Abstract
With multifactorial etiologies, combined with disease heterogeneity and a lack of suitable diagnostic markers and therapy, endometriosis remains a major reproductive health challenge. Extracellular vesicles (EVs) have emerged as major contributors of disease progression in several conditions, including a variety of cancers; however, their role in endometriosis pathophysiology has remained elusive. Using next-generation sequencing of EVs obtained from endometriosis patient tissues and plasma samples compared with controls, we have documented that patient EVs carry unique signatures of miRNAs and long noncoding RNAs (lncRNAs) reflecting their contribution to disease pathophysiology. Mass spectrophotometry–based proteomic analysis of EVs from patient plasma and peritoneal fluid further revealed enrichment of specific pathways, as well as altered immune and metabolic processes. Functional studies in endometriotic epithelial and endothelial cell lines using EVs from patient plasma and controls clearly indicate autocrine uptake and paracrine cell proliferative roles, suggestive of their involvement in endometriosis. Multiplex cytokine analysis of cell supernatants in response to patient and control plasma–derived EVs indicate robust signatures of important inflammatory and angiogenic cytokines known to be involved in disease progression. Collectively, these findings suggest that endometriosis-associated EVs carry unique cargo and contribute to disease pathophysiology by influencing inflammation, angiogenesis, and proliferation within the endometriotic lesion microenvironment.
Keywords: Reproductive Biology
Keywords: Expression profiling, Fertility, Molecular diagnosis
Next generation sequencing of extracellular vesicles from endometriosis patient tissues and plasma samples reveals a unique miRNA-lncRNA profile.
Introduction
Endometriosis is a major reproductive pathology that is defined by the benign growth of endometrial tissue in ectopic sites (1). Endometriosis places a major economic and clinical burden on the industrialized world (2), accounting for more than $95 billion and $1.8 billion dollars in annual societal costs in the United States and Canada, respectively (3, 4). The etiology is ultimately unknown; however, the widely accepted Sampson’s theory of retrograde menstruation suggests that menstrual effluent is refluxed into the peritoneal cavity by uterine contractions. Once in the peritoneum, this endometrial tissue can adhere to peritoneal structures, develop a blood supply, and develop into endometriotic lesions. However, 78%–90% of women experience retrograde menstruation, while only 5%–10% of women end up developing the disease (1, 2). This suggests that the women who develop endometriosis potentially have dysfunctional immune responses to the refluxed endometrial debris, which prevents their clearance and ultimately facilitates an inflammatory, angiogenic, and pathologic environment. Nevertheless, the early molecular mechanisms underlying immune system regulation, as well as early lesion establishment and proliferation, remain poorly understood.
Angiogenesis and inflammation are 2 key biological events that have been implicated in the pathogenesis and pathophysiology of endometriosis (5, 6). Indeed, increased proinflammatory cytokines persist in the peritoneal fluid (PF) of endometriosis patients (5). Previous findings from our group demonstrate that endometriotic lesions possess unique immune-inflammation gene signatures (7) and are critical promoters of systemic inflammation in endometriosis patients (8). Furthermore, we have shown the potential for aberrant endometriosis-associated cytokines and regulators of cytokine production to promote inflammation and stimulate the production of angiogenic cytokines in vitro (9, 10). However, it is not currently clear which regulators may mediate the perpetuation of inflammation and angiogenesis in the early endometriotic-endothelial cross-talk.
Extracellular vesicles (EVs), particularly exosomes, are bioactive nanovesicles that range in size, between 30–150 nm, and are released from the cell surface via exocytosis. EVs can be released by the vast majority of cell types that make up the human body, including the uterine fluid and endometrium (11, 12). EVs contain various contents, specific to the cell type, but have been broadly found to include RNAs, proteins, lipids, and small noncoding RNAs including microRNAs (miRNAs) and long noncoding RNAs (lncRNAs) (13–15). Indeed, immunomodulatory functions of exosomes are now widely recognized in several physiological conditions, including pregnancy or pathological scenarios such as various cancers and chronic inflammatory diseases with potential implications in development of novel diagnostic and therapeutic modalities (reviewed in ref. 16). In the context of endometriosis, there are no reports describing exosomal contents from endometriosis patients or their potential role in disease pathophysiology. To our knowledge, only 1 report so far suggests that exosomes derived from endometrial stromal cells of endometriotic lesions have proangiogenic effects on HUVECs in vitro (17).
With this important knowledge gap in mind, the primary objective of this study was to delineate unique small RNA and proteomic signatures in EVs from matched patient endometriotic and normal endometrial tissue, as well as matched patient plasma and peritoneal fluid compared to menstrual stage matched normal women. This study also sought to reveal the functional role of EVs in the endometrial epithelial-endothelial cross talk using a potentially novel in vitro coculturing-based approach. Ultimately, our study is the first to our knowledge to reveal unique exosomal/EV signatures in endometriosis patients and demonstrate increased angiogenesis when cocultured with HUVECs, and it provides dimensions in the complexities of endometriosis pathophysiology.
Results
Characterization of EVs, including exosomes, in endometriosis patient samples and human umbilical vein endothelial, endometrial and endometriotic epithelial cells.
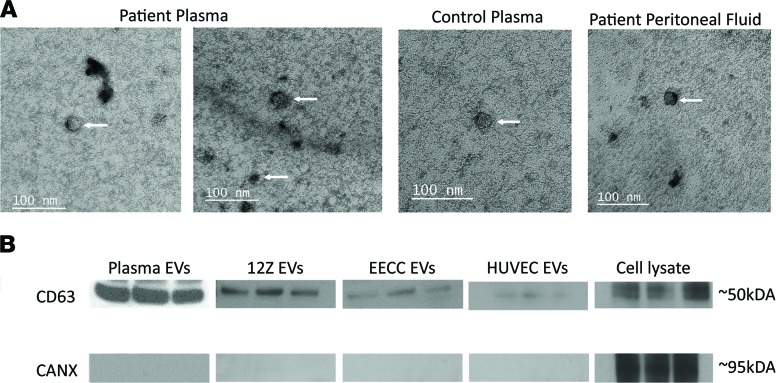
We isolated EVs, including exosomes, from endometriosis patients and healthy, fertile control subject tissue, PF, and plasma samples and verified them using transmission electron microscopy (TEM) (Figure 1A). TEM detected EVs ranging from 30–150 nm in size, further confirming the presence of exosomes (Figure 1A). We then probed for the presence of CD63, which is a tetraspanin family member commonly found in exosomes (Figure 1B). The endoplasmic reticulum–specific protein calnexin was not detected in any of our isolated EV samples (Figure 1B), suggesting purified EV isolation containing exosomes with very minimal to no detection of other cellular contaminants normally present in cell lysates. We also isolated EVs from the patient-derived endometriotic epithelial (12Z) cell line, as well as endometrial epithelial carcinoma (EECC) and HUVEC cell lines that serve as in vitro models to examine EV uptake and their functions relevant in the endometrial and endometriotic microenvironment.
Figure 1. Characterization of Extracellular vesicles, including exosomes, in endometriosis patient samples, HUVEC, and endometrial and endometriotic epithelial cells.
Transmission electron microscopy scans of purified extracellular vesicles (EVs), including exosomes, derived from endometriosis patient plasma, control plasma, and peritoneal fluid. (A). Extracellular vesicles, including exosomes, ranging in size from approximately 30–120 nm were detected in all 3 groups of patient samples (arrows point to isolated EVs in each group), and all group EVs displayed a cup-shaped morphology — a key signature of intact EV morphology. (B) Exosomal marker probing using Western blotting illustrated positivity of tetraspanin exosomal marker CD63 in all EV groups, as well as in cell lysate from 12Z cultured cells. The endoplasmic reticulum marker CANX was negative for all EV groups and positive for the cell lysate.
Small RNA species analysis using a next-generation sequencing platform revealed unique miRNA signatures implicated in endometriosis pathophysiology.
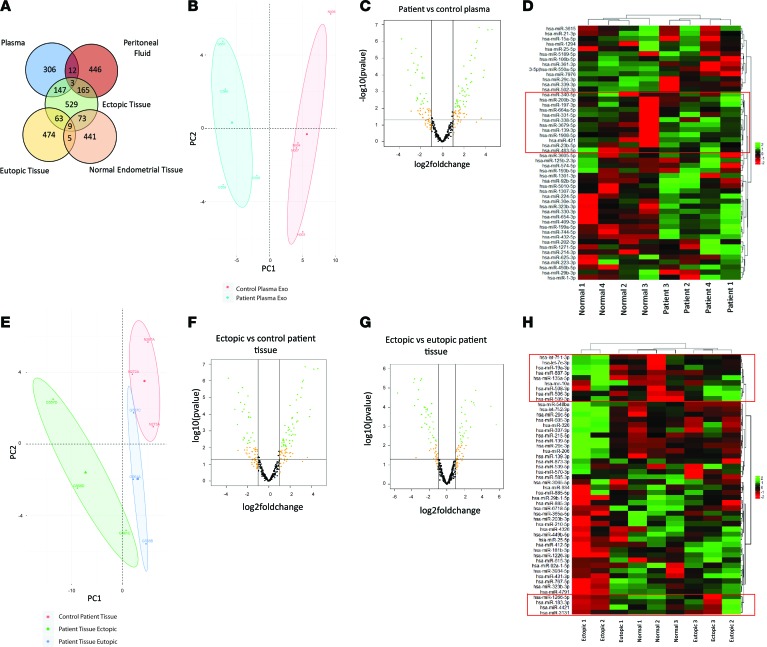
To unravel the unique EV, including exosomes, profiles in a well-defined and matched patient sample cohort, we analyzed the small RNA content in matched eutopic and ectopic endometriotic lesions, as well as from the matched patient peripheral blood and PF EVs. Biotype distribution within the next-generation sequencing (NGS) library revealed approximately 1 × 106 reads for miRNA, 1.8 × 105 reads for transfer RNA (tRNA), and approximately 1 × 104 reads for lncRNAs (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.128846DS1). Of the 2196 miRNAs detected across all samples, 529 belonged to the endometriotic lesion group, 446 to the PF, and 309 to the patient plasma groups (Figure 2A). Importantly, 14 miRNAs (miR-206, –29c-3p, –139-3p, –let-7a-3p, –95-3p, –29b-3p, –495-3p, –136-3p, –887-3p, –381-3p, –100-5p, –193b-3p, –335-5p, –411-5p) were differentially expressed (DE) between EVs isolated from ectopic endometriotic lesions and eutopic endometrium compared with endometrium from normal healthy, fertile women (Figure 2A). Principle component analysis (PCA) of control vs. patient plasma–derived EVs revealed distinct demarcation between each group (Figure 2B) and 14 DE miRNAs (Figure 2, C and D).
