Adenoid cystic carcinoma is a rare but aggressive type of salivary gland malignancy. This article addresses the need for more effective, biomarker‐informed therapies in rare cancers, focusing on clinical utility and financial sustainability of integrated next‐generation sequencing in routine practice.
Keywords: Salivary glands, Adenoid cystic carcinoma, Molecular diagnostics, NOTCH1, MYB
Abstract
Background.
Adenoid cystic carcinoma (ACC) is an aggressive salivary gland malignancy without effective systemic therapies. Delineation of molecular profiles in ACC has led to an increased number of biomarker‐stratified clinical trials; however, the clinical utility and U.S.‐centric financial sustainability of integrated next‐generation sequencing (NGS) in routine practice has, to our knowledge, not been assessed.
Materials and Methods.
In our practice, NGS genotyping was implemented at the discretion of the primary clinician. We combined NGS‐based mutation and fusion detection, with MYB break‐apart fluorescent in situ hybridization (FISH) and MYB immunohistochemistry. Utility was defined as the fraction of patients with tumors harboring alterations that are potentially amenable to targeted therapies. Financial sustainability was assessed using the fraction of global reimbursement.
Results.
Among 181 consecutive ACC cases (2011–2018), prospective genotyping was performed in 11% (n = 20/181; n = 8 nonresectable). Testing identified 5/20 (25%) NOTCH1 aberrations, 6/20 (30%) MYB‐NFIB fusions (all confirmed by FISH), and 2/20 (10%) MYBL1‐NFIB fusions. Overall, these three alterations (MYB/MYBL1/NOTCH1) made up 65% of patients, and this subset had a more aggressive course with significantly shorter progression‐free survival. In 75% (n = 6/8) of nonresectable patients, we detected potentially actionable alterations. Financial analysis of the global charges, including NGS codes, indicated 63% reimbursement, which is in line with national (U.S.‐based) and international levels of reimbursement.
Conclusion.
Prospective routine clinical genotyping in ACC can identify clinically relevant subsets of patients and is approaching financial sustainability. Demonstrating clinical utility and financial sustainability in an orphan disease (ACC) requires a multiyear and multidimensional program.
Implications for Practice.
Delineation of molecular profiles in adenoid cystic carcinoma (ACC) has been accomplished in the research setting; however, the ability to identify relevant patient subsets in clinical practice has not been assessed. This work presents an approach to perform integrated molecular genotyping of patients with ACC with nonresectable, recurrent, or systemic disease. It was determined that 75% of nonresectable patients harbor potentially actionable alterations and that 63% of charges are reimbursed. This report outlines that orphan diseases such as ACC require a multiyear, multidimensional program to demonstrate utility in clinical practice.
Introduction
Adenoid cystic carcinoma (ACC) is, despite its deceptively low‐grade histological appearance, an aggressive type of salivary gland malignancy arising in major and minor salivary glands. In the U.S., there are ∼15,000 patients with ACC alive today, and ACC thereby fulfills the criteria of the “rare (orphan) disease act” of 2002 (affecting <200,000 people in the U.S.). ACC grows slowly, and the disease has a seemingly indolent, yet relentless, course. Disease‐specific survival at 5 years is 89% and drops to 40% at 15 years [1]. Complete surgical resection of ACC is, whenever possible, the treatment of choice [2]; however, ACC frequently recurs, and treatment failure is often characterized by distant metastasis [1]. Radiation therapy can lower the rate of local recurrence (so‐called “local control”) but does not improve overall survival [3]. Currently, there is no effective chemotherapy [4], clinical trial data are sparse [5], [6], and standard off‐label use of cytotoxic chemotherapy is so ineffective that current guidelines state that enrollment in clinical trials is preferred [7].
Numerous studies have delineated key molecular profiles of ACC [8], [9], [10], [11], [12], [13] and collectively indicate pathways for genotype‐stratified therapies [14], [15], [16], [17], [18]. The concept, to match each patient according to the tumor pathway profile to the most effective (biologically sound) and least toxic treatment, rests on the prevalence of actionable alterations [14], [19], [20], [21]. Assessment of established alterations in clinical practice requires comprehensive genotyping efforts, for example, accomplished via next‐generation sequencing (NGS) technologies. For routine patient management in the U.S., tests are required to be clinical grade (i.e., derived from a Clinical Laboratory Improvements Amendments (CLIA)‐certified laboratory) and, because of the increased quality standards and documentation efforts, are typically 2–3 times more expensive than the same test in the research setting [22], [23]. Although drug costs are a major burden in precision medicine, test cost coverage and financial sustainability are substantial challenges to clinical genomics—in particular for orphan diseases.
Through institutional prioritization, we have initiated pan‐cancer genetic testing in 2008, transitioned to NGS‐based testing in 2011, and adopted specific NGS billing codes in 2015. In common cancers, the identification of rare subsets of actionable targets has been established (e.g., National Comprehensive Cancer Network [NCCN], College of American Pathologists guidelines); however, the need for more effective, biomarker‐informed therapies in a disease as rare as ACC poses unique challenges to demonstrate clinical utility and U.S.‐centric financial sustainability [14], [19], [21], [23].
Here we assessed clinical utility by determining the prevalence and relevance of molecular alterations. In addition, we assessed financial sustainability by determining reimbursement rates in comparison with local, national, and international rates. By combining clinical utility and financial sustainability, we outline an integrated approach to identify clinically relevant subsets of patients with ACC in an economically sustainable manner.
Materials and Methods
Study Design
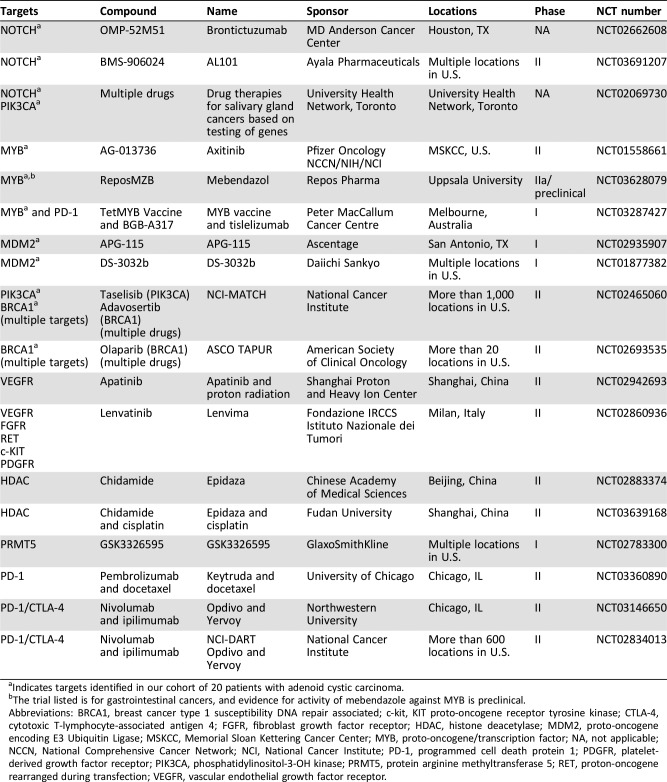
Samples were identified retrospectively (2011–2018), and we included only patients with a histologically confirmed diagnosis of ACC. We obtained demographics, anatomic location, date of diagnosis, time to progression, and/or last follow‐up by review of the electronic medical record (Qpid Health, Boston, MA). Appropriate institutional review board approval was obtained, and the research was performed in accordance with the Declaration of Helsinki. We defined “relevant” as having diagnostic and/or prognostic value and “actionable” as having potential therapeutic relevance [24]—assessed via review of relevant trials (Table 1).
