This article reports results of a study that examined the clinicopathological characteristics of patients with metastatic colorectal cancer displaying amplification or overexpression of the HER2 oncogene, focusing on response to anti‐EGFR treatment.
Keywords: Colorectal cancer, HER2, ERBB2, Anti‐EGFR monoclonal antibodies
Abstract
Background.
HER2 amplification is detected in 3% of patients with colorectal cancer (CRC), making tumors in the metastatic setting vulnerable to double pharmacological HER2 blockade. Preclinical findings show that it also might impair response to anti‐epidermal growth factor receptor (EGFR) treatment.
Subjects and Methods.
Patients with KRAS exon 2 wild‐type metastatic CRC underwent molecular screening of HER2 positivity by HERACLES criteria (immunohistochemistry 3+ or 2+ in ≥50% of cells, confirmed by fluorescence in situ hybridization). A sample of consecutive HER2‐negative patients was selected as control. A regression modeling strategy was applied to identify predictors explaining the bulk of HER2 positivity and the association with response to previous anti‐EGFR treatment.
Results.
From August 2012 to April 2018, a total of 100 HER2‐positive metastatic CRC tumors were detected out of 1,485 KRAS exon 2 wild‐type screened patients (6.7%). HER2‐positive patients show more frequently lung metastases (odds ratio [OR], 2.04; 95% confidence interval [CI], 1.15–3.61; p = .014) and higher tumor burden (OR, 1.48; 95% CI, 1.10–2.01; p = .011), and tumors were more likely to be left sided (OR, 0.50; 95% CI, 0.22–1.11; p = .088). HER2‐positive patients who received treatment with anti‐EGFR agents (n = 79) showed poorer outcome (objective response rate, 31.2% vs. 46.9%, p = .031; progression‐free survival, 5.7 months vs. 7 months, p = .087).
Conclusion.
Testing for HER2 should be offered to all patients with metastatic CRC because the occurrence of this biomarker is unlikely to be predicted based on main clinicopathological features. Patients with HER2‐amplified metastatic CRC are less likely to respond to anti‐EGFR therapy.
Implications for Practice.
Patients with HER2‐amplified/overexpressed metastatic colorectal cancer (mCRC) harbor a driver actionable molecular alteration that has been shown in preclinical models to hamper efficacy of the anti‐epidermal growth factor receptor (EGFR) targeted therapies. The present study confirmed that this molecular feature was associated with worse objective tumor response and shorter progression‐free survival in response to previous anti‐EGFR therapies. Moreover, it was found that the occurrence of this biomarker is unlikely to be predicted based on main clinicopathological features. Therefore, HER2 status assessment should be included in the molecular diagnostic workup of all mCRC for speedy referral to clinical trials encompassing HER2‐targeted double blockade independently of previous anti‐EGFR treatment.
摘要
背景 3% 的结直肠癌 (CRC) 患者中检测到 HER2 扩增,这使得转移灶中的肿瘤容易受到双重药物 HER2 阻断。临床前研究表明,HER2 还可能损害对抗表皮生长因子受体 (EGFR) 治疗的反应。
受试者和方法 KRAS 外显子 2 野生型转移性CRC患者采用 HERACLES 标准进行 HER2 阳性分子筛查(在 ≥ 50% 的细胞中免疫组化 3 + 或 2+,经荧光原位杂交证实)。选择连续HER2 阴性患者作为对照。采用回归建模策略确定预测因素,对主要 HER2 阳性以及与先前抗 EGFR 治疗反应的相关性进行解析。
结果 2012 年 8 月至 2018 年 4 月期间,在 1 485 例 KRAS 外显子 2 野生型筛查患者中,共检测出 100 例 (6.7%) 转移性 CRC 肿瘤 HER2 阳性患者。HER2 阳性患者肺转移更频繁 [比值比 (OR), 2.04; 95% 可信区间 (CI), 1.15‐3.61; p = 0.014],肿瘤负荷较高(OR, 1.48; 95% CI, 1.10‐2.01;p = 0.011),肿瘤位于左侧的可能性更高(OR, 0.50; 95% CI,0.22‐1.11; p = 0.088)。接受抗 EGFR 药物治疗的 HER2 阳性患者 (n = 79) 预后较差(客观有效率 31.2% vs. 46.9%, p = 0.031;无进展生存期,5.7 个月 vs. 7 个月,p = 0.087)。
结论 HER2 这种生物标志物的发生不太可能根据主要的临床病理特征进行预测,故而所有转移性 CRC 患者均应接受 HER2 检测。HER 2 扩增转移性CRC 患者对抗 EGFR 治疗不太容易产生反应。
实践意义:HER2 扩增/过表达转移性结直肠癌 (mCRC) 患者存在一种驱动可操作的分子改变,临床前模型中显示出,这种改变会阻碍抗表皮生长因子受体 (EGFR) 靶向治疗的疗效。本研究证实,这一分子特征与客观肿瘤反应较差和针对之前的抗 EGFR 治疗的无进展生存时间较短有关。此外,我们还发现这种生物标志物的发生不太可能根据主要的临床病理特征进行预测。因此,所有 mCRC 的分子诊断工作都应纳入 HER2 状态评估,以便在不依赖以往抗 EGFR 治疗的情况下,快速转介到包含 HER2 靶向双重阻断的临床试验中。
Introduction
Amplification and/or overexpression of human epidermal growth factor receptor 2 (HER2) encoded by the ERBB2 gene is a negative prognostic biomarker and a positive predictor of response to anti‐HER2 therapies in metastatic breast [1] and gastroesophageal junction adenocarcinoma [2]. In metastatic colorectal cancer (mCRC), the rate of HER2 amplification or overexpression ranges from 1.3% to 3.2% of cases in all stages, molecularly unselected [3], up to 5.2% in KRAS exon 2 wild‐type patients in the metastatic setting [4]. In a recent study performed in a large cohort of colorectal cancer (CRC) samples from stage IV patients by next‐generation sequencing (NGS) of a panel of 315 cancer‐related genes, amplification of ERBB2 was found in 2.8%, and KRAS mutations were found to occur less frequently in ERBB2‐amplified samples [5].
