Abstract
The transformational impact of bioorthogonal chemistries has inspired new strategies for the in vivo synthesis of bioactive agents through non-natural means. Among these, palladium (Pd) catalysts have played a prominent role in the growing subfield of bioorthogonal catalysis by producing xenobiotics and uncaging biomolecules in living systems. However, delivering catalysts selectively to specific cell types still lags behind catalyst development. Here we have developed a bio-artificial device consisting of cancer-derived exosomes loaded with Pd catalysts by a method that enables the controlled assembly of Pd nanosheets directly inside the vesicles. This hybrid system mediates Pd-triggered dealkylation reactions in vitro and inside cells and displays preferential tropism for their progenitor cells. The use of Trojan exosomes to deliver abiotic catalysts into designated cancer cells creates the opportunity for a new targeted therapy modality: exosome-directed catalyst prodrug therapy, whose first steps are presented herein with the cell-specific release of the anticancer drug panobinostat.
Introduction
In recent years, transition metals conventionally confined to the chemistry lab (e.g. Pd, Ru, Au) have been shuttled into living cells, tissues and animals in pursuit of tools capable of catalysing first-in-life reactions.1–31 From organometallic complexes,1–13 artificial metalloenzymes14–16 and metal-loaded nanocarriers17–24 to larger-than-cells implantable devices,25–31 a selected number of agents have demonstrated catalytic activity and maintained their functional compatibility with the biological milieu. Although the development of effective intracellular catalysts based on abiotic metals remains challenging, various examples reported in the literature have tested the feasibility of this concept.4–9,16–24,32 One of the most exciting opportunities of this active field of research is, however, still in its infancy: the development of devices that not only display bioorthogonal catalytic activity but are also precisely delivered to specific anatomical locations, e.g. inside tumors.21,23,30 First attempts to achieve this have explored physical (EPR effect, magnetism)21,23 and surgical30 approaches, but intercellular trafficking pathways have not yet been investigated. In principle, natural cell-specific delivery vectors such as exosomes33 could be ideally suited to carry miniaturized catalytic reactors to individual cancerous cells.
Exosomes are membrane-enclosed vesicles released by cells to the extracellular space to mediate intercellular exchange of biomolecules and thereby regulate a variety of physiological functions.33–35 The biochemical composition of the exosomal membrane is reflective of the donor cell, thus conferring on the exosomes a preferential tropism for cells that are parented to their cell of origin.34,35 In the tumour microenvironment, exosomes are believed to play a fundamental role in the communication between cancer cells to orchestrate cancer progression, invasion, angiogenesis and immune regulation.35–37 Thanks to their long circulating half-life, small size, low immunogenicity and ability to preferentially target particular cell types, exosomes exhibit ideal characteristics as vectors to deliver a therapeutic cargo (such as drugs, biomolecules or hyperthermia-capable nanoparticles) to specific cells.38,39 However, introducing large molecules or nanoparticles inside exosomes is challenging because the methods often used (e.g. electroporation, sonication, etc.) can disrupt the exosomal membrane and compromise their targeting ability.
Here, motivated by the possibility of using exosomes as Trojan horses,34,38–41 we aimed to hijack this ancient intercellular communication system to protect and deliver a catalytic cargo into a specific type of cancer cells. To implement this strategy, a mild chemical methodology was first developed to enable the controlled assembly of Pd nanostructures directly inside cancer cell-derived exosomes without compromising their integrity and functionality. The resulting bioartificial vesicles (Pd-Exo) are highly catalytic under physiological conditions, capable of preferentially delivering their catalytic cargo to their target cells and activate an anticancer prodrug inside them by Pd-mediated dealkylation. The concept of using exosome-directed catalyst delivery into designated cell types to mediate bioorthogonal processes in the inner cell space opens up a plethora of possibilities in the fields of chemical biology and biomedicine.
Results
Design, fabrication and characterization of Pd-Exo
The use of exosomes as drug delivery carriers has attracted increasing attention from academics and industry in recent years.42 Various methods have been developed to ship a diverse range of therapeutic agents inside exosomes, including small molecule drugs, proteins, RNA molecules, and more complex cargoes such as modified viruses and gene-editing tools.43–50 Rather than loading exosomes with a limited amount of a bioactive agent, we rationalized that the incorporation of transition metal-based catalysts into such a targeted delivery system could be used to build a deliverable nanoreactor able to direct a bioorthogonal catalytic payload towards cancer cells and facilitate amplified production of therapeutic agents directly at the cancer site. The first challenge was to develop a method that allows loading a metal of choice into these vesicles without affecting the exosome targeting properties and preserves the metal’s catalytic activity, since current biophysical techniques for loading exosomes with metallic nanostructures51–55 can induce detrimental defects on the integrity of the exosome membrane.56,57
Encouraged by prior success on the controlled growth of Pd nanosheets using carbon monoxide (CO) as a gaseous reducing agent under wet conditions,58,59 a chemical methodology was investigated to generate Pd nanostructures directly inside exosomes. Figure 1A illustrates the stepwise production of Pd-ExoA549 (see full procedure in the Methods). After purification by ultracentrifugation from non-small cell lung carcinoma (NSCLC) A549 cell culture (see Suppl. Fig. 1), cancer-derived exosomes ExoA549 were incubated with K2PdCl4 (water soluble Pd2+ reagent) in PBS at physiological conditions for 12 h. This was then followed by a second ultracentrifugation step (to remove extracellular Pd2+ species) and 40-min treatment under a CO atmosphere (6 bar) and gentle stirring at low temperature (30 °C, to prevent protein denaturation) to mediate the in situ reduction of Pd2+ to Pd0 and self-assembly into stable Pd nanostructures. Procedural conditions (temperature, incubation times, CO pressure, etc.) were optimized for each step of the protocol to enhance the diffusion of Pd2+ species and reductant gas through the exosome membrane, promoting Pd nanoparticle formation without causing functional defects to the exosomes. The choice of a mild antioxidant such as CO was addressed to minimize side reactions, since most reductants capable of reducing Pd2+ species can potentially react with functional groups present in most biomolecules, including proteins and lipids of the exosome membrane.
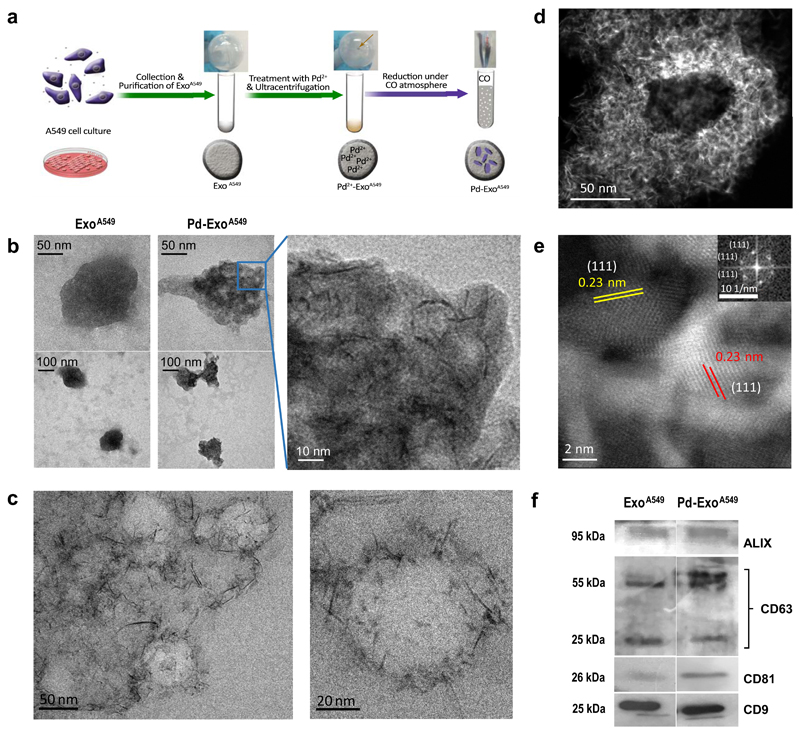
Figure 1. Preparation and characterization of Pd-functionalised exosomes.
