Abstract
Natalizumab (NZM), a humanized monoclonal IgG4 antibody to α4 integrins, is used to treat patients with relapsing-remitting multiple sclerosis (MS)1,2, but in about 6% of the cases persistent neutralizing anti-drug antibodies (ADAs) are induced leading to therapy discontinuation3,4. To understand the basis of the ADA response and the mechanism of ADA-mediated neutralization, we performed an in-depth analysis of the B and T cell responses in two patients. By characterizing a large panel of NZM-specific monoclonal antibodies, we found that, in both patients, the response was polyclonal and targeted different epitopes of the NZM idiotype. The neutralizing activity was acquired through somatic mutations and correlated with a slow dissociation rate, a finding that was supported by structural data. Interestingly, in both patients, the analysis of the CD4+ T cell response, combined with mass spectrometry-based peptidomics, revealed a single immunodominant T cell epitope spanning the FR2-CDR2 region of the NZM light chain. Moreover, a CDR2-modified version of NZM was not recognized by T cells, while retaining binding to α4 integrins. Collectively, our integrated analysis identifies the basis of T-B collaboration that leads to ADA-mediated therapeutic resistance and delineates an approach to design novel deimmunized antibodies for autoimmune disease and cancer treatment.
The therapeutic use of monoclonal antibodies and other biopharmaceutical products can result in an immune response to the drug that, in some cases, affects its efficacy due to the production of neutralizing ADAs5. Several clinical studies have measured ADA levels in sera of selected cohorts of patients and concluded that not all antibody responses lead to drug neutralization6–11. However, an explanation for these heterogeneous responses and an integrated characterization of the B and T cell responses to the drug are still missing. In this study, we isolated NZM-specific B and T cell clones from memory cells of two MS patients, who had a hypersensitivity reaction following drug infusion and developed high titers of ADAs (Supplementary Table 1 and Supplementary Fig. 1).
To identify NZM-specific antibodies, we screened supernatants of immortalized B cells12 and isolated 30 and 10 anti-NZM monoclonal antibodies from patients A and B, respectively (Fig. 1). Most antibodies from patient A showed high affinity for NZM (KD values 1-6,790 pM, median 6.1 pM), while antibodies from patient B showed lower affinity (KD values 0.4-22.7 nM, median 2.3 nM) (Supplementary Table 2). Sixty percent (18/30) of the antibodies from patient A potently inhibited binding of NZM to α4 integrins on the surface of T cells (IC90 values 17-271 ng/ml, defined as NAbs, neutralizing antibodies), while the remaining showed reduced or no inhibitory capacity (IC90 values > 1,000 ng/ml, defined as BAbs, binding antibodies) (Fig. 2a and Supplementary Table 2). Interestingly, patient B developed only non-neutralizing BAbs, a finding that may be related to the lower number of NZM infusions received.
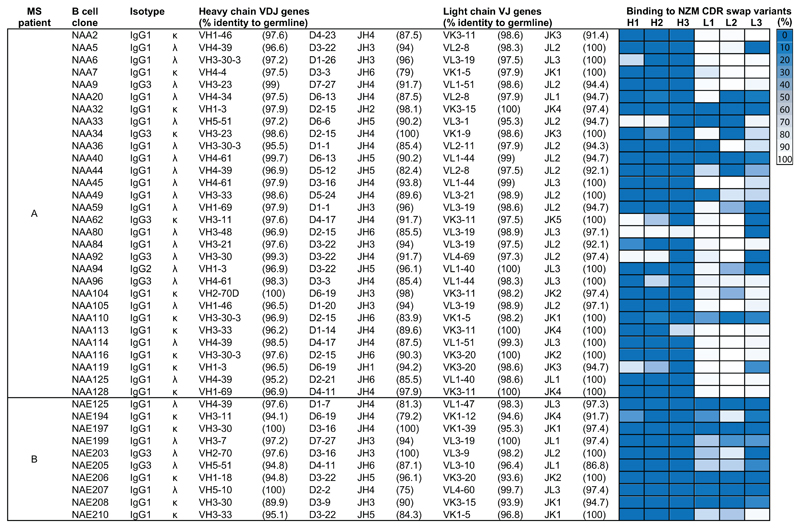
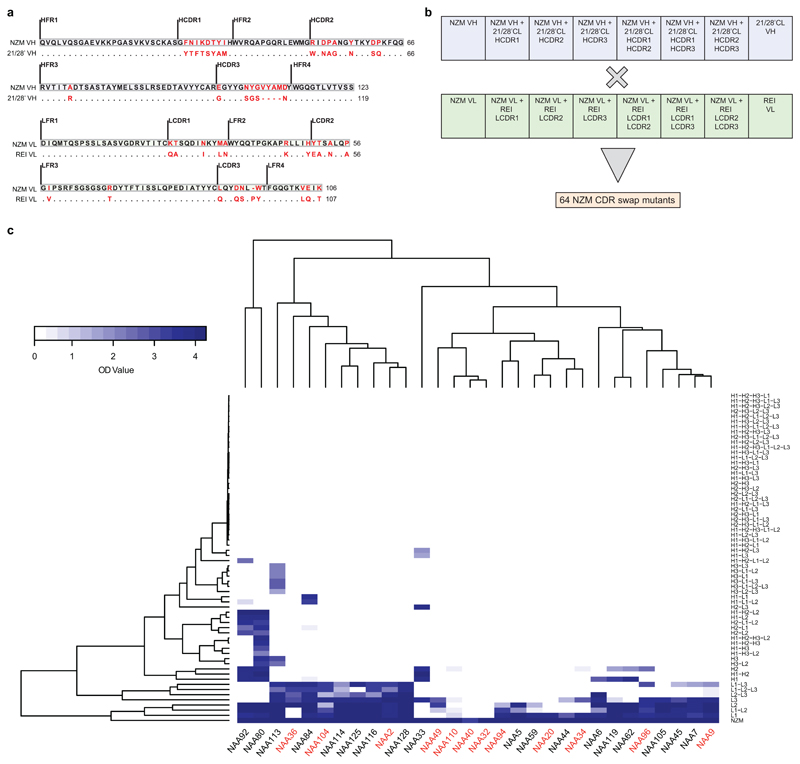
Fig. 1. V(D)J gene usage and epitope mapping of 40 anti-natalizumab monoclonal antibodies.
The colored cells show the binding of the antibodies to six selected NZM variants in which individual CDRs were swapped with the counterparts of the human scaffold antibody used for NZM humanization (H, heavy chain; L, light chain; 1, CDR1; 2, CDR2; 3, CDR3). Shown is the percentage of binding of the antibodies to the NZM CDR swap variants relative to NZM, as tested by ELISA, with a three-color gradation scale from minimum (0%, blue) to maximum (100%, white). OD values and exact % binding values are shown in Source Data 1.
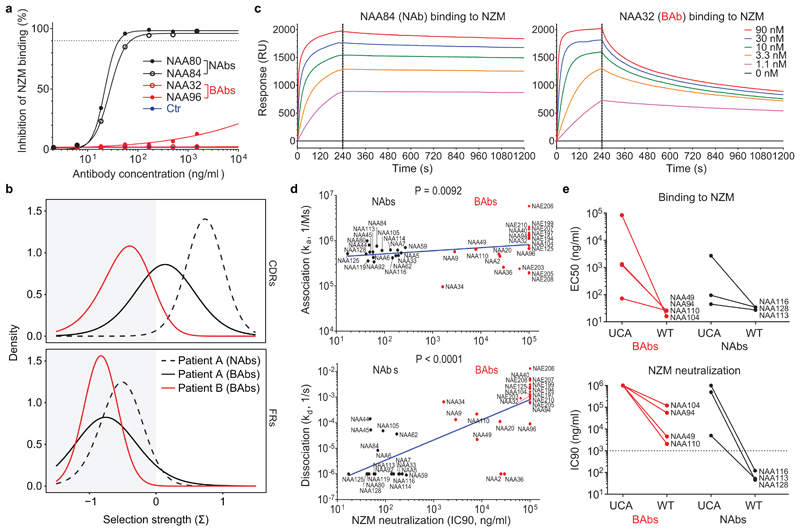
Fig. 2. The neutralizing activity of ADAs is acquired through somatic mutations and correlates with a slow dissociation rate.