Although the role of HER2 as a biomarker for prognosis in CRC remains uncertain [6], its value as a therapeutic target has been recently established [7], [8], [9]. The HERACLES A phase II trial of dual HER2‐targeted therapy (trastuzumab plus lapatinib) was conducted in patients with KRAS exon 2 wild‐type, HER2‐positive (defined according to the HERACLES diagnostic criteria [10]) mCRC who were refractory to standard‐of‐care treatments, making HER2 the first specific oncogenic target that can be successfully actioned at the clinical level in this tumor type. Durable responses were indeed observed in 30% of patients, with high ERBB2 copy number variation being a further potential positive biomarker for treatment refinement [11]. These results have been subsequently confirmed in the MyPathway basket study, in which patients with HER2‐amplified or HER2‐overexpressed mCRC who had exhausted standard treatment options received a combination of trastuzumab and pertuzumab with an objective response rate of 38% [12]. Interestingly, it has been pointed out by preclinical models and analysis of retrospective limited cohorts of patients that HER2 amplification or overexpression can be also a negative predictor of response to epidermal growth factor receptor (EGFR)‐targeted treatments [7], [11], [13], [14]. Hence, patients with HER2‐positive mCRC harbor a driver actionable molecular alteration that, at the same time, might hamper efficacy of a standard treatment option, with relevant implications for the treatment algorithm of stage IV patients. Here we present the largest clinically annotated series to date of HER2‐positive CRCs, including clinical outcome of EGFR‐targeted therapies, to identify the clinicopathological features associated with this driver genetic tumor abnormality and test the impact on anti‐EGFR therapies.
Patients and Methods
Study Design and Cohort Selection
This is a retrospective longitudinal cohort study examining clinicopathological characteristics of patients with mCRC displaying amplification and/or overexpression of the HER2 oncogene. In the frame of HERACLES studies, patients affected by KRAS exon 2 wild‐type mCRC and refractory to standard of care were screened at Niguarda Cancer Center, Grande Ospedale Metropolitano Niguarda, Milano, Italy [10], [11], [15], [16]. HERACLES Diagnostic screened 256 patients retrospectively, whereas in the therapeutic studies, 1,229 patients with KRAS exon 2 wild‐type mCRC from four Italian institutions were screened centrally at Pathology Department of the Niguarda Cancer Center according to HERACLES diagnostic criteria [10]. All patients with HER2‐positive mCRC were included in the analysis, and a sample of consecutive KRAS exon 2 wild‐type patients without HER2 amplification or overexpression was selected as control. The nine clinicopathological features selected for present study and retrospectively collected from electronical charts were age, gender, site of primary tumor (i.e., right sided, from cecum up to transverse colon included, vs. left sided, from splenic flexure to rectum; colon vs. rectum), stage of disease at diagnosis, time to progression before recurrence, histological grade of the primary tumor, and number of metastatic sites at diagnosis in case of synchronous metastases or at progression to stage IV in case of metachronous metastases. In addition, objective response and progression‐free survival achieved for each previous anti‐EGFR treatment were collected. All patients treated with anti‐EGFR in both the HER2‐positive and HER2‐negative cohorts were pan‐RAS wild‐type as per label for receiving cetuximab or panitumumab in Italy. Date of death or last follow‐up was acquired to establish overall survival. Date of HER2 status assessment, site of assessment (primary tumor or metastatic site), immunohistochemistry (IHC) score, and percent of tumor cells stained with HER2 IHC and ERBB2‐centromere of chromosome 17 (CEN17) ratio at fluorescence in situ hybridization (FISH) were also obtained.
HER2 Status Characterization
The pathological methods to define HER2 amplification or overexpression (HER2 positivity) in CRC have been described elsewhere [10]. Briefly, HER2‐positive CRCs were defined by immunohistochemistry 3+ or 2+ in at least 50% of cells, confirmed by FISH. Samples not meeting these criteria were considered negative.
Study Objectives and Endpoints
The primary study objective was to assess the predictive role of HER2 positivity for sensitivity and resistance to anti‐EGFR therapy measured in terms of objective response rate and progression‐free survival. The secondary objective was to identify clinicopathological features predictive of presence of HER2 positivity and to develop a diagnostic model.
Statistical Analysis
Logistic and Cox regression models were used to detect and estimate the association between HER2 positivity, objective response rate, and progression‐free survival in response to anti‐EGFR treatment and to develop and internally (i.e., using resampling techniques) validate a diagnostic model predicting HER2 status. The logistic regression model was used to evaluate the following aims:
To test and estimate the statistical association between clinicopathological features and HER2 status (amplified vs. not amplified).
To develop and internally validate a multivariable model predicting HER2 positive status. Based on the multivariable model, a nomogram calculating probabilities for HER2 positive status was also obtained.
