Abstract
BACKGROUND
Fecal microbiota transplantation (FMT) is an effective treatment for recurrent Clostridioides difficile infection (rCDI) in adults and children, but donor stool samples are currently screened for only a limited number of potential pathogens. We sought to determine whether putative procarcinogenic bacteria (enterotoxigenic Bacteroides fragilis, Fusobacterium nucleatum, and Escherichia coli harboring the colibactin toxin) could be durably transmitted from donors to patients during FMT.
METHODS
Stool samples were collected from 11 pediatric rCDI patients and their respective FMT donors prior to FMT as well as from the patients at 2–10 weeks, 10–20 weeks, and 6 months after FMT. Bacterial virulence factors in stool DNA extracts and stool cultures were measured by quantitative PCR: Bacteroides fragilis toxin (bft), Fusobacterium adhesin A (fadA), and Escherichia coli colibactin (clbB).
RESULTS
Four of 11 patients demonstrated sustained acquisition of a procarcinogenic bacteria. Whole genome sequencing was performed on colony isolates from one of these donor/recipient pairs and demonstrated that clbB+ E. coli strains present in the recipient after FMT were identical to a strain present in the donor, confirming strain transmission. Conversely, 2 patients exhibited clearance of procarcinogenic bacteria following FMT from a negative donor.
CONCLUSION
Both durable transmission and clearance of procarcinogenic bacteria occurred following FMT, suggesting that additional studies on appropriate screening measures for FMT donors and the long-term consequences and/or benefits of FMT are warranted.
FUNDING
Crohn’s & Colitis Foundation, the Bloomberg~Kimmel Institute for Cancer Immunotherapy at Johns Hopkins University School of Medicine, the National Cancer Institute, and the Canadian Institutes of Health Research.
Keywords: Gastroenterology, Microbiology
Keywords: Bacterial infections, Colorectal cancer, Inflammatory bowel disease
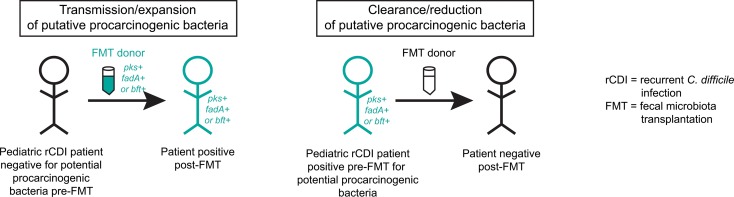
Transmission and/or clearance of putative procarcinogenic bacteria may be a frequent, unintended consequence of fecal microbiota transplantation in pediatric patients with recurrent Clostridioides difficile infection.
Introduction
Infection with Clostridioides difficile (formerly Clostridium difficile) is a significant cause of diarrheal illness in both adults and children (1), particularly in patients with inflammatory bowel disease (IBD) (2). Fecal microbiota transplantation (FMT) is a life-saving therapeutic option for patients with recurrent C. difficile infection (rCDI), with successful treatment outcomes in more than 80% of adult cases compared with traditional vancomycin treatment alone (~30%) (3–5). Similar success rates have also been reported in pediatric rCDI cohorts, including a multicenter cohort of 335 children (6–16). Given its successes in rCDI, FMT is being increasingly explored as a therapeutic option for a number of gastrointestinal disorders, as well as other conditions associated with microbial dysbiosis, with recent promising results in clinical trials in ulcerative colitis (UC) (17–19) and immune checkpoint inhibitor–associated colitis in cancer patients (20). Despite the promising potential of FMT for these disease indications, the long-term consequences of FMT are not well understood.
Growing evidence supports a role for specific microbes in the etiology of a number of disorders, including colorectal cancer (CRC). Enterotoxigenic Bacteroides fragilis (ETBF), Fusobacterium nucleatum, and Escherichia coli harboring the polyketide synthase island that synthesizes the genotoxin colibactin (clbB) are tumorigenic in murine models of CRC and are more prevalent in CRC and IBD patients compared with healthy controls (21–28), suggesting a potential link between these organisms and procarcinogenic/proinflammatory mechanisms. These bacteria are not presently included in current FMT donor screening protocols. Though large prospective cohort studies have yet to be performed to directly demonstrate causality versus association of these bacteria with CRC, small cohort studies (<50 patients) have revealed that these organisms can be frequently detected even in healthy adult populations using sensitive culture-based techniques, with prevalences of up to 67% for ETBF (26), 26.5% for F. nucleatum (28), and 21% for clbB+ E. coli (21) in healthy controls. The presence of ETBF is associated with diarrhea in children aged 1–5 years, but is otherwise rare in stool samples from healthy children in the United States (culture-positive rates of 3%–6%) (29–31). Pediatric colonization rates for F. nucleatum and clbB+ E. coli are not established. We thus hypothesized that some otherwise healthy FMT donors might harbor these potential pathogens, and that transmission of these organisms to recipients could be a hidden risk of FMT in a pediatric rCDI cohort.