a, Dose-dependent inhibition of NZM binding to T cells of four representative antibodies (two NAbs in black and two BAbs in red) compared to an unrelated antibody used as a control (representative of n = 2 independent experiments). The dotted line indicates the 90% of inhibition. b, Positive and negative selection strengths (Σ) in CDRs and FRs of 18 NAb and 12 BAb heavy chain sequences from patient A (black and dotted black lines) and 10 BAb heavy chain sequences from patient B (red line) estimated using the Bayesian estimation of Antigen-driven SELectIoN (BASELINe), which compares the observed frequencies of replacement and silent mutations with the expected mutations. c, Kinetics of binding of two representative antibodies (NAA84 and NAA32) to different doses of NZM as measured by SPR (representative of n = 2 independent experiments). The dotted line separates the association and the dissociation phase. RU, resonance units. d, Correlation of NZM neutralization (expressed as IC90, i.e. the antibody concentration required for 90% inhibition of NZM binding to T cells) with association constant (ka, upper panel) and dissociation constant (kd, lower panel) of n = 40 antibodies measured through SPR (representative of n = 2 experiments). BAbs and NAbs are shown as red and black symbols (circle for patient A and diamond for patient B), respectively. A two-tailed Spearman’s correlation was performed; r coefficient, 0.4070 (ka) and 0.7673 (kd); 95% confidence interval, 0.09993 to 0.6433 (ka) and 0.5928 to 0.8730 (kd). e, Comparison of binding to NZM expressed as half-maximal effective concentration, EC50 (upper panel) and NZM neutralization expressed as IC90 (lower panel) between the unmutated common ancestor (UCA) and the wild type (WT) versions of seven representative antibodies (four BAbs and three NAbs) (representative of n = 2 independent experiments). The dotted line indicates the threshold of neutralization set at 1,000 ng/ml.
The antibodies isolated were mostly IgG1-λ, used different V(D)J genes and showed moderate levels of somatic mutations (Fig. 1). NAbs from patient A carried a high load of replacement mutations in the complementary determining regions (CDRs), consistent with an antigen-driven selection (Fig. 2b), while BAbs, from both patients, carried few replacement mutations in the CDRs. The antibody fine specificity was tested using 64 NZM variants generated by swapping the hypervariable CDR loops with the counterparts of the human scaffold antibody used for NZM humanization (Extended Data Fig. 1a-b). The antibodies recognized epitopes comprising one to six NZM CDRs, with preferential recognition of the heavy chain CDRs (Fig. 1 and Extended Data Fig. 1c). These findings demonstrate that NZM induces a neutralizing polyclonal anti-idiotypic antibody response that targets multiple epitopes located primarily in the heavy chain CDRs.
The difference in neutralizing activity of the anti-NZM antibodies isolated may be due to the binding to distinct epitopes or, alternatively, to different affinity or kinetics of binding. When compared for their capacity to bind to the 64 NZM CDR swap variants, BAbs and NAbs did not cluster separately (Extended Data Fig. 1c), indicating that the difference is not related to epitope specificity. However, BAbs and NAbs showed different binding kinetics, as assessed by surface plasmon resonance (SPR). In particular, while the association constant (ka) was comparable, NAbs showed a lower dissociation constant (kd) that significantly correlated with NZM neutralization ability (Fig. 2c-d and Supplementary Table 2). The role of somatic mutations was addressed by comparing the antibodies to the unmutated common ancestors (UCAs). In three out of four BAbs tested, the UCAs had low binding affinity for NZM, which was increased by somatic mutations. In contrast, in two out of three NAbs tested, the UCAs showed already high affinity, but required somatic mutations to gain full neutralizing activity (Fig. 2e). Collectively, these findings indicate a critical role for somatic mutations in the generation of high-affinity antibodies and highlight the importance of dissociation rate for ADA neutralizing activity.