The statistical procedure for the development and internal validation of a predictive model was applied to two samples. The first sample (sample A) included all patients enrolled in the study. The second sample (sample B) included patients enrolled who had been treated with anti‐EGFR therapy. Candidate predictors were the same of sample A, with the addition of response to anti‐EGFR therapy (complete response or partial response [CR/PR] vs. stable disease or progressive disease [SD/PD]) and line of anti‐EGFR therapy. The presence of heavy overfitting was evident from the low value of the total likelihood ratio predictors (6.256 for sample A; 11.091 for sample B) and the high number of predictors (5 degrees of freedom [d.f.] for sample A; 7 d.f. for sample B). The Van Houwelingen‐Le Cessie heuristic shrinkage estimate was 0.20 for sample A and 0.37 for sample B, indicating that these models would validate on new data respectively about 80% and 63% worse than on these data sets. In order to control overfitting, we attempted to reduce the number of predictors, but no predictor variable explained the bulk of HER2 amplification. Thus, the full models were preserved, and the overfitting was controlled by the estimation of true regression parameters using a penalized maximum likelihood approach. Candidate predictors were considered to be primary tumor grading, primary tumor location, time to metastatic presentation (synchronous vs. metachronous), and burden of metastatic disease (liver limited disease, liver and other organs, only other organs). The second sample (sample B) included patients who had been treated with anti‐EGFR compounds. Candidate predictors were the same as sample A, with the addition of response to anti‐EGFR therapy (CR/PR vs. SD/PD) and line of anti‐EGFR therapy. The statistical procedure was conducted as follows:
Categories with fewer than 15 patients were merged.
A single conditional imputation of missing values for all candidate predictors was performed. This used the R “transcan” function with the following options: imputed = T, transformed = T, impact = “score.”
A full multivariable model with a linear combination of predictors on the original scale was fitted. The Van Houwelingen‐Le Cessie heuristic shrinkage estimate was used to ascertain the likely amount of overfitting.
To address the overfitting of multivariable model and to identify the predictors that explain the bulk of HER2 mutational status, a fast‐backward step‐down procedure with total residual Akaike information criterion as the stopping rule was investigated. The R “fastbw” function was used.
The final model was internally validated for calibration and discrimination ability. In the presence of overfitting, true regression parameters were estimated using a penalized maximum likelihood approach. The receiver operating characteristic area was considered to be the summary measure of discrimination; it was fairly estimated using bootstrapping. The R “validate” function with B = 50 as option was used.
Regression modeling strategies and nomogram plotting were performed using R statistical software, version 3.3.2 [17]. Clinicopathological features were summarized using descriptive statistics (median and range for continuous variables; absolute and percentage frequencies for categorical variables). We investigated the influence of HER2 status on the following study outcomes: response rate (RR), disease control rate (DCR), progression‐free survival (PFS), and overall survival (OS). The logistic regression model was used to detect and estimate the association between HER2 positive status and RR and DCR. PFS was calculated by the Kaplan‐Meier method from the date of starting anti‐EGFR treatment until disease progression or death, whichever occurred first. OS was calculated by the Kaplan‐Meier method from the date of diagnosis of metastatic disease until death from cancer or death for any cause. The reverse Kaplan‐Meier method was used to estimate median and interquartile range (IQR) follow‐up. Clark's C index was used to estimate the follow‐up completeness [18]. Because of imbalances in follow‐up, the log‐rank test was used to assess differences between subgroups within the first 5 years (i.e., survival times longer than 5 years were right‐censored at 5 years). Because of the descriptive nature of this study, hypothesis testing was applied qualitatively and not formally (e.g., no threshold for statistical significance level was defined). Statistical analysis was performed using SAS software, version 9.4 (SAS Institute Inc., Cary, NC). Survival functions were plotted using Stata software, version 12.1 (StataCorp, College Station, TX).
Results
Clinicopathological Features of Patients with HER2‐Positive Metastatic Colorectal Cancer
From August 2012 to April 2018, 1,485 patients were tested for HER2 in primary or metastatic tumor tissue or both. A total of 100 positive cases were detected at molecular screening, and all were considered in the present study (supplemental online Fig. 1). Of these, 63 were eligible and, consequently, enrolled into clinical studies encompassing HER2‐directed therapies: 33 in HERACLES A [19] and 30 in HERACLES B trials [16]. One hundred sixteen consecutive patients without HER2 amplification or overexpression were selected as controls among those diagnosed at our institution from January 2012 to March 2017 based on availability of medical records for retrieving selected clinicopathological features. As shown in Table 1, median age at diagnosis of mCRC was 58.9 versus 58.4 years for patients with HER2‐positive versus HER2‐negative tumors, respectively; most patients were males (75% vs. 68.1%) and had stage IV disease at diagnosis of CRC (62.5 vs. 59.5%). The primary tumor site was left sided in 89.5% versus 80.9% of patients. Supplemental online Table 1 shows cancer treatments received before study entry for the HER2‐positive cohort of patients.
Table 1. Comparison of clinicopathological features between patients with HER2‐positive and HER2‐negative metastatic colorectal cancer.
