Abstract
Cetuximab, an EGFR-blocking antibody, is currently approved for treatment of metastatic head and neck squamous cell carcinoma (HNSCC), but its response rate is limited. In addition to blocking EGFR-stimulated cell signaling, cetuximab can induce endocytosis of ASCT2, a glutamine transporter associated with EGFR in a complex, leading to glutathione biosynthesis inhibition and cellular sensitization to ROS. Pyruvate dehydrogenase kinase-1 (PDK1), a key mitochondrial enzyme overexpressed in cancer cells, redirects glucose metabolism from oxidative phosphorylation toward aerobic glycolysis. In this study, we tested the hypothesis that targeting PDK1 is a rational approach to synergize with cetuximab through ROS overproduction. We found that combination of PDK1 knockdown or inhibition by dichloroacetic acid (DCA) with ASCT2 knockdown or with cetuximab treatment induced ROS overproduction and apoptosis in HNSCC cells, and this effect was independent of effective inhibition of EGFR downstream pathways but could be lessened by N-acetyl cysteine, an anti-oxidative agent. In several cetuximab-resistant HNSCC xenograft models, DCA plus cetuximab induced marked tumor regression, whereas either agent alone failed to induce tumor regression. Our findings call for potentially novel clinical trials of combining cetuximab and DCA in patients with cetuximab-sensitive EGFR-overexpressing tumors and patients with cetuximab-resistant EGFR-overexpressing tumors.
Keywords: Metabolism, Therapeutics
Keywords: Head and neck cancer
Combining ASCT2 glutamine transporter-targeting through cetuximab and pyruvate dehydrogenase kinase 1 inhibition induced tumor regression in preclinical head and neck squamous cell carcinoma models.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a prevalent cancer worldwide. In the United States, HNSCC accounts for approximately 3% of all cancers, and it is estimated that there will be 53,000 new cases and 10,860 deaths from this disease in 2019 (1). Recently, there have been encouraging improvements in survival in patients with recurrent or metastatic HNSCC after treatment with the the anti–programmed cell death protein-1 antibodies nivolumab and pembrolizumab (2, 3). However, despite notable progress in treatment of HNSCC with surgery, radiotherapy, chemotherapy, targeted therapy, and immunotherapy, the 5-year survival rate remains low in patients with advanced metastatic HNSCC (4, 5).
HNSCC cells frequently exhibit overexpression of the EGFR. Cetuximab, an EGFR-blocking antibody, is currently the only approved EGFR-targeting therapy for patients with metastatic HNSCC (6, 7). The efficacy of cetuximab is limited by intrinsic and acquired drug resistance, which is due mainly to tumor heterogeneity and genetic instability; these lead to constitutive activation of EGFR downstream signaling pathways and functional redundancy or compensatory activation of alternative growth factor receptor pathways (8–14). The current response rate to cetuximab as a single agent in HNSCC is only approximately 13% (15).
Cancer cells, including HNSCC cells, metabolize glucose by aerobic glycolysis in the cytoplasm (i.e., the Warburg effect) to meet the critical biosynthetic and bioenergetic requirements for cancer cell growth and proliferation (16, 17). In contrast, normal cells metabolize glucose by oxidative phosphorylation via the tricarboxylic acid cycle in the mitochondria. The mitochondrial enzyme pyruvate dehydrogenase kinase-1 (PDHK1 or PDK1, hereafter referred to as PDK1) is an emerging target for cancer treatment owing to its key role in switching glucose metabolism toward aerobic glycolysis in cancer cells (18, 19). In HNSCC, overexpression of PDK1 has been reported to be associated with a poor outcome in patients (20). PDK1 promotes aerobic glycolysis by phosphorylating and inactivating pyruvate dehydrogenase (PDH), which is the gate-keeping enzyme that controls the flux of glucose into the mitochondria by converting pyruvate to acetyl-CoA for oxidative phosphorylation through the tricarboxylic acid cycle (21–23). Inhibition of PDK1 leads to redirection of glucose metabolism from aerobic glycolysis to oxidative phosphorylation, resulting in overproduction of ROS in cancer cells. Although an elevated level of ROS can activate the protumorigenic signaling that is common in many types of cancer cells, overproduction of ROS can also lead to cancer cell death via apoptosis (24).
Cancer cells, however, are often intrinsically resistant to ROS-induced apoptosis, and one of the underlying mechanisms is cancer cells’ ability to detoxify ROS (25). Glutathione (GSH), a small tripeptide consisting of glutamate, cysteine, and glycine, is the major nonenzymatic cellular antioxidant when in its reduced state. There are 2 key determinants of glutathione biosynthesis: availability of cysteine, the sulfur amino acid precursor of glutathione, and activity of the rate-limiting enzyme glutamate cysteine ligase (26). Because of the oxidizing environment in the extracellular space, approximately 90% of cysteine is oxidized as the dimeric cystine. Cells therefore mainly use the stable cystine as a precursor for glutathione synthesis (26). Cystine is exclusively transported into cells by the xC– cystine/glutamate antiporter, which is a heterodimer consisting of xCT, the regulatory subunit of the system, and 4F2hc/CD98, the functional subunit of the system (27, 28). Once inside the cell, cystine is rapidly reduced to cysteine because of the reducing intracellular milieu (28–30).
Whereas xCT regulates the transport of cystine into cells, the alanine-serine-cysteine transporter 2 (ASCT2) regulates the transport of glutamine, cysteine, and a few other neutral amino acids into cells (27, 31). Glutamine has broad roles in cell metabolism; it is involved not only in the biosynthesis of glutathione and other macromolecules required for growth and proliferation of cancer cells, but also in several other important metabolic processes, including glutaminolysis, which provides biofuels through catabolizing glutamine to generate ATP and lactate (32, 33). Many cancer cells are addicted to glutamine (34–38). Thus, ASCT2 is an appealing target for cancer treatment (32–40). However, ASCT2 also plays an important role in normal cell metabolism, and direct targeting of ASCT2 is technically challenging.
We recently reported that ASCT2 is physically associated with EGFR on the plasma membrane of HNSCC cells in a molecular complex involving AP1G1 and that the EGFR-AP1G1-ASCT2 complex can be coendocytosed upon cetuximab binding to EGFR (41, 42). This potentially novel activity of cetuximab that downregulates ASCT2 is independent of whether EGFR downstream cell signaling is inhibited by cetuximab (41, 42). In this study, we sought to test the hypotheses that (a) the combination of cetuximab with PDK1 inhibition is a rational strategy for enhancing overall therapeutic response in HNSCC and (b) an underlying mechanism is that cetuximab mediates downregulation of ASCT2 and consequent inhibition of glutamine uptake and intracellular glutathione biosynthesis, thereby sensitizing HNSCC cells to PDK1 inhibition, which exerts antitumor activity via increasing ROS production in cancer cells.
Results
Both PDK1 and SLC1A5 (ASCT2) are overexpressed in HNSCC and in other cancers, and their dual silencing is synthetically lethal.
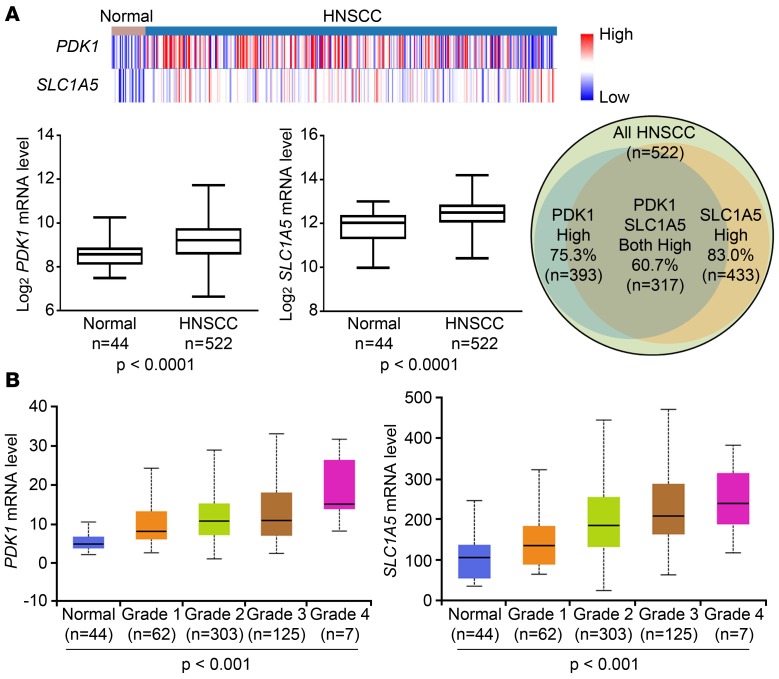
We queried The Cancer Genome Atlas (TCGA) database (https://xena.ucsc.edu/) and found that the mRNA levels of PDK1 and SLC1A5 (ASCT2) were both significantly higher in primary human HNSCC tissues (n = 522) than in the adjacent normal tissues (n = 44) (Figure 1A). We found that, of the 522 HNSCC samples, 393 (75.3%) had a higher level of PDK1 mRNA, 433 (83.0%) had a higher level of SLC1A5 mRNA, and 317 (60.7%) had higher levels of both PDK1 mRNA and SLC1A5 mRNA than the mean values of these gene expression levels in normal tissues (Figure 1). The mRNA levels of PDK1 and SLC1A5 in the HNSCC samples in the TCGA database also individually correlated with tumor grade (Figure 1B), which is linked to tumor recurrence, metastasis, and patient mortality (43). Furthermore, we found that the mRNA levels of PDK1 and SLC1A5 were elevated not only in HNSCC, but also in other types of cancers in a pancancer cohort consisting of 12 datasets, including bladder urothelial carcinoma, breast invasive carcinoma, colon adenocarcinoma, glioblastoma multiforme, HNSCC, kidney renal clear cell carcinoma, acute myeloid leukemia, lung adenocarcinoma, lung squamous cell carcinoma, ovarian serous cystadenocarcinoma, rectum adenocarcinoma, and uterine corpus endometrioid carcinoma (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.131106DS1). High mRNA levels of PDK1 and SLC1A5 individually correlated with poor survival of patients in the cohort (Supplemental Figure 1, C and D).