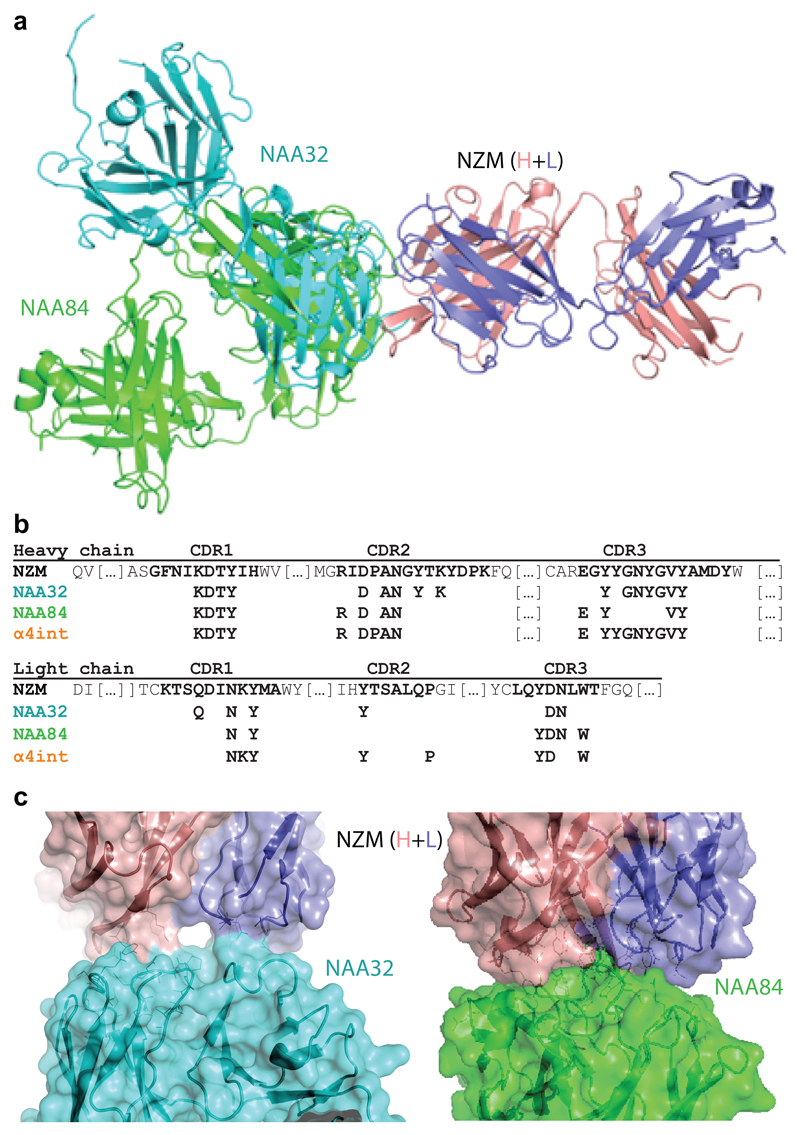
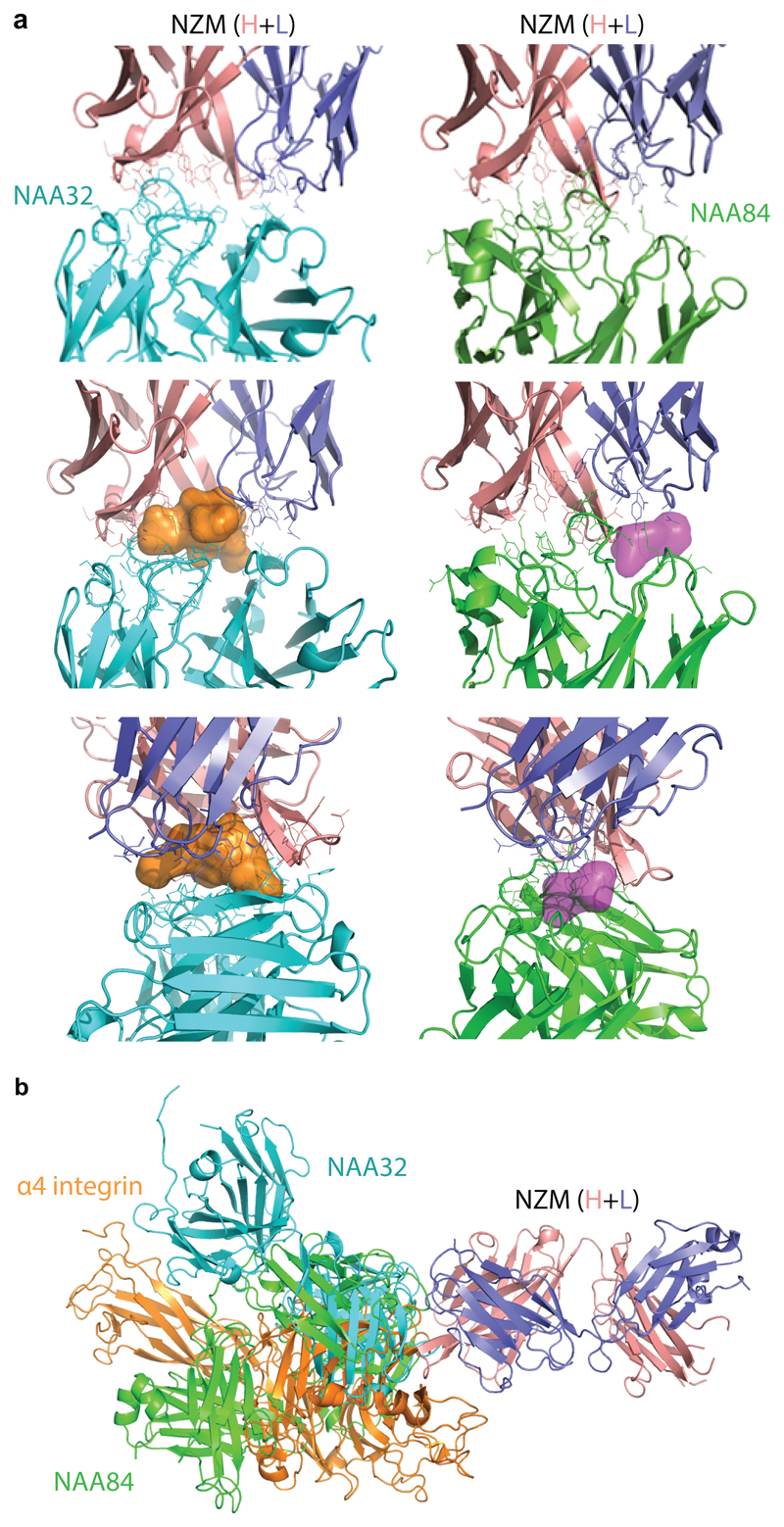
We next determined the crystal structure of a BAb (NAA32) and a NAb (NAA84) in complex with NZM. With 2.8 Å resolution, the structures revealed that both antibodies interacted with the same surface area on NZM but engaged the molecule with different orientation (Fig. 3a and Supplementary Table 3). NAA32 and NAA84 recognized 22 and 18 residues of NZM CDRs, respectively, which were mostly located in the heavy chain, a finding consistent with the CDR swap variant specificity (Fig. 3b). In particular, 14 of these residues were recognized by both antibodies, highlighting potential immunodominant B-cell epitopes on NZM. The contact surface area between NAA32 and NZM was 757Å2, although the main interactions occurred at two separate contact points leaving an empty space in between the two interfaces (Fig. 3c and Extended Data Fig. 2a). In contrast, the contact surface area between NAA84 and NZM was 617Å2, but interactions were much tighter, with deeper residue contacts and contiguous surface complementarity that could account for its slower dissociation rate from NZM (Fig. 3c and Extended Data Fig. 2a). Importantly, the NZM surface area engaged by NAA32 and NAA84 overlaps with the area engaged by α4 integrin13 with 13 shared residues (Fig. 3b). While both NAA32 and NAA84 antibodies occluded α4 integrin binding site, neither of them provided molecular mimicry of the integrin, which exhibited a different binding orientation (Extended Data Fig. 2b). Together with the functional data, these structural data suggest that the difference between BAbs and NAbs cannot be explained by the fine epitope specificity, but rather by the strength and the fit of the interaction with NZM.
Fig. 3. Structural features of the interaction of NZM with a NAb and a BAb.
a, Superimposition of the antigen-binding fragment of NZM in complex with NAA84 (NAb, green) and NAA32 (BAb, cyan). NZM heavy and light chains are shown in salmon and slate blue, respectively. Proteins are displayed in ribbon diagram. b, Alignment of the NZM residues that are recognized by NAA32, NAA84 and α4 integrin. c, Detailed visualization of the interacting interfaces of NZM and NAA32 (left) and NAA84 (right). The antibodies are shown as ribbon diagrams with overlapping surfaces. The Sc values were 0.696 and 0.707 for NAA32/NZM and NAA84/NZM complexes, respectively.
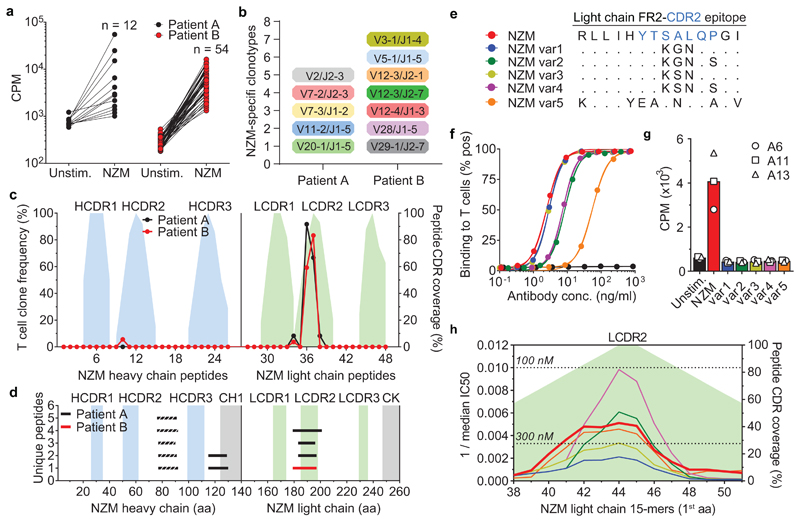
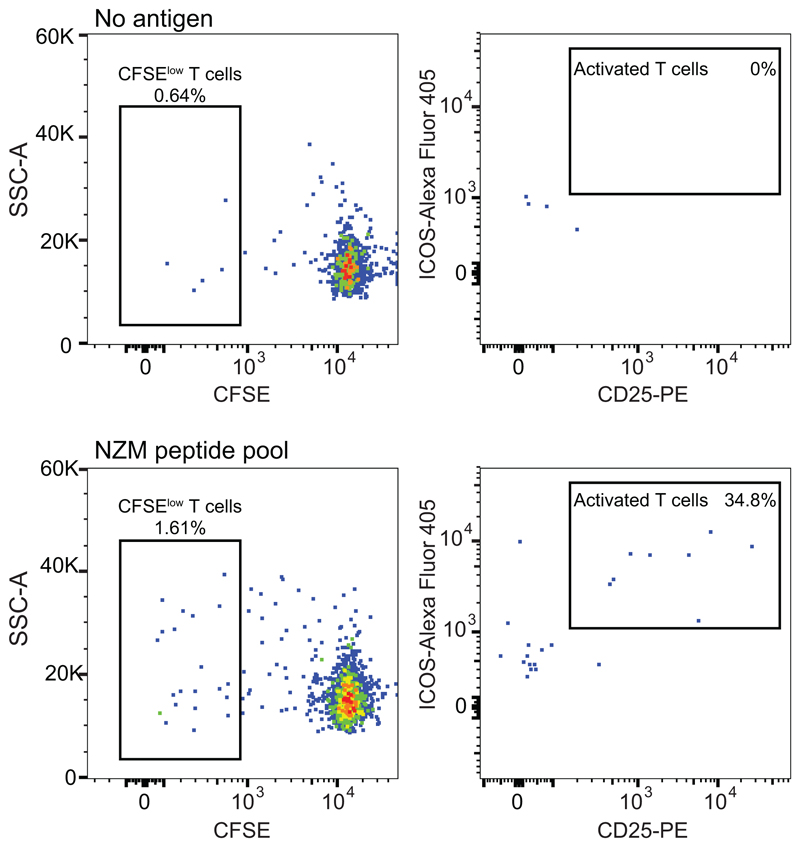
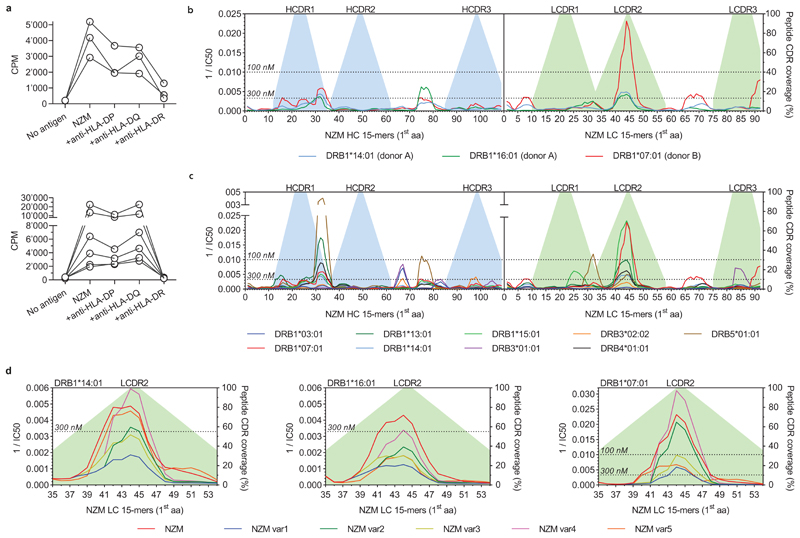
The generation of neutralizing ADAs through somatic mutations is consistent with an affinity maturation driven by CD4+ T cells targeting non-self regions of the NZM idiotype. To address this hypothesis and to characterize the specificity of the T-cell response, we stimulated CFSE-labeled memory CD4+ T cells from both patients with overlapping peptides covering the variable regions of NZM (Extended Data Fig. 3). Cloning of activated CFSElow T cells resulted in the isolation of several NZM-reactive T cell clones (12 from patient A and 54 from patient B) (Fig. 4a). T cell receptor (TCR) Vβ-gene sequencing revealed the presence of 5 and at least 7 distinct T cell clonotypes in patient A and B, respectively (Fig. 4b and Supplementary Table 4). Strikingly, most of the T cell clones from both patients recognized two overlapping peptides spanning a region comprising the end of framework region 2 (FR2) and the CDR2 of the NZM light chain (GKAPRLLIHYTSALQPGI, named NZM-LCFR2-CDR2) (Fig. 4c). Of note, T cell recognition of NZM-LCFR2-CDR2 was restricted by HLA-DRB1*14/16 in patient A and by DRB1*07/07 in patient B (Extended Data Fig. 4a and Supplementary Table 1 and 4). Taken together, these findings suggest that, in these patients, a single T cell epitope in the NZM molecule induced an HLA-DR-restricted CD4+ T-cell response that was sufficient to sustain a strong polyclonal B cell response to the NZM idiotype.