Table 1. Clinical trials for patients with adenoid cystic carcinoma by molecular target.
Indicates targets identified in our cohort of 20 patients with adenoid cystic carcinoma.
The trial listed is for gastrointestinal cancers, and evidence for activity of mebendazole against MYB is preclinical.
Abbreviations: BRCA1, breast cancer type 1 susceptibility DNA repair associated; c‐kit, KIT proto‐oncogene receptor tyrosine kinase; CTLA‐4, cytotoxic T‐lymphocyte‐associated antigen 4; FGFR, fibroblast growth factor receptor; HDAC, histone deacetylase; MDM2, proto‐oncogene encoding E3 Ubiquitin Ligase; MSKCC, Memorial Sloan Kettering Cancer Center; MYB, proto‐oncogene/transcription factor; NA, not applicable; NCCN, National Comprehensive Cancer Network; NCI, National Cancer Institute; PD‐1, programmed cell death protein 1; PDGFR, platelet‐derived growth factor receptor; PIK3CA, phosphatidylinositol‐3‐OH kinase; PRMT5, protein arginine methyltransferase 5; RET, proton‐oncogene rearranged during transfection; VEGFR, vascular endothelial growth factor receptor.
Morphology
Pathological TNM staging followed the American Joint Committee on Cancer (AJCC) staging system, 8th edition [25]. At least three board‐certified pathologists reviewed and followed the diagnostic criteria proposed by the World Health Organization [26]. ACC was subclassified by growth pattern into cribriform, tubular, solid, or mixed (defined as containing a solid component). Mixed tumors containing several growth patterns, but with one predominant (>60%), were classified according to the predominant pattern.
Next‐Generation Sequencing
Genotyping at our institution is performed in a clinical (CLIA‐certified) molecular diagnostics laboratory and includes NGS‐based testing and reporting of results into the medical record. Briefly, isolated nucleic acids from tumor specimens were analyzed using anchored multiplex polymerase chain reaction (AMP) to detect single‐nucleotide variants and insertions/deletions in a target set of cancer‐related genes. To detect fusion transcripts, we also applied the AMP technology in a separate RNA‐based NGS assay [27]. We sequenced on Illumina NextSeq (for details, see supplemental online data).
FISH Assay and Immunohistochemistry
MYB gene rearrangements were analyzed using a break‐apart probe strategy (supplemental online data), and Myb protein expression was assessed using immunohistochemistry. A detailed description of both methods is provided in supplemental online material and methods.
Economic Analysis
We reviewed reimbursement (defined as amount refunded from payors) by current procedural terminology (CPT) code and claim adjustment codes (if applicable). We standardized across payors using the national values provided via the clinical laboratory fee schedules/physician fee schedule from the Center for Medicare and Medicaid Services (CMS). In 2015, the American Medical Association introduced specific NGS panel codes, and we calculated the overall reimbursement rate by dividing the total sum of obtained payments through the total sum of charges using CMS values as a reference. For comparison of our reimbursement rates with local, national, and international rates, we reviewed reimbursement data from selected publications [28], [29], [30], [31], [32], [33], [34], [35], [36].
Results
Clinically Integrated Molecular‐Genetic Testing in ACC
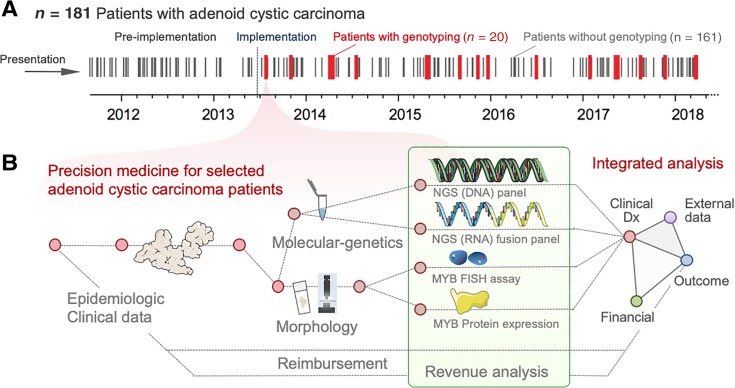
In total, 181 patients with the diagnosis of an ACC were identified between 2011 and 2018. Twenty of these ACC cases were genotyped clinically (Fig. 1A) by combining NGS‐based DNA mutation assessment, RNA‐based fusion detection, MYB immunohistochemistry, and MYB break‐apart fluorescent in situ hybridization (FISH). The clinicopathological characteristics of the study cohort are summarized in Table 2. We refer to this approach as clinically integrated molecular‐genetic testing, and an overview is provided in Figure 1B. In our clinical practice, providers order molecular testing in patients with recurrent, progressive, or nonresectable tumors (see below) in order to identify clinically relevant and potentially actionable alterations (i.e., genetic alterations expected to change management or treatment; Table 1) [24].
Figure 1.
Clinically integrated diagnostics and analytical workflow. (A): Timeline in days, indicating the diagnosis of adenoid cystic carcinoma (ACC) between 2011 and 2018 by black lines (patients without genotyping) as well as 20 ACC cases, that were sequenced within that timeframe (red lines, patients with genotyping). (B): Analysis of 20 clinically performed formalin‐fixed paraffin‐embedded tissue samples and corresponding clinical data. We integrated five separate diagnostic components including (a) histomorphology, (b) MYB immunohistochemistry, (c) MYB break‐apart FISH, (d) an NGS DNA‐based gene panel, and (e) an RNA‐based NGS panel for fusion detection. For outcome assessment, we employed 201 publicly available (external) data.
Abbreviations: Clinical Dx, clinical diagnostics; FISH, fluorescent in situ hybridization; NGS, next‐generation sequencing.
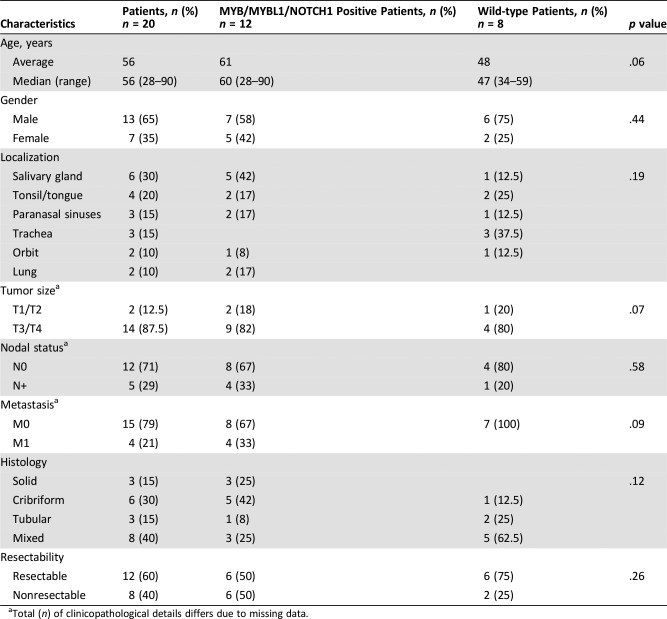
Table 2. Clinicopathological characteristics of the study cohort according to mutational status of MYB, MYBL1, and/or NOTCH1.
Total (n) of clinicopathological details differs due to missing data.