Figure 2. Small RNA species analysis using next-generation sequencing platform revealed unique miRNA signatures implicated in endometriosis pathogenesis and unique miRNA expression across tissue types and plasma samples compared with healthy control samples.
(A) Of the 2196 miRNAs detected across all samples using our small RNA–seq, 529 belonged to the ectopic tissue group, 446 to the peritoneal fluid, and 306 to the patient plasma groups. Importantly, 14 microRNAs (miR-206, –29c-3p, –139-3p, –let-7a-3p, –95-3p, –29b-3p, –495-3p, –136-3p, –887-3p, –381-3p, –100-5p, –193b-3p, –335-5p, –411-5p) were differentially expressed between EVs isolated from ectopic endometriotic lesions and eutopic endometrium compared with endometrium from normal healthy, fertile women. (B–D) Principle component analysis (PCA) of control vs. patient plasma–derived EVs (PC1, 40.7%; PC2, 15.9%) reveal demarcations between each group (B) and 14 differentially expressed (DE) miRNAs as indicated in the box plot and listed (C and D). (E) A multigroup comparison was also employed between eutopic, ectopic, and normal endometrium tissues. PCA (PC1, 50.4%; PC2, 17%) demonstrate primary overlap between eutopic and control patient tissue miR profiles and miR profiles for ectopic tissues from endometriosis patients. (F and G) A total of 78 and 63 miRs were differentially expressed in ectopic vs. control tissue and ectopic vs. eutopic patient tissues, respectively. (H) Ectopic vs. eutopic tissue differentially expressed miRs. n = 4 biological sample/group (B–D) and n = 3 biological sample/group (E–H).
In our second comparison, we employed a multigroup comparison between eutopic, ectopic, and normal endometrial tissues. Biotype distribution within each sample indicates highest that miRNA is the most abundant biotype among all endometriosis lesion and eutopic endometrium from endometriosis patients and normal endometrium (Supplemental Figure 2). Plasma samples differed and had an overall abundance of tRNA compared with miRNA, compared with other patient samples (Supplemental Figure 2). PCA revealed primary overlap between eutopic tissues from endometriosis patients and endometrial samples from healthy, fertile control tissue miRNA profiles, with a fully distinct miRNA profile for ectopic lesions from endometriosis patients (Figure 2E). A total of 78 and 63 miRNAs were DE in ectopic vs. control tissue and ectopic vs. eutopic (matched) patient tissues, respectively (Figure 2, F and G). Figure 2H lists ectopic vs. eutopic tissue DE miRNAs. We compared DE miRNAs between eutopic and ectopic tissues; miR-451a and miR–144-5p were significantly upregulated compared with normal endometrial tissue from fertile healthy controls (Supplemental Table 1). In control endometrial tissue and ectopic endometriotic lesions, miR–1266-5p, –200c-3p, –200a-3p, –20b-5p, –200a-5p, and –96-5p were significantly upregulated compared with matched eutopic tissues (Supplemental Table 1). Additionally, miR-451a and miR–144-5p were significantly upregulated in endometriotic lesions and matched eutopic endometrium from patients compared with normal endometrial tissues (Supplemental Table 1). To gain insights into the localized peritoneal vs. systemic microenvironment and to establish whether EVs from PF carry unique cargo compared with plasma EVs, we compared DE miRNAs between patient PF and plasma (Supplemental Figure 3, A–C). With the exception of 1 PF sample (no. 1; Supplemental Figure 3C), the groupings were profound, and 2 clusters of DE miRNAs were identified (Supplemental Figure 3C). The top and bottom clusters included 10 and 21 DE miRNAs, respectively.
Validation of miRNAs DE in multiple sample and comparison groups using quantitative PCR (qPCR).
Eleven miRNAs (miR-27a, -30d, -100, -136, -144, -193b, -200c, –200a-5p, -206, -375, –let-7a) DE in multiple sample groups from the small RNA–seq comparisons (from Supplemental Table 1) were selected for validation purposes (Supplemental Figure 4). Of the 11, 6 were validated (miR-27a, -200c, –200a-5p, -375, -144, –let-7a), which were composed of at least 1 miRNA from each group of DE miRNAs (Supplemental Table 1). Notably, miR-27a and -375 from our network analysis were significantly different for all group comparisons, and miR-30d, while not significant, demonstrated similar expression patterns as miR-375 (P = 0.09 for comparisons).
Unique miRNA expression across tissue types and plasma samples compared with healthy controls samples.
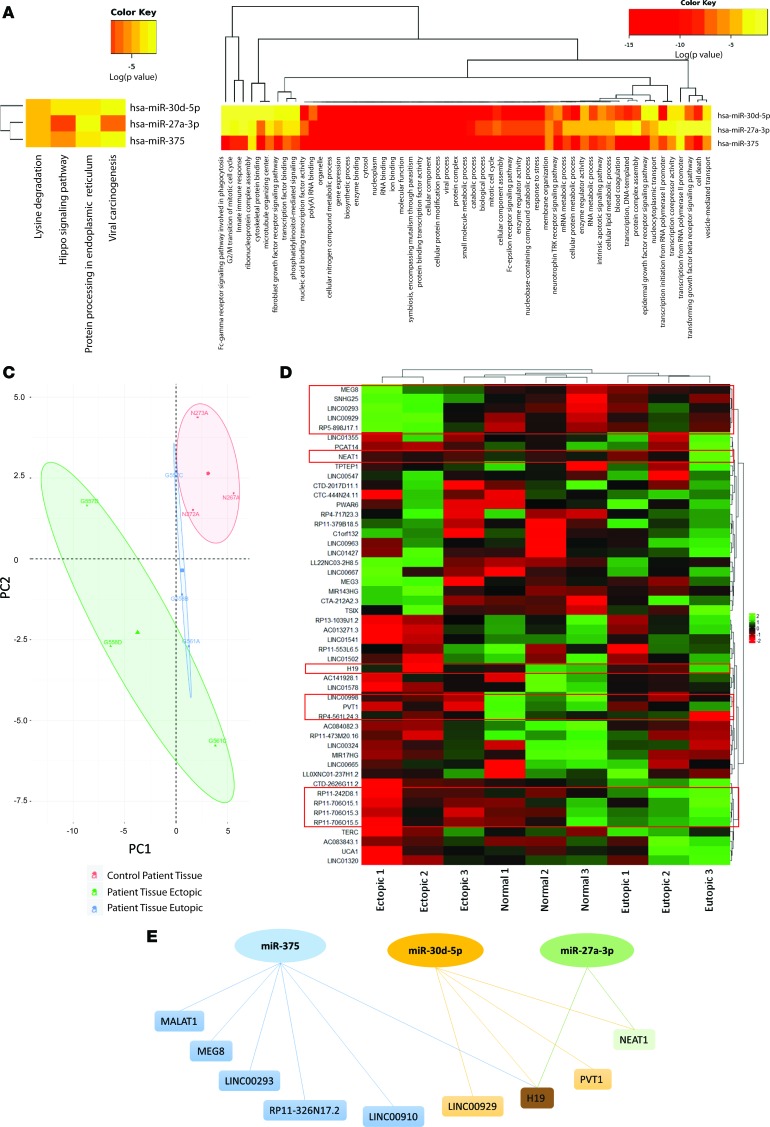
We further curated the miRNA data from EVs to determine some of the unique miRNA signatures DE across all comparisons. In terms of patient vs. control plasma–derived EVs, 21 miRNAs were DE (Supplemental Table 1), with miR-375, miR–27a-3p, and miR–30d-5p as examples of being significantly downregulated in patient plasma compared with healthy, fertile control plasma EVs. Two of the 3 miRNAs (miR-375 and miR–30d-5p) were significantly downregulated in endometriosis patients (ectopic endometriotic lesions compared with normal endometrium; Supplemental Table 1) and also upregulated in ectopic lesions when compared with matched eutopic endometrium (Supplemental Table 1). miR–27a-3p, on the other hand, was significantly upregulated in endometriosis patient tissues (both eutopic and ectopic compared with normal endometrium) and displayed significantly higher expression in the eutopic endometrium compared with ectopic lesion. These 3 miRNAs may be highly relevant to the disease state, as they were also found to be DE in both patient plasma- and tissue-derived EVs. This unique miRNA signature was subjected to DIANA miRpath software tools for miRNA pathway union analysis. This analysis revealed that lysine degradation, hippo signaling pathway, protein processing in endoplasmic reticulum, and viral carcinogenesis are the 4 molecular processes implicated with this miRNA signature (Figure 3A). Additionally, miRpath analysis listed vesicle-mediated transport with these miRNAs, particularly hsa-miR-27a-3p and miR–30d-5p, as well as small molecular metabolic processes, RNA binding, and protein complexes (Figure 3B).