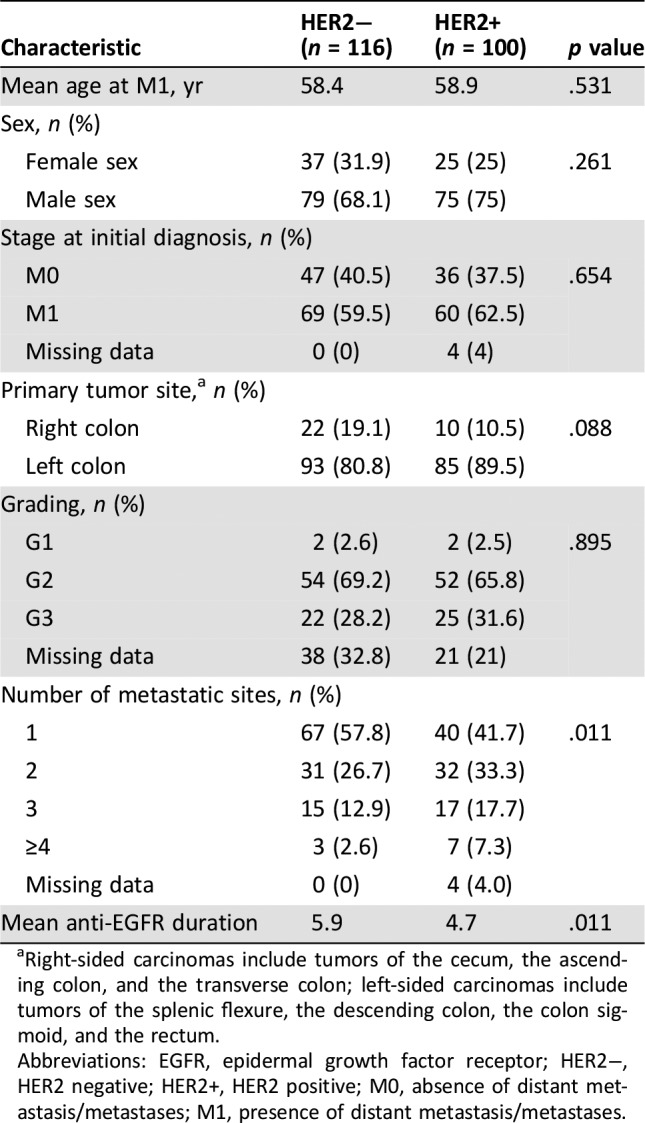
Right‐sided carcinomas include tumors of the cecum, the ascending colon, and the transverse colon; left‐sided carcinomas include tumors of the splenic flexure, the descending colon, the colon sigmoid, and the rectum.
Abbreviations: EGFR, epidermal growth factor receptor; HER2−, HER2 negative; HER2+, HER2 positive; M0, absence of distant metastasis/metastases; M1, presence of distant metastasis/metastases.
Association of HER2 Positivity and Clinicopathological Characteristics
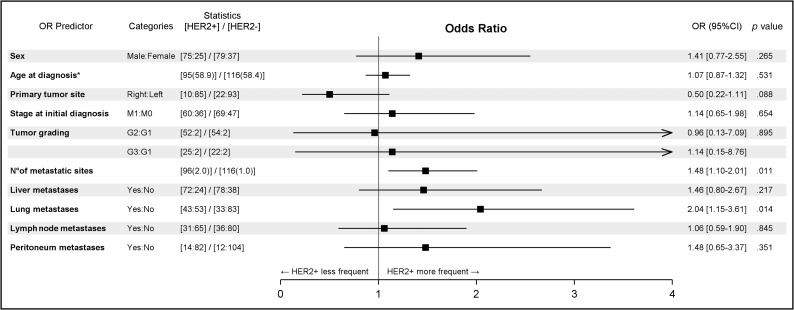
Among selected clinicopathological variables, only the presence of lung metastases (odds ratio [OR], 2.04; 95% confidence interval [CI], 1.15–3.61; p = .014) and of a higher metastatic burden defined as number of metastatic sites at diagnosis (OR, 1.48; 95% CI, 1.10–2.01; p = .011) were associated with HER2 positivity (Table 1). It should be noted that right‐sided mCRCs were 50% less likely to be HER2 positive than left‐sided mCRCs (OR, 0.50; 95% CI, 0.22–1.11; p = .088). Figure 1 shows a forest plot with the odds ratio for selected clinicopathological variables of HER2‐positive tumors.
Figure 1.
Forest plot showing odds ratio and 95% confidence intervals of HER2‐positive metastatic colorectal cancers for selected clinicopathological variables. *, Age at diagnosis: OR increases every 10 years.
Abbreviations: CI, confidence interval; M0, absence of distant metastasis/metastases; M1, presence of distant metastasis/metastases; OR, odds ratio.
Prediction of HER2 Positivity by Clinicopathological Characteristics
Full multiple logistic regression models were fitted for both the whole cohort and the subset of patients who have been treated with anti‐EGFR therapy. Penalized maximum likelihood estimates of the full models for both cohorts were plotted in two nomograms (see supplemental online Statistical Methods for details). The two resulting nomograms are shown in supplemental online Figure 3. In nomograms developed in this study, the regression coefficients have been summarized into the patient's probability to be HER2 positive. Referring to the whole cohort, the likelihood of carrying an HER2‐positive tumor ranged from 2% to 8%. The area under the curve (AUC) of the full model, which is a summary index of discrimination, was 0.59. Using bootstrapping (internal validation), a good estimate of the AUC that would be obtained by a future independent data set was 0.54. Referring to the subset of patients treated with anti‐EGFR therapy, the likelihood of carrying an HER2‐positive tumor ranged from 2% to 9%. The AUC of the full model was 0.65. Using bootstrapping, a good estimate of AUC that would be obtained by a future independent data set was 0.59.
Overall, because of the low discrimination power and the very limited range of probabilities detected, both nomograms do not appear clinically useful for enriching the population of patients who are candidates for HER2 amplification testing.