Results
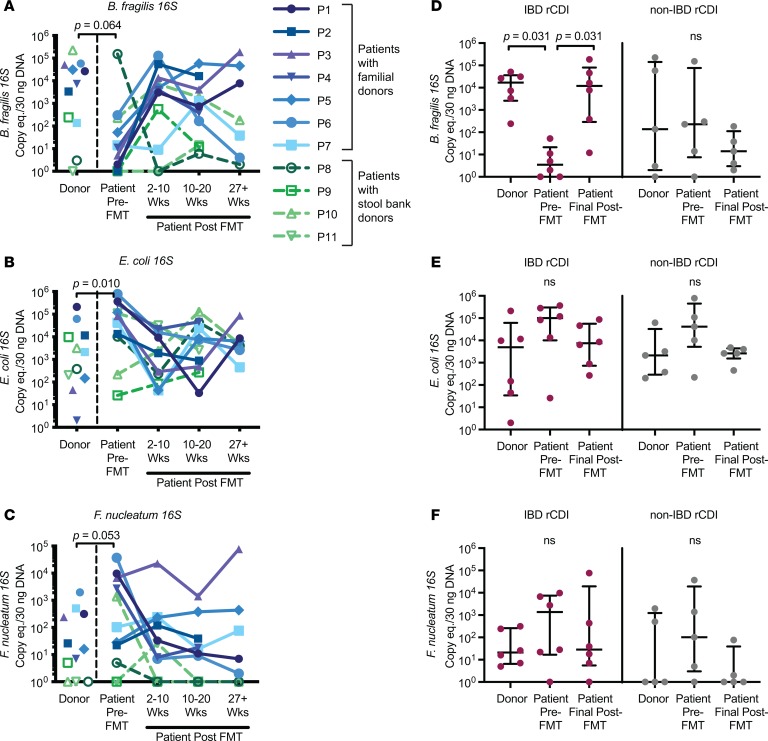
A flow chart of the study design is shown in Figure 1. Only stool samples from patients for whom we had donor stool, patient stool pre-FMT, and patient stool at least 2 time points after FMT were analyzed. We first examined levels of the species-specific 16S rRNA–encoding genes in the donors and recipients by quantitative PCR (qPCR) on DNA extracted from stool samples. The pediatric rCDI patients prior to FMT tended to have lower levels of B. fragilis 16S (P = 0.064) and higher levels of both E. coli (P = 0.010) and F. nucleatum 16S (P = 0.053) in stool compared with their donors, levels that converged toward donor levels after FMT (Figure 2, A–C; paired-sample Wilcoxon’s test). These findings are consistent with previous data indicating that rCDI-associated dysbiosis is marked by high levels of Proteobacteria and low levels of commensals/symbiotes from the Bacteroidetes phylum (6, 15). The trend toward lower levels of B. fragilis 16S rDNA genes in the patients before FMT was largely driven by the IBD rCDI patients (Table 1), who had significantly lower levels of this bacteria compared with their donors (Figure 2D, P = 0.031, paired-sample Wilcoxon’s test). In contrast, the significant elevation in E. coli 16S and the trend toward elevated F. nucleatum 16S prior to FMT were similar in IBD and non-IBD patients (Figure 2, E and F).
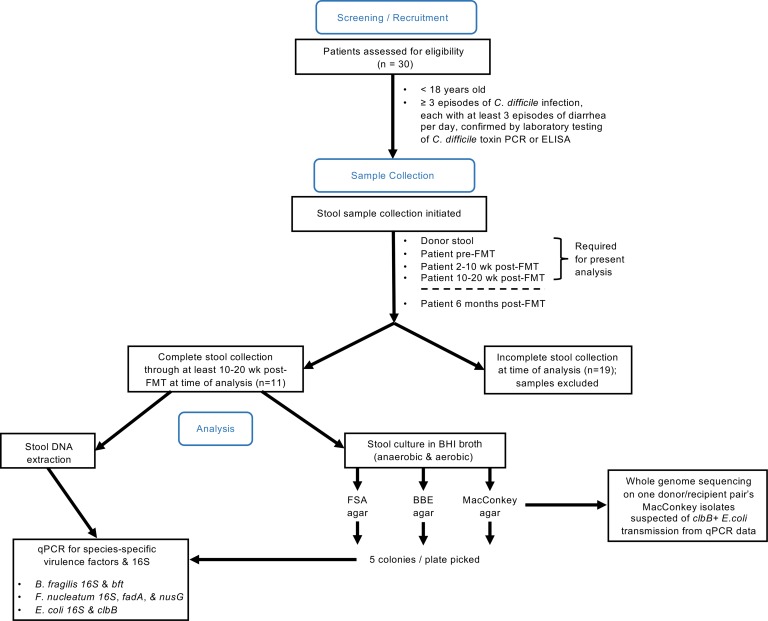
Figure 1. Flow diagram of study design.
Screening/recruitment: pediatric patients (<18 years old) with at least 3 episodes of C. difficile infection were recruited (n = 30 donor/patient pairs). Each episode consisted of at least 3 episodes of diarrhea per day and was confirmed by laboratory test of C. difficile toxin by PCR or ELISA. Sample collection: stool samples were collected from donors and patients at the designated time points. Analysis: at the time of analysis, only patient/donor pairs with a complete set of stool samples collected through at least the 10–20 weeks post-FMT time point were included for further study (n = 11 donor/patient pairs).
Figure 2. Longitudinal dynamics of bacterial species by 16S qPCR.
(A–C) Detection of species-specific 16S rRNA–encoding genes in stool DNA by qPCR for FMT donors, patients prior to FMT, and patients at various time points after FMT (2–10 weeks, 10–20 weeks, and >27 weeks) (n = 11 patient/donor pairs). Colors denote the source of the FMT donor stool, with familial donors in shades of blue/purple (P1–P7) and commercial stool bank donors shown in shades of green (P8–P11). The paired-sample Wilcoxon’s test P value shown refers to the levels of the donors compared with the patients prior to FMT. eq., equivalents. (D–F) 16S rRNA–encoding gene levels are divided according to whether the rCDI patient had IBD (red, left panels, n = 6 patient/donor pairs) or not (gray, right panels, n = 5 patient/donor pairs). The final time point available for each patient is presented as the patient’s final post-FMT value, corresponding to either the 10–20 Wks or the 27+ Wks time point. Error bars represent medians with interquartile ranges. For all data, undetectable data points were assigned a value of 1 for display purposes on the log10 scale; statistical analyses (paired-sample Wilcoxon’s test) were performed on the raw data.
Table 1. Patient demographics.