Figure 3. lncRNA-miRNA–seq analysis demonstrate intricate endometriosis-specific lncRNA miR-375, –30d-5p, –27a-3p axis network.
(A) miRpath analysis revealed that lysine degradation, hippo signaling pathway, protein processing in ER, and viral carcinogenesis are the 4 molecular processes implicated with this miRNA signature. (B) Vesicle-mediated transport is associated with miR–27a-3p and –30d-5p, as well as small molecular metabolic processes, RNA binding, and protein complexes. (C) PCA for PC1 (46.7%) and PC2 (23.2%) grouping for control patient tissue EVs and moderate overlapping between eutopic tissue EVs and control patient tissue EVs, with groupings of ectopic tissue EVs. (D) DE analysis for small RNAs, including lncRNAs, revealed key signatures between matched ectopic and eutopic tissues, as well as when compared with normal endometrial tissue from patients. Heatmap analysis of DE lncRNAs between ectopic and eutopic plus normal control endometrium show a cluster of lncRNAs, including MEG8, SNHG25, LINC00293, LINC00929, and RP5-898J17.1, which are reduced in ectopic endometriotic lesion EVs (top red outline). The lncRNAs NEAT1 and H19 cluster in ectopic endometriotic lesions (second and third red outlines from top). The lncRNAs LINC00998, PVT1, and RP4-561L24.3 also significantly cluster in ectopic endometriotic lesion EVs (fourth red outline from top). Further, lncRNAs RP11-242D6.1, RP11-706O15.1, -.3, -.5 also significantly cluster in ectopic endometriotic lesion EVs (fifth red outline from top). (E) lncRNAs NEAT1 and H19 were the only lncRNAs that shared predicted binding sites with 2 or more miRNAs from this axis, with H19 being the only that had predicted binding sites with all 3 miRNAs. n = 3 biological sample/group.
lncRNA-miRNA–seq analysis demonstrate intricate lncRNA-miR375, miR-30d-5p, miR-27a-3p axis network.
Small RNA species including lncRNAs were further analyzed using both small RNA–seq and in silico LncBase software to examine predicted lncRNA-miRNA interactions (Supplemental Table 2). Since lncRNA read counts were low for plasma, these samples were excluded from analysis, and only tissue EVs were analyzed. PCA revealed high percentage grouping for healthy, fertile control tissue EVs and moderate overlapping between eutopic tissue EVs and healthy, fertile control tissue EVs, with distinct grouping of ectopic tissue EVs (Figure 3C). Differential expression analysis for small RNAs, including lncRNAs, revealed key signatures between matched ectopic and eutopic tissues, as well as when compared with endometrial tissue from healthy, fertile controls (Figure 3D). Heatmap analysis of DE lncRNAs between ectopic, eutopic, and healthy, fertile control endometrium show a cluster of lncRNAs including MEG8, SNHG25, LINC00293, LINC00929, and RP5-898J17.1, which are downregulated in ectopic endometriotic lesion EVs compared with both matched eutopic lesions and normal endometrium from healthy, fertile controls (Figure 3D, top red outline). Additionally, the lncRNAs NEAT1 and H19 were significantly upregulated in ectopic endometriotic lesions (Figure 3D, second and third red outline from top) compared with normal endometrium from healthy, fertile controls. The lncRNAs LINC00998, PVT1, and RP4-561L24.3 also significantly cluster and are significantly upregulated in ectopic endometriotic lesion EVs compared with normal endometrium from healthy, fertile controls (Figure 3D, fourth red outline from top). These lncRNA clusters all share predicted miRNA binding sites between miR–30d-5p, miR–27a-3p, and miR-375; however, 1 cluster of lncRNAs that are significantly upregulated in endometriotic lesions does not (Figure 3D, bottom red outline). Using a combination of software including LncBase, a custom predictive binding network map using the lncRNA clusters and miR-375, miR–30d-5p, and miR-27a-3p was created (Figure 3E). LncRNAs NEAT1 and H19 shared predicted binding sites with 2 or more miRNAs from this axis, with H19 being the only lncRNA that had predicted binding sites with all 3 miRNAs (Figure 3E). Additionally, TargetScan for the proteomic signatures implicated in crucial angiogenesis and inflammatory-related pathways obtained using mass spectrometry (MS) in endometriosis patients were analyzed for conserved miRNA binding sites. Thrombospondin-1 (TSP1/THBS1), TSP2/THBS2, pigment epithelium-derived factor (SERPINF1), α-1-antitrypsin (SERPINA1), angiopoietin-related protein 6 (ANGPTL6), PDGF-α (PDGFA), and IFN regulatory factor 4 (IRF4) were all detected from the proteomic database and share binding sites with the unique miRNA signatures. Our TargetScan for conserved binding sites indicated that miR-27a has binding sites for SERPINA1, THBS1, and PDGFA; miR-30d has binding sites for THBS2, SERPINF2, and IRF4; and miR-375 has binding sites for PDGFA. We also used the bioinformatics tool miRNet, a network-based visual analysis of miRNAs, targets, and functions, to develop further functional interpretations from our small RNA–seq data (18). We compiled and developed a node of annotated common genes that have binding sites shared among miR–30d-5p, miR–27a-3p, and miR-375, and several key genes responsible for proinflammatory and proangiogenic signaling, such as ZFP36L2 (antiinflammatory), HOXB3 (proangiogenic), and the tumor suppressor gene TP53 were identified (Supplemental Figure 5A). Additionally, significant interacting genes (P < 0.05) of miR–30d-5p, miR–27a-3p, and miR-375 were computed from the Reactome database (19) and highlighted (Supplemental Figure 5B). Many cytokines and signaling pathway genes implicated in endometriosis, as well as inflammation and angiogenesis, were shared among 2 or all 3 miRNAs in the network (IL-1A, IFNG, EGFR, ERBB2, MAP3K8, JAK2, STAT3, NOTCH1, YAP1, IRF3; Supplemental Figure 5B). Furthermore, genes heavily implicated in cancer were also detected (MYC, TP53, KRAS; Supplemental Figure 5B). It is important to note that mutations in KRAS have also been previously identified as cancer-associated mutations in endometriosis without cancer (20), and activation of KRAS contributes to the pathogenesis of endometriosis (21).
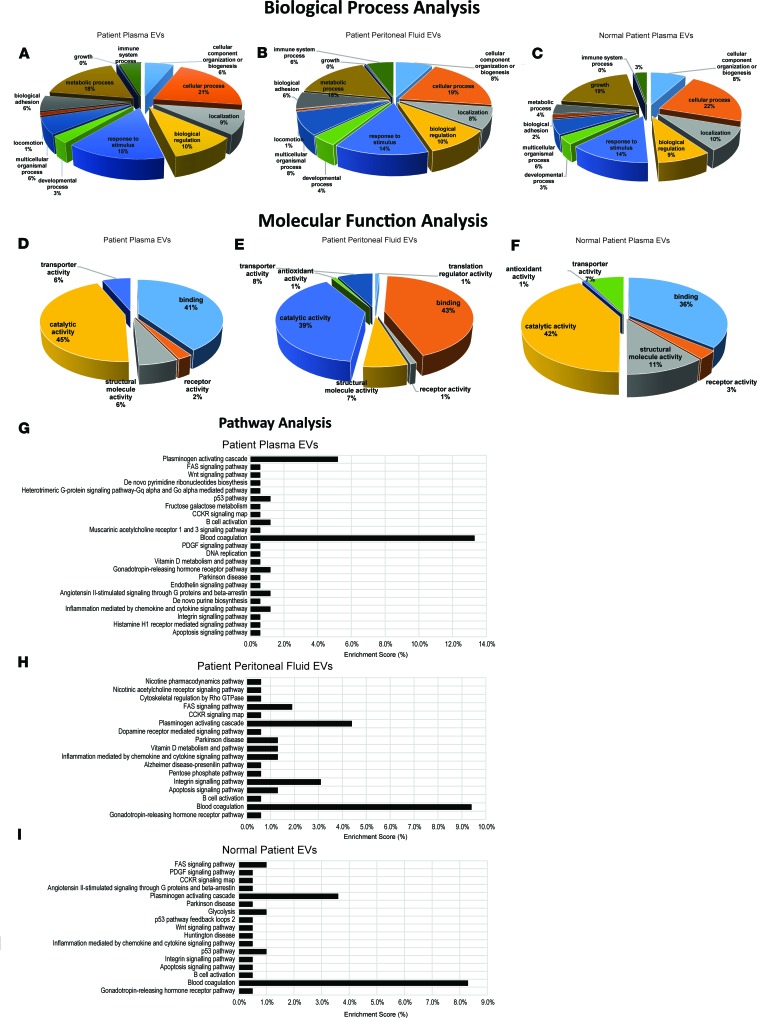
Proteomiccharacterization of matched ectopic endometriotic lesion, eutopic endometrium, patient plasma, and PF EVs, including exosomes, using MS. In order to understand the vast protein array networks implicated both in EV biology and endometriosis pathophysiology, we conducted MS in the same matched patient and control sample cohorts that were analyzed by RNA-seq. We also conducted MS of 2 representative epithelial cell lines, including 12Z endometriotic epithelial and EECC EVs, and of the primary endothelial cells, HUVECs (Supplemental Figure 6). To delineate the categories of proteins contained in different EV subtypes, we conducted PantherDB gene ontology (GO) analysis and separated our findings based on biological process, molecular function, and pathways implicated (Figure 4). Biological process analysis of patient plasma and PF EVs vs. healthy, fertile control plasma EVs revealed elevated immune system and metabolic processes in both endometriotic plasma and PF EVs compared with healthy, fertile control plasma (Figure 4, A–C). The same analysis in cell lines revealed that HUVECs have a unique biological process profile, compared with the 2 epithelial-based cell lines, displaying higher developmental process, lower biological regulation, higher biological adhesion, and higher immune system process (Supplemental Figure 6, A–C). 12Zs display zero-growth biological process (vs. 2% in EECC and HUVECs; Supplemental Figure 6, D–F). EECCs display higher metabolic process compared with both HUVECs and EECCs (Supplemental Figure 6, D–F).