Impact of HER2 Positivity on Clinical Outcome of EGFR‐Targeted Treatments
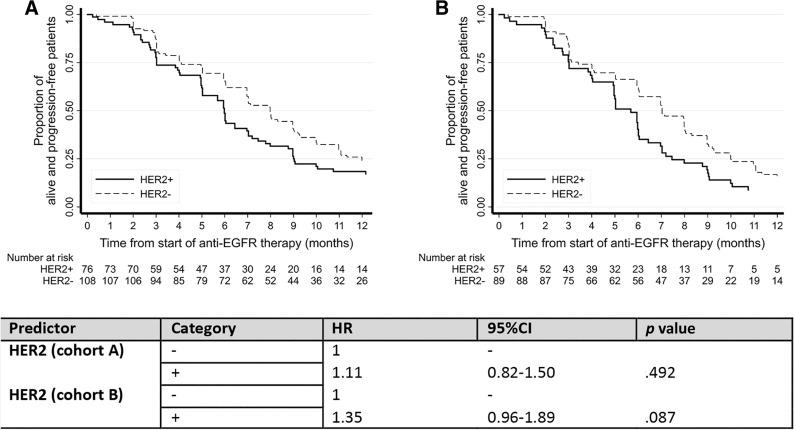
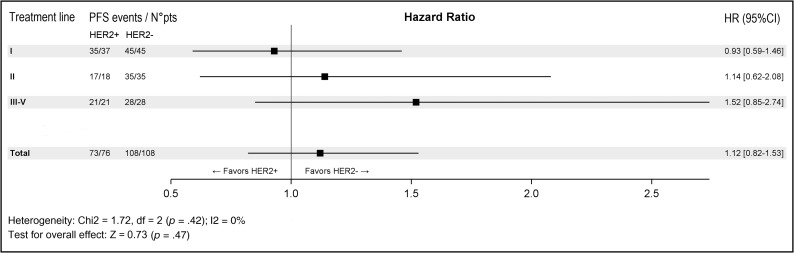
A comparative analysis of the subgroups of patients with RAS wild‐type tumors treated with an anti‐EGFR monoclonal antibody (n = 79/100, 79% among HER2‐positive patients; and 113/116, 97% among HER2‐negative patients; see supplemental online Table 2 for details of anti‐EGFR treatment received) showed that patients with HER2‐positive tumors were 50% less likely to achieve a complete or partial response when treated with anti‐EGFR therapy (OR, 0.51; 95% CI, 0.28–0.94; p = .031). This result was confirmed by a multivariate analysis adjusted for the number of metastatic sites, tumor sidedness, and line of anti‐EGFR treatment (OR, 0.52; 95% CI, 0.27–0.98; p = .044; Table 2). Progression‐free survival analysis was also performed, showing that HER2‐positive patients displayed a trend toward a worse outcome: the median progression‐free survival in response to anti‐EGFR treatment was 5.7 months (95% CI, 4.9–6.0) in HER2‐positive patients and 7 months (95% CI, 6.0–8.0) in HER2‐negative patients (hazard ratio [HR], 1.35; 95% CI, 0.96–1.89; p = .087; Fig. 2A). Patients who underwent liver resection after an anti‐EGFR‐based neoadjuvant treatment (HER2 positive, n = 43; HER2 negative, n = 27) were excluded from this analysis because of the confounding effect of surgery (Fig. 2B). Anti‐EGFR treatment was administered mostly in the first line as combination with a backbone of FOLFIRI chemotherapy (supplemental online Table 1). Figure 3 displays a forest plot for the HR of progression in response to anti‐EGFR treatment according to the line of treatment, showing an overall negative impact on clinical outcome of HER2 positivity that is stronger in more advanced lines in which the confounding impact of chemotherapy decreases.
Table 2. Univariate and multivariate analysis of objective response of anti‐EGFR therapy in patients with HER2‐negative and HER2‐positive metastatic colorectal cancer.
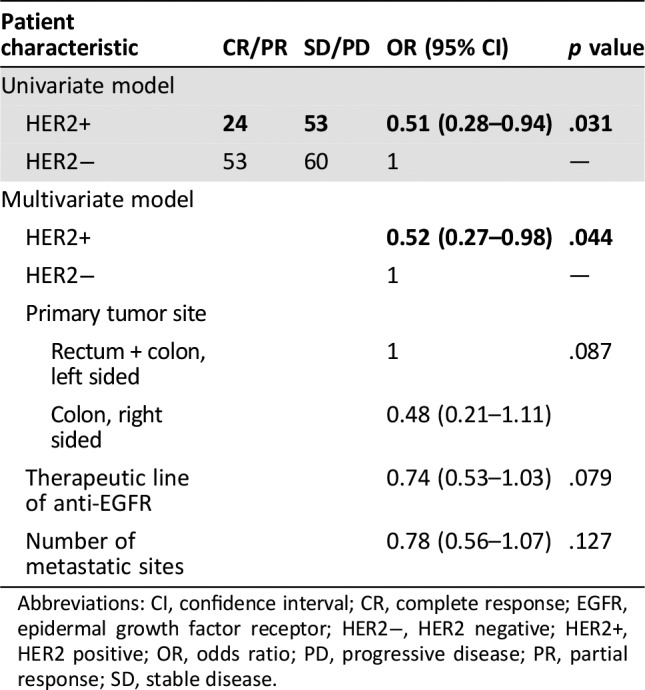
Abbreviations: CI, confidence interval; CR, complete response; EGFR, epidermal growth factor receptor; HER2−, HER2 negative; HER2+, HER2 positive; OR, odds ratio; PD, progressive disease; PR, partial response; SD, stable disease.
