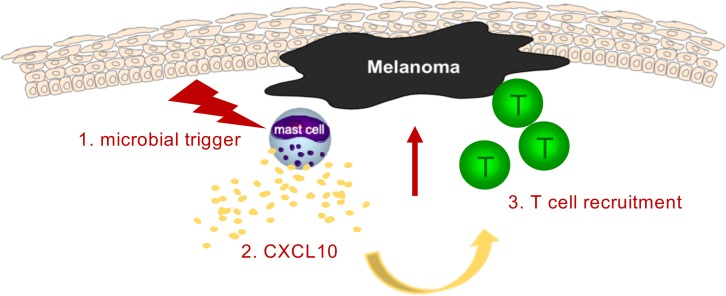
Abstract
Immune checkpoint blockade has revolutionized cancer treatment. Patients developing immune mediated adverse events, such as colitis, appear to particularly benefit from immune checkpoint inhibition. Yet, the contributing mechanisms are largely unknown. We identified a systemic LPS signature in melanoma patients with colitis following anti–cytotoxic T lymphocyte–associated antigen 4 (anti–CTLA-4) checkpoint inhibitor treatment and hypothesized that intestinal microbiota–derived LPS contributes to therapeutic efficacy. Because activation of immune cells within the tumor microenvironment is considered most promising to effectively control cancer, we analyzed human and murine melanoma for known sentinels of LPS. We identified mast cells (MCs) accumulating in and around melanomas and showed that effective melanoma immune control was dependent on LPS-activated MCs recruiting tumor-infiltrating effector T cells by secretion of CXCL10. Importantly, CXCL10 was also upregulated in human melanomas with immune regression and in patients with colitis induced by anti–CTLA-4 antibody. Furthermore, we demonstrate that CXCL10 upregulation and an MC signature at the site of melanomas are biomarkers for better patient survival. These findings provide conclusive evidence for a “Trojan horse treatment strategy” in which the plasticity of cancer-resident immune cells, such as MCs, is used as a target to boost tumor immune defense.
Keywords: Immunology, Oncology
Keywords: Chemokines, Mast cells, Melanoma
Targeted activation of melanoma-associated mast cells initiates effective tumor immune defense by CXCL10-induced recruitment of tumor-infiltrating T cells.
Introduction
Targeting the immune checkpoint inhibitors cytotoxic T lymphocyte–associated antigen 4 (CTLA-4) and programmed cell death 1 (PD-1) prolongs the overall survival of patients with melanoma by activating T cell–mediated antitumor immune responses (1). Yet, a substantial number of patients with melanoma progress because of innate or acquired resistance. To increase the response rates and to sustain treatment success, combined targeting of different molecules and pathways is increasingly studied, with initial data demonstrating the superiority of dual strategies compared with single-target strategies (2). These approaches aim to promote the effectiveness of tumor-infiltrating effector T cells (TILs) by reversing immune suppression within the tumor microenvironment (TME) (3, 4). Predictors for the therapeutic success of immune checkpoint inhibitors include the abundance of preexisting tumor-specific T cells at tumor sites (5, 6) and immune-mediated adverse events such as colitis, which are associated with tumor regression (7–10). So far, little is known about mechanisms contributing to the favorable patient outcome and adverse events after checkpoint inhibition. Based on data demonstrating a role for the intestinal microbiota in the responses to cancer therapy (11–15), we hypothesized that checkpoint inhibitor–associated colitis results in systemic exposure to microbial products like LPS, which contribute to immune modulation by targeting previously suppressive immune cells within the TME. Important suppressive noncancerous cells in the TME are tumor-associated macrophages, myeloid-derived suppressor cells, tumor-infiltrating DCs, and cancer-associated fibroblasts (16, 17). In addition, mast cells (MCs) might have thus-far-underestimated therapeutic potential because they are long-lived (18), frequently detected in the TME (19), and characterized by functional plasticity (20–22). MCs can serve as important innate immune sentinels such as for LPS (23) and have the ability to enhance T cell–mediated immune reactions but were also shown to suppress immune responses under other circumstances (22, 24–28). Consistent with their functional plasticity, MC numbers in the TME were reported to correlate with cancer progression as well as with improved patient survival (29).
Indeed, we identified a serum response signature indicative of exposure to intestinal microbiota–derived LPS in melanoma patients with colitis after anti–CTLA-4 melanoma treatment. Furthermore, we detected MCs in and around melanomas, and our in vivo models provide evidence that exposing these immune cells adjacent to melanoma to LPS initiates melanoma immune defense. Importantly, effective melanoma treatment was dependent on LPS-activated melanoma-resident MCs, which recruited TILs by secretion of CXCL10. Our data provide strong evidence for a new treatment opportunity targeting MCs within the TME by taking advantage of their plasticity. Additionally, a mast cell signature as well as CXCL10 expressed in excised human tissues of primary melanomas proved to be positive biomarkers for patient survival.
Results
LPS signature in melanoma patients treated with α–CTLA-4 antibody.
We analyzed 38 patients with stage IV melanoma treated with anti–CTLA-4 and anti–PD-1 antibodies (ipilimumab and nivolumab) who developed immune-related adverse events (irAEs) in terms of best overall response and overall survival. The results showed that patients diagnosed with immune-related enterocolitis had a significantly better overall response (Figure 1A; P = 0.0116) and significantly better survival (Figure 1B; P = 0.0042) compared with patients who developed other irAEs. Complete response (CR) was observed in 25% of the patients with immune-related colitis compared with 4.55% of patients with other irAEs. Likewise, progressive disease (PD) was less frequent in patients with immune-related colitis (25%) versus patients with other irAEs (59%). These findings confirm and extend previous analyses on melanoma therapy with the anti–CTLA-4 antibody ipilimumab. Here, enterocolitis, a leading irAE in melanoma therapy with the anti–CTLA-4 antibody ipilimumab, is associated with tumor regression and now considered an independent predictor of improved survival (30). Because immune-mediated enterocolitis leads to increased intestinal permeability, allowing systemic exposure to intestinal microbiota–derived components, we wanted to identify dominant biomarkers and selected serum samples from patients with stage IV melanoma before (“pre α–CTLA-4”) and after (“post α–CTLA-4”) (Figure 1C) ipilimumab treatment and in the absence or presence of therapy-associated enterocolitis (“w/o colitis” or “colitis,” respectively). Multiplex protein arrays from samples before therapy for proinflammatory cytokines showed little difference between the 2 groups (pre α–CTLA-4, Figure 1C). In sharp contrast, a dominant increase in proinflammatory cytokines and chemokines in the colitis group compared with the “w/o colitis” group was observed following ipilimumab treatment (post α–CTLA-4, Figure 1C). Strikingly, the LPS-induced key cytokines TNF-α, IL-8, and IL-6 (31) were increased only in the sera of patients with ipilimumab-triggered colitis (Figure 1D), indicating systemic exposure to intestinal microbiota–derived LPS following enterocolitis-associated barrier defects in the gut. The interpretation of these findings as “LPS signature” was confirmed by the detection of increased serum levels of the LPS-specific marker LPS binding protein (LPS-BP) (32) in the colitis group only, with significantly higher intraindividual upregulation of LPS-BP following α–CTLA-4 administration compared with the patient group without colitis (Figure 1, E and F; Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125057DS1). Analysis of sCD14, another LPS-inducible protein, provided similar data (Supplemental Figure 1B). Importantly, analyzing 458 patients with melanoma from the data portal The Cancer Genome Atlas (TCGA) of the National Institutes of Health (http://cancergenome.nih.gov) for the expression of the LPS receptor TLR4 revealed a significantly longer median overall survival (P < 0.0001) for the 50th percentile of patients with high compared with low TLR4 expression. The LPS signature and the beneficial role of the presence of its receptor, TLR4, underline the potential role of TLR4 activation in melanoma-specific immune defense (Supplemental Figure 2). This finding led us to investigate whether local exposure to the microbial cell wall component LPS functions as an initiator of antitumor immune responses in melanoma.