Figure 4. Proteomic characterization of matched ectopic endometriotic lesion, eutopic endometrium, patient plasma, and peritoneal fluid EVs, including exosomes, using mass spectrometry.
(A–I) PantherDB gene ontology analysis was performed on EVs from endometriosis patients and separated our findings based on biological process (A–C), molecular function (D–F), and pathways implicated (G–I). Biological process analysis of patient plasma and peritoneal fluid EVs vs. normal plasma EVs revealed higher immune system and metabolic processes in both endometriotic plasma and PF EVs compared with normal patient plasma (A–C). Molecular function analysis of patient plasma and peritoneal fluid EVs vs. normal plasma EVs revealed higher binding and lower structural molecule activity in patient plasma and PF EVs compared with normal patient plasma EVs (D–F). Pathway analysis revealed that pathways implicated in patient plasma EVs were more abundant than in normal patient plasma EVs (24 total pathways compared with 17 and 18 peritoneal fluid and normal patient plasma EVs, respectively; G–I). Proteins implicated in blood coagulation pathway were markedly increased in patient plasma compared with normal patient plasma EVs (G vs. I). Both patient plasma and peritoneal fluid EVs also had an increase in inflammation mediated by chemokine and cytokine signaling pathway and B cell activation compared with normal patient plasma EVs (G–I). n = 6 biological sample/group.
Molecular function analysis of patient plasma and PF EVs vs. healthy, fertile control plasma EVs revealed higher binding and lower structural molecule activity in patient plasma and PF EVs compared with healthy, fertile control plasma EVs (Figure 4, D–F). In the cell lines, EECCs and 12Z endometriotic epithelial cells possess similar molecular function profiles, with lower or no receptor activity compared with HUVECs (Supplemental Figure 6, D–F). HUVECs displayed the presence of antioxidant activity compared with the absence of this function in both 12Zs and EECCs (Supplemental Figure 6, D–F).
Pathway analysis revealed that pathways implicated in patient plasma EVs were more multifactorial than in healthy, fertile control plasma EVs (24 total pathways compared with 17 and 18 PF and healthy, fertile control plasma EVs, respectively; Figure 4, G–I). Proteins implicated in blood coagulation pathways were markedly increased in patient plasma EVs compared with healthy, fertile control plasma EVs (Figure 4, G–I). Both patient plasma and PF EVs also had an increase in inflammation mediated by the chemokine and cytokine signaling pathway and B cell activation compared with normal control plasma EVs (Figure 4, G–I). In the cell lines, HUVEC EVs displayed a higher-grade integrin signaling pathway compared with 12Z and EECC EVs; EECCs displayed medium and 12Zs displayed the lowest integrin signaling pathways (Supplemental Figure 6, G–I). Additionally, 12Z EVs have markedly higher cytoskeletal regulation by Rho GTPase compared with HUVECs and EECCs, and the CCKR signaling map pathway is present only in 12Z EVs (Supplemental Figure 6G). HUVECs have a higher plasminogen activating cascade pathway compared with EECC and 12Z EVs (Supplemental Figure 6, G–I).
Uptake of endometriosis patient plasma and PF EV, as well as endometriotic and endometrial epithelial–derived EV uptake in human endothelial cells.
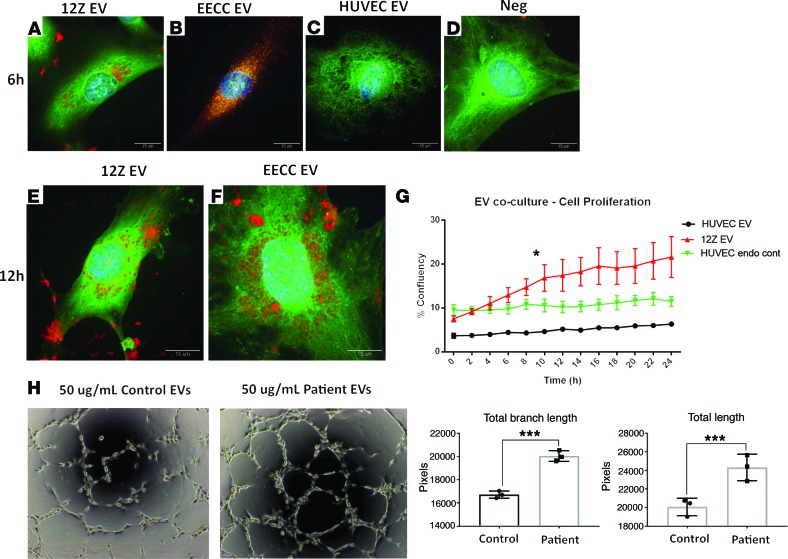
To gain further insight as to the early pathogenesis of endometriosis, specifically as the endometriotic epithelial cells establish a communication with endothelial cells to gain vascularity via angiogenic cytokines, we examined EV uptake using confocal microscopy and the live cell imaging platform, Incucyte. We cocultured fluorescently labeled EVs derived from 12Zs (endometriotic epithelial), EECCs, and HUVECs. We observed similar EV uptake in endothelial cells from both 12Z- and EECC-derived EV cocultured groups at 6 hours (Figure 5, A and B), compared with endogenous and negative controls (Figure 5, C and D). Interestingly, we observed an accumulation of fluorescently labeled 12Z- and EECC-derived EVs in the perinuclear and cytoplasm of endothelial cells after 12 hours (Figure 5, E and F). The same phenomenon was observed using the Incucyte platform, and we detected continued accumulation of patient-derived EVs in endothelial cells (Supplemental Video 1 and Supplemental Figure 7). EV uptake was confirmed using the video-codec package from ImageJ to compile videos, and it was observed that EVs were indeed taken up by the cell and not overlaid when imaged (Supplemental Video 1).
Figure 5. Uptake of endometriosis patient plasma and peritoneal fluid EVs, as well as endometriotic and endometrial epithelial–derived EV uptake in human endothelial cells and extracellular vesicle, including exosomes, coculture with HUVECs results in increased angiogenesis.
(A–D) Confocal microscopy images of labeled EVs from 12Z (A), EECC (B), HUVEC (C) and negative (raw HUVEC supernatant labeled; no EV purification) (D) at 6 hours in HUVECs. 12Z-labeled EVs were found in moderate to high quantities in the cytoplasm of endothelial cells (A) at 6 hours. EECC-labeled exosomes were detected in high to very high levels in the cytoplasm and perinuclear space in endothelial cells at 6 hours (B). HUVEC-labeled exosomes were detected in low quantities in the cytoplasm and nucleus of endothelial cells at 6 hours (C), and no exosomes were detected in endothelial cells from negative group (D). (E and F) 12Z- and EECC-labeled exosomes accumulated in high quantities in the cytoplasmic, perinuclear, and basal locations of endothelial cells at 12 hours. (G) Confluencies were calculated using the cell proliferation algorithm (Incucyte software) and plotted over 24 hours for each group, with 12Z EVs displaying increased cell proliferation at 10 hours (2-way RM ANOVA). (H) Bright-field images of endometriosis patient plasma EVs cocultured with HUVECs results in increased angiogenesis compared with healthy, fertile control plasma EVs (t test). Branch length and total lengths were plotted and compared between endometriosis patient plasma EVs cocultured with HUVECs compared with healthy, fertile control plasma EVs. Expression data illustrated as mean ± SEM. *P < 0.05. Green dye indicate cytoplasm, blue dye indicate nucleus (DAPI), and red dye indicate fluorescently labeled exosomes. Images representative of n = 3 independent experiments. Scale bar: 15 μm.
EV, including exosome coculture with human umbilical vein endothelial cells, results in increased angiogenesis.
To investigate functional effects and uptake in the epithelial-endothelial cross-talk, we cocultured EVs derived from 12Zs (endometriotic epithelial), EECCs, and HUVECs. We fluorescently labeled EVs and cocultured with HUVECs to subsequently monitor EV uptake and cell proliferation via the Incucyte live cell imaging platform. Additionally, supernatants from HUVECs were collected and fluorescent label was added without the EV purification step in order to control for endogenous HUVEC EV uptake. Endothelial cells in all treated groups took up EVs as early as 0–2 hours upon initial EV addition (Supplemental Figure 7, A–L). This was confirmed via fluorescent histograms, which illustrated significant increases in the red fluorescent (EV/exosome labeled) channel, which matches data presented illustrating EV uptake using confocal microscopy (Figure 5, A, B, E, and F). Red fluorescent intensity detected in the endothelial cells continued to increase for all time points (Supplemental Figure 7, M–P). Significantly increased cellular proliferation (P < 0.05) was observed in endothelial cells that took up EECC- and 12Z-derived EVs (Figure 5G). Additionally, HUVECs treated with patient plasma–derived EVs had significantly elevated tube formation, suggesting increased angiogenesis compared with normal plasma-derived EVs (Figure 5H). Both branch length (P = 0.0005) and total lengths (P = 0.0012) were significantly different (Figure 5H).