(a) Overview of the step-wise protocol to generate Pd-ExoA549 from A549 cells. (b) Representative TEM images of ExoA549 and Pd-ExoA549 at different magnifications. (c) Representative cryo-TEM images of Pd-ExoA549 at two magnifications. (d) HRSTEM-HAADF image of a representative Pd-ExoA459. (e) HRSTEM-HAADF zoomed-in image showing highly-crystalline Pd nanostructures with a lattice spacing of 0.23 nm, which correspond to Pd (1 1 1)-surfaces. Inset shows the FFT spectrum generated from the image. (f) Western blots of exosome-specific biomarkers ALIX, CD63, CD81 and CD9 of ExoA549 and Pd-ExoA549.
Exosomes were characterized before and after treatment by different procedures, including physicochemical (transmission electron microscopy (TEM), cryo-TEM, high-resolution scanning TEM-high angle annular dark field (HRSTEM-HAADF), inductively coupled plasma mass spectrometry (ICP-MS), X-ray spectroscopy, and zeta potential nanoparticle tracking analysis (NTA)) as well as biological (Western Blot assays against different protein markers, total protein content) techniques. Rounded vesicles with an average diameter of 100-140 nm were observed from both samples, whereas dark nanostructures (Pd) with laminar shape were only visible inside Pd-ExoA549 (Figure 1B). Importantly, all Pd-ExoA549 contained such nanostructures and minimal alterations were observed in the size and morphology of the vesicles. Due to the benefits of vesicle cryopreservation, cryo-TEM permitted visualization of the native spherical morphology of Pd-ExoA549 and identified the presence of numerous Pd-nanosheets that can be observed mainly clustered around the exosomal membrane. HRSTEM-HAADF was then employed to study the crystallinity of the Pd nanosheets inside exosomes (Figure 1D,E), revealing lattice fringes with d-spacing of 0.23 nm that match with metallic Pd (1 1 1)-surfaces.60 The Fast Fourier Transform spectrum generated from this image (see inset of Fig 1E) corroborated this observation. This agrees with the expected dual role of CO as an electron donor to reduce Pd2+ ions to Pd0 atoms and as a capping agent to control the confined growth of Pd structures into a planar shape due to its preferential adsorption to basal {111}.58 High resolution imaging showed the Pd nanostructures displayed an oriented 2D configuration (nanosheets) with a thickness of approximately 1.4 nm. The amount of Pd inside the vesicles was quantified by ICP-MS (ElanDRC-e, PerkinElmer) and normalized to the total protein amount of Pd-ExoA549, providing an average Pd content of 0.64 μg of Pd / μg of Pd-ExoA549 protein. UV-VIS spectral analysis showed a maximal absorbance peak in the near infrared range (Supp. Fig. 3), which is characteristic of Pd nanosheets58, while energy-dispersive X-ray spectroscopy analysis (Suppl. Fig. 4) and X-ray photoelectron spectroscopy (XPS, see Suppl. Fig. 5)24 confirmed the presence of Pd0 inside the exosomes. Analysis of the atomic environment for N 1s detected no changes between ExoA549 and Pd-ExoA549, suggesting that the employed methodology does not alter protein composition.
To further study if membrane proteins had been altered during the chemical procedure, the presence of exosome-specific antigens CD9, CD63, CD81 and ALIX were characterized by western blot. As shown in Figure 1F, similar protein levels were observed before and after Pd loading, indicating that the mild reductive conditions employed to reduce Pd2+ species do not degrade membrane proteins. Under native (non-denaturing) conditions, same bands were observed for both samples, but their migration rate showed clear differences (see Suppl. Fig. 6). The protein bands from Pd-ExoA549 migrated at a reduced rate and slightly towards the left, indicating a change in charge and mass. This artefact may be consequence of the Pd-ExoA549 lysing step during sample processing and attributed to coordination with free Pd species.61 The low impact of the process to the protein composition of the exosomes is further supported by the XPS data (Suppl. Fig. 6) and the zeta potential analysis (Suppl. Fig. 7). Also, bicinchoninic acid assay showed that the CO treatment did not affect total exosomal protein content. Protein concentrations of 101.45 and 102.95 μg/mL were respectively obtained for ExoA549 and CO-treated ExoA549.
The stability of this hybrid system was studied by analysis of particle size and zeta potential of ExoA549 and Pd-ExoA549 at different time points using a Nanosight NS500 (Malvern Instruments Ltd, UK) and a Zeta Plus (Brookhaven Instruments Corp., USA), respectively. As shown in Suppl. Fig. 7, average particle diameters of the exosomes before and after NP generation were equivalent (146.5 and 155 nm, respectively) and the size and zeta potential of Pd-ExoA549 remain relatively stable after 72 h at room temperature, indicating that Pd-loaded exosomes are sufficiently stable to perform reproducible in vitro and cell-based studies.
Pd-ExoA549 as catalytic nanoreactors
The catalytic properties of Pd-ExoA549 were evaluated with the Pd-sensitive off-on sensor 1, which was designed to release the membrane-permeant red-fluorescent dye resorufin (2) upon a single O-depropargylation reaction (see Figure 2A). The lipophilicity of both 1 and 2 facilitates back-and-forth diffusion across the exosome membrane before and after activation, thus facilitating reaction monitoring by fluorometric analysis of the reaction medium. Compound 1 was synthesized by reacting 2 with propargyl bromide and DBU in DMF at room temperature. As shown in Suppl. Fig 8, 1 shows negligible fluorescence emission under white LED excitation (>200-fold difference compared to that of dye 2 at 590 nm), a consequence of blocking the electronic conjugation of the system by alkylation of the phenolic OH.
