Abstract
Mitochondrial genes are placed on one molecule, which implies that they should carry consistent phylogenetic information. Following this advantage, we present a well-supported phylogeny based on mitochondrial genomes from almost 300 representatives of Passeriformes, the most numerous and differentiated Aves order. The analyses resolved the phylogenetic position of paraphyletic Basal and Transitional Oscines. Passerida occurred divided into two groups, one containing Paroidea and Sylvioidea, whereas the other, Passeroidea and Muscicapoidea. Analyses of mitogenomes showed four types of rearrangements including a duplicated control region (CR) with adjacent genes. Mapping the presence and absence of duplications onto the phylogenetic tree revealed that the duplication was the ancestral state for passerines and was maintained in early diverged lineages. Next, the duplication could be lost and occurred independently at least four times according to the most parsimonious scenario. In some lineages, two CR copies have been inherited from an ancient duplication and highly diverged, whereas in others, the second copy became similar to the first one due to concerted evolution. The second CR copies accumulated over twice as many substitutions as the first ones. However, the second CRs were not completely eliminated and were retained for a long time, which suggests that both regions can fulfill an important role in mitogenomes. Phylogenetic analyses based on CR sequences subjected to the complex evolution can produce tree topologies inconsistent with real evolutionary relationships between species. Passerines with two CRs showed a higher metabolic rate in relation to their body mass.
Keywords: concerted evolution, control region, duplication, mitochondrial DNA, Passeriformes, phylogeny
Introduction
Phylogeny and Evolution of Passeriformes
Among avian orders, Passeriformes were subjected to the most explosive radiation, which resulted in at least 6,321 currently described species (Clements et al. 2018). They constitute as many as 60% of all bird species. According to recent analyses, passerines originated and diversified in Australo-Pacific at the beginning of Cenozoic (Mayr 2013; Jarvis et al. 2014; Claramunt and Cracraft 2015; Gibb et al. 2015; Prum et al. 2015; Selvatti et al. 2015, 2017; Oliveros et al. 2019), later than the end of Cretaceous, as was previously assumed (Ericson et al. 2002; Barker et al. 2004; Aggerbeck et al. 2014; Ericson et al. 2014)—see supplementary material, Supplementary Material online, for a more detailed description and discussion about the evolution of Passeriformes. Many climatic and geologic events, for example, Oligocene glaciation and inundation of New Zealand, influenced the dispersal and diversification of passerines (Oliveros et al. 2019). Acanthisitti (or Acanthisittia), an endemic New Zealand group, was the first lineage diverged from all other passerines classified into Eupasseres, which next split into two suborders, the Suboscines (or Tyranni) and the Oscines (or Passeri). The former group evolved in Western Gondwana including Antarctica and South America, whereas the latter in the Australian region.
Suboscines diversified into two infraorders: Tyrannides, inhabiting Central and South America (Ohlson et al. 2013), and Eurylaimides, widespread in Africa, Madagascar and Southeast Asia, with one family known from South America and fossils found in Europe (Mayr and Manegold 2006; Moyle et al. 2006). Oscines, representing nearly 80% of all passerine diversity, are typically divided into two parvorders: Corvida and Passerida (or Eupasseri) (Sibley and Ahlquist 1990; Barker et al. 2002, 2004; Ericson et al. 2002), but other groups are also distinguished: Basal Oscines (the earliest diverged lineages), Corvoidea (or core Corvoidea, the main group of Corvida), and Transitional Oscines (or Basal Passerida) (Jonsson et al. 2011; Gibb et al. 2015; Selvatti et al. 2015). Passerida is a monophyletic group related with paraphyletic Corvida. Basal and Transitional Oscines (also paraphyletic and restricted to the Australian-Oceanian region) are early diverged lineages of all Oscines and those related to Passerida, respectively. Corvoidea and Passerida expanded most probably from Australasia to Eurasia and next to Africa and Americas (Selvatti et al. 2015; Moyle et al. 2016; Oliveros et al. 2019).
The rapid radiation of passerine lineages caused that the deep relationships in phylogenetic trees are poorly or inconsistently resolved across various approaches. The most extensive global avian phylogenies including a substantial number of Passeriformes were based on 259 nuclear genes and 44 passerine taxa (Prum et al. 2015) as well as 1,156 nuclear genes and 99 passerine taxa (Claramunt and Cracraft 2015). A comprehensive genome-scale analysis included 4,155 ultraconserved loci from 104 oscine passerines (Moyle et al. 2016) and 4,060 also ultraconserved nuclear genes from 209 suboscine and oscine passerines (Oliveros et al. 2019). Phylogenetic analyses focused on Passeriformes were carried out also with various combinations of markers and taxa, for example, 2 nuclear genes and 144 taxa (Barker et al. 2004), 7 nuclear genes and 55 taxa (Ericson et al. 2014), and 4 nuclear and 5 mitochondrial genes from 1,119 taxa (Selvatti et al. 2015). Two studies involved 125 (Barker 2014) and 102 complete mitochondrial (mtDNA) genomes (Gibb et al. 2015).
The latter approach seems especially promising because the mitochondrial markers show a variable evolutionary rate, which is between slowly evolving exons and rapidly changing introns (Gibb et al. 2015). Moreover, the relatively rapid evolutionary rate of mitochondrial sequences may lead to accumulation of mutations along short internodes in phylogeny, where relevant information from nuclear markers may be limited (Tamashiro et al. 2019). It was shown that mitogenomes retain important phylogenetic information in the case of passerines and performed well in recovering relationships at multiple hierarchical levels (Barker 2014). Only transitions in the third codon position revealed some level of saturation. However, their influence on the phylogenetic tree reconstruction was negligible, probably due to compensating effect of the applied partitioning model (Barker 2014). In fact, more complex substitution models often improve phylogenetic trees based on mitochondrial sequences and produce accurate trees (Braun and Kimball 2002; Leavitt et al. 2013; Tamashiro et al. 2019).
Moreover, the mtDNA genes are located on one molecule and are inherited together (Moore 1995; Berlin and Ellegren 2001). Thus, the individual markers should bear the same phylogenetic signal. It contrasts with nuclear genes which are more susceptible to incomplete lineage sorting and hidden gene paralogy (Moore 1995; Maddison 1997; Page 2000; Gribaldo and Philippe 2002; Martin and Burg 2002; Feiner et al. 2009; Kuraku 2013, 2010). Such nonorthologous genes can cause disagreement between gene and species trees. Thereby, it is reasonable to include mtDNA markers in phylogenetic analyses because they can provide consistent trees with well-resolved deep nodes as well as between later diverged clades. Much better results are obtained when concatenated alignments of many mitochondrial genes are used than those based on single markers due to a greater probability of stochastic errors in the latter case (Russo et al. 1996; Miya and Nishida 2000; Rohland et al. 2007; San Mauro et al. 2009; Willerslev et al. 2009; Duchene et al. 2011; Talavera and Vila 2011; Lambret-Frotte et al. 2012; Havird and Santos 2014). In agreement with that, quite well-resolved phylogenies based on avian mitogenomes were also received by many authors (Slack et al. 2007; Pacheco et al. 2011; Powell et al. 2013; Meikejohn et al. 2014; Urantowka et al. 2017b; Tamashiro et al. 2019).
Duplications in Mitochondrial Genomes
Mitochondrial genomes have become interesting also because of their compact structure and organization as well as their relation to main transitions in animal evolution (Lavrov 2011, 2007). Contrary to the previous view assuming that vertebrate mitogenomes are highly conserved in terms of the gene content and order (Clayton 1991; Boore 1999), more and more data, also about Aves, indicate that the control region (CR) with neighboring genes can be subjected to tandem duplications and subsequent degenerations of the duplicated copies. The mitogenomes containing such duplication were found in many avian orders: Accipitriformes (Gibb et al. 2007), Bucerotiformes (Sammler et al. 2011), Coraciiformes (Huang et al. 2016), Cuculiformes (Pratt et al. 2009; Pacheco et al. 2011; Wang, Liang, et al. 2016), Falconiformes (Gibb et al. 2007; Ryu et al. 2012; Dou et al. 2016; Wang, Zhang, et al. 2016; Sveinsdottir et al. 2017), Gruiformes (Akiyama et al. 2017), Pelecaniformes (Zhou et al. 2014), Phoenicopteriformes (Morgan-Richards et al. 2008; Luo et al. 2016), Piciformes (Gibb et al. 2007), Procellariiformes (Abbott et al. 2005; Gibb et al. 2007, 2013, 2015; Lounsberry et al. 2015), Psittaciformes (Eberhard et al. 2001; Schirtzinger et al. 2012; Eberhard and Wright 2016; Urantowka et al. 2017a, 2018), Strigiformes (Hanna et al. 2017), Suliformes (Morris-Pocock et al. 2010) as well as Passeriformes (Mindell et al. 1998; Bensch and Harlid 2000; Alstrom et al. 2006; Singh et al. 2008; Cerasale et al. 2012; Cooke et al. 2012; Wang et al. 2015; Caparroz et al. 2018).
It is commonly assumed that the mitogenomic duplications are derived states and occur independently in many lineages of a given order, for example, parrots and passerines (Mindell et al. 1998; Alstrom et al. 2006; Schirtzinger et al. 2012; Eberhard and Wright 2016; Caparroz et al. 2018). However, such a view can result from the usage of the standard polymerase chain reaction strategy that misses the duplication. Therefore, an appropriate methodology should be applied and gene orders should be reassigned in many mitogenomes (Gibb et al. 2007; Singh et al. 2008). In agreement with that, reanalyses of some crane, passerine and ardeid mitogenomes (Singh et al. 2008; Zhou et al. 2014; Gibb et al. 2015; Akiyama et al. 2017) showed the existence of duplicated regions, although previously the single version was found. Moreover, the appropriate approach revealed that duplication was the ancestral rather than derived state for parrots (Urantowka et al. 2018). Taking into account this finding as well as that Psittaciformes and Passeriformes are sister groups (Ericson et al. 2006; Hackett et al. 2008; Suh et al. 2011; Jarvis et al. 2014; Claramunt and Cracraft 2015; Prum et al. 2015), we can assume that the mitogenomic duplication was also plesiomorphic for passerines. Therefore, the aim of this study was to obtain a robust phylogeny of Passeriformes based on mitochondrial genomes and mapping on it the presence and absence of the mitogenomic duplications to verify the concept about their ancestry and reconstruct the evolution of the duplicated regions. We also included the mitogenomes of Falconiformes in this study, because this avian group constitutes an order related to Passeriformes (Ericson et al. 2006; Hackett et al. 2008; Suh et al. 2011; Jarvis et al. 2014; Claramunt and Cracraft 2015; Prum et al. 2015), so is important in the determination of ancestral state in the latter. Psittaciformes and Passeriformes are considered sister to each other and form a group named Psittacopasserae, which in turn is sister to Falconiformes, so all three orders are lumped together under the name Eufalconimorphae (Suh et al. 2011). Thanks to the comparison of passerine mitogenomes with those in parrots and falcons, we were able to present the evolution of duplications in these genomes in much broader context. The phylogenetic analyses based on almost 300 passerine mitogenomes also enabled us to obtain better resolved relationships with taxonomic implications.
Materials and Methods
Phylogenetic Analyses
Phylogenetic relationships between passerines were inferred using all available 294 complete mitochondrial genomes of Passeriformes (supplementary table S1, Supplementary Material online) and 5 representatives of Psittaciformes used as an outgroup: Coracopsis vasa (KM611468.1), Myiopsitta monachus (KM611471.1), Nestor notabilis (MH133967.1), Nymphicus hollandicus (MH133968.1), and Psittacus erithacus (KM611474.1). The sequences were downloaded from the GenBank database. From the mitogenomic records, we selected sequences of 13 protein coding genes (PCGs), 12S and 16S rRNAs, as well as 22 tRNAs. Phylogenetic analyses of CR comprised five sets: Suboscines, Turdidae, Petroicidae, Sylvioidea, and Falconiformes (supplementary table S2, Supplementary Material online). The sequences were aligned in MAFFT using a slow and accurate algorithm L-INS-i with 1,000 cycles of iterative refinement (Katoh and Standley 2013). The resulted alignments were edited manually in JalView (Waterhouse et al. 2009) and sites suitable for phylogenetic study were selected in GBlocks (Talavera and Castresana 2007). The concatenated alignment of mitochondrial genes consisted of 15,307 bp, whereas the CR alignments contained 694–1,175 bp (supplementary table S2, Supplementary Material online).
Three phylogenetic approaches were applied to infer evolutionary relationships between passerines: the maximum likelihood (ML) method in IQ-TREE (Nguyen et al. 2015), as well as two Bayesian analyses, in MrBayes (Ronquist et al. 2012) and PhyloBayes (Lartillot and Philippe 2004). In order to check the necessity of using separate nucleotide substitution models for the mitogenomic data, we considered 63 potential partitions, that is, 3 codon positions for each individual PCG and separate partitions for each of the RNA genes (supplementary table S3, Supplementary Material online).
The ModelFinder program associated with IQ-TREE (Chernomor et al. 2016; Kalyaanamoorthy et al. 2017) provided three substitution models for the defined partitions of the mitochondrial gene set (supplementary table S3, Supplementary Material online). For the CR alignments, we selected the best one-partition model (supplementary table S2, Supplementary Material online). In IQ-TREE, we applied Shimodara–Hasegawa-like approximate likelihood ratio test (SH-aLRT) assuming 10,000 replicates and nonparametric bootstrap with 1,000 replicates. In the tree search, we used more thorough and slower NNI search.
In MrBayes analyses of the mitogenomic set, we assumed 26 substitution models for the appropriate partitions according to the results of PartitionFinder (Lanfear et al. 2012) using BIC criterion for the model selection (supplementary table S3, Supplementary Material online). However, we applied mixed models rather than fixed ones to specify appropriate substitution models across the large parameter space (Huelsenbeck et al. 2004), but the models describing heterogeneity rate across sites were adopted according to PartitionFinder. In the case of CR sets, we assumed the mixed models and the heterogeneity rate across sites described by the discrete gamma model according to results of jModelTest 2.1 (Darriba et al. 2012). Two independent runs starting from random trees, each using 32 Markov chains, were applied for the mitogenomic set. For the largest set of CRs from Sylvioidea, we assumed eight Markov chains and for the rest CR sets, four chains. The trees were sampled every 100 generations for 40,000,000 generations for the mitogenomic and Sylvioidea CR set, and for 10,000,000 generations in the case of the other CR sets. In the final analysis, we selected trees from the last 4,285,000 to 20,749,000 generations (depending on the alignment set) that reached the stationary phase and convergence, that is, when the standard deviation of split frequencies stabilized and was much below the proposed 0.01 threshold.
In PhyloBayes, we applied the CAT + GTR + Γ model and the number of components, weights, and profiles of the applied model were inferred from the data. Two independent Markov chains were run for 100,000 generations with 1 tree sampled for each generation. The last 75,000–95,000 trees (depending on the alignment set) from each chain were collected to compute posterior consensus trees after reaching convergence, when the largest discrepancy observed across all bipartitions (maxdiff) was below the recommended threshold 0.1.