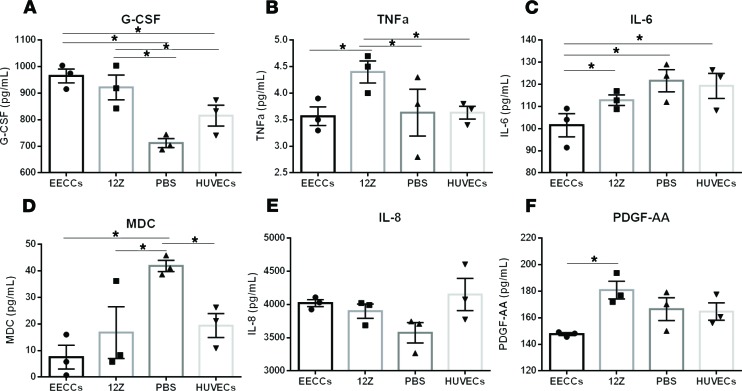
Endometriotic-specific proangiogenic and proinflammatory EV signatures exist in endothelial cells cocultured with EVs from 12Z and EECCs.
Endometriotic epithelial cells have been previously established to be highly invasive and able to establish a blood supply via increased angiogenesis in a highly inflammatory microenvironment (22). To investigate the early effects of endometriotic and endometrial-endothelial cross-talk in the perpetuation of inflammation and potential angiogenesis, we analyzed proangiogenic and proinflammatory cytokines using a 42-plex cytokine array. Of the 42 cytokines analyzed, granulocyte CSF (G-CSF) was significantly upregulated in both endothelial cell groups treated with EECC- and 12Z-derived EVs compared with both HUVEC-derived EVs and HUVEC endogenous control groups (P < 0.05; Figure 6A). Additionally, the proinflammatory cytokine TNFα was also significantly upregulated in endothelial cells treated with 12Z-derived EVs compared with cells treated with EECC, HUVEC-derived EVs, and HUVEC endogenous control (P < 0.05; Figure 6B), which suggests a proinflammatory microenvironment stimulated by 12Z endometriotic EV contents. We observed significant downregulation of the proinflammatory cytokine IL-6 in endothelial cells treated with EECC-derived EVs when compared with other groups (P < 0.05; Figure 6C). With the exception of HUVEC endogenous control, we also observed a significant downregulation of macrophage-derived chemokine (MDC), which is primarily produced by macrophages and DCs and is downregulated by Th1-type cytokines; upon stimulation, it is involved in the recruitment of DCs and Th2-type cytokines, in all treatment groups of EVs, including HUVEC EVs treated back onto endothelial cells (P < 0.05) (Figure 6D). We observed no significant differences in the inflammatory cytokine IL-8 (Figure 6E). Finally, significantly elevated levels of PDGF, a key proangiogenic factor, were observed in endothelial cells treated with 12Z-derived EVs compared with cells treated with EECC-derived EVs (P < 0.05; Figure 6F).
Figure 6. Endometriotic-specific proangiogenic and proinflammatory EV signatures exist in endothelial cells cocultured with EVs from 12Z and EECCs.
(A) Of the 42 cytokines analyzed with protein multiplexing, granulocyte CSF (G-CSF) was significantly upregulated in both endothelial cell groups treated with EECC and 12Z-derived EV compared with both HUVECs-derived EV and HUVEC endogenous control groups (P < 0.05). (B) TNFα was also significantly upregulated in endothelial cells treated with 12Z-derived EV compared with cells treated with EECC, HUVECs-derived EV, and HUVEC endogenous control (P < 0.05). (C) IL-6 was significantly downregulated in endothelial cells treated with EECC-derived EVs when compared with other groups (P < 0.05). (D) With the exception of HUVEC endogenous control, macrophage-derived chemokine (MDC) was downregulated in all treatment groups of EVs, including HUVEC EVs treated back onto endothelial cells. (E) No significant differences in the inflammatory cytokine IL-8. (F) PDGF-AA was significantly higher in endothelial cells treated with 12Z-derived EVs compared with cells treated with EECCs (P < 0.05). All comparisons were made using a 1-way ANOVA. n = 3 independent experiments. Expression data illustrated as mean ± SEM. *P < 0.05.
Discussion
In this study, we sought to determine unique exosomal contents and their potential role in endometriosis pathophysiology using patient samples and representative cell lines. Purified EV populations containing exosomes have been previously isolated and characterized in the uterine microenvironment and have been shown to participate as a form of intercellular communication (23). However, in the context of endometriosis, there are no reports of non–endometriotic stromal cell (ESC) patient-derived exosomes (17). One of the core findings of this study is that EVs obtained from endometriotic lesions carry a unique miRNA signature compared with EVs derived from matched patient eutopic endometrium and normal healthy endometrium. We discovered miRNAs within EVs that have been previously established to be functionally relevant in the pathophysiology of endometriosis (24), such as let-7a (miR–let-7a) (downregulated in ectopic compared with eutopic tissues), miR-23a (upregulated in ectopic compared with eutopic tissues), miR-143, and miR-320a. Furthermore, we sought to determine if we could capture some of these miRNA alterations in the EVs obtained from the peripheral blood of endometriosis patients. Indeed, we found that patient plasma–derived EVs carry unique miRNAs compared with healthy control EVs, such as miR–30d-5p, miR–16-5p, and miR–27a-3p. When we accounted for the sample-type comparisons, we found selected miRNAs including miR–30d-5p, miR–27a-3p, and miR-375 that are unique to endometriosis, and these miRNAs were corroborated with findings from other groups implicating them in endometrial pathologies, including endometriosis (25–27).
Previous work in endometriosis has also corroborated our findings, with a report indicating that let-7a is downregulated in serum in a mouse model of endometriosis (28), miR-23a is upregulated in ectopic compared with eutopic tissues (29), and differential expression of miR-143 and -320a in endometriomas is shown compared with endometrium (24). Our qPCR-based validation studies involved additional DE miRNAs from Supplemental Table 1 of commonly grouped DE miRNAs such as miR-29c, which has been reported to be downregulated in ectopic endometrium (30), as well as miR-451, which is elevated in endometriotic tissue compared with paired eutopic endometrium from women with disease in the patient cohort from (29) (Ectopic > Normal and Eutopic > Normal tissues as detected in the small RNA-seq data). These findings corroborate with the literature and point toward the potential role of EVs in modulating disease pathophysiology. With respect to plasma EVs, miR-27a, which we found to be downregulated in plasma EVs from endometriosis patients compared with control, was also found in the plasma samples from endometriosis patients compared with controls free of endometriosis (31). This miR has been demonstrated in the literature to be implicated both in autophagy and in regulation of inflammatory response by targeting IL-10 (32). A literature scan of miR-30d, another DE miRNA specifically downregulated in plasma from endometriosis patients compared with control, did not reveal any association. This miR has been shown to be secreted by the human endometrium into exosomes/EVs (33) and is a known plasma circulating inflammation–related miR (34). When put together, our findings of these EVs in diseased vs. normal plasma is in direct agreement with the endometriosis literature, and we hypothesize that these miRs are likely deficient in endometriosis patient plasma EVs, consequential to the dysregulated chronic inflammation. Since endometriosis lesion microenvironment is complex and dynamic, future functional studies should be targeted to establish disease-specific exosome signatures and determine individual stromal, epithelial, endothelial, and immune EV contents from the lesion microenvironment.
While the field of lncRNA research is in its infancy, we attempted to ascertain important potential lncRNA-miRNA interactions in endometriosis. Using our discovery patient cohort, our analysis revealed an intricate network of lncRNAs that share binding sites with our DE miRNAs from all comparison groups, with lncRNAs H19 and NEAT1 being the only 2 that yielded more than 1 miRNA predicted binding site. Both of these lncRNAs have been previously implicated in endometrial pathologies involving cell proliferation of stromal cells via insulin growth factor signaling, as well as invasive and migratory abilities (35, 36), with H19 mRNA levels being significantly downregulated in eutopic endometrium from women with endometriosis compared with healthy controls (35). Our lncRNA-seq results are in accordance with these findings, and notably, we did not see this difference in the eutopic endometrium from endometriosis patients. We believe that these lncRNAs, as well as the clusters that we discovered, will likely yield diagnostic benefit, since there was a stark contrast between the matched ectopic and eutopic endometrial tissue from these patients, as well as a contrast compared with endometrium from normal, fertile, healthy control women. Additionally, future studies investigating lncRNA-miRNA interactor signatures in primary stromal cells will further elucidate the epithelial-stromal EV roles in endometriosis. Furthermore, the data generated from these analyses will prove beneficial as future algorithms and software are developed to better infer pathology with lncRNA-miRNA interactions.
A primary focus was to identify functional effects of EVs derived from endometriotic and endometrial epithelial cells on endothelial cell uptake. We report for the first time to our knowledge increased endothelial cell proliferation resulting from treatment with endometriotic and endometrial epithelial–derived EVs. We also report a stimulation of inflammatory and angiogenic factors in endothelial cells cocultured with the aforementioned EVs, with higher levels of angiogenic and inflammatory cytokines being present in the cells cocultured with the endometriotic epithelial EVs. These EVs exhibit alternative modes of paracrine cell proliferative and endocrine communication roles via their functions based on their unique cargo, which are able to modify the activity or properties of the recipient endothelial target cells. Based on the chronic inflammatory microenvironment both in the ectopic endometriotic lesion and eutopic endometrium in patients with endometriosis and the unique cargo that the EVs from these types carry compared with normal endometrium, there exists the paradigm that these EVs further perpetuate the inflammatory state via their paracrine roles. Similar paradigms have been previously hypothesized in different human pathologies (37, 38). Previous reports have demonstrated that miR-16 and -30d specifically can regulate and promote angiogenesis by targeting the VEGF and MYPT1/cJUN/VEGFA pathway, respectively (39, 40), and we also found differential expression of miR-16 and -30d in the patient plasma–derived EVs compared with healthy control EVs. These findings are in direct agreement of our hypothesis and suggest that angiogenic factors released from these EVs, including exosomes, are responsible for the increased endothelial cell proliferation and, although speculative, that the inflammatory cytokine profiles reflect an inherent return to inflammatory phenotype for the 12Z endometriotic cells. These cells have been previously established to be highly invasive and can, thus, thrive in inflammatory conditions, due to their lineage and immortalized generation from endometriotic epithelial cells (22). Indeed, further mechanistic work using luciferase assays will be required to examine and validate these properties in vitro with respect to the endometriotic epithelial-endothelial microenvironment.