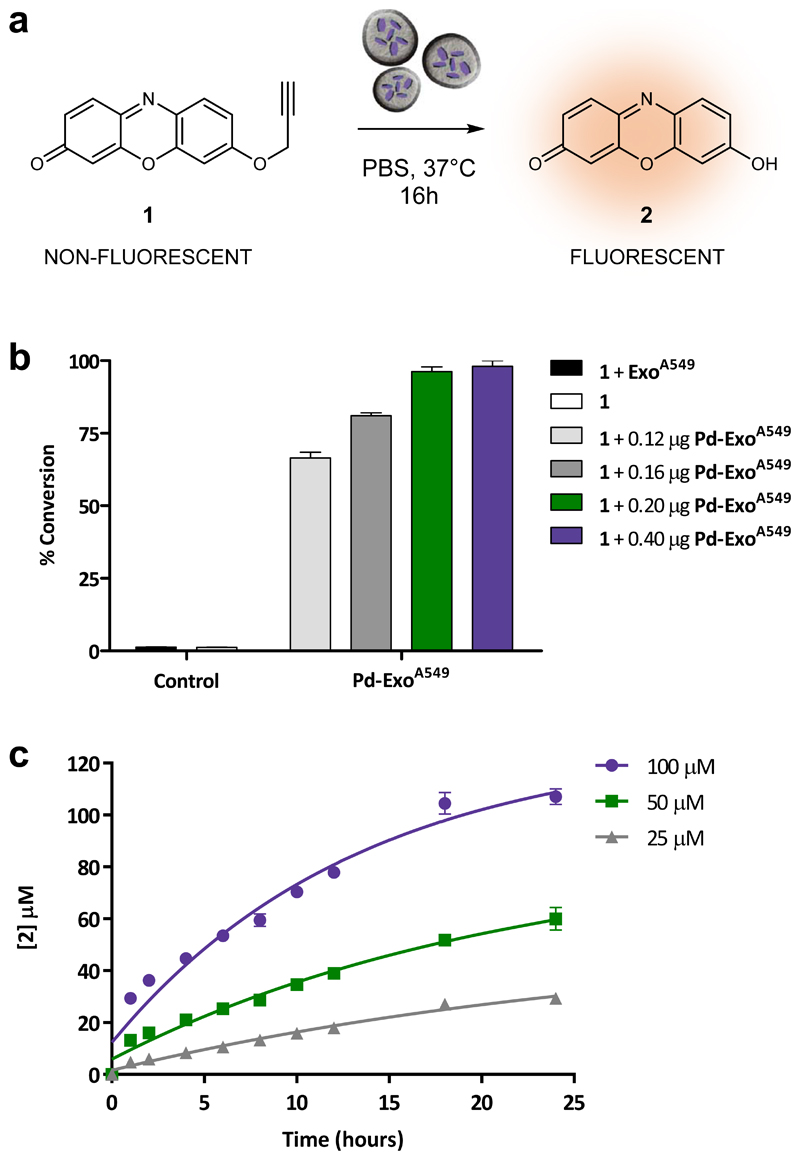
Figure 2. Study of the catalytic properties of Pd-ExoA549.
(a) Conversion of non-fluorescent 1 into highly red-fluorescent resorufin 2 mediated by Pd-ExoA549 under biocompatible conditions (37 °C, PBS, pH 7.4, isotonicity). (b) Conversion efficiency after 16 h incubation of 1 (100 μM) with Pd-ExoA549 (0.12, 0.16, 0.2 and 0.4 μg / 100 μL). Conversion values (%) were calculated from fluorescence intensity measurements at λex/em= 540/590 nm using the fluorescence intensity of 2 (100 μM) as 100 %. Negative controls: 1 (100 μM) with or without ExoA549 (0.2 μg/100 μL). Error bars: ±SD, n=3. (c) Kinetic study of the reaction of Pd-ExoA549 (0.2 μg/100μL) with different concentrations of 1 (25, 50 and 100 μM) in PBS at 37 °C. Concentration values of product 2 were calculated from fluorescence intensity measurements at λex/em= 540/590 nm. Curves fit: non-linear exponential equation. Error bars: ±SD, n=3.
Pd-ExoA549 were incubated at different concentrations (0.12, 0.16, 0.2 and 0.4 μg/100μL) with non-fluorescent compound 1 (100 μM) in PBS and 37 °C for 16 h. Note that, to be able to study and compare the activity of vesicles loaded or not with Pd, the concentration values used in the experiments are expressed as μg of exosome protein /100 μL. The fluorescence intensity of the samples was measured with a spectrofluorometer (Ex/Em: 540/590 nm) and values compared to that of 2 at 100 μM (= 100% conversion). Incubation of sensor 1 (100 μM) alone or in the presence of ExoA549 (0.2 μg / 100μL) were used as negative controls. Treatment of 1 with Pd-ExoA549 resulted in a dramatic increase of fluorescent emission, with the top concentrations of 0.2 and 0.4 μg / 100μL yielding near to complete conversion (>97% yield after 16 h, see Figure 2B). In contrast, sensor 1 was stable on its own and in the presence of ExoA549 (no fluorescence detected). Time lapse imaging of the reaction illustrates the fast conversion rate of non-fluorescent 1 into highly red fluorescent 2 in the presence of Pd-loaded exosomes (see Supp. Video 1).
To further study the kinetics of the process, three concentrations of substrate 1 (25, 50 and 100 μM) were treated with Pd-ExoA549 (0.2 μg/100 μL) at the same conditions and the formation of dye 2 monitored by fluorometry at different timepoints. As shown in Figure 2C, the rate of product formation followed a classical Michaelis Menten growth curve and all reactions were completed before 24 h. Kinetic parameters were calculated by plotting the natural log of the concentration of substrate ([1]) versus time (Suppl. Fig. 9), displaying pseudo first-order kinetics with a K = 0.1228 ±0.0049 s-1 and half-life of 5.64 h at the range of concentrations used. These results agree with the O-depropargylation rate found for previously-reported Pd-based heterogeneous catalysts.25–30 The reusability of Pd-ExoA549 was then tested by recycling (centrifugation) the vesicles after reaction with 1, followed by re-treatment with 1. Repetition of this process 3 times verified the recyclability of these catalytic vesicles (Supp. Fig. 10), although a moderate reduction of activity (45% and 20% reduction yield in cycles 2 and 3, respectively) was observed after each cycle, which we attribute to a loss of material due to incomplete recovery by the centrifugation process.
Pd-ExoA549 biocompatibility and intracellular entry
The tolerability of human cells to exposure to Pd-loaded exosomes was determined by incubating increasing quantities of Pd-ExoA549 with lung cancer A549 cells for 5 d. Cell viability assays showed that Pd-ExoA549 induce minimal effect on cell proliferation even at the top dose of 0.6 μg / 100μL (see Supp. Fig. 11).
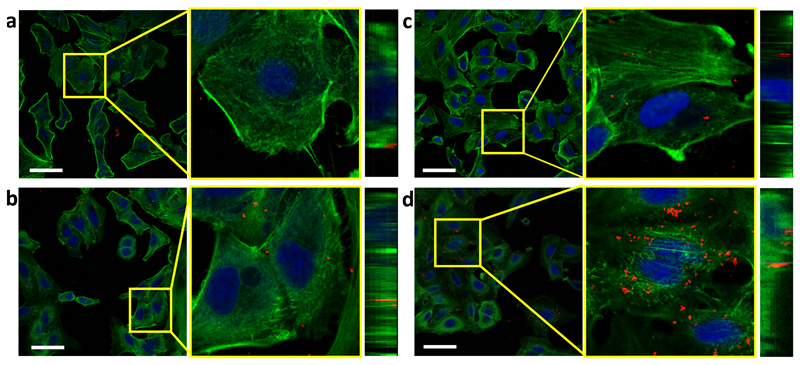
Next, the internalization of Pd-ExoA549 into their parental cells A549 was studied by confocal microscopy in search for the optimal incubation time to perform intracellular catalytic experiments. After treating A549 cells with Pd-ExoA549 under standard cell culture conditions, cells were fixed and labelled with phaloidin-488 (actin fibres) and Draq5 (nuclei). Due to the inherent reflective optical properties of the Pd nanosheets, clusters of Pd-ExoA549 were directly observed by reflection of the incident light in the confocal microscope at 488/490 nm excitation/emission wavelengths (identified in Figure 3 as red dots). Representative images from A549 cells after treatment with Pd-ExoA549 for 1, 2, 4 and 6 h are shown in Figure 3 (see study at 24 h in Suppl. Fig. 12). Quantitative analysis of the red fluorescent signal confirmed that the highest cytoplasmic presence of Pd-loaded vesicles was reached after 6 h incubation of cells with Pd-ExoA549 (Suppl. Fig. 13). Z-stack sections of the images at 6 h confirmed the intracellular localization of Pd-ExoA549 aggregates (Figure 3), while co-localization studies revealed that the majority of Pd-nanosheets were not in lysosomal compartments at that time point (Suppl. Fig. 14).