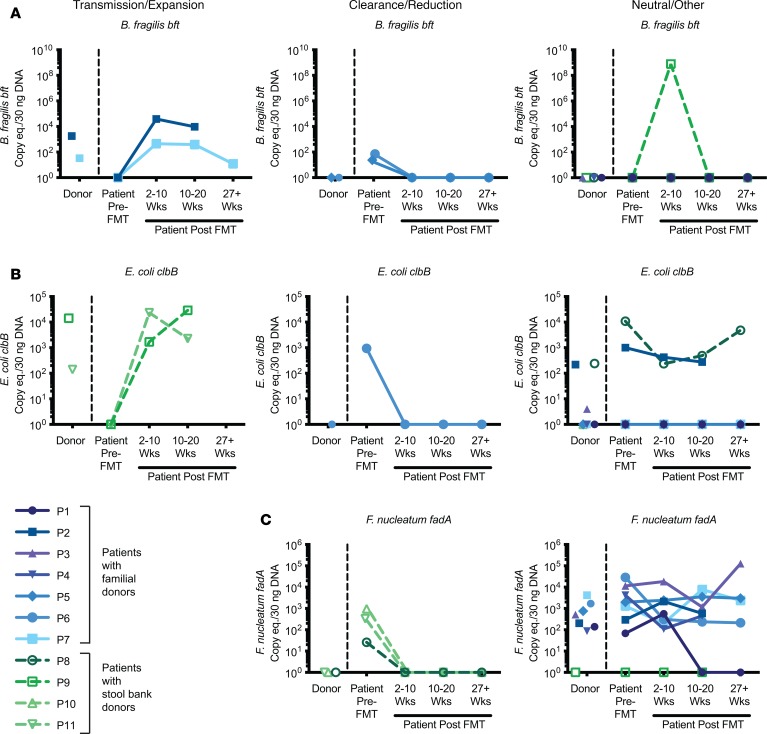
However, whereas nearly all F. nucleatum strains harbor the fadA adhesin factor, only a subset of B. fragilis and E. coli strains harbor the putative CRC-associated toxin genes bft and clbB, respectively (32–38). We therefore examined levels of the bacterial virulence factors in the FMT rCDI cohort. Transmission/expansion events were defined as patients who were undetectable for a given virulence factor prior to FMT but rose to donor levels after FMT and remained detectable throughout the time course. Clearance/reduction events were defined as patients who were positive prior to FMT for a given virulence factor but had undetectable levels at all post-FMT time points by total stool qPCR following FMT from a donor negative for that particular virulence factor. Strikingly, we found several instances of potential transmission events of putative procarcinogenic bacteria from donors to their respective FMT recipients. Two of these cases involved bft (Figure 3A, left panel, patients 2 and 7) and 2 involved clbB (Figure 3B, left panel, patients 9 and 11).
Figure 3. Longitudinal transmission and remission of bacteria expressing CRC-associated virulence factors.
Detection of virulence factors in stool DNA by qPCR are shown for FMT donors, patients prior to FMT, and patients at various time points after FMT (2–10 weeks, 10–20 weeks, and >27 weeks) (n = 11 patient/donor pairs). Virulence factors B. fragilis bft (A), E. coli clbB (B), and F. nucleatum fadA (C). Left panels indicate whether there was evidence for transmission/expansion; middle panels, clearance/reduction; right panels, neutral/other changes in the levels of bacterial virulence factors. Colors denote the source of the FMT donor stool, with familial donors in shades of blue/purple (P1–P7) and commercial stool bank donors shown in shades of green (P8–P11). Undetectable data points were assigned a value of 1 for display purposes on the log10 scale.
No F. nucleatum transmissions were observed, because all of the recipients were positive for F. nucleatum before FMT, with the exception of patient 9, whose donor was also F. nucleatum negative. However, we did observe multiple cases of F. nucleatum clearance/reduction following FMT (Figure 3C, middle panel, patients 8, 10, and 11). Reductions in patients 10 and 11 were confirmed by qPCR using an additional F. nucleatum–specific marker gene, nusG, though nusG analyses overall were less sensitive than the other markers examined (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.130848DS1). Notably, the donor in each of the F. nucleatum clearance/reduction cases was a nonfamilial, commercial stool bank donor. Indeed, while all 7 of the familial donors had high levels of F. nucleatum, none of the 4 commercial stool bank donors were positive by qPCR. Two additional patients demonstrated loss of other procarcinogenic bacteria to undetectable levels following FMT from a negative donor: one for bft (Figure 3A, middle panel, patient 5) and one for both bft and clbB (Figure 3, A and B, middle panels, patient 6).
All remaining patients either maintained their positive status for a given virulence factor following FMT from a positive donor or remained negative following FMT from a negative donor, with the exception of 2 patients who demonstrated abrupt changes: patient 9 had a temporary spike in bft at 2–10 weeks after FMT that disappeared by 10–20 weeks (Figure 3A, right panel), while patient 1 had a spontaneous clearance/reduction of fadA at 10–20 weeks (Figure 3C, right panel) and remained undetectable at the 6-month time point. Otherwise, changes in detection of virulence factors in total stool by qPCR consistently followed the trajectory of the donor.
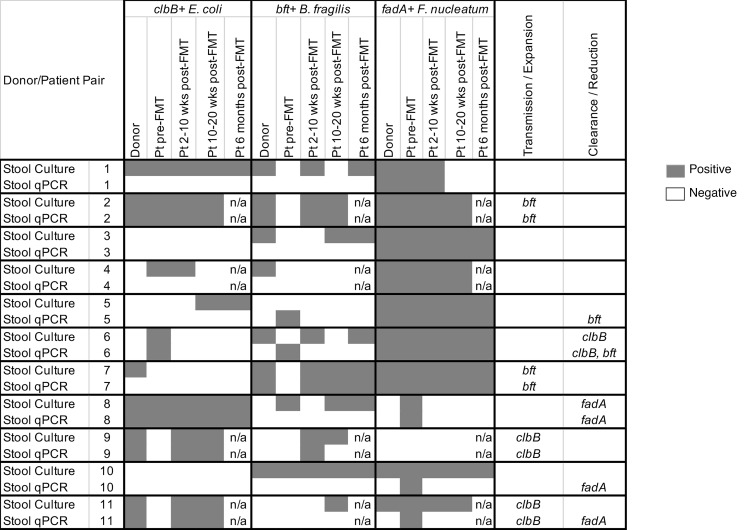
To confirm the transmission/clearance cases, we cultured all of the donor and recipient samples on selective media and tested 5 colonies per plate for the presence of procarcinogenic bacteria and their virulence factors. In general, detection rates by culture were similar to or higher than for the total stool qPCR (Supplemental Tables 2–4) but also displayed greater variability in patients at time points after FMT (Figure 4 and Supplemental Table 5). Only patients who were consistently positive (or negative) following FMT were considered to have validated transmission/clearance events. Overall, all 4 transmission cases originally identified by qPCR (2 bft, 2 clbB) were confirmed by stool culture, while only 2 of the 5 clearance cases were confirmed by culture (1 clbB, 1 fadA) (Figure 4, patients 6 and 8). The remaining clearance cases originally identified by total stool qPCR were subsequently found to harbor viable bft+ B. fragilis or F. nucleatum strains by culture for at least one time point after FMT (Figure 4, patients 5, 10, and 11).
Figure 4. Comparison of virulence factor detection by stool culture versus total stool DNA qPCR.
