Abstract
Occurrence of Candida nivariensis and Candida bracarensis, two species phenotypically similar to Candida glabrata sensu stricto, in human clinical samples from different geographical settings remains unknown. This study developed a low-cost multiplex PCR (mPCR) and three species-specific singleplex PCR assays. Reference strains of common Candida species were used during development and the performance of mPCR and singleplex PCR assays was evaluated with 440 clinical C. glabrata sensu lato isolates. The internal transcribed spacer (ITS) region of rDNA was also sequenced from 85 selected isolates and rDNA sequence variations were used for determining genetic relatedness among the isolates by using MEGA X software. Species-specific amplicons for C. glabrata (~360 bp), C. nivariensis (~250 bp) and C. bracarensis (~180 bp) were obtained in mPCR while no amplicon was obtained from other Candida species. The three singleplex PCR assays also yielded expected results with reference strains of Candida species. The mPCR amplified ~360 bp amplicon from all 440 C. glabrata sensu lato isolates thus identifying all clinical isolates in Kuwait as C. glabrata sensu stricto. The results of mPCR were confirmed for all 440 isolates as they yielded an amplicon only in C. glabrata sensu stricto-specific singleplex PCR assay. The rDNA sequence data identified 28 ITS haplotypes among 85 isolates with 18 isolates belonging to unique haplotypes and 67 isolates belonging to 10 cluster haplotypes. In conclusion, we have developed a simple, low-cost mPCR assay for rapid differentiation of C. glabrata sensu stricto from C. nivariensis and C. bracarensis. Our data obtained from a large collection of clinical C. glabrata sensu lato isolates show that C. nivariensis and C. bracarensis are rare pathogens in Kuwait. Considerable genetic diversity among C. glabrata sensu stricto isolates was also indicated by rDNA sequence analyses.
Introduction
Candida spp. are identified as the fourth most frequent cause of bloodstream infections in hospitalized patients and third common cause of central-line associated bloodstream infections in patients admitted to intensive care units (ICUs) [1, 2]. Invasive candidiasis is associated with mortality rates of ~50% [1, 2]. Nearly 90% of Candida infections are caused by only four species/species complexes comprising Candida albicans, Candida glabrata, Candida parapsilosis and Candida tropicalis [3]. Although C. albicans is the most commonly isolated species, >50% of all Candida infections are now caused by non-albicans species of Candida [2, 4–6]. Candida glabrata sensu stricto has emerged as the second or third most frequently isolated Candida species from patients with bloodstream, vulvovaginal and oral infections [2, 6–8].
The emergence of C. glabrata as a major cause of invasive fungal infections is worrisome due to high mortality rates associated with these infections, particularly in immunocompromised elderly patients requiring major surgery and neutropenic patients [2, 9]. The higher mortality rates have been attributed to the intrinsic and/or rapidly acquired resistance of C. glabrata to extended-spectrum triazoles, particularly fluconazole, as a consequence of widespread use of these relatively safer antifungal drugs and the haploid nature of this organism [9–12]. Although echinocandins are now preferred as first-line agents for treatment of invasive Candida infections [13], resistance to echinocandins in Candida spp. has also appeared in recent years and breakthrough invasive C. glabrata infections have been reported in patients on micafungin therapy [6, 14–19]. Resistance to polyenes has also been described in clinical C. glabrata isolates [20–24]. A multidrug-resistant C. glabrata phenotype (resistant to azoles and echinocandins) occurring in ICU and non-ICU settings has also been noted in recent years [25, 26]. Two other closely related species: Candida nivariensis and Candida bracarensis, with similar niches in humans, share many phenotypic characteristics with C. glabrata sensu stricto. Candida nivariensis and C. bracarensis are usually misidentified as C. glabrata based on phenotypic identification methods alone and are generally less susceptible to azoles and amphotericin B [9, 27–30]. Accurate species-specific identification of all clinical C. glabrata sensu lato isolates is thus warranted.
The three species belonging to C. glabrata complex can be accurately identified only by molecular methods including matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) [9, 30, 31]. The occurrence of C. nivariensis and C. bracarensis among phenotypically identified clinical C. glabrata sensu lato isolates from different geographical settings remains unknown. This study developed a simple, low-cost multiplex PCR (mPCR) assay for rapid detection and differentiation of C. nivariensis and C. bracarensis from C. glabrata sensu stricto isolates. Furthermore, three species-specific singleplex PCR assays were also developed and the protocols were tested on a large collection of Candida glabrata sensu lato isolates obtained from various clinical specimens in Kuwait. The internal transcribed spacer (ITS) region of rDNA was also sequenced and rDNA sequence variations were used for determining genetic relatedness among selected isolates.
Materials and methods
Reference strains and clinical isolates
Reference strains or well characterized clinical isolates of C. glabrata sensu stricto (ATCC 90030 and CBS 138), C. nivariensis (CBS 9983), C. bracarensis (CBS 10154), C. albicans (ATCC 76615), C. parapsilosis (ATCC 22019), C. tropicalis (ATCC 750) and C. dubliniensis (CBS 7987), C. lusitaniae (CBS 4413), C. guilliermondii (CBS 6021), C. kefyr (ATCC 28838), C. famata (CBS 796), Saccharomyces cerevisiae (Kw1336/19) and Trichosporon asahii (Kw1728/19) were used as reference Candida or other yeast species. The clinical isolates used in this study were obtained from different specimens, including blood samples, from patients after obtaining verbal consent at various hospitals across Kuwait. The isolation and identification of fungal pathogens was performed as part of routine patient care and diagnostic work-up. The results obtained from the clinical isolates used in this study are presented on deidentified samples. The clinical isolates were initially identified at the hospital microbiology laboratory as C. glabrata sensu lato by the Vitek2 yeast identification system (bioMerieux, Marcy-lEtoile, France). The cultured isolates were sent to Mycology Reference Laboratory (MRL), Department of Microbiology, Faculty of Medicine, Kuwait University for further identification and antifungal susceptibility testing. A total of 440 C. glabrata sensu lato isolates, collected during 2006 to 2015, were randomly selected from the stock cultures maintained at MRL for this study. The clinical specimens yielding 440 C. glabrata sensu lato isolates used in this study are listed in S1 Table.
Phenotypic identification
All 440 C. glabrata sensu lato isolates, initially identified by Vitek2 ID-YST system (bioMerieux, Marcy-lEtoile, France) were grown on CHROMagar Candida (Becton Dickinson, Bootle, UK) for phenotypic identification and the results were interpreted according to manufacturer’s instructions. The typical pink color of C. glabrata sensu stricto and white color of C. nivariensis and C. bracarensis [30] was used for species-specific identification of these isolates.
Template DNA preparation and molecular identification
The genomic DNA from reference strains and clinical C. glabrata sensu lato isolates was extracted by using Gentra Puregene Yeast DNA extraction kit (Qiagen, Hilden Germany) according to kit instructions or by the rapid boiling method using Chelex-100 as described previously [32].