Figure 1. PDK1 and SLC1A5 are both overexpressed in HNSCC tumors, and their mRNA levels are associated with tumor grade in HNSCC.
(A) The mRNA levels of PDK1 and SLC1A5 in HNSCC and adjacent normal tissues were retrieved from the TCGA database (hosted at https://xena.ucsc.edu/). Heatmaps of PDK1 and SLC1A5 mRNA levels in HNSCC and normal tissues were created (top), and their expression levels were plotted and analyzed by Student’s t test (bottom). Blue, less than the median; red, greater than the median. The Venn diagram at right shows the numbers of patients who had higher mRNA expression of PDK1, SLC1A5, or both in HNSCC tissues than the mean value of expression of these genes in the adjacent normal tissues. (B) The mRNA levels of PDK1 and SLC1A5 were compared among HNSCC tumors of different grades and corresponding adjacent normal tissue. The data were analyzed by 1-way ANOVA and are presented as box-and-whisker plots; plots show median values (line), 25th–75th percentiles (box outline), and minimum and maximum values (whiskers). Grade 1, well differentiated; grade 2, moderately differentiated; grade 3, poorly differentiated; grade 4, undifferentiated. See also Supplemental Figure 1.
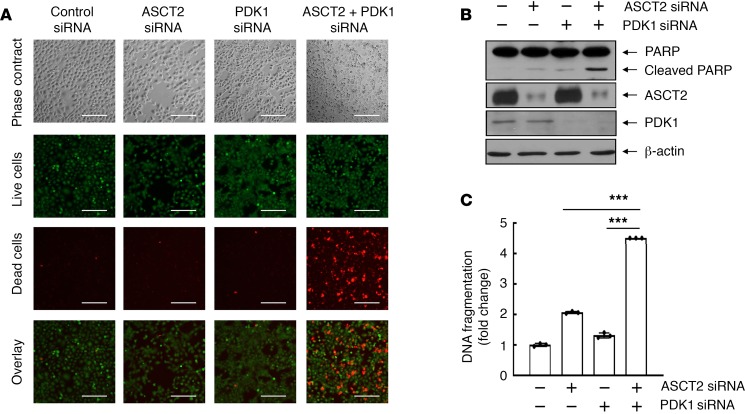
We next investigated the impact of PDK1 and ASCT2 levels on survival of HNSCC cells using siRNA-mediated expression silencing to knock down PDK1 and ASCT2 alone and together. As shown in Figure 2A, knockdown of PDK1 or ASCT2 expression alone had no marked effect on cell survival of HN5 cells, an HNSCC cell line that expresses a very high level of EGFR (44, 45); however, dual knockdown of PDK1 and ASCT2 expression led to massive cell death, measured by a fluorescence-based LIVE/DEAD cell viability assay. Apoptosis assays showed much greater poly (ADP-ribose) polymerase (PARP) cleavage cleavage detected by Western blotting (Figure 2B) and DNA fragmentation measured by an apoptosis ELISA (Figure 2C) following dual knockdown of PDK1 and ASCT2 than following individual knockdown of PDK1 or ASCT2. Similar results were observed in another HNSCC cell line, FaDu, which expresses a moderately high level of EGFR (Supplemental Figure 2).
Figure 2. Dual silencing of ASCT2 and PDK1 is synthetically lethal to HNSCC cells.
HN5 cells were transfected with control siRNA, ASCT2 siRNA, PDK1 siRNA, or ASCT2 siRNA plus PDK1 siRNA for 72 hours. (A) HN5 cells were subjected to LIVE/DEAD cell viability assay as described in Methods and then observed under a fluorescence microscope. Scale bars: 200 μm. (B) The cell lysates were subjected to Western blotting with the indicated antibodies. (C) The cell lysates were subjected to quantitative apoptosis ELISA as described in Methods. Error bars indicate ± SD. ***P < 0.001 (2-way ANOVA, n = 3). See also similar data on FaDu cells in Supplemental Figure 2.
Together, these results indicate that both PDK1 and ASCT2 are overexpressed in the tumors of HNSCC patients and are associated with tumor grade and poor clinical outcomes and that dual knockdown of PDK1 and ASCT2 can be synthetically lethal, suggesting that cotargeting of PDK1 and ASCT2 may represent a rational combination approach for treatment of HNSCC.
Cetuximab mediates coendocytosis of EGFR-associated ASCT2 and sensitizes HNSCC cells to PDK1 knockdown–induced or PDK1 inhibition–induced ROS overproduction and apoptosis.
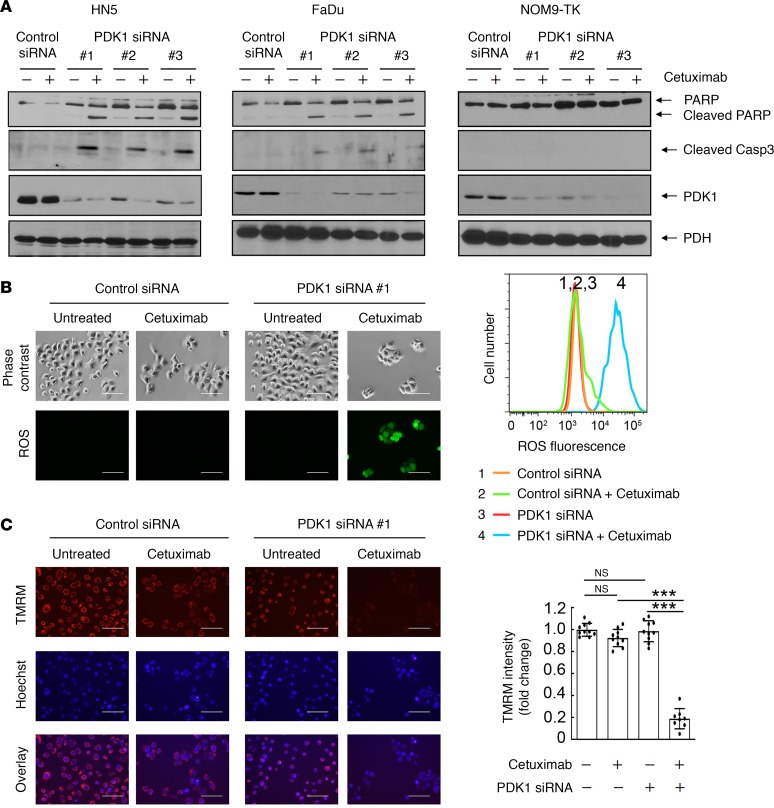
Unlike common druggable targets, such as receptor tyrosine kinases, ASCT2 is technically challenging to target by a pharmacological approach; however, at least a fraction of ASCT2 is associated in a molecular complex with EGFR on the cell surface and can be downregulated by cetuximab (41, 42). Importantly, this action is mediated through cetuximab-induced EGFR endocytosis that is independent of cetuximab-mediated inhibition of EGFR downstream cell signaling (41, 42). We thus sought to examine whether the combination of cetuximab with PDK1 silencing or inhibition can induce cell death in representative HNSCC cells. Although HN5 and FaDu cells are sensitive to cetuximab-induced growth inhibition (a cytostatic effect), their treatment with cetuximab alone or PDK1 knockdown with any of 3 different siRNAs had no effect on induction of apoptosis; in contrast, the combination of PDK1 knockdown with cetuximab induced apoptosis, as indicated by detection of cleaved PARP and caspase 3 on Western blotting (Figure 3A), in both HN5 and FaDu cells, but not in NOM9-TK cells, an immortalized nonmalignant head and neck epithelial cell line (46–48). We further found that induction of apoptosis after treatment with the combination of PDK1 knockdown with cetuximab was accompanied by overproduction of ROS (Figure 3B) and rapid depolarization of inner mitochondrial membrane potential caused by ROS-induced oxidative stress (Figure 3C) in HN5 cells. Similar results were observed in FaDu cells (Supplemental Figure 3).
Figure 3. Cetuximab sensitizes HNSCC cells to PDK1-silencing–induced apoptosis, ROS overproduction, and mitochondria depolarization.