Stool samples from 11 patient/donor pairs were amplified in broth and then cultured on selective media for E. coli (MacConkey agar), B. fragilis (BBE agar), and F. nucleatum (FSA agar), with up to 5 colonies analyzed per plate. Data are compared with the qPCR results from direct stool DNA extractions. Dark gray shading represents positive samples, equating to at least 1 colony being positive for the respective virulence factor for the culture-based studies or at least 10 copy equivalents per 30 ng stool DNA for the stool qPCR studies. Pt, patient; n/a, stool samples were not available for the time point for analysis.
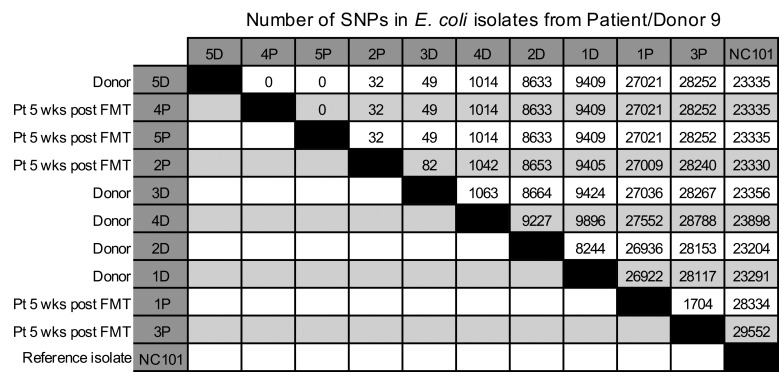
Finally, to determine whether the putative strain transmission events observed in the patients were in fact from the donor and not from environmental sources or from low-abundance strains in the patients themselves, we recultured the stool samples from a clbB+ E. coli transmission case from a commercial stool bank donor (donor/patient pair 9) and amplified individual MacConkey isolates in LB to generate sufficient DNA for single-colony whole genome sequencing (WGS). qPCR analyses on these additional colonies confirmed our earlier findings of transmission (Supplemental Table 5), with all of the additional colonies from the recipient prior to FMT being clbB– (0 of 5), and the donor (5 of 5) and recipient after FMT (3 of 5) consistently harboring clbB+ colonies. We then performed WGS on DNA extracted from these isolates, using a cutoff of 10 SNPs as our threshold for identifying transmitted strains based on prior literature (39). WGS on these colony isolates revealed that one of the donor strains (colony 5D) was closely related to 2 of the patient’s strains after FMT (4P and 5P), with 0 SNPs detected between these isolates using the core genome alignment tool Parsnp (Figure 5). Donor strain 3D and patient strain 2P were also similar to 5D, harboring only 49 and 32 SNP differences, respectively, likely reflecting the presence of additional clbB+ strains in the donor that were transmitted to the recipient. These related strains (5D, 4P, 5P, 3D, and 2P) were all clbB+ by qPCR. In contrast, all of the patient’s pre-FMT strains from MacConkey agar were clbB– by qPCR and were classified as Klebsiella or Morganella species by sequencing (data not shown; these reads were too divergent to be analyzed alongside the other strains in Parsnp). These data confirmed that the strain transmission likely occurred directly from the donor and not from an environmental source.
Figure 5. Number of SNPs identified via WGS.
MacConkey agar isolates from patient/donor pair 9 underwent WGS and core genome alignment analysis with Parsnp. Data are ordered according to their similarity with donor isolate 5D. Murine clbB+ isolate NC101 is shown for comparison. D, donor isolates; P, patient isolates. Five donor (D) and 5 patient post-FMT (P) isolates are shown. Five isolates from the patient pre-FMT were also sequenced but were too divergent to be aligned alongside the E. coli isolates, as the pre-FMT isolates were all Klebsiella or Morganella species.
Discussion
Overall, our data suggest that FMT in pediatric rCDI results in a remarkable degree of gut microbiome reconstitution in patients that closely matches donor microbiomes at a quantitative level for E. coli, B. fragilis, F. nucleatum, and their associated virulence factors. These changes were stable by qPCR through the 6-month time point, the latest time point that our study captured. Four of the 11 patients in our study demonstrated potential engraftment after FMT of donor strains harboring virulence factors (2 ETBF, 2 clbB+ E. coli) by qPCR analysis. All 4 of these transmission cases were confirmed by culture, with one clbB+ E. coli transmission from a commercial stool bank donor definitively proven by WGS. Our strain data therefore represent a proof of concept that transmission of putative procarcinogenic organisms can occur, although additional WGS studies would be needed to similarly prove transmission in the other 3 cases. Our strain data are also in line with recent publications on strain tracking in FMT using stool whole genome shotgun sequencing, in which strains were transmitted in 85% of adult rCDI FMT cases in one study (40) and approximately 38% of donor strains remained in metabolic syndrome FMT recipients at 3 months after FMT in another study (41). Additional clinical studies using 16S rRNA amplicon sequencing have similarly demonstrated that the microbiota of recipients consistently shifts toward that of the donor after FMT in rCDI, even in pediatric cohorts (6, 7). Thus, sustained donor strain engraftment is not unexpected during FMT and indeed is quite frequent.
Conversely, we also observed several cases of clearance/reduction of putative procarcinogenic organisms, particularly for F. nucleatum, for which 3 patients demonstrated a loss of detection by qPCR following FMT from F. nucleatum–negative donors, all of whom were commercial stool bank donors. Two additional patients exhibited loss of other pathogenic bacteria (one ETBF, one ETBF and clbB+ E. coli). However, of the 5 total potential clearance cases observed by total stool qPCR, samples from only 2 patients were also consistently negative by culture (patient 8 for F. nucleatum, patient 6 for clbB+ E. coli). In general, the culture-based and total stool qPCR–based analyses were complementary but only approximately 75% concordant overall. Culture was more sensitive but also more inconsistent longitudinally, likely due to limitations in the number of colonies screened or the viability of individual microbes, whereas qPCR afforded a more quantitative view.
The striking difference in the rates of F. nucleatum positivity by qPCR in the parental donors (7 of 7) versus the commercial stool bank donors (0 of 4) was unexpected. F. nucleatum is a common oral pathogen and is enriched in CRC specimens, but is detected in only approximately 25% of healthy individuals (28). While our study was limited in size, the high prevalence in the patients prior to FMT (91%) and in familial donors (100%) by qPCR suggests that F. nucleatum colonization of the gut may be associated with recurrent C. difficile infection, as either a cause or consequence of dysbiosis. Additionally, these data suggest that F. nucleatum may be readily transmitted in family settings. However, as the familial and commercial stool bank donors were processed in different facilities, and the stool from stool bank donors underwent an additional freeze/thaw prior to testing, differences in the way in which the samples were prepared could contribute to the differences seen in F. nucleatum levels in the donors. Future prospective studies would be necessary to determine any potential contributing roles of F. nucleatum to familial transmissions and/or rCDI.