A simple, low-cost mPCR assay was developed for rapid molecular identification of C. glabrata sensu stricto, C. nivariensis and C. bracarensis isolates. For this purpose, three different forward primers targeting specific sequences within ITS-1 region of rDNA of the three species and one common reverse primer targeting 5.8S rRNA gene were synthesized (S2 Table). The primer sequences were selected based on multiple sequence alignment of ITS region sequences of multiple strains of all commonly encountered Candida species that are available from the GenBank. The mPCR should yield amplicons of ~360 bp, ~250 bp and ~180 bp (due to slight variations in the length of ITS region of rDNA) from C. glabrata sensu stricto, C. nivariensis and C. bracarensis respectively, while no amplicon is expected from other Candida or other yeast species. The species specificity of the combination of forward primers mCGLF, mCNIF and mCBRF together with a common reverse primer (mCGCR) for C. glabrata sensu stricto, C. nivariensis and C. bracarensis, respectively, was also indicated by BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi?). The mPCR amplification was performed in a final volume of 50 μl containing 1x AmpliTaq DNA polymerase buffer I and 1 unit of AmpliTaq DNA polymerase (Applied Biosystems, Brachburg, NJ, USA), 10 pmol of mCGLF, mCNIF, mCBRF and mCGCR primers, 2 μl of template DNA and 100 μM of each dNTP. Cycling conditions included an initial denaturation at 95°C for 5 min followed by 30 cycles of 95°C for 1 min, 52°C for 30 s and 72°C for 1 min and a final extension at 72°C for 10 min. PCR products (20 μl) were run on 2% (w/v) agarose gels, as described previously [33].
Primer design for three species-specific singleplex PCR assays
The results of mPCR were confirmed for all isolates by developing three species-specific singlelex PCR assays. For this purpose, three pairs of forward and reverse primers, each pair specific for C. glabrata sensu stricto, C. nivariensis or C. bracarensis were designed (S2 Table). Again, primer sequences were selected based on multiple sequence alignment of ITS region sequences of multiple strains of common Candida species available from the GenBank. Primers CGLF + CGLR, CNIF + CNIR and CBRF + CBRR should yield amplicons of ~360 bp, ~288 bp and ~299 bp (due to slight variations in the length of ITS region of rDNA) from C. glabrata sensu stricto, C. nivariensis and C. bracarensis, respectively, while no amplicon is expected from other Candida or other yeast species. The species specificity of CGLF + CGLR, CNIF + CNIR and CBRF + CBRR primer pairs for C. glabrata sensu stricto, C. nivariensis and C. bracarensis, respectively, was also indicated by BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi?). PCR amplification was performed and the amplicons were detected as described above except that CGLF + CGLR or CNIF + CNIR or CBRF + CBRR primer pairs were used.
PCR-sequencing of ITS region of rDNA and identification of major ITS haplotypes
The ITS region of rDNA was amplified by using panfungal ITS1 and ITS4 primers and the amplicons were sequenced by using internal sequencing (ITS1FS, ITS2, ITS3 or ITS4RS) primers, as described previously [34, 35]. The ITS region sequence for each isolate was assembled and used for BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi?). Sequence identity >99% with the corresponding sequence from reference strains of Candida species was used for species identification. The genotypic relationship among the isolates was studied by comparing ITS region sequences. Pairwise comparisons and multiple sequence alignments were performed with CLUSTAL W2. Phylogenetic tree was constructed with MEGA software version X using the neighbor-joining method with pair-wise deletion of gaps option and the maximum composite likelihood model and the robustness of tree branches was assessed by bootstrap analysis with 1,000 replicates, as described previously [36].
Results
Phenotypic identification of C. glabrata complex isolates
The reference strains of C. glabrata sensu stricto (ATCC 90030), C. nivariensis (CBS 9983) and C. bracarensis (CBS 10154) yielded purple, white and white colored colonies on CHROMagar Candida, respectively, as expected. However, when 440 clinical isolates were streaked on CHROMagar candida medium, only 42 isolates produced typical purple color colonies, while 321 isolates produced light purple colonies with white tinge and the remaining 77 isolates formed completely white colonies with no noticeable purple color.
Establishment of a multiplex PCR (mPCR) assay and analysis of clinical C. glabrata sensu lato isolates
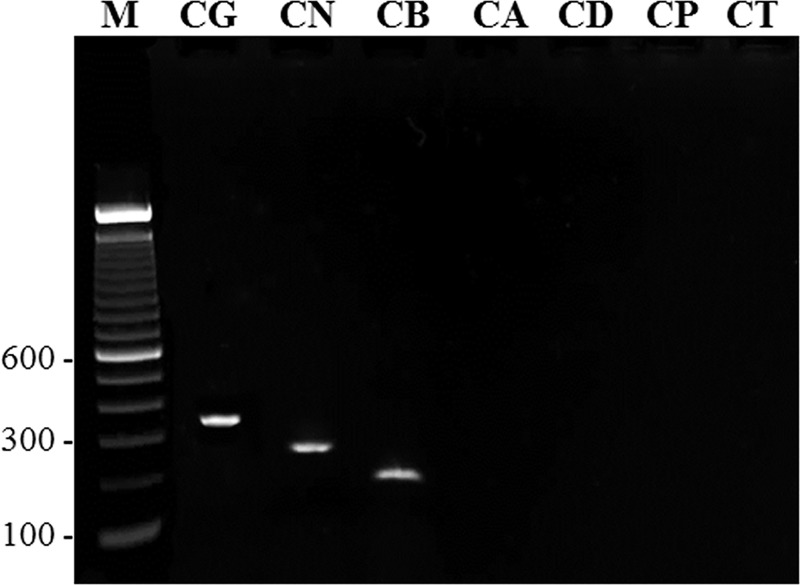
The mPCR amplification performed with mCGLF + mCNIF + mCBRF + mCGCR primers yielded an expected size amplicon of nearly 360 bp, 250 bp and 180 bp with genomic DNA from the reference strains of C. glabrata sensu stricto (ATCC 90030), C. nivariensis (CBS 9983) and C. bracarensis (CBS 10154), respectively (Fig 1). No amplification was obtained in mPCR with DNA from reference strains of C. albicans (ATCC 76615), C. dubliniensis (CBS 7987), C. parapsilosis (ATCC 22019) or C. tropicalis (ATCC 750) (Fig 1). No amplification was also obtained in mPCR with DNA prepared from Candida lusitaniae (CBS 4413), Candida guilliermondii (CBS 6021), Candida kefyr (ATCC 28838), Candida famata (CBS 796), Saccharomyces cerevisiae (Kw1336/19), Trichosporon asahii (Kw1728/19) or from human cells, as expected. Ten-fold serial dilutions were made and the minimum detection limit for a positive test in the mPCR assay was found to be 960 pg, 500 pg and 512 pg of genomic DNA from C. glabrata, C. nivariensis and C. bracarensis, respectively. When DNA prepared from clinical C. glabrata sensu lato isolates was used, all 440 isolates yielded an amplicon of ~360 bp only which is characteristic of C. glabrata sensu stricto strains. None of the clinical isolates yielded an amplicon of 250 bp (characteristic of C. nivariensis) or 180 bp (characteristic of C. bracarensis). Thus mPCR data showed lack of detection of C. nivariensis and C. bracarensis among 440 clinical C. glabrata sensu lato isolates in Kuwait.
Fig 1. An agarose gel of amplified products obtained in mPCR using C. glabrata sensu stricto-specific (mCGLF), C. nivariensis-specific (mCNIF) and C. bracarensis-specific (mCBRF) forward and C. glabrata complex-specific (mCGCR) reverse primer with genomic DNA from reference strain of C. glabrata sensu stricto ATCC 90030 (lane CG), C. nivariensis CBS 9983 (lane CN), C. bracarensis CBS 10154 (lane CB), C. albicans ATCC 76615 (lane CA), C. dubliniensis CBS 7987 (lane CD), C. parapsilosis ATCC 22019 (lane CP) and C. tropicalis ATCC750 (lane CT).
Lane M is 100 bp DNA ladder and the position of migration of 100 bp, 300 bp and 600 bp fragments are marked.