Comutational Landscape of ACC in Clinical Practice
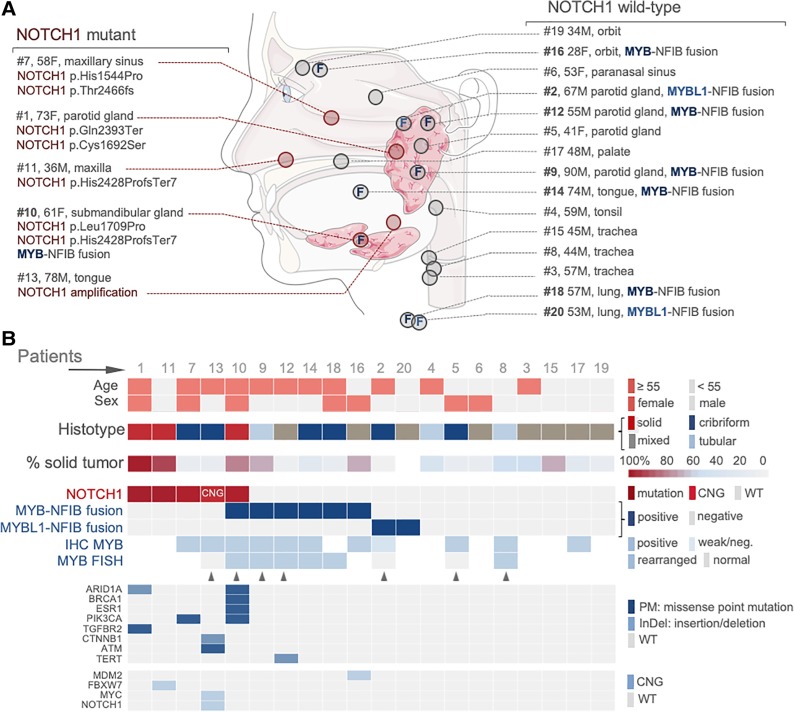
NGS identified 27 alterations in 12/20 (60%) patients in our study cohort (Fig. 2), and 8/20 patients (40%) were fusion positive. Fusions for MYB‐NFIB or MYBL1‐NFIB (40%) and NOTCH1 mutations (25%) were the most common alterations. Across all cancer types tested in our institution (n = 2,701 cases were genotyped between 2013 and 2018) [27], [37], we have only seen MYB and MYBL1 fusions in adenoid cystic carcinomas (specificity: 100%) [38]. We modeled that even if a non‐ACC would harbor one MYB/MYBL1 fusion, it would take ∼14 cases of non‐ACC for the specificity to drop below 99.5%. Therefore, ACC should be the top differential diagnosis when encountering an MYB/MYBL1 fusion (= diagnostic relevance).
Figure 2.
Anatomic location and mutational landscape. (A): Anatomic location of each case shown with salient clinical and key mutational findings (i.e., NOTCH1/MYB/MYBL1 findings). (B): Mutational landscape provided for each sample (column) along with clinical and molecular features (rows). Triangles mark cases with additional details provided in the Supplement.
Abbreviations: CNG, copy number gain; F, female; InDel, insertion/deletion; M, male; NGS, next‐generation sequencing; weak/neg., weak/negative; PM, point (missense) mutation; WT, wild‐type.
Anatomical distribution and gender were not correlated with fusion‐ or mutational status (Fig. 2A). However, as previously noted [16], NOTCH1 mutations were solely found in solid and cribriform subtypes (Fig. 2B) and occurred predominantly in older patients (i.e., ≥55 years; p = .17; Fisher's exact). Additional (and in part recurrent) mutations were present in ARID1A, BRCA1, phosphatidylinositol‐3‐OH kinase (PIK3CA), TGFBR2, CTNNB1, ATM, TERT, MDM2, FBXW7, and MYC; for details, see supplemental online Table 1. Notably, in case 13, we identified coamplification of NOTCH1 and MYC genes (supplemental online Fig. 1A), whereas case 12 was remarkable for a canonical TERT promoter mutation at position −124 (supplemental online Fig. 1B) [39]. Importantly, these additionally detected genetic aberrations occurred in tumors with MYB/MYBL1/NOTCH1 alterations, whereas the remaining “wild‐type” tumors showed no additional changes by our targeted gene panel (p = .002, Fisher's exact).
MYB‐Fusion Status and Correlation with Protein Expression
Interestingly, the occurrence of MYB/MYBL1/NOTCH1 gene alterations was, with one exception (case 10), mutually exclusive [40]. Review of this comutated case 10 showed a geographically distinct MYB protein expression pattern (supplemental online Fig. 2A), possibly reflecting intratumoral heterogeneity [41] with respect to the underlying MYB‐NFIB and NOTCH1 gene alterations. Unless limited by tissue availability, we tested NGS‐positive MYB‐NFIB cases by MYB break‐apart FISH and confirmed all cases as MYB rearranged (Fig. 2B). Correlation of the three methods identified (a) NGS‐concordant protein expression (supplemental online Fig. 3) [10], [17], (b) strong overexpression of MYB protein (supplemental online Fig. 2B) despite being fusion‐ and FISH negative (supplemental online Fig. 2C), or (c) unusual “green‐only” MYB probe pattern by FISH (supplemental online Fig. 3E, circles) with MYB protein expression (supplemental online Fig. 3F). These findings are compatible with loss of the corresponding exons (supplemental online Fig. 4A–C) and/or a locus disruption outside the MYB coding region affecting the “red” probe binding site. One way to improve sensitivity of the FISH assay would be to modify FISH probe position (supplemental online Fig. 4A–C). In other words, relying on a single method may be inferior because our findings underscore the complementary nature of various testing modalities [38].
Outcome Analysis and Clinical Utility
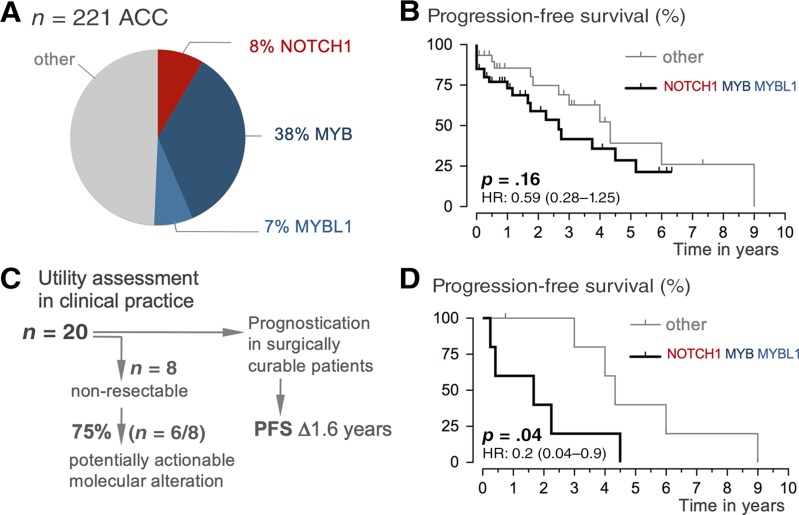
To assess prognostic relevance in our setting, we first integrated our 20 (internal) samples/patients with 201 publicly available (external; www.cbioportal.org) data (total case number = 221). Notably, when comparing progression‐free survival periods, we found significantly shorter progression‐free survival in our internal population as compared with publicly available data (supplemental online Fig. 5; p = .0003, log‐rank). The significant difference is likely due to specific order‐practice in our setting, where providers use genotyping in patients with locally advanced or systemic/palliative disease settings (Table 2; 87.5% pT3/4 internal vs. 55% external). A striking 40% of the tested patients in our setting experienced immediate tumor progression (supplemental online Fig. 5). Comparing the prevalence of NOTCH1 mutations and MYB/MYBL1 fusions revealed a similar frequency in external and internal data sets (not shown), and both cohorts were pooled for further analysis (n = 221). The group of cases with NOTCH1, MYB, and MYBL1 alterations makes up half of all ACC (Fig. 3A), and when compared with the wild‐type group, the NOTCH1/MYB/MBL1 group (= signature) does not demonstrate significantly different clinicopathological features (Table 2). Although the median progression‐free survival in the “signature” group was only 2.6 years (vs. 4.3 years in the wild‐type group), the difference did not reach statistical significance (p = .16, log‐rank; Fig. 3B). Prior studies have reported a more aggressive clinical course of NOTCH1/MYB/MYBL1 patients [10], [15], [16]; however, the direct clinical application as a prognosticator remains to be explored [42], [43]. For assessment of clinical utility, we considered two key clinical management questions regarding (a) identification of treatment options (= actionability) and (b) prognostication. First, we examined the subset of patients with initially nonresectable or systemic tumors (n = 8/20, 40% of the cohort) and determined the fraction of identified potentially actionable molecular alterations. When taking into account the shifting landscape of clinical trials (including “basket trials” and those listed in Table 1), our integrated molecular workup revealed potentially actionable alterations in 6 of 8 patients (75%; Fig. 3C). Second, when initial complete surgical resection can be achieved, we examined whether the NOTCH1/MYB/MYBL1 signature can serve as a prognosticator. We found that median progression‐free survival in the signature group was 1.6 years versus 4.3 years in the wild‐type group (p = .04, log‐rank; Fig. 3D). Although conclusions will inevitably suffer from the very small sample size, considering that ACC is an orphan disease, these data outline clinical utility by distinguishing relevant subgroups of patients with ACC in clinical practice.