Limitations of this study include the small sample size and follow-up through only 6 months. Larger and longer-term prospective studies are necessary to demonstrate whether engraftment of putative procarcinogenic strains is (a) truly stable over the span of years, not just months; and (b) leads to an actual greater risk of CRC in patients, a salient question given emerging early-onset CRC (42). In terms of colonization stability of these microbes, a study by Zitomersky et al. of B. fragilis strains cultured from healthy adult stools demonstrated that detection (or not) of bft+ B. fragilis strains over the course of a year was remarkably consistent. Of 13 patients evaluated, 5 remained bft+ throughout, 7 were bft– throughout, and only 1 changed status from bft– to bft+ (43). No bft+ participants lost bft in that study, although 2 subjects carried both bft+ and bft– strains simultaneously. Longitudinal colonization studies are lacking for the other bacteria of interest. In our study, we observed only 2 sudden colonization changes that occurred seemingly independently of the donor status: patient 9 transiently gained bft+ B. fragilis at 2–10 weeks after FMT despite having a bft– donor, and patient 1 lost F. nucleatum at 10–20 wks after FMT despite having an F. nucleatum+ donor. Both of these cases were confirmed by culture and qPCR. These data suggest that there may be some colonization instability in these microbes in the post-FMT period, although all the other microbes and cases were largely stable over the 6-month period of examination. Future studies would need to examine these patients over a longer period of time (years, not months) to determine true persistence of any donor-derived strains.
The potential role of these microbes in CRC warrants further study. CRC is a complex disease with well-established risk factors, including host genetics, diet, smoking, and other environmental factors, of which the microbiota is just one component. Thus, transmission of one of these putative procarcinogenic bacteria may not impact CRC risk at all, or perhaps only slightly increase the risk. The mechanisms pointing to a procarcinogenic role for ETBF, clbB+ E. coli, and F. nucleatum are largely based on animal models but include directly genotoxic mechanisms (clbB), induction of ROS (bft), and induction of Wnt signaling (bft and fadA) (44). Mechanistic and longitudinal data linking colonization with these microbes to actual CRC risk in patients are lacking for each of these bacteria, although a combination of 16S-, metagenomics-, and qPCR-based approaches have established an enrichment of these bacteria in case-control studies. For example, meta-analyses of 16S rRNA gene amplicon sequencing have demonstrated enrichment of F. nucleatum in both stool (n = 506–752 patients) and mucosa (n = 551 patients) of CRC patients compared with healthy controls (27, 45). 16S rRNA gene–based approaches are not suitable for determining the association of bft or clbB with CRC, as only a subset of B. fragilis and E. coli bacteria harbor these virulence factors. However, in a recent meta-analysis of shotgun metagenomic data on fecal samples from 768 patients, fadA and pks were found to be significantly enriched in CRC versus healthy controls (46). Numerous studies involving smaller cohorts have found an enrichment of bft in CRC using culture-based methods followed by qPCR in both the colon mucosa (85% CRC vs. 53% healthy controls) (26) and stool (38% CRC vs. 12% controls, and 27% vs. 10%, respectively) (47, 48). Proving mechanistically that any of these 3 bacteria are truly causally involved (or not) in CRC progression would therefore likely require both sequencing and culture-based analyses of longitudinal stool samples.
Whether the putative procarcinogenic bacteria are also readily transmitted (or cleared) in non-rCDI FMT cohorts remains to be studied. The potential for transmission of procarcinogenic bacteria during FMT may be lower in non-rCDI FMT cohorts such as those with UC, where multiple iterations of FMT are frequently necessary, suggesting that the microbiota in UC patients is more resistant and/or resilient to change (17–19). Alternatively, more frequent FMTs may provide more opportunities for individual bacteria to colonize. Thus, while our data suggest that the transfer of putative procarcinogenic strains may be relevant in FMT for pediatric rCDI, heightened scrutiny of donor selection procedures may be warranted in other FMT settings as well.
Conversely, should future mechanistic studies confirm that these putative procarcinogenic bacteria are causally involved in human CRC or other disorders, our data suggest that FMT from donors who are negative for these organisms could be explored as a potential preventive/therapeutic approach. FMT has already shown some promise in non-rCDI patients as a tool to prevent or clear other individual pathogenic bacteria including antibiotic-resistant organisms, although with variable success (49–51). However, given that FMT itself is not risk free (e.g., there are risks associated with anesthesia for colonoscopy-delivered FMT), any such investigational studies should not be explored lightly, as the risks involved could very likely exceed any benefit. The recent report of transmission of an extended-spectrum β-lactamase–producing (ESBL-producing) E. coli from a donor (who had not been screened for multidrug-resistant organisms [MDROs]) to 2 immunocompromised FMT recipients (one of whom died) highlights the very real risks posed by this still investigational procedure. These severe adverse reaction events led the US FDA to institute immediate changes in donor screening for Investigational New Drug (IND) holders, including mandatory testing for the MDROs ESBL-producing Enterobacteriaceae, vancomycin-resistant enterococci, carbapenem-resistant Enterobacteriaceae, and methicillin-resistant Staphylococcus aureus. MDRO transmission was not examined in the present study.
Conceptually, an engineered consortium of human-derived strains or other approaches may ultimately be safer and preferred by patients over traditional FMT, and would alleviate concerns over transmission of potential pathogens such as the ones observed in our study as well as established or emerging MDROs such as that in the ESBL–E. coli event. Quality-controlled microbial-based alternatives to FMT and novel ways to protect the gut microbiota from the action of antibiotics are under active investigation (52–56). However, at this time there is no evidence of an engineered microbial product having superiority over FMT, and FMT remains a potentially life-saving therapeutic option for rCDI (57). Overall, the durable changes that we observed in our study out to 6 months after FMT suggest that long-term consequences — both promising and deleterious — should be considered when screening FMT donors and recipients, especially in the context of pediatric rCDI.
Methods
Patients.