Fig. 4. Identification of a single immunodominant T cell epitope that can be engineered to deimmunize NZM.
a, Proliferative response of NZM-reactive T cell clones isolated after ex-vivo stimulation of memory CD4+ T cells, upon re-stimulation with the overlapping peptide pool covering the entire sequences of the variable regions of the NZM heavy and light chains. Proliferation was assessed on day 3 after 16 h pulse with [3H]-thymidine. Data are expressed as counts per minute (cpm). Unstim., unstimulated T cells. b, TCR Vβ gene repertoire of NZM-reactive CD4+ T cell clones isolated from MS patients A and B. The y-axis indicates the number of unique clonotypes identified. c, Epitope mapping of NZM-specific CD4+ T cell clones isolated from patients A and B. The epitopes were identified by screening the T cell clones with overlapping peptides spanning the NZM heavy and light chain variable region. The left y-axis indicates the frequency of T cell clones reactive to each peptide. The right y-axis indicates the percentage of CDR residues within each peptide shown as filled areas in the background (representative of n = 2 experiments). d, Mass spectrometry-based MHC peptidomics of NZM-specific B cell clones pulsed with NZM. Each line represents a unique MHC-II-bound peptide identified in n = 2 independent experiments. Dashed lines indicates peptides belonging to both NZM and B cell receptor of the antigen-presenting cells. The y-axis indicates the number of unique peptides overlapping in the same NZM region. e, Comparison of the amino acid sequences of the LCFR2-CDR2 epitope of NZM and the five variants engineered for the drug deimmunization. f, Binding of NZM variants to α4 integrin expressed on T cells analyzed by flow cytometry. The NZM variants are color coded as depicted in panel e. An antibody with different specificity was used a negative control (black curve). g, Proliferation of three NZM-LCFR2-CDR2-reactive T cell clones (A6, A11 and A13) after stimulation with autologous B cells pulsed with NZM and the five engineered variants (representative of n = 2 independent experiments). The bars show the mean proliferation. h, Predicted binding affinities of 15mer peptides spanning the light chain CDR2 region of NZM variants to a reference set of nine DRB1 and DRB3/4/5 alleles (DRB1*0301, DRB1*0701, DRB1*1301, DRB1*1401, DRB1*1501, DRB3*0101, DRB3*0202, DRB4*0101, DRB5*0101)15,16. The NZM variants are color coded as depicted in panel e. The affinities are shown as reciprocal median IC50 (nM) values. The dotted lines define the thresholds of high-affinity binding set at 100 nM and low-affinity binding set at 300 nM.
To investigate the mechanism that leads to the presentation of the NZM T cell epitope, we pulsed NZM-specific B cell clones with NZM and identified the naturally processed MHC-II-bound peptides by mass spectrometry-based peptidomics. Three sets of nested peptides that mapped to the variable regions of NZM were identified (Fig. 4d and Supplementary Table 5). Two of these sets covered the heavy chain FR3 and the FR4-CH1 region, which are in the human germline configuration and therefore are not expected to induce an immune response, a notion consistent with our failure to isolate specific T cell clones. Remarkably, the third set of peptides mapped to the FR2-CDR2 region of the NZM light chain (consensus, TPGKAPRLLIHYTSALQPGIPSR) that spanned the immunodominant NZM-LCFR2-CDR2 epitope (GKAPRLLIHYTSALQPGI) recognized by NZM-reactive T cell clones (Fig. 4d and Supplementary Table 5). The NetMHCIIpan algorithm14 applied to predict binding to the HLA-DR alleles carried by the two patients identified the same three peptides, as well as several other peptides mapping to CDRs and FRs of NZM. (Extended Data Fig. 4b). In addition, this algorithm predicted the binding of the NZM-LCFR2-CDR2 epitope to a reference panel of nine DRB1 and DRB3/4/5 alleles15,16 (Extended Data Fig. 4c), suggesting its potential immunodominance also in individuals with a diverse HLA background. Collectively, these findings demonstrate that only one of the potentially immunogenic peptides encoded by the six CDRs of NZM was a naturally presented T cell epitope able to generate a polyclonal CD4+ T-cell response.
The identification of the immunodominant NZM-LCFR2-CDR2 epitope prompted us to use a structure-guided design to engineer a “deimmunized” version of NZM. First, we identified residues of NZM light chain CDR2 that were not engaging α4 integrin binding and modelled different mutants with the constraint to preserve the conformation of the CDR2 and the specificity of NZM. Four NZM variants (var1-4) were retained for experimental testing and validation together with a fifth variant (var5) in which the CDR2 of the light chain was reverted to the germline sequence of the human antibody scaffold (Fig. 4e). Two variants, var1 and var3, retained binding to α4 integrins, while var2 and var4 showed partial loss of binding that was considerably reduced in the case of var5 (Fig. 4f). Remarkably, none of the five NZM variants was able to trigger proliferation of T cell clones specific for the naturally presented NZM-LCFR2-CDR2 peptide (Fig. 4g), a finding consistent with either absence of TCR cross-reactivity or with a reduced binding to class II molecules. Prediction of binding to a reference set of nine DRB1 and DRB3/4/5 alleles15,16, as well as to the DRB1 alleles of both patients, showed a reduction of predicted binding affinity for the var1 and var3 peptides compared to the original NZM peptide (Fig. 4h and Extended Data Fig. 4d). These results provide two deimmunized versions of NZM that can be tested in vivo.
This study integrates, for the first time, clonal analysis of B and T cell repertoires and mass spectrometry-based peptidomics to identify the factors that underpin the neutralizing antibody response to a humanized therapeutic antibody. The large number of monoclonal antibodies isolated recognize multiple epitopes spanning different NZM CDRs and therefore represent classical anti-idiotypic antibodies17,18 rather than internal image of the antigen. Interestingly, neutralizing antibodies showed high-level replacement mutations in the CDRs and low dissociation rate, suggesting that B cell selection was driven by decreased kd rather than increased ka19.
The highly diverse anti-idiotypic response is consistent with the presence of multiple B cell epitopes recognized by naïve B cells and contrasts with the T cell response that is largely limited to a single epitope that we mapped to the FR2-CDR2 region of NZM light chain. This finding highlights the merit of the humanization technology in limiting the T cell immunogenicity of therapeutic antibodies, since chimeric antibodies were found to elicit T cell responses against multiple epitopes in the FRs and CDRs of both heavy and light chains20. As expected, the residual immunogenicity is primarily in the CDR regions21, but it is still limited by HLA restriction and processing by antigen-presenting cells.
Previous studies reported a positive association of NZM-related hypersensitivity reactions with DRB1*13 and 14 alleles16, but did not investigate any T cell response. The finding that the NZM-LCFR2-CDR2 peptide is naturally presented in the context of the HLA-DRB1*07 and DRB1*14/16 alleles of the two patients, together with the prediction of its binding to different alleles, including DRB1*13, suggests that this peptide is a major source of T cell help driving the anti-idiotypic B cell response to NZM. This may explain the high frequency of MS patients producing ADAs to NZM3,4 and offers the possibility for the deimmunization of the drug for a more safe treatment. Structured-guided mutagenesis and experimental tests suggest that a little amendment of only three residues of the CDR2 of the NZM light chain may be sufficient to remove the immunogenic T cell epitope without interfering with the interaction of NZM with its target.