Establishment of three species-specific singleplex PCR assays and analysis of clinical C. glabrata sensu lato isolates
The three singleplex PCR assays (S1 Fig, panels A-C) correctly identified the three target species and did not show any cross-reaction with other Candida/yeast species, as expected. When DNA isolated from 440 clinical C. glabrata sensu lato isolates was tested, an amplicon of ~360 bp was obtained in C. glabrata sensu stricto-specific singleplex PCR assay only and thus confirmed mPCR data.
ITS haplotypes among C. glabrata isolates
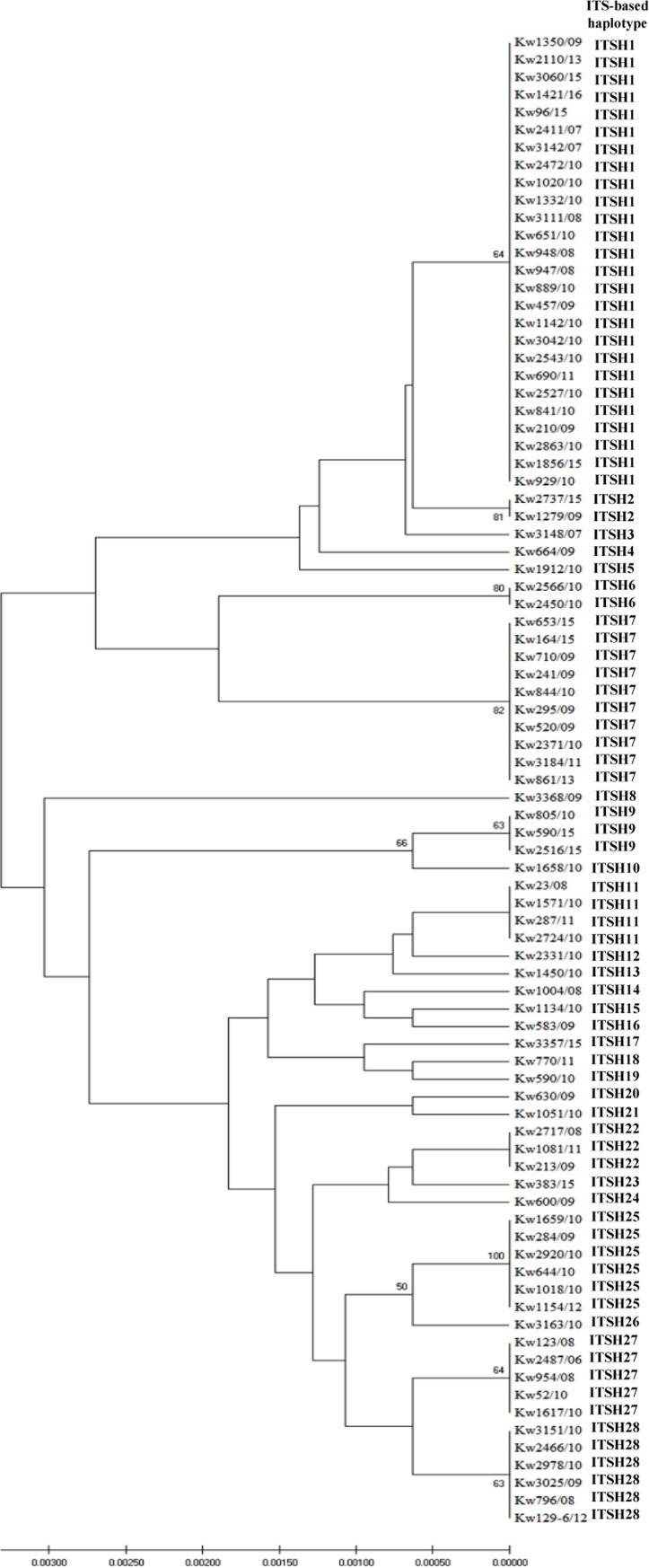
DNA sequencing data for the ITS region of rDNA from 85 randomly selected clinical C. glabrata isolates from Kuwait showed >99% similarity with sequences from reference C. glabrata sensu stricto strains ATCC 90030 and CBS 138 and were distinct from sequences of reference C. nivariensis and C. bracarensis strains. Multiple sequence alignment was carried out to compare the ITS region sequences. Based on ITS region sequence data, 28 distinct haplotypes (arbitrarily assigned as ITSH1 to ITSH28) were identified among 85 clinical C. glabrata isolates from Kuwait The discriminatory power for the ITS-based fingerprinting method was 0.88. Eighteen of 85 (21%) isolates belonged to unique ITS haplotypes while 67 isolates belonged to 10 cluster haplotypes with each cluster containing 2–26 isolates (Fig 2). The most common haplotype was ITSH1 shared among 26 (31%) isolates followed by ITSH7 among 10 (12%) isolates and ITSH25 and ITSH28 shared among six (7%) isolates each (Fig 2). The ITS-based fingerprinting data thus showed considerable genotypic heterogeneity among clinical C. glabrata sensu stricto isolates in Kuwait. There was no association of specific/predominant ITS haplotypes with the specimen types of C. glabrata isolates. The ITS region sequence data have been submitted to European Molecular Biology Laboratory (EMBL) database under accession numbers LS398112 to LS398120, LS398122, LS398123, LT837719 to LT837724 and LT837726 to LT837744.
Fig 2. Neighbor-joining tree based on DNA sequence data for the ITS region of rDNA from 85 C. glabrata sensu stricto isolates.
The bootstrap frequencies (>50%) on the node branches are shown. The ITS haplotype (ITSH) arbitrarily assigned for each isolate is also shown in the right hand column.
Discussion
Candida nivariensis and C. bracarensis generally exhibit lower susceptibility to triazoles and amphotericin B than C. glabrata sensu stricto isolates warranting species-specific identification of all clinical C. glabrata sensu lato isolates [9, 27–30, 37]. All 440 C. glabrata isolates used in this study were first tested on CHROMagar Candida. Although reference strains of C. glabrata sensu stricto (purple), C. nivariensis (creamy white) and C. bracarensis (creamy white) produced expected results as described previously [27], production of creamy white colonies on CHROMagar Candida is not restricted for C. nivariensis and C. bracarensis strains only as Candida norvegensis, C. inconspicua and some strains of C. glabrata also produce creamy white colonies on this medium [30, 38]. Similar to the results reported by Bishop et al. [38], only 363 of 440 (82.5%) isolates produced completely or partially purple colonies while 77 (17.5%) isolates produced creamy white colonies. The occurrence of creamy white colonies was not restricted to any specific specimen type. Lockhart et al [30] also reported that 11 of 14 isolates producing creamy white colonies in their study were subsequently identified as C. glabrata sensu stricto strains by two molecular tests. Thus creamy white colonies on CHROMagar Candida is not diagnostic for the identification of C. nivariensis and C. bracarensis strains.
In this study, we have also developed a simple, low-cost mPCR assay for accurate identification of C. glabrata sensu stricto, C. nivariensis and C. bracarensis isolates in a single PCR assay. The accuracy of mPCR assay was tested by using reference strains of several Candida/other yeast species. All 440 clinical C. glabrata sensu lato isolates analyzed in this study were identified as C. glabrata sensu stricto by mPCR. The mPCR-based identification of all isolates was also confirmed by a C. glabrata sensu stricto-specific singleplex PCR assay. The mPCR assay developed here can be completed within 4 hours using basic PCR and gel electrophoresis equipment that are readily available in clinical microbiology laboratories and will only cost ~2 US$ per sample (using Chelex-100-based boiling method for DNA extraction and excluding the cost of culture and personnel time).