(A) HN5, FaDu, and NOM9-TK cells were transfected with control siRNA or each of 3 different PDK1 siRNAs for 72 hours. Cetuximab (20 nM) was added or not added during the last 24 hours of siRNA transfection as indicated. Cell lysates were then prepared and subjected to Western blotting with the indicated antibodies. (B and C) HN5 cells were transfected with control siRNA or PDK1 siRNA #1 for 72 hours. Cetuximab (20 nM) was added or not added during the last 24 hours of siRNA transfection as in A. In B, the cells were stained with Enzo Life Sciences’ ROS detection kit and then observed under a fluorescence microscope (left) or subjected to FACS analysis after staining (right). Scale bars: 100 μm. In C, the cells were stained with mitochondrial membrane potential–sensitive dye tetramethyl rhodamine methyl ester (TMRM) and then observed under a fluorescence microscope (left) or analyzed with a fluorescence plate reader (right). Scale bars: 100 μm. Error bars ± SD. ***P < 0.001 (2-way ANOVA, n = 10). Similar results were observed with 2 other PDK1 siRNAs. See also similar data on FaDu cells in Supplemental Figure 3.
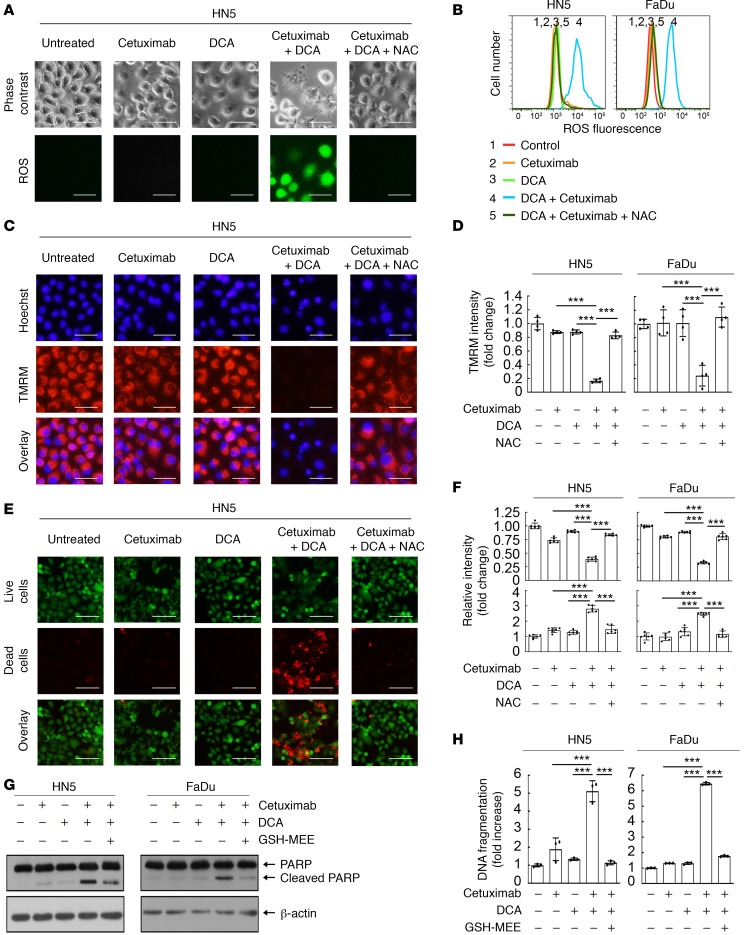
Similar to the results observed with PDK1 knockdown, the combination of cetuximab with pharmacological inhibition of PDK1 enzyme activity with dichloroacetic acid (DCA) resulted in overproduction of ROS (Figure 4, A and B), mitochondrial depolarization (Figure 4, C and D), and massive cell death detected by the fluorescence-based LIVE/DEAD cell viability assay (Figure 4, E and F) in HN5 cells. Similar results were observed in FaDu cells (Figure 4 and Supplemental Figure 4). Apoptosis assays detected marked PARP cleavage and DNA fragmentation following treatment with DCA plus cetuximab in both HN5 and FaDu cells (Figure 4, G and H). Importantly, addition of N-acetyl cysteine (NAC), a glutathione precursor, or glutathione monoethyl ester (GSH-MEE), a membrane-permeable form of glutathione, into the culture medium — which tempered overproduction of ROS and mitochondrial depolarization — decreased the extent of cell death induced by the combination treatment. This important observation confirms that overproduction of ROS causally contributed to the induction of cell death by the combination of cetuximab with DCA.
Figure 4. Cetuximab sensitizes HNSCC cells to DCA-induced ROS overproduction, mitochondria depolarization, and apoptosis.
HN5 cells (A–H) and FaDu cells (B, D, F, and H) were left untreated or treated with 20 nM cetuximab, 10 mM DCA, or both with or without 10 mM NAC as indicated for 24 hours. (A and B) The cells were stained with Enzo Life Sciences’ ROS detection kit and then observed under a microscope (A) and subjected to FACS analysis (B). Scale bars: 50 μm. (C and D) The cells were stained with mitochondrial membrane potential–sensitive dye tetramethyl rhodamine methyl ester (TMRM) and then observed under a microscope (C) or analyzed with a fluorescence plate reader (D). Scale bars: 50 μm. (E and F) The cells were subjected to LIVE/DEAD cell viability assay and then observed under a microscope (E) or analyzed with a fluorescence plate reader (F). Scale bars: 100 μm. (G and H) Lysates of cells treated as indicated were subjected to Western blotting with the indicated antibodies (G) and to an apoptosis ELISA (H). All error bars indicate ± SD. ***P < 0.001 (2-way ANOVA, n = 4 in D, n = 6 in F, n = 3 in H). See similar results for FaDu cells in Supplemental Figure 4.
To further address the extent to which the combination of cetuximab with DCA induces apoptosis across different HNSCC cell lines and across different types of cancer cell lines, we examined the effect of combination treatment in a panel including 9 additional types of HNSCC cells and several other types of cancer cells. We found that the combination treatment induced apoptosis in all the cancer cell lines examined (Supplemental Figure 5, A and B). DCA inhibited PDK1 enzymatic activity as expected, shown by a decreased level of S239-phosphorylated PDH (which leads to PDH activation) but no change in the level of total PDH in HN5 and FaDu HNSCC cells and in the immortalized nonmalignant NOM9-TK cells (Supplemental Figure 5C).
Taken together, these results suggest that HNSCC cells and other types of cancer cells exhibit intrinsic resistance to induction of apoptosis following overproduction of ROS, as a result of PDK1 expression knockdown–induced or PDK1 enzyme inhibition–induced overactivation of PDH, and they suggest that this resistance can be overcome by cotreatment of the cells with cetuximab.
Cetuximab sensitizes HNSCC cells to PDK1 targeting–induced apoptosis via diminishing ASCT2-mediated glutamine uptake.
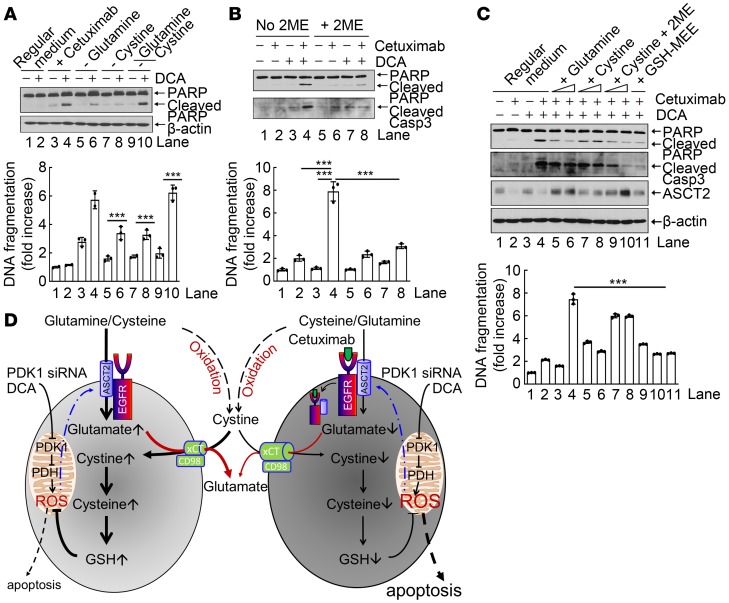
To further determine the extent to which inhibition of glutamine uptake following cetuximab-induced downregulation of ASCT2 contributes to cetuximab-mediated sensitization of HNSCC cells to PDK1 targeting–induced overproduction of ROS and subsequent apoptosis, we first mimicked the consequence of an attenuated transporter function of ASCT2 by directly removing glutamine, the consequence of an attenuated transporter function of xCT by directly removing cystine, or the consequence of directly removing both glutamine and cystine from culture medium. Removal of glutamine, cystine, or both had no clear impact on induction of apoptosis in cells cultured for 24 hours, but removal of glutamine or cystine — and particularly removal of both glutamine and cystine — sensitized the cells to DCA-induced apoptosis (Figure 5A), indicating that both glutamine and cystine were required to protect against DCA-induced apoptosis. We next tested the impact of chemical conversion of cystine (the oxidized form of cysteine present in regular medium) to cysteine on DCA plus cetuximab-induced apoptosis by addition of the reducing agent 2-mercaptoethanol (2ME) in culture medium. Conversion of cystine to cysteine reduced the degree to which cetuximab sensitized cells to DCA-induced apoptosis (Figure 5B), indicating a more critical role of cysteine than of cystine in counteracting the apoptotic effect of DCA in the presence of cetuximab. Addition into culture medium of 2- to 4-fold excess amounts of glutamine or of cystine plus 2ME (i.e., converted to cysteine), both of which require ASCT2 for transport into cells, upregulated ASCT2 and reduced the apoptotic effect of DCA plus cetuximab (Figure 5C), consistent with the general knowledge that both glutamine and cysteine can protect against ROS-induced apoptosis after transport via ASCT2. In contrast, addition of an excess amount of cystine alone did not upregulate ASCT2 and only minimally reduced the apoptotic effect of DCA plus cetuximab (Figure 5C), strongly supporting the role of ASCT2 level in regulating DCA plus cetuximab–induced apoptosis.