Collectively, our results demonstrate how the integration of peptidomics, structural data, in silico predictions and dissection of the specific B and T cell responses represents a powerful approach to define the immunogenic landscape of therapeutic antibodies and to guide the deimmunization strategies of next-generation biological therapeutics for autoimmune and cancer disease. Currently used fully human antibodies, including checkpoint inhibitors, may also benefit from the deimmunization strategy. Reciprocally, this approach could be used to improve the immunogenicity of vaccines through the engineering of dominant T cell epitopes driving neutralizing antibody responses.
Online Methods
Patients and sample collection
Blood samples were collected from two MS patients (A and B) who had a hypersensitivity reaction following infusion of NZM and developed high titers of ADAs. Patient A was treated with NZM at the University Hospital for Neurology in Innsbruck, Austria, in 2014, while patient B was treated at the Mondino Foundation in Pavia, Italy, in 2018. Both patients provided written informed consent for this study. The study was approved by the Ethical committees of Innsbruck (UN2013-0040_LEK) and Pavia (P-20170027756). Blood samples were processed to obtain serum and peripheral blood mononuclear cells (PBMCs). For serum preparation, whole blood was collected in Vacutainer tubes (BD Biosciences) containing clot activators and kept at room temperature until a clot was formed. The tube was centrifuged at 2,000g for 10 min at 22°C, and the serum fraction was stored at −80°C. PBMCs were isolated from whole blood through Ficoll density gradient centrifugation and were resuspended in freezing medium for long-term storage in liquid nitrogen.
Production of NZM Fc-variants and CDR swap variants
A variant of NZM in which the human Fc was replaced with the murine counterpart (NZM-mFc) was produced by molecular cloning and used in ELISA and FACS assays to avoid reactivity with secondary anti-human Fcγ-specific antibodies. Briefly, synthetic genes expressing the NZM heavy chain and light chain variable regions (KEGG DRUG Database entry: D06886) were produced by Genscript and sublconed into vectors for expression of chimeric human CH1-murine IgG2a heavy chain (mFc) and human Igκ, respectively. The chains were expressed following transient transfection of these vectors into Expi293F cells (ThermoFisher Scientific) using polyethylenimine. Cell lines were routinely tested for mycoplasma contamination. NZM CDR swap variants were designed by aligning the sequences of NZM with those of the human antibodies (21/28’CL and REI) used for humanization (GenBank accession numbers AAA52825 and 751419A), synthetized by Genscript and subcloned into the mFc vector. NZM deimmunized variants were synthetized by Genscript and subcloned into vectors for expression of full human IgG4. The antibodies were purified by protein A or protein G chromatography (GE Healthcare) and concentrated by Amicon Ultra filter units (100K, Millipore). For SPR, mass-spectrometry and T cell experiments, NZM IgG was purified from TYSABRI® drug solution for infusion (Biogen). Total IgGs were quantified by Pierce BCA protein assay (ThermoFischer).
B cell and T cell sorting
Monocytes were isolated from PBMCs by positive selection using CD14 magnetic microbeads (Miltenyi Biotech). CD14-depleted fractions were stained on ice for 15–20 min with the following fluorochrome-labeled mouse monoclonal antibodies: CD8-PE–Cy5 (clone B9.11; cat. no. A07758), CD14-PE–Cy5 (clone RMO52; cat. no. A07765), CD16-PE–Cy5 (clone 3G8; cat. no. A07767), CD45RA-FITC (clone ALB11; cat. no. A07786) from Beckman Coulter, CD19-PE–Cy7 (clone SJ25C1; cat. no. 341113), CD25–PE (clone M-A251; cat. no. 555432) from BD Biosciences, CD4–PE–Texas Red (clone S3.5; cat. no. MHCD0417) from ThermoFisher Scientific, CCR7–BV421 (clone G043H7; cat. no. 353208) from BioLegend, Alexa Fluor 647-conjugated goat anti-human IgG (cat. no. 109-606-170) from Jackson ImmunoResearch. IgG+ memory B cells were sorted from CD19+ B cells to over 98% purity on a FACSAria III (BD). Memory CD4+ T cells were sorted to over 98% purity after exclusion of naïve CD4+ T cells (CD45RA+CCR7+) and CD8+, CD14+, CD16+, CD19+, CD25bright cells.
B cell immortalization and isolation of monoclonal antibodies
B cells were cultured in RPMI 1640 medium supplemented with 2 mM glutamine, 1% (v/v) nonessential amino acids, 1% (v/v) sodium pyruvate, penicillin (50 U/ml), streptomycin (50 μg/ml) (all from Invitrogen) and 10% fetal bovine serum (HyClone, characterized, GE Healthcare Life Science). Sorted IgG+ memory B cells were immortalized with Epstein–Barr virus (EBV) and plated in single-cell cultures in the presence of CpG-DNA (2.5 μg/ml) and irradiated PBMC-feeder cells, as previously described12. Two weeks post immortalization, the culture supernatants were tested by ELISA for binding to NZM-mFc as well as to a control antibody of an irrelevant specificity. B cell cultures that tested positive only for NZM-mFc were isolated and expanded.
Sequence analysis of antibody cDNAs and production of recombinant antibodies
cDNA was synthesized from selected B cell cultures, and both the VH and VL genes were sequenced as previously described22. The genes that encoded the VH and VL and the number of somatic mutations were determined by analyzing the homology between the genes encoding the VH and VL sequences of the monoclonal antibodies and the known genes encoding human V, D and J regions that are present in the international immunogenetics information system (IMGT) database (version 3.4.17)23. Antibody-coding sequences were amplified and sequenced with primers specific for the V and J regions of the given antibody. Sequences of the unmutated common ancestor (UCA) of the VH- and VL-coding genes were constructed using IMGT/V-QUEST23 and synthetized by Genscript. To calculate the frequency of mutations, the entire sequence of each heavy chain variable region was compared to the germline sequence to identify replacement (R) and silent (S) mutations. The selection strength was estimated using BASELINe framework which compares the observed frequencies of replacement and silent mutations with the expected ones24. All calculations were done using the “calcBaseline” and “groupBaseline” functions from SHazaM (version 0.1.11)25. Sequences encoding antibody heavy and light chains were cloned into vectors for expression of human IgG1, Igκ and Igγ, and the chains were expressed following transient transfection of these vectors into Expi293F cells, as described above. Selected antibodies were also expressed as antigen-binding fragments for X-ray crystallography analysis.
ELISA assays for screening and characterization of anti-NZM antibodies
Total IgGs were quantified using 96-well MaxiSorp plates (Nunc) coated with 10 μg/ml goat anti–human IgG (SouthernBiotech, cat. no. 2040-01). Plates were then blocked with PBS with 1% BSA and incubated with titrated monoclonal antibodies, using Certified Reference Material 470 (ERMs-DA470, Sigma-Aldrich) as a standard. Plates were then washed and incubated with 1/500 alkaline phosphatase (AP)-conjugated goat anti–human IgG (Southern Biotech, cat. no. 2040-04). Substrate (para-nitrophenyl phosphate (p-NPP), Sigma) was added and plates were read at wavelength of 405 nm to determine optical density (OD) values. To test specific antibody binding, ELISA plates were coated with 1 μg/ml of NZM-mFc or a control antibody to test for nonspecific binding. Plates were blocked with PBS with 1% BSA and incubated with titrated sera or monoclonal antibodies, followed by 1/2,500 AP-conjugated goat anti-human IgG, Fcγ fragment specific antibody (Jackson ImmunoResearch, cat. no. 109-056-098), goat anti-human serum IgA, α chain specific (Jackson ImmunoResearch, cat. no. 109-055-011), or goat anti-human IgM, Fc5μ fragment specific (Jackson ImmunoResearch, cat. no. 109-055-129). In some dissociation assays, the samples were supplemented with 25% (vol/vol) of an alkaline dissociation buffer (2.5% Triton X-100, 2M ethanolamine, 0.15M NaCl, pH 11.6) one minute before the end of incubation. To test antibody binding to NZM CDR swap variants, ELISA plates were coated with 2 μg/ml goat anti–mouse IgG, human adsorbed (Southern Biotech, cat. no. 1030-01) and blocked with PBS with 1% BSA. After washing, the plates were incubated with 2 μg/ml of NZM CDR swap variants, followed by monoclonal antibodies at 2 μg/ml concentration and 1/2,500 AP-conjugated goat anti-human IgG, Fcγ fragment specific antibody. A heat map of the patterns of antibody binding to the 64 NZM CDR swap variants was computed by using the default clustering parameters of the heatmap.2 function from ‘gplots’ package in R in which the Euclidean metric and complete aggregation method were selected.