Several molecular methods have been described previously for species-specific identification of C. glabrata sensu stricto, C. nivariensis and/or C. bracarensis isolates [39–44]. Of these, the mPCR assays described by Romeo et al. [41] and Arastehfar et al. [43] are very similar to our assay except that longer amplicons were obtained in their study. The PCR protocols yielding smaller amplicons are generally preferred [45, 46]. The larger amplicon size (~450 bp) for C. glabrata in SeptiFast real-time PCR assays yielded false-nagative results from candidemia patients in some studies even when the blood cultures yielded C. glabrata [47–49]. Singleplex PCR assays have also been described for the detection of C. glabrata sensu stricto and C. nivariensis [39, 40], however, they (including those developed in this study) are time consuming as they require three separate PCR reactions for each isolate. Although the singleplex PCR assay described by Enache-Angoulvant et al., [42] can potentially detect all the three species in a single test, the primers are not specific as amplicons were also obtained from S. cerevisiae and few other close relatives [42]. Additionally, poor resolution in agarose gels due to small (~100 bp) differences among larger (~1000 bp) amplicons and variations in the length of intronic sequences used as target among the isolates may lead to misidentification in some cases. The PNA-FISH is labor intensive and time consuming and species-specific probes have to be tested for all phenotypically identified C. glabrata sensu lato isolates [44]. Although rapid identification of C. nivariensis and C. bracarensis is possible with MALDI-TOF MS analysis, the requirement for fresh cultures often necessitates sub-culturing for obtaining interpretable results and these two species are included in the database of only the Bruker Daltonics Biotyper system but not in the database of the Biomerieux Vitek MS system [31, 50, 51].
The ITS-based fingerprinting data showed considerable genotypic heterogeneity as 28 distinct haplotypes were identified among 85 isolates. Eighteen isolates belonged to unique haplotypes while 67 isolates clustered in 10 haplotypes. The discriminatory power for the ITS-based fingerprinting method was 0.88 which is nearly same as the discriminatory power of the six-loci-based multilocus sequence typing (MLST) scheme for C. glabrata [52]. Fingerprinting of a global collection of 109 C. glabrata isolates by MLST yielded only 30 sequence types (STs) [52]. Fingerprinting studies for other Candida species have yielded variable results. While the ITS region sequences of 24 C. parapsilosis sensu stricto isolates analyzed so far from Kuwait were identical, three haplotypes were identified among 19 isolates of C. orthopsilosis, a species very closely related to C. parapsilosis sensu stricto [32, 53]. Likewise, the ITS region sequences among C. albicans are highly similar as only four haplotypes have been identified among a large collection of isolates so far, nine haplotypes have been detected in clinical isolates of the sister species, C. dubliniensis [36, 54–56]. The highest variability among ITS region sequences has been noted recently among C. lusitaniae strains as nine haplotypes were detected among 13 clinical isolates [57]. Taken together, the ITS region of rDNA presents as an attractive target and has the potential for improving the discriminatory power of the MLST scheme [52] for routine fingerprinting of clinical C. glabrata sensu stricto isolates.
Although we analyzed a large (n = 440) and diverse collection of clinical C. glabrata sensu lato isolates, C. nivariensis and C. bracarensis were not detected implying that these two cryptic species are not clinically important in Kuwait. The findings are consistent with a recent study that reported a low prevalence of 0.12% for C. nivariensis and 0.01% for C. bracarensis among phenotypically identified C. glabrata strains [9]. A search of published studies was carried out to determine which clinical specimens or ethnic background of patients may be more suitable for the isolation of these rare yeast pathogens and the results are summarized in Table 1. In total, 11 case reports and 18 research studies were identified which either reported the isolation or lack of isolation of C. nivariensis and/or C. bracarensis. C. nivariensis has a wider global distribution as it has been isolated from several countries in Europe, Asia (including Iran in the Middle East), Australia and South America while C. bracarensis has mainly been isolated from Canada, USA and Mexico in North America, some European countries, China and Argentina [27–30, 37, 44, 51, 58–79]. Surprisingly, both species were isolated simultaneously from only two (China and Argentina) countries [37, 51, 76]. Altogether, eight countries/geographical locations (including Kuwait) involving 2560 C. glabrata sensu lato isolates failed to identify the presence of either C. nivariensis or C. bracarensis indicating that these species are rare yeast pathogens in some countries/geographical locations [30, 68, 69, 71, 77].
Table 1. Summary of published studies/case reports and screening methods used for the identification of C. nivariensis and C. bracarensis among C. glabrata sensu lato isolates.
| No. | Type of study | Year of publication | Country/Region | No. of isolates screened | No. (%) of isolates identified as | Identification method(s) | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| CG | CN | CB | Preliminary test | Confirmatory test | ||||||
| 1 | Case reports | 2005 | Spain | NA | NA | 3 | - | CHROMagar | DNA sequencing | Alcoba-Flo´rez et al., 2005 [27] |
| 2 | Case report | 2006 | Portugal/UK | NA | NA | - | 2 | - | DNA sequencing | Correia et al., 2006 [28] |
| 3 | Case report | 2007 | Japan | NA | NA | 1 | - | - | DNA sequencing | Fujita et al., 2007 [58] |
| 4 | Case report | 2008 | Indonesia | NA | NA | 1 | - | CHROMagar/Auxacolor2 | DNA sequencing | Wahyuningsih et al., 2008 [59] |
| 5 | Case reports | 2008 | UK | NA | NA | 16 | - | CHROMagar/Auxacolor2/ API 20C |
DNA sequencing | Borman et al., 2008 [29] |
| 6 | Case report | 2010 | Canada | NA | NA | - | 2 | CHROMagar | DNA sequencing | Warren et al., 2010 [60] |
| 7 | Case report | 2013 | Spain | NA | NA | 1 | - | CHROMagar | DNA sequencing | Lopez-Soria et al., 2013 [61] |
| 8 | Case reports | 2013 | UK | NA | NA | 5 | - | CHROMagar | DNA sequencing/MALDI-TOF | Gorton et al., 2013 [62] |