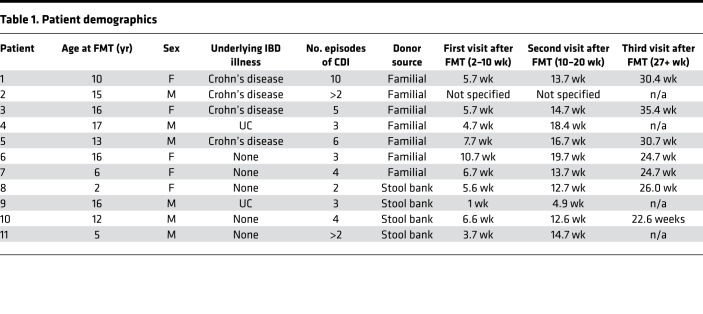
The study cohort included 30 pediatric patients from Johns Hopkins Hospital who had at least 3 episodes of CDI (initial infection and 2 recurrences) and underwent an FMT procedure between August 2013 and December 2015. Parents of any child scheduled to receive a clinically indicated fecal transplant for rCDI were approached for recruitment into the study in the pediatric gastroenterology clinic. All rCDI patients had clinical resolution of C. difficile following FMT. rCDI patients included both those with and those without IBD (Crohn’s disease or UC). Patients in this cohort have previously been described (6). At the time of the present analysis, 11 patient/donor pairs had both pre-FMT stool samples for the donor and patient as well at least 2 post-FMT stool samples for the patient (Table 1). Stool donations for the FMT patients were either familial or from a commercial stool bank (OpenBiome). Familial donors were screened for potential blood and stool pathogens, completed a health questionnaire, and underwent a personal history and physical examination by their primary physician as previously described (6). The anonymous OpenBiome donors underwent a rigorous screening protocol that supplements clinical history evaluations with blood and stool testing. Patients were instructed to stop antibiotics treatment for their rCDI 48 hours prior to the FMT procedure. Pre-FMT stool samples were collected from patients 24–48 hours prior to the FMT procedure. Fresh familial donor stools were collected within 12 hours of the FMT procedure. One hundred grams of fresh stool from familial donors or thawed stool from stool bank donors were vortexed with 400 mL of nonbacteriostatic saline, filtered, and delivered to patients via colonoscopy by a pediatric gastroenterologist, with the majority of the slurry being delivered in the cecum and a small amount delivered throughout the rest of the colon as the colonoscope was withdrawn. Patients were required to lie down in a flat position for 2 hours after the procedure and received loperamide (weight-based standard dosing) to promote retention of the FMT product. Patients returned at 3 follow-up time points within 2–10 weeks, 10–20 weeks, and 6 months after FMT for monitoring of adverse effects and collection of post-FMT stool samples. All stools were aliquoted and stored at –80° prior to screening. One patient (patient 2) was evaluated clinically at Johns Hopkins Hospital but underwent FMT at Sinai Hospital in Baltimore. Both patient 2 and patient 11 met the criteria for inclusion in the study (initial infection and at least 2 rCDI events), but the exact number of rCDI episodes was unknown.
DNA extraction from total stool.
DNA was extracted from stool using the QIAamp DNA Stool Mini Kit (QIAGEN) and stored at –80° prior to qPCR testing.
Stool cultures.
Approximately 50 mg of stool from each patient was amplified in liquid broth at 37°C for 24–48 hours in brain-heart infusion (BHI) broth in an anaerobic chamber for F. nucleatum and B. fragilis or aerobically in LB for E. coli until growth was observed. Amplified cultures were then plated onto the following selective plates for an additional 24–48 hours: Fusobacterium Selective Agar (FSA, Anaerobe Systems) for F. nucleatum, Bacteroides Bile Esculin Agar (BBE, Anaerobe Systems) for B. fragilis, and MacConkey Agar (MilliporeSigma) for E. coli. Up to 5 individual colonies were selected from each plate and boiled for 10 minutes to lyse bacteria. In some cases, fewer than 5 colonies grew on the respective plates within 48 hours. Boiled colonies were centrifuged to remove cellular debris, and supernatants containing crude DNA extracts were harvested.
qPCR of extracted DNA.
Amplification was performed on a 7500 ABI instrument with 30 ng stool DNA and TaqMan Gene Expression Mix (Thermo-Fisher Scientific) supplemented with 0.1 μg/μL BSA using the following cycling parameters: 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds and 60°C or 58°C for 1 minute (see Supplemental Table 1 for annealing temperatures and primer/probe sequences; refs. 21, 23, 25, 26, 58, 59). Primers and probes were synthesized by Integrated DNA Technologies except for the bft probe, which was synthesized by Applied Biosystems.
To quantitate copy numbers of bacteria and their virulence factors, standard curves were generated from pure colonies containing each respective virulence factor (enterotoxigenic B. fragilis strain 086-5443-2-2 for bft; a clinical isolate of F. nucleatum from Johns Hopkins Hospital for fadA; and the murine E. coli strain NC101 for the colibactin gene clbB). The threshold used for detection was 10 copies/30 ng DNA, with the exception of nusG (SYBR green), for which the limit of quantitation was 1000 copies/30 ng DNA. All no-template controls were below this threshold.
Microbial standards.
The clbB+ E. coli NC101 strain used for qPCR standard curves was a gift from the laboratory of Christian Jobin (University of Florida College of Medicine, Gainesville, Florida, USA). The clinical F. nucleatum isolate used for qPCR standard curves was a gift from Brandon Ellis (Division of Medical Microbiology, Johns Hopkins Hospital).
Illlumina MiSeq WGS.
DNA from candidate E. coli isolates from donor/recipient pair 9 were obtained by reculturing fecal matter on MacConkey plates and harvesting up to 5 colonies per sample. Colony DNA was extracted using the Quick-DNA Fecal/Soil Microbe Kit (Zymo Research) and quantitated using a Qubit Fluorometer (Thermo Fisher Scientific). Sequencing libraries were generated using the Nextera DNA Flex Library Prep Kit (Illumina). Samples were sequenced on a MiSeq instrument using v3 2x75 chemistry. E. coli reads were assembled into contigs using SPAdes v3.10.1 (https://github.com/ablab/spades/releases/tag/v3.10.1) and aligned using the core genome alignment software Parsnp (https://harvest.readthedocs.io/en/latest/content/parsnp.html) (60), which generated 2,466 alignment blocks with an average cluster length of 1,611 bp. The final SNP counts represent SNPs per approximately 4 Mb of the E. coli genome. Sequencing reads from isolates too divergent to align with E. coli in Parsnp were classified using Centrifuge (61) with the NCBI database of archaea, bacteria, and virus genomes. WGS sequencing data are available through the NCBI Sequence Read Archive (SRA; PRJNA529243). The code used to perform the whole genome sequencing analysis is available at https://github.com/timplab/drewes_fmt
Statistics.