One of the major limitations facing this study is that all of the mechanistic evidence was performed in vitro. Although this study elucidated many altered molecular pathways involved in this challenging disease, future studies in an in vivo model will be beneficial in determining whether this complex intercellular communication modality fully or partially can be recapitulated in the pathogenesis of endometriotic lesions. Another confounding factor may be an inherent database bias used to obtain network analyses, as these databases are modeled with a propensity for cancer research using large patient cohorts, such as The Cancer Genome Atlas. Additionally, it is uncertain whether all exosome subsets in the EV population will be uptaken or if there is selective sorting that occurs in the presence of local factors in vivo. Techniques including fluorescent tagging of differential EV subsets will be required to further delineate this modality.
To the best of our knowledge, this report is the first to demonstrate functional effects of both patient-derived endometriotic epithelial and highly characterized and established immortalized endometriotic epithelial and endometrial epithelial–derived EVs, including exosomes, in the endometrial-endothelial cross-talk. Overall, we demonstrate the following: (a) potentially novel miRNA-lncRNA proteomic signatures and networks inherent in endometriosis patient EVs compared with controls; (b) unique endometriotic and endometrial epithelial-endothelial uptake and cross-talk in endothelial cells; (c) direct angiogenic and inflammatory effects exerted from endometriotic epithelial EVs on endothelial cells; (d) increased endothelial cell angiogenesis (tube formation) by endometriosis patient EVs, with a profound increase in cellular growth occurring from endometriotic epithelial EV uptake.
Currently, there are no effective treatments to either cure or provide remission of severe symptoms from endometriosis. Surgery is the only available approach for advanced cases, which comprise the majority of patients with this disease, due to a lack of biomarkers to diagnose early cases. Indeed, over half of the patients who undergo invasive laparoscopic surgery to remove lesions have recurrence of endometriosis (41). Our findings from this report reveal many suitable candidates to potentially serve as biomarkers. While conducting this study, Kojima and colleagues published on designer exosomes, which had been custom-tagged to allow for preferential uptake and to deliver therapeutic cargo for Parkinson’s disease (42). Although a different pathology, this body of work can prove to be a unique avenue of future research for therapeutic treatments in peritoneal-related reproductive pathologies, including endometriosis.
Methods
Sample collection from endometriosis patients and control women.
Matched human endometrium (n = 6), endometriotic lesions (n = 6), PF (n = 6), and plasma (n = 6) from stage III–IV endometriosis patients were obtained from the Greenville Hospital Systems (Greenville, South Carolina, USA), upon written informed consent. The eutopic endometrium samples were obtained by Pipelle sampling during the time of laparoscopic surgery for removal of endometriotic lesions. All patient subjects from Greenville Hospital were free from hormonal therapy for 3 months before the collection of samples. Endometriosis staging was determined based on the American Society of Reproductive Medicine (ASRM) criteria previously established (43). Patient baseline characteristics accounted for included: age (24–39), race (Caucasian), pathology, other pathologies, stage of endometriosis (III–IV), and location of endometriosis (deep infiltrating nodules). Control women enrolled in this study were free from hormonal therapy for 3 months before the collection of samples and did not have any indicators of endometriosis including infertility and/or gynecological malignancies. All patients and controls were matched for the secretory phase of the menstrual cycle when samples were obtained. Samples were snap-frozen in liquid nitrogen and then stored at –80°C until further use.
Cell culture.
Endometriotic epithelial 12Z, EEC-1 (ATCC CRL-2923), and HUVEC (ATCC CRL-2873) cells were all grown in antibiotic-free media in T75 flasks. Cell lines were previously checked for mycoplasma detection with STR profiling, and only passages 3–15 were used for EEC and 12Z cells. 12Z cells were grown as per a previously published protocol (22). Human umbilical vein cell passages between 2–5 were used, and all downstream experiments were designed to use matched passage HUVECs. Cellular morphology and proliferative properties were monitored during each passage to ensure cell lines maintained their phenotype and cellular integrity.
Isolation of EVs, including exosomes, from patient plasma, tissue, PF, and cell culture supernatants.
EVs, including exosomes, were isolated from plasma, tissue, and PF from patients with endometriosis and normal fertile healthy controls, as well as cell culture supernatants using our hybrid protocol as previously described (44). All EV collection followed MISEV2018 guidelines (45). Briefly, miRCURY exosome isolation kit (300102; Exiqon Inc.) was used with an addition of a 100,000 g ultracentrifuge step prior to precipitation. Patient tissue, PF, and cell culture supernatants were then spun 2 times at 10,000 g for 5 minutes to remove cellular debris, and precipitation buffer was added to the remaining mixture and incubated overnight. Samples were subjected to additional filtration for coagulants and other potential clinical contaminants using a 0.22-μM filter. Quality of EVs, including exosomes, isolated from this method were compared with those from ultracentrifugation protocols and were found to be comparable. For patient plasma–derived EVs, including exosomes, thrombin was added and incubated for 5 minutes prior to the cellular debris–filtering steps. For patient tissue–derived EVs, including exosomes, an additional filtration with a 0.22-μM filter was performed following the initial filtration, followed by further vortexing of resuspension buffer and EV pellet for 15 minutes, with intermittent aspirations with a pipette to further mix the sample. All samples with resuspension buffer were spun 2 times at 10,000 g for 30 minutes at 20°C, and supernatants were removed. EV pellets were resuspended in 100 μl of resuspension buffer and were immediately used for downstream applications.
TEM analysis of EVs/exosomes.
EVs, including exosomes, derived from patient plasma and PF, as well as 12Z, EECC, and HUVEC cells, were imaged as per our previously established protocol (44). Briefly, EVs were mixed with 4% paraformaldehyde (Sigma Aldrich) for 30 minutes, counterstained with Alcian blue (Sigma Aldrich) for 30 minutes and subjected to a series of PBS (Thermo Fisher Scientific) washes. Pellets were obtained via centrifugation at 15,000 g and fixed in 2% glutaraldehyde for 5 minutes. Resuspended EV mixtures were transferred to 300-mesh formvar nickel grids and incubated for 40 minutes. All grids were negatively stained with 4% saturated aqueous uranyl acetate for 15 minutes. Grids were analyzed using a FEI Tecnai Osiris transmission electron microscope at 200 kV (FEI) by trained EM specialists at the Reactor Materials Testing Laboratory at Queen’s University. All EVs detected were subjected to morphometric analysis.
Detection of EVs, including exosomes, using Western blotting.
Exosomal marker CD63 and Calnexin (CANX) were probed in isolated EVs from patient plasma, 12Z, EECC, and HUVECs, as per our previously published protocol (44) and followed MISEV2018 guidelines (45). Briefly, EV protein concentrations were determined using a bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) as per manufacturer’s instructions. All samples were normalized to a protein concentration of 5 μg/μl, and samples were denatured at 99.9°C for 10 minutes. A total volume of 15 μl containing 10 μl of normalized EV protein and 5 μl of loading dye was pipetted to appropriate wells of 4–20% Tris-glycine precast gels (Bio-Rad) and separated at 120 V for 1 hour. Transferring was performed onto PVDF membranes (Thermo Fisher Scientific) and run at 100 V for 2 hours. Membranes were blocked in 5% skim milk TBS-T solution overnight. Rabbit polyclonal anti-CD63 (1 μg/ml; Ab134045, Abcam) and 1.5 μg/ml rabbit polyclonal anti-CANX (NB100-1965, Novus Biologicals) antibodies were added as primary antibodies. Membranes were rinsed with TBS-T solution. HRP-conjugated goat anti–rabbit IgG 1:2000 (R&D, HAF008) secondary antibody was added in 5% skim milk TBS-T solution to each membrane and incubated at room temperature (RT) for 2 hours. Enhanced chemiluminescence detection was completed with Clarity chemiluminescent substrate solution (Bio-Rad) and imaged on a Kodak X-ray film processor. All films were scanned on a flatbed scanner at 600 dpi grayscale, and images were analyzed using ImageJ software (NIH).
RNA extraction.
Total RNA was collected in triplicate from patient tissue, plasma, PF, and cell culture–purified EVs (12Z, EECC, HUVECs) using Norgen purification kit (Norgen Biotek). RNA quality and concentration was determined using both Nanodrop UV-Vis 2000C (Thermo Fisher Scientific, ) and Agilent 2100 Bioanalyzer machines with Picochip Bioanalyzer RNA kit using Eukaryote Total RNA microfluidic chips (Agilent).
Small RNA–seq analysis of EVs from patient peripheral blood, PF, and ectopic and eutopic endometriotic lesions.