The gamma-distributed rate variation across the sites was approximated by five discrete rate categories in Bayesian approaches. Tip-to-root distances for Sylvioidea and Paroidea were calculated in TempEst (Rambaut et al. 2016). Trees were edited in FigTree (Rambaut 2012) and TreeGraph (Stover and Muller 2010).
The data about the presence and absence of duplication in the passerine mitogenomes were mapped on the MrBayes tree using Mesquite (Maddison and Maddison 2017). The lack of data about the duplication was coded as missing data. We applied maximum parsimony (MP) and ML reconstruction methods. In the latter case, we used Mk1 model (Markov k-state 1 parameter model) because it fit the data better according to AIC criterion than the alternative AsymmMk model (asymmetrical Markov k-state 2 parameter model).
Other Bioinformatic and Statistical Analyses
The normalized differences in complementary nucleotides were measured as AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) (Perna and Kocher 1995; Reyes et al. 1998). These parameters were calculated for the L-strand sequence of complete genomes, as well as for all PCGs and 4-fold degenerated (4FD) sites. The composition of sites in the ND6 gene, whose sense strand is located in the genomic H-strand, was determined from the complementary L-strand. We analyzed in this way complete mitogenomes from 10 Falconiformes, 271 Passeriformes (71 with the duplicated region and 200 with the single version), and 47 Psittaciformes (23 with the duplicated region and 24 with the single version). The data for parrots were taken from Urantowka et al. (2018).
The program water from EMBOSS package (Rice et al. 2000) based on the Smith–Waterman algorithm was used to calculate the local alignment of two CR sequences.
The data on body mass (M) and basal metabolic rate (BMR) for Passeriformes were collected from AnAge database (http://genomics.senescence.info/species) and various references (Hails 1983; Daan et al. 1990; Tieleman and Williams 2000; Rezende et al. 2002; McKechnie et al. 2006; White et al. 2007; Wiersma et al. 2007; Makarieva et al. 2008; McNab 2009; Sabat et al. 2010; Londono et al. 2015; Bech et al. 2016; Bushuev et al. 2018). We averaged values from various sources and expressed BMR in watt (W). After extensive literature survey, we found information for 11 passerines containing the duplicated region in their mitogenome and for 53 passerines without this region, 38 of them have M in the range of the first set (supplementary table S4, Supplementary Material online). The relationship between BMR and M was modeled using the linear model log10(BMR) = a × log10(M) + b.
In order to check if the analyzed variables are normally distributed, we used the Shapiro–Wilk test. The homogeneity of variance across the studied groups was verified with the Levene test. Because these assumptions were not fulfilled, the nonparametric unpaired Wilcoxon–Mann–Whitney tests was applied in the comparison of the length of single versus duplicated CRs, body mass, and BMR of the two groups of passerines as well as tip-to-root distances in the MrBayes subtree between Sylvioidea (with duplicated CR) and Paroidea (without duplicated CR), whereas the Dunn’s test following Kruskal–Wallis test was used in the multiple comparisons of the CR lengths as well as AT- and GC-skew between three bird orders. In this multiple testing, P values were corrected using the Benjamini and Hochberg method. Body mass-specific metabolic rate (BMR/M) as well as the ratio of the logarithm from average maximum life span to the logarithm from body mass of the passerines with duplicated and single CR was compared in t-test. The paired Wilcoxon signed rank test with continuity correction was applied in the comparison of the length distribution between two duplicated CRs within a given avian order. The null hypotheses were rejected at the 0.05 level. The statistical analyses were carried out in R package 3.5.1 (R Core Team 2018).
Results and Discussion
Phylogenetic Relationships between Passerines
Quality of Phylogenetic Inferring and High-Level Relationships
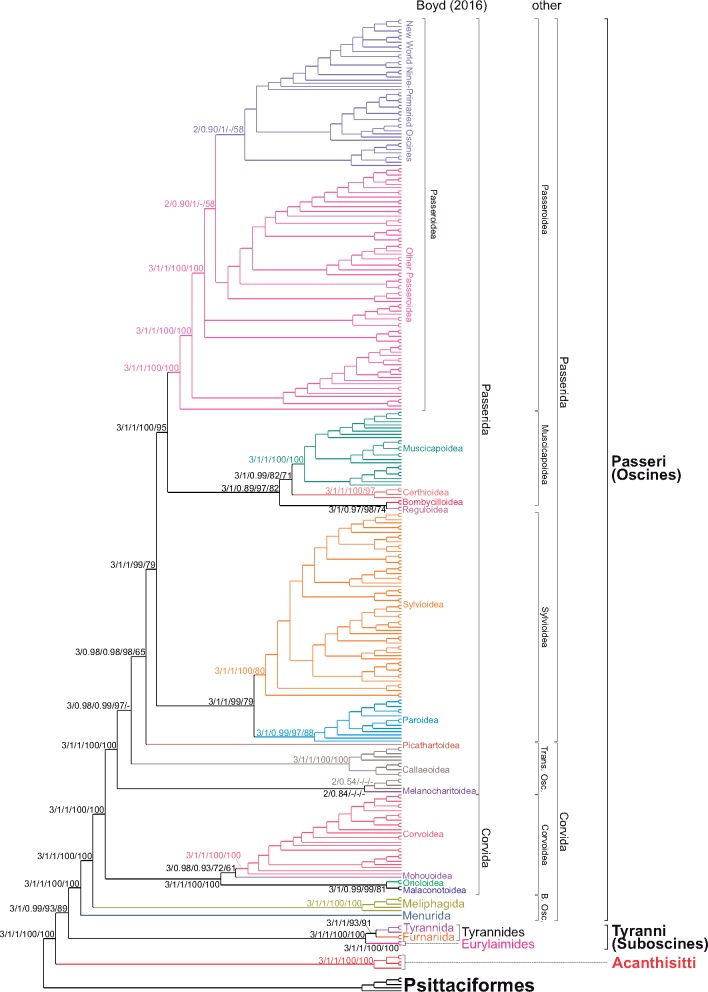
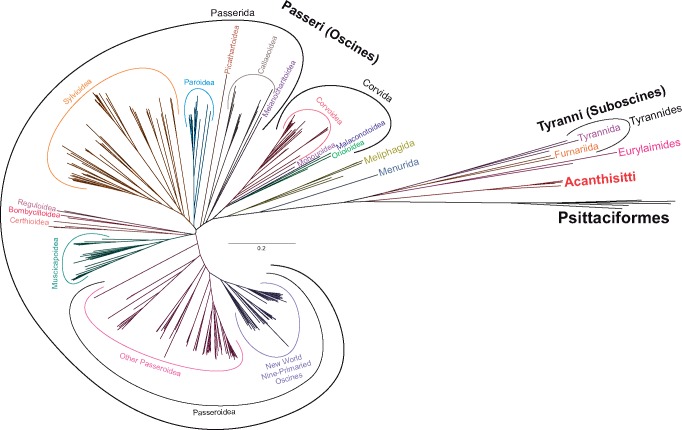
Despite the huge data set consisting of 299 taxa and 15,307 characters, we applied partitioned substitution models and 3 advanced approaches, 2 Bayesian in MrBayes and PhyloBayes as well as the ML in IQ-TREE. We decided to use the latter instead of faster and commonly used RAxML/ExaML or FastTree because IQ-TREE turned out to outperform the other programs in various tests (Zhou et al. 2018). The three approaches produced very consistent well-resolved tree topologies. The consensus cladogram of three methods is presented in figure 1 and supplementary figure S1, Supplementary Material online, whereas the phylogram calculated in MrBayes in figure 2 and supplementary figure S2, Supplementary Material online. As many as 90% and 88% out of all 296 nodes obtained posterior probability (PP) >0.95 in MrBayes and PhyloBayes, respectively. Only two and seven nodes were supported with PP smaller than 0.5, respectively. In consequence, the average PP was 0.98 and 0.97 for the resolved nodes in MrBayes and PhyloBayes. In the case of IQ-TREE, 82% of nodes received support larger than 95 in SH-aLRT approach and 81% of nodes were supported by bootstrap percentage (BP) >75. Only 20 and 28 nodes obtained values smaller than 50. The average values of SH-aLRT and BP were 97 and 94, respectively. The applied methods reproduced the same deep nodes, most of which obtained very high support values.
Fig. 1.
—The simplified consensus cladogram of three trees obtained in MrBayes, PhyloBayes, and IQ-TREE based on passerine mitogenomic genes. Representatives of Psittaciformes were used as an outgroup. The values at nodes, in the following order N/MB/PB/SH/BP, indicate the number of trees containing a given node (N), posterior probabilities found in MrBayes (MB), and PhyloBayes (PB) as well as SH-aLRT (SH) and nonparametric bootstrap (BP) percentages calculated in IQ-TREE. The posterior probabilities <0.5 and the percentages <50% were omitted or indicated by a dash “-.” Two distinct classifications of Passeriformes at the medium taxonomic level were presented. B. Osc., Basal Oscines; Trans. Osc., Transitional Oscines. See supplementary figure S1, Supplementary Material online, for the full tree with species and family names as well as support values for all nodes.
Fig. 2.
—The phylogram obtained in MrBayes based on passerine mitogenomic genes. Representatives of Psittaciformes were used as an outgroup. See supplementary figure S2, Supplementary Material online, for the full tree with species and family names as well as support values.
The relationships between the high-level groups of Passeriformes, that is, Acanthisitti, Suboscines, and Oscines, and their main subgroups are consistent with the results of other authors (Barker et al. 2004; Barker 2014; Ericson et al. 2014; Claramunt and Cracraft 2015; Gibb et al. 2015; Prum et al. 2015; Selvatti et al. 2015; Moyle et al. 2016; Oliveros et al. 2019), but we received much higher support by more methods in many cases. See figures 1 and 2 and supplementary figures S1 and S2, Supplementary Material online, for phylogenetic trees based on the complete mitochondrial genomes and supplementary material, Supplementary Material online, for a more detailed description and discussion of the obtained phylogenies. Below we focused on differences and controversial groupings.
Relationships among Corvides and Transitional Oscines
Earlier diverged lineages of Corvides, that is, Campephagidae, Oriolidae, and Vireonidae, were significantly grouped in all three methods applied by us (PPMrBayes = 1, PPPhyloBayes = 0.99, aLRT = 99, BP = 81). These relationships differ from other phylogenies in the position of Campephagidae, which created a separate lineage (Jonsson et al. 2016; Moyle et al. 2016; Oliveros et al. 2019). Oriolidae and Vireonidae were separated in our analyses, as in Barker et al. (2004) and Claramunt and Cracraft (2015), but unlike others (Jonsson et al. 2011, 2016; Aggerbeck et al. 2014; Ericson et al. 2014; Selvatti et al. 2015; Moyle et al. 2016; Oliveros et al. 2019). Mohoua clustered in all three trees (PPMrBayes = 0.98, PPPhyloBayes = 0.93, aLRT = 72, BP = 61) with the highly supported clade including Rhipiduridae, Monarchidae, Laniidae, and Corvidae (supplementary fig. S1, Supplementary Material online). According to other authors, Mohoua diverged after Campephagidae and before the divergence of the mentioned families (Moyle et al. 2016; Oliveros et al. 2019), or even earlier (Aggerbeck et al. 2014; Claramunt and Cracraft 2015).
The position of the Transitional Oscines depends on the method used. The earliest diverged lineage included Melanocharitidae as well as the clade of Notiomystidae and Callaeidae in IQ-TREE and MrBayes, but in the PhyloBayes tree, Callaeidae branched off first. Unfortunately, these relationships were not highly supported (supplementary fig. S1, Supplementary Material online) but were also obtained for selected taxa in most approaches applied by Barker (2014). Notiomystidae and Callaeidae were also grouped in the trees received by Selvatti et al. (2015) and Oliveros et al. (2019), but Melanocharitidae was sister to the rest Transitional Oscines and Passerida. However, all three methods used by us showed with high support (PP ≥ 0.98 and aLRT ≥ 97) that Petroicidae diverged before Picathartidae, as in Selvatti et al. (2015). It contrasts with other results, in which the order was reversed (Barker et al. 2004; Beresford et al. 2005; Jonsson et al. 2011) or these families were grouped together (Jonsson et al. 2007; Moyle et al. 2016; Oliveros et al. 2019).
The closer relationship of Picathartidae with Passerida was interpreted that the ancestor of the latter evolved in Africa (Beresford et al. 2005; Jonsson and Fjeldsa 2006; Jonsson et al. 2007), because the majority of picathartes live on this continent. The opposite view assumes that Australasian Petroicidae is sister to Passerida, which then inhabited continental Asia from Australia (Barker et al. 2002, 2004; Johansson et al. 2008). However, a member of Picathartidae, Eupetes macrocerus, inhabits Southeast Asia, which suggests that picathartes could be originally widespread in Asia and migrated to Africa later. Thereby, the dispersal of the Passerida ancestor from Australia to Asia is also possible under the tree topology obtained in this study. It agrees with the recent estimations of divergence time and biogeographic analyses (Selvatti et al. 2015; Moyle et al. 2016; Oliveros et al. 2019), see supplemental material, Supplementary Material online, for discussion.
Relationships among Passerida
Among Passerida, four main groups can be distinguished: Passeroidea, Muscicapoidea, Sylvioidea (Sibley and Ahlquist 1990; Ericson and Johansson 2003), and Paroidea (Nabholz et al. 2010; Alstrom et al. 2014), but relationships between them depend on the data and methods used.
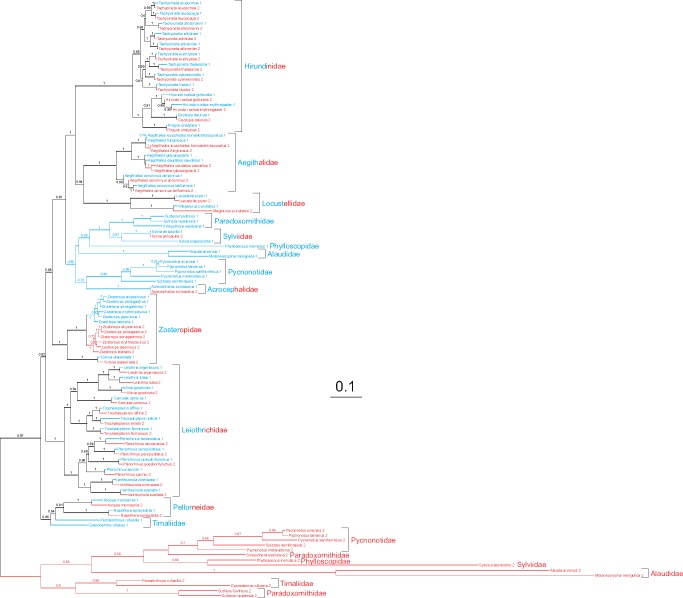
Our results are consistent with most other analyses based on big data sets (Barker et al. 2004; Barker 2014; Ericson et al. 2014; Claramunt and Cracraft 2015; Gibb et al. 2015; Prum et al. 2015; Selvatti et al. 2015; Moyle et al. 2016; Oliveros et al. 2019) indicating that alternative topologies are less probable (Johansson et al. 2008; Nabholz et al. 2010; Alstrom et al. 2014; Wu et al. 2015). Trees obtained by us are also better resolved than those by other authors (figs. 1, 2, and supplementary fig. S1, Supplementary Material online). Sylvioidea (sensu stricto) and Paroidea create one significant clade with PPs = 1, aLRT = 99, and BP = 79. Sylvioidea and Paroidea are also very well supported individually (PPs ≥ 0.99, aLRT ≥ 97, BP ≥ 80). The two other groups, Passeroidea and Muscicapoidea (sensu lato) are also significantly clustered with PPs = 1, aLRT = 100, and BP = 95. The former received all four support values maximal and the latter had PPMrBayes = 1, PPPhyloBayes = 0.89, aLRT = 97, and BP = 82.