Figure 3.
Outcome analysis and clinical utility. (A): Distribution of NOTCH1, MYB, and MYBL1 alterations in a cohort of 221 cases. (B): Progression‐free survival of patients by NOTCH1/MYB/MYBL1 status (altered vs. nonaltered). Note: The grouping is based on prognostic impact, and we acknowledge that MYB/MYBL1 and NOTCH1 signaling is functionally distinct. Kaplan‐Meier survival plots demonstrate shorter time to metastasis or local tumor progression for patients whose tumors showed NOTCH1/MYB/MYBL alterations versus those who did not; the difference did not reach statistical significance (p, log‐rank test). (C): Utility assessment of molecular findings is twofold: (a) identification of potential actionable alterations in patients with nonresectable tumors (see also Table 1) and (b) prognostication in surgically curable patients. (D): Progression‐free survival of patients by NOTCH1/MYB/MYBL1 status (altered vs. nonaltered). Kaplan‐Meier plots show shorter progression‐free survival time for patients with surgically resectable tumors in the subgroup with NOTCH1/MYB/MYBL alterations (p, log‐rank test).
Abbreviation: ACC, adenoid cystic carcinoma; HR, hazard ratio; PFS, progression‐free survival.
Assessment of Financial Sustainability
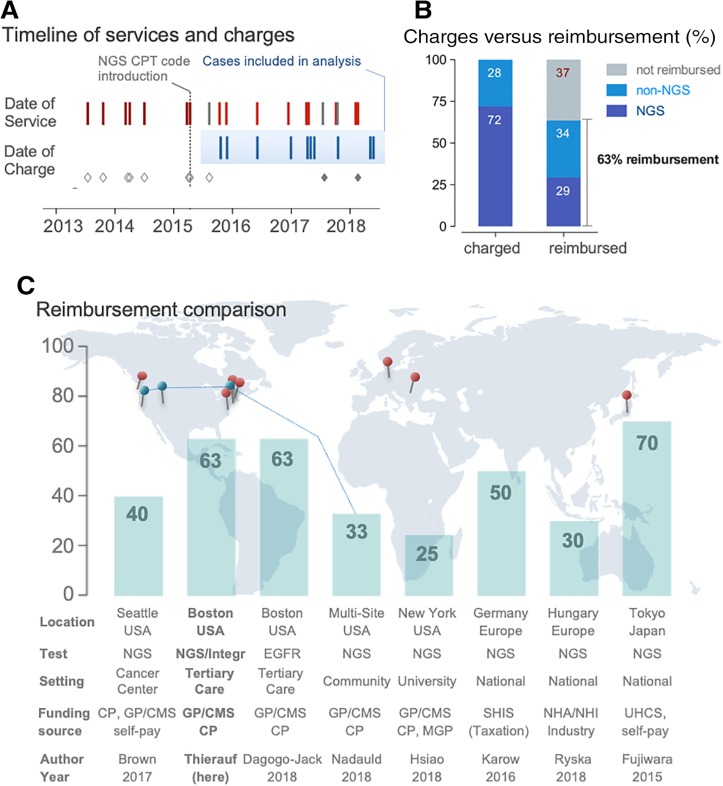
A key factor in maintaining an integrated genotyping program in clinical practice is achieving financial sustainability. In contrast to cancers with higher prevalence, the rarity in orphan diseases poses additional hurdles [14], [21], [44], particularly when genotyping is applied only to the meaningful subsets of patients. During the initial phase (2013–2015; Fig. 4A), we applied the miscellaneous CPT code 81479 “Unlisted molecular pathology procedure” for NGS testing because specific NGS codes were not available at the time. We excluded data from this time frame because of the nonspecific nature of the CPT code and high variability in payments. Of the 12 cases charged after NGS code introduction, we excluded 2 cases because of payment through alternative sources. Review of the remaining 10 cases included in the analysis (Fig. 4A) showed that 72% of the total amount charged derived from NGS panel codes whereas 28% derived from non‐NGS codes. Reimbursement analysis indicates that 29% of payments derive from NGS codes whereas 34% of payments derive from non‐NGS codes. In summary, the fraction of reimbursement for NGS and non‐NGS tests sums up to a total of 63% (Fig. 4B). When comparing this fraction of reimbursement with published local, national, and international reimbursement rates, the overall fraction is similar (Fig. 4C). Notably, these comparison data are obtained in nonorphan disease settings and include multisite studies including large community centers or even national initiatives [28], [29], [30], [31], [32], [33], [34], [35], [36]. Nonetheless, obtaining reimbursement requires carefully designed workflows to account for prior authorization, policy tailoring, and appeals processes [45], [46], [47]. Review of the claim adjustment codes in our cohort indicates “charge exceeds maximum allowable” (n = 6 of 10) as the most frequent reason for adjusted payments.
Figure 4.
Financial sustainability of adenoid cystic carcinoma testing in clinical practice. (A): Timeline shows dates of service and dates of charges. In 2015, CPT codes for NGS‐based panel testing were available and introduced (vertical line). We excluded cases with miscellaneous CPT codes (applied before availability of NGS codes indicated by open gray diamonds) and those payed for from alternative sources (indicated by filled gray diamonds). The cases included in the reimbursement analysis span 2.5 years. (B): Bar graphs comparing charged (left) and reimbursed amounts (right) separated by NGS‐ and non‐NGS related fractions. Note: The lower rate in the fraction of reimbursement of NGS codes when compared with non‐NGS codes is likely related to a combination of (a) lower rates of payments, (b) denials, and/or (c) a relative larger fraction of payments related to non‐NGS codes. (C): Bar graphs indicating reimbursement in percent; the location of the eight references (see Results) is provided on the world map.
Abbreviations: Author, first author of the publication; CP, commercial plans; CPT, current procedural terminology; EGFR, epidermal growth factor receptor (single gene testing); GP/CMS, government plans/Center for Medicare and Medicaid services; MGP, Managed government plans (i.e., Medicaid health maintenance organizations and managed Medicaid/Medicare plans); NGS, next‐generation sequencing; NHA/NHI, national health authority/national health insurance; SHIS, Germany's statutory health insurance system; UHCS, Japanese universal health care system.
Therefore, our data indicate that integrated genotyping of selected patients with ACC in our practice is approaching financially sustainability. In conjunction with the ability to identify potentially (therapeutically) actionable subsets of patients, our data indicate that achieving financial sustainability requires a multiyear and multidimensional program.