Figure 5. Cetuximab sensitizes HNSCC cells to DCA-induced apoptosis via diminishing ASCT2-mediated glutamine uptake.
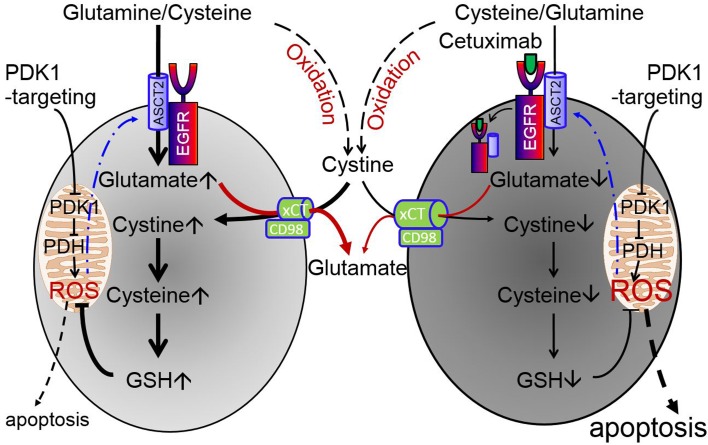
(A–C) HN5 cells were treated as indicated, and cell lysates were subjected to Western blotting with the indicated antibodies (upper panels) and to ELISA for quantification of apoptosis (lower panels). (A) The cells were cultured in regular medium or glutamine- and/or cystine-deficient medium with or without 10 mM DCA for 24 hours. (B) The cells were left untreated or treated with cetuximab, DCA, or both as indicated for 24 hours in medium with or without addition of 10 μM 2-mercaptoethanol (2ME). (C) The cells were left untreated or treated with cetuximab, DCA, or both as indicated for 24 hours in regular medium; regular medium supplemented with additional glutamine (final concentrations 5 mM and 10 mM in lanes 5 and 6, respectively) or cystine (final concentrations 0.2 mM and 0.4 mM in lanes 7 and 8 or final concentrations 0.2 mM and 0.4 mM plus 10 μM 2ME in lanes 9 and 10, respectively); or regular medium supplemented with 10 mM glutathione monoethyl ester (GSH-MEE). All error bars indicate ± SD. ***P < 0.001 (2-way ANOVA, n = 3). (D) Proposed working model in which cetuximab diminishes intracellular glutathione via downregulation of the ASCT2-EGFR complex, thereby sensitizing cells to PDK siRNA– or DCA-induced apoptosis.
Taken together with our recently reported findings that cetuximab downregulates EGFR-associated ASCT2 via inducing endocytosis of the complex, which leads to decreased uptake of glutamine and a lower intracellular level of glutamate (41, 42), our results in Figure 5, A–C, suggest the validity of the working model depicted in Figure 5D, wherein the left and right sides of the model illustrate the process of glutathione biosynthesis in the absence and presence of cetuximab, respectively. In this model, cetuximab diminishes glutamine uptake via downregulation of the ASCT2-EGFR complex, leading to a low intracellular glutamate level; the low intracellular glutamate level decreases xC– cystine/glutamate antiporter-mediated glutamate-cystine exchange, which leads to a lower intracellular level of cysteine; and the low intracellular level of cysteine leads to reduced glutathione biosynthesis and, consequently, increased sensitivity to ROS-induced apoptosis.
Combination of cetuximab with PDK1 inhibition induces apoptosis and tumor regression in cetuximab-resistant HNSCC cells.
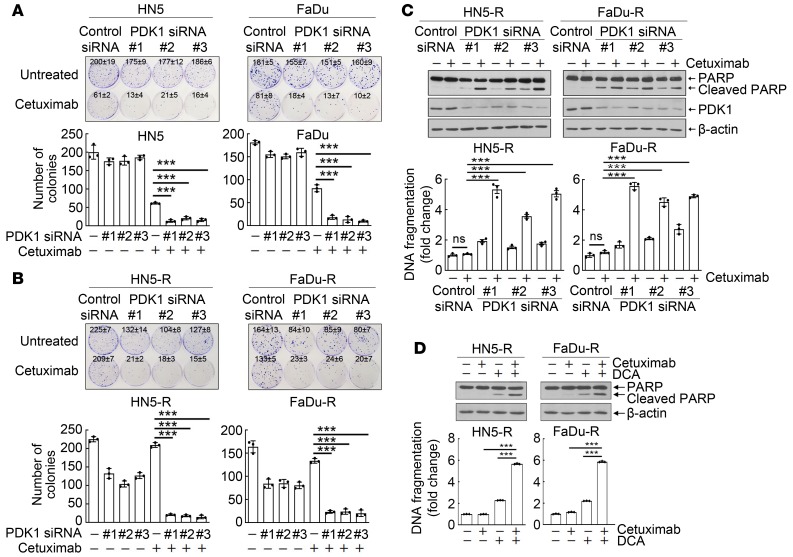
To determine if inhibition of EGFR downstream signaling by cetuximab plays a role in the induction of apoptosis by treatment with the combination of cetuximab with DCA, we compared responses to cetuximab plus PDK1 knockdown between parental cetuximab-sensitive HN5 and FaDu cells and HN5-R and FaDu-R cells, sublines of HN5 and FaDu cells with acquired resistance to cetuximab (48). Knockdown of PDK1 with any of 3 validated PDK1 siRNAs (as shown in Figure 3A) had minimal inhibitory effect on clonogenic growth of cetuximab-sensitive HN5 and FaDu cells (Figure 6A). Cetuximab strongly inhibited clonogenic growth of cetuximab-sensitive HN5 and FaDu cells, and the effect was further enhanced by PDK1 knockdown (Figure 6A). Cetuximab-resistant HN5-R and FaDu-R cells exhibited considerable resistance to the anticlonogenic growth activity of cetuximab, as expected (Figure 6B). Interestingly, the cetuximab-resistant sublines were more sensitive to PDK1 silencing than were their respective parental cells (Figure 6B). Most importantly, the anticlonogenic growth effects of the combination of cetuximab with PDK1 knockdown in cetuximab-resistant HN5-R and FaDu-R cells (Figure 6B) were as strong as the effects observed in the cetuximab-sensitive parental cells (Figure 6A). This effect of the combination treatment was further supported by our observation of induction of apoptosis in HN5-R and FaDu-R cells by the combination of cetuximab with PDK1 knockdown (Figure 6C) or cetuximab with PDK1 inhibition (Figure 6D), at levels similar to those in HN5 and FaDu cells (Figure 3A and Figure 4G). These results indicated that an antiproliferative response to cetuximab treatment is not essential for the induction of apoptosis by the combination of cetuximab with PDK1 knockdown or PDK1 inhibition.
Figure 6. Cetuximab plus PDK1 knockdown or inhibition decreases clonogenic survival of cetuximab-sensitive HNSCC cells and HNSCC cells with acquired resistance to cetuximab.
(A and B) Parental cetuximab-sensitive HN5 and FaDu cells (A), and their cetuximab-resistant sublines HN5-R and FaDu-R cells (B), were transfected with a control siRNA or each of 3 different PDK1 siRNAs for 72 hours; then, the siRNA-treated cells were seeded at low density and cultured with or without 2 nM cetuximab for 13 days for HN5 and HN5-R and 22 days for FaDu and FaDu-R cells. Upper panels, photographs of culture dishes. Lower panels, mean colony numbers ± SDs of triplicate wells. (C and D) HN5-R and FaDu-R cells were transfected with a control siRNA or each of 3 different PDK1 siRNAs for 72 hours (C) or were treated with DCA for 24 hours (D). Cetuximab (20 nM) was added or not during the last 24 hours of siRNA transfection or DCA treatment as indicated. Cell lysates were then prepared and subjected to Western blotting with the indicated antibodies (upper panels) and to quantitative apoptosis ELISA (bottom panels). All error bars indicate ± SD. ***P < 0.001 (2-way ANOVA, n = 3).