Figure 1. LPS signature in melanoma patients treated with α–CTLA-4.
(A) Best overall response to immunotherapy with α–CTLA-4 and α–PD-1 in patients with stage IV melanoma as assessed by staging and the immune-related adverse events (irAEs) and (B) Kaplan-Meier analyses comparing patients with checkpoint treatment–related colitis (n = 16) and patients with other irAEs (n = 22). (C–F) Serum samples before and after α–CTLA-4 treatment of patients with stage IV melanoma were divided into the following 2 groups: patients without (“w/o colitis”) or with (“colitis”) therapy-provoked enterocolitis (n = 6 per group). (C) Heatmap of different protein levels as determined by multiplex immunoassays of the sera. (D) Pie charts of protein serum levels of 3 LPS-induced key cytokines shown in C, where the size of the pie pieces correlates with the expression level of the cytokines. The schematic on the top shows the arrangement of the different conditions; the colors in the pie charts are the same as in the heatmap. (E) Serum LPS-BP levels of the “colitis” group measured by quantitative ELISA. (F) Changes in LPS-BP serum concentration with α–CTLA-4 treatment. P values were calculated with log-rank test (B), Wilcoxon’s test (E), or Mann-Whitney U test (F) using GraphPad Prism software. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease. *P < 0.05; **P < 0.005.
LPS initiates melanoma immune defense.
It is well known that antitumor immune responses are mediated primarily by tumor-specific T cells. In melanoma, spontaneous regression occurs and is mediated by tumor-specific TILs, although the stimuli initiating this spontaneous regression are still unknown (33–35). To investigate whether local exposure to the microbial cell wall component LPS functions as an initiator of antitumor immune responses, we established a mouse melanoma model. Ovalbumin-expressing (OVA-expressing) B16 melanomas were grafted into the skin of mice, and adoptive transfer of OVA- and thus tumor-specific OT-I and OT-II T cells was carried out 1 week later. Subsequently, the tissue surrounding the tumors in these mice was complemented by 3 consecutive peritumoral LPS injections or vehicle control (Figure 2A). In sham-treated mice, the transfer of tumor-specific T cells significantly reduced the tumor volume by approximately 50% at day 14 (Figure 2B). In contrast, mice that received T cell transfer and treatment with LPS completely controlled the tumor (Figure 2C). This effect was dependent on tumor-specific T cells because their presence determines disease outcome in Tcrb–/– Tcrd–/– mice, which lack functional endogenous T cells (Figure 2D), possibly mediating bystander effects in WT mice.
Figure 2. Melanoma immune control by exposure to LPS.
(A) Protocol for the mouse melanoma model: 7 days after intradermal injection of B16-OVA melanoma cells, adoptive transfer of tumor-specific OT-I and OT-II T cells (TCs) was performed, followed by 3 consecutive peritumoral injections of LPS or PBS (control). (B–D) Tumor volume (mm3) of cutaneous melanomas in mice as referred to in the protocol in A over time (left) and at the endpoint, shown as bars (right) with or without adoptive TC transfer and without (B) or with exposure to LPS (C) as indicated by arrows. (D) Tumor volume of melanomas in Tcrb–/– Tcrd–/– mice lacking functional TCs. Data are presented as the mean ± SEM (n = 6–9 per group). Differences were assessed by 2-way ANOVA and Sidak’s multiple-comparisons test. *P < 0.05; ****, ####P < 0.0001.
LPS selectively targets melanoma-resident MCs to initiate tumor defense.
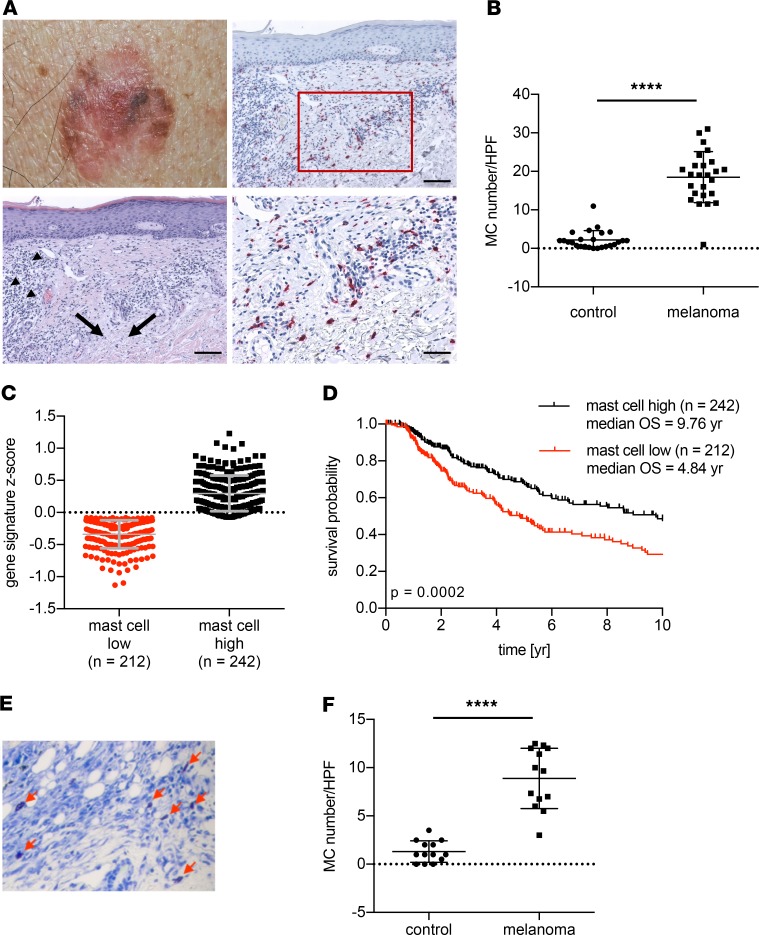
To analyze the underlying mechanisms of LPS-induced tumor immune control, different cell types and pathways as possible targets of LPS were considered. Therefore, we next investigated the natural initiation of local tumor immune control as it is found in human melanomas with immune regression (Figure 3A). H&E staining showed infiltrating lymphocytes and fibrosis as signs of immune regression (Figure 3A; arrowheads and black arrows, respectively; Supplemental Figure 4A). Already in the H&E staining, increased numbers of cells with a morphology indicative of MCs and of lymphocytes were detected (Supplemental Figure 3). Anti-CD3 immunohistochemistry characterized the infiltrating lymphocytes as T cells (Supplemental Figure 4B). MC-specific immunohistochemistry with antitryptase antibody confirmed significantly increased MC numbers within these areas of immune regression compared with the intraindividual areas of adjacent normal skin (Figure 3, A and B, and Supplemental Figure 4C). To explore whether MC infiltration may represent a meaningful biomarker in melanoma patients, we generated Z scores based on the expression of 55 MC signature genes (36) in cutaneous melanoma and defined groups as MC low and MC high (Figure 3C). Correlating them to patient survival, we found a significantly better outcome for patients with a high MC Z score (Figure 3D, P < 0.0002). This also suggests that MC accumulation at the site of melanoma is beneficial for patients with melanoma. To allow investigations regarding the functional consequences of MCs, we next analyzed our melanoma model. Similar to human melanomas, MC accumulation was also observed next to melanomas in the mouse models (Figure 3, E and F). To investigate a potential involvement of MCs in the LPS-induced initiation of anti-melanoma immune response, we applied our melanoma model with adoptive T cell transfer (Figure 2A) to 2 MC-deficient mouse lines. Both B6.Cg-kitW–sh/W–sh mice (hereafter referred to as “KitW–sh/W–sh”) and Kit-independent MC-deficient Mcpt5-cre+ R-DTAfl/fl mice failed to initiate melanoma immune defense upon LPS exposure (Figure 4, A and B, respectively). In contrast, reconstituting the skin of KitW–sh/W–sh mice with in vitro–generated bone marrow–derived MCs (BMMCs) 6 weeks before tumor implantation (Supplemental Figure 5, A–C) completely restored LPS-induced melanoma immune control (Figure 4A). Of note, in some experiments, accelerated melanoma growth led to half-maximal tumor volume at the start of immunotherapy. However, even under these conditions, LPS exposure and MCs were able to establish immune control of melanomas (Figure 4B). LPS is recognized by TLR4, which is also expressed by MCs (37, 38) (Supplemental Figure 5D). To address whether MCs are directly targeted by LPS, the skin of TLR4-deficient mice was populated with either WT or Tlr4–/– MCs. The latter showed unchanged responsiveness to different stimuli in vitro, except to LPS (Supplemental Figure 5E). In TLR4-deficient mice, only reconstitution with WT MCs, but not with Tlr4–/– MCs, allowed initiation of tumor immune control equivalent to that in WT C57BL/6 mice following peritumoral LPS exposure (Figure 4C). This result indicates that targeting innate immune pathways in MCs is sufficient to initiate melanoma immune defense. To further support this hypothesis, we analyzed melanoma immune control in Mcpt5-cre+ IKBfl/fl mice in which the signal transducer downstream of TLR4, NF-κB, (39), is endogenously activated because of Cre-mediated deletion of its negative regulator, IκB, selectively in MCs. Indeed, active NF-κB in MCs (“MC NF-κB*”) was sufficient to initiate an antitumor immune response comparable to that induced by exposure to LPS in littermates without IκB deletion and in WT mice (Figure 4D and Supplemental Figure 6). Thus, LPS administration selectively targets MCs to initiate effective tumor immune defense.