| 9 | Case reports | 2016 | Spain | NA | NA | 4 | - | CHROMagar | DNA sequencing/MALDI-TOF | Aznar-Marin et al., 2016 [63] |
| 10 | Case report | 2016 | Brazil/Rio de Janeiro | NA | NA | 1 | - | CHROMagar/Vitek2 | Figueiredo-Carvalho et al., 2016 [64] | |
| 11 | Case report | 2018 | Mexico | NA | NA | - | 1 | API 20C | DNA sequencing | Treviño-Rangel et al., 2018 [65] |
| 12 | Research/screening | 2008 | USA/Baltimore | 137 | 134 (98) | 0 | 3 (2) | CHROMagar/Vitek2 | PNA FISH/DNA sequencing | Bishop et al., 2008 [44] |
| 13 | Research/screening | 2009 | North America | 838 | 836 (99.8) | 0 | 2 (0.2) | CHROMagar | PCR/PNA FISH | Lockhart et al., 2009 [30] |
| South America | 133 | 133 (100) | 0 | 0 | CHROMagar | PCR/PNA FISH | ||||
| Europe | 400 | 400 (100) | 0 | 0 | CHROMagar | PCR/PNA FISH | ||||
| Asia | 111 | 111 (100) | 0 | 0 | CHROMagar | PCR/PNA FISH | ||||
| Australia | 42 | 41 (97.6) | 1 (2.4) | 0 | CHROMagar | PCR/PNA FISH | ||||
| Africa | 71 | 71 (100) | 0 | 0 | CHROMagar | PCR/PNA FISH | ||||
| 14 | Research/screening | 2010 | India | 363 | 361 (99.5) | 2 (0.5) | 0 | CHROMagar | DNA sequencing | Chowdhary et al., 2010 [66] |
| 15 | Research/screening | 2011 | Spain | 143 | 140 (98) | 0 | 3 (2) | NS | DNA sequencing | Cuenca-Estrella et al., 2011 [67] |
| 16 | Research/screening | 2011 | Denmark | 133 | 133 (100) | 0 | 0 | NS | PCR-RFLP/PNA FISH | Mirhendi et al., 2011 [68] |
| 17 | Research/screening | 2012 | Spain | 158 | 158 (100) | 0 | 0 | NS | PCR/DNA sequencing | Pemán et al, 2012 [69] |
| 18 | Research/screening | 2013 | India | 100 | 96 (96) | 4 (4) | 0 | CHROMagar/Vitek2 | PCR/DNA sequencing | Sharma et al., 2013 [70] |
| 19 | Research/screening | 2013 | Italy | 1000 | 1000 (100) | 0 | 0 | CHROMagar/ID32C | mPCR | Esposto et al., 2013 [71] |
| 20 | Research/screening | 2014 | China/Shenzhen | 301 | 293 (97.3) | 7 (2.3) | 1 (0.3) | CHROMagar/API 20C | DNA sequencing | Li et al., 2014 [37] |
| 21 | Research/screening | 2014 | Poland | 224 | 211 (94) | 13 (6) | 0 | CHROMagar/ID32C | DNA sequencing | Swoboda-Kopec et al., 2014 [72] |
| 22 | Research/screening | 2014 | Malaysia | 185 | 183 (99) | 2 (1) | 0 | CHROMagar | PCR (RPL31 gene)/DNA sequencing | Tay et al., 2014 [73] |
| 23 | Research/screening | 2015 | China/Shanghai | NA | NA | 1 | - | CHROMagar/API 20C | DNA sequencing | Feng et al., 2015 [74] |
| 24 | Research/screening | 2016 | Argentina/Buenos Aires | 117 | 114 (97) | 3 (3) | 0 | CHROMagar | PCR (RPL31 gene)/DNA sequencing | Morales-López et al., 2016 [75] |
| 25 | Research/screening | 2017 | Argentina/Buenos Aires | 122 | 114 (93.5) | 5 (4) | 3 (2.5) | NS | mPCR/DNA sequencing | Morales-López et al., 2017 [76] |
| 26 | Research/screening | 2017 | China/Beijing | 960 | 947 (98.6) | 12 (1.3) | 1 (0.1) | Vitek2 | DNA sequencing | Hou et al., 2017 [51] |
| 27 | Research/screening | 2018 | Spain | 114 | 114 (100) | 0 | 0 | CHROMagar/ID32C | mPCR | Miranda-Cadena et al., 2018 [77] |
| 28 | Research/screening | 2018 | Poland | 353 | 352 (99.7) | 0 | 1 (0.3) | CHROMagar/Vitek2 | mPCR/DNA sequencing | Małek et al., 2018 [78] |
| 29 | Research/screening | 2019 | Iran | 213 | 209 (98) | 4 (2) | 0 | CHROMagar/mPCR | DNA sequencing/MALDI-TOF | Arastehfar et al., 2019 [79] |
| 30 | Research/screening | 2019 | Kuwait | 440 | 440 (100) | 0 | 0 | CHROMagar/Vitek2 | PCR/mPCR/DNA sequencing | This study |
CG, C. glabrata sensu stricto; CN, C. nivariensis; CB, C. bracarensis; mPCR, multiplex PCR; PNA-FISH, peptide nucleic acid-fluorescent in situ hybridization; NS, not specified; NA, not applicable
The clinical specimens yielding C. nivariensis or C. bracarensis among ~6500 C. glabrata sensu lato isolates screened so far in various studies are listed in Table 2. The data show that bloodstream, vaginal specimens and respiratory specimens are more likely to yield C. nivariensis or C. bracarensis.
Table 2. Number of C. nivariensis and/or C. bracarensis isolates identified in various clinical specimens among ~6500 C. glabrata sensu lato isolates screened so far.
| Clinical specimens | No. of C. nivariensis | No. of C. bracarensis |
|---|---|---|
| screened | isolates detected | isolates detected |
| Blood | 22 | 5 |
| Vaginal swab/exudate | 12 | 3 |
| Respiratory tract/oral cavity/sputum | 14 | 2 |
| Ascitic fluid | 3 | 0 |
| Urine/renal sample | 2 | 0 |
| Stool | 0 | 2 |
| Abscess/pus | 2 | 2 |
| Pleural fluid | 1 | 1 |
| Cerebrospinal fluid | 1 | 0 |
| Catheter exudate/exit site swab | 1 | 1 |
| Surgical site swab | 1 | 0 |
| CAPB dialysis fluid | 1 | 0 |
| Unspecified | 6 | 0 |
| Total | 66 | 16 |
CAPB, continuous ambulatory peritoneal bag
In conclusion, we have described a simple, cost effective mPCR assay by using highly conserved rDNA sequences for rapid detection and differentiation of C. nivariensis and C. bracarensis from C. glabrata sensu stricto isolates. The method involves single-tube PCR amplification followed by agarose gel electrophoresis for detection of species-specific amplicons. Only basic PCR and gel electrophoresis equipment are needed which are readily available in most mycology laboratories and the requirement for a minimal amount of genomic DNA allows the whole procedure to be completed within 4 hours. The mPCR accurately identified all 440 C. glabrata sensu stricto isolates. Furthermore, our study highlights lack of detection of C. nivariensis and C. bracarensis from Kuwait further supporting that C. nivariensis and C. bracarensis are rare yeast pathogens in some geographical locations. Considerable genotypic diversity among C. glabrata sensu stricto isolates was also indicated by rDNA sequence analyses.
Our study has few limitations. Only one reference strain was available for C. nivariensis and C. bracarensis during specificity testing studies and an internal PCR control was not included. Also, misidentification of one of the Candida species may occur in the event of mixed colonies as it will not be amplified by mPCR.
Supporting information
(DOCX)
(DOCX)
Agarose gel of amplified products obtained in PCR using C. glabrata-specific (panel A), C. nivariensis-specific (panel B) and C. bracarensis-specific (panel C) primers with genomic DNA from reference strain of C. glabrata (lane CG), C. nivariensis (lane CN), C. bracarensis (lane CB), C. albicans (lane CA), C. dubliniensis (lane CD), C. parapsilosis (lane CP) and C. tropicalis (lane CT). Lane M is 100 bp DNA ladder.