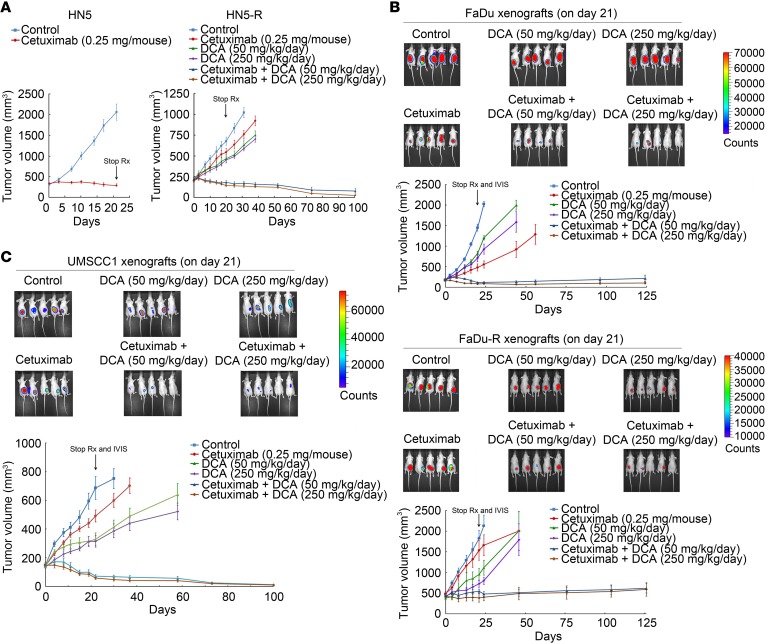
Lastly, we assessed the antitumor activity of the combination strategy in vivo. Compared with HN5 xenografts, which were strongly inhibited by cetuximab alone, HN5-R xenografts were resistant to cetuximab alone but moderately susceptible to DCA alone at 50 mg/kg body weight/day (equivalent to the recommended clinical dose of DCA for patients; ref. 49) and 250 mg/kg body weight/day (Figure 7A). However, the growth of HN5-R xenografts was strongly inhibited by DCA plus cetuximab. The mice in the combination-treatment groups were tumor free or had barely noticeable tumor nodules almost 80 days after treatment was stopped on day 21 (Figure 7A). FaDu xenografts were sensitive to cetuximab alone (Figure 7B, upper), although not as sensitive as HN5 xenografts (Figure 7A). Compared with FaDu xenografts, FaDu-R xenografts were resistant to cetuximab alone but responsive to DCA alone, as shown by both tumor size measurements and the findings on in vivo imaging of bioluminescence-positive xenografts on day 21 (Figure 7B, lower). FaDu xenografts inhibited by cetuximab and FaDu-R xenografts inhibited by DCA quickly resumed growth after the treatments were stopped, indicating that the effect of either treatment was only cytostatic. In contrast, the FaDu and FaDu-R xenografts treated with DCA plus cetuximab did not resume growth after treatment was stopped. These mice were tumor free or had only a small tumor nodule over 100 days after treatment was stopped on day 21, suggesting that the tumor cells underwent apoptosis or senescence. We further confirmed the effect of the cetuximab-DCA combination using a third type of HNSCC xenograft, UMSCC1 xenografts, which are less sensitive to cetuximab than HN5 and FaDu xenografts (50). Similar to the responses of HN5-R and FaDu-R xenografts, the growth of UMSCC1 xenografts was only minimally inhibited by cetuximab 0.25 mg/mouse twice a week but strongly inhibited by DCA plus cetuximab (Figure 7C). All the mice were tumor free for over 80 days after treatment was stopped on day 21.
Figure 7. Cetuximab plus PDK1 inhibition induces apoptosis in HNSCC cells with acquired resistance to cetuximab and inhibits their growth in vivo when used in combination with DCA.
(A) Left, established HN5 xenografts (~250 mm3) were untreated or treated with cetuximab (0.25 mg/mouse, twice a week for 3 weeks). Right, HN5-R xenografts (~250 mm3) were untreated or treated with the same dose of cetuximab, DCA (50 or 250 mg/kg/day), or DCA plus cetuximab for 3 weeks. (B) Established FaDu (upper) and FaDu-R (lower) xenografts (~250 mm3) were untreated or treated as described in A for HN5-R xenografts. After completion of treatments, the mice were subjected to IVIS imaging and then observed for additional days for tumor progression or regression. The recording of tumor sizes was stopped when fewer than 4 mice remained in any group because of sacrifice of mice with large tumor burden or morbid or moribund status. (C) Established UMSCC1 xenografts (~150 mm3) were untreated or treated with cetuximab (0.25 mg/mouse, twice a week for 3 weeks), DCA (50 or 250 mg/kg/day), or DCA plus cetuximab for 3 weeks. After completion of treatments on day 21, the mice were subjected to IVIS imaging and then observed for additional days for tumor progression or regression. The recording of tumor sizes was stopped when fewer than 4 mice remained in any group because of sacrifice of mice with large tumor burden or morbid or moribund status. Error bars indicate ± SEM. Rx, treatment.
In summary, our results from these preclinical studies indicate that the combination of cetuximab with DCA is a potentially novel approach, sensitizing cetuximab-sensitive and cetuximab-resistant EGFR-overexpressing HNSCC cells to PDK1 inhibition–induced ROS overproduction and apoptosis.
Discussion
In this paper, we report our findings from exploring a therapeutic strategy for overcoming cetuximab resistance in preclinical models of HNSCC. Our findings support a strategy of a combination of cetuximab with DCA, which has been approved by the US Food and Drug Administration (FDA) for a noncancer indication (a rare hereditary lactate metabolism disorder in children) for over 30 years (51, 52). In this combination strategy, cetuximab downregulates the EGFR-AP1G1-ASCT2 complex via cetuximab-induced EGFR endocytosis independently of EGFR downstream signaling inhibition (41, 42) and inhibits glutathione synthesis through inhibition of ASCT2-mediated glutamine uptake; DCA induces oxidative stress via inhibition of PDK1 and subsequent activation of PDH; and the combination of cetuximab with DCA leads to induction of apoptosis.
This combination strategy is compelling for 2 main reasons. First, unlike previously proposed strategies to overcome cetuximab resistance, our strategy does not require functional inhibition of EGFR downstream cell signaling pathways by cetuximab in order to be effective. Therefore, several known mechanisms of cetuximab resistance, including constitutive activation of EGFR downstream signaling pathways and functional redundancy or compensatory activation of alternative growth factor receptor pathways, such as HER2/HER3 or c-MET (13, 14), are unlikely to influence the outcome of our approach. Second, the combination of cetuximab with DCA not only induces cell death, but more importantly can induce cell death selectively in cancer cells. This selectivity lies in the known differences in metabolism and in EGFR expression level between normal cells and cancer cells. Only in cancer cells, which heavily rely on the Warburg effect, will ROS be overproduced upon treatment with the combination of PDK1 inhibition and ASCT2 downregulation. Our approach downregulates ASCT2 via targeting EGFR, which is often overexpressed in HNSCC cells compared with normal cells. Our data showing lack of toxicity of the combination treatment in NOM9-TK, a normal oral mucosa cell line, support this explanation (Figure 3A and Supplemental Figure 5C).
Elevated ASCT2 expression has been reported in many types of cancer cells (53); however, it is challenging to develop a therapy targeting ASCT2 directly because such a therapy would have to target the ASCT2 on cancer cells but not normal cells, and because amino acid transporters are generally not easily druggable (54). Glutamine analogs can block ASCT2 but also block several other glutamine transporters, including SNAT1, SNAT2, and LAT1 (55, 56). Targeting ASCT2 alone is also unlikely to exert durable therapeutic effects against cancer or cure cancer, given that the role of ASCT2 can be compensated for by the other glutamine transporters (57) and given that cancer metabolism can be easily reprogrammed when intracellular glutamine level is deficient.
Cancer treatment based on PDK1 as a metabolic target of the FDA-approved DCA has been investigated in several types of preclinical cancer models (24, 58–66) and in several clinical trials (https://clinicaltrials.gov/), including in patients with HNSCC (NCT01163487 and NCT01386632). In this study, we found that targeting PDK1 alone via PDK1 expression knockdown or enzyme inhibition with DCA at the doses reported in the literature did not produce significant antitumor effects against several HNSCC models either in vitro or in vivo, unless PDK1 targeting was combined with ASCT2 knockdown or cetuximab.
We found that the degree to which cetuximab enhanced PDK1-targeted therapy was substantially reduced in the presence of excess NAC or GSH. This finding strongly supports a functional role of ROS in the induction of apoptosis by the combination of cetuximab with PDK1 inhibition. Our conclusion that cetuximab-mediated ASCT2 downregulation contributes to the induction of apoptosis by the combination of cetuximab with DCA, as illustrated in the model in Figure 5D, is corroborated by our finding that dual silencing of PDK1 and ASCT2 was synthetically lethal to cancer cells (Figure 2). EGFR is known to physically associate with multiple partner proteins (67, 68). Whether our approach reveals a new paradigm of strategically developing therapies based on cetuximab-mediated coendocytosis of EGFR-associated proteins is worth further exploring through study of various other combinations.
As mentioned in the introduction, glutamine is an important metabolic fuel that helps rapidly proliferating cells to meet the increased demand of cancer cells for ATP, biosynthetic precursors, and reducing agents. Thus, the impact of cetuximab-mediated downregulation of ASCT2 and subsequent inhibition of glutamine uptake on cell growth and proliferation may not be due only to inhibition of glutathione synthesis. However, because ASCT2 silencing alone failed to produce noticeable effects on cell survival but ASCT2 silencing was synthetically lethal to cells when combined with PDK1 silencing — and because the combination of cetuximab with DCA induced apoptosis in both cetuximab-sensitive and cetuximab-resistant HNSCC cells and obliterated well-established HNSCC xenografts that were resistant to treatment with cetuximab or DCA alone — our results strongly support a dominant role of decreased glutathione synthesis and enhanced ROS sensitization resulting from downregulation of ASCT2 by cetuximab. It may also be worth testing the combination of cetuximab with PDK1 inhibition in other types of EGFR-overexpressing cancers because cetuximab may downregulate ASCT2 in other types of cancers beyond HNSCC; metastatic colorectal cancer may be a good candidate, given that cetuximab is currently approved for treatment of this disease. Our findings are highly translational, given that cetuximab is currently approved for treatment of metastatic HNSCC and DCA has been tested in patients with HNSCC. Our findings call for novel clinical trials of combining cetuximab and DCA in patients with cetuximab-sensitive EGFR-overexpressing tumors and patients with cetuximab-resistant EGFR-overexpressing tumors.