Discussion
Here we report that clinically integrated molecular‐genetic diagnostics can identify a significant subset of potentially actionable molecular alterations in patients with nonresectable ACC. Identification of MYB/MYBL1 fusions is 100% specific for ACC and can serve as a diagnostic biomarker. Molecular genotyping can furthermore serve as a prognostication tool for progression‐free survival in initially surgically curable patients with ACC. Finally, we present reimbursement data, indicating that approaching financial sustainability requires a multiyear and multidimensional program.
Generally, ACC is being portrayed as an indolent disease; however, it has become clear that this is not always the case [48]. We fully acknowledge that the rarity and the small number of patients represent a significant limitation of our study. However, the demand for larger cohorts and studies does not solve day‐to‐day clinical management questions: for example, how to identify the subset of patients with ACC with a more aggressive clinical course. Notably, despite various studies outlining the prognostic relevance of molecular markers [5], [9], [15], [16], [42], [43], [48], [49], [50], [51], [52], [53], [54], clinical guidelines currently rely on staging, margin status [55], perineural invasion [56], and resectability for treatment stratification (NCCN guidelines Version 2/2018; www.nccn.org, last accessed 12/22/2018).
Molecular genotyping in ACC is primarily performed in research settings, and numerous studies have outlined relevant mutational profiles [17], [57], [58], [59]. In addition, various biological [12], [17], [60], diagnostic [60], [61], [62], [63], [64], prognostic [16], [54], [64], and therapeutic advances [14], [15], [59] have been proposed. It is important to note that our study did not focus on identification or description of novel biomarkers. In contrast, our aim was to specifically address clinical utility of established alterations in clinical practice. In other words, our integrated precision medicine program explicitly prioritizes the individualization of care and focuses attention on unique characteristics of a particular patient subset. The approach thereby differs from traditional evidence‐based medicine, which seeks to determine the best course of action for a patient with an appeal to generalizable knowledge gained from population‐based studies [24], [65]. In the case of the orphan disease ACC, this specifically means to demonstrate utility in the small subset of meaningful patients. Demonstrating utility of previously described molecular alterations in the relevant subset of patients in clinical practice is the next logical and important step toward identifying when expanded or integrated genotyping should be considered standard of care.
The MYB translocation t(6;9)(q21‐24;p13‐23) results in a fusion protein of two transcription factors (MYB and NFIB). The MYB‐NFIB fusion results in an increased expression of the protein Myb [43]. Myb is important for the growth of multiple solid tumors, and MYB protein overexpression in ACC is associated with a more aggressive clinical course [33], [47], [48], [49]. For example, Mitani et al. [40] reported that the level of MYB expression in identical histological subtypes of ACC is associated with a shorter survival time. Recently, xenografting [66] and the first MYB‐NFIB‐positive cell line (UM‐HACC‐2A) have been established and represent interesting tools to explore novel therapeutic possibilities [67]. Therapeutically, MYB‐targeted antisense oligodeoxynucleotides (G4460; INX‐3001; LR3001) [68] have not been effective in clinical trials [69]; however, several lines of evidence suggest that MYB is a promising therapeutic target [70], [71], [72], [73]. Currently, the first clinical trial of a therapy that directly targets MYB, in combination with an anti‐PD‐1 immune checkpoint inhibitor, opened (MYPHISMO, NCT03287427), and the first patient with ACC received her first infusion of the MYB DNA vaccine in September 2018 [74]. Collectively, these data suggest that identification of MYB alterations represents a promising approach to identify a subset of patients with ACC for molecularly targeted therapies.
NOTCH1 alterations also identify a subset of patients with an aggressive disease course [50] that are potential candidates for clinical trial enrollment. Importantly, NOTCH1 knockdown results in effective suppression of metastasis in vivo [51], and the efficacy of gamma‐secretase inhibitors targeting the canonical NOTCH1 signaling pathway [19] is currently being explored (NCT03422679). Ferrarotto et al. [75] described activating NOTCH1 mutations that define a distinct subgroup of patients with adenoid‐cystic carcinoma who have poor prognosis, propensity to bone and liver metastasis, and potential responsiveness to Notch1 inhibitors in a patient‐derived xenograft model. Furthermore, an index patient with NOTCH1‐mutant ACC showed partial response to brontictuzumab [16]. Those results led to a clinical trial for patients with ACC treated with brontictuzumab (OMP‐52 M51), a humanized monoclonal antibody directed against the Notch‐1 receptor with potential antineoplastic activity (NCT02662608). Moreover, CB‐103, a small molecule inhibitor, is being tested in a phase IIA trial in up to 140 patients with preselected cancer indications with tumor cells characterized by NOTCH overactivation (NCT03422679). In addition, the first phase II clinical trial of a NOTCH inhibitor recently opened (NCT03691207). These data collectively indicate NOTCH1 and MYB genes as promising therapeutic targets. Most recently a clinical trial for NOTCH‐mutant ACCs has started at our institution (ACCURACY trial; NCT03691207).
Additional cancer‐relevant alterations co‐occurred within the set of MYB/MYBL1/NOTCH1 aberrations that may contribute to the more aggressive clinical course in this group of ACC. Several other aberrations may lead to new therapeutic strategies (Table 1). For example, PARP inhibitors such as rucaparib and other agents such as trabectedin are actively tested in tumors with BRCA1 mutations (NCT01989546). PIK3CA are responsible for coordinating a diverse range of cell functions including proliferation and survival. Novel therapeutics targeting different components of the PI3K pathway showed preclinical efficacy in an array of human cancer, and several compounds as well as dual PI3K‐mTOR inhibitors, PI3K inhibitors (that do not inhibit mTOR), AKT inhibitors, and mTOR catalytic site inhibitors are moving toward clinical trials (Table 1) [76], [77], [78]. Interestingly, ESR1 has also been linked to PIK3CA variants in metastatic breast cancer [53], [79], and one of our ACC cases with a PIK3CA mutation (number 10) also harbored an ESR1 aberration. Strategies to block estrogen signaling have shown extraordinary success in the prevention and treatment of breast cancer, and the development of therapeutic approaches that directly target ESR1‐mutated clones is an appealing concept.