Relationships among families in our analyses are generally the same as in phylogenomic studies by Moyle et al. (2016) and Oliveros et al. (2019) except for 1) Hyliotidae, which was sister to other Paroidea and Sylvioidea representatives, but in our trees, this family significantly clustered with Paridae and Remizidae (PPMrBayes = 1, PPPhyloBayes = 0.99, aLRT = 97, BP = 88) and 2) Regulidae, which grouped with Certhioidea, but in our phylogenies, it was sister to Bombycilloidea (PPMrBayes = 1, PPPhyloBayes = 0.97, aLRT = 98, BP = 74).
Taxonomic Implications
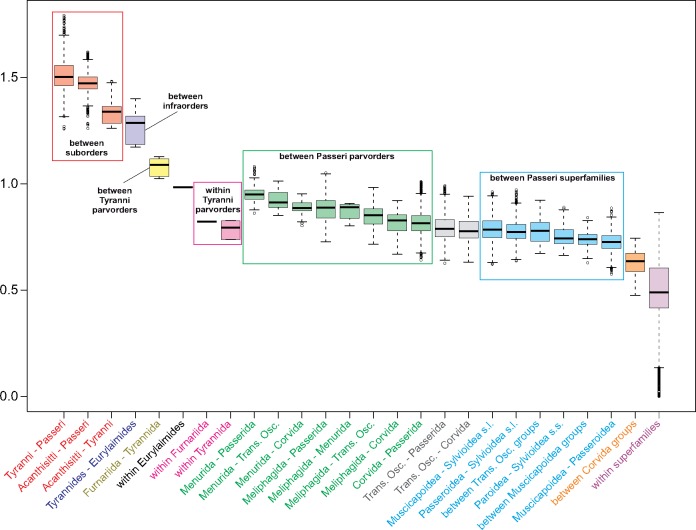
Since the influential work by Sibley and Ahlquist (1990), many classification schemes of Passeriformes have been proposed. Some of them were mentioned in the discussion of the phylogenetic analyses and presented in figure 1 and supplementary figure S1, Supplementary Material online. Some lineages with the same taxonomic rank are characterized by different genetic differentiation level, and groups with different taxonomic rank can have the same genetic diversity. To assess the lineage variation, we calculated phylogenetic distances (measured as the average number of nucleotide substitutions per site) based on the MrBayes tree between pairs of various lineages and within them (fig. 3). As expected, the divergence between suborders is the largest, whereas between two infraorders of Suboscines slightly smaller. Tyranni parvorders show a larger distance between themselves than parvorders of Passeri, but intraparvordes distances of Tyranni are of the same order as those of Passeri. The distances between and within Passeri parvorders can be arranged in a set of overlapping distributions from the largest to the smallest values. The intergroup distances are located in the first half of this arrangement, whereas the intragroup distances in the second half. As expected, the smallest values are for comparisons within superfamilies.
Fig. 3.
—Box-plots of phylogenetic distances (measured as the average number of nucleotide substitutions per site) based on the MrBayes tree between pairs of various passerine lineages and within them. The thick line indicates median, the boxes show quartile range, and the whiskers denote the range without outliers. Trans. Osc., Transitional Oscines; Corvida is considered as in Boyd (2016), that is, does not include Basal and Transitional Oscines; Muscicapoidea is considered as in the traditional classification, that is, it includes groups, which have the level of superfamily in Boyd (2016): Bombycilloidea, Certhioidea, core Muscicapoidea, and Reguloidea; Passerida is considered as in the traditional classification, that is, does not include Transitional Oscines; Sylvoidea s.l. (sensu lato) is considered as in the traditional classification, that is, it includes also Paroidea; Sylvoidea s.s. (sensu stricto) is considered as in Boyd (2016), that is, it does not include Paroidea.
In the light of our and previous studies, for example, Selvatti et al. (2015) and Oliveros et al. (2019), Basal Oscines and Transitional Oscines are undoubtedly paraphyletic groups. Therefore, Boyd’s classification excluding these lineages from Corvida seems reasonable (Boyd 2016). Thereby, Corvida becomes monophyletic, whereas the individual lineages of Basal Oscines obtain the rank of parvorders, for example, Menurida and Meliphagida. In this classification, the parvorder Corvida would consist of only one superfamily Corvoidea, if we assume the traditional classification (Sibley and Ahlquist 1990). Alternatively, according to Boyd (2016), Corvida is split into several superfamilies, for example, Malaconotoidea, Orioloidea, Mohouoidea, and Corvoidea. We compared the phylogenetic distances calculated between these groups with inter- and intra-superfamilies distances (fig. 3). If these groups represent the superfamily rank, the distances should be of the same order as between other well-established superfamilies of Passeri. The results, however, are not unambiguous because the distribution of distances between the Corvida groups is located between the distributions of inter- and intra-superfamilies distances. Nevertheless, the median value of the distances between the Corvida groups is closer to that calculated for the distances between other superfamilies. Thus, the superfamily level of these groups could be justified.
In order to avoid the paraphyly of Transitional Oscines, it seems sensible and concordant with the phylogenetic analyses to extend parvorder Passerida by these lineages as in Boyd’s classification (Boyd 2016), which makes the whole Passerida monophyletic. The lineages of Transitional Oscines are represented in this classification as superfamilies, which corresponds to the phylogenetic distances calculated between them. These distances are comparable to those between other superfamilies of Passeri (fig. 3).
In terms of the phylogenetic distances as well as tree topology, it is also justified to recognize Paroidea and Sylvoidea as two superfamilies, which was proposed by Alstrom et al. (2014) and Boyd (2016). The separation of these groups is also substantiated by their mitogenome organization. All known mitogenomes of Sylvoidea comprise a duplicated region, but none of the Paroidea members (see below). Alstrom et al. (2014) and Boyd (2016) elevated clades formerly classified into one group Muscicapoidea also to the level of superfamily. This proposition is supported by our comparisons because the phylogenetic distances between these clades (Bombycilloidea, Certhioidea, core Muscicapoidea, and Reguloidea) are in the same range as those between other superfamilies (fig. 3). The median of these distances is even larger than that calculated for the distances between Passeroidea and these clades of Muscicapoidea considered as a whole.
Duplications in Passerine Mitogenomes
General Characteristic of Rearrangement Types
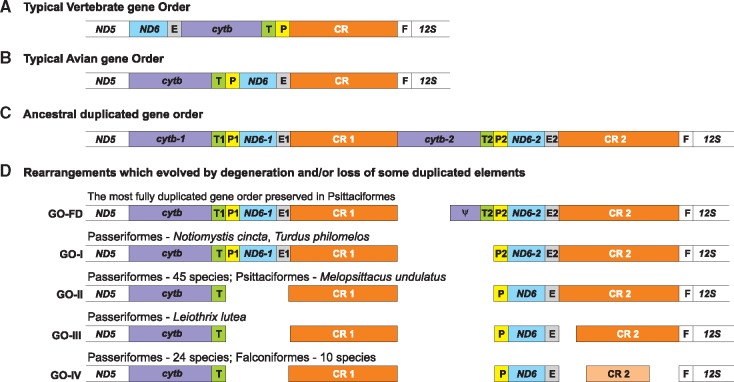
Mitochondrial genomes with duplicated regions have been found so far in 71 passerines (supplementary table S1, Supplementary Material online). Two hundred mitogenomes do not have such regions reported and 22 mitochondrial genomes are not completely sequenced, so it is not possible to classify them to one of these groups. In the case of Acanthisitta chloris, it is highly probable that duplication exists in its mitogenome because the sequencing of the region from tRNA-Phe gene to the CR proved difficult without the cloning of DNA fragments, on account of the presence of repeats and heteroplasmy (Harrison et al. 2004). The same explanation refers also to the parrot mitogenome of Strigops (Urantowka et al. 2018). Four types of gene orders can be recognized in Passeriformes. They are presented in figure 4 and compared with other gene rearrangements, typical of vertebrates and birds, as well as the most fully duplicated gene order preserved in Psittaciformes and a rearrangement reported in Falconiformes. These two latter avian orders occurred closely related with Passeriformes in many analyses (Ericson et al. 2006; Hackett et al. 2008; Suh et al. 2011; Jarvis et al. 2014; Claramunt and Cracraft 2015; Prum et al. 2015).
Fig. 4.
—The comparison of mitochondrial gene orders between ND5 and 12S rRNA for a typical vertebrate gene order (A), a typical avian gene order (B), an ancestral duplicated gene order assuming the tandem duplication of the cytb to CR segment (C), and rearrangements which evolved by degeneration and/or loss of some duplicated elements in Psittaciformes, Passeriformes, and Falconiformes (D). ND5, NADH dehydrogenase subunit 5 gene; cytb, cytochrome b gene; T, tRNA gene for threonine; P, tRNA gene for proline; ND6, NADH dehydrogenase subunit 6; E, tRNA gene for glutamic acid; CR, control region; F, tRNA gene for phenylalanine; 12S, 12S rRNA gene. Pseudogenes are marked by ψ and colored correspondingly to their functional gene copy.
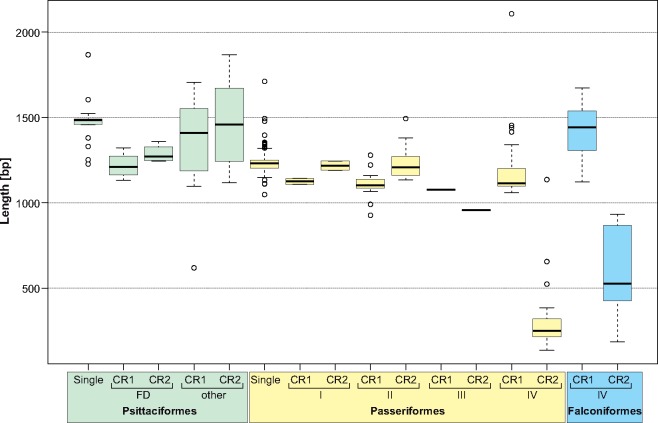
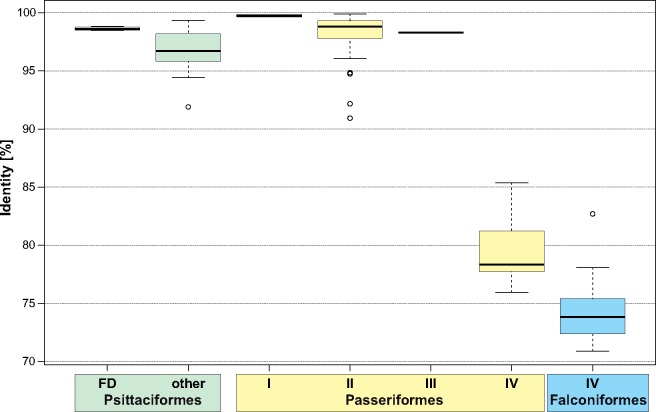
The region that is most often subjected to duplication in Aves contains genes: cytb, tRNA-Thr, tRNA-Pro, Nd6, and tRNA-Glu, as well as the CR (fig. 4). However, the complete duplication is very rarely preserved in the considered avian groups. The most complete version with the second copy of cytb pseudogenized was found only in four parrots (Urantowka et al. 2018) and is named in figure 4 as GO-FD. The second copy of the control region (CR2) is always slightly longer than the first one (CR1) in these mitogenomes (fig. 5). The average and the range are 1,287 bp (1,243–1,360 bp) and 1,218 bp (1,131–1,322 bp), respectively. The CR sequences show on average 98.6% (98.5–98.8%) identity and 3.9% (1.2–7.9%) gaps in the local alignment (fig. 6). The more reduced versions of gene rearrangements reported in other Psittaciformes retained the similar parameters for the CRs. The average length of CR2 is 1,452 bp (1,117–1,867 bp) and CR1 is 1,344 bp (619–1,705 bp). The CRs show slightly weaker sequence conservation: 97% (91.9–99.3%) identity and 9% (0.1–58.1%) gaps.
Fig. 5.
—Box-plots of the control region (CR) length for Psittaciformes, Passeriformes, and Falconiformes. The thick line indicates median, the boxes show quartile range, and the whiskers denote the range without outliers. Results for various types of CRs and gene orders were shown. Single, CR present in one copy; CR1 and CR2, two copies in one mitogenome; FD, the most fully duplicated gene order preserved in Psittaciformes; I–IV, gene orders found in Passeriformes shown in figure 4.
Fig. 6.
—Box-plots of the identity percent between two copies of control region in one mitogenome for Psittaciformes, Passeriformes, and Falconiformes. Other explanations as in figure 5.
In two passerine mitogenomes, Notiomystis cincta and Turdus philomelos (Gibb et al. 2015), the second copies of cytb gene and tRNA-Thr were finally lost but other elements were maintained (GO-I in fig. 4). The CR2 is also slightly longer (1,245 and 1,191 bp) than the CR1 (1,143 and 1,108 bp), respectively (fig. 5). Both regions maintained a very high identity, 99.6% and 99.8% in these two mitogenomes, respectively (fig. 6).
The majority of Passeriformes, that is, 45 species, have lost in addition to cytb and tRNA-Thr, the first adjacent copies of tRNA-Pro, Nd6, and tRNA-Glu (GO-II in fig. 4). In these mitogenomes, the second copies of these genes have been preserved and seem to be functional. The length of CR2 is a little larger than that of CR1: 1,230 bp (1,133–1,493 bp) versus 1,111 bp (927–1,279 bp)—figure 5. In only one case, Tachycineta thalassina, the CR2 was shorter than the CR1 by 18 bp. The CRs in the GO-II type are also highly conserved with 98% (90.9–99.9%) identity and 2% (0.1–7.7%) gaps (fig. 6). Such rearrangement type was also found in the parrot Melopsittacus undulates with 1,307-bp CR1 and 1,378-bp CR2, showing 95.6% identity and 10.8% gaps.
In the passerine Leiothrix lutea, the reduction went further because the second CR lost almost 200 nucleotides at the beginning (GO-III in fig. 4). However, the preserved region maintained a high sequence identity, that is, 98.3% with 10.4% gaps (fig. 6). The more advanced level of degradation was reached by 24 mitogenomes of Passeriformes, in which the second copy of CR significantly shortened by 900 bp on average and diverged at the sequence level from the first one (GO-IV in fig. 4). The average length and ranges of CR2 and CR1 in this type are 312 bp (136–1,136 bp) and 1,207 bp (1,058–2,107 bp), whereas the identity percent is 79% (76.0–85.4%) and gaps is 39% (17.3–50.4%) (figs. 5 and 6). This stage is also represented by all ten known mitogenomes of Falconiformes. The CR2 is on average 800 bp shorter than CR1: 599 bp (186–932 bp) versus 1,427 bp (1,122–1,673 bp). These regions show even greater sequence divergence, that is,75% (70.9–82.7%) identity with 48% (42–54.7%) gaps in the local alignment.