Surface plasmon resonance (SPR) assay
To study the kinetics of interaction of NZM-specific monoclonal ADAs, NZM IgG (50 nM) was stabilized in 10 mM acetate buffer, pH 4.5, and immobilized onto a EDC/NHS pre-activated ProteOn sensor chip (Biorad) through amine coupling; unreacted groups were blocked by injection of ethanolamine HCl (1 M). HEPES buffered saline (HBS) (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant Tween-20) was used as running buffer. All injections were made at flow rate of 100 μl/min. Monoclonal ADAs were diluted to 30 nM and injected onto the NZM coated chip; one channel of the chip was injected with HBS and used as reference for the analysis. Injection time and dissociation time were 240 s and 600 s, respectively. The binding interaction of each antibodies with NZM was assessed using a ProteON XPR36 instrument (BioRad). SPR data were processed with ProteOn Manager Software and ka (1/Ms), kd (1/s) and KD (M) parameters were calculated applying the Langmuir fit model.
NZM binding and inhibition of NZM binding assays
Serial dilutions of NZM and NZM variants IgG4 were prepared in MACS buffer (PBS 1% FBS, 2mM EDTA). T cells isolated from healthy donors were used as source of the cell adhesion molecule α4-integrin and added (50,000 cell/well) to the plates for 30 min, 4°C. T cells were washed and stained with 3.75 μg/ml Alexa Fluor 647–conjugated goat anti-human IgG (Jackson ImmunoResearch, cat. no. 109-606-170) for 30 min, 4°C. Cells were washed and analyzed by FACS. NZM binding was calculated as percentage of IgG+ stained cells. To study NZM neutralization, NZM-mFc was diluted to 5 ng/ml (final concentration) in MACS buffer (PBS 1% FBS, 2mM EDTA) and incubated with titrated monoclonal antibodies for 1 h, 37°C. T cells were added to the plates for 30 min, 4°C, then washed and stained with secondary goat anti-mouse-IgG-PE (SouthernBiotech, cat. no. 1030-09) at 1 μg/ml for 30 min, 4°C. Cells were washed and analyzed by FACS. NZM neutralization was calculated for each well as percentage of inhibition of binding of NZM-mFc to T cells with the following formula: 1 – % of cells stained by NZM-mFc. Gates were defined based on negative and positive controls.
X-ray crystallography and structure-guided deimmunization
NZM, NAA32 and NAA84 antibodies were produced as antigen-binding fragments and purified on IMAC (HitrapTM IMAC HP, GE Healthcare). Stable complexes of NZM-NAA32 and NZM-NAA84 were prepared in D-PBS buffer (PBS Dulbecco’s Gibco 14190-094) at 1.5:1 molar ratio, purified by size-exclusion chromatography (Superdex 200, GE healthcare) and concentrated to 10 mg/ml. Crystal were grown by vapor diffusion with a well solution containing 1.7 M sodium malonate pH 6 at 292 K (NZM-NAA32) or 35 % PEG400, 200 mM NaCl, 4 % MPD, 100 mM MES pH 6 (NZM-NAA84), and they typically appeared within a week. Crystals were flash frozen and x-ray data were collected at the European Synchrotron Radiation Facility (Beamline ID30A-1, Grenoble) using MxCube2 software. Data collection and processing statistics are outlined in Supplementary Table 3. Structure solution was performed by molecular replacement using MOLREP (version 11.6.03)26 through the CCP4 suite (version 7.0.058)27 and crystallographic refinement was carried out using BUSTER (version 2.11.7)28. The shape correlation statistic Sc of protein interface was calculated with CCP4. Superimposition was performed using the structure of the NZM-α4β7 integrin complex as a reference (4IRZ, https://www.rcsb.org/). Surface area is calculated based on Van der Waals surfaces of atoms of NZM lying at 4Å distance from any atom of NAA32 or NAA84. For deimmunization of NZM, the CDR2 of the light chain of the antibody was modelled by mutating residues that were positioned at more than 4.0 Å distance from α4-integrin as observed in the 4IRZ structure and did not affect the conformation of the antibody CDR region. A non-exhaustive search was performed and a number of potential mutations were selected for mutagenesis.
Ex-vivo stimulation of memory CD4+ T cells
T cells were cultured in RPMI 1640 medium supplemented with 2 mM glutamine, 1% (v/v) nonessential amino acids, 1% (v/v) sodium pyruvate, penicillin (50 U/ml), streptomycin (50 μg/ml) (all from Invitrogen) and 5% heat-inactivated human serum (Swiss Red Cross). Sorted memory CD4+ T cells were labelled with 5-(and 6)-carboxyfluorescein diacetate succinimidyl ester (CFSE, ThermoFisher) and cultured at a ratio of 2:1 with irradiated autologous monocytes untreated or pre-pulsed for 2-3 h with a peptide pool (15mers overlapping of 10) covering the entire sequence of the variable region of the NZM heavy and light chains (NZM peptide pool, 3 μM per peptide, produced by A&A Labs). After 12 days, cells were stained with antibodies to CD25–PE (BD Biosciences, clone M-A251, cat. no. 555432) and ICOS–Pacific Blue (Biolegend, clone C398.4A, cat. no. 313522).
Isolation of NZM-specific T cell clones
Proliferating activated T cells from ex-vivo stimulated cultures were sorted as CFSElowCD25+ICOS+ and cloned by limiting dilution. T cell clones reactivity was determined by stimulation with irradiated autologous monocytes or B cells, untreated or pre-pulsed for 2-3 h with NZM peptide pool (3 μM per peptide) or, in some experiments, with recombinant NZM (5 μg/ml). T cell clones proliferation was measured on day 3 after 16 h incubation with 1 μCi/ml [methyl-3H]thymidine (Perkin Elmer). Positive T cell clones were selected based on a cut-off value of (i) counts per minute (cpm) with antigen and antigen-presenting cells (APCs) ≥1000, and (ii) stimulation index ≥1.5 (cpm with antigen and APCs / cpm with APCs only). To determine MHC restriction, stimulation assay was performed in the absence or presence of blocking anti-MHC-II monoclonal antibodies produced in house from hybridoma cell lines (anti-HLA-DR, clone L243 from ATCC, cat. no. HB-55; anti-HLA-DQ, clone SPVL329; anti-HLA-DP, clone B7/2130). Epitope mapping was performed by stimulation of T cell clones with irradiated EBV-immortalized B-cell (EBV-B) clones, untreated or pre-pulsed for 2-3 h with individual peptides (15mers overlapping of 10) covering the entire sequence of the variable region of the NZM heavy and light chains (3 μM per peptide).
Sequence analysis of TCR Vβ genes
Sequence analysis of rearranged TCR Vβ genes of NZM-specific T cell clones was performed as previously described31. Briefly, cDNA from individual T cell clones was obtained by reverse transcription of total RNA from 103-104 cells per reaction. Rearranged TCR Vβ genes were PCR amplified using forward primer pool targeting Vβ genes, and reverse primer pairing to C1–C2 β-chain constant region. Sequence amplification was assessed through agarose gel electrophoresis; successfully amplified fragments were sequenced by Sanger method, and TCR sequence annotation was carried out by using IMGT/V-QUEST algorithm23.
HLA typing and peptide-MHC-II binding affinity predictions
HLA genotype of the patients was determined by reverse sequence-specific oligonucleotides probes (revPCR-SSO) DNA typing (LABType, One Lambda Inc.) performed at the IRCCS San Matteo Hospital Foundation, Pavia, Italy. Predicted IC50 (nM) binding values of all theoretical NZM-derived peptides (15mers overlapping of 14) to HLA-DRB1 alleles carried by the two patients and to a reference set of nine HLA-DRB1 and HLA-DRB3/4/5 alleles including DRB1*0301, DRB1*0701, DRB1*1501, DRB3*0101, DRB3*0202, DRB4*0101, DRB5*010115 and DRB1*13:01/14:0116 were calculated using NetMHCIIpan 3.2 server (http://www.cbs.dtu.dk/services/NetMHCIIpan-3.2/)14.