Small RNA species-based RNA-seq was performed to detect and quantify NGS libraries containing small RNA species, with a focus specifically on miRNAs and lncRNAs (Norgen BioTek Corp.). Briefly, small RNA libraries were prepared using a company-specific Small RNA library prep kit (catalog 63600, Norgen Biotek). Samples were used to conduct NGS on an Illumina NextSeq 500 (Illumina). NextSeq 500/550 High output kit v2 was used as a sequencing platform reagent (Norgen Biotek). Data were indexed and saved as Fastq raw files. The raw Fastq files were normalized and analyzed by 2 trained bioinformaticians by the company using excerpt small RNA–seq Pipeline (v4.6.2) available on Genboree. Small RNA reference sequence databases used were as follows: miRNAs, miRBase version 21; tRNAs, gtRNAdb; piRNAs, RNAdb; and Genome, Gencode version 24 (hg38) which includes lncRNA. CPM cut-off values for lncRNA were set to 76 to filter at a minimum of 1 count. The lncRNA-miRNA interaction network was generated using LncBase Experimental v.2 (DIANA tools). All DE miRNAs were inputted, and lncRNAs with binding affinity were matched with predicted scores (miRNA target gene score; miTG score). The DE lncRNAs were cross-referenced to confirm lncRNA-miRNA predicted binding and recorded. lncRNA-miRNA binding sites were evaluated using lncBase; http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex-experimental Filtration was at a minimum of 1 count, and CPM cut-off values were set to 2227 in 3 or more replicates for all analyses. Network nodes of genes interacting with miR–30d-5p, miR–27a-3p, and miR-375 were developed using miRNet network–based visual analysis of miRNAs, targets, and functions. All compiled nodes were imported into Cytoscape (Version 3.6.1) and sorted based on Reactome detections or shared miRNA binding genes.
Validation of miRNAs DE in multiple sample and comparison groups using qPCR.
Validation of eleven miRNAs (miR-27a, -30d, -100, -136, -144, -193b, -200c, –200a-5p, -206, -375, –let-7a) DE in multiple sample groups from the small RNA–seq comparisons (Supplemental Table 1) was conducted for independent validation using custom miScript qPCR assays (Qiagen). Briefly, purified RNA from EVs were reverse transcribed using miScript II RT Kit (Qiagen) as per manufacturer’s protocol. Real-time PCR was conducted using plate-based Roche LC480 Platform (Roche Diagnostics) and custom miScript primer assays (miR-27a: MS00004231; miR-30d: MS00009387; miR-375: MS00031829; miR-100: MS00031234; miR-136: MS00008645; miR-144: MS00020328; miR-193b: MS00031549; miR-200c: MS00003752; miR–200a-5p: MS00009009; miR-206: MS00003787; miR–let-7a: MS00008274) as per manufacturer’s instructions. Relative quantification was performed using U6 as a control. Pooled control plasma cDNA was used as a calibrator. Expression of U6 did not differ across groups by 1-way ANOVA. All samples were run in triplicates. The run protocol was the following: denaturation, 95°C, 15 minutes; amplification, 45 cycles, 95°C for 15 seconds, 55°C for 30 seconds, 70°C for 30 seconds; melting curve, 70°C–95°C, at a rate of 0.1°C per second. Data were analyzed using the ΔΔCt method.
Proteomic analysis of EVs by MS.
Total protein from all sample type EVs was extracted using total exosome protein isolation kit (4478545; Invitrogen) according to manufacturer’s instructions. The sample was desalted with a dialysis in 10 mM ammonium bicarbonate solution and subsequently digested with trypsin (20 μg) at 37°C for 18 hours. The peptide mixture was further analyzed by liquid chromotagraphy–tandem MS (LC-MS/MS) on a nLC-Orbitrap Velos Pro (Thermo Scientific), and MS/MS spectra were collected. The data were searched using Thermo Proteome Discoverer 1.4.0.288.
EV proteome analysis.
MS/MS-identified EV proteins were subjected to enrichment analysis by PANTHER GO analysis software (Protein Analysis Through Evolutionary Relationships; http://www.pantherdb.org). This software specializes in predictions and classification of proteins in order to facilitate high-throughput analysis. Using this software, the classified proteins were further classified according to their biological, cellular, and molecular function. Protein IDs were also subjected to further classification using available reference or background lists based on the Illumina probe list, as well as a custom reference list including only genes that are detected in the experimental protein database as a background. Conserved miRNA binding sites from proteomic signature genes were assessed using Targetscan v 7.2 software (http://www.targetscan.org/vert_72/). Findings from PANTHER were compared with 3 other GO platforms (gProfiler, BiNGO, Gorilla), in order to determine if there is heterogeneity in the output results, and all platforms maintained similarity. This process yielded families and subfamilies of proteins that were annotated with ontology terms, and sequences were assigned to PANTHER pathway cell signaling pathways.
Confocal microscopy analysis of EV shuttling and uptake.
EVs derived from 12Z, EECC, or HUVECs were stained with CellTracker CM-Dil tracking dye that binds to plasma membranes (CellTracker, C7000, Thermo Fisher Scientific) as per manufacturer. Fluorescently labeled exosomes were reconstituted in cell growth medium and added onto HUVECs grown on coverslips in 12-well culture plates. Cells were fixed at varying time points with 100% ice cold methanol for 5 minutes and rinsed with ice cold PBS 3 times for 5 minutes. Cells were dyed with CellTracker Green BODIPY (1:1000, Thermo Fisher Scientific) and incubated at 37°C for 30 minutes. Cells were washed and mounted with Prolong Gold Antifade DAPI using coverslips. All coverslips were immediately examined under a confocal microscope, and images were acquired for further image processing using ImageJ software (NIH).
EV exosomal coculturing and cell proliferation analysis using quantitative live-cell analysis.
Fluorescently labeled EVs derived from patient samples (peripheral blood and PF) — as well as EVs derived from 12Z, EECC, HUVECs as described above — were incubated with fluorescently labeled HUVECs in 12-well plates that were dyed with CellTracker Green BODIPY (1:1000) and were monitored with the Incucyte real-time quantitative live-cell analysis ZOOM system (Essen Instruments) according to the supplier’s protocols. Briefly, cells cocultured with exosomes were incubated for 24 hours, and the Incucyte software obtained images from 4 individual areas of each well at 2-hour time points. Cell proliferation was measured and EV uptake videos were rendered using the built-in ZOOM analysis software suite as per manufacturer’s guidelines.
Endothelial tube formation assay.
The μ-slide angiogenesis tube formation assay (Ibidi, catalog 81506) was performed as per the manufacturer’s protocol. Growth factor reduced, phenol red free matrigel (10 μl; Corning, catalog 356221) was put into each well of a μ-slide and incubated at 37°C for 30 minutes. HUVECs were harvested with 1× Trypsin-EDTA and seeded at 1 × 104 cells per well. A total of 50 μg/ml plasma-derived EVs from patient samples and healthy, fertile controls were added to the plate. Cells were incubated for 4 hours at 37°C. Images were taken with Olympus CKX41 microscope and Infinity 1 image analyzer. Tube formation was quantified using NIH ImageJ with the angiogenesis analyzer plugin (46).
Multiplex cytokine analysis.
A human 42-plex commercially available multiplex assay was conducted from Eve Technologies using Luminex xMAP laser bead platform as per their workflow (Bio-Rad). This multiplex assay include cytokines involved in inflammation and angiogenesis pathways such as: EGF, Eotaxin-1, FGF-2, Flt-3L, Fractalkine, G-CSF, GM-CSF, GROα, IFNα2, IFNγ, IL-1α, IL-1β, IL-1ra, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-17A, IL-18, IP-10, MCP-1, MCP-3, MDC, MIP-1α, MIP-1β, PDGF-AA, PDGF-AB/BB, RANTES, sCD40L, TGFα, TNFα, TNFβ, and VEGF-A. Color-coded polystyrene beads were coupled with capture antibodies for each cytokine.
Statistics.
GraphPad Prism 6.05 software was used for statistical analysis. Data throughout the paper are expressed as mean ± SEM. Cellular proliferation analysis using Incucyte was analyzed using repeated measures (RM) 2-way ANOVA. This analysis was subjected to Tukey’s multiple comparisons test for main row effect and simple effects within rows to compare time and interaction groups. miRNA validation experiments using qPCR were analyzed by 1-way ANOVA. Tube formation assay experiments were analyzed by 2-tailed Student’s t test. All data were assessed for equal variance and normality before conducting statistical tests. In vitro cytokine protein multiplexing data were analyzed by 1-way ANOVA. P < 0.05 was considered as statistically significant.
Study approval.
Ethics approval was provided by the Health Sciences Research Ethics Board, Queen’s University, Kingston, Ontario, Canada. Human plasma from endometriosis patients and healthy, fertile women were obtained after written informed consent with the use of a protocol approved by the Institutional Review Committees at Greenville Health Systems (Greenville, South Carolina, USA) and the University of North Carolina (Chapel Hill, North Carolina, USA; IRB protocol no. Pro00000993).
Author contributions
KK and CT conceived the experiments. SLY and BAL collected patient samples. ATF and CT contributed reagents. KK, JEM, and HL conducted the experiments. KK and JEM analyzed the results. KK wrote the manuscript. CT supervised data analysis and provided critical suggestions for manuscript writing. CT and MK provided funding. All authors reviewed the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Christina Peixoto for assistance in EM sample processing, as well as Ryan Marks and Lindsey Symons for valuable editorial comments for manuscript preparation. The authors acknowledge financial support by Queen’s Graduate Award, R. Samuel McLaughlin and Robert Wilson Fellowships (KK), NIH (ATF, SLY, and BAL), and Canadian Institutes of Health Research (CT and MK).
Version 1. 09/19/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(18):e128846.https://doi.org/10.1172/jci.insight.128846.