Generally, the CRs of Passeriformes (the average 1,215 bp, excluding the highly reduced CR2 in GO-IV) are significantly shorter (P < 0.0002) than in Psittaciformes (1,407 bp) and Falconiformes (1,427 bp, excluding the highly reduced CR2)—figure 5. The length of the single versions of CR in parrot mitogenomes (1,473 bp) is also significantly greater than CR1 (1,321 bp, P = 0.042) but not CR2 (1,422 bp, P = 0.073). In the case of passerines, the single CRs (1,238 bp) are significantly longer (P < 0.00002) from CR1 in the gene orders I/II (1,112 bp) and III/IV (1,201 bp) as well as CR2 in GO-III/IV (338 bp) but its length is comparable with CR2 in GO-I/II (1,229 bp, P = 0.21). There is a common trend in the shortening of CR1 compared with CR2 in Psittaciformes (1,321 bp vs. 1,422 bp, P = 0.0011) as well as in Passeriformes mitogenomes containing the gene orders I and II (1,112 bp vs. 1,229 bp, P = 4 × 10−9). On the other hand, the falcon mitogenomes and passerine mitogenomes with the gene orders III and IV have a significantly shorter CR2 than CR1: 599 bp versus 1,427 bp (P = 0.002), and 338 bp versus 1,201 bp (P = 6 × 10−8), respectively.
Phylogenetic Distribution of Rearrangement Types
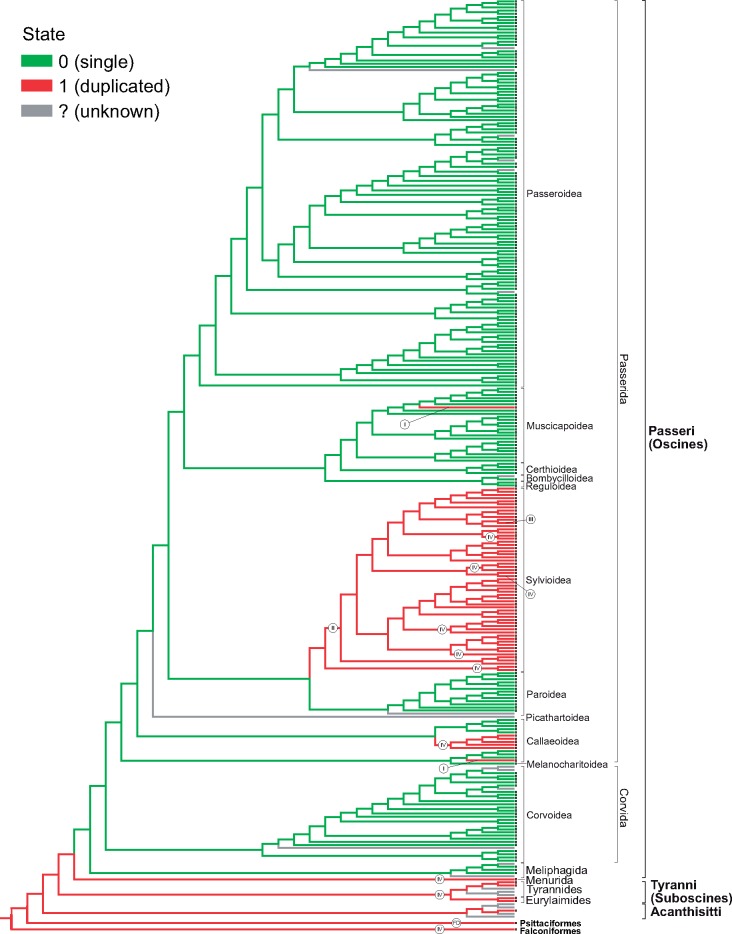
Using the phylogenetic relationships inferred from mitogenomes, we mapped onto it the presence and absence of the mitogenome duplication (fig. 7 and supplementary figs. S3 and S4, Supplementary Material online). The four types of rearrangements described in Passeriformes were differently distributed in the tree. The GO-I type could occur independently because it was found in two unrelated lineages: one known representative of Notiomystidae (Callaeoidea, Transitional Oscines), that is, N.cincta, and one out of seven species of Turdus (Turdidae, core Muscicapoidea), that is, T. philomelos. The relative completeness of their duplicated region suggests that its duplication could occur quite recently.
Fig. 7.
—The MP reconstruction of ancestral states and mapping of mitogenomic duplications onto the phylogenetic tree of passerines with verified presence or absence of the duplication. The parrot and falcon lineages are also included. Gene orders I–IV were marked at appropriate lineages. See supplementary figure S3, Supplementary Material online, for the full tree with species and family names.
The GO-II is more common and exclusively present in many members of Sylvioidea, sensu Alstrom et al. (2014) and Boyd (2016), namely all representatives of the families: Acrocephalidae, Aegithalidae, Hirundinidae, Locustellidae, Pellorneidae, and Zosteropidae (fig. 7 and supplementary figs. S3 and S4, Supplementary Material online). This gene order occurs also in the mitogenomes of all Leiothrichidae, except for Leiothrix lutea, which has the GO-III. One member of Sylvidae, Sylvia atricapilla has also the GO-II type, whereas the second, that is, Sylvia crassirostris has GO-IV (Singh et al. 2008). The latter type is more widespread because it was also found in the mitogenomes of other Sylvioidea families: Alaudidae, Paradoxornithidae, Phylloscopidae, Pycnonotidae, and Timaliidae, as well as earlier diverged passerine lineages: genus Petroica (Callaeoidea, Transitional Oscines) (Cooke et al. 2012), Menura novaehollandiae (Menurida, Basal Oscines), and Suboscines. Not all these lineages are directly related and are separated by groups with the single CR, so it means that this gene order could occur independently. The presence of the duplicated region in all known Sylvioidea mitogenomes indicates that its common ancestor already had this region. However, species including mitogenomes with one type of duplication do not form one monophyletic clade. Taking into account such distribution as well as that the GO-III and -IV represent more reduced versions, we can assume that they derived independently from the GO-II type due to CR2 reduction, which originally was longer than CR1.
Using ML and MP methods, we deduced the ancestral states for the mitogenome based on the assumed tree topology (fig. 7 and supplementary figs. S3 and S4, Supplementary Material online). In the tree, we also included the lineages of Psittaciformes and Falconiformes, which are sister to Passeriformes. For both parrots and falcons, we assumed that the mitogenomic duplication was a plesiomorphic state. In the case of parrots, such assumption is based on the recent analyses of their mitogenomes, especially the discovery of the mitogenomic duplications in the early evolved lineages (Urantowka et al. 2018). In turn, the supposition of the ancestral mitogenomic duplication in Falconiformes is supported by the presence of the duplication in all known mitochondrial genomes of this order (Gibb et al. 2007; Ryu et al. 2012; Dou et al. 2016; Wang, Zhang, et al. 2016; Sveinsdottir et al. 2017).
The two methods consistently showed that the common ancestor of subsequently diverged lineages Eufalconimorphae, Psittacopasserae, Passeriformes, Eupasseres, and Suboscines contained duplication (fig. 7 and supplementary figs. S3 and S4, Supplementary Material online). The ML method provided the probability P > 0.9999 for this state. The mitogenome of Oscines ancestor also contained a duplicated region with P = 0.991. The assumption of an unknown state for A.chloris did not change the high probability for the ancestral duplication for Eufalconimorphae, Psittacopasserae, Eupasseres, Suboscines, and Oscines. After the separation of Menurida, the ancestor of its sister lineage most likely lost the duplicated region (P = 0.991) and such state was kept in the ancestors of later evolved lineages because the next deep nodes of passerine groups have a larger probability of the single mitogenomic region. However, this region could duplicate again independently in some groups at least four times, that is, Notiomystidae, the ancestor of Petroica (P = 0.991), the ancestor of Sylvioidea (P = 0.991), and one Turdus species.
Our results differ from those by Caparroz et al. (2018), who proposed at least six independent duplication events. This discrepancy results from disregarding the data about the groups Falconiformes and Psittaciformes, which are sister to Passeriformes. The authors also obtained in phylogenetic analyses an unexpected position of A.chloris, which is not placed basal to the rest of Passeriformes, that is, Suboscines + Oscines, but only to Oscines. Therefore, their analyses showed that the ancestor of passerines had no duplication in its mitogenome. Our conclusions about the evolution of mitogenomic duplications are the same when we take into account the newest tree topology received by Oliveros et al. (2019).
Phylogeny of Duplicated Regions in Individual Lineages
Analyses of many avian mitochondrial genomes showed that the duplicated regions are subjected to concerted evolution (Arndt and Smith 1998; Kumazawa et al. 1996, 1998; Eberhard et al. 2001; Abbott et al. 2005; Shao et al. 2005; Gibb et al. 2007; Kurabayashi et al. 2008; Cadahia et al. 2009; Eda et al. 2010; Morris-Pocock et al. 2010; Sammler et al. 2011; Schirtzinger et al. 2012). Under this process, some parts of duplicated regions became very similar or identical, and other parts diverged or degenerated. Predominantly, CRs are prone to homogenization, which is manifested in phylogenetic trees by the common clustering of two paralogous CR copies from the same species (CR1 and CR2). Alternatively, in the case of an ancient duplication, inheritance, and independent evolution of the two copies, we should expect that the corresponding CR regions from different species (CR1s or CR2s), that is, orthologous, are grouped together.
In order to analyze evolutionary relationships between the duplicated CRs in a bigger collection of passerines, we performed a phylogenetic analysis for five selected groups, in which duplications were reported. MrBayes phylograms, including information on the substitution number in individual lineages, are presented in figures 8–10, whereas consensus cladograms of three methods are shown in supplementary figure S5, Supplementary Material online. Interestingly, we found two scenarios on CR evolution in passerines.
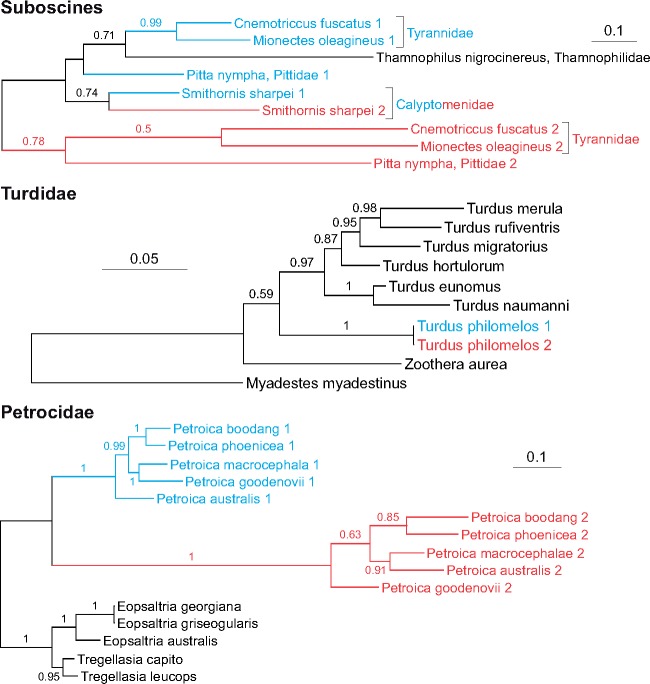
Fig. 8.
—The phylograms obtained in MrBayes based on control regions for Suboscines, Turdidae, and Petroicidae. The blue and red colors correspond to the first and the second copy of CR, respectively. The values at nodes indicate posterior probabilities. The values <0.5 were omitted. See supplementary figure S5, Supplementary Material online, for consensus cladograms of three methods.
Fig. 10.
—The phylogram obtained in MrBayes based on control regions for Falconiformes. Other explanations as in figure 8.
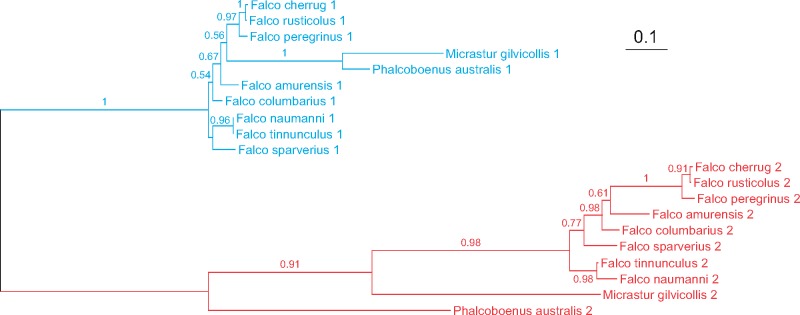
In Suboscines (fig. 8), the first and the second CR copies from Cnemotriccus fuscatus, Mionectes oleaginous, and Pitta nympha are separated in the trees. These species represent all two infraorders recognized in Suboscines, Tyrannides, and Eurylaimides, which indicates that the duplication occurred before the divergence of these lineages. The CR2s revealed much greater divergence, that is, the average 1.25 nucleotide substitutions per site (sub/site) in pairwise comparisons, than their CR1s counterparts, that is, the average 0.48 sub/site. However, a concerted evolution must have happened in the Smithornis sharpie mitogenome, because its two CRs are clustered together and show the average 0.49 sub/site. However, the CR2 has accumulated two times more substitutions than CR1 since their divergence, that is, 0.34 and 0.16 sub/site, respectively. The single CR of Thamnophilus nigrocinereus groups only with the CR1 sequences which suggests that the second copy was lost or was not identified yet in its genome.
The recent duplication of CR occurred in T.philomelos but not in other species of this genus (fig. 8). These two copies are almost identical (99.8% identity). This duplication is also found in N.cincta, whose CRs have 99.6% identity (Gibb et al. 2015). However, it is the only representative of Notiomystidae, so further consideration in the context of a wider group is not possible. In turn, the analysis of Petroicidae phylogeny (fig. 8) showed that the duplication of CR occurred before the differentiation of the current species of Petroica. The CR2 copies of these taxa also showed the average number of substitutions per site (0.31) almost two times higher than CR1 versions (0.18). The branch leading from the common ancestor of the two CR copies to the ancestor of CR2 in these species is even more than four times longer than that referring to the ancestor of CR1 (0.58 sub/site vs. 0.13 sub/site).
The evolution of CRs in Sylvioidea is more complicated. The CR phylogenetic tree includes a clearly separated clade grouping exclusively CR2 sequences (fig. 9). They come from various families: Paradoxornithidae, Phylloscopidae, Pycnonotidae, Sylviidae, Timaliidae as well as Alaudidae, which is the earliest diverged family of Sylvioidea (supplementary fig. S1, Supplementary Material online). These sequences showed more than two and half times higher accumulation of substitutions (1.33 sub/site) than their paralogous CR1 copies (0.50 sub/site). The second clade contains, besides these first copies, also CR1 and CR2 sequences from other members of Sylvioidea. In 35 species or subspecies from many families: Acrocephalidae, Aegithalidae, Hirundinidae, Leiothrichidae, Locustellidae, Pellorneidae, Sylviidae, and Zosteropidae, their paralogous copies of CRs are clustered together (fig. 9). There are also cases in which CRs of the same type (1 or 2) are more closely related than the copies in the same mitogenome. It concerns two pairs of some species of Aegithalos: Ae. fuliginosus with Ae. iouschistos, and Ae. caudatus with Ae. glaucogularis. Similarly, CRs from all species of Zosterops are split into two clades including the first and the second copy. However, the evolution of CRs can be more complex because, as shown for Aegithalos, most of the duplicated regions remain identical within an individual and evolve under concerted evolution, whereas the 5′ and 3′ end of the duplicated CRs are more variable and evolve independently (Wang et al. 2015).