Figure 3. Confocal study of Pd-ExoA549 internalization in A549 cells.
Merged confocal images of A549 cells after treatment with Pd-ExoA549 for 1 h (a), 2 h (b), 4 h (c) and 6 h (d). Actin fibres (phaloidin-488) are shown in green, cell nuclei (Draq5) in blue and clusters of Pd-ExoA549 in red. Scale bar= 50 μm. The middle and right images show the amplified and z-stacks sections, respectively.
Targeted intracellular prodrug activation mediated by Pd-ExoA549
The pan-histone deacetylase (HDAC) inhibitor panobinostat (3) received clinical approval to treat multiple myeloma in 2015,62 and is currently in clinical trials for the treatment of a variety of cancers including NSCLC. Despite its broad activity against many cancer types, dose-limiting systemic toxicities including diarrhoea, cardiac ischemic events, arrhythmias and myelosuppression, limit the use of panobinostat and other HDAC inhibitors against solid tumours. Selective delivery and localized activation of this class of epigenetic targeting drugs at cancer sites have been proposed to enhance their clinical tolerability and antitumor efficacy, with different prodrug strategies for medically-used vorinostat and belinostat being recently showcased in the literature.63,64 However, prodrugs of 3 have not yet been reported, perhaps due to its higher structural complexity.
At this point of the investigation, Pd-ExoA549 had shown to be catalytic under physiological conditions, harmless to cells and capable of entering A549 cells. To further challenge the multifunctional capacities of the nanodevices, it was essential to determine if Pd-ExoA549 were also able to mediate the bioorthogonal uncaging of a bioactive substance inside their target cells. To test this, a Palladium-activatable prodrug of 3 was developed (see Figure 4A). Given the essential role of the Zn-chelating hydroxamate group of 3 in its cytotoxic mode of action,64 the O-alkyl hydroxamate derivative 4 was designed to reduce its capacity to inhibit HDACs while, at the same time, making it sensitive to Pd chemistry.
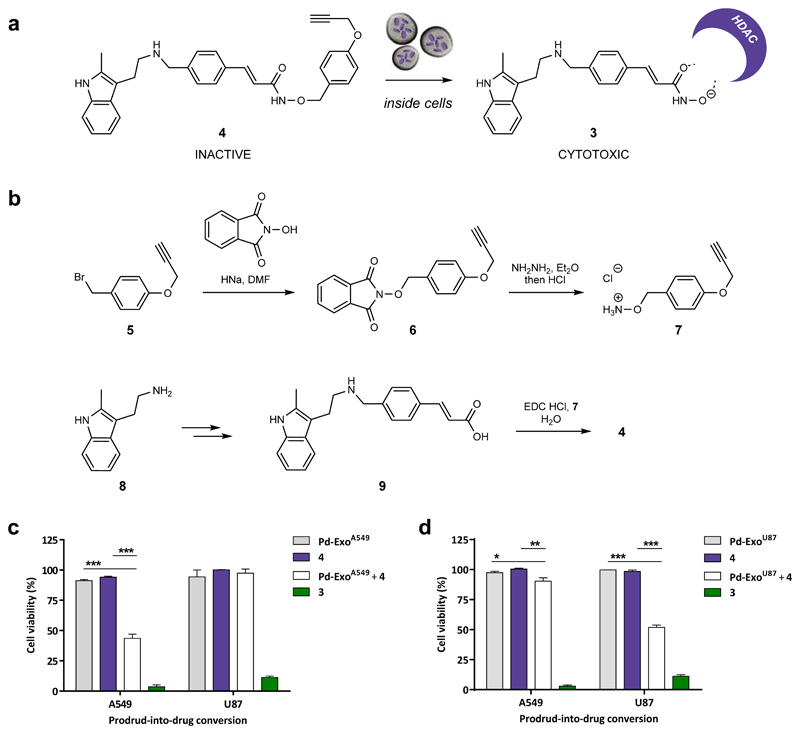
Figure 4. Design and synthesis of prodrug 4 and targeted intracellular activation mediated by Pd-ExoA549 and Pd-ExoU87 in A549 and U87 cells.
(a) Intracellular Pd-ExoA549-mediated conversion of prodrug 4 into clinically-approved HDAC inhibitor 3. (b) Total synthesis of compound 4. (c) Pd-ExoA549-mediated uncaging of 4 inside cells. A549 and U87 cells were incubated for 6 h with 0.4 and 0.53 μg / 100 μL, respectively. Cells were thoroughly washed to eliminate extracellular vesicles followed by addition of prodrug 4 (0.2 μM). Controls: Pd-ExoA549 only (–ve control, grey); prodrug 4 only (–ve control, purple); 3 (+ve control, green). Cell viability was measured at day 5 using PrestoBlue. Error bars: ± SEM, n = 3. (d) Pd-ExoU87-mediated uncaging of 4 inside cells. A549 and U87 cells were incubated for 6 h with 0.4 and 0.53 μg / 100 μL, respectively. Cells were thoroughly washed to eliminate extracellular vesicles followed by addition of prodrug 4 (0.2 μM). Controls: Pd-ExoU87 only (–ve control, grey); prodrug 4 only (–ve control, purple); 3 (+ve control, green). Cell viability was measured at day 5 using PrestoBlue. Error bars: ± SEM, n = 3.
Direct attempts to alkylate the OH group of 3 with p-(propargyloxy)benzyl bromide64 were unsuccessful due the presence of various nucleophilic groups in the molecule. Consequently, compound 4 was prepared by total synthesis following the scheme summarized in Figure 4B. Briefly, p-(propargyloxy)benzyl bromide (5) was reacted with N-hydroxyphthalimide in the presence of HNa to afford intermediate 6, which was converted into hydroxylamine 7 by hydrazinolysis. β-Substituted-acrylic acid 9 was prepared by reductive amination of 3-(4-formyl-phenyl)-acrylic acid methyl ester with 2-methyltryptamine (8)65 followed by basic hydrolysis. Carbodiimide-mediated coupling of 7 and 9 resulted in the generation of prodrug 4 in moderate yield.
To evaluate the efficiency of the masking strategy, the antiproliferative properties of prodrug 4 and unmodified drug 3 were assessed against NSCLC A549 and glioblastoma U87 cells. Cells were treated for 5 d with compounds at concentrations ranging from 0.003 to 100 μM and cell viability determined with the PrestoBlue reagent. Comparison between the dose response curves of 3 and 4 evidenced that the alkylation of the hydroxamate group leads to a significant reduction of the bioactivity of the compound in both cell lines (see Suppl. Fig. 15).