Contributor Information
Kasra Khalaj, Email: k.khalaj@queensu.ca.
Harshavardhan Lingegowda, Email: h.lingegowda@queensu.ca.
Steven L. Young, Email: youngs@med.unc.edu.
Bruce A. Lessey, Email: blessey@wakehealth.edu.
Madhuri Koti, Email: madhuri.koti@queensu.ca.
References
- 1.Giudice LC. Clinical practice. Endometriosis. N Engl J Med. 2010;362(25):2389–2398. doi: 10.1056/NEJMcp1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramer DW, Missmer SA. The epidemiology of endometriosis. Ann N Y Acad Sci. 2002;955:11–22. doi: 10.1111/j.1749-6632.2002.tb02761.x. [DOI] [PubMed] [Google Scholar]
- 3.Simoens S, et al. Endometriosis cost assessment (the EndoCost study): a cost-of-illness study protocol. Gynecol Obstet Invest. 2011;71(3):170–176. doi: 10.1159/000316055. [DOI] [PubMed] [Google Scholar]
- 4.Levy AR, et al. Economic burden of surgically confirmed endometriosis in Canada. J Obstet Gynaecol Can. 2011;33(8):830–837. doi: 10.1016/S1701-2163(16)34986-6. [DOI] [PubMed] [Google Scholar]
- 5.Gazvani R, Templeton A. Peritoneal environment, cytokines and angiogenesis in the pathophysiology of endometriosis. Reproduction. 2002;123(2):217–226. doi: 10.1530/rep.0.1230217. [DOI] [PubMed] [Google Scholar]
- 6.Fainaru O, et al. Dendritic cells support angiogenesis and promote lesion growth in a murine model of endometriosis. FASEB J. 2008;22(2):522–529. doi: 10.1096/fj.07-9034com. [DOI] [PubMed] [Google Scholar]
- 7.Ahn SH, Khalaj K, Young SL, Lessey BA, Koti M, Tayade C. Immune-inflammation gene signatures in endometriosis patients. Fertil Steril. 2016;106(6):1420–1431.e7. doi: 10.1016/j.fertnstert.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monsanto SP, et al. Surgical removal of endometriotic lesions alters local and systemic proinflammatory cytokines in endometriosis patients. Fertil Steril. 2016;105(4):968–977.e5. doi: 10.1016/j.fertnstert.2015.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn SH, Edwards AK, Singh SS, Young SL, Lessey BA, Tayade C. IL-17A Contributes to the Pathogenesis of Endometriosis by Triggering Proinflammatory Cytokines and Angiogenic Growth Factors. J Immunol. 2015;195(6):2591–2600. doi: 10.4049/jimmunol.1501138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khalaj K, et al. A balancing act: RNA binding protein HuR/TTP axis in endometriosis patients. Sci Rep. 2017;7(1):5883. doi: 10.1038/s41598-017-06081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burns G, Brooks K, Wildung M, Navakanitworakul R, Christenson LK, Spencer TE. Extracellular vesicles in luminal fluid of the ovine uterus. PLoS One. 2014;9(3):e90913. doi: 10.1371/journal.pone.0090913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns GW, Brooks KE, Spencer TE. Extracellular Vesicles Originate from the Conceptus and Uterus During Early Pregnancy in Sheep. Biol Reprod. 2016;94(3):56. doi: 10.1095/biolreprod.115.134973. [DOI] [PubMed] [Google Scholar]
- 13.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 14.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahadi A, Brennan S, Kennedy PJ, Hutvagner G, Tran N. Long non-coding RNAs harboring miRNA seed regions are enriched in prostate cancer exosomes. Sci Rep. 2016;6:24922. doi: 10.1038/srep24922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Toro J, Herschlik L, Waldner C, Mongini C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol. 2015;6:203. doi: 10.3389/fimmu.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harp D, et al. Exosomes derived from endometriotic stromal cells have enhanced angiogenic effects in vitro. Cell Tissue Res. 2016;365(1):187–196. doi: 10.1007/s00441-016-2358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet - dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44(W1):W135–W141. doi: 10.1093/nar/gkw288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabregat A, et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anglesio MS, et al. Cancer-Associated Mutations in Endometriosis without Cancer. N Engl J Med. 2017;376(19):1835–1848. doi: 10.1056/NEJMoa1614814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo JY, et al. KRAS Activation and over-expression of SIRT1/BCL6 Contributes to the Pathogenesis of Endometriosis and Progesterone Resistance. Sci Rep. 2017;7(1):6765. doi: 10.1038/s41598-017-04577-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banu SK, Lee J, Starzinski-Powitz A, Arosh JA. Gene expression profiles and functional characterization of human immortalized endometriotic epithelial and stromal cells. Fertil Steril. 2008;90(4):972–987. doi: 10.1016/j.fertnstert.2007.07.1358. [DOI] [PubMed] [Google Scholar]
- 23.Ng YH, et al. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS One. 2013;8(3):e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawkins SM, et al. Functional microRNA involved in endometriosis. Mol Endocrinol. 2011;25(5):821–832. doi: 10.1210/me.2010-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laudanski P, Charkiewicz R, Tolwinska A, Szamatowicz J, Charkiewicz A, Niklinski J. Profiling of Selected MicroRNAs in Proliferative Eutopic Endometrium of Women with Ovarian Endometriosis. Biomed Res Int. 2015;2015:760698. doi: 10.1155/2015/760698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canlorbe G, et al. Identification of microRNA expression profile related to lymph node status in women with early-stage grade 1-2 endometrial cancer. Mod Pathol. 2016;29(4):391–401. doi: 10.1038/modpathol.2016.30. [DOI] [PubMed] [Google Scholar]
- 27.Di Pietro C, et al. MiR-27a-3p and miR-124-3p, upregulated in endometrium and serum from women affected by Chronic Endometritis, are new potential molecular markers of endometrial receptivity. Am J Reprod Immunol. 2018;80(3):e12858. doi: 10.1111/aji.12858. [DOI] [PubMed] [Google Scholar]
- 28.Seifer BJ, Su D, Taylor HS. Circulating miRNAs in Murine Experimental Endometriosis. Reprod Sci. 2017;24(3):376–381. doi: 10.1177/1933719116667228. [DOI] [PubMed] [Google Scholar]
- 29.Nothnick WB. MicroRNAs and Endometriosis: Distinguishing Drivers from Passengers in Disease Pathogenesis. Semin Reprod Med. 2017;35(2):173–180. doi: 10.1055/s-0037-1599089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long M, Wan X, La X, Gong X, Cai X. miR-29c is downregulated in the ectopic endometrium and exerts its effects on endometrial cell proliferation, apoptosis and invasion by targeting c-Jun. Int J Mol Med. 2015;35(4):1119–1125. doi: 10.3892/ijmm.2015.2082. [DOI] [PubMed] [Google Scholar]
- 31.Jia S-Z, Yang Y, Lang J, Sun P, Leng J. Plasma miR-17-5p, miR-20a and miR-22 are down-regulated in women with endometriosis. Hum Reprod. 2013;28(2):322–330. doi: 10.1093/humrep/des413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie N, et al. miR-27a regulates inflammatory response of macrophages by targeting IL-10. J Immunol. 2014;193(1):327–334. doi: 10.4049/jimmunol.1400203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vilella F, et al. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development. 2015;142(18):3210–3221. doi: 10.1242/dev.124289. [DOI] [PubMed] [Google Scholar]
- 34.Caserta S, Kern F, Cohen J, Drage S, Newbury SF, Llewelyn MJ. Circulating Plasma microRNAs can differentiate Human Sepsis and Systemic Inflammatory Response Syndrome (SIRS) Sci Rep. 2016;6:28006. doi: 10.1038/srep28006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.et al. H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Mol Med. 2015;7(8):996–1003. doi: 10.15252/emmm.201505245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.et al. Overexpression of long noncoding RNA, NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother. 2016;84:244–251. doi: 10.1016/j.biopha.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 37.Huang-Doran I, Zhang CY, Vidal-Puig A. Extracellular Vesicles: Novel Mediators of Cell Communication In Metabolic Disease. Trends Endocrinol Metab. 2017;28(1):3–18. doi: 10.1016/j.tem.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Forterre A, et al. Proteomic analysis of C2C12 myoblast and myotube exosome-like vesicles: a new paradigm for myoblast-myotube cross talk? PLoS One. 2014;9(1):e84153. doi: 10.1371/journal.pone.0084153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun CY, et al. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis. 2013;34(2):426–435. doi: 10.1093/carcin/bgs333. [DOI] [PubMed] [Google Scholar]
- 40.Lin ZY, et al. MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol Cancer. 2017;16(1):48. doi: 10.1186/s12943-017-0615-x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Guo SW. Recurrence of endometriosis and its control. Hum Reprod Update. 2009;15(4):441–461. doi: 10.1093/humupd/dmp007. [DOI] [PubMed] [Google Scholar]
- 42.Kojima R, et al. Designer exosomes produced by implanted cells intracerebrally deliver therapeutic cargo for Parkinson’s disease treatment. Nat Commun. 2018;9(1):1305. doi: 10.1038/s41467-018-03733-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.[No authors listed] Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67(5):817–821. doi: 10.1016/S0015-0282(97)81391-X. [DOI] [PubMed] [Google Scholar]
- 44.Bidarimath M, Khalaj K, Kridli RT, Kan FW, Koti M, Tayade C. Extracellular vesicle mediated intercellular communication at the porcine maternal-fetal interface: A new paradigm for conceptus-endometrial cross-talk. Sci Rep. 2017;7:40476. doi: 10.1038/srep40476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Théry C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.