to incubate for an additional 30 min at 4°C with rotation. Beads were washed four times in buffer DG and bound proteins were eluted by boiling in 1× SDS-PAGE sample buffer. Total RNA was extracted with the TRIzol reagent.
Electrophoretic mobility shift assays
Binding reactions were prepared in 10 µL total volume and contained 10 nM 5′-Cy5-labeled U1-SL4 RNA (Integrated DNA Technologies), 2.2 mM MgCl2, 60% buffer DG, and varying concentrations of GST-UBL protein. After incubation for 1 h at RT, the binding reactions were loaded onto a horizontal native-PAGE gel (6% 29:1 Bis-Acrylamide, 0.1% ammonium persulfate, and 1:1000 dilution of TEMED in 0.5× TBE buffer) and run at 100 V for 2 h at 4°C (Dowdle et al. 2017). The gels were visualized using the Typhoon FLA 9500 Imager and bands were quantified with ImageQuant Software (GE Healthcare). For competition EMSAs, first, binding reactions containing 10 nM 5′-Cy5-U1-SL4 and 500 nM GST-UBL protein were prepared (Ryder et al. 2008). Then the competitor RNAs were added at varying concentrations and incubation was continued at RT for 1 h. The complexes were separated and visualized as above. Dose-response curves for fraction of Cy5-U1-SL4 bound versus the log of protein or competitor RNA concentrations were generated by a nonlinear regression analysis of the data and assuming one binding site using GraphPad Prism Software v8.1.0.
Surface plasmon resonance
SPR experiments were performed on the Pioneer FE Surface Plasmon Resonance System (ForteBio) and largely based on parameters described by Katsamba et al. (2002). 5′-biotinylated RNAs (Integrated DNA Technologies) were immobilized on streptavidin-coated biosensor chips (SADH Biosensors). Prior to immobilization, the RNAs were diluted to 1 µM in base SPR buffer (10 mM Tris-HCL, pH 8.0, 150 mM NaCl) heated to 65°C for 5 min, and cooled slowly to RT to allow folding. Folded RNA substrates were diluted 100-fold in SPR running buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5% glycerol, 62.5 µg/mL bovine serum albumin, 125 µg/mL tRNA, 1 mM dithiothreitol, and 0.05% tween-20), and injected at 10 µL/min until 45–50 response units of RNA were captured on the SADH biosensor. Protein stocks were diluted to a 500 nM concentration in SPR running buffer and 10 to 12, twofold serial dilutions were prepared and injected using the One-Step Kinetics assay setting. Binding experiments were performed at 20°C at a flow rate of 150 µL/min. Under the conditions used, >95% of the protein was removed from the surface during the dissociation phase and therefore a regeneration step was not required. Background signal from a streptavidin-only reference flow cell was subtracted from all data sets. Data were fit assuming a 1:1 stoichiometry to the Langmuir binding model to obtain kinetic parameters and dissociation constant.
Sequence analysis
Annotated human UBL domains and/or proteins were collected from the integrated annotations for ubiquitin and UBL conjugations database (iUUCD) as well as from the InterPro application from EMBL-EBI (Zhou et al. 2017; Mitchell et al. 2019). A total of 955 proteins were manually scanned for the presence of a conserved tyrosine at positions analogous to 772 and/or 773, the carboxy-terminal RGG motif, and carboxy-terminal positively charged residues. This search narrowed down the number of analyzed UBL domains to approximately 146 proteins. From this initial group, redundant protein sequences were purged, and sequences representative of UBL families were selected, yielding 38 UBL domains, which were aligned to SF3A1-UBL. The alignment of this final collection of UBL domains is displayed in Figure 8 and UniProt accession numbers are reported in Supplemental Table S2. All protein sequence alignments were performed using ClustalW (Thompson et al. 1994). Electrostatic surface potential analysis for SF3A1-UBL domain (PDB ID: 1ZKH) was performed in PyMOL (DeLano 2002). RNA secondary structure predictions were performed in the mfold online web server (Zuker 2003).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds to S.S. from the National Institutes of Health, National Institute of General Medical Sciences (R01GM127464) and the American Cancer Society (Institutional Research grant 74-001-34-IRG), and to S.S. and W.M. from the Valley Research Partnership Program (P1-4009). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.072256.119.