Next, a cell-based assay was performed to determine if the devices have preferential tropism for their progenitor cells66 by comparing their capacity to mediate bioorthogonal uncaging reactions in different cell types. Two cell lines were used for the experiment: the parental cancer cells A549 (target cells) and unparented glioblastoma cells U87 (non-target cells). To maximise cell entry, A549 and U87 cells were incubated with Pd-ExoA549 for 6 h at 0.4 or 0.53 μg / 100μL, respectively. Note that higher Pd-ExoA549 concentration was used for U87 cells to normalise the quantity of vesicles per number of seeded cells. After several washing steps to remove non-internalised vesicles, cells were treated with prodrug 4 for 5 d (0.2 μM; activation assay), aiming to observe a reduction in cell viability resulting from the bioorthogonal release of cytotoxic 3 in cells containing catalytically-active Pd-ExoA549. Cells separately treated with 4 (0.2 μM) or Pd-ExoA549 were used as negative controls, and treatment with 3 (0.2 μM) used as positive control. As observed in Figure 4C, A549 cells sequentially treated with Pd-ExoA549 and 4 exhibited significantly lower cell viability than the negative controls, evidence of the generation of cytotoxic levels of 3 inside cells and, therefore, of the presence of catalytically-active Pd-ExoA549. Additional negative controls further confirmed that the observed cytotoxic activity was driven by Pd mediated drug uncaging (Suppl. Fig. 16 and 17). In contrast, no effect was observed on the viability of U87 cells after the same treatment, indicating that the lower uptake or the reduced catalytic activity of Pd-ExoA549 in U87 cells results in insufficient intracellular production of drug 3.
To study if the reverse situation was also true, i.e. superior uptake of catalytically-active Pd-ExoU87 by U87 cells relative to A549 cells leads to higher prodrug-into-drug conversion levels and cell death, Pd-ExoU87 were generated from U87 cells following the methodology described before. After ICP-MS analysis and testing their catalytic properties in vitro (see Suppl. Fig. 19), A549 and U87 cells were sequentially treated with Pd-ExoU87 and 4 using the same conditions and controls above described, and cell viability measured at day 5. As shown in Fig 4D, results followed the opposite trend to those found with Pd-ExoA549, further supporting that the preferential tropism of exosomes for their cancer cells of origin is important to deliver sufficient Pd nanosheets into cells and produce bioactive concentrations of drug 3.
Additionally, this study was carried out in a third cell line: the monocyte/macrophage-like cell line RAW 264.7. These phagocytic cells were independently treated with Pd-ExoA549 or Pd-ExoU87, followed by treatment with 4. Suppl. Fig. 20 shows that both Pd-Exo / 4 combinations were capable of reducing cell proliferation in this cell line, evidence of in situ drug 3 synthesis, although at lower levels than those achieved in their cell of origin, even if RAW 264.7 cells are much more sensitive to 3. It is important to note that while the higher Pd-Exo internalisation levels would be consistent with the results observed, they are not the only factor in play. Thus, differences in intracellular trafficking routes could also have a significant effect in the exosome-mediated delivery of catalytically-active Pd nanostructures into progenitor cells.
Last, in situ production of 3 was further confirmed by studying intracellular target displacement of coumarin–SAHA, a blue-emitting fluorescent inhibitor of HDAC proteins whose fluorescent properties are quenched when bound to their targets.64 Displacement from their targets by other competitive inhibitor results in an increase of intracellular fluorescence intensity that can be measured by flow cytometry (λex/em = 405/450 nm) and thereby be used as evidence of in situ drug synthesis and target engagement. As shown in Suppl. Fig. 21, cells pre-treated with coumarin-SAHA and Pd-ExoA549 followed by washing and treatment with prodrug 4 resulted in significantly higher fluorescent intensity than in the absence of Pd-ExoA549, further corroborating the intracellular generation of biologically-effective levels of 3 by Palladium chemistry.
Conclusions
The nanoscale bio-hybrid system developed in this work brings together the targeting capabilities of exosomes, a robust intercellular trafficking pathway evolved in nature across millions of years, and the bioorthogonal concept developed by Bertozzi and others67–69 in the last two decades. Cell-targeting catalytic devices can be generated from cancer-derived exosomes using a methodology based on CO-mediated reduction at low temperature to generate ultrathin Pd nanosheets directly inside the vesicles. The mildness of the protocol yielded catalytically active Pd nanostructures that maintained the targeting capabilities of the exosomes. This methodology can be considered —in its own right— as a bioorthogonal process suitable to load metallic structures into biological vesicles.
The development of Pd nanodevices that can perform uncaging chemistries inside cells require robust vectors able to enter cells with high efficacy and compatible with the complex intracellular environment (redox, pH, etc.). Herein we show that Pd-Exo not only display the capacity of entering the cancer cells from where they originated and perform bioorthogonal uncaging reactions, but also the ability to discriminate over other cell types. The intracellular catalytic properties of Pd-ExoA549 were demonstrated by the in situ activation of the anticancer drug panobinostat in lung cancer A549 cells, but not in U87 (a glioma cell line), indicating preferential tropism for their progenitor cells. Opposite results were observed in the reverse situation, i.e. Pd-loaded exosomes originated from U87 cells displayed superior drug-mediated cytotoxic effect in U87 cells than in A549 cells. This proof-of-concept study illustrates the therapeutic potential of combining exosome-mediated delivery of catalysts and bioorthogonal uncaging chemistries to activate bioactive substances in a spatiotemporal selective manner.
Methods
ExoA549 purification and characterization
A549 cells (kind gift from Dr S. Wilkinson; ATTC, CCL-185) or glioma U87 cells (a kind gift from Dr N. Gammoh; ATTC, HTB-14) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10 % fetal bovine serum (FBS), 1 % penicillin/streptomycin and 1 % amphotericin, under normoxic conditions. Neither A549 nor U87 cell lines are included in the register of misidentified cell lines (source: ICLAC database v9, updated on October 2018). Cells were checked for mycoplasma before use. Ultracentrifuged serum (Ultracen medium, 100,000 g, 8 h, 4 ºC) was employed to guarantee exosomes free medium. Exo were collected and purified by successive ultracentrifugation cycles from supernatants of cell culture at confluency (see detailed protocol in the Supplementary Methods).
Preparation of Pd-Exo
The purified exosome fraction was then dispersed in a PBS solution and treated with K2PdCl4 (0.06 mM) at 40 °C for 12 h to maximize the internalization of Pd2+ ions. The mixture was subsequently ultracentrifuged at 35,000 g and 4ºC for 2 h to discard non-internalized Pd species and avoid contamination with extra-exosomal metal nanoparticles during the reduction step. Pd2+-Exo were resuspended in PBS and treated for 40 min to a 6 bar CO atmosphere using a high-pressure Teflon lined autoclave under gentle stirring and 30 °C to reduction of Pd2+ species into Pd0. A light dark colour was immediately observed after CO treatment, inferring formation of Pd nanoparticles. After treatment, CO was desorbed with air to keep the resulting Pd-Exo in a neat environment.
Characterization of ExoA549 and Pd-ExoA549
Phosphotungstic acid (3 %) was used as contrast agent to evaluate the morphology, size and shape of ExosA549 and Pd-ExoA549 by TEM operated at 200 kV with a LaB6 electron source fitted with a SuperTwin® objective lens allowing a point-to-point resolution of 2.4 Å. Analysis of the content specific exosome surface protein was performed by western blot. In brief, 10 μg of exosomes were lysed in RIPA buffer and the protein fraction precipitated with cold acetone (1:1 w/w) at -20 ºC for 2 h. The precipitated fraction was resuspended in Laemmly buffer and boiled at 90 ºC for 10 min. Subsequently, proteins were separated by 12 % SDS-polyacrylamide gel electrophoresis at 100 V for 2 h and transferred to a nitrocellulose membrane at 4 ºC for 4 h. Blots were blocked overnight with TBS-5% milk at 4 ºC and incubated with CD9; 1:2000 (Abcam, UK), CD63; 1:1000 (BD Bioscience, United States), CD81; 1:500 (Santa Cruz Biotechnology, United States) and ALIX; 1:1000 (Cell Signaling Technologies, United States) for 1.5 h. Membranes were washed 3 times with TBS-Tween (TTBS) followed by incubation of the secondary antibody (anti-HRP, Sigma Aldrich). Last, membranes were washed extensively and imagined by chemiluminescence.