Figure 3. Mast cell accumulation adjacent to melanomas.
(A) Representative photomicrograph of a human melanoma with spontaneous immune regression (upper left) and histological sections thereof. Lower left: H&E staining of melanoma regression area with infiltrating lymphocytes and fibrosis as indicated by arrowheads and arrows, respectively (scale bar: 100 μm). Upper right: immunohistochemistry identifying MCs with antitryptase antibody (red stain; scale bar: 100 μm) and magnification thereof as indicated by a rectangle (lower right, scale bar: 50 μm). (B) Numbers of MCs within the melanoma immune regression area compared with MCs in control healthy skin regions at the rim of the excised margins. MC numbers/high-power field (HPF) are shown; n = 25; Wilcoxon’s test was used for P value calculation. (C) MC signature Z scores of 452 patients with melanoma derived from the TCGA data set as dichotomized values and (D) in correlation with overall survival data of the patients. (E) Representative example of toluidine blue–stained sections of mouse B16-OVA melanoma (arrows refer to MCs) and (F) MC quantification (MC numbers/HPF). Statistical difference was determined by paired t test (n = 13). ****P < 0.0001.
Figure 4. LPS targets melanoma-resident MCs to initiate tumor immune defense.
(A–D) Tumor volume (mm3) in mice over time according to the protocol shown in Figure 2A with TC transfer. Right: Graphs depict the tumor volume at the endpoint comparing LPS- and PBS-exposed mice. Skin reconstitution with MCs was performed 6 weeks before the tumor protocol. Statistical analysis was done by 2-way ANOVA and Tukey’s multiple-comparisons test. (A) Melanoma growth in MC-deficient KitW–sh/W–sh mice with and without WT MC reconstitution (n = 8 per group). (B) Melanoma growth in MC-deficient mice because of MC-specific (Mcpt5) expression of diphtheria toxin compared with WT mice (n = 16–20 per group). (C) Tlr4–/– mice reconstituted with WT or Tlr4–/– MCs compared with WT mice reconstituted with WT MC (right) (n = 5–10 per group). (D) Melanoma in Mcpt5-cre+ IKBfl/fl mice with MC-specific endogenous activation of NF-κB (MC NF-κB*) compared with LPS-induced exogenous NF-κB activation and PBS controls in Mcpt5-cre+ IKBwt/wt or IKBfl/wt mice (n = 8–22 per group). Means ± SEM are shown. *, #P < 0.05; ##, ‡‡P < 0.005; ***, P < 0.0005; ****, ####, ‡‡‡‡P < 0.0001.
MC-derived CXCL10 mediates tumor control by recruiting tumor-infiltrating T cells.
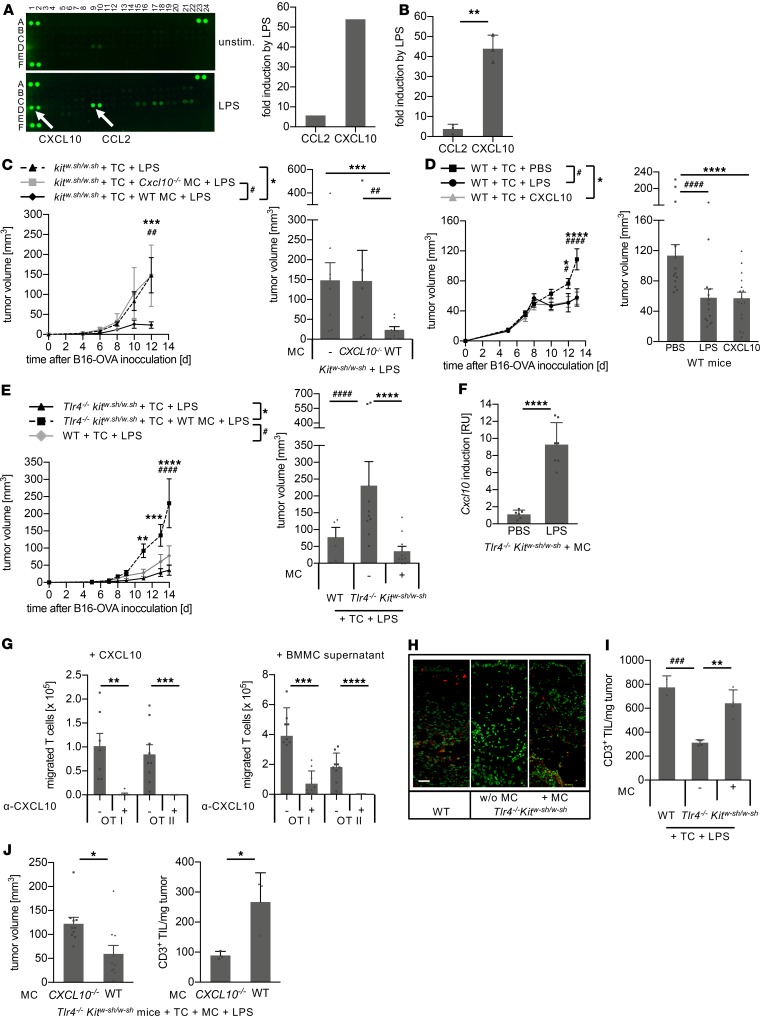
To identify the underlying mechanisms of MC-mediated tumor immune defense, MC supernatants from BMMC cultures either untreated or exposed to LPS were analyzed by a cytokine array detecting 40 secretory proteins. Surprisingly, 2 MC-derived chemokines were selectively upregulated by LPS treatment. CCL2 (monocyte chemotactic protein 1) was increased almost 6-fold, and CXCL10 (IFN-inducible protein 10) was induced by more than 50-fold (Figure 5A and Supplemental Figure 7). Both findings were supported by quantitative ELISAs (Figure 5B). Notably, among the CXCR3 ligands, only CXCL10, but not CXCL9 and CXCL11, was induced by LPS in MCs (Figure 5A and Supplemental Figure 7). Subsequently, we investigated the role of MC-derived CXCL10 in tumor immune control and reconstituted the skin of MC-deficient KitW–sh/W–sh mice with MCs from different genotypes before establishing cutaneous melanomas. In contrast with WT MCs, reconstitution with TLR4-responsive Cxcl10-deficient MCs (Supplemental Figure 5F) resulted in the complete loss of LPS-induced tumor immune control (Figure 5C), demonstrating that MC-derived CXCL10 is crucial for the LPS-initiated tumor defense. Indeed, in WT mice peritumoral administration of CXCL10 instead of LPS resulted in the same extent of tumor control as did exposure to LPS (Figure 5D). To exclude the effects mediated by LPS-reactive cells other than MCs, we next generated a mouse deficient in both TLR4 and MCs (Tlr4–/– KitW–sh/W–sh). As expected, Tlr4–/– KitW–sh/W–sh mice failed to establish LPS-induced tumor immune control (Supplemental Figure 8A). However, reconstitution of the skin with WT MCs completely rescued LPS-induced tumor immune defense (Figure 5E), with strong induction of cutaneous Cxcl10 upon LPS exposure (Figure 5F).