(DOCX)
Acknowledgments
We thank A. Theyyathel and L. Joseph for technical assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
The study was supported by College of Graduate Studies and Department of Microbiology, Faculty of Medicine, Kuwait University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 2007; 20:133–163. 10.1128/CMR.00029-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diekema D, Arbefeville S, Boyken L, Kroeger J, Pfaller M. The changing epidemiology of healthcare-associated candidemia over three decades. Diagn Microbiol Infect Dis 2012; 73:45–48. 10.1016/j.diagmicrobio.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 3.Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 2014; 20(Supp. 6):5–10. [DOI] [PubMed] [Google Scholar]
- 4.Pfaller MA, Moet GJ, Messer SA, Jones RN, Castanheira M. Candida bloodstream infections: comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY Antimicrobial Surveillance Program, 2008–2009. Antimicrob Agents Chemother 2011; 55:561–566. 10.1128/AAC.01079-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khan Z, Ahmad S, Joseph L and Chandy R. Candida dubliniensis: an appraisal of its clinical significance as a bloodstream pathogen. PLoS One 2012; 7:e32952 10.1371/journal.pone.0032952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lockhart SR, Iqbal N, Cleveland AA, Farley MM, Harrison LH, Bolden CB, et al. Species identification and antifungal susceptibility testing of Candida bloodstream isolates from population-based surveillance studies in two U.S. cities from 2008 to 2011. J Clin Microbiol 2012; 50:3435–3442. 10.1128/JCM.01283-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleveland AA, Farley MM, Harrison LH, Stein B, Hollick R, Lockhart SR, et al. Changes in incidence and antifungal drug resistance in candidemia: results from population-based laboratory surveillance in Atlanta and Baltimore, 2008–2011. Clin Infect Dis 2012; 55:1352–1361. 10.1093/cid/cis697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombo AL, Guimarães T, Sukienik T, Pasqualotto AC, Andreotti R, Queiroz-Telles F, et al. Prognostic factors and historical trends in the epidemiology of candidemia in critically ill patients: an analysis of five multicenter studies sequentially conducted over a 9-year period. Intensive Care Med 2014; 40:1489–1498. 10.1007/s00134-014-3400-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angoulvant A, Guitard J, Hennequin C. Old and new pathogenic Nakaseomyces species: epidemiology, biology, identification, pathogenicity and antifungal resistance. FEMS Yeast Res 2016; 16:fov114 10.1093/femsyr/fov114 [DOI] [PubMed] [Google Scholar]
- 10.Pfaller MA. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am J Med 2012; 125:S3–S13. 10.1016/j.amjmed.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 11.Sanguinetti M, Posteraro B, Lass-Flörl C. Antifungal drug resistance among Candida species: mechanisms and clinical impact. Mycoses 2015; 58 Suppl 2:2–13. [DOI] [PubMed] [Google Scholar]
- 12.Kołaczkowska A, Kołaczkowski M. Drug resistance mechanisms and their regulation in non-albicans Candida species. J Antimicrob Chemother 2016; 71:1438–1450. 10.1093/jac/dkv445 [DOI] [PubMed] [Google Scholar]
- 13.Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, et al. Clinical Practice Guideline for the Management of Candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis 2016; 62:e1–e50. 10.1093/cid/civ933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katiyar S, Pfaller M, Edlind T. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob Agents Chemother 2006; 50:2892–2894. 10.1128/AAC.00349-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desnos-Olivier M, Dromer F, Dannaoui E. Detection of capofungin resistance in Candida spp. by Etest. J Clin Microbiol 2008; 46:2389–2392. 10.1128/JCM.00053-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dannaoui E, Desnos-Ollivier M, Garcia-Hermoso D, Grenouillet F, Cassaing S, Baixench MT, et al. Candida spp. with acquired echinocandin resistance, France, 2004–2010. Emerg Infect Dis 2012; 8:86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander BD, Johnson MD, Pfeiffer CD, Jiménez-Ortigosa C, Catania J, Booker R, et al. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 2013; 56:1724–1732. 10.1093/cid/cit136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saraya T, Tanabe K, Araki K, Yonetani S, Makino H, Watanabe T, et al. Breakthrough invasive Candida glabrata in patients on micafungin: a novel FKS gene conversion correlated with sequential elevation of MIC. J Clin Microbiol 2014; 52:2709–2712. 10.1128/JCM.03593-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan Z, Ahmad S, Mokaddas E, Meis JF, Joseph L, Abdullah A, et al. Development of echinocandin resistance in Candida tropicalis following short-term exposure to caspofungin for empiric therapy. Antimicrobial Agents and Chemotherapy 2018; 62:e01926–17. 10.1128/AAC.01926-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan ZU, Ahmad S, Al-Obaid I, Al-Sweih NA, Joseph L, Farhat D. Emergence of resistance to amphotericin B and triazoles in Candida glabrata vaginal isolates in a case of recurrent vaginitis. J Chemother 2008; 20:488–491. 10.1179/joc.2008.20.4.488 [DOI] [PubMed] [Google Scholar]
- 21.Vandeputte P, Tronchin G, Larcher G, Ernoult E, Bergès T, Chabasse D, et al. A nonsense mutation in the ERG6 gene leads to reduced susceptibility to polyenes in a clinical isolate of Candida glabrata. Antimicrob Agents Chemother 2008; 52:3701–3709. 10.1128/AAC.00423-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hull CM, Bader O, Parker JE, Weig M, Gross U, Warrilow AG, et al. Two clinical isolates of Candida glabrata exhibiting reduced sensitivity to amphotericin B both harbor mutations in ERG2. Antimicrob Agents and Chemother 2012; 56:6417–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan Z, Ahmad S, Joseph L, Al-Obaid K. Isolation of cholesterol-dependent, multidrug-resistant Candida glabrata strains from blood cultures of a candidemia patient in Kuwait. BMC Infect Dis 2014;14:188 10.1186/1471-2334-14-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad S, Joseph L, Parker JE, Asadzadeh M, Kelly SL, Meis JF, Khan Z. ERG6 and ERG2 are major targets conferring reduced susceptibility to amphotericin B in clinical Candida glabrata isolates in Kuwait. Antimicrob Agents Chemother 2019; 63:e01900–18. 10.1128/AAC.01900-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, et al. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun 2016; 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Healey KR, Perlin DS. Fungal Resistance to Echinocandins and the MDR Phenomenon in Candida glabrata. J fungi 2018; 4:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alcoba-Flórez J, Méndez-Alvarez S, Cano J, Guarro J, Pérez-Roth E, del Pilar Arévalo M. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J Clin Microbiol 2005; 43:4107–4111. 10.1128/JCM.43.8.4107-4111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Correia A, Sampaio P, James S, Pais C. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int J Syst Evol Microbiol 2006; 56:313–317. 10.1099/ijs.0.64076-0 [DOI] [PubMed] [Google Scholar]