Figure 2.
Progression‐free survival in response to anti‐EGFR‐based‐therapeutic treatment in patients with HER2‐positive and HER2‐negative metastatic colorectal cancer. (A): All patients. (B): Subset of patients with exclusion of those who underwent surgery (metastasectomy of liver or lung).
Abbreviations: CI, confidence interval; EGFR, epidermal growth factor receptor; HR, hazard ratio.
Figure 3.
Forest plot showing progression‐free survival in response to anti‐EGFR therapy in patients with HER2‐positive and HER2‐negative metastatic colorectal cancer, stratified for line of treatment.
Abbreviations: CI, confidence interval; df, degrees of freedom; HR, hazard ratio; PFS, progression‐free survival.
HER2 Positivity and Overall Survival
At the time of data analysis, 54 patients with HER2‐positive mCRC out of 94 (57.4%) and 89 patients with HER2‐negative mCRC out of 116 (76.7%) were dead. The median overall survival was 44.6 (95% CI, 35.2–49.4) months in patients with HER2‐positive tumors and 43.7 (95% CI, 34.3–49.7) months in patients with HER2‐negative tumors. Because the follow‐up time was different between groups (lower in HER2‐positive patients with a median of 50.1 months [IQR 35.2–87.0] vs. 83.7 [IQR 59.8–110.9], respectively) and this could have influenced the comparison, survival distribution was formally compared in the first 5 years. A 5 years’ threshold was chosen because the completeness of follow‐up was moderate (Clark's index 76% in HER2‐positive patients and 90% in HER2‐negative patients). The log‐rank test at 5 years did not show association between HER2 status and survival (p = .851; supplemental online Fig. 2). It should be noticed that among patients with HER2‐positive tumors, 64 out of 100 (64%) were eventually enrolled in at least one of the HERACLES trials during the continuum of care and thus received a targeted HER2‐directed treatment that could likely have impacted on overall survival, making this piece of data of uncertain interpretation.
Discussion
HER2 overexpression or amplification is an emerging genetic alteration that can be screened for with established diagnostic tools and acted on at the therapeutic level in tumors without mutations in KRAS exon 2 [20]. Here we present the largest clinically annotated series to date of HER2‐positive CRCs (n = 100), demonstrating that this molecular feature is also associated with worse objective tumor response and shorter progression‐free survival in response to anti‐EGFR therapies.
In the present analysis, we observed a 6.7% incidence of HER2 positivity, in line with studies highlighting a higher incidence of HER2 positivity among patients without KRAS mutations than in the unselected population [5]. We found that HER2 positivity was associated with the presence of lung metastases and a higher tumor burden. However, a regression logistic model taking into account the main clinicopathological features did not allow us to identify predictors explaining the bulk of HER2 positivity and therefore obtain a meaningful statistical tool to enrich patient selection for screening. Although the limited sample size could have increased sampling variability, the predictive performance of the full models was unsatisfactory and unlikely to improve even after increasing the sample size. Therefore, recommendations for enriching the population to be tested for HER2 should not be made according to the selected clinicopathological characteristics, and testing should be offered to all patients with mCRC.
Our data suggest that right‐sided tumors are less likely to be HER2 positive, in line with previous reports showing that chromosomal regions hosting receptor tyrosine kinases are more often amplified in distal than in proximal carcinomas [21], [22]. On one hand, however, this should not preclude testing for HER2 based on sidedness, as some positive cases might be missed; on the other hand, the predominant presence of HER2 positivity in the left colon carries therapeutic implications by mitigating the clinical paradigm according to which left‐sided CRCs as a whole are more sensitive to EGFR‐targeted treatment [23].
This study confirmed previous preclinical data and hypothesis‐generating retrospective analyses in limited case series [13], [24], [25] showing that HER2 positivity is associated with worse outcome in response to anti‐EGFR therapies. Our findings have a strong biological rationale, as overexpression of receptor tyrosine kinases other than EGFR has been shown to obviate the need for activated EGFR signaling and to be responsible for resistance to anti‐EGFR therapies in preclinical models of CRC and other cancers [7].
Our analysis has several limitations, as the lack of association between the clinicopathological features selected and HER2 status could be due to dilution of real HER2‐driven mCRCs by HER2‐overexpressing tumors in which the biomarker does not play a central role in tumorigenesis and/or tumor progression. In the phase II HERACLES A clinical trial, patients with higher ERBB2 gene copy number (i.e., ≥9.6 copies) had indeed significantly longer time to progression and overall survival, indicating that further discrimination at the molecular level among HER2‐positive cases according to the level of amplification is warranted. However, in the present study this piece of data was not available for all patients and therefore only IHC and FISH scoring were used without knowledge of the actual ERBB2 gene dosage of all samples. Furthermore, not having tested all samples by NGS limited our knowledge of the wider range of genome abnormalities that could have impacted, concomitantly with HER2, on the association with main clinicopathological features of tumors.