Therapeutic strategies, such as the one we describe in this paper, that target alterations in cancer cell metabolism rather than alterations in cell signaling pathways may hold promise for improving the therapeutic effects of targeted cancer therapy in HNSCC and other types of cancers (69–71). Future directions to further develop this strategy would include identifying markers for predicting response to the combination treatment and further improving therapeutic efficacy through treatment optimization.
Methods
TCGA data retrieval and analysis.
TCGA data were retrieved from the TCGA data coordination center through https://xena.ucsc.edu/ The TCGA data we analyzed included gene expression data from the following datasets: bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), Head-Neck Squamous Cell Carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), acute myeloid leukemia (LAML), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), rectum adenocarcinoma (READ), and uterine corpus endometrioid carcinoma (UCEC). SLC1A5 (ASCT2) and PDK1 mRNA expression profiling data measured using the Illumina HiSeq 2000 RNA sequencing platform were downloaded and were classified on the basis of sample types (primary tumor vs. adjust normal tissue) or HNSCC tumor grade. Expression of each transcript was shown as log2(x+1) transformed normalized count.
Cell lines and cell culture.
The HN5 cell line was originally obtained from Helmout Modjtahedi (Kingston University London, London, United Kingdom). Other HNSCC cell lines (FaDu, HN30, MDA1986, UMSCC1, UMSCC2, UMSCC17A, UMSCC22B, OSC19, SqCC/Y1, and TU167) were originally obtained from Kian Ang and Luka Milas (MD Anderson Cancer Center). The cells were maintained in DMEM/F12 medium supplemented with 10% FBS, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin under conditions of 5% CO2 at 37°C in an incubator, unless otherwise specified. Generation and characterization of the HN5-R and FaDu-R cetuximab–resistant sublines were recently described (48). Immortalized nonmalignant NOM9-TK human head and neck epithelial cells were maintained in serum-free keratinocyte basal growth medium supplemented with components in the KGM SingleQuots kit, including bovine pituitary extract, recombinant human EGF, insulin, hydrocortisone, and gentamicin sulfate (Lonza) (46–48).
Reagents.
Cetuximab was obtained from ImClone Systems, an Eli Lilly company. Glutathione monoethyl ester (membrane-permeable GSH-MEE) was purchased from Santa Cruz Biotechnology Inc. All other chemicals, including DCA and NAC, were purchased from MilliporeSigma unless otherwise specified.
cDNA construct, siRNA duplexes, and transfection.
siRNA oligonucleotide duplexes for ASCT2 and PDK1 were purchased from MilliporeSigma. The DNA targeting sequences of siRNAs were as follows: ASCT2: sequence 1, 5′ - GTCAGCAGCCTTTCGCTCA - 3′; sequence 2, 5′ - CCAAGCACATCAGCCGTTT - 3′. PDK1: sequence 1, 5′ - GGATGAAATTGCACCTATT - 3′; sequence 2, 5′ - GTCCAGGAGACTGTGTCAT - 3′; sequence 3, 5′ - TGCTAGGCGTCTGTGTGAT - 3′. The siRNA oligonucleotides were transfected into the targeted cells with Lipofectamine 2000 according to the manufacturer’s instructions (Invitrogen).
Western blotting and immunoprecipitation.
Cultured cells were washed twice with cold PBS and harvested with a rubber scraper. Cell pellets were lysed and kept on ice for at least 10 minutes in a buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 0.5% IGEPAL CA-630, 50 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 25 μg/ml leupeptin, and 25 μg/ml aprotinin (72, 73). The lysates were cleared by centrifugation at 15,600 g for 30 minutes at 4°C, and the supernatants were collected. Lysate proteins were quantified using the Pierce Coomassie Plus colorimetric protein assay (Thermo Fisher Scientific). Whole-cell lysates were separated by SDS-PAGE and subjected to Western blotting analysis with various primary antibodies as follows: anti-PDH (catalog 3205, Cell Signaling Technology), anti-PARP (catalog 9542, Cell Signaling Technology), anti-ASCT2 (H-52) (catalog sc-99002, Santa Cruz Biotechnology Inc.), anti-PDK1 (catalog ADI-KAP-PK112-F, Enzo Life Sciences), anti–S293-phosphorylated PDH (catalog NB110-93479, Novus Biologicals), and anti–β-actin (catalog A2066, MilliporeSigma). The signals were visualized using the enhanced chemiluminescence detection kit (Amersham Biosciences) after incubation with HRP-labeled secondary antibodies.
Quantitative apoptosis ELISA.
A colorimetric Cell Death Detection ELISA kit (Roche Diagnostics) was used to detect induction of apoptosis by quantitatively measuring the level of cytoplasmic histone–associated DNA fragments (mononucleosomes and oligonucleosomes).
ROS detection.
A total ROS detection kit (Enzo Life Sciences) was used to detect intracellular ROS (24). After indicated treatment of cells in a 12-well plate at around 40%–60% confluency, the cells were washed with a buffer provided in the kit and were stained directly with ROS detection solution at 37°C for 1 hour and then observed under a fluorescence microscope or analyzed with an LSRFortessa cell analyzer (BD Biosciences) after trypsinization and resuspension in Eppendorf tubes.
Mitochondrial membrane potential assay.
The fluorescent dye TMRM (Invitrogen) was used to stain the cells for measuring mitochondrial membrane potential (24). Cells were seeded in a 12-well plate at around 60%–80% confluency. After indicated treatments, cells were stained with TMRM and counterstained with Hoechst 33258 at 37°C for 30 minutes and then observed under a fluorescence microscope equipped with appropriate filters for TMRM and Hoechst 33258. For quantitative analysis, cells were seeded in clear-bottom, opaque-wall 96-well plates at around 60%–80% confluency. Relative TMRM staining intensity was determined as the ratio of the reading at 590 nm (for TMRM) to the reading at 460 nm (for Hoechst 33258) using a fluorescence microplate reader.
LIVE/DEAD cell viability assay.
The fluorescence-based LIVE/DEAD cell viability assay kit (Invitrogen) was used to detect live or dead cells (73). Following treatments, the cells were incubated with 4 μM calcein acetoxymethyl ester (calcein AM) and 2 μM ethidium homodimer-1 (Eth-D) together in a 37°C, 5% CO2 incubator for 45 minutes. The cells were then rinsed gently with PBS and observed for cell viability under a fluorescence microscope. Live cells were identified by green fluorescence, and dead cells were identified by bright red fluorescence. For quantification of the ratio of live to dead cells, cells were seeded in a clear-bottom, opaque-wall 96-well plate. After indicated treatments, cells stained with calcein AM and Eth-D as described above and were analyzed with a fluorescence microplate reader with emission wavelength at 610 nm for Eth-D and at 520 nm for calcein AM. The relative values of live and dead cells in the treated groups were expressed as a percentage of the fluorescence reading of the corresponding group of untreated cells.
Cell culture with glutamine and cystine deprivation and supplementation.
For cell culture with glutamine and cystine deprivation, glutamine- and cystine-deficient DMEM/F12 medium was used. Cells were cultured in the deficient medium or in the deficient medium with freshly added 2.5 mM glutamine (i.e., cystine deprivation), 0.1 mM cystine (i.e., glutamine deprivation), or both (i.e., regular medium) for 24 hours.
For cell culture with glutamine and cystine supplementation (without or with 10 μM 2ME), excess amounts of glutamine and/or cystine were freshly added into the regular medium so that the final concentrations of glutamine and cystine were 2 and 4 times as high as their concentrations in regular medium (5 mM and 10 mM glutamine and 0.2 mM and 0.4 mM cystine).
Clonogenic survival assay.
Cells were seeded in triplicate into 6-cm dishes at densities varying from 250–1,000 cells/dish. The cells were cultured in a 37°C, 5% CO2 incubator for 2–3 weeks (13–22 days) depending on the proliferation rate of the cells. Surviving clones in the dishes were fixed and stained with a solution containing 0.2% crystal violet in 10% ethanol for 30 minutes, and individual colonies (>50 cells/colony) were counted (74, 75).
Animal studies and bioluminescence tumor imaging.
Male and female Swiss nude mice (4–6 weeks old, colony maintained by the Department of Experimental Radiation Oncology, MD Anderson Cancer Center) were used for s.c. inoculation of HN5, HN5-R, FaDu, FaDu-R, or UMSCC1 cells (1 × 107 cells/mouse in 100 μl of serum-free medium) on the right flanks. When xenograft volume reached 150–250 mm3, the mice were randomly divided into groups (6–7 mice in each group) with similar average tumor volume for starting treatments. DCA was added in drinking water at 62.5 mg/l and 312.5 mg/l, which converts to 50 mg/kg/day and 250 mg/kg/day based on our observation that each mouse consumes an average of 20 ml of water per day and the average body weight of the mice we used in our study was 25 g. Cetuximab was administered i.p. at 0.25 mg/mouse, twice a week, alone and in combination with DCA. All treatments were stopped on day 21 after initiation of treatment.