Figure 5. MC-derived CXCL10 mediates melanoma immune control by TIL recruitment.
(A) Cytokine array performed with supernatants from untreated (upper left) and LPS-stimulated bone marrow–derived MCs (BMMCs) (lower left) and the 2 cytokines with the strongest LPS induction as determined by densitometric quantification (right). (B) Induction of CCL2 and CXCL10 in LPS-exposed BMMCs compared with untreated BMMCs as determined by quantitative ELISA. Means ± standard deviation of 3 independent experiments are shown; P value was calculated with unpaired Student’s t test. (C–E) Tumor volume over time (left) and at the endpoint (right) according to the protocol outlined in Figure 2A. (C) KitW–sh/W–sh mice with or without MC reconstitution demonstrating loss of tumor immune control with Cxcl10–/– MCs (n = 6–8 per group). (D) According to the protocol in Figure 2A, WT C57BL/6 mice were exposed to LPS or PBS or CXCL10 by peritumoral injections (n = 14–16 per group). (E) Tumor analysis in Tlr4–/– KitW–sh/W–sh mice with or without MC reconstitution compared with WT C57BL/6 mice (n = 6–10 per group). (F) LPS-induced Cxcl10 expression in the skin of Tlr4–/– KitW–sh/W–sh mice reconstituted with WT MCs. (Means ± standard deviation; n = 6 per group; unpaired Student’s t test.) (G) In vitro Transwell migration assay with OT-I and OT-II TCs in the presence of either CXCL10 (left) or supernatant from LPS-exposed BMMCs (right) used as chemoattractants. Blocking anti-CXCL10 antibody was used as a specificity control (means ± SEM; n = 7–9 per group; unpaired Student’s t test). (H and I) CD3+ TC infiltration (TIL) adjacent to and within melanomas of the experiment depicted in E. (H) Representative immunofluorescence tissue sections oriented with the epidermis to the top and the tumor to the bottom (red, TCs; green, nuclear stain; scale bar: 50 μm). (I) Flow cytometry analysis showing TILs (means ± standard deviation). (J) According to the protocol in Figure 2A, 14 days after B16-OVA inoculation, tumor volume (left) and TILs were determined by flow cytometry (right) in Tlr4–/– KitW–sh/W–sh mice reconstituted with Cxcl10–/– MCs or WT MCs. P values calculated with 2-way ANOVA and Tukey’s test (C–E and I) or with unpaired Student’s t test (J). *, #P < 0.05; **, ##P < 0.005; ***, ###P < 0.0005; ****, ####P < 0.0001.
CXCL10 acts as a chemoattractant via CXCR3 found on the surface of activated T cells (40). Tumor-specific OT-I and OT-II T cells, as used in this model, express CXCR3 (Supplemental Figure 9), and CXCR3+ OT-I and OT-II T cells migrated in response to CXCL10 and to supernatants from LPS-exposed MCs (Figure 5G). Importantly, migration of OT-I and OT-II T cells to CXCL10 and supernatants from LPS-exposed MCs was completely abrogated by the addition of blocking anti-CXCL10 antibodies. This demonstrates that the CXCL10 within MC supernatants is responsible for the migration of tumor-specific T cells (Figure 5G). Indeed, immunofluorescence analysis revealed an infiltration of T cells in melanomas from Tlr4–/– KitW–sh/W–sh mice reconstituted with WT MCs and exposed to LPS, comparable to C57BL/6 WT mice (Figure 5H). In contrast, very few T cells were detected in melanomas of LPS-exposed Tlr4–/– KitW–sh/W–sh mice without MC reconstitution (Figure 5H). Analysis of tumor-infiltrating T cells (TILs) of these melanomas by flow cytometry showed that TILs in and adjacent to tumor tissues in MC-competent mice (Figure 5I) inversely correlated with the tumor volume (Figure 5E). TIL numbers per milligram of tumor were significantly lower in the absence of MCs (Figure 5I). Skin reconstitution with Cxcl10–/– MCs in Tlr4–/– KitW–sh/W–sh mice also failed to restore LPS-induced tumor immune control (Supplemental Figure 8B). Further analyses of TIL recruitment in LPS-exposed melanomas demonstrated that only MCs secreting CXCL10 recruited TILs and mediated tumor immune control, whereas reconstitution of Tlr4–/– KitW–sh/W–sh mice with Cxcl10–/– MCs failed to do so (Figure 5J). Together with our findings that tumor-specific T cells are required for LPS-induced and MC-mediated tumor immune defense (Figure 2D), these data demonstrate that effective melanoma control following LPS exposure is mediated by MC-derived CXCL10 via the recruitment of T cells to the tumor site.
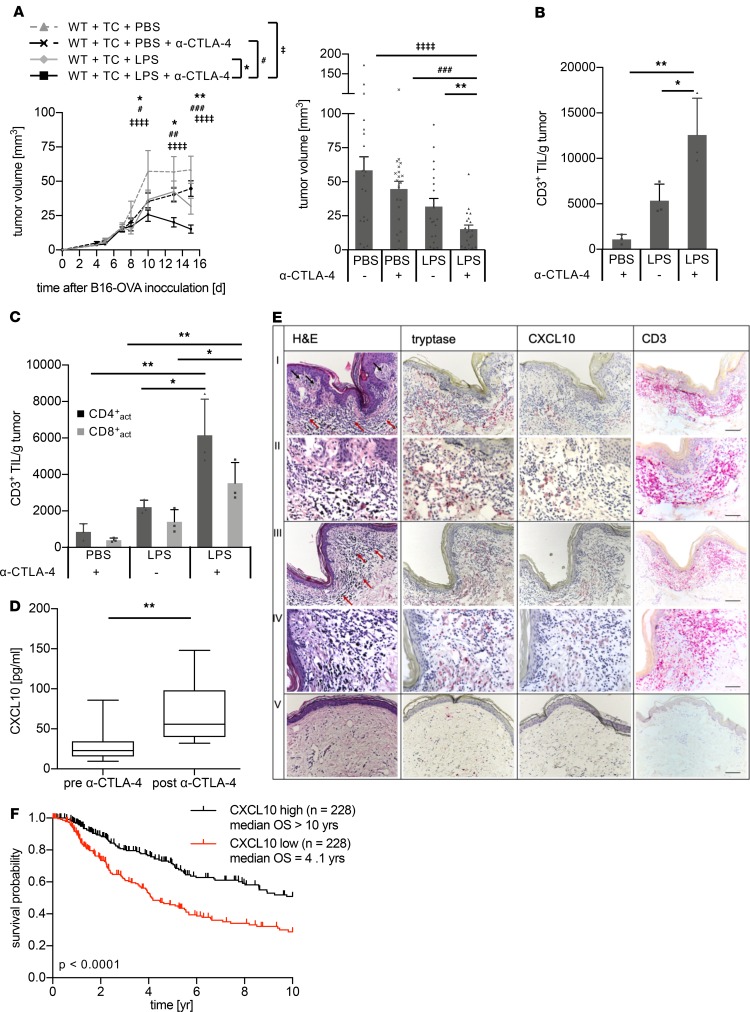
In patients with melanoma, an LPS signature was detected following enterocolitis induced by anti–CTLA-4 antibody. Therefore, we investigated whether anti–CTLA-4 antibody treatment and LPS exposure boost melanoma immune control in our murine model without inducing enterocolitis. In fact, the combination of anti–CTLA-4 antibodies and LPS exposure initiated the most effective antitumor immune defense, as demonstrated by strongly reduced tumor volume (Figure 6A, Supplemental Figure 10A) and increased numbers of TILs (Figure 6, B and C), while none of the mice showed evidence of colitis. No significant differences were found for NK cells (Supplemental Figure 10B) and macrophages (data not shown). Similar results were obtained using a combination of anti–CTLA-4 antibodies and CXCL10 (Supplemental Figure 11). Thus, immune checkpoint blockade by systemic anti–CTLA-4 antibodies and local LPS exposure orchestrated effective melanoma immune defense, which is indicative of a new treatment strategy. Indeed, patients with therapy-evoked enterocolitis had significantly upregulated CXCL10 levels following anti–CTLA-4 antibody treatment (Figure 6D). Moreover, immunohistochemistry of human melanoma revealed CXCL10 staining colocalizing with MCs, particularly within areas of inflammation merging into areas of immune regression characterized by infiltrating lymphocytes. In contrast, bordering healthy skin of the same sections did not show any CXCL10 staining, indicating that, when triggered by LPS, only activated MCs release CXCL10, leading to infiltration of lymphocytes and finally to regression (Figure 6E). Additionally, analysis of melanoma patient data sets from TCGA data portal revealed a significantly longer survival time for patients with high compared with low CXCL10 expression (Figure 6F, P < 0.0001). This indicates that CXCL10 is also a biomarker for long-term survival.