Conclusion
The knowledge presented in this study suggests that HER2 status assessment should be included in the molecular diagnostic workup of all mCRC for speedy referral to clinical trials encompassing HER2‐targeted double blockade independently of previous anti‐EGFR treatment.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
We are grateful to Drs. Andrea Ardizzoni, Salvatore Artale, Stefano Cordio, Antonio Cubillo, Carlo Garufi, Filippo Pietrantonio, Alberto Sobrero, and Maria Giulia Zampino for referral of patients with HER2‐positive mCRC. This work was partially presented as a poster at the 2018 American Society of Clinical Oncology Gastrointestinal Cancers Symposium [J Clin Oncol 2018;36(suppl 4): abstract 581]. The study has been supported by grants from Associazione Italiana Ricerca Cancro (AIRC) grant AIRC 5 x mille [Project ID 51000] Special Program Molecular Clinical Oncology and AIRC Special Program 5 per mille metastases [Project ID 21091]; CORDIS Community Research and Development Information Service, Horizon 2020 [Project ID 635342] grant Molecularly Guided Trials with Specific Treatment Strategies in Patients with Advanced Newly Molecular Defined Subtypes of Colorectal Cancer (MoTriColor); Fondazione Oncologia Niguarda Onlus, grant Terapia Molecolare dei Tumori.
Contributed equally.
Footnotes
For Further Reading: Devarati Mitra, Jeffrey W. Clark, Helen A. Shih et al. Enrichment of HER2 Amplification in Brain Metastases from Primary Gastrointestinal Malignancies. The Oncologist 2019;24:193–201.
Implications for Practice: HER2 amplification is a well‐known driver of oncogenesis in breast cancer, with associated increased risk of brain metastases and response to HER2‐directed therapy. In nongastric gastrointestinal (GI) cancers, HER2 amplification is not common and consequently is infrequently tested. The current study shows that brain metastases in patients with GI primary malignancies have a relatively high likelihood of being HER2 positive despite HER2 amplification or overexpression being less commonly found in matched tissue from prior sites of disease. This suggests that regardless of prior molecular testing, patients with GI cancer with brain metastases who have tissue available are likely to benefit from HER2 assessment to identify potential novel therapeutic options.
Author Contributions
Conception/design: Andrea Sartore‐Bianchi, Alessio Amatu, Salvatore Siena
Provision of study material or patients: Andrea Sartore‐Bianchi, Alessio Amatu, Sara Lonardi, Francesco Leone, Francesca Bergamo, Elisabetta Fenocchio, Erika Martinelli, Beatrice Borelli, Federica Tosi, Patrizia Racca, Emanuele Valtorta, Emanuela Bonoldi, Giovanna Marrapese, Fortunato Ciardiello, Vittorina Zagonel, Salvatore Siena
Collection and/or assembly of data: Andrea Sartore‐Bianchi, Alessio Amatu, Luca Porcu, Silvia Ghezzi, Sara Lonardi, Francesco Leone, Francesca Bergamo, Elisabetta Fenocchio, Erika Martinelli, Beatrice Borelli, Federica Tosi, Patrizia Racca, Emanuele Valtorta, Emanuela Bonoldi, Cosimo Martino, Caterina Vaghi, Giovanna Marrapese, Fortunato Ciardiello, Vittorina Zagonel, Alberto Bardelli, Livio Trusolino, Valter Torri, Silvia Marsoni, S. Siena
Data analysis and interpretation: Andrea Sartore‐Bianchi, Alessio Amatu, Luca Porcu, Silvia Ghezzi, Sara Lonardi, Francesco Leone, Francesca Bergamo, Elisabetta Fenocchio, Erika Martinelli, Beatrice Borelli, Federica Tosi, Patrizia Racca, Emanuele Valtorta, Emanuela Bonoldi, Cosimo Martino, Caterina Vaghi, Giovanna Marrapese, Fortunato Ciardiello, Vittorina Zagonel, Alberto Bardelli, Livio Trusolino, Valter Torri, Silvia Marsoni, Salvatore Siena
Manuscript writing: Andrea Sartore‐Bianchi, Alessio Amatu, Luca Porcu, Silvia Ghezzi, Sara Lonardi, Francesco Leone, Francesca Bergamo, Elisabetta Fenocchio, Erika Martinelli, Beatrice Borelli, Federica Tosi, Patrizia Racca, Emanuele Valtorta, Emanuela Bonoldi, Cosimo Martino, Caterina Vaghi, Giovanna Marrapese, Fortunato Ciardiello, Vittorina Zagonel, Alberto Bardelli, Livio Trusolino, Valter Torri, Silvia Marsoni, Salvatore Siena
Final approval of manuscript: Andrea Sartore‐Bianchi, Alessio Amatu, Luca Porcu, Silvia Ghezzi, Sara Lonardi, Francesco Leone, Francesca Bergamo, Elisabetta Fenocchio, Erika Martinelli, Beatrice Borelli, Federica Tosi, Patrizia Racca, Emanuele Valtorta, Emanuela Bonoldi, Cosimo Martino, Caterina Vaghi, Giovanna Marrapese, Fortunato Ciardiello, Vittorina Zagonel, Alberto Bardelli, Livio Trusolino, Valter Torri, Silvia Marsoni, Salvatore Siena
Disclosures
Andrea Sartore‐Bianchi: Amgen, Bayer, Sanofi (SAB); Alessio Amatu: Amgen, Bayer (SAB); Sara Lonardi: Amgen, Bayer, Merck Serono, Eli Lilly & Co., Bristol‐Myers Squibb (SAB), Amgen, Merck Serono (RF); Fortunato Ciardiello: Roche, Merck Serono, Bayer, Amgen, Pfizer, Bristol‐Myers Squibb, (C/A), Roche, Merck Serono, Bayer, Amgen, Ipsen (RF); Livio Trusolino: Servier, Merus, Symphogen, Pfizer (RF); Salvatore Siena: Amgen, Roche, Bayer, Sanofi, Merck, Merus, Ignyta, Eli Lilly & Co., Novartis (SAB). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Slamon DJ, Leyland‐Jones B, Shak S et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783–792. [DOI] [PubMed] [Google Scholar]