REFERENCES
- Afroz T, Hock EM, Ernst P, Foglieni C, Jambeau M, Gilhespy LAB, Laferriere F, Maniecka Z, Plückthun A, Mittl P, et al. 2017. Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation. Nat Commun 8: 45 10.1038/s41467-017-00062-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, Allain FHT. 2007. Solving the structure of PTB in complex with pyrimidine tracts: an NMR study of protein–RNA complexes of weak affinities. J Mol Biol 367: 174–186. 10.1016/j.jmb.2006.12.053 [DOI] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Lührmann R, Stark H. 2017. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170: 701–713.e11. 10.1016/j.cell.2017.07.011 [DOI] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Lührmann R. 2010. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16: 2384–2403. 10.1261/rna.2456210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesler C, Rigo N, Anokhina MM, Tauchert MJ, Agafonov DE, Kastner B, Urlaub H, Ficner R, Will CL, Lührmann R. 2016. A spliceosome intermediate with loosely associated tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat Commun 7: 11997 10.1038/ncomms11997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R, Gröning K, Behrens S, Lührmann R, Krämer A. 1993. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kDa subunit to yeast PRP9. Science 262: 102–105. 10.1126/science.8211112 [DOI] [PubMed] [Google Scholar]
- Chang CK, Wu TH, Wu CY, Chiang MH, Toh EKW, Hsu YC, Lin KF, Liao YH, Huang TH, Huang JJT. 2012. The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem Biophys Res Commun 425: 219–224. 10.1016/j.bbrc.2012.07.071 [DOI] [PubMed] [Google Scholar]
- Charenton C, Wilkinson ME, Nagai K. 2019. Mechanism of 5′ splice site transfer for human spliceosome activation. Science 364: 362–367. 10.1126/science.aax3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisci A, Raleff F, Bagdiul I, Raabe M, Urlaub H, Rain JC, Krämer A. 2015. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res 43: 10456–10473. 10.1093/nar/gkv952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R. 2000. Functional association of U2 snRNP with the ATP-independent splicecosomal complex E. Mol Cell 5: 779–787. 10.1016/S1097-2765(00)80318-4 [DOI] [PubMed] [Google Scholar]
- DeLano WL. 2002. The PyMOL molecular graphics system, v2.3.2. DeLano Scientific, San Carlos, CA. [Google Scholar]
- Dowdle ME, Imboden SB, Park S, Ryder SP, Sheets MD. 2017. Horizontal gel electrophoresis for enhanced detection of protein-RNA complexes. J Vis Exp e56031 10.3791/56031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilebrecht S, Smet-Nocca C, Wieruszeski JM, Benecke A. 2010. SUMO-1 possesses DNA binding activity. BMC Res Notes 3: 146 10.1186/1756-0500-3-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei DL, Motowski H, Chatrikhi R, Prasad S, Yu J, Gao S, Kielkopf CL, Bradley RK, Varmus H. 2016. Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet 12: e1006384 10.1371/journal.pgen.1006384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, Nagai K. 2017. Structure of a spliceosome remodelled for exon ligation. Nature 542: 377–380. 10.1038/nature21078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Bartel B, Varshavsky A. 1989. The tails of ubiquitin precursors are ribosomal protein whose fusion to ubiquitin facilitates ribosome biogenesis. Nature 338: 394–400. 10.1038/338394a0 [DOI] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Potashkin J, Reed R. 1998. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18: 4752–4760. 10.1128/MCB.18.8.4752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselbach D, Komarov I, Agafonov DE, Hartmuth K, Graf B, Dybkov O, Urlaub H, Kastner B, Lührmann R, Stark H. 2018. Structure and conformational dynamics of the human spliceosomal Bact complex. Cell 172: 454–464.e11. 10.1016/j.cell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Huang CJ, Ferfoglia F, Raleff F, Krämer A. 2011. Interaction domains and nuclear targeting signals in subunits of the U2 small nuclear ribonucleoprotein particle-associated splicing factor SF3a. J Biol Chem 286: 13106–13114. 10.1074/jbc.M110.201491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsamba PS, Myszka DG, Laird-Offringa IA. 2001. Two functionally distinct steps mediate high affinity binding of U1A protein to U1 hairpin II RNA. J Biol Chem 276: 21476–21481. 10.1074/jbc.M101624200 [DOI] [PubMed] [Google Scholar]
- Katsamba PS, Park S, Laird-Offringa IA. 2002. Kinetic studies of RNA–protein interactions using surface plasmon resonance. Methods 26: 95–104. 10.1016/S1046-2023(02)00012-9 [DOI] [PubMed] [Google Scholar]
- Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC-W, Ramakrishnan A, Li Y, Chung YR, Micol JB, Murphy ME, et al. 2015. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 27: 617–630. 10.1016/j.ccell.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner LS, Stratakis CA. 2000. Structure of the human ubiquitin fusion gene Uba80 (RPS27a) and one of its pseudogenes. Biochem Biophys Res Commun 270: 1106–1110. 10.1006/bbrc.2000.2568 [DOI] [PubMed] [Google Scholar]