GraphPad Prism (version 8) was used for all statistical tests. A paired-sample Wilcoxon’s test was used to analyze data in Figure 2 comparing donor values with their respective patient pre-FMT or post-FMT values. P < 0.05 was considered statistically significant. No other statistical analyses were performed.
Study approval.
This study was reviewed and approved by the Institutional Review Board at Johns Hopkins Hospital. All subjects or, when applicable, their parents or guardians, provided informed consent prior to their participation in the study.
Author contributions
JLD and CLS had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. JLD, MOH, and CLS conceived and designed the study. JLD, AC, US, YF, SKH, MW, SDS, PJS, HW, WT, MOH, and CLS acquired, analyzed, and interpreted data. JLD, AC, US, YF, SKH, MW, SDS, PJS, HW, WT, MOH, and CLS engaged in critical revision of the manuscript for important intellectual content. JLD and HW were responsible for statistical analysis. MOH and CLS obtained funding. JLD, WT, MOH, and CLS supervised the work.
Supplementary Material
Acknowledgments
This study was funded by the Crohn’s & Colitis Foundation (CCFA 1853; to CLS), the Bloomberg~Kimmel Institute for Cancer Immunotherapy at Johns Hopkins University School of Medicine (JHU SOM), funds from the Bloomberg-Kimmel Chair in Immunotherapy (CLS), a Doctoral Foreign Study Award from the Canadian Institutes of Health Research (to YF), and a K99 award from the National Cancer Institute (K99CA230192 to JLD).
Version 1. 10/03/2019
Electronic publication
Footnotes
US’s current address is: Stanford University School of Medicine, Stanford, California, USA.
SKH’s current address is: Inova Children’s Hospital, Falls Church, Virginia, USA.
MW’s current address is: Department of Pediatric Gastroenterology and Nutrition, Rutgers Robert Wood Johnson Medical School, New Brunswick, New Jersey, USA.
Conflict of interest: WT has received research funding from Merck & Co. WT has 2 patents (8,748,091 and 8,394,584) licensed to Oxford Nanopore Technologies.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(19):e130848.https://doi.org/10.1172/jci.insight.130848.
Contributor Information
Julia L. Drewes, Email: jdrewes2@jhmi.edu.
Alina Corona, Email: 6acorona@gmail.com.
Uriel Sanchez, Email: usanchez@stanford.edu.
Yunfan Fan, Email: yfan7@jhmi.edu.
Melissa Weidner, Email: mweidner@rwjms.rutgers.edu.
Hao Wang, Email: hwang76@jhmi.edu.
Winston Timp, Email: wtimp@jhu.edu.
Maria Oliva-Hemker, Email: moliva@jhmi.edu.
References
- 1.Lessa FC, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372(9):825–834. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hourigan SK, Oliva-Hemker M, Hutfless S. The prevalence of Clostridium difficile infection in pediatric and adult patients with inflammatory bowel disease. Dig Dis Sci. 2014;59(9):2222–2227. doi: 10.1007/s10620-014-3169-4. [DOI] [PubMed] [Google Scholar]
- 3.van Nood E, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368(5):407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 4.Cammarota G, et al. Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment Pharmacol Ther. 2015;41(9):835–843. doi: 10.1111/apt.13144. [DOI] [PubMed] [Google Scholar]
- 5.Li YT, Cai HF, Wang ZH, Xu J, Fang JY. Systematic review with meta-analysis: long-term outcomes of faecal microbiota transplantation for Clostridium difficile infection. Aliment Pharmacol Ther. 2016;43(4):445–457. doi: 10.1111/apt.13492. [DOI] [PubMed] [Google Scholar]
- 6.Hourigan SK, et al. Microbiome changes associated with sustained eradication of Clostridium difficile after single faecal microbiota transplantation in children with and without inflammatory bowel disease. Aliment Pharmacol Ther. 2015;42(6):741–752. doi: 10.1111/apt.13326. [DOI] [PubMed] [Google Scholar]
- 7.Li X, et al. Clinical efficacy and microbiome changes following fecal microbiota transplantation in children with recurrent clostridium difficile infection. Front Microbiol. 2018;9:2622. doi: 10.3389/fmicb.2018.02622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kronman MP, et al. Fecal microbiota transplantation via nasogastric tube for recurrent clostridium difficile infection in pediatric patients. J Pediatr Gastroenterol Nutr. 2015;60(1):23–26. doi: 10.1097/MPG.0000000000000545. [DOI] [PubMed] [Google Scholar]
- 9.Russell GH, et al. Fecal transplant for recurrent Clostridium difficile infection in children with and without inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2014;58(5):588–592. doi: 10.1097/MPG.0000000000000283. [DOI] [PubMed] [Google Scholar]
- 10.Pierog A, Mencin A, Reilly NR. Fecal microbiota transplantation in children with recurrent Clostridium difficile infection. Pediatr Infect Dis J. 2014;33(11):1198–1200. doi: 10.1097/INF.0000000000000419. [DOI] [PubMed] [Google Scholar]
- 11.Walia R, et al. Efficacy of fecal microbiota transplantation in 2 children with recurrent Clostridium difficile infection and its impact on their growth and gut microbiome. J Pediatr Gastroenterol Nutr. 2014;59(5):565–570. doi: 10.1097/MPG.0000000000000495. [DOI] [PubMed] [Google Scholar]
- 12.Kahn SA, Young S, Rubin DT. Colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection in a child. Am J Gastroenterol. 2012;107(12):1930–1931. doi: 10.1038/ajg.2012.351. [DOI] [PubMed] [Google Scholar]
- 13.Rubin TA, Gessert CE, Aas J, Bakken JS. Fecal microbiome transplantation for recurrent Clostridium difficile infection: report on a case series. Anaerobe. 2013;19:22–26. doi: 10.1016/j.anaerobe.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Kelly CR, et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol. 2014;109(7):1065–1071. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fareed S, et al. Applying fecal microbiota transplantation (FMT) to treat recurrent Clostridium difficile infections (rCDI) in children. PeerJ. 2018;6:e4663. doi: 10.7717/peerj.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Outryve M, Debongnie JC. GLEM/LOK report on proctology practice in Belgium. Results, comments and recommendations. Acta Gastroenterol Belg. 2006;69(1):25–30. [PubMed] [Google Scholar]