Fig. 9.
—The phylogram obtained in MrBayes based on control regions for Sylvioidea. Other explanations as in figure 8.
Such phylogenetic relationships between CRs suggest that the duplication occurred before the Sylvioidea radiation or very early in its evolution (Singh et al. 2008). Next, CR2 evolved from CR1 within many individual species lineages at a frequency higher than the rate of speciation during the process of concerted evolution. However, such quick changes have not yet occurred in few cases. For example, the homogenization of CRs proceeded before the emergence of the current Zosterops species but not later within these species lineages. An interesting case is represented by Sylvia. In one of its species, Sylvia atricapilla, the concerted evolution already happened but Sylvia crassirostris still has two copies generated in the ancient duplication event, see also Singh et al. (2008).
We also compared the number of substitutions per site in the branches leading to the paralogous sequences of CRs present in the same mitogenome and clustered together in the tree. The number was greater in CR2 than CR1 in 17 cases (members of Locustellidae, Pellorneidae, Zosteropidae, and the majority of Leiothrichidae), whereas in 16 cases (members of Aegithalidae, Acrocephalidae, Sylviidae, and the majority of Hirundinidae), it was the opposite. However, there were 15 cases in which the number of substitutions in CR2 was more than twice that in CR1, and only 8 cases of the opposite observation. CR1s and CR2s of Zosterops showed a comparable average number of substitutions in pairwise comparison: 0.038 and 0.037 sub/site, respectively. In turn, the CR2s of Ae. fuliginosus and Ae. iouschistos were diverged to a greater extent than the CR1s as can be seen by the different branch lengths. The branch leading to the CR2 ancestor has 0.0138 sub/site and to the CR1 ancestor, 0.0031 sub/site.
In Falconiformes, we found only one type of CR evolution. The CR sequences created two clades, separate for CR1s and CR2s (fig. 10). The analyzed falcons represent all two subfamilies recognized in this order, Polyborinae and Falconinae, so we can assume that the duplication occurred before the divergence of the current Falconiformes lineages, see also Cadahia et al. (2009). The CR2 sequences accumulated 2.7 times more substitutions than CR1 sequences. The average for CR2s is 0.70 sub/site and for CR1s 0.26 sub/site.
Relationships between Mitogenomic Duplications and DNA Strand Asymmetry
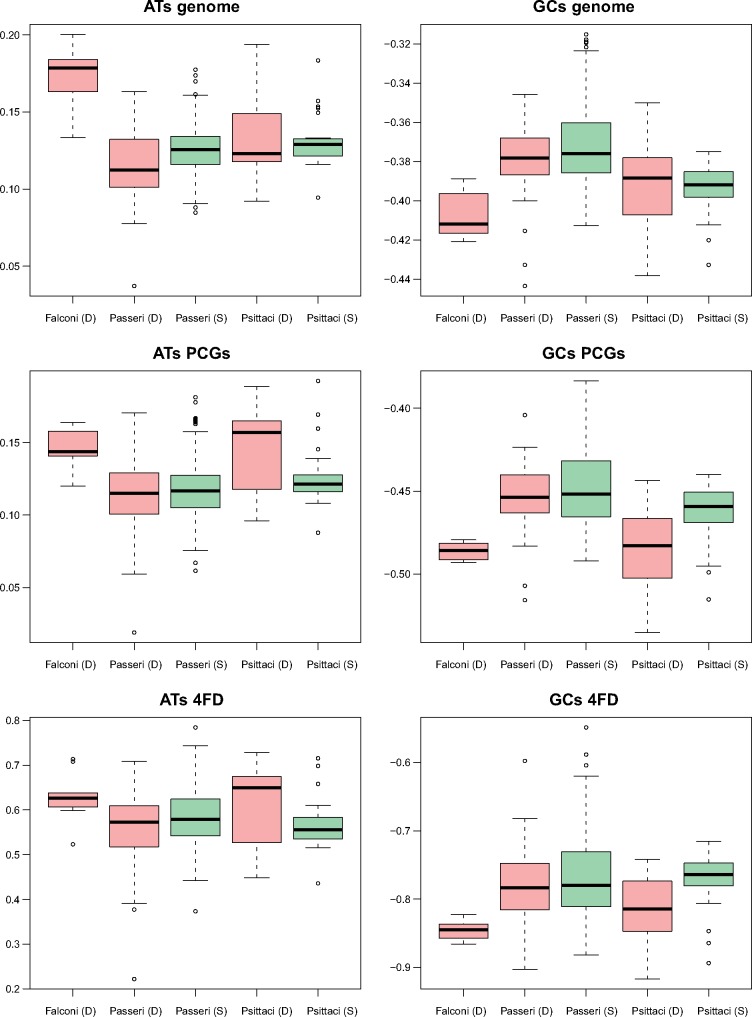
Recent analyses of parrot mitogenomes showed that the presence of duplicated CR regions is related with an elevated compositional asymmetry between DNA strands, which reflects differences in directional mutational pressure associated with replication between the heavy and light strands (Eberhard and Wright 2016; Urantowka et al. 2018). In order to check this relationship for Passeriformes and Falconiformes, we calculated normalized differences in complementary nucleotides, that is, AT- and GC-skew for the whole genomes, as well as for all PCGs and 4FD sites (fig. 11).
Fig. 11.
—Box-plots of AT-skew (ATs) and GC-skew (GCs) calculated for the entire genome as well as all sites in protein coding sequences (PCGs) and 4-fold degenerate (4FD) sites in mitogenomes with duplicated (D) or single (S) control regions for Falconiformes (Falconi), Passeriformes (Passeri), and Psittaciformes (Psittaci). The thick line indicates median, the boxes show quartile range, and the whiskers denote the range without outliers.
In the case of falcon mitogenomes, we did not have any sample with a single CR region for comparison. However, when compared with parrot mitogenomes comprising the duplicated region, Falconiformes PCGs and 4FD skews were comparable, whereas their genome AT-skew was even significantly higher (P = 0.0008) and GC-skew significantly lower (P = 0.046) than in the parrot genomes (fig. 11). All falcon skew parameters, except for the genome GC-skew, were also significantly higher in absolute value than those in parrot mitogenomes without the duplicated CR (P < 0.04). On the other hand, Passeriformes mitogenomes revealed a significantly weaker DNA asymmetry (fig. 11). It concerned all measures, except for 4FD AT-skew, calculated for the passerine mitogenomes without the duplication and compared with the falcon mitogenomes (P < 0.0002). The difference was also statistically significant (P < 0.013) in the comparison of such Passeriformes genomes with the parrot mitogenomes containing the duplication (in PCGs AT-skew, and all types of GC-skews) and parrot mitogenomes without the duplication (in genome and PCGs GC-skews). The passerine mitogenomes even with the duplicated region were characterized by a weak DNA asymmetry. All skew measures for such mitogenomes were significantly lower in absolute value than those obtained for falcon and parrot mitogenomes containing the second CR (P < 0.043). The genome skews of these passerine genomes showed also smaller absolute values than those in parrot mitogenomes without the duplication (P < 0.0025).
Interestingly, we did not observe any significant difference (P > 0.072) in five out of six skew measures between two types of Passeriformes mitogenomes, with and without the duplication (fig. 11). Unexpectedly, the mitogenomes with the single CR characterized by a significantly (P = 0.0008) larger genome AT-skew than the genomes with the duplication. The conclusions about the passerine mitogenomes did not change when we analyzed separately the groups with unreduced (GO-I and -II) and reduced (GO-III and -IV) CRs (data not shown). The parrot mitogenomes showed expected results because the genomes with the duplication had significantly higher DNA asymmetry than the genomes without the additional CR, in 4FD AT-skew (P = 0.049) as well as PCGs (P = 0.009) and 4FD GC-skew (P = 0.008).
Although we cannot exclude that among the passerine mitogenomes without the reported duplication, there are cases with a hidden duplication. They could change the unexpected results for the genome AT-skew. However, the general similarity of the skew measures between the two genome types implies that a potential relationship between the compositional asymmetry and the additional CR copy does not exist in the passerine mitogenomes as it does in parrots.
Because a stronger DNA asymmetry is expected in genomes with slower replication when their H-strand remains exposed to mutations for a longer time (Reyes et al. 1998), we can assume that the passerine mitogenomes replicate in a rather similar time irrespective of the additional CR copy.
The compositional strand bias in the mitochondrial genome is mainly caused by a higher rate of substitutions occurring in the parental H-strand during the mitogenome replication. According to the strand-asynchronous or strand-displacement model of mtDNA replication (Robberson et al. 1972; Clayton 1982), this strand stays for a longer time in a single-stranded state until the replication of L-strand, proceeding on this H-strand as template, is completed. In this state, the H-strand is prone to deamination of C and A. However, additional studies have challenged this conventional replication scheme both in mammals (Holt et al. 2000; Bowmaker et al. 2003) and birds (Reyes et al. 2005). According to the new strand-coupled model, DNA replication can start from multiple origins and proceeds bidirectionally until termination occurs in the D-loop region. This replication type still introduces the compositional asymmetry between DNA strands due to the different synthesis mode between leading and lagging strands as in bacterial genomes (Frank and Lobry 1999; Kowalczuk et al. 2001), but the template for the newly synthetized lagging strand stays exposed for a shorter time than the H-strand in the strand-displacement replication. Thus, we can expect a smaller compositional bias in the former case than in the latter. Consequently, the significantly smaller AT- and GC-skew values of passerine mitogenomes can suggest that these genomes replicate according to the strand-coupled model, whereas mtDNA in Falconiformes characterized by a much larger compositional bias is synthetized in accordance with the strand-displacement model. It cannot be excluded that different replication mechanisms operate in various tissues and cell types as well as physiological conditions to ensure appropriate mtDNA copy number, mitochondrial gene expression and, in consequence, energy production (Pohjoismaki and Goffart 2011; McKinney and Oliveira 2013).
Relationships of Mitogenomic Duplications with Body Mass and BMR
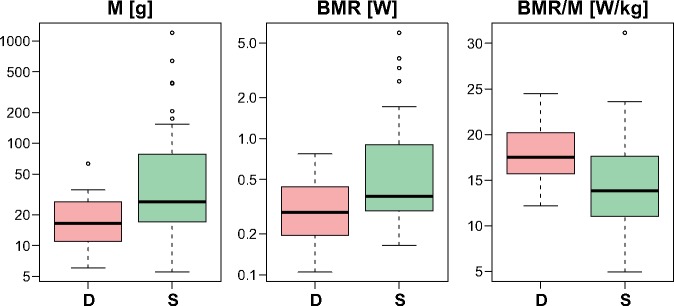
The presence of two CRs suggests that they can increase the overall number of genome copies per mitochondrion and, in consequence, efficiency of energy production. Therefore, we tested how possessing mitogenomes with a single or duplicated region is associated with BMR as well as body mass (M), with which BMR is scaled as a power function (Kleiber 1932; West et al. 1997; Dodds et al. 2001; White and Kearney 2014). The comparisons showed that passerines with a mitogenomic duplicated region are characterized by significantly smaller (P = 0.024) body mass (median 16.40, average 21.24) than the passerines without such region (median 26.90, average 91.07)—figure 12. Due to a positive relationship between BMR and M, the passerines with a duplicated region showed also smaller BMR (median 0.287, average 0.354) than those without the region (median 0.377, average 0.790), but the difference was not statistically significant (P = 0.057). However, the difference in body mass-specific metabolic rate (BMR/M) was already significant (P = 0.014). The passerines with a duplication have a higher rate (median 17.50, average 18.02) than passerines without duplication (median 13.851, average 14.354).
Fig. 12.
—Box-plots of body mass (M), basal metabolic rate (BMR), and mass-specific metabolic rate (BMR/M) calculated for passerines with duplicated (D) or single (S) control regions in their mitogenomes. M and BMR are shown in logarithmic scale. The thick line indicates median, the boxes show quartile range, and the whiskers denote the range without outliers.
Such relations could be expected because BMR shows negative allometric relationship with M (Kleiber 1932; West et al. 1997; Dodds et al. 2001; White and Kearney 2014). Therefore, we compared linear modeling of log10(BMR) ∼ log10(M) separately for passerines with and without duplicated regions. This analysis demonstrated that the linear function for the first group of birds has a slope by 20% larger than that in the second group, that is, 0.859 ± 0.093 (standard error) versus 0.716 ± 0.022. The intercept was −1.582 ± 0.117 and −1.423 ± 0.037, whereas adjusted R2 were 0.894 and 0.952, respectively. All coefficients were significant with P < 7E-6. The slope of the function for passerines with two CRs became larger by almost 29%, when was compared with the slope in the function log10 BMR = 0.667(±0.065) × log10 M − 1.362(±0.087) with adjusted R2 = 0.739, calculated for the species with the single CR and body mass in the range of passerines containing the mitogenomic duplication. The results might suggest that passerines with the mitogenomic duplication can have generally a higher metabolic rate in relation to their body mass than passerines without the duplication. However, additional analyses on much larger data set are necessary to prove this view, because avian BMR can be related with many various factors: phylogeny, season, temperature, habitat, environment, behavior, and migratory tendency, for example, McNab (2009, 2012).
Recently, Berv and Field (2018) demonstrated for birds that body mass and metabolic rate may increase the overall substitution rate, when there exists a selection for small body and/or population size. It may be associated with greater production of mutagenic oxygen radicals in smaller-bodied organism with higher body mass-specific metabolic rates (Gillooly et al. 2005, 2007). If duplicated regions in mitogenomes increase copy numbers of these genomes due to additional replication initiation site, and consequently intensify the production of energy by mitochondria and free radicals as by-products, we could expect higher mutation and substitution rates in such genomes. Interestingly, when we compared tip-to-root distances between two closely related groups, containing the mitogenomic duplications (Sylvioidea) and lacking them (Paroidea), we found that the former group was characterized by a significantly (P = 1.3E-7) higher median substitution rate than the latter, that is, 0.396 (lower and upper quartiles, 0.384–0.434) and 0.339 (0.327–0.353), respectively.
Possible Scenario and Mechanisms for Evolution of Duplicated Regions in Mitogenomes and Their Selective Importance
Because the duplicated CRs are present in some phylogenetic lineages and absent from others, there are two theoretical scenarios for their evolution: 1) the region was duplicated in a common ancestor and next inherited or lost in its descendants or 2) the duplication occurred independently in some lineages. The first possibility could be supported by a common grouping of the corresponding (orthologous) CR regions from different lineages, whereas in the second case, two paralogous CR copies from the same species should be clustered together. However, the duplicated regions can be often subjected to homogenization via concerted evolution, which erases the ancient phylogenetic signal accumulated between the copies since their duplication in the past. In consequence, phylogenetic trees of the duplicated regions inherited from the common ancestor can resemble trees produced under the second scenario and imply multiple origins of the duplication. Studying individual groups of passerines, we actually observed these two ways of CR grouping.