Purification of MHC-II presented peptides
NZM-specific EBV-B clones isolated from the two patients were pulsed overnight with 5 μg/ml NZM at a cellular density of 5x106 cells/ml. MHC-II complexes were purified from about 109 NZM-pulsed EBV-B cells with a protocol adapted from Bassani-Sternberg M. et al32. Briefly, the B cells were lysed with 0.25% sodium deoxycholate, 1% octyl-β-D glucopyranoside (Sigma), 0.2 mM iodoacetamide, 1 mM EDTA, and Complete Protease Inhibitor Cocktail (Roche) in PBS at 4°C for 1 h. The lysates were cleared by 20 min centrifugation at 18,000 g at 4°C, and MHC-II complexes were purified by immunoaffinity chromatography with the anti-HLA-DR/DP/DQ HB-145 monoclonal antibody produced in house from hybridoma cell line IVA12 (ATCC, cat. no. HB-145) and covalently bound to Protein-A Sepharose beads (Thermo Fisher Scientific). In detail, the cleared lysates were loaded 3 times into the affinity columns at 4°C, and subsequently washed at 4°C with 10 column volumes of 150 mM NaCl, 20 mM Tris•HCl, pH 8 (buffer A); 10 column volumes of 400 mM NaCl, 20 mM Tris•HCl, pH 8; 10 column volumes of buffer A; and finally 10 column volumes of 20 mM Tris•HCl, pH 8. The HLA-II complexes were eluted at room temperature by addition of 500 µl of 0.1 M acetic acid, in total five elutions for each sample. Small aliquots of each eluted fraction were analyzed by 12% SDS-PAGE to evaluate yield and purity of MHC-II complexes. Sep-Pak tC18 (Waters, Milford, MA) cartridges were used for further separation of peptides from MHC-II subunits. The cartridges were prewashed with 80% acetonitrile (AcN) in 0.5% formic acid, followed by 0.2% trifluoroacetic acid (TFA), and subsequently loaded 3 times with each fraction eluted from the immunoaffinity column. After loading, the cartridges were washed with 0.2% TFA, and the peptides were separated from the more hydrophobic MHC-II chains by elution with 30% AcN in 0.2% TFA. The peptides were further purified using a Silica C18 column tip (Harvard Apparatus, Holliston, MA) and eluted again with 30% AcN in 0.2% TFA. Finally, the peptides were concentrated by vacuum centrifugation, and resuspended in 2% AcN, 0.1% TFA, 0.5% formic acid for MS analysis.
Liquid chromatography–mass spectrometry (LC-MS/MS) and data analysis
MHC-II peptides were separated on an EASY-nLC 1200 HPLC system coupled online to a Q Exactive mass HF spectrometer via a nanoelectrospray source (Thermo Fisher Scientific). Peptides were loaded in buffer A (0.1% formic acid) on in-house packed columns (75 μm inner diameter, 50 cm length, and 1.9 μm C18 particles from Dr. Maisch GmbH) and eluted with a non-linear 120 min gradient of 5%–60% buffer B (80% ACN, 0.1% formic acid) at a flow rate of 250 nl/min and a column temperature of 50°C. The Q Exactive was operated in a data dependent mode with a survey scan range of 300-1650 m/z and a resolution of 60,000 at m/z 200. Up to 10 most abundant isotope patterns with a charge ≥ 1 were isolated with a 1.8 Th wide isolation window and subjected to higher-energy C-trap dissociation (HCD) fragmentation at a normalized collision energy of 27. Fragmentation spectra were acquired with a resolution of 15,000 at m/z 200. Dynamic exclusion of sequenced peptides was set to 30 s to reduce the number of repeated sequences. Thresholds for the ion injection time and ion target values were set to 80 ms and 3E6 for the survey scans and 120 ms and 1E5 for the MS/MS scans, respectively. Data were acquired using the Xcalibur software (Thermo Scientific). MaxQuant software was used to analyze mass spectrometry raw files. MS/MS spectra were searched against the full-length NZM heavy and light chains sequences, the human Uniprot FASTA database, and a common contaminants database (247 entries) by the Andromeda search engine33. N-terminal acetylation and methionine oxidation were set as variable modifications; no fixed modifications were selected; the enzyme specificity was set to “unspecific” with a minimum peptide length of 8 amino acids. A false discovery rate (FDR) of 1% was required for peptides. Peptide identification was performed with an allowed precursor mass deviation of up to 4.5 ppm and an allowed fragment mass deviation of 20 ppm; “match between runs” option was disabled.
Statistical analysis
GraphPad Prism 7 software was used for to perform all the statistical analyses. EC50 (ng/ml) and IC90 (ng/ml) values were calculated for every antibody tested with the different ELISA assays and the inhibition of NZM binding assay, respectively, by nonlinear regression analysis using the GraphPad Prism 7 software. A two-tailed Spearman’s correlation was performed to correlate NZM neutralization (IC90) with association constant (ka) and dissociation constant (kd). “n” indicates the number of antibodies tested. With 17 or more pairs, GraphPad Prism 7 software computes an approximate P value for nonparametric correlation. An extremely significant P value is indicated as P < 0.0001.
Extended Data
Extended Data Fig. 1. Epitope mapping of NZM-specific antibodies.
a, Alignment of NZM heavy and light chain variable regions (NZM VH and NZM VL) to the human scaffold antibody counterparts (21/28’CL and REI) used for NZM humanization. Mutated residues are shown in red. Dots indicate the same residue. b, Scheme of the 8 heavy and 8 light chains variants of NZM that were combined in an 8x8 matrix to express 64 different NZM CDR swap variants. c, Cluster analysis of binding of 30 antibodies isolated from patient A to the 64 NZM swap variants by ELISA. BAbs and NAbs are indicated on the x-axis in red and black, respectively. The NZM swap variants are shown on the right y-axis (H, heavy chain; L, light chain; 1, CDR1; 2, CDR2; 3, CDR3). Optical density (OD) values are shown with a two-color gradation scale from minimum (white) to maximum (blue).
Extended Data Fig. 2. Structural details of the interaction of NZM with a NAb, a BAb and α4-integrin.
a, Closer view of the interaction interface between NZM and NAA32 (left) and NAA84 (right). Epitope and paratope residues are shown in solid sticks. Proteins are displayed in ribbon diagram. The empty space in the interface between the NZM and NAA32 or NAA84 is represented as orange or purple surface, respectively, in two different orientations b, Superimposition of the antigen-binding fragment of NZM in complex with NAA84 (NAb, green), NAA32 (BAb, cyan) and α4-integrin (orange). NZM heavy and light chains are shown in salmon and slate blue, respectively. Proteins are displayed in ribbon diagram.
Extended Data Fig. 3. Sorting of NZM-activated memory CD4+ T cells from MS patients.
Flow cytometry analysis of memory CD4+ T cells at day 12 after ex-vivo stimulation with irradiated autologous monocytes untreated (upper panels) or pre-pulsed with NZM peptide pool (lower panels). CFSElowCD25+ICOS+ T cells reactive to NZM peptide pool were FACS-sorted and cloned by limiting dilution (representative of n = 2 biologically independent samples).
Extended Data Fig. 4. MHC restriction of NZM-reactive CD4+ T cell clones and peptide-MHC-II binding affinity predictions of NZM and deimmunized variants.
a, MHC restriction of NZM-reactive T cell clones. NZM-specific CD4+ T cell clones isolated from patient A (upper panel) and patient B (lower panel) were stimulated with antigen-pulsed autologous APCs in the absence or presence of blocking anti-MHC-II antibody (anti-HLA-DR, clone L243; anti-HLA-DQ, clone SPVL3; anti-HLA-DP, clone B7/21). Proliferation was measured on day 3 after a 16-h pulse with [3H]-thymidine, and is expressed as counts per minute (cpm). Inhibition of T cell proliferation was >80% only in the presence of the anti-HLA-DR antibody. b and c, Predicted binding affinities of all theoretical 15mer peptides derived from NZM heavy chain (HC) and light chain (LC) to HLA-DRB1 alleles carried by the two patients (b), or to a reference set of nine HLA-DRB1 and HLA-DRB3/4/5 alleles (c). The affinities are shown as reciprocal IC50 (nM) values. The dotted lines define the thresholds of high-affinity binding set at 100 nM and low-affinity binding set at 300 nM. d, Predicted binding affinities of 15mer peptides spanning the light chain CDR2 region of NZM variants to HLA-DRB1 alleles carried by patient A (DRB1*14:01 and DRB1*16:01) and patient B (DRB1*0701). The affinities are shown as reciprocal median IC50 (nM) values. The dotted lines define the thresholds of high-affinity binding set at 100 nM and low-affinity binding set at 300 nM.
Supplementary Material
Acknowledgments
The authors would like to thank the patients for their participation in the study. We would like to thank M. Nussenzweig (Rockefeller University) for providing reagents for antibody cloning and expression, and Servizio Tipizzazione of the IRCCS San Matteo Hospital Foundation, Pavia, Italy, for HLA typing. This work was supported by the Swiss National Science Foundation (grant no. 176165 to A.L.) and by the Innovative Medicines Initiative Joint Undertaking ABIRISK (Anti-Biopharmaceutical Immunization: Prediction and analysis of clinical relevance to minimize the risk) project under grant agreement no. 115303, resources of which are composed of financial contribution from the European Union's Seventh Framework Program (FP7/2007-2013) and EFPIA Companies. A.L. and F.S. are supported by the Helmut Horten Foundation.