Figure 6. Immune checkpoint inhibitor treatment and complementation with LPS orchestrate effective melanoma immune defense.
(A) The protocol shown in Figure 2A was extended by additional application of α–CTLA-4 antibodies. Tumor volume in WT C57BL/6 mice over time (left) and as bars at the endpoint (right) (n = 20 per group). (B and C) Flow cytometry analysis of melanomas from WT C57BL/6 mice treated with α–CTLA-4 and/or LPS. (B) TILs determined as CD45+CD3+ cells, (C) activated CD4+ TCs (CD45+CD3+CD4+CD44+CD62L–, black bars), and CD8+ TCs (CD45+CD3+CD8+CD44+CD62L–, gray bars) (n = 3 per group). P values were calculated with 2-way ANOVA (A) or with 1-way ANOVA (B and C) followed by Tukey’s test. Means ± SEM are shown. (D) Changes in CXCL10 serum levels in patients with melanoma before (“pre α–CTLA-4”) and with ipilimumab-provoked enterocolitis (“post α–CTLA-4”). P value was calculated with Wilcoxon’s test; n = 8. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range. (E) Representative example of human melanoma with areas of inflammation (first and second rows) merging into immune regression (second and fourth rows) stained with H&E (first column) and immunohistochemically (red dye) for tryptase identifying MCs (second column), CXCL10 (third column) and CD3+ marking TCs (fourth column). Fifth row shows healthy skin at the rim of the excision. Infiltrating lymphocytes are marked by red arrows; black arrows refer to melanoma cells. Scale bars: 100 μm (first, third, and fifth rows), 50 μm (second and fourth rows). (F) Kaplan-Meier analyses of TCGA data set of 454 melanoma patients with high (black curve) and low (red curve) CXCL10 levels based on the 50th percentile of gene expression showed significantly better patient survival in patients with high CXCL10 levels (P < 0.001). Statistical analysis was done with log-rank test using R software. *, #P < 0.05; **, ##P < 0.005; ###P < 0.0005; ‡‡‡‡P < 0.0001.
Current immunotherapies using checkpoint inhibitors depend on (a) preexisting effector T cells recognizing tumor (neo-) antigens and (b) a permissive TME allowing adaptive immune responses against the tumors. Here, we show that targeting the MCs within the TME is a strategy to initiate tumor immune defense, taking advantage of the localization of these cells, their plasticity, and our increasing understanding on innate immune modulation. We provide evidence that microbiota-derived components can function as a source of adjuvants affecting the TME with consequences for MC immune behavior (see graphical abstract).
Discussion
The combination of treatment modalities, including immune checkpoint inhibition, targeting the BRAF mutation, and tumor radiation, is promising and trials are ongoing (41–44). Among these strategies are concepts that are focusing on targeting mediators of immune suppression within the TME (16, 17). In particular, there is growing evidence that innate immune cells, such as MCs, and especially their plasticity are relevant in cancer progression (45). MCs, known for their detrimental effects in allergy and anaphylaxis (46), are in fact multifunctional cells and play a key role in certain immune responses (22, 47). They are located at strategically important sites, predominantly at interfaces to the microenvironment, and possess phenotypic and functional heterogeneity as they achieve their terminal differentiation and maturation dependent on the tissue’s microenvironment (48). Equipped with a broad range of receptors and costimulatory molecules, MCs are able to rapidly respond to incoming signals and to secrete a variety of stored and newly synthesized mediators (49). These properties allow MCs to modulate innate and adaptive immune responses through amplification or suppression (50). This plasticity of MCs leads to an ambiguous role in immune responses and can contribute to cancer progression or control (22). MCs may support tumor progression by enhancing angiogenesis, by the release of growth factors and mediators for tissue remodeling, such as metalloproteinases activating tryptase (22). In addition, MC-derived cytokines have been thought to orchestrate immune evasion (51). On the other hand, MCs can contribute to controlling cancer by release of antitumorigenic heparin, histamine, IL-6, and TNF-α (52–55). Once recruited, MCs are tissue resident, and their phenotypes and effector functions depend on the shaping capacity of factors of the tissue micromilieu (48). This plasticity of MCs might open up new avenues to therapeutically reshape MCs’ phenotype toward tumor defense by appropriate stimulation.
Growing evidence indicates a pivotal role of the microbiome in orchestrating immune responses of defense and tolerance in health and disease (56). Also, in cancer patients it has been shown that microbes and microbial substances affect the efficacy of anticancer therapies (57).
Herein, we demonstrate a thus-far-unrecognized mechanism of MC-mediated tumor immune defense. As important immune cells of body surfaces (50), MCs are prototypic innate cells responding to various stimuli, including signals and components derived from human microbiota, and could therefore function as modulators of suppressive immune responses, initiating tumor immune control (22). We show that targeting MCs with the microbial adjuvant LPS is effective to boost T cell–mediated tumor cell clearance. This strategy takes advantage of the localization of MCs in the vicinity of the tumor, the plasticity of MC regarding their effector functions, and the potentiating antitumor effects generated by the recruitment of effector T cells. Furthermore, our findings demonstrate that CXCL10 plays a crucial role in melanoma immune defense. It has been previously postulated that repression of CXCL10 because of enhanced production of NO by melanoma-derived inducible NOS (iNOS) favors a protumorigenic microenvironment (58). Our data demonstrate that MCs are an important source of CXCL10 that might be harnessed for future therapeutic strategies. Targeting tumor-resident immune cells, as shown here for MCs, has substantial aptitude to improve current cancer therapies and warrants further investigation. Additionally, findings of this study highlight the crucial role of CXCL10 as an important chemokine for T cell recruitment and both MCs and CXCL10 as positive biomarkers of overall survival in melanoma patients.
Methods
Patient data analysis.
Serum samples of patients from the Department of Dermatology, Eberhard Karls University, Tübingen, Germany, who had stage IV melanoma and were treated with ipilimumab were included in the study. The serum samples of ipilimumab-treated patients were obtained between April 2008 and January 2014 (Ethics Committee approval number 567/2012BO2). Another retrospective analysis included 38 stage IV melanoma patients with irAEs treated between January 2015 and December 2017, in the Section of Dermato-oncology of the Department of Dermatology, Eberhard Karls University, Tübingen, Germany (Ethics Committee approval number 053/2019BO2). Best response was determined by staging with computed tomography and classified as CR, PR, SD, and PD according to the Response Evaluation Criteria in Solid Tumors (version 1.1) (59).
Mice.