Mapping the presence and absence of duplications onto the phylogenetic tree can help to solve the issue. The occurrence of duplicated regions in the mitogenomes of early diverged passerine lineages and two closely related groups of other avian orders indicates that the common ancestor of Passeriformes could contain a duplication. Otherwise, we should assume at least three independent duplication events in the passerine lineages. The single origin of the duplicated region in a common ancestor was also proposed not only for falcons (Cadahia et al. 2009) and parrots (Urantowka et al. 2018) but also for sulids (Morris-Pocock et al. 2010), ardeids (Zhou et al. 2014), and cranes (Akiyama et al. 2017). The single occurrence of a duplicated region and its inheritance from a common ancestor is also consistent with the view that similar gene arrangements are likely to be shared only as a result of common ancestry because the great number of potentially possible arrangements makes convergence of gene orders less likely (Rokas and Holland 2000). Accordingly, independent duplications in separate lineages would require a number of identical evolutionary events involving the same pattern of recombination and replication errors as well as duplication and degeneration (Shao et al. 2005; Morris-Pocock et al. 2010; Zhou et al. 2014; Akiyama et al. 2017).
Taking into account the distribution of duplication in other passerines, five events are the most parsimonious, that is, one loss and four independent occurrences in separate lineages (N.cincta, Petroica, Sylvioidea, and T.philomelos). Alternatively, we could suppose that the duplications found in these four lineages were inherited from an ancestor, whereas other passerine lineages lost the duplicated region. This opposite possibility would require more than ten losses and therefore, it seems statistically less probable based on the current data. However, the presented model postulates that the occurrence and loss of mitogenomic duplication are equally probable. It does not have to be true because of different probabilities of duplications and deletions of DNA fragments in the genomes due to molecular mechanisms. Moreover, the duplication can be favored or disapproved due to selective reasons.
Because the CR seems to be responsible for initiation of transcription and replication (Boore 1999), this region and adjacent genes could be especially prone to duplication. The mitochondrial tandem duplications are mainly caused by replication errors, such as slipped-strand mispairing or asynchrony in the points of initiation and termination of DNA synthesis (Stanton et al. 1994; Broughton and Dowling 1997; Mueller and Boore 2005; San Mauro et al. 2006; Fujita et al. 2007). The replication slippage is often promoted by repetitive sequences. Actually, many CRs in bird mitogenomes contain such types of sequences known as microsatellites (Berg et al. 1995; Roques et al. 2004; He et al. 2013). However, it is not clear whether these repeats can stimulate the duplication of a larger genome portion including not only the microsatellite region but also the whole CR and neighboring genes.
More certain is the importance of CR in increasing recombination frequency, which finally leads to homogenization of duplicated CRs (Kumazawa et al. 1996, 1998; Kurabayashi et al. 2008; Morris-Pocock et al. 2010; Sammler et al. 2011; Zhou et al. 2014; Wang et al. 2015). It is postulated that in the 3′ side of the CR, that is, D-loop region, the replication fork stalls (Bowmaker et al. 2003), which causes that the 3′ end of the nascent strand is exposed for a long time until the replication restarts. It makes the nascent strands susceptible to recombination. The strands can be exchanged between two mitogenome molecules or within a single one if two independent replication forks occur, for example, from two CRs. Repair of resultant heteroduplex DNA or secondary replication lead to homogenization of CRs and flanking genes. This mechanism can explain the concerted evolution in many mitogenomes (Kurabayashi et al. 2008; Sammler et al. 2011; Zhou et al. 2014; Wang et al. 2015). Alternatively, the homogenized sequences can occur due to tandem duplication involving replication slippage, but the sequences of the duplicated region would need to be sequentially homogenized in this case (Kumazawa et al. 1998).
Because CRs can be “hot spots” of recombination (Kurabayashi et al. 2008), the homogenized CRs are present in many mitogenomes, where the homogenization occurs within individual species lineages at a frequency higher than the rate of speciation or even in every generation (Sammler et al. 2011). The identical or almost identical CRs copies were found in species of many avian groups: Psittaciformes (Eberhard et al. 2001; Eberhard and Wright 2016; Urantowka et al. 2018), Procellariiformes (Abbott et al. 2005; Eda et al. 2010), Pelecaniformes (Cho et al. 2009; Zhou et al. 2014), Charadriiformes (Verkuil et al. 2010), Suliformes (Morris-Pocock et al. 2010; Gibb et al. 2013), Bucerotiformes (Sammler et al. 2011), Gruiformes (Akiyama et al. 2017), Strigiformes (Kang et al. 2018), and also Passeriformes (Gibb et al. 2015; Caparroz et al. 2018). We also reported the concerted evolution of CRs for 35 passerine species. The generation of two identical or very similar copies of CR can be favored by selection because then the replication and transcription started from the CRs can be regulated more effective by the same factors. However, it is interesting that CRs are not always very quickly homogenized because we observed their independent evolution for Suboscines, Petrocidae, and 12 genera belonging to Sylvioidea as well as Falconiformes. It should be noted that the nonhomogenized CRs can be preserved longer than the emergence of a new family. It may be associated with a different mechanism of replication in which the D-loop region is not such a strong barrier for the replication fork.
However, not the entire CR must be subjected to concerted evolution, at least with the same rate. It was found for several avian groups (Eberhard et al. 2001; Abbott et al. 2005; Cadahia et al. 2009; Morris-Pocock et al. 2010; Verkuil et al. 2010; Sammler et al. 2011), including passerines (Wang et al. 2015), that the central parts of paralogous CR copies are usually identical or at least more similar in a given species than flanking 5′ and 3′ sections, showing greater variability. It implies that the former parts evolve in concert and the latter independently.
It is generally assumed that many rearrangements in mitochondrial genomes evolve according to the tandem duplication and random loss model, also in birds (Moritz and Brown 1987; Bensch and Harlid 2000; Boore 2000; San Mauro et al. 2006; Gibb et al. 2007). It postulates that, after duplication, one of the copied elements (gene or CR) becomes a pseudogene or is eliminated from the genome due to mutation accumulation. This process is assumed to be random, so various gene rearrangement can occur. However, nonrandom loss was reported in some insect mitogenomes (Lavrov et al. 2002). Therefore, it is possible that parallel degenerations of duplicated regions result in the same pattern, which does not have to be explained by independent multiple origins. Such convergent evolution of initially complete gene orders leading to the reduced versions (GO-III and -IV) could occur in at least eight separate passerine lineages. The parallel occurrence of the same derived gene arrangement was also postulated for frog, reptile, and bird mitogenomes (Bensch and Harlid 2000; Macey et al. 2004; Kurabayashi et al. 2008, 2010; Verkuil et al. 2010).
The existence of duplicated regions in mitogenomes may depend on whether they prove beneficial or unfavorable to an organism. On the one hand, the duplications can be a burden for a genome, because the extra copy can prolong the replication time and increase the energy consumed in the synthesis of the longer genome. Thus, elimination of the duplicated regions should be favored. On the other hand, selection can accept mitogenomes with duplicated regions due to several reasons. Additional CRs can increase the number of replication and transcription events per unit time and efficiency of these processes, and thereby the overall number of genomic and transcript copies per mitochondrion (Kumazawa et al. 1996; Arndt and Smith 1998; Tang et al. 2000; Umeda et al. 2001; Jiang et al. 2007). This view is supported by the experiment which showed that human mitogenomes with two CRs reached a higher number than those with one CR in a culturing cell population (Tang et al. 2000). Other study showed that the extra origin of replication does not disturb the function of another and is competent for DNA synthesis (Umeda et al. 2001). Thus, this additional CR can give a replicative advantage by increasing twice the opportunity to initiate the replication.
As a result of the more effective replication and the larger number of genome copies, mitochondria can produce energy more intensively. A certain premise to this may be our finding that passerines with two CRs are characterized by a higher metabolic rate in relation to their body mass, but additional biochemical and physiological studies are necessary to confirm the direct relationship between the mitogenomic duplications and more efficient energy production. Nevertheless, Urantowka et al. (2018) found another relationship between duplications in the mitogenome and phenotype, namely parrots keeping two copies of the CR showed morphological features related to more active flight, which is connected with consuming a lot of energy. These parrots were also characterized by larger body mass, which is opposite to that found for passerines. In the latter case, species with the duplication were more lightweight. The difference may be associated with distinct physiology of these avian groups and needs further study.
Global investigations of 92 bird families (Skujina et al. 2016) and Psittaciformes as one group (Urantowka et al. 2018) revealed that species living longer have mitogenomes with the duplicated CR. It was proposed that the additional copies may protect the mitochondrion from loss of function resulting from age related deletions as well as can increase flexibility of mitochondrial response to environmental changes associated with the elevated metabolism (Skujina et al. 2016). Based on the data collected by Skujina et al. (2016) for Passeriformes, we calculated the ratio of the logarithm from average maximum life span to the logarithm from body mass and found that this parameter was larger for the species containing duplicated CR than those with the single one. Mean and standard deviation were 0.84±0.17 (the species number, N = 13) and 0.75±0.13 (N = 23) respectively, but the difference was not statistically significant (P value = 0.14), probably due to poor sampling. Additional analyses are necessary to verify if the relationship between the age and mitochondrial duplications is also valid for passerines.
In summary, the conducted analyses of duplicated regions in mitogenomes of Passeriformes indicate that the primary duplication occurred in the passerine ancestor and independent duplication events were not so frequent in individual passerine lineages, but once they occurred, they were maintained during evolutionary time. In contrast to Boore et al. (2005) and consistent with our findings, other authors also postulated that the copied CRs with flanking genes are maintained by recombination for a long time, in the case of mantellid frogs for at least 33 Ma (Kurabayashi et al. 2008), cranes for about 34 Ma (Akiyama et al. 2017), and snakes for at least 70 Ma (Kumazawa et al. 1996). Assuming that the duplication state occurred in the ancestor of Passeriformes and was inherited to the present by the early diverged lineages, we can assume that this state existed in these groups for about 50 Ma, according to the recent estimations of passerine radiation by Oliveros et al. (2019). However, the duplication state could last even 10 Ma longer if we take into account the presence of the duplication in the common ancestor of Falconiformes, Psittaciformes, and Passeriformes. In the preservation of the repeated regions, an important role was fulfilled by recombination, which often unified the duplicated sequences. The mitogenomes with extra CR could be additionally favored by selection because these CRs increased the replication and transcription efficiency, and consequently the energy production. Nevertheless, it should be emphasized that the scenario suggested by us for the passerine mitogenome evolution is valid for the current data and may change when additional data about the mitogenomes appears. It is important to verify the already sequenced mitogenomes in terms of a duplicated region by using appropriate methods for their detection (Gibb et al. 2007).
Conclusions
The mitochondrial gene analyses have generated a well-supported phylogeny of Passeriformes, from the level of suborder to species. The analyses confirmed the higher-taxonomic classification of passerines and resolved deep relationships including early evolved lineages. In terms of phylogenetic relationships and variation, the Basal Oscines can represent groups in the rank of parvorders, whereas the lineages of Transitional Oscines can be included as superfamilies into parvorder Passerida. The other two sister groups of Passerida, supported in phylogenetic analyses, are Sylvioidea with Paroidea and Passeroidea with Muscicapoidea. The mitogenomes of Passeriformes have four types of rearrangements in respect to the duplication of CR and neighboring genes. Comparison of the mitogenome organization and phylogenetic relationships showed that the common ancestor of not only Passeriformes but also Falconiformes and Psittaciformes had a duplication of a CR with adjacent genes in the mitochondrial genome. This state was retained in early diverged lineages of passerines but the duplication disappeared after separation of Menurida. Other duplicated regions that are observed in Notiomystis, Petroica, Turdus, and the ancestor of Sylvioidea might result from independent duplications or inheritance from an ancestor. In some lineages, both duplicated regions were preserved but the second copy showed a greater tendency to shortening and accumulation of larger number of nucleotide substitutions. However, when the second copy was not degenerated, it usually lasted longer than its paralog. In other groups, the two CR copies became very similar due to concerted evolution when this homogenization occurred at the frequency higher than the speciation rate. The presence of the two CR copies in passerine mitogenomes is not associated with an increased mutational pressure in the mitogenome, for example, due to slower replication, as in parrots, and may be associated with a different replication mode of the mitogenome. The complex evolution of CRs means that phylogenetic trees based on this marker will show incorrect relationships between taxa, when only one CR copy is preserved or used in the reconstruction. The preservation of the second CR copy for a long time implies that both regions can play an important role in initiation and regulation of replication and transcription. They might also influence metabolic rate in Passeriformes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are very grateful to the Associate Editor and anonymous reviewers for their insightful comments and remarks, which significantly improved the manuscript. This work was supported by the National Science Centre Poland (Narodowe Centrum Nauki, Polska) under grant no. 2015/17/B/NZ8/02402. Some computations were carried out at the Wrocław Center for Networking and Supercomputing under the grant nos. 307 and 442.