Elemental analysis of Pd-ExoA549 was assessed with an EDS (EDAX) detector which allows performing EDS experiments in the scanning mode (FEI Inspect F30). The amount of Pd inside the vesicles was quantified by ICP-MS (ElanDRC-e, PerkinElmer) and normalized to the total protein amount of Pd-ExoA549. For that, samples were digested with 10 % Aqua regia (HNO3 + 3HCl) in 1.5 mL of distilled water. Digestion was performed at rt for 1 h. Calibrations were carried out using Pd standards in 10 % Aqua regia ranging from 0 to 10 ppm. Characterization of the absorbance properties of Pd-ExoA549 was determined by UV-VIS spectroscopy (Jasco V670) to identify the characteristic plasmon at the NIR range of Pd nanosheets. For Cryo-TEM imaging, the grid preparation of cryogenically immobilized Pd-ExoA549 samples required an extremely rapid sample freezing to achieve a vitreous state. A thin film vitrified specimen was prepared with Vitrobot (FEI) in melting ethane. Afterwards, the sample holder with the specimen grid was maintained under cryogenic conditions with liquid nitrogen. Cryo-TEM observations were carried using a T20-FEI microscope with a LaB6 electron source fitted with a SuperTwin® objective lens allowing a point-to-point resolution of 2.4 Å. Total protein amount of native ExoA549 and ExoA549 treated with CO was quantified by Pierce BCA protein assay (Thermo Fisher Scientific, USA) following manufacturer instructions.
Fluorogenic studies
0.6, 0.8, 1 and 2 μg of Pd-ExoA549 were added to a 500 μL solution of reagent 1 (100 μM) in PBS, to obtain a final concentration of 0.12, 0.16, 0.20 and 0.40 μg of Pd-ExoA549/100 μL, respectively. The mixtures were shaken at 700 rpm and 37 °C in a Thermomixer, and reactions were monitored after 16 h using a PerkinElmer EnVision 2101 multilabel reader (Ex/Em: 540 /590 nm). Sensor 1 alone (100 μM) or in the presence of ExoA549 (1 μg of exosomes in 500 μL of PBS; 0.20 μg of ExoA549/100 μL) were used as negative controls and fluorescent dye 2 at 100 μM as reference control. Reactions were done in triplicates and the conversion % calculated according to the reference control.
Time-lapse imaging
The Pd-ExoA549–mediated activation of 1 into 2 was also visualized in real time by time lapse microscopy with a 20x objective (Ex/Em: 560/630 nm, Leica AF6000 LX, Germany), using non-fluorescent 1 alone as negative control and resorufin 2 alone as positive control. 0.5 μg of Pd-ExoA549 were added to a 20 μM solution of 1 in PBS (0.5 mL) in a 24-well plate and incubated for 24 h. Frames were taken every 15 min for 24 h in DIC mode and under fluorescence emission. Videos were created using ImageJ software.
Reusability study
After performing the reaction above described, Pd-ExoA549 were recovered by centrifugation (13,000 rpm, 30 min) and a fresh solution of 1 (100 μM) in PBS added to the recovered fraction of Pd-ExoA549. The mixtures were shaken at 700 rpm and 37 °C in a Thermomixer, and reactions monitored after 16 h using a PerkinElmer EnVision 2101 multilabel reader (Ex/Em: 540 /590 nm). This cycle was repeated 3 times in total.
Study of Pd-ExoA549 biocompatibility
A549 cells were cultured in DMEM supplemented with 10 % of FBS and L-glutamine (2 mM) and maintained in a tissue culture incubator at 37 °C and 5% CO2. Experiments were performed in DMEM supplemented with exosome-depleted FBS (Gibco™) and L-glutamine (2 mM). A549 cells were seeded in a 96-well plate format (at 1,500 cells / well) and incubated for 24 h before treatment. Each well was then replaced with a suspension of Pd-ExoA549 in culture media depleted of exosomes at 0.2, 0.4 and 0.6 μg per 100 μL and incubated for 6 h. Cells were then washed with PBS buffer and fresh media added. After 5 d, PrestoBlue™ cell viability reagent (10 % v/v) was added to each well and the plate incubated for 90 min. Fluorescence emission was detected using a PerkinElmer EnVision 2101 multilabel reader (Ex / Em: 540 / 590 nm). Experiments were performed in triplicates.
Confocal microscopy study of Pd-ExoA549 cell internalization
A549 cells were seeded at a density of 1.5 x 104 onto 20 mm cover slips (in a 24-well plate) and incubated under standard culture conditions for 24 h. 1 μg of Pd-ExoA549 was then added to each well and incubated for 1, 2, 4, 6 and 24 h. Cells were fixed with 4 % paraformaldehyde and stained with phalloidin-Alexa488 (Invitrogen) and Draq-5. Cells were imaged by confocal microscopy (Spectral Confocal Microscope Leica TCA SP2) with a 63x oil immersed N.A. 1.40 objective. Reflection of the incident light at 488/490 was used to directly visualized Pd-ExoA549. Z-stack orthogonal projections were also carried out to enable visualization of exosomes inside cells cytosol.
Cell viability study: 4 vs 3
Antiproliferative activity of 3 and 4 was compared by performing dose-response studies against A549 and U87 cells. Both cell lines were seeded in a 96-well plate format (at 1,500 cells / well for A549 and 2,000 cells / well for U87) and incubated for 24 h before treatment. Each well was then replaced with fresh media, containing compounds 3 and 4 (0.003-100 μM). Untreated cells were incubated with DMSO (0.1 % v/v). After 5 d of incubation, cell viability was determined as described above. All conditions were normalized to the untreated cells (100 %) and curves fitted using GraphPad Prism using a sigmoidal variable slope curve. Experiments were performed in triplicates.
Intracellular prodrug activation study
A549 and U87 cells were seeded in a 96-well plate format (at 1,500 cells / well for A549 and 2,000 cells / well for U87) and incubated for 24 h before treatment. The corresponding wells were then replaced with a suspension of Pd-ExoA549 or Pd-ExoU87 in culture media depleted of exosomes (at 0.4 μg / 100 μL for A549 and 0.53 μg / 100 μL for U87, in order to normalize exosome quantity per number of seeded cell). After 6 h of incubation, these wells were washed with PBS buffer to remove extracellular vesicles and treated with fresh media containing either DMSO (0.1% v/v) or 4 (0.2 μM). Cells treated with 3 and 4 at 0.2 μM were used as positive and negative controls, respectively. After 5 d of treatment, cell viability was determined as described above. Experiments were performed in triplicates.