- 29.Borman AM, Petch R, Linton CJ, Palmer MD, Bridge PD, Johnson EM. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J Clin Microbiol 2008; 46:933–938. 10.1128/JCM.02116-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lockhart SR, Messer SA, Gherna M, Bishop JA, Merz WG, Pfaller MA, et al. Identification of Candida nivariensis and Candida bracarensis in a large global collection of Candida glabrata isolates: comparison to the literature. J Clin Microbiol 2009; 47:1216–1217. 10.1128/JCM.02315-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamal WY, Ahmad S, Khan ZU, Rotimi VO. Comparative evaluation of two matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems for the identification of clinically significant yeasts. Int J Infect Dis 2014; 26:167–170. 10.1016/j.ijid.2014.05.031 [DOI] [PubMed] [Google Scholar]
- 32.Asadzadeh M, Ahmad S, Hagen F, Meis JF, Al-Sweih N, Khan Z. Simple, low-cost detection of Candida parapsilosis complex isolates and molecular fingerprinting of Candida orthopsilosis strains in Kuwait by ITS region sequencing and amplified fragment length polymorphism analysis. PLoS One 2015; 10:e0142880 10.1371/journal.pone.0142880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmad S, Mustafa AS, Khan Z, Al-Rifaiy AI, Khan ZU. PCR-enzyme immunoassay of rDNA in the diagnosis of candidemia and comparison with amplicon detection by agarose gel electrophoresis. Int J Med Microbiol 2004; 294:45–51. 10.1016/j.ijmm.2004.01.002 [DOI] [PubMed] [Google Scholar]
- 34.Khan ZU, Ahmad S, Mokaddas E, Chandy R, Cano J, Guarro J. Actinomucor elegans var. kuwaitiensis isolated from the wound of a diabetic patient. Antonie van Leeuwoenhoek 2008; 94:343–352. [DOI] [PubMed] [Google Scholar]
- 35.Khan ZU, Ahmad S, Hagen F, Fell JW, Kowshik T, Chandy R, et al. Cryptococcus randhawai sp. nov., a novel anamorphic basidiomycetous yeast isolated from tree trunk hollow of Ficus religiosa (peepal tree) from New Delhi, India. Antonie Van Leeuwenhoek 2010; 97:253–259. 10.1007/s10482-009-9406-8 [DOI] [PubMed] [Google Scholar]
- 36.Asadzadeh M, Ahmad S, Al-Sweih N, Khan Z. Population structure and molecular genetic characterization of 5-flucytosine-susceptible and -resistant clinical Candida dubliniensis isolates from Kuwait. PLoS One 2017; 12:e0175269 10.1371/journal.pone.0175269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, Shan Y, Fan S, Liu X. Prevalence of Candida nivariensis and Candida bracarensis in vulvovaginal candidiasis. Mycopathologia 2014; 178:279–283. 10.1007/s11046-014-9800-2 [DOI] [PubMed] [Google Scholar]
- 38.Bishop JA, Chase N, Lee R, Kurtzman CP, Merz WG. Production of white colonies on CHROMagar Candida medium by members of the Candida glabrata clade and other species with overlapping phenotypic traits. J Clin Microbiol 2008; 46:3498–3500. 10.1128/JCM.00982-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo G & Mitchell TG. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J Clin Microbiol 2002; 40:2860–2865. 10.1128/JCM.40.8.2860-2865.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcoba-Flórez J, Arévalo Mdel P, González-Paredes FJ, Cano J, Guarro J, Pérez-Roth E, et al. PCR protocol for specific identification of Candida nivariensis, a recently described pathogenic yeast. J Clin Microbiol 2005; 43:6194–6196. 10.1128/JCM.43.12.6194-6196.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Romeo O, Scordino F, Pernice I, Lo Passo C, Criseo G. A multiplex PCR protocol for rapid identification of Candida glabrata and its phylogenetically related species Candida nivariensis and Candida bracarensis. J Microbiol Methods 2009; 79:117–120. 10.1016/j.mimet.2009.07.016 [DOI] [PubMed] [Google Scholar]
- 42.Enache-Angoulvant A, Guitard J, Grenouillet F, Martin T, Durrens P, Fairhead C, et al. Rapid discrimination between Candida glabrata, Candida nivariensis, and Candida bracarensis by use of a singleplex PCR. J Clin Microbiol 2011; 49:3375–3379. 10.1128/JCM.00688-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arastehfar A, Fang W, Pan W, Liao W, Yan L, Boekhout T. Identification of nine cryptic species of Candida albicans, C. glabrata, and C. parapsilosis complexes using one-step multiplex PCR. BMC Infect Dis. 2018; 18:480 10.1186/s12879-018-3381-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bishop JA, Chase N, Magill SS, Kurtzman CP, Fiandaca MJ, Merz WG. Candida bracarensis detected among isolates of Candida glabrata by peptide nucleic acid fluorescence in situ hybridization: susceptibility data and documentation of presumed infection. J Clin Microbiol 2008; 46:443–446. 10.1128/JCM.01986-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber JA, Morrison HG, Huse SM, Neal PR, Sogin ML, Mark Welch DB. Effect of PCR amplicon size on assessments of clone library microbial diversity and community structure. Environ Microbiol 2009; 11:1292–1302. 10.1111/j.1462-2920.2008.01857.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Debode F, Marien A, Janssen E, Bragard C, Berben G. The influence of amplicon length on real-time PCR results. Biotechnol Agron Soc Environ 2017; 21:3–11. [Google Scholar]
- 47.Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, et al. A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol 2008; 197:313–324. 10.1007/s00430-007-0063-0 [DOI] [PubMed] [Google Scholar]
- 48.Lucignano B, Ranno S, Liesenfeld O, Pizzorno B, Putignani L, Bernaschi P, et al. Multiplex PCR allows rapid and accurate diagnosis of bloodstream infections in newborns and children with suspected sepsis. J Clin Microbiol 2011; 49:2252–2258. 10.1128/JCM.02460-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grif K, Fille M, Würzner R, Weiss G, Lorenz I, Gruber G, et al. Rapid detection of bloodstream pathogens by real-time PCR in patients with sepsis. Wien Klin Wochenschr 2012; 124:266–270. 10.1007/s00508-012-0159-4 [DOI] [PubMed] [Google Scholar]
- 50.Galán F, García-Agudo L, Guerrero I, Marín P, García-Tapia A, García-Martos P, et al. Evaluation of mass spectrometry for the identification of clinically interesting yeasts. Enferm Infecc Microbiol Clin 2015; 33:372–378. 10.1016/j.eimc.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 51.Hou X, Xiao M, Chen SC, Wang H, Yu SY, Fan X, et al. Identification and Antifungal Susceptibility Profiles of Candida nivariensis and Candida bracarensis in a Multi-Center Chinese Collection of Yeasts. Front Microbiol 2017; 8:5 10.3389/fmicb.2017.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dodgson AR, Pujol C, Denning DW, Soll DR, Fox AJ. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J Clin Microbiol 2003; 41:5709–5717. 10.1128/JCM.41.12.5709-5717.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asadzadeh M, Ahmad S, Al-Sweih N, Khan ZU. Rapid molecular differentiation and genotypic heterogeneity among Candida parapsilosis and Candida orthopsilosis strains isolated from clinical specimens in Kuwait. J Med Microbiol 2009; 58:745–752. 10.1099/jmm.0.008235-0 [DOI] [PubMed] [Google Scholar]
- 54.Gee SF, Joly S, Soll DR, Meis JF, Verweij PE, Polacheck I, et al. Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo. J Clin Microbiol 2002; 40:556–574. 10.1128/JCM.40.2.556-574.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmad S, Khan ZU, Joseph L, Asadzadeh M, Theyyathel A. Genotypic heterogeneity and molecular basis of 5-flucytosine resistance among Candida dubliniensis isolates recovered from clinical specimens in Kuwait. Med Mycol 2012; 50:244–251. 10.3109/13693786.2011.597446 [DOI] [PubMed] [Google Scholar]