- 17.Paramsothy S, et al. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389(10075):1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 18.Moayyedi P, et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology. 2015;149(1):102–109.e6. doi: 10.1053/j.gastro.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Costello SP, et al. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321(2):156–164. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. 2018;24(12):1804–1808. doi: 10.1038/s41591-018-0238-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arthur JC, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castellarin M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boleij A, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60(2):208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drewes JL, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes. 2017;3:34. doi: 10.1038/s41522-017-0040-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss J, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17(9):1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 29.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22(2):349–69. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.San Joaquin VH, Griffis JC, Lee C, Sears CL. Association of Bacteroides fragilis with childhood diarrhea. Scand J Infect Dis. 1995;27(3):211–215. doi: 10.3109/00365549509019011. [DOI] [PubMed] [Google Scholar]
- 31.Sack RB, et al. Enterotoxigenic Bacteroides fragilis: epidemiologic studies of its role as a human diarrhoeal pathogen. J Diarrhoeal Dis Res. 1992;10(1):4–9. [PubMed] [Google Scholar]
- 32.Han YW, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187(15):5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu P, Liu Y, Wang J, Guo Y, Zhang Y, Xiao S. Detection of fusobacterium nucleatum and fadA adhesin gene in patients with orthodontic gingivitis and non-orthodontic periodontal inflammation. PLoS ONE. 2014;9(1):e85280. doi: 10.1371/journal.pone.0085280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shetab R, et al. Detection of Bacteroides fragilis enterotoxin gene by PCR. J Clin Microbiol. 1998;36(6):1729–1732. doi: 10.1128/jcm.36.6.1729-1732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pantosti A, Malpeli M, Wilks M, Menozzi MG, D’Ambrosio F. Detection of enterotoxigenic Bacteroides fragilis by PCR. J Clin Microbiol. 1997;35(10):2482–2486. doi: 10.1128/jcm.35.10.2482-2486.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claros MC, et al. Characterization of the Bacteroides fragilis pathogenicity island in human blood culture isolates. Anaerobe. 2006;12(1):17–22. doi: 10.1016/j.anaerobe.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Claros MC, et al. Occurrence of Bacteroides fragilis enterotoxin gene-carrying strains in Germany and the United States. J Clin Microbiol. 2000;38(5):1996–1997. doi: 10.1128/jcm.38.5.1996-1997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Putze J, et al. Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae. Infect Immun. 2009;77(11):4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24(4):350–354. doi: 10.1016/j.cmi.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Smillie CS, et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe. 2018;23(2):229–240.e5. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li SS, et al. Durable coexistence of donor and recipient strains after fecal microbiota transplantation. Science. 2016;352(6285):586–589. doi: 10.1126/science.aad8852. [DOI] [PubMed] [Google Scholar]
- 42.Siegel RL, et al. Colorectal cancer incidence patterns in the United States, 1974-2013. J Natl Cancer Inst. 2017;109(8):djw322. doi: 10.1093/jnci/djw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zitomersky NL, Coyne MJ, Comstock LE. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun. 2011;79(5):2012–2020. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15(3):317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah MS, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67(5):882–891. doi: 10.1136/gutjnl-2016-313189. [DOI] [PubMed] [Google Scholar]
- 46.Wirbel J, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–689. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toprak NU, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12(8):782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 48.Keenan JI, Aitchison A, Purcell RV, Greenlees R, Pearson JF, Frizelle FA. Screening for enterotoxigenic Bacteroides fragilis in stool samples. Anaerobe. 2016;40:50–53. doi: 10.1016/j.anaerobe.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Singh R, et al. Donor feces infusion for eradication of extended spectrum beta-lactamase producing Escherichia coli in a patient with end stage renal disease. Clin Microbiol Infect. 2014;20(11):O977–O978. doi: 10.1111/1469-0691.12683. [DOI] [PubMed] [Google Scholar]
- 50.Dinh A, et al. Clearance of carbapenem-resistant Enterobacteriaceae vs vancomycin-resistant enterococci carriage after faecal microbiota transplant: a prospective comparative study. J Hosp Infect. 2018;99(4):481–486. doi: 10.1016/j.jhin.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 51.Huttner BD, et al. A 5-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant Enterobacteriaceae: a randomized clinical trial. Clin Microbiol Infect. 2019;25(7):830–838. doi: 10.1016/j.cmi.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 52.Orenstein R, et al. Safety and durability of RBX2660 (microbiota suspension) for recurrent Clostridium difficile infection: results of the PUNCH CD study. Clin Infect Dis. 2016;62(5):596–602. doi: 10.1093/cid/civ938. [DOI] [PubMed] [Google Scholar]
- 53.Khanna S, et al. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. J Infect Dis. 2016;214(2):173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- 54.Bristol A, Hubert S, Hofmann F, Baer H. Formulation development of SYN-004 (ribaxamase) oral solid dosage form, a β-lactamase to prevent intravenous antibiotic-associated dysbiosis of the colon. Int J Pharm. 2017;534(1-2):25–34. doi: 10.1016/j.ijpharm.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 55.de Gunzburg J, et al. Protection of the human gut microbiome from antibiotics. J Infect Dis. 2018;217(4):628–636. doi: 10.1093/infdis/jix604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Petrof EO, et al. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating’ the gut. Microbiome. 2013;1(1):3. doi: 10.1186/2049-2618-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDonald LC, et al. Clinical practice guidelines for clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin Infect Dis. 2018;66(7):987–994. doi: 10.1093/cid/ciy149. [DOI] [PubMed] [Google Scholar]
- 58.Tong J, Liu C, Summanen P, Xu H, Finegold SM. Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe. 2011;17(2):64–68. doi: 10.1016/j.anaerobe.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 59.Kato N, et al. A new subtype of the metalloprotease toxin gene and the incidence of the three bft subtypes among Bacteroides fragilis isolates in Japan. FEMS Microbiol Lett. 2000;182(1):171–176. doi: 10.1111/j.1574-6968.2000.tb08892.x. [DOI] [PubMed] [Google Scholar]
- 60.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15(11):524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26(12):1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.