Although primarily designed to identify therapeutically actionable alterations (Table 1), our testing platform also showed prognostically relevant signatures (Fig. 3). Many head and neck cancer centers will not have local access to molecular testing that integrates DNA and RNA NGS, FISH, and IHC data. Furthermore, establishing such testing practices is challenging when cost‐coverage for molecular tests is restricted to settings that have been shown to directly influence therapeutic decision‐making. In some diagnostically challenging cases, there is value in detecting a fusion that is highly specific for ACC; however, our testing approach was not specifically built for ACC. We combined and repurposed several existing infrastructures from former initiatives [80], [81], [82], [83] to illustrate clinical utility in ACC. At first glance, this cross‐functionality seems straightforward; however, we want to underscore that this platform is not a “one‐time research project” but represents a fully integrated clinical‐grade workflow. Two components are noteworthy: first, a dedicated clinical data science team that maintains the computational infrastructure; and second, adopting up‐to date U.S.‐billing practices to approach financial sustainability. With only 10 cases over multiple years, any economic argument can only be anecdotal and numerous limitations apply (e.g., variance by payor, coinsurances, copayments, deductibles, etc.). Furthermore, some may argue that CMS charges can only be regarded as an imperfect reference cost and do not account for additional indirect cost components, laboratory maintenance cost, etc. Importantly, when taking into account that our approach requires navigation of at least four distinct CPT codes from various payors, different negotiated hospital contracts (that changed over time), and the various components and tools of the U.S.‐centric revenue cycle, it becomes clear that achieving financial sustainability in an orphan disease is a multiyear endeavor and that the integrated testing approach has to entail a highly specialized and policy‐tailored billing process [45], [46], [47], an arguably overlooked aspect of personalized medicine. Demonstrating financial sustainability is important; however, in other settings, the implementation of integrated NGS testing in an orphan cancer will require modification(s). Despite these limitations, our comparison with other molecular testing initiatives (Fig. 4C) indicates largely compatible rates, and it is critical to understand the underlying motivation of the reimbursement or funding source. In our setting—a U.S.‐based, CLIA‐certified diagnostic laboratory in a tertiary care (academic) medical center—we were able to align the goals of private payors for medically meaningful testing (and reduction of unnecessary testing) with the needs of our providers and the aims of the health care organization. When considering other variables, for example, single‐payer systems with national quality improvement initiatives, or initiatives that aim to tackle multiple diseases or even disease groups, financial sustainability can only be achieved through meticulous planning and the combination of mural and extramural funding strategies. Simply put, the (financial) sustainability of a novel initiative is a direct function of how well the core value‐proposition of the program is aligned with the aims of the funding source (supplemental online Fig. 6). For orphan cancers, there is currently no generalizable “cost‐benefit approach” that can serve as a one‐size‐fits‐all approach for the plethora of possible settings. Therefore, managing the variable economic burden of NGS testing programs remains context dependent, and we have summarized the paradigm for achieving financial sustainability in modern medicine by funding source (supplemental online Fig. 6). Interestingly, several centers recently established rare tumor clinics [14], [21], [44], and we wanted to share our financial data because these additional layers of complexity in sustaining clinical genotyping are largely ignored, particularly when relevant subsets of patients with recurrent or metastatic ACC only present 2–3 times per year.
Conclusion
Clinically integrated workup of patients with ACC can identify a significant subset of potentially actionable molecular alterations in nonresectable tumors and serves as a prognostication tool for progression‐free survival in initially surgically curable patients. The full potential of molecularly informed, personalized medicine relies on the availability of testing, access to molecularly matched therapeutic strategies, and ultimately demonstration of outcome differences. In our practice, a significant fraction of patients could benefit from genotype‐stratified and targeted therapies—which matches the experience by others. However, clinical implementation requires at least initial investment until financial sustainability is achieved, especially at times of intense economic pressure and demand to manage rapidly escalating health care costs.
See http://www.TheOncologist.com for supplemental material available online.
Acknowledgments
We thank all members of the Center for Integrated Diagnostics for the excellent technical and administrative support. We specifically acknowledge discussions with J. Batten on achieving financial sustainability.
Footnotes
For Further Reading: Zviadi Aburjania, Samuel Jang, Jason Whitt et al. The Role of Notch3 in Cancer. The Oncologist 2018;23:900–911.
Implications for Practice: The Notch family is a highly conserved gene group that regulates cell‐cell interaction, embryogenesis, and tissue commitment. This review summarizes the existing data on the third subtype of the Notch family, Notch3. The role of Notch3 in different types of cancers is discussed, as well as implications of its modification and new strategies to affect Notch3 signaling activity.
Author Contributions
Conception/design: Julia Thierauf, Jochen K. Lennerz
Provision of study material or patients: Julia Thierauf, Nisha Ramamurthy, Vickie Y. Jo, Hayley Robinson, Ryan P. Frazier, Jonathan Gonzalez, Maciej Pacula, Enrique Dominguez Meneses, Vania Nose, Dora Dias‐Santagata, Long P. Le, Derrick T. Lin, William C. Faquin, A. John Iafrate, Jochen K. Lennerz
Collection and/or assembly of data: Julia Thierauf, Nisha Ramamurthy, Vickie Y. Jo, Hayley Robinson, Ryan P. Frazier, Jonathan Gonzalez, Maciej Pacula, Enrique Dominguez Meneses, Jochen Hess, Jochen K. Lennerz
Data analysis and interpretation: Julia Thierauf, Ryan P. Frazier, Jonathan Gonzalez, Valentina Nardi, Dora Dias‐Santagata, Jochen Hess, Jochen K. Lennerz
Manuscript writing: Julia Thierauf, Jochen K. Lennerz
Final approval of manuscript: Julia Thierauf, Nisha Ramamurthy, Vickie Y. Jo, Hayley Robinson, Ryan P. Frazier, Jonathan Gonzalez, Maciej Pacula, Enrique Dominguez Meneses, Vania Nose, Valentina Nardi, Dora Dias‐Santagata, Long P. Le, Derrick T. Lin, William C. Faquin, Lori J. Wirth, Jochen Hess, A. John Iafrate, Jochen K. Lennerz
Disclosures
Vickie Y. Jo: Merck (E [spouse], OI [spouse]); Long P. Le: ArcherDx (C/A, OI, IP); Lori J. Wirth: Ayala, Bayer, Eisai, Loxo, Merck (C/A); Jochen Hess: Merck Sharp & Dohme, Bristol‐Myers Squibb (H, SAB); A. John Iafrate: ArcherDx (IP). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1.Coca‐Pelaz A, Rodrigo JP, Bradley PJ et al. Adenoid cystic carcinoma of the head and neck–An update. Oral Oncol 2015;51:652–661. [DOI] [PubMed] [Google Scholar]
- 2.Trope M, Triantafillou V, Kohanski MA et al. Adenoid cystic carcinoma of the sinonasal tract: A review of the national cancer database. Int Forum Allergy Rhinol 2019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 3.Buchsenschutz K, Veit JA, Schuler PJ et al. Molecular approaches to systemic therapy of adenoid cystic carcinoma of the head and neck area [in German]. Laryngorhinootologie 2014;93:657–664. [DOI] [PubMed] [Google Scholar]
- 4.Laurie SA, Ho AL, Fury MG et al. Systemic therapy in the management of metastatic or locally recurrent adenoid cystic carcinoma of the salivary glands: A systematic review. Lancet Oncol 2011;12:815–824. [DOI] [PubMed] [Google Scholar]
- 5.Till BG, Martins RG. Response to paclitaxel in adenoid cystic carcinoma of the salivary glands. Head Neck 2008;30:810–814. [DOI] [PubMed] [Google Scholar]