Footnotes
Author contributions
A.C. characterized the T cell response, performed the peptidomics, analyzed the data and wrote the manuscript; V.M. performed structural analyses, modeling, deimmunization and supervision of structural studies; T.B. determined the crystal structures; S.P. performed crystallization and characterization of antibody complexes; J.L.P. cloned the antigen-binding fragments for crystallization; P.F. purified antibodies; J.D. expressed the antigen-binding fragments for crystallization; M.A. collected clinical data and samples; F.D. collected clinical data and provided supervision; M.G. collected clinical data and samples; D.F. collected clinical data and provided supervision; C.S.-F. immortalized memory B cells and performed screenings; B.F.R. sequenced and expressed antibodies; I.G.-S. analyzed antibody sequences; M.F. performed bioinformatics analyses; D.J. performed cell sorting; R.G. analyzed mass-spectrometry data; F.S. provided supervision and wrote the manuscript; A.L. provided supervision, analyzed the data and wrote the manuscript; L.P. provided overall supervision, designed the experiments, characterized the antibodies, analyzed the data and wrote the manuscript.
Competing interests. V.M., T.B., S.P., J.L.P., P.F. and J.D. are all employees of Sanofi. F.D. has participated in meetings sponsored by or received honoraria for acting as an advisor/speaker for Biogen Idec, Celgene, Genzyme-Sanofi, Merck, Novartis Pharma, and Roche. His institution has received research grants from Biogen and Genzyme Sanofi. He is section editor of the MSARD Journal (Multiple Sclerosis and Related Disorders). A.L. is a Senior Vice President and Senior Research Fellow at Vir Biotechnology, Inc.
Reporting Summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary linked to this article.
Data availability. All requests for raw and analyzed data and materials will be promptly reviewed by the Institute for Research in Biomedicine to verify if the request is subject to any intellectual property or confidentiality obligations. Patient-related data not included in the paper may be subject to patient confidentiality. Any data and materials that can be shared will be released via a Material Transfer Agreement. Source data of Fig. 1 are provided. Sequence data of the monoclonal antibodies isolated in this study have been deposited in GenBank (MN044260-MN044339). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE34 partner repository with the dataset identifier PXD013599. The X-ray structure factors and coordinates have been deposited in the Protein Data Bank (access numbers are 6FG1 and 6FG2).
References
- 1.Hao L, Fang-Hong S, Shi-Ying H, Shun-Guo Z, Min-Ling C. A Review on Clinical Pharmacokinetics, Pharmacodynamics, and pharmacogenomics of natalizumab: A Humanized Anti-alpha4 Integrin Monoclonal Antibody. Curr Drug Metab. 2018 doi: 10.2174/1389200219666180427165841. [DOI] [PubMed] [Google Scholar]
- 2.Chataway J, Miller DH. Natalizumab therapy for multiple sclerosis. Neurotherapeutics. 2013;10:19–28. doi: 10.1007/s13311-012-0171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calabresi PA, et al. The incidence and significance of anti-natalizumab antibodies: results from AFFIRM and SENTINEL. Neurology. 2007;69:1391–1403. doi: 10.1212/01.wnl.0000277457.17420.b5. [DOI] [PubMed] [Google Scholar]
- 4.Bachelet D, et al. Occurrence of Anti-Drug Antibodies against Interferon-Beta and Natalizumab in Multiple Sclerosis: A Collaborative Cohort Analysis. PLoS One. 2016;11:e0162752. doi: 10.1371/journal.pone.0162752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rup B, et al. Standardizing terms, definitions and concepts for describing and interpreting unwanted immunogenicity of biopharmaceuticals: recommendations of the Innovative Medicines Initiative ABIRISK consortium. Clin Exp Immunol. 2015;181:385–400. doi: 10.1111/cei.12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Link J, et al. Clinical practice of analysis of anti-drug antibodies against interferon beta and natalizumab in multiple sclerosis patients in Europe: A descriptive study of test results. PLoS One. 2017;12:e0170395. doi: 10.1371/journal.pone.0170395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn N, et al. Rituximab in multiple sclerosis: Frequency and clinical relevance of anti-drug antibodies. Mult Scler. 2018;24:1224–1233. doi: 10.1177/1352458517720044. [DOI] [PubMed] [Google Scholar]
- 8.Quistrebert J, et al. Incidence and risk factors for adalimumab and infliximab anti-drug antibodies in rheumatoid arthritis: A European retrospective multicohort analysis. Semin Arthritis Rheum. 2018 doi: 10.1016/j.semarthrit.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Jensen PEH, et al. Detection and kinetics of persistent neutralizing anti-interferon-beta antibodies in patients with multiple sclerosis. Results from the ABIRISK prospective cohort study. J Neuroimmunol. 2019;326:19–27. doi: 10.1016/j.jneuroim.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Zare N, Zarkesh-Esfahani SH, Gharagozloo M, Shaygannejad V. Antibodies to interferon beta in patients with multiple sclerosis receiving CinnoVex, rebif, and betaferon. J Korean Med Sci. 2013;28:1801–1806. doi: 10.3346/jkms.2013.28.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murdaca G, et al. Immunogenicity of infliximab and adalimumab: what is its role in hypersensitivity and modulation of therapeutic efficacy and safety? Expert Opin Drug Saf. 2016;15:43–52. doi: 10.1517/14740338.2016.1112375. [DOI] [PubMed] [Google Scholar]
- 12.Traggiai E, et al. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat Med. 2004;10:871–875. doi: 10.1038/nm1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu Y, Schurpf T, Springer TA. How natalizumab binds and antagonizes alpha4 integrins. J Biol Chem. 2013;288:32314–32325. doi: 10.1074/jbc.M113.501668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen KK, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul S, et al. Development and validation of a broad scheme for prediction of HLA class II restricted T cell epitopes. J Immunol Methods. 2015;422:28–34. doi: 10.1016/j.jim.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Hera B, et al. Natalizumab-related anaphylactoid reactions in MS patients are associated with HLA class II alleles. Neurol Neuroimmunol Neuroinflamm. 2014;1:e47. doi: 10.1212/NXI.0000000000000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan Y, Yuhasz SC, Amzel LM. Anti-idiotypic antibodies: biological function and structural studies. FASEB J. 1995;9:43–49. doi: 10.1096/fasebj.9.1.7821758. [DOI] [PubMed] [Google Scholar]
- 18.van Schie KA, et al. Neutralizing capacity of monoclonal and polyclonal anti-natalizumab antibodies: The immune response to antibody therapeutics preferentially targets the antigen-binding site. J Allergy Clin Immunol. 2017;139:1035–1037 e1036. doi: 10.1016/j.jaci.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Foote J, Milstein C. Kinetic maturation of an immune response. Nature. 1991;352:530–532. doi: 10.1038/352530a0. [DOI] [PubMed] [Google Scholar]
- 20.Hamze M, et al. Characterization of CD4 T Cell Epitopes of Infliximab and Rituximab Identified from Healthy Donors. Front Immunol. 2017;8:500. doi: 10.3389/fimmu.2017.00500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding FA, Stickler MM, Razo J, DuBridge RB. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. MAbs. 2010;2:256–265. doi: 10.4161/mabs.2.3.11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiller T, et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yaari G, Uduman M, Kleinstein SH. Quantifying selection in high-throughput Immunoglobulin sequencing data sets. Nucleic Acids Res. 2012;40:e134. doi: 10.1093/nar/gks457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta NT, et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 27.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bricogne G, et al. BUSTER version 2.11.7. Cambridge, United Kingdom: Global Phasing Ltd; 2017. [Google Scholar]
- 29.Spits H, et al. Characterization of monoclonal antibodies against cell surface molecules associated with cytotoxic activity of natural and activated killer cells and cloned CTL lines. Hybridoma. 1983;2:423–437. doi: 10.1089/hyb.1983.2.423. [DOI] [PubMed] [Google Scholar]
- 30.Watson AJ, DeMars R, Trowbridge IS, Bach FH. Detection of a novel human class II HLA antigen. Nature. 1983;304:358–361. doi: 10.1038/304358a0. [DOI] [PubMed] [Google Scholar]
- 31.Latorre D, et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature. 2018;562:63–68. doi: 10.1038/s41586-018-0540-1. [DOI] [PubMed] [Google Scholar]
- 32.Bassani-Sternberg M, Pletscher-Frankild S, Jensen LJ, Mann M. Mass spectrometry of human leukocyte antigen class I peptidomes reveals strong effects of protein abundance and turnover on antigen presentation. Mol Cell Proteomics. 2015;14:658–673. doi: 10.1074/mcp.M114.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.