Literature Cited
- Abbott CL, Double MC, Trueman JW, Robinson A, Cockburn A.. 2005. An unusual source of apparent mitochondrial heteroplasmy: duplicate mitochondrial control regions in Thalassarche albatrosses. Mol Ecol. 14(11):3605–3613. [DOI] [PubMed] [Google Scholar]
- Aggerbeck M, Fjeldså J, Christidis L, Fabre P-H, Jønsson KA.. 2014. Resolving deep lineage divergences in core corvoid passerine birds supports a proto-Papuan island origin. Mol Phylogenet Evol. 70:272–285. [DOI] [PubMed] [Google Scholar]
- Akiyama T, et al. 2017. Gene duplication and concerted evolution of mitochondrial DNA in crane species. Mol Phylogenet Evol. 106:158–163. [DOI] [PubMed] [Google Scholar]
- Alstrom P, Ericson PG, Olsson U, Sundberg P.. 2006. Phylogeny and classification of the avian superfamily Sylvioidea. Mol Phylogenet Evol. 38:381–397. [DOI] [PubMed] [Google Scholar]
- Alstrom P, et al. 2014. Discovery of a relict lineage and monotypic family of passerine birds. Biol Lett. 10:20131067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt A, Smith MJ.. 1998. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol Biol Evol. 15(8):1009–1016. [DOI] [PubMed] [Google Scholar]
- Barker FK. 2014. Mitogenomic data resolve basal relationships among passeriform and passeridan birds. Mol Phylogenet Evol. 79:313–324. [DOI] [PubMed] [Google Scholar]
- Barker FK, Barrowclough GF, Groth JG.. 2002. A phylogenetic hypothesis for passerine birds: taxonomic and biogeographic implications of an analysis of nuclear DNA sequence data. Proc R Soc Lond B 269(1488):295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J.. 2004. Phylogeny and diversification of the largest avian radiation. Proc Natl Acad Sci U S A. 101(30):11040–11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech C, Chappell MA, Astheimer LB, Londoño GA, Buttemer WA.. 2016. A ‘slow pace of life’ in Australian old-endemic passerine birds is not accompanied by low basal metabolic rates. J Comp Physiol B 186(4):503–512. [DOI] [PubMed] [Google Scholar]
- Bensch S, Harlid A.. 2000. Mitochondrial genomic rearrangements in songbirds. Mol Biol Evol. 17(1):107–113. [DOI] [PubMed] [Google Scholar]
- Beresford P, Barker FK, Ryan PG, Crowe TM.. 2005. African endemics span the tree of songbirds (Passeri): molecular systematics of several evolutionary ‘enigmas’. Proc R Soc B 272(1565):849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Moum T, Johansen S.. 1995. Variable numbers of simple tandem repeats make birds of the order Ciconiiformes heteroplasmic in their mitochondrial genomes. Curr Genet. 27(3):257–262. [DOI] [PubMed] [Google Scholar]
- Berlin S, Ellegren H.. 2001. Evolutionary genetics. Clonal inheritance of avian mitochondrial DNA. Nature 413(6851):37–38. [DOI] [PubMed] [Google Scholar]
- Berv JS, Field DJ.. 2018. Genomic signature of an avian Lilliput effect across the K-Pg extinction. Syst Biol. 67(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boore JL. 2000. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals In: Sankoff D, Nadeau JH, editors. Comparative genomes: empirical and analytical approaches to gene order dynamics, map alignment and the evolution of gene families. Dordrecht (the Netherlands: ): Kluwer Academic Publishers; p. 133–148. [Google Scholar]
- Boore JL, Macey JR, Medina M.. 2005. Sequencing and comparing whole mitochondrial genomes of animals. Methods Enzymol. 395:311–348. [DOI] [PubMed] [Google Scholar]
- Bowmaker M, et al. 2003. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J Biol Chem. 278(51):50961–50969. [DOI] [PubMed] [Google Scholar]
- Boyd J. 2016. Taxonomy in Flux Checklist 3.08. Available from: http://jboyd.net/Taxo/
- Braun EL, Kimball RT.. 2002. Examining Basal avian divergences with mitochondrial sequences: model complexity, taxon sampling, and sequence length. Syst Biol. 51(4):614–625. [DOI] [PubMed] [Google Scholar]
- Broughton RE, Dowling TE.. 1997. Evolutionary dynamics of tandem repeats in the mitochondrial DNA control region of the minnow Cyprinella spiloptera. Mol Biol Evol. 14(12):1187–1196. [DOI] [PubMed] [Google Scholar]
- Bushuev A, Tolstenkov O, Zubkova E, Solovyeva E, Kerimov A.. 2018. Basal metabolic rate in free-living tropical birds: the influence of phylogenetic, behavioral, and ecological factors. Curr Zool. 64(1):33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadahia L, et al. 2009. Repeated sequence homogenization between the control and pseudo-control regions in the mitochondrial genomes of the subfamily Aquilinae. J Exp Zool Part B Mol Dev Evol. 312B:171–185. [DOI] [PubMed] [Google Scholar]
- Caparroz R, et al. 2018. Mitogenomes of two neotropical bird species and the multiple independent origin of mitochondrial gene orders in Passeriformes. Mol Biol Rep. 45(3):279.. [DOI] [PubMed] [Google Scholar]
- Cerasale DJ, Dor R, Winkler DW, Lovette IJ.. 2012. Phylogeny of the Tachycineta genus of New World swallows: insights from complete mitochondrial genomes. Mol Phylogenet Evol. 63(1):64–71. [DOI] [PubMed] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ.. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HJ, et al. 2009. Tandem duplication of mitochondrial DNA in the black-faced spoonbill, Platalea minor. Genes Genet Syst. 84(4):297–305. [DOI] [PubMed] [Google Scholar]
- Claramunt S, Cracraft J.. 2015. A new time tree reveals Earth history’s imprint on the evolution of modern birds. Sci Adv. 1(11):e1501005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA. 1982. Replication of animal mitochondrial DNA. Cell 28(4):693–705. [DOI] [PubMed] [Google Scholar]
- Clayton DA. 1991. Replication and transcription of vertebrate mitochondrial-DNA. Annu Rev Cell Biol. 7(1):453–478. [DOI] [PubMed] [Google Scholar]
- Clements JF, et al. 2018. The eBird/Clements checklist of birds of the world: v2018. Available from: http://www.birds.cornell.edu/clementschecklist/download/
- Cooke GM, King AG, Johnson RN, Boles WE, Major RE.. 2012. Rapid characterization of mitochondrial genome rearrangements in Australian songbirds using next-generation sequencing technology. J Hered. 103(6):882–886. [DOI] [PubMed] [Google Scholar]
- Daan S, Masman D, Groenewold A.. 1990. Avian basal metabolic rates: their association with body composition and energy expenditure in nature. Am J Physiol. 259:R333–R340. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PS, Rothman DH, Weitz JS.. 2001. Re-examination of the “3/4-law” of metabolism. J Theor Biol. 209(1):9–27. [DOI] [PubMed] [Google Scholar]
- Dou HS, et al. 2016. Complete mitochondrial genome of the Merlin (Falco columbarius). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1547–1548. [DOI] [PubMed] [Google Scholar]
- Duchene S, Archer FI, Vilstrup J, Caballero S, Morin PA.. 2011. Mitogenome phylogenetics: the impact of using single regions and partitioning schemes on topology, substitution rate and divergence time estimation. PLoS One 6:e27138.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard JR, Wright TF.. 2016. Rearrangement and evolution of mitochondrial genomes in parrots. Mol Phylogenet Evol. 94:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard JR, Wright TF, Bermingham E.. 2001. Duplication and concerted evolution of the mitochondrial control region in the parrot genus Amazona. Mol Biol Evol. 18(7):1330–1342. [DOI] [PubMed] [Google Scholar]
- Eda M, Kuro-o M, Higuchi H, Hasegawa H, Koike H.. 2010. Mosaic gene conversion after a tandem duplication of mtDNA sequence in Diomedeidae (albatrosses). Genes Genet Syst. 85(2):129–139. [DOI] [PubMed] [Google Scholar]
- Ericson PG, et al. 2006. Diversification of Neoaves: integration of molecular sequence data and fossils. Biol Lett. 2(4):543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson PGP, Johansson US.. 2003. Phylogeny of Passerida (Aves: Passeriformes) based on nuclear and mitochondrial sequence data. Mol Phylogenet Evol. 29(1):126–138. [DOI] [PubMed] [Google Scholar]
- Ericson PGP, Klopfstein S, Irestedt M, Nguyen JMT, Nylander JAA.. 2014. Dating the diversification of the major lineages of Passeriformes (Aves). BMC Evol Biol. 14(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson PGP, et al. 2002. A Gondwanan origin of passerine birds supported by DNA sequences of the endemic New Zealand wrens. Proc R Soc Lond B 269(1488):235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner N, Begemann G, Renz AJ, Meyer A, Kuraku S.. 2009. The origin of bmp16, a novel Bmp2/4 relative, retained in teleost fish genomes. BMC Evol Biol. 9(1):277.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank AC, Lobry JR.. 1999. Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238(1):65–77. [DOI] [PubMed] [Google Scholar]
- Fujita MK, Boore JL, Moritz C.. 2007. Multiple origins and rapid evolution of duplicated mitochondrial genes in parthenogenetic geckos (Heteronotia binoei; Squamata, Gekkonidae). Mol Biol Evol. 24(12):2775–2786. [DOI] [PubMed] [Google Scholar]
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D.. 2007. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 24(1):269–280. [DOI] [PubMed] [Google Scholar]
- Gibb GC, Kennedy M, Penny D.. 2013. Beyond phylogeny: pelecaniform and ciconiiform birds, and long-term niche stability. Mol Phylogenet Evol. 68(2):229–238. [DOI] [PubMed] [Google Scholar]
- Gibb GC, et al. 2015. New Zealand Passerines help clarify the diversification of major songbird lineages during the Oligocene. Genome Biol Evol. 7(11):2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly JF, Allen AP, West GB, Brown JH.. 2005. The rate of DNA evolution: effects of body size and temperature on the molecular clock. Proc Natl Acad Sci U S A. 102(1):140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly JF, McCoy MW, Allen AP.. 2007. Effects of metabolic rate on protein evolution. Biol Lett. 3(6):655–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S, Philippe H.. 2002. Ancient phylogenetic relationships. Theor Popul Biol. 61(4):391–408. [DOI] [PubMed] [Google Scholar]
- Hackett SJ, et al. 2008. A phylogenomic study of birds reveals their evolutionary history. Science 320(5884):1763–1768. [DOI] [PubMed] [Google Scholar]
- Hails CJ. 1983. The metabolic-rate of tropical birds. Condor 85(1):61–65. [Google Scholar]
- Hanna ZR, et al. 2017. Complete mitochondrial genome sequences of the northern spotted owl (Strix occidentalis caurina) and the barred owl (Strix varia; Aves: Strigiformes: Strigidae) confirm the presence of a duplicated control region. PeerJ 5:e3901.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison GL, et al. 2004. Four new avian mitochondrial genomes help get to basic evolutionary questions in the late Cretaceous. Mol Biol Evol. 21(6):974–983. [DOI] [PubMed] [Google Scholar]
- Havird JC, Santos SR.. 2014. Performance of single and concatenated sets of mitochondrial genes at inferring metazoan relationships relative to full mitogenome data. PLoS One 9(1):e84080.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XL, Ding CQ, Han JL.. 2013. Lack of structural variation but extensive length polymorphisms and heteroplasmic length variations in the mitochondrial DNA control region of highly inbred crested ibis, Nipponia nippon. PLoS One 8(6):e66324.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt IJ, Lorimer HE, Jacobs HT.. 2000. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100(5):515–524. [DOI] [PubMed] [Google Scholar]
- Huang R, et al. 2016. Complete mitochondrial genome and phylogenetic relationship analysis of Garrulax affinis (Passeriformes, Timaliidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3502–3503. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Larget B, Alfaro ME.. 2004. Bayesian phylogenetic model selection using reversible jump Markov chain Monte Carlo. Mol Biol Evol. 21(6):1123–1133. [DOI] [PubMed] [Google Scholar]
- Jarvis ED, et al. 2014. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346(6215):1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang ZJ, et al. 2007. Comparative mitochondrial genomics of snakes: extraordinary substitution rate dynamics and functionality of the duplicate control region. BMC Evol Biol. 7(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson US, Fjeldsa J, Bowie RC.. 2008. Phylogenetic relationships within Passerida (Aves: Passeriformes): a review and a new molecular phylogeny based on three nuclear intron markers. Mol Phylogenet Evol. 48(3):858–876. [DOI] [PubMed] [Google Scholar]
- Jonsson KA, Fabre PH, Ricklefs RE, Fjeldsa J.. 2011. Major global radiation of corvoid birds originated in the proto-Papuan archipelago. Proc Natl Acad Sci U S A. 108(6):2328–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson KA, Fjeldsa J.. 2006. Determining biogeographical patterns of dispersal and diversification in oscine passerine birds in Australia, Southeast Asia and Africa. J Biogeogr. 33(7):1155–1165. [Google Scholar]
- Jonsson KA, Fjeldsa J, Ericson PGP, Irestedt M.. 2007. Systematic placement of an enigmatic Southeast Asian taxon Eupetes macrocerus and implications for the biogeography of a main songbird radiation, the Passerida. Biol Lett. 3:323–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson KA, et al. 2016. A supermatrix phylogeny of corvoid passerine birds (Aves: Corvides). Mol Phylogenet Evol. 94:87–94. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS.. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Li B, Ma X, Xu Y.. 2018. Evolutionary progression of mitochondrial gene rearrangements and phylogenetic relationships in Strigidae (Strigiformes). Gene 674:8–14. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiber M. 1932. Body size and metabolism. Hilgardia 6(11):315–353. [Google Scholar]
- Kowalczuk M, et al. 2001. DNA asymmetry and the replicational mutational pressure. J Appl Genet. 42(4):553–577. [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T.. 1996. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control-region-like sequences duplicated and inserted into a tRNA gene cluster. Mol Biol Evol. 13(9):1242–1254. [DOI] [PubMed] [Google Scholar]
- Kumazawa Y, Ota H, Nishida M, Ozawa T.. 1998. The complete nucleotide sequence of a snake (Dinodon semicarinatus) mitochondrial genome with two identical control regions. Genetics 150(1):313–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurabayashi A, et al. 2008. Phylogeny, recombination, and mechanisms of stepwise mitochondrial genome reorganization in mantellid frogs from Madagascar. Mol Biol Evol. 25(5):874–891. [DOI] [PubMed] [Google Scholar]
- Kurabayashi A, et al. 2010. Complete mitochondrial DNA sequence of the endangered frog Odorrana ishikawae (family Ranidae) and unexpected diversity of mt gene arrangements in ranids. Mol Phylogenet Evol. 56(2):543–553. [DOI] [PubMed] [Google Scholar]
- Kuraku S. 2010. Palaeophylogenomics of the vertebrate ancestor-impact of hidden paralogy on hagfish and lamprey gene phylogeny. Integr Comp Biol. 50(1):124–129. [DOI] [PubMed] [Google Scholar]
- Kuraku S. 2013. Impact of asymmetric gene repertoire between cyclostomes and gnathostomes. Semin Cell Dev Biol. 24(2):119–127. [DOI] [PubMed] [Google Scholar]
- Lambret-Frotte J, Perini FA, Russo CAD.. 2012. Efficiency of nuclear and mitochondrial markers recovering and supporting known amniote groups. Evol Bioinform Online 8:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S.. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Philippe H.. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21(6):1095–1109. [DOI] [PubMed] [Google Scholar]
- Lavrov DV. 2007. Key transitions in animal evolution: a mitochondrial DNA perspective. Integr Comp Biol. 47(5):734–743. [DOI] [PubMed] [Google Scholar]
- Lavrov DV. 2011. Key transitions in animal evolution: a mitochondrial DNA perspective In: Desalle R, Schierwater B, editors. Key transitions in animal evolution. Boca Raton (FL: ): CRC Press; p. 34–53. [Google Scholar]
- Lavrov DV, Boore JL, Brown WM.. 2002. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss. Mol Biol Evol. 19(2):163–169. [DOI] [PubMed] [Google Scholar]