C57BL/6, OT-I, OT-II, Tcrbtm1Mom Tcrdtm1Mom, Kitw–sh/w–sh, and Cxcl10–/– mice were purchased from Charles River or The Jackson Laboratory. Tlr4–/– mice were provided by S. Akira (Frontiers Research Center, Osaka University, Osaka, Japan). Mcpt5-cre+ (60) and Mcpt5-cre+ R-DTA (25) were given by A. Roers (Institute for Immunology, Technical University Dresden, Dresden, Germany), and IKBfl/fl mice (61) came from R. Rupec (Department of Dermatology and Allergology, Ludwig-Maximilians University, Munich, Germany) and were crossed with Mcpt5-cre+ mice. The Tlr4–/– Kitw–sh/w–sh mice were generated by crossing Tlr4–/– with Kitw–sh/w–sh mice, which were bred to homozygosity. All mice were kept and bred under specific pathogen–free conditions in accordance with the guidelines of the Federation of European Laboratory Science Association.
Cell culture.
OVA-expressing B16 melanoma cells were a gift from R. Dutton (Trudeau Institute, Saranac Lake, New York, USA). OVA expression was confirmed by PCR (primers OVA forward: 5′-CGTGGTGCTCTTTTGCAC-3′; OVA reverse: 5′-TCAGAGTTCACCATTGGGCTC-3′, both from Eurofins, Munich, Germany). Femoral bone marrow was cultured in RPMI medium with 20% FCS, 1% X63Ag8-653mIL-3–conditioned medium as a source of IL-3, medium conditioned with 1% CHO–murine stem cell factor (CHO–mSCF) as a source of SCF, and 50 U/mL penicillin/streptomycin for 3 to 4 weeks. Conditioned media were obtained from cell culture supernatants of the above cell lines. After flow cytometry analysis, cultures with 90% or more CD117+FcεRI+ cells were used for in vitro or in vivo studies. For BMMC stimulation, 10 ng Salmonella minnesota R595 LPS/mL (Alexis Biochemicals) was used. OVA-specific CD4+ and CD8+ T cells were isolated from OT-II and OT-I mice, respectively, and expanded in vitro with OVA323-339 or SIINFKL peptide (both from EMC) in the presence of antigen-presenting cells as previously described (62). Migration assays were performed in Transwell chambers with 5-μm pore inserts. RPMI medium with 0.5% BSA was used, and CXCL10 (Peprotech), anti-CXCL10 antibody (Peprotech), or supernatants from cultured BMMCs were added as indicated. After 3 hours, the cells in the lower chamber were stained with trypan blue and the viable cells were counted. All cultured cells were tested regularly for mycoplasma and no contamination was detected.
MC reconstitution and melanoma model with adoptive T cell transfer.
Mouse treatment was performed as outlined in Figure 2A: 7.5 × 104 B16-OVA cells were injected, and after 1 week, 1.2 × 107 OT-I CD8+ and 0.4 × 107 OT-II CD4+ T cells were adoptively transferred by injection into the tail vein. Where stated, 1 μg of LPS, 1 μg of CXCL10 (Peprotech) or only PBS (control) was injected peritumorally on days 8, 10, and 12. Skin MC repopulation was achieved by intradermally injecting 1 × 106 BMMCs 6–8 weeks (63) before B16-OVA melanoma injection. α–CTLA-4 antibodies were purified from the supernatant of UC10-4F10-11 hybridoma cells (ATCC) and i.p. injected (3× 200 μg). Tumor size was measured with a caliper.
Histology.
Histological sections from paraffin-embedded tissues were H&E or toluidine stained. Immunohistochemistry was performed with anti–human tryptase antibody (clone AA1; DakoCytomation), anti–human CD3 antibody (BRB063; Zytomed Systems), and anti–human CXCL10 antibody (ab9807; Abcam). For murine immunofluorescence, sections were blocked using donkey serum (MilliporeSigma) and incubated with goat anti–mouse CD3ε antibody (sc-1127; Santa Cruz Biotechnology). Bound antibody was visualized using donkey anti–goat Cy3 antibody (112-165-167; Dianova GmbH). For nuclear staining, we used Yopro (1:2000; Invitrogen). Sections were analyzed using a Leica TCS-SP confocal laser scanning microscope (Leica Mikrosysteme Wetzlar), a PL Fluotar ×25/0.75 oil lens (Leica), and Mowiol medium (Hoechst). Images were processed with Leica Confocal Software LCS (version 2.61). Original magnification was ×250.
Analysis of gene expression.
Six hours after intracutaneous injection of LPS into BMMC-reconstituted Tlr4–/– Kitw–sh/w–sh mice, total RNA was extracted from the skin using an RNA kit (MACHEREY-NAGEL). RNA was reverse-transcribed to cDNA using an iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instructions. Quantitative real-time PCR was carried out with a Light-Cycler LC480 (Roche) using SYBR Green Supermix (Roche). Values were normalized to the housekeeping gene B-actin. The primers used were as follows: Cxcl10 forward: 5′-GGATGGCTGTCCTAGCTCTG-3′, Cxcl10 reverse: 5′-ATAACCCTTGGGAAGATGG-3′; B-actin forward: 5′-CTAAGGCCAACCGTGAAAAG-3′; B-actin reverse: 5′-ACCAGAGGCATACAGGGACA-3′ (all from Eurofins). The MC signature score was defined as the average Z score of the Z score–transformed data set comprising 55 genes from Dwyer et al. (36). The Z scores were generated using the R2: Genomics Analysis and Visualization Platform. Kaplan-Meier survival analysis of patients with melanoma from the TCGA melanoma data set was done with log-rank test using GraphPad Prism 6.0 software.
ELISA and flow cytometry.
Human sera were analyzed using Luminex assay (R&D Systems), Legendplex human inflammation panel (BioLegend), sCD13 ELISA (R&D Systems), and LPS-BP ELISA (Biometeo). Mouse cell culture supernatants were analyzed by proteome profiler cytokine array panel A (R&D Systems) and by ELISA kits for CXCL10 (R&D Systems), CCL2 (PeproTech), and IL-6 (BD Biosciences).
Tumor preparation and flow cytometry.
Melanoma samples were explanted from mouse skin, dissected with scissors, and digested with collagenase A (Serva) for 30 minutes at 37°C. The tissues were homogenized with a gentleMACS dissociator (Miltenyi Biotec), passed through a nylon mesh cell strainer, and washed 3 times. The following antibodies were used: (a) from BioLegend, anti–CCR3-PE (catalog 144506), anti–CD117-APC (catalog 105812), anti–CD3-BV421 (catalog 100336), anti–CD3-PerCP (catalog 553067), anti–CD4-PE-Cy7 (catalog 100422), anti–CD44-PerCP-Cy5.5 (catalog 103032), anti–CD45.2-APC-Cy7 (catalog 109824), anti–CD49b-PE-Cy7 (catalog 108921), anti–CD62L-FITC (catalog 104406), anti–CD8-APC (catalog 100712), anti–NK1.1-FITC (catalog 108705), and Zombie Aqua fixable viability dye; and (b) from eBioscience, anti–CD45-eF450 (catalog 48-0451-80), anti–FcεRI-PE (catalog 12-5898-81), and fixable viability dye eFluor 780. Flow cytometry data were acquired using an LSRII or FACSCanto II (BD Biosciences) and analyzed with FACSDIVA software (BD Biosciences) or FlowJo software (Tree Star, BD Biosciences).
Statistics.
If not otherwise stated, data are presented as the mean ± SEM. Mouse experiments are representative of at least 2 experiments. Statistical analysis of human data was performed with Fisher’s exact test, Mann-Whitney U, or Wilcoxon’s test. Murine data were analyzed with unpaired Student’s t test (2 tailed) or with 2-way repeated-measures ANOVA and Tukey’s multiple-comparisons test. Analysis was performed with GraphPad Prism software. P values less than 0.05 were considered statistically significant and are marked as follows: *, #P < 0.05; **, ##, ‡‡P < 0.005; ***, ###, ‡‡‡P < 0.0005; and ****, ####, ‡‡‡‡P < 0.00005.