Tumor size was measured twice a week with calipers. Tumor volume was calculated using the formula π/6 × ab2 (a, length; b, width; a > b) and was plotted as a function of the days without or with indicated treatments. Mice were euthanized when their tumor size was greater than 1.5 cm in diameter or when the animals became morbid or moribund. When fewer than 4 mice remained in any group, all mice in the group were euthanized. Mice implanted with luciferase-positive FaDu, FaDu-R, and UMSCC1 cells were subjected to bioluminescent imaging after termination of various treatments on day 21. Bioluminescent imaging of xenografts was performed with the Xenogen in vitro imaging system (IVIS) in living animals after i.p. injection of D-luciferin (3.3 mg/100 μl) and induction of anesthesia by inhalation of 2.5% isoflurane (IsoSol; Vedco Inc.).
Statistics.
Differential expression between primary cancer tissues and their respective adjacent normal tissues was performed by using 2-tailed Student’s t test. For Kaplan-Meier survival analysis, a log-rank (Mantel-Cox) test was used to compare each of the arms. For all other assays, differences between 2 groups were analyzed by using 2-tailed Student’s t test, whereas differences between multiple groups were analyzed by using 1-way or 2-way ANOVA with Bonferroni post hoc test. A P value less than 0.05 was considered significant for all analyses.
Study approval.
All animal studies were approved by the IACUC of MD Anderson Cancer Center.
Author contributions
HL and ZF conceptualized the hypothesis and designed the study. HL performed most of the experiments. YL, SQ, and XL helped to develop the methods and performed some experiments. YX helped with TCGA data analysis and data plotting. ZF supervised the study. HL and ZF analyzed all the results and wrote the manuscript. All authors reviewed and commented on the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by a US NIH R01 award (CA179015) and an NIH R21 award (DE021883) to ZF. The work was also supported in part by the NIH through MD Anderson’s Cancer Center Support Grant, CA016672. We thank Stephanie Deming in Scientific Publications, Research Medical Library, at MD Anderson Cancer Center for editing this manuscript. This work is dedicated to the memory of John Mendelsohn, a pioneer in targeted cancer therapy whose groundbreaking work led to the development of cetuximab.
Version 1. 10/03/2019
Electronic publication
Version 2. 10/04/2019
A missing comma was added to the Results section.
Version 3. 10/04/2019
The word median was changed to mean in the Results section.
Footnotes
HL’s present address is: Institute for Cell Engineering, The Johns Hopkins University School of Medicine, Baltimore, Maryland, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(19):e131106.https://doi.org/10.1172/jci.insight.131106.
Contributor Information
Haiquan Lu, Email: lvhaiquan@gmail.com.
Yang Lu, Email: ylufan@yahoo.com.
Yangyiran Xie, Email: yxie28@jhu.edu.
Songbo Qiu, Email: sqiu@mdanderson.org.
Xinqun Li, Email: xinquli@mdanderson.org.
Zhen Fan, Email: zfan@mdanderson.org.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Ferris RL, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375(19):1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seiwert TY, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956–965. doi: 10.1016/S1470-2045(16)30066-3. [DOI] [PubMed] [Google Scholar]
- 4.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371(9625):1695–1709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malone E, Siu LL. Precision Medicine in Head and Neck Cancer: Myth or Reality? Clin Med Insights Oncol. 2018;12:1179554918779581. doi: 10.1177/1179554918779581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Specenier P, Vermorken JB. Cetuximab: its unique place in head and neck cancer treatment. Biologics. 2013;7:77–90. doi: 10.2147/BTT.S43628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho C. Cetuximab in locally advanced head-and-neck cancer: defining the population. Curr Oncol. 2010;17(4):48–51. doi: 10.3747/co.v17i4.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkins G, O’Byrne KJ, Panizza B, Richard DJ. Genome stability pathways in head and neck cancers. Int J Genomics. 2013;2013:464720. doi: 10.1155/2013/464720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riaz N, Morris LG, Lee W, Chan TA. Unraveling the molecular genetics of head and neck cancer through genome-wide approaches. Genes Dis. 2014;1(1):75–86. doi: 10.1016/j.gendis.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung CH, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5(5):489–500. doi: 10.1016/S1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 11.Lui VW, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3(7):761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozawa H, et al. SMAD4 Loss Is Associated with Cetuximab Resistance and Induction of MAPK/JNK Activation in Head and Neck Cancer Cells. Clin Cancer Res. 2017;23(17):5162–5175. doi: 10.1158/1078-0432.CCR-16-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollock NI, et al. Increased Expression of HER2, HER3, and HER2:HER3 Heterodimers in HPV-Positive HNSCC Using a Novel Proximity-Based Assay: Implications for Targeted Therapies. Clin Cancer Res. 2015;21(20):4597–4606. doi: 10.1158/1078-0432.CCR-14-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pollock NI, Grandis JR. HER2 as a therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res. 2015;21(3):526–533. doi: 10.1158/1078-0432.CCR-14-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vermorken JB, et al. Open-label, uncontrolled, multicenter phase II study to evaluate the efficacy and toxicity of cetuximab as a single agent in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck who failed to respond to platinum-based therapy. J Clin Oncol. 2007;25(16):2171–2177. doi: 10.1200/JCO.2006.06.7447. [DOI] [PubMed] [Google Scholar]
- 16.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134(5):703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329(5998):1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W, Zhang SL, Hu X, Tam KY. Targeting Tumor Metabolism for Cancer Treatment: Is Pyruvate Dehydrogenase Kinases (PDKs) a Viable Anticancer Target? Int J Biol Sci. 2015;11(12):1390–1400. doi: 10.7150/ijbs.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuy F, et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015;22(4):577–589. doi: 10.1016/j.cmet.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Wigfield SM, Winter SC, Giatromanolaki A, Taylor J, Koukourakis ML, Harris AL. PDK-1 regulates lactate production in hypoxia and is associated with poor prognosis in head and neck squamous cancer. Br J Cancer. 2008;98(12):1975–1984. doi: 10.1038/sj.bjc.6604356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hitosugi T, et al. Tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase 1 is important for cancer metabolism. Mol Cell. 2011;44(6):864–877. doi: 10.1016/j.molcel.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathania D, Millard M, Neamati N. Opportunities in discovery and delivery of anticancer drugs targeting mitochondria and cancer cell metabolism. Adv Drug Deliv Rev. 2009;61(14):1250–1275. doi: 10.1016/j.addr.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Roche TE, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci. 2007;64(7-8):830–849. doi: 10.1007/s00018-007-6380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnet S, et al. A mitochondria-K+ channel axis is suppressed in cancer and its normalization promotes apoptosis and inhibits cancer growth. Cancer Cell. 2007;11(1):37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 25.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8(7):579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 26.Lu SC. Glutathione synthesis. Biochim Biophys Acta. 2013;1830(5):3143–3153. doi: 10.1016/j.bbagen.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch. 2004;447(5):469–479. doi: 10.1007/s00424-003-1146-4. [DOI] [PubMed] [Google Scholar]
- 28.Lo M, Wang YZ, Gout PW. The x(c)- cystine/glutamate antiporter: a potential target for therapy of cancer and other diseases. J Cell Physiol. 2008;215(3):593–602. doi: 10.1002/jcp.21366. [DOI] [PubMed] [Google Scholar]
- 29.Liu R, et al. Cystine-glutamate transporter SLC7A11 mediates resistance to geldanamycin but not to 17-(allylamino)-17-demethoxygeldanamycin. Mol Pharmacol. 2007;72(6):1637–1646. doi: 10.1124/mol.107.039644. [DOI] [PubMed] [Google Scholar]
- 30.Okuno S, et al. Role of cystine transport in intracellular glutathione level and cisplatin resistance in human ovarian cancer cell lines. Br J Cancer. 2003;88(6):951–956. doi: 10.1038/sj.bjc.6600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGivan JD, Bungard CI. The transport of glutamine into mammalian cells. Front Biosci. 2007;12:874–882. doi: 10.2741/2109. [DOI] [PubMed] [Google Scholar]
- 32.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29(3):313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang CV. Glutaminolysis: supplying carbon or nitrogen or both for cancer cells? Cell Cycle. 2010;9(19):3884–3886. doi: 10.4161/cc.9.19.13302. [DOI] [PubMed] [Google Scholar]