Supplementary Material
Acknowledgements
Financial support from EPSRC (Healthcare Technology Challenge Award EP/N021134/1) and the ERC Advanced Grant CADENCE (ERC-2016-ADG-742684) are gratefully acknowledged. We also thank Dr Silvia Irusta at the University of Zaragoza for performing XPS measurements. MSA thanks the Spanish Government for an FPU PhD research fellowship. BRR thanks the EC (H2020-MSCA-IF-2014-658833, ChemoBOOM). MA thanks the financial support of the ERC Consolidator Grant program (ERC-2013-CoG-614715). PMD thanks AECC Fund and Refbio-Lactodermal for financial support.
Footnotes
Competing Interests
The authors declare that compound 4 is protected under patent application PCT/GB2017/051379.
Author Contributions
M.S.-A., B.R.-R., A.M.P.-L. and V.S. prepared and characterised the materials, planned and performed the experiments, analysed the data and wrote the Methods; V.S., P.M.-D. and M.A. planned and supervised the research, analysed the data and contributed to the manuscript writing; J.S. and A.U.-B. designed, coordinated and supervised the research, analysed the data and wrote the paper. All the authors checked the manuscript.
Data availability statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Streu C, Meggers E. Ruthenium-induced allylcarbamate cleavage in living cells. Angew Chem Int Ed. 2006;45:5645–5648. doi: 10.1002/anie.200601752. [DOI] [PubMed] [Google Scholar]
- 2.Li N, Lim RK, Edwardraja S, Lin Q. Copper-free sonogashira cross-coupling for functionalization of alkyne-encoded proteins in aqueous medium and in bacterial cells. J Am Chem Soc. 2011;133:15316–15319. doi: 10.1021/ja2066913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spicer CD, Triemer T, Davis BG. Palladium-mediated cell-surface labeling. J Am Chem Soc. 2012;134:800–803. doi: 10.1021/ja209352s. [DOI] [PubMed] [Google Scholar]
- 4.Michel BW, Lippert AR, Chang CJ. A reaction-based fluorescent probe for selective imaging of carbon monoxide in living cells using a palladium-mediated carbonylation. J Am Chem Soc. 2012;134:15668–15671. doi: 10.1021/ja307017b. [DOI] [PubMed] [Google Scholar]
- 5.Yang M, Jia S, Zhang X, Chen P. Palladium-triggered deprotection chemistry for protein activation in living cells. Nat Chem. 2014;6:352–361. doi: 10.1038/nchem.1887. [DOI] [PubMed] [Google Scholar]
- 6.Mascareñas JL, Sánchez MI, Penas C, Vázquez ME. Metal-catalyzed uncaging of DNA-binding agents in living cells. Chem Sci. 2014;5:1901. [PMC free article] [PubMed] [Google Scholar]
- 7.Völker T, Dempwolff F, Graumann PL, Meggers E. Angew Chem Int Ed. 2014;53:10536. doi: 10.1002/anie.201404547. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, et al. Chemical remodeling of cell-surface sialic acids through a palladium-triggered bioorthogonal elimination reaction. Angew Chem Int Ed. 2015;54:5364–5368. doi: 10.1002/anie.201409145. [DOI] [PubMed] [Google Scholar]
- 9.Tomás-Gamasa M, Martínez-Calvo M, Couceiro JR, Mascareñas JL. Transition metal catalysis in the mitochondria of living cells. Nat Commun. 2016;7:12538. doi: 10.1038/ncomms12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsubokura K, et al. In Vivo Gold Complex Catalysis within Live Mice. Angew Chem Int Ed. 2017;56:3579–3584. doi: 10.1002/anie.201610273. [DOI] [PubMed] [Google Scholar]
- 11.Destito P, Couceiro JR, Faustino H, López F, Mascareñas JL. Ruthenium-catalyzed azide-thioalkyne cycloadditions in aqueous media: a mild, orthogonal, and biocompatible chemical ligation. Angew Chem Int Ed. 2017;56:10766–10770. doi: 10.1002/anie.201705006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vidal C, Tomás-Gamasa M, Destito P, López F, Mascareñas JL. Concurrent and orthogonal gold(I) and ruthenium(II) catalysis inside living cells. Nat Commun. 2018;9:1913. doi: 10.1038/s41467-018-04314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenton BJ, Oliveira BL, Matos MJ, Sinatra L, Bernardes GJL. A thioether-directed palladium-cleavable linker for targeted bioorthogonal drug decaging. Chem Sci. 2018;9:4185–4189. doi: 10.1039/c8sc00256h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee A, et al. An enantioselective artificial Suzukiase based on the biotin–streptavidin technology. Chem Sci. 2016;7:673–677. doi: 10.1039/c5sc03116h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeschek M, et al. Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature. 2016;537:661–665. doi: 10.1038/nature19114. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto Y, et al. A cell-penetrating artificial metalloenzyme regulates a gene switch in a designer mammalian cell. Nat Commun. 2018;9:1943. doi: 10.1038/s41467-018-04440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yusop RM, Unciti-Broceta A, Johansson EMV, Sánchez-Martín RM, Bradley M. Palladium-mediated intracellular chemistry. Nat Chem. 2011;3:239–243. doi: 10.1038/nchem.981. [DOI] [PubMed] [Google Scholar]
- 18.Unciti-Broceta A, Johansson EMV, Yusop RM, Sánchez-Martín RM, Bradley M. Synthesis of polystyrene microspheres and functionalization with Pd(0) nanoparticles to perform bioorthogonal organometallic chemistry in living cells. Nat Protocols. 2012;7:1207–1218. doi: 10.1038/nprot.2012.052. [DOI] [PubMed] [Google Scholar]
- 19.Tonga GY, Jeong Y, Duncan B, Mizuhara T, Mout R, et al. Supramolecular regulation of bioorthogonal catalysis in cells using nanoparticle-embedded transition metal catalysts. Nat Chem. 2015;7:597–603. doi: 10.1038/nchem.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller MA, et al. Nano-palladium is a cellular catalyst for in vivo chemistry. Nat Commun. 2017;8:15906–15919. doi: 10.1038/ncomms15906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clavadetscher J, Indrigo E, Chankeshwara SV, Lilienkampf A, Bradley M. In-cell dual drug synthesis by cancer-targeting palladium catalysts. Angew Chem Int Ed. 2017;56:6864–6868. doi: 10.1002/anie.201702404. [DOI] [PubMed] [Google Scholar]
- 22.Hoop M, et al. Mobile magnetic nanocatalysts for bioorthogonal targeted cancer therapy. Adv Funct Mater. 2018;28:1705920. [Google Scholar]
- 23.Liu Y, et al. Catalytically active single-chain polymeric nanoparticles: exploring their functions in complex biological media. J Am Chem Soc. 2018;140:3423–3433. doi: 10.1021/jacs.8b00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F, Zhang Y, Du Z, Ren J, Qu X. Designed heterogeneous palladium catalysts for reversible light-controlled bioorthogonal catalysis in living cells. Nat Commun. 2018;9:1209. doi: 10.1038/s41467-018-03617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss JT, et al. Extracellular palladium-catalysed dealkylation of 5-fluoro-1-propargyl-uracil as a bioorthogonally activated prodrug approach. Nat Commun. 2014;5:3277. doi: 10.1038/ncomms4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss JT, et al. Development and bioorthogonal activation of palladium-labile prodrugs of gemcitabine. J Med Chem. 2014;57:5395–5404. doi: 10.1021/jm500531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss JT, Carragher NO, Unciti-Broceta A. Palladium-mediated dealkylation of N-propargyl-floxuridine as a bioorthogonal oxygen-independent prodrug strategy. Sci Rep. 2015;5:9329. doi: 10.1038/srep09329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clavadetscher J, et al. Copper catalysis in living systems and in situ drug synthesis. Angew Chem Int Ed. 2016;55:15662–15666. doi: 10.1002/anie.201609837. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-López AM, et al. Gold-triggered uncaging chemistry in living systems. Angew Chem Int Ed. 2017;56:12548–12552. doi: 10.1002/anie.201705609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray TL, et al. Bright insights into palladium-triggered local chemotherapy. Chem Sci. 2018;9:7354–7361. doi: 10.1039/c8sc02291g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adam C, et al. Bioorthogonal uncaging of the active metabolite of irinotecan by palladium-functionalized microdevices. Chem Eur J. 2018;24:16783–16790. doi: 10.1002/chem.201803725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eda S, et al. Biocompatibility and therapeutic potential of glycosylated albumin artificial metalloenzymes. Nat Catal. 2019 doi: 10.1038/s41929-019-0317-4. [DOI] [Google Scholar]