Study approval.
All animal experiments were performed in compliance with both European Union and German law and approved by local authorities (Regierungspräsidium Tübingen, HT1/05, HT3/09, HT5/14; Regierungspräsidium Oberbayern 55.2-1-54-2532-19-2016). Human studies were approved by the Ethics Committee, University of Tübingen (567/2012BO2 and 053/2019BO2). Human participants provided informed consent before their participation.
Author contributions
SK and TB designed the study, analyzed the data, and wrote the manuscript. SK performed the experiments. YS, FW, MK, and TV assisted in experiments and participated in the manuscript preparation. WEK supported the acquisition and analysis of flow cytometry data. BW, CG, GM, MS, SM, and AY cooperated with regard to human samples and KH with regard to mouse strains. TS and TA performed parts of the patient data and the survival analyses, and HGR and MR contributed to the development of the project by fruitful discussion.
Supplementary Material
Acknowledgments
We thank Ulrike Schmidt, Stefanie Müller, Synia Haub, Natalie Mucha, Birgit Fehrenbacher, Renate Nordin, Eva Müller-Hermelink, and Susanne Bogner for excellent technical assistance. We gratefully acknowledge Phil Cheng and Mitch Levesque for providing the UZH Cancer Browser. We thank A. Siegel from the Department of Internal Medicine IV, University of Tübingen, for measuring the Luminex assay; Axel Roers for providing the Mcpt5-cre+ and Mcpt5-cre+-RDTA mice; and Christian Posch for critically reading the manuscript. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 685 A06, SFB 824 B10, BI696/10-1, SPP1394 BI696/5-2, SFB156 B06, SFB 1335 P17, SFB1371 P06), the Helmholtz Gesellschaft (KKG “Einheit klinische Allergologie”), the Wilhelm Sander-Stiftung 2012.056.3, and the European Research Council (AdG 339842 MUTAEDITING to HGR).
Version 1. 10/03/2019
Electronic publication
Footnotes
Conflict of interest: MR reports grant funding from AB Science, Abbott, AbbVie, Alcedis, Almirall Hermal, Amgen, AnaptysBio, argenx, AstraZeneca, Bayer, Biogen Idec, Boehringer Ingelheim, Celgene, CureVac, DelArrivo, Deutsche Forschungsgemeinschaft, Deutsche Krebshilfe, Dynavax Technologies, Eli Lilly, GSK, Hoffmann-La Roche, Hokusai, Idera Pharmaceuticals, Ilkos Therapeutic, Immatics Biotechnologies, Incyte, Iovance Biotherapeutics, Janssen-Cilag, Johnson & Johnson, LEO Pharma, MSD Sharp & Dohme, Novartis Pharmaceuticals, PellePharm, Pfizer, Philogen, Regeneron Pharmaceuticals, Sanofi-Aventis, Schering-Plough, Sun Pharma, UCB, Wilhelm Sander-Stiftung, and 4SC; grant funding from and being a shareholder of Bristol-Myers Squibb and Merck; travel support from Deutsche Dermatologische Gesellschaft, European Academy of Dermatology and Venereology, and International League of Dermatological Societies; grant support from and serving as a consultant for Galderma; and honoraria as a consultant for Institut für medizinische und pharmazeutische Prüfungsfragen.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(19):e125057.https://doi.org/10.1172/jci.insight.125057.
Contributor Information
Susanne Kaesler, Email: susanne.kaesler@tum.de.
Florian Wölbing, Email: florian.woelbing@tum.de.
Yuliya Skabytska, Email: yuliya.skabytska@tum.de.
Martin Köberle, Email: martin.koeberle@tum.de.
Thomas Volz, Email: thomas.volz@tum.de.
Tobias Sinnberg, Email: Tobias.Sinnberg@med.uni-tuebingen.de.
Teresa Amaral, Email: teresa.amaral@med.uni-tuebingen.de.
Gisela Metzler, Email: gisela.metzler@med.uni-tuebingen.de.
Martin Schaller, Email: Martin.Schaller@med.uni-tuebingen.de.
Karin Hartmann, Email: karin.hartmann@usb.ch.
Benjamin Weide, Email: benjamin.weide@med.uni-tuebingen.de.
Claus Garbe, Email: Claus.Garbe@med.uni-tuebingen.de.
Martin Röcken, Email: martin.roecken@med.uni-tuebingen.de.
Tilo Biedermann, Email: tilo.biedermann@tum.de.
References
- 1.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14(8):561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 2.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14(8):463–482. doi: 10.1038/nrclinonc.2017.43. [DOI] [PubMed] [Google Scholar]
- 3.Callahan MK, Postow MA, Wolchok JD. Targeting T cell co-receptors for cancer therapy. Immunity. 2016;44(5):1069–1078. doi: 10.1016/j.immuni.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 4.Lin WM, Fisher DE. Signaling and immune regulation in melanoma development and responses to therapy. Annu Rev Pathol. 2017;12:75–102. doi: 10.1146/annurev-pathol-052016-100208. [DOI] [PubMed] [Google Scholar]
- 5.Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Attia P, et al. Autoimmunity correlates with tumor regression in patients with metastatic melanoma treated with anti-cytotoxic T-lymphocyte antigen-4. J Clin Oncol. 2005;23(25):6043–6053. doi: 10.1200/JCO.2005.06.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beck KE, et al. Enterocolitis in patients with cancer after antibody blockade of cytotoxic T-lymphocyte-associated antigen 4. J Clin Oncol. 2006;24(15):2283–2289. doi: 10.1200/JCO.2005.04.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camacho LH, et al. Phase I/II trial of tremelimumab in patients with metastatic melanoma. J Clin Oncol. 2009;27(7):1075–1081. doi: 10.1200/JCO.2008.19.2435. [DOI] [PubMed] [Google Scholar]
- 10.Dubin K, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaput N, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 12.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivan A, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vétizou M, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viaud S, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976. doi: 10.1126/science.1240537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 18.Padawer J. Mast cells: extended lifespan and lack of granule turnover under normal in vivo conditions. Exp Mol Pathol. 1974;20(2):269–280. doi: 10.1016/0014-4800(74)90059-8. [DOI] [PubMed] [Google Scholar]
- 19.Maciel TT, Moura IC, Hermine O. The role of mast cells in cancers. F1000Prime Rep. 2015;7:09. doi: 10.12703/P7-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galli SJ, Borregaard N, Wynn TA. Phenotypic and functional plasticity of cells of innate immunity: macrophages, mast cells and neutrophils. Nat Immunol. 2011;12(11):1035–1044. doi: 10.1038/ni.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marichal T, Tsai M, Galli SJ. Mast cells: potential positive and negative roles in tumor biology. Cancer Immunol Res. 2013;1(5):269–279. doi: 10.1158/2326-6066.CIR-13-0119. [DOI] [PubMed] [Google Scholar]
- 22.Dudeck A, et al. Mast cells as protectors of health. J Allergy Clin Immunol. 2018;S0091-6749(18):31605-1. doi: 10.1016/j.jaci.2018.10.054. [DOI] [PubMed] [Google Scholar]
- 23.Piliponsky AM, Romani L. The contribution of mast cells to bacterial and fungal infection immunity. Immunol Rev. 2018;282(1):188–197. doi: 10.1111/imr.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biedermann T, et al. Mast cells control neutrophil recruitment during T cell-mediated delayed-type hypersensitivity reactions through tumor necrosis factor and macrophage inflammatory protein 2. J Exp Med. 2000;192(10):1441–1452. doi: 10.1084/jem.192.10.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dudeck A, et al. Mast cells are key promoters of contact allergy that mediate the adjuvant effects of haptens. Immunity. 2011;34(6):973–984. doi: 10.1016/j.immuni.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 26.Echtenacher B, Männel DN, Hültner L. Critical protective role of mast cells in a model of acute septic peritonitis. Nature. 1996;381(6577):75–77. doi: 10.1038/381075a0. [DOI] [PubMed] [Google Scholar]