- 2.Bang Y‐J, Van Cutsem E, Feyereislova A et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): A phase 3, open‐label, randomised controlled trial. Lancet 2010;376:687–697. [DOI] [PubMed] [Google Scholar]
- 3.Ingold Heppner B, Behrens H‐M, Balschun K et al. HER2/neu testing in primary colorectal carcinoma. Br J Cancer 2014;111:1977–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richman SD, Southward K, Chambers P et al. HER2 overexpression and amplification as a potential therapeutic target in colorectal cancer: Analysis of 3256 patients enrolled in the QUASAR, FOCUS and PICCOLO colorectal cancer trials. J Pathol 2016;238:562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross JS, Fakih M, Ali SM et al. Targeting HER2 in colorectal cancer: The landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer 2018;124:1358–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siena S, Sartore‐Bianchi A, Marsoni S et al. Targeting the human epidermal growth factor receptor 2 (HER2) oncogene in colorectal cancer. Ann Oncol 2018;29:1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertotti A, Migliardi G, Galimi F et al. A molecularly annotated platform of patient‐derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab‐resistant colorectal cancer. Cancer Discov 2011;1:508–523. [DOI] [PubMed] [Google Scholar]
- 8.Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartore‐Bianchi A, Loupakis F, Argiles G et al. Challenging chemoresistant metastatic colorectal cancer: Therapeutic strategies from the clinic and from the laboratory. Ann Oncol 2016;27:1456–66. [DOI] [PubMed] [Google Scholar]
- 10.Valtorta E, Martino C, Sartore‐Bianchi A et al. Assessment of a HER2 scoring system for colorectal cancer: Results from a validation study. Mod Pathol 2015;28:1481–1491. [DOI] [PubMed] [Google Scholar]
- 11.Sartore‐Bianchi A, Trusolino L, Martino C et al. Dual‐targeted therapy with trastuzumab and lapatinib in treatment‐refractory, KRAS codon 12/13 wild‐type, HER2‐positive metastatic colorectal cancer (HERACLES): A proof‐of‐concept, multicentre, open‐label, phase 2 trial. Lancet Oncol 2016;17:738–746. [DOI] [PubMed] [Google Scholar]
- 12.Hainsworth JD, Meric‐Bernstam F, Swanton C et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: Results from MyPathway, an open‐label, phase IIa multiple basket study. J Clin Oncol 2018;36:536–542. [DOI] [PubMed] [Google Scholar]
- 13.Yonesaka K, Zejnullahu K, Okamoto I et al. Activation of ERBB2 signaling causes resistance to the EGFR‐directed therapeutic antibody cetuximab. Sci Transl Med 2011;3:99ra86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin V, Landi L, Molinari F et al. HER2 gene copy number status may influence clinical efficacy to anti‐EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 2013;108:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siena S, Bardelli A, Sartore‐Bianchi A et al. HER2 amplification as a ‘molecular bait’ for trastuzumab‐emtansine (T‐DM1) precision chemotherapy to overcome anti‐HER2 resistance in HER2 positive metastatic colorectal cancer: The HERACLES‐RESCUE trial. J Clin Oncol 2016;34(suppl 4):TPS774A. [Google Scholar]
- 16.Marsoni S, Bertotti A, Sartore‐Bianchi A et al. Dual anti‐HER2 treatment of patients with HER2‐positive metastatic colorectal cancer: The HERACLES trial (HER2 Amplification for Colo‐rectaL Cancer Enhanced Stratification). J Clin Oncol 2013;31(suppl 15):TPS3648A. [Google Scholar]
- 17.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2017. Available at https://www.R-project.org/. Accessed March 29, 2019. [Google Scholar]
- 18.Clark TG, Altman DG, De Stavola BL. Quantification of the completeness of follow‐up. Lancet 2002;359:1309–1310. [DOI] [PubMed] [Google Scholar]
- 19.Siena S, Sartore‐Bianchi A, Trusolino L et al. Final results of the HERACLES trial in HER2‐amplified colorectal cancer. Cancer Res 2017;77:CT005A. [Google Scholar]
- 20.Sartore‐Bianchi A, Marsoni S, Siena S. Human epidermal growth factor receptor 2 as a molecular biomarker for metastatic colorectal cancer. JAMA Oncol 2018;4:19–20. [DOI] [PubMed] [Google Scholar]
- 21.Missiaglia E, Jacobs B, D'Ario G et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol 2014;25:1995–2001. [DOI] [PubMed] [Google Scholar]
- 22.Nam SK, Yun S, Koh J et al. BRAF, PIK3CA, and HER2 oncogenic alterations according to KRAS mutation status in advanced colorectal cancers with distant metastasis. PloS One 2016;11:e0151865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holch JW, Ricard I, Stintzing S et al. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta‐analysis of first‐line clinical trials. Eur J Cancer 2017;70:87–98. [DOI] [PubMed] [Google Scholar]
- 24.Martin V, Landi L, Molinari F et al. HER2 gene copy number status may influence clinical efficacy to anti‐EGFR monoclonal antibodies in metastatic colorectal cancer patients. Br J Cancer 2013;108:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raghav KPS, Overman MJ, Yu R et al. HER2 amplification as a negative predictive biomarker for anti‐epidermal growth factor receptor antibody therapy in metastatic colorectal cancer. J Clin Oncol 2016;34:3517–3517. [DOI] [PubMed] [Google Scholar]