- 33.Valadi H, et al. Exosome mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 34.Gould SJ, Booth AM, Hildreth JE. The Trojan exosome hypothesis. Proc Natl Acad Sci USA. 2003;100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gourlay J, et al. The emergent role of exosomes in glioma. J Clin Neurosci. 2017;35:13–23. doi: 10.1016/j.jocn.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 36.Hornick NI, et al. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci Signal. 2016;9:ra88. doi: 10.1126/scisignal.aaf2797. [DOI] [PubMed] [Google Scholar]
- 37.Huan J, et al. Coordinate regulation of residual bone marrow function by paracrine trafficking of AML exosomes. Leukemia. 2015;29:2285–2295. doi: 10.1038/leu.2015.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Erviti L, et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 39.Li X, et al. Exosomes in cancer: Small transporters with big functions. Cancer Lett. 2018;435:55–65. doi: 10.1016/j.canlet.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 40.Meckes DG., Jr Exosomal communication goes viral. J Virol. 2015;89:5200–5203. doi: 10.1128/JVI.02470-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrami L, et al. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Rep. 2013;5:986–996. doi: 10.1016/j.celrep.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cross R. Meet the exosome, the rising star in drug delivery. Chem Eng News. 2018;96:22–23. [Google Scholar]
- 43.Zitvogel L, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell-derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 44.Munoz JL, et al. Delivery of functional anti-mir-9 by mesenchymal stem cell-derived exosomes to glioblastoma multiforme cells conferred chemosensitivity. Mol Ther Nucleic Acids. 2013;2:e126. doi: 10.1038/mtna.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smyth T, et al. Surface functionalization of exosomes using click chemistry. Bioconjug Chem. 2014;25:1777–1784. doi: 10.1021/bc500291r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang T, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. 2015;32:2003–2014. doi: 10.1007/s11095-014-1593-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.György B, et al. Rescue of hearing by gene delivery to inner-ear hair cells using exosome-associated AAV. Mol Ther. 2017;25:379–391. doi: 10.1016/j.ymthe.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SM, et al. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release. 2017;266:8–16. doi: 10.1016/j.jconrel.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, et al. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat Commun. 2018;9:960. doi: 10.1038/s41467-018-03390-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hood JL, Scott MJ, Wickline SA. Maximizing exosomes colloidal stability following electroporation. Anal Biochem. 2014;448:41–49. doi: 10.1016/j.ab.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alhasan AH, Patel PC, Choi CHJ, Mirkin CA. Exosome encased spherical nucleic acid gold nanoparticle conjugates as potential microRNA regulation agents. Small. 2014;10:186–192. doi: 10.1002/smll.201302143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Busato A, et al. Magnetic resonance imaging of ultrasmall superparamagnetic iron oxide-labeled exosomes from stem cells: a new method to obtain labeled exosomes. Int J Nanomedicine. 2016;11:2481–2490. doi: 10.2147/IJN.S104152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Illes B, et al. Exosome-Coated Metal–Organic Framework Nanoparticles: An Efficient Drug Delivery Platform. Chem Mater. 2017;29:8042–8046. [Google Scholar]
- 55.Betzer O, et al. In vivo neuroimaging of exosomes using gold Nanoparticles. ACS Nano. 2017;11:10883–10893. doi: 10.1021/acsnano.7b04495. [DOI] [PubMed] [Google Scholar]
- 56.Ha D, Yang N, Nadithe V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: current perspectives and future challenges. Acta Pharm Sin B. 2016;6:287–296. doi: 10.1016/j.apsb.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luan X, et al. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38:754–763. doi: 10.1038/aps.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sebastian V, Smith CD, Jensen KF. Shape-controlled continuous synthesis of metal nanostructures. Nanoscale. 2016;8:7534–7543. doi: 10.1039/c5nr08531d. [DOI] [PubMed] [Google Scholar]
- 59.Herrer L, et al. High surface coverage of a self-assembled monolayer by in situ synthesis of palladium nanodeposits. Nanoscale. 2017;9:13281–13290. doi: 10.1039/c7nr03365f. [DOI] [PubMed] [Google Scholar]
- 60.Zhang D, et al. An In situ TEM study of the surface oxidation of palladium nanocrystals assisted by electron irradiation. Nanoscale. 2017;9:6327–6333. doi: 10.1039/c6nr08763a. [DOI] [PubMed] [Google Scholar]
- 61.Kikot P, Polat A, Achilli E, Fernandez Lahore M, Grasselli M. Immobilized palladium(II) ion affinity chromatography for recovery of recombinant proteins with peptide tags containing histidine and cysteine. J Mol Recognit. 2014;27:659–668. doi: 10.1002/jmr.2389. [DOI] [PubMed] [Google Scholar]
- 62.Raedler LA. Farydak (Panobinostat): First HDAC Inhibitor Approved for Patients with Relapsed Multiple Myeloma. Am Health Drug Benefits. 2016;9:84–87. [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng S, et al. Biocompatible Boron-Containing Prodrugs of Belinostat for the Potential Treatment of Solid Tumors. ACS Med Chem Lett. 2018;9:149–154. doi: 10.1021/acsmedchemlett.7b00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubio-Ruiz B, Weiss JT, Unciti-Broceta A. Efficient palladium-triggered release of vorinostat from a bioorthogonal precursor. J Med Chem. 2016;59:9974–9980. doi: 10.1021/acs.jmedchem.6b01426. [DOI] [PubMed] [Google Scholar]
- 65.Wang H, et al. Discovery of (2E)-3-{2-Butyl-1-[2-(diethylamino)ethyl]-1H-benzimidazol-5-yl}-N-hydroxyacrylamide (SB939), an Orally Active Histone Deacetylase Inhibitor with a Superior Preclinical Profile. J Med Chem. 2011;54:4694–4720. doi: 10.1021/jm2003552. [DOI] [PubMed] [Google Scholar]
- 66.Sancho-Albero M, et al. Exosome origin determines cell targeting and the transfer of therapeutic nanoparticles towards target cells. J Nanobiotechnol. 2019;17:16. doi: 10.1186/s12951-018-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, Chen PR. Development and application of bond cleavage reactions in bioorthogonal chemistry. Nat Chem Biol. 2016;12:129–137. doi: 10.1038/nchembio.2024. [DOI] [PubMed] [Google Scholar]
- 69.Devaraj NK. The Future of Bioorthogonal Chemistry. ACS Cent Sci. 2018;4:952–959. doi: 10.1021/acscentsci.8b00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request.