- 27.Malaviya R, Ikeda T, Ross E, Abraham SN. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996;381(6577):77–80. doi: 10.1038/381077a0. [DOI] [PubMed] [Google Scholar]
- 28.Kneilling M, et al. Targeted mast cell silencing protects against joint destruction and angiogenesis in experimental arthritis in mice. Arthritis Rheum. 2007;56(6):1806–1816. doi: 10.1002/art.22602. [DOI] [PubMed] [Google Scholar]
- 29.Rigoni A, Colombo MP, Pucillo C. The role of mast cells in molding the tumor microenvironment. Cancer Microenviron. 2015;8(3):167–176. doi: 10.1007/s12307-014-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, et al. Immune-checkpoint inhibitor-induced diarrhea and colitis in patients with advanced malignancies: retrospective review at MD Anderson. J Immunother Cancer. 2018;6(1):37. doi: 10.1186/s40425-018-0346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat Rev Immunol. 2003;3(2):169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- 32.Brass DM, Savov JD, Whitehead GS, Maxwell AB, Schwartz DA. LPS binding protein is important in the airway response to inhaled endotoxin. J Allergy Clin Immunol. 2004;114(3):586–592. doi: 10.1016/j.jaci.2004.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Kalialis LV, Drzewiecki KT, Klyver H. Spontaneous regression of metastases from melanoma: review of the literature. Melanoma Res. 2009;19(5):275–282. doi: 10.1097/CMR.0b013e32832eabd5. [DOI] [PubMed] [Google Scholar]
- 34.Mackensen A, et al. Evidence for in situ amplification of cytotoxic T-lymphocytes with antitumor activity in a human regressive melanoma. Cancer Res. 1993;53(15):3569–3573. [PubMed] [Google Scholar]
- 35.McGovern VJ. Spontaneous regression of melanoma. Pathology. 1975;7(2):91–99. doi: 10.3109/00313027509092702. [DOI] [PubMed] [Google Scholar]
- 36.Dwyer DF, Barrett NA, Austen KF, Immunological Genome Project Consortium Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat Immunol. 2016;17(7):878–887. doi: 10.1038/ni.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsushima H, Yamada N, Matsue H, Shimada S. TLR3-, TLR7-, and TLR9-mediated production of proinflammatory cytokines and chemokines from murine connective tissue type skin-derived mast cells but not from bone marrow-derived mast cells. J Immunol. 2004;173(1):531–541. doi: 10.4049/jimmunol.173.1.531. [DOI] [PubMed] [Google Scholar]
- 38.Kulka M, Metcalfe DD. TLR3 activation inhibits human mast cell attachment to fibronectin and vitronectin. Mol Immunol. 2006;43(10):1579–1586. doi: 10.1016/j.molimm.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 39.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11(5):373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 40.Liu M, et al. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22(3):121–130. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiniker SM, et al. A prospective clinical trial combining radiation therapy with systemic immunotherapy in metastatic melanoma. Int J Radiat Oncol Biol Phys. 2016;96(3):578–588. doi: 10.1016/j.ijrobp.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodi FS, et al. Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2-year overall survival outcomes in a multicentre, randomised, controlled, phase 2 trial. Lancet Oncol. 2016;17(11):1558–1568. doi: 10.1016/S1470-2045(16)30366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long GV, et al. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N Engl J Med. 2017;377(19):1813–1182. doi: 10.1056/NEJMoa1708539. [DOI] [PubMed] [Google Scholar]
- 44.Wolchok JD, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2017;377(14):1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hölzel M, Bovier A, Tüting T. Plasticity of tumour and immune cells: a source of heterogeneity and a cause for therapy resistance? Nat Rev Cancer. 2013;13(5):365–376. doi: 10.1038/nrc3498. [DOI] [PubMed] [Google Scholar]
- 46.Galli SJ, Tsai M. IgE and mast cells in allergic disease. Nat Med. 2012;18(5):693–704. doi: 10.1038/nm.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cildir G, Pant H, Lopez AF, Tergaonkar V. The transcriptional program, functional heterogeneity, and clinical targeting of mast cells. J Exp Med. 2017;214(9):2491–2506. doi: 10.1084/jem.20170910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurish MF, Austen KF. Developmental origin and functional specialization of mast cell subsets. Immunity. 2012;37(1):25–33. doi: 10.1016/j.immuni.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 49.Moon TC, Befus AD, Kulka M. Mast cell mediators: their differential release and the secretory pathways involved. Front Immunol. 2014;5:569. doi: 10.3389/fimmu.2014.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elieh Ali Komi D, Grauwet K. Role of mast cells in regulation of T cell responses in experimental and clinical settings. Clin Rev Allergy Immunol. 2018;54(3):432–445. doi: 10.1007/s12016-017-8646-z. [DOI] [PubMed] [Google Scholar]
- 51.Visciano C, et al. Mast cells induce epithelial-to-mesenchymal transition and stem cell features in human thyroid cancer cells through an IL-8-Akt-Slug pathway. Oncogene. 2015;34(40):5175–5186. doi: 10.1038/onc.2014.441. [DOI] [PubMed] [Google Scholar]
- 52.Samoszuk M, Kanakubo E, Chan JK. Degranulating mast cells in fibrotic regions of human tumors and evidence that mast cell heparin interferes with the growth of tumor cells through a mechanism involving fibroblasts. BMC Cancer. 2005;5:121. doi: 10.1186/1471-2407-5-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lázár-Molnár E, et al. Inhibition of human primary melanoma cell proliferation by histamine is enhanced by interleukin-6. Eur J Clin Invest. 2002;32(10):743–749. doi: 10.1046/j.1365-2362.2002.01020.x. [DOI] [PubMed] [Google Scholar]
- 54.Shikotra A, Ohri CM, Green RH, Waller DA, Bradding P. Mast cell phenotype, TNFα expression and degranulation status in non-small cell lung cancer. Sci Rep. 2016;6:38352. doi: 10.1038/srep38352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oldford SA, Haidl ID, Howatt MA, Leiva CA, Johnston B, Marshall JS. A critical role for mast cells and mast cell-derived IL-6 in TLR2-mediated inhibition of tumor growth. J Immunol. 2010;185(11):7067–7076. doi: 10.4049/jimmunol.1001137. [DOI] [PubMed] [Google Scholar]
- 56.Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. 2017;46(4):562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271–285. doi: 10.1038/nrc.2017.13. [DOI] [PubMed] [Google Scholar]
- 58.Tanese K, Grimm EA, Ekmekcioglu S. The role of melanoma tumor-derived nitric oxide in the tumor inflammatory microenvironment: its impact on the chemokine expression profile, including suppression of CXCL10. Int J Cancer. 2012;131(4):891–901. doi: 10.1002/ijc.26451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eisenhauer EA, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 60.Scholten J, et al. Mast cell-specific Cre/loxP-mediated recombination in vivo. Transgenic Res. 2008;17(2):307–315. doi: 10.1007/s11248-007-9153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rebholz B, et al. Crosstalk between keratinocytes and adaptive immune cells in an IkappaBalpha protein-mediated inflammatory disease of the skin. Immunity. 2007;27(2):296–307. doi: 10.1016/j.immuni.2007.05.024. [DOI] [PubMed] [Google Scholar]
- 62.Kaesler S, et al. Effective T-cell recall responses require the taurine transporter Taut. Eur J Immunol. 2012;42(4):831–841. doi: 10.1002/eji.201141690. [DOI] [PubMed] [Google Scholar]
- 63.Grimbaldeston MA, Chen CC, Piliponsky AM, Tsai M, Tam SY, Galli SJ. Mast cell-deficient W-sash c-kit mutant Kit W-sh/W-sh mice as a model for investigating mast cell biology in vivo. Am J Pathol. 2005;167(3):835–848. doi: 10.1016/S0002-9440(10)62055-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.