- 56.Borman AM, Szekely A, Linton CJ, Palmer MD, Brown P, Johnson EM. Epidemiology, antifungal susceptibility, and pathogenicity of Candida africana isolates from the United Kingdom. J Clin Microbiol 2013; 51:967–972. 10.1128/JCM.02816-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan Z, Ahmad S, Al-Sweih N, Khan S, Joseph L. Candida lusitaniae in Kuwait: prevalence, antifungal susceptibility and role in neonatal fungemia. PLoS One 2019; 14: e0213532 10.1371/journal.pone.0213532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fujita S, Senda Y, Okusi T, Ota Y, Takada H, Yamada K, Kawano M. Catheter-related fungemia due to fluconazole-resistant Candida nivariensis. J Clin Microbiol 2007; 45:3459–3461. 10.1128/JCM.00727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wahyuningsih R, SahBandar IN, Theelen B, Hagen F, Poot G, Meis JF, et al. Candida nivariensis isolated from an Indonesian human immunodeficiency virus-infected patient suffering from oropharyngeal candidiasis. J Clin Microbiol 2008; 46:388–391. 10.1128/JCM.01660-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warren TA, McTaggart L, Richardson SE, Zhang SX. Candida bracarensis bloodstream infection in an immunocompromised patient. J Clin Microbiol 2010; 48:4677–4679. 10.1128/JCM.01447-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.López-Soria LM, Bereciartua E, Santamaría M, Soria LM, Hernández-Almaraz JL, Mularoni A, et al. First case report of catheter-related fungemia by Candida nivariensis in the Iberian Peninsula. Rev Iberoam Micol 2013; 30:69–71. 10.1016/j.riam.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 62.Gorton RL, Jones GL, Kibbler CC, Collier S. Candida nivariensis isolated from a renal transplant patient with persistent candiduria-Molecular identification using ITS PCR and MALDI-TOF. Med Mycol Case Rep 2013; 2:156–158. 10.1016/j.mmcr.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aznar-Marin P, Galan-Sanchez F, Marin-Casanova P, García-Martos P, Rodríguez-Iglesias M. Candida nivariensis as a new emergent agent of vulvovaginal candidiasis: description of cases and review of published studies. Mycopathologia 2016; 181:445–449. 10.1007/s11046-015-9978-y [DOI] [PubMed] [Google Scholar]
- 64.Figueiredo-Carvalho MH, Ramos Lde S, Barbedo LS, Chaves AL, Muramoto IA, Santos AL, et al. First description of Candida nivariensis in Brazil: antifungal susceptibility profile and potential virulence attributes. Mem Inst Oswaldo Cruz 2016; 111:51–58. 10.1590/0074-02760150376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Treviño-Rangel RJ, Espinosa-Pérez JF, Villanueva-Lozano H, Montoya AM, Andrade A, Bonifaz A, et al. First report of Candida bracarensis in Mexico: hydrolytic enzymes and antifungal susceptibility pattern. Folia Microbiol (Praha) 2018; 63:517–523. [DOI] [PubMed] [Google Scholar]
- 66.Chowdhary A, Randhawa HS, Khan ZU, Ahmad S, Juneja S, Sharma B, et al. First isolations in India of Candida nivariensis, a globally emerging opportunistic pathogen. Med Mycol 2010; 48:416–420. 10.1080/13693780903114231 [DOI] [PubMed] [Google Scholar]
- 67.Cuenca-Estrella M, Gomez-Lopez A, Isla G, Rodriguez D, Almirante B, Pahissa A, et al. Prevalence of Candida bracarensis and Candida nivariensis in a Spanish collection of yeasts: comparison of results from a reference centre and from a population-based surveillance study of candidemia. Med Mycol 2011; 49:525–529. 10.3109/13693786.2010.546373 [DOI] [PubMed] [Google Scholar]
- 68.Mirhendi H, Bruun B, Schønheyder HC, Christensen JJ, Fuursted K, Gahrn-Hansen B, et al. Differentiation of Candida glabrata, C. nivariensis and C. bracarensis based on fragment length polymorphism of ITS1 and ITS2 and restriction fragment length polymorphism of ITS and D1/D2 regions in rDNA. Eur J Clin Microbiol Infect Dis 2011; 30:1409–1416. 10.1007/s10096-011-1235-9 [DOI] [PubMed] [Google Scholar]
- 69.Pemán J, Cantón E, Quindós G, Eraso E, Alcoba J, Guinea J, et al. Epidemiology, species distribution and in vitro antifungal susceptibility of fungaemia in a Spanish multicentre prospective survey. J Antimicrob Chemother 2012; 67:1181–1187. 10.1093/jac/dks019 [DOI] [PubMed] [Google Scholar]
- 70.Sharma C, Wankhede S, Muralidhar S, Prakash A, Singh PK, Kathuria S, et al. Candida nivariensis as an etiologic agent of vulvovaginal candidiasis in a tertiary care hospital of New Delhi, India. Diagn Microbiol Infect Dis 2013; 76:46–50. 10.1016/j.diagmicrobio.2013.02.023 [DOI] [PubMed] [Google Scholar]
- 71.Esposto MC, Prigitano A, Romeo O, Criseo G, Trovato L, Tullio V, et al. Looking for Candida nivariensis and C. bracarensis among a large Italian collection of C. glabrata isolates: results of the FIMUA working group. Mycoses 2013; 56:394–396. 10.1111/myc.12026 [DOI] [PubMed] [Google Scholar]
- 72.Swoboda-Kopeć E, Sikora M, Golas M, Piskorska K, Gozdowski D, Netsvyetayeva I. Candida nivariensis in comparison to different phenotypes of Candida glabrata. Mycoses 2014; 57:747–753. 10.1111/myc.12264 [DOI] [PubMed] [Google Scholar]
- 73.Tay ST, Lotfalikhani A, Sabet NS, Ponnampalavanar S, Sulaiman S, Na SL, et al. Occurrence and characterization of Candida nivariensis from a culture collection of Candida glabrata clinical isolates in Malaysia. Mycopathologia 2014; 178:307–314. 10.1007/s11046-014-9778-9 [DOI] [PubMed] [Google Scholar]
- 74.Feng X, Ling B, Yang X, Liao W, Pan W, Yao Z. Molecular Identification of Candida Species Isolated from Onychomycosis in Shanghai, China. Mycopathologia 2015; 180:365–371. 10.1007/s11046-015-9927-9 [DOI] [PubMed] [Google Scholar]
- 75.Morales-López SE, Taverna CG, Bosco-Borgeat ME, Maldonado I, Vivot W, Szusz W, et al. Candida glabrata species complex prevalence and antifungal susceptibility testing in a culture collection: First description of Candida nivariensis in Argentina. Mycopathologia 2016; 181:871–878. 10.1007/s11046-016-0052-1 [DOI] [PubMed] [Google Scholar]
- 76.Morales-López S, Dudiuk C, Vivot W, Szusz W, Córdoba SB, Garcia-Effron G. Phenotypic and Molecular Evaluation of Echinocandin Susceptibility of Candida glabrata, Candida bracarensis, and Candida nivariensis Strains Isolated during 30 Years in Argentina. Antimicrob Agents Chemother 2017; 61:e00170–17. 10.1128/AAC.00170-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miranda-Cadena K, Marcos-Arias C, Mateo E, Aguirre JM, Quindós G, Eraso E. Prevalence and antifungal susceptibility profiles of Candida glabrata, Candida parapsilosis and their close-related species in oral candidiasis. Arch Oral Biol 2018; 95:100–107. 10.1016/j.archoralbio.2018.07.017 [DOI] [PubMed] [Google Scholar]
- 78.Malek M, Mrowiec P, Klesiewicz K, Skiba-Kurek I, Szczepański A, Białecka J, et al. Prevalence of human pathogens of the clade Nakaseomyces in a culture collection-the first report on Candida bracarensis in Poland. Folia Microbiol (Praha) 2018. October 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arastehfar A, Daneshnia F, Salehi MR, Zarrinfar H, Khodavaisy S, Haas PJ, et al. Molecular characterization and antifungal susceptibility testing of Candida nivariensis from blood samples—an Iranian multicentre study and a review of the literature. J Med Microbiol 2019; 68:770–777. 10.1099/jmm.0.000963 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Agarose gel of amplified products obtained in PCR using C. glabrata-specific (panel A), C. nivariensis-specific (panel B) and C. bracarensis-specific (panel C) primers with genomic DNA from reference strain of C. glabrata (lane CG), C. nivariensis (lane CN), C. bracarensis (lane CB), C. albicans (lane CA), C. dubliniensis (lane CD), C. parapsilosis (lane CP) and C. tropicalis (lane CT). Lane M is 100 bp DNA ladder.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.