- 34.Erickson JW, Cerione RA. Glutaminase: a hot spot for regulation of cancer cell metabolism? Oncotarget. 2010;1(8):734–740. doi: 10.18632/oncotarget.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu W, Pelicano H, Huang P. Cancer metabolism: is glutamine sweeter than glucose? Cancer Cell. 2010;18(3):199–200. doi: 10.1016/j.ccr.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajagopalan KN, DeBerardinis RJ. Role of glutamine in cancer: therapeutic and imaging implications. J Nucl Med. 2011;52(7):1005–1008. doi: 10.2967/jnumed.110.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10(9):671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 39.Jones NP, Schulze A. Targeting cancer metabolism--aiming at a tumour’s sweet-spot. Drug Discov Today. 2012;17(5-6):232–241. doi: 10.1016/j.drudis.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 40.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu H, Li X, Lu Y, Qiu S, Fan Z. ASCT2 (SLC1A5) is an EGFR-associated protein that can be co-targeted by cetuximab to sensitize cancer cells to ROS-induced apoptosis. Cancer Lett. 2016;381(1):23–30. doi: 10.1016/j.canlet.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tao X, Lu Y, Qiu S, Wang Y, Qin J, Fan Z. AP1G1 is involved in cetuximab-mediated downregulation of ASCT2-EGFR complex and sensitization of human head and neck squamous cell carcinoma cells to ROS-induced apoptosis. Cancer Lett. 2017;408:33–42. doi: 10.1016/j.canlet.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roland NJ, Caslin AW, Nash J, Stell PM. Value of grading squamous cell carcinoma of the head and neck. Head Neck. 1992;14(3):224–229. doi: 10.1002/hed.2880140310. [DOI] [PubMed] [Google Scholar]
- 44.Modjtahedi H, Affleck K, Stubberfield C, Dean C. EGFR blockade by tyrosine kinase inhibitor or monoclonal antibody inhibits growth, directs terminal differentiation and induces apoptosis in the human squamous cell carcinoma HN5. Int J Oncol. 1998;13(2):335–342. doi: 10.3892/ijo.13.2.335. [DOI] [PubMed] [Google Scholar]
- 45.Modjtahedi H, et al. Targeting of cells expressing wild-type EGFR and type-III mutant EGFR (EGFRvIII) by anti-EGFR MAb ICR62: a two-pronged attack for tumour therapy. Int J Cancer. 2003;105(2):273–280. doi: 10.1002/ijc.11055. [DOI] [PubMed] [Google Scholar]
- 46.Chakravarti N, et al. Differential inhibition of protein translation machinery by curcumin in normal, immortalized, and malignant oral epithelial cells. Cancer Prev Res (Phila) 2010;3(3):331–338. doi: 10.1158/1940-6207.CAPR-09-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao M, et al. Assembly and initial characterization of a panel of 85 genomically validated cell lines from diverse head and neck tumor sites. Clin Cancer Res. 2011;17(23):7248–7264. doi: 10.1158/1078-0432.CCR-11-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu H, Li X, Luo Z, Liu J, Fan Z. Cetuximab reverses the Warburg effect by inhibiting HIF-1-regulated LDH-A. Mol Cancer Ther. 2013;12(10):2187–2199. doi: 10.1158/1535-7163.MCT-12-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stacpoole PW, et al. Controlled clinical trial of dichloroacetate for treatment of congenital lactic acidosis in children. Pediatrics. 2006;117(5):1519–1531. doi: 10.1542/peds.2005-1226. [DOI] [PubMed] [Google Scholar]
- 50.Luo J, et al. Acetyl-CoA carboxylase rewires cancer metabolism to allow cancer cells to survive inhibition of the Warburg effect by cetuximab. Cancer Lett. 2017;384:39–49. doi: 10.1016/j.canlet.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stacpoole PW. The pharmacology of dichloroacetate. Metab Clin Exp. 1989;38(11):1124–1144. doi: 10.1016/0026-0495(89)90051-6. [DOI] [PubMed] [Google Scholar]
- 52.Stacpoole PW, Nagaraja NV, Hutson AD. Efficacy of dichloroacetate as a lactate-lowering drug. J Clin Pharmacol. 2003;43(7):683–691. doi: 10.1177/0091270003254637. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y, Zhao T, Li Z, Wang L, Yuan S, Sun L. The role of ASCT2 in cancer: A review. Eur J Pharmacol. 2018;837:81–87. doi: 10.1016/j.ejphar.2018.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Bröer A, Fairweather S, Bröer S. Disruption of Amino Acid Homeostasis by Novel ASCT2 Inhibitors Involves Multiple Targets. Front Pharmacol. 2018;9:785. doi: 10.3389/fphar.2018.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiu M, et al. GPNA inhibits the sodium-independent transport system L for neutral amino acids. Amino Acids. 2017;49(8):1365–1372. doi: 10.1007/s00726-017-2436-z. [DOI] [PubMed] [Google Scholar]
- 56.Cormerais Y, et al. The glutamine transporter ASCT2 (SLC1A5) promotes tumor growth independently of the amino acid transporter LAT1 (SLC7A5) J Biol Chem. 2018;293(8):2877–2887. doi: 10.1074/jbc.RA117.001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bröer A, Rahimi F, Bröer S. Deletion of Amino Acid Transporter ASCT2 (SLC1A5) Reveals an Essential Role for Transporters SNAT1 (SLC38A1) and SNAT2 (SLC38A2) to Sustain Glutaminolysis in Cancer Cells. J Biol Chem. 2016;291(25):13194–13205. doi: 10.1074/jbc.M115.700534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br J Cancer. 2008;99(7):989–994. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michelakis ED, et al. Metabolic modulation of glioblastoma with dichloroacetate. Sci Transl Med. 2010;2(31):31ra34. doi: 10.1126/scitranslmed.3000677. [DOI] [PubMed] [Google Scholar]
- 60.Madhok BM, Yeluri S, Perry SL, Hughes TA, Jayne DG. Dichloroacetate induces apoptosis and cell-cycle arrest in colorectal cancer cells. Br J Cancer. 2010;102(12):1746–1752. doi: 10.1038/sj.bjc.6605701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun RC, Fadia M, Dahlstrom JE, Parish CR, Board PG, Blackburn AC. Reversal of the glycolytic phenotype by dichloroacetate inhibits metastatic breast cancer cell growth in vitro and in vivo. Breast Cancer Res Treat. 2010;120(1):253–260. doi: 10.1007/s10549-009-0435-9. [DOI] [PubMed] [Google Scholar]
- 62.Subramanian K, Ramaian AS. Development of a less toxic dichloroacetate analogue by docking and descriptor analysis. Bioinformation. 2010;5(2):73–76. doi: 10.6026/97320630005073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dunbar EM, et al. Phase 1 trial of dichloroacetate (DCA) in adults with recurrent malignant brain tumors. Invest New Drugs. 2014;32(3):452–464. doi: 10.1007/s10637-013-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garon EB, et al. Dichloroacetate should be considered with platinum-based chemotherapy in hypoxic tumors rather than as a single agent in advanced non-small cell lung cancer. J Cancer Res Clin Oncol. 2014;140(3):443–452. doi: 10.1007/s00432-014-1583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sutendra G, Michelakis ED. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front Oncol. 2013;3:38. doi: 10.3389/fonc.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strum SB, Adalsteinsson O, Black RR, Segal D, Peress NL, Waldenfels J. Case report: Sodium dichloroacetate (DCA) inhibition of the “Warburg Effect” in a human cancer patient: complete response in non-Hodgkin’s lymphoma after disease progression with rituximab-CHOP. J Bioenerg Biomembr. 2013;45(3):307–315. doi: 10.1007/s10863-012-9496-2. [DOI] [PubMed] [Google Scholar]
- 67.Foerster S, et al. Characterization of the EGFR interactome reveals associated protein complex networks and intracellular receptor dynamics. Proteomics. 2013;13(21):3131–3144. doi: 10.1002/pmic.201300154. [DOI] [PubMed] [Google Scholar]
- 68.Tong J, Taylor P, Moran MF. Proteomic analysis of the epidermal growth factor receptor (EGFR) interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Mol Cell Proteomics. 2014;13(7):1644–1658. doi: 10.1074/mcp.M114.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandulache VC, Myers JN. Altered metabolism in head and neck squamous cell carcinoma: an opportunity for identification of novel biomarkers and drug targets. Head Neck. 2012;34(2):282–290. doi: 10.1002/hed.21664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kamarajan P, Rajendiran TM, Kinchen J, Bermúdez M, Danciu T, Kapila YL. Head and Neck Squamous Cell Carcinoma Metabolism Draws on Glutaminolysis, and Stemness Is Specifically Regulated by Glutaminolysis via Aldehyde Dehydrogenase. J Proteome Res. 2017;16(3):1315–1326. doi: 10.1021/acs.jproteome.6b00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X, et al. Modulators of Redox Metabolism in Head and Neck Cancer. Antioxid Redox Signal. 2018;29(16):1660–1690. doi: 10.1089/ars.2017.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X, Lu Y, Liang K, Pan T, Mendelsohn J, Fan Z. Requirement of hypoxia-inducible factor-1alpha down-regulation in mediating the antitumor activity of the anti-epidermal growth factor receptor monoclonal antibody cetuximab. Mol Cancer Ther. 2008;7(5):1207–1217. doi: 10.1158/1535-7163.MCT-07-2187. [DOI] [PubMed] [Google Scholar]
- 73.Li X, Lu Y, Lu H, Luo J, Hong Y, Fan Z. AMPK-mediated energy homeostasis and associated metabolic effects on cancer cell response and resistance to cetuximab. Oncotarget. 2015;6(13):11507–11518. doi: 10.18632/oncotarget.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 75.Lu H, Liang K, Lu Y, Fan Z. The anti-EGFR antibody cetuximab sensitizes human head and neck squamous cell carcinoma cells to radiation in part through inhibiting radiation-induced upregulation of HIF-1α. Cancer Lett. 2012;322(1):78–85. doi: 10.1016/j.canlet.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.