- Leavitt JR, Hiatt KD, Whiting MF, Song H.. 2013. Searching for the optimal data partitioning strategy in mitochondrial phylogenomics: a phylogeny of Acridoidea (Insecta: Orthoptera: Caelifera) as a case study. Mol Phylogenet Evol. 67(2):494–508. [DOI] [PubMed] [Google Scholar]
- Londono GA, Chappell MA, Castaneda MD, Jankowski JE, Robinson SK.. 2015. Basal metabolism in tropical birds: latitude, altitude, and the ‘pace of life’. Funct Ecol. 29:338–346. [Google Scholar]
- Lounsberry ZT, et al. 2015. Next-generation sequencing workflow for assembly of nonmodel mitogenomes exemplified with North Pacific albatrosses (Phoebastria spp.). Mol Ecol Resour. 15(4):893–902. [DOI] [PubMed] [Google Scholar]
- Luo X, Kang XB, Zhang DY.. 2016. Complete mitochondrial genome of the American flamingo, Phoenicopterus ruber (Phoenicopteriformes, Phoenicopteridae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3519–3520. [DOI] [PubMed] [Google Scholar]
- Macey JR, Papenfuss TJ, Kuehl JV, Fourcade HM, Boore JL.. 2004. Phylogenetic relationships among amphisbaenian reptiles based on complete mitochondrial genomic sequences. Mol Phylogenet Evol. 33(1):22–31. [DOI] [PubMed] [Google Scholar]
- Maddison WP. 1997. Gene trees in species trees. Syst Biol. 46(3):523–536. [Google Scholar]
- Maddison WP, Maddison DR.. 2017. Mesquite: a modular system for evolutionary analysis. Version 3.31. Available from: https://www.mesquiteproject.org/
- Makarieva AM, et al. 2008. Mean mass-specific metabolic rates are strikingly similar across life’s major domains: evidence for life’s metabolic optimum. Proc Natl Acad Sci U S A. 105(44):16994–16999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP, Burg TM.. 2002. Perils of paralogy: using HSP70 genes for inferring organismal phylogenies. Syst Biol. 51(4):570–587. [DOI] [PubMed] [Google Scholar]
- Mayr G. 2013. The age of the crown group of passerine birds and its evolutionary significance—molecular calibrations versus the fossil record. Syst Biodivers. 11(1):7–13. [Google Scholar]
- Mayr G, Manegold A.. 2006. A small suboscine-like passeriform bird from the early oligocene of France. Condor 108(3):717–720. [Google Scholar]
- McKechnie AE, Freckleton RP, Jetz W.. 2006. Phenotypic plasticity in the scaling of avian basal metabolic rate. Proc R Soc B 273(1589):931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EA, Oliveira MT.. 2013. Replicating animal mitochondrial DNA. Genet Mol Biol. 36(3):308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab BK. 2009. Ecological factors affect the level and scaling of avian BMR. Comp Biochem Physiol A Mol Integr Physiol. 152:22–45. [DOI] [PubMed] [Google Scholar]
- McNab BK. 2012. Extreme measures: the ecological energetics of birds and mammals. Chicago (IL: ): University of Chicago Press. [Google Scholar]
- Meikejohn KA, et al. 2014. Incongruence among different mitochondrial regions: a case study using complete mitogenomes. Mol Phylogenet Evol. 78:314–323. [DOI] [PubMed] [Google Scholar]
- Mindell DP, Sorenson MD, Dimcheff DE.. 1998. Multiple independent origins of mitochondrial gene order in birds. Proc Natl Acad Sci U S A. 95(18):10693–10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya M, Nishida M.. 2000. Use of mitogenomic information in teleostean molecular phylogenetics: a tree-based exploration under the maximum-parsimony optimality criterion. Mol Phylogenet Evol. 17(3):437–455. [DOI] [PubMed] [Google Scholar]
- Moore WS. 1995. Inferring phylogenies from mtDNA variation—mitochondrial-gene trees versus nuclear-gene trees. Evolution 49:718–726. [DOI] [PubMed] [Google Scholar]
- Morgan-Richards M, et al. 2008. Bird evolution: testing the Metaves clade with six new mitochondrial genomes. BMC Evol Biol. 8(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz C, Brown WM.. 1987. Tandem duplications in animal mitochondrial DNAs: variation in incidence and gene content among lizards. Proc Natl Acad Sci U S A. 84(20):7183–7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Pocock JA, Taylor SA, Birt TP, Friesen VL.. 2010. Concerted evolution of duplicated mitochondrial control regions in three related seabird species. BMC Evol Biol. 10(1):14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle RG, Chesser RT, Prum RO, Schikler P, Cracraft J.. 2006. Phylogeny and evolutionary history of Old World suboscine birds (Aves: Eurylaimides). Am Mus Novit. 3544(1):1–22. [Google Scholar]
- Moyle RG, et al. 2016. Tectonic collision and uplift of Wallacea triggered the global songbird radiation. Nat Commun. 7(1):12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RL, Boore JL.. 2005. Molecular mechanisms of extensive mitochondrial gene rearrangement in plethodontid salamanders. Mol Biol Evol. 22(10):2104–2112. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Jarvis ED, Ellegren H.. 2010. Obtaining mtDNA genomes from next-generation transcriptome sequencing: a case study on the basal Passerida (Aves: Passeriformes) phylogeny. Mol Phylogenet Evol. 57(1):466–470. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson JI, Irestedt M, Ericson PGP, Fjeldsa J.. 2013. Phylogeny and classification of the New World Suboscines (Aves, Passeriformes). Zootaxa 3613(1):1–35. [DOI] [PubMed] [Google Scholar]
- Oliveros CH, et al. 2019. Earth history and the passerine superradiation. Proc Natl Acad Sci U S A. 116(16):7916–7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco MA, et al. 2011. Evolution of modern birds revealed by mitogenomics: timing the radiation and origin of major orders. Mol Biol Evol. 28(6):1927–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. 2000. Extracting species trees from complex gene trees: reconciled trees and vertebrate phylogeny. Mol Phylogenet Evol. 14(1):89–106. [DOI] [PubMed] [Google Scholar]
- Perna NT, Kocher TD.. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41(3):353–358. [DOI] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S.. 2011. Of circles, forks and humanity: topological organisation and replication of mammalian mitochondrial DNA. BioEssays 33:290–299. [DOI] [PubMed] [Google Scholar]
- Powell AF, Barker FK, Lanyon SM.. 2013. Empirical evaluation of partitioning schemes for phylogenetic analyses of mitogenomic data: an avian case study. Mol Phylogenet Evol. 66(1):69–79. [DOI] [PubMed] [Google Scholar]
- Pratt RC, et al. 2009. Toward resolving deep Neoaves phylogeny: data, signal enhancement, and priors. Mol Biol Evol. 26(2):313–326. [DOI] [PubMed] [Google Scholar]
- Prum RO, et al. 2015. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature 526(7574):569–573. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing; Available form: https://www.R-project.org/ [Google Scholar]
- Rambaut A. 2012. FigTree v1.4.0. Available from: http://tree.bio.ed.ac.uk/software/figtree/.
- Rambaut A, Lam TT, Max Carvalho L, Pybus OG.. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2(1):vew007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Pesole G, Saccone C.. 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 15(8):957–966. [DOI] [PubMed] [Google Scholar]
- Reyes A, Yang MY, Bowmaker M, Holt IJ.. 2005. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J Biol Chem. 280(5):3242–3250. [DOI] [PubMed] [Google Scholar]
- Rezende EL, Swanson DL, Novoa FF, Bozinovic F.. 2002. Passerines versus nonpasserines: so far, no statistical differences in the scaling of avian energetics. J Exp Biol. 205(Pt 1):101–107. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Robberson DL, Kasamatsu H, Vinograd J.. 1972. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci U S A. 69(3):737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, et al. 2007. Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup. PLoS Biol. 5(8):e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A, Holland PW.. 2000. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 15(11):454–459. [DOI] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roques S, Godoy JA, Negro JJ, Hiraldo F.. 2004. Organization and variation of the mitochondrial control region in two vulture species, Gypaetus barbatus and Neophron percnopterus. J Hered. 95(4):332–337. [DOI] [PubMed] [Google Scholar]
- Russo CAM, Takezaki N, Nei M.. 1996. Efficiencies of different genes and different tree-building methods in recovering a known vertebrate phylogeny. Mol Biol Evol. 13(3):525–536. [DOI] [PubMed] [Google Scholar]
- Ryu SH, Lee JH, Hwang UW.. 2012. Complete mitochondrial genome of the peregrine falcon Falco peregrinus (Aves, Falconiformes, Falconidae): genetic differences between the two individuals. Mitochondrial DNA 23(2):139–141. [DOI] [PubMed] [Google Scholar]
- Sabat P, Ramirez-Otarola N, Barcelo G, Salinas J, Bozinovic F.. 2010. Comparative basal metabolic rate among passerines and the food habit hypothesis. Comp Biochem Physiol A Mol Integr Physiol. 157:35–40. [DOI] [PubMed] [Google Scholar]
- Sammler S, Bleidorn C, Tiedemann R.. 2011. Full mitochondrial genome sequences of two endemic Philippine hornbill species (Aves: Bucerotidae) provide evidence for pervasive mitochondrial DNA recombination. BMC Genomics. 12(1):35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Mauro D, Gower DJ, Zardoya R, Wilkinson M.. 2006. A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Biol Evol. 23(1):227–234. [DOI] [PubMed] [Google Scholar]
- San Mauro D, et al. 2009. Experimental design in caecilian systematics: phylogenetic information of mitochondrial genomes and nuclear rag1. Syst Biol. 58(4):425–438. [DOI] [PubMed] [Google Scholar]
- Schirtzinger EE, et al. 2012. Multiple independent origins of mitochondrial control region duplications in the order Psittaciformes. Mol Phylogenet Evol. 64(2):342–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvatti AP, Galvão A, Pereira AG, Pedreira Gonzaga L, Russo CA.. 2017. An African origin of the eurylaimides (Passeriformes) and the successful diversification of the ground-foraging pittas (Pittidae). Mol Biol Evol. 34(2):483–499. [DOI] [PubMed] [Google Scholar]
- Selvatti AP, Gonzaga LP, Russo CA.. 2015. A Paleogene origin for crown passerines and the diversification of the Oscines in the New World. Mol Phylogenet Evol. 88:1–15. [DOI] [PubMed] [Google Scholar]
- Shao R, Barker SC, Mitani H, Aoki Y, Fukunaga M.. 2005. Evolution of duplicate control regions in the mitochondrial genomes of metazoa: a case study with Australasian Ixodes ticks. Mol Biol Evol. 22(3):620–629. [DOI] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE.. 1990. Phylogeny and classification of birds: a study in molecular evolution. New Haven (CT: ): Yale University Press. [Google Scholar]
- Singh TR, Shneor O, Huchon D.. 2008. Bird mitochondrial gene order: insight from 3 warbler mitochondrial genomes. Mol Biol Evol. 25(3):475–477. [DOI] [PubMed] [Google Scholar]
- Skujina I, McMahon R, Lenis VPE, Gkoutos GV, Hegarty M.. 2016. Duplication of the mitochondrial control region is associated with increased longevity in birds. Aging 8(8):1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack KE, Delsuc F, McLenachan PA, Arnason U, Penny D.. 2007. Resolving the root of the avian mitogenomic tree by breaking up long branches. Mol Phylogenet Evol. 42(1):1–13. [DOI] [PubMed] [Google Scholar]
- Stanton DJ, Daehler LL, Moritz CC, Brown WM.. 1994. Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics 137(1):233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover BC, Muller KF.. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh A, et al. 2011. Mesozoic retroposons reveal parrots as the closest living relatives of passerine birds. Nat Commun. 2(1):443.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveinsdottir M, Guomundsdottir L, Magnusson KP.. 2017. Complete mitochondrial genome of the gyrfalcon Falco rusticolus (Aves, Falconiformes, Falconidae). Mitochondrial DNA A DNA Mapp Seq Anal. 28:370–371. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Talavera G, Vila R.. 2011. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol Biol. 11(1):315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamashiro RA, et al. 2019. What are the roles of taxon sampling and model fit in tests of cyto-nuclear discordance using avian mitogenomic data? Mol Phylogenet Evol. 130:132–142. [DOI] [PubMed] [Google Scholar]
- Tang Y, Manfredi G, Hirano M, Schon EA.. 2000. Maintenance of human rearranged mitochondrial DNAs in long-term cultured transmitochondrial cell lines. Mol Biol Cell 11(7):2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman BI, Williams JB.. 2000. The adjustment of avian metabolic rates and water fluxes to desert environments. Physiol Biochem Zool. 73(4):461–479. [DOI] [PubMed] [Google Scholar]
- Umeda S, et al. 2001. Both heavy strand replication origins are active in partially duplicated human mitochondrial DNAs. Biochem Biophys Res Commun. 286(4):681–687. [DOI] [PubMed] [Google Scholar]
- Urantowka AD, Kroczak A, Mackiewicz P.. 2017a. Complete mitochondrial genome of bronze-winged parrot (Pionus chalcopterus chalcopterus, Psittaciformes). Mitochondrial DNA B Resour. 2:744–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urantowka AD, Kroczak A, Mackiewicz P.. 2017b. The influence of molecular markers and methods on inferring the phylogenetic relationships between the representatives of the Arini (parrots, Psittaciformes), determined on the basis of their complete mitochondrial genomes. BMC Evol Biol. 17(1):166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urantowka AD, et al. 2018. New insight into parrots’ mitogenomes indicates that their ancestor contained a duplicated region. Mol Biol Evol. 35:2989–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkuil YI, Piersma T, Baker AJ.. 2010. A novel mitochondrial gene order in shorebirds (Scolopacidae, Charadriiformes). Mol Phylogenet Evol. 57(1):411–416. [DOI] [PubMed] [Google Scholar]
- Wang H-W, Zhang H-F, et al. 2016. The whole mitochondrial genome of the Lesser Kestrel (Falco naumanni). Mitochondrial DNA A DNA Mapp Seq Anal. 27:2385–2386. [DOI] [PubMed] [Google Scholar]
- Wang N, Liang B, Huo J, Liang W.. 2016. Complete mitochondrial genome and the phylogenetic position of the Lesser Cuckoo, Cuculus poliocephalus (Ayes: Cuculiformes). Mitochondrial DNA A DNA Mapp Seq Anal. 27:4409–4410. [DOI] [PubMed] [Google Scholar]
- Wang XY, Huang Y, Liu N, Yang J, Lei FM.. 2015. Seven complete mitochondrial genome sequences of bushtits (Passeriformes, Aegithalidae, Aegithalos): the evolution pattern in duplicated control regions. Mitochondrial DNA 26(3):350–356. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ.. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West GB, Brown JH, Enquist BJ.. 1997. A general model for the origin of allometric scaling laws in biology. Science 276(5309):122–126. [DOI] [PubMed] [Google Scholar]
- White CR, Blackburn TM, Martin GR, Butler PJ.. 2007. Basal metabolic rate of birds is associated with habitat temperature and precipitation, not primary productivity. Proc R Soc B 274(1607):287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CR, Kearney MR.. 2014. Metabolic scaling in animals: methods, empirical results, and theoretical explanations. Compr Physiol. 4(1):231–256. [DOI] [PubMed] [Google Scholar]
- Wiersma P, Munoz-Garcia A, Walker A, Williams JB.. 2007. Tropical birds have a slow pace of life. Proc Natl Acad Sci U S A. 104(22):9340–9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerslev E, et al. 2009. Analysis of complete mitochondrial genomes from extinct and extant rhinoceroses reveals lack of phylogenetic resolution. BMC Evol Biol. 9(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, et al. 2015. A phylogeny of the Passerida (Aves: Passeriformes) based on mitochondrial 12S ribosomal RNA gene. Avian Res. 6:1. [Google Scholar]
- Zhou X, Lin Q, Fang W, Chen X.. 2014. The complete mitochondrial genomes of sixteen ardeid birds revealing the evolutionary process of the gene rearrangements. BMC Genomics. 15(1):573.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Shen XX, Hittinger CT, Rokas A.. 2018. Evaluating fast maximum likelihood-based phylogenetic programs using empirical phylogenomic data sets. Mol Biol Evol. 35(2):486–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.