- 6.van Herpen CM, Locati LD, Buter J et al. Phase II study on gemcitabine in recurrent and/or metastatic adenoid cystic carcinoma of the head and neck (EORTC 24982). Eur J Cancer 2008;44:2542–2545. [DOI] [PubMed] [Google Scholar]
- 7.Colevas AD, Yom SS, Pfister DG et al. NCCN Guidelines Insights: Head and Neck Cancers, Version 1.2018. J Natl Compr Canc 2018;16(5). [DOI] [PubMed] [Google Scholar]
- 8.Agrawal N, Frederick MJ, Pickering CR et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011;333:1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arruga F, Gizdic B, Serra S et al. Functional impact of NOTCH1 mutations in chronic lymphocytic leukemia. Leukemia 2014;28:1060–1070. [DOI] [PubMed] [Google Scholar]
- 10.Brayer KJ, Frerich CA, Kang H et al. Recurrent fusions in MYB and MYBL1 define a common, transcription factor‐driven oncogenic pathway in salivary gland adenoid cystic carcinoma. Cancer Discov 2016;6:176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmaier RJ, Albacker LA, Chmielecki J et al. High‐throughput genomic profiling of adult solid tumors reveals novel insights into cancer pathogenesis. Cancer Res 2017;77:2464–2475. [DOI] [PubMed] [Google Scholar]
- 12.Sant DW, Tao W, Field MG et al. Whole exome sequencing of lacrimal gland adenoid cystic carcinoma. Invest Ophthalmol Vis Sci 2017;58:BIO240–BIO6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wysocki PT, Izumchenko E, Meir J et al. Adenoid cystic carcinoma: Emerging role of translocations and gene fusions. Oncotarget 2016;7:66239–66254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chahal M, Pleasance E, Grewal J et al. Personalized oncogenomic analysis of metastatic adenoid cystic carcinoma: Using whole‐genome sequencing to inform clinical decision‐making. Cold Spring Harb Mol Case Stud 2018;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrarotto R, Heymach JV, Glisson BS. MYB‐fusions and other potential actionable targets in adenoid cystic carcinoma. Curr Opin Oncol 2016;28:195–200. [DOI] [PubMed] [Google Scholar]
- 16.Ferrarotto R, Mitani Y, Diao L et al. Activating NOTCH1 mutations define a distinct subgroup of patients with adenoid cystic carcinoma who have poor prognosis, propensity to bone and liver metastasis, and potential responsiveness to Notch1 inhibitors. J Clin Oncol 2017;35:352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho AS, Kannan K, Roy DM et al. The mutational landscape of adenoid cystic carcinoma. Nat Genet 2013;45:791–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoeck A, Lejnine S, Truong A et al. Discovery of biomarkers predictive of GSI response in triple‐negative breast cancer and adenoid cystic carcinoma. Cancer Discov 2014;4:1154–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chabner BA, Ellisen LW, Iafrate AJ. Personalized medicine: Hype or reality. The Oncologist 2013;18:640–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson DB, Dahlman KH, Knol J et al. Enabling a genetically informed approach to cancer medicine: A retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next‐generation sequencing panel. The Oncologist 2014;19:616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato S, Kurasaki K, Ikeda S et al. Rare tumor clinic: The University of California San Diego Moores Cancer Center experience with a precision therapy approach. The Oncologist 2018;23:171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanagal‐Shamanna R, Singh RR, Routbort MJ et al. Principles of analytical validation of next‐generation sequencing based mutational analysis for hematologic neoplasms in a CLIA‐certified laboratory. Expert Rev Mol Diagn 2016;16:461–472. [DOI] [PubMed] [Google Scholar]
- 23.Madhavan S, Ritter D, Micheel C et al. ClinGen Cancer Somatic Working Group ‐ Standardizing and democratizing access to cancer molecular diagnostic data to drive translational research. Pac Symp Biocomput 2018;23:247–258. [PMC free article] [PubMed] [Google Scholar]
- 24.Carr TH, McEwen R, Dougherty B et al. Defining actionable mutations for oncology therapeutic development. Nat Rev Cancer 2016;16:319–329. [DOI] [PubMed] [Google Scholar]
- 25.Amin MB, Edge S, Greene F et al. AJCC Cancer Staging Manual. 8th ed. New York: Springer International Publishing, 2017. [Google Scholar]
- 26.El‐Naggar AK, Chan JKC, Grandis JR et al. WHO Classification of Head and Neck Tumours. Lyon, France: IARC Publications, 2017. [Google Scholar]
- 27.Zheng Z, Liebers M, Zhelyazkova B et al. An chored multiplex PCR for targeted next‐generation sequencing. Nat Med 2014;20:1479–1484. [DOI] [PubMed] [Google Scholar]
- 28.Brown TD, Tameishi M, Liu X et al. Analysis of reimbursement (R) for next generation sequencing (NGS) on patients' tumors in the context of a personalized medicine program. J Clin Oncol 2017;35(suppl 15):6506a. [Google Scholar]
- 29.Dagogo‐Jack I, Azzolli CG, Fintelmann F et al. Clinical utility of rapid EGFR genotyping in advanced lung cancer. JCO Precis Oncol 10.1200/PO.17.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujiwara Y. Evolution of frameworks for expediting access to new drugs in Japan. Nat Rev Drug Discov 2016;15:293–294. [DOI] [PubMed] [Google Scholar]
- 31.Hsiao SJ, Mansukhani MM, Carter MC. The history and impact of molecular coding changes on coverage and reimbursement of molecular diagnostic tests: Transition from stacking codes to the current molecular code set including genomic sequencing procedures. J Mol Diagn 2018;20:177–183. [DOI] [PubMed] [Google Scholar]
- 32.Karow J. German Health Insurance Allows Reimbursement of NGS Tests Up to Limit; Excludes cfDNA analysis, PGx. Available at https://www.genomeweb.com/molecular‐diagnostics/german‐health‐insurance‐allows‐reimbursement‐ngs‐tests‐limit‐excludes‐cfdna#.XB6NSc9KjmE. Accessed December 22, 2018. [Google Scholar]
- 33.Kohno T. Implementation of “clinical sequencing” in cancer genome medicine in Japan. Cancer Sci 2018;109:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadauld LD, Ford JM, Pritchard D et al. Strategies for clinical implementation: Precision oncology at three distinct institutions. Health Aff (Millwood) 2018;37:751–756. [DOI] [PubMed] [Google Scholar]
- 35.Ryska A, Berzinec P, Brcic L et al. NSCLC molecular testing in Central and Eastern European countries. BMC Cancer 2018;18:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunami K, Takahashi H, Tsuchihara K et al. Clinical practice guidance for next‐generation sequencing in cancer diagnosis and treatment (Edition 1.0). Cancer Sci 2018;109:2980–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zomnir MG, Lipkin L, Pacula M et al. Artificial intelligence approach for variant reporting. JCO Clin Cancer Inform 10.1200/CCI.16.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Togashi Y, Dobashi A, Sakata S et al. MYB and MYBL1 in adenoid cystic carcinoma: Diversity in the mode of genomic rearrangement and transcripts. Mod Pathol 2018;31:934–946. [DOI] [PubMed] [Google Scholar]
- 39.Huang FW, Bielski CM, Rinne ML et al. TERT promoter mutations and monoallelic activation of TERT in cancer. Oncogenesis 2015;4:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitani Y, Liu B, Rao PH et al. Novel MYBL1 gene rearrangements with recurrent MYBL1‐NFIB fusions in salivary adenoid cystic carcinomas lacking t(6;9) translocations. Clin Cancer Res 2016;22:725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu B, Mitani Y, Rao X et al. Spatio‐temporal genomic heterogeneity, phylogeny, and metastatic evolution in salivary adenoid cystic carcinoma. J Natl Cancer Inst 2017;109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He S, Li P, Zhong Q et al. Clinicopathologic and prognostic factors in adenoid cystic carcinoma of head and neck minor salivary glands: A clinical analysis of 130 cases. Am J Otolaryngol 2017;38:157–162. [DOI] [PubMed] [Google Scholar]