- Krämer A, Mulhauser F, Wersig C, Gröning K, Bilbe G. 1995. Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of Saccharomyces cerevisiae. RNA 3: 260–272. [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu S, Jiang J, Zhang L, Espinosa S, Hill RC, Hansen KC, Zhou ZH, Zhao R. 2017. CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing. Nat Commun 8: 1035 10.1038/s41467-017-01241-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Xu RM. 2012. Structure and assembly of the SF3a splicing factor complex of U2 snRNP. EMBO J 31: 1579–1590. 10.1038/emboj.2012.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19: 1512–1517. 10.1101/gad.1302205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang HY, El-Gebali S, Fraser MI, et al. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47: D351–D360. 10.1093/nar/gky1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D, Krämer A. 2002. Domains in human splicing factors SF3a60 and SF3a66 required for binding to SF3a120, assembly of the 17S U2 snRNP, and prespliceosome formation. Mol Cell Biol 21: 6406–6417. 10.1128/MCB.21.19.6406-6417.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, Ding L, Ketkar-Kulkarni S, Hundal J, Laird KM, et al. 2015. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia 29: 909–917. 10.1038/leu.2014.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano JM, Clingman CC, Ryder SP. 2011. Quantitative approaches to monitor protein–nucleic acid interactions using fluorescent probes. RNA 17: 14–20. 10.1261/rna.2428111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. 2016. Genomic classification and prognosis in acute myeloid leukemia. J New Engl Med 374: 2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin PC, Nagai K. 2017. Structure of a pre-catalytic spliceosome. Nature 546: 617–621. 10.1038/nature22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin P-C, Charenton C, Nagai K. 2018. Prespliceosome structure provides insights into spliceosome assembly and regulation. Nature 559: 419–422. 10.1038/s41586-018-0323-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Oubridge C, Leung AKW, Li J. 2009. Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature 458: 475–480. 10.1038/nature07851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Lim L-Z, Wei Y, Song J. 2014. TDP-43 N terminus encodes a novel ubiquitin-like fold and its unfolded form in equilibrium that can be shifted by binding to ssDNA. Proc Natl Acad Sci 111: 18619–18624. 10.1073/pnas.1413994112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder SP, Recht MI, Williamson JR. 2008. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol Biol 488: 99–115. 10.1007/978-1-60327-475-3_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Maris C, Allain FH-T, Black DL. 2011. U1 snRNA directly interacts with polypyrimidine tract-binding protein during splicing repression. Mol Cell 41: 579–588. 10.1016/j.molcel.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Wongpalee SP, Vashisht A, Wohlschlegel JA, Black DL. 2014. Stem–loop 4 of U1 snRNA is essential for splicing and interacts with the U2 snRNP-specific SF3A1 protein during spliceosome assembly. Genes Dev 28: 2518–2531. 10.1101/gad.248625.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smet-Nocca C, Wieruszeski JM, Léger H, Eilebrecht S, Benecke A. 2011. SUMO-1 regulates the conformational dynamics of thymine-DNA glycosylase regulatory domain and competes with its DNA binding activity. BMC Biochem 12: 4 10.1186/1471-2091-12-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanackovic G, Krämer A. 2004. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol Biol Cell 16: 1366–1377. 10.1091/mbc.e04-11-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-kumar S, Bugg CE, Cook WJ. 1987. Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol 194: 531–544. 10.1016/0022-2836(87)90679-6 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Schneider C, Hossbach M, Ublaur H, Rauhut R, Elbashir S, Tuschl T, Lührmann R. 2004. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10: 929–941. 10.1261/rna.7320604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying L, Lau A, Alvira CM, West R, Cann GM, Zhou B, Kinnear C, Jan E, Sarnow P, Van de Rijn M, et al. 2009. LC3-mediated fibronectin mRNA translation induces fibrosarcoma growth by increasing connective tissue growth factor. J Cell Sci 9: 1441–1451. 10.1242/jcs.025957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. 10.1038/nature10496 [DOI] [PubMed] [Google Scholar]
- Zhan X, Yan C, Zhang X, Lei J, Shi Y. 2018. Structures of the human pre-catalytic spliceosome and its precursor spliceosome. Cell Res 28: 1129–1140. 10.1038/s41422-018-0094-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lieu YK, Ali AM, Penson A, Reggio KS, Rabadan R, Raza A, Mukherjee S, Manley JL. 2015. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci 112: E4726–E4734. 10.1073/pnas.1514105112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yan C, Zhan X, Li L, Lei J, Shi Y. 2018. Structure of the human activated spliceosome in three conformational states. Cell Res 28: 307–322. 10.1038/cr.2018.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Boudreau N, Coulber C, Hammarback J, Rabinovitch M. 1997. Microtubule-associated protein 1 light chain 3 is a fibronectin mRNA-binding protein linked to mRNA translation in lamb vascular smooth muscle cells. J Clin Invest 12: 3070–3082. 10.1172/JCI119862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Xu Y, Lin S, Guo Y, Deng W, Zhang Y, Guo A, Xue Y. 2017. iUUCD 2.0: an update with rich annotations for ubiquitin and ubiquitin-like conjugations. Nucleic Acids Res 46: D447–D453. 10.1093/nar/gkx1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.