- 43.Xu B, Drill E, Ho A et al. Predictors of outcome in adenoid cystic carcinoma of salivary glands: A clinicopathologic study with correlation between MYB fusion and protein expression. Am J Surg Pathol 2017;41:1422–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Choi M. Ultra‐rare disease and genomics‐driven precision medicine. Genomics Inform 2016;14:42–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deverka PA, Kaufman D, McGuire AL. Overcoming the reimbursement barriers for clinical sequencing. JAMA 2014;312:1857–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lennerz JK, McLaughlin HM, Baron JM et al. Health care infrastructure for financially sustainable clinical genomics. J Mol Diagn 2016;18:697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sireci AN, Aggarwal VS, Turk AT et al. Clinical genomic profiling of a diverse array of oncology specimens at a large academic cancer center: Identification of targetable variants and experience with reimbursement. J Mol Diagn 2017;19:277–287. [DOI] [PubMed] [Google Scholar]
- 48.van Weert S, Reinhard R, Bloemena E et al. Differences in patterns of survival in metastatic adenoid cystic carcinoma of the head and neck. Head Neck 2017;39:456–463. [DOI] [PubMed] [Google Scholar]
- 49.Cross RS, Malaterre J, Davenport AJ et al. Therapeutic DNA vaccination against colorectal cancer by targeting the MYB oncoprotein. Clin Transl Immunology 2015;4:e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao J, Aksoy BA, Dogrusoz U et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mitani Y, Li J, Rao PH et al. Comprehensive analysis of the MYB‐NFIB gene fusion in salivary adenoid cystic carcinoma: Incidence, variability, and clinicopathologic significance. Clin Cancer Res 2010;16:4722–4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rettig EM, Tan M, Ling S et al. MYB rearrangement and clinicopathologic characteristics in head and neck adenoid cystic carcinoma. Laryngoscope 2015;125:E292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spoerke JM, Gendreau S, Walter K et al. Heterogeneity and clinical significance of ESR1 mutations in ER‐positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 2016;7:11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie M, Wu X, Zhang J et al. The prognostic significance of Notch1 and fatty acid binding protein 7 (FABP7) expression in resected tracheobronchial adenoid cystic carcinoma‐A multicenter retrospective study. Cancer Res Treat 2018;50:1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garden AS, Weber RS, Morrison WH et al. The influence of positive margins and nerve invasion in adenoid cystic carcinoma of the head and neck treated with surgery and radiation. Int J Radiat Oncol Biol Phys 1995;32:619–626. [DOI] [PubMed] [Google Scholar]
- 56.Jang S, Patel PN, Kimple RJ et al. Clinical outcomes and prognostic factors of adenoid cystic carcinoma of the head and neck. Anticancer Res 2017;37:3045–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris LG, Chandramohan R, West L et al. The molecular landscape of recurrent and metastatic head and neck cancers: Insights from a precision oncology sequencing platform. JAMA Oncol 2016. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stephens PJ, Davies HR, Mitani Y et al. Whole exome sequencing of adenoid cystic carcinoma. J Clin Invest 2013;123:2965–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ross JS, Wang K, Rand JV et al. Comprehensive genomic profiling of relapsed and metastatic adenoid cystic carcinomas by next‐generation sequencing reveals potential new routes to targeted therapies. Am J Surg Pathol 2014;38:235–238. [DOI] [PubMed] [Google Scholar]
- 60.Drier Y, Cotton MJ, Williamson KE et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat Genet 2016;48:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pusztaszeri MP, Faquin WC. Update in salivary gland cytopathology: Recent molecular advances and diagnostic applications. Semin Diagn Pathol 2015;32:264–274. [DOI] [PubMed] [Google Scholar]
- 62.Pusztaszeri MP, Faquin WC. MYB is a helpful diagnostic marker for adenoid cystic carcinoma in fine‐needle aspiration biopsy. Arch Pathol Lab Med 2015;139:157–158. [DOI] [PubMed] [Google Scholar]
- 63.Pusztaszeri MP, Sadow PM, Ushiku A et al. MYB immunostaining is a useful ancillary test for distinguishing adenoid cystic carcinoma from pleomorphic adenoma in fine‐needle aspiration biopsy specimens. Cancer Cytopathol 2014;122:257–265. [DOI] [PubMed] [Google Scholar]
- 64.Sajed DP, Faquin WC, Carey C et al. Diffuse staining for activated NOTCH1 correlates with NOTCH1 mutation status and is associated with worse outcome in adenoid cystic carcinoma. Am J Surg Pathol 2017;41:1473–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tonelli MR, Shirts BH. Knowledge for precision medicine: Mechanistic reasoning and methodological pluralism. JAMA 2017;318:1649–1650. [DOI] [PubMed] [Google Scholar]
- 66.Cornett A, Athwal HK, Hill E et al. Serial patient‐derived orthotopic xenografting of adenoid cystic carcinomas recapitulates stable expression of phenotypic alterations and innervation. EBioMedicine 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Warner KA, Oklejas AE, Pearson AT et al. UM‐HACC‐2A: MYB‐NFIB fusion‐positive human adenoid cystic carcinoma cell line. Oral Oncol 2018;87:21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.National Cancer Institute. NCI Drug Dictionary. Available at https://www.cancer.gov/publications/dictionaries/cancer-drug. Accessed December 22, 2018.
- 69.Liu X, Xu Y, Han L et al. Reassessing the potential of Myb‐targeted anti‐cancer therapy. J Cancer 2018;9:1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ho AL, Dunn L, Sherman EJ et al. A phase II study of axitinib (AG‐013736) in patients with incurable adenoid cystic carcinoma. Ann Oncol 2016;27:1902–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uttarkar S, Dasse E, Coulibaly A et al. Targeting acute myeloid leukemia with a small molecule inhibitor of the Myb/p300 interaction. Blood 2016;127:1173–1182. [DOI] [PubMed] [Google Scholar]
- 72.Uttarkar S, Frampton J, Klempnauer KH. Targeting the transcription factor Myb by small‐molecule inhibitors. Exp Hematol 2017;47:31–35. [DOI] [PubMed] [Google Scholar]
- 73.Walf‐Vorderwulbecke V, Pearce K, Brooks T et al. Targeting acute myeloid leukemia by drug‐induced c‐MYB degradation. Leukemia 2018;32:882–889. [DOI] [PubMed] [Google Scholar]
- 74.Adenoid Cystic Carcinoma Research Foundation. Clinical Trials ACCRF Research Update November 2018. Available at https://myemail.constantcontact.com/ACCRF‐Research‐Update.html?soid=1102247034976&aid=KPrur38UlCI. Accessed December 22, 2018.
- 75.Ferrarotto R, Heymach JV. Taking it up a NOTCH: A novel subgroup of ACC is identified. Oncotarget 2017;8:81725–81726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bowles DW, Kochenderfer M, Cohn A et al. A randomized, phase II trial of cetuximab with or without PX‐866, an irreversible oral phosphatidylinositol 3‐kinase inhibitor, in patients with metastatic colorectal carcinoma. Clin Colorectal Cancer 2016;15:337–344.e2. [DOI] [PubMed] [Google Scholar]
- 77.Mahadevan D, Chiorean EG, Harris WB et al. Phase I pharmacokinetic and pharmacodynamic study of the pan‐PI3K/mTORC vascular targeted pro‐drug SF1126 in patients with advanced solid tumours and B‐cell malignancies. Eur J Cancer 2012;48:3319–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papadopoulos KP, Tabernero J, Markman B et al. Phase I safety, pharmacokinetic, and pharmacodynamic study of SAR245409 (XL765), a novel, orally administered PI3K/mTOR inhibitor in patients with advanced solid tumors. Clin Cancer Res 2014;20:2445–2456. [DOI] [PubMed] [Google Scholar]
- 79.Simoncini T, Hafezi‐Moghadam A, Brazil DP et al. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol‐3‐OH kinase. Nature 2000;407:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dias‐Santagata D, Akhavanfard S, David SS et al. Rapid targeted mutational analysis of human tumours: A clinical platform to guide personalized cancer medicine. EMBO Mol Med 2010;2:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dienstmann R, Dong F, Borger D et al. Standardized decision support in next generation sequencing reports of somatic cancer variants. Mol Oncol 2014;8:859–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nardi V, Sadow PM, Juric D et al. Detection of novel actionable genetic changes in salivary duct carcinoma helps direct patient treatment. Clin Cancer Res 2013;19:480–490. [DOI] [PubMed] [Google Scholar]
- 83.Sequist LV, Heist RS, Shaw AT et al. Implementing multiplexed genotyping of